Abstract
Background
Early diagnosis of coronavirus disease 2019 (COVID-19) is of the utmost importance but remains challenging. The objective of the current study was to characterize exhaled breath from mechanically ventilated adults with COVID-19.
Methods
In this prospective observational study, we used real-time, online, proton transfer reaction time-of-flight mass spectrometry to perform a metabolomic analysis of expired air from adults undergoing invasive mechanical ventilation in the intensive care unit due to severe COVID-19 or non-COVID-19 acute respiratory distress syndrome (ARDS).
Findings
Between March 25th and June 25th, 2020, we included 40 patients with ARDS, of whom 28 had proven COVID-19. In a multivariate analysis, we identified a characteristic breathprint for COVID-19. We could differentiate between COVID-19 and non-COVID-19 ARDS with accuracy of 93% (sensitivity: 90%, specificity: 94%, area under the receiver operating characteristic curve: 0•94-0•98, after cross-validation). The four most prominent volatile compounds in COVID-19 patients were methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal.
Interpretation
The real-time, non-invasive detection of methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal in exhaled breath may identify ARDS patients with COVID-19.
Funding
The study was funded by Agence Nationale de la Recherche (SoftwAiR, ANR-18-CE45-0017 and RHU4 RECORDS, Programme d'Investissements d'Avenir, ANR-18-RHUS-0004), Région Île de France (SESAME 2016), and Fondation Foch.
Keywords: COVID-19, Intensive care, Mechanical ventilation, Breath analysis, Metabolomics
Research in context.
Evidence before this study
Early diagnosis of coronavirus disease 2019 (COVID-19) is of the utmost importance but remains challenging. Around 5% of patients with COVID-19 will develop acute respiratory distress syndrome (ARDS), septic shock and/or multiple organ failure; ideally, these patients should be identified as soon as possible. Breath analysis is an innovative, non-invasive, real-time, point-of-care technique for detecting volatile organic compounds (VOCs) in expired breath. It has potential for use in diagnosis and large-scale screening. However, it was not previously known whether patients with COVID-19 have a breath “signature” (also known as a “breathprint”).
Added value of this study
Here, we show that breath analysis can discriminate between COVID-19 ARDS and non-COVID-19 ARDS. We characterized a VOC breathprint that was able to identify COVID-19 ARDS patients requiring invasive mechanical ventilation with high sensitivity and specificity. The four most prominent volatile compounds in the patients’ breath were methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal. The COVID-19 breathprint did not depend on the severity of the ARDS or the patient's viral load.
Implications of all the available evidence
All the available evidence suggest that real-time, non-invasive breath analysis could enable the large-scale screening and thus earlier treatment of patients likely to develop severe forms of COVID-19.
Alt-text: Unlabelled box
Introduction
As of November 21st, 2020, about 57 million of people worldwide had been infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and about 1•4 million had died from coronavirus disease 2019 (COVID-19) [1]. Approximately 5% of patients with COVID-19 will develop acute respiratory distress syndrome (ARDS), septic shock, or multiple organ dysfunction [2]. Around the world, unprecedented research efforts are being focused on the prevention, early detection, diagnosis and management of this lethal disease. To date, only one antiviral drug (remdesivir) has been approved for the treatment of patients hospitalized for COVID-19 [3]. More recently, a large trial showed that dexamethasone at a daily dose of 6 mg for 10 days substantially reduced the risk of 28 day death (age-adjusted rate ratio [95% confidence interval (CI)]: 0•83 [0•75 to 0•93], particularly in patients with severe disease requiring invasive mechanical ventilation (rate ratio: 0•64 [0•51 to 0•81]) [4]. Although the early immune response may not depend on the severity of the illness, the most severely ill patients show persistent elevations of blood inflammatory markers (such as IL-1α, IL-1β, IL-6, IL-10, IL-18 and TNF-α) 10 or so days after SARS-CoV-2 infection, with a very high risk of subsequent organ injury 5, 6, 7. Proteomic and metabolomic studies of serum have described a COVID-19-specific molecular signature; severe and non-severe forms of COVID-19 differ with regard to amino acid metabolism and the expression of acute phase proteins [8]. Breath analysis is an innovative, non-invasive, real-time point-of-care technique for detecting volatile organic compounds (VOCs) with potential for use in diagnosis and large-scale screening [9,10]. Thousands of VOCs have been identified in human breath following infectious, inflammatory or pathological events [11,12]. It has been suggested that the analysis of exhaled breath can be used to diagnose tuberculosis, invasive fungal infections, and bacterial colonization of the respiratory tract 13, 14, 15, 16, together with ARDS and ventilator-associated pneumonia in patients in the intensive care unit (ICU) 17, 18, 19, 20, 21, 22. Likewise, previous studies have suggested that VOC analysis is of value in the diagnosis of viral infections in patients with chronic obstructive pulmonary disease and of influenza infections in a swine model [23,24]. The airway and lung damage caused by SARS-CoV-2 [25] might conceivably result in the release of characteristic VOCs in the exhaled breath. To test this hypothesis, we determined the metabolomic breath signature in a group of ARDS patients with or without COVID-19 and who required invasive mechanical ventilation.
