See Cerri and Valente (doi:10.1093/brain/awaa324) for a scientific commentary on this article.
Ubiquinol-cytochrome c reductase core protein I (UQCRC1) is a complex III core protein of the mitochondrial electron transport chain. Lin et al. identify rare missense mutations in mitochondrial UQCRC1 that segregate with Parkinson’s disease, and show that they are functionally pathogenic.
Keywords: Parkinson’s disease, mitochondria, respiratory chain complex III, UQCRC1, ubiquinol-cytochrome c reductase core protein 1
Abstract
Parkinson’s disease is a neurodegenerative disorder with a multifactorial aetiology. Nevertheless, the genetic predisposition in many families with multi-incidence disease remains unknown. This study aimed to identify novel genes that cause familial Parkinson’s disease. Whole exome sequencing was performed in three affected members of the index family with a late-onset autosomal-dominant parkinsonism and polyneuropathy. We identified a novel heterozygous substitution c.941A>C (p.Tyr314Ser) in the mitochondrial ubiquinol-cytochrome c reductase core protein 1 (UQCRC1) gene, which co-segregates with disease within the family. Additional analysis of 699 unrelated Parkinson’s disease probands with autosomal-dominant Parkinson’s disease and 1934 patients with sporadic Parkinson’s disease revealed another two variants in UQCRC1 in the probands with familial Parkinson’s disease, c.931A>C (p.Ile311Leu) and an allele with concomitant splicing mutation (c.70-1G>A) and a frameshift insertion (c.73_74insG, p.Ala25Glyfs*27). All substitutions were absent in 1077 controls and the Taiwan Biobank exome database from healthy participants (n = 1517 exomes). We then assayed the pathogenicity of the identified rare variants using CRISPR/Cas9-based knock-in human dopaminergic SH-SY5Y cell lines, Drosophila and mouse models. Mutant UQCRC1 expression leads to neurite degeneration and mitochondrial respiratory chain dysfunction in SH-SY5Y cells. UQCRC1 p.Tyr314Ser knock-in Drosophila and mouse models exhibit age-dependent locomotor defects, dopaminergic neuronal loss, peripheral neuropathy, impaired respiratory chain complex III activity and aberrant mitochondrial ultrastructures in nigral neurons. Furthermore, intraperitoneal injection of levodopa could significantly improve the motor dysfunction in UQCRC1 p.Tyr314Ser mutant knock-in mice. Taken together, our in vitro and in vivo studies support the functional pathogenicity of rare UQCRC1 variants in familial parkinsonism. Our findings expand an additional link of mitochondrial complex III dysfunction in Parkinson’s disease.
See Cerri and Valente (doi:10.1093/brain/awaa324) for a scientific commentary on this article.
Introduction
Parkinson’s disease is a common neurodegenerative disorder affecting 7 to 10 million individuals worldwide (Pringsheim et al., 2014). The aetiology is multifactorial, arising from the interplay of genetic and environmental factors. While most patients appear to be sporadic, ∼10% have a family history of parkinsonism and numerous monogenic causes have now been identified (Hardy et al., 2009). The identification of genes causing rare familial forms of parkinsonism provides molecular insight into the underlying disease process (Trinh and Farrer, 2013). For example, missense and genomic multiplication mutations in SNCA lead to intra-neuronal misfolded α-synuclein that accumulates as Lewy bodies, whereas mutations in LRRK2, VPS35, Rab39B, and DNAJC family members have been linked to aberrant intracellular vesicle trafficking and protein recycling pathways (Ferreira and Massano, 2017). PINK1 is a mitochondrial-targeted ubiquitin kinase that regulates mitophagy together with parkin (PRKN) and also modulates neuroinflammation (Lazarou et al., 2015; Matheoud et al., 2019). Several other genes linked to familial parkinsonism include PARK7 (DJ1), CHCHD2 and VPS13A are also involved in mitochondrial function (Ferreira and Massano, 2017).
To extend our knowledge of the genetic architecture and pathophysiology of Parkinson’s disease, we performed whole exome sequencing and comparative analysis of three affected family members in a multi-incident pedigree with parkinsonism. Mutations in known genes for Parkinson’s disease were excluded but a novel, putatively pathogenic, substitution in mitochondrial ubiquinol-cytochrome c reductase core protein 1 (UQCRC1) was identified (Lin et al., 2019). Two additional potentially pathogenic variants were further found from 699 unrelated autosomal dominant familial Parkinson’s disease but none in 1077 ethnically matched control subjects. UQCRC1 is a subunit of complex III in the mitochondrial respiratory chain (Hoffman et al., 1993). The functional effect of heterozygous UQCRC1 mutations were subsequently examined in knock-in cell lines, Drosophila and mouse models, to assess their potential pathogenicity in the disease process.
Materials and methods
Human study
Participants
We enrolled 10 individuals from one Taiwanese multiplex family carrying a molecularly unassigned autosomal-dominant inherited Parkinson’s disease. Five of these individuals had Parkinson’s disease and five did not (Family A in Fig. 1A). An additional 699 unrelated families with molecularly unassigned inherited Parkinson’s disease (200 Taiwanese Parkinson’s disease families and 499 Japanese Parkinson’s disease families), 1934 patients with sporadic Parkinson’s disease (782 Taiwanese and 1152 Korean patients), and 1077 control subjects (597 Taiwanese and 480 Korean subjects) were recruited for candidate gene analysis. Pathogenic variants in 40 genes known to be associated with parkinsonism were excluded in the index patients by a targeted next-generation sequencing panel as previously described (Lin et al., 2019). All patients fulfilled the UK Brain Bank diagnostic criteria for Parkinson’s disease (Gelb et al., 1999). Parkinson’s disease symptom severity was evaluated using the Unified Parkinson’s Disease Rating Scale (UPDRS), and Hoehn and Yahr staging. Genomic DNA was isolated from peripheral blood following a standard protocol. The Ethical Review Boards of the participating institutions approved this study, including National Taiwan University Hospital, Seoul National University Hospital, and Jutendo University Hospital. All patients provided signed informed consent prior to enrolment.
Figure 1.
