Abstract
Cluster of differentiation (CD) 166 or activated leukocyte cell adhesion molecule (ALCAM) is a transmembrane molecule known to be an intercellular adhesion factor. The expression and function of ALCAM in medulloblastoma (MB), a pediatric brain tumor with highly advanced molecular genetics, remains unclear. Therefore, this study aimed to clarify the significance and functional role of ALCAM expression in MB. ALCAM expression in 45 patients with MB was evaluated by immunohistochemical analysis of formalin-fixed paraffin-embedded clinical specimens and the relationship between ALCAM expression and pathological type/molecular subgroup, such as WNT, SHH, Group 3, and Group 4, was examined. Eight ALCAM positive (18%), seven partially positive (16%), and 30 negative (67%) cases were detected. All seven cases of the WNT molecular subgroup were ALCAM positive and ALCAM expression strongly correlated with this subgroup (P < 0.0001). In addition, functional studies using MB cell lines revealed ALCAM expression affected proliferation and migration as a positive regulator in vitro. However, ALCAM silencing did not affect survival or the formation of leptomeningeal dissemination in an orthotopic mouse model, but did induce a malignant phenotype with increased tumor cell invasion at the dissemination sites (P = 0.0029). In conclusion, our results revealed that ALCAM exhibited highly specific expression in the WNT subgroup of MB. Furthermore, we demonstrated that the cell kinetics of MB cell lines can be altered by the expression of ALCAM.
Introduction
Medulloblastoma (MB) is the most common pediatric malignant brain tumor of the cerebellum. Histologically, MB is an embryonal tumor that may differentiate into cells of neural lineages and is classified as a grade IV tumor by the World Health Organization (WHO). There are several MB histological subtypes, including classic, desmoplastic/nodular, MB with extensive nodularity (MBEN), and large cell/anaplastic. Patients with MB are clinically stratified into average or high-risk groups according to their age, metastatic status, and the presence of residual tumor following resection [1]. However, these classifications do not account for MB heterogeneity. Recently, integrated genomic analysis of MB showed that MB consists of at least four distinct molecular subgroups, WNT, SHH, Group 3, and Group 4 [2–4]. This molecular classification of MB reflects distinct demographics and clinical features, including prognosis, transcriptomes, and genetics [2–5], and have been incorporated into the WHO classification of tumors of the central nervous system as revised in 2016 [6]. This new classification also provides for additional clinical risk stratification [5, 7]. However, a further understanding of disease based on the molecular subgroups and the subsequent development of treatment strategies is necessary.
Activated leukocyte cell adhesion molecule (ALCAM) is a transmembrane glycoprotein that belongs to the immunoglobulin superfamily. ALCAM has been identified in a wide variety of tissues and cells, such as selected epithelia, lymphoid and myeloid cells, fibroblasts, neurons, and hepatocytes and is also referred to as CD166, CD6 ligand, MEMD, SB10 antigen, and HCA [8]. ALCAM is involved in cell-cell adhesion, either by homophilic (ALCAM-ALCAM) or heterophilic (ALCAM-CD6) interaction, and is also involved in organ development, neurogenesis, hematopoiesis, and immune responses [8]. In cancer, ALCAM expression is a prognostic marker for various tumor types [9]. For example, membranous ALCAM expression and ALCAM overexpression are independent markers of poor prognosis in colorectal carcinoma and pancreatic cancer, respectively [10, 11]. Contrarily, decreased ALCAM expression in breast cancer and loss of ALCAM membrane expression in ovarian cancer have been correlated with poor prognosis [12, 13]. Therefore, the impact of ALCAM expression on prognosis seems to depend on the cancer type, and in some types of cancer, membranous versus cytoplasmic ALCAM expression should also be considered [9]. In vitro and in vivo studies have demonstrated that ALCAM is involved in migration, invasion, and stemness in several cancers [14–19]. However, the functional significance of ALCAM in cancer is not consistent, with the differences depending on both the cancer type and tumor microenvironment.
The expression of ALCAM in MB has been previously reported, but its relevance to the molecular subgroups or histological classification has not been examined [20]. Furthermore, no study has analyzed the functional role of ALCAM in MB. In the current study, we retrospectively evaluated ALCAM expression in samples from patients with MB and examined the correlation between ALCAM expression and the MB molecular subgroups and histological subtypes. In addition, we investigated the functional role of ALCAM in MB using in vitro assays and an in vivo orthotopic mouse model.
Materials and methods
Clinical samples and patient characteristics
We retrospectively recruited sample cases that a) had available specimens surgically removed between 1996 and 2020 and b) were diagnosed with MB at the original treating institute. All cases were then centrally reviewed by a senior board-certified neuropathologist (Y.K.) for inclusion in the study. Forty-five case specimens of MB and their clinicopathologic information were obtained from 11 collaborating institutions (Osaka University Graduate School of Medicine, Osaka National Hospital, Kansai Medical University, Wakayama Medical University School of Medicine, Ehime University School of Medicine, Kitano Hospital, Takatsuki General Hospital, Kansai Rosai Hospital, Osaka City University Graduate School of Medicine, Osaka City General Hospital, and Hyogo College of Medicine) in the Kansai Molecular Diagnosis Network for CNS Tumors [21]. Approval of the study was obtained from the Institutional Review Boards (IRBs) of Osaka University Graduate School of Medicine (approval number: 13244), Osaka National Hospital (approval number: 713), and all the collaborative institutes. For all cases, either written informed consent was obtained or its requirement was waived by the IRB with a public announcement on the institution website. Immunohistochemistry and data analysis were performed at Osaka University Graduate School of Medicine and genetic analysis was performed at Osaka National Hospital and The Hospital for Sick Children. MB was histologically classified based on hematoxylin-eosin (HE) and reticulin silver staining as classic, desmoplastic/nodular, MBEN, or large cell/anaplastic subtype according to the 2016 WHO classification.
Immunohistochemistry
Six-micrometer sections of formalin-fixed paraffin-embedded (FFPE) tissues were used for immunohistochemistry. Heat-induced antigen retrieval was performed using a pressure cooker in 0.01 M citrate buffer (pH 6.0) for 10 min. Sections were incubated with a primary antibody against CD166/ALCAM [EPR2759(2); Abcam, Cambridge, MA, USA; 1:100 dilution] and β-catenin (BD Bioscience, San Jose, CA, USA; 1:100 dilution) at 4 °C overnight. Histofine Simple Stain MAX-PO (Nichirei, Tokyo, Japan) was used as a secondary antibody. The antibody complexes were visualized using the Dako Liquid DAB + Substrate Chromogen System (Dako, Carpinteria, CA, USA) and the sections were then counterstained with hematoxylin.
