Abstract
GOLM1, a type II transmembrane protein, is associated with tumor progression, metastasis and immunosuppression. However, the relationship between GOLM1 and the immunosuppressive molecule PD-L1 in HCC remains largely unclear. Here, we revealed that GOLM1 acts as a novel positive regulator of PD-L1, whose abnormal expression plays a crucial role in cancer immune evasion and progression. We found that GOLM1 is overexpressed and positively correlated with PD-L1 expression in HCC. Mechanistically, we found that GOLM1 promotes the phosphorylation of STAT3 by enhancing the level of EGFR, which in turn upregulates the transcriptional expression of PD-L1. Taken together, we demonstrated that GOLM1 acts as a positive regulator of PD-L1 expression via the EGFR/STAT3 signaling pathway in human HCC cells. This study provides a new insight into the regulatory mechanism of PD-L1 expression in HCC, which may provide a novel therapeutic target for HCC immunotherapy.
Keywords: GOLM1, hepatocellular carcinoma, PD-L1, EGFR, STAT3
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the second leading cause of cancer-related deaths in the world [1]. Although great progress has been made in the diagnosis and treatment of HCC, its overall prognosis is still poor, with a 5-year survival rate of approximately 12% [2]. At present, treatments for early HCC include surgical resection, liver transplantation and local radiofrequency (RF) ablation [3], but their efficacy is still limited. Molecular targeted therapies, such as the small-molecule multikinase inhibitor sorafenib (first-line use) [4], regorafenib (second-line use) [5] and lenvatinib (first-line use) [6] have been used for the treatment of advanced HCC. However, these drugs only extend the median overall survival of patients with advanced HCC by no longer than 4 months [7]. Immune checkpoint blockade (ICB) has brought considerable clinical benefits in the treatment of different tumors [8-11]. Among these immune checkpoints, the research on programmed cell death protein 1 and its ligand (PD-1/PD-L1) has attracted the most widespread attention [12]. However, the anti-PD-1 therapy approved for HCC treatment only achieved a 20% response rate [13]. Treatment with pembrolizumab or nivolumab failed to reach the primary endpoints of the KEYNOTE-240 and CheckMate-459 HCC clinical trials [14,15]. Thus, exploring the regulatory mechanism of immune checkpoints and identifying novel therapies to improve HCC patients’ long-term outcome is urgently needed.
Golgi membrane protein 1 (GOLM1, also known as Golgi protein 73, GP73 or Golgi phosphoprotein 2, GOLPH2), a type II transmembrane protein, is associated with tumor progression [16,17], metastasis [18-21] and immunosuppression [22,23]. It has been reported that the expression level of GOLM1 can be increased not only in viral infections [24-27] but also in several types of cancer, such as HCC, lung adenocarcinoma and prostate cancer [28-30]. Our previous study revealed that GOLM1 is one of the leading genes associated with HCC metastasis. We found that GOLM1 expression was closely related to early cancer recurrence, metastasis and poor prognosis in HCC patients. Detailed studies revealed that GOLM1, as a specific mediator (cargo adaptor), selectively interacts with epidermal growth factor receptor (EGFR) to assist EGFR/RTK anchoring on the trans-Golgi network (TGN) and recycling back to the plasma membrane, leading to the continued activation of downstream kinases, and finally mediating HCC metastasis [18]. However, the relationship between GOLM1 and the immunosuppressive molecule PD-L1 in HCC remains largely unclear.
Studies in various tumor types have demonstrated that levels of PD-L1 expression can predict therapeutic responses to monotherapies blocking the PD-1/PD-L1 axis [31-33]. These levels of PD-L1 expression are regulated in highly complicated manners and can be affected by transcriptional control and post-translational regulation [34]. One of the most critical transcription factors in human cancer cells is the signal transducer and activator of transcription 3 (STAT3), which can directly act on the PD-L1 promoter to enhance PD-L1 expression [35]. Some studies have suggested that EGFR is involved in the activation of STAT3 [36-38]. Activated EGFR recruits and phosphorylates STAT3 at Tyr705, then phosphorylated STAT3 enters the nucleus to promote the expression of some cancer-related genes [39-41]. Due to the important regulation of GOLM1 on EGFR, we speculate that there might be a regulatory relationship in the GOLM1/EGFR/STAT3/PD-L1 axis.
Herein, we found that GOLM1 promoted PD-L1 transcriptional expression by enhancing STAT3 Tyr705 phosphorylation. Specifically, we found that GOLM1 enhanced levels of EGFR, hence the levels of phosphorylated EGFR, which phosphorylated STAT3, promoting the expression of PD-L1 in HCC. Our results identified that GOLM1 is a positive regulator of PD-L1 expression, which provides a new insight in the relationship between GOLM1 and immune response.
Materials and methods
Antibodies and reagents
The antibodies listed below were used in Western blotting, immunohistochemical: anti-GOLM1 (ab109628, Abcam; ab92612, Abcam), anti-PD-L1 (#13684T, Cell Signaling Technology; #64988S, Cell Signaling Technology), anti-STAT3 (#9139S, Cell Signaling Technology), anti-p-STAT3 (Tyr705) (#9145S, Cell Signaling Technology), anti-EGFR (ab52894, Abcam), anti-p-EGFR (phospho Y1068) (ab40815, Abcam). BP-1-102 (#T3708) was purchased from Topscience. Gefitinib (ab142052) was purchased from Abcam.
HCC cell lines
The HCC cell lines HCC-LM3, Huh7, Hep3B, Bel-7402 and Hepa1-6 were obtained from Liver Cancer Institute and Zhongshan Hospital, Fudan University, Shanghai, China. These cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco). All cell lines were incubated in a humidified incubator containing 5% CO2 at 37°C.
Tissue microarray and immunohistochemistry
Tissue microarrays containing 239 matched pairs of primary HCC samples and adjacent nontumor liver tissues were constructed as described previously [42,43]. In brief, tumor specimens were collected from HCC patients who underwent surgical resection from January 2005 to December 2006 in Liver Surgery Department of Zhongshan Hospital, Fudan University, Shanghai, China. The Research Ethics Committee of Zhongshan Hospital approved the research protocol and all patients signed the informed consent forms. Paraffin-embedded implanted tumors were cut into 5-μm sections. Immunohistochemical (IHC) staining of the HCC samples was performed as described previously [44]. Briefly, each sample was stained with the indicated antibodies and then incubated with an avidin-biotin-peroxidase complex. The chromogen 3-amino-9-ethylcarbazole was used to visualize the target protein. The expression levels of GOLM1 and PD-L1 in tissues were evaluated by the H-score method through combining the value of the immunoreaction intensity and the percentage of stained cells [45]. H-score = ∑π (i + 1), where i is the intensity of the stained cells (0 for negative staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining) and π is the value corresponding to the percentage of stained cells with different intensities (0, < 5% positive cells; 1, 6-25% positive cells; 2, 26-50% positive cells; 3, 51-75% positive cells; 4, > 75% positive cells). Tumor tissues with H-scores greater than the median of all scored tumor tissues were classified as high expression.
