Abstract
Background
N6-methyladenosine (m6A) is the most abundant modification known in mRNAs. It participates in a variety of physiological and pathological processes, such as metabolism, inflammation, and apoptosis.
Aims
To explore the mechanism of m6A in cisplatin-induced acute kidney injury (AKI) and berberine alleviation in mouse.
Methods
This study investigated the N6-methyladenosine (m6A) methylome of kidneys from three mouse groups: C57 mice (controls), those with CI-AKI (injury group, IG), and those pretreated with berberine (treatment group, TG). Methylated RNA Immunoprecipitation Next Generation Sequencing (MeRIP-seq) and RNA-seq were performed to identify the differences between the injury group and the control group (IvC) and between the treatment group and the injury group (TvI). Western blotting was performed to identify the protein levels of candidate genes.
Results
In IvC, differentially methylated genes (DMGs) were enriched in metabolic processes and cell death. In TvI, DMGs were enriched in tissue development. Several genes involved in important pathways related to CI-AKI showed opposite methylation and expression trends in the IvC and TvI comparisons.
Conclusion
m6A plays an important role in cisplatin induced AKI and berberine may alleviate this process.
Keywords: M6A, cisplatin induced nephrotoxicity, berberine, FGA, SLC12A1, Havcr1
Introduction
Cisplatin is an anticancer drug widely used for the treatment of various solid tumors, but it is also known for its nephrotoxicity. It exerts its function by interacting with and disrupting DNA and mitochondrial function. During drug metabolism, cisplatin accumulates in renal tubular cells, causing cell death and resulting in acute kidney injury (AKI) (Lebwohl and Canetta, 1998; Cummings and Schnellmann, 2002; Ozkok and Edelstein, 2014; George et al., 2018; Holditch et al., 2019; Yimit et al., 2019). Recent studies have revealed that apoptosis, necrosis, inflammation, and other mechanisms play significant roles in cisplatin-induced AKI (Sahu et al., 2015; Zuk and Bonventre, 2016; Humanes et al., 2017; Long et al., 2017).
Berberine is the principal component of many popular medical plants, such as Coptis chinensis, Rhizoma coptidis, Hydrastis canadensis, Berberis aquifolium, and Berberis vulgaris (Wang et al., 2017; Duan et al., 2018; Fan et al., 2019). As a promising drug, various pharmacological activities of berberine have been reported, including antimicrobial, antiemetic, antipyretic, anti-pruritic, antioxidant, anti-inflammatory, hypotensive, anti-arrhythmic, and sedative activities (Shamsa et al., 1999; Chen et al., 2012; Caliceti et al., 2016). Several reports have mentioned that berberine shows nephroprotective effects against cisplatin-induced kidney damage (Domitrović et al., 2013; Teng et al., 2015; Ojha et al., 2016; Ahmad et al., 2019). However, the mechanism of alleviating CI-AKI is still unclear.
m6A has been implicated in all aspects of posttranscriptional RNA metabolism, such as regulating reversible modifications, alternative splicing, stability, and translation. It prefers to modify sequences identified as RRACH, where R is adenine or guanine, A is the m6A site, and H is adenine, cytosine, or uracil. In addition, m6A modifications exhibit enrichment in the 3’ UTR near mRNA stop codons and within long internal exons. The writing of m6A is accomplished via a complex composed of methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associated protein (Ping et al., 2014; Wang et al., 2014; Spitale et al., 2015). Two m6A demethylases, fat mass and obesity-associated protein (FTO) and AlkB homolog 5, have been discovered as “erasers” (Jia et al., 2011). While proteins containing the YT521-B homology (YTH) domain, such as YTHDC1, YTHDF1, YTHDF2, and YTHDF3, directly bind m6A sites and act as readers of the m6A signal (Zheng et al., 2013).
Several studies have focused on the correlation between m6A methylation and kidney injury. Wang J. et al. (2019) reported that METTL3 overexpression plays a protective role against colistin-induced oxidative stress and apoptosis in renal tubular epithelial cells in mice. Alteration of m6A regulators was associated with pathologic stage in patients with clear cell renal cell carcinoma (Zhou J. C. et al., 2019). Another study reported that cisplatin treatment reduced FTO expression and increased m6A levels in vivo and in vitro. They also found that inhibiting FTO by meclofenamic acid aggravated renal damage and increased apoptosis in cisplatin-treated kidneys (Zhou P. et al., 2019). METTL14 is elevated in people with AKI (Xu et al., 2020). However, few studies have investigated the m6A methylome in cisplatin-induced AKI and the potential mechanism of berberine alleviation.
In this study, we found that berberine significantly alleviated cisplatin-induced AKI in a reliable mouse model. To further investigate the role of m6A in this process, MeRIP-seq was used to establish the first known transcriptome-wide m6A methylome profiles of kidneys from normal, CI-AKI, and berberine-pretreated mice. RNA-seq was performed to detect differentially expressed genes (DEGs) among the groups. Based on our results, we speculate that berberine may alleviate CI-AKI by regulating m6A methylation.
Materials and Methods
Animals and Tissue Collection
Male C57BL/6 mice (aged 8 weeks) were randomly assigned to the control group, injury group (IG), and treatment group (TG), with four mice per group. All mice were housed under a 12 h light/dark schedule with free access to food and water. Control and IG mice were subjected to daily intraperitoneal (i.p) injections with vehicle (normal saline), while TG mice were injected daily with berberine (Sigma, St. Louis, MO, United States, 20 mg/kg) (Ruan et al., 2017). After 14 days of drug treatment, the IG and TG were injected intravenously with cisplatin (20 mg/kg), while controls were injected intravenously with a vehicle. After cisplatin injection, berberine pretreatment was stopped and all mice were housed as usual. All mice in the three groups were sacrificed at 72 h postinjection by cervical dislocation after CO2-induced narcosis (Zhang et al., 2016; Dutta et al., 2017). Immediately afterward, the kidneys were collected.
Serum Levels of Creatinine and Blood Levels of Urea Nitrogen
Before the mice were sacrificed, blood was drawn from their tail veins. Serum samples were collected. Serum levels of creatinine (Scr) and urea nitrogen (BUN) were analyzed using a standard spectrophotometric assay (Roche Diagnostics GmbH, Mannheim, Germany).
Histopathology Analyses
Renal tissue harvested from animals was washed with 0.9% saline, fixed in 10% neutral buffered formalin, and then embedded in 10% paraffin. Sections (5 μm thick) were stained with hematoxylin eosin for further microscopic analyses. A tubular injury score was calculated (Leemans et al., 2005). The percentage of damaged tubules in the corticomedullary junction was estimated by a nephropathologist using a 5-point scale according to the following criteria: tubular dilation, cast deposition, brush border loss, and necrosis in eight randomly chosen, non-overlapping fields (×400 magnification). Lesions were graded on a scale from 0 to 5: 0 = normal; 1 = mild, involvement of less than 10% of the cortex; 2 = moderate, involvement of 10–25% of the cortex; 3 = severe, involvement of 25–50% of the cortex; 4 = very severe, involvement of 50–75% of cortex; 5 = extensive damage, involvement of more than 75% of the cortex.
RNA MeRIP-seq and Data Analyses
In accordance with the manufacturer’s instructions, TRIzol reagent (Invitrogen Corporation, CA, United States) was used to extract total RNA from kidney tissue. Ribosomal RNA was removed from total RNA with a Ribo-Zero rRNA Removal Kit (Illumina, Inc., CA, United States). Then, fragmentation buffer (Illumina, Inc.) was used to split the RNA into fragments of approximately 100 nucleotides in length. MeRIP-Seq was performed by Cloudseq Biotech Inc. (Shanghai, China). In brief, total RNA was extracted from kidney tissue using TRIzol Reagent (Life Technologies CA, United States). RNA was tested for quality via NanoDrop (Thermo Fisher Scientific, MA, United States). RNA integrity was assessed using a denaturing agarose gel. mRNA was isolated from total RNA using a Seq-StarTM poly(A) mRNA Isolation Kit (Arraystar, MD, United States). The GenSeqTM m6A RNA IP Kit (GenSeq Inc., China) was used to perform m6A RNA immunoprecipitation. A NEBNext® Ultra II Directional RNA Library Prep Kit (New England Biolabs, Inc., MA, United States) was used to construct both the input samples without immunoprecipitation and the m6A IP samples. All samples were subjected to 150 bp paired-end sequencing on an Illumina HiSeq instrument (Illumina, Inc.).
