Abstract
Cancer is one of the leading causes of morbidity and mortality in the globe. Microbiological infections account for up to 20% of the total global cancer burden. The human microbiota within each organ system is distinct, and their compositional variation and interactions with the human host have been known to attribute detrimental and beneficial effects on tumor progression. With the advent of next generation sequencing (NGS) technologies, data generated from NGS is being used for pathogen detection in cancer. Numerous bioinformatics computational frameworks have been developed to study viral information from host-sequencing data and can be adapted to bacterial studies. This review highlights existing popular computational frameworks that utilize NGS data as input to decipher microbial composition, which output can predict functional compositional differences with clinically relevant applicability in the development of treatment and prevention strategies.
Keywords: Cancer microbiome, Computational frameworks, NGS
Introduction
Cancer is one of the leading causes of morbidity and mortality in the globe. Annually an estimated 14.1 million are diagnosed, and 8.2 million die from cancers around the world. In the United States alone, 1.7 million cases are diagnosed, and about six hundred thousand die from the disease [1–3]. Cancer is a multifactorial disease with known genetic and environmental etiologies. Microbiological infections account for up to 20% of the total global cancer burden [4, 5]. Viruses are commonly attributed and are responsible for at least 10% of all human cancers [6]. Multiple studies have evaluated viral content and its influence on cancer pathogenesis utilizing advanced technologies and bioinformatics approaches.
Meanwhile, recent limited evidence exists proposing relationships between bacterial species and disease either as effector or consequence of tumorigenesis. While much effort has gone into characterizing cavity organs microbiota, that of solid tumors is less explored. The characterization of tissue-associated microbiota is challenging as well as computationally intensive. Next-generation sequencing technologies provide an opportunity to explore better bioinformatics approaches to detect microbial agents and can assist in the interpretation of not only viral but bacterial species impact in tumor tissue. The examination of microbial species is pivotal to developing new prevention and treatment strategies.
Relationship of microbiota with cancer pathogenesis
The human microbiome, defined as the aggregation of microorganisms that live in and on our bodies, contributes to our broader genetic portrait [7, 8]. The microbiota within each organ system is distinct, which can drive functionally relevant inter-individual variations and determinants of disease [7, 9–12]. Microbial community variations, production of bacterial metabolites, and microbial interactions with the human host have been attributed to detrimental and beneficial tumoral effects since the eighteenth century [13, 14]. This highlights the unique agonistic and antagonistic effects of the human microbiome in cancer progression and has become an area of intense exploration. While contribution by some viral pathogens is firmly established, the role of the bacterial community remains controversial. The mechanisms by which viral agents contribute to pathogenesis have been reviewed in detail and are not covered here [15–18]. Mechanisms by which bacteria contribute to the alterations and the carcinogenic process are not all well understood. It is known, however, that similar to viruses, persistent and chronic infections may initiate the process or promote established cancers [14, 19–22]. Alteration of the bacterial community could also result in beneficial effects on the tumor microenvironment. In fact, according to the literature, any agent capable of stimulating host immune defenses can minimize the incidence and be advantageous to established tumors. Modification of the immune cascade in response to infection or dysbiosis is one of the most critical aspects of tumor-microenvironment cross-talk [23, 24]. Altered host-dynamics can increase bacterial translocation as a direct consequence of changes in microbial composition, resulting in increased inflammation. Bacterial products and bacterial metabolites may have protective effects on survival, reduced growth of cancer cells, or modulate anticancer immunosurveillance at local or distant sites [10]. Butyrate for example, which has anti-inflammatory properties, is thought to be protective while secondary bile acids are considered carcinogenic [25, 26]. These variations in the microbial composition may be directly or indirectly responsible for the carcinogenic process in susceptible populations, alter the course of established cancer, or influence therapeutic response and can assist in understanding patient inter-variability [27, 28].
New microbial (viral, bacterial, and other pathogens) contributions to cancer, whether beneficial or detrimental, are being discovered. Improved techniques and integrated data networks facilitate discoveries and have become the focus of multiple studies [29–37]. Recent studies have found that specific bacterial taxa are consistently identified in tumor tissue [38]. Compared to adjacent or control tissue, Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia, and Methanobacteriales are found at increased levels in tumor, while Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium, Roseburia, and Treponema are at decreased levels [38–46] (Table 1). Also, viral and bacterial co-occurrence is thought to modulate tumor aggressiveness [47–49]. Based on epidemiological and geographic correlations analyses, it is suggested that viral agents interact with bacteria resulting in more aggressive tumors. For example, stomach tumors infected with Epstein Barr virus are recognized to be molecularly distinct. Meanwhile, Epstein Barr virus is thought to interact with Helicobacter pylori driving aggressiveness, however insufficient evidence exists. In hepatocellular carcinoma viral co-infection with HBV or HCV and the interaction between the proteins, HBx HCV core and NS5a, can also lead to more aggressive tumors. Interaction with other exposures, alcohol consumption, smoking, co-morbidities, betel nut chewing can act as co-factors altering the tumor microenvironment in cancers of the head and neck [50].
Table 1.
