SUMMARY
Paternal DNA demethylation in mammalian zygotes is achieved through Tet3-mediated iterative oxidation of 5-methylcytosine (5mC) coupled with replication-dependent dilution. Tet3-mediated paternal DNA demethylation is believed to play important roles in mouse development given that Tet3 heterozygous embryos derived from Tet3-deficient oocytes exhibit embryonic sublethality. Here we demonstrate that the sublethality phenotype of the Tet3 maternal KO mice is caused by haploinsufficiency, but not by defective paternal 5mC oxidation. We found that Tet3 heterozygous progenies derived from heterozygous father or mother also exhibit sublethality. Importantly, wild-type embryos reconstituted with paternal pronuclei that bypassed 5mC oxidation develop and grow to adulthood normally. Genome-scale DNA methylation analysis demonstrated that hypermethylation in maternal Tet3 KO embryos is largely diminished by the blastocyst stage. Our study thus reveals that Tet3-mediated paternal 5mC oxidation is dispensable for mouse development and suggests the existence of a compensatory mechanism for defective 5mC oxidation in preimplantation embryos.
INTRODUCTION
DNA methylation, the addition of a methyl group to the fifth position of cytosine (5-methylcytosine, 5mC), plays important roles in gene silencing and genome stability and is essential for mammalian development (Sasaki and Matsui, 2008; Smith and Meissner, 2013). DNA methylation is established by the de novo DNA methyltransferases, DNMT3A and DNMT3B, and is maintained by DNMT1. While the DNA methylation pattern is faithfully maintained throughout generations in somatic cells, it is globally erased during preimplantation development (Saitou et al., 2011; Sasaki and Matsui, 2008). After fertilization, both paternal and maternal genomes become hypomethylated and reach the lowest levels at the blastocyst stage, despite of the hypomethylated status being established differentially between the parental genomes. Maternal 5mC is mostly diluted in a DNA replication-dependent manner (Rougier et al., 1998) likely due to the limited availability of DNMT1 in the early embryos (Hirasawa et al., 2008). In contrast, the paternal genome is subjected to global active demethylation (Mayer et al., 2000; Oswald et al., 2000). We and others have found that Tet3 oxidizes paternal 5mC into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine, and 5-carboxylcytosine in mouse zygotes (Gu et al., 2011; Inoue et al., 2011; Inoue and Zhang, 2011; Wossidlo et al., 2011), and that the 5mC oxidation products are gradually lost during preimplantation development through DNA replication-dependent passive dilution (Inoue et al., 2011; Inoue and Zhang, 2011). Recent genome-scale analysis has also revealed that a large proportion of paternal 5mCs undergo passive dilution without oxidation (Guo et al., 2014a; Shen et al., 2014).
In contrast to the great progress in understanding the mechanism of paternal DNA demethylation, the biological significance of this process still remains poorly understood. The importance of this event for mammalian development has been suggested by previous studies demonstrating that paternal DNA demethylation is conserved in certain species including rabbit, bovine, and human (Guo et al., 2014b; Lepikhov et al., 2008; Reis Silva et al., 2012; Wossidlo et al., 2011). However, a similar phenomenon has not been observed in sheep, pig, and goat, questioning a general role for this event in mammalian development (Beaujean et al., 2004; Jeong et al., 2007; Park et al., 2011). In addition, a previous study has demonstrated that although paternal DNA demethylation was impaired in mouse zygotes derived from round spermatid injection, the development of these zygotes was normal (Polanski et al., 2008). Taken together, these studies have generated conflicting interpretations regarding the role of paternal DNA demethylation in mammalian development.
Nevertheless, it is generally believed that Tet3-mediated paternal DNA demethylation plays an important role in mouse development (Kohli and Zhang, 2013; Messerschmidt et al., 2014; Pastor et al., 2013; Seisenberger et al., 2013; Wu and Zhang, 2014). This notion is based on the observation that loss of maternal Tet3 protein prevents paternal 5mC oxidation in zygotes and leads to embryonic sublethality (Gu et al., 2011). In the study, heterozygous (Het) embryos derived from crosses of germ cell conditional knockout (CKO) females with wild-type (WT) males exhibited delayed expression of Oct4 gene from the paternal allele during preimplantation development, and importantly, ~40% of the Het embryos degenerated after midgestation (Gu et al., 2011). This sublethal phenotype can be interpreted in at least two ways: First, Tet3-mediated paternal DNA demethylation is required for mouse development. The delayed paternal Oct4 expression is unlikely to be the cause of the sublethality because it is recovered by the blastocyst-stage, a stage beyond which the embryos can develop normally (Gu et al., 2011). The second possibility is Tet3 haploinsufficiency due to deletion of the maternal allele. This possibility should be considered given that 42% of mouse genes previously screened exhibit haploinsufficiency (White et al., 2013). Evaluation of these two possibilities is thus necessary for addressing whether Tet3-mediated paternal DNA demethylation is required for mouse development.
In this study, we demonstrated that the sublethality of Tet3 maternal KO mice is caused by Tet3 haploinsufficiency, but not by defective paternal 5mC oxidation. Furthermore, genome-scale DNA methylation analysis revealed that hypermethylation in maternal KO zygotes is largely reset by the blastocyst stage, suggesting the existence of a compensatory demethylation pathway in preimplantation embryos.
RESULTS
Tet3 maternal KO causes neonatal sublethality
As reported recently (Shen et al., 2014), we generated a Tet3 conditional KO (CKO) mouse that allows deletion of Tet3 in oocytes expressing Zp3Cre. We refer 2 loxP and 1 loxP (deleted) alleles as “f” and “–”, respectively. Immunostaining with an anti-Tet3 antibody confirmed that Tet3 is depleted in zygotes derived from oocytes of [Zp3Cre, Tet3f/f] females fertilized with WT sperm (maternal KO zygotes). In contrast, Tet3 is readily detectable in paternal pronuclei of control zygotes derived from oocytes of [Tet3f/f] females (Fig. 1A). Consistent with the notion that 5hmC in paternal pronuclei is generated by maternally-deposited Tet3, 5hmC is completely lost in maternal KO zygotes, while the 5mC signal in paternal pronuclei is slightly increased (Fig. 1B).
