Recombinant human erythropoietin (rhEPO) and its derivatives are some of the most important treatment options against anemia associated with chronic kidney disease, chemotherapy, and low-risk myelodysplastic syndrome.1 On the other hand, it has also gained fame as a doping agent in endurance sports. All this became possible by translating basic research findings that started more than 100 years ago with the recognition of an increase in red blood cell number in animals living at high altitude (i.e., at a decreased pO2) and that went, via purification, partial sequencing, cloning, and basic understanding of EPO gene regulation, into clinics (for excellent reviews of EPO history see2-5).
With its cloning and the fast-improving molecular biological tools, findings from hypoxia-inducible EPO gene regulation also became a paradigm to understand basic principles of how cells sense and adapt to oxygen availability. The discovery of those principles were acknowledged by part of the 2019 Nobel prize in Physiology or Medicine.6
The major production sites of EPO are peritubular fibroblasts in the kidney as well as hepatocytes and hepatic stellate cells (about 90% of circulating EPO stems from kidney and 10% from liver). But EPO expression can also be found in brain, testis, uterus, and osteoblasts.7-10 Apart from its major function as a driver of erythropoiesis, EPO was also found to exert organ- and tissuerestricted protective functions in the brain, cardiovascular system, adipose tissue, and bones.11,12
Experiments with transgenic mice revealed that critical parts of the EPO gene that are located away from the proper EPO coding exons in the distal 5’- and 3’-flanking regions are responsible for tissue-specific and hypoxiadriven EPO gene expression. Up to then, it had been known that an array consisting of the kidney-inducible element (KIE; between -14 kb to -9.5 kb 5’ from the promoter) and a negative regulatory element (NRE, between -6kb to -0.4kb) in the 5’-part seem to be of special importance for EPO expression in kidney (Figure 1). By contrast, in liver, hypoxia-inducible EPO expression appeared to depend largely on an enhancer in the so-called liverinducible element (LIE) that was found 3’-distal from the EPO polyadenylation side (Figure 1). That area contained, among other functional sites, an HRE (hypoxia response element) that served as a binding site for a hypoxiainducible factor (HIF).13-16 From the three HIFs known, HIF-2α has been defined as a major part of the hypoxiainducible EPO gene activating complex.17,18
While the role of the HRE in the 3’-enhancer is well established and found to be necessary and sufficient to confer liver-specific EPO gene expression, the DNA element responsible for kidney-specific EPO gene expression is far less well characterized and nothing was known about the presence and function of HREs in EPO-producing none-kidney tissues such as neurons of the brain. This problem has been tackled by Orlando and co-workers, and the outcome of their studies is described in an article in the current issue of Haematologica.19
In their studies, the authors built on findings where they discovered a distal 5’-HRE within a DNaseI hypersensitive site -9.2 kb upstream of the EPO transcriptional start site which was supposed to contribute to oxygenregulated EPO expression in the kidney.20 As the EPOproducing cells in the kidney derive from neural crest and neuroepithelial cells,21 the authors went on to compare the function of this part containing the 5’ HRE and the established 3’-HRE in human neuroblastoma cells and liver hepatocellular carcinoma cells; both cell types are able to express EPO in response to hypoxia.22
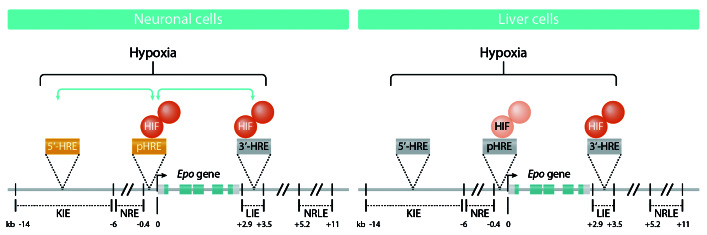
Figure 1.
Hypoxia-regulated human erythropoietin (EPO) gene expression in neuronal and liver cells. Hypoxia leads to stabilization of hypoxia-inducible factor (HIF), and mainly HIF-2 containing complexes are able to induce EPO gene expression. In neuronal cells, HIF-containing complexes bind most strongly to the newly identified hypoxia response element (HRE) in the promoter and to a lesser extent to the 3'-HRE. Although HIF-complexes appear not to bind the 5'-HRE, the 5´-HRE co-operates in the hypoxic induction process. In liver cells, HIF-complexes bind mainly to the 3´-HRE, weakly to the pHRE, and not to the 5´- HRE. Lighter colors indicate less HIF binding; green arrows indicate co-operation between the HREs. KIE: kidney-inducible element; NRE: negative regulatory element; LIE: liver-inducible element; HRE: hypoxia response element; NRLE: negative regulatory liver element; Prom: promoter; pHRE: promoter HRE functioning as one entity but consisting of a tandem dimeric repeat with two HRE sequences.