Known and suspected microbial association with cancer pathogenesis
| Cancer type | Known microbial associations | Suspected agents | References |
|---|---|---|---|
|
Breast Triple-negative, HER2+, ER+ |
None |
Epstein–Barr virus, human papillomaviruses Alistipes spp. Bacteroides fragilis, Sphingobium yanoikuyae, Microbial dysbiosis |
[35, 36, 39, 40] |
|
Prostate Prostate adenocarcinoma |
None |
Cutibacterium acnes Bacteroides massiliensis Streptococcus spp. Staphylococcus spp. Microbial dysbiosis |
[37, 41, 42] |
|
Stomach Stomach adenocarcinoma |
Helicobacter pylori, Epstein Barr Virus |
Microbial dysbiosis | [57, 70] |
|
Liver Liver and intrahepatic bile duct |
Hepatitis viruses, Parasitic infections |
Helicobacter pylori | [43] |
|
Cervical Cervical squamous cell and endometrial carcinoma |
Human papillomaviruses | Chlamydia trachomatis, microbiome dysbiosis | [63] |
|
Head and Neck Oropharyngeal and laryngeal |
Epstein Barr Virus, Human papillomaviruses |
Fusobacterium nucleatum, microbiome dysbiosis | [56, 58] |
|
Colon and rectum Colorectal adenocarcinoma |
Microbial dysbiosis Fusobacterium nucleatum |
Human papillomavirus Helicobacter pylori, Streptococcus bovis, E. Escherichia coli, E. Bacteroides fragilis, Campylobacter spp. |
[10, 31, 32, 55] |
|
Kidney Renal cell carcinoma and clear cell carcinoma |
None |
Hepatitis C virus Epstein Barr Virus Urinary tract infection-associated pathogens |
[44] |
|
Lung Lung squamous cell and adenocarcinomas |
None |
Epstein Barr Virus Molluscum Contagiosum virus Microbial dysbiosis Chlamydia pneumoniae |
[45] |
|
Bladder Bladder squamous cell carcinoma |
Schistosoma haematobium |
Human papillomavirus Epstein–Barr Virus |
[46] |
Common cancer types listing known and suspected microbial (viral, bacterial, and other) agents associated with cancer pathogenesis or that have been identified as common causes of infection in cancer patients, which may play a role in patient inter-variability
Competitive interaction between viral-bacterial species and other exposures may be more apparent at broader taxonomic levels. Taxonomic level analyses of the gut, oral, and other cavity organ microbiomes reveal bacterial candidates associated with pathology of disease [33, 35, 51]. These findings could be applied to preventive or complementary therapies. Questions remain, whether microbial composition findings derived from surrogate material, like stool and saliva within these cavity organs, directly relate to the microbial composition within the solid tumor tissue and surrounding tumor microenvironment. Further, whether the tissue-associated tumor microbial composition can be consistently derived from existing human sequencing data and how to best discern microbial roles in inter-population variability. Identification of microbial composition directly from tumor tissue human sequences enables not only the study of microbial changes and cancer pathogenesis but microbial genomic integration [34]. Integration of microbial DNA into the human genome may prove key in the identification of passager versus driver bacteria in cancer pathogenesis.
Microbiome detection in high throughput sequencing data
Next-generation sequencing (NGS) technologies, also known as high-throughput, provide a powerful tool for the evaluation of the role of microbes in cancer development and progression as well as differences across populations. NGS is a useful and unbiased tool that can be used for the identification of previously undetected or unsuspected causative microorganisms in molecular diagnostics [52]. It has become vital and necessary for the integrative analysis of cancer biology, enabling description of the mutational and molecular landscape of cancer for both direct and indirect taxonomic studies [53]. These techniques take advantage of NGS production of short reads and the predominance of host-derived sequences to examine pathogen-host interaction, including their correlation with metabolic and regulatory mechanisms in cancer [30, 32, 54–58]. Although the establishment of a causal relationship requires a more detailed characterization of the tumor microbiota and microbial population dynamics, integration of host sequencing data with clinical and epidemiological data can provide valuable information to the understanding of the role bacteria play in cancer pathogenesis and population differences. Given the close interaction between microbes and the host responses, it is essential to identify the compositional structure and clinically relevant functional pathways with an integrated approach.