Figure 1. Tet3 maternal KO progenies show neonatal sublethality.
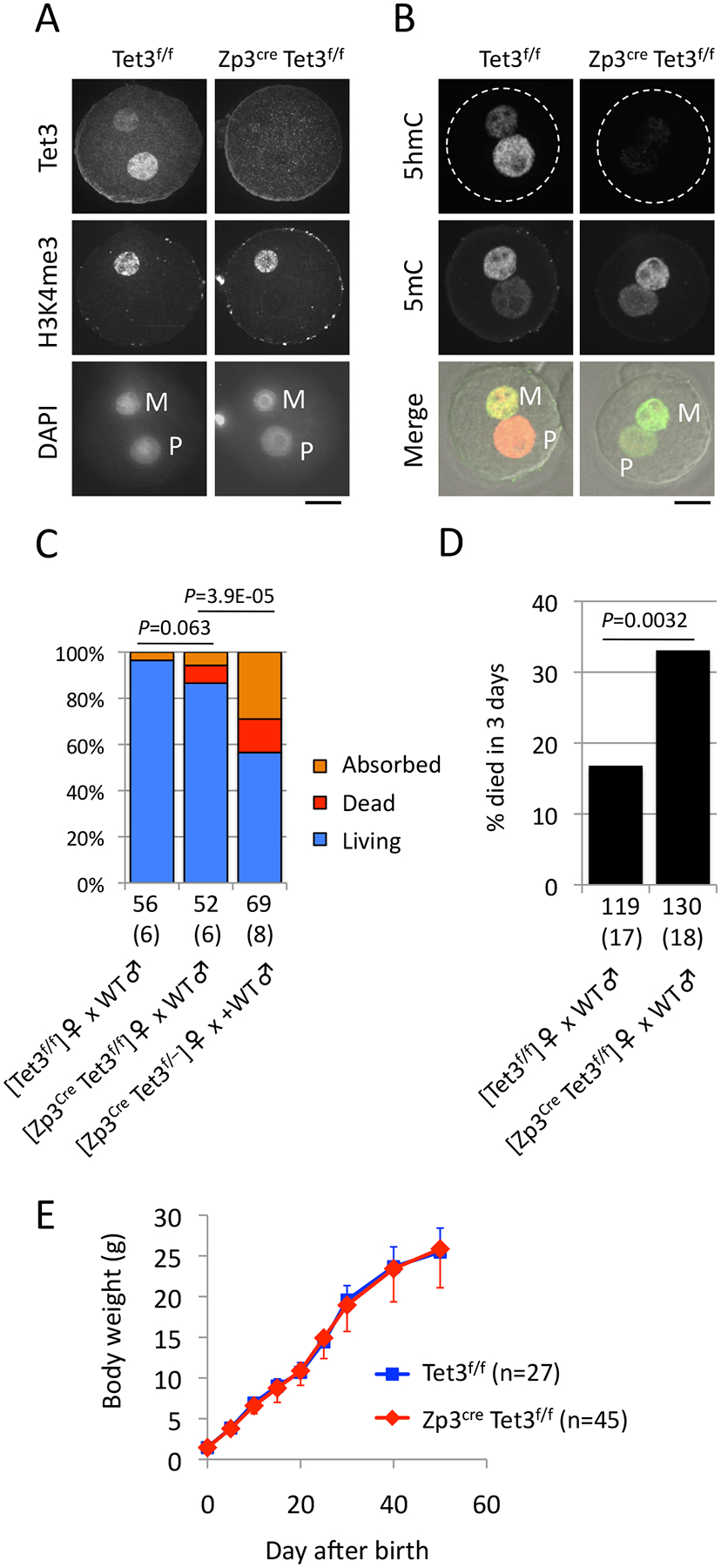
(A) Representative images of zygotes stained with anti-Tet3 antibody. Maternal chromatin is marked by H3K4me3. M, maternal pronucleus; P, paternal pronucleus. Scale bar, 20 μm.
(B) Representative images of zygotes stained with anti-5hmC (red in merge) and anti-5mC (green in merge).
(C) Percentage of living (blue), dead (red), and absorbed (orange) embryos at E19.5. Implantation sites without visible embryos were counted as “Absorbed”. Dead bodies at birth were counted as “Dead”. C57BL/6J males were used for mating. The total number of embryos obtained is indicated below the bars. The total number of litters examined is indicated within the parentheses. P, χ2-test for ”Living”.
(D) Percentage of progenies that died within 3 days after birth. C57BL/6J males were used for mating. The total numbers of progenies examined are indicated below the bars. The total number of deliveries during mating term for 3–4 months is indicated within parentheses. A total of 7 mating pairs were examined in both groups. P, χ2-test.
(E) Growth curve of progenies from the indicated females. The numbers of progenies examined are indicated in parentheses. Error bar, SD.
