Abstract
Background & Aims
There is a considerable degree of variation in bone mineral density (BMD) within populations. Use of plasma metabolomics may provide insight into established and novel determinants of BMD variance, such as nutrition and gut microbiome composition, to inform future prevention and treatment strategies for loss of BMD. Using high-resolution metabolomics (HRM), we examined low-molecular weight plasma metabolites and nutrition-related metabolic pathways associated with BMD.
Methods
This cross-sectional study included 179 adults (mean age 49.5 ± 10.3 yr, 64% female). Fasting plasma was analyzed using ultra-high-resolution mass spectrometry with liquid chromatography. Whole body and spine BMD were assessed by dual energy x-ray absorptiometry and expressed as BMD (g/cm2) or Z-scores. Multiple linear regression, pathway enrichment, and module analyses were used to determine key plasma metabolic features associated with bone density.
Results
Of 10,210 total detected metabolic features, whole body BMD Z-score was associated with 710 metabolites, which were significantly enriched in seven metabolic pathways, including linoleic acid, fatty acid activation and biosynthesis, and glycerophospholipid metabolism. Spine BMD was associated with 970 metabolites, significantly enriched in pro-inflammatory pathways involved in prostaglandin formation and linoleic acid metabolism. In module analyses, tryptophan- and polyamine-derived metabolites formed a network that was significantly associated with spine BMD, supporting a link with the gut microbiome.
Conclusions
Plasma HRM provides comprehensive information relevant to nutrition and components of the microbiome that influence bone health. This data supports pro-inflammatory fatty acids and the gut microbiome as novel regulators of postnatal bone remodeling.
Keywords: Inflammation, nutrition, omega-6, fats, PUFA, skeletal health
1. INTRODUCTION
Numerous factors contribute to postnatal skeletal remodeling, peak bone mineral density (BMD), and age-related skeletal involution. These include non-modifiable influences such as sex, age, genetics, and ethnicity, as well as modifiable factors such as dietary intake, physical activity, tobacco use, and hormonal status (1). As lifestyle behaviors determine an estimated 20–40% of peak bone mass (2), strategies to optimize post-natal skeletal development or prevent metabolic bone diseases often focus on understanding these influences to identify interventions that can maximize accrual of bone mass in children and young adults or minimize bone loss throughout aging.
Adequate nutrient intake plays an integral role in optimizing bone health. Knowledge of the relationships between bone metabolism and diet has expanded beyond classical nutrients such as vitamin D and calcium to include additional critical minerals, vitamins, bioactive compounds, and macronutrient intake (1). More recently, investigators have examined dietary patterns, rather than single nutrients, and linked more healthful dietary patterns to improved measures of bone health (3).
Dietary intake, including fiber and other compounds, also influences the composition and diversity of the gut microbiome (4–6). The intestinal microbiome is a diverse environment that hosts trillions of microorganisms, which interact with metabolism and digestion and impact the host’s immune system (7). It is now clear that a significant number of plasma metabolites are products of gut microbiome metabolism (8). Emerging research suggests that microbiome-derived metabolites may have effects beyond the gastrointestinal tract such as regulating bone formation and resorption, though few studies have investigated this relationship in humans (4, 9–11).
While adequate intake of nutrients in the diet is needed for optimal bone mass accrual and health, rigorous data on these relationships are conflicting (2). It is therefore necessary to increase our understanding of interactions between endogenous nutrition-related metabolism and bone health to inform new approaches. High-resolution metabolomics (HRM) is a novel and rapidly developing tool that enables complex biosystems research by integrating metabolic pathways with environmental determinants and health outcomes (12). HRM assessment provides a global, unbiased measure of metabolism by profiling thousands of low molecular weight chemicals that are derived from food and beverage consumption, the gut microbiome, drugs, environmental chemicals, and biological compounds (12). Thus, utilizing HRM as a state-of-the-art tool can provide innovative understanding of the interactions between diet and the gut microbiome with bone metabolism. Given the limited knowledge about these relationships, we utilized HRM to interrogate the plasma metabolome related to bone health, as this novel platform can identify metabolites and metabolic pathways derived from both macronutrient metabolism and the gut microbiome.
2. MATERIALS AND METHODS
2.1. Participants and Study Design
This study population included 179 adults (mean age 49.5 ± 10.3 yr., 64% female) from a metropolitan area cohort. Participants were primarily Emory University or Healthcare employees enrolled in the Emory-Georgia Tech Center for Health Discovery and Well-Being Cohort in whom baseline plasma HRM was measured. Participants were invited to join the study and enrolled from 2007–2013 (13). Subjects participated in extensive health assessments involving clinical laboratories, health and medical history questionnaires, body composition analysis, and physical tests. A complete description of the study and included assessment procedures is available (14). Major exclusion criteria included current pregnancy, new prescription medications within the previous year other than blood pressure or diabetic medications, history of severe psychosocial disorder, history of substance or alcohol abuse, acute illness within two weeks of the study visit, or uncontrolled or poorly controlled autoimmune, cardiovascular, endocrine, gastrointestinal, hematologic, infectious, inflammatory, musculoskeletal, neurologic, psychiatric or respiratory disease (14). The study was approved by the Emory University Institutional Review Board, and all participants provided written informed consent prior to study participation.
2.2. Demographic, Clinical, and Lifestyle Measures
Sex and race were determined by self-report. Current antibiotic, probiotic, supplement, and medication use were assessed by reviewing self-reported medication logs. Participants were categorized as having a chronic disease if they reported a current diagnosis of hypertension, diabetes mellitus, and/or hyperlipidemia, or if they were taking a medication to treat those conditions at the time of enrollment. Peripheral blood samples were collected following an overnight fast, and plasma and serum were stored at −80°C until ready for assay analyses. Serum 25-hydroxyvitamin D [25(OH) D] concentrations were measured commercially (Quest Diagnostics Nichols Valencia, Valencia, CA) by liquid chromatography/tandem mass spectrometry. Tobacco use was collected by self-report. Physical activity was assessed using the Cross-Cultural Activity Participation Survey (CAPS) (15). Participants were then categorized into meeting or not meeting the 2007 American College of Sports Medicine/American Heart Association physical activity and strength guidelines if they reported completing 30 minutes of moderate aerobic activity five days a week or 20 minutes of vigorous aerobic activity three times a week and two days a week of strength training, respectively (16). Alcohol consumption was calculated from Block food frequency questionnaire data. The daily reported consumption of alcoholic drinks was converted to grams of ethanol per day and categorized into light (<11 g/day), moderate (11–30 g/day), or heavy (>30 g/day) alcohol consumption. Dietary intake was estimated using Block food frequency questionnaires (FFQ) (17, 18). Dietary data that was considered implausible (<500 or >5,000 kcal/day) were excluded from analysis, and all reported dietary intake data were adjusted per 1000 kcal consumed.