Computational frameworks and tissue-associated bacteria detection in cancer
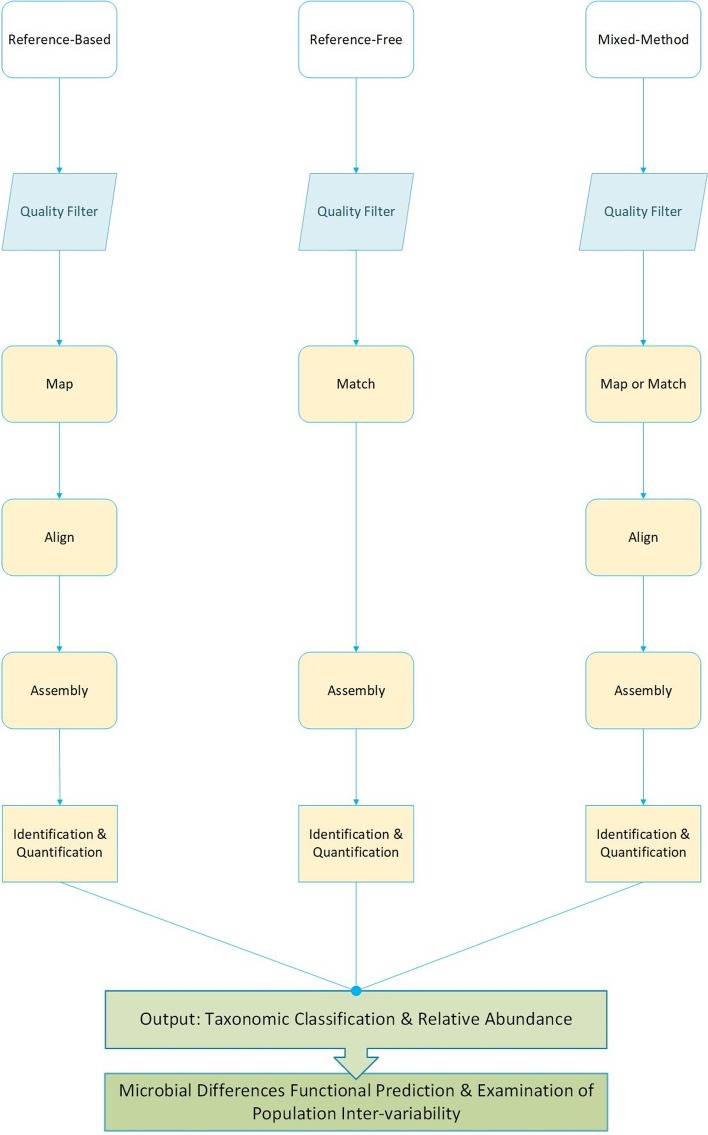
Bioinformatics computational frameworks are methods and pipelines able to accommodate user-defined parameters and deliverables to understand the basis of biological concepts [59]. Mining NGS data using bioinformatics computational frameworks provide great opportunities in understanding the role of bacteria in cancer pathogenesis. Numerous state-of-the-art bioinformatics tools and methods are available today that support the identification of microbial novel targets in cancer diagnostics, treatment, prevention, and control. Several studies have demonstrated that pathogenic and commensal bacteria composition can be derived from human tumor tissue utilizing various bioinformatics computational approaches by sequential filtering and matching steps [52, 60–63]. Pathogen detection derived from human sequences has been primarily completed by computational subtraction with one of three approaches, reference-based, reference-free, or mixed methods with one primary core pipeline involving the removal of human-host sequences to characterize remaining sequencing reads (Fig. 1). Pathogen detection algorithms may be classified by (1) their methodology, (2) the order in which human sequencing reads are identified and removed, and (3) what happens with the remaining sequences (whether these go through de-novo assembly or are filtered out). Here, we discuss ten computational frameworks, PathSeq, SRSA, CaPSID, PathoScope 2.0, SURPI, VirusScan, MetaShot, ConStrains, RINS, and GRAMMY, designed to identify microbiota (virus, bacteria, and other) derived from human sequences with applications in human cancer (Table 2). Computational frameworks that strictly match sequencing reads to pathogen libraries or those designed for direct metagenomics analyses are not included (see Nooij et al. 2018 for a recent in-depth review of these tools [64]).
Fig. 1.
Generic pipeline comparing three basic computational frameworks designed to identify microbial reads from human sequences. Generic pipelines can be summarized into three general stages, pre-processing (blue), processing (yellow), and analyses post-processing (green). During pre-process, most methodologies trim and quality filter sequencing reads. Quality reads are mapped and aligned during the processing steps to either human or pathogen reference sequences or key identifying factors before making a final identification call. Once species have been identified, their composition is characterized in detail, depending on the methodology being used. Finally, having taxonomic classification and compositional structure permits downstream correlation analyses and functional-relevant identification of molecular pathways. Differential functional prediction and patient inter-variability aid in the identification of novel microbe based prevention and treatment strategies
Table 2.