Methods
Study design and oversight
This prospective study was part of the observational phase of the ongoing RECORDS trial (NCT04280497) and was conducted at the ICU of Raymond Poincaré Hospital (Garches, France). The RECORDS study protocol was approved by an ethics commitee (Comite de Protection des Personnes EST I, Dijon, France; reference 20.03.10.51415) and the French National Agency for Healthcare Product Safety (ANSM, Paris, France). The study was registered with the European Union Drug Regulating Authorities Clinical Trials Database (EudraCT 2020-000296-21). Whenever possible, participants or their legally authorized next of kin provided written, informed consent before inclusion. In the remaining cases, patients provided their deferred, written, informed consent. This investigator-led study was publicly funded. All the authors had full and independent access to all data and vouch for the integrity, accuracy, and completeness of the data and analysis and for the adherence of the trial to the protocol.
Study participants
Adult patients (aged 18 or over) in ICUs were eligible for inclusion if they had ARDS and required invasive mechanical ventilation. ARDS was defined as all of the following: (i) acute onset, i.e., within one week of an apparent clinical insult, followed by progression of the respiratory syndrome, (ii) bilateral opacities on chest imaging not explained by another lung disease (e.g., pleural effusion, atelectasis, nodules etc.), (iii) no evidence of heart failure or volume overload, and (iv) PaO2/FiO2 ≤ 300 mm Hg, and positive end expiratory pressure (PEEP) ≥ 5 cm H2O [26]. The main exclusion criteria were pregnancy, an expectation of death within 48 h, and the withholding or withdrawal of treatment.
Study measurements and procedures
Variables recorded at baseline were patient demographics and anthropometrics, the source of infection, and the severity of illness (according to the Simplified Acute Physiology Score (SAPS) II and the Sequential Organ Failure Assessment (SOFA)) [27,28]. The following variables were recorded at baseline and daily during the hospital stay: core body temperature, vital signs, central hemodynamic data, standard laboratory data, microbiological and virologic data. Samples for routine surveillance of lower respiratory tract colonization were obtained every 72 h until the patient had been weaned off mechanical ventilation or had died. A nonbronchoscopic bronchoalveolar lavage was performed with three 20 mL aliquots of sterile 0•9% saline solution, with a view to collect at least 5–10 mL of effluent per sample. Samples of blood and nasopharyngeal, bronchial or bronchoalveolar lavage fluids were assayed for SARS-CoV-2 and other respiratory viruses with a PCR test, as described by the French National Reference Center for Respiratory Viruses (Institut Pasteur, Paris, France). We also recorded life-supportive therapies including mechanical ventilation, renal replacement therapy, intravenous fluids bolus and the administration of vasopressors, and adjunct therapies including corticosteroids, thiamine, vitamin C, other vitamins, nutritional supplements, blood products, anticoagulants, sedatives, stress ulcer prophylaxis, and anti-infective drugs.
Breath analysis
Each patient's expired air was analyzed daily in the morning until discharge. Measurements were made with a proton-transfer-reaction quadrupole time-of-flight mass spectrometer (Ionicon Analytik GmbH, Innsbruck, Austria) placed outside the patient room. Samples were obtained via a heated transfer line (length: 1.6 m) connected directly to the end of the endotracheal tube (i.e., without disconnection from the mechanical ventilator) and with an air flow of 50 mL/min. To eliminate the dependency on the oxygen concentration in the sample matrix, recordings were performed in patients with a fraction of inspired oxygen of 100% for at least 3 min [29]. The acquisition took 2 min. H3O+ was used as the primary ion and the instrument settings were as follows: source voltage, 120 V; drift tube pressure, 3•8 mbar; drift tube temperature, 60 °C; and drift tube voltage, 959 V. The mass spectrum was acquired up to m/z = 392, with a time resolution of 0•1 s.
Data and statistical analysis
Patient characteristics were expressed as the median [interquartile range (IQR)] for continuous variables and the frequency (percentage) for categorical variables. Patients with and without COVID-19 were compared using Fisher's exact test for categorical variables, and a t-test or the Mann-Whitney test for normally and non-normally distributed continuous variables (as evaluated with the d'Agostino-Pearson test), respectively.