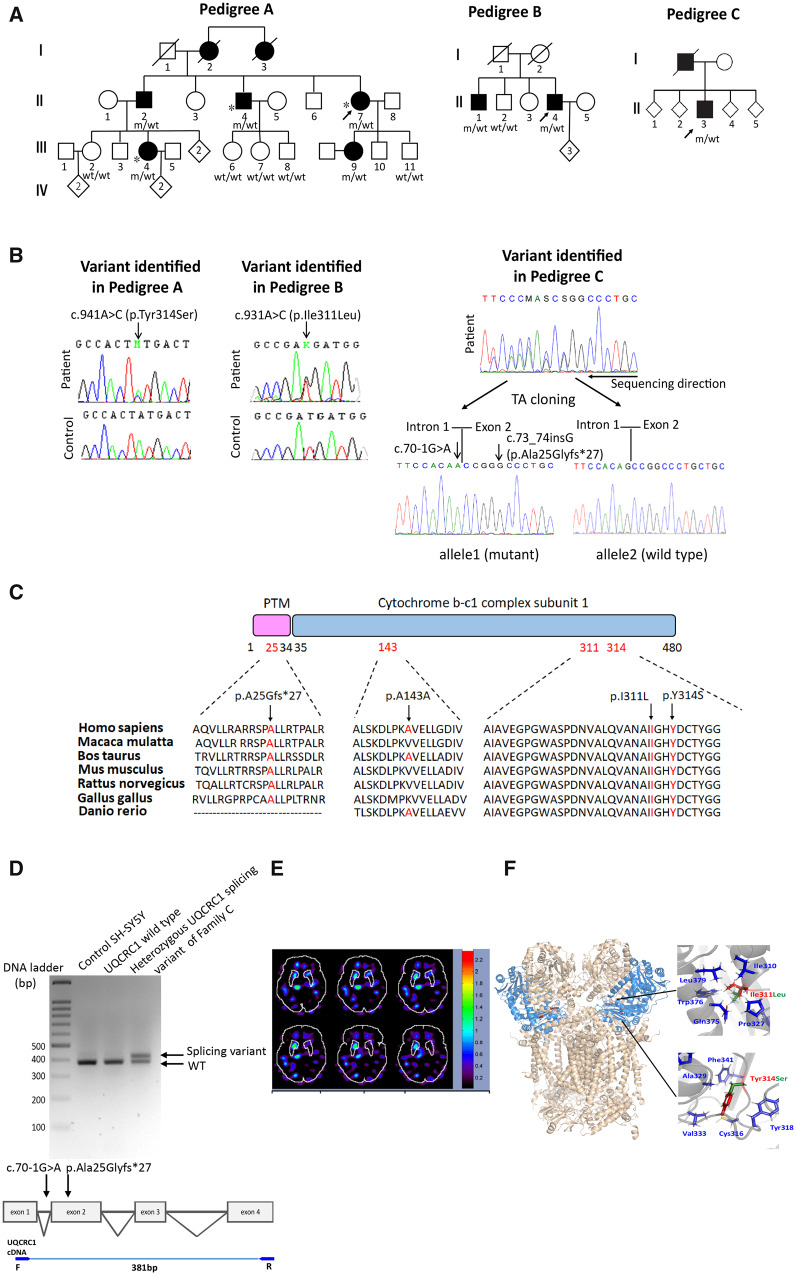
UQCRC1 mutations in families with Parkinson’s disease. (A) Pedigrees of three families with rare missense or splicing variants identified in the UQCRC1 gene. m/wt = heterozygous carriers of the UQCRC1 mutation; wt/wt = non-carriers; open symbols = unaffected; filled symbols = affected; symbol with a diagonal line = deceased; diamond = total number of children, unknown sex; arrow = proband. Asterisks indicate patients whose whole exomes were sequenced. (B) Sanger sequencing traces confirming the c.941A>C (p.Tyr314Ser), c.931A>C (p.Ile311Leu) and the allele with concomitant c.70-1G>A and c.73_73insG (p.Ala25Glyfs*27) variants identified in the proband of Family C (RefSeq NM_003365.2). (C) Alignment of multiple UQCRC1 orthologues shows conservation of the Tyr314, Ile311 and Ala25 residues across species. (D) RT-PCR showed aberrant splicing of UQCRC1 in mutant knock-in SH-SY5Y cells expressing the allele with concomitant c.70-1G>A and c.73_73insG (p.Ala25Glyfs*27) variants identified in the proband of Family C (top). The PCR was performed with the primers targeting cDNA of UQCRC1 (bottom). (E) Tc-99m TRODAT SPECT image shows dopamine transporter activity (red-orange regions) in the brain of Patient II : 4 of Family A. Note the asymmetric uptake due to reduced uptake in the left basal ganglia. The asymmetry of specific uptake ratio (SUR) of caudate, putamen and striatum were 58.3%, 39.1% and 43.8%, respectively (left to right ratio). (F) Left: The predicted protein structural model of human respiratory chain complex III protein. The UQCRC1 proteins are marked in blue and the Ile311 and Tyr314 residues are marked in red. Right: The relationships between the Ile311 (top) and Tyr314 (bottom) residues and the surrounding residues.
Genetic analysis
We performed whole exome sequencing in three affected members from Family A using the Ion Torrent™ Next-Generation Sequencing Exon v2 kit and platform (Life Technologies) for exome enrichment and sequencing. As parkinsonism in Family A appears to be inherited in an autosomal dominant manner, we identified heterozygous variants shared by all three patients and selected these for further analyses. We excluded genetic variants that did not alter coding sequences and variants with a minor allele frequency of >0.001 in control subjects in one or more reference databases, specifically the Single Nucleotide Polymorphism Database dbSNP144; https://www.ncbi.nlm.nih.gov/snp); 1000 Genomes Project (1000 Genomes Project Consortium et al., 2012); Exome Aggregation Consortium (ExAC version 0.3) (Lek et al., 2016); The Genome Aggregation Database (gnomAD, n = 123 136 exome and 15 496 whole-genome sequences; http://gnomad.broadinstitute.org/); and The Taiwan biobank (https://taiwanview.twbiobank.org.tw/index, n = 1517 exomes). Next, we performed Sanger sequencing on the remaining variants to determine whether any completely co-segregated with parkinsonism in Family A. Combined Annotation Dependent Depletion (CADD; https://cadd.gs.washington.edu/) scores were used to summarize how deleterious amino acid substitutions were to protein function (Adzhubei et al., 2010).
We then performed Sanger sequencing of all exons and exon-intron boundary junctions in the potential candidate gene in 699 unrelated probands with familial parkinsonism. We further screened for mutations in the potential candidate gene in an additional 1934 patients with sporadic Parkinson’s disease and 1077 controls to confirm the pathogenic role of the potential candidate gene in familial parkinsonism.
In vitro study
Generation and characterization of mutant UQCRC1 knock-in SH-SY5Y cell lines
We computationally identified the CRISPR/Cas91.1 target sites at the Zhang Lab website (http://crispr.mit.edu/) to generate mutant UQCRC1 knock-in human neuroblastoma SH-SY5Y cells having human mutations as previously described (Lin et al., 2019). Neurite lengths for differentiated SH-SY5Y cells stimulated by 24-h treatment of retinoic acid with individual genotype were manually quantified with ImageJ software (National Institutes of Health, Bethesda, Maryland, USA) (Lin et al., 2016).
Reverse transcription PCR
Reverse transcription PCR (RT-PCR) was performed in mutant knock-in SH-SY5Y cells expressing the allele with concomitant c.70-1G>A and c.73_73insG (p.Ala25Glyfs*27) variants identified in the proband of Family C. The primer sequences targeting cDNA of UQCRC1 were: F: 5′-GCCGGGGCACAAGTGCTAT-3′ and R: 5′-CTTGGACAGCGCCTTGATGT-3′.
Measurement of cellular respiration and individual mitochondrial respiratory chain complex activity
General mitochondrial respiration and activities of individual mitochondrial respiratory chain complex I–IV were measured in live wild-type or mutant SH-SY5Y knock-in cells in a Seahorse XFe24 extracellular flux analyzer (Seahorse Bioscience) as previously described (Salabei et al., 2014).
Measurement of reactive oxygen species production
Cells were washed twice with prewarmed phosphate-buffered saline (PBS) and incubated with 25 μM CM-H2DCFDA (Invitrogen™ C6827) for 20 min. Cells were then rinsed twice with prewarmed PBS and were examined by flow cytometry (BD FACSCalibur™).
In vivo study
Generation of mutant UQCRC1 knock-in Drosophila models
All Drosophila stocks were grown on standard culture medium at 22°C, under a natural light-dark cycle. The V5-tagged human transgenes UAS-UQCRC1WT-V5 and UAS-UQCRC1Y314S-V5 were generated with the PhiC31 integrase-mediated transgenesis system with an insertion at an estimated position of 25C6, at the attP40 site (BestGene, Inc.) (Markstein et al., 2008).
Human UQCRC1 constructs and CRISPR/Cas9-based fly UQCRC1 null mutant
To mimic heterozygous variants of UQCRC1 in patients, we used CRISPR/Cas9-based genome editing to knock-out an allele of endogenous fly UQCRC1 gene (CG3731) and created a heterozygous UQCRC1 null mutant. We then introduced either the human wild-type or UQCRC1 p.Tyr314Ser mutant cDNA to this heterozygous UQCRC1 null mutant background to establish a heterozygous UQCRC1 mutant fly model to mimic the human heterozygous mutation state.
