Abstract
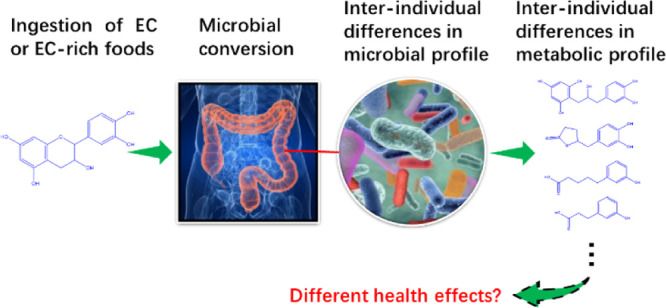
To quantify interindividual differences in the human intestinal microbial metabolism of (−)-epicatechin (EC), in vitro anaerobic incubations with fecal inocula from 24 healthy donors were conducted. EC-derived colonic microbial metabolites were qualitatively and quantitively analyzed by liquid chromatography triple quadrupole mass spectrometry (LC-TQ-MS) and liquid chromatography time-of-flight mass spectrometry (LC-TOF-MS). Quantitative microbiota characterization was achieved by 16S rRNA analysis. The results obtained show 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-dihydroxyphenyl)-2-propanol (3,4-diHPP-2-ol) and 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone (3,4-diHPV) to be key intermediate microbial metabolites of EC and also revealed the substantial interindividual differences in both the rate of EC conversion and the time-dependent EC metabolite pattern. Furthermore, substantial differences in microbiota composition among different individuals were detected. Correlations between specific microbial phylotypes and formation of certain metabolites were established. It is concluded that interindividual differences in the intestinal microbial metabolism of EC may contribute to interindividual differences in potential health effects of EC-abundant dietary foods or drinks.
Keywords: (−)-epicatechin, phenyl-γ-valerolactones, microbiota, microbial metabolism, interindividual differences
Introduction
Epicatechin (EC) is one of the important dietary catechins that belong to the group of flavan-3-ols and widely exists in plant-based foods, for example, cocoa, apple, green tea, berries, chocolates, grape seeds, and so forth.1−3 The ingestion of these EC-rich foods is often reported to be associated with beneficial health effects, including anti-inflammatory, cardioprotective, anticancer activities, and so forth.4−7 However, the bioavailability of catechins has been reported to be low because of the fact that upon ingestion in the small intestine, only a limited amount of the parent compounds will be absorbed with subsequent phase II biotransformation reactions, that is, conjugated in glucuronidation, methylation, and/or sulfation reactions.8,9 A substantial part of the catechins is known to be passed on to the large intestine, where they are subjected to intense microbial metabolism to form a battery of smaller phenolic molecules.10,11 For example, although EC possesses the highest bioavailability among other major catechins, for example, (−)-epigallocatechin (EGC), (−)-epicatechin-3-O-gallate (ECG), and (−)-epigallocatechin-3-O-gallate (EGCG), only about 20% of the intake was reported to pass into the systemic circulation via absorption in the small intestine, while over 70% of the ingested EC appeared to end up in the systemic circulation in the form of colonic phenolic metabolites.11,12 These intestinal microbial metabolites thus contribute substantially to the systemic metabolites of EC.13
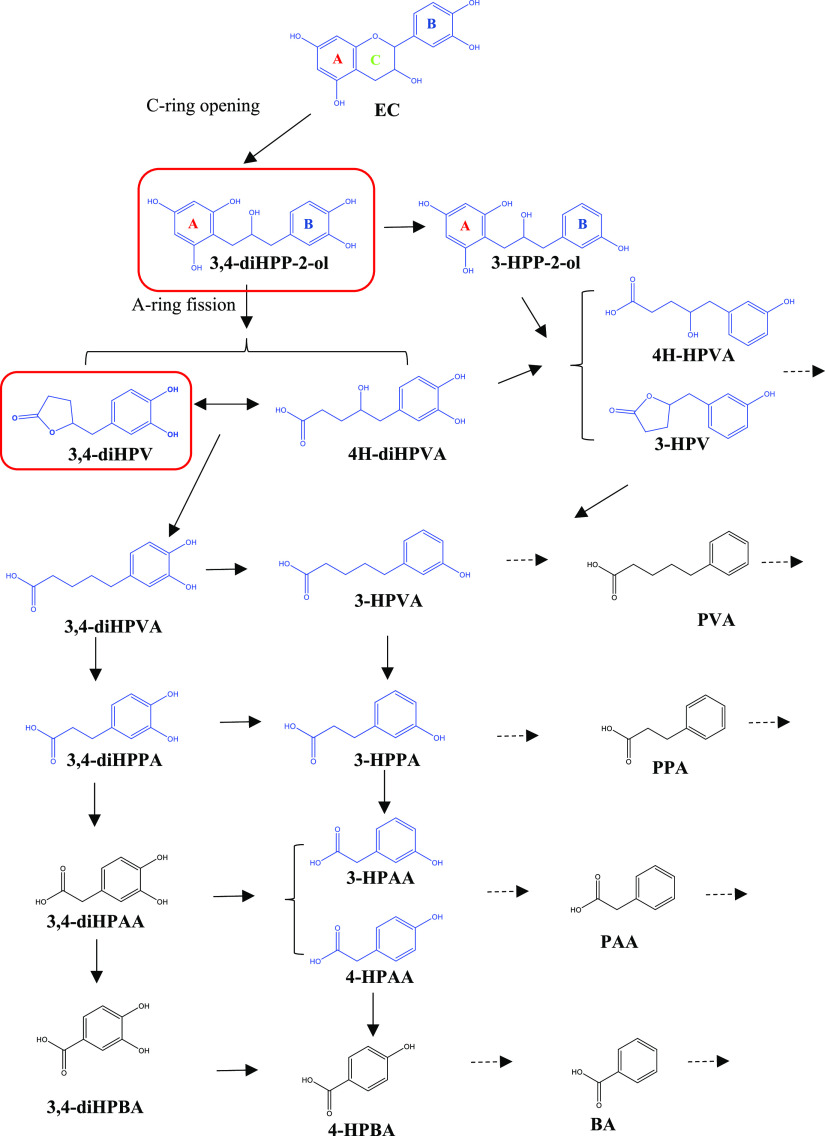
An overview of the major metabolic pathways in the potential intestinal microbial degradation of EC is presented in Figure 1. Given the many potential metabolites that can be formed and the substantial differences in the interindividual composition of the human microbiota,14,15 the microbial metabolic routes of EC may be expected to show interindividual differences, which have not been characterized so far.9 Additionally, knowledge on the microbiota-mediated conversion of catechins is limited.
Figure 1.
Proposed pathways for the colonic metabolism of EC. Structures in blue color were detected in the present study. The chemicals in the red boxes [i.e., 1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-2-propanol (3,4-diHPP-2-ol) and 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone (3,4-diHPV)] were the major metabolites formed in the time-dependent in vitro fecal incubation.
The aim of the present study is to obtain insights into the potential interindividual human differences in this intestinal microbial metabolism of EC. This insight will also contribute to a better understanding of the causal relationship between EC intake and potential beneficial effects because it has been hypothesized that these are due to the microbial metabolites of EC.16−18
Materials and Methods
Chemicals and Reagents
EC, catechol, gallic acid, 3′-hydroxyphenylacetic acid (3-HPAA), 4′-hydroxyphenylacetic acid (4-HPAA), 3-(3′-hydroxyphenyl)propionic acid (3-HPPA), 3-(3′,4′-dihydroxyphenyl)propionic acid (3,4-diHPPA), 5-(3′-hydroxyphenyl)valeric acid (3-HPVA), 5-(3′,4′-dihydroxyphenyl)valeric acid (3,4-diHPVA), and glycerol were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). 5-(3′-hydroxyphenyl)-γ-valerolactone (3-HPV) and 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone (3,4-diHPV) were purchased from Enamine (Kyiv, Ukraine). Methanol and acetonitrile (ACN) were obtained from Biosolve BV (Valkenswaard, The Netherlands). Phosphate buffer saline (PBS) was purchased from Gibco (Paisley, UK). Acetic acid was ordered from Merck KGaA (Darmstadt, Germany), and formic acid was purchased from VWR CHEMICA (Amsterdam, The Netherlands).
Fecal Slurry Preparation
Feces were collected from 24 healthy individuals without dietary restrictions. They were of different genders, ages, and ethnicities. All of them had not taken antibiotics for at least 3 months prior to the study. After volunteers’ donations, fecal samples were collected, weighed, and then immediately transferred to the anaerobic operating chamber BACTRON300 (Cornelius, USA) with an atmosphere of 85% N2, 10% CO2, and 5% H2 within 5 min. Subsequently, fecal samples were diluted with an anaerobic solution of 10% (v/v) glycerol in PBS to obtain a final fecal concentration of 20% (w/v). Samples were filtered in Para-Pak SpinCon tubes (New Hampshire, USA) by centrifugation (21,500g for 5 min at 4 °C) to remove the larger particles, and the resulting fecal suspension was collected and aliquoted. A pooled sample was obtained by mixing equal volumes of the first 14 fecal suspensions and was subsequently aliquoted. All the abovementioned steps were carried out under anaerobic conditions. In the end, all samples were stored at −80 °C until use. The effect of freezing, storing, and thawing on the microbial metabolism of EC by the fecal samples was tested and shown not to significantly affect the activity (Figure S1 in Supporting Information). The experimental protocol was approved not to require further evaluation by the Medical Ethical Reviewing Committee of Wageningen University (METC-WU) based on the Dutch Medical Research Involving Human Subjects Act. All participants gave their written consent.
Fecal Batch Culture Incubation
The experiments were performed in 1.5 mL Eppendorf tubes containing 39.5 μL of the anaerobic PBS, 10 μL of the fecal slurries (final fecal concentration: 40 mg/mL), and 0.5 μL of EC (added from a 100 times concentrated stock solution in methanol; final concentration 100 μM) or methanol as control (referred to as “negative control”). The final concentration of 100 μM EC allows for detection of the substrate and its metabolites without affecting the microbiota.19 Incubations of EC without fecal slurries were included as blank control. All fecal incubations were carried out in the BACTRON300 anaerobic chamber with an atmosphere of 85% N2, 10% CO2, and 5% H2 at 37 °C. Aliquots of the fermented fecal samples (50 μL) were collected after 0, 1, 2, 3, 4, 5, 6, and 24 h of incubation, and the reaction was terminated by adding 1 volume of ice-cold methanol. For studies on interindividual differences, a time point of 2 h was selected. Subsequently, samples were put on ice for at least 15 min, followed by centrifugation at 21,500g for 15 min at 4 °C to precipitate proteins, microorganisms, and so forth. The supernatant of each sample was collected and stored immediately at −80 °C until liquid chromatography triple quadrupole mass spectrometry (LC-TQ-MS) and LC-TOF-MS analyses. The incubation of each individual and pooled fecal sample was performed in triplicate.
