Abstract
Objectives
The aim of this study was to develop a single blood test that could determine gestational age and estimate the risk of preterm birth by measuring serum metabolites. We hypothesised that serial metabolic modelling of serum analytes throughout pregnancy could be used to describe fetal gestational age and project preterm birth with a high degree of precision.
Study design
A retrospective cohort study.
Setting
Two medical centres from the USA.
Participants
Thirty-six patients (20 full-term, 16 preterm) enrolled at Stanford University were used to develop gestational age and preterm birth risk algorithms, 22 patients (9 full-term, 13 preterm) enrolled at the University of Alabama were used to validate the algorithms.
Outcome measures
Maternal blood was collected serially throughout pregnancy. Metabolic datasets were generated using mass spectrometry.
Results
A model to determine gestational age was developed (R2=0.98) and validated (R2=0.81). 66.7% of the estimates fell within ±1 week of ultrasound results during model validation. Significant disruptions from full-term pregnancy metabolic patterns were observed in preterm pregnancies (R2=−0.68). A separate algorithm to predict preterm birth was developed using a set of 10 metabolic pathways that resulted in an area under the curve of 0.96 and 0.92, a sensitivity of 0.88 and 0.86, and a specificity of 0.96 and 0.92 during development and validation testing, respectively.
Conclusions
In this study, metabolic profiling was used to develop and test a model for determining gestational age during full-term pregnancy progression, and to determine risk of preterm birth. With additional patient validation studies, these algorithms may be used to identify at-risk pregnancies prompting alterations in clinical care, and to gain biological insights into the pathophysiology of preterm birth. Metabolic pathway-based pregnancy modelling is a novel modality for investigation and clinical application development.
Keywords: health informatics, risk management, obstetrics
Strengths and limitations of this study.
The insensitivity of the prediction model to gestational age (GA) window of sample collection increases its flexibility and opportunity for potential clinical use.
This study is among the first to propose a pathway-based computational methodology to estimate GA and predict preterm birth.
The overall cohort size is modest, and the distribution of sampling time is different between patients and cohorts.
It is a retrospective study, a larger prospective cohort study is necessary before applying the estimates and prediction to a broader population for clinical utility.
Introduction
Gestational age (GA) dating is a core element of standard prenatal care.1–4 Prenatal ultrasound (US) is an established modality for estimating GA, monitoring fetal growth and screening for fetal anomalies.5 According to the policy statement of the Committee on Obstetric Practice, the American Institute of Ultrasound in Medicine and the Society for Maternal-Fetal Medicine, a pregnancy is considered optimally dated through a combination of last menstrual period (LMP) and an accurate US obtained prior to 22 0/7 weeks.6 Accordingly, LMP is dependent on maternal recall and many pregnancies do not present for a first prenatal US evaluation until the second or third trimester. Thus, there is a need for a molecular method that would complement the potential shortcomings of LMP recall and US dating outside the first trimester. Moreover, it is possible that molecular pregnancy dating will provide greater resolution to pregnancy risk then current information based on calendar dating (LMP) and anthropometrics (US). Although experience is accumulating with the use of second and third trimester US for an estimation of risk of preterm birth (PTB),7–9 to date these measures have not been widely adopted, are subject to user experience and have reported variable performance characteristics. The availability and expertise of US in disadvantaged areas is limited.10 Therefore, there is a need to develop an alternative measure of fetal progression to estimate GA and pregnancy risk in a variety of settings and especially when US and LMP dates are unavailable or unreliable.
Compared with imaging methodologies, blood-based molecular testing may provide a more reproducible and precise modality in clinical applications for the frequent monitoring of health status and detection of early signs of disease. Genomic, gene expression, protein and metabolite profiles measured in human blood have been increasingly used for the determination of disease risk and to gain disease-specific pathophysiology insight. Attempts at estimating GA using molecular adaptations have included modelling of RNA, protein or immune cell changes, and most recently metabolites in maternal or newborn blood.11–17 Similarly, risk prediction of PTB in clinical settings is currently primarily based on maternal history. Biomarkers have been suggested from genetic and proteomic analyses, but less effort has been focused on understanding maternal metabolic signatures of pregnancy.18–24
In this study, we hypothesised that longitudinal metabolic profiling of pregnancy reflects the temporal progression of fetal development with a high degree of precision. Moreover, we posited that if a normal pregnancy progression profile could be defined in metabolic terms, then aberrations from the normal profile may identify a pregnancy at risk for PTB. Our findings suggest that composite metabolic panel modelling may serve as a reproducible and precision approach to GA dating of pregnancy and prediction of PTB.
Materials and methods
Definition
In this study, a full-term pregnancy was defined as a pregnancy ending with a delivery at ≥37 weeks. PTB was defined by delivery at <35 weeks GA in order to make a complete separation from the full-term subjects.