To examine the developmental potential of maternal KO embryos, WT [Tet3f/f] or CKO [Zp3Cre, Tet3f/f] females were crossed with WT males. Caesarian (C)-section at E19.5 revealed that the maternal KO embryos do not show a significant increase of developmental failure that is evident by implantation sites and dead bodies (Fig 1C and Table S1). We confirmed that all of the maternal KO embryos genotyped were Het (29/29), indicating efficient deletion of the Tet3 allele by Zp3Cre. This result indicates that defective paternal 5mC oxidation does not significantly compromise embryonic development. This seems to be in conflict with a previous study reporting that ~40% of Tet3 maternal KO embryos ([TNAP Cre, Tet3f/–] female × WT male) die before birth (Gu et al., 2011). This phenotypic difference could be potentially caused by the difference in maternal genotype as we used CKO mice with WT background ([Tet3f/f]) while the previous study used mice with Het background ([Tet3f/–]). To test this possibility, we generated CKO females with Het background [Zp3Cre, Tet3f/–] and crossed with WT males. C-section at E19.5 revealed that, consistent with the previous report, ~40% of embryos showed lethality during embryogenesis (Fig 1C and Table S1).
We next assessed neonatal and postnatal growth of the maternal KO progenies that were derived from [Zp3Cre, Tet3f/f] females naturally mated with WT males. Daily checking of delivery and counting the number of surviving pups revealed that a significantly larger population (33%) of pups died within 3 days after birth in maternal KO progenies than in control (17%) (Fig. 1D and Table S2). The surviving progenies at day 3 were viable and grew normally (Fig. 1E). Thus, these results indicate that Tet3 maternal KO progenies display neonatal sublethality.
Tet3 heterozygous mice exhibit neonatal sublethality
Because maternal KO progenies are Het, and Tet3-null mice are known to be neonatal lethal (Gu et al., 2011; Wang et al., 2013), the observed sublethality can be caused by Tet3 haploinsufficiency. To examine this possibility, we asked whether Tet3 Het progenies derived from Het females exhibit neonatal sublethality similarly to the maternal KO progenies. Mating of Het females with WT males followed by C-section at E19.5 showed no significant developmental failure in the embryos (Fig. 2A and Table S3). As expected, about half (49%) of the embryos were Het (Fig. 2B). Analysis of neonatal and postnatal growth indicated that a significant population (31%) of the progenies died within 3 days after birth (Fig. 2C and Table S4). All the progenies that survived the first 3 days were viable at least until 20 days after birth. Genotyping of the living pups at 20 days after birth revealed that the Het population (38%) was markedly less than that of WT (Fig. 2D). These results indicate that Tet3 Het progenies exhibit neonatal sublethality in a similar fashion to those of maternal KO progenies.
Figure 2. Tet3 heterozygous mice show neonatal sublethality.
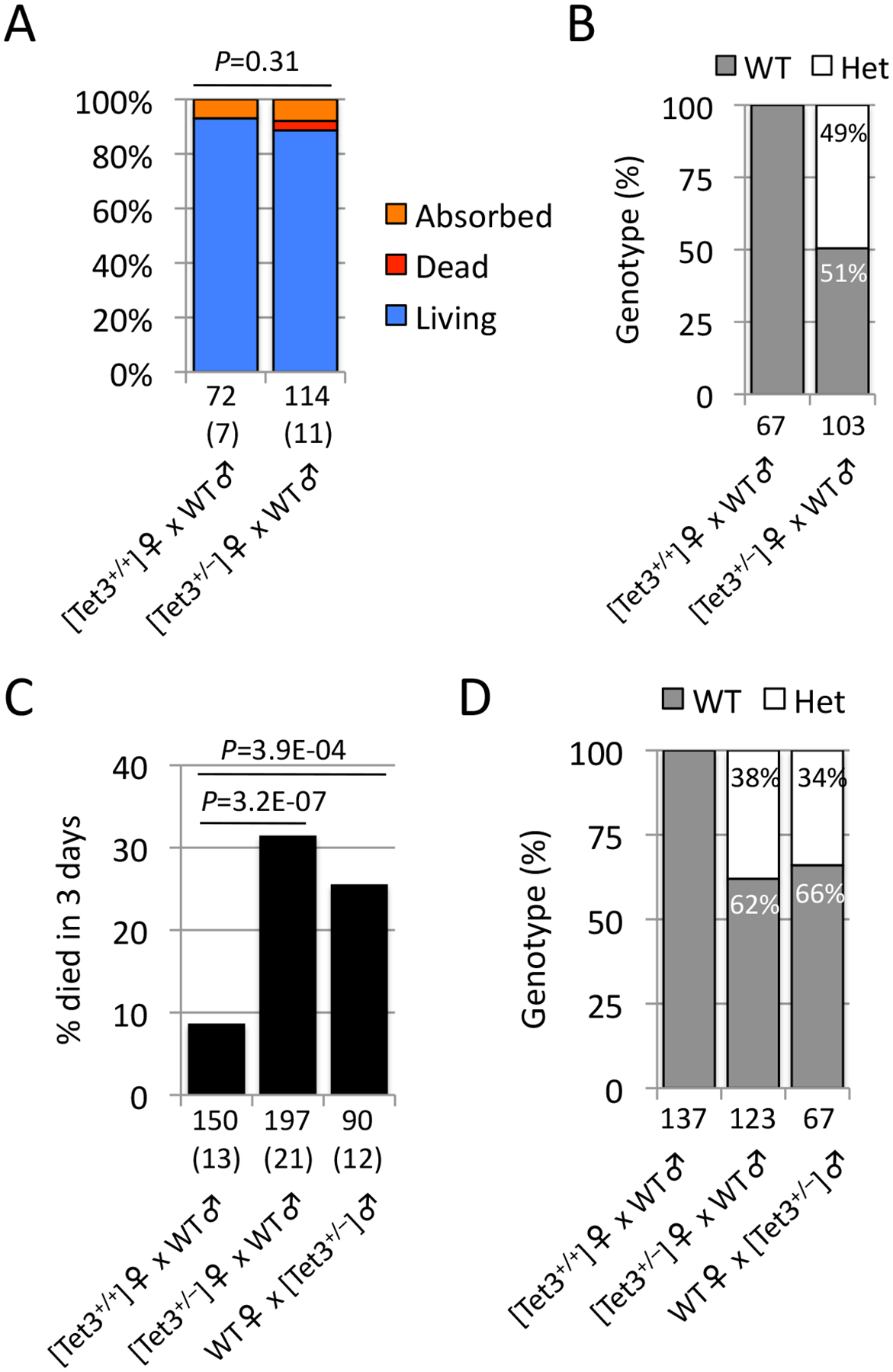
(A) Percentage of living (blue), dead (red), and absorbed (orange) embryos at E19.5. Implantation sites without visible embryos were counted as “Absorbed”. Dead bodies at birth were counted as “Dead”. C57BL/6J males were used for mating. The total numbers of embryos obtained are indicated below the bars. The total number of litters examined is indicated within parentheses. P, χ2-test for “Living”.