LC-TQ-MS Analysis
A Shimadzu LC-TQ-MS 8045 was used to quantify the concentration of EC and its metabolites. This equipment consisted of an ultrahigh-performance liquid chromatography system coupled to a tandem triple quadrupole mass spectrometer containing an ESI source (s-Hertogenbosch, The Netherlands). The ESI source was operated in negative ion mode, and fragment ions m/z were obtained. Chromatographic separation was performed on a Waters Acquity UPLC BEH C18 column (2.1 × 50 mm; 1.7 μm) at 40 °C. Solvent A was composed of water: acetic acid (999 : 1, v/v), and solvent B was methanol absolute. The eluents were delivered at a flow rate of 0.4 mL/min, and the following gradient was used: 0–0.5 min: 5% B, 0.5–5 min: 5–25% B, 5–6 min: 25–100% B, 6–7 min: 100% B, 7–8 min: 100–5% B, and 8–13 min: 5% B. The ESI parameters were as follows: nebulizing gas flow, 3.0 L/min; drying gas flow and heating gas flow, 10.0 L/min; interface temperature, 300 °C; desolvation temperature, 250 °C; and heat block temperature, 400 °C. Chromatographic peaks were identified by comparing the retention time and the ion values of the samples with those of the commercial authentic standards. The identification information for EC and its potential colonic metabolites are summarized in Table 1. These compounds were included in the triple quadrupole based on several in vitro and in vivo studies reporting their formation.3,8,9,11,20−22 To quantify the metabolites formed during in vitro incubations, a set of mixed authentic standards were used to build calibration curves as appropriate using 14 concentrations ranging between 0.01 and 50 μM (the area of peak against the concentration was obtained to define the linear regression equation). To guarantee the accuracy of each quantification, a set of calibrators to define the calibration curve was included in each measurement series. Concentrations of the formed metabolites were calculated correcting for values of parallel negative controls incubated without EC. Each experimental replicate was corrected with the mean value of three negative controls obtained for the respective fecal sample.
Table 1. LC-TQ-MS and LC-TOF-MS Identification of EC and Its Potential Colonic Metabolites, Based on Relative Retention Time and MS (or MS Fragmentation) Values.
| analytical platform | chemical | parent ion (m/z) | product ion (m/z) | collision energy (V) | retention time |
|---|---|---|---|---|---|
| LC-TQ-MS | EC | 289.1 | 245.10, 109.10, 203.00 | 14, 25, 18 | 5.72 |
| 3,4-diHPVA | 209.0 | 135.10, 191.05, 122.10 | 18, 15, 21 | 7.60 | |
| 3,4-diHPV | 207.1 | 163.05, 122.15, 161.10 | 16, 21, 21 | 5.00 | |
| 3-HPVA | 193.1 | 106.15, 149.05, 175.05 | 23, 17, 15 | 7.88 | |
| 3-HPV | 191.1 | 147.05, 102.90, 135.00 | 8, 15, 15 | 6.93 | |
| 3,4-diHPPA | 181.1 | 137.10, 59.00, 109.00 | 14, 17, 15 | 3.95 | |
| 3-HPPA | 165.1 | 121.20, 106.10, 119.15 | 13, 22, 16 | 6.33 | |
| 3-HPAA | 151.0 | 107.10, 65.00 | 12, 25 | 4.30 | |
| 4-HPAA | 151.0 | 107.10, 79.15, 93.20 | 10, 20, 22 | 3.82 | |
| catechol | 109.2 | 90.95, 81.10, 53.15 | 22, 21, 20 | 1.85 | |
| LC-TOF-MS | 3,4-diHPP-2-ol | 291.09 | 19.6 | ||
| 3-HPP-2-ol | 275.09 | 23.1 | |||
| 4H-diHPVA | 225.08 | 15.7 | |||
| 4H-HPVA | 209.08 | 15.2 |
LC-TOF-MS Analysis
An Agilent 1200 series high-performance liquid chromatography system coupled with a Bruker micrOTOF was applied to qualitatively and quantitively detect 3,4-diHPP-2-ol, 1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-2-propanol (3-HPP-2-ol), 4-hydroxy-5-(3′,4′-dihydroxyphenyl)-valeric acid (4H-diHPVA), and 4-hydroxy-5-(3′-hydroxyphenyl)-valeric acid (4H-HPVA). Chromatographic separation was performed on the same column as used in the LC-TQ-MS analysis. Solvent A was composed of water/formic acid (999:1, v/v), and solvent B was ACN/water (999:1, v/v). The eluents were delivered at a flow rate of 0.2 mL/min, and the following gradient was used: 0–2.5 min: 100% A, 2.5–40 min: 0–40% B, 40–45 min: 40–100% B, 45–50 min: 100% B, 50–55 min: 100–0% B, and 55–90 min: 100% A. The mass detection was operated in negative mode. Relative retention time and M–H values were used to identify these three EC colonic metabolites. Specifically, molecules such as 3,4-diHPP-2-ol, 3-HPP-2-ol, and 4H-diHPVA have high M–H values (i.e., 291.09, 275.09, and 225.08), which are unique in the EC microbial conversion pathway. Furthermore, their time-dependent formations (e.g., the first two metabolites were largely detected at early hours of incubation) were taken into consideration. Additionally, chemical standards used in LC-TQ-MS were also analyzed in LC-TOF-MS, which helped to distinguish compounds sharing the same M–H values. For example, although 4H-HPVA and 3,4-diHPVA share the same M–H value in negative mode, the measurement of the 3,4-diHPVA standard in LC-TOF-MS ensured the unequivocal identification of both 3,4-diHPVA and 4H-HPVA. Altogether, this approach and the combination of LC-TQ-MS and LC-TOF-MS also ensured the unequivocal identification of those metabolites for which standards were not available. Quantification was carried out through the calibration curves of EC (for quantifications of 3,4-diHPP-2-ol and 3-HPP-2-ol) or 3,4-diHPV (for quantifications of 4H-diHPVA and 4H-HPVA). Detailed identification information is summarized in Table 1.
Microbial Taxonomic Profiling and Total Bacterial Load
Aliquots of all 24 fecal samples were sent to an accredited commercial laboratory (IMGM Laboratories GmbH, Martinsried, Germany) for DNA extraction, PCR, library preparation, and sequencing. Additionally, quantification of the bacterial load was carried out by real-time qPCR. PCR products were generated by amplification using 16S V3–V4 primers (F-NXT-Bakt-341F: 5′-CCTACGGGNGGCWGCAG-3′ and R-NXT-Bakt-805R: 5′-GACTACHVGGGTATCTAATCC-3′). During an index PCR, barcodes for multiplexed sequencing were introduced using overhang tags. A sequencing library was prepared from the barcoded PCR products and sequenced on the Illumina MiSeq next-generation sequencing system (Illumina Inc.). Signals were processed to *.fastq-files, and the resulting 2 × 250 bp reads were demultiplexed. Microbiota identification was carried out by clustering the operational taxonomic units (OTUs).
Data Analysis
Metabolic profile data acquisition and processing were carried out using the Labsolutions software in the LC-TQ-MS system and HyStar software in the LC-TOF-MS system. Graphics were drawn by using Graphpad Prism 8.2 (San Diego, USA). ChemDraw 18.0 (PerkinElmer, Waltham, USA) was used to draw chemical structures. Results are shown as mean ± standard deviation (SD). Subsequently, principal component analysis (PCA) was applied to study the differences and relatedness of metabolic profiles following fecal incubations of EC (at t = 2 h) between different individuals. Using this approach, the multivariate metabolic profile data derived from interindividual fecal incubations were summarized, simplified, and then transformed into a smaller data set called principal components (PCs) of PCA. In addition to PCA, a heatmap was used to visualize the hierarchic clustering of different individuals based on their metabolic profiles and EC residuals. Both PCA and heatmap development were done using XLSTAT (Addinsoft, New York, USA). Spearman’s rank correlation coefficients ρ (−1 ≤ ρ ≤ 1) were calculated between the concentration of the six most relevant metabolites present after 2 h of incubation and the relative abundance of the phyla (range between 0 and 100%). To display the correlations, a heatmap was constructed. Statistically significant correlation was tested by Student t-test (p < 0.05).
Results
LC-TQ-MS- and LC-TOF-MS-Based Identification of Colonic Metabolites of EC
Upon time-dependent incubation of EC with fecal samples, various metabolites were formed. These metabolites were identified and quantified based on chromatography of standard reference compounds and coupled with mass spectrometric fragmentation patterns. The analytical details of the LC-TQ-MS- and LC-TOF-MS-based detection of EC and its potential colonic metabolites are summarized in Table 1. In total, during the 8 h incubation time, 12 metabolites, of which the chemical structures are highlighted in Figure 1 in blue color, were observed. These metabolites were 3,4-diHPP-2-ol, 3-HPP-2-ol, 4H-diHPVA, 3,4-diHPV, 4H-HPVA, 3-HPV, 3,4-diHPVA, 3-HPVA, 3,4-diHPPA, 3-HPPA, 3-HPAA, and 4-HPAA.