Study design
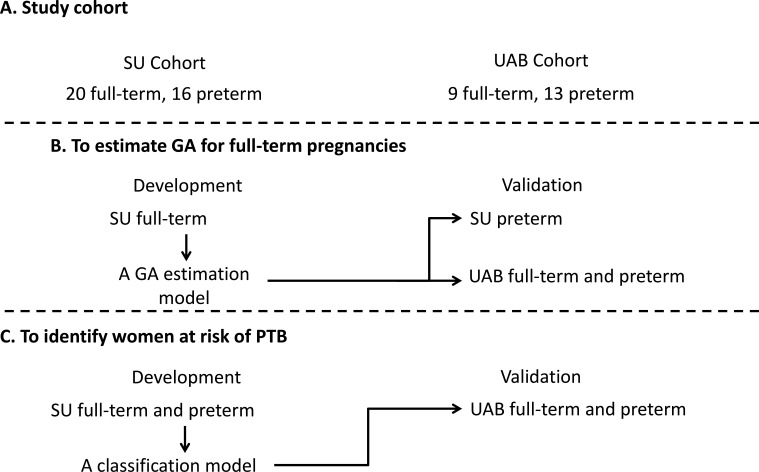
The study was conducted in two phases: (1) modelling to devise a metabolite-based estimation of GA during full-term pregnancies and (2) modelling to devise a metabolic panel predictive of PTB (figure 1). In this study, the ‘gold’ standard of GA was US measurement based on the crown-rump length at the first trimester.25 Serum samples were collected in the first, second or third trimester during pregnancy for each individual woman. Each participant had one to four time-points collected prior to delivery. Samples were provided by Stanford Hospital and Clinics (SU) and the University of Alabama (UAB). Metabolic concentrations in each sample were measured by targeted and untargeted mass spectrometry (MS) analysis. Models that estimated GA or predicted PTB were developed using the SU cohort and validated using the UAB cohort. All samples were collected after informed consent was obtained. All statistical analyses were done in R software.
Figure 1.
Study design. Models were developed separately to estimate gestational age during full-term pregnancy, and to predict the risk of preterm birth. Both models were developed with the Stanford Hospital and Clinics (SU) cohort and validated with the University of Alabama (UAB) cohort.
Targeted and global MS analysis
Samples of full-term and preterm patients as well as quality control (QC) samples were injected into the MS. Targeted MS analysis was done through flow injection methods by using Ultimate 3000 Ultra-High-Performance Liquid Chromatography (UHPLC) system and Quantiva Triple Quadrupole Mass Spectrometer. Global (ie, untargeted) MS analysis was done by using a Vanquish UHPLC system coupled to a Q Exactive plus mass spectrometer and Q Exactive HF hybrid Quadrupole-Orbitrap mass spectrometer.
Data preprocessing and metabolic identification
A data preprocessing procedure was conducted to convert the raw data generated by MS analysis into a matrix of relative concentrations of metabolites versus samples.26 This procedure was done by R package. Metabolic values in each sample were then normalised by the median values measured with QC samples to reduce the batch effects.
Compounds detected by untargeted analyses were matched to metabolites in the Human Metabolome Database by putative identification.27 Accurate mass was used for the mapping. Metabolites were mapped to pathways using Kyoto Encyclopedia of Genes and Genomes and Human Metabolome Database. Only endogenous pathways were considered.
Metabolic compound selection, pathway computation and model development
Metabolites measured by targeted and untargeted MS were aggregated and filtered. The remaining metabolites were mapped to pathways. The value of each pathway was calculated as the weighted sum of the normalised concentrations of metabolites on the pathway divided by the number of metabolites. An XGBoost model was developed with the pathway values of samples from full-term patients to estimate the GA. R-squared (R2; goodness-of-fit of the model), root-mean-square error (RMSE) and error distribution were calculated to evaluate the model performance. A second XGBoost model was developed to predict PTB. To evaluate the model performance, Mann-Whitney U tests were used to compare the distribution of final predictive estimates, that is, XGBoost model values, on full-term and PTB samples. Additional details of model development are described in online supplemental appendix text A1. ELISA tests were conducted on the SU and UAB cohorts to evaluate the insulin-like growth factor-binding protein 4 (IBP4)/sex hormone-binding globulin (SHBG) signature, a predictor that was validated in a prospective study as a predictor of spontaneous PTB.19 Serum concentrations were measured using commercial kits Human IGFBP4 ELISA Kit (Abcam, Burlingame, California, USA) and Human SHBG Quantikine ELISA Kit (R&D System). Results were compared with our metabolic model.
bmjopen-2020-040647supp001.pdf (9.2MB, pdf)
Patient and public involvement statement
This retrospective research was done without patient involvement. Patients were not invited to comment on the study design and were not consulted to develop patient relevant outcomes or interpret the results. Patients were not invited to contribute to the writing or editing of this document for readability or accuracy.
Results
Samples
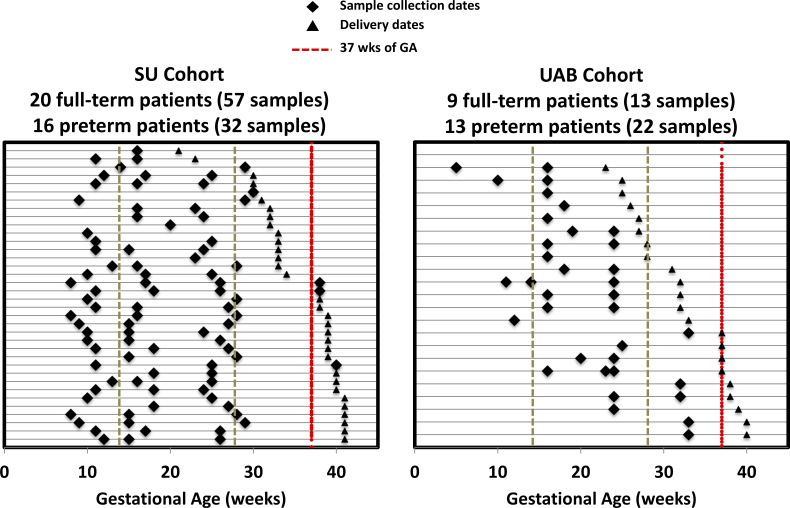
As shown in figure 2, the SU cohort had 20 full-term pregnancies with 57 blood samples (17, 32 and 8 collected in the first, second and third trimesters, respectively) and 16 preterm pregnancies with 32 blood samples (9, 19 and 4 collected in the first, second and third trimesters, respectively). The UAB cohort had 9 full-term pregnancies with 13 blood samples (8 and 5 in the second and third trimesters, respectively) and 13 preterm pregnancies with 22 blood samples (4 and 18 in the first and second trimesters, respectively). In the SU cohort, two (12.5%) were extremely preterm (<28 weeks) and five (31.3%) were very preterm (28–31 weeks). In the UAB cohort, six (46.2%) were extremely preterm, and three (23.1%) were very preterm. Our SU and UAB cohorts were assembled: no complications of pregnancy were included; all deliveries were singleton and all PTB were spontaneous. Demographics of the two cohorts are shown in table 1.