To identify tumor cells in the orthotopic mouse model, a primary antibody to human STEM121 (Takara Bio Inc., Shiga, Japan; 1:1,000 dilution) was used with POD Conjugate Set Anti Mouse, For Mouse Tissue reagent (Takara Bio Inc.).
When we analyzed ALCAM expression, we also evaluated whether the membrane or cytoplasm of the tumor cells were stained. The immunostaining of ALCAM was evaluated as the proportion of ALCAM-positive tumor cells in a representative area of tumor in the section. The cutoff values for the subdivision of the ALCAM staining was set at < 1% for negative staining, 1–25% for partially positive staining, and > 25% for positive staining. β-catenin immunostaining of tumor cells was considered positive only in cases of nuclear staining.
For ALCAM immunohistochemical staining, the identification of optimal cutoff points for the proportion of ALCAM-positive tumor cells in the WNT molecular subgroup was evaluated using receiver operating characteristic (ROC) curves and assessment of the area under the ROC curve (AUC).
Molecular subgrouping and genetic analysis
All samples were analyzed with molecular diagnostic techniques using the nanoString nCounter system (NanoString Technologies Inc., Seattle, WA, USA) [3, 4, 22] and by Sanger sequencing. The CTNNB1 mutation hotspot region in exon 3 was amplified and sequenced using forward primer 5’-TGGAACCAGACAGAAAAGCG-3’ and reverse primer 5’-ACAGGACTTGGGAGGTATCC-3’. Cases classified by the nanoString nCounter analysis or that presented with the CTNNB1 mutation and nuclear staining of β-catenin were defined as WNT subtype.
Quantitative PCR (qPCR)
Total RNA was extracted from cultured cells and clinical samples using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA) or QIAzol Lysis Reagent (Qiagen) and reverse transcribed using PrimeScript RT Master Mix (Takara Bio Inc.) following the manufacturers’ protocols. qPCR was performed using TB Green Premix Ex Taq II (Takara Bio Inc.) and the Applied Biosystems ViiA7 Real-Time PCR System (Thermo Fisher Scientific Inc., Waltham, MA, USA). To measure ALCAM expression, the following primer pair was used: 5’-TCCTGCCGTCTGCTCTTCT-3’ (forward) and 5’-TTCTGAGGTACGTCAAGTCGG-3’ (reverse) [19]. As an internal reference for normalization, ACTB expression was measured using the following primer pair: 5’-CACCAACTGGGACGACAT-3’ (forward) and 5’-ACAGCCTGGATAGCAACG-3’ (reverse). Expression was measured relative to Human Brain, Cerebellum Total RNA (Takara Bio) and relative quantification analyses were performed using the ΔΔCT method [23].
R2: Genomics analysis and visualization platform dataset analysis
To validate ALCAM expression in MB, the Cavalli-763 MB dataset [24] from the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) was used. ALCAM gene expression, patient age, histological variant, and molecular subgroup data were extracted from the dataset.
Cell lines
Four established human MB cell lines were used in this study, Daoy, D341, ONS-76, and D283. Daoy, D341, and D283 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). ONS-76 was purchased from the Japanese Collection of Research Bioresources Cell Bank. Daoy and ONS-76 were cultured as adherent cells and D341 and D283 were cultured as cell suspensions. Daoy cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS); D341 cells were cultured in Minimum Essential Medium (MEM) supplemented with 20% FBS and 1% Non-Essential Amino Acids Solution (Gibco, Grand Island, NY, USA); ONS-76 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, and D283 cells were cultured in MEM supplemented with 10% FBS. Cultured cells were maintained at 37°C in a 5% CO2 atmosphere.
Flow cytometric analysis
Cells from subconfluent monolayer cultures were suspended in phosphate-buffered saline (PBS) and incubated with phycoerythrin (PE)-conjugated mouse anti-human CD166/ALCAM antibody (3A6; BD Bioscience). The labeled cells were then analyzed on a FACS Aria III flow cytometer (BD Bioscience) according to the manufacturer’s instructions. Analysis of the flow cytometry data was performed using FlowJo, version 10 software (BD Bioscience). ALCAM fluorescence intensity is reported as the intensity ratio (IR) obtained as follows: IR = ALCAM mean fluorescence intensity / negative control mean fluorescence intensity.
Knockdown of ALCAM expression
To generate cell lines in which ALCAM expression had been stably knocked down, we used the MISSION® RNAi system and lentiviral pLKO-puro vector (Sigma Aldrich, St Louis, MO, USA). The sequences of the shRNAs were as follows: shALCAM1, 5’-CCGGCAGCCATGATAATAGGTCATACTCGAGTATGACCTATTATCATGGCTGTTTTTG-3’ and shALCAM2, 5’-CCGGCTTCGATCTAGCCCGTCATTTCTCGAGAAATGACGGGCTAGATCGAAGTTTTTG-3’. Non-target shRNA Control (Sigma Aldrich) was used as negative control shRNA. Lentivirus was produced by transfection of the lentiviral vector and the Lentiviral Packaging Mix (Sigma Aldrich) in 293T cells. Daoy and ONS-76 cells were then infected with the lentivirus expressing the shRNAs. Knockdown of ALCAM was confirmed by flow cytometry and qPCR after puromycin selection. Before performing the in vitro and in vivo assays, the 10–15% cells with lower ALCAM expression were sorted from the heterogeneous Daoy cell population in which ALCAM had been knocked down (expressing different levels of the protein) by FACS using PE-conjugated mouse anti-human ALCAM antibody.
ALCAM overexpression
The expression vector ALCAM cDNA ORF clone pCMV3-C-GFPSpark (Sino Biological, Beijing, China) was used for ALCAM overexpression. D341 cells were transfected with the ALCAM cDNA plasmid using a lipofection method and Sinofection (Sino Biological). pCMV3-C-GFPSpark Control Vector (Sino Biological) was used as negative control. Overexpression of ALCAM was confirmed by qPCR after hygromycin B selection.
Cell proliferation assays
For cell proliferation assays, Daoy and ONS-76 cells were seeded into 6-well plates (2.0 × 104 cells per well). After incubation for 48 h and 96 h, live cells were counted using a OneCell Counter (BMS, Tokyo, Japan). For D341 cell proliferation assays, cells were seeded into 12-well plates (2.0 x 105 cells in 2 ml of medium per well) and live cells counted after incubation for 96 h and 192 h.