To do this, they used gene editing by CRISPR/Cas9 to specifically mutate the -9.2 kb 5’-HRE and the +3.0 kb 3’- HRE in the EPO gene locus, and combined this with subsequent HIF-DNA interaction studies, RNA and protein expression measurements and reporter gene assays.
Intriguingly, when analyzing EPO mRNA and secreted EPO protein levels upon exposure of the engineered cells to hypoxia (0.2% O2) for 24 hours, the authors found that the neuroblastoma cells with mutations in both the 5’- HRE and the 3’-HRE did not increase EPO mRNA and protein levels to the same extent as wild-type cells. By contrast, only mutation of the 3’-HRE reduced the hypoxic EPO induction in hepatoma cells.
When the authors expanded these experiments by transiently transfecting cells with minimal EPO promoter-driven reporter genes, enhanced by various DNA fragments containing the 5’- and/or 3’-HRE, they found that EPO promoter-driven reporter gene activity in neuroblastoma cells was best when a 100 bp 5’-region containing the HRE was combined with a 126 bp HRE containing 3’- fragment.
Further experiments with different EPO promoter HRE constructs indicated that the most efficient hypoxiadependent EPO induction requires co-operation between the EPO 5’- and 3’-HRE specifically in neuronal cells, and that additional distal and proximal 5’-flanking elements further contribute to tissue-specific and conditional EPO regulation.
In addition, the minimal EPO gene promoter alone was able to promote hypoxia-dependent reporter gene activation in neuroblastoma cells and hepatoma cells indicating that this part may contain previously unidentified HRE. Indeed, the authors found a tandem dimeric repeat with two HRE sequences that had not previously been reported. Importantly, mutation of each single HRE reduced reporter gene activity, but simultaneous mutation of both HREs did not have an additional effect, suggesting that both HREs behave as an entity.
Next, the authors were able to link the observed findings to HIF binding. Chromatin immunoprecipitation assays in neuroblastoma cells revealed that none of the HIF subunits bound to the 5’-HRE, whereas a significant hypoxic increase in HIF-2α/HIF-b binding to the 3’-HRE could be detected. By contrast, in hepatocellular carcinoma cells, HIF-2α/HIF-b binding could be detected with the promoter region and the 3’-HRE. Importantly, non- EPO producing cells did not show any HIF binding to the examined regions of the EPO gene. Further experiments with the CRISPR/Cas9 engineered cells showed that mutation of the 3’-HRE also decreased HIF binding to the promoter region; vice versa, mutation of the 5’-HRE impaired HIF binding to the 3’-HRE.
Taken together, the study shows that hypoxic EPO expression in hepatic cells appears only to depend on HIF interaction with the 3'-HRE and not on the 5'-HRE or the HIF binding EPO promoter. By contrast, neuronal EPO expression seems to require co-operation with intact 5'- and 3’-HRE, with HIF interacting strongly with the new HRE in the EPO promoter, and to a lesser extent with the 3' HRE.
Another important aspect of this study is that hypoxiadependent gene expression is not a uniform process and needs to be seen and analyzed in a cell type-specific context. This is because some cells’ HIF may not bind to all HREs, although they may confer oxygen sensitivity. This may be of importance in gene-wide association studies where different SNPs or disease-associated polymorphisms may represent HREs which may, depending on the context, gain or lack their function.
Overall, the investigation by Orlando et al.19 presents novel and interesting findings highlighting that a complex interplay between various HREs, HIFs and other factors at the EPO locus contribute to its hypoxia-dependent and organ-specific expression.
Supplementary Material
Acknowledgments
TK was supported by the Academy of Finland SA296027, the Jane and Aatos Erkko Foundation, the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, and Biocenter Oulu.