Figure 2.
Cohort construction. Each line represents an individual patient. Diamond and triangle markers indicate sample collection dates and delivery dates, respectively. The red dashed line represents 37 weeks’ gestational age.
Table 1.
Maternal characteristics in SU and UAB cohorts
| Characteristic | Full-term (n=20) | SU | P value | Full-term (n=9) | UAB | P value | SU vs UAB |
| Preterm (n=16) | Preterm (n=13) | P value | |||||
| Race, n (%) | <0.001*** | 0.5 | <0.001*** | ||||
| Asian | 0 | 1 (6.3) | 0 | 0 | |||
| White | 20 (100) | 5 (31.3) | 0 | 2 (15.4) | |||
| Black | 0 | 1 (6.3) | 9 (100) | 10 (76.9) | |||
| American Indian | 0 | 2 (12.5) | 0 | 0 | |||
| Pacific Islander | 0 | 1 (6.3) | 0 | 0 | |||
| Other/Unknown | 0 | 6 (37.5) | 0 | 1 (7.7) | |||
| Hispanic, n (%) | 0 | 8 (50) | <0.001*** | 0 | 1 (7.7) | 0.9 | 0.1 |
| Maternal age, year, mean (SD) | 31.9 (4.8) | 29.8 (7.5) | 0.3 | 25.6 (5.0) | 27.5 (4.5) | 0.4 | 0.008** |
| Gestational age at delivery, weeks, median (IQR) | 39.5 (39, 41) | 32 (30, 33) | <0.001*** | 38 (37, 39) | 28 (26, 32) | <0.001*** | 0.01* |
| Having previous pregnancy, n (%) | 9 (45) | 6 (37.5) | 0.7 | 9 (100) | 13 (100) | 0.4 | <0.001*** |
| BMI, kg/m2, median (IQR) | 22.3 (20.2, 24.7) | 27.6 (23.4, 33.9) | 0.003*** | 30.4 (22.3, 33.1) | 26.5 (22.6, 36.5) | 0.8 | 0.06 |
| History of PTB, n (%) | 3 (15) | 8 (50) | 0.03* | 7 (77.8) | 13 (100) | 0.2 | <0.001*** |
*P<0.05; **P<0.01; ***P<0.005.
BMI, body mass index; PTB, preterm birth; SU, Stanford Hospital and Clinics; UAB, University of Alabama.
LC-MS/MS metabolomics
The study targeted 315 metabolites by LC-MS/MS, including 13 categories: acyl-carnitine (11, 3.5%), amino acid (9, 2.9%), fatty acid (6, 1.9%), ceramide (12, 3.8%), ceramide 1-phosphate (8, 2.5%), galactosylceramide (5, 1.6%), phosphatidyl acid (15, 4.8%), phosphatidylethanolamine (52, 16.5%), phosphatidylglycerol (5, 1.6%), phosphatidylinositol (11, 3.5%), phophatidylcholine (130, 41.3%), cholesteryl ester (16, 5.1%) and sphingomyelin (35, 11.1%). The study also identified 1627 positively and 295 negatively charged compounds through untargeted analyses. Together these formed the initial set of 2237 compounds.
Feature selection of GA estimation modelling
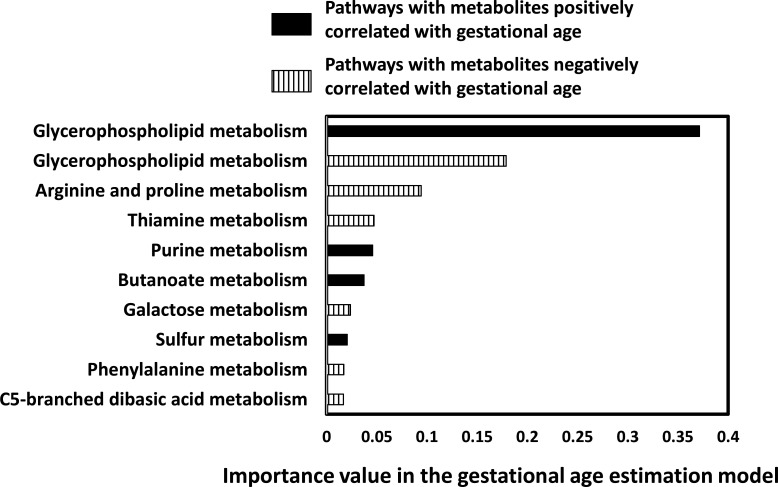
Of the 2237 compounds, 118 had an absolute Pearson’s correlation coefficient of >0.35 with GA. The cut-off of ±0.35 was selected based on the false discovery rate (FDR) values of the mapped pathways <1% (online supplemental appendix figure A1). The 118 compounds were mapped to 89 pathways, 33 of which were selected by the XGBoost model. The normalised value of each pathway varied over the course of gestation (online supplemental appendix figure A2). Univariate analysis of the 33 pathways is shown in online supplemental appendix figure A3, and the top 10 pathways in the model is depicted in figure 3. The top 10 pathways included those associated in the metabolisms of: glycerophospholipid, arginine and proline, thiamine, purine, butanoate, galactose, sulfur, phenylalanine and C5-branched dibasic acid.
Figure 3.
The importance of the top 10 metabolic pathways in the gestational age estimation model. Pathways either positively or negatively correlated gestational age.