Computational frameworks designed to detect microbiota from human sequences by subtractive, filtration, or mixed methods
| Framework | Approach | Dependencies | Input | output | Advantages/disadvantages | Cancer validation | Refs. |
|---|---|---|---|---|---|---|
| PathSeq | Alignment and de novo assembly |
BLAST BLASTN BLASTX MAQ MegaBLAST RepeatMasker Velvet |
Input: RNA-seq or DNA-seq Output: Pathogen presence/absence |
Scalable cloud computing Feasible for known and novel pathogen identification Two-pass subtraction with increased filtering costs |
Cervical cancer (cell line and simulated data) TCGA ovarian |
[63, 68] |
| SRSA | Alignment and de novo assembly |
Velvet MegaBLAST BLAST BWA TopHat |
Input: RNA-seq Output: Species-level taxonomy characterization (prevalence) |
Incorporates sample pre-processing, quality filtering, sequence mapping, and assembly Not freely available No known updates Original work validation was limited to cell line |
HIV-1 cell line | [60] |
| CaPSID | Mix-method, simultaneous alignment, filtration and de novo assembly |
BioPython Bowtie2 Trinity |
Input: RNA-seq or DNA-seq Output: Top-hit pathogen genome identification ranked by maximum gene coverage |
Web-based, open-source and scalable application; Modular analyses; Single pass filtering, which may fail to subtract host reads |
Ovarian cancer TCGA stomach |
[67] |
| SURPI | Dual scanning mode; Known pathogens identification or de novo assembly |
SNAP RAPSearch BWA BLASTN Bowtie2 DUST in PRINSEQ |
Input: Paired-end metagenomic Output: Species-level taxonomic classification and coverage map |
Scalable to cloud or standalone servers Capacity to incorporate reference database Dual-mode: quantitative and semi-quantitative pathogen identification |
Prostate cancer (cell line, tissue biopsies) Colorectal cancer (tissue biopsies) |
[71] |
| PathoScope 2.0 | Penalized probabilistic identification; Modular filtration, alignment and assignment |
SAMtools BLASTX Bowtie2 thetaPrior |
Input: Metagenomic or genomic (RNA-seq or DNA-seq) Output: Strain level pathogen relative abundance |
Modular detailed result reporting with Designed for low abundance strain-level identification MySQL server required; no connection to the population structure of relevant species |
TCGA stomach | [69, 70] |
| VirusScan | Identification of known viral and integration sites |
BWA BLAST MegaBLAST Pindel RepeatMasker PHYLIP |
Input: RNA-seq Output: Viral read abundance and integration sites |
Designed for viral identification; Abundance and integration sites analyses |
TCGA cancer cohorts | [72] |
| MetaShot | Two-step similarity filtering and taxonomic assessment |
Bowtie2 TANGO STAR Bash |
Input: RNA-Seq or DNA-Seq Output: Assigned read report and Krona plot with relative abundance |
Extracts unassigned reads; Allow for functional annotations; Slower than other applications |
None | [73] |
| ConStrains |
Marker-based (SNP patterns) Strain-level prediction |
MetaPhlAn PhyloPhlAn Bowtie2 SAMtools Metropolis-Hasting Monte-Carlo |
Input: Metagenomics (RNA-seq) Output: Strain-level prediction and relative abundance |
Single reference strain collection; Facilitates functional analyses when combined with reference genome-based gene coverage metadata |
None | [74] |
| RINS | Intersection based identification and removal |
Bowtie BLAST BLAT Trinity |
Input: Mate-paired RNA-seq unmapped reads Output: Pathogen contigs |
Requires prior knowledge of reference; Detection limited to user-defined parameters |
Prostate cancer (cell line) |
[66] |
| GRAMMy | Mix- model Bayesian, Expectation–Maximization and maximum likelihood estimation |
BLAST BLAT MAQ Bowtie PerM BLASY |
Input: Metagenomics reads Output: Genomic relative abundance as numerical vectors |
User flexibility Probabilistic handling of ambiguous hits Computational efficiency |
None | [76] |
Comparison of computational workflows designed to derive microbial content from human sequences by subtractive and filtering methods, broadly categorized as reference-based, reference-free, and mixed methods approaches. Data requirements to run the pipeline, output information, as well as advantages and disadvantages for each, are summarized. Most have been validated with large cancer datasets, including TCGA sequencing data. ConStrains is based on reference-free, while all other approaches are reference-based or mixed-methods
In NGS, about 10% of the sequencing reads are flagged unmapped to the human genome after alignment [65]. Under the assumption that the sequenced tissue contains both host and microbial information, the bacterial composition can then be detected after the computational subtraction of human content [61–63]. Computational subtraction methods for microbial identification and discovery derived from human tissue were first introduced by Weber et al. and Xu et al. [61, 62]. These early approaches were computationally intensive and involved creation of a cDNA library with subsequent subtraction of human-expressed sequence tags [61, 62]. Newer methods take advantage of NGS data repositories’ unmapped-to-human sequences and have lower computational requirements. Frameworks that consider unmapped-to-human sequencing reads as input data can lower computational costs while facilitating novel discoveries.