(B) Genotype of embryos living at E19.5. The total number of pups examined is indicated below the bars.
(C) Percentage of progenies that died within 3 days after birth. C57BL/6J males were used for mating as WT. The total number of progenies examined is indicated below the bars. The total number of deliveries during mating term for 3–4 months is indicated within parentheses. A total of 5, 9, and 4 mating pairs were examined for [Tet3+/+] females, [Tet3+/−] females, and [Tet3+/−] males, respectively. P, χ2-test.
(D) Genotype of living pups at 20 days after birth. The total numbers of pups examined are indicated below the bars.
It is assumed that oocytes from Het females may have only half the amount of Tet3 protein, which may cause incomplete oxidation of paternal 5mC in zygotes. This assumption makes it unclear whether the sublethality of Tet3 Het progenies was caused by haploinsufficiency at the neonatal stage or caused by potential compromise of paternal 5mC oxidation at the zygote stage. To distinguish between these possibilities, we generated Tet3 Het males and crossed them with WT females. Given that paternal 5mC oxidation is solely dependent on maternally deposited Tet3 protein (Gu et al., 2011; Guo et al., 2014a; Shen et al., 2014), zygotes derived from these mating pairs should undergo normal 5mC oxidation. Notably, natural mating of Het males with WT females also resulted in neonatal sublethality (Fig. 2C and Table S4). Genotyping of the surviving pups at 20 days after birth revealed that the Het population (34%) was substantially lower than that of WT mice (Fig. 2D), similar to that observed in crosses of Het females with WT males. Taken together, these results demonstrate that Tet3 Het mice exhibit haploinsufficiency defects in neonatal development.
Tet3-mediated paternal 5mC oxidation is dispensable for mouse development
Although the above results suggest that the Tet3 gene shows haploinsufficiency, it is still unclear whether Tet3-mediated paternal 5mC oxidation is developmentally important. Thus, it is necessary to distinguish the potential effect of defective paternal 5mC oxidation from the effect of haploinsufficiency. We therefore attempted to reconstruct genetically-WT zygotes that bypass paternal 5mC oxidation through pronuclear transfer (NT). First, we removed paternal pronuclei from WT zygotes at the late-zygotic (PN5) stage and used the remaining cytoplasm containing maternal pronuclei as recipients (Fig. 3A). We then isolated paternal pronuclei from the same zygotic stage of maternal KO zygotes ([Zp3Cre, Tet3f/f] females × WT males) as donor pronuclei. Importantly, while the donor paternal pronuclei are genetically WT, they escape Tet3-mediated 5mC oxidation. Fusion with the recipients allowed the reconstruction of genetically-WT zygotes with paternal pronuclei bypassing 5mC oxidation (Fig. 3A, NT-KO). As a control, we used paternal pronuclei from WT zygotes ([Tet3f/f] females × WT males) that had gone through 5mC oxidation as donors (Fig. 3A, NT-WT).
Figure 3. Tet3-mediated paternal 5mC oxidation is dispensable for mouse development.
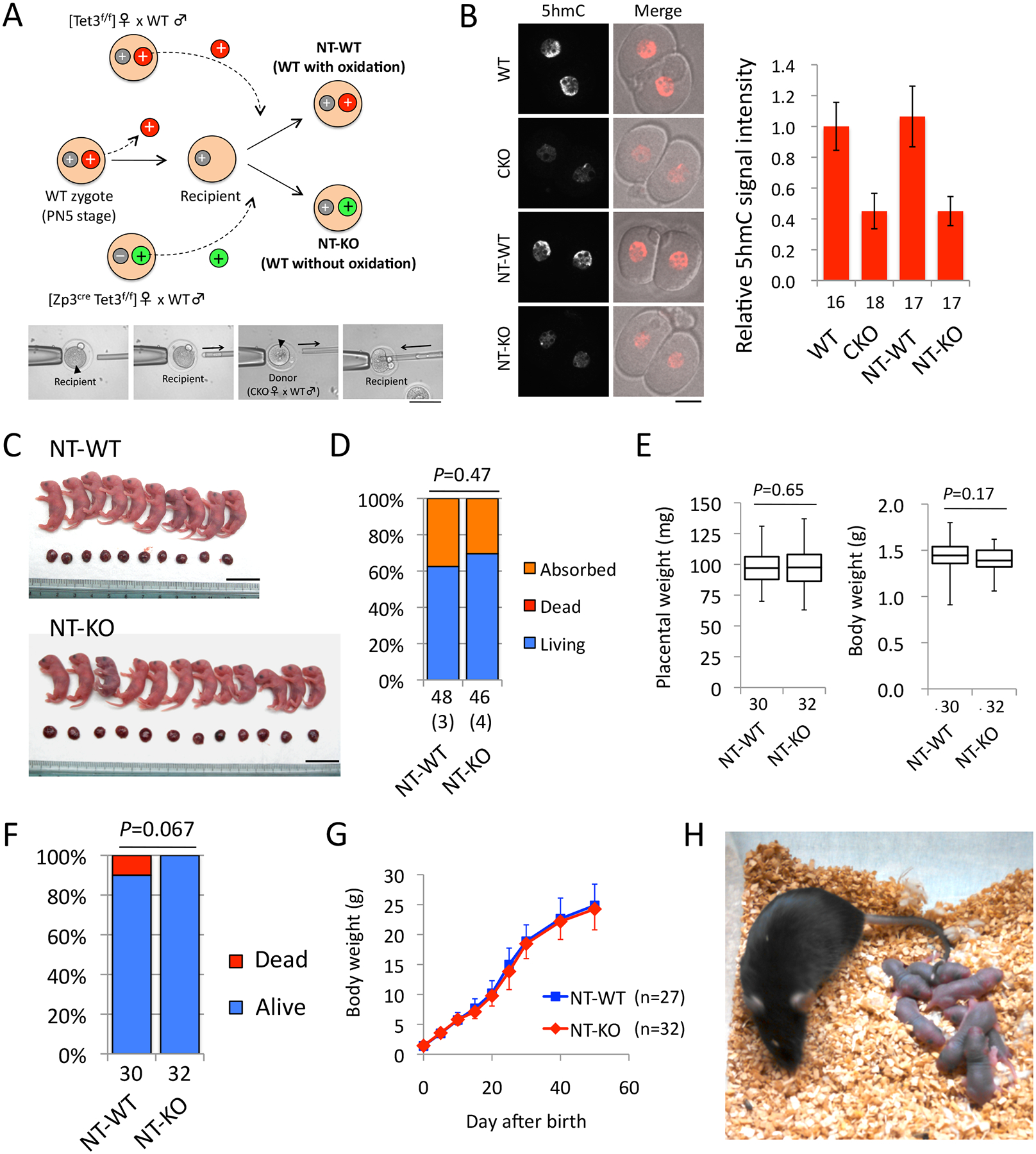