References
- 1.Rizzo JD, Brouwers M, Hurley P, et al. American Society of Hematology/American Society of Clinical Oncology clinical practice guideline update on the use of epoetin and darbepoetin in adult patients with cancer. Blood. 2010;116(20):4045-4059. [DOI] [PubMed] [Google Scholar]
- 2.Fandrey J. Oxygen-dependent and tissue-specific regulation of erythropoietin gene expression. Am J Physiol Regul Integr Comp Physiol. 2004;286(6):R977-988. [DOI] [PubMed] [Google Scholar]
- 3.Wenger RH, Kurtz A. Erythropoietin. Compr Physiol. 2011;1(4):1759-1794. [DOI] [PubMed] [Google Scholar]
- 4.Bunn HF. Erythropoietin. Cold Spring Harb Perspect Med. 2013;3(3):a011619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jelkmann W. Erythropoietin. Front Horm Res. 2016;47:115-127. [DOI] [PubMed] [Google Scholar]
- 6.Kietzmann T. The air that we breeze: from 'Noble' discoveries of a general oxygen-sensing principle to its clinical use. Acta Physiol (Oxf). 2020;228(2):e13416. [DOI] [PubMed] [Google Scholar]
- 7.Marti HH. Erythropoietin and the hypoxic brain. J Exp Biol. 2004;207(Pt 18):3233-3242. [DOI] [PubMed] [Google Scholar]
- 8.Magnanti M, Gandini O, Giuliani L, et al. Erythropoietin expression in primary rat Sertoli and peritubular myoid cells. Blood. 2001;98(9):2872-2874. [DOI] [PubMed] [Google Scholar]
- 9.Yasuda Y, Masuda S, Chikuma M, Inoue K, Nagao M, Sasaki R. Estrogen-dependent production of erythropoietin in uterus and its implication in uterine angiogenesis. J Biol Chem. 1998;273(39):25381-25387. [DOI] [PubMed] [Google Scholar]
- 10.Rankin EB, Wu C, Khatri R, et al. The HIF signaling pathway in osteoblasts directly modulates erythropoiesis through the production of EPO. Cell. 2012;149(1):63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suresh S, Rajvanshi PK, Noguchi CT. The many facets of erythropoietin physiologic and metabolic response. Front Physiol. 2020;10:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng B, Kong G, Yang C, Ming Y. Erythropoietin and its derivatives: from tissue protection to immune regulation. Cell Death Dis. 2020;11(2):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL, Traystman MD, Gearhart JD, Antonarakis SE. Polycythemia in transgenic mice expressing the human erythropoietin gene. Proc Natl Acad Sci U S A. 1989;86(7):2301-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenza GL, Dureza RC, Traystman MD, Gearhart JD, Antonarakis SE. Human erythropoietin gene expression in transgenic mice: multiple transcription initiation sites and cis-acting regulatory elements. Mol Cell Biol. 1990;10(3):930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madan A, Lin C, Hatch SL 2nd, Curtin PT. Regulated basal, inducible, and tissue-specific human erythropoietin gene expression in transgenic mice requires multiple cis DNA sequences. Blood. 1995;85(10):2735-2741. [PubMed] [Google Scholar]
- 16.Madan A, Curtin PT. A 24-base-pair sequence 3' to the human erythropoietin gene contains a hypoxia-responsive transcriptional enhancer. Proc Natl Acad Sci U S A. 1993;90(9):3928-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imeri F, Nolan KA, Bapst AM, et al. Generation of renal Epo-producing cell lines by conditional gene tagging reveals rapid HIF-2 driven Epo kinetics, cell autonomous feedback regulation, and a telocyte phenotype. Kidney Int. 2019;95(2):375-387. [DOI] [PubMed] [Google Scholar]
- 18.Kapitsinou PP, Liu Q, Unger TL, et al. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood. 2010;116(16):3039-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlando IMC, Lafleur VN, Storti F, et al. Distal and proximal hypoxia response elements co-operate to regulate organ-specific erythropoietin gene expression. Haematologica. 2020;105(12):2774-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storti F, Santambrogio S, Crowther LM, et al. A novel distal upstream hypoxia response element regulating oxygen-dependent erythropoietin gene expression. Haematologica. 2014;99(4):e45-e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano I, Suzuki N. The neural crest as the first production site of the erythroid growth factor erythropoietin. Front Cell Dev Biol. 2019;7:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stolze I, Berchner-Pfannschmidt U, Freitag P, et al. Hypoxiainducible erythropoietin gene expression in human neuroblastoma cells. Blood. 2002;100(7):2623-2628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.