Paired-end reads were quality controlled by Q30. Trimming of the 3’ adaptor and low-quality read removal were performed using Cutadapt software (v1.9.3) (Kechin et al., 2017). Hisat2 software (v2.0.4) (Kim et al., 2015) was used to align clean reads of all libraries to the reference genome (mm10). Then, methylated sites on RNAs (peaks) were identified using Model-based Analysis of ChIP-Seq (MACS) software (Zhang et al., 2008; Li et al., 2017; Huang et al., 2019). Identified m6A peaks were subjected to motif enrichment analyses using Hypergeometric Optimization of Motif EnRichment software (Heinz et al., 2010), and metagene m6A distribution was characterized using R package MetaPlotR (Olarerin-George and Jaffrey, 2017). Differentially methylated sites with a fold change cutoff of ≥ 2 and P < 0.05 were identified using the diffReps differential analysis package (Shen et al., 2013; Wang Y. et al., 2019; Wang et al., 2020). The peaks identified by MACS and diffReps overlapping with exons of mRNA were selected for further analyses. Gene ontology (GO) and pathway enrichment analyses were performed on the differentially methylated proteins for using the GO1 and Kyoto Encyclopedia of Genes and Genomes (KEGG)2 databases.
RNA-Seq and Data Analyses
Total RNA was extracted from kidney samples using TRIzol reagent (Life Technologies) according to the manufacturer’s protocol. Denaturing agarose gel electrophoresis was used to evaluate the integrity of total RNA. A Seq-Star TM poly(A) mRNA Isolation Kit (Arraystar, MD, United States) was utilized to purify mRNA from total RNA after measuring the quantity and quality on a NanoDrop ND-2,000. Then, a BGISEQ-500 platform was used to subject fragmented mRNA to 50 bp single-end sequencing. Adapter and low-quality reads were trimmed using SOAPnuke (Chen et al., 2018), and those trimmed reads were aligned to the reference genome using bowtie2 (Langmead and Salzberg, 2012). Finally, cuffdiff was use to analyze differential expressed genes (DEGs) (Trapnell et al., 2012).
Western Blotting
Mouse tissues were lysed using a protein lysis buffer containing 20 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 25 mM sodium pyrophosphate, and 2 mM sodium orthovanadate aprotinin. All denatured proteins were separated on an SDS-PAGE gel and then transferred to polyvinylidene difluoride membranes (Roche, Netley, NJ, United States). The membranes were blocked with 5% skimmed milk in Tris-buffered saline and then were incubated with 1:500 dilutions of primary antibodies as follows: anti-FGA (Abcam, Cambridge, MA, United States), anti-Slc12a1 (Abcam, Cambridge, MA, United States), and anti-Havcr1 (Abcam, Cambridge, MA, United States). Then, the samples were incubated with a horseradish peroxidase-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch, PA, United States). The bands were visualized using an ECL Western Blotting Kit (Biovision, Milpitas, CA, United States) and were quantified by Quantity One software (Bio-Rad, Hercules, CA, United States).
Statistical Analyses
Data are expressed as mean ± standard deviation (SD). Student’s t-test was used to compare two groups. ANOVA with Tukey’s post-test was used to assess the statistical significance between-group means for comparisons between multiple groups. The differences were considered statistically significant at P < 0.05.
Results
Establishment of a Reliable CI-AKI Mouse Model
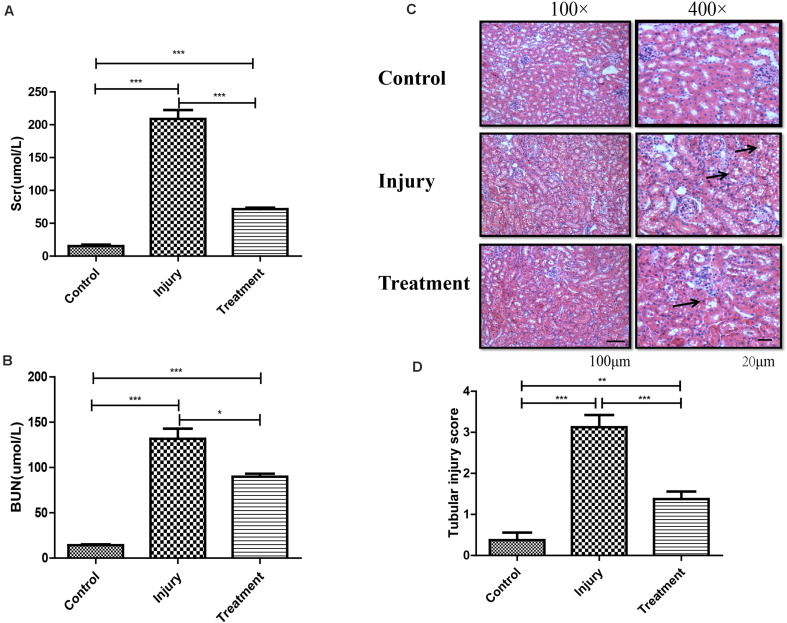
We found that 20 mg/kg cisplatin injection resulted in an approximately 18–20-fold increase in Scr and BUN relative to the control group, but significantly lower levels in TG than in IG (Figures 1A,B). Hematoxylin–eosin staining indicated that cisplatin induced remarkable renal structure damage in IG tissue, including extensive tubular vacuolization, tubular epithelial cell exfoliation, and thickening of glomerular basement membrane (P < 0.001), while these changes were notably alleviated in TG mice (P < 0.001) (Figures 1C,D). This indicated successful and reliable establishment of a CI-AKI mouse model and relief of the injury through berberine pretreatment.
FIGURE 1.
Establishment of cisplatin-induced acute kidney injury model in C57 mouse. (A) Analysis of Scr level in mice following different treatments. Error bars represent the standard deviation. ***p < 0.001, Student’s t-test. (B) Analysis of BUN level in mice following different treatments. Error bars represent the standard deviation. *p < 0.05, ***p < 0.001, Student’s t-test. (C) Represents the image of hematoxylin and eosin staining in kidney (black arrows indicating the injury). (D) Score for characteristic histologic signs of renal injury. **p < 0.01, ***p < 0.001, Student’s t-test.
General Features of m6A Methylation
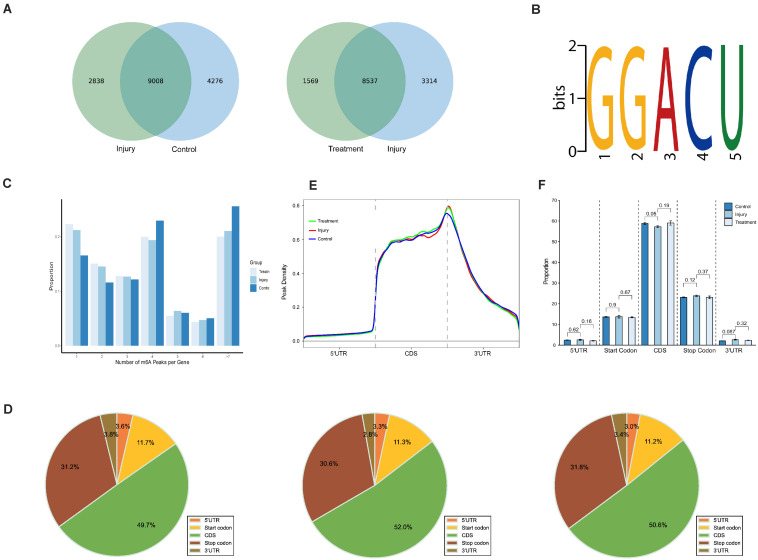
MeRIP-seq analyses of mRNA derived from kidneys revealed 13,284 m6A peaks within 7,942 coding gene transcripts in controls; the values were 11,846 within 7,422 mRNAs in IG and 10,106 within 6,481 mRNAs in TG. Overall, 7,337 peaks overlapped among the three groups (Supplementary Figure 1). Pairwise comparisons showed that 9,008 peaks (more than 55.8% of all peaks) overlapped in IG versus control (IvC) comparisons and 8,537 peaks (more than 63.6% of all peaks) overlapped in TG versus IG (TvI) comparisons (Figure 2A). Approximately 40% of all peaks were non-overlapping, suggesting differences among those groups.
FIGURE 2.