Performance of GA estimation
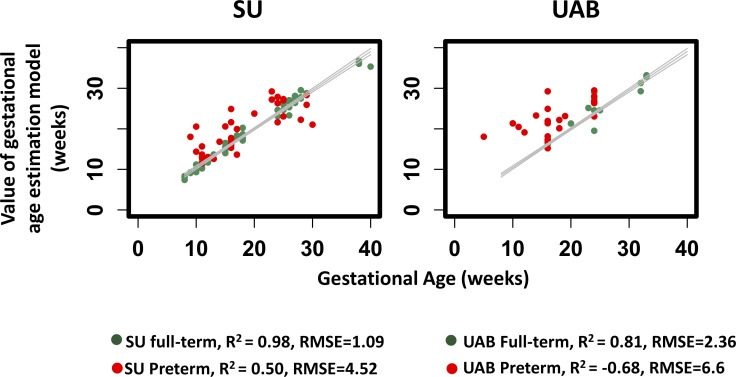
The performance of GA estimates on full-term samples was similar in the development phase (SU cohort, R2=0.98, RMSE=1.09) and the validation phase (UAB cohort, R2=0.81, RMSE=2.36) (figure 4). In our validation testing, 66.7% of the estimates were within ±1 week of the US results (online supplemental appendix figure A4).
Figure 4.
Gestational age estimates of the gestational age model with the Stanford Hospital and Clinics (SU) (R2=0.98, root-mean-square error (RMSE)=1.09 weeks) and University of Alabama (UAB) cohorts (R2=0.81, RMSE=2.36 weeks).
Intriguingly, model performance significantly deteriorated when applied to samples from PTB pregnancies (R2=−0.68 and RMSE=6.6 in validation; see figure 4). It suggested that the relationships between metabolic parameters and full-term pregnancies were not maintained in PTB pregnancies. Furthermore, such disruptions were notable as early as 10 weeks’ GA (figure 4) or early to mid-gestation. These findings prompted the development of a metabolic-based model of PTB estimation.
Performance of PTB prediction
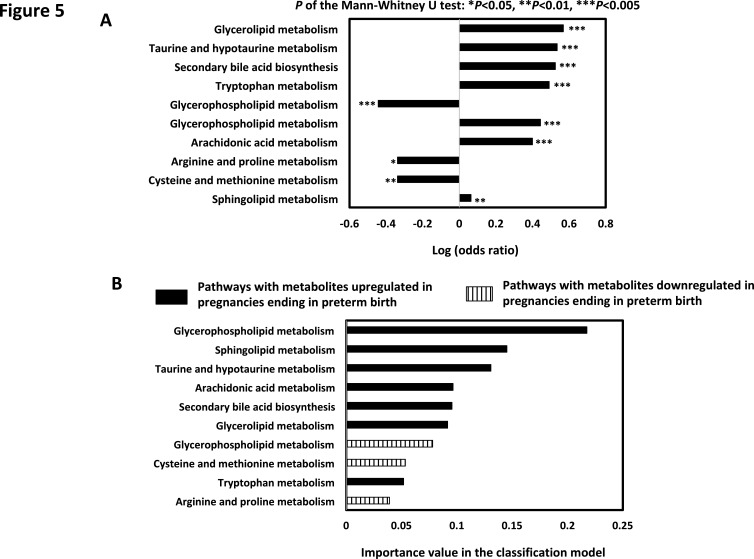
Samples collected before 35 weeks’ GA were used to develop a model that differentiated PTB pregnancies from those full-term. As before, the model was developed with the SU cohort that had 20 full-term (54 samples) and 16 preterm (32 samples) pregnancies, and was validated with the UAB cohort that had 9 full-term (13 samples) and 13 preterm (22 samples) pregnancies. In total, 148 metabolic compounds (with Mann-Whitney U test p<0.05) were mapped to 66 pathways (FDR <1.5%; see online supplemental appendix figure A5). Further model development selected 10 pathways as strong predictors covering the metabolisms of glycerophospholipid, sphingolipid, taurine and hypotaurine, arachidonic acid, secondary bile acid biosynthesis, glycerolipid, cysteine and methionine, tryptophan and arginine and proline (figure 5).
Figure 5.
(A) Univariate analysis of the 10 metabolic pathways in the preterm birth prediction model. OR of each pathway was calculated. (B) The importance of the metabolic pathways in the preterm birth prediction model. Pathways were either upregulated or downregulated in relation to preterm birth.