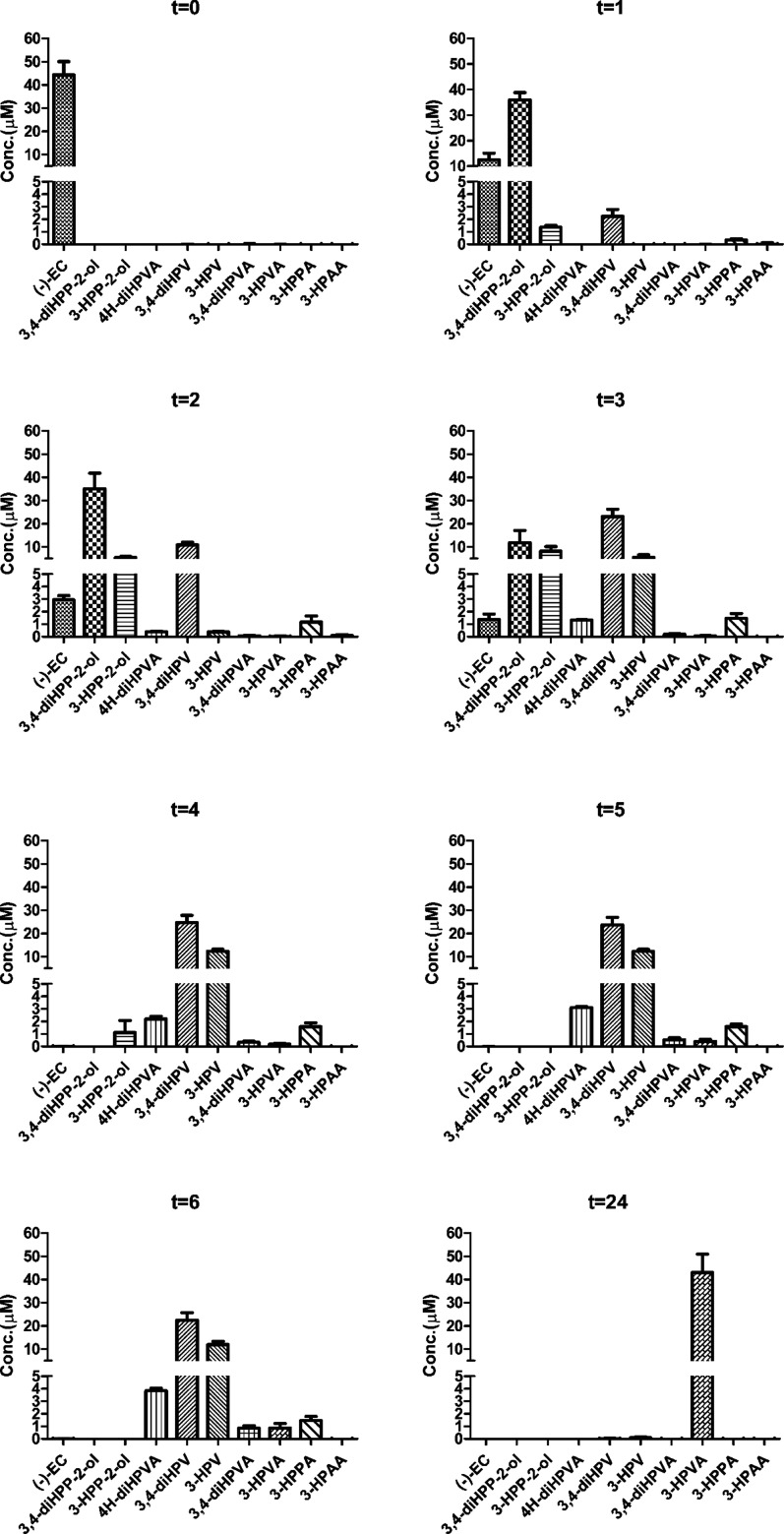
Quantification was achieved via calibration curves made using commercially available reference compounds, except for 3,4-diHPP-2-ol, 3-HPP-2-ol, 4H-diHPVA, and 4H-HPVA, for which authentic references were not available. For these compounds, quantification was achieved using the calibration curves of EC for the first two and of 3,4-diHPV for the last two based on their structural similarity. However, the amount of 4H-HPVA observed appeared to be below the levels required for quantification. The limit of detection (LOD) and the limit of quantification (LOQ) of these compounds are listed in Table S1 in Supporting Information. Figure 2 presents the metabolite concentrations detected at increasing time of incubation with the pooled fecal samples. Concentrations of the metabolites formed were calculated, including a correction for the amounts detected in corresponding negative controls incubated without adding EC. Six polyphenols were detected in the negative controls, albeit in limited amounts (generally <11.5% of the total mass recovery) (Figure S2 in Supporting Information). The results obtained show total molar mass recoveries that ranged between 82.9 and 112.9%, being somewhat lower at the later time points (Table 2). Comparison of time-dependent metabolite profiles reveals similar patterns with 3,4-diHPP-2-ol being the first (detectable) and most abundant metabolite formed within the first 2 h of incubation, representing more than 70% of the initial amount of EC. Upon prolonged incubation, its relative abundance dropped to 23.7 ± 10.4% of EC equivalents at 3 h, while this metabolite was no longer detected at and beyond 4 h incubation. On the contrary, 3,4-diHPV was detected in large amounts at later time points (3–6 h) and appeared to be the dominant metabolite in the metabolite patterns at and beyond 3 h of incubation. Its relative abundance appeared to be relatively constant from 3 to 6 h, ranging between 45.0 and 49.2% of the total metabolite pattern. In blank control incubations where EC was incubated without fecal slurries, no metabolite formation was observed and EC was stable with an average mass recovery at all incubation time points of 95.3%. (Figure S4 in Supporting Information).
Figure 2.
Time-dependent quantitative analysis of EC and its microbial metabolites formed during 24 h of anaerobic incubation with a pooled fecal sample from 14 donors. Results are shown as mean ± SD from three independent incubations.
Table 2. Molar Mass Recoveries for EC and Its Colonic Metabolites (%) during 24 Hours of Anaerobic Incubation with a Pooled Fecal Sample from 14 Donors.
| chemical | 0 (h) | 1 (h) | 2 (h) | 3 (h) | 4 (h) | 5 (h) | 6 (h) | 24 (h) |
|---|---|---|---|---|---|---|---|---|
| ECb | 88.54 ± 11.51 | 24.92 ± 5.21 | 5.86 ± 0.68 | 2.74 ± 0.86 | 0.04 ± 0.03 | 0.01 ± 0.01 | 0.03 ± 0.03 | n.d.d |
| 3,4-diHPP-2-olc | n.d. | 71.92 ± 5.61 | 70.13 ± 13.47 | 23.74 ± 10.42 | n.d. | n.d. | n.d. | n.d. |
| 3-HPP-2-olc | n.d. | 2.72 ± 0.27 | 10.86 ± 0.84 | 16.32 ± 3.89 | 2.22 ± 1.93 | n.d. | n.d. | n.d. |
| 4H-diHPVAc | n.d. | n.d. | 0.79 ± 0.10 | 2.64 ± 0.04 | 4.39 ± 0.42 | 6.20 ± 0.14 | 7.68 ± 0.39 | n.d. |
| 3,4-diHPVb | n.d. | 4.48 ± 1.08 | 21.78 ± 2.19 | 46.19 ± 6.37 | 49.19 ± 6.48 | 47.48 ± 6.37 | 45.01 ± 6.25 | 0.04 ± 0.08 |
| 3-HPVb | n.d. | n.d. | 0.75 ± 0.12 | 11.03 ± 2.15 | 24.68 ± 1.62 | 24.51 ± 1.80 | 23.76 ± 2.75 | 0.17 ± 0.14 |
| 3,4-diHPVAb | n.d. | n.d. | 0.14 ± 0.07 | 0.39 ± 0.10 | 0.68 ± 0.16 | 1.08 ± 0.29 | 1.69 ± 0.40 | n.d. |
| 3-HPVAb | n.d. | 0.02 ± 0.02 | 0.06 ± 0.06 | 0.09 ± 0.09 | 0.36 ± 0.18 | 0.79 ± 0.34 | 1.73 ± 0.71 | 86.14 ± 15.61 |
| 3-HPPAb | n.d. | 0.66 ± 0.22 | 2.34 ± 0.95 | 2.94 ± 0.75 | 3.14 ± 0.65 | 3.16 ± 0.40 | 2.95 ± 0.62 | n.d. |
| 3-HPAAb | n.d. | 0.03 ± 0.23 | 0.16 ± 0.18 | n.d. | n.d. | n.d. | n.d. | n.d. |
| total | 88.54 | 104.75 | 112.88 | 106.08 | 84.70 | 83.23 | 82.85 | 86.35 |
Results are shown as mean ± SD from three independent incubations.
Qualitatively and quantitatively detected by LC-TQ-MS.
Qualitatively and quantitatively detected by LC-TOF-MS.
Not detected.
Comparison of EC In Vitro Metabolite Profiles to the Reported In Vivo Plasma or Urinary EC Metabolite Data
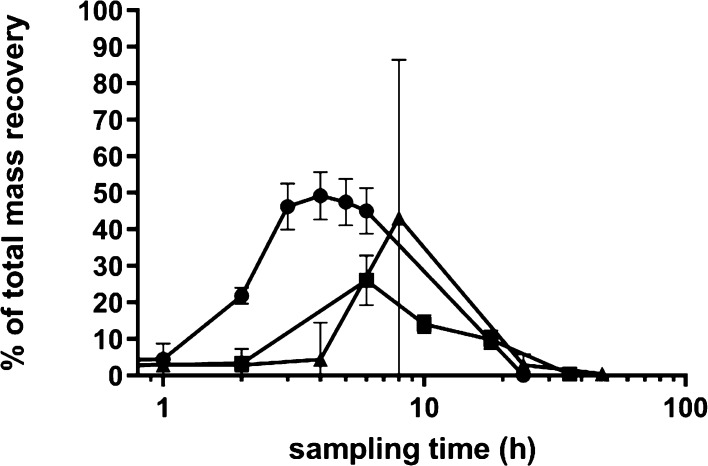
To enable the evaluation of the in vitro metabolite profiles obtained, the human intestinal microbial metabolite patterns obtained in the anaerobic fecal incubations in the present study were compared to the available literature data on EC metabolite patterns in human plasma and urine upon oral dosing.8,11 Wiese et al. (2015) reported that after the ingestion of EC by six volunteers, four major microbial metabolites could be identified in the pharmacological kinetic course in human plasma samples including 4-HPAA, ferulic acid, 3,4-diHPV, and 4H-diHPVA.8 In the fecal incubations, three of these four metabolites were detected, that is, 3,4-diHPV, 4H-diHPVA, and 4-HPAA, with the latter being present in trace amounts only. The formation of ferulic acid takes place in the liver and kidney;9 hence, it was not targeted in the in vitro incubation. Ottaviani et al. (2016) reported time-dependent EC urinary metabolites from eight volunteers after the intake of the 14C-EC test drink. They reported that about 70% of the ingested EC was absorbed in the large intestine.11 In both studies, 3,4-diHPV conjugates were the most abundant EC metabolites. Figure 3 presents a comparison of the time-dependent relative abundance of 3,4-diHPV formed in the in vitro fecal incubation model and in the in vivo plasma and urinary metabolite patterns (summing up unconjugated and conjugated forms) after the ingestion of EC. Similar kinetics were observed. Moreover, the total excretion of 3,4-diHPV conjugates accounted for 53.5% of the total EC that passed into the large intestine, which is comparable with the large and stable amount (45.0–49.2% of the total metabolite pattern) produced in the in vitro fecal incubations.