(A) A schematic presentation of the pronuclear transfer (NT) procedure. The recipient cytoplasm was prepared by removing the paternal pronucleus from PN5-stage WT zygotes. Donor pronuclei were isolated from zygotes obtained from CKO [Zp3Cre, Tet3f/f] or WT [Tet3f/f] females mated with WT males. Fusion with the recipients yields genetically-WT zygotes with defective paternal 5mC oxidation (NT-KO) or control zygotes (NT-WT). Gray, maternal pronuclei. Green, paternal pronuclei with defective 5mC oxidation. Red, paternal pronuclei with oxidized 5mC. +, Tet3 WT allele. –, Tet3 KO allele. Images at bottom represent manipulation of pronuclear transfer. Black arrowheads indicate paternal pronuclei.
(B) Representative images of 2-cell embryos stained with anti-5hmC antibody. WT and CKO embryos were prepared as positive and negative controls, respectively. Scale bar, 20 μm. The graph at right indicates quantification of the 5hmC signal. The value of WT embryos was set as 1. The numbers of embryos examined were indicated below the bars. Error bars, SD.
(C) Representative images of embryos and placentae from a single litter of NT-WT and NT-KO at E19.5. Scale bars, 20 mm.
(D) Percentage of living (blue) and absorbed (orange) embryos at E19.5. Implantation sites without visible embryos were counted as “Absorbed”. Dead bodies were not observed in both groups. The total number of embryos examined is indicated below the bars. The total number of litters is indicated within parentheses. P, χ2-test.
(E) Box plot representations of body and placental weight. Middle lines in the boxes indicate the medians. Box edges and whiskers indicate the 25th/75th and 0th/100th percentiles, respectively. P, two-tailed Student’s t-test.
(F) Percentage of progenies that were alive (blue) or dead within 3 days after birth (red). The total numbers of progenies examined are indicated below the bars. P, χ2-test.
(G) Growth curve of progenies. The number of pups examined is indicated in parentheses. Error bar, SD.
(H) Representative image of an adult NT-KO female with its pups after crossing with a WT male.
Immunostaining with anti-5hmC antibody at the 2-cell stage (20 hrs after NT) confirmed that 5hmC level in the NT-KO embryos is as low as that in maternal KO embryos, indicating that Tet3-mediated paternal 5mC oxidation does not occur in NT-KO embryos (Fig. 3B). This suggests that Tet3 protein retained in the recipient cytoplasm is not sufficient to trigger massive 5mC oxidation in the reconstructed embryos. This is plausible as Tet3 mainly localizes to the paternal pronuclei that have been removed from the recipients (Gu et al., 2011; Guo et al., 2014a; Inoue et al., 2012; Shen et al., 2014). Additionally, since Tet3 is no longer localized to nuclei after the first mitosis (Gu et al., 2011), the time window during which the remaining Tet3 can function is very limited due to the quick entry of the embryo into the first mitosis (within 2 hrs after fusion). Thus, we successfully created genetically-WT zygotes without paternal 5mC oxidation.
To examine development of the reconstructed embryos, we transplanted them into pseudopregnant females. C-section at E19.5 revealed that NT-KO embryos could develop to term at a ratio similar to NT-WT (Fig. 3C and 3D and Table S5). Furthermore, no significant difference in the weights of embryos and placentae were observed (Fig. 3E). Importantly, no NT-KO pups showed neonatal lethality, and all the mice developed normally to adulthood (Fig. 3F and 3G). Mating of adult NT-KO mice with WT mice confirmed that both NT-KO males and females are fertile (Fig. 3H). Taken together, these results demonstrate that Tet3-mediated paternal 5mC oxidation is dispensable for mouse development.