The encyclopedia of RNA interactomes (ENCORI) database
Co-Expression Analysis of GOLM1 and PD-L1 in 374 HCC patients in The Encyclopedia of RNA Interactomes (ENCORI) database is available from the Web site of (https://starbase.sysu.edu.cn/panGeneCoExp.php#).
Western blotting
Total proteins extracted from different HCC cell lines with cell lysis buffer supplemented with protease inhibitors were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After being blocked with 5% nonfat milk in TBST for 1 h at room temperature, the membranes were incubated with the primary antibody at 4°C overnight and then incubated with HRP-conjugated goat anti-mouse or anti-rabbit secondary antibodies for 2 h at room temperature after being washed 3 times with TBST. And then the membranes were washed 3 times and detected with an ECL detection kit (Thermo Fisher Scientific, Waltham, MA). The band intensity was quantified with Image J (National Institutes of Health, Bethesda, MD, USA).
RNA extraction and real-time quantitative reverse-transcription polymerase chain reaction
Total RNA was isolated by using Trizol reagent (Invitrogen, Carlsbad, CA, USA). According to the manufacturer’s protocol (Takara), the same amount of RNA was reverse transcribed into cDNA. Real-time quantitative reverse-transcription polymerase chain reaction (RT-qPCR) was performed using SYBR-Green PCR Master mix (Yeasen Biotechnology Co., Ltd.) according to the manufacturer’s instructions. The primers’ sequences used were as follows: human GOLM1 forward, 5’-CCGGAGCCTCGAAAAGAGATT-3’ and reverse, 5’-ATGATCCGTGTCTGGAGGTC-3’; human PD-L1 forward, 5’-TCACTTGGTAATTCTGGGAGC-3’ and reverse, 5’-CTTTGAGTTTGTATCTTGGATGCC-3’; human GAPDH forward, 5’-ACAGTCCATGCCATCACTGCC-3’ and reverse, 5’-GCCTGCTTCACCACCTTCTTG-3’.
Lentivirus transfection
Cells were seeded into the 6-well cell culture plates and were cultured for 24 hours to 50% confluence in DMEM containing 10% FBS at 37°C. Then the medium was changed to serum-free medium. According to the pre-experimental infection conditions, the lentiviruses (GeneChem, Shanghai, China) were added to the 6-well cell culture plates. After 12 hours, the cell culture medium was replaced with complete medium. The efficiency of transfection was measured under the fluorescence microscope. After 72 hours, the cells were cultured in medium containing 5 μg/ml puromycin until the fluorescence efficiency reached nearly 100%. Then the concentration of puromycin was reduced to 1 μg/ml to select stably transfected cells. The expression level of GOLM1 was evaluated by Western blotting and RT-qPCR.
Cell proliferation assay
Cell proliferation was assessed with cell counting kit-8 (CCK-8) method (Dojindo, Kumamoto, Japan). Briefly, cells were added into 96-well plates at the initial density of 5 × 103 cells/well. At time points of 24, 48 and 72 hours, 10 μL of CCK-8 reagent was added to the cells in high-glucose DMEM medium for 2 hours. And then the absorbance values were measured at wavelength of 450 nm. Experiments were repeated three times under the same conditions.
Cell migration assay
Cell migration was measured with a Transwell migration apparatus (Becton Dickinson Labware, Franklin Lakes, NJ) of 8-μm pore-size. HCC cells (5 × 104 cells for Hep3B, 2 × 104 cells for Huh7) mixed with 100 µl of serum-free DMEM were added into the upper chamber, and 500 µl of DMEM containing 10% FBS were added into the lower chamber. Then, cells were cultured at 37°C and in a humidified atmosphere with 5% CO2. After 36 h, cells in the upper chamber without migration were removed with a cotton swab. The membranes were subsequently washed with phosphate buffered saline (PBS), and cells adhered to the membranes were then fixed with 4% paraformaldehyde for 15 min, and stained with Giemsa dye for 10 min. The stained cells were washed with PBS three times and counted under a microscope. The data were presented as mean counts of stained cells in 12 random fields/chamber. Experiments were repeated three times under the same conditions.
In vivo experiment
Hepa1-6 cells with stable knockdown of GOLM1 (Hepa1-6-GOLM1-KD) were constructed through lentiviral transfection. Hepa1-6-NC cells were used as negative control. The C57BL/6 mice (male, 5-6 weeks old, weighing 20-22 g) were randomly divided into 2 groups (Hepa1-6-NC and Hepa1-6-GOLM1-KD, 5 mice per group). Cells were injected (1 × 107 cells, suspended in 0.2 mL PBS, transplanted subcutaneously) to grow into tumors. This study was approved by the Shanghai Medical Experimental Animal Care Committee and performed according to the National Institutes of Health “Guide for the Care and Use of Laboratory Animals”. Subcutaneous tumors were measured using a caliper every 3 days. Tumor volumes were calculated using the formula: tumor volume = length × width2/2. At the end of the experiment, the mice were euthanized by cervical dislocation, and the tumors were weighed and then used for subsequent histological analyses.
Statistical analysis
The data were presented as the means ± standard deviation (SD). All statistical analyses were carried out with GraphPad Prism 7 (GraphPad Software, CA, USA). Student’s t test was performed to analyze the differences between two groups. Survival curves were estimated by the Kaplan-Meier method, and the log-rank test was applied to compare two groups. The association between GOLM1 or PD-L1 expression and clinicopathological variables was analyzed by the χ2 test or the Fisher’s exact test. The correlation between the expression levels of GOLM1 and PD-L1 was analyzed by Pearson correlation analysis and linear regression analysis. P < 0.05 was considered statistically significant.
Results
GOLM1 is overexpressed in human HCC and associated with tumor progression and poor survival
We conducted the IHC analysis on tissue microarrays including tumor samples and corresponding adjacent normal liver tissues from 239 patients. The representative GOLM1 IHC staining images of cancer tissues and corresponding adjacent normal tissues are shown in the Figure 1A and 1B. The expression level of GOLM1 in cancer tissues was significantly higher than that in adjacent normal tissues (P < 0.0001) (Figure 1C). The correlation between clinicopathological features and GOLM1 expression is shown in Table 1. A high expression level of GOLM1 was related to tumor size (P = 0.0007), tumor number (P = 0.0269), cancer thrombus (P = 0.0102) and tumor recurrence (P = 0.0027) (Table 1), indicating that the high expression of GOLM1 promotes the progression of HCC. We also found that high expression of GOLM1 was closely related to HBV infection (P = 0.0129) (Table 1). Survival analysis showed that the overall survival (OS) of patients with high GOLM1 expression was poorer than those with low GOLM1 expression (P = 0.0011) (Figure 1F). These findings suggest that the high expression of GOLM1 is correlated with the progression of HCC and poor survival.
Figure 1.