Figure 3.
Comparison of EC microbial metabolite 3,4-diHPV from in vivo human urine samples (square, Ottaviani et al., 2016), human plasma samples (triangle, Wiese et al., 2015), and the in vitro data from the present study (circle).
Interindividual Differences of EC Bioconversion by Human Colonic Microbiota
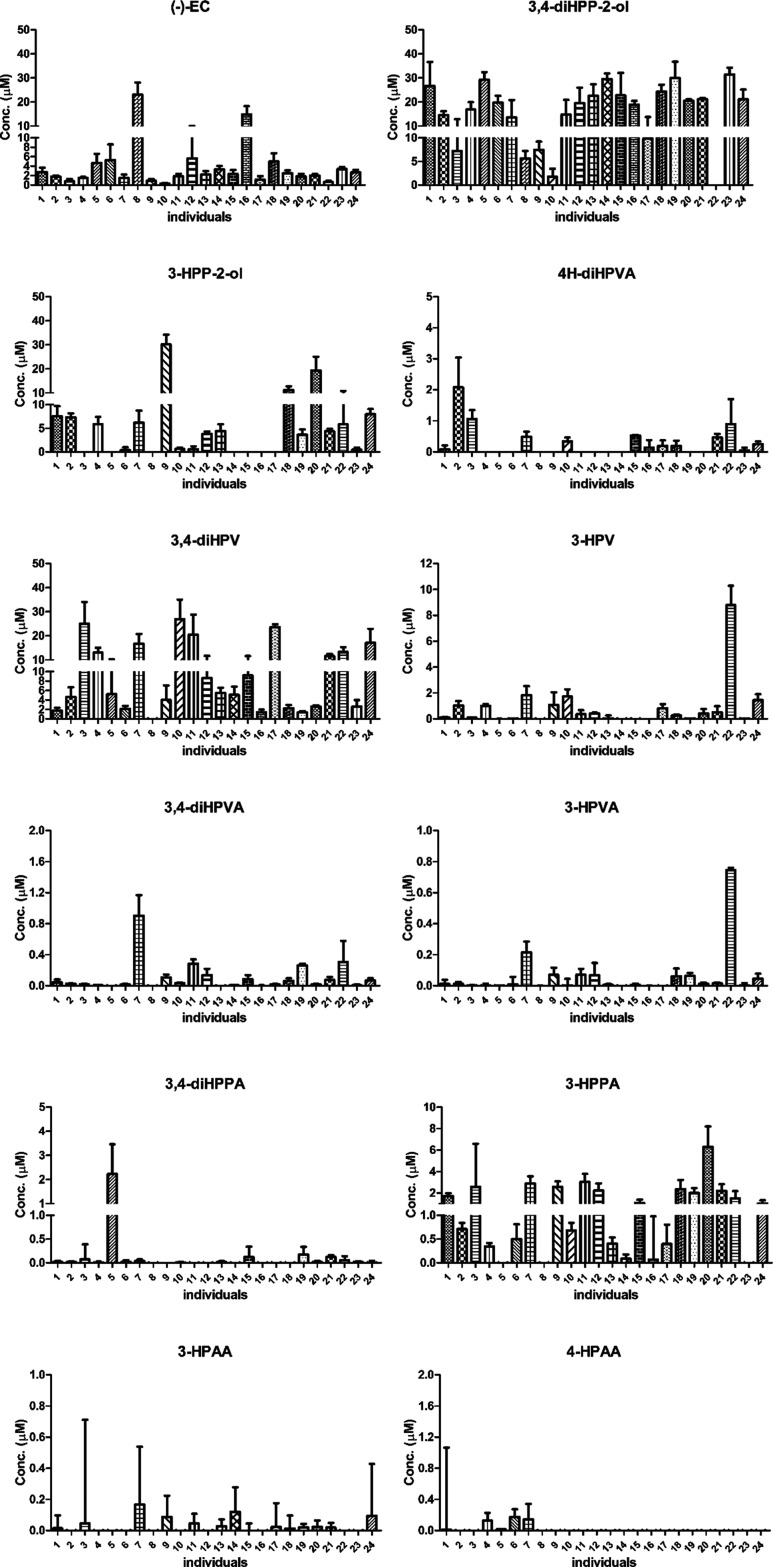
Based on the results obtained for the time-dependent in vitro microbial EC conversion in incubations with a pooled fecal sample, a period of 2 h was selected to quantify potential human interindividual differences in the EC intestinal microbial metabolism. This time point was selected, as the residual EC was still present, while also showing substantial metabolite formation. At 1 h, limited colonic metabolites were detected, while at 3 h, the residual EC was limited (less than 3% of the original amount). Furthermore, the number of colonic metabolites detected at t = 3 h is the same as at t = 2 h. Figure 4 provides an overview of the metabolite profiles detected after 2 h incubation of EC with fecal slurries from 24 individual human donors. Phenolic compounds quantified in different individuals in the negative controls are presented in Figure S3 in the Supporting Information. All phenolics were present in the negative controls at levels <12.8% of the total mass recovery with the exception of 3-HPPA (82.5% of the total mass recovery) in individual 3 and 3,4-diHPPA (22.5% of the total mass recovery) in individual 5. Data from Figure 4 clearly elucidate that the metabolite patterns obtained upon 2 h incubation with the fecal slurries from different individuals show marked differences in terms of both the types and amounts of metabolites formed as well as in the level of EC conversion. For instance, in the incubations with fecal slurry from individuals 8 and 16, 23.0 and 14.8 μM of EC (46.1 and 29.6% of the amount added) remained, respectively, while in incubations with fecal slurry from individuals 10 and 22, this amounted to less than 1 μM (<1.3%) of EC (0.3 and 0.6 μM, respectively). Meanwhile, substantial differences were also observed for the major metabolite 3,4-diHPV. For instance, 26.9 μM (53.8% of original EC equivalents) of 3,4-diHPV was formed in the incubations with fecal slurry from individual 10, while only 1.8 μM (3.5% of original EC equivalents) was detected in incubations with fecal slurry from individual 1 and none for individual 8. On the basis of the data obtained, it can be concluded that the amount of the residual EC after the first 2 h of incubation differed up to 76.7-fold (calculated by using the highest EC residual from individual 8, i.e., 23.0 μM, divided by the lowest EC residual from individual 10, i.e., 0.3 μM) between 24 individuals (Figure 4). It appears that individuals 8 and 16 can be classified as slow EC metabolizers (<70.4% EC conversion within the first 2 h), and the other individuals were defined as fast EC metabolizers (>88.7% EC conversion within the first 2 h). Setting 3,4-diHPV as the biomarker of EC colonic microbial bioconversion, individuals 3, 10, and 17 can be classified as fast 3,4-diHPV producers (>47.3% of original EC equivalents of 3,4-diHPV formed within the first 2 h). Individual 22 was detected to form the highest level of 3-HPV (17.5% of original EC equivalents) among others, which is likely because of the fast dehydroxylation of 3,4-diHPV. The other individuals showed lower formation of 3,4-diHPV either because of a relatively low EC clearance rate (individuals 8 and 16) or because of the accumulation of the precursor 3,4-diHPP-2-ol.
Figure 4.
EC metabolite patterns formed in incubations with fecal slurries from 24 individuals at 2 h. Molar mass recoveries for EC and its colonic metabolites of each individual ranged from 53.3 to 103.1% (Figure S7 in Supporting Information).
Besides, differences in the rate of metabolic conversion and also the types of metabolites observed in the metabolite profiles formed by different individuals varied from one another. For example, besides EC, only 3,4-diHPP-2-ol was detected in incubations with fecal slurry from individual 8, while 11 different metabolites were observed in incubations with fecal slurry from individuals 1 and 7. Additionally, for individual 5, only dihydroxyphenols were detected in the metabolic profile, which may indicate that this individual is in favor of producing dihydroxyphenolics rather than monohydroxyphenolics. Moreover, unlike most individuals, individual 9 tended to produce 3-HPP-2-ol (60.2% of original EC equivalents) rather than 3,4-diHPP-2-ol (14.9% of original EC equivalents).