Wound healing assays
For wound healing assays, 1.2 × 105 Daoy cells or 1.6 × 105 ONS-76 cells were seeded into 12-well plates and incubated until confluency (24 h). A wound was made in the cell monolayer by scraping with a CELL Scratcher (AGC Techno Glass Co., Shizuoka, Japan). The wound gap width was measured at 0, 24, and 48 h for the Daoy cells and 0, 12, and 24 h for the ONS-76 cells using a Nikon TMS inverted microscope (Nikon, Tokyo, Japan) and WRAYCAM camera (WRAYMER INC., Osaka, Japan). Images were analyzed using Photoshop CC (Adobe Systems Co., San Jose, CA, USA).
Orthotopic mouse model
Male NOD/Shi-scid, IL-2RγKO Jic mice (7–8 weeks; In-Vivo Science Inc., Tokyo, Japan) were used for xenograft implantation. All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee at the Osaka University Medical School (approval number: 29–041). All procedures involving animals were performed according to the animal use guidelines of the Animal Experiment Committee of Osaka University. The mice were anesthetized via intraperitoneal injection with three mixed sedatives (midazolam, butorphanol tartrate, and buprenorphine) and 2 × 105 stably transduced Daoy shControl or Daoy shALCAM1 cells in 2 μL of PBS were injected (n = 11 per group) through a burr hole into the right cerebellar hemisphere using a stereotactic injector (Stoelting, Wood Dale, IL). The mice were monitored every 1–2 days and immediately euthanized by overdose-anesthesia when severe neurological symptoms such as weight loss, loss of mobility, and severe paralysis were observed. The brains and spinal cords of the mice were subsequently dissected and formalin-fixed for histopathological analysis. For each mouse, the brain was sliced using a Mouse Brain Slicer (Muromati Kikai Co., Tokyo, Japan) to obtain three cerebral and three cerebellar coronal sections and the spinal cord was sliced to obtain five coronal sections. All dissected tissues were stained with HE and immunostained for CD166/ALCAM. STEM121 was used as a cytoplasm marker. Leptomeningeal dissemination and invasion at dissemination sites to the cerebrum, brain stem, and spinal cord of each section were evaluated histologically. Invasion was defined as a lesion that was continuous from the disseminated lesion and infiltrated into the subpial parenchyma. The histological evaluation of each section was performed blindly by three independent observers and the scores obtained were averaged.
Transwell assay
The invasion ability of ALCAM-depleted Daoy cells was assessed using a BioCoat Matrigel Invasion Chamber (BD Bioscience) according to the manufacturer’s instructions. Briefly, 1.0 × 105 Daoy cells in serum-free DMEM were seeded onto the Matrigel insert membrane and control insert membrane. DMEM containing 10% fetal bovine serum was added to the lower chamber as a chemoattractant and the chambers were incubated for 22 h. Afterward, the cells on the lower surface of the membrane were stained with Diff-Quik (Sysmex Co., Hyogo, Japan) and counted. The invasion percentage was calculated as follows:
Statistical analysis
All statistical analyses were performed using JMP, version 13 software (SAS Institute, Cary, NC, USA). Analysis of the relationship between ALCAM expression and the clinicopathological and molecular genetic parameters was tested using univariate analysis with the Fisher’s exact test. Comparison of ALCAM expression between groups in the R2: Genomics Analysis and Visualization Platform was performed using Student’s t-test and Tukey-Kramer test. For in vitro and in vivo studies, the distribution of data was verified with the Shapiro–Wilk test and parametric (One-way ANOVA and Dunnet’s post hoc test) and non-parametric (Wilcoxon rank sum test) methods were used for statistical analysis of the data. Survival data from the mouse model is presented in Kaplan-Meier plots and was analyzed with the log-rank test. Data are expressed as means ± SE. For all analyses, a P value < 0.05 was considered statistically significant.
Results
ALCAM expression correlated with the WNT molecular subgroup of MB
We investigated ALCAM expression in 45 human FFPE MB specimens using immunohistochemistry. We detected eight cases positive for ALCAM staining (18%), seven partially positive cases (16%), and 30 negative cases (67%). Interestingly, ALCAM expression showed a strong significant correlation to MB molecular subgroups (P < 0.0001; Table 1). All seven WNT cases and one SHH case, which histologically belongs to the MBEN subgroup, were ALCAM positive (Table 2). There was no clear correlation between ALCAM expression and the histologic variant of MB (P = 0.32; Table 1). Factors related to the WNT subgroup, such as nuclear β-catenin expression and CTNNB1 mutation status, correlated with ALCAM expression (P < 0.0001; Table 1).
Table 1. Correlation between clinicopathological/molecular data and ALCAM expression.
| ALCAM positive (n = 8) | ALCAM negative (n = 30) & partially positive (n = 7) | P value | |
|---|---|---|---|
| Age group | 0.022 | ||
| Infant (< 4 years) | 1 | 7 | |
| Child (4–16 years) | 3 | 27 | |
| Adult (> 16 yeas) | 4 | 3 | |
| Gender | 0.19 | ||
| Male | 4 | 29 | |
| Female | 4 | 8 | |
| Molecular subgroup | < 0.0001 | ||
| WNT | 7 | 0 | |
| SHH | 1 | 7 | |
| Group 3 | 0 | 7 | |
| Group 4 | 0 | 21 | |
| N/A | 0 | 2 | |
| Histological variant | 0.32 | ||
| classic | 7 | 29 | |
| desmoplastic/nodular | 0 | 5 | |
| MBEN | 1 | 0 | |
| LCA | 0 | 1 | |
| CTNNB1 status | < 0.0001 | ||
| mutation | 7 | 1a | |
| wild type | 1 | 36 | |
| Nuclear β-catenin expression | < 0.0001 | ||
| positive | 6 | 0 | |
| negative | 2 | 37 |
ALCAM, activated leukocyte cell adhesion molecule; N/A, data not available; MBEN, medulloblastoma with extensive nodularity; LCA, large cell/anaplastic.
aThis case had a CTNNB1 silent mutation. Statistically significant findings are in bold.