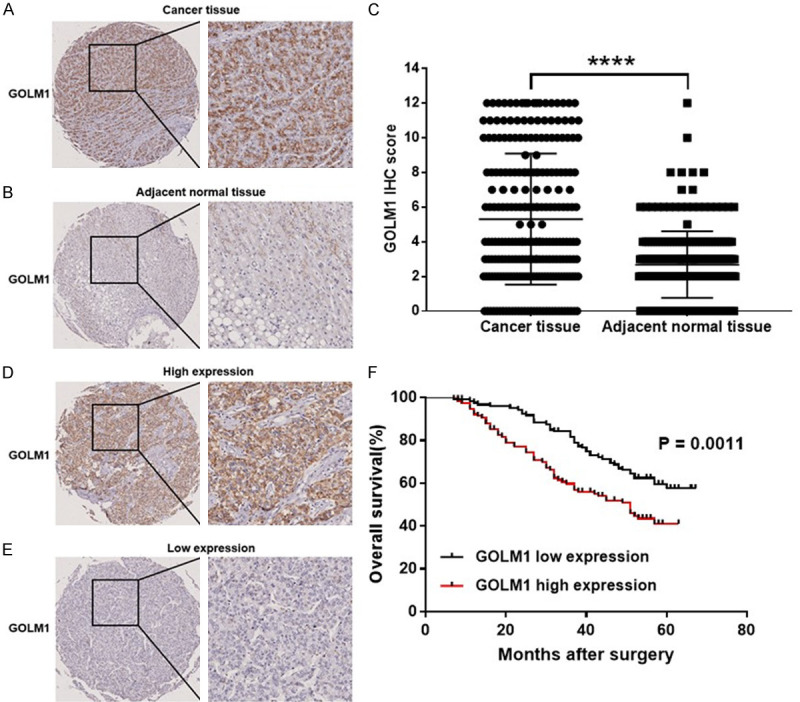
Expression of GOLM1 in HCC. GOLM1 IHC staining of (A) cancer tissues and (B) corresponding adjacent normal tissues (Scale bar, 100 μm). (C) H-score of GOLM1 expression of cancer tissues and adjacent normal tissues. (D) High GOLM1 expression in human HCC tissues (Scale bar, 100 μm). (E) Low GOLM1 expression in human HCC tissues (Scale bar, 100 μm). (F) Survival curves of HCC patients with GOLM1 high expression and GOLM1 low expression. Significance was evaluated using the Log-rank test (P = 0.0011). Tumor tissues with H-scores greater than the median of all scored tumor tissues were classified as high GOLM1 expression. ****P < 0.0001.
Table 1.
Correlation of GOLM1 or PD-L1 expression with clinicopathological factors
| Parameters | n | GOLM1 | P value | PD-L1 | P value | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Low | High | Low | High | ||||
| Age (yr) | |||||||
| ≤ 50 | 89 | 49 | 40 | 0.7203 | 45 | 44 | 0.5387 |
| > 50 | 150 | 79 | 71 | 82 | 68 | ||
| Gender | |||||||
| Male | 197 | 103 | 94 | 0.3931 | 104 | 93 | 0.8163 |
| Female | 42 | 25 | 17 | 23 | 19 | ||
| HBsAg | |||||||
| Positive | 225 | 116 | 109 | 0.0129* | 117 | 108 | 0.1575 |
| Negative | 14 | 12 | 2 | 10 | 4 | ||
| AFP (ng/mL) | |||||||
| ≤ 20 | 94 | 55 | 39 | 0.2163 | 59 | 35 | 0.0163* |
| > 20 | 145 | 73 | 72 | 68 | 77 | ||
| Tumor size (cm) | |||||||
| ≤ 5 | 156 | 96 | 60 | 0.0007*** | 87 | 69 | 0.2638 |
| > 5 | 83 | 32 | 51 | 40 | 43 | ||
| Tumor number | |||||||
| Solitary | 197 | 112 | 85 | 0.0269* | 108 | 89 | 0.2584 |
| Multiple | 42 | 16 | 26 | 19 | 23 | ||
| Cancer thrombus | |||||||
| Yes | 81 | 34 | 47 | 0.0102* | 37 | 44 | 0.0980 |
| No | 158 | 94 | 64 | 90 | 68 | ||
| Tumor capsule | |||||||
| Yes | 132 | 72 | 60 | 0.7335 | 72 | 60 | 0.6282 |
| No | 107 | 56 | 51 | 55 | 52 | ||
| Tumor recurrence | |||||||
| Yes | 90 | 37 | 53 | 0.0027** | 39 | 51 | 0.0182* |
| No | 149 | 91 | 58 | 88 | 61 | ||
| Clinical stage | |||||||
| I+II | 236 | 127 | 109 | 0.4797 | 125 | 111 | 0.6365 |
| III | 3 | 1 | 2 | 2 | 1 | ||
P < 0.05;
P < 0.01;
P < 0.001.
GOLM1 is positively correlated with PD-L1 expression in human HCC tissue
GOLM1 was reported to play an important role in tumor progression [16,17], metastasis [18-21] and immunosuppression [22,23], and its overexpression has been associated with cancer recurrence, metastasis and poor prognosis in different tumors [16,18-21,46], but the effect of GOLM1 on the immunosuppressive molecule PD-L1 remains largely unknown. To determine the relation between GOLM1 and PD-L1 expression, we measured their expression in HCC and corresponding adjacent normal tissues by tissue microarray. The representative PD-L1 IHC staining images of cancer tissues and corresponding adjacent normal tissues are shown in the Figure 2A and 2B. We found that the expression level of PD-L1 in cancer tissues was significantly higher than that in adjacent nontumor tissues (Figure 2C). The correlation between PD-L1 and the clinicopathological factors was summarized in Table 1. A high expression level of PD-L1 was associated with alpha fetoprotein (AFP) level (P = 0.0163) and tumor recurrence (P = 0.0182) (Table 1). Survival analysis showed the OS of patients with high PD-L1 expression was poorer than those with low PD-L1 expression (P = 0.0326) (Figure 2F). Representative pictures of IHC staining of HCC for GOLM1 and PD-L1 are shown in the Figure 3A. We then investigated the correlation between GOLM1 and PD-L1 using Pearson correlation and linear regression analysis and found that PD-L1 expression was positively correlated with GOLM1 expression (r = 0.3919, P < 0.0001) (Figure 3B). This finding was further confirmed using the ENCORI database, which showed a positive correlation between GOLM1 and PD-L1 mRNA expression (r = 0.224, P = 0.0000128, Figure 3C), implying that the regulation between GOLM1 and PD-L1 may occur at the transcriptional level.
Figure 2.
