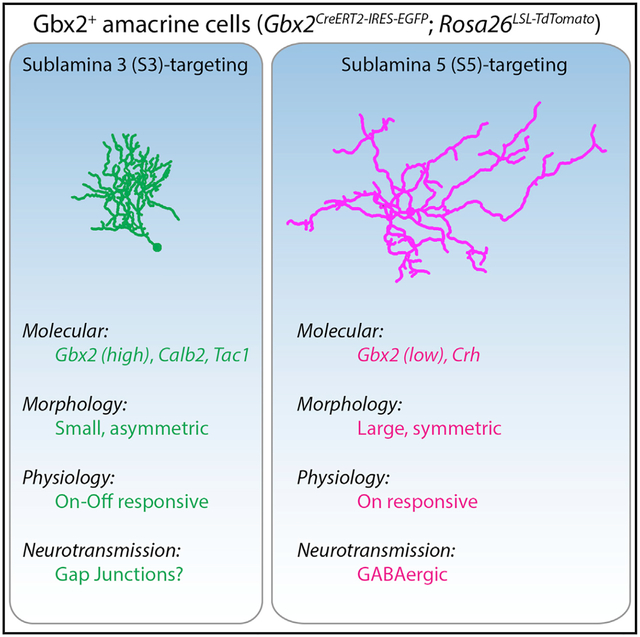
SUMMARY
Our understanding of nervous system function is limited by our ability to identify and manipulate neuronal subtypes within intact circuits. We show that the Gbx2CreERT2-IRES-EGFP mouse line labels two amacrine cell (AC) subtypes in the mouse retina that have distinct morphological, physiological, and molecular properties. Using a combination of RNA-seq, genetic labeling, and patch clamp recordings, we show that one subtype is GABAergic that receives excitatory input from On bipolar cells. The other population is a non-GABAergic, non-glycinergic (nGnG) AC subtype that lacks the expression of standard neurotransmitter markers. Gbx2+ nGnG ACs have smaller, asymmetric dendritic arbors that receive excitatory input from both On and Off bipolar cells. Gbx2+ nGnG ACs also exhibit spatially restricted tracer coupling to bipolar cells (BCs) through gap junctions. This study identifies a genetic tool for investigating the two distinct AC subtypes, and it provides a model for studying synaptic communication and visual circuit function.
Graphical Abstract
In Brief
Investigations into neural circuit development and function are limited by the lack of genetic tools to label and perturb individual neuronal subtypes. Using the Gbx2CreERT2 mouse line, Kerstein et al. identify two amacrine cell subtypes in the mouse retina and explore their distinct molecular, morphological, and physiological characteristics.
INTRODUCTION
The mammalian nervous system comprises hundreds of distinct neuronal subtypes that form precise connections with one another. Neuronal subtypes can be defined by a combination of their morphological, physiological, and molecular properties (Zeng and Sanes, 2017). Recent single-cell RNA sequencing (scRNA-seq) approaches have greatly expanded the catalog of neuronal subtypes based on transcriptional profiles (Macosko et al., 2015; Saunders et al., 2018; Tasic et al., 2018). However, linking the morphological and physiological properties of neuronal subtypes to their molecular profile and identifying their function within neural circuits remains a major challenge.
The retina is an ideal system to address such questions. It contains a complete neural circuit organized in a highly stereotyped manner within a compact space. Three classes of excitatory neurons, photoreceptors, bipolar cells (BCs), and retinal ganglion cells (RGCs), connect in sequence to sense light and transmit this sensory information to the brain. Two classes of inhibitory neurons, horizontal cells and amacrine cells, increase the feature selectivity of these sensory signals by providing spatial and temporal regulation of excitatory cell activity (Diamond, 2017). Within these 5 classes, there are >120 distinct neuronal subtypes (Macosko et al., 2015; Rheaume et al., 2018; Sanes and Masland, 2015; Shekhar et al., 2016; Tran et al., 2019; Yan et al., 2020). This high level of diversity reflects the enormous amount of computation necessary to encode up to 40 distinct representations of the visual field (Baden et al., 2016).
Amacrine cells (ACs) exhibit the greatest diversity in number and variance between subtypes. Morphological analysis of ACs predicts there are ~45 AC subtypes (Badea and Nathans, 2004; Helmstaedter et al., 2013; Lin and Masland, 2006; MacNeil et al., 1999), while recent single-cell transcriptomic analysis predicts >60 distinct AC types (Peng et al., 2019; Yan et al., 2020). The data from these scRNA-seq studies can provide potential markers for identifying neuronal subtypes in mouse (Macosko et al., 2015; Rheaume et al., 2018; Shekhar et al., 2016; Tran et al., 2019) and primate retinas (Peng et al., 2019). AC subtypes display characteristic specializations evident in their selective synaptic connectivity and neurotransmitter release. The dendritic morphology and stratification of an AC subtype determines its receptive field size and dictates the potential pre- and postsynaptic partners within the inner plexiform layer (Diamond, 2017; MacNeil and Masland, 1998). Two broad groups of AC subtypes are defined by their expression of either glycine or GABA. In addition to the inhibitory neurotransmitter, some AC subtypes co-release an excitatory neurotransmitter—for example, glycine and glutamate (Haverkamp and Wässle, 2004; Johnson et al., 2004; Lee et al., 2014) or GABA and acetylcholine (Brecha et al., 1988; Vaney and Young, 1988). Other ACs also release neuromodulators such as dopamine (Newkirk et al., 2013) or neuropeptides (Zalutsky and Miller, 1990). Furthermore, in addition to neurochemical signaling, AC subtypes can form electrical synapses via gap junctions with BCs, RGCs, and other ACs (Bloomfield and Völgyi, 2009; Vaney and Weiler, 2000). Despite this broad functional and morphological diversity, most AC subtypes have not been thoroughly characterized due to a lack of genetic tools to prospectively identify and manipulate them.
Here, we identify two AC subtypes that are genetically labeled by a mouse line expressing tamoxifen-inducible Cre recombinase from the endogenous locus of the transcription factor Gbx2 (Gbx2CreERT2-IRES-EGFP). We use this genetic tool to define the distinct morphological, physiological, and molecular properties of the two AC subtypes. S5-Gbx2+ ACs have larger dendritic arbors that stratify in S5, receive inputs exclusively from On type BCs, and are GABAergic. In contrast, S3-Gbx2+ ACs have smaller, dense, asymmetric dendritic arbors that stratify in S3, receive input from both On and Off BCs, and are non-GABAergic non-glycinergic (nGnG) ACs. S3-Gbx2+ AC subtypes also exhibit exclusively heterotypic tracer coupling to BCs, suggesting that they communicate in part through electrical synapses.
RESULTS
Gbx2CreERT2-IRES-EGFP Marks Two Distinct AC Subtypes
To begin unraveling the neuronal subtype complexity in the retina, we sought to identify Cre or CreERT2 mouse lines that could be used as genetic tools to selectively label and manipulate single neuronal subtypes in the retina. Using scRNA-seq datasets and transgenic mouse databases as a guide (Macosko et al., 2015; Siegert et al., 2009), we identified Gbx2CreERT2-IRES-EGFP as a mouse line predicted to label a sparse population of neurons in the retina (Chen et al., 2009). Since the CreERT2-IRES-EGFP cassette is knocked into the Gbx2 locus, labeled neurons are expected to faithfully recapitulate its endogenous expression pattern. Crossing the Gbx2CreERT2-IRES-EGFP line to multiple Cre-dependent reporter lines labeled neurons in the inner nuclear layer (INL; Figure 1A) and the ganglion cell layer (GCL; Figure 1B). Low doses of tamoxifen (<0.02 mg) labeled neurons in both the INL and GCL with dendrites that selectively stratify in sublamina 3 (S3) of the inner plexiform layer (IPL). When we used a saturating dose of tamoxifen (2.0 mg/day for 2 days; Figures S1D–S1I), we labeled a second neuronal subtype with dendrites that stratify in sublamina 5 (S5) of the IPL (Figures 1C and 1G). Importantly, the same two neuronal populations were labeled whether tamoxifen was administered at embryonic (E16), postnatal (P0–P2), or adult (P28) ages (Figures S1A–S1C), demonstrating that the Gbx2CreERT2-IRES-EGFP line provides a consistent and stable tool for genetically labeling the same neuronal populations from embryonic to adult stages.
Figure 1. Gbx2CreERT2-IRES-EGFP Selectively Labels a Subtype of Amacrine Cells (ACs) in the Mouse Retina.
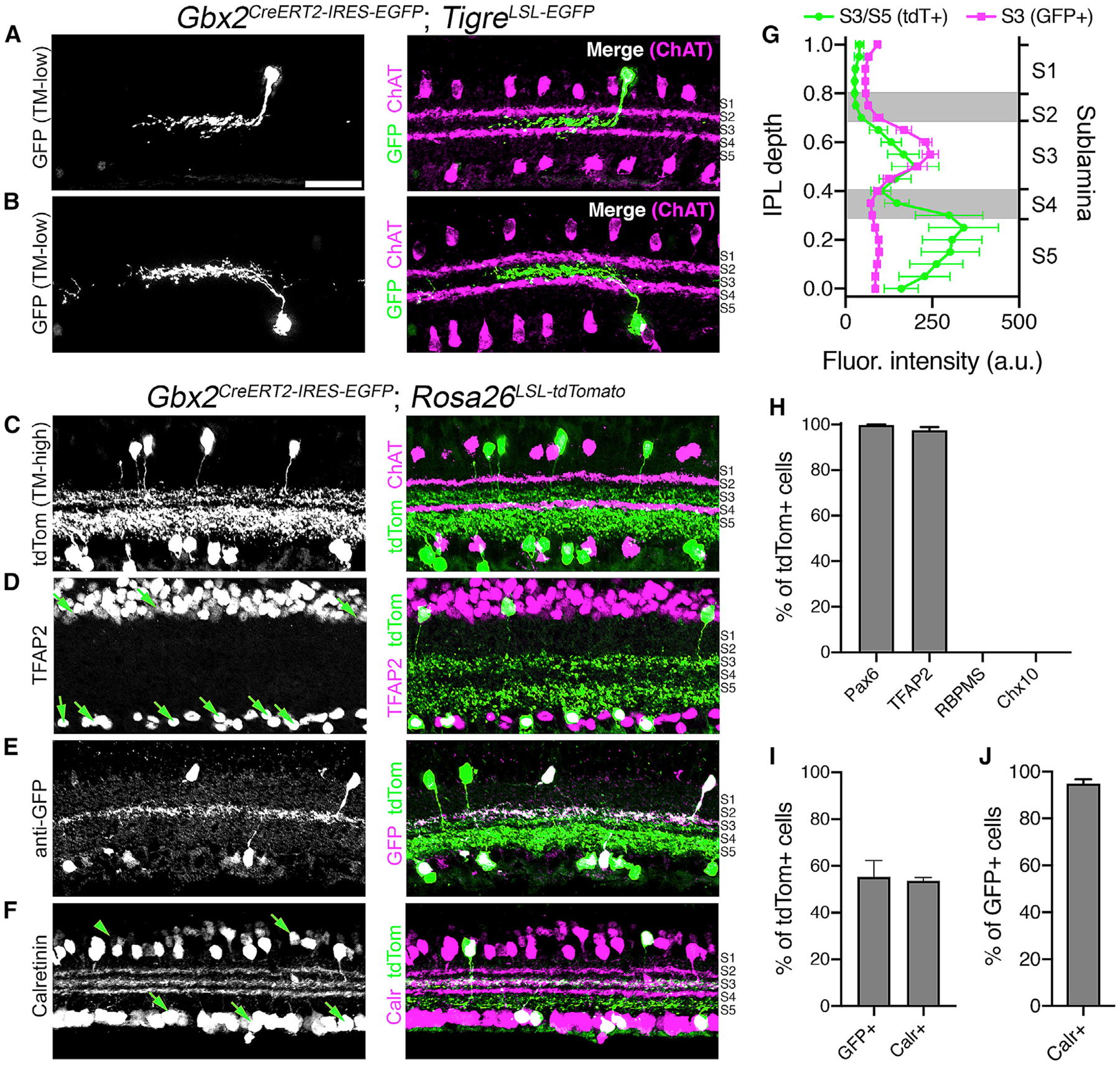
(A and B) Left: cross-sections of adult retinas from a Gbx2CreERT2-IRES-EGFP; TIGRETIT2L-GFP-ICL-tTA2 mouse labeling single Gbx2+ ACs (low-TM, 0.02 mg tamoxifen) in the (A) INL and (B) GCL. Right: merged image, choline acetyltransferase (ChAT; magenta, S2, S4), Gbx2-GFP (green, S3).
(C) Cross-sections of an adult retina from a Gbx2CreERT2; Rosa26LSL-tdTomato mouse labeling the total Gbx2+ AC population (high-TM, 2.0 mg tamoxifen). Right: merged image, ChAT (magenta) and Gbx2-tdTom (green, S3 and S5).
(D–F) Left, retinal sections from a Gbx2CreERT2-IRES-EGFP; Rosa26LSL-tdTomato mouse immunolabeled with (D) an AC marker TFAP2, (E) GFP, and (F) calretinin co-labeled with Gbx2+ tdTomato+ retinal neurons (green, right).
(G) Fluorescent intensity plotted in IPL depth using ChAT bands in S2 and S4 as reference points. Magenta: Gbx2+ ACs detected by EGFP immunohistochemistry (n = 6 measurements, 3 retinas); green: Gbx2+ ACs detected by tdTomato labeling (n = 6 measurements, 3 retinas).
(H) Quantification of the percentage of tdTomato+ cell co-labeling with AC markers Pax6 and TFAP2, retinal ganglion cell marker RBPMS, and bipolar cell maker Chx10.
(I) Percentage of tdTomato+ cells co-labeling with GFP or calretinin.