Each subject underwent a total body composition scan assessed by dual energy x-ray absorptiometry (DXA, Lunar iDXA densitometer, GE Healthcare, Madison, WI, USA) using the Lunar reference population. For the assessment, subjects laid in the supine position with arms at their sides and were instructed to wear clothes that did not contain any metal. Whole body BMD and spine BMD were expressed in Z-scores or as gm/cm2.
2.3. High-Resolution Metabolomics (HRM)
Plasma HRM analyses followed a published workflow in the Emory University Clinical Biomarkers Laboratory (19–22). Briefly, 65 μL of plasma was added to 130 μL acetonitrile along with a mixture of internal standard stable isotopes (19). All samples were analyzed in triplicate in batches of 20 samples. Quality control samples were included at the beginning and end of each batch. Samples were analyzed on a Fourier transform mass spectrometer (Dionex Ultimate 3000, Q-Exactive HF, Thermo Fisher) with C18 chromatography in positive electrospray ionization (ESI) mode and 70,000 resolution (19). Data extraction of raw files was completed using the validated R programs apLCMS (23) and xMSanalyzer (24) with subsequent batch correction by ComBat (25). Triplicate values for each sample were averaged, filtered for less than 50% non-missing values, log10 transformed, and mean-centered. Metabolite annotation was performed by matching accurate mass to charge (m/z) ratios of metabolic features to previously confirmed identities (19, 20) (26), which are equivalent to a Metabolomics Standard Initiative (MSI) level 1 metabolite identification (27). Additionally, select metabolic features were confirmed using ion dissociation spectra (MS/MS) that were obtained using a Thermo Scientific Fusion Mass Spectrometer for MS/MS spectral library matching using the mzCloud database (www.mzcloud.org). When level 1 metabolite identification was not available, annotation was completed using the R package xMSannotator (28). xMSannotator provides a confidence score for metabolite annotation from 0 to 3, with 3 being the highest confidence according to four orthogonal criteria. Annotations from xMSannotator with a high confidence score (≥2) were considered a level 2 annotation according to MSI. Low confidence xMSannotator annotations were considered an MSI level 3 or 4 annotation. Metabolite databases, including Human Metabolome Database (www.hmdb.ca) and Metlin (metlin.scripps.edu), were searched using common adducts for positive mode data and a threshold of 10 ppm. Metabolic features without matches to common adducts within a 10 ppm window were labeled as “unknown.”
2.4. Statistical Analyses and Bioinformatics
Descriptive statistics were performed for demographic and clinical variables using JMP Pro (Version 14, SAS Institute Inc, Cary, NC). Continuous variables were summarized as mean ± standard deviation and categorical variables were summarized as count (percentage). Bioinformatics analyses for HRM data were performed using R. The metabolome wide association studies (MWAS) utilized multiple linear regression analyses to determine the associations of metabolic features (metabolites) with whole body BMD Z-score and spine BMD, adjusting for age, race, sex, and body mass index (BMI) as a priori covariates. False discovery rate (FDR) correction was applied using the Benjamini-Hochberg method (q=0.2) (29). Additional variables considered for confounding were exogenous hormone use (estrogen or testosterone), antibiotic use, probiotic use, and tobacco smoking but due to very small proportions of the population using these products, the variables were not included in analyses. Physical activity and alcohol intake were also considered as confounders but were not related to the measures of bone density, so were not included in statistical models. History of diabetes, hypertension, or dyslipidemia were also evaluated for confounding, however, any statistically significant differences in BMD outcomes between groups with and without history of disease were mediated by a higher BMI, which was included in the final models of all analyses. For specific metabolic features of interest, MetabNet (30) was used to examine networks of significantly associated metabolic features using Spearman correlations. Pathway and module analyses were performed using mummichog (31). Mummichog utilizes computational algorithms to provide pathway enrichment and module analyses of metabolic features without requiring initial chemical identity. The pathway enrichment and module analyses differ in that pathway enrichment analyses include known biological pathways, while module analyses are unbiased and build modules of significantly correlated metabolic features.
3. RESULTS
Demographic, clinical, and lifestyle characteristics of the 179 participants are shown in Table 1. The average age of participants was 49.5 years. The majority of the cohort was female and Caucasian. Among the 116 females, 41 (23%) were classified as post-menopausal. The average BMI was in the overweight range (27.3 ± 5.5). Almost one-third of subjects reported a current diagnosis of diabetes mellitus, hypertension, and/or hyperlipidemia, or were taking medications to treat glucose intolerance (n=9), hypertension (n=7) or hyperlipidemia (n=8). Only 4 participants fasting plasma glucose values within the hyperglycemic range (>125 mg/dL). The majority of the cohort (89%) had adequate levels of serum 25(OH)D. A small proportion of participants reported taking a vitamin D (11%) or calcium (18%) supplement. Two individuals reported daily use of a probiotic, and two participants reported use of an antibiotic. Eight participants (4%) reported use of testosterone, and one participant reported use of estrogen. Eight participants (4%) reported taking bisphosphonates. Nearly one-half of participants met the moderate or vigorous physical activity guidelines (42%), while only one-fourth of the participants met the strength exercises guideline (23%). A small proportion of the population were current tobacco smokers (4%). The majority of the cohort reported light alcohol consumption (67%). Whole body BMD Z-score and spine BMD were positively correlated (r= 0.59, p< 0.001, Supplemental Figure 1).
Table 1.
Demographic, clinical, and lifestyle characteristics of all participants (n=179)
| Demographic characteristics | |
| Age, years | 49.5 ± 10.3 |
| Female, n (%) | 116 (65) |
| Post-menopausal, n (%) | 41 (23) |
| Race | |
| White, n (%) | 137 (77) |
| Black, n (%) | 34 (19) |
| Asian, n (%) | 8 (4) |
| Clinical characteristics | |
| Body mass index, kg/m2 | 27.3 ± 5.5 |
| History of chronic disease, n (%) | 49 (27) |
| History of type 2 diabetes mellitus, n (%)a | 33 (18) |
| History of hypertension, n (%)a | 14 (8) |
| History of dyslipidemia, n (%)a | 35 (20) |
| 25(OH)D, ng/mLb | 34.2 ± 13.5 |
| 25(OH)D <20 ng/mL, n (%) | 20 (11) |
| Supplements and Medications | |
| Vitamin D supplementation, n (%) | 20 (11) |
| Calcium supplementation, n (%) | 33 (18) |
| Current probiotics use, n (%)c | 2 (1) |
| Current antibiotics prescription, n (%) | 2 (1) |
| Testosterone prescription, n (%) | 1 (<1) |
| Estrogen prescription, n (%) | 8 (4) |
| Bisphosphonate prescription, n (%) | 8 (4) |
| Lifestyle Characteristics | |
| Smoking, n (%) | 8 (4) |
| Met MVPA guidelines, n (%) | 75 (42) |
| Met strength guidelines, n (%) | 41 (23) |
| Light alcohol consumption (<11 g/day), n (%) | 119 (66) |
| Moderate alcohol consumption (11–30 g/day), n (%) | 46 (26) |
| Heavy alcohol consumption (>30 g/day), n (%) | 14 (8) |
| Bone density measures | |
| Spine BMD, g/cm | 1.1 ± 0.2 |
| Whole Body BMD Z-Score | 0.5 ± 1.2 |
Data are presented as mean ± SD or n (%).