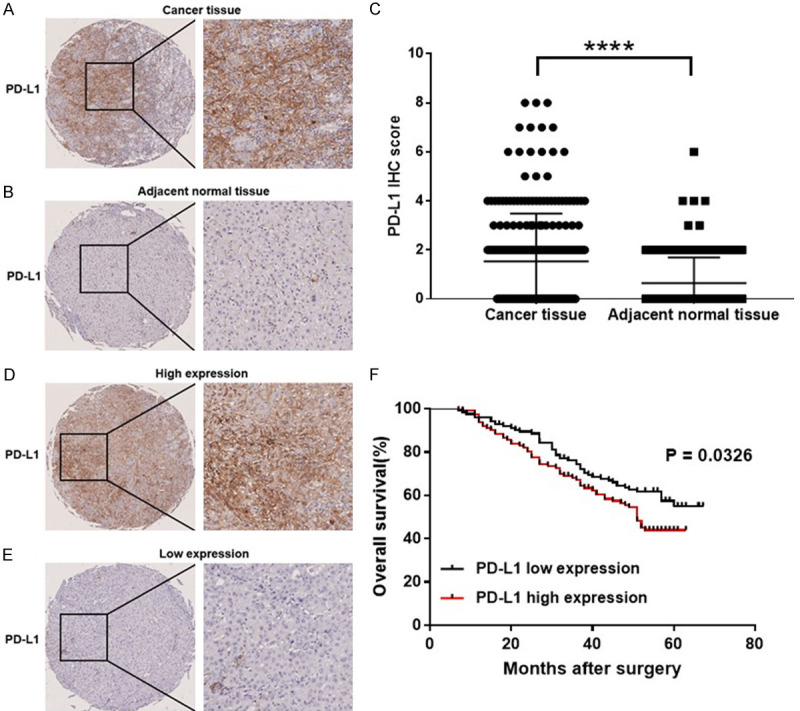
Expression of PD-L1 in HCC. PD-L1 IHC staining of (A) cancer tissues and corresponding (B) adjacent normal tissues (Scale bar, 100 μm). (C) H-score of PD-L1 expression of cancer tissues and adjacent normal tissues. (D) High PD-L1 expression in human HCC tissues (Scale bar, 100 μm). (E) Low PD-L1 expression in human HCC tissues (Scale bar, 100 μm). (F) Survival curves of HCC patients with PD-L1 high expression and PD-L1 low expression. Significance was evaluated using the Log-rank test (P = 0.0326). Tumor tissues with H-scores greater than the median of all scored tumor tissues were classified as high PD-L1 expression. ****P < 0.0001.
Figure 3.
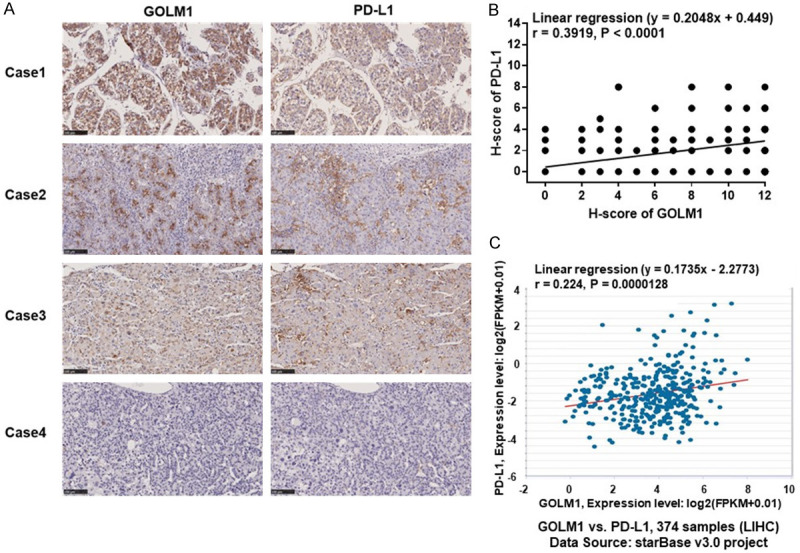
Correlation of GOLM1 and PD-L1. (A) Patient tissue samples were stained for GOLM1 and PD-L1. Representative pictures of IHC staining of HCC tumors for GOLM1 and PD-L1. (B) Correlation analysis of GOLM1 and PD-L1 expression in human HCC tissue microarray (n = 239, Linear regression (y = 0.2048x+0.449), r = 0.3919, P < 0.0001). (C) Correlation of GOLM1 and PD-L1 gene expression in the ENCORI database (C) from the ENCORI database.
GOLM1 upregulates PD-L1 mRNA expression in HCC cell lines
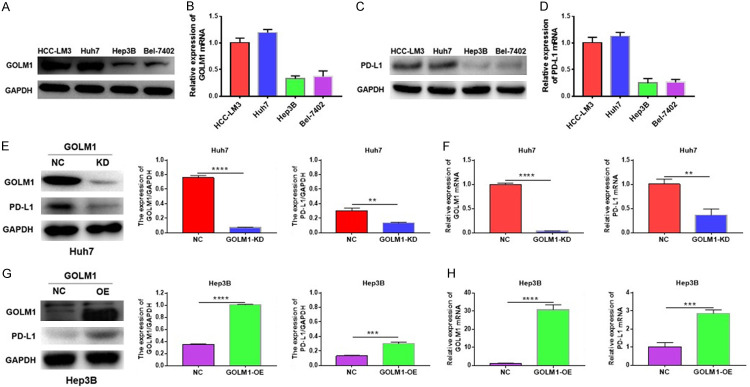
Our previous study found that compared with HCC cell lines with lower metastatic potentials, HCC cells with higher invasion and metastasis capabilities have significantly increased GOLM1 levels [18]. To determinate whether GOLM1 regulates PD-L1 expression, we first detected the expression of GOLM1 and PD-L1 in HCC cell lines: HCC-LM3, Huh7, Hep3B and Bel-7402 cells (HCC-LM3 and Huh7 cells with higher invasion and metastasis capabilities; Hep3B and Bel-7402 cells with lower invasion and metastasis potentials). We found that the expression levels of GOLM1 and PD-L1 were relatively high in HCC-LM3 and Huh7 cell lines and relatively low in Hep3B and Bel7-402 cell lines at both protein (Figure 4A, 4C) and mRNA levels (Figure 4B, 4D). Among these four cell lines, the expression of GOLM1 was highest in Huh7 cells and lowest in Hep3B cells. We then 1) knocked down GOLM1 expression in Huh7 cells and 2) overexpressed GOLM1 in Hep3B cells by the lentivirus transfection (Supplementary Figure 1A, 1B). We found that PD-L1 expression was significantly decreased when knocking down GOLM1 expression in Huh7 cells (Figure 4E, 4F), while its expression was significantly up-regulated when GOLM1 expression was overexpressed in Hep3B cells (Figure 4G, 4H). These results indicate that GOLM1 may positively regulate PD-L1 mRNA expression.
Figure 4.
GOLM1 upregulates PD-L1 expression in HCC cell lines. A and B. Expression levels of GOLM1 in different HCC cell lines: HCC-LM3, Huh7, Hep3B and Bel-7402. C and D. Expression levels of PD-L1 in different HCC cell lines: HCC-LM3, Huh7, Hep3B and Bel-7402. Protein expression was detected by Western blotting. mRNA expression was detected by RT-qPCR. E and F. Western blot and RT-qPCR analysis of PD-L1 levels in GOLM1-knockdown Huh7 cells. Quantitative analysis of GOLM1 and PD-L1 expression after GOLM1 was knockdown in Huh7 cells through Image J intensity measurement. G and H. Western blot and RT-qPCR analysis of PD-L1 levels in GOLM1-overexpressed Hep3B cells. Quantitative analysis of GOLM1 and PD-L1 expression after GOLM1 was overexpressed in Hep3B cells through Image J intensity measurement. **P < 0.01, ***P < 0.001, ****P < 0.0001.