(J) Percentage of GFP+ cells co-labeling with calretinin.
n = 4 measurements, 3 animals for each experiment in (H)–(J). Data represented as means ± SEMs. Scale bar, 25 μm.
See also Figures S1–S3.
Both the S3- and S5-stratifying Gbx2+ populations colocalized with pan-AC markers TFAP2 and Pax6 (Figures 1D, 1H, and S2A) and lacked expression of the retinal ganglion cell marker RBPMS and BC marker Chx10 (Figure 1H, S2B, and S2C). GFP expression could be detected in 55.3% of the total Gbx2/tdTomato+ population (Figures 1E and 1I). A second marker, calretinin, labeled 53.6% of the total Gbx2+ ACs (Figures 1F and 1I), and nearly all (94.9%) of the GFP+ Gbx2+ ACs were co-labeled by calretinin (Figure 1J). Both GFP and calretinin label the S3 sublamina of the IPL, whereas S5 is negative for both GFP and calretinin (Figures 1E–1G). Some retinal neuron subtypes, such as starburst ACs, organize in regularly spaced mosaics, while many others do not (Keeley et al., 2020). Both S3- and S5-stratifying Gbx2+ populations fall into this latter category, forming irregular mosaics in the INL and GCL (Figures S3A–S3K). The GFP+/calretinin+ S3-stratifying subtype was present at a slightly higher density than the GFP−/calretinin− S5-stratifying subtype (Figure S3I). The cell density of both subtypes was consistent across the different areas of the retina (Figures S3L–S3N). These data suggest that Gbx2CreERT2-IRES-GFP labels two distinct AC subtypes: a GFP+/calretinin+ population that stratifies in S3 and a GFP−/calretinin− population that stratifies in S5 of the IPL.
S3- and S5-Gbx2+ ACs Have Distinct Molecular Profiles
The transcriptomic profiles of related neuronal subtypes can provide clues to the molecular basis of the morphology, development, and function, as well as identify markers for distinguishing individual subtypes. We used genetic labeling with the Gbx2CreERT2-IRES-EGFP line to identify the molecular differences between S3- and S5-Gbx2+ ACs by performing bulk RNA-seq on Gbx2+ ACs isolated from P8 retinas of Gbx2CreERT2-IRES-EGFP; R26LSL-tdTomato mice administered 25 μg tamoxifen at P1. We used fluorescence-activated cell sorting (FACS) to separate the S3-Gbx2+ AC (GFP+/tdTomato+) and S5-Gbx2+ AC (GFP−/tdTomato+) populations (Figure S4). RNA-seq was performed on these isolated populations and identified 18 and 67 differentially expressed genes (DEGs) enriched in S3- and S5-Gbx2+ ACs, respectively (Figure 2A) (GEO: GSE157271). Gbx2 was expressed 13-fold higher in the S3-Gbx2+ ACs versus the S5-Gbx2+ ACs (Figure 2B). Consistent with this result, GFP (driven from the endogenous Gbx2 locus) was expressed 10-fold higher in the S3-Gbx2+ ACs compared to the S5 subtype (Figure 2B). Calretinin (Calb2), which distinguishes S3- and S5-Gbx2+ ACs by immunohistochemistry (Figures 1F–1J), was expressed 19-fold higher in S3-Gbx2+ ACs (Figure 2B).
Figure 2. Transcriptomic Profiling of S3- and S5-Stratifying Gbx2+ AC Subtypes Identifies Molecular Differences.
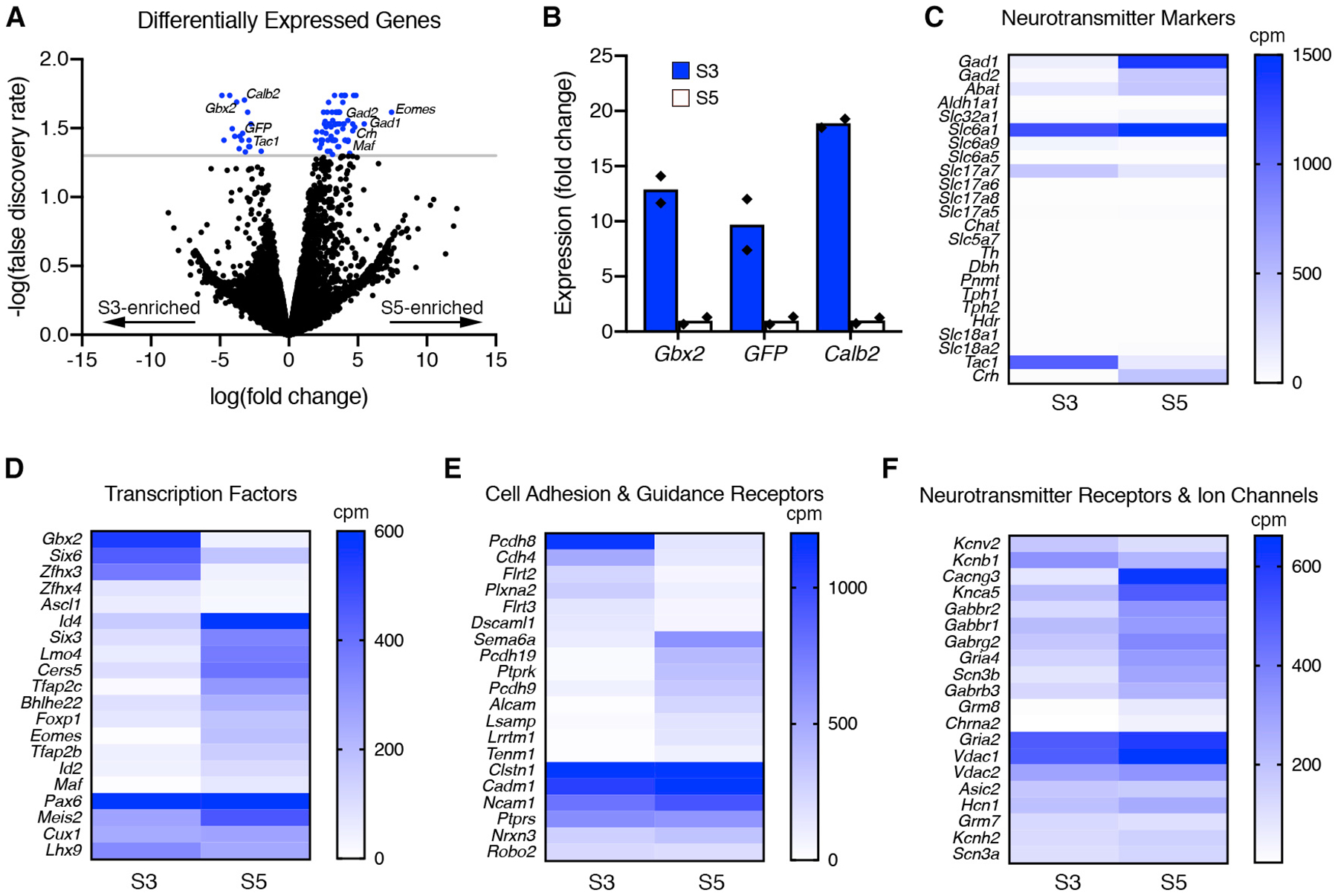
(A) Differentially expressed genes (DEGs) identified by RNA-seq between S3- and S5-Gbx2+ ACs are displayed in a volcano plot (negative fold change, S3 enriched; positive fold change, S5 enriched). The blue points represent genes that have significant differences in between the S3- and S5- Gbx2+ AC subtypes (p < 0.05, false discovery rate [FDR], gray line).
(B) Fold change in expression determined by RNA-seq for known S3-specific markers in S3 (blue bars) and S5 (white bars) Gbx2+ AC subtypes. Each point (black diamond) represents a single sample/animal.
(C) The mean expression of neurotransmitter markers from RNA-seq between S3- and S5-Gbx2+ ACs displayed in a heatmap. Neurotransmitter markers are subdivided into GABA synthesis (Gad1, Gad2, Abat, Aldh1a1), GABA transport (Slc32a1, Slc6a1), glycine transport (Slc6a9, Slc6a5), glutamate transport (Slc17a7, Slc17a6, Slc17a8, Slc17a5), cholinergic synthesis and transport (Chat, Slc5a7), catecholamine synthesis (Th, Dbh, Pnmt), serotonin synthesis (Tph1, Tph2), histamine synthesis (Hdc), monoamine transport (Slc18a1, Slc18a2), and neuropeptides (Tac1, Crh).
(D and E) Heatmaps comparing the mean expression of highly and differentially expressed (D) transcription factors, (E) cell adhesion and axon guidance receptors, and (F) neurotransmitter receptors and ion channels. Cpm, counts per million reads.
The data are represented as means.
See also Figure S4.
The majority of AC subtypes are categorized by their neurotransmitter profile as either GABAergic or glycinergic (Kay et al., 2011). Based on the RNA-seq results, the GABAergic enzymes Gad1 and Gad2 are highly expressed in S5-Gbx2+ ACs (Figure 2C). In contrast, S3-Gbx2+ ACs were negative for all of the standard neurotransmitter markers, including the GABAergic markers Gad1 and Gad2 and the glycinergic markers Slc6a9 and Slc6a5 (Figure 2C). Both S3 and S5 subtypes expressed low levels of the glutamatergic markerSlc17a7; however, this is likely due to photoreceptor contamination. The neuropeptides Tachykinin1 (Tac1) and Corticotropin-releasing hormone (Crh) were highly expressed in the S3- and S5-Gbx2+ subtypes, respectively (Figure 2C).
To gain additional insight into the differences between S3- and S5-Gbx2+ ACs, we expanded our analysis to include transcription factors (Figure 2D), cell adhesion and axon guidance receptors (Figure 2E), and neurotransmitter receptors and ion channels (Figure 2F). The S3-Gbx2+ AC subtype expressed the transcription factors Gbx2 and Zfhx3 significantly higher than in the S5 subtype, whereas the S5 subtype selectively expressed Id4, Lmo4, Tfap2c, Eomes, and Maf (Figure 2D). Both S3 and S5-Gbx2+ AC subtypes expressed high levels of the transcription factors Pax6, Meis2, Cux1, and Lhx9 (Figure 2D). The cell adhesion molecule Pcdh8 was expressed significantly higher in the S3-Gbx2+ ACs, whereas Pcdh19, Alcam, Lrrtm1, and Tenm1 were higher in the S5 subtype (Figure 2E). Both shared high expression of the adhesion receptor genes Clstn1, Cadm1, Ncam1, Ptprs, Nrxn3, and Robo2 (Figure 2E). Unlike transcription factors and adhesion receptors, neurotransmitter receptors and ion channels showed little difference in expression between the two subtypes, with the exception of Cacng3, Grm8, and Chrna2 genes showing significant enrichment in the S5 subtype (Figure 2F). These differentially expressed genes identify markers to distinguish the two Gbx2+ AC subtypes, and provide insight into the function of these cells, particularly with respect to neurotransmitter release.
S3-Gbx2+ ACs Are Non-GABAergic, Non-glycinergic (nGnG)
The majority of ACs (~85%) release either GABA or glycine to inhibit downstream postsynaptic neurons (Kay et al., 2011). Based on the RNA-seq results, S5-Gbx2+ ACs express both Gad1 and Gad2 and are therefore GABAergic (Figure 2C). We confirmed this with immunohistochemistry, which showed that28.4% of Gbx2+ ACs in the INL co-labeled with the GABAergic marker GAD67, while the remaining Gbx2+ ACs were negative for GAD67 (Figures 3A–3C, and 3H). The GAD67+/Gbx2+ ACs are likely S5-Gbx2+ ACs, as ~37% of all Gbx2+ ACs in the INL are the S5 subtype (calretinin−/GFP−) (Figure S3). Neither Gbx2+ AC subtype colocalized with the glycinergic marker GlyT1/2 (Figures 3D, 3E, and 3H). Therefore, S3-Gbx2+ ACs are a subtype of non-glycinergic, non-GABAergic (nGnG) AC. These results are consistent with a previous scRNA-seq analysis that showed that a cluster of ACs identified by their expression of Gbx2 lacked the expression of Slc6a9 or Gad1/2 (Macosko et al., 2015; Yan et al., 2020). When we examined other neurotransmitter markers known to be expressed in specific AC subtypes, we found little (<1%) or no co-localization with Gbx2+ ACs (Figures 3H and S5), consistent with our RNA-seq results (Figure 2C).
Figure 3. S3-Gbx2+ Neurons Are a Subtype of non-GABAergic Non-glycinergic ACs.