Includes participants who reported a history of disease or current prescription to treat respective disease state.
n=175
Reported daily use of probiotic supplements containing 1 billion colony-forming units (CFU)
Abbreviations: MVPA, moderate to vigorous physical activity; 25(OH)D, 25-hydroxyvitamin D; BMD, bone mineral density
3.1. Whole Body BMD Z-Score MWAS
A total of 10,210 metabolic features were detected by HRM in plasma samples. Whole body BMD Z-score was significantly associated with 22 metabolic features at FDR q=0.2, including metabolites related to linoleic acid (α-linolenoyl ethanoliamide), phenylacetic acid, and a leukotriene metabolite (Supplementary Table 1). There were 710 metabolites significantly associated with whole body BMD Z-score at p<0.05, which were utilized as input for pathway enrichment and module analyses in mummichog (Figure 1A). Pathway analysis identified seven significantly enriched pathways (all p<0.05), primarily comprised of fatty acid related-metabolic pathways (Figure 1B). Enriched metabolic pathways associated with whole body BMD Z-score included linoleic acid metabolism, fatty acid activation and biosynthesis, glycosphingolipid metabolism, glycerophospholipid metabolism, saturated fatty acids beta-oxidation, and fatty acid metabolism. Independent of pathway analyses, module analyses identified a group of fatty acid-related metabolites that were all significantly, inversely associated with whole body BMD Z-score (p=0.002), including linoleic acid, gamma-linolenic acid, oleic acid, palmitic acid, and the linoleic acid oxylipins hydroperoxy-octadecadienoic acid (HPODE), hydroxyoctadecadienoic acid (HODE), and epoxyoctadecenoic acid (EpOME) (Figure 1C). Linoleic acid, HPODE, and HODE/EpOME metabolites were all confirmed using MS/MS methods and are considered a level 1 identification. The inverse relationships between linoleic acid, HPODE, and HODE/EpOME with whole body BMD Z-score are shown in Figure 2 (p<0.05 for all).
Figure 1.
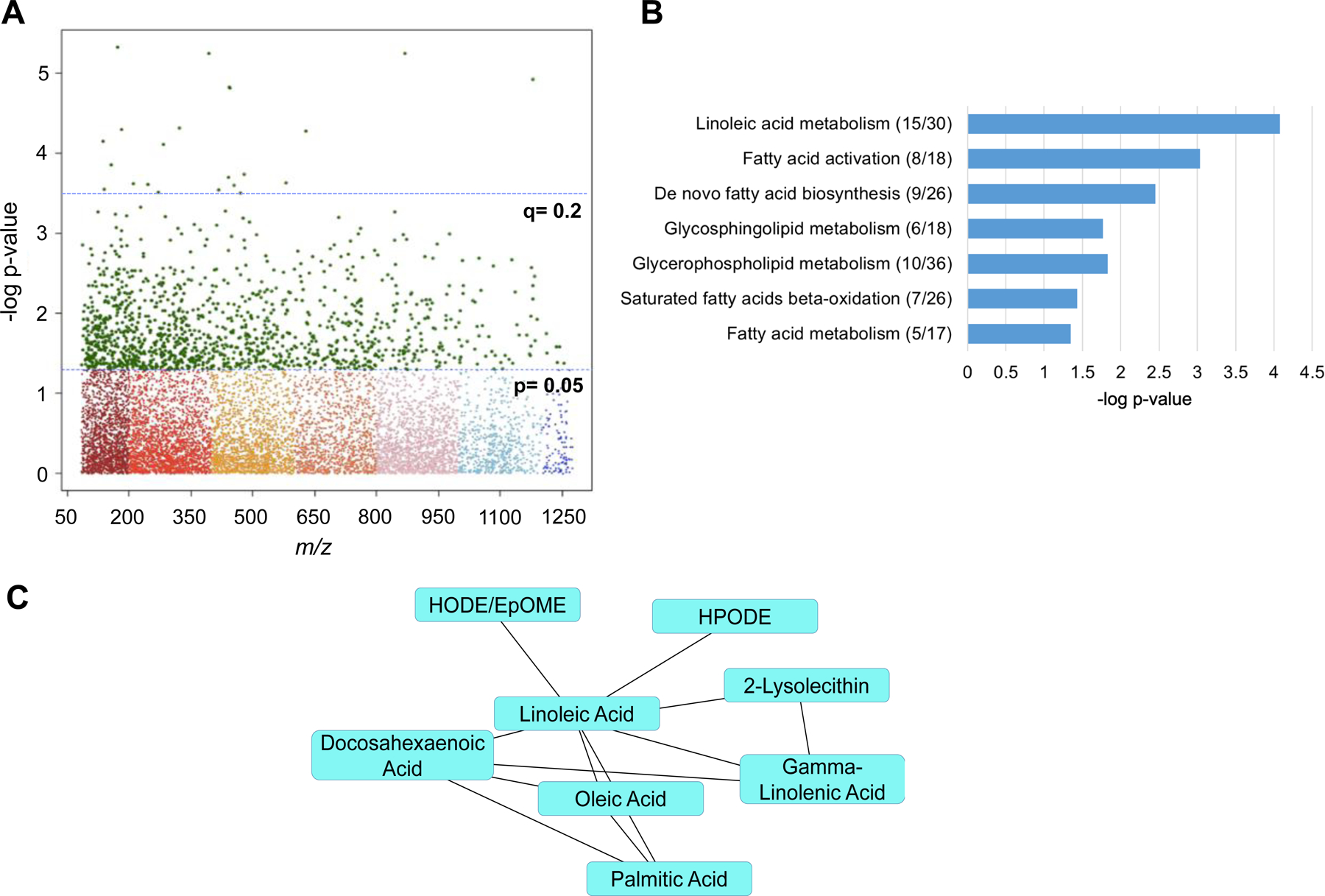
A) Manhattan plot denoting 1,267 significant metabolic features at p<0.05, and 22 metabolic features significant at q<0.2. B) Significantly enriched metabolic pathways associated with whole body BMD Z-score. Significant metabolites over total pathway metabolites are shown in parentheses. C) Module analysis of the relationship between metabolites significantly, inversely associated with whole body BMD Z-score. Linoleic acid (C18:2), HPODE, and HODE/EpOME were all confirmed using MS/MS methods. Abbreviations: HPODE, hydroperoxy-octadecadienoic acid; HODE, hydroxyoctadecadienoic acid; EpOME, epoxyoctadecenoic acid.