We injected Drosophila UQCRC1-targeted single guide RNA (sgRNA) into a vas-Cas9(X) embryo as previously described to generate a CRISPR/Cas9-based targeted gene deletion (Bassett et al., 2013). We identified one knock-out allele of Drosophila UQCRC1 with a 4-bp deletion that caused a frameshift and early termination at amino acid 111 in the Drosophila UQCRC1 protein (Supplementary Fig. 2A). The pUASTattB-UQCRC1WT-V5 construct was generated by amplifying human UQCRC1 wild-type cDNA from a pGEM-UQCRC1 cDNA clone (HG16163-G, Sino Biological Inc.) by PCR. The primers were engineered to place EcoRI/KpnI restriction sites at both ends and a V5 tag on the C-terminal and the product was subcloned into a pUASTattB vector (New England Biolabs) (Groth et al., 2004). We then used the Actin-GAL4 driver to ubiquitously express wild-type human UQCRC1 (Act>UQCRC1WT) or the mutant UQCRC1 p.Tyr314Ser (Act>UQCRC1Y314S) in a heterozygous UQCRC1 null knock-out mutant genetic background (UQCRC1null/+) to serve as a wild-type or heterozygous mutant UQCRC1 knock-in Drosophila model.
Behavioural assays
Negative geotaxis and jumping assays
Climbing (negative geotaxis) and jumping assays were used to assess locomotor ability as previously described (Lin et al., 2010). Cohorts of 50 flies from each experimental group were subjected to the assay weekly. In brief, flies were placed in a vertical plastic column (length 25 cm, diameter 1.5 cm) for the climbing assay and tapped to the bottom of the column to start climbing. Climbing ability was defined as the ability to climb 8 cm in 5 s. For the jumping assay, flies were placed in a petri dish and flies that landed upside down when the dish was tapped had to right themselves. Jumping ability was defined as the ability of the fly to right itself by jumping. Both climbing and jumping activities were expressed as the percentage of flies that could climb or jump as defined.
Immunostaining of whole-mount dissected brains and neuromuscular junctions
Adult fly brains were dissected, whole-mounted, and immunostained with fluorescently labelled anti-tyrosine hydroxylase (TH) antibodies at 4 weeks of age for measuring the number of dopaminergic neurons as previously described (Lin et al., 2010). The third instar larvae from individual genotypes were dissected and immunostained for measuring the presynaptic and postsynaptic morphology of neuromuscular junctions (NMJs), as previously described (Tsai et al., 2018).
Generation of mutant UQCRC1 knock-in mice
For details on the generation of UQCRC1 knock-in mice, see the Supplementary material. PCR genotyping used the following primers: VF1: 5′-CCACGTGGCCATTGCAGTAGA-3′ and VR1: 5′- TCGATACTCATGGCATCACAGAC-3′, digesting the amplified product with BsrGI as the mutant allele has this restriction site (Fig. 4C).
Figure 4.
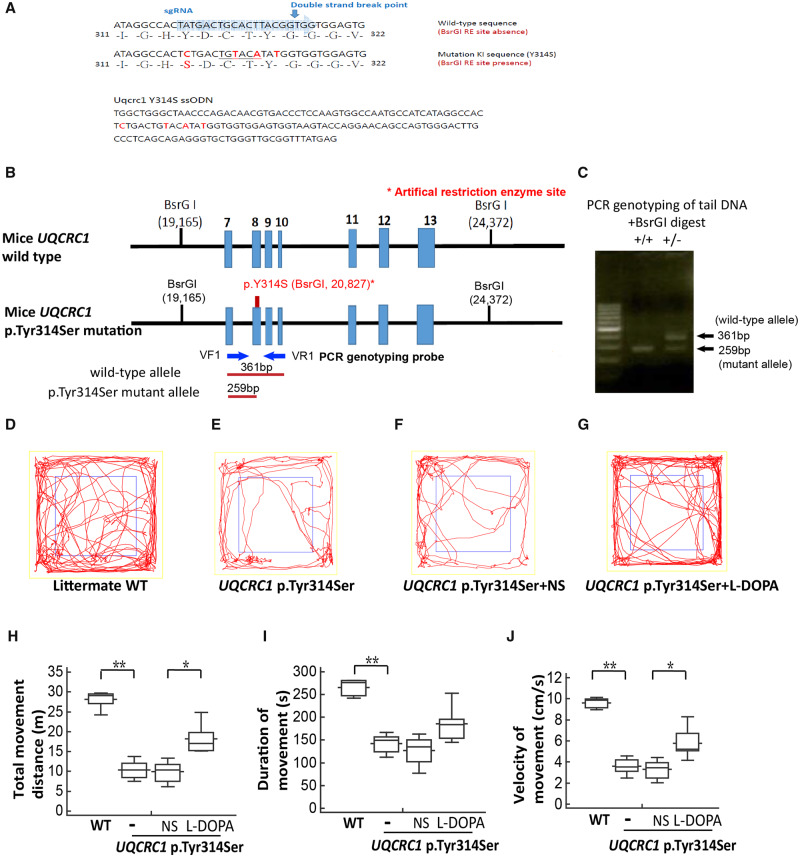
Genotype analysis and locomotor behaviour assay of UQCRC1 p.Y314 knock-in mice. (A) Single-stranded oligodeoxynucleotide for targeting mice UQCRC1 and Sanger sequencing of UQCRC1 p.Tyr314Ser sequence in knock-in mice. Top: The wild-type target mouse UQCRC1 gene sequence (blue); Bottom: The p.Tyr314Ser substitution sequence of the single-stranded oligodeoxynucleotide (ssODN) used for targeted mutagenesis in mice. Silent mutations were included to generate BsrGI restriction enzyme recognition sequences. (B) Gene targeting strategy. Top: Region of the mouse wild-type UQCRC1 gene that encompasses exons 7–13. Middle: Exons 7–13 are shown with the c.941A>C mutation (p.Tyr314Ser). The 5′ and 3′ diagnostic probes (blue arrows) were used in the PCR genotyping analysis. The artificial restriction enzyme sites recognized by the BsrGI enzyme are shown (black ticks) with their locations (bp) in the gene. Bottom: BsrGI restriction fragments predicted to be unique to the wild-type (361bp) and targeted (259 bp) alleles. (C) PCR genotyping of tail-blood DNA from mice of two genotypes, a mutant homozygote (+/+, left) and heterozygote (+/−; right), with a molecular weight marker. DNA samples were digested with BsrGI enzyme and PCR was performed with the following primers: VF1: 5′-CCACGTGGCCATTGCAGTAGA-3′ and VR1: 5′- TCGATACTCATGGCATCACAGAC-3′. (D–G) Trace plots of the open field locomotor tests of age and gender-matched littermate wild-type (WT) control (D), UQCRC1 p.Tyr314Ser knock-in mice (E), and UQCRC1 p.Tyr314Ser knock-in mice receiving normal saline (NS) (F) and l-DOPA treatment (G) at the age of 12 months. (H–J) Parameters of wild-type and UQCRC1 p.Tyr314Ser knock-in mouse locomotor activities with and without normal saline or l-DOPA treatment. (H) Total distance travelled of UQCRC1 p.Tyr314Ser knock-in mice versus control mice was 10.9 ± 0.8 m versus 28.9 ± 2.4 m; F(1,11) = 72.3, P < 0.01 by one-way ANOVA. The total movement distance of UQCRC1 p.Tyr314Ser knock-in mouse after intraperitoneal injection of l-DOPA compared to vehicle (saline) was 18.3 ± 0.9 m versus 9.9 ± 0.8 m, F(1,10) = 6.4, P = 0.03 by one-way ANOVA. (I) Total movement time of UQCRC1 p.Tyr314Ser knock-in mice versus control mice; 142.3 ± 11.8 s versus 265.2 ± 28.7 s; F(1,11) = 21.3, P < 0.01 by one-way ANOVA. The total movement duration of UQCRC1 p.Tyr314Ser knock-in mouse after intraperitoneal injection of l-DOPA compared to vehicle (saline) was 175.9 ± 38.5 s versus 127.2 ± 33.0 s, F(1,10) = 2.1, P = 0.07 by one-way ANOVA. (J) Movement velocity of UQCRC1 p.Tyr314Ser knock-in mice versus control mice was 3.6 ± 0.3 cm/s versus 9.6 ± 0.8 cm/s; F(1,11) = 59.1, P < 0.01 by one-way ANOVA. Movement velocity of UQCRC1 p.Tyr314Ser knock-in mouse after intraperitoneal injection of l-DOPA compared to vehicle (saline) was 5.6 ± 1.7 cm/s versus 3.3 ± 0.9 cm/s, F(1,10) = 8.7, P = 0.01 by one-way ANOVA. n = 6 for each group; *P < 0.05; ** P < 0.01.