Overview of N6-methyladenosine methylation within mRNAs in the control, injury, and treatment groups. (A) Venn diagram showing the overlap of m6A peaks within mRNAs between IvC (left) and TvI (right). (B) The top motif enriched across m6A peaks identified from three groups. (C) Proportion of genes harboring different numbers of m6A peaks in three groups. The majority of genes harboring more than one m6A peak. (D) Pie charts showing the percentage of m6A peaks in five segments of transcripts. m6A peaks were most enriched in the coding sequence segment. (E) Distributions of fold enrichment of m6A peaks in five segments. The mean fold enrichment in the stop codon segments was the highest in injury group, while that value in CDS was the largest in treatment group. (F) Analysis of m6A peaks in different groups. Error bars represent the standard deviation. Student’s t-test.
To determine whether the m6A peaks contained the RRACH conserved sequence motif, we detected all samples in three groups. One thousand peaks with the highest scores (−10∗log10, p-value) were analyzed using DREME software. The results showed that the same sequence motif (RRACH) was necessary for m6A methylation in kidney mRNAs in each group (Figure 2B), consistent with other studies (Dominissini et al., 2012; Meyer et al., 2012; Luo et al., 2019).
Further analyses demonstrated that 16.59, 21.21, and 22.29% of the genes with m6A-methylation sites in controls, IG, and TG contained one m6A peak (Figure 2C). More than 75% of the genes contained two or more peaks (Figure 2C). This result is different from other reports on mouse liver (Luo et al., 2019) and brain (Dominissini et al., 2012), suggesting that the kidney is unique.
To understand the preferential locations of m6A peaks, all peaks were categorized into five transcript segments: the 5’ UTR, start codon segment, coding sequence (CDS), stop codon segment, and 3’ UTR. We found that m6A was mostly enriched in the CDS, and there was some enrichment near stop codons (Figure 2D). These results are also similar previous studies (Dominissini et al., 2012; Meyer et al., 2012).
Furthermore, m6A peaks in all groups had the highest density in stop codons (Figure 2E). Comparisons between different groups showed no significant difference in the volume of m6A peaks, which indicates that although many variants in m6A methylation were detected in different segments, the total proportion of m6A peaks did not significantly change (Figure 2F).
Distribution of Differentially Methylated m6A Sites
In IvC, we found 2,981 differentially methylated m6A sites (DMMSs) within 2,227 genes, of which 48.17% (1,436/2,981) exhibited significant increases in methylation. Table 1 displays the top 20 differentially methylated m6A sites.
TABLE 1.
The top 20 differently methylated m6A peaks in IvC.
| Gene name | Gene ID | Fold change | Regulation | Chromosome | Peak start | Peak end | Peak length | p-value |
| BC061237 | 385138 | 1489 | Up | chr14 | 44504106 | 44504291 | 185 | 1.48521E-12 |
| BC061237 | 385138 | 816.8 | Up | chr14 | 44500121 | 44500197 | 76 | 3.23828E-13 |
| Krt20 | 66809 | 498.7531381 | Up | chr11 | 99430752 | 99430920 | 168 | 9.96013E-10 |
| Krt20 | 66809 | 498.0274348 | Up | chr11 | 99429021 | 99429078 | 57 | 9.08594E-10 |
| Krt20 | 66809 | 483.5165816 | Up | chr11 | 99432181 | 99432343 | 162 | 2.59398E-09 |
| 1700001F09Rik | 71826 | 383.8 | Up | chr14 | 43346701 | 43346790 | 89 | 5.98092E-15 |
| Ccdc85b | 240514 | 328.5877193 | Up | chr19 | 5454141 | 5454580 | 439 | 3.35868E-11 |
| Gm3543 | 100041849 | 318 | Up | chr14 | 41982201 | 41982290 | 89 | 1.38739E-12 |
| Serpina3n | 20716 | 316.3 | Up | chr12 | 104414261 | 104414329 | 68 | 7.77029E-14 |
| Gm3543 | 100041849 | 254 | Up | chr14 | 41982133 | 41982180 | 47 | 4.03611E-10 |
| Alms1 | 236266 | 503.6 | Down | chr6 | 85694833 | 85694951 | 118 | 4.2803E-10 |
| Tas2r119 | 57254 | 385.6 | Down | chr15 | 32177288 | 32177620 | 332 | 9.86777E-14 |
| Ctnna2 | 12386 | 220.8 | Down | chr6 | 77600041 | 77600380 | 339 | 1.36972E-13 |
| Afm | 280662 | 195.1 | Down | chr5 | 90518931 | 90519060 | 129 | 1.23171E-10 |
| Slc5a4a | 64452 | 126.7 | Down | chr10 | 76163688 | 76163769 | 81 | 7.19208E-10 |
| Nat1 | 17960 | 123.5 | Down | chr8 | 67490861 | 67491460 | 599 | 2.54765E-10 |
| Pzp | 11287 | 111.1290323 | Down | chr6 | 128526621 | 128526720 | 99 | 8.47489E-15 |
| Dpf3 | 70127 | 99.2 | Down | chr12 | 83215461 | 83215800 | 339 | 3.4897E-12 |
| Dgkg | 110197 | 95.3 | Down | chr16 | 22479365 | 22479426 | 61 | 1.61986E-10 |
| Dpf3 | 70127 | 94.3 | Down | chr12 | 83214541 | 83214840 | 299 | 1.03615E-10 |
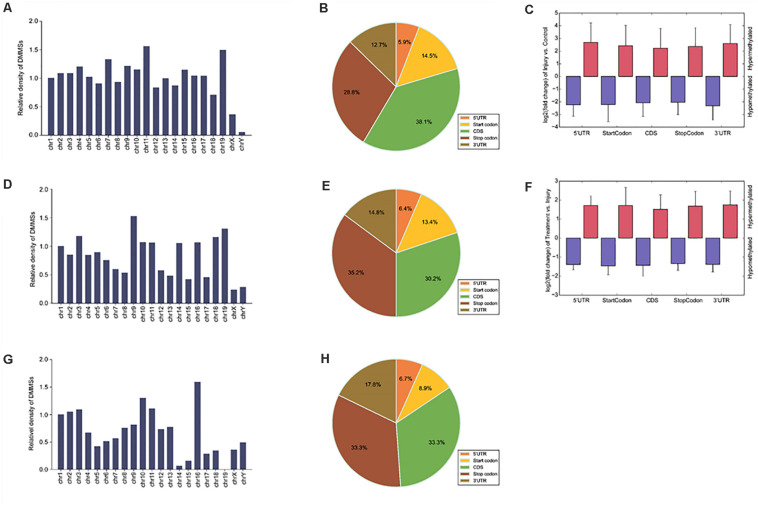
To understand the DMMS distribution profiles in those two groups, we mapped them to chromosomes. Chromosomes 4, 2, 7, and 11 were the top four chromosomes, harboring more than 200 DMMSs (Supplementary Figure 2A). However, when the number of DMMSs was normalized by the length of chromosomes, the top four with the highest relative densities were 11, 19, 7, and 4 (Figure 3A). Meanwhile, most DMMSs were within a CDS (Figure 3B). Further analyses showed that at sites with increased methylation, DMMSs within the 5’ UTR had the highest fold change, while among the sites with decreased methylation, DMMSs within the 3’ UTR had the highest fold change (Figure 3C), demonstrating the location preferences of methylation and demethylation within the genome.
FIGURE 3.
Distribution of differentially methylated N6-methyladenosine sites. (A) Relative occupancy of differentially methylated m6A sites in each chromosome normalized by length in IvC. (B) Pie chart showing the percentage of DMM peaks in five non-overlapping segments in IvC. (C) Statistics of fold change of DMM peaks in five segments. The histogram shows the mean of the fold change in IvC. Error bars represent the standard error of the mean. (D) Relative occupancy of differentially methylated m6A sites in each chromosome normalized by length in TvI. (E) Pie chart showing the percentage of DMM peaks in five non-overlapping segments in TvI. (F) Statistics of fold change of DMM peaks in five segments. The histogram shows the mean of the fold change in TvI. Error bars represent the standard error of the mean. (G) Relative occupancy of differentially methylated m6A sites in each chromosome normalized by length. (H) Pie chart showing the percentage of DMM peaks in five non-overlapping segments.
In TvI comparisons, 526 DMMSs were identified within 420 genes, of which 66.73% (351/526) were sites with increased methylation. The top 20 m6A sites with the most increased and decreased methylation are shown in Table 2.
TABLE 2.