Class Signatures of Metabolic Profiles and Hierarchic Clustering of 24 Individuals
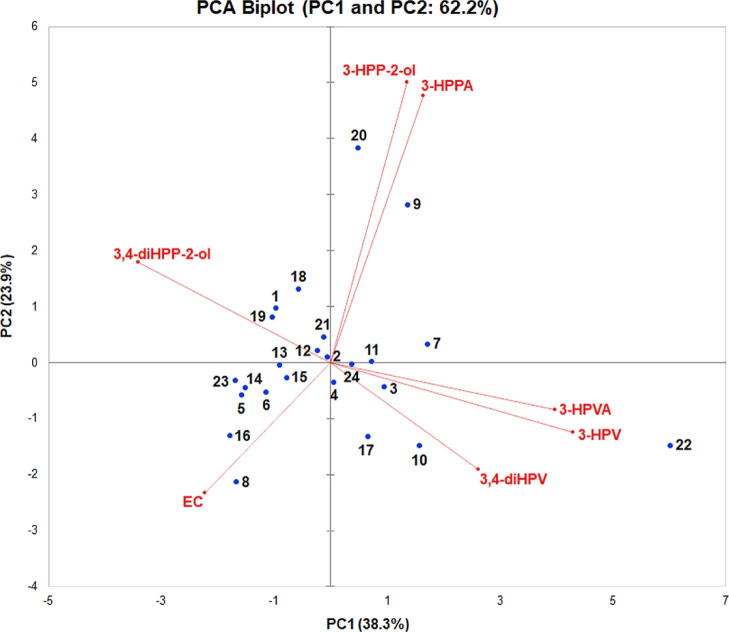
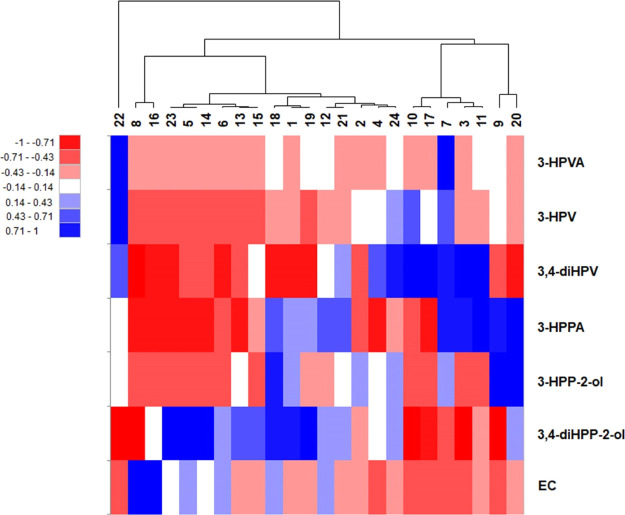
To further evaluate the interindividual differences in the intestinal microbial metabolic patterns and the possible existence of metabotypes among 24 individuals, a PCA analysis was performed. The quantified amounts of the residual EC and all the 11 metabolites detected at t = 2 h (Figure 4) were used as the data input for XLSTAT. The PCA results thus obtained revealed that only 43.8% of all variance in the metabolite patterns could be explained by two PCs (i.e., PC1 and PC2; Figure S5 in Supporting Information). For further evaluation, the quantified amounts of the residual EC and only its six major metabolites detected at t = 2 h were used to define the multivariate data set for the PCA analysis. These six major metabolites included 3,4-diHPP-2-ol, 3-HPP-2-ol, 3,4-diHPV, 3-HPV, 3-HPVA, and 3-HPPA. The PCA analysis thus obtained (Figure 5) could explain 62.2% of the variation in the metabolite profiles using only two PCs (PC1 and PC2). PC1 accounts for the largest possible variance in the data set (38.3%), where PC2 captures 23.9% of the remaining variation. In order to visualize the hierarchic clustering of different individuals based on their metabolic profiles, a heatmap was produced (Figure 6). From the heatmap, it appears that individuals 18, 1, and 19 cluster together mainly because of the similar amounts of the residual EC and 3,4-diHPP-2-ol in their metabolite patterns at 2 h incubation, while the individuals 23, 5, and 14 cluster together because of similar amounts in these two compounds as well. In the PCA biplot, these individuals also locate close to one another driven by these similar metabolite levels. Individuals 9 and 20 cluster together and are clearly separated from other individuals both in the PCA plot as well as in the heatmap because of substantially higher amounts of 3-HPP-2-ol and 3-HPPA formed. Individual 22 was fully separated from other individuals both in the PCA plot and the heatmap mainly because of a high level of 3-HPV. Furthermore, individuals 8 and 16 cluster together in the heatmap because of the high residual amount of EC in line with their earlier identification as slow metabolizers. Individuals 10 and 17 cluster together and are also clearly separated from other individuals in the heatmap and PCA plot because of the substantially higher level of 3,4-diHPV in the metabolite profile at 2 h incubation time.
Figure 5.
PCA based on the concentration of the residual EC and the six major EC microbial metabolites present in incubations with fecal slurries from 24 individuals at 2 h.
Figure 6.
Hierarchic clustering illustrated by a heatmap composed based on the concentration of the residual EC and the six major EC microbial metabolites present in incubations with fecal slurries from 24 individuals at 2 h.
Quantitative Microbial Profile and Correlation with Metabolite Formation
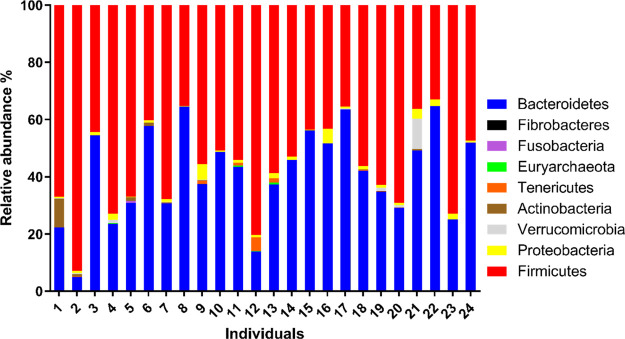
In Figure 7, the relative microbial profile at the phylum level of the 24 individual fecal samples is reported. The bacteria fall into nine different phyla, in particular, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, Tenericutes, Euryarchaeota, Fusobacterial, and Fibrobacteres. Interindividual differences in the microbial relative abundance were observed. However, bacteria belonging to Firmicutes and Bacteroidetes were consistently found to be the dominant phyla in all samples. Detailed information of the relative/absolute taxon abundances and diversity at the phylum/genus level is included in the Supporting Information File1.
Figure 7.
Relative microbial profile at the phylum level of the 24 human fecal samples.
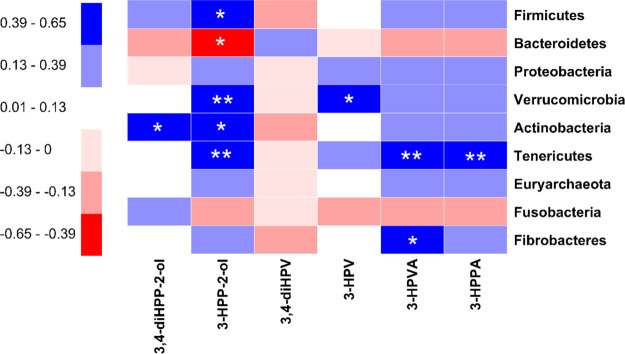
In order to reveal the interplay between the microbial taxonomic profile of the fecal samples of the 24 individuals and their intestinal microbial metabolite profiles, a Spearman correlation study was performed between the relative abundance of the bacterial phyla and the concentrations of the most relevant metabolites present after 2 h of incubation in the fecal slurry incubations (correlations at the bacterial genus level are presented in Figure S6 in Supporting Information). Figure 8 illustrates the Spearman correlation coefficients by a heatmap. The statistical correlation is significant (p < 0.05) with ρ ≤ −0.39 or ρ ≥ 0.39. The results thus obtained reveal several significant correlations for 3-HPP-2-ol, an early metabolite in the metabolic pattern of EC (Figure 1), showing a negative correlation with the amount of Bacteroidetes and positive correlations with the amount of Firmicutes, Verrucomicrobia, Actinobacteria, and Tenericutes. A significant positive correlation was also found for 3,4-diHPP-2-ol with the amount of Actinobacteria.
Figure 8.
Spearman correlations between the taxon abundances of nine bacterial phyla and the concentration of six major EC microbial metabolites present in incubations with fecal slurries from 24 individuals at 2 h. The sign and strength of the Spearman correlation coefficients are represented as colors (blue: positive; red: negative). The significances of the correlations are indicated by asterisks: *P < 0.05, **P < 0.01.
Discussion
In the present study, it was shown that in vitro anaerobic fecal incubations can be used to characterize in vivo colonic metabolism and related human plasma metabolites of the model catechin EC. During pooled human fecal incubations, nine colonic metabolites of EC could be detected and quantified. Most of these intermediates are also reported in previous studies using human or animal in vitro fecal incubation models21,23−25 or in plasma, urine, and feces in human or animal interventions.8,26,27 The relative abundance of these microbial metabolites varied between different studies. For instance, 3-HPPA was reported to be the predominant metabolite in two in vitro human fecal incubation studies,28,29 while it was detected in our in vitro model to only a limited extent. These different results could result from different materials used (e.g., different fecal sources) or different experimental conditions (e.g., different extraction methods) applied. The results obtained in our study showed that 3,4-diHPV and its precursor, 3,4-diHPP-2-ol, were the two most dominant metabolites formed in the fecal incubations with pooled human fecal samples. The formation of 3,4-diHPV as a major EC colonic metabolite is in line with previous studies.30−32 The latter intermediate is often suggested to be a transient catabolite,23,25,33 which was also confirmed in this study. Its formation is the result of the first step in the EC colonic microbial metabolism, formed by the cleavage of the C ring of EC. This intermediate would be rapidly metabolized into 3,4-diHPV, which seemed to be more stable because phenyl-γ-valerolactone was present at a high abundance at 3 h and beyond (around 45–50% of the total mass balance). In the present study, we investigated the use of an in vitro anaerobic fecal incubation model to study the colonic metabolism of EC. This model does not account for the uptake of EC in the small intestine and its subsequent conjugation. However, under in vivo conditions, the percentage of EC ending up in the large intestine is known to be substantial (∼70%), although it may vary with the individual.20 This importance of colonic metabolism is corroborated by the fact that the absorption of EC in the small intestine was reported to be only up to ∼20%.11 The importance also follows from the fact that among all colonic metabolites, 3,4-diHPV-derived metabolites were reported to account for 42–60% of the EC intake,11,34 which is in agreement with the results derived from the in vitro incubations in the present study. This emphasizes the importance of the intestinal microbiome in improving the bioavailability of catechins or rather of their intestinal metabolites.
Furthermore, the results of the present study also revealed that 3-HPVA was the terminal metabolite upon 24 h anaerobic incubations when EC was completely degraded. This metabolite may result from the further conversion of 3-HPV, which is believed to be formed by reverse Claisen-driven degradation of the A-ring of EC.35 3-HPVA accounted for 86.1% of the total molar mass at 24 h [and even at 48 h (data not shown)], whereas at these time points, the residual amounts of other phenolic metabolites were negligible, pointing at their further degradation by the colonic bacteria of these other phenolic metabolites, while the degradation of 3-HPVA is apparently limited. This result is in line with what was reported by Stoupi et al. (2010), demonstrating that 3-HPVA, 3-HPPA, and phenyl acetic acid were dominant metabolites after 24 h or longer incubations with human fecal samples.28 However, this 3-HPVA metabolite may have very limited efficacy in the host because the phenylcarboxylic acids are hydrophilic with poor solubility within biological membranes, which restrict their bioavailability resulting in limited bioefficacies.36 This is important when studying the physiological consequences of the microbial metabolism of EC.