Behavioural assays and l-DOPA challenge test
Two behavioural tests for motor function assay were used, the open field test and CatWalk XT gait analysis. Experimenters were blinded to genotypes during behavioural testing as previously described (Angeby-Moller et al., 2008).
l-DOPA (25 mg/kg, i.p.; Sigma) or saline vehicle (0.9%, i.p.) was administered 30 min before each session in a counterbalanced manner. The peripheral decarboxylase inhibitor benserazide (12.5 mg/kg, i.p.; Sigma) was administered 20 min before l-DOPA while saline-treated animals received an additional injection of saline at the same time point.
Measurement of striatal dopamine content with 18F-fluoro-l-DOPA PET and HPLC analysis
18F-Fluoro-l-DOPA (FDOPA) PET in live animals was performed as previously described (Lee et al., 2013). Post-mortem mouse brains were frozen in liquid nitrogen and subsequently sectioned on a freezing microtome at a thickness of 900 μm. Punches (2 mm diameter) of striatal tissue were weighed, homogenized and centrifuged at 10 000 rpm for 20 min at 4°C. The supernatant was filtered and was then analysed for dopamine and metabolites using high-performance liquid chromatography (HPLC). Data were collected and analysed using ChromPerfect software (Justice Innovations, Mountain View, CA).
Additional methods
See the Supplementary material for details on TH, phospho-α-synuclein, hippocampal Nissl staining and glutamic acid decarboxylase immunostaining, as well as nerve conduction studies and sural nerve fibre analysis in adult mice.
Transmission electron microscopy of mitochondria in mice nigral neurons
Sections of substantia nigra were cut with a vibratome (VT1000S, Leica Microsystems) after perfusion and then fixed in 1% osmium tetroxide for 2 h at 4°C. The sections were dehydrated by immersion in a series of increasing ethanol concentrations and were then embedded using the Spurr’s resin kit (cat-14300; EMS) overnight at room temperature. Embedded tissues were sectioned into 70-µm thick sections with an ultramicrotome (EM-UC7, Leica Microsystems) and mounted on copper grids. Ultramicrographs were acquired with a transmission electron microscope (JEM-1400; JEOL) at 100 kV.
Measurement of mitochondrial complex III activity
The complex III enzyme activity of mitochondrial fractionations from mouse substantia nigra and striatal homogenates was measured using a mitochondrial complex III activity assay (BioVision) according to the manufacturer's instructions (Kirby et al., 2007).
Statistical analysis
Sample sizes estimates used the resource equation method but were refined using G power software on effect size (mean/standard deviation within groups) for 1 − beta = 0.8 and alpha = 0.05 (Faul et al., 2007). Data are expressed as the mean ± standard error of the mean (SEM), except when otherwise stated. The number of animals is indicated as n. The Mann-Whitney U-test was used for statistical comparisons between two groups. The one-way ANOVA post hoc Tukey test was performed for comparisons among multiple groups (adjustment applied), and the chi-square test was used for comparing behavioural test results. Statistical tests (two-sided) were performed using Stata (StataCorp LP, College Station, Texas) and a two-sided P < 0.05 was considered significant.
Data availability
The data that support the findings of this study are available from the corresponding author.
Results
Exome sequencing, clinical features, and in silico analysis
We initially examined a three-generations Taiwanese family with autosomal dominant parkinsonism and 10 family members, including five affected and five asymptomatic, were enrolled in the study (Family A in Fig. 1A). We previously applied whole exome sequencing in three affected patients (Patients II-4, II-7, and III-4 of Family A) (Fig. 1A and Supplementary Table 1) identified one potentially pathogenic missense variant, c.941A>C (p.Tyr314Ser) in UQCRC1 (RefSeq NM_003365.2) segregating with the parkinsonism phenotype (Fig. 1A) (Lin et al., 2019). The DNA from the fourth generation of Family A were not sampled as they were younger than 18 years old.
We proceeded to sequence all exons and exon-intron boundary junctions of UQCRC1 in 699 unrelated probands with familial parkinsonism and identified two more UQCRC1 variants, including c.931A>C (p.Ile311Leu) (Family B in Fig. 1A), and an allele with concomitant c.70-1G>A and c.73_74insG, p.Ala25Glyfs*27 variations (Family C in Fig. 1A). All variants were confirmed by Sanger sequencing (Fig. 1B) and were absent from the public databases. Among these two variants, UQCRC1 c.931A>C (p.Ile311Leu) also co-segregated with parkinsonism within the family (Family B in Fig. 1A). However, additional DNA samples from Family C were not available for segregation analysis (Family C in Fig. 1A). In addition, there were 14 coding missense variants in UQCRC1 with a minor allele frequency <0.001 present in the Taiwan biobank from 1517 control exomes (Supplementary Table 2). We did not find any of these 14 rare variants in our 699 enrolled probands by Sanger sequencing.
The identified three non-synonymous variants in UQCRC1, c.931A>C (p.Ile311Leu), c.941A>C (p.Tyr314Ser) and p.Ala25Glyfs*27, are all conserved across species (Fig. 1C). The variants identified in Family C are within splice junctions (Fig. 1B) and are bioinformatically predicted to affect exon splicing by Human Splicing Finder (HSF, Version 3.1; http://www.umd.be/HSF). Furthermore, we performed TaqMan® genotyping of UQCRC1 c.931A>C (p.Ile311Leu) and c.941A>C (p.Tyr314Ser) in 1934 unrelated patients with sporadic Parkinson’s disease but identified no additional heterozygotes.
Most affected UQCRC1 heterozygotes carriers presented with asymmetrical tremor-predominant parkinsonism, consistent with idiopathic Parkinson’s disease. The onset ages ranged from 40 to 60 years (Table 1). All patients have a good response to treatment of levodopa. We treated one affected member (Patient II-4 of Family A) with levodopa (600 mg/day) and ropinirole (2 mg/day). The UPDRS part III score improved from 54/108 to 27/108 after dopaminergic treatment. Nearly all affected members had anxiety as well. Asymmetric loss of the dopamine transporter was detected by Tc-99m TRODAT single-photon emission computed tomography (SPECT) in one affected member (Patient II-4) from Family A, mimicking idiopathic Parkinson’s disease. The asymmetry of specific uptake ratio (SUR) of caudate, putamen and striatum were 58.3%, 39.1% and 43.8%, respectively, which were compatible with the patient’s right-side predominant symptoms (Fig. 1E). In addition to parkinsonism, patients having UQCRC1 p.Tyr314Ser heterozygotes also had slowly progressive onset of numbness and weakness of feet followed by hand muscles involvement beginning in their early twenties. Numbness were noted in the feet and ascended to the mid-calf or knee levels and then followed by palms numbness. Patients II-2, II-4 and II-7 needed a cane to assist walk when they were in their fifties. Patients III-4 and III-9 had similar ascending weakness and numbness symptoms beginning in their early twenties. Both patients had right side parkinsonism features by their early forties and their daily activities do not need assistance at their current age (mid-forties). Neurological examinations of all affected members, even those without receiving levodopa, had variable degrees of distal legs and hands muscle atrophy and weakness accompanied with absent deep tendon reflexes. Sensory examinations showed decreased pinprick, temperature and vibration sensation over lower limbs to the knees level and hands. Nerve conduction studies of all affected members revealed axonal type-predominant sensorimotor polyneuropathy; EMG studies revealed neurogenic polyphasic waves in sampled distal muscles (Table 1). Somatosensory evoked potentials (SSEPs) exhibited abnormal tibial and median nerve SSEPs, as evidenced by prolongation of peripheral potentials (N9-median nerve, and N20-tibial nerve). None of the family members have diabetes mellitus or renal dysfunction. Patients II-2, II-4 and II-7 received cardiac echo examinations in their sixties and the results were normal. As the phenotype of sensorimotor neuropathy also co-segregated with UQCRC1 p.Tyr314Ser substitution in the index Family A, we examined the raw data of WES for all the causative genes contributing to either demyelinating type of axonal type of Charcot-Marie-Tooth (CMT) diseases, including PMP22, GJB1, MPZ, MFN2 and other rare genes for CMT disease (Rossor et al., 2013) and no pathogenic variants for CMT were identified. We also further screened UQCRC1 in a cohort of 32 CMT families without known mutations (mean age at examination was 36.6 ± 20.9 years old; 46.8% were male). All affected family members had electrophysiological evidence of either demyelinating or axonal type of polyneuropathy. All exon and intron-exon boundary junctions of UQCRC1 were Sanger sequenced in all probands of CMT families and we did not identify potential pathogenic variants among these patients.