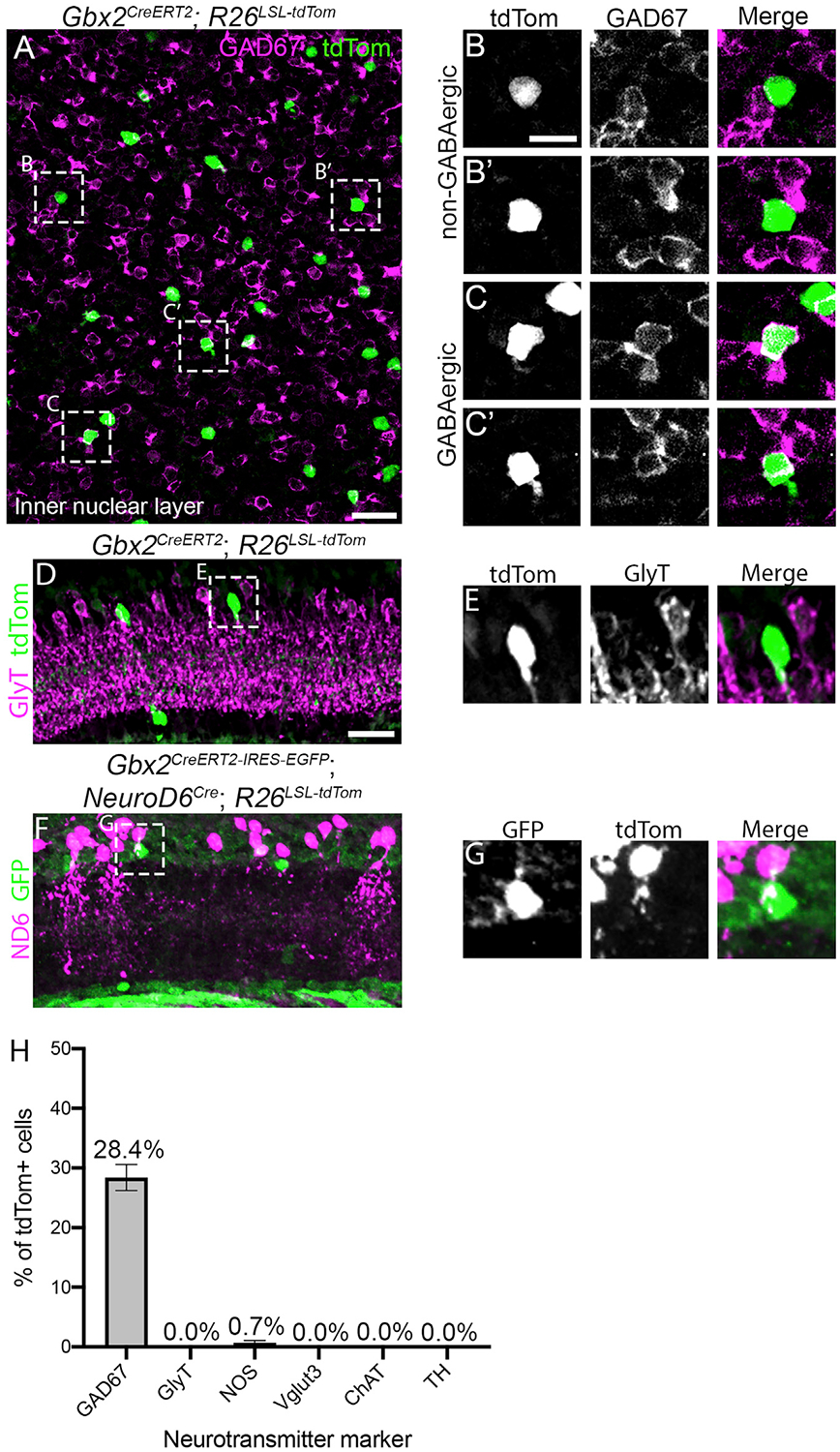
(A) Inner nuclear layer of an adult retina en face from a Gbx2CreERT2; R26LSL-tdTom mouse immunolabeled for tdTomato (green) and GAD67 (magenta).
(B and C) Magnified images of Gbx2+ ACs from (A) that do not colocalize with GAD67 (B–B’) and GABAergic Gbx2+ ACs that colocalize with GAD67 (C–C’).
(D and E) A retinal cross-section from a Gbx2CreERT2; R26LSL-tdTom mouse shows no colocalization between Gbx2+ ACs with GlyT1/2.(F) Gbx2+ ACs (GFP, green) do not colocalize with NeuroD6+ nGnG ACs (tdTom, magenta) in retinal sections from a Gbx2CreER-ires-EGFP; NeuroD6Cre; Rosa26LSL-tdTomto mouse.
(G) Magnified image from (F) labeled with GFP (Gbx2, left), tdTomato (NeuroD6, center), and a merge image (right).
(H) Quantification of the percentage of tdTomato+ cells that colocalize with neurotransmitter markers. GAD67 quantification was performed in INL only. n > 125 neurons and 3 mice for each condition.
The data represented are as means ± SEMs. Scale bar, 25 μm in (A), (D), and (F), 10 μm in (B), (C), (E), and (G).
See also Figure S5.
Previous studies have hinted at the presence of at least two nGnG AC subtypes (Cherry et al., 2009; Macosko et al., 2015; Yan et al., 2020). One nGnG AC subtype is defined by the expression of the transcription factor NeuroD6 (Kay et al., 2011). NeuroD6+ nGnG ACs have a narrow dendritic arbor that stratifies diffusely through multiple layers of the IPL, in contrast to the dendritic arbors of S3-Gbx2+ ACs, which have a medium-size arbor that is monostratified. To confirm that Gbx2 and NeuroD6 label different populations of nGnG ACs, we generated NeuroD6Cre; Gbx2CreERT2-IRES-EGFP; R26LSL-tdTomato mice and found no overlap between the NeuroD6+ (tdTomato+) and Gbx2+ (GFP+) AC subtypes (Figures 3F and 3G). Therefore, S3-Gbx2+ ACs are a distinct subtype of the nGnG AC.
S3- and S5-Gbx2 ACs Have Distinct Morphological Properties
The dendritic morphology of a neuronal subtype is directly related to its function (Lefebvre et al., 2015). Dendritic shape and size are particularly important for the computations performed by AC subtypes, as dendrites contain both pre- and postsynaptic sites. We used the Gbx2CreERT2-IRES-EGFP line to define the morphological properties of S3- and S5-stratifying Gbx2+ ACs by 2 different sparse-labeling techniques: a genetic approach using low-dose tamoxifen (0.05 mg) or single-cell fills of Gbx2+ ACs with neurobiotin (Figures 4A–4D). S3-Gbx2+ ACs had an average dendritic arbor area of 10,331 ± 629 μm2 and a total dendrite length of 1,766 ± 109 μm. S5-Gbx2+ ACs had significantly larger dendritic arbors, with an area of 44,643 ± 11,699 μm2 and length of 2,800 ± 401 μm (Figures 4E and S6A). S3-Gbx2 ACs had a significantly higher average dendrite and branch density of 117.8 ± 16.9 branches/100 μm2, compared to 34.3 ± 12.9 in S5-Gbx2+ ACs (Figures 4F and 4G). We did not observe any difference between S3- and S5-Gbx2+ ACs in total number of branches (Figure S6B), branch crossovers (Figure S6C), or coverage factor (Figure S6D).
Figure 4. Distinct Morphological Features of S3- and S5-Gbx2+ ACs.
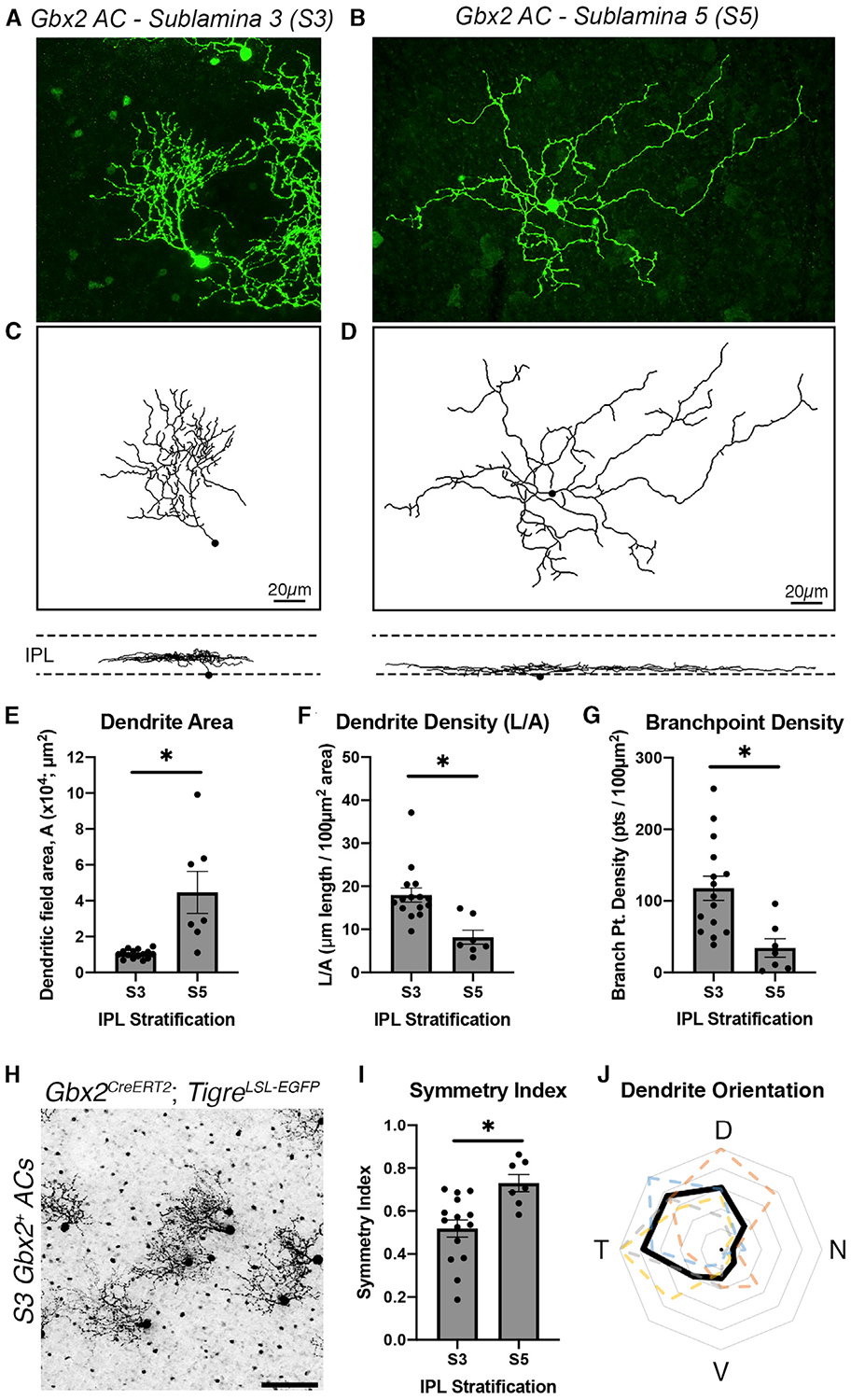
(A and B) Single-cell labeling of (A) S3 stratifying and (B) S5 stratifying Gbx2+ ACs using a Gbx2CreERT2-IRES-EGFP; TIGRETIT2L-GFP-ICL-tTA2 mouse and neurobiotin cell fill.
(C and D) Dendrite morphology traces constructed using the Filaments plugin in Imaris software for the Gbx2+ ACs in (A) and (B), respectively. Below are the traces in the orthogonal view to display the stratifications of each neuron in the IPL.
(E–G) S3- and S5-Gbx2+ AC morphology in (E) dendrite area (p = 0.0003), (F) dendrite density (p =0.0015), and (G) branchpoint density (p = 0.0009). Morphological data were collected from 15 S3-Gbx2+ ACs (3 animals) and 7 S5-Gbx2+ ACs (4 animals).
(H) Gbx2+ ACs sparsely labeled in a flat-mounted P21 retinainaGbx2CreERT2-IRES-EGFP;TIGRETIT2L-GFP-ICL-tTA2 mouse.
(I) Quantification of dendritic arbor symmetry between S3- and S5-stratifying ACs (p = 0.0039).
(J) A polar plot of dendrite orientation of S3-targeting Gbx2+ ACs; black trace represents the mean and colored dashed traces represent neurons quantified from a single retina (n > 20 neurons per retina, n = 4 retinas). The concentric gray rings represent the average percentage of neurons with dendrites extending in the specific orientation (outermost ring = 100%, 20% increments).
The data are represented as means ± SEMs. *p < 0.05 by an unpaired t test with Welch’s correction. Scalebar, 20 μm in (C) and (D), 50 μm in (G).
See also Figure S6.
Sparse labeling revealed that the dendritic arbors of S3-Gbx2+ ACs exhibited a clear asymmetry, with the arbor rarely exceeding 180° (Figure 4H). As a population, S3-Gbx2+ ACs had a significantly lower symmetry index than S5-Gbx2+ ACs (Figure 4I). We analyzed the dendrite orientation of S3-Gbx2+ ACs at a population level and found that their asymmetric dendrites were consistently oriented in a dorsal-temporal direction (Figure 4J). Both the dendrite asymmetry and orientation of S3-Gbx2+ ACs were consistent across all quadrants of the retina (Figures S6E and S6F and S6G–S6J, respectively). Overall, these data show that S3- and S5-Gbx2+ ACs exhibit significantly different morphological features, supporting the hypothesis that they are distinct subtypes.
S3-Gbx2+ ACs Are Tracer Coupled to Bipolar Cells
Many ACs show selective electrical coupling to other neurons, including bipolar cells (BCs), ganglion cells, and other ACs (Bloomfield and Völgyi, 2009). Electrical synapses play a major role in the transmission of visual information in the retina (O’Brien and Bloomfield, 2018) and are typically revealed by filling cells with neurobiotin, a small tracer molecule that is permeable through most gap junctions (Vaney, 1991). When we filled genetically labeled S3-Gbx2+ AC somas in the ganglion cell layer with neurobiotin, we found that the tracer spread into a sparse number of neighboring retinal neurons with somas in the INL (Figures 5A and 5B). The coupled cells could be identified as BCs based on their morphology in orthogonal optical sections (Figure 5G). At higher magnification, we also observed tracer-filled BC axon terminals overlapping S3-Gbx2+ AC dendrites in the same optical section en face (Figure 5H). In contrast to the S3-Gbx2 ACs, S5-Gbx2+ ACs did not show consistent tracer coupling with other retinal cells (Figure 5J).
Figure 5. Gbx2+ ACs Are Electrically Coupled to Neighboring Bipolar Cells.