Figure 2.
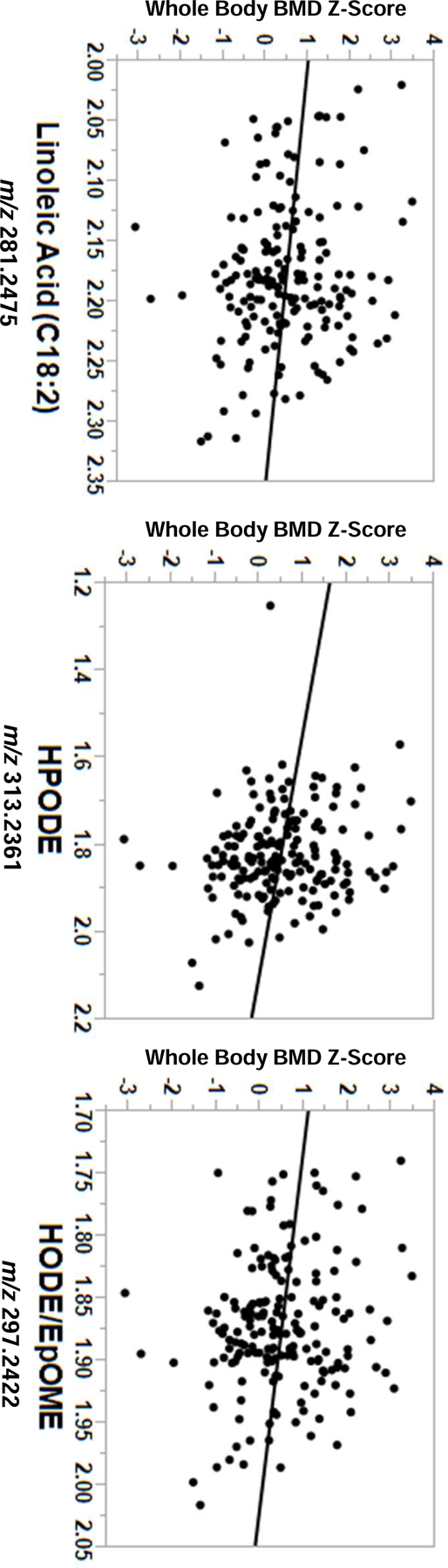
Inverse relationships between linoleic acid (C18:2) and linoleic acid-derived oxylipins with whole body BMD Z-score (p<0.05 for all), adjusting for age, race, sex, and body mass index. Metabolite intensity values are x10−4. Abbreviations: HPODE, hydroperoxy-octadecadienoic acid; HODE, hydroxyoctadecadienoic acid; EpOME, epoxyoctadecenoic acid.
3.2. Spine BMD MWAS
Spine BMD was significantly associated with nine metabolites at q=0.2 (Supplemental Table 1) and 970 metabolites at p<0.05. All metabolites significant at a raw p-value were used as input for pathway enrichment analysis (Figure 3A). Three pathways were significantly enriched (all p<0.05), including prostaglandin formation from arachidonic acid, linoleic acid metabolism, and glycolysis and gluconeogenesis (Figure 3B). Module analyses grouped several metabolites related to tryptophan and polyamine metabolism into an overarching module that was significantly associated with spine BMD (p=0.001) and contained the microbiome-related metabolites, indoleacetylaldehyde and N-acetylputrescine, which were positively and inversely correlated with spine BMD, respectively (Figure 3C).
Figure 3.
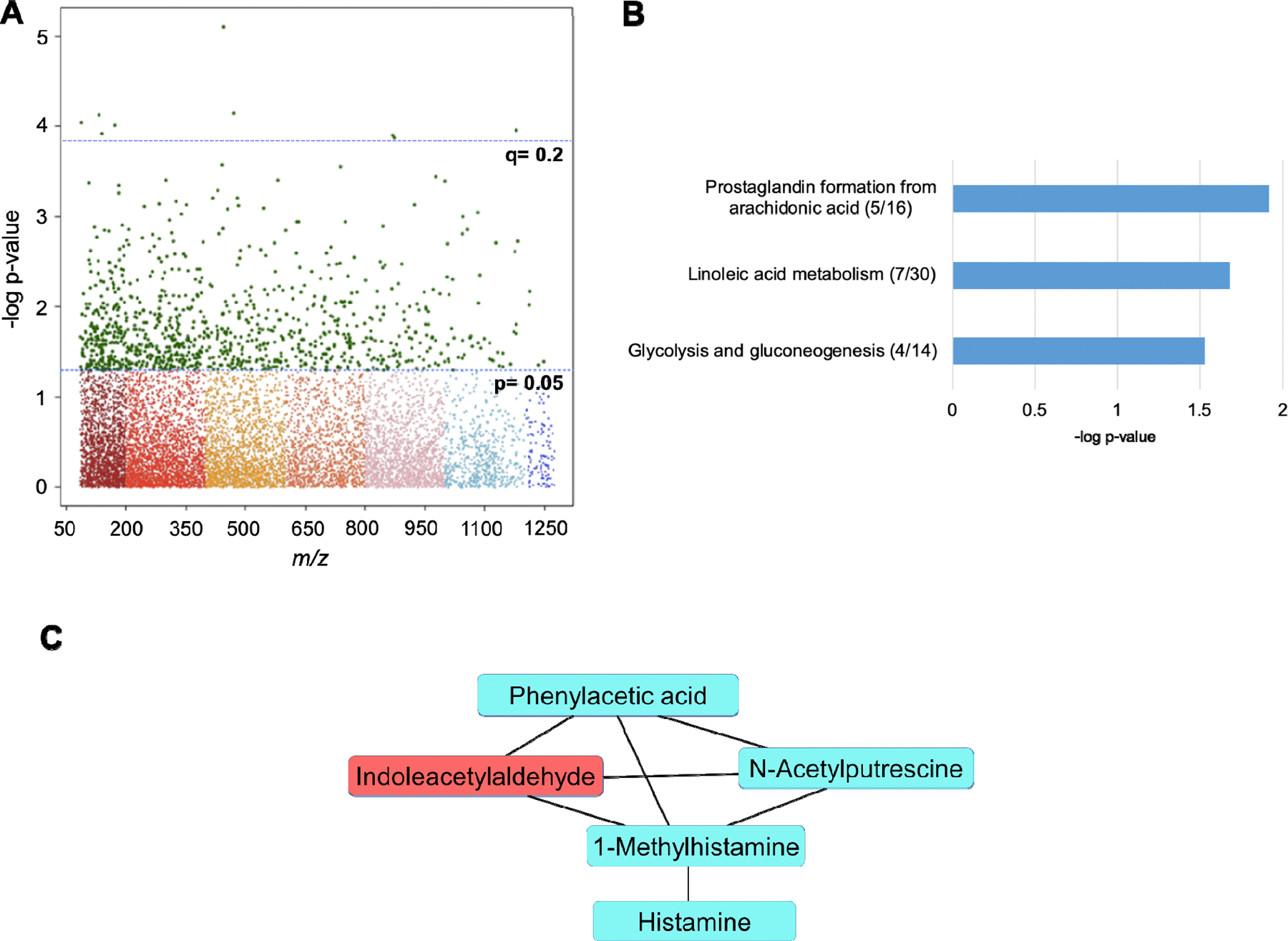
A) Manhattan plot denoting 970 significant metabolic features at p<0.05, and nine metabolic features significant at q<0.2. B) Significantly enriched metabolic pathways associated with spine BMD. Significant metabolites over total pathway metabolites detected are shown in parentheses. C) Module analysis of the relationship between metabolites significantly associated with spine BMD, where blue metabolites are inversely associated and red metabolites are positively associated with spine BMD, respectively. All metabolites included in the module are annotated with an MSI level 1 or 2 confidence.