Table 1.
Clinical features of patients with UQCRC1 mutations
| Family | A |
B |
C |
|||||
|---|---|---|---|---|---|---|---|---|
| II : 2 | II : 4 | II : 7 | III : 4 | III : 9 | II : 1 | II : 4 | III : 1 | |
| Current age, years | 77 | 74 | 69 | 49 | 42 | 76 | 71 | 69 |
| Onset age, years | 62 | 60 | 59 | 48 | 42 | 69 | 66 | 54 |
| Sex | M | M | F | F | F | M | M | M |
| Initial symptoms | ||||||||
| Asymmetry | + | + | + | + | + | + | + | + |
| Rest tremor | ++ | ++ | ++ | − | − | ++ | ++ | + |
| Rigidity | + | + | + | + | + | + | + | + |
| Bradykinesia | + | + | + | + | + | + | + | + |
| UPDRS scores (on time) | ||||||||
| Part I | 1 | 0 | 2 | 1 | 0 | N/A | N/A | N/A |
| Part II | 12 | 9 | 15 | 6 | 3 | N/A | N/A | N/A |
| Part III | 35 | 27 | 39 | 12 | 9 | N/A | N/A | N/A |
| Hoehn-Yahr stage (on time) | III | III | IV | I | I | III | II | V |
| Cognition | ||||||||
| MMSE | 29 | 30 | 28 | 30 | 30 | N/A | N/A | N/A |
| Associated symptoms | ||||||||
| Anxiety | + | + | + | + | + | N/A | N/A | N/A |
| Depression | − | − | + | + | − | N/A | N/A | N/A |
| Polyneuropathy | + | + | + | + | − | N/A | N/A | N/A |
| Drug response | ||||||||
| Levodopa | Well | Well | Well | Well | Well | Well | Well | Well |
| Anticholinergics | Well | Well | Well | N/A | N/A | Well | Well | Well |
| Brain MRI study | Mild diffuse atrophy | Mild diffuse atrophy | Mild diffuse atrophy | Unremarkable | Unremarkable | N/A | N/A | N/A |
| Nerve conduction study | ||||||||
| Ulnar nerve MNCV, m/s (normal: >50 m/s) | 45↓ | 46↓ | 45↓ | 48↓ | 52 | N/A | N/A | N/A |
| Ulnar nerve cMAP, mV (normal: >6.0 mV) | 3.2↓ | 3.0↓ | 3.7↓ | 5.2↓ | 5.8↓ | N/A | N/A | N/A |
| Ulnar nerve SAP, mV (normal: >10.0 mV) | 2.8↓ | 2.9↓ | 3.2↓ | 9.5↓ | 9.2↓ | N/A | N/A | N/A |
| Sural nerve SAP, mV (normal: >5.0 mV) | Absent | Absent | Absent | Absent | Absent | N/A | N/A | N/A |
| EMG study | Neurogenica | Neurogenica | Neurogenica | Neurogenica | N/A | N/A | N/A | N/A |
cMAP = compound muscle action potential; F = female; M = male; MMSE = Mini-Mental State Examination; MNCV = motor nerve conduction velocity; N/A = not assessed or not available from medical records; SAP = sensory action potential; UPDRS = Unified Parkinson's Disease Rating Scale, ‘on time’ refers to periods of adequate control of Parkinson’s disease motor symptoms and was usually within 3 h after taking levodopa medications. Upward arrow indicates increase; downwards arrow indicates decrease; −= absent; + = mild; ++ = moderate.
Neurogenic polyphasic waves in bilateral first dorsal interosseous muscles and tibialis anterior muscles.
The structure of human respiratory complex III at 3.4 Å resolution using cryo-electron microscopy has been reported. Monomers of UQCRC1 Ile311 and Tyr314 are required in complex III homodimerization (Fig. 1F, left) (Guo et al., 2017). We studied the structural changes induced by p.Ile311Leu and p.Tyr314Ser substitutions using in silico protein modelling. The side chain of Ile311 points into the hydrophobic core of the protein and forms contacts with Ile310, Pro327, Gln375, Trp376 and Leu379 (Fig. 1F, top right). A substitution of Ile (CH3-) to Leu (CH3-CH2-), although a modest aliphatic change, alters the packing of nearby side chains in the structure and potentially influences the activity of respiratory complex III. Similarly, Tyr314 has a bulky, rigid ring structure and hydrophobic interactions with Cys317, Tyr318, Ala329, Val333, and Phe341 (Fig. 1F, bottom right). Substitution from Tyr (-C6H5OH) to Ser (-CH2-OH) disrupts the stacking of the aromatic rings and would disturb side-chain-to-side-chain van der Waals packing. Both substitutions may therefore lead to destabilization of complex III protein structure, which is a homodimer, and have the potential to influence mitochondrial respiratory chain complex III activity.
As the identified genetic variants in UQCRC1 in autosomal-dominant Parkinson’s disease are very rare and the pathogenicity of the splicing variant in Family C remains uncertain due to lack of DNA samples of family members for co-segregation analysis, it is critical to prove the pathogenicity of UQCRC1 mutations in Parkinson’s disease by functional analyses focused on the two missense variants shown to co-segregate with parkinsonism in Families A and B, c.931A>C (p.Ile311Leu) and c.941A>C (p.Tyr314Ser).
Mutant UQCRC1 expression leads to neurite retraction and mitochondrial respiratory chain dysfunction
UQCRC1 is widely expressed in tissues including brain, heart, kidney, liver, and skeletal muscles. Notably, UQCRC1 is abundant in the substantia nigra and the striatum, which are predominantly affected in Parkinson’s disease (Supplementary Fig. 1A). Mitochondrial respiratory chain complex III is required for proper mitochondrial function and ATP synthesis and is essential for neuronal morphogenesis and function (Oruganty-Das et al., 2012).
To characterize the impact of the UQCRC1 carrying individual substitutions of Families A–C on human dopaminergic neurons, we established mutant UQCRC1 knock-in SH-SY5Y cell lines with CRISPR-Cas9 technology with sequence confirmation (Supplementary Fig. 1B) (Ran et al., 2013). As the variants identified in Family C are within splice junctions between exons 1 and 2, we examined the cDNA expression of SH-SY5Y cell with the heterozygous splicing variant of Family C by RT-PCR. We observed an aberrant splicing of UQCRC1 transcript (Fig. 1D), indicating the variations of Family C affect exon splicing of UQCRC1. Neurite morphology was examined in retinoic acid-stimulated differentiated SH-SY5Y cells with similar UQCRC1 protein expression levels, except for the aberrant splicing variant of Family C which showed decreased full-length UQCRC1 protein expression (Supplementary Fig. 1C). Confocal microscopy showed that cells expressing UQCRC1 with either the p.Tyr314Ser or p.Ile311Leu substitution, or the aberrant splicing variation consistently exhibited shortened neurites compared to wild-type neurons (SH-SY5Y wild-type control: 82.6 ± 10.8 μm; UQCRC1 p.Tyr314Ser: 42.6 ± 7.3 μm; UQCRC1 p.Ile311Leu: 55.1 ± 9.5 μm; UQCRC1 concomitant c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant: 54.1 ± 10.2 μm; P = 0.008 for wild-type versus UQCRC1 p.Tyr314Ser; P = 0.03 for wild-type versus UQCRC1 p.Ile311Leu; P = 0.01 for wild-type versus UQCRC1 aberrant splicing variant) (Supplementary Fig. 1D–G; statistics in Supplementary Fig. 1H). These results demonstrate mutant UQCRC1 proteins impair neurite morphogenesis.