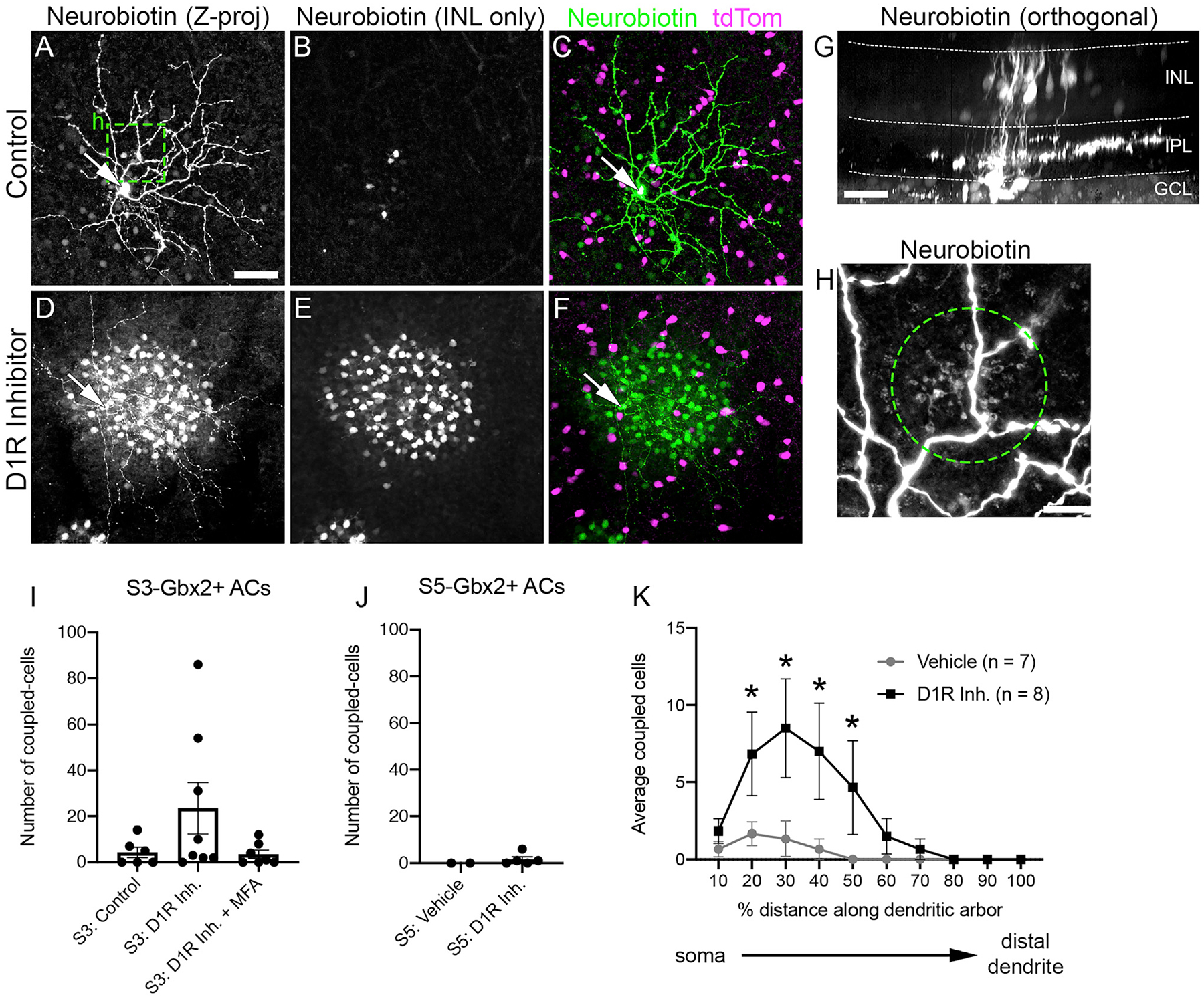
(A–F) Neurobiotin-filled S3-stratifying Gbx2+ AC (arrows, cell bodies) incubated in (A–C) vehicle and (D–F) D1 receptor inhibitor, 50 μM SCH23390, in flat-mounted retinas. (A and D) Z projection through the entire retina. (B and E) Z projection through the INL only. (C and F) Merged Z projection with neurobiotin (green) and Gbx2+ ACs (magenta).
(G) Orthogonal view of neurobiotin-filled Gbx2+ AC (arrow) and dye-coupled BCs.
(H) Magnified image (box in A) of bipolar cell (BC) axon terminals (circle) colocalizing with a dendritic branch of neurobiotin-filled Gbx2+ ACs.
(I and J) Mean number of neurons dye coupled to a single (I) S3- and (J) S5-stratifying Gbx2+ AC (S3-vehicle, n = 6 cell-fills; S3-D1R inhibitor [Inh.], n = 8; S3-D1R Inh. + MFA, n = 7; S5-vehicle, n = 2; S5-D1R Inh., n = 3; Welch’s t test).
(K) The spatial distribution of dye-coupled cells to a single S3-stratifying Gbx2+ AC along its dendritic arbor. *p < 0.05 calculated by a 2-way ANOVA with multiple comparisons.
The data are represented as means ± SEMs. Scale bar, 50 μm in (A)–(G), 10 μm in (H).
The extent and strength of coupling in the retina is linked to the ambient light level through the release of the neuromodulator dopamine. Dopamine release increases as the background light level increases (Hampson et al., 1992). The increased concentration of dopamine activates D1 receptors in all AII ACs, reducing their coupling to BCs. To determine whether S3-Gbx2+ AC to BC coupling is also modulated by dopamine, we measured coupling in the presence of the D1 receptor antagonist SCH23390 (50 μM). The inhibition of D1 receptors dramatically increased coupling to BCs in 3 of 8 S3-Gbx2+ ACs, while coupling in the remaining S3-Gbx2+ ACs was unaffected (Figures 5D–5I). The specificity of tracer coupling was confirmed by bath application of the gap junction antagonist meclofenamic acid (MFA, 100 μM), which blocked coupling in both baseline and D1 receptor antagonist conditions (Figure 5I).
The coupling between S3-Gbx2+ ACs and BCs shows unusual spatial features. In other ACs, coupling typically occurs homo-typically and spreads laterally to serve a signal averaging function (Bloomfield and Völgyi, 2009). In contrast, we never observed S3-Gbx2+ ACs coupled to neighboring S3-Gbx2+ ACs, and their coupling to BCs appeared to be spatially selective. BCs coupled to S3-Gbx2+ ACs were clustered toward the center of the dendritic arbor (Figure 5K). Even with the addition of the D1R antagonist, BC coupling to Gbx2+ ACs never extended into the distal 20% of the dendrite arbor (Figure 5J). Inhibition of the D1 receptors did not unmask coupling between the S5-Gbx2+ ACs and other retinal neurons (Figure 5J).
Input Differences between S3- and S5-Gbx2+ ACs
We next sought to characterize the intrinsic physiological properties of S3- and S5-Gbx2+ ACs. S3-Gbx2+ ACs cells stratify at the border between the On and Off sublamina of the IPL and thus potentially receive input from both On- and Off-type BCs. The S5-Gbx2+ ACs cells stratify within the On-sublamina, and are therefore likely to receive input from On-type BCs. To test these predictions, we recorded light-evoked postsynaptic potentials (PSPs) from S3- and S5-Gbx2+ ACs. Genetically labeled Gbx2+ Acs were stimulated with light spots of 50% to 80% contrast, square-wave modulated at 2 Hz, centered on the receptive field. The background intensity was set at a photopic level (103 photons/μm2/s). Consistent with the stratification levels of their dendrites, S3-Gbx2+ ACs cells showed strong depolarization during both the On phase (increase in luminance) and the Off phase (decrease in luminance) of the stimulus (black trace, Figure 6A, left). In contrast, S5-Gbx2+ ACs cells were depolarized only during the On phase and are hyperpolarized during the Off phase (black trace, Figure 6A, right). Thus, the physiological inputs are consistent with the stratification level of the dendrites for both S3- and S5-Gbx2+ ACs.
Figure 6. S3- and S5-Gbx2+ ACs Exhibit Distinct Spatial Receptive Field Properties and On-Off Inputs.
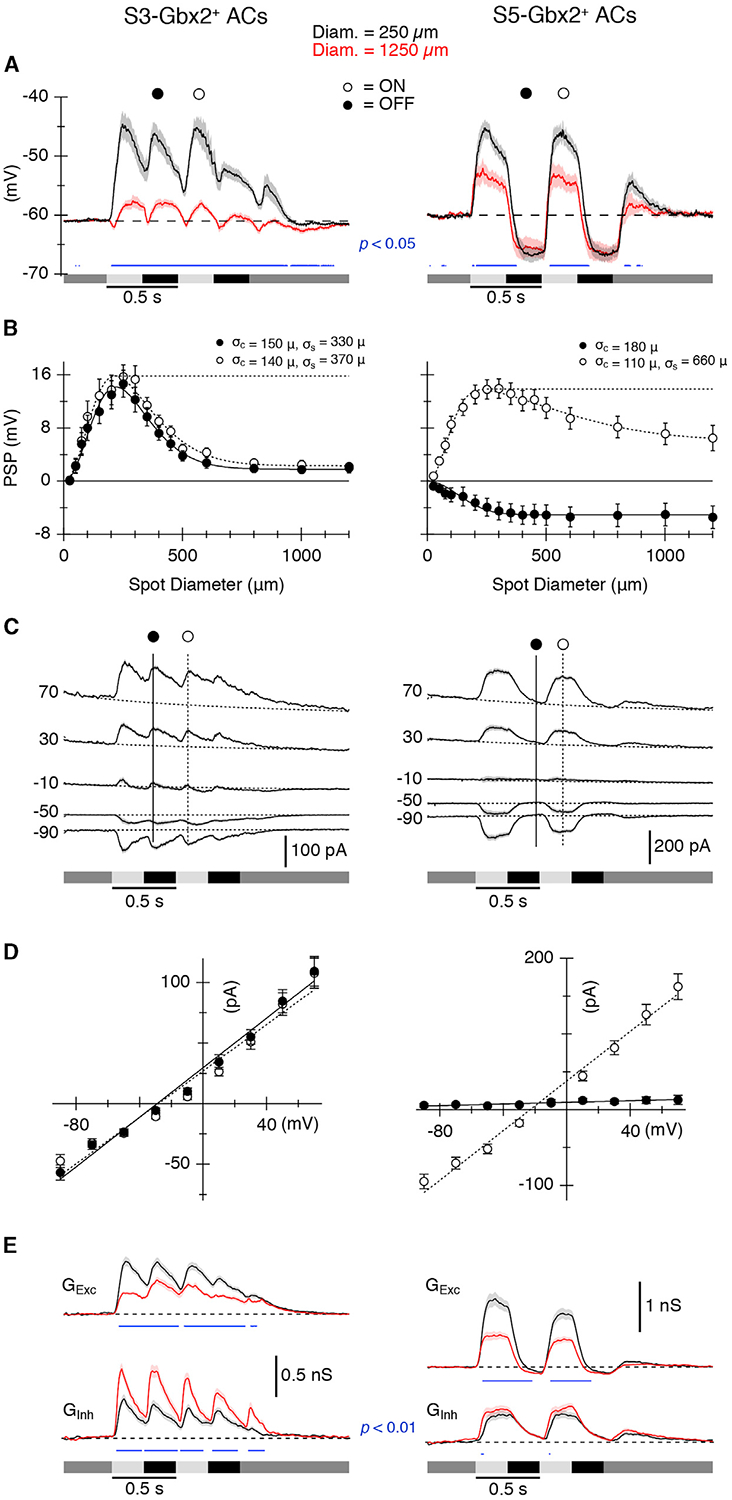
(A) Average light-evoked postsynaptic potentials (PSPs) in S3 (n = 12) (left) and S5 (n = 17) (right) Gbx2+ ACs in response to a 250-μm (black) or 1,200-μm (red) diameter spot stimulus. Luminance indicated by shaded bars underneath traces.
(B) Area response functions from S3 (left) and S5 (right) cells measured as the amplitude of the PSPs versus stimulus diameter at indicated time points in (A). Surround suppression of PSPs was 86% for S3 and 53% for S5 cells.
(C) Average light-evoked postsynaptic currents (PSCs) in (n = 56) (left) S3 and (n = 45) (right) S5 cells for center spot stimuli during a series of voltage steps from −90 to +70 mV.
(D) Current-voltage (I-V) relations for the PSCs at the time points indicated in (C). The dotted and solid lines show average least-squares fits to the I-V relations.
(E) Average synaptic conductance traces calculated from fits to I-V relations measured every 10 ms during the light stimulus (see Method Details). The traces and symbols represent the average responses. The shading and error bars represent SEMs.
Blue traces in (A) and (E) indicate significant differences (paired t test) between the solid and dashed data. p values are indicated in the panels.
S3- and S5-Gbx2+ ACs Receive Center and Surround Inhibition
In many retinal neurons, the spatial structure of the receptive field is shaped by center/surround organization, in which illumination outside the excitatory center receptive field of a neuron elicits an antagonistic response. We presented spots with a range of diameters to test for differences between the spatial extent of the receptive fields of the two cell types. The largest spots suppressed center responses in both cell types. For S3-Gbx2+ ACs, maximal surround stimulation suppressed On and Off PSPs equally by 86% (Figures 6A and 6B, left), whereas surround suppression was weaker in the S5-Gbx2+ ACs, reducing center PSPs by ~53% (Figures 6A and 6B, right). Activation of the surround had little effect on the amplitude of the hyperpolarization evoked during the Off phase of the stimulus (Figure 6B, left, filled symbols). The spatial extent of the center and surround were estimated at fixed time points during the On and Off phases of the stimulus by fitting a difference of Gaussians (DOG) function to the amplitude of the responses. The On and Off responses of the S3 ACs showed essentially identical spatial tuning profiles, with the extent of the surround being ~2.5-fold wider than the center (Figure 6B, left). The maximum dendritic diameter averaged 181 ± 8 μm (n = 17), as estimated from the morphological analysis, which compares well with the physiological center size of ~145 μm, estimated as the space constant from the DOG fit. The S5 ACs were more divergent. The maximum dendritic diameter averaged 258 ± 12 μm (n = 27), while the physiological center size estimated during the On phase was only ~110 μm (Figure 6B, right), and was smaller than the center size measured during the Off phase (~180 μm). A similar difference was apparent in a previous study (Park et al., 2018).