The in vitro time-dependent formation of the major metabolite 3,4-diHPV was compared with in vivo plasma and urinary data. The result indicates that the in vitro metabolite patterns matched the in vivo patterns quite well and that the fecal incubations may provide an adequate mean to obtain insights into the intestinal microbial conversions. This conclusion is in line with that reported by Lagkouvardos et al. (2017), who performed a thorough review on the cultivation of bacteria from the intestine of mammals, concluding that up to 65% of molecular species detected by sequencing have corresponding strains in culture.37 It is also in line with a study that compared microbial-related in vivo metabolic changes in the gut tissue, cecum content, and feces of rats treated with antibiotics concluding that “as a non-invasive sampling method, feces provide a suitable matrix for studies on metabolism by the gut microbiota”.38 Thus, using fecal incubations to describe intestinal microbial metabolism appears to be a useful approach. In addition, it is noted that an in vivo study with the same donors as the ones used for donation of the fecal samples remains of interest for future studies to enable further evaluation of this in vitro incubation model. Also, comparison with more than the currently available in vivo studies and/or similar studies with other substrates for colonic metabolism would be of use.
In the next step, the in vitro fecal incubation method was used to study interindividual differences in EC microbial metabolism. Substantial interindividual differences were observed in EC microbial metabolism both in terms of type and level of metabolites formed among 24 volunteers. To obtain insights into the potential underlying explanation, microbial taxonomy and relative abundance of the 24 fecal samples were characterized by 16S rRNA analysis. The results obtained indicate that in line with the incubations with the pooled human fecal sample, 3,4-diHPP-2-ol and 3,4-diHPV were the two dominant colonic metabolites of EC. Interindividual differences in the rate of clearance of EC were observed, pointing at the existence of slow metabolizers, of which 2 were identified among 24 subjects. The difference in the rate of metabolism was also found previously for the conversion of 3-(4′-hydroxyphenyl)propionic acid to 3-phenylpropionic acid, further indicating the differences in the ability of the intestinal microbiota of individuals to metabolize phenolics.29 Furthermore, differences in subsequent metabolic conversions resulted in different EC metabolite patterns among the individuals, with 2 out of 24 volunteers showing relatively high intermediate levels of accumulation of 3-HPP-2-ol and 3 individuals showing relatively high levels of 3,4-diHPV, pointing at potentially slow metabolizers for phenyl-γ-valerolactone formation or degradation. The high interindividual variations in metabolism (or formation) of EC and phenyl-γ-valerolactones were in agreement with the reported in vivo human intervention studies.32 For instance, Hollands and colleagues reported a 21-fold and 60-fold interindividual difference in urinary excretion of EC and phenyl-γ-valerolactones, respectively, after 28 days of daily ingestion of high dose EC mixed with oligomeric procyanidins capsules.39 Anesi and colleagues found a 7.5-fold variation in human urinary excretion of conjugated phenyl-γ-valerolactones after the consumption of apples.2 These in vivo studies already pointed at interindividual differences in EC metabolism based on two of its major urinary metabolites. The results of the present study reveal that interindividual differences in the microflora-derived production of EC metabolites may result in differences in colonic metabolites formed beyond differences in the formation of phenyl-γ-valerolactones. It is worth noting that some individuals might be able to form metabolites, which were not included in the set of metabolites included in the present MS analytical methods, reflected by relatively lower mass recoveries, for example, for individuals 8 and 6, a molar mass recovery of less than 60% was obtained (Figure S7 in Supporting Information). We could hypothesize here that different individuals may have different metabolic pathways or different pathways could coexist at the same time, but the host may have its priorities from one another. However, for most individuals, the molecular mass balance of the metabolite patterns amounted to >75%, indicating that 11 metabolites now identified represent the majority of the patterns obtained.
There are a number of factors that can contribute to the interindividual metabotype variations observed. Of all the factors, the host microbial profile is most obvious.3,40,41 There are 1011 to 1012 bacteria/g in the colon content, the highest amount of all parts of the gastrointestinal tract of the host.42 The individual microbiota composition patterns can be different because of different ages, genders, ethnics, dietary and living habits, and so forth. Mena and colleagues were the first to describe three urinary metabotypes after daily ingestion of green tea extract tablets for 8 weeks.32,40 The three putative metabotypes were characterized by different amounts of tri- and dihydroxyphenyl-γ-valerolactones and 3-(hydroxyphenyl)propionic acid produced among 11 volunteers. Given that in this study green tea, and not EC, was dosed, the differences may in part also have been caused by differences in the colonic metabolism of other green tea or dietary constituents because in addition to EC, catechins, gallocatechins, procyanidins, and prodelphinidins have also been reported to be metabolized to phenyl-γ-valerolactones. Because the gut microbial compositions of these individuals were not studied, a link between the metabotypes and host microbial abundance was not established. In this sense, the microbial taxonomic abundance of 24 fecal samples was analyzed in the present study; correlations between microbial taxonomic abundance and metabolic abundance could be defined. The bacterial load results highlighted indeed rather a small variation, where the loads of slow metabolizers (individuals 8 and 16) were not the lowest two among all individuals (Figure S8 in Supporting Information). The gene copy numbers per gram of fecal slurry were comparable to the ones of healthy human individuals reported in literature.43 However, differences were observed both in the relative abundances and in the composition of the microbiota. To the best of our knowledge, Eubacterium SDG-2, Lactobacillus plantarum, Eggerthella lenta, and Adlercreutzia equolifaciens are the only bacteria reported so far to be capable of producing the metabolite diphenylpropan-2-ol from epicatechins (or catechins), a reaction comparable to the formation of 3,4-diHPP-2-ol from EC detected in the present study. The first two belong to the phylum Firmicutes and the latter two to the phylum Actinobacteria.44−46 Accordingly, in the present study, a statistically significant positive correlation was found between the abundance of Actinobacteria and the quantity of 3,4-diHPP-2-ol. In addition, we found statistically significant positive correlations between the relative abundance of the Lachnospiraceae NC2004 group and the formation of 3,4-diHPP-2-ol, suggesting this bacteria to be capable of cleaving the C-ring of EC, which was not reported before (Figure S6 in Supporting Information). The levels of the dehydroxylated metabolite 3-HPP-2-ol were positively correlated with Firmicutes, Verrucomicrobia, Actinobacteria, and Tenericutes, suggesting that the dehydroxylation reaction of phenolics is not a strain-specific microbial conversion. In the next metabolic step, diphenylpropan-2-ol is transformed into 3,4-diHPV and 4H-diHPVA, reported to be mediated by Flavonifractor plautii, which belongs to the family of Ruminococcace, phylum Firmicutes.44 In the present study, we found that several bacteria from this family, Ruminococcace, were positively correlated with the formation of 3-HPP-2-ol, phenyl-γ-valerolactones, and phenylvaleric acids (Figure S6 in Supporting Information), indicating that these bacteria could catalyze the conversion of diphenylpropan-2-ol to downstream metabolites and have the ability to perform dehydroxylation reaction. Besides, Phascolarctobacterium was newly found to be significantly positively correlated with the concentration of 3,4-diHPV (Figure S6 in Supporting Information). The relative abundance of Prevotella 7 and Bacteroides was positively and negatively associated with the production of 4H-diHPVA, phenyl-γ-valerolactones, phenylvaleric acids, and 3-HPPA, which is in line with the conclusion derived from the human intervention study carried out by Trost and colleagues.3 Additionally, several bacteria from other genera (e.g., Barnesiella and Akkermansia) also showed positive correlations with the formation of phenyl-γ-valerolactones, phenylvaleric acids, and 3-HPPA; however, their relative abundances were generally lower than 1% in most individuals. Altogether, the correlations obtained support that in anaerobic fecal incubations provide a novel way to investigate the potential link between bacterial abundance and colonic metabolite patterns. Nevertheless, more work on screening and isolation of catechin-catalyzing bacteria is needed in order to explain the observed different metabotypes after consumption of dietary flavan-3-ols.47,48 Moreover, intestinal bacteria from different species also have different metabolic abilities in catechin biotransformation, which cannot be simply extrapolated from one species to another, further pointing at the relevance of developing and evaluating fecal incubations as an adequate model to obtain human data.26
It is interesting to note that the two major intermediates detected in our study, 3,4-diHPP-2-ol and 3,4-diHPV, both have a catechol moiety that possesses the potential to oxidize them to quinones or semiquinones.49 According to previous studies, catechins (or other flavonoids) were reported to activate the Keap1-Nrf2 pathway by means of their (semi)quinone formation.50,51 The (semi)quinone-type metabolites were considered to play a role in the induction of detoxifying enzymes via activation of the Keap1-Nrf2 pathway.51 Based on these considerations, we hypothesized that EC colonic metabolites (especially the catechol-type phenolic EC metabolites), such as 3,4-diHPP-2-ol and 3,4-diHPV, would be able to trigger the activation of the Keap1-Nrf2 pathway in vivo after EC-rich foods consumption and may thus contribute to the observed health-promoting effects, as shown for other catechol-containing phenolics.52 The role of these EC colonic metabolites in inducing the beneficial effects of EC may also be relevant because these phenolic/catechol intestinal metabolites appear to be better bioavailable than the parent EC.53 Although EC and its phenolic metabolites also have antioxidative properties, it should be noted that the capacity to activate the Keap1-Nrf2 pathway may prove more important for their beneficial health effects than their antioxidant activity because, given their low micromolar systemic concentrations,53−55 EC and its metabolites may not be able to effectively compete with the antioxidant activity of tocopherols and/or ascorbate present in substantially higher concentrations in vivo. For instance, the in vivo concentrations of EC and its colonic metabolites were reported with a Cmax of ∼1.2 μM in plasma, while α-tocopherol and ascorbic acid were reported to be around 24 μM and 70 μM, respectively, in the serum or plasma in Western populations.11,56
In conclusion, the microbial metabolism of EC is crucial in producing potential bioactive phenolics. Among all the colonic metabolites, 3,4-diHPV seems to be the most important metabolite, considering its abundance, stability, and catechol structural moiety. Striking interindividual differences were found in the formation of EC intestinal microbial metabolites, including 3,4-diHPV, both in terms of type and level, which is likely because of substantial variations of host microbiota composition. Altogether, our results suggest that substantial interindividual differences in the gut microbial metabolism of EC may contribute to interindividual differences in the potential beneficial effects of EC-rich foods.