Most computational subtraction frameworks are reference-based approaches [60, 63, 66, 67]. Reference-based, by definition, requires mapping to a reference, in this case, human host genome, then allocating all leftover unmapped-to-human reads to pathogen target genomes. PathSeq, for example, combines alignment and de novo assembly with a two-pass subtraction process [63]. It aligns the sequencing reads to target genomes and quantify their abundance based on the total number of aligned sequencing reads and the genome coverage, enabling identification of both commensals and pathogens whether known or novel. However, the two-pass filtration process may eliminate a high number of sequences, which may increase filtration costs and limit identification. PathSeq has been utilized in pathogen identification for various infection-associated and inflammation-associated cancers, notably the emerging association of Fusobacterium nucleatum in colorectal cancer [68]. SRSA, short RNA subtraction, and assembly utilize short RNA mapping and assembly to identify pathogens in relation to host-sequencing reads [60]. SRSA has the capability for use in microbial identification in infection-associated cancers. However, initial work was limited to mycoplasma detection in HIV-1 cell lines, and its computational methods are also not freely available. Unlike SRSA, CaPSID (computational pathogen sequence identification) is a web-based open-source platform that similar to PathSeq, performs mapping and de novo assembly [67]. CaPSID differs in its single-pass alignment and filtration process, where both human and pathogen reads are aligned to reference genomes while separating those that do not match either for de novo assembly simultaneously. Its potential in cancer was demonstrated by Borozan et al. in stomach adenocarcinoma samples from TCGA and other cancer networks [49]. Borozan et al. evaluated human herpesvirus 4 (HHV-4) variants to determine oncogenic potential differences among samples from different country origins providing evidence of the potential of such frameworks in future population studies [49]. Unlike PathSeq, SRSA, and CaPSID, PathoScope 2.0 does not perform de novo assembly; instead, it utilizes penalized statistical mix-model and probabilistic pathogen identification [69]. It also provides detailed reports with core and optional module format that enable user customization. On the downside, the target reference genome must be present for precise identification of microbes. PathoScope 2.0 is designed to identify low abundant strains, making it an ideal tool for host-derived microbial analyses due to the low abundance of microbial reads in relation to host reads found in sequencing data. Zhang et al. incorporated PathoScope 2.0 methods with its WGS PathSeq-based methods for microbial relative abundance estimation of gastric cancer clinical samples and existing sequencing data [70]. SURPI, sequence-based ultra-rapid pathogen identification, was also designed for pathogen detection from clinical samples for surveillance similar to PathoScope 2.0. One of the advantages of SURPI is the capacity for quantitative and semi-quantitative simultaneous identification, meaning it can perform mapping and de novo assembly for divergent microbial analyses [71]. SURPI has been validated against samples from colon and prostate cancer-derived datasets. Unlike those before mentioned that were designed to identify various microorganisms, VirusScan is a referenced-based computational subtraction approach designed to profile the viral composition. It also calculates abundance and integration sites within human tumors utilizing unmapped-to-humans and poorly mapped to human genome reads [72]. This approach was used to identify population viral differences in TCGA’s liver and stomach cancer cohorts [72]. The inclusion of bacterial libraries could assist in future co-occurrence and tumor microbiome analyses. MetaShot is similar to prior mentioned reference-based approaches in that it shares a two-step filtration method to identify candidate pathogens; however, it is a bit more stringent in its taxonomic assignment [73]. This feature enables functional annotation with great potential in tissue-associated bacterial composition analyses. On the other hand, its rigorous approach comes with higher computational costs and has yet to be validated in cancer datasets.
Other methods may utilize pre-defined target genomic markers like k-mers, single nucleotide polymorphisms (SNP), or unique sequence tag libraries to identify and retain pathogen information while removing human host sequences from further consideration. These approaches can be described as marker-based methods and are mostly considered reference-free. Reference-free, marker-based approaches such as ConStrains, conspecific strains rely on the creation of SNP profiles to predict pathogen strains contained within the sequencing sample [74]. However, methods such as this are not wholly reference-free, rather minimally reference-dependent [74]. ConStrains works by inferring microbial abundance of conspecific strains utilizing SNP patterns and de novo assembly with microbial prediction estimation based on Metropolis-Hasting Markov Chain Monte-Carlo model. Although ConStrains has not been used in cancer genomic data, it has the capability for functional analyses, which are pivotal in understanding different microbial effects in cancer pathogenesis, particularly those of infectious etiology.
Computational frameworks may also take advantage of mixed approaches which can be reference-free or reference-based. Reference-Free Mixed or mixture-model approach utilizes intersection analyses, while mixture-model approaches take advantage of both reference and marker-based methods. RINS, rapid identification of non-human sequences, uses intersection analysis. Similar to ConStrain is not completely reference-free. It employs a pre-defined query reference that includes genomes of viruses, bacteria, or other pathogens to find the intersect, rather than mapping and subtracting the human reference genome [66]. RINS has been validated in prostate cancer and has low computational requirements. However, it can only detect pathogens that are explicitly defined within the query reference [66]. By only being able to identify defined references expressly, it risks the removal of unknown sequences, hindering novel pathogen discovery. Mixture-model approaches differ from traditional computational subtraction in that these either maps against a pre-determined pathogen reference in series [66, 73], against both human and pathogen in parallel [75], or some combination of these before filtering out human-host sequences. Mixture-model approaches like GRAMMy, genome relative abundance estimation framework using mixture model theory, utilize expectation–maximization algorithms to calculate microbial genome relative abundance at different taxonomic levels [76]. GRAMMy is designed to use either mapping or de novo assembly in the absence of a reference genome [76].
Computational pipelines and functional prediction of microbial differences
Recent works in the gut microbiome revealed the utility of taxonomic differences, epigenetic, heritable, and co-occurrence patterns in the understanding of cancer pathogenesis [77]. Microbial compositional differences and population variations have been thoroughly reviewed in [78]. From these and other works, we understand that accurate interpretation of microbial impact cancer pathogenesis involves more than compositional differences. Functional annotation and prediction of molecular processes are equally important in the identification of clinically relevant microbial interactions within the human host.