Table 2. Overview of clinicopathological/molecular characteristics and ALCAM expression in the MB cases examined.
| Case No. | Age group | molecular subgroup | Histological type | CTNNB1 mutation status | Nuclear β-catenin expression | ALCAM expression (positive cell proportion) | ALCAM staininglocalization |
|---|---|---|---|---|---|---|---|
| MB22 | adulta | WNT | classic | c.94G>A, p.D32N | Positive | Positive (> 75%) | Cytoplasm + partially membrane |
| MB29 | adulta | WNT | classic | c.98C>T, p.S33F | Positive | Positive (> 50%) | Cytoplasm |
| MB32 | adulta | WNT | classic | c.134C>T, p.S45F | Positive | Positive (> 75%) | Cytoplasm |
| MB34 | child | WNT | classic | c.101G>A, p.G34E | Negative | Positive (> 75%) | Cytoplasm |
| MB43 | adult | WNT | classic | c.98C>G, p.S33C | Positive | Positive (>75%) | Cytoplasm |
| MB9 | child | WNTb | classic | c.95A>G, p.D32G | Positive | Positive (> 75%) | Cytoplasm + partially membrane |
| MB30 | child | WNTb | classic | c.98C>T, p.S33F | Positive | Positive (> 75%) | Cytoplasm + partially membrane |
| MB12 | child | SHH | classic | NM | Negative | Negative | N/A |
| MB40 | adult | SHH | classic | NM | Negative | Negative | N/A |
| MB45 | child | SHH | classic | NM | Negative | Negative | N/A |
| MB7 | adult | SHH | desmoplastic/nodular | NM | Negative | Negative | N/A |
| MB25 | infant | SHH | desmoplastic/nodular | NM | Negative | Negative | N/A |
| MB36 | child | SHH | desmoplastic/nodular | NM | Negative | Negative | N/A |
| MB37 | infant | SHH | desmoplastic/nodular | NM | Negative | Negative | N/A |
| MB16 | infant | SHH | MBEN | NM | Negative | Positive (> 75%) | Membrane + partially cytoplasm |
| MB5 | child | Group 3 | classic | NM | Negative | Partially positive | Cytoplasm |
| MB13 | infant | Group 3 | classic | NM | Negative | Partially positive | Cytoplasm |
| MB11 | child | Group 3 | classic | NM | Negative | Negative | N/A |
| MB20 | infant | Group 3 | classic | NM | Negative | Negative | N/A |
| MB21 | child | Group 3 | classic | NM | Negative | Negative | N/A |
| MB35 | adult | Group 3 | classic | c.91C>T, p.L31Lc | Negative | Negative | N/A |
| MB39 | child | Group 3 | classic | NM | Negative | Negative | N/A |
| MB3 | child | Group 4 | classic | NM | Negative | Partially positive | Cytoplasm |
| MB17 | child | Group 4 | classic | NM | Negative | Partially positive | Cytoplasm |
| MB1 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB2 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB6 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB8 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB15 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB23 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB24 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB26 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB27 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB28 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB31 | infant | Group 4 | classic | NM | Negative | Negative | N/A |
| MB33 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB14 | infant | Group 4 | classic | NM | Negative | Negative | N/A |
| MB19 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB38 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB41 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB42 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB44 | child | Group 4 | classic | NM | Negative | Negative | N/A |
| MB18 | child | Group 4 | LCA | NM | Negative | Partially positive | Cytoplasm |
| MB10 | child | N/A | classic | NM | Negative | Partially positive | Cytoplasm |
| MB4 | infant | N/A | desmoplastic/nodular | NM | Negative | Partially positive | Cytoplasm |
Adult: > 16 years, child: 4–16 years, infant: < 4 years.
ALCAM, activated leukocyte cell adhesion molecule; IHC, immunohistochemistry; M, male; F, female; N/A, data not available; MBEN, medulloblastoma with extensive nodularity; LCA, large cell/anaplastic; NM, no mutation.
a17–21 years.
bThese cases were defined as WNT according to the mutation of CTNNB1 and nuclear staining of β-catenin.
cSilent mutation.
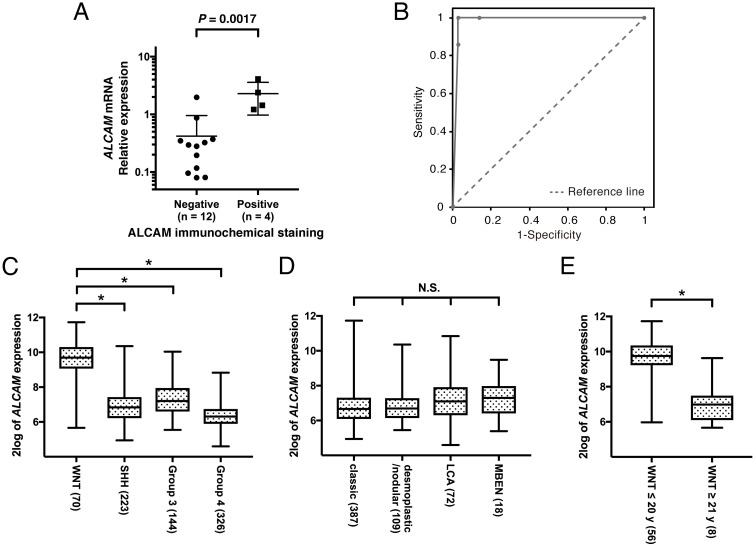
To validate the correlation between the immunohistochemical staining data and ALCAM expression, we performed qPCR analysis of four ALCAM-positive and 13 ALCAM-negative cases. The mean relative expression level in the ALCAM-positive cases was 10 times higher than that in the ALCAM-negative cases (P = 0.0017; Fig 1A). These findings showed that ALCAM expression was higher in the WNT subgroup of MB than in the non-WNT subgroup. Furthermore, the correlation between ALCAM protein level determined by immunohistochemical staining and ALCAM mRNA expression was confirmed.
Fig 1.
(A) Correlation between ALCAM immunohistochemical staining and ALCAM mRNA expression. ALCAM-positive medulloblastoma (MB) cases showed increased levels of ALCAM compared with the ALCAM-negative cases (P = 0.0017). The relative expression in human brain (cerebellum) was arbitrarily set at 1.0. (B) Receiver operating characteristic (ROC) curve for positive immunohistochemical expression of ALCAM in WNT subgroup. Area under the curve (AUC), 0.984. (C–E) ALCAM Expression in the Cavalli-763 MB cohort from the R2 Genomics Platform. (C) The WNT subgroup of MB strongly expressed ALCAM as compared with the other subgroups. (D) There was no clear correlation between ALCAM expression levels and histologic MB subtypes. (E) In the WNT subgroup of MB cases, ALCAM expression differed between the ages ≤ 20 y and ≥ 21 y. The box-and-whisker plots show the medians (thick horizontal lines) and interquartile ranges (IQRs; boundaries of the box) and ranges. *: P < 0.0001; N.S.: not significant.
To evaluate the reliability of the WNT subgroup of MB using ALCAM immunohistochemical staining, ROC curves were prepared using the proportion of ALCAM-positive tumor cells (0, 1, 25, 50, 75, 100%), from representative tumor areas, as the independent variable (Fig 1B). Analyzing the seven cases in WNT subgroup and the 36 cases in the non-WNT subgroup, the AUC was 0.984, indicating a high accuracy, and the optimal cutoff was > 25% or > 50%.