To examine the potential synaptic mechanisms providing surround antagonism, we recorded light-evoked postsynaptic currents (PSCs) in response to center-only and full-field visual stimuli, and calculated the component excitatory and inhibitory inputs (Figures 6C–6E). Excitation and inhibition were activated during the On and Off phases of the stimulus for S3-Gbx2+ ACs and appeared to have very similar temporal dynamics (Figure 6E, left). The S5 ACs showed a similar pattern, with excitation and inhibition showing similar temporal waveforms. However, while inhibition was slow to turn off during the Off phase, excitation declined more rapidly and dipped below zero (Figure 6E, right), presumably driving the hyperpolarization seen during the Off phase in Figure 6A. Comparison of the center-only and full-field responses provides insight into the mechanisms of surround antagonism. For the S3 ACs, excitation was suppressed and inhibition enhanced for full-field stimuli relative center-only stimuli (Figure 6E, red versus black, left), suggesting that the antagonistic surround shown in Figure 6A results from complementary changes in both excitation and inhibition. For S5 ACs, by contrast, activation of the surround suppressed only the excitation, and had no significant effect on the inhibitory input (Figure 6E, red versus black, right).
S3- and S5-Gbx2+ ACs Receive Distinct On and Off Pathway Excitation and Inhibition
These data suggest that excitation and inhibition may arise through both the On and Off pathways for S3-Gbx2+ ACs, but only the On pathway for S5-Gbx2+ ACs. Consistent with this hypothesis, when the On pathway signaling was blocked by using mGluR6 agonist L-AP4 to prevent depolarization of On BCs, light-evoked synaptic input to S5-Gbx2+ ACs was abolished (Figure 7A, right). In contrast, the light-evoked synaptic inputs to S3-Gbx2+ ACs during the On phase of the stimulus were strongly suppressed, while the Off pathway excitation and inhibition were largely unaffected. (Figure 7A, left). Inhibition is in-phase with excitation during On and Off responses in S3-Gbx2+ ACs and during On responses in S5-Gbx2+ ACs, suggesting that it could serve to regulate the gain of signaling by counter-balancing excitation.
Figure 7. S3- and S5-Gbx2+ AC Subtypes Have Distinct Excitatory and Inhibitory Inputs.
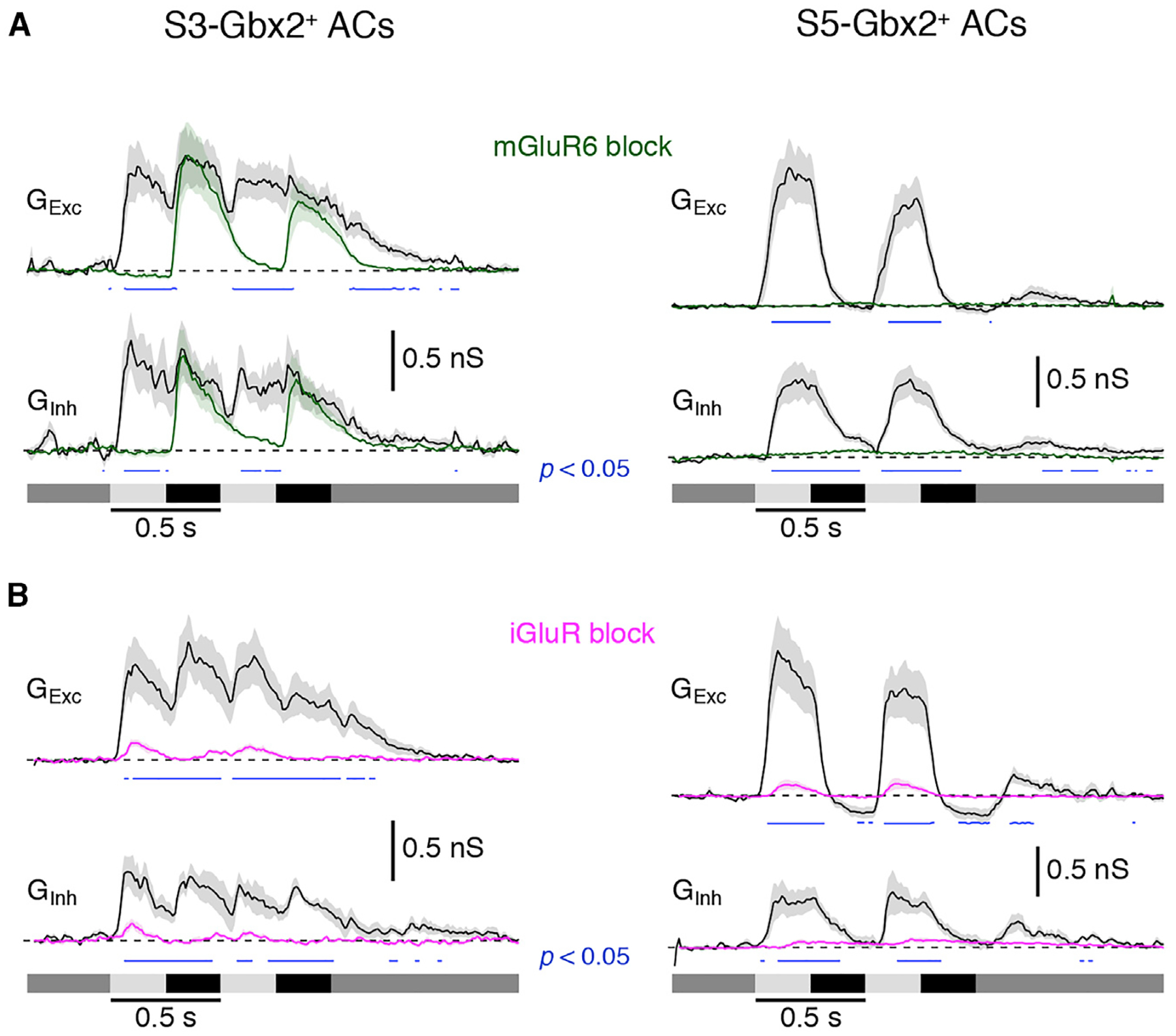
(A) Conductance measurements from (n = 7) S3 (left) and S5 (right) cells in response to a center spot stimulus in the presence (green) or absence (black) of mGluR6 receptor agonist 10 μM L-AP4 (On pathway block).
(B) Conductance measurements from (n = 6) S3 and S5 cells in response to a center spot stimulus in the presence (magenta) or absence (black) of a cocktail of iGluR receptor agonists: 1 μM ACET, 20 μM GYKI, and 50 μM D-AP5.
The traces represent the average responses. The shading represents SEM. Blue traces indicate significant differences (paired t test) between the drug and control conditions. p values indicated on graphs (p < 0.05).
The S3- and S5-Gbx2+ ACs show distinct neurobiotin-coupling patterns to peri-somatic BCs (Figure 5). To test whether these gap junction connections support significant, light-evoked electrical synaptic input to the ACs, we blocked glutamatergic inputs using a cocktail of antagonists, including the NMDA receptor antagonist D-AP5 (50 μm), and the AMPA and kainate receptor antagonists ACET and GYKI (1 and 50 μM). Under these conditions, mGluR6-mediated transmission in On-type BCs is preserved, and if gap junctions mediate electrical transmission to S3- or S5-Gbx2+ ACs, excitatory On responses in these cells should also be preserved. However, light-evoked responses using stimulus spots large enough to activate all of the coupled BCs are almost completely blocked (Figure 7B, magenta traces), suggesting that excitatory drive through any electrical synapses from the coupled BCs is functionally insignificant under these recording conditions. These experiments do not rule out the possible presence of functionally significant electrical synapses between Off BCs and S3-Gbx2+ ACs because AMPA/kainate antagonists block cone input to Off BCs. However, current-voltage relations of excitatory synaptic inputs during light-off responses, recorded with inhibition blocked, were linear and reversed near 0 mV, which is inconsistent with a major input through gap junctions (data not shown).
DISCUSSION
We have identified the Gbx2CreERT2-IRES-EGFP mouse line as a tool that can be used to genetically label two AC subtypes in the mouse retina that are morphologically, physiologically, and molecularly distinct from each other. S3-Gbx2+ ACs are On-Off-type cells (Figure 5), have smaller, asymmetric dendrites that stratify in sublamina 3 of the IPL (Figure 4), and exhibit heterotypic gap junction coupling (Figure 6). S5-Gbx2+ ACs are On-type cells, have larger, more symmetric dendrites that stratify in sublamina 5 of the IPL, and lack consistent gap junction coupling. S3-Gbx2+ ACs lack expression of any of the standard inhibitory or excitatory neurotransmitters, identifying them as a subtype of nGnG ACs, whereas the S5-Gbx2+ ACs are GABAergic. RNA-seq revealed the distinct molecular profiles of these two Gbx2+ AC subtypes, which confirms their identification as separate subtypes and informs future analyses of the cell-specific mechanisms that underlie their distinct morphology and physiological features.
Categorization of AC Subtypes
Previous studies have used Golgi staining, cell fills, and sparse genetic labeling to categorize many AC subtypes based on their morphology (Badea and Nathans, 2004; Lin and Masland, 2006; MacNeil et al., 1999). Reconstruction of a 114 × 80-μm volume of mouse retina by serial block-face electron microscopy indicated the presence of >40 subtypes of ACs (Helmstaedter et al., 2013). However, these approaches may undersample rare neurons, and do not provide a means to prospectively identify and manipulate individual subtypes. A recent single-cell transcriptomic study provides evidence for the existence of 63 molecularly distinct subtypes, indicating that there is greater subtype diversity than previously predicted (Yan et al., 2020). One of the challenges going forward will be to match these molecularly defined subtypes with their morphological counterparts and begin to tease out their connectivity patterns with visual circuits.
Single-cell transcriptomic studies of AC subtypes initially identified two different nGnG AC subtypes, one that is molecularly similar to glycinergic ACs and one that is similar to GABAergic ACs (Cherry et al., 2009; Macosko et al., 2015). The glycinergic-like nGnG AC subtype is marked by the specific expression of NeuroD6 (Kay et al., 2011). A recent study identified two additional nGnG subtypes closely related to NeuroD6+ nGnG ACs (Yan et al., 2020). In contrast, the S3-Gbx2+ AC subtype is the “GABAergic-like” nGnG subtype cluster 4 in the Macosko dataset (Macosko et al., 2015) and nGnG 4, cluster 36 in the Yan dataset (Yan et al., 2020). S3-Gbx2+ nGnG ACs appear to be conserved in primates based on cross-species analysis of single-cell transcriptomics (Peng et al., 2019).
S5-Gbx2+ ACs closely resemble cluster 7 from the Macosko study based on the selective expression of Maf, Cxcl14, Id4, and Lmo4 in both transcriptomic datasets (Figure 2A). S5-Gbx2+ ACs also share morphological, molecular, and physiological properties with the previously described CRH-1 AC (Jacoby et al., 2015; Park et al., 2018; Zhu et al., 2014). S5-Gbx2+ ACs and CRH-1 ACs have dendritic arbors of a similar size and shape that stratify in S5 of the IPL, they both express the neuropeptide Crh, and have similar physiological properties in response to On light stimuli (Jacoby et al., 2015; Park et al., 2018). The CRH-IRES-Cre line used in these studies labels at least three different AC subtypes, which can be distinguished based on their morphological differences. An intersectional approach pairing the Gbx2CreERT2-IRES-EGFP with a recently developed Crh-IRES-FlpO line (Jackson Labs strain 031559) and a dual-recombinase reporter line should selectively label S5-Gbx2+ ACs and allow the direct testing of whether they are the same subtype as CRH-1 ACs.
Dendrite Morphology and Gbx2+ AC Function
The receptive field properties of ACs are dependent on the morphologies of their dendritic arbors. By using a combination of genetic labeling and electrophysiological recordings, we show that the morphologies and the spatial receptive field properties of the S3- and S5-Gbx2+ ACs are quite different from one another (Figures 6A and 6B). Although both cells display a center/surround organization, with fairly similar center diameters, the extent of the surround is much smaller for the S3 than for the S5 cells. For S3-Gbx2+ ACs, responses during both the On and Off phases of the stimulus were strongly suppressed by the surround, with space constants of ~350 μm (Figure 6B). The S5-Gbx2+ ACs showed much weaker surround suppression, with a space constant exceeding 650 μm. With stronger surround suppression, activated with a shorter space constant, the S3 cells will be tuned to higher spatial frequencies than the S5 cells. It seems likely that the narrow surround inhibition of the S3 cells arises in the IPL, while the broader surround of the S5 ACs probably reflects, at least in part, the center-surround organization of the cone photoreceptor output, generated by horizontal cell feedback in the OPL. The lack of any surround suppression during the Off phase of the response (Figure 6B) is expected if the response were produced by turning off presynaptic On BCs that provide a tonic excitatory input to the S5 ACs. The disparity in the center size estimates for the On and Off phases of the stimulus may be explained by inputs from two populations of On BCs with different spatial distributions across the dendritic arbor. Further work will be required to test this hypothesis. Unlike S5 ACs, CRH-1 ACs have been shown to lack any surround suppression during the On phase of the response (compare Figure 3 of Park et al., 2018 with Figure 6A). This discrepancy probably reflects differences in the physiological conditions of the preparations rather than evidence that S5 Gbx2+ ACs and CRH-1 ACs are different subtypes.
Unlike the S5-Gbx2+ ACs, the dendritic arbors of the S3-Gbx2+ ACs are asymmetric along the dorsal-temporal axis, with the soma offset relative to the dendritic arbor. Such asymmetries can influence the response properties of neurons. For example, J-RGCs display a similar asymmetry, which appears to enhance directional responses to ventral motion (Kim et al., 2008; Liu and Sanes, 2017). Examples of functional asymmetry are also found outside the retina, such as in Drosophila proprioceptors, which are spatially tuned for the direction of muscle movement (He et al., 2019). It remains to be determined whether the asymmetric dendrites of S3-Gbx2+ ACs result in anisotropic physiological receptive field structure. The developmental mechanisms that instruct dendrite asymmetry and orientation are poorly understood, but likely involve a combination of selective directional growth and pruning of developing arbors (Liu and Sanes, 2017).