Post-processing pipelines have been developed to translate microbial composition outputs into predicted mechanisms through which bacteria may influence host immune responses, gene, and protein expression within the tumor microenvironment. For example, pipelines such as PICRUSt [79], Tax4Fun [80], and ShortBRED [81] can assist in the identification of functional annotations and subtle differences across populations within and across tumor types. Although these pipelines are designed to predict functional profiles derived from 16S rRNA sequencing data, they have application in host-derived microbial profiles when used in integrated approaches. For example, PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) infers microbial community host-associated functional composition based on gene annotation databases such as the Kyoto Encyclopaedia of Genes and Genomes (KEGG) or the Clusters of Orthologous Group (COGs) [82]. Tax4Fun (Taxonomy functional community profiling) on the other hand, predicts the functional capabilities of microbial communities based on 16S rRNA datasets. Tax4Fun provides an excellent approximation to functional profiles obtained from metagenomic shotgun sequencing approaches and has been successfully used to identify signs of ethnic acculturation in oral microbiota [80]. Both methods, in combination with computational frameworks designed to determine the microbial composition, provide insight into tumor-microbial associations and enable the discovery of new associations, the identification of patterns of co-occurrence, and possible host interaction effects. Gene and protein expression within the tumor and surrounding tissue information in conjunction with microbial composition may provide much-needed information on differential analyses. ShortBRED (Short, Better REad Dataset) is one that quantifies the abundance of functional gene families to predict protein profiles within the sample [81]. It can predict antibiotic resistance genes and virulence factors protein families that are pivotal in understanding therapeutic response. A combination of microbial detection and functional prediction approaches is critical, especially given the potential use in microbe-based prevention strategies and targeted therapies.
Conclusions
There is a great diversity present in the human tumor microenvironment that makes identification of the microbial community challenging. Next generation sequencing technologies and the use of these computational tools permit the discovery of new microbes that are non-culturable and would otherwise remain undiscovered [83]. Profiling and characterization of the bacterial community and functional annotations can provide information on the effects of microbiota on colonized tissue, the progression of inflammation, alteration of cellular processes, and impact on tumor-promoting genes within the tumor microenvironment. Computational frameworks for microbial detection evaluated here are broadly classified as reference-based or reference-free, or mixed methods and mainly utilize computational subtraction that has been used or have the potential for such microbial diversity evaluations. These methodologies could help shed light on the role of the microbiota in cancer pathogenesis. Further, the output from these workflows combined with phylogenetic and protein-functional predictions from bioinformatics pipelines such as PICRUSt, Tax4Fun, and ShortBRED, among others, provide important clues in the understanding of microbial differences and commonalities and the potential impact on differential outcomes, therapeutic response, and population inter-variability. Recent works [84–86] demonstrate the utility of tissue-associated microbial detection derived from existing human sequencing data and the computational tools to characterize them. Differences may highlight effectors that impact the treatment decision making process and potential for targeted therapies. Their use should be promoted as first approach to the identification or confirmation of known, suspected, and novel pathogen associations in cancer.
Acknowledgements
Not applicable.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 21 Supplement 9, 2020: Selected Articles from the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP 2019). The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-21-supplement-9.
Abbreviations
- CaPSID
Computational Pathogen Sequence Identification workflow
- cDNA
Complementary DNA
- COG
Cluster of Orthologous Group
- ConStrains
Conspecific strain workflow
- GRAMMy
Genome Relative Abundance Estimation framework
- HBV
Human hepatitis virus B
- HBx
Hepatitis viral X protein
- HCV
Human hepatitis virus C
- HCV core
Hepatitis viral core protein
- HHV-4
Human herpes virus 4
- HIV-1
Human immunodeficiency virus 1
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- MetaShot
Metagenomics Taxon Classification workflow
- NGS
Next generation sequencing
- NS5a
Non-structural protein 5A
- PathoScope
Pathogen Identification and Quantitation modular workflow
- PathSeq
Pathogen sequence workflow
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- RINS
Rapid Identification of Non-human Sequence workflow
- ShortBRED
Short Better Read Dataset
- SNP
Single nucleotide polymorphism
- SRSA
Short-RNA subtraction and assembly workflow
- SURPI
Sequence Based Ultra Rapid Pathogen Identification workflow
- Tax4Fun
Taxonomy functional community profiling
- TCGA
The Cancer Genome Atlas
- VirusScan
Viral sequence scanner workflow
- WGS
Whole genome sequencing
Authors’ contributions
RMR and VSK contributed to the conceptualization and wrote the original draft. MM, BYH and YD contributed to the conceptualization, review and editing of manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Institute on Minority Health and Health Disparities and the Health and Wellness Achieved by Impacting Inequalities [Grant Number 2U54MD007601-32].
The Population Sciences in the Pacific Epidemiology Program at the University of Hawaii Cancer supported RMR. The Bioinformatics Core is supported in part by the National Institute of Health [Grant Numbers P20GM103466, U54MD007584, 5R01CA223490 and 5R01CA230514)].
The contents of this work are solely the responsibility of the authors and do not necessarily represent the views of the funding bodies. Funding bodies played no part in the design, collection, analyses, interpretation of data, or writing of this manuscript. Publication cost was funded by INBRE.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rebecca M. Rodriguez and Vedbar S. Khadka contributed equally and should be considered co-first authors
Contributor Information
Rebecca M. Rodriguez, Email: rmr4@hawaii.edu
Vedbar S. Khadka, Email: vedbar@hawaii.edu
Mark Menor, Email: mmenor@hawaii.edu.