GOLM1 upregulates PD-L1 expression via the EGFR/STAT3 pathway
Given STAT3 can directly act on the PD-L1 promoter to increase PD-L1 expression in human cancer cells [35], EGFR is suggested to be involved in STAT3 activation [36-38], and GOLM1 can modulate EGFR levels [18], we hypothesized that GOLM1 regulates PD-L1 mRNA expression by activating EGFR/STAT3 pathway.
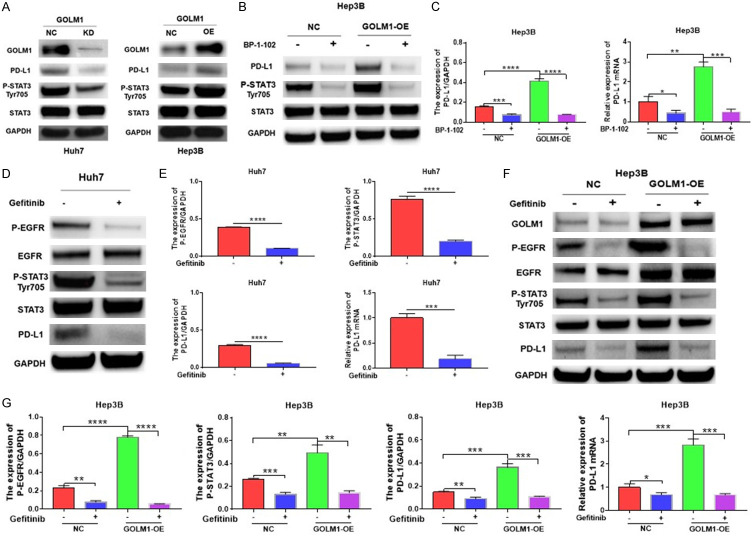
First, we found that STAT3 Tyr705 phosphorylation was inhibited after knocking down GOLM1 in Huh7 cells, while it was enhanced after overexpressing GOLM1 in Hep3B cells (Figure 5A).
Figure 5.
GOLM1 promotes PD-L1 expression via the EGFR/STAT3 pathway. A. Western blot analysis of PD-L1 and phosphorylation of STAT3 Tyr705 in GOLM1-knockdown Huh7 cells and in GOLM1-overexpressed Hep3B cells. B. GOLM1 upregulates the expression of PD-L1 through promoting phosphorylation of STAT3 Tyr705 (P-STAT3). The expression of PD-L1, P-STAT3 and STAT3 was detected by Western blot after incubation with BP-1-102 (5 μM) for 12 hours in Hep3B cells. C. Quantitative analysis of PD-L1 expression after treatment with BP-1-102 through Image J intensity measurement. The relative expression of PD-L1 mRNA in Hep3B cells and GOLM1-overexpressed Hep3B cells was detected by RT-qPCR after incubation with BP-1-102 (5 μM) for 12 hours. D. The expression of EGFR, the phosphorylation of EGFR (P-EGFR), STAT3, P-STAT3 and PD-L1 was detected by Western blot after treatment with Gefitinib (4 μM) for 6 hours in Huh7 cells. E. Quantitative analysis of P-EGFR, P-STAT3 and PD-L1 expression after treatment with Gefitinib through Image J intensity measurement. The relative expression of PD-L1 mRNA was detected by RT-qPCR after treatment with Gefitinib in Huh7 cells. F. GOLM1 enhances the phosphorylation of STAT3 Tyr705 through the regulation of P-EGFR and EGFR. The expression of GOLM1, EGFR, P-EGFR, STAT3, and P-STAT3 was detected by Western blot in Hep3B cells and GOLM1-overexpressed Hep3B cells after incubation with Gefitinib. G. Quantitative analysis of P-EGFR, P-STAT3 and PD-L1 expression after treatment with Gefitinib through Image J intensity measurement. The relative expression of PD-L1 mRNA in Hep3B cells and GOLM1-overexpressed Hep3B cells was detected by RT-qPCR after treatment with Gefitinib. Cells were incubated with EGF (50 ng/ml).
To investigate whether the GOLM1-mediated upregulation of PD-L1 expression is dependent on STAT3 Tyr705 phosphorylation in HCC, we further inhibited STAT3 with selective small-molecule inhibitors (BP-1-102) in GOLM1-overexpressed Hep3B cells. PD-L1 expression was decreased when STAT3 Tyr705 phosphorylation was inhibited (Figure 5B, 5C), suggesting that the upregulation of PD-L1 expression induced by GOLM1 is dependent on STAT3 Tyr705 phosphorylation.
To determine whether the GOLM1-mediated upregulation of STAT3 Tyr705 phosphorylation is dependent on EGFR phosphorylation in HCC, we investigated the effect of GOLM1 on the phosphorylation of EGFR. We found that the phosphorylated EGFR were significantly increased after overexpressing GOLM1 in Hep3B cells. We then blocked the phosphorylation of EGFR with the inhibitor Gefitinib and found that the phosphorylation of STAT3 was significantly inhibited in Huh7 cells (Figure 5D, 5E). Furthermore, we found that expression of PD-L1 and the phosphorylation of STAT3 were simultaneously suppressed after Gefitinib treatment in GOLM1 overexpressed Hep3B cells (Figure 5F, 5G), indicating that GOLM1 regulates phosphorylation of STAT3 and expression of PD-L1 by promoting phosphorylation of EGFR. Taken together, the above data suggestes that GOLM1 upregulates the expression of PD-L1 via the EGFR/STAT3 pathway.
GOLM1 promotes the proliferation and migration abilities of HCC cells in vitro
We conducted the CCK-8 assay to evaluate the effects of GOLM1 on cell proliferation in vitro. We observed that GOLM1 knockdown decreased the proliferation of Hep3B cells and GOLM1 overexpression promoted the proliferation of Huh7 cells significantly in vitro compared to their respective negative controls at the time point of 72 h (Supplementary Figure 2A, 2B). Transwell migration assay was conducted to investigate the modulation of GOLM1 on cell migration in vitro. As shown in Supplementary Figure 2C-F, GOLM1 overexpression promoted the migration of Hep3B cells compared with the negative control group. In contrast, the knockdown of GOLM1 decreased the migration ability of Huh7 cells.
GOLM1 knockdown inhibits tumor growth and downregulates the expression of PD-L1 in Hepa1-6 cells in vivo
The subcutaneous tumor model was used to evaluate the effect of GOLM1 on HCC cell proliferation in vivo. Mice were randomly divided into 2 groups (Hepa1-6-NC and Hepa1-6-GOLM1-KD, 5 mice per group). Comparison of the two groups of mice showed that the tumors in GOLM1-KD group were significantly smaller than those in NC group (Figure 6A-C). IHC staining of PD-L1 showed that GOLM1 knockdown downregulates PD-L1 expression. PD-L1 expression in the GOLM1-KD group was significantly lower than that in the NC group (Figure 6D, 6E). Thus, GOLM1 plays an important role in promoting tumor growth and regulating PD-L1 expression.