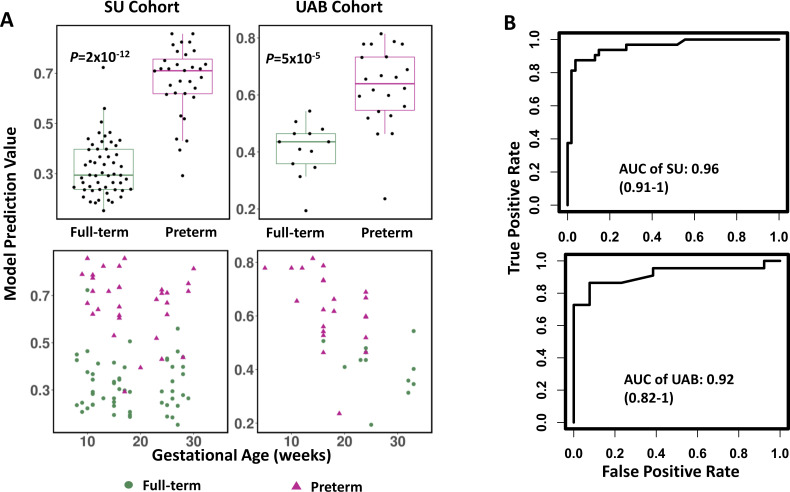
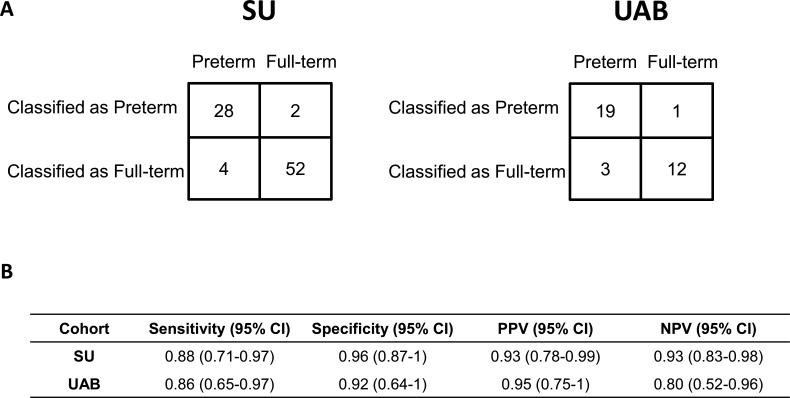
The level of prediction accuracy was maintained in the validation cohort (p=5×10−5, area under the curve (AUC)=0.92; see figure 6). The prevalence-corrected positive predictive values (PPVs) across model values (ie, scores) were plotted based on the PTB prevalence in Alabama in 2018 (12.5%; see online supplemental appendix figure A6). A threshold value of 0.52 was selected as a high-risk threshold for PTB, which was associated with a PPV of 0.70, a relative risk (RR) of 5.6 compared with the US population baseline (=0.70/12.5%), a sensitivity of 0.86 (19 of 22) and a specificity of 0.92 (12 of 13; figure 7). The sensitivities and specificities with cut-off values are shown in online supplemental table A1.
Figure 6.
(A) Prediction of preterm birth risk grouped by full-term and preterm birth patients (top) and over the course of gestation (bottom). (B) Area under the curve (AUC) performance of the prediction in Stanford Hospital and Clinics (SU) and University of Alabama (UAB) cohorts. P value was calculated using Mann-Whitney U test.
Figure 7.
Performance of the preterm birth prediction model. (A) A contingency table showing the number of samples in each category. (B) Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) together with the 95% CIs. SU, Stanford Hospital and Clinics; UAB, University of Alabama.
In the validation cohort, 12 of 13 full-term samples and 19 of 22 preterm samples were classified correctly. The misclassified full-term sample was from a mother that delivered at 37 weeks’ GA. The 19 correctly classified PTB samples were from 13 PTB pregnancies. Of the 13 pregnancies, 9 were identified as high risk at or earlier than 16 weeks’ GA. The median gap between the time of identification and the delivery was 11 weeks’ GA (IQR: 8, 15.5).
To determine the performance of our metabolic model against existing models, a comparison between the metabolic PTB risk model and the commercially available IBP4/SHBG PTB test was performed and is summarised in online supplemental appendix text A2 and online supplemental appendix figure A7.
Metabolite-based model and pathway-based model: a comparison
To determine the effectiveness of model performance based on robustness of biological features, we compared model performance using pathway or individual metabolite as selected features in estimating GA and predicting PTB. The performance of the pathway-based models were significantly better than the metabolite-based models, with a lower RMSE (Student’s t-test p=4×10−3; online supplemental appendix figure A8) and a larger AUC (DeLong test p=0.03; online supplemental appendix figure A9).
Discussion
Principal findings
In this study, we report a panel of metabolic pathways measured in maternal serum that provides an estimation of GA over the course of a full-term pregnancy. A second and distinct set of metabolic pathways was also identified in maternal serum that could distinguish pregnancies ending with PTB (<35 weeks) from full-term (≥37 weeks) with a high degree of precision. The models were developed and validated using two independent cohorts from two different institutions in order to test the robustness of the biological features driving the classifications. Intriguingly, PTB pregnancies do not demonstrate the same temporal relationship as term pregnancies on metabolic modelling across gestation (figure 4). Indeed, PTB pregnancies demonstrate a marked departure from the term metabolic profile (figure 4) that is dramatic (R2=0.98 train and 0.81 test for term model; compared with R2=0.50 train and −0.68 test for PTB pregnancy in term model), and is recognisable as early as 10 weeks’ GA as determined by the current standard of US dating. Recognising the metabolic pathway aberration of PTB pregnancies, a second model was developed using metabolic pathway analyses to quantify the risk of PTB prior to 35 weeks’ GA. Once again, metabolic profiling proved to be robust in identifying PTB pregnancies with a high degree of sensitivity (AUC 0.96 training; AUC 0.92 testing) and precision (training PPV 0.93 (95% CI 0.78 to 0.99); testing PPV 0.95 (95% CI 0.75 to 1). Taken together, this study demonstrated a powerful new, reproducible methodology for monitoring pregnancy progression and identifying abnormal pregnancies.
Clinical and research implications
The potential clinical utility of developing a test for pregnancy monitoring is appealing. There is a need to develop a more robust method than LMP and an alternative to first trimester US that captures pregnancy progression, a complex relationship of fetal and placental growth, development and function. To support these processes, there is a need for energy transfer between mother and fetus throughout gestation. We therefore reasoned that metabolic phenotyping would be ideally suited to capture this relationship. Despite a modest cohort size, the results of metabolic modelling demonstrate a high degree of concordance with clinical standard US dating performed by experts as reflected by 66.7% of model estimates falling within ±1 week of US results (online supplemental appendix figure A4). Moreover, unlike the deterioration experienced with US dating of pregnancy, metabolic modelling was shown to achieve near equivalent performance in the first, second and third trimesters, indicating the potential for broad clinical applicability that might achieve independence of reliance on accuracy of LMP or concordance among modality testing. The result of PTB prediction is equally robust demonstrating a high degree of precision. Beyond relying on clinical histories or self-reported symptoms, the model proposed here provides a molecular classification that may be more accurate than current methods and further reflect a comprehensive measure of aberrant pregnancy based on metabolic changes. In practice, clinicians could use the PTB prediction model to differentiate high-risk from low-risk patients. Low-risk patients would then be subject to GA estimation panel testing, all from the same blood draw.