Brenda Y. Hernandez, Email: brenda@cc.hawaii.edu
Youping Deng, Email: dengy@hawaii.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 5.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609–616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 6.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15:465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 11.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser MJ. Understanding microbe-induced cancers. Cancer Prev Res. 2008;1:15–20. doi: 10.1158/1940-6207.CAPR-08-0024. [DOI] [PubMed] [Google Scholar]
- 13.Nauts HC. Bacteria and cancer–antagonisms and benefits. Cancer Surv. 1989;8:713–723. [PubMed] [Google Scholar]
- 14.Nauts HC. Bacterial products in the treatment of cancer: past, present and future. London and New York: Academic Press; 1982. [Google Scholar]
- 15.Burnett-Hartman AN, Newcomb PA, Potter JD. Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev. 2008;17:2970–2979. doi: 10.1158/1055-9965.EPI-08-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hattori N, Ushijima T. Epigenetic impact of infection on carcinogenesis: mechanisms and applications. Genome Med. 2016 doi: 10.1186/s13073-016-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Flora S, Bonanni P. The prevention of infection-associated cancers. Carcinogenesis. 2011;32:787–795. doi: 10.1093/carcin/bgr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248:171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 19.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Biological agents. Volume 100 B. A review of human carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100 Pt B:1–441. [PMC free article] [PubMed]
- 20.Chang AH, Parsonnet J. Role of bacteria in oncogenesis. Clin Microbiol Rev. 2010;23:837–857. doi: 10.1128/CMR.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci USA. 2013;110:9862–9867. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 24.Beuth J. Microorganisms and Cancer. In: From Friends to Foes; Old Herborn University. Germany: Herborn Literature; 2005.
- 25.Parsonnet J. Bacterial infection as a cause of cancer. Environ Health Perspect. 1995;103(Suppl 8):263–268. doi: 10.1289/ehp.95103s8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bordonaro M, Lazarova DL, Sartorelli AC. Butyrate and Wnt signaling: a possible solution to the puzzle of dietary fiber and colon cancer risk? Cell Cycle. 2008;7:1178–1183. doi: 10.4161/cc.7.9.5818. [DOI] [PubMed] [Google Scholar]
- 27.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/AEM.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goyal S, Nangia-Makker P, Farhana L, Yu Y, Majumdar AP. Racial disparity in colorectal cancer: Gut microbiome and cancer stem cells. World J Stem Cells. 2016;8:279–287. doi: 10.4252/wjsc.v8.i9.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas AM, Jesus EC, Lopes A, Aguiar S, Begnami MD, Rocha RM, et al. Tissue-associated bacterial alterations in rectal carcinoma patients revealed by 16S rRNA community profiling. Front Cell Infect Microbiol. 2016 doi: 10.3389/fcimb.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, et al. Towards the human colorectal cancer microbiome. PLoS ONE. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Thotakura PL, Tiwary BK, Krishna R. Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions. BMC Microbiol. 2016;16:84. doi: 10.1186/s12866-016-0700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riley DR, Sieber KB, Robinson KM, White JR, Ganesan A, Nourbakhsh S, et al. Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput Biol. 2013;9:e1003107. doi: 10.1371/journal.pcbi.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan AA, Bashir M, Rivas MN, Duvall K, Sieling PA, Pieber TR, et al. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS ONE. 2017;12:e0188873. doi: 10.1371/journal.pone.0188873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yow MA, Tabrizi SN, Severi G, Bolton DM, Pedersen J, Giles GG, et al. Characterisation of microbial communities within aggressive prostate cancer tissues. Infect Agent Cancer. 2017 doi: 10.1186/s13027-016-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3:130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xuan C, Shamonki JM, Chung A, Dinome ML, Chung M, Sieling PA, et al. Microbial dysbiosis is associated with human breast cancer. PLoS ONE. 2014;9:e83744. doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, Wei Z, Tan F, Peck KN, Shih N, Feldman M, et al. Distinct microbiological signatures associated with triple negative breast cancer. Sci Rep. 2015;5:15162. doi: 10.1038/srep15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golombos DM, Ayangbesan A, O’Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: a prospective. Pilot Study Urol. 2018;111:122–128. doi: 10.1016/j.urology.2017.08.039. [DOI] [PubMed] [Google Scholar]
- 42.Cavarretta I, Ferrarese R, Cazzaniga W, Saita D, Lucianò R, Ceresola ER, et al. The microbiome of the prostate tumor microenvironment. Eur Urol. 2017;72:625–631. doi: 10.1016/j.eururo.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 43.Grąt M, Wronka KM, Krasnodębski M, Masior Ł, Lewandowski Z, Kosińska I, et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant Proc. 2016;48:1687–1691. doi: 10.1016/j.transproceed.2016.01.077. [DOI] [PubMed] [Google Scholar]