Mass spectrometry data were processed with the ptairMS R package (https://github.com/camilleroquencourt/ptairMS) and included mass calibration, expiratory phase detection on the CO2 extracted ion chromatogram, peak detection and quantification with background subtraction, normalization, alignment, isotope identification, and imputation of missing values. All concentration values were quoted in ppb [30]. After aligning each individual peak, ions detected in more than 70% of at least one group (COVID vs. non-COVID-19 ARDS) were kept; this resulted in 81 features. Missing values (corresponding to ions in exhaled breath that were not detected by the preprocessing algorithm) were imputed with the ptairMS package, which returns to the raw data and integrates the noise at the exact missing m/z. Data were then log2-transformed and standardized. Outliers (patients with a z-score >3 for at least five features) were deleted. In the remaining patients, saturated ions (acetone, H3O+, H2O-H3O+, oxygen) and isotopes were deleted to leave a final table of 65 features. For the univariate analysis, a Wilcoxon test was performed and p-values were adjusted to control for the false discovery rate [31]. For multivariate analysis, data were analyzed first with principal component analysis and then with machine learning algorithms with different mathematical backgrounds (orthogonal partial least-squares discriminant analysis (OPLS-DA), linear support vector machine (SVM), elastic net, and random forest (RF); summarized in Table S1) with the R packages ropls, e1071, and caret 32, 33, 34, 35. A 10-fold, stratified cross-validation was repeated four times (in order to avoid overfitting the small number of data points), and features were selected with the elastic net and RF approaches. The models’ parameters were tuned to optimize the accuracy of cross-validation. Features were ranked according to the specific metrics of each modeling method (p-values from the Wilcoxon test, absolute loading values from PCA, the variable importance in projection from OPLS-DA, the coefficient values from the elastic net and SVM models, and the feature importance from the RF model). An aggregated ranking was then computed by maximizing the sum of the Spearman correlation with each of the metric rankings (RankAggreg R package) [36]. The correlations between the metric rankings and the aggregated rank are shown in Fig. S3. To limit the risk of overfitting, we aggregated several metrics from statistical models with different mathematical backgrounds. The effects of tidal volume, serum C-reactive protein (CRP) level, body temperature, and the number of days spent in the ICU were investigated in a correlation test with the three first components of the PCA (using a Pearson's test for continuous variables and a chi-squared test for categorical variables) to detect putative factors with a strong impact on the VOC concentrations which may interfere with the prediction of the COVID-19 status (Fig. S1). For the positive end-expiratory pressure (PEEP) and respiratory rate (the median levels of which differed for each COVID-19 status), we performed a Pearson correlation test within each group (as described in the Supplementary Material and Fig. S2). No significant correlations were detected by any of these tests.
A longitudinal univariate analysis of the most important features was performed with a mixed effects model. The fixed effect represents the change in the VOC concentration as a function of the period of mechanical ventilation, with only one measurement per patient per day. We chose a spline function (sum of four b-spline functions basis of degree three uniformly distributed over time) for the fixed effect and an intercept per patient for the random effect. Intergroup differences in trends and means were assessed with an F-test (p-value <0•05) adjusted for the false discovery rate. The test compares the residuals of models with and without COVID status as a predictor. Correlations between VOC concentrations, the SAPS II, the SOFA score, and the viral load were analyzed using Pearson's correlation test, after adjustment for the false discovery rate.
Role of the funding source
The funding source had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. The corresponding author confirms that he had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Patients
Between March 25th and June 25th, 2020, 40 patients (of whom 28 had confirmed COVID-19-related ARDS) were included in the study and a total of 303 measurements were made. Compared with the patients with non-COVID-19 ARDS, the patients with COVID-19 ARDS had (i) a higher respiratory rate, FiO2, PEEP and CRP on admission, (ii) a higher incidence of treatment with hydroxychloroquine and a lower incidence of treatment with fludrocortisone after admission, and (iii) a greater likelihood of renal replacement therapy (Table 1).
Table 1.