A distinct advantage of the PTB risk prediction developed in this study is that it has a wide window of sampling. Samples were collected broadly before 35 weeks’ GA, which is wider than the window of other well-established biomarkers such as fetal fibronectin (between 24 and 34 weeks’ GA),20 IBP4/SHBG (19–21 weeks)19 and inter-alpha-trypsin inhibitor heavy chain 4 protein (24 and 28 weeks).18 Relatively stable AUC levels were maintained throughout the diagnostic window (online supplemental taxt A2). The insensitivity of the prediction model to GA at testing increases its flexibility and opportunity for potential clinical use. An additional advantage of the model herein is the ability for early identification of high-risk women. Although there is no standardised guideline for early gestation management of patients at risk of PTB delivery, metabolic modelling for PTB risk may provide a not previously possible opportunity for early gestation risk mitigation. Clinical trials have suggested that hormone treatment and maternal physical activity modifications applied between 16 and 37 weeks’ GA reduced the PTB rate of women who were deemed at high risk due to a history of prior PTB delivery.28 29 In many cases, PTB cannot be prevented, however any opportunity is deemed highly desirable for even a modest delay (1–2 weeks) in PTB or an enhanced ability to more accurately triage for delivery to centres with the capability to manage profoundly premature neonates.30–32
This study is among the first to propose a pathway-based computational methodology to estimate GA and predict PTB. Metabolic pathways are linked to chemical functions, and the alteration or disruption of specific functions participate in disease phenotypes, facilitating the use of pathways to function as higher-level biomarkers of diseases.33 The role of metabolic pathways in disease diagnosis has been explored in several preliminary clinical studies.34 35 Pathway performance in differentiating patients with disease from healthy controls has been found to be effective compared with using individual metabolites.35 Similarly, we found the pathway-based models had less variability and higher sensitivity than metabolite-based models that were developed using the same population. One plausible explanation for this observation may be attributed to the calculation of pathway values, which represents the sum of individual metabolites and thus may amplify association to outcome relationships. This hypothesis is supported by the FDR comparison (online supplemental appendix figure A8 and A9): pathway-based analysis had lower FDR values than metabolite models. This study adds to the exploration of the feasibility of using pathways for health monitoring and prediction.
In this study, glycerophospholipid metabolism was identified as the most significant contributing pathway for both GA estimation and preterm birth prediction. Glycerophospholipids consist of fatty acid chains and have been previously cited as strong correlates to birth weight, pregnancy duration and risk of preterm birth.36 These same authors also found different polyunsaturated fatty acid components of glycerophospholipid had differential effects on fetal growth. Gao et al has reported a potential association between glycerophospholipid and labour timing in rodent models.37 38 The current study extends those prior observations through a quantitative assessment of the relationship between glycerophospholipid metabolism, GA and the risk of preterm birth. The leading effect of glycerophospholipid pathway metabolism in the current study was positive in both the assessment of GA and risk of preterm birth. These findings add further insight into the role of glycerophospholipid metabolism in human pregnancy. Other contributing pathways for preterm birth prediction such as sphingolipid metabolism, arachidonic acid metabolism and arginine and proline metabolism were also found associated to preterm. Alterations in plasma sphingolipids were found in women who had spontaneous PTB.39 Increase of arachidonic acid metabolism might correlate to bacteria activities that led to preterm labour.40 Plasma level of arginine and citrulline was significantly lowered in preterm babies.41
Taken together, the analysis of the leading pathways found to significantly contribute to the metabolic pregnancy modelling herein provide ample insights to deepen our understanding of pregnancy progression and may facilitate the identification and interpretation of potential therapeutic targets. Furthermore, we speculate that the platform and approaches outlined herein may be extended to the interrogation of additional conditions of pregnancy including abnormalities of placentation, gestational diabetes and fetal growth disturbances among others.
Limitations
This study has several limitations. First, the overall cohort size was modest, and pregnancies with delivery at 35 or 36 weeks were not included in the study. Second, blood samples were collected in a non-uniform manner with respect to GA timing and time of day. The time between two adjacent samples corresponding to the same patient varied. Third, the distribution of samples throughout pregnancy were different between patients and cohorts. In the SU cohort, none of the full-term patients had samples collected between 30 and 37 weeks. In the UAB cohort, none of the full-term patients had sampling in the first trimester, and none of the PTB patients had sampling in the third trimester. Fourth, for methodologic reasons, not all serum analytes could be identified and mapped to known metabolites. Fifth, baseline characteristics of patients were not included in the analysis. Sixth, the study was retrospective, and the participants were solely from California and Alabama. A larger prospective cohort study with a reasonable ratio of full-term to preterm is necessary before applying the estimates and prediction to a broader population for clinical utility.
Conclusion
The present study demonstrates that maternal serum-based metabolic profiling is a highly sensitive and accurate method for determining GA and prediction of PTB. The pathway-based analysis supports the hypothesis of the orderly metabolic progression of pregnancy that can be reproducibly captured using metabolic profiling. The robustness of the modelling reinforces the potential appeal for further clinical development and as a platform to investigate the pathophysiology associated with aberrant fetal development and pregnancy progression. This study is the first to report a single blood test for metabolic pathway-based determination of GA dating, and early detection of PTB risk.