- 44.Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi J, et al. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol. 2013 doi: 10.3389/fcimb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Tong H, Brindley PJ, Meyer CG, Velavan TP. Parasite infection carcinogenesis and human malignancy. EBioMedicine. 2016;15:12–23. doi: 10.1016/j.ebiom.2016.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huo Q, Zhang N, Yang Q. Epstein-Barr virus infection and sporadic breast cancer risk: a meta-analysis. PLoS ONE. 2012;7:e31656. doi: 10.1371/journal.pone.0031656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilar R, Casabonne D, O’Callaghan-Gordo C, Vidal M, Campo JJ, Mutalima N, et al. Assessment of the combined effect of Epstein–Barr Virus and Plasmodium falciparum infections on endemic Burkitt lymphoma using a multiplex serological approach. Front Immunol. 2017 doi: 10.3389/fimmu.2017.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borozan I, Zapatka M, Frappier L, Ferretti V. Analysis of Epstein–Barr Virus genomes and expression profiles in gastric adenocarcinoma. Journal of Virology. 2018 doi: 10.1128/JVI.01239-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hernandez BY, Zhu X, Goodman MT, Gatewood R, Mendiola P, Quinata K, et al. Betel nut chewing, oral premalignant lesions, and the oral microbiome. PLoS ONE. 2017;12:e0172196. doi: 10.1371/journal.pone.0172196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie G, Wang X, Liu P, Wei R, Chen W, Rajani C, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget. 2016;7:19355–19366. doi: 10.18632/oncotarget.8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daly GM, Leggett RM, Rowe W, Stubbs S, Wilkinson M, Ramirez-Gonzalez RH, et al. Host subtraction, filtering and assembly validations for novel viral discovery using next generation sequencing data. PLoS ONE. 2015;10:e0129059. doi: 10.1371/journal.pone.0129059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Contreras AV, Cocom-Chan B, Hernandez-Montes G, Portillo-Bobadilla T, Resendis-Antonio O. Host-microbiome interaction and cancer: potential application in precision medicine. Front Physiol. 2016 doi: 10.3389/fphys.2016.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5:4724. doi: 10.1038/ncomms5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz ELS, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cristescu R, Lee J, Nebozhyn M, Kim K-M, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Funchain P, Bebek G, Altemus J, Zhang H, Niazi F, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. 2017 doi: 10.1186/s13073-017-0405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leipzig J. A review of bioinformatic pipeline frameworks. Brief Bioinform. 2017;18:530–536. doi: 10.1093/bib/bbw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isakov O, Modai S, Shomron N. Pathogen detection using short-RNA deep sequencing subtraction and assembly. Bioinformatics. 2011;27:2027–2030. doi: 10.1093/bioinformatics/btr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Identification of foreign gene sequences by transcript filtering against the human genome. Nat Genet. 2002;30:141–142. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Stange-Thomann N, Weber G, Bo R, Dodge S, David RG, et al. Pathogen discovery from human tissue by sequence-based computational subtraction. Genomics. 2003;81:329–335. doi: 10.1016/S0888-7543(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 63.Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RGW, Getz G, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nooij S, Schmitz D, Vennema H, Kroneman A, Koopmans MPG. Overview of virus metagenomic classification methods and their biological applications. Front Microbiol. 2018;9:749. doi: 10.3389/fmicb.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tae H, Karunasena E, Bavarva JH, McIver LJ, Garner HR. Large scale comparison of non-human sequences in human sequencing data. Genomics. 2014 doi: 10.1016/j.ygeno.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhaduri A, Qu K, Lee CS, Ungewickell A, Khavari PA. Rapid identification of non-human sequences in high-throughput sequencing datasets. Bioinformatics. 2012;28:1174–1175. doi: 10.1093/bioinformatics/bts100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borozan I, Wilson S, Blanchette P, Laflamme P, Watt SN, Krzyzanowski PM, et al. CaPSID: a bioinformatics platform for computational pathogen sequence identification in human genomes and transcriptomes. BMC Bioinform. 2012;13:206. doi: 10.1186/1471-2105-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C, Cleveland K, Schnoll-Sussman F, McClure B, Bigg M, Thakkar P, et al. Identification of low abundance microbiome in clinical samples using whole genome sequencing. Genome Biol. 2015 doi: 10.1186/s13059-015-0821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cao S, Wendl MC, Wyczalkowski MA, Wylie K, Ye K, Jayasinghe R, et al. Divergent viral presentation among human tumors and adjacent normal tissues. Sci Rep. 2016;6:28294. doi: 10.1038/srep28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fosso B, Santamaria M, D’Antonio M, Lovero D, Corrado G, Vizza E, et al. MetaShot: an accurate workflow for taxon classification of host-associated microbiome from shotgun metagenomic data. Bioinformatics. 2017;33:1730–1732. doi: 10.1093/bioinformatics/btx036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naeem R, Rashid M, Pain A. READSCAN: a fast and scalable pathogen discovery program with accurate genome relative abundance estimation. Bioinformatics. 2013;29:391–392. doi: 10.1093/bioinformatics/bts684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia LC, Cram JA, Chen T, Fuhrman JA, Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS ONE. 2011;6:e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018;16:e2006842. doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31:2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol. 2015;11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Relman DA. Detection and identification of previously unrecognized microbial pathogens. Emerg Infect Dis. 1998;4:382–389. doi: 10.3201/eid0403.980310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodriguez RM, Hernandez BY, Menor M, Deng Y, Khadka VS. The landscape of bacterial presence in tumor and adjacent normal tissue across 9 major cancer types using TCGA exome sequencing. Comput Struct Biotechnol J. 2020;18:631–641. doi: 10.1016/j.csbj.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Livyatan I, Nejman D, Shental N, Straussman R. Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. OncoImmunology. 2020;9:1800957. doi: 10.1080/2162402X.2020.1800957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.