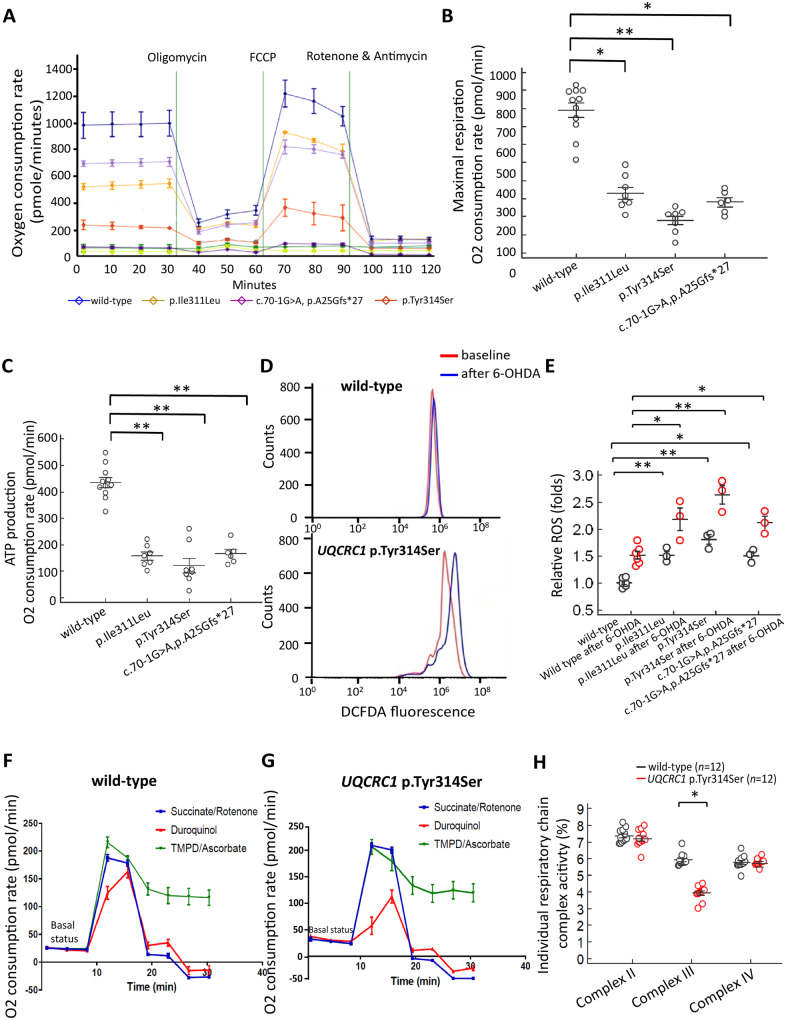
We next examined whether variants of UQCRC1 affects the cellular respiration of neurons using a Seahorse extracellular flux analyzer. We measured oxygen consumption rates (OCRs) at regular intervals with the addition of compounds to test mitochondrial function (Fig. 2A). We found that the maximal respiration activity and ATP production were markedly decreased in neurons having UQCRC1 p.Ile311Leu, p.Tyr314 or the aberrant splicing variation when compared to wild-type neurons (Fig. 2B and C). Consistently, the production of reactive oxygen species was increased in UQCRC1 mutants compared to wild-type neurons, either during basal state or after stressed with treatment of 6-OHDA (Fig. 2D; statistics in Fig. 2E). These effects were more significant for UQCRC1 p.Tyr314Ser neurons (Fig. 2A–E).
Figure 2.
Variants of UQCRC1 alter mitochondrial respiratory chain function of SH-SY5Y cells. (A) The oxygen consumption rate (OCR; pmol/min) was measured with a Seahorse Extracellular Flux Analyzer in 8 × 105 cultured wild-type SH-SY5Y cells (blue line), UQCRC1 p.Ill311Leu (yellow line), UQCRC1 p.Tyr314Ser (orange line) and UQCRC1 concomitant c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant knock-in SH-SY5Y cells (purple line). OCRs were measured at regular intervals with the addition of compounds to test mitochondrial function. (B) Maximal respiration is decreased in all mutant neurons compared to wild-type neurons. Each cell line was assayed in at least three independent experiments, and means were calculated. Error bars are SEM. F(1,12) = 17.55, P = 0.006, one-way ANOVA for wild-type versus UQCRC1 p.Tyr314Ser cells; F(1,13) = 10.12, P = 0.013, for wild-type versus UQCRC1 p.Ile311Leu cells; F(1,12) = 11.35, P = 0.018, for wild-type versus UQCRC1 concomitant c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant. (C) ATP production through oxidative phosphorylation is decreased in neurons having all human UQCRC1 variants compared to wild-type neurons. F(1,12) = 18.21, P = 0.004, one-way ANOVA for wild-type versus UQCRC1 p.Tyr314Ser cells; F(1,13) = 15.73, P = 0.008, for wild-type versus UQCRC1 p.Ile311Leu cells; F(1,15) = 17.29, P = 0.009, for wild-type versus UQCRC1 concomitant c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant. (D) Representative flow cytometry profiles of reactive oxygen species production by DCFDA fluorescence showing reactive oxygen species production in wild-type cells and UQCRC1 p.Tyr314Ser cells before (baseline, red lines) and after exposure to 6-OHDA for 3 h (blue lines). (E) Amount of reactive oxygen species produced in individual genotypes of cells with or without treatment with 6-OHDA normalized to wild-type cells without exposure to 6-OHDA is presented as mean ± SEM. Data are representative of three experiments. F(1,4) = 26.73, P = 0.007, one-way ANOVA for wild-type versus UQCRC1 p.Ile311Leu cells; F(1,4) = 6.38, P = 0.065, for UQCRC1 p.Ile311Leu versus UQCRC1 p.Tyr314Ser cells; F(1,4) = 55.03, P = 0.002, one-way ANOVA for wild-type versus UQCRC1 p.Tyr314Ser cells; F(1,4) = 10.21, P = 0.012, one-way ANOVA for wild-type versus UQCRC1 c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant cells; F(1,4) = 6.88, P = 0.06, for wild-type versus UQCRC1 p.Ile311Leu cells after treatment with 6-OHDA; F(1,4) = 25.55, P = 0.007, one-way ANOVA for wild-type versus UQCRC1 p.Tyr314Ser cells after treatment with 6-OHDA. F(1,4) = 9.98 P = 0.016, one-way ANOVA for wild-type versus UQCRC1 c.70-1G>A and p.Ala25Glyfs*27 aberrant splicing variant cells after treatment with 6-OHDA. (F–H) The individual activity of mitochondrial respiratory chain complex I–IV was examined further in wild-type cells and UQCRC1 p.Tyr314Ser knock-in SH-SY5Y cells. Representative seahorse profiles for the individual activity of mitochondrial respiratory chain complexes I–IV are illustrated in wild-type cells (F) and UQCRC1 p.Tyr314Ser knock-in SH-SY5Y cells (G) by measuring the OCRs before (basal respiration) and after sequential additions of oligomycin (ATP synthase inhibitor), CCCP (uncoupling protonophore), and then rotenone (complex I inhibitor) and succinate (complex II inhibitor) (blue lines); duroquinol (complex III inhibitor) (red lines); and TMPD and ascorbate (complex IV inhibitors) (green lines). This strategy allowed for the determination of the contribution of each component of respiration chain complexes. (H) The percentages of OCR attributable to the activities of complexes I–IV. In each experiment, data were collected and averaged from four separate wells for each individual cell line. Each cell line was assayed in at least three independent experiments, and means were calculated. Error bars are SEM. F(1,22) = 31.22, P = 0.018, one-way ANOVA for wild-type versus UQCRC1 p.Tyr314Ser for complex III activity. *P < 0.05. **P < 0.01.