Patient characteristics and treatments
| COVID-19 ARDS | Non-COVID-19 ARDS | p value | |
|---|---|---|---|
| Number of patients (n) | 28 | 12 | - |
| Males/females (n) | 20/8 | 6/6 | 0•28 |
| Age (years) | 61 [55-72] | 72 [54-79] | 0•75 |
| Body weight (kg) | 80•0 [66•6-87•6] | 86•5 [65•3-94•1] | 0•71 |
| Height (cm) | 170 [164-175] | 173 [169-175] | 0•55 |
| Body mass index (kg/m²) | 26•3 [23•7-32•4] | 28•9 [23•0-30•9] | 0•79 |
| SAPS II score in the first 24 hours | 62 [49-68] | 46 [40-57] | 0•051 |
| SOFA score in the first 24 hours | 11 [7-12] | 8 [5-12] | 0•37 |
| Comorbidities: (n (%)) | |||
|
11 (39) | 6 (50) | 0•73 |
|
2 (7) | 1 (8) | >0•99 |
|
5 (18) | 3 (25) | 0•68 |
|
2 (7) | 3 (25) | 0•15 |
| Treatments before admission: (n (%)) | |||
|
1 (4) | 3 (25) | 0•073 |
|
5 (18) | 1 (8) | 0•54 |
|
2 (7) | 2 (16) | 0•57 |
| Interventions after admission: (n (%)) | |||
|
17 (61) | 4 (33) | 0•17 |
|
9 (32) | 0 (0) | 0•038 |
| Treatments after admission: (n (%)) | |||
|
27 (96) | 1 (8) | <0•0001 |
|
2 (7) | 0 (0) | >0•99 |
|
7 (25) | 0 (0) | 0•081 |
|
11 (39) | 6 (50) | 0•73 |
|
1 (4) | 4 (33) | 0•022 |
|
12 (43) | 4 (33) | 0•73 |
| Body temperature at first sample (°C) | 37•4 [36•5-38•3] | 37•3 [36•8-37•8] | 0•84 |
| Respiratory rate at first sample (breaths per min) | 26 [25-28] | 20 [18-23] | <0•0001 |
| Tidal volume at first sample (mL) | 420 [400-475] | 438 [400-490] | 0•99 |
| Fraction of inspired oxygen at first sample (%) | 80 [50-100] | 48 [31-68] | 0•007 |
| Positive end-expiratory pressure at first sample (cm H2O) | 10 [8-13] | 5•5 [5-8] | 0•0002 |
| Serum creatinine at first sample (µM) | 74 [56-137] | 67 [44-86] | 0•30 |
| Serum C-reactive protein at first sample (mg/L) | 195 [175-268] | 76 [23-119] | 0•002 |
Continuous data are presented as the median [IQR].
Metabolomic analysis of exhaled breath
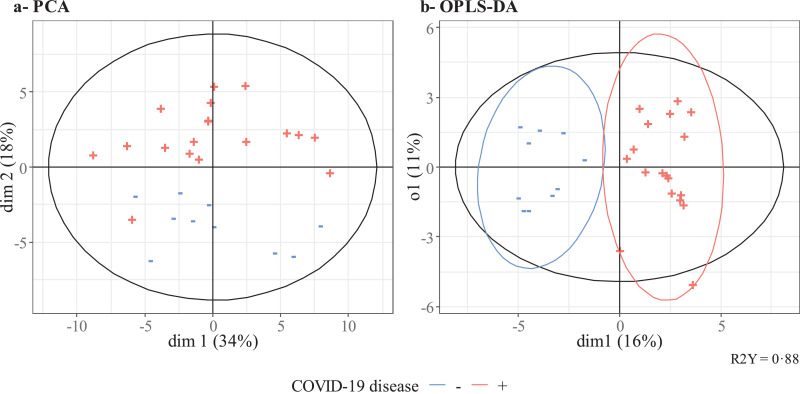
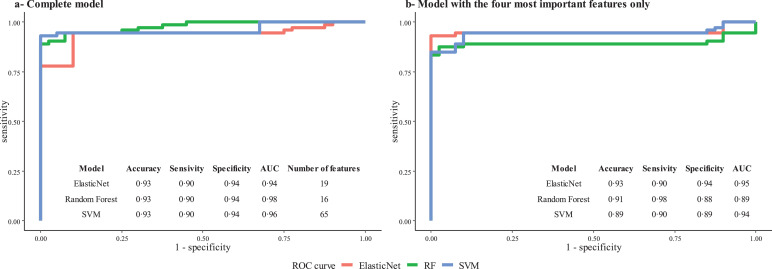
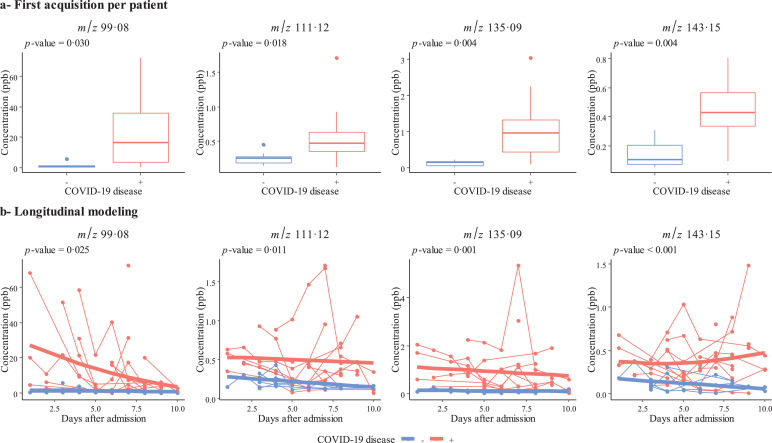
We first used an untargeted metabolomic strategy to discover the signature associated with COVID-19 ARDS. To this end, we used the first breath sample collected after admission. Twelve of the 40 participants had been hospitalized for more than 10 days at the start of the sampling period and so were excluded from this first part of the study. Hence, we analyzed 18 patients with COVID-19 ARDS and 10 with non-COVID-19 ARDS. The study groups’ demographic characteristics are summarized in Table S2. A principal component analysis and an orthogonal partial least-squares discriminant analysis showed that COVID-19 was associated with a specific signature in the expired air, i.e., the breathprint could discriminate between COVID-19 ARDS and non-COVID-19 ARDS cases (Fig. 1). The use of three machine learning algorithms yielded an accuracy of 93% for all three classifiers, based on the selection of 19, 16 or 65 features for the elastic net, random forest, and support vector machine algorithms, respectively (in a 10-fold stratified cross-validation, repeated four times). The corresponding receiver operating characteristic curves are shown in Fig. 2a. A Wilcoxon test with p-value correction for the false discovery rate highlighted VOCs that significantly distinguished between the