The top 20 differently methylated m6A peaks in TvI.
| Gene name | Gene ID | Fold change | Regulation | Chromosome | Peak start | Peak end | Peak length | p-value |
| Syt15 | 319508 | 63.6 | Up | chr14 | 34228038 | 34228320 | 282 | 1.59789E-06 |
| Celf5 | 319586 | 48.7 | Up | chr10 | 81469443 | 81469523 | 80 | 1.58151E-06 |
| Cav3 | 12391 | 46.9 | Up | chr6 | 112459821 | 112459925 | 104 | 3.0379E-06 |
| Npffr2 | 104443 | 44.1 | Up | chr5 | 89582741 | 89583200 | 459 | 5.67547E-06 |
| Kcnf1 | 382571 | 19.08571429 | Up | chr12 | 17174961 | 17175260 | 299 | 3.61832E-06 |
| Amy2a4 | 100043684 | 18.27027027 | Up | chr3 | 113279836 | 113279860 | 24 | 1.19296E-06 |
| Metap2 | 56307 | 18.03125 | Up | chr10 | 93858488 | 93858660 | 172 | 2.64599E-06 |
| Tbc1d10c | 108995 | 17.81081081 | Up | chr19 | 4184361 | 4184740 | 379 | 4.61772E-06 |
| Cx3cr1 | 13051 | 16.29090909 | Up | chr9 | 120051221 | 120051460 | 239 | 1.12074E-09 |
| Bhlhe22 | 59058 | 16.16216216 | Up | chr3 | 18055801 | 18056180 | 379 | 1.39035E-06 |
| Fgg | 99571 | 12.77879342 | Down | chr3 | 83008647 | 83008741 | 94 | 2.1945E-09 |
| Fgg | 99571 | 11.67778106 | Down | chr3 | 83010063 | 83010180 | 117 | 1.6791E-09 |
| Dvl3 | 13544 | 7.87254902 | Down | chr16 | 20522461 | 20522660 | 199 | 8.52622E-06 |
| Fgb | 110135 | 7.239726027 | Down | chr3 | 83049701 | 83049803 | 102 | 2.426E-08 |
| Slc2a5 | 56485 | 6.895348837 | Down | chr4 | 150143641 | 150143960 | 319 | 9.64308E-09 |
| Gm21994 | 102637277 | 6.525504152 | Down | chr2 | 150254517 | 150255160 | 643 | 3.39444E-07 |
| Fga | 14161 | 5.909136445 | Down | chr3 | 83031021 | 83031620 | 599 | 1.55539E-08 |
| Fga | 14161 | 5.251931475 | Down | chr3 | 83026152 | 83026260 | 108 | 8.18498E-12 |
| Mpped1 | 223726 | 5.03943662 | Down | chr15 | 83856361 | 83856840 | 479 | 6.39334E-11 |
| Calcb | 116903 | 4.906439854 | Down | chr7 | 114721974 | 114722161 | 187 | 6.45098E-12 |
Chromosomes 3, 1, 9, and 4 were the top four chromosomes harboring the most DMMSs (Supplementary Figure 2B). The top four with the highest relative densities were 9, 19, 18, and 3 (Figure 3D). Most DMMSs were within a CDS (Figure 3E). DMMSs within the 3’ UTR had the highest fold change among sites with increased methylation, while those near the start codon had the highest fold change among sites with decreased methylation (Figure 3F).
We further analyzed DMMSs with contrary methylation trends in IvC and TvI. In total, 94 DMMSs showed opposite trends. Of these, 42.55% (40/94) exhibited significantly increased methylation in IvC but decreased methylation in TvI.
Chromosomes 1, 2, 11, and 10 were the top four chromosomes harboring the most DMMSs (Supplementary Figure 3). The top four with the highest relative densities were 16, 10, 11, and 3 (Figure 3G). Most DMMSs were within a CDS (Figure 3H).
Differentially Methylated RNAs Are Involved in Important Biological Pathways
To explore the role of m6A in different groups, GO enrichment analyses and KEGG pathway analyses were used to analyze all protein coding genes containing DMMSs.
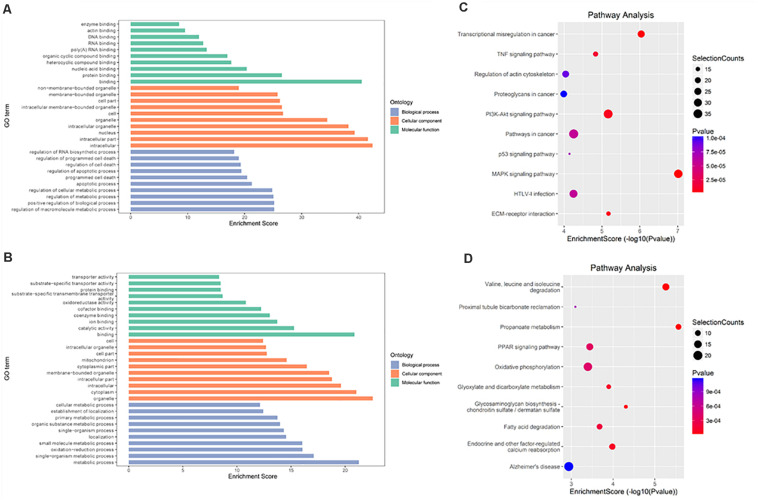
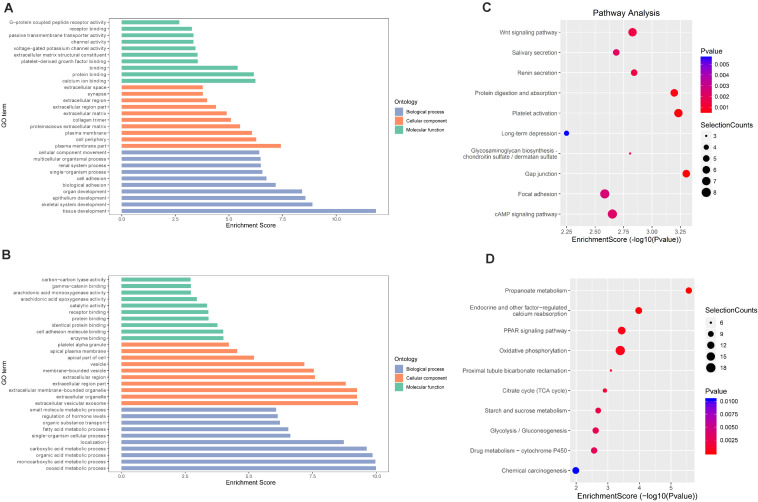
In IvC (Figure 4), for the BP category, genes with increased methylation of m6A sites were significantly (p < 0.05) enriched in the regulation of metabolic processes and cell death processes, such as macromolecule metabolic, cellular metabolic, and apoptotic processes (Figure 4A); genes with decreased methylation were highly enriched in metabolic processes, oxidation–reduction processes, transport, transmembrane transport, and others (Figure 4B). Regarding pathways, the former were significantly involved in apoptosis-associated pathways (e.g., TNF, MAPK, P53 signaling pathways) while the latter were involved in propanoate metabolism, oxidative phosphorylation, and others (Figures 4C,D). For the CC category, genes containing DMMSs were mainly enriched in the intracellular organelles, cytoplasm, and nucleus intracellular membrane-bound organelles. For the MF category, upregulation of m6A was notably enriched in genes involved in enzyme binding, actin binding, DNA binding, and RNA compound binding, while loss of m6A methylation was enriched in genes involved in catalytic activity, ion binding, coenzyme binding, and cofactor binding (Figures 4A,B). These results suggest that m6A has complicated roles in CI-AKI, with primary roles in metabolism, various pathways related to cell death, and oxidation.
FIGURE 4.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses of coding genes containing DMMSs in IvC. (A) Major gene ontology terms were enriched for the genes containing up-regulated m6A sites in IvC. (B) Major gene ontology terms were enriched for the genes containing down-regulated m6A sites in IvC. (C) Major enriched pathways for the genes containing up-regulated m6A sites in IvC. (D) Major enriched pathways for the genes containing down-regulated m6A sites in IvC.