Similar results were observed in the large cohorts of the Cavalli-763 MB dataset [24] from the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). Specifically, ALCAM expression was significantly higher in the WNT subgroup of MB compared to that in the SHH, Group 3, and Group 4 subgroups (P < 0.0001; Fig 1C). On the other hand, no correlation was found between ALCAM expression and histologic subtypes of MB (Fig 1D). ALCAM expression was generally higher in the WNT subgroup, but this feature was not observed in cases of patients older than 21 y (mean, 33.2; range, 23–56 y; P < 0.0001; Fig 1E).
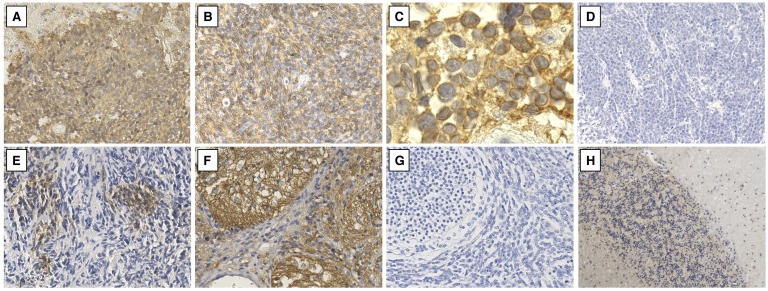
In the seven WNT molecular subgroup cases, the tumor cells of the whole section were diffusely stained for ALCAM (Fig 2A and 2B). Most of the ALCAM-positive tumor cells stained predominantly in the cytoplasm, but a few of them showed some staining in the cell membrane (Fig 2C). In the majority of the SHH, Group 3, and Group 4 subgroup cases, no expression of ALCAM was observed in the tumor cells (Fig 2D). We also observed seven partially positive cases in which ALCAM-positive cells with cytoplasmic staining were observed in some areas of the tissue section, two Group 3 cases, three Group 4 cases, and two cases in the N/A group (Fig 2E). The MBEN histological subtype, observed in only one case, showed a characteristic staining pattern of neuropil-like tissue and differentiated neurocytic cells in the expanded lobular architecture stained positive for ALCAM (mainly in the membrane), while the small round neurocytic cells in the internodular areas were negative (Fig 2F). In contrast, in five desmoplastic/nodular cases, the tumor cells showing signs of variable neurocytic maturation in pale islands failed to show ALCAM staining, similar to the internodular areas (Fig 2G). However, in one desmoplastic/nodular case, weak ALCAM staining was observed in the tumor cells of a few pale islands. In the normal cerebellum, ALCAM staining was absent in the molecular layer, while the granular layer and white matter stained weakly (Fig 2H).
Fig 2. Immunohistochemical staining of ALCAM in medulloblastoma (MB) specimens.
(A, B) ALCAM-positive cases: the WNT subgroup showed diffuse positive staining of ALCAM. (C) Most of the ALCAM-positive cells stained predominantly in the cytoplasm. (D) An ALCAM-negative case. (E) A partially positive case of ALCAM staining: few scattered ALCAM-positive cells can be observed. (F) The MBEN histological variant demonstrated a different ALCAM expression profile: positive staining in the lobular architectural region and negative in the internodular region. (G) The desmoplastic/nodular variant showed no ALCAM expression in most of the pale nodular areas and internodular areas. (H) In normal cerebellar tissue, the expression of ALCAM is not observed in the molecular layer, but is weakly detected in the granular layer and white matter. Original magnification, ×200 (A, B, D–G), ×1,000 (C) and ×100 (H).
Expression of ALCAM and its effect on cell proliferation and migration in MB cell lines
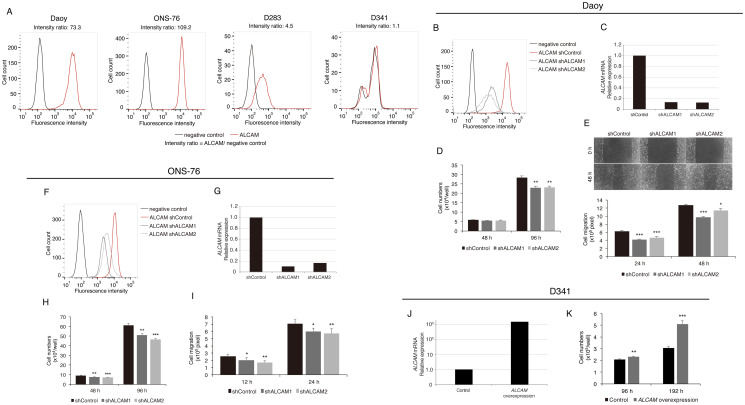
To evaluate the expression levels of ALCAM, we performed flow cytometry analysis of four human MB cell lines. ALCAM expression differed in each of the cell lines (Fig 3A) with the lowest levels being found in D341 cells (IR = 1.1) and the highest levels in Daoy and ONS-76 cells (IR = 73.3 and 109.2, respectively). Based on these results, we selected Daoy cells as a representative cell line with high ALCAM expression to investigate the functional role of ALCAM in MB. Two ALCAM-depleted Daoy cell lines were generated by RNA interference and the reduction of ALCAM expression was confirmed by flow cytometry analysis and qPCR (Fig 3B and 3C, respectively). The comparison between ALCAM-depleted and control Daoy cells showed that ALCAM depletion decreased cell proliferation and migration (Fig 3D and 3E, respectively). Specifically, there was no difference in cell proliferation at 48 h, while a difference was observed at 96 h, with the mean numbers (± SE) of the shControl, shALCAM1, and shALCAM2 Daoy cells being 28.3 ± 0.879, 22.9 ± 0.970, and 23.1 ± 0.798 × 104 cells, respectively (Fig 3D). Additionally, in wound healing assays, shALCAM1 and shALCAM2 Daoy cells showed 32% and 26% reduced migration at 24 h, respectively, and 24% and 11% at 48 h, respectively, compared to that of the shControl Daoy cells (Fig 3E). Similar results were observed with the ALCAM-depleted ONS-76 cell lines (Fig 3F–3I). These results demonstrated that ALCAM acted as a positive regulator of proliferation and migration in Daoy and ONS-76 cells in vitro.
Fig 3. Expression of ALCAM in medulloblastoma (MB) cell lines and in vitro study of its function in Daoy and ONS-76 cells.
(A) Four MB cell lines exhibited different ALCAM expression as measured by flow cytometry. (B, C, F, G) Knockdown of ALCAM in Daoy and ONS-76 cells was verified using flow cytometry (B, F) and qPCR (C, G). (D, E, H, I) ALCAM silencing inhibits the proliferation (D, H) and migration (E, I) of Daoy and ONS-76 cells. (J) Overexpression of ALCAM in D341 cells was confirmed by qPCR. (K) ALCAM overexpression promotes the proliferation of D341 cells. Data are reported as the mean ± SE. *: P < 0.05; **: P < 0.01; ***: P < 0.0001.