As UQCRC1 p.Tyr314Ser knock-in neurons express the most prominent phenotypes, the individual activities of composite respiratory chain complexes and further functional assay in in vivo Drosophila and mouse models were examined. Treatment with individual respiratory chain complex inhibitors demonstrated that UQCRC1 p.Tyr314Ser expression had no obvious influence on mitochondrial complex I, II, or IV activity (Fig. 2F and G; statistics in Fig. 2H), but remarkably reduced the oxygen consumption of complex III activity (Fig. 2F and G; statistics in Fig. 2H). This finding suggests that UQCRC1 p.Tyr314Ser disturbs mitochondrial respiratory chain complex III activity in SH-SY5Y cells.
UQCRC1 p.Tyr314Ser expression in flies causes age-dependent locomotor defects and loss of TH-positive neurons
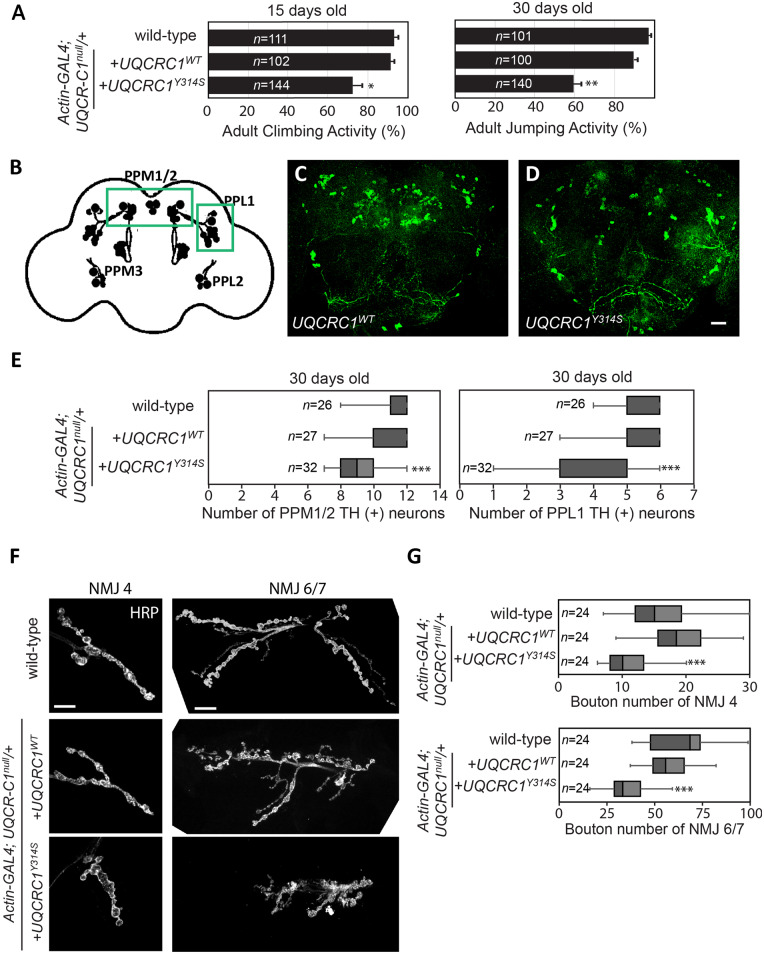
The age-dependent locomotor phenotypes of behaviour assays, decreased life span and the number of dopaminergic neurons were comparable between wild-type Drosophila and the introduction of the UQCRC1WT allele in a heterozygous UQCRC1 null allele background (Fig. 3A, E and Supplementary Fig. 2E), indicating UQCRC1 has a species-conserved function in both humans and flies. We next examined whether introducing UQCRC1 p.Tyr314Ser into neurons would cause parkinsonism-like phenotypes in in vivo fly models. We found that adult UQCRC1Y314S flies displayed significant impairments in locomotion at 15 days after eclosion and more prominent decline at 30 days after eclosion, compared to UQCRC1WT flies (Fig. 3A). Furthermore, adult UQCRC1Y314S flies also exhibited an age-dependent decreased life span compared to wild-type control or UQCRC1WT flies (Supplementary Fig. 2E). In addition, we observed a significant loss of dopaminergic neurons in the paired protocerebral posterior medial 1/2 (PPM1/2) and protocerebral posterior lateral clusters (PPL1) in aged UQCRC1Y314S adult brains at the fourth week after eclosion (Fig. 3B–D) compared to UQCRC1WT brains from flies of the same age (Fig. 3E, P < 0.01). We did not observe any abnormal phospho-α-synuclein immunostaining in dopaminergic neurons of aged UQCRC1Y314S adult fly brains (data not shown). These results suggest that the UQCRC1 p.Tyr314Ser substitution induces progressive loss of dopaminergic neurons and compromises motor activity in an age-dependent manner.
Figure 3.
UQCRC1 p.Tyr314Ser expression in Drosophila causes age-dependent locomotor defects, loss of TH-positive neurons and presynaptic defects in NMJs. (A) Climbing and jumping assays for wild-type control and flies that expressed either the wild-type UQCRC1 (UQCRC1WT) or mutant UQCRC1 p.Tyr314Ser (UQCRC1YS314S). The number of tested flies was shown in the bar graph and assays were repeated three times for each genotype. For the climbing assay, wild-type versus UQCRC1WT was 95.38 ± 6.21% versus 93.24 ± 5.13%, F(1, 81) = 4.8, P = 0.43; UQCRC1WT versus UQCRC1YS314S was 93.24 ± 5.13% versus 72.35 ± 7.38%, F(1,84) = 5.7, P = 0.021. For the jumping assay, wild-type versus UQCRC1WT was 98.25 ± 3.51% versus 90.13 ± 4.88%, F(1,81) = 6.7, P = 0.10; UQCRC1WT versus UQCRCY314S was 90.13 ± 4.88% versus 60.79 ± 6.42%, F(1,98) = 45.2, P = 0.006; all were based on one-way ANOVA. **P < 0.01. (B–D) Whole-mount adult brains were immunostained with anti-TH (green) to label individual dopaminergic neuronal clusters in 30-day-old flies. Images are representative of (B) schematic representation of the distribution of dopaminergic neurons in the Drosophila adult brain. Dopaminergic neurons are grouped in small clusters arranged with bilateral symmetry. PPL = protocerebral posterior lateral; PPM = protocerebral posterior medial. (C) UQCRC1WT files, and (D) UQCRC1Y314S flies. Scale bar = 20 µm. (E) Quantification of neurons that stained anti-TH-positive in the PPM1/2 and PPL1 clusters in individual genotypes at 30 days old [for PPM1/2, wild-type versus UQCRC1WT was 11.75 ± 0.38 versus 10.58 ± 0.23, F(1,45) = 6.8, P = 0.23; UQCRC1WT versus UQCRC Y314S was 10.58 ± 0.23 versus 8.12 ± 0.19, F(1,57) = 27.8, P = 0.007; For PPL1, wild-type versus UQCRC1WT was 5.57 ± 0.11 versus 5.61 ± 0.14, F(1,45) = 5.3, P = 0.49; UQCRC1WT versus UQCRC1 Y314S was 5.61 ± 0.14 versus 4.08 ± 0.22, F(1,57) = 20.3, P = 0.009 by one-way ANOVA]. (F) Representative confocal microscopy images show NMJs in third-instar larvae. Left: NMJ segment 4 (NMJ4) and right: NMJ segment 6/7 (NMJ6/7) are labelled with the membrane marker HRP. Fly genotypes are: (top row) wild-type control, (middle row) wild-type human UQCRC1 (UQCRC1WT), and (bottom row) human UQCRC1 p.Tyr314Ser mutant (UQCRC1Y314S). Scale bars = 20 µm. (G) NMJ sizes were quantified as the bouton number divided by the muscle area of NMJ4 or NMJ 6/7, expressed as bouton number × 104/muscle area (µm2). Average NMJ sizes (mean ± SEM) were compared with the one-way ANOVA post hoc Tukey test; ***P < 0.001. Sample numbers (n) are shown inside the box-and-whiskers plots.