In TvI (Figure 5), for the BP category, genes with increased methylation of m6A sites were significantly (p < 0.05) enriched in some development-associated processes, such as tissue, skeletal system, epithelium, and organ development (Figure 5A), while those with decreased methylation were highly enriched in acid metabolism processes, such as the oxoacid, organic acid, and carboxylic acid metabolic processes (Figure 5B). Regarding pathways, the former were significantly involved in gap junctions, protein digestion and absorption, and the Wnt pathway, while the latter were involved in metabolic processes, such as the PPAR signaling pathway (Figures 5C,D). For the CC category, genes containing DMMSs induced by cisplatin-induced AKI were mainly enriched in plasma membrane, cell periphery extracellular vesicular exosome, and extracellular organelles. For the MF category, increased methylation of m6A was notably enriched in calcium ion binding, protein binding, and channel activity, while decreased methylation of m6A was enriched in enzyme binding, cell adhesion molecule binding, and identical protein binding (Figures 5A,B). These results suggest that m6A methylation is involved in berberine alleviating CI-AKI in mouse kidneys.
FIGURE 5.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses of coding genes containing DMMSs in TvI. (A) Major gene ontology terms were enriched for the genes containing up-regulated m6A sites in TvI. (B) Major gene ontology terms were enriched for the genes containing down-regulated m6A sites in TvI. (C) Major enriched pathways for the genes containing up-regulated m6A sites in TvI. (D) Major enriched pathways for the genes containing down-regulated m6A sites in TvI.
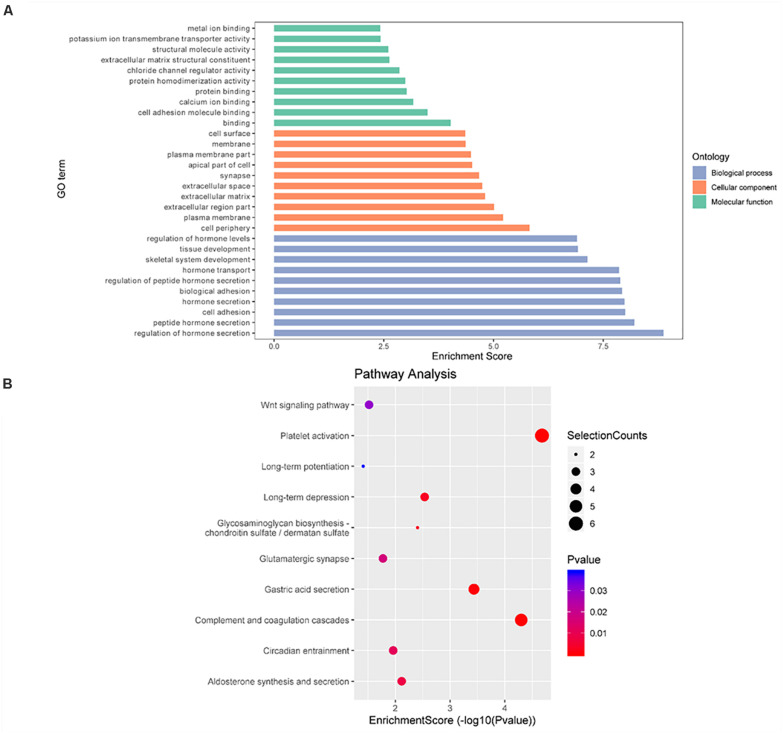
Finally, GO enrichment and KEGG pathway analyses were conducted on genes containing m6A sites showing opposite methylation trends in IvC and TvI (Figure 6). For the BP category, genes with DMMSs were significantly (p < 0.05) enriched in hormone secretion processes, such as regulation of hormone secretion and peptide hormone secretion (Figure 6A). Pathway analyses demonstrated that genes with DMMSs were involved in platelet activation, complement and coagulation cascades, and gastric acid secretion (Figure 6B). For the CC category, genes containing DMMSs were mainly enriched in the cell periphery, plasma membrane, and extracellular matrix. For the MF category, genes were enriched in binding, cell adhesion molecule binding, and calcium ion binding (Figure 6A). These results suggest that berberine might resist the nephrotoxicity of cisplatin through different pathways.
FIGURE 6.
Gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses of coding genes containing DMMSs. (A) Major gene ontology terms were enriched for the genes containing m6A sites with reverse methylation trend between IvC and TvI. (B) Major enriched pathways for the genes containing m6A sites with reverse trend between IvC and TvI.
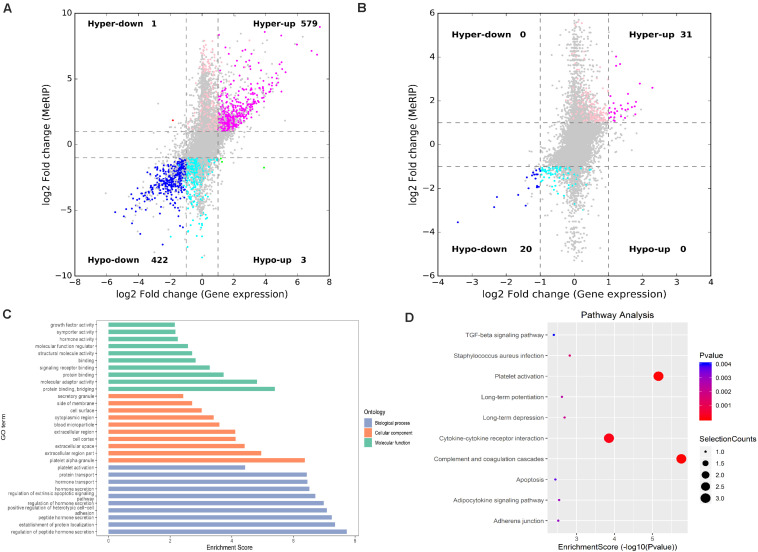
Conjoint Analyses of m6A Modification and Gene Regulation
RNA-seq was used to detect DEGs among the groups. In IvC, 4,469 genes were differentially expressed (fold change ≥ 2 and p < 0.05), including 1,655 downregulated genes and 2,814 upregulated genes (Figure 7). Conjoint analyses of DMGs and DEGs resulted in four groups of genes: 576 hypermethylated and upregulated genes, 422 hypomethylated and downregulated genes, 1 hypermethylated but downregulated gene, and 3 hypomethylated but upregulated genes (Figure 7A). In TvI, 350 genes were differentially expressed, including 84 downregulated genes and 266 upregulated genes. Conjoint analyses revealed 31 hypermethylated and upregulated genes, 20 hypermethylated but downregulated genes, and no hypomethylated upregulated or hypomethylated downregulated genes (Figure 7B).
FIGURE 7.
Conjoint analysis of differentially methylated genes and differentially expressed genes. (A) Four-quadrant graph exhibiting the DEGs containing differentially methylated m6A peaks in IvC. (B) Four-quadrant graph exhibiting the DEGs containing differentially methylated m6A peaks in TvI. (C) Major gene ontology terms were significantly enriched for the genes containing m6A sites with reverse methylation and expression trends between IvC and TvI. (D) Major enriched pathways for the genes containing m6A sites with reverse methylation and expression trends between IvC and TvI.
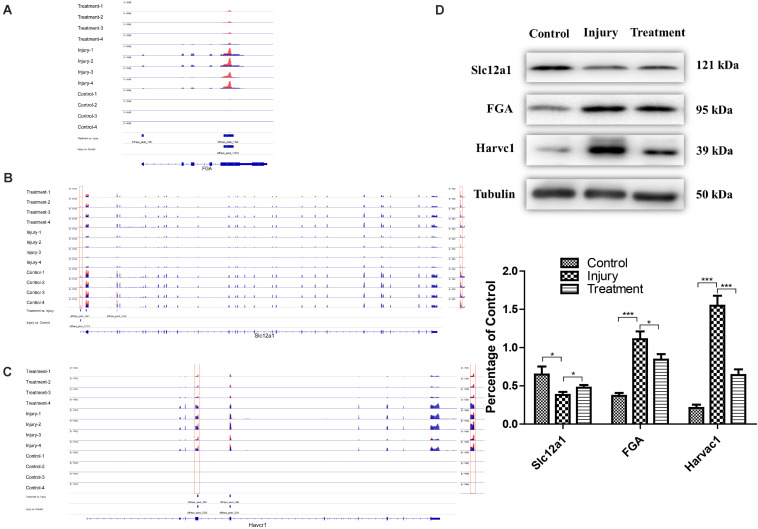
To further analyze the role of m6A in alleviating the action of berberine, genes with opposite methylation and expression trends in IvC and TvI were selected (Table 3). Overall, 9 were hypermethylated and upregulated in IvC but hypomethylated and downregulated in TvI, and 12 were hypomethylated and downregulated in IvC but hypermethylated and upregulated in TvI. GO and pathway analyses were performed to uncover the biological processes associated with these genes (Figure 7C). These genes were found to be highly enriched in complement and coagulation cascades, platelet activation, and cytokine–cytokine receptor interaction (Figure 7D). For instance, FGA/FGB/FGG genes encoding fibrinogen (Martini et al., 2019; Vilar et al., 2020) were hypermethylated and upregulated in IvC but were hypomethylated and downregulated in TvI (Figure 8A). Slc12a1, the key gene that mediates the electroneutral movement of Na(+) and K(+) across cell membranes (Schiessl et al., 2013; Xue et al., 2014; Hao et al., 2018; Kawaguchi et al., 2018), was hypomethylated and downregulated in IvC but hypermethylated and upregulated in TvI (Figure 8B). Havcr1, also known as Kim-1, is a well-known biomarker for kidney injury (Lippi et al., 2018; Seibert et al., 2018; Song et al., 2019; Zdziechowska et al., 2020). Cisplatin induced hypermethylation and upregulation of Havcr1, while berberine reversed these effects (Figure 8C).