two groups (p<0•05). We checked that none of the other external covariates impacted the VOC concentrations and interfered with the model's predictions (see the Supplementary Material). To determine which VOCs were most discriminant for COVID-19 ARDS, we performed a rank aggregation based on the various metrics from the previously mentioned models and the hypothesis tests. The four most relevant features in the rank aggregation were at m/z 99•08, 111•12, 135•09, and 143•15 (Fig. 3a). Using these four features only, the elastic net, random forest, and support vector machine algorithms yielded an accuracy of between 89% and 93% (Fig. 2b). We therefore investigated the expression of these VOCs in the whole study population throughout the period of mechanical ventilation (Fig. 3b). We observed that the VOC concentrations (i) were significantly higher in the breath of patients with COVID-19 ARDS than in the breath of patients with non-COVID-19 ARDS, and (ii) tended to decrease over the first 10 days of hospitalization. The putative annotations for the four compounds at m/z 99•08, 111•12, 135•09, and 143•15 were respectively methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal.
Fig. 1.
Multivariate analysis. Principal component analysis (left) and orthogonal partial least squares - discriminant analysis (right) of the breath signature in intubated, mechanically ventilated ICU patients with a positive (red) or negative (blue) PCR test for SARS-CoV-2.
Fig. 2.
Receiver operating characteristic curves for models classifying patients with COVID-19 vs. non-COVID-19 ARDS. a. Complete model. The use of three machine learning algorithms (elastic net, support vector machine (SVM), and random forest (RF)) yielded an accuracy of up to 93%, with a 10-fold cross validation repeated four times and based on the selection of 19 features (elastic net), 16 features (random forest) or all 65 features (support vector machine) from the full dataset. After internal cross-validation, the sensitivity was 90% and the specificity was 94%. b. Model with the four most important features only. After internal cross-validation, the sensitivity ranged from 90% to 98% and the specificity ranged from 88% to 94%.
Fig. 3.
Longitudinal analysis of VOCs in expired breath. The four features (m/z 99•08, 111•12, 135•09, and 143•15) contributing the most to the models were assessed in the first sample available for each patient (a) and over time (b) during the ICU stay for intubated, mechanically ventilated patients with COVID-19 ARDS (in red, n = 28) or non-COVID-19 ARDS (in blue, n = 12). All the points for a given patient are connected, and the bold lines correspond to the fixed effect of the mixed model for each group. p-values come from a Wilcoxon test (a) and an F-test (b).
Correlation with viral load and severity scores
The viral load in bronchoalveolar fluid was measured for 18 patients. The median [IQR] value in the first sample was 7•2 [6•2–8•4] log eq. copies/mL. The VOC concentrations in the first sample were not significantly correlated with the bronchoalveolar fluid viral load or with the severity of illness (i.e., the SAPS II and SOFA score) [27,28] measured during the first 24 h in the ICU (Table 2, |r| < 0•4).
Table 2.