Figure 6.
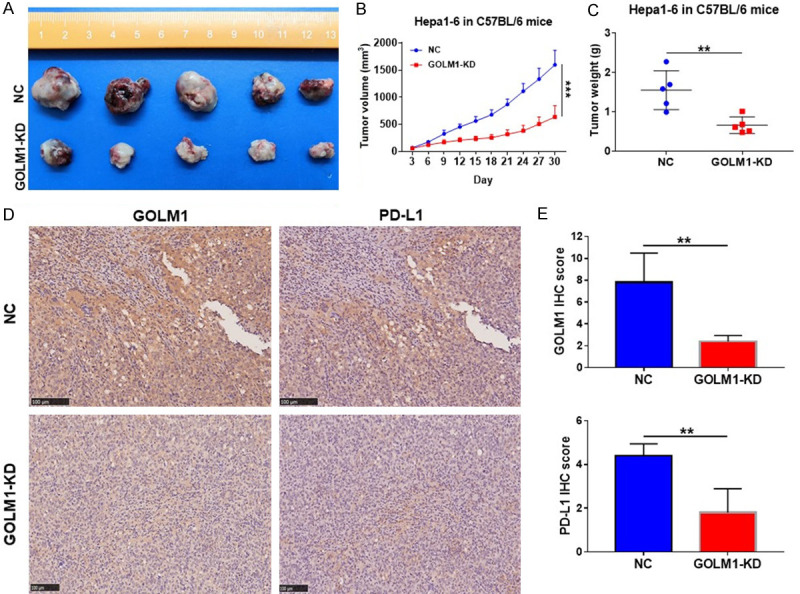
GOLM1 knockdown inhibits tumor growth and downregulates the expression of PD-L1 in Hepa1-6 cells in vivo. A. Images of Hepa1-6 subcutaneous HCC tumors from each group (n = 5). B. Tumor growth curves in C57BL/6 mice from the Hepa1-6-NC and -GOLM1-KD groups. C. Tumor weight from the Hepa1-6-NC and -GOLM1-KD groups. D. Representative IHC images of GOLM1 and PD-L1 expression in Hepa1-6 subcutaneous HCC tumors. Scale bar = 100 μm. E. Graphic representation of IHC score of GOLM1 and PD-L1 expression in Hepa1-6 subcutaneous HCC tumors from NC and GOLM1-KD groups. **P < 0.01, ***P < 0.001.
Discussion
GOLM1 was reported to play an important role in tumor progression [16,17], metastasis [18-21] and immunosuppression [22,23], and its overexpression was associated with cancer recurrence, metastasis, and poor prognosis in different tumors [16,18-21,46]. Especially, in our previous study, we found that GOLM1 could lead to the continued activation of downstream kinases by stabilizing EGFR expression, and finally mediating HCC metastasis [18]. In addition, GOLM1 was reported inducing immunosuppression, but the effect of GOLM1 on the immunosuppressive molecule PD-L1 remains largely unknown. In this study, we unexpectedly found that GOLM1 upregulated PD-L1 transcriptional expression by promoting STAT3 Tyr705 phosphorylation. Furthermore, we found that GOLM1 enhanced the level of EGFR, further leading to upregulation of phosphorylation of STAT3, and finally promoted the expression of PD-L1 in HCC. In conclusion, our results suggest a novel molecular regulatory mechanism - GOLM1/EGFR/STAT3/PD-L1 signaling pathway (Figure 7), which may serve as a novel target axis in searching of a new and effective HCC immunotherapy.
Figure 7.
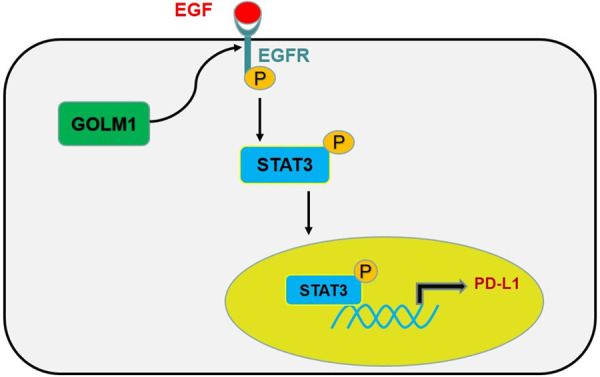
A brief illustration of the proposed working model. GOLM1 can upregulate the level of EGFR. After binding to its ligand EGF, EGFR is phosphorylated, which then promotes phosphorylation of STAT3. Phosphorylated STAT3 plays an important role in promoting the expression of PD-L1.
STAT3 mediates a variety of cellular biological processes, including tumor cell proliferation, metastasis and immune response [47-49]. In malignant tumors, excessive activation of STAT3 leads to inflammation-driven repair, thereby promoting drug resistance or tumor progression [50-53]. STAT3 was reported to induce immunosuppression in cancer by upregulating PD-L1 and overexpression of PD-L1 significantly associates with the level of phosphorylated STAT3 [54,55]. PD-L1 is an important immunomodulatory molecule that can inhibit the immune response by binding to its receptor, PD-1 [56]. In several human cancers, including melanoma, non-small cell lung cancer, hepatocellular carcinoma, gastric cancer, esophageal cancer, and urothelial carcinoma, high PD-L1 expression is associated with tumor immunosuppression and poor prognosis [57-59]. In the current study, we found that GOLM1 upregulated PD-L1 transcriptional expression by promoting STAT3 Tyr705 phosphorylation. This positive regulation effect is attenuated by STAT3 inhibitor due to decreased Tyr705 phosphorylation of STAT3. Our findings reveal GOLM1 as an upstream positive regulator of STAT3 and uncover a new mechanism that regulates the expression of PD-L1 in HCC.
Regarding the mechanism, we found that upon GOLM1 overexpression in HCC cells, EGFR phosphorylation levels were significantly increased, thereby enhancing STAT3 phosphorylation at Tyr705, which finally led to upregulation of PD-L1 in HCC cells. Together this suggests that the phosphorylation level of EGFR regulated by GOLM1 not only affects tumor metastasis but also participates in immune escape in HCC. EGFR is a member of the EGFR/ErbB subfamily of receptor tyrosine kinases (RTKs), which plays an important role in the development of various cancers, cell proliferation, survival and metastasis [60,61]. Furthermore, we previously showed that EGFR recycle regulated by GOLM1 promotes HCC metastasis [18]. Our present study reveals another mechanism by which GOLM1 affects the prognosis of HCC patients.
In summary, we demonstrated that GOLM1 promoted the expression of PD-L1 by regulating the EGFR/STAT3 signaling pathway. Our data reveals a novel regulatory mechanism of PD-L1 expression in HCC, which may provide a novel therapeutic target for HCC immunotherapy.