Furthermore, D341 cells, which had the lowest levels of ALCAM expression, were used to establish an ALCAM-overexpressing D341 cell line using cDNA transduction. Increased ALCAM expression was confirmed by qPCR analysis (Fig 3J). Comparison of ALCAM-overexpressing D341 cells and control D341 cells revealed that enhanced ALCAM expression resulted in increased cell proliferation (Fig 3K).
Depletion of ALCAM affects invasion at dissemination sites in an orthotopic mouse model of MB
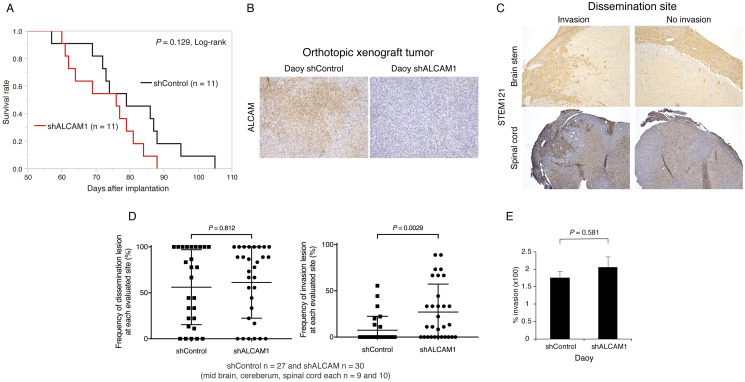
Next, we assessed the effects of ALCAM on tumor progression and dissemination in vivo. Stably transfected ALCAM-depleted or control Daoy cells were inoculated into the right cerebellar hemisphere of mice. Tumors developed in both groups of mice. During the experiment, 9 mice in the shControl were euthanized and two were found dead while 10 mice in the shALCAM group were euthanized and one mouse was found dead. Although there was no significant difference in the survival rates between the shControl and shALCAM groups (P = 0.129, log-rank test), the survival of mice injected with ALCAM-depleted cells appeared to be shorter than that of the control mice (Fig 4A). Upon pathological evaluation of the brains and spinal cords of the mice, the primary tumors were found in the cerebellum and leptomeningeal disseminated lesions were found on the surface of the cerebrum, brain stem, and spinal cord. Immunostaining of ALCAM confirmed that depletion of ALCAM was maintained in the shALCAM group (Fig 4B). In the disseminated lesions, several foci of invasive tumor cells were found into the subpial parenchyma (Fig 4C). The frequency of sections with dissemination lesions in the two groups was similar (P = 0.812; Fig 4D). However, invasion at the dissemination sites was observed more frequently in the shALCAM group compared to that in the shControl group (P = 0.0029; Fig 4D). These results indicated that ALCAM depletion may be associated with a more invasive tumor-cell phenotype in vivo. On the other hand, there was no significant difference in the invasiveness of shControl and shALCAM1 Daoy cells as per the results of the in vitro Transwell assay. The invasion percentage (± SE) of the shControl and shALCAM1 Daoy cells was 176 ± 18.5 and 206 ± 29.8, respectively (P = 0.581; Fig 4E).
Fig 4. Functional analysis of ALCAM in an orthotopic mouse model and in vitro transwell assay in Daoy cells.
(A) Kaplan–Meier survival curves of mice injected with ALCAM-silenced and control Daoy cells showed no significant difference between the two groups (P = 0.129, log-rank test). (B) Representative immunohistochemical staining of ALCAM in primary tumors in the cerebellum. (C) Representative STEM121 staining showing invasive and noninvasive tumor cells in disseminated lesions. (D) The frequency of dissemination in the ALCAM-silenced and control group was similar (P = 0.812). The frequency of invasion observed at the dissemination sites was significantly higher in the ALCAM-silenced group than that in the control group (P = 0.0029). (E) The invasion percentage of ALCAM-silenced and control Daoy cells was not significantly different, as per the results of the transwell assay (P = 0.581). Original magnification, ×100 (B, C).
Discussion
In the current study, we first showed that ALCAM expression was strongly correlated to the WNT molecular subgroup of MB. Few studies have investigated the expression of ALCAM in MB; among them, one study assessed ALCAM levels in various primary central nervous system tumors, including MB [20]. In that study, Allmendinger et al [20] showed that the positive rate of ALCAM in MB is 31%, similar to that obtained in our study. However, the authors did not examine the relation between ALCAM expression and the molecular subgroup or histological subtypes [20].
Recent studies have focused on the molecular classification of MB and the expression of several genes related to molecular subgroups has been determined in MB. However, there is no report investigating the relationship between ALCAM and the WNT subgroup. Immunohistochemically, the nuclear staining of β-catenin resulting from a CTNNB1 mutation is associated with the WNT subgroup and is a good diagnostic tool for clinical applications [25]. However, immunohistological diagnosis of the WNT subgroup of MB using β-catenin nuclear staining alone is considered insufficient [26]. ALCAM has the potential of being a new WNT-related biomarker for improving the reliability of a WNT subgroup diagnosis of MB.
Interestingly, it has been recently reported that there is a strong correlation between nuclear staining of β-catenin and ALCAM levels in adamantinomatous craniopharyngioma [27], which is closely related to the Wnt signal pathway [28]. This report and our current results suggest a relationship between the Wnt signal pathway and ALCAM expression. It is known that embryonic expression of ALCAM occurs through non-canonical Wnt/JNK signaling [29, 30]. However, it is unclear whether the activation of the Wnt pathway caused by the CTNNB1 mutation is associated with ALCAM expression.
Despite the fact that ALCAM was originally identified as a membrane protein, the localization of ALCAM staining in MB was mainly cytoplasmic and only partially observed in the membrane. This is consistent with a previous report by Allmendinger et al [20]. In fact, ALCAM is known to be expressed in the cytoplasm of many carcinomas and the pattern of expression seems to vary depending on the particular carcinoma [11, 13, 19, 20]. There is also a report that cell function is altered by the translocation of ALCAM from the membrane to the cytoplasm [13].
Regarding the immunohistochemical evaluation of ALCAM, it is necessary to understand ALCAM expression in the normal brain. As we have shown, ALCAM was slightly expressed in the granular layer of the cerebellum. Furthermore, Allmendinger et al [20] have reported that ALCAM is expressed in the normal cerebral tissue, such as hippocampus and basal ganglia, and also in reactive glial cells. In the current study, we performed Immunohistochemical evaluation of ALCAM in representative tumor areas.