In targeted MetabNet analyses of microbiome-related metabolic features, there were 1,429 metabolic features significantly associated with indoleacetylaldehyde and 1,867 features significantly associated with N-acetylputrescine at p<0.05. In subsequent mummichog analyses of the significantly enriched metabolic features, indoleacetylaldehyde was associated with 17 significantly enriched pathways including branched chain amino acid metabolism, pathways related to polyamine metabolism involving methionine and cysteine metabolism and arginine and proline metabolism, butanoate (butyrate) metabolism, tryptophan metabolism, and energy generating pathways (Table 2). N-acetylputrescine was associated with three significantly enriched pathways including bile acid synthesis, arginine and proline metabolism, and carnitine shuttle (Table 2).
Table 2.
Enriched metabolic pathways associated with microbiome-related metabolic features
| Targeted metabolic feature | Metabolic pathway | Overlap size | Pathway size | p-value |
|---|---|---|---|---|
| Indoleacetylaldehyde | Lysine metabolism | 7 | 22 | <0.001 |
| m/z 160.0761 | Valine, leucine, and isoleucine degradation | 7 | 23 | <0.001 |
| TCA cycle | 5 | 13 | <0.001 | |
| Methionine and cysteine metabolism | 8 | 39 | 0.001 | |
| Carnitine shuttle | 6 | 27 | 0.001 | |
| Arginine and proline metabolism | 7 | 34 | 0.001 | |
| Purine metabolism | 7 | 39 | 0.003 | |
| Aspartate and asparagine metabolism | 9 | 55 | 0.004 | |
| Butanoate metabolism | 4 | 20 | 0.005 | |
| Bile acid biosynthesis | 7 | 42 | 0.005 | |
| Pyrimidine metabolism | 6 | 36 | 0.006 | |
| Urea cycle/amino group metabolism | 7 | 47 | 0.01 | |
| Fatty acid activation | 4 | 24 | 0.01 | |
| Glycine, serine, alanine and threonine metabolism | 6 | 40 | 0.01 | |
| Tryptophan metabolism | 7 | 51 | 0.02 | |
| Pentose phosphate pathway | 4 | 32 | 0.04 | |
| Glycolysis and gluconeogenesis | 4 | 33 | 0.04 | |
| N-Acetylputrescine | Bile acid biosynthesis | 6 | 42 | 0.009 |
| m/z 131.1181 | Arginine and proline metabolism | 5 | 34 | 0.01 |
| Carnitine shuttle | 4 | 27 | 0.02 |
3.3. Dietary Intake
A summary of dietary intake data from all participants is shown in supplemental table 2. Post-hoc analyses of the relationships between dietary polyunsaturated fatty acid (PUFA) intake and bone mineral density are shown in Table 3. Reported dietary intake of omega-6 (n-6) fatty acids (predominantly from linoleic acid) was significantly, inversely related to whole body BMD z-score (−0.14 ± 0.06, p=0.02) and trended towards a negative relationship with spine BMD (−0.01 ± 0.008, p=0.07). Intake of omega-3 (n-3) fatty acids was not significantly related to BMD, and the ratio of n-6 to n-3 intake was inversely related to bone, although the relationships did not reach statistical significance.
Table 3.
Multiple linear regression analyses of dietary intake of omega-6 (n-6) fatty acids, omega-3 (n-3) fatty acids, and ratio of n-6 to n-3 intake related to bone mineral density
| Dietary variable | Whole body BMD Z score β ± SE | Spine BMD β ± SE |
|---|---|---|
| Dietary n-6, 18:2 FA Percent total calories, %1 | −0.14 ± 0.06 (0.02) | −0.01 ± 0.008 (0.07) |
| Dietary n-3, 18:3 FA Percent total calories, %1 | −0.35 ± 0.38 (0.37) | −0.05 ± 0.05 (0.32) |
| Ratio 18:2 FA to 18:3 FA | −0.07 ± 0.04 (0.13) | −0.006 ± 0.006 (0.30) |
Results are consistent for gm/1000 kcal intake; FA, fatty acids; BMD, bone mineral density
All analyses are adjusted for age, race, sex, and body mass index.
Intake of dietary omega-6, 18:2 fatty acids and omega-3, 18:3 fatty acids includes intake from supplements
4. DISCUSSION
Pathways related to fatty acid metabolism, particularly the n-6 fatty acid linoleic acid, were significantly associated with BMD. The metabolic features associated with adverse bone health in this study have pro-inflammatory characteristics and may be indicative of increased inflammation and altered cellular signaling that negatively affects bone remodeling (32). Metabolites linked to the gut microbiome and related to tryptophan and polyamine metabolism were also highly associated with BMD. Findings for whole body BMD Z-score and spine BMD reflected overlapping fatty acid-related metabolism and inflammatory pathways. The differences found between whole body BMD Z-score and spine BMD may reflect differences in site-specific bone tissue metabolism.
While previous metabolomics-based studies of bone health have reported associations with amino acids and assessment of bone health (33–35), our findings implicated plasma lipids as predominant correlates of BMD. The discrepancies in findings may be related to the different metabolomics platforms used or populations studied. In our heterogeneous population, altered metabolism of linoleic acid was associated with whole body BMD Z-score and spine BMD. Metabolic products of linoleic acid, namely arachidonic acid (ARA), HODE, and EpOME/HPODE, were inversely associated with bone health. ARA is the primary precursor of the pro-inflammatory eicosanoid pathway (36–38) and is suggested, via prostaglandin E2, to induce osteoclast formation and bone resorption in human bone marrow cultures (39, 40). Linoleic acid-derived oxylipins, including 9-HODE and 13-HODE, are peroxisome proliferator-activated receptor gamma (PPARγ) agonists that promote mesenchymal stem cell differentiation to adipocytes at the expense of osteoblast differentiation (41, 42). In agreement with other reports (43, 44), we found alterations of other membrane lipid pathways, such as glycosphingolipids and glycerophospholipids, that were adversely linked to BMD. Glycosphingolipids have been shown to have pro-osteoclastogenic effects in experimental models (45). Thus, our findings support the concept that pro-inflammatory lipid molecules have adverse effects on bone health that may stem from increased bone resorption and impaired bone formation.