Glossary
Abbreviations Used
- LC-TQ-MS
liquid chromatograph triple quadrupole mass spectrometry
- LC-TOF-MS
liquid chromatograph time-of-flight mass spectrometry
- EC
(−)-epicatechin
- ECG
(−)-epicatechin-3-O-gallate
- EGC
(−)-epigallocatechin
- EGCG
(−)-epigallocatechin-3-O-gallate
- 3,4-diHPP-2-ol
1-(3′,4′-dihydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-2-propanol
- 3-HPP-2-ol
1-(3′-hydroxyphenyl)-3-(2″,4″,6″-trihydroxyphenyl)-2-propanol
- 4H-diHPVA
4-hydroxy-5-(3′,4′-dihydroxyphenyl)-valeric acid
- 4H-HPVA
4-hydroxy-5-(3′-hydroxyphenyl)-valeric acid
- 3,4-diHPV
5-(3′,4′-dihydroxyphenyl)-γ-valerolactone
- 3-HPV
5-(3′-hydroxyphenyl)-γ-valerolactone
- 3,4-diHPVA
5-(3′,4′-dihydroxyphenyl)valeric acid
- 3-HPVA
5-(3′-hydroxyphenyl)valeric acid
- PVA
5-phenylpentanoic acid
- 3,4-diHPPA
3-(3′,4′-dihydroxyphenyl)propionic acid
- 3-HPPA
3-(3′-hydroxyphenyl)propionic acid
- PPA
3-phenylpropionic acid
- 3,4-diHPAA
3′,4′-dihydroxyphenylacetic acid
- 3-HPAA
3′-hydroxyphenylacetic acid
- 4-HPAA
4′-hydroxyphenylacetic acid
- PAA
phenylacetic acid
- 3,4-diHPBA
3′,4′-dihydroxybenzoic acid
- 4-HPBA
4′-hydroxybenzoic acid
- BA
hydroxybenzoic acid
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.0c05890.
LOD and LOQ of EC and its major metabolites; fresh and frozen fecal incubations; negative controls of pooled fecal incubations; negative controls of individual fecal incubations; EC contents of blank control; PCA of EC and its 11 metabolites; Spearman correlations between the taxon abundances of bacterial genera and the concentration of residual EC and eight EC microbial metabolites; total mass recoveries of 24 individuals; bacterial load of 24 individuals; and absolute and relative bacteria in phylum and genus levels (PDF)
Author Present Address
§ Wageningen Food Safety Research, PO Box 230, 6700 AE Wageningen, The Netherlands.
Chen Liu is grateful for the financial support of the China Scholarship Council (CSC). Grant number: 201803250053.
The authors declare no competing financial interest.
Supplementary Material
References
- Rodriguez-Mateos A.; Cifuentes-Gomez T.; Gonzalez-Salvador I.; Ottaviani J. I.; Schroeter H.; Kelm M.; Heiss C.; Spencer J. P. E. Influence of age on the absorption, metabolism, and excretion of cocoa flavanols in healthy subjects. Mol. Nutr. Food Res. 2015, 59, 1504–1512. 10.1002/mnfr.201500091. [DOI] [PubMed] [Google Scholar]
- Anesi A.; Mena P.; Bub A.; Ulaszewska M.; Del Rio D.; Kulling S. E.; Mattivi F. Quantification of urinary phenyl-γ-valerolactones and related valeric acids in human urine on consumption of apples. Metabolites 2019, 9, 254. 10.3390/metabo9110254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trošt K.; Ulaszewska M. M.; Stanstrup J.; Albanese D.; De Filippo C.; Tuohy K. M.; Natella F.; Scaccini C.; Mattivi F. Host: Microbiome co-metabolic processing of dietary polyphenols–An acute, single blinded, cross-over study with different doses of apple polyphenols in healthy subjects. Food Res. Int. 2018, 112, 108–128. 10.1016/j.foodres.2018.06.016. [DOI] [PubMed] [Google Scholar]
- Ruijters E. J. B.; Weseler A. R.; Kicken C.; Haenen G. R. M. M.; Bast A. The flavanol (-)-epicatechin and its metabolites protect against oxidative stress in primary endothelial cells via a direct antioxidant effect. Eur. J. Pharmacol. 2013, 715, 147–153. 10.1016/j.ejphar.2013.05.029. [DOI] [PubMed] [Google Scholar]
- Morrison M.; van der Heijden R.; Heeringa P.; Kaijzel E.; Verschuren L.; Blomhoff R.; Kooistra T.; Kleemann R. Epicatechin attenuates atherosclerosis and exerts anti-inflammatory effects on diet-induced human-CRP and NFκB in vivo. Atherosclerosis 2014, 233, 149–156. 10.1016/j.atherosclerosis.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Claude S.; Boby C.; Rodriguez-Mateos A.; Spencer J. P. E.; Gérard N.; Morand C.; Milenkovic D. Flavanol metabolites reduce monocyte adhesion to endothelial cells through modulation of expression of genes via p38-MAPK and p65-Nf-kB pathways. Mol. Nutr. Food Res. 2014, 58, 1016–1027. 10.1002/mnfr.201300658. [DOI] [PubMed] [Google Scholar]
- Saha A.; Kuzuhara T.; Echigo N.; Suganuma M.; Fujiki H. New role of (−)-epicatechin in enhancing the induction of growth inhibition and apoptosis in human lung cancer cells by curcumin. Canc. Prev. Res. 2010, 3, 953–962. 10.1158/1940-6207.capr-09-0247. [DOI] [PubMed] [Google Scholar]
- Wiese S.; Esatbeyoglu T.; Winterhalter P.; Kruse H.-P.; Winkler S.; Bub A.; Kulling S. E. Comparative biokinetics and metabolism of pure monomeric, dimeric, and polymeric flavan-3-ols: A randomized cross-over study in humans. Mol. Nutr. Food Res. 2015, 59, 610–621. 10.1002/mnfr.201400422. [DOI] [PubMed] [Google Scholar]
- Monagas M.; Urpi-Sarda M.; Sánchez-Patán F.; Llorach R.; Garrido I.; Gómez-Cordovés C.; Andres-Lacueva C.; Bartolomé B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- Fraga C. G.; Oteiza P. I.; Galleano M. Plant bioactives and redox signaling:(−)-Epicatechin as a paradigm. Mol. Aspect. Med. 2018, 61, 31–40. 10.1016/j.mam.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Ottaviani J. I.; Borges G.; Momma T. Y.; Spencer J. P.; Keen C. L.; Crozier A.; Schroeter H. The metabolome of [2-14 C](−)-epicatechin in humans: Implications for the assessment of efficacy, safety, and mechanisms of action of polyphenolic bioactives. Sci. Rep. 2016, 6, 29034. 10.1038/srep29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmach A.; Troufflard S.; Serafini M.; Crozier A. Absorption, metabolism and excretion of Choladi green tea flavan-3-ols by humans. Mol. Nutr. Food Res. 2009, 53, S44–S53. 10.1002/mnfr.200800169. [DOI] [PubMed] [Google Scholar]
- Del Rio D.; Calani L.; Cordero C.; Salvatore S.; Pellegrini N.; Brighenti F. Bioavailability and catabolism of green tea flavan-3-ols in humans. Nutrition 2010, 26, 1110–1116. 10.1016/j.nut.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Guarner F.; Malagelada J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. 10.1016/s0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- Eckburg P. B.; Bik E. M.; Bernstein C. N.; Purdom E.; Dethlefsen L.; Sargent M.; Gill S. R.; Nelson K. E.; Relman D. A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel N.; Rekdal V. M.; Balskus E. P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdal T.; Sela D. A.; Xiao J.; Boyacioglu D.; Chen F.; Capanoglu E. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016, 8, 78. 10.3390/nu8020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson G.; Clifford M. N. Role of the small intestine, colon and microbiota in determining the metabolic fate of polyphenols. Biochem. Pharmacol. 2017, 139, 24–39. 10.1016/j.bcp.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Puupponen-Pimiä R.; Nohynek L.; Meier C.; Kähkönen M.; Heinonen M.; Hopia A.; Oksman-Caldentey K.-M. Antimicrobial properties of phenolic compounds from berries. J. Appl. Microbiol. 2001, 90, 494–507. 10.1046/j.1365-2672.2001.01271.x. [DOI] [PubMed] [Google Scholar]
- Borges G.; Ottaviani J. I.; van der Hooft J. J. J.; Schroeter H.; Crozier A. Absorption, metabolism, distribution and excretion of (−)-epicatechin: A review of recent findings. Mol. Aspect. Med. 2018, 61, 18–30. 10.1016/j.mam.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Roowi S.; Stalmach A.; Mullen W.; Lean M. E. J.; Edwards C. A.; Crozier A. Green tea flavan-3-ols: colonic degradation and urinary excretion of catabolites by humans. J. Agric. Food Chem. 2010, 58, 1296–1304. 10.1021/jf9032975. [DOI] [PubMed] [Google Scholar]
- Chen H.; Sang S. Biotransformation of tea polyphenols by gut microbiota. J. Funct. Foods 2014, 7, 26–42. 10.1016/j.jff.2014.01.013. [DOI] [Google Scholar]
- Takagaki A.; Nanjo F. Catabolism of (+)-catechin and (−)-epicatechin by rat intestinal microbiota. J. Agric. Food Chem. 2013, 61, 4927–4935. 10.1021/jf304431v. [DOI] [PubMed] [Google Scholar]
- Schantz M.; Erk T.; Richling E. Metabolism of green tea catechins by the human small intestine. Biotechnol. J. 2010, 5, 1050–1059. 10.1002/biot.201000214. [DOI] [PubMed] [Google Scholar]
- Meselhy M. R.; Nakamura N.; Hattori M. Biotransformation of (-)-epicatechin 3-O-gallate by human intestinal bacteria. Chem. Pharm. Bull. 1997, 45, 888–893. 10.1248/cpb.45.888. [DOI] [PubMed] [Google Scholar]
- Meng X.; Sang S.; Zhu N.; Lu H.; Sheng S.; Lee M.-J.; Ho C.-T.; Yang C. S. Identification and characterization of methylated and ring-fission metabolites of tea catechins formed in humans, mice, and rats. Chem. Res. Toxicol. 2002, 15, 1042–1050. 10.1021/tx010184a. [DOI] [PubMed] [Google Scholar]