In our functional study using MB cell lines, we demonstrated that in vitro silencing of ALCAM in Daoy and ONS-76 cells inhibited cell proliferation and migration. Meanwhile, the overexpression of ALCAM in D341 cells promoted cell proliferation, although cell migration could not be evaluated due to the fact that the cells grow in suspension and are not adherent cells. Our findings indicate that ALCAM may act as a positive regulator of cell proliferation in MB. However, we obtained unexpected results in the orthotopic mouse model, where the survival of mice injected with ALCAM-silenced Daoy cells appeared to be shorter than that of the control mice, even though the difference between the two groups was not statistically significant. We also found that ALCAM depletion in Daoy cells increased invasion at disseminated sites in the orthotopic mouse model. On the other hand, the increased invasiveness of ALCAM-depleted Daoy cells was not observed in the context of an in vitro transwell assay. These results seem contradictory, but the difference between ALCAM function in vitro and in vivo indicates that ALCAM may be affected by the surrounding microenvironment. Notably, our results are consistent with those from previous reports regarding other carcinomas in which depletion of ALCAM suppresses cell proliferation and migration in vitro, but the cells demonstrate invasiveness in vivo [14, 16, 19, 31]. For example, in cell lines of malignant mesothelioma and endometrioid endometrial cancer, proliferation and migration are inhibited in vitro by ALCAM silencing [16, 19]. Nevertheless, ALCAM-negativity at the invasive front of the tumor has been reported as a marker of myometrial invasion in tissue of endometrioid endometrial cancer [31]. Moreover, in a metastatic melanoma cell line, interfering with endogenous ALCAM through the expression of an amino-terminally truncated ALCAM, which disrupts ALCAM–ALCAM interactions, increases cell invasive growth in skin reconstructions [14]. The attenuation of ALCAM enhances the invasive abilities of the tumor, which is arguably caused by the reduction of ALCAM-mediated adhesion, resulting in less adhesive and more mobile tumor cells [14, 31]. Therefore, the functional role of ALCAM might vary depending on the surrounding microenvironment.
ALCAM has been shown to be a prognostic marker in several types of cancers; however, it is both a marker of good and poor prognosis depending on the cancer type [9–13]. In our study, the expression of ALCAM was observed particularly concentrated in the WNT subgroup of MB, which typically shows a good long-term prognosis in comparison to the other subgroups. Therefore, ALCAM may be regarded as a good prognostic marker in MB. Conversely, a lack of ALCAM may contribute to poor prognosis. Differences in ALCAM expression levels might be related to a factor regulating the cell kinetics between WNT MB and non-WNT MB. Accordingly, we showed that ALCAM silencing enhanced the invasiveness of disseminated lesions in the orthotopic mouse model. The invasion ability at disseminated lesions is an important negative prognostic factor of MB as most MB recurrence is caused by leptomeningeal dissemination.
A limitation of our current study was the relatively small sample size of our cohort compared to that of other large cohorts. The small sample size used for ALCAM immunohistochemical staining may have resulted in inevitable statistical bias. However, there was no significant difference in our molecular subgrouping results compared to that using another cohort [4]. It is noteworthy that our study lacked a validation study of the ALCAM immunohistochemical staining, and thus our immunohistochemical findings may be insufficient to assert ALCAM as a biomarker for the WNT subgroup of MB. However, we did perform a comparative analysis using a large cohort (Cavalli-763 MB dataset) [24] of the R2: Genomics Analysis and Visualization Platform. This comparative analysis was based on gene expression and showed a strong correlation between ALCAM expression and the WNT subgroup of MB. Consistent with this, our study showed a correlation between ALCAM protein levels based on immunohistochemical staining and ALCAM mRNA gene expression levels analyzed by RT-qPCR. As evaluation of patient age and pathological type was insufficient in our cohort, we examined this using expression data from a large cohort of the R2 database. Analysis of the R2 database cases revealed ALCAM expression was low in patients 21 y and older, even in the WNT subgroup with high ALCAM expression. Therefore, we believe that limiting the evaluation of cases to patients 20 years or younger improves the strength of the correlation between ALCAM and the WNT subgroup.
According to the results from the analysis of clinical sample in which ALCAM was strongly expressed in the WNT subgroup, it may be more appropriate to use a WNT subgroup cell line. However, cell lines belonging to the WNT subgroup are not readily available [32]. Daoy and ONS-76 cells used in our study strongly express ALCAM and have been considered to be of the SHH subgroup, while D341 and D231 cells weakly express ALCAM and are considered to be of the Group 3 and Group 3/4 subgroups, respectively [32]. In our current study, positive ALCAM immunostaining was observed in only one of the eight cases of the SHH subgroup and ALCAM expression was low. Further studies are required to determine whether ALCAM is strongly expressed in the SHH subgroup cell line, despite low expression of ALCAM in the SHH subgroup clinical sample.
Conclusions
Clinical sample analysis performed in our current study suggested that ALCAM expression in MB was strongly correlated to the molecular subgroup. Specifically, ALCAM was highly expressed in the WNT subgroup of MB compared to that in non-WNT subgroups of MB. Furthermore, the cell kinetics of MB cell lines were altered by ALCAM expression. ALCAM appeared to be involved as a positive regulator of proliferation and migration of MB tumor cells in vitro. However, in vivo studies using an orthotopic mouse model demonstrated that the attenuation of ALCAM enhanced cell invasiveness in disseminated lesions.
Acknowledgments
The authors thank Ai Takada (Institute for Clinical Research, Osaka National Hospital) for her generous assistance in collecting the clinical samples and Tomomi Inai (Center for Medical Research and Education, Osaka University Graduate School of Medicine) for flow cytometric analysis and cell sorting. The authors also thank Editage (www.editage.jp) for English language editing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
YF Japan Society for the Promotion of Science (JSPS) KAKENHI https://www.jsps.go.jp/english/ Grant Number JP16K10755 YK Japan Agency for Medical Research and Development (AMED) https://www.amed.go.jp/en/ Grant Number JP18ck0106330 and JP18gm0810011.