The relationships of total and individual dietary polyunsaturated fatty acid (PUFA) intake with bone health is complex (44, 46, 47) and may be influenced by nutrient-nutrient interactions, as well as the ratio of n-6 to omega-3 fatty acid (n-3) intake. In this cohort, higher intake of n-6 fatty acids was inversely related to bone density, particularly whole body BMD Z-score. As an essential fatty acid, adequate amounts of dietary linoleic acid are necessary for normal health and functioning, but excessive intake, which is typical in Western-style diets, may be deleterious for bone health (1, 38). These negative effects largely stem from n-6 fatty acid promotion of inflammation and oxidative stress and suppression of the predominantly anti-inflammatory effects of n-3 PUFAs and related metabolites (48). A high dietary intake of n-6 linoleic acid is reflected by higher amounts of pro-inflammatory n-6 derived oxylipins (36), which were observed in this cohort. Recently, consumption of an omega-3 fatty acid supplement with eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) was shown to increase n-3 derived oxylipins and suppress the effects of n-6 oxylipins (49). Additional experimental and clinical trials are required to determine if higher intakes of n-3 fatty acids and/or lower intakes of n-6 fatty acids promote bone health.
Emerging evidence supports the gut microbiome as a potent modulator of bone metabolism (4, 11, 50). Cytokines produced by interactions with the gut microbiome may promote the expression of receptor activated NF-κB ligand (RANKL), TNF-α, IL-17 and other factors, which stimulate bone resorption (50), while the microbial-derived short chain fatty acid (SCFA) butyrate stimulates bone formation and bone resorption (4). In our study, several metabolites linked to the gut microbiome were significantly associated with spine BMD. Indoleacetylaldehyde was positively associated with spine BMD, and the polyamine N-acetylputrescine was negatively associated with spine BMD. Both endogenous (e.g. kyurenines) and bacterial-derived (e.g., indoleacetylaldehyde) tryptophan metabolites contribute to the maintenance of gut barrier integrity and reduce inflammatory signaling (6, 51). A study of Caucasian women also identified a tryptophan-related metabolite, serum formylkyurenine, as positively associated with BMD (34). Polyamines are also important for gut microbiome homeostasis and cellular processes (52). Few studies have investigated the relationship between polyamines and bone health, although experimental animal studies have indicated that the polyamines spermine and spermidine prevent bone loss (53), and excess polyamine catabolism impairs osteoblastogenesis (54). Elevated N-acetylated polyamines, such as N-acetylputrescine detected in the current study, may be reflective of increased polyamine catabolism (55). An emerging area of research suggests that probiotic supplementation may influence gut microbiome activity that favorably impacts biomarkers relevant to bone health (9, 56). Thus, our findings are in line with the current field of osteomicrobiology (57). Additional mechanistic and clinical studies are required to fully elucidate the relationship between the gut microbiome and bone health.
It is possible that the plasma metabolome also reflects energy metabolism of the bone itself. Bone resorption and formation processes require significant amounts of adenosine triphosphate (ATP)(58). Glucose metabolism is thought to be the dominant energy generating pathway for osteoblasts (59). Here, pathways linked to BMD were predominantly comprised of fatty acid metabolism, although glycolysis and gluconeogenesis were found to be a significantly enriched pathway related to spine BMD. Metabolism of fatty acids also produces large amounts of ATP via beta-oxidation, which may be utilized to meet the high-energy needs of osteoclasts (59). Findings in this study cannot distinguish if plasma metabolites are directly related to bone tissue metabolism, however, future metabolomic investigations of bone turnover markers, as well as osteoblast-derived metabolic hormones such as osteocalcin, will provide valuable insight into metabolism specific to bone tissue resorption and formation.
This study provides detailed information on plasma nutrition-related metabolic pathways associated with bone density. These findings provide insight into possible mechanisms contributing to insufficient bone accrual or age-related bone loss and potential targets for therapeutic interventions in a heterogeneous population. Limitations of this study include the cross-sectional nature, which limits our ability to establish causality in the reported relationships. The participants were not specifically recruited to assess bone accrual or age-related bone loss; therefore, prospective studies with well-curated populations specifically designed and powered to address these questions will be needed. The DXA assessments of BMD utilized in this study are a composite measure of cortical and trabecular bone; however, these tissues have unique responses to disease, medications, physical activity, and hormonal changes, which we were not able to assess. Site-specific measures of bone density such as lumbar spine and femoral neck, which were not available from the total body composition DXA scans, may yield different results. As previously published (60), this cohort reported relatively high income and education levels, which may affect the generalizability of our findings. Small sample sizes limited our ability to assess the influence of specific disease states (such as type 2 diabetes and dyslipidemia) or medications on our findings. Finally, the composition of the gut microbiome was not directly characterized in this cohort. Future studies should link data from the plasma metabolome with gut microbiome composition and bone turnover markers in longitudinal studies of disease specific or at-risk cohorts to increase understanding of these multifaceted relationships.
5. CONCLUSIONS
Novel findings in this adult cohort highlight the pro-inflammatory linoleic acid pathway and other membrane lipid-related pathways, defined using plasma HRM, as inversely associated with bone health. This study also demonstrated that plasma HRM allows for profiling of gut microbiome-related molecules and implicated such circulating metabolites as associated with BMD in humans. Further investigation is needed to study the interactions between dietary fats, gut microbiome indices and their associated metabolites, and mechanistic influence on bone turnover and maintenance of bone health.
Supplementary Material
Highlights.
A variety of modifiable factors, including diet, influence bone health
Metabolomics is a novel tool to explore factors related to bone health
Plasma linoleic acid and oxidized metabolites were inversely related to bone density
Gut microbiome-related metabolites were also associated with bone density
Metabolomics is a valuable method for examining regulators of bone metabolism
Acknowledgements:
We would like to thank the reviewers of this manuscript, whose questions and insight greatly contributed to creating a stronger manuscript.
Sources of Support: This work is based on information from the Emory Predictive Health Institute and Center for Health Discovery and Well Being Database supported by the National Center for Advancing Translational Sciences of the NIH under award number UL1 TR002378. Additional grant support included NIH K01 DK102851 (JAA), R03 DK117246 (JAA), U54 AG062334 (JAA, MNW), K24 DK096574 (TRZ), P30 ES019776 (Health and Exposome Research Center at Emory; DPJ, TRZ), R01 DK112946 (RP), R01 DK108842 (RP), S10 RR028009 (RP), R01 AR068157 (MNW), and R01 AR070091 (MNW). MNW was also supported by a grant from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (5I01BX000105).