Correlations between VOC concentrations and the SAPS II, SOFA score and viral load.
| SAPS II score | SOFA score | Viral load | ||||
|---|---|---|---|---|---|---|
| VOC (m/z) | r | p-value | r | p-value | r | p-value |
| 99•08 | 0•04 | 0•88 | 0•36 | 0•13 | 0•08 | 0•70 |
| 111•12 | 0•02 | 0•93 | 0•28 | 0•25 | -0•14 | 0•48 |
| 135•09 | 0•05 | 0•85 | 0•35 | 0•14 | -0•0004 | 1•00 |
| 143•15 | 0•12 | 0•62 | 0•27 | 0•25 | -0•23 | 0•24 |
r: Pearson's correlation coefficient.
Discussion
This study provided proof of concept for the measurement of VOCs and the determination of a specific VOC breathprint in the exhaled breath from patients with COVID-19-related ARDS requiring invasive mechanical ventilation in the ICU. This breathprint was independent of the severity of illness and the viral load. Four VOCs (methylpent-2-enal, 2,4-octadiene 1-chloroheptane, and nonanal) may discriminate between COVID-19 and non-COVID-19 ARDS.
We applied a highly sensitive, rapid, non-invasive, real-time mass spectrometry breath analysis [37,38]. This contrasts with offline technologies, which require a sampling step and remote, time-consuming analytical steps [21,22]. Implementation of a non-targeted strategy (as described here) is mandatory for the discovery of novel biomarkers. The subsequent diagnostic validation and clinical implementation can be based on less cumbersome technologies, such as mass spectrometers dedicated to targeted analyses or portable “electronic noses” with a set of sensors that are relatively selective for different families of VOCs (as previously used in patients with ARDS) [20].
The first (cross-sectional) part of the present study enabled us to identify a specific signature. We then performed a longitudinal analysis of expired air in ARDS patients, which allowed us to confirm the VOC signature and to track the changes over time in the VOC concentrations. Two of the four prominent VOCs (methylpent-2-enal and nonanal) are aldehydes, while 2,4-octadiene is an alkadiene. These three compounds are known to be expressed in breath [39,40], while 1-chloroheptane is probably not endogenous. Nonanal is a sub-product of the destruction of the cell membrane as a result of oxidative stress; reactive oxygen species may be generated by various type of inflammatory, immune and structural cell in the airways [41]. In studies of expired air from patients with ARDS, Schubert et al. found abnormally low isoprene concentrations and Bos et al. reported abnormally high concentrations of octane, acetaldehyde and 3-methylheptane [21,22]. Differences in study populations (non-COVID-19 vs. COVID-19 ARDS) and analytical methods (offline vs. online) might explain the differences between the VOCs identified in the present study and those identified in previous studies of ARDS [21,22]. Although there may be an association between VOCs and disease, the underlying biochemistry has not been fully characterized.
In line with previous reports, the VOC concentrations measured here were not correlated with the severity of illness (as judged by the SAPS II and the SOFA score) [21]. This finding suggest that the exhaled breath signature is a marker of COVID-19 per se, rather than of the severity of illness. Likewise, the VOC concentrations were not correlated with viral load, suggesting that this signature may be a marker of the disease related to SARS-CoV-2 rather than of virus carriage.
Our interpretation of the present data may have been limited by differences between the COVID-19 and non- COVID-19 ARDS subgroups. Patients with COVID-19 ARDS cohort had higher respiratory rate, FiO2, PEEP and CRP values on admission. The respiratory rate, PEEP and CRP were not found to interfere with the VOC predictive signature, and all the patients were sampled when breathing 100% FiO2 (to avoid mass spectrometry interference by oxygen) [29]. Similarly, patients with COVID-19 ARDS were more likely to have been treated with hydroxychloroquine. However, this drug was administered to the patients after their first sample had been analyzed. Although the VOC concentrations decreased over time, the treatments did not change, and there was no correspondence between the VOCs described in the present study and the molecular masses of the known metabolites of hydroxychloroquine. Lastly, the sample size of this pilot study was limited and these observations will require confirmation with an external validation cohort.
In conclusion, we determined a COVID-19-specific breath metabolomic signature in patients with ARDS requiring invasive mechanical ventilation. Knowledge of this specific breathprint might enable the development of rapid, non-invasive, point-of-care tests for large-scale COVID-19 screening.
Acknowledgments
Acknowledgements
The authors would like to thank all the staff members at the intensive care unit at Raymond Poincaré Hospital for their collaboration, Professor Marie-Anne Welti and Professor Elyanne Gault for providing virological data, and Dr David Fraser (Biotech Communication SARL, Ploudalmézeau, France) for copy-editing assistance. The study was funded by Agence Nationale de la Recherche (SoftwAiR, ANR-18-CE45-0017 and RHU4 RECORDS, Programme d'Investissements d'Avenir, ANR-18-RHUS-0004), Région Île de France (SESAME 2016), and Fondation Foch.