References
- 1.Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7: 813–820. 10.1016/S1470-2045(06)70867-1 [DOI] [PubMed] [Google Scholar]
- 2.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29: 1408–1414. 10.1200/JCO.2009.27.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho Y-J, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123: 465–472. 10.1007/s00401-011-0922-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kool M, Korshunov A, Remke M, Jones DTW, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123: 473–484. 10.1007/s00401-012-0958-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, et al. Medulloblastoma. Nat Rev Dis Primers. 2019;5: 11 10.1038/s41572-019-0063-6 [DOI] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, Deimling von A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131: 803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy V, Remke M, Bouffet E, Bailey S, Clifford SC, Doz F, et al. Risk stratification of childhood medulloblastoma in the molecular era: the current consensus. Acta Neuropathol. 2016;131: 821–831. 10.1007/s00401-016-1569-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swart GWM. Activated leukocyte cell adhesion molecule (CD166/ALCAM): developmental and mechanistic aspects of cell clustering and cell migration. Eur J Cell Biol. 2002;81: 313–321. 10.1078/0171-9335-00256 [DOI] [PubMed] [Google Scholar]
- 9.Weidle UH, Eggle D, Klostermann S, Swart GWM. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7: 231–243. [PubMed] [Google Scholar]
- 10.Weichert W, Knösel T, Bellach J, Dietel M, Kristiansen G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57: 1160–1164. 10.1136/jcp.2004.016238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahlert C, Weber H, Mogler C, Bergmann F, Schirmacher P, Kenngott HG, et al. Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br J Cancer. 2009;101: 457–464. 10.1038/sj.bjc.6605136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King JA, Ofori-Acquah SF, Stevens T, Al-Mehdi A-B, Fodstad O, Jiang WG. Activated leukocyte cell adhesion molecule in breast cancer: prognostic indicator. Breast Cancer Res. 2004;6: R478–87. 10.1186/bcr815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mezzanzanica D, Fabbi M, Bagnoli M, Staurengo S, Losa M, Balladore E, et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin Cancer Res. 2008;14: 1726–1733. 10.1158/1078-0432.CCR-07-0428 [DOI] [PubMed] [Google Scholar]
- 14.van Kempen LCLT, Meier F, Egeblad M, Kersten-Niessen MJF, Garbe C, Weidle UH, et al. Truncation of activated leukocyte cell adhesion molecule: a gateway to melanoma metastasis. J Invest Dermatol. 2004;122: 1293–1301. 10.1111/j.0022-202X.2004.22531.x [DOI] [PubMed] [Google Scholar]
- 15.Dalerba P, Dylla SJ, Park I-K, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104: 10158–10163. 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiguro F, Murakami H, Mizuno T, Fujii M, Kondo Y, Usami N, et al. Activated leukocyte cell-adhesion molecule (ALCAM) promotes malignant phenotypes of malignant mesothelioma. J Thorac Oncol. 2012;7: 890–899. 10.1097/JTO.0b013e31824af2db [DOI] [PubMed] [Google Scholar]
- 17.Kijima N, Hosen N, Kagawa N, Hashimoto N, Nakano A, Fujimoto Y, et al. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro Oncol. 2012;14: 1254–1264. 10.1093/neuonc/nor202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara K, Ohuchida K, Sada M, Horioka K, Ulrich CD, Shindo K, et al. CD166/ALCAM expression is characteristic of tumorigenicity and invasive and migratory activities of pancreatic cancer cells. PLoS ONE. 2014;9: e107247 10.1371/journal.pone.0107247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devis L, Moiola CP, Masia N, Martinez-Garcia E, Santacana M, Stirbat TV, et al. Activated leukocyte cell adhesion molecule (ALCAM) is a marker of recurrence and promotes cell migration, invasion, and metastasis in early-stage endometrioid endometrial cancer. J Pathol. 2017;241: 475–487. 10.1002/path.4851 [DOI] [PubMed] [Google Scholar]
- 20.Allmendinger O, Trautmann K, Mittelbronn M, Waidelich J, Meyermann R, Tatagiba M, et al. Activated leukocyte cell adhesion molecule is expressed in neuroepithelial neoplasms and decreases with tumor malignancy, matrix metalloproteinase 2 expression, and absence of IDH1R132H mutation. Hum Pathol. 2012;43: 1289–1299. 10.1016/j.humpath.2011.10.007 [DOI] [PubMed] [Google Scholar]
- 21.Sasaki T, Fukai J, Kodama Y, Hirose T, Okita Y, Moriuchi S, et al. Characteristics and outcomes of elderly patients with diffuse gliomas: a multi-institutional cohort study by Kansai Molecular Diagnosis Network for CNS Tumors. J Neurooncol. 2018;57: 1–11. 10.1007/s11060-018-2957-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Northcott PA, Shih DJH, Remke M, Cho Y-J, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123: 615–626. 10.1007/s00401-011-0899-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31: 737–754.e6. 10.1016/j.ccell.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23: 7951–7957. 10.1200/JCO.2005.01.5479 [DOI] [PubMed] [Google Scholar]
- 26.Goschzik T, Mühlen zur A, Kristiansen G, Haberler C, Stefanits H, Friedrich C, et al. Molecular stratification of medulloblastoma: comparison of histological and genetic methods to detect Wnt activated tumours. Neuropathol Appl Neurobiol. 2015;41: 135–144. 10.1111/nan.12161 [DOI] [PubMed] [Google Scholar]
- 27.Guadagno E, de Divitiis O, Solari D, Borrelli G, Bracale UM, Di Somma A, et al. Can recurrences be predicted in craniopharyngiomas? β-catenin coexisting with stem cells markers and p-ATM in a clinicopathologic study of 45cases. J Exp Clin Cancer Res. 2017;36: 95 10.1186/s13046-017-0562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekine S, Shibata T, Kokubu A, Morishita Y, Noguchi M, Nakanishi Y, et al. Craniopharyngiomas of adamantinomatous type harbor beta-catenin gene mutations. Am J Pathol. 2002;161: 1997–2001. 10.1016/s0002-9440(10)64477-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cizelsky W, Tata A, Kühl M, Kühl SJ. The Wnt/JNK signaling target gene alcam is required for embryonic kidney development. Development. 2014;141: 2064–2074. 10.1242/dev.107938 [DOI] [PubMed] [Google Scholar]
- 30.Seigfried FA, Cizelsky W, Pfister AS, Dietmann P, Walther P, Kühl M, et al. Frizzled 3 acts upstream of Alcam during embryonic eye development. Dev Biol. 2017;426: 69–83. 10.1016/j.ydbio.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 31.Devis L, Martinez-Garcia E, Moiola CP, Quiles MT, Arbos MA, Stirbat TV, et al. ALCAM shedding at the invasive front of the tumor is a marker of myometrial infiltration and promotes invasion in endometrioid endometrial cancer. Oncotarget. 2018;9: 16648–16664. 10.18632/oncotarget.24625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanov DP, Coyle B, Walker DA, Grabowska AM. In vitro models of medulloblastoma: Choosing the right tool for the job. J Biotechnol. 2016;236: 10–25. 10.1016/j.jbiotec.2016.07.028 [DOI] [PubMed] [Google Scholar]