TABLE 3.
Genes with contrary methylation and expression trends between IvC and TvI.
| Gene name | Gene ID | m6A regulation & gene regulation in IvC | m6A regulation & gene regulation in TvI | Chromosome | Peak start | Peak end | P-value in IvC | FDR in IvC | P-value in TvI | FDR in TvI |
| Akap12 | 83397 | Up | Down | chr10 | 4353141 | 4353240 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Amica1 | 270152 | Up | Down | chr9 | 45107681 | 45108040 | 0.00335 | 0.052867 | 2.00E-04 | 0.000571614 |
| Fga | 14161 | Up | Down | chr3 | 83031021 | 83031620 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Fgb | 110135 | up | Down | chr3 | 83042246 | 83042340 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Fgg | 99571 | Up | Down | chr3 | 83010063 | 83010180 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Havcr1 | 171283 | Up | Down | chr11 | 46756128 | 46756260 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Inhbb | 16324 | Up | Down | chr1 | 119422161 | 119422248 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Lep | 16846 | Up | Down | chr6 | 29073261 | 29073500 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Mlph | 171531 | Up | Down | chr1 | 90950081 | 90950420 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Egfl6 | 54156 | Down | Up | chrX | 166538413 | 166538734 | 5.00E-05 | 0.000154677 | 5.00E-05 | 0.00196624 |
| Gbp4 | 17472 | Down | Up | chr5 | 105118221 | 105118499 | 5.00E-05 | 0.000154677 | 0.00195 | 0.0357303 |
| Gnaz | 14687 | Down | Up | chr10 | 75015621 | 75015980 | 2.00E-04 | 0.00635985 | 0.00045 | 0.00121049 |
| Ppp1r1a | 58200 | Down | Up | chr15 | 103537841 | 103537992 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Ranbp3l | 223332 | Down | Up | chr15 | 8967948 | 8968040 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Slc12a1 | 20495 | Down | Up | chr2 | 125152504 | 125152633 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Slc16a7 | 20503 | Down | Up | chr10 | 125230591 | 125230700 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Snai2 | 20583 | Down | Up | chr16 | 14708181 | 14708720 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Tfap2b | 21419 | Down | Up | chr1 | 19212053 | 19212140 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Tmem207 | 100043057 | Down | Up | chr16 | 26504161 | 26504620 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Tnfsf10 | 22035 | Down | Up | chr3 | 27342154 | 27342427 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
| Tril | 66873 | Down | Up | chr6 | 53818361 | 53819000 | 5.00E-05 | 0.00196624 | 5.00E-05 | 0.000154677 |
FIGURE 8.
Representative genes with opposite m6A methylation and expression trends between IvC and TvI. (A) Visualization of m6A-modified genes FGA in the control group, injury group, and treatment group. (B) Visualization of m6A modified genes Slc12a1 in the control group, injury group, and treatment group. (C) Visualization of m6A-modified genes Havcr1 in the control group, injury group, and treatment group. (D) Western blot images of FGA, Slc12a1, and Havcr1 in control group, injury group, and Havcr1 group. Error bars represent the standard deviation. *p < 0.05, ***p < 0.001, Student’s t-test.
Western Blotting Analyses of Protein Expression in FGA, Slc12a1, and Havcr1
Western blotting was conducted to detect the protein levels of FGA, Slc12a1, and Havcr1 in each group. Cisplatin induced an increase in Slc12a1 protein levels and a decrease in FGA and Havcr1 protein levels. However, berberine pretreatment reversed these effects (Figure 8D).
Discussion
m6A methylation is considered a reversible dynamic modification in many species. Previous studies have shown that m6A modification can regulate cellular responses to stimuli by affecting mRNA transcription, splicing, localization, translation, stability, and posttranscriptional regulation of gene expression at the RNA level (Zhou et al., 2015; Fry et al., 2017). Evidence (Anders et al., 2018; Wang Y. et al., 2019; Zhou P. et al., 2019; Li et al., 2020; Liu et al., 2020; Xu et al., 2020) also suggests a strong relationship between m6A modification and kidney disease, thereby revealing an important biological role for m6A in the regulation of kidney injury.
In this study, a reliable cisplatin-induced AKI model was established in mice, and the model was tested by analyzing Scr, BUN, and kidney section images. The kidney injury resulted in dynamic m6A modifications and gene expression in the kidneys. Differentially methylated mRNAs were found to be involved in many biological pathways. m6A methylation was enriched in the CDS in all groups. In IvC, GO, and KEGG analyses of coding genes harboring DMMSs demonstrated that genes with increased methylation were primarily enriched in the pathways related to metabolic processes and cell death process, such as macromolecule metabolic processes, cellular metabolic processes, the TNF signaling pathway, and the MAPK signaling pathway, while decreases in methylation were mainly enriched in metabolic processes, oxidation, and transport, indicating that cisplatin induced complicated variation in m6A methylation. Several pathways have been thoroughly studied as factors affecting CI-AKI. However, we found that variation in metabolic processes constituted the foundation of the biological and pathological changes in CI-AKI.
Different m6A methylation sites were also detected in TvI. GO, and KEGG analyses of coding genes with DMMSs revealed that the genes with increases in methylation were primarily enriched in pathways associated with tissue, the skeletal system, and epithelium development, such as gap junctions and the Wnt signaling pathway. Genes with decreases in methylation were mainly enriched in the oxoacid metabolic process and organic metabolic process. These results provide evidence of a link between m6A and berberine-mediated regulation of kidney injury, showing that berberine attenuated CI-AKI by increasing the methylation of genes associated with tissue, the skeletal system, and epithelium development. These results should prove useful for directing future research into the underlying medical value of berberine in CI-AKI.
Furthermore, 94 DMMSs showed opposite methylation trends in IvC and TvI. The genes with those DMMSs were primarily enriched in pathways associated with hormone secretion and complement and coagulation cascades, which implies that berberine probably relieves CI-AKI by directly reversing some changes in m6A methylation.
Finally, conjoint analyses of m6A modifications and gene regulation provided a broad picture of CI-AKI and the impacts of berberine pretreatment. In total, 21 genes were found to show opposite m6A methylation and expression trends in IvC and TvI. Among these candidate genes, several drew our attention. For instance, FGA/FGB/FGG are the core genes encoding fibrinogen. Cisplatin may cause endothelial injury and inflammation, which can activate coagulation cascades (Ozkok and Edelstein, 2014). Thus, FGA/FGB/FGG were hypermethylated and upregulated in CI-AKI. However, berberine pretreatment significantly alleviated these trends. This result is consistent with one previous study (Riccioni et al., 2018). Slc12a1, also known as NKCC2, is the molecular target of loop diuretics, as it is expressed on the apical membrane of the thick ascending limb of Henle epithelial cells (Hao et al., 2018). It mediates the electroneutral movement of Na(+) and K(+), which is tightly coupled to the movement of Cl(–) across cell membranes (Schiessl et al., 2013; Xue et al., 2014; Kawaguchi et al., 2018). In our study, Slc12a1 was hypomethylated and downregulated by cisplatin. Berberine pretreatment maintained transmembrane ion exchange by hypermethylating and upregulating Slc12a1. Havcr1, also known as KIM-1, is a promising biomarker for predicting kidney injury (Lippi et al., 2018; Seibert et al., 2018; Song et al., 2019; Zdziechowska et al., 2020). It may reduce acute injury by mediating phagocytosis (Yang et al., 2015). In our study, it was hypermethylated and upregulated in CI-AKI. However, berberine reversed these trends. Consistently, the protein expression levels of FGA, Slc12a1, and KIM-1 were similar to the m6A methylation and gene expression levels. These results suggest that berberine has a protective effect in CI-AKI through different pathways.