As a transcription factor, Gbx2 is poised to regulate the cellular identity and development of Gbx2+ ACs. Subtype-specific transcription factors in retinal neurons have an important role establishing correct cell body location, dendrite morphology, and synaptic connectivity (Kay et al., 2011; Liu et al., 2018; Peng et al., 2017, 2020; Whitney et al., 2014). Gbx2 regulates the specification, migration, and axon guidance of several neuronal populations in the brain and spinal cord (Chatterjee et al., 2012; Chen et al., 2010; Luu et al., 2011; Mallika et al., 2015). Therefore, Gbx2 may regulate the development of Gbx2+ ACs through the regulation of effector genes specific to this AC subtype, but its exact function in AC identity and development remains to be determined.
Modes of Neurotransmission in S3-Gbx2+ ACs
Without GABA, glycine, and other chemical neurotransmitters, how do S3-Gbx2+ ACs communicate with other neurons? One possibility is that these neurons are in fact GABAergic, and the very low levels of Gad1 and Gad2 in these cells is sufficient for low levels of GABA synthesis. There are also non-canonical pathways for GABA synthesis, although S3-Gbx2+ ACs do not express appreciable levels of Abat (GABA transaminase) or Aldh1a1 (aldehyde dehydrogenase 1a1), the enzymes required for these pathways (Kim et al., 2015; Tritsch et al., 2012, 2014). Alternatively, S3-Gbx2+ ACs could release GABA without de novo synthesis. Midbrain dopaminergic lack expression of Gad1 and Gad2 expression, yet release GABA that is taken up from the extracellular environment by plasma membrane GABA transporters (Tritsch et al., 2014). S3-Gbx2+ ACs express relatively high levels of Slc6a1, which encodes the plasma membrane GABA transporter Gat1 (Figure 2C).
In addition to chemical neurotransmitters, many retinal neurons also express neuromodulators and neuropeptides. Our RNA-seq results revealed that the S3-Gbx2+ ACs highly express the neuropeptide gene Tachykinin1 (Tac1), that encodes several neuropeptides, the most common one being substance P (Figure 2C). Release of substance P from ACs can modulate the excitability of downstream retinal ganglion cells, at a slower timescale than most neurotransmitters, however (Zalutsky and Miller, 1990). The genetic identity of substance P-releasing ACs in the mouse retina is unknown, and several other AC subtypes also express Tac1 in addition to S3-Gbx2+ ACs (Yan et al., 2020). Ultimately, directly testing whether S3-Gbx2+ ACs use standard chemical neurotransmitters and/or neuromodulators will require identifying their downstream synaptic partners.
Many ACs exhibit homotypic coupling with neighboring ACs of the same subtype, as seen with AII and neuronal nitric oxide synthase-2 (nNOS-2) AC subtypes (Jacoby and Schwartz, 2018; Vaney, 1991). Coupling can spread signals laterally and may serve a “signal averaging” function in coupled cells. However, such signal averaging seems unlikely in S3-Gbx2+ ACs because we never observed homotypic coupling despite significant overlap in their dendritic arbors (Figure 7C). Rather, S3-Gbx2+ ACs are heterotypically coupled with neighboring BCs. Heterotypic coupling between ACs and BCs has been previously observed in glycinergic AII and A8 ACs with cone BCs (Vaney, 1991; Yadav et al., 2019). The spatial pattern of the S3-Gbx2+ AC:BC tracer coupling is unusual, as it never extends beyond the central 80% of the dendritic arbor of the S3-Gbx2+ AC. Although the physiological recordings (Figure 7B) indicate that electrical synapses contribute minimal excitatory input to S3-Gbx2+ ACs at photopic light levels, it remains possible that signaling between S3-Gbx2+ ACs and cone BCs through electrical synapses could become prominent under specific conditions. Tracer coupling to BCs increased in a subset of S3-Gbx2+ ACs during the application of a D1 receptor antagonist, which mimics low dopamine release that occurs under scotopic conditions (Figures 5D–5F and 5I). These results demonstrate that tracer coupling can be modulated and suggest that the physiological role of gap junctions may depend on the adaptation level. It is also possible that gap junction connections may mediate S3-Gbx2+ AC to excite ganglion cells indirectly by depolarizing BCs. In this situation, electrical synapses could amplify local signaling between S3-Gbx2+ ACs and BCs. While the BC subtype coupled to S3-Gbx2+ ACs is unknown, based on the stratification of coupled-BC axon terminals in the IPL, it is likely to include type 5 and possibly types 6 and 7a BCs (Ghosh et al., 2004). Future studies should be able to determine this connectivity with a combination of immunohistochemistry or genetic markers for BC subtypes. In addition, the identity of the connexins that mediate coupling between S3-Gbx2+ ACs and BC remains unknown. In our RNA-seq data, Gjd2 (Cx36), Gjc1 (Cx45), and Gje1 (Cx23) are expressed in S3-Gbx2+ ACs, implicating a number of potential targets.
Potential Visual Modalities Requiring Gbx2+ ACs
What are the functions of Gbx2+ ACs in the retina? While this remains an open question, the stratification pattern of Gbx2+ AC dendrites to specific lamina in the IPL restricts their potential synaptic partners and allows for some speculation. Each sublamina in the IPL contains the dendrites of a specific subset of RGC subtypes and responds to a specific set of visual modalities (Roska and Werblin, 2001). If S5-Gbx2 ACs are the CRH-1 subtype, as hypothesized above, we would expect that they provide inhibition onto “sustained” suppressed-by-contrast (SbC) RGCs and On aRGCs (also known as the M4 subtype of intrinsically photosensitive RGCs [ipRGCs]) (Estevez et al., 2012; Jacoby and Schwartz, 2018; Jacoby et al., 2015; Park et al., 2018). Sublamina 5 also contains the dendrites of the M2, M3, and M5 subtypes of ipRGCs, which are involved in visual modalities involved in light avoidance, pupillary reflex, and circadian rhythm behaviors (Schmidt et al., 2011; Sonoda et al., 2020).
In contrast to the S5-Gbx2+ ACs, the S3-Gbx2+ ACs do not resemble any previously described AC subtype. With dendrites that ramify in sublamina 3 of the IPL (Figure 1), the S3-Gbx2+ ACs could be connected to W3B RGCs that are involved in object motion detection (Krishnaswamy et al., 2015; Zhang et al., 2012) or other small-field RGCs involved in spatial vision (Jacoby and Schwartz, 2018). However, it is important to note that not all ACs and RGCs that co-stratify in an IPL sublamina are synaptically connected (Krishnaswamy et al., 2015). Furthermore, we currently lack a complete accounting of the dendritic stratification of the ≥40 RGC subtypes that have been defined molecularly, suggesting that there are other potential synaptic partners (Rheaume et al., 2018; Tran et al., 2019). Ultimately, the presence of functional connections between S3- and S5-Gbx2+ ACs and their potential postsynaptic targets will require paired recordings, which should be feasible now that we have genetic tools to prospectively identify Gbx2+ ACs.
Over the past few years, single-cell transcriptomics has greatly increased the cellular inventory of molecularly distinct neuronal subtypes. Going forward, we will need genetic tools to interrogate specific neuronal subtypes within a given neural circuit, as we have done in this study. The Gbx2CreERT2-IRES-EGFP line has allowed us to define the molecular, morphological, and physiological properties of two AC subtypes. In particular, the unusual properties of the S3-Gbx2+ AC subtype raise a number of intriguing questions about their form and function within the retina for future studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Requests for additional information and resources should be directed to the Lead Contact, Kevin Wright (wrighke@ohsu.edu).
Materials Availability
No mice or reagents were generated in this study. The mouse lines used in the study will be provided with permission from the investigator that generated them (see Key Resources Table). Questions and inquiries should be directed to the Lead Contact.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP (1:1000, IHC) | Abcam | Cat# AB6556; RRID: AB_305564 |
| Rabbit anti-DsRed (1:500, IHC) | Clontech | Cat# 632496; RRID: AB_10013483 |
| Goat anti-ChAT (1:500, IHC) | Millipore | Cat# AB144P; RRID: AB_11214092 |
| Rabbit anti-TFAP2 (1:1000, IHC) | DSHB (Univ. of Iowa) | Cat# 3B5; RRID: AB_528084 |
| Mouse anti-Pax6 (1:500, IHC) | DSHB (Univ. of Iowa) | Cat# PAX6; RRID: AB_528427 |
| Guinea pig anti-RBPMS (1:500, IHC) | PhosphoSolutions | Cat# 1832-RBPMS; RRID: AB_2492226 |
| Goat anti-Chx10 (1:500, IHC) | Santa Cruz Biotech | Cat# sc-21690; RRID: AB_2216006 |
| Rabbit anti-Calretinin (1:2000, IHC) | Swant | Cat# CG 1; RRID: AB_2619710 |
| Rabbit anti-GlyT1 (1:500, IHC) | Antibodies-Online | Cat# ABIN1841935; RRID: AB_2876781 |
| Mouse anti-GAD67 (1:500, IHC) | Millipore | Cat# MAB5406; RRID: AB_2278725 |
| Rabbit anti-NOS (1:1000, IHC) | ThermoFisher | Cat# 61–7000; RRID: AB_2313734 |
| Guinea pig anti-Vglut3 (1:2000, IHC) | Millipore | Cat# AB5421; RRID: AB_2187832 |
| Rabbit anti-Tyrosine hydroxylase (1:1000, IHC) | Millipore | Cat# AB152; RRID: AB_390204 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-AP4 (20 μM) | Tocris Bioscience | Cat# 0103 |
| D-AP5 (50 μM) | Abcam Biochemicals | Cat# 120003 |
| GYKI-53655 (50 μM) | Tocris Bioscience | Cat# 2555 |
| ACET (1 μM) | Tocris Bioscience | Cat# 2728 |
| Neurobiotin Tracer (60mM) | Vector Labs | Cat# SP-1120 |
| Streptavidin, Alexa Fluor 488 (1:500) | Fisher Scientific | Cat# S11223A |
| Papain (10U) | Roche | Cat# 10108014001 |
| Critical Commercial Assays | ||
| Absolutely RNA Nanoprep Kit | Agilent | Cat# 400753 |
| SMART-Seq Ultra Low Input RNA Kit | Takara | Cat# 634888 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Gbx2CreERT2-IRES-EGFP | Jackson Laboratory | JAX: 022135, MGI: 3840448 |
| Mouse: TIGRElsl-gfp (Ai140D, TIT2L-GFP-ICL-tTA2) | Jackson Laboratory | JAX: 030220, MGI: 5904001 |
| Mouse: Rosa26LSL-tdTomato (Ai9) | Jackson Laboratory | JAX: 007909, MGI: 3809523 |
| Mouse: NeuroD6Cre (NEXCre) | (Schwab et al., 2000) | MGI: 2668659 |
| Deposited Data | ||
| RNA-sequencing data | This manuscript | GEO (https://www.ncbi.nlm.nih.gov/geo/) Accession number: GSE157271 |
| Software/Algorithms | ||
| ZEN Blue | Zeiss | RRID: SCR_013672 |
| ImageJ/FIJI | (Schindelin et al., 2012) | https://fiji.sc/; RRID: SCR_002285 |
| WinDRP | (Rodieck, 1991) | N/A |
| Imaris | Bitplane | RRID: SCR_007370 |
| Prism 8 | Graphpad Software, Inc. | RRID: SCR_002798 |
| Igor Pro 8.02 | WaveMetrics, Inc. | RRID: SCR_000325 |
| FastQC | Simon Andrews (Babraham Institute) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/; RRID: SCR_014583 |
| MultiQC | (Ewels et al., 2016) | https://multiqc.info/; RRID: SCR_014982 |
| Trimmomatic | (Bolger et al., 2014) | https://github.com/timflutre/trimmomatic RRID: SCR_011848 |
| STAR (v2.7.3a) | (Dobin et al., 2013) | RRID: SCR_015899 |
| RNA-SeQC | (DeLuca et al., 2012) | RRID: SCR_005120 |
| R and R Studio | R | RRID: SCR_000432 |
Data and Code Availability
RNA-sequencing data generated in this manuscript are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), GEO accession number: GSE157271. No new code was generated in this study.
EXPERIMENTAL MODEL AND SUJECT DETAILS
Animals and Animal Procedures
Gbx2CreERT2-IRES-EGFP (Chen et al., 2009), NeuroD6Cre (NEXCre) (Schwab et al., 2000), Ai9/Rosa26LSL-tdTomato (Madisen et al., 2010), and Ai140D/TIGRETRE2-LSL-GFP, CAG-LSL-tTA2 (Daigle et al., 2018) mice were maintained on a C57BL/6J background. Mice of both sexes were used for all experiments. Tamoxifen was administered at E16, P0, and adults (> P28) ages. For E16 time points, 200μL of 5mg/mL tamoxifen and 2.5mg/mL progesterone dissolved in sunflower seed oil was administered to pregnant dam by oral gavage. Pregnancies were timed by the presence of a vaginal plug marked as E0.5. At P0 time points, 50 μL of 0.5mg/mL tamoxifen was injected with into the milk pouch of the mouse pups, as previously described (Pitulescu et al., 2010). At adult ages, 200μL of 5mg/mL tamoxifen was administered by oral gavage for at least two consecutive days to ensure complete recombination of the Cre-dependent reporter (Figure S1). All animal procedures were approved by Oregon Health and Science University Institutional Animal Care and Use Committee, the Institutional Animal Care and Use Committee of University of California (Berkeley, CA), and conformed to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Immunohistochemistry
Adult retinas were prepared for immunolabeling by removing the eyes from the head of a recently euthanized mouse. The cornea was removed and the eyes were fixed in 4% EM-grade paraformaldehyde (PFA) for 30 min at room temperature. Eyes were washed in PBS for 30 min post-fixation. Tissue for cryosections was cryo-protected in 10% and then 20% sucrose in PBS for 1 h each at 4°C. The lenses were removed and the eye cups were placed in cryomold with Optimal Cutting Temperature media and frozen. Retinas were sectioned at 20μm using a cryostat. Slide mounted retinal sections were washed for 10 mins in PBS and blocked with 2% normal donkey serum, 0.2% Triton X-100 in PBS for 30 mins. Sections were incubated in primary antibody in blocking buffer overnight at 4°C. Primary antibodies were used at the dilutions listed in the Key Resources Table. Sections were washed 3 times for 10 mins in PBS and incubated in secondary antibodies in blocking buffer for 2 hr at room temperature. All secondary antibodies were used at a 1:500 dilution. Retinal sections were washed three times in PBS for 10 mins with DAPI (1:5000) included in the first wash step. Tissue was mounted in Fluoromount-G (Southern Biotech) and coverslipped for imaging.