Acknowledgements
We would like to thank Nadia Alatrakchi and Christopher Oetheimer for their excellent work in correcting spelling/grammar errors and improving scientific writing. This work was supported in part by the National Natural Science Foundation of China (81572301, 81502487, 81802893, 81871924), Shanghai Sailing Program (19YF1407600) and the State Scholarship Fund of China (201906100143).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Nault JC. The end of almost 10 years of negative RCTs in advanced hepatocellular carcinoma. Lancet. 2017;389:4–6. doi: 10.1016/S0140-6736(16)32480-1. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, Gerolami R, Masi G, Ross PJ, Song T, Bronowicki JP, Ollivier-Hourmand I, Kudo M, Cheng AL, Llovet JM, Finn RS, LeBerre MA, Baumhauer A, Meinhardt G, Han G RESORCE Investigators. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 6.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, Baron A, Park JW, Han G, Jassem J, Blanc JF, Vogel A, Komov D, Evans TRJ, Lopez C, Dutcus C, Guo M, Saito K, Kraljevic S, Tamai T, Ren M, Cheng AL. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 7.Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, Cicin I, Merle P, Chen Y, Park JW, Blanc JF, Bolondi L, Klumpen HJ, Chan SL, Zagonel V, Pressiani T, Ryu MH, Venook AP, Hessel C, Borgman-Hagey AE, Schwab G, Kelley RK. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379:54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, Dronca R, Gangadhar TC, Patnaik A, Zarour H, Joshua AM, Gergich K, Elassaiss-Schaap J, Algazi A, Mateus C, Boasberg P, Tumeh PC, Chmielowski B, Ebbinghaus SW, Li XN, Kang SP, Ribas A. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol. 2018;18:153–167. doi: 10.1038/nri.2017.108. [DOI] [PubMed] [Google Scholar]
- 10.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, Ready N, Gainor J, Aren Frontera O, Havel L, Steins M, Garassino MC, Aerts JG, Domine M, Paz-Ares L, Reck M, Baudelet C, Harbison CT, Lestini B, Spigel DR. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, Choueiri TK, Gurney H, Donskov F, Bono P, Wagstaff J, Gauler TC, Ueda T, Tomita Y, Schutz FA, Kollmannsberger C, Larkin J, Ravaud A, Simon JS, Xu LA, Waxman IM, Sharma P CheckMate 025 Investigators. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X, Wang J, Deng X, Xiong F, Ge J, Xiang B, Wu X, Ma J, Zhou M, Li X, Li Y, Li G, Xiong W, Guo C, Zeng Z. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, Meyer T, Kang YK, Yeo W, Chopra A, Anderson J, Dela Cruz C, Lang L, Neely J, Tang H, Dastani HB, Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finn RS, Ryoo BY, Merle P, Kudo M, Bouattour M, Lim HY, Breder V, Edeline J, Chao Y, Ogasawara S, Yau T, Garrido M, Chan SL, Knox J, Daniele B, Ebbinghaus SW, Chen E, Siegel AB, Zhu AX, Cheng AL KEYNOTE-240 investigators. Pembrolizumab as second-line therapy in patients with advanced hepatocellular carcinoma in keynote-240: a randomized, double-blind, phase III trial. J. Clin. Oncol. 2020;38:193–202. doi: 10.1200/JCO.19.01307. [DOI] [PubMed] [Google Scholar]
- 15.Zhou B, Yan J, Guo L, Zhang B, Liu S, Yu M, Chen Z, Zhang K, Zhang W, Li X, Xu Y, Xiao Y, Zhou J, Fan J, Hung MC, Li H, Ye Q. Hepatoma cell-intrinsic TLR9 activation induces immune escape through PD-L1 upregulation in hepatocellular carcinoma. Theranostics. 2020;10:6530–6543. doi: 10.7150/thno.44417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Zhang Y, He F, Li J, Wei X, Li Y, Liao X, Sun J, Yi W, Niu D. Expression of GOLPH2 is associated with the progression of and poor prognosis in gastric cancer. Oncol Rep. 2014;32:2077–2085. doi: 10.3892/or.2014.3404. [DOI] [PubMed] [Google Scholar]
- 17.Duan J, Li X, Huang S, Zeng Y, He Y, Liu H, Lin D, Lu D, Zheng M. GOLPH2, a gene downstream of ras signaling, promotes the progression of pancreatic ductal adenocarcinoma. Mol Med Rep. 2018;17:4187–4194. doi: 10.3892/mmr.2018.8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye QH, Zhu WW, Zhang JB, Qin Y, Lu M, Lin GL, Guo L, Zhang B, Lin ZH, Roessler S, Forgues M, Jia HL, Lu L, Zhang XF, Lian BF, Xie L, Dong QZ, Tang ZY, Wang XW, Qin LX. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;30:444–458. doi: 10.1016/j.ccell.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XM, Cao LL. Identification of GOLM1 as a positively regulator of tumor metastasis by regulating MMP13 in gastric carcinoma. Cancer Biomark. 2019;26:421–430. doi: 10.3233/CBM-190301. [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, Zhu Z, Shen W, Li X, Dhoomun DK, Tian Y. Golgi Membrane Protein 1 (GOLM1) promotes growth and metastasis of breast cancer cells via regulating matrix metalloproteinase-13 (MMP13) Med Sci Monit. 2019;25:847–855. doi: 10.12659/MSM.911667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song YX, Xu ZC, Li HL, Yang PL, Du JK, Xu J. Overexpression of GP73 promotes cell invasion, migration and metastasis by inducing epithelial-mesenchymal transition in pancreatic cancer. Pancreatology. 2018;18:812–821. doi: 10.1016/j.pan.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Zhu C, Wang T, Jiang H, Ren Y, Zhang Q, Wu K, Liu F, Liu Y, Wu J. GP73 represses host innate immune response to promote virus replication by facilitating MAVS and TRAF6 degradation. PLoS Pathog. 2017;13:e1006321. doi: 10.1371/journal.ppat.1006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang QF, Ji Q, Tang Y, Hu SJ, Bao YJ, Peng W, Yin PH. Golgi phosphoprotein 2 down-regulates the Th1 response in human gastric cancer cells by suppressing IL-12A. Asian Pac J Cancer Prev. 2013;14:5747–5751. doi: 10.7314/apjcp.2013.14.10.5747. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, Liu L, Pan X, Wei K, Wei M, Liu L, Yang H, Liu Q. Serum Golgi protein 73 (GP73) is a diagnostic and prognostic marker of chronic HBV liver disease. Medicine (Baltimore) 2015;94:e659. doi: 10.1097/MD.0000000000000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei H, Zhang J, Li H, Ren H, Hao X, Huang Y. GP73, a new marker for diagnosing HBV-ACLF in population with chronic HBV infections. Diagn Microbiol Infect Dis. 2014;79:19–24. doi: 10.1016/j.diagmicrobio.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Wei H, Hao X, Li B, Li X, Hou J, Qiao Y, Zhang R, Li X. GP73 is a potential marker for evaluating AIDS progression and antiretroviral therapy efficacy. Mol Biol Rep. 2013;40:6397–6405. doi: 10.1007/s11033-013-2754-5. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Zhang Y, Wang Y, Xu L, Xu W. Alpha-fetoprotein-L3 and Golgi protein 73 may serve as candidate biomarkers for diagnosing alpha-fetoprotein-negative hepatocellular carcinoma. Onco Targets Ther. 2016;9:123–129. doi: 10.2147/OTT.S90732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu WJ, Guo BL, Han YG, Shi L, Ma WS. Diagnostic value of alpha-fetoprotein-L3 and Golgi protein 73 in hepatocellular carcinomas with low AFP levels. Tumour Biol. 2014;35:12069–12074. doi: 10.1007/s13277-014-2506-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Gu Y, Li X, Wang W, He J, Peng T. Up-regulated Golgi phosphoprotein 2 (GOLPH2) expression in lung adenocarcinoma tissue. Clin Biochem. 2010;43:983–991. doi: 10.1016/j.clinbiochem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Kristiansen G, Fritzsche FR, Wassermann K, Jager C, Tolls A, Lein M, Stephan C, Jung K, Pilarsky C, Dietel M, Moch H. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. Br J Cancer. 2008;99:939–948. doi: 10.1038/sj.bjc.6604614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, Savage KJ, Hernberg MM, Lebbe C, Charles J, Mihalcioiu C, Chiarion-Sileni V, Mauch C, Cognetti F, Arance A, Schmidt H, Schadendorf D, Gogas H, Lundgren-Eriksson L, Horak C, Sharkey B, Waxman IM, Atkinson V, Ascierto PA. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Pan Y, Cao W, Xia F, Liu B, Niu J, Alfranca G, Sun X, Ma L, de la Fuente JM, Song J, Ni J, Cui D. A tumor microenvironment responsive biodegradable CaCO3/MnO2- based nanoplatform for the enhanced photodynamic therapy and improved PD-L1 immunotherapy. Theranostics. 2019;9:6867–6884. doi: 10.7150/thno.37586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, Wasik MA. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci U S A. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T, Zhang C, Zhao G, Zhang X, Hao M, Hassan S, Zhang M, Zheng H, Yang D, Liu L, Mehraein-Ghomi F, Bai X, Chen K, Zhang W, Yang J. IGFBP2 regulates PD-L1 expression by activating the EGFR-STAT3 signaling pathway in malignant melanoma. Cancer Lett. 2020;477:19–30. doi: 10.1016/j.canlet.2020.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi HI, Kim DH, Park JS, Kim IJ, Kim CS, Bae EH, Ma SK, Lee TH, Kim SW. Peroxiredoxin V (PrdxV) negatively regulates EGFR/Stat3-mediated fibrogenesis via a Cys48-dependent interaction between PrdxV and Stat3. Sci Rep. 2019;9:8751. doi: 10.1038/s41598-019-45347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhat AA, Lu H, Soutto M, Capobianco A, Rai P, Zaika A, El-Rifai W. Exposure of Barrett’s and esophageal adenocarcinoma cells to bile acids activates EGFR-STAT3 signaling axis via induction of APE1. Oncogene. 2018;37:6011–6024. doi: 10.1038/s41388-018-0388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao H, Cheng HY, Cook RG, Tweardy DJ. Identification and characterization of signal transducer and activator of transcription 3 recruitment sites within the epidermal growth factor receptor. Cancer Res. 2003;63:3923–3930. [PubMed] [Google Scholar]
- 40.Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14:6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci U S A. 1996;93:13704–13708. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Li CW, Li X, Ding Q, Guo L, Liu S, Liu C, Lai CC, Hsu JM, Dong Q, Xia W, Hsu JL, Yamaguchi H, Du Y, Lai YJ, Sun X, Koller PB, Ye Q, Hung MC. MET inhibitors promote liver tumor evasion of the immune response by stabilizing PDL1. Gastroenterology. 2019;156:1849–1861. e1813. doi: 10.1053/j.gastro.2019.01.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, Xu Y, Li YW, Tang ZY. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J. Clin. Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- 44.Zhou B, Guo L, Zhang B, Liu S, Zhang K, Yan J, Zhang W, Yu M, Chen Z, Xu Y, Xiao Y, Zhou J, Fan J, Li H, Ye Q. Disulfiram combined with copper induces immunosuppression via PD-L1 stabilization in hepatocellular carcinoma. Am J Cancer Res. 2019;9:2442–2455. [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Huang T, Zou Q, Liu D, Wang Y, Tan X, Wei Y, Qiu H. FGFR2 promotes expression of PD-L1 in colorectal cancer via the JAK/STAT3 signaling pathway. J Immunol. 2019;202:3065–3075. doi: 10.4049/jimmunol.1801199. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Chen L, Zhang T. Increased GOLM1 Expression independently predicts unfavorable overall survival and recurrence-free survival in lung adenocarcinoma. Cancer Control. 2018;25:1073274818778001. doi: 10.1177/1073274818778001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Li H, Lin HJ, Yang S, Lin J, Liang G. Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Zheng H, Hong H, Zhang L, Cai X, Hu M, Cai Y, Zhou B, Lin J, Zhao C, Hu W. Nifuratel, a novel STAT3 inhibitor with potent activity against human gastric cancer cells. Cancer Manag Res. 2017;9:565–572. doi: 10.2147/CMAR.S146173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 51.Jarnicki A, Putoczki T, Ernst M. Stat3: linking inflammation to epithelial cancer - more than a “gut” feeling? Cell Div. 2010;5:14. doi: 10.1186/1747-1028-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lankadasari MB, Aparna JS, Mohammed S, James S, Aoki K, Binu VS, Nair S, Harikumar KB. Targeting S1PR1/STAT3 loop abrogates desmoplasia and chemosensitizes pancreatic cancer to gemcitabine. Theranostics. 2018;8:3824–3840. doi: 10.7150/thno.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, Chang LL, Yan FJ, Hu Y, Zeng CM, Zhou TY, Yuan T, Ying MD, Cao J, He QJ, Yang B. AKR1C1 activates STAT3 to promote the metastasis of non-small cell lung cancer. Theranostics. 2018;8:676–692. doi: 10.7150/thno.21463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bu LL, Yu GT, Wu L, Mao L, Deng WW, Liu JF, Kulkarni AB, Zhang WF, Zhang L, Sun ZJ. STAT3 induces immunosuppression by upregulating PD-1/PD-L1 in HNSCC. J Dent Res. 2017;96:1027–1034. doi: 10.1177/0022034517712435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning W, Zeng H, Zhang N, Du W, Chen C, Huang JA. PD-L1 induced by IFN-gamma from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol. 2017;22:1026–1033. doi: 10.1007/s10147-017-1161-7. [DOI] [PubMed] [Google Scholar]
- 56.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, Okazaki T, Tokura Y. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer. 2010;116:1757–1766. doi: 10.1002/cncr.24899. [DOI] [PubMed] [Google Scholar]
- 58.Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Krejci KG, Lobo JR, Sengupta S, Chen L, Zincke H, Blute ML, Strome SE, Leibovich BC, Kwon ED. Costimulatory B7-H1 in renal cell carcinoma patients: indicator of tumor aggressiveness and potential therapeutic target. Proc Natl Acad Sci U S A. 2004;101:17174–17179. doi: 10.1073/pnas.0406351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med. 2015;21:24–33. doi: 10.1016/j.molmed.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 61.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.