Conclusion
We characterized the differential m6A methylome in normal, CI-AKI, and berberine-pretreated CI-AKI in mice. Our results suggest a strong association between m6A methylation and the regulation of CI-AKI. In addition, the candidate genes identified reveal the possible pathways by which berberine alleviates kidney injury. Altogether, our results provide a fundamental contribution to research into the mechanisms of and novel therapies for CI-AKI.
Data Availability Statement
The datasets generated for this study can be found in (US National Center for Biotechnology Information Gene Expression Omnibus). The accession ID is GSE157261 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157261).
Ethics Statement
The animal study was reviewed and approved by The Institutional Animal Care and Use Committee of Shanghai Jiaotong University affiliated Renji Hospital.
Author Contributions
ZN and XC contributed to design the research. JS, WW, and XS conducted the experiments. JS and WW analyzed and interpreted the data, and drafted the manuscript. JW and SL helped to polish the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to BioDataStudio (Shanghai) for the assistance of drawing figures. We are also grateful to Drs. Chaojun Qi and Wenyan Zhou for the assistance in pathobiology.
Abbreviations
- m6A
N6-methyladenosine
- CI-AKI
cisplatin-induced acute kidney injury
- IvC
injury group vs. control group
- TvI
treatment group vs. injury group
- YTH
YT521-B homology
- DEGs
differentially expression genes, negative control; i.p. injected intraperitoneally
- Scr
serum creatinine
- BUN
blood urea nitrogen
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- CDS
coding sequence
- MACS
Model-based Analysis of ChIP-Seq
- DMMSs
differentially methylated m6A sites.
- H-U
hyper-methylated and upregulated, 422 hypo-methylated and downregulated (hypo-down) genes, one hyper-methylated but down-regulated (hyper-down) genes and 3 hypo-methylated but up-regulated (hypo-up).
Funding. This work was supported in part by grants from the National Natural Science Foundation of China (No. 81700586); Key Shanghai Laboratory of Nucleic Acid Chemistry and Nanomedicine (2020ZYB009); Shanghai Municipal Planning Commission of science and Research [No. ZY(2018-2020)-FWTX-1001)]; Medical Scientific Research Project of Health and Family Planning Commission of Jiangsu Province (No. Z2018026).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.584460/full#supplementary-material
Venn diagram showing the overlap of m6A peaks within mRNAs in three groups.
Chromosomal distribution of all DMM sites. (A) Chromosomal distribution of all DMM sites in IvC. (B) Chromosomal distribution of all DMM sites in TvI.
Chromosomal distribution of DMM sites presenting contrary methylation trend between IvC and TvI.
References
- Ahmad S., Hussain A., Hussain A., Abdullah I., Ali M. S., Froeyen M., et al. (2019). Quantification of berberine in Berberis vulgaris L. root extract and its curative and prophylactic role in cisplatin-induced in vivo toxicity and in vitro cytotoxicity. Antioxidants 8:185. 10.3390/antiox8060185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders M., Chelysheva I., Goebel I., Trenkner T., Zhou J., Mao Y., et al. (2018). Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1:e201800113. 10.26508/lsa.201800113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliceti C., Franco P., Spinozzi S., Roda A., Cicero A. F. (2016). Berberine: new insights from pharmacological aspects to clinical evidences in the management of metabolic disorders. Curr. Med. Chem. 23 1460–1476. 10.2174/0929867323666160411143314 [DOI] [PubMed] [Google Scholar]
- Chen H. Y., Ye X. L., Cui X. L., He K., Jin Y. N., Chen Z., et al. (2012). Cytotoxicity and antihyperglycemic effect of minor constituents from Rhizoma Coptis in HepG2 cells. Fitoterapia 83 67–73. 10.1016/j.fitote.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 7 1–6. 10.1093/gigascience/gix120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings B. S., Schnellmann R. G. (2002). Cisplatin-induced renal cell apoptosis: caspase 3-dependent and -independent pathways. J. Pharmacol. Exp. Ther. 302 8–17. 10.1124/jpet.302.1.8 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Domitrović R., Jakovac H., Marchesi V. V., Blažeković B. (2013). Resolution of liver fibrosis by isoquinoline alkaloid berberine in CCl4-intoxicated mice is mediated by suppression of oxidative stress and upregulation of MMP-2 expression. J. Med. Food 16 518–528. 10.1089/jmf.2012.0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y., Liu T., Zhou Y., Dou T., Yang Q. (2018). Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine. J. Biol. Chem. 293 15429–15438. 10.1074/jbc.RA118.004351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R. K., Kondeti V. K., Sharma I., Chandel N. S., Quaggin S. E., Kanwar Y. S. (2017). Beneficial effects of myo-inositol oxygenase deficiency in cisplatin-induced AKI. J. Am. Soc. Nephrol. 28 1421–1436. 10.1681/asn.2016070744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Liu L., Wu Z., Cao M. (2019). Combating neurodegenerative diseases with the plant alkaloid berberine: molecular mechanisms and therapeutic potential. Curr. Neuropharmacol. 17 563–579. 10.2174/1570159x16666180419141613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry N. J., Law B. A., Ilkayeva O. R., Holley C. L., Mansfield K. D. (2017). N(6)-methyladenosine is required for the hypoxic stabilization of specific mRNAs. RNA 23 1444–1455. 10.1261/rna.061044.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George B., Joy M. S., Aleksunes L. M. (2018). Urinary protein biomarkers of kidney injury in patients receiving cisplatin chemotherapy. Exp. Biol. Med. 243 272–282. 10.1177/1535370217745302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S., Hao M., Ferreri N. R. (2018). Renal-specific silencing of TNF (tumor necrosis factor) unmasks salt-dependent increases in blood pressure via an NKCC2A (Na(+)-K(+)-2Cl(-) cotransporter isoform A)-dependent mechanism. Hypertension 71 1117–1125. 10.1161/hypertensionaha.117.10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., et al. (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holditch S. J., Brown C. N., Lombardi A. M., Nguyen K. N., Edelstein C. L. (2019). Recent advances in models, mechanisms, biomarkers, and interventions in cisplatin-induced acute kidney injury. Int. J. Mol. Sci. 20:3011. 10.3390/ijms20123011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Keren Zhou K., Wu T., Zhao B., Sun M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 567 414–419. 10.1038/s41586-019-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humanes B., Camaño S., Lara J. M., Sabbisetti V., González-Nicolás M., Bonventre J. V., et al. (2017). Cisplatin-induced renal inflammation is ameliorated by cilastatin nephroprotection. Nephrol. Dial. Transplant. 32 1645–1655. 10.1093/ndt/gfx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K., Hatano R., Matsubara M., Asano S. (2018). Internalization of NKCC2 is impaired in thick ascending limb of Henle in moesin knockout mice. Pflugers Arch. 470 1055–1068. 10.1007/s00424-018-2134-z [DOI] [PubMed] [Google Scholar]
- Kechin A., Boyarskikh U., Kel A., Filipenko M. (2017). cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24 1138–1143. 10.1089/cmb.2017.0096 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebwohl D., Canetta R. (1998). Clinical development of platinum complexes in cancer therapy: an historical perspective and an update. Eur. J. Cancer 34 1522–1534. 10.1016/s0959-8049(98)00224-x [DOI] [PubMed] [Google Scholar]
- Leemans J. C., Stokman G., Claessen N., Rouschop K. M., Teske G. J., Kirschning C. J., et al. (2005). Renal-associated TLR2 mediates ischemia/reperfusion injury in the kidney. J. Clin. Invest. 115 2894–2903. 10.1172/JCI22832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Tong J., Zhu S., Batista P., Duffy E., Zhao J., et al. (2017). m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 338–342. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang J., Huang C., Shen M., Zhan H., Xu K. (2020). RNA N6-methyladenosine: a promising molecular target in metabolic diseases. Cell. Biosci. 10:19 10.1186/s13578-020-00385-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi I., Perondi F., Meucci V., Bruno B., Gazzano V., Guidi G. (2018). Clinical utility of urine kidney injury molecule-1 (KIM-1) and gamma-glutamyl transferase (GGT) in the diagnosis of canine acute kidney injury. Vet. Res. Commun. 42 95–100. 10.1007/s11259-018-9711-7 [DOI] [PubMed] [Google Scholar]