Contributors
S.G.D. and D.A. conceived the study. S.G.D., P.M. and C.R. defined parameters for mass spectrometry breath analysis. S.G.D., G.S., S.C., J.F., H.S., E.N., L-J.C., P.D., P.M., D.A. performed the experiments and analyzed and/or interpreted results. S.G.D., P.M., N.H., D.A. collected epidemiological and clinical data. P.M. and N.H. assisted in patient recruitment. C.R. and E.T. developed software and analyzed the data. S.G.D. and C.R. checked the underlying data. S.G.D., C.R. and D.A. drafted the manuscript. E.T., P.M., P.D., L-J.C., H.S, E.N., G.S., S.C., N.H., J.F. revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Competing Interests
DA has received a grant from Agence Nationale de la Recherche to conduct the RECORDS program, of which this study is part (ANR-18-RHUS-0004). S.G.D, C.R., E.T. and D.A. are named as inventors on a patent application covering breath analysis in COVID-19. The authors declare no other conflicts of interest.
Data sharing statement
The study protocol and the datasets generated during and/or analysed during the current study, including deidentified participant data will be available with publication from the corresponding author on reasonable request. The ptairMS R package used for data analysis is publicly available at https://github.com/camilleroquencourt/ptairMS
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103154.
Appendix. Supplementary materials
References
- 1.COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html (accessed October 19th, 2020).
- 2.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 3.Beigel JH, Tomashek KM, Dodd LE. Remdesivir for the treatment of COVID-19 - final report. N Engl J Med. 2020 doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 4.Recovery_Collaborative_Group. Horby P, Lim WS. Dexamethasone in hospitalized patients with COVID-19 - preliminary report. N Engl J Med. 2020 [Google Scholar]
- 5.Lucas C, Wong P, Klein J. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadjadj J, Yatim N, Barnabei L. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuri-Cervantes L, Pampena MB, Meng W. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5(49) doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen B, Yi X, Sun Y. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1) doi: 10.1016/j.cell.2020.05.032. 59-72 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataoka H, Saito K, Kato H, Masuda K. Noninvasive analysis of volatile biomarkers in human emanations for health and early disease diagnosis. Bioanalysis. 2013;5(11):1443–1459. doi: 10.4155/bio.13.85. [DOI] [PubMed] [Google Scholar]
- 10.Rattray NJ, Hamrang Z, Trivedi DK, Goodacre R, Fowler SJ. Taking your breath away: metabolomics breathes life in to personalized medicine. Trends Biotechnol. 2014;32(10):538–548. doi: 10.1016/j.tibtech.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Amann A, de Lacy Costello B, Miekisch W. The human volatilome: volatile organic compounds (VOCs) in exhaled breath, skin emanations, urine, feces and saliva. J Breath Res. 2014;8(3) doi: 10.1088/1752-7155/8/3/034001. [DOI] [PubMed] [Google Scholar]
- 12.de Lacy Costello B, Amann A, Al-Kateb H. A review of the volatiles from the healthy human body. J Breath Res. 2014;8(1) doi: 10.1088/1752-7155/8/1/014001. [DOI] [PubMed] [Google Scholar]
- 13.Koo S, Thomas HR, Daniels SD. A breath fungal secondary metabolite signature to diagnose invasive aspergillosis. Clin Infect Dis. 2014;59(12):1733–1740. doi: 10.1093/cid/ciu725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakhleh MK, Jeries R, Gharra A. Detecting active pulmonary tuberculosis with a breath test using nanomaterial-based sensors. Eur Respir J. 2014;43(5):1522–1525. doi: 10.1183/09031936.00019114. [DOI] [PubMed] [Google Scholar]
- 15.Coronel Teixeira R, Rodriguez M, Jimenez de Romero N. The potential of a portable, point-of-care electronic nose to diagnose tuberculosis. J Infect. 2017;75(5):441–447. doi: 10.1016/j.jinf.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Suarez-Cuartin G, Giner J, Merino JL. Identification of pseudomonas aeruginosa and airway bacterial colonization by an electronic nose in bronchiectasis. Respir Med. 2018;136:111–117. doi: 10.1016/j.rmed.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Schnabel R, Fijten R, Smolinska A. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci Rep. 2015;5:17179. doi: 10.1038/srep17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipiak W, Beer R, Sponring A. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: a prospective pilot study. J Breath Res. 2015;9(1) doi: 10.1088/1752-7155/9/1/016004. [DOI] [PubMed] [Google Scholar]