Abbreviations:
- BMD
bone mineral density
- HRM
high-resolution metabolomics
- BMI
body mass index
- ARA
arachidonic acid
- n-6
omega-6
- n-3
omega-3
- PUFA
polyunsaturated fatty acid
- DHA
Docosahexaenoic acid
- EPA
eicosapentaenoic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors have no conflicts of interest to declare.
References
- 1.Cashman KD. Diet, nutrition, and bone health. J of Nutr 2007;137(11):2507S–12S. doi: 10.1093/jn/137.11.2507s. [DOI] [PubMed] [Google Scholar]
- 2.Weaver CM, Gordon CM, Janz KF, Kalkwarf HJ, Lappe JM, Lewis R, O’Karma M, Wallace TC, Zemel BS. The National Osteoporosis Foundation’s position statement on peak bone mass development and lifestyle factors: a systematic review and implementation recommendations. Osteoporos Int 2016;27(4):1281–386. doi: 10.1007/s00198-015-3440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Movassagh EZ, Vatanparast H. Current evidence on the association of dietary patterns and bone health: a scoping review. Adv Nutr 2017;8(1):1–16. doi: 10.3945/an.116.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaiss MM, Jones RM, Schett G, Pacifici R. The gut-bone axis: how bacterial metabolites bridge the distance. J Clin Invest 2019;129(8):3018–28. doi: 10.1172/jci128521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 2014;146(6):1564–72. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “Western-lifestyle” inflammatory diseases. Immunity 2014;40(6):833–42. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Tuddenham S, Sears CL. The intestinal microbiome and health. Curr Opin Infect Dis 2015;28(5):464–70. doi: 10.1097/qco.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Nat Acad of Sci USA 2009;106(10):3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlsson C, Engdahl C, Fak F, Andersson A, Windahl SH, Farman HH, Moverare-Skrtic S, Islander U, Sjogren K. Probiotics protect mice from ovariectomy-induced cortical bone loss. PLoS One 2014;9(3):e92368. doi: 10.1371/journal.pone.0092368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez CJ, Guss JD, Luna M, Goldring SR. Links between the microbiome and bone. J Bone Miner Res 2016;31(9):1638–46. doi: 10.1002/jbmr.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlsson C, Sjögren K. Osteomicrobiology: A new cross-disciplinary research field. Calcif Tissue Int 2018;102(4):426–32. doi: 10.1007/s00223-017-0336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones DP, Park Y, Ziegler TR. Nutritional metabolomics: progress in addressing complexity in diet and health. Ann Rev Nutr 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rask KJ, Brigham KL, Johns MM. Integrating comparative effectiveness research programs into predictive health: A unique role for academic health centers. Acad Med 2011;86(6):718–23. doi: 10.1097/ACM.0b013e318217ea6c. [DOI] [PubMed] [Google Scholar]
- 14.Brigham KL. Predictive health: the imminent revolution in health care. J Amer Geriatr Soc 2010;58 Suppl 2:S298–302. doi: 10.1111/j.1532-5415.2010.03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ainsworth BE, Irwin ML, Addy CL, Whitt MC, Stolarczyk LM. Moderate physical activity patterns of minority women: the Cross-Cultural Activity Participation Study. J Womens Health Gend Based Med 1999;8(6):805–13. doi: 10.1089/152460999319129. [DOI] [PubMed] [Google Scholar]
- 16.Haskell WL, Lee IM, Pate RR, Powell KE, Blair SN, Franklin BA, Macera CA, Heath GW, Thompson PD, Bauman A. Physical activity and public health: Updated recommendation for adults from the American College of Sports Medicine and the American Heart Association. Circulation 2007;116(9):1081–93. doi: 10.1161/circulationaha.107.185649. [DOI] [PubMed] [Google Scholar]
- 17.NutritionQuest. Our Research. Internet: https://www.nutritionquest.com/company/our-research-questionnaires/(accessed October 30 2019).
- 18.Block G, Woods M, Potosky A, Clifford C. Validation of a self-administered diet history questionnaire using multiple diet records. J Clin Epi 1990;43(12):1327–35. doi: 10.1016/0895-4356(90)90099-b. [DOI] [PubMed] [Google Scholar]
- 19.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013;9(1 Suppl):S132–s43. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, Strobel F, Quyyumi AA, Ziegler TR, Pennell KD, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol Sci 2015;148(2):531–43. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones DP, Walker DI, Uppal K, Rohrbeck P, Mallon CT, Go YM. Metabolic pathways and networks associated with tobacco use in military personnel. J Occup Med 2016;58(8 Suppl 1):S111–6. doi: 10.1097/jom.0000000000000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE, Wilson ME, Sutliff RL, Mansfield KG, Wachtman LM, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicol 2012;295(1–3):47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25(15):1930–6. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, Jones DP. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC Bioinformatics 2013;14:15. doi: 10.1186/1471-2105-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Go YM, Liang Y, Uppal K, Soltow QA, Promislow DE, Wachtman LM, Jones DP. Metabolic characterization of the common marmoset (Callithrix jacchus). PLoS One 2015;10(11):e0142916. doi: 10.1371/journal.pone.0142916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3(3):211–21. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uppal K, Walker DI, Jones DP. xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal Chem 2017;89(2):1063–7. doi: 10.1021/acs.analchem.6b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y and Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 1995;57(1):289–300. [Google Scholar]
- 30.Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, Jones DP. MetabNet: an R package for metabolic association analysis of high-resolution metabolomics data. Front Bioeng Biotech 2015;3:87. doi: 10.3389/fbioe.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Bio 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naughton SS, Mathai ML, Hryciw DH, McAinch AJ. Linoleic acid and the pathogenesis of obesity. Prostaglandins Other Lipid Mediat 2016;125:90–9. doi: 10.1016/j.prostaglandins.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto T, Hirayama A, Sato Y, Koboyashi T, Katsuyama E, Kanagawa H, Fujie A, Morita M, Watanabe R, Tando T, et al. Metabolomics-based profiles predictive of low bone mass in menopausal women. Bone Rep 2018;9:11–8. doi: 10.1016/j.bonr.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Q, Shen H, Su KJ, Zhang JG, Tian Q, Zhao LJ, Qiu C, Zhang Q, Garrett TJ, Liu J, et al. Metabolomic profiles associated with bone mineral density in US Caucasian women. Nutr Metab 2018;15:57. doi: 10.1186/s12986-018-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You YS, Lin CY, Liang HJ, Lee SH, Tsai KS, Chiou JM, Chen YC, Tsao CK, Chen JH. Association between the metabolome and low bone mineral density in Taiwanese women determined by (1)H NMR spectroscopy. J Bone Miner Res 2014;29(1):212–22. doi: 10.1002/jbmr.2018. [DOI] [PubMed] [Google Scholar]