- Stalmach A.; Mullen W.; Steiling H.; Williamson G.; Lean M. E. J.; Crozier A. Absorption, metabolism, and excretion of green tea flavan-3-ols in humans with an ileostomy. Mol. Nutr. Food Res. 2010, 54, 323–334. 10.1002/mnfr.200900194. [DOI] [PubMed] [Google Scholar]
- Stoupi S.; Williamson G.; Drynan J. W.; Barron D.; Clifford M. N. A comparison of the in vitro biotransformation of (−)-epicatechin and procyanidin B2 by human faecal microbiota. Mol. Nutr. Food Res. 2010, 54, 747–759. 10.1002/mnfr.200900123. [DOI] [PubMed] [Google Scholar]
- Gross G.; Jacobs D. M.; Peters S.; Possemiers S.; van Duynhoven J.; Vaughan E. E.; Van de Wiele T. In vitro bioconversion of polyphenols from black tea and red wine/grape juice by human intestinal microbiota displays strong interindividual variability. J. Agric. Food Chem. 2010, 58, 10236–10246. 10.1021/jf101475m. [DOI] [PubMed] [Google Scholar]
- Appeldoorn M. M.; Vincken J.-P.; Aura A.-M.; Hollman P. C. H.; Gruppen H. Procyanidin dimers are metabolized by human microbiota with 2-(3, 4-dihydroxyphenyl) acetic acid and 5-(3, 4-dihydroxyphenyl)-γ-valerolactone as the major metabolites. J. Agric. Food Chem. 2009, 57, 1084–1092. 10.1021/jf803059z. [DOI] [PubMed] [Google Scholar]
- Das N. P. Studies on flavonoid metabolism. Biochem. Pharmacol. 1971, 20, 3435–3445. 10.1016/0006-2952(71)90449-7. [DOI] [PubMed] [Google Scholar]
- Mena P.; Bresciani L.; Brindani N.; Ludwig I. A.; Pereira-Caro G.; Angelino D.; Llorach R.; Calani L.; Brighenti F.; Clifford M. N.; Gill C. I. R.; Crozier A.; Curti C.; Del Rio D. Phenyl-γ-valerolactones and phenylvaleric acids, the main colonic metabolites of flavan-3-ols: Synthesis, analysis, bioavailability, and bioactivity. Nat. Prod. Rep. 2019, 36, 714–752. 10.1039/c8np00062j. [DOI] [PubMed] [Google Scholar]
- Groenewoud G.; Hundt H. K. L. The microbial metabolism of (+)-catechin to two novel diarylpropan-2-ol metabolites in vitro. Xenobiotica 1984, 14, 711–717. 10.3109/00498258409151469. [DOI] [PubMed] [Google Scholar]
- Ottaviani J. I.; Fong R.; Kimball J.; Ensunsa J. L.; Britten A.; Lucarelli D.; Luben R.; Grace P. B.; Mawson D. H.; Tym A. Evaluation at scale of microbiome-derived metabolites as biomarker of flavan-3-ol intake in epidemiological studies. Sci. Rep. 2018, 8, 9859. 10.1038/s41598-018-28333-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J. F.; Maier C. S. The chemistry of gut microbial metabolism of polyphenols. Phytochem. Rev. 2016, 15, 425–444. 10.1007/s11101-016-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semalty A.; Semalty M.; Singh D.; Rawat M. S. M. Phyto-phospholipid complex of catechin in value added herbal drug delivery. J. Inclusion Phenom. Macrocyclic Chem. 2012, 73, 377–386. 10.1007/s10847-011-0074-8. [DOI] [Google Scholar]
- Lagkouvardos I.; Overmann J.; Clavel T. Cultured microbes represent a substantial fraction of the human and mouse gut microbiota. Gut Microb. 2017, 8, 493–503. 10.1080/19490976.2017.1320468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr C.; Sperber S.; Jiang X.; Strauss V.; Kamp H.; Walk T.; Herold M.; Beekmann K.; Rietjens I. M. C. M.; Van Ravenzwaay B. Microbiome-related metabolite changes in gut tissue, cecum content and feces of rats treated with antibiotics. Toxicol. Appl. Pharmacol. 2018, 355, 198–210. 10.1016/j.taap.2018.06.028. [DOI] [PubMed] [Google Scholar]
- Hollands W. J.; Philo M.; Perez-Moral N.; Needs P. W.; Savva G. M.; Kroon P. A. Monomeric Flavanols Are More Efficient Substrates for Gut Microbiota Conversion to Hydroxyphenyl-γ-Valerolactone Metabolites Than Oligomeric Procyanidins: A Randomized, Placebo-Controlled Human Intervention Trial. Mol. Nutr. Food Res. 2020, 64, 1901135. 10.1002/mnfr.201901135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena P.; Ludwig I. A.; Tomatis V. B.; Acharjee A.; Calani L.; Rosi A.; Brighenti F.; Ray S.; Griffin J. L.; Bluck L. J.; Del Rio D. Inter-individual variability in the production of flavan-3-ol colonic metabolites: Preliminary elucidation of urinary metabotypes. Eur. J. Nutr. 2019, 58, 1529–1543. 10.1007/s00394-018-1683-4. [DOI] [PubMed] [Google Scholar]
- Eker M. E.; Aaby K.; Budic-Leto I.; Rimac Brnčić S.; El S. N.; Karakaya S.; Simsek S.; Manach C.; Wiczkowski W.; de Pascual-Teresa S. A review of factors affecting anthocyanin bioavailability: Possible implications for the inter-individual variability. Foods 2020, 9, 2. 10.3390/foods9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S.; Grootaert C.; Vermeiren J.; Gross G.; Marzorati M.; Verstraete W.; de Wiele T. The Intestinal Environment in Health and Disease - Recent Insights on the Potential of Intestinal Bacteria to Influence Human Health. Curr. Pharm. Des. 2009, 15, 2051–2065. 10.2174/138161209788489159. [DOI] [PubMed] [Google Scholar]
- Brukner I.; Longtin Y.; Oughton M.; Forgetta V.; Dascal A. Assay for estimating total bacterial load: relative qPCR normalisation of bacterial load with associated clinical implications. Diagn. Microbiol. Infect. Dis. 2015, 83, 1–6. 10.1016/j.diagmicrobio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Kutschera M.; Engst W.; Blaut M.; Braune A. Isolation of catechin-converting human intestinal bacteria. J. Appl. Microbiol. 2011, 111, 165–175. 10.1111/j.1365-2672.2011.05025.x. [DOI] [PubMed] [Google Scholar]
- Takagaki A.; Nanjo F. Bioconversion of (−)-epicatechin,(+)-epicatechin,(−)-catechin, and (+)-catechin by (−)-epigallocatechin-metabolizing bacteria. Biol. Pharm. Bull. 2015, 38, 789–794. 10.1248/bpb.b14-00813. [DOI] [PubMed] [Google Scholar]
- Sánchez-Patán F.; Tabasco R.; Monagas M.; Requena T.; Peláez C.; Moreno-Arribas M. V.; Bartolomé B. Capability of Lactobacillus plantarum IFPL935 To Catabolize Flavan-3-ol Compounds and Complex Phenolic Extracts. J. Agric. Food Chem. 2012, 60, 7142–7151. 10.1021/jf3006867. [DOI] [PubMed] [Google Scholar]
- Cortés-Martín A.; Selma M. V.; Tomás-Barberán F. A.; González-Sarrías A.; Espín J. C. Where to look into the puzzle of polyphenols and health? The postbiotics and gut microbiota associated with human metabotypes. Mol. Nutr. Food Res. 2020, 64, 1900952. 10.1002/mnfr.201900952. [DOI] [PubMed] [Google Scholar]
- Espín J. C.; González-Sarrías A.; Tomás-Barberán F. A. The gut microbiota: A key factor in the therapeutic effects of (poly) phenols. Biochem. Pharmacol. 2017, 139, 82–93. 10.1016/j.bcp.2017.04.033. [DOI] [PubMed] [Google Scholar]
- Awad H. M.; Boersma M. G.; Boeren S.; van Bladeren P. J.; Rietjens I. M. C. M.; Rietjens I. M. Structure– activity study on the quinone/quinone methide chemistry of flavonoids. Chem. Res. Toxicol. 2001, 14, 398–408. 10.1021/tx000216e. [DOI] [PubMed] [Google Scholar]
- Lee-Hilz Y. Y.; Boerboom A.-M. J. F.; Westphal A. H.; van Berkel W. J. H.; Rietjens I. M. C. M.; Rietjens I. M. Pro-oxidant activity of flavonoids induces EpRE-mediated gene expression. Chem. Res. Toxicol. 2006, 19, 1499–1505. 10.1021/tx060157q. [DOI] [PubMed] [Google Scholar]
- Muzolf-Panek M.; Gliszczyńska-Świgło A.; de Haan L.; Aarts J. M. M. J. G.; Szymusiak H.; Vervoort J. M.; Tyrakowska B.; Rietjens I. M. C. M. Role of catechin quinones in the induction of EpRE-mediated gene expression. Chem. Res. Toxicol. 2008, 21, 2352–2360. 10.1021/tx8001498. [DOI] [PubMed] [Google Scholar]
- Bolton J. L.; Dunlap T. L.; Dietz B. M. Formation and biological targets of botanical o-quinones. Food Chem. Toxicol. 2018, 120, 700–707. 10.1016/j.fct.2018.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford M. N.; van der Hooft J. J.; Crozier A. Human studies on the absorption, distribution, metabolism, and excretion of tea polyphenols. Am. J. Clin. Nutr. 2013, 98, 1619S–1630S. 10.3945/ajcn.113.058958. [DOI] [PubMed] [Google Scholar]
- van Duynhoven J.; van der Hooft J. J. J.; van Dorsten F. A.; Foltz M.; Gomez-Roldan V.; Vervoort J.; de Vos R. C. H.; Jacobs D. M.; Jacobs D. M. Rapid and sustained systemic circulation of conjugated gut microbial catabolites after single-dose black tea extract consumption. J. Proteome Res. 2014, 13, 2668–2678. 10.1021/pr5001253. [DOI] [PubMed] [Google Scholar]
- Manach C.; Williamson G.; Morand C.; Scalbert A.; Rémésy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005, 81, 230S–242S. 10.1093/ajcn/81.1.230s. [DOI] [PubMed] [Google Scholar]
- Schwedhelm E.; Maas R.; Troost R.; Böger R. H. Clinical pharmacokinetics of antioxidants and their impact on systemic oxidative stress. Clin. Pharmacokinet. 2003, 42, 437–459. 10.2165/00003088-200342050-00003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.