- 19.Schnabel RM, Boumans ML, Smolinska A. Electronic nose analysis of exhaled breath to diagnose ventilator-associated pneumonia. Respir Med. 2015;109(11):1454–1459. doi: 10.1016/j.rmed.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Bos LD, Schultz MJ, Sterk PJ. Exhaled breath profiling for diagnosing acute respiratory distress syndrome. BMC Pulm Med. 2014;14:72. doi: 10.1186/1471-2466-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bos LD, Weda H, Wang Y. Exhaled breath metabolomics as a noninvasive diagnostic tool for acute respiratory distress syndrome. Eur Respir J. 2014;44(1):188–197. doi: 10.1183/09031936.00005614. [DOI] [PubMed] [Google Scholar]
- 22.Schubert JK, Muller WP, Benzing A, Geiger K. Application of a new method for analysis of exhaled gas in critically ill patients. Intensive Care Med. 1998;24(5):415–421. doi: 10.1007/s001340050589. [DOI] [PubMed] [Google Scholar]
- 23.van Geffen WH, Bruins M, Kerstjens HA. Diagnosing viral and bacterial respiratory infections in acute COPD exacerbations by an electronic nose: a pilot study. J Breath Res. 2016;10(3) doi: 10.1088/1752-7155/10/3/036001. [DOI] [PubMed] [Google Scholar]
- 24.Traxler S, Bischoff AC, Sass R. VOC breath profile in spontaneously breathing awake swine during Influenza A infection. Sci Rep. 2018;8(1):14857. doi: 10.1038/s41598-018-33061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackermann M, Verleden SE, Kuehnel M. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Force ADT, Ranieri VM, Rubenfeld GD. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307(23):2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 27.Le Gall JR, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA. 1993;270(24):2957–2963. doi: 10.1001/jama.270.24.2957. [DOI] [PubMed] [Google Scholar]
- 28.Vincent JL, Moreno R, Takala J. The SOFA (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the european society of intensive care medicine. Intensive Care Med. 1996;22(7):707–710. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- 29.Trefz P, Pugliese G, Brock B, Schubert JK, Miekisch W. Effects of elevated oxygen levels on VOC analysis by means of PTR-ToF-MS. J Breath Res. 2019;13(4) doi: 10.1088/1752-7163/ab28ec. [DOI] [PubMed] [Google Scholar]
- 30.Hansel A, Jordan A, Holzinger R, Prazeller P, Vogel W, Lindinger W. Proton transfer reaction mass spectrometry: on-line trace gas analysis at the ppb level. Int J Mass Spectrom Ion Process. 1995;149-150:609–619. [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 32.Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 33.Thevenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res. 2015;14(8):3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- 34.Weston J, Mukherjee S, Chapelle O, Pontil M, Poggio T, Vapnik V. Feature selection for SVMs. Adv Neural Inform Process Syst. 2000;13 [Google Scholar]
- 35.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B. 2005;67(2):301–320. [Google Scholar]
- 36.Pihur V, Datta S, Datta S. RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics. 2009;10:62. doi: 10.1186/1471-2105-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trefz P, Schmidt M, Oertel P. Continuous real time breath gas monitoring in the clinical environment by proton-transfer-reaction-time-of-flight-mass spectrometry. Anal Chem. 2013;85(21):10321–10329. doi: 10.1021/ac402298v. [DOI] [PubMed] [Google Scholar]
- 38.Brock B, Kamysek S, Silz J, Trefz P, Schubert JK, Miekisch W. Monitoring of breath VOCs and electrical impedance tomography under pulmonary recruitment in mechanically ventilated patients. J Breath Res. 2017;11(1) doi: 10.1088/1752-7163/aa53b2. [DOI] [PubMed] [Google Scholar]
- 39.van de Kant KD, van Berkel JJ, Jobsis Q. Exhaled breath profiling in diagnosing wheezy preschool children. Eur Respir J. 2013;41(1):183–188. doi: 10.1183/09031936.00122411. [DOI] [PubMed] [Google Scholar]
- 40.Corradi M, Pignatti P, Manini P. Comparison between exhaled and sputum oxidative stress biomarkers in chronic airway inflammation. Eur Respir J. 2004;24(6):1011–1017. doi: 10.1183/09031936.04.00002404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman I. Oxidative stress, chromatin remodeling and gene transcription in inflammation and chronic lung diseases. J Biochem Mol Biol. 2003;36(1):95–109. doi: 10.5483/bmbrep.2003.36.1.095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.