The paternal genome of maternal Tet3 KO embryos is hypomethylated by the blastocyst-stage
After the first wave of DNA demethylation in zygotes, the embryonic genome becomes further demethylated during preimplantation development and reaches its lowest point of methylation at the blastocyst stage (Guo et al., 2014b; Smith et al., 2014; Smith et al., 2012). To examine the effect the maternal loss of Tet3 on DNA methylation at the blastocyst stage, we performed genome-scale methylome analysis at 1-cell and blastocyst-stage embryos generated using Tet3 CKO oocytes (C57BL/6J × 129/Sv background) and CAST/EiJ sperm (Table S6). Due to the limited cell numbers of the samples, we used the reduced representative bisulfite sequencing (RRBS) method, a technique that samples ~5% of total CpG of the mouse genome (Smith et al., 2012). Using single-nucleotide polymorphism (SNP) information unique to the CAST strain, we dissociated the methylation state of paternal genome from that of maternal genome. In total, we identified 83,172 SNP-tracked CpGs commonly covered in all samples, among which 17,442 CpGs undergo dramatic DNA demethylation in zygotes [Methylation Level in sperm (MLsp)≥80%, and Relative Demethylation Level in WT zygotes (RDLWT)≥0.3, where RDLWT is defined as [(MLSp–MLWT)/MLSp]. We then focused on Tet3-dependent demethylated CpGs, which showed lower RDL values in maternal KO zygotes than in WT (RDLCKO/RDLWT ≤0.6, n=10,559), and then examined the methylation levels of blastocyst embryos (Figure 4A). Interestingly, the hypermethylated CpG sites in maternal KO zygotes become drastically hypomethylated at the blastocyst stage, and the extent of methylation difference between WT and maternal KO is much less obvious (Figure 4A and B), suggesting that demethylation of these loci takes place independently of maternal Tet3. These results imply that, even in the absence of Tet3-mediated oxidation at zygotes, the paternal genome can be globally hypomethylated by the blastocyst stage, which may explain why Tet3-mediated paternal 5mC oxidation is dispensable for later development.
Figure 4. Effect of maternal Tet3 KO on the paternal methylome.
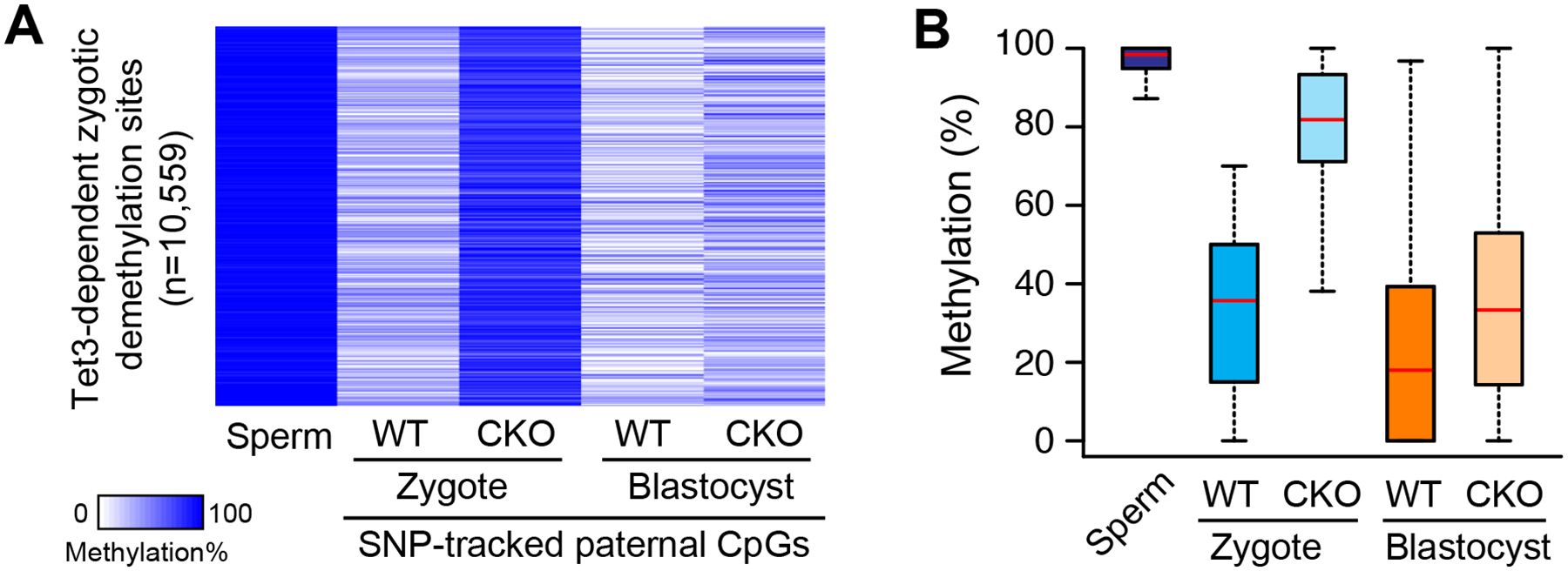
(A) Heatmap of 10,559 SNP-tracked paternal CpG sites that are methylated in sperm (MLSp≥80%) and demethylated after fertilization in a Tet3-dependent manner (RDLWT Zygote ≥ 0.3 and RDLCKO Zygote/RDLWT Zygote ≤0.6). ML, Methylation Level; RDL, Relative Demethylation Level, defined as [(MLSp–MLWT Zygote)/MLSp]. CKO, Tet3 maternal KO.
(B) Box plot of methylation levels. Red line represents the median. Boxes and whiskers represent the 25th and 75th, and 2.5th and 97.5th percentiles, respectively.