Supplementary Material
Acknowledgments
The authors would like to thank colleagues at the Stanford University Pediatric Proteomics group and the March of Dimes Prematurity Research Center at Stanford University for critical discussions.
Footnotes
Contributors: XL, KGS and HJC contributed to concept development and design. JY, RJW and DKS contributed to the acquisition of data. KGS, SH, LZ, XY, LT, LM, SL, RJW, GMS, DKS, JCW and DBM contributed to the analysis and interpretation of data. KGS and SH drafted the manuscript. JY, LZ, LT, XY, LM, SL, RJW, GMS, DKS, HJC, JCW, DBM and XL critically revised the manuscript. All the authors gave final approval of the version to be submitted and agreed to be accountable for all aspects of the work.
Funding: This work was supported in part by the March of the Dimes Prematurity Research Center at Stanford University, and Stanford Child Health Research Institute. Grant numbers are N/A.
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The study was approved by the Institutional Review Board of both sites (Protocol #21956).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The datasets used and/or analysed in this study are available on reasonable request to the corresponding author. Once published, data will also be uploaded at the laboratory website (http://translationalmedicine.stanford.edu) and shared with March of Dimes Database.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Brownfoot FC, Gagliardi DI, Bain E, et al. Different corticosteroids and regimens for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst Rev 2013;8:CD006764. 10.1002/14651858.CD006764.pub3 [DOI] [PubMed] [Google Scholar]
- 2.Raju TNK, Mercer BM, Burchfield DJ, et al. Periviable birth: executive summary of a joint workshop by the Eunice Kennedy Shriver National Institute of child health and human development, Society for Maternal-Fetal medicine, American Academy of pediatrics, and American College of obstetricians and Gynecologists. J Perinatol 2014;34:333–42. 10.1038/jp.2014.70 [DOI] [PubMed] [Google Scholar]
- 3.Vohr B. Long-term outcomes of moderately preterm, late preterm, and early term infants. Clin Perinatol 2013;40:739–51. 10.1016/j.clp.2013.07.006 [DOI] [PubMed] [Google Scholar]
- 4.Pereira APE, Dias MAB, Bastos MH, et al. Determining gestational age for public health care users in Brazil: comparison of methods and algorithm creation. BMC Res Notes 2013;6:60. 10.1186/1756-0500-6-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peek MJ, Devonald KJ, Beilby R, et al. The value of routine early pregnancy ultrasound in the antenatal Booking clinic. Aust N Z J Obstet Gynaecol 1994;34:140–3. 10.1111/j.1479-828X.1994.tb02676.x [DOI] [PubMed] [Google Scholar]
- 6.Committee on Obstetric Practice, the American Institute of Ultrasound in Medicine, and the Society for Maternal-Fetal Medicine Committee opinion no 700: methods for estimating the due date. Obstet Gynecol 2017;129:e150–4. 10.1097/AOG.0000000000002046 [DOI] [PubMed] [Google Scholar]
- 7.Roman A, Saccone G, Dude CM, et al. Midtrimester transvaginal ultrasound cervical length screening for spontaneous preterm birth in diamniotic twin pregnancies according to chorionicity. Eur J Obstet Gynecol Reprod Biol 2018;229:57–63. 10.1016/j.ejogrb.2018.08.006 [DOI] [PubMed] [Google Scholar]
- 8.Erkamp JS, Voerman E, Steegers EAP, et al. Second and third trimester fetal ultrasound population screening for risks of preterm birth and small-size and large-size for gestational age at birth: a population-based prospective cohort study. BMC Med 2020;18:63. 10.1186/s12916-020-01540-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziadosz M, Bennett T-A, Dolin C, et al. Uterocervical angle: a novel ultrasound screening tool to predict spontaneous preterm birth. Am J Obstet Gynecol 2016;215:376.e1–7. 10.1016/j.ajog.2016.03.033 [DOI] [PubMed] [Google Scholar]
- 10.Jehan I, Zaidi S, Rizvi S, et al. Dating gestational age by last menstrual period, symphysis-fundal height, and ultrasound in urban Pakistan. Int J Gynaecol Obstet 2010;110:231–4. 10.1016/j.ijgo.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson K, Hawken S, Potter BK, et al. Accurate prediction of gestational age using newborn screening analyte data. Am J Obstet Gynecol 2016;214:513.e1–e9. 10.1016/j.ajog.2015.10.017 [DOI] [PubMed] [Google Scholar]
- 12.Knight AK, Craig JM, Theda C, et al. An epigenetic clock for gestational age at birth based on blood methylation data. Genome Biol 2016;17:206. 10.1186/s13059-016-1068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jelliffe-Pawlowski LL, Norton ME, Baer RJ, et al. Gestational dating by metabolic profile at birth: a California cohort study. Am J Obstet Gynecol 2016;214:511 e1–3. 10.1016/j.ajog.2015.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aghaeepour N, Ganio EA, Mcilwain D, et al. An immune clock of human pregnancy. Sci Immunol 2017;2:eaan2946 10.1126/sciimmunol.aan2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang L, Rasmussen M-LH, Piening B, et al. Metabolic dynamics and prediction of gestational age and time to delivery in pregnant women. Cell 2020;181:1680–92. 10.1016/j.cell.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngo TTM, Moufarrej MN, Rasmussen M-LH, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 2018;360:1133–6. 10.1126/science.aar3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aghaeepour N, Lehallier B, Baca Q, et al. A proteomic clock of human pregnancy. Am J Obstet Gynecol 2018;218:347.e1–14. 10.1016/j.ajog.2017.12.208 [DOI] [PubMed] [Google Scholar]