- Liu P., Zhang B., Chen Z., He Y., Du Y., Liu Y., et al. (2020). m(6)A-induced lncRNA MALAT1 aggravates renal fibrogenesis in obstructive nephropathy through the miR-145/FAK pathway. Aging 12 5280–5299. 10.18632/aging.102950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Zhen X., Zhu F., Hu Z., Lei W., Li S., et al. (2017). Hyperhomocysteinemia exacerbates cisplatin-induced acute kidney injury. Int. J. Biol. Sci. 13 219–231. 10.7150/ijbs.16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zhang Z., Tai L., Zhang L., Sun Z., Zhou L. (2019). Comprehensive analysis of differences of N(6)-methyladenosine RNA methylomes between high-fat-fed and normal mouse livers. Epigenomics 11 1267–1282. 10.2217/epi-2019-0009 [DOI] [PubMed] [Google Scholar]
- Martini F., Cecconi N., Paolicchi A., Galimberti S., Cervetti G., Buda G., et al. (2019). Interference of monoclonal gammopathy with fibrinogen assay producing spurious dysfibrinogenemia. TH Open 3 e64–e66. 10.1055/s-0039-1683969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha S., Venkataraman B., Kurdi A., Mahgoub E., Sadek B., Rajesh M. (2016). Plant-derived agents for counteracting cisplatin-induced nephrotoxicity. Oxid. Med. Cell Longev. 2016:4320374. 10.1155/2016/4320374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarerin-George A. O., Jaffrey S. R. (2017). MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics 33 1563–1564. 10.1093/bioinformatics/btx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkok A., Edelstein C. L. (2014). Pathophysiology of cisplatin-induced acute kidney injury. Biomed. Res. Int. 2014:967826. 10.1155/2014/967826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccioni G., Gammone M. A., Currenti W., D’Orazio N. (2018). Effectiveness and safety of dietetic supplementation of a new nutraceutical on lipid profile and serum inflammation biomarkers in hypercholesterolemic patients. Molecules 23:1168. 10.3390/molecules23051168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H., Zhan Y. Y., Hou J., Xu B., Chen B., Tian Y., et al. (2017). Berberine binds RXRα to suppress β-catenin signaling in colon cancer cells. Oncogene 36 6906–6918. 10.1038/onc.2017.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B. D., Mahesh Kumar J., Sistla R. (2015). Baicalein, a bioflavonoid, prevents cisplatin-induced acute kidney injury by up-regulating antioxidant defenses and down-regulating the MAPKs and NF-κB pathways. PLoS One 10:e0134139. 10.1371/journal.pone.0134139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl I. M., Rosenauer A., Kattler V., Minuth W. W., Oppermann M., Castrop H. (2013). Dietary salt intake modulates differential splicing of the Na-K-2Cl cotransporter NKCC2. Am. J. Physiol. Renal Physiol. 305 F1139–F1148. 10.1152/ajprenal.00259.2013 [DOI] [PubMed] [Google Scholar]
- Seibert F. S., Sitz M., Passfall J., Haesner M., Laschinski P., Buhl M., et al. (2018). Prognostic value of urinary calprotectin, NGAL and KIM-1 in chronic kidney disease. Kidney Blood Press. Res. 43 1255–1262. 10.1159/000492407 [DOI] [PubMed] [Google Scholar]
- Shamsa F., Ahmadiani A., Khosrokhavar R. (1999). Antihistaminic and anticholinergic activity of barberry fruit (Berberis vulgaris) in the guinea-pig ileum. J. Ethnopharmacol. 64 161–166. 10.1016/s0378-8741(98)00122-6 [DOI] [PubMed] [Google Scholar]
- Shen L., Shao N. Y., Liu X., Maze I., Feng J., Nestler E. J. (2013). diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One 8:e65598. 10.1371/journal.pone.0065598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Yu J., Prayogo G. W., Cao W., Wu Y., Jia Z., et al. (2019). Understanding kidney injury molecule 1: a novel immune factor in kidney pathophysiology. Am. J. Transl. Res. 11 1219–1229. [PMC free article] [PubMed] [Google Scholar]
- Spitale R. C., Flynn R. A., Zhang Q. C., Crisalli P., Lee B., Jung J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519 486–490. 10.1038/nature14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Z. Y., Cheng X. L., Cai X. T., Yang Y., Sun X. Y., Xu J. D., et al. (2015). Ancient chinese formula qiong-yu-gao protects against cisplatin-induced nephrotoxicity without reducing anti-tumor activity. Sci. Rep. 5:15592. 10.1038/srep15592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar R., Fish R. J., Casini A., Neerman-Arbez M. (2020). Fibrin(ogen) in human disease: both friend and foe. Haematologica 105 284–296. 10.3324/haematol.2019.236901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ishfaq M., Xu L., Xia C., Chen C., Li J. (2019). METTL3/m(6)A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front. Pharmacol. 10:517. 10.3389/fphar.2019.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Feng X., Chai L., Cao S., Qiu F. (2017). The metabolism of berberine and its contribution to the pharmacological effects. Drug Metab. Rev. 49 139–157. 10.1080/03602532.2017.1306544 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z., Zhao J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16 191–198. 10.1038/ncb2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun J., Lin Z., Zhang W., Wang S., Wang W., et al. (2020). m6A mRNA methylation controls functional maturation in neonatal murine β-cells. Diabetes Metab. Res. Rev. 69 1708–1722. 10.2337/db19-0906 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zeng L., Liang C., Zan R., Ji W., Zhang Z., et al. (2019). Integrated analysis of transcriptome-wide m 6 A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics 11 693–1715. 10.2217/epi-2019-0262 [DOI] [PubMed] [Google Scholar]
- Xu Y., Yuan X. D., Wu J. J., Chen R. Y., Xia L., Zhang M., et al. (2020). The N6-methyladenosine mRNA methylase METTL14 promotes renal ischemic reperfusion injury via suppressing YAP1. J. Cell. Biochem. 121 524–533. 10.1002/jcb.29258 [DOI] [PubMed] [Google Scholar]
- Xue H., Zhang Z. J., Li X. S., Sun H. M., Kang Q., Wu B., et al. (2014). Localization and vasopressin regulation of the Na+-K+-2Cl– cotransporter in the distal colonic epithelium. World J. Gastroenterol. 20 4692–4701. 10.3748/wjg.v20.i16.4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Brooks C. R., Xiao S., Sabbisetti V., Yeung M. Y., Hsiao L. L., et al. (2015). KIM-1-mediated phagocytosis reduces acute injury to the kidney. J. Clin. Invest. 125 1620–1636. 10.1172/jci75417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimit A., Adebali O., Sancar A., Jiang Y. (2019). Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Nat. Commun. 10:309. 10.1038/s41467-019-08290-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdziechowska M., Gluba-Brzózka A., Franczyk B., Rysz J. (2020). Biochemical markers in the prediction of contrast-induced acute kidney injury. Curr. Med. Chem. [Epub ahead of print]. 10.2174/0929867327666200502015749 [DOI] [PubMed] [Google Scholar]
- Zhang J., Rudemiller N. P., Patel M. B., Wei Q., Karlovich N. S., Jeffs A. D., et al. (2016). Competing actions of type 1 angiotensin II receptors expressed on T lymphocytes and kidney epithelium during cisplatin-induced AKI. J. Am. Soc. Nephrol. 27 2257–2264. 10.1681/asn.2015060683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S. R., Qian S. B. (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. C., Wang J. Y., Hong B. A., Ma K. F., Xie H. B., Li L., et al. (2019). Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging 11 1633–1647. 10.18632/aging.101856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Wu M., Ye C., Xu Q., Wang L. (2019). Meclofenamic acid promotes cisplatin-induced acute kidney injury by inhibiting fat mass and obesity-associated protein-mediated m(6)A abrogation in RNA. J. Biol. Chem. 294 16908–16917. 10.1074/jbc.RA119.011009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk A., Bonventre J. V. (2016). Acute kidney injury. Annu. Rev. Med. 67 293–307. 10.1146/annurev-med-050214-013407 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn diagram showing the overlap of m6A peaks within mRNAs in three groups.
Chromosomal distribution of all DMM sites. (A) Chromosomal distribution of all DMM sites in IvC. (B) Chromosomal distribution of all DMM sites in TvI.
Chromosomal distribution of DMM sites presenting contrary methylation trend between IvC and TvI.
Data Availability Statement
The datasets generated for this study can be found in (US National Center for Biotechnology Information Gene Expression Omnibus). The accession ID is GSE157261 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157261).