DISCUSSION
Based on the previous report that maternal depletion of Tet3 blocks paternal 5mC oxidation and leads to embryonic sublethality (Gu et al., 2011), it has been believed that Tet3-mediated paternal 5mC oxidation plays an important role in mouse development (Kohli and Zhang, 2013; Messerschmidt et al., 2014; Pastor et al., 2013; Seisenberger et al., 2013; Wu and Zhang, 2014). However, because the progenies of Tet3 CKO females are heterozygous, it remained to be determined whether the sublethality was due to Tet3 haploinsufficiency or defective paternal 5mC oxidation. In this study, we explored these two possibilities and found that haploinsufficiency, but not defective paternal 5mC oxidation, is the cause of this phenotype. The notion that proper expression of Tet3 is required for neonatal growth is consistent with previous findings that Tet3-null mice exhibit neonatal lethality (Gu et al., 2011; Wang et al., 2013). Future studies should reveal why Tet3 is required for neonatal growth.
The mammalian zygote is one of the best in vivo models for studying the mechanism of DNA demethylation. Interestingly, a recent study suggested that 5mCs within certain genomic loci are converted to unmodified cytosines in mouse zygotes in a Tet3-dependent but thymine DNA glycosylase (TDG)-independent manner, implying the existence of an undefined demethylation pathway (Guo et al., 2014a). To reveal such mechanism, a hypothesis-driven candidate approach will be required, as genome-wide screening is difficult to perform in zygotes (Gkountela and Clark, 2014). However, there may be many candidate factors, including deaminase, base excision repair enzymes, decarboxylases, and the elongator, that can be involved in active DNA demethylation (Messerschmidt et al., 2014; Wu and Zhang, 2014; Wu and Zhang, 2010). Since Tet3-mediated 5mC oxidation had been thought to be required for development, only genes known to be relevant to development might have been listed as candidates. Nevertheless, our study indicates that such factors are not necessarily essential for development, and thus non-essential genes should also be considered.
We and others have recently reported that Tet3 oxidizes not only the paternal genome but also the maternal genome, although to a lesser extent (Guo et al., 2014a; Shen et al., 2014). Because our pronuclear transfer experiment could not evaluate the role of maternal 5mC oxidation, we cannot exclude the possibility that Tet3-mediated maternal 5mC oxidation may play a role in development. Nevertheless, such possibility is less likely given that phenotypes of maternal KO embryos, in which both paternal and maternal 5mC oxidations are defective, are not more severe than those of the Het embryos, in which 5mC oxidation takes place in both genomes normally, derived from Het males or females crossed with WT (compare Figure 1D and 2C).
We found that the paternal genome can be largely hypomethylated by the blastocyst stage in the absence of maternal Tet3 (Figure 4). This demethylation might be achieved by DNA replication-dependent passive dilution of 5mC during preimplantation development. This notion is supported by our recent observation that Tet3-dependent demethylated regions can partially undergo replication-coupled demethylation at the 1-cell stage (Shen et al., 2014). Thus, consecutive passive dilutions during preimplantation development might compensate for the loss of maternal Tet3, leading to hypomethylation of the paternal genome. Alternatively, Tet1 and Tet2 might also contribute to DNA demethylation, as both begin to be expressed after the 2-cell stage (Iqbal et al., 2011). It is also possible that a Tet-independent demethylation pathway may be involved in the observed compensation (Wang et al., 2014). Further studies will be needed to identify all the players involved in DNA demethylation, and to address the biological significance of global DNA demethylation in preimplantation embryos.
EXPERIMENTAL PROCEDURES
Mice
All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School. Tet3 CKO mice were generated as described previously (Shen et al., 2014). Tet3 Het and WT mice were obtained by crossing CKO ([Zp3Cre, Tet3f/f] or [Zp3Cre, Tet3f/–]) females with C57BL/6J males, followed by crosses of the Het progenies with C57BL/6J mice. Genotyping primer sequences were reported previously (Shen et al., 2014). The day when a vaginal plug appears at noon is defined as embryonic day (E) 0.5. At E19.5, the progenies were collected by dissecting pregnant females that had been injected with 0.2 ml of 10 mg/ml progesterone (Sigma-Aldrich) at E17.5 and E18.5. For neonatal and postnatal growth, we took daily recordings of the delivery and viability from natural mating pairs with C57BL/6J mice.
Pronuclear transfer
For preparation of donor zygotes, 8 week-old CKO [Zp3Cre, Tet3f/f] or WT [Tet3f/f] females were superovulated by injecting 7.5 I.U of PSMG (Millipore) and hCG (Millipore) followed by mating with C57BL/6J males. For preparation of recipient zygotes, 8 week-old B6D2F1 females were superovulated similarly and mated with C57BL/6J males. At noon of day E0.5, PN2–3 zygotes were collected and cultured in KSOM (Millipore) in a humidified atmosphere of 5% CO2/95% air at 37.8°C. Five hours later, zygotes reached the PN5 stage and were then transferred into M2 media containing 5 μM cytochalasin B (Sigma-Aldrich). Zona pellucidae were cut by a Piezo impact-driven micromanipulator (Prime Tech Ltd., Ibaraki, Japan). The paternal pronuclei were removed from the recipient zygotes, and the remaining cytoplasms containing maternal pronuclei were served as recipients. Parental pronuclei can be distinguished by the distance from the second polar body and by the pronuclear size. Next, paternal pronuclei were isolated from PN5-stage WT or CKO zygotes and fused with the recipients by using sendai virus (HVJ, Cosmo-bio) as described previously (Inoue et al., 2008).
Two-cell stage embryos were transferred to the oviducts of pseudopregnant (E0.5) ICR females. The pups were recovered by C-section on the day of delivery (E19.5) and nursed by lactating ICR females. After they grew to the adulthood, each of three NT-KO males and females were mated with C57BL/6J mice for fertility test, and all gave 7–11 pups.
Immunostaining
Tet3, 5mC, and 5hmC staining were performed as described previously (Shen et al., 2014).