- 18.Esplin MS, Merrell K, Goldenberg R, et al. Proteomic identification of serum peptides predicting subsequent spontaneous preterm birth. Am J Obstet Gynecol 2011;204:391.e1–8. 10.1016/j.ajog.2010.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saade GR, Boggess KA, Sullivan SA, et al. Development and validation of a spontaneous preterm delivery predictor in asymptomatic women. Am J Obstet Gynecol 2016;214:633.e1–24. 10.1016/j.ajog.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 20.Peaceman AM, Andrews WW, Thorp JM, et al. Fetal fibronectin as a predictor of preterm birth in patients with symptoms: a multicenter trial. Am J Obstet Gynecol 1997;177:13–18. 10.1016/S0002-9378(97)70431-9 [DOI] [PubMed] [Google Scholar]
- 21.Strauss JF. 3Rd, Romero R, Gomez-Lopez N, et al. spontaneous preterm birth: advances toward the discovery of genetic predisposition. Am J Obstet Gynecol 2018;218:294–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virgiliou C, Gika HG, Witting M, et al. Amniotic fluid and maternal serum metabolic signatures in the second trimester associated with preterm delivery. J Proteome Res 2017;16:898–910. 10.1021/acs.jproteome.6b00845 [DOI] [PubMed] [Google Scholar]
- 23.Li J, Lu YP, Reichetzeder C, et al. Maternal PCaaC38:6 is Associated With Preterm Birth - a Risk Factor for Early and Late Adverse Outcome of the Offspring. Kidney Blood Press Res 2016;41:250–7. 10.1159/000443428 [DOI] [PubMed] [Google Scholar]
- 24.Hawdon JM. Ward Platt Mp Fau - Aynsley-Green A, Aynsley-Green A. Patterns of metabolic adaptation for preterm and term infants in the first neonatal week. (1468-2044 (Electronic)). [DOI] [PMC free article] [PubMed]
- 25.Robinson HP. Sonar measurement of fetal crown-rump length as means of assessing maturity in first trimester of pregnancy. (0007-1447 (print)). [DOI] [PMC free article] [PubMed]
- 26.Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011;6:1060–83. 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- 27.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis chemical analysis Working Group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2007;3:211–21. 10.1007/s11306-007-0082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meis PJ, Klebanoff M, Thom E, et al. Prevention of recurrent preterm delivery by 17 alpha-hydroxyprogesterone caproate. N Engl J Med 2003;348:2379–85. 10.1056/NEJMoa035140 [DOI] [PubMed] [Google Scholar]
- 29.Evenson KR, Siega-Riz AM, Savitz DA, et al. Vigorous leisure activity and pregnancy outcome. Epidemiology 2002;13:653–9. 10.1097/00001648-200211000-00009 [DOI] [PubMed] [Google Scholar]
- 30.McIntire DD, Leveno KJ. Neonatal mortality and morbidity rates in late preterm births compared with births at term. Obstet Gynecol 2008;111:35–41. 10.1097/01.AOG.0000297311.33046.73 [DOI] [PubMed] [Google Scholar]
- 31.Henderson-Smart DJ. The effect of gestational age on the incidence and duration of recurrent apnoea in newborn babies. Aust Paediatr J 1981;17:273–6. 10.1111/j.1440-1754.1981.tb01957.x [DOI] [PubMed] [Google Scholar]
- 32.Khashu M, Narayanan M, Bhargava S, et al. Perinatal outcomes associated with preterm birth at 33 to 36 weeks' gestation: a population-based cohort study. Pediatrics 2009;123:109–13. 10.1542/peds.2007-3743 [DOI] [PubMed] [Google Scholar]
- 33.Lee D-S, Park J, Kay KA, et al. The implications of human metabolic network topology for disease comorbidity. Proc Natl Acad Sci U S A 2008;105:9880–5. 10.1073/pnas.0802208105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumgartner C, Böhm C, Baumgartner D, et al. Supervised machine learning techniques for the classification of metabolic disorders in newborns. Bioinformatics 2004;20:2985–96. 10.1093/bioinformatics/bth343 [DOI] [PubMed] [Google Scholar]
- 35.Huang S, Chong N, Lewis NE, et al. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med 2016;8:34. 10.1186/s13073-016-0289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grootendorst-van Mil NH, Tiemeier H, Steenweg-de Graaff J, et al. Maternal plasma n-3 and n-6 polyunsaturated fatty acids during pregnancy and features of fetal health: fetal growth velocity, birth weight and duration of pregnancy. Clin Nutr 2018;37:1367–74. 10.1016/j.clnu.2017.06.010 [DOI] [PubMed] [Google Scholar]
- 37.Menon R, Bonney EA, Condon J, et al. Novel concepts on pregnancy clocks and alarms: redundancy and synergy in human parturition. Hum Reprod Update 2016;22:535–60. 10.1093/humupd/dmw022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, Rabbitt EH, Condon JC, et al. Steroid receptor coactivators 1 and 2 mediate fetal-to-maternal signaling that initiates parturition. J Clin Invest 2015;125:2808–24. 10.1172/JCI78544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morillon A-C, Yakkundi S, Thomas G, et al. Association between phospholipid metabolism in plasma and spontaneous preterm birth: a discovery lipidomic analysis in the cork pregnancy cohort. Metabolomics 2020;16:19. 10.1007/s11306-020-1639-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett PR, Rose MP, Myatt L, et al. Preterm labor: stimulation of arachidonic acid metabolism in human amnion cells by bacterial products. Am J Obstet Gynecol 1987;156:649–55. 10.1016/0002-9378(87)90070-6 [DOI] [PubMed] [Google Scholar]
- 41.Contreras MT, Gallardo MJ, Betancourt LR, et al. Correlation between plasma levels of arginine and citrulline in preterm and full-term neonates: therapeutical implications. J Clin Lab Anal 2017;31:e22134. 10.1002/jcla.22134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-040647supp001.pdf (9.2MB, pdf)