Retina flat-mounts were fixed and washed as described above. The retinas were isolated from the eye cup and flattened by making 3–4 equally spaced incisions from the edge of the retina. Retina flat-mounts were post-fixed in 4% PFA in PBS for 10 min to help maintain their shape. Retinas were washed in blocking buffer (4% normal donkey serum, 0.2% Triton X-100) 3 times for 30 min. Retinas were incubated in primary antibody in blocking buffer for 3 days at room temperature. Following incubation of primary antibody, retinas were washed 3 times for 30 min in blocking buffer. Retinas were then incubated in secondary antibody overnight at room temperature. Next, retinas were washed in PBS three times for 30 min, flattened on slides, and coverslipped in Fluoromount-G for imaging.
Fluorescence Image Acquisition
All retinal sections were imaged on a Zeiss Axio Imager M2 upright microscope equipped with an ApoTome2 using a 20× objective. Retinal flatmounts for mosaic analysis, single-cell morphology, and tracer coupling experiments were imaged on a Zeiss LSM 880 confocal microscope using a 40× objective. Images were acquired using the Zeiss Zen Imaging software for both microscopes.
Electrophysiology
Adult mice of either sex were dark adapted for 1–2 h. Animals were anesthetized with isoflurane before being euthanized via cervical dislocation. Subsequent to enucleation, all procedures were performed under infrared illumination. The retina and attached pigment epithelium were dissected free from the sclera and placed in a recording chamber under a microscope and continuously perfused (5ml/min) with Ames medium maintained at 34°C. Cells were visualized though a 40× water-immersion objective and Dodt contrast illumination. Fluorescent Gbx2+ ACs were targeted under 2-photon guidance (excitation wavelength: 920 nm) with a Ti:sapphire laser (Chameleon ultra II; Coherent). Correct targeting was confirmed by visualizing the Alexa dye in intracellular solution fill the soma and processes of the target cell.
Patch electrodes were pulled from borosilicate glass to a final resistance of 8–12 MΩ. For voltage-clamp recordings, pipettes were filled with an intracellular solution containing the following (in mM): 125 Cs-methanesulphonate, 7 CsCl, 10 Na-HEPES, 3 phosphocreatine-Na2, 1 EGTA, 2 Mg-ATP, 1 Na-GTP, 0.1 Alexa Fluor 488 hydrazide, and 3 QX-314 chloride. The solution was adjusted to pH7.35 using CsOH. Cesium was included in place of potassium to block voltage-gated potassium currents, thereby improving the voltage clamp at positive potentials. QX-314 was included to block voltage-gated sodium channels. For current-clamp recordings, all solution components were the same except potassium was used in place of cesium, and QX-314 was not included. Currents were sampled at 10 kHz and filtered at 2 kHz through the four-pole Bessel filter in an EPC-10 patch clamp amplifier (HEKA). Voltages were corrected for a liquid junction potential of −10mV.
Visual stimuli were produced using custom software based on PsychoPy routines (Peirce, 2007). The stimuli, generated on a Texas Instruments digital light projector (DLP; Lightcrafter 4500), were projected onto the photoreceptor layer through a 10× water immersion objective (0.3 NA, Olympus). The DLP intensity was linearized using a calibrated lookup table. DLP intensity was attenuated using neutral density filters to produce a gray adapting background flux of ~3.4 × 105 photons/μm2/s. Stimuli were first aligned to the receptive field center of each cell using a series of 100 × 1000 mm vertical and horizontal bars to locate the cell’s maximal response. All subsequent stimuli were centered on the coordinate of maximum response. Receptive field sizes were estimated from area-response data and fit to a difference of Gaussians function:
where R is the peak response evoked by a stimulus of diameter d, Kc and Ks are the amplitudes of the excitatory and inhibitory components, respectively, and σc and σs are their space constants.
Pharmacological Agents
Drugs were added to the perfusion solution. The following agents were used: L-(+)-2-amino-4-phophonobutyric acid (L-AP4; 20 μM; Tocris Bioscience, catalog #0103), D-(−)-2-amino-5-phophonopentanoic acid (D-AP5; 50 μM; Abcam Biochemicals, catalog #120003), 1-(4-aminophenyl)-3-methylcarbamyl-4-methyl-3,4-dihydro-7,8-meth- ylenedioxy-5H-2,3-benzodiazepine hydrochloride [GYKI-53655 (GYKI); 50 μM;Tocris Bioscience catalog #2555), (S)-1-(2-Amino-2-carboxyethyl)-3-(2-carboxy-5-phenylthiophene-3-yl-methyl)-5-methylpyrimidine-2,4-dione (ACET; 1 μM; Tocris Bioscience, catalog #2728).
Data Analysis
Light-evoked synaptic conductances were calculated as described previously (Taylor and Vaney, 2002). Briefly, current–voltage (I–V) relations were measured at 10 ms intervals over a range of voltage steps from −90 +50 mV in 20mV increments. The total light-evoked conductance was calculated as the difference between the I–V relation at each time point and the “leak” I–V relation measured just before the onset of the light stimulus. To avoid errors in calculating the net light-evoked currents due to a sloping baseline during positive voltage steps, a single exponential trend was subtracted from the current traces for each voltage step before t. he leak subtraction. The excitatory and inhibitory conductances could then be calculated at each time point using the observed I–V reversal potential along with the cation and chloride reversal potentials.
Tracer Coupling
Gbx2CreERT2-IRES-EGFP; Rosa26LSL-tdTomato mice were used to target tdTomato positive neurons in the GCL for electroporation of Neurobiotin, as previously described (Kanjhan and Vaney, 2008; Sivyer and Vaney, 2010). In isolated preparations of retina bathed in Ames medium at room temperature, tdTomato positive neurons were lose seal patched with an intracellular solution containing (in mM): 120 K-gluconoate; 6 KCl;10 HEPES Na;10 Phosphocreatine-Na2; 60 Neurobiotin-Cl; 2 ATP; 0.5 GTP pH balanced to 7.2 with KOH. Once a seal was established the HEKA EPC-800 amplifier was switched to current clamp mode and ACs were electro-porated by applying 0.5–1nA pulses for 0.5 s at 1 Hz. Following electroporation, retinae were incubated in Ames medium for 45 mins at 35 C before being transferred to room temperature Ames, mounted on nitrocellulose filter paper and fixed in cold paraformaldehyde in PBS for 30 mins. Retinas were then wash twice with 0.2% Triton in 1× PBS and incubated with 10 μg/mL Streptavidin Alexa Fluor 488 (Thermo Fisher) at room temperature overnight. Before imaging, stained retinas were washed three times in 1× PBS before being mounted on a glass coverslip.
Retina Dissociation and FACS
Retinas from P6 or P7 mouse pups were dissected and isolated in 1× Hank’s Balanced Salt Solution (HBSS). Cells were dissociated into a single-cell suspension by incubating the retinas in 1mL of HBSS containing 10U of papain (Roche) and 200μM cysteine. Retinal tissue was incubated at 37°C for 30 mins. After papain incubation, the tissue was pelleted using a bench top microfuge and washed twice with 1mL of HBSS. Cells were gently dissociated in 500μL of HBSS by flicking the tube. Dissociated cells were passed through a cell strainer (35μm nylon mesh) to remove any cell aggregates. DNase was added to the solution and the samples were placed on ice until sorted. Gbx2+ amacrine cells were sorted into two groups S3-cells that were both GFP+ and tdTomato+ and S5-cells that were only tdTomato+. All fluorescence activated cell sorting (FACS) was completed in the OHSU Flow Cytometry Core using a BD InFlux equipped with 488nm and 561nm laser lines. All cells were sorted directly into cell lysis buffer and RNA was isolated with Agilent Absolutely RNA Nanoprep Kit.
RNaseq Library Preparation and Sequencing
The cDNA libraries for used for sequencing of 4 total RNA samples were synthesized using a SMART-Seq Ultra Low Input RNA kit (Takara) in the OHSU Massively Parallel Sequencing Shared Resource Core Facility. Two of the cDNA libraries contained tdTomato+/GFP+ cells (S3-Gbx2+ ACs) and two samples contained the cDNA libraries for tdTomato+ only cells (S5-Gbx2+ ACs). Quality and quantity of the cDNA libraries was determined on a Bioanalyzer. For multiplex sequencing, all four cDNA sample libraries were loaded on a single lane on a HiSeq 2500 sequencer (Illumina). Libraries were sequenced to a depth of 45–55 million reads per sample. Alignment rate of total reads was >97% across all samples.
QUANTIFICATION AND STATISTICAL ANALYSIS
Co-localization Analysis
All co-localization experiments used retinal sections from the Gbx2CreERT2-IRES-EGFP; Rosa26LSL-tdTomato mouse line. Co-localization was determined by antibody labeling within the soma of the Gbx2+ amacrine cell bodies. Images were collected and analyzed as z stacks to confirm the antibody labeling was within the soma of the correct z-depth. Offline analysis was completed using FIJI (Schindelin et al., 2012).
Mosaic Cell Spacing Analysis
To analyze cell density and mosaic cell spacing, we used a 500μm × 500μm area of tissue from 4–8 locations within a single retina. Retina orientation was maintained to make spatial measurements in dorsal, ventral, temporal, and nasal areas of the retina. In addition, measurements in the peripheral and central regions of the retina made in ROIs ~200μm from the peripheral edge or optic nerve head respectively. Cell counts and X-Y coordinates were measured offline in FIJI (Schindelin et al., 2012) and density recovery profiles were obtained by analysis completed in WinDRP (Rodieck, 1991).
Neuron Morphology Analysis
For analysis of dendrite stratification in cross-section, we made measurements of fluorescent intensity along IPL depth using FIJI. These values were binned into 5% increments along the IPL depth using the FIJI plugin, IPLaminator (Li et al., 2016). For analysis of dendritic arbor morphology in retinal flatmounts we analyzed isolated single cells labeled using either sparse expression in Gbx2CreERT2-IRES-EGFP; TigreLSL-GFP mice or by targeted cell fills using Alexa Fluor 488 hydrazide (Fisher Scientific). Offline tracing and analysis of dendritic arbors were made using the Filaments plugin in Imaris (Bitplane). Dendrite density (L/A) was calculated by dividing dendrite length (L) over dendrite area (A). Coverage factor was calculated by dividing the dendrite area over the cell density of Gbx2+ ACs in the INL and GCL for each subtype. Symmetry Index was calculated by subtracting the sums of missing dendrite coverage from 360 and then divided by 360, as previously described (Sun et al., 2013). Dendrite orientation was determined by the direction of the longest dendritic branch of a single cell and values were binned into 8 different groups based on the cardinal directions.
RNA-seq Analysis
Differential expression analysis was performed by the ONPRC Bioinformatics and Biostatistics Core. The quality of the raw sequencing files was evaluated using FastQC combined with MultiQC (https://multiqc.info/) (Ewels et al., 2016). Trimmomatic was used to remove any remaining Illumina adapters (Bolger et al., 2014). Reads were aligned to Ensembl’s GRCm38 along with its corresponding annotation, release 99. The program STAR (v2.7.3a) was used to align the reads to the genome (Dobin et al., 2013). STAR has been shown to perform well compared to other RNA-seq aligners (Engström et al., 2013). Since STAR utilizes the gene annotation file, it also calculated the number of reads aligned to each gene. RNA-SeQC (DeLuca et al., 2012) and another round of MultiQC were utilized to ensure alignments were of sufficient quality.
Gene-level differential expression analysis was performed in open source software R (R Core Team, https://www.R-project.org/). Gene-level raw counts were filtered to remove genes with extremely low counts in many samples following the published guidelines (Chen et al., 2016), normalized using the trimmed mean of M-values method (TMM) (Oshlack et al., 2010), and transformed to log-counts per million with associated sample wise quality weight and observational precision weights using voom method (Law et al., 2014). Gene-wise linear models comparing the cell types (tdTom+ versus GFP+/tdTom+) were employed for differential expression analyses using limma with empirical Bayes moderation (Ritchie et al., 2015) and false discovery rate (FDR) adjustment (Benjamini and Hochberg, 1995).
RNaseq datasets were organized and displayed into gene families with R Studio and the START app (Nelson et al., 2017); and using the reference gene groups determined by the HUGO Gene Nomenclature Committee at the European Bioinformatics Institute (https://www.genenames.org). All graphs displaying RNaseq data were made using the Prism 8 Software (Graphpad Software, Inc.).
Statistics
For each experiment and time point a minimum of 3 retinas from three different mice were analyzed. For analysis of neuron morphology and tracer coupling, at least 5 neurons were analyzed from at least 3 animals. For all datasets, the variance was reported as mean ± SEM. Each dataset was first tested for normality. Analysis between two groups was completed by using unpaired Student’s t test (parametric) or Mann-Whitney U test (nonparametric). For analysis between more than two groups, we used either a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test (parametric) or Kruskal-Wallis with Dunn’s multiple comparison test (nonparametric). All statistical significance tests were completed using Prism 8 Software (Graphpad Software, Inc.) and Igor Pro 8.02 (WaveMetrics, Inc.) for electrophysiological analyses.