- 36.Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr 2015;6(5):513–40. doi: 10.3945/an.114.007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Innes JK, Calder PC. Omega-6 fatty acids and inflammation. Prostaglandins Leukot Essent Fatty Acids 2018;132:41–8. doi: 10.1016/j.plefa.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Zhu J, Lyu F, Panigrahy D, Ferrara KW, Hammock B, Zhang G. omega-3 polyunsaturated fatty acids-derived lipid metabolites on angiogenesis, inflammation and cancer. Prostaglandins Other Lipid Mediat 2014;113–115:13–20. doi: 10.1016/j.prostaglandins.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flanagan AM, Stow MD, Kendall N, Brace W. The role of 1,25-dihydroxycholecalciferol and prostaglandin E2 in the regulation of human osteoclastic bone resorption in vitro. Int J Exp Pathol 1995;76(1):37–42. [PMC free article] [PubMed] [Google Scholar]
- 40.Lader CS, Flanagan AM. Prostaglandin E2, interleukin 1alpha, and tumor necrosis factor-alpha increase human osteoclast formation and bone resorption in vitro. Endocrinology 1998;139(7):3157–64. doi: 10.1210/endo.139.7.6085. [DOI] [PubMed] [Google Scholar]
- 41.Lecka-Czernik B, Moerman EJ, Grant DF, Lehmann JM, Manolagas SC, Jilka RL. Divergent effects of selective peroxisome proliferator-activated receptor-gamma 2 ligands on adipocyte versus osteoblast differentiation. Endocrinology 2002;143(6):2376–84. doi: 10.1210/endo.143.6.8834. [DOI] [PubMed] [Google Scholar]
- 42.Lin SJ, Yang DR, Yang G, Lin CY, Chang HC, Li G, Chang C. TR2 and TR4 orphan nuclear receptors: An overview. Curr Top Dev Biol 2017;125:357–73. doi: 10.1016/bs.ctdb.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Cabrera D, Kruger M, Wolber FM, Roy NC, Totman JJ, Henry CJ, Cameron-Smith D, Fraser K. Association of plasma lipids and polar metabolites with low bone mineral density in Singaporean-Chinese menopausal women: a pilot study. Int J Environ Res Public Health 2018;15(5). doi: 10.3390/ijerph15051045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farina EK, Kiel DP, Roubenoff R, Schaefer EJ, Cupples LA, Tucker KL. Plasma phosphatidylcholine concentrations of polyunsaturated fatty acids are differentially associated with hip bone mineral density and hip fracture in older adults: the Framingham Osteoporosis Study. J Bone Miner Res 2012;27(5):1222–30. doi: 10.1002/jbmr.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ersek A, Xu K, Antonopoulos A, Butters TD, Santo AE, Vattakuzhi Y, Williams LM, Goudevenou K, Danks L, Freidin A, et al. Glycosphingolipid synthesis inhibition limits osteoclast activation and myeloma bone disease. J Clin Invest 2015;125(6):2279–92. doi: 10.1172/jci59987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longo AB, Ward WE. PUFAs, bone mineral density, and fragility fracture: findings from human studies. Adv Nutr 2016;7(2):299–312. doi: 10.3945/an.115.009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahni S, Mangano KM, McLean RR, Hannan MT, Kiel DP. Dietary approaches for bone health: lessons from the Framingham Osteoporosis Study. Current Osteoporos Rep 2015;13(4):245–55. doi: 10.1007/s11914-015-0272-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DiNicolantonio JJ, O’Keefe JH. Importance of maintaining a low omega-6/omega-3 ratio for reducing inflammation. Open Heart 2018;5(2):e000946. doi: 10.1136/openhrt-2018-000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostermann AI, West AL, Schoenfeld K, Browning LM, Walker CG, Jebb SA, Calder PC, Schebb NH. Plasma oxylipins respond in a linear dose-response manner with increased intake of EPA and DHA: results from a randomized controlled trial in healthy humans. Am J Clin Nutr 2019;109(5):1251–63. doi: 10.1093/ajcn/nqz016. [DOI] [PubMed] [Google Scholar]
- 50.Li JY, Yu M, Tyagi AM, Vaccaro C, Hsu E, Adams J, Bellido T, Weitzmann MN, Pacifici R. IL-17 Receptor signaling in osteoblasts/osteocytes mediates PTH-induced bone loss and enhances osteocytic RANKL production. J Bone Miner Res 2019;34(2):349–60. doi: 10.1002/jbmr.3600. [DOI] [PubMed] [Google Scholar]
- 51.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, Yin Y. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tofalo R, Cocchi S, Suzzi G. Polyamines and gut microbiota. Front Nutr 2019;6. doi: 10.3389/fnut.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto T, Hinoi E, Fujita H, Iezaki T, Takahata Y, Takamori M, Yoneda Y. The natural polyamines spermidine and spermine prevent bone loss through preferential disruption of osteoclastic activation in ovariectomized mice. Br J Pharmacol 2012;166(3):1084–96. doi: 10.1111/j.1476-5381.2012.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pirnes-Karhu S, Maatta J, Finnila M, Alhonen L, Uimari A. Overexpression of spermidine/spermine N1-acetyltransferase impairs osteoblastogenesis and alters mouse bone phenotype. Transgenic Res 2015;24(2):253–65. doi: 10.1007/s11248-014-9836-6. [DOI] [PubMed] [Google Scholar]
- 55.Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J 2009;421(3):323–38. doi: 10.1042/bj20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Britton RA, Irwin R, Quach D, Schaefer L, Zhang J, Lee T, Parameswaran N, McCabe LR. Probiotic L. Reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J Cell Physiol 2014;229(11):1822–30. doi: 10.1002/jcp.24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones RM, Mulle JG, Pacifici R. Osteomicrobiology: the influence of gut microbiota on bone in health and disease. Bone 2018;115:59–67. doi: 10.1016/j.bone.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Lee WC, Guntur AR, Long F, Rosen CJ. Energy metabolism of the osteoblast: implications for osteoporosis. Endo Rev 2017;38(3):255–66. doi: 10.1210/er.2017-00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dirckx N, Moorer MC, Clemens TL, Riddle RC. The role of osteoblasts in energy homeostasis. Nat Rev Endocrinol 2019;15(11):651–65. doi: 10.1038/s41574-019-0246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellissimo MP, Cai Q, Ziegler TR, Liu KH, Tran PH, Vos MB, Martin GS, Jones DP, Yu T, Alvarez JA. Plasma high-resolution metabolomics differentiates adults with normal weight obesity from lean individuals. Obesity 2019;27(11):1729–37. doi: 10.1002/oby.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