RRBS
MII oocytes were collected from 8 week-old superovulated females. They were transferred into HTF medium supplemented with 10 mg/ml bovine serum albumin (BSA; Sigma-Aldrich) and inseminated with activated spermatozoa collected from the caudal epididymides of adult CAST/EiJ males. Five hours after fertilization, zygotes were transferred into KSOM. Zygotes and blastocysts were collected at 13 and 96 hours postfertilization, respectively. Biological duplicates for each sample were collected, with each sample containing 40–50 zygotes or 4 blastocysts. Sperm genomic DNA was extracted from CAST males as described previously (Weyrich, 2012).
RRBS analyses were performed as described previously (Shen et al., 2014), and sequencing reads were mapped to the mouse genome (mm9) using Bismark v0.10.1 (Babraham Bioinformatics) after adapter trimming by Trim Galore (Babraham Bioinformatics) with the “--rrbs” option. Paternal reads were extracted from the total mapped reads by SNPs between CAST and C57BL/6J mice. The methylation level of each covered cytosine in CpG context was calculated by dividing the number of reported C with the total number of reported C and T. Only CpG sites that were commonly covered by at least five reads in all samples were used for the subsequent analyses.
Supplementary Material
HIGHLIGHTS.
Tet3 maternal KO causes neonatal sublethality
The sublethality is caused by Tet3 haploinsufficiency
Tet3-mediated paternal 5mC oxidation is dispensable for mouse development
The paternal genome of maternal KO embryos is hypomethylated at blastocyst stage
ACKNOWLEDGEMENTS
We thank L.M. Tuesta and S. Yamaguchi for critical reading of the manuscript, and G.L. Xu for the Tet3 antibody. The work was partly supported by NIH grant U01-DK089565. S.M. is a research fellow for Research Abroad of the Japan Society for the Promotion of Science. Y.Z. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Beaujean N, Hartshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, Meehan R, and Young L (2004). Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol 14, R266–267. [DOI] [PubMed] [Google Scholar]
- Gkountela S, and Clark A (2014). A big surprise in the little zygote: the curious business of losing methylated cytosines. Cell Stem Cell 15, 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. (2011). The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610. [DOI] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. (2014a). Active and passive demethylation of male and female pronuclear DNA in the Mammalian zygote. Cell Stem Cell 15, 447–458. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. (2014b). The DNA methylation landscape of human early embryos. Nature 511, 606–610. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, and Sasaki H (2008). Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev 22, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Matoba S, and Zhang Y (2012). Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation. Cell Res 22, 1640–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Nakajima R, Nagata M, and Aoki F (2008). Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod 23, 1377–1384. [DOI] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, and Zhang Y (2011). Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21, 1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, and Zhang Y (2011). Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin S, Pfeifer G, and Szabó P (2011). Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A 108, 3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Yeo S, Park J, Koo D, Chang W, Lee K, and Kang Y (2007). DNA methylation state is preserved in the sperm-derived pronucleus of the pig zygote. Int J Dev Biol 51, 707–714. [DOI] [PubMed] [Google Scholar]
- Kohli R, and Zhang Y (2013). TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepikhov K, Zakhartchenko V, Hao R, Yang F, Wrenzycki C, Niemann H, Wolf E, and Walter J (2008). Evidence for conserved DNA and histone H3 methylation reprogramming in mouse, bovine and rabbit zygotes. Epigenetics Chromatin 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, and Haaf T (2000). Demethylation of the zygotic paternal genome. Nature 403, 501–502. [DOI] [PubMed] [Google Scholar]
- Messerschmidt D, Knowles B, and Solter D (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28, 812–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, and Walter J (2000). Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10, 475–478. [DOI] [PubMed] [Google Scholar]
- Park J, Lee D, Cho S, Shin S, and Kang Y (2011). Active loss of DNA methylation in two-cell stage goat embryos. Int J Dev Biol 54, 1323–1328. [DOI] [PubMed] [Google Scholar]
- Pastor W, Aravind L, and Rao A (2013). TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski Z, Motosugi N, Tsurumi C, Hiiragi T, and Hoffmann S (2008). Hypomethylation of paternal DNA in the late mouse zygote is not essential for development. Int J Dev Biol 52, 295–298. [DOI] [PubMed] [Google Scholar]
- Reis Silva A, Adenot P, Daniel N, Archilla C, Peynot N, Lucci C, Beaujean N, and Duranthon V (2012). Dynamics of DNA methylation levels in maternal and paternal rabbit genomes after fertilization. Epigenetics 6, 987–993. [DOI] [PubMed] [Google Scholar]
- Rougier N, Bourc’his D, Gomes D, Niveleau A, Plachot M, Pàldi A, and Viegas-Péquignot E (1998). Chromosome methylation patterns during mammalian preimplantation development. Genes Dev 12, 2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, and Kurimoto K (2011). Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139, 15–31. [DOI] [PubMed] [Google Scholar]
- Sasaki H, and Matsui Y (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nature reviews Genetics 9, 129–140. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Peat J, and Reik W (2013). Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Current opinion in cell biology 25, 281–288. [DOI] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, and Zhang Y (2014). Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell 15, 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z, Chan M, Humm K, Karnik R, Mekhoubad S, Regev A, Eggan K, and Meissner A (2014). DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, and Meissner A (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013). DNA methylation: roles in mammalian development. Nature reviews Genetics 14, 204–220. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. (2014). Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyrich A (2012). Preparation of genomic DNA from mammalian sperm. Current protocols in molecular biology / edited by Frederick M Ausubel [et al. ] Chapter 2, Unit 2 13 11–13. [DOI] [PubMed] [Google Scholar]
- White J, Gerdin A, Karp N, Ryder E, Buljan M, Bussell J, Salisbury J, Clare S, Ingham N, Podrini C, et al. (2013). Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 154, 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques C, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, and Walter J (2011). 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2, 241. [DOI] [PubMed] [Google Scholar]
- Wu H, and Zhang Y (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, and Zhang Y (2010). Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