Supplementary Material
Highlights.
Gbx2 identifies two amacrine cell interneurons in the mouse retina
The Gbx2+ subtypes are molecularly, morphologically, and physiologically distinct
One Gbx2+ subtype lacks classical neurotransmitters but has electrical synapses
ACKNOWLEDGMENTS
We would like to thank the members of the Wright, Taylor, and Sivyer laboratories for their assistance and discussion throughout the course of this study. We also thank Dr. Kelly Monk and Dr. Martin Riccomagno for their comments on the manuscript. This work was supported by NIH grant R01 NS091027, a Whitehall Foundation research grant, and an OHSU University Shared Resources Grant, to K.M.W.; NIH grant F32 EY029974, the Collins Medical Trust, and the Knights Templar Eye Foundation, to P.C.K.; NIH grant T32 NS007466, to J.L.; NIH grants R01 EY022070 and P30 EY003176, to W.R.T.; and NIH grant P30 EY010572 and unrestricted departmental funding from Research to Prevent Blindness (New York, NY) and support from the donors of National Glaucoma Research, a program of BrightFocus Foundation, to B.S. Confocal microscopy and analysis was performed in the OHSU Advance Light Microscopy Core supported by NIH grant P30 NS061800. Cell sorting was performed in the OHSU Flow Cytometry Shared Resource, and the core is supported by the Knight NCI Cancer Center Support Grant. Short-read sequencing assays were performed by Dr. Amy Carlos and Dr. Robert Searles of the OHSU Massively Parallel Sequencing Shared Resource. The authors acknowledge the support of Dr. Suzanne Fei and Dr. Lina Gao of the Oregon National Primate Research Center Bioinformatics and Biostatistics Core, which is funded in part by NIH grant OD P51 OD011092.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108382.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Badea TC, and Nathans J (2004). Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J. Comp. Neurol 480, 331–351. [DOI] [PubMed] [Google Scholar]
- Baden T, Berens P, Franke K, Román Rosón M, Bethge M, and Euler T (2016). The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- Bloomfield SA, and Völgyi B (2009). The diverse functional roles and regulation of neuronal gap junctions in the retina. Nat. Rev. Neurosci 10, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecha N, Johnson D, Peichl L, and Wassle H (1988). Cholinergic amacrine cells of the rabbit retina contain glutamate decarboxylase and gamma-aminobutyrate immunoreactivity. Proc. Natl. Acad. Sci. USA 85, 6187–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee M, Li K, Chen L, Maisano X, Guo Q, Gan L, and Li JY (2012). Gbx2 regulates thalamocortical axon guidance by modifying the LIM and Robo codes. Development 139, 4633–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guo Q, and Li JY (2009). Transcription factor Gbx2 acts cell-non-autonomously to regulate the formation of lineage-restriction boundaries of the thalamus. Development 136, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chatterjee M, and Li JY (2010). The mouse homeobox gene Gbx2 is required for the development of cholinergic interneurons in the striatum. J. Neurosci 30, 14824–14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lun AT, and Smyth GK (2016). From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 5, 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry TJ, Trimarchi JM, Stadler MB, and Cepko CL (2009). Development and diversification of retinal amacrine interneurons at single cell resolution. Proc. Natl. Acad. Sci. USA 106, 9495–9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, and Getz G (2012). RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JS (2017). Inhibitory Interneurons in the Retina: Types, Circuitry, and Function. Annu. Rev. Vis. Sci 3, 1–24. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström PG, Steijger T, Sipos B, Grant GR, Kahles A, Rätsch G, Goldman N, Hubbard TJ, Harrow J, Guigó R, and Bertone P; RGASP Consortium (2013). Systematic evaluation of spliced alignment programs for RNA-seq data. Nat. Methods 10, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez ME, Fogerson PM, Ilardi MC, Borghuis BG, Chan E, Weng S, Auferkorte ON, Demb JB, and Berson DM (2012). Form and function of the M4 cell, an intrinsically photosensitive retinal ganglion cell type contributing to geniculocortical vision. J. Neurosci 32, 13608–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Käller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Bujan S, Haverkamp S, Feigenspan A, and Wassle H (2004). Types of bipolar cells in the mouse retina. J Comp Neurol 469, 70–82. [DOI] [PubMed] [Google Scholar]
- Hampson EC, Vaney DI, and Weiler R (1992). Dopaminergic modulation of gap junction permeability between amacrine cells in mammalian retina. J. Neurosci 12, 4911–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, and Wässle H (2004). Characterization of an amacrine cell type of the mammalian retina immunoreactive for vesicular glutamate transporter 3. J. Comp. Neurol 468, 251–263. [DOI] [PubMed] [Google Scholar]
- He L, Gulyanon S, Mihovilovic Skanata M, Karagyozov D, Heckscher ES, Krieg M, Tsechpenakis G, Gershow M, and Tracey WD Jr. (2019). Direction Selectivity in Drosophila Proprioceptors Requires the Mechanosensory Channel Tmc. Curr. Biol 29, 945–956.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Turaga SC, Jain V, Seung HS, and Denk W (2013). Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174. [DOI] [PubMed] [Google Scholar]
- Jacoby J, and Schwartz GW (2018). Typology and Circuitry of Suppressed-by-Contrast Retinal Ganglion Cells. Front. Cell. Neurosci 12, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby J, Zhu Y, DeVries SH, and Schwartz GW (2015). An Amacrine Cell Circuit for Signaling Steady Illumination in the Retina. Cell Rep. 13, 2663–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J, Sherry DM, Liu X, Fremeau RT Jr., Seal RP, Edwards RH, and Copenhagen DR (2004). Vesicular glutamate transporter 3 expression identifies glutamatergic amacrine cells in the rodent retina. J. Comp. Neurol 477, 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjhan R, and Vaney DI (2008). Semi-loose seal Neurobiotin electroporation for combined structural and functional analysis of neurons. Pflugers Arch. 457, 561–568. [DOI] [PubMed] [Google Scholar]
- Kay JN, Voinescu PE, Chu MW, and Sanes JR (2011). Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate. Nat. Neurosci 14, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Eglen SJ, and Reese BE (2020). From random to regular: variation in the patterning of retinal mosaics. J. Comp. Neurol 528, 2135–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, and Sanes JR (2008). Molecular identification of a retinal cell type that responds to upward motion. Nature 452, 478–482. [DOI] [PubMed] [Google Scholar]
- Kim JI, Ganesan S, Luo SX, Wu YW, Park E, Huang EJ, Chen L, and Ding JB (2015). Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science 350, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, and Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen L, Chen M, Ye M, Seal RP, and Zhou ZJ (2014). An unconventional glutamatergic circuit in the retina formed by vGluT3 amacrine cells. Neuron 84, 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Sanes JR, and Kay JN (2015). Development of dendritic form and function. Annu. Rev. Cell Dev. Biol 31, 741–777. [DOI] [PubMed] [Google Scholar]
- Li S, Woodfin M, Long SS, and Fuerst PG (2016). IPLaminator: an ImageJ plugin for automated binning and quantification of retinal lamination. BMC Bioinformatics 17, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, and Masland RH (2006). Populations of wide-field amacrine cells in the mouse retina. J. Comp. Neurol 499, 797–809. [DOI] [PubMed] [Google Scholar]
- Liu J, and Sanes JR (2017). Cellular and Molecular Analysis of Dendritic Morphogenesis in a Retinal Cell Type That Senses Color Contrast and Ventral Motion. J. Neurosci 37, 12247–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Reggiani JDS, Laboulaye MA, Pandey S, Chen B, Rubenstein JLR, Krishnaswamy A, and Sanes JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nat. Neurosci 21, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu B, Ellisor D, and Zervas M (2011). The lineage contribution and role of Gbx2 in spinal cord development. PLOS ONE 6, e20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil MA, and Masland RH (1998). Extreme diversity among amacrine cells: implications for function. Neuron 20, 971–982. [DOI] [PubMed] [Google Scholar]
- MacNeil MA, Heussy JK, Dacheux RF, Raviola E, and Masland RH (1999). The shapes and numbers of amacrine cells: matching of photofilled with Golgi-stained cells in the rabbit retina and comparison with other mammalian species. J. Comp. Neurol 413, 305–326. [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. (2015). Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallika C, Guo Q, and Li JY (2015). Gbx2 is essential for maintaining thalamic neuron identity and repressing habenular characters in the developing thalamus. Dev. Biol 407, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sklenar J, Barnes AP, and Minnier J (2017). The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics 33, 447–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk GS, Hoon M, Wong RO, and Detwiler PB (2013). Inhibitory inputs tune the light response properties of dopaminergic amacrine cells in mouse retina. J. Neurophysiol 110, 536–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, and Bloomfield SA (2018). Plasticity of Retinal Gap Junctions: Roles in Synaptic Physiology and Disease. Annu. Rev. Vis. Sci 4, 79–100. [DOI] [PubMed] [Google Scholar]
- Oshlack A, Robinson MD, and Young MD (2010). From RNA-seq reads to differential expression results. Genome Biol 11, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJH, Pottackal J, Ke JB, Jun NY, Rahmani P, Kim IJ, Singer JH, and Demb JB (2018). Convergence and Divergence of CRH Amacrine Cells in Mouse Retinal Circuitry. J. Neurosci 38, 3753–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JW (2007). PsychoPy–Psychophysics software in Python. J. Neurosci. Methods 162, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron 95, 869–883.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Shekhar K, Yan W, Herrmann D, Sappington A, Bryman GS, van Zyl T, Do MTH, Regev A, and Sanes JR (2019). Molecular Classification and Comparative Taxonomics of Foveal and Peripheral Cells in Primate Retina. Cell 176, 1222–1237.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, James RE, Yan W, Kay JN, Kolodkin AL, and Sanes JR (2020). Binary Fate Choice between Closely Related Interneuronal Types Is Determined by a Fezf1-Dependent Postmitotic Transcriptional Switch. Neuron 105, 464–474.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu ME, Schmidt I, Benedito R, and Adams RH (2010). Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc 5, 1518–1534. [DOI] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, Sun L, Robson P, and Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun 9, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW (1991). The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis. Neurosci 6, 95–111. [DOI] [PubMed] [Google Scholar]
- Roska B, and Werblin F (2001). Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature 410, 583–587. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The types of retinal ganglion cells: current status and implications for neuronal classification. Annu. Rev. Neurosci 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, Chen SK, and Hattar S (2011). Intrinsically photosensitive retinal ganglion cells: many subtypes, diverse functions. Trends Neurosci. 34, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, and Nave KA (2000). Neuronal basic helix-loop-helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J. Neurosci 20, 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. (2016). Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, and Roska B (2009). Genetic address book for retinal cell types. Nat. Neurosci 12, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Sivyer B, and Vaney DI (2010). Dendritic morphology and tracer-coupling pattern of physiologically identified transient uniformity detector ganglion cells in rabbit retina. Vis. Neurosci 27, 159–170. [DOI] [PubMed] [Google Scholar]
- Sonoda T, Okabe Y, and Schmidt TM (2020). Overlapping morphological and functional properties between M4 and M5 intrinsically photosensitive retinal ganglion cells. J. Comp. Neurol 528, 1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, Yau KW, Feller MB, and Kolodkin AL (2013). On and off retinal circuit assembly by divergent molecular mechanisms. Science 342, 1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, and Vaney DI (2002). Diverse synaptic mechanisms generate direction selectivity in the rabbit retina. J. Neurosci 22, 7712–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, et al. (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 104, 1039–1055.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Ding JB, and Sabatini BL (2012). Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature 490, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Oh WJ, Gu C, and Sabatini BL (2014). Midbrain dopamine neurons sustain inhibitory transmission using plasma membrane uptake of GABA, not synthesis. eLife 3, e01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney DI (1991). Many diverse types of retinal neurons show tracer coupling when injected with biocytin or Neurobiotin. Neurosci. Lett 125, 187–190. [DOI] [PubMed] [Google Scholar]
- Vaney DI, and Weiler R (2000). Gap junctions in the eye: evidence for heteromeric, heterotypic and mixed-homotypic interactions. Brain Res. Brain Res. Rev 32, 115–120. [DOI] [PubMed] [Google Scholar]
- Vaney DI, and Young HM (1988). GABA-like immunoreactivity in cholinergic amacrine cells of the rabbit retina. Brain Res. 438, 369–373. [DOI] [PubMed] [Google Scholar]
- Whitney IE, Keeley PW, St John AJ, Kautzman AG, Kay JN, and Reese BE (2014). Sox2 regulates cholinergic amacrine cell positioning and dendritic stratification in the retina. J. Neurosci 34, 10109–10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SC, Tetenborg S, and Dedek K (2019). Gap Junctions in A8 Amacrine Cells Are Made of Connexin36 but Are Differently Regulated Than Gap Junctions in AII Amacrine Cells. Front. Mol. Neurosci 12, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Laboulaye MA, Tran NM, Whitney IE, Benhar I, and Sanes JR (2020). Mouse Retinal Cell Atlas: Molecular Identification of over Sixty Amacrine Cell Types. J. Neurosci 40, 5177–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalutsky RA, and Miller RF (1990). The physiology of substance P in the rabbit retina. J. Neurosci 10, 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, and Sanes JR (2017). Neuronal cell-type classification: challenges, opportunities and the path forward. Nat. Rev. Neurosci 18, 530–546. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kim IJ, Sanes JR, and Meister M (2012). The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proc. Natl. Acad. Sci. USA 109, E2391–E2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Xu J, Hauswirth WW, and DeVries SH (2014). Genetically targeted binary labeling of retinal neurons. J. Neurosci 34, 7845–7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data generated in this manuscript are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), GEO accession number: GSE157271. No new code was generated in this study.