UQCRC1 p.Tyr314Ser substitution causes presynaptic defects at the neuromuscular junction
As heterozygous UQCRC1 p.Tyr314Ser patients also present with mild signs of peripheral neuropathy, this observation suggests mutant UQCRC1 may also lead to peripheral neurodegeneration in addition to dopaminergic neuronal degeneration. We therefore examined the morphology of NMJs in late, third-instar larvae of wild-type (UQCRC1WT) and mutant (UQCRC1Y314S) flies to elucidate the effects of mutant UQCRC1 on peripheral motor nerves. Axonal processes were labelled with anti-HRP antibody. Notably, the NMJs were significantly smaller and had markedly reduced numbers of boutons in larvae expressing UQCRC1Y314S compared to those in UQCRC1WT larvae (Fig. 3F, statistics in Fig. 3G, P < 0.001 in either NMJ 4 or NMJ 6/7 segments).
Several other changes in NMJ morphology were obvious in larvae expressing mutant UQCRC1Y314S. We observed that the presynaptic compartments were unevenly enlarged in UQCRC1Y314S mutant NMJs compared to those in wild-type UQCRC1WT larvae (Supplementary Fig. 2B). The distribution of the presynaptic marker Bruchpilot (Brp), a component of T-bars at active zones, appeared to form clusters in the enlarged boutons in the mutant UQCRCY314S larvae (Supplementary Fig. 2B) and the total Brp expression levels were decreased (statistics in Supplementary Fig. 2D), suggesting a presynaptic defect at NMJs. Additionally, in wild-type UQCRC1WT larvae, the NMJ is organized into a series of synaptic boutons that can be directly visualized using the membrane marker HRP and another presynaptic vesicle marker cysteine string protein (CSP) (Supplementary Fig. 2B). However, the presynaptic nerve terminals in UQCRC1Y314S larvae lacked discrete synaptic boutons and there was no characteristic regular inter-bouton spacing (Supplementary Fig 2B). In contrast, immunostaining for discs large (Dlg), a postsynaptic marker of morphology, was comparable in mutant UQCRC1Y314S larvae and controls, and total Dlg expression at the NMJ was equivalent (Supplementary Fig. 2C and D). These findings suggest UQCRC1 p.Tyr314Ser results in specific presynaptic nerve defects at the NMJs, consistent with clinical phenotypes of peripheral neuropathy observed in human patients with a heterozygous UQCRC1 p.Tyr314Ser substitution.
Establishment of a heterozygous UQCRC1 p.Tyr314Ser knock-in mouse
We generated a CRISPR/Cas9-based knock-in mouse with the human p.Tyr314Ser variation in the mouse UQCRC1 genome (UQCRC1 p.Tyr314Ser knock-in mouse) (Fig. 4A) to validate our observations in the fly models. Given the human clinical phenotypes are observed in heterozygous patients, we used heterozygous UQCRC1 p.Tyr314Ser knock-in mice to assess the functional consequences of this mutation.
The gene targeting strategy and PCR-based genotyping for the knock-in mice are summarized in Fig. 4B. PCR analyses of tail DNA (Fig. 4C) from the offspring of heterozygous parents showed that the mouse UQCRC1 gene p.Tyr314Ser variation was correctly mutated and Sanger sequencing of the PCR product confirmed the c.941A>C (p.Tyr314Ser) substitution in the mutant allele (Supplementary Fig. 2F).
UQCRC1 p.Tyr314Ser knock-in mice show age-dependent locomotor defects
Heterozygous UQCRC1 p.Tyr314Ser knock-in mice grew normally and for the first 9 months of age were indistinguishable from wild-type mice in outward appearance, size, and body weight, but then began to exhibit behavioural phenotypes. At 12 months, open field test to assess spontaneous locomotor activity of UQCRC1 p.Tyr314Ser knock-in mice revealed profound deficits in distance travelled (Fig. 4D and E, statistics in Fig. 4H, P < 0.01), total movement time (Fig. 4I, P < 0.01), and movement velocity (Fig. 4J, P < 0.01) compared to wild-type age and gender-matched littermate controls.
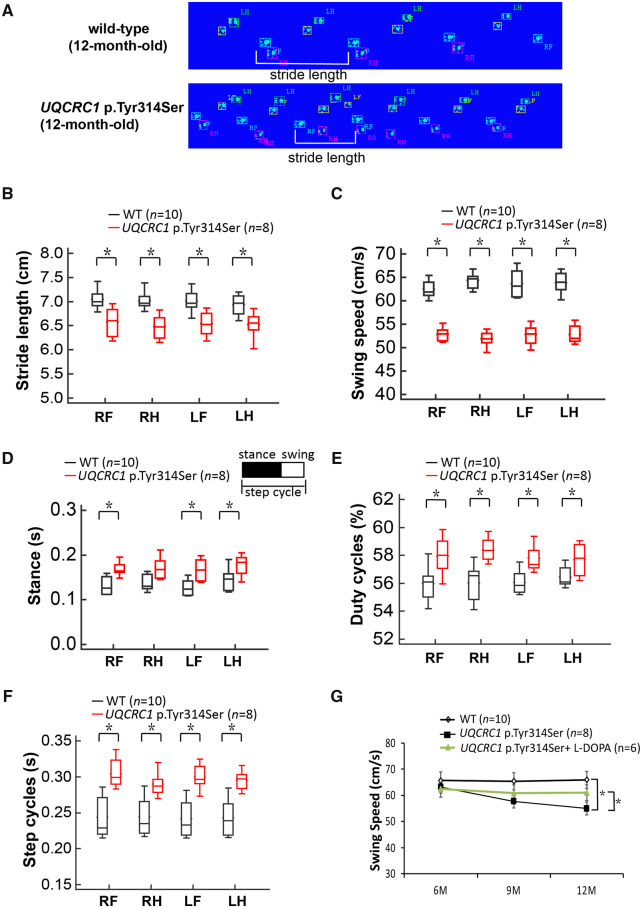
We then evaluated gait in both wild-type and heterozygous UQCRC1 p.Tyr314Ser knock-in mice with a CatWalk system at 6, 9, and 12 months of age (Fig. 5A). The time taken to walk a specified distance was significantly longer for heterozygous mice than for their wild-type littermates at 9 and 12 months of age. We also noted that the UQCRC1 p.Tyr314Ser knock-in mice exhibited a significantly shorter stride length compared to littermate controls (Fig. 5A and B, P < 0.01) as well as a slower walking swing speed (Fig. 5C, P < 0.01), which mimicked the small-stepped gait of Parkinson’s disease patients. Consequently, the stances (the time duration of paws in contact with the glass plate in seconds), duty cycles (the percentage of stance over the sum of stance and swing duration), and paw step cycles (the time in seconds between two consecutive initial contacts of the same paw) increased substantially in UQCRC1 p.Tyr314Ser knock-in mice compared to controls at 12 months of age (Fig. 5D–F), which recapitulated the shuffling and freezing gait of Parkinson’s disease patients. Analysis at 6–12 months of age showed a significant age-dependent reduction in swing speed in UQCRC1 p.Tyr314Ser knock-in mice compared to littermate controls (Fig. 5G, P = 0.01). These findings indicated an age-dependent slowing of movement as well as locomotor defects in UQCRC1 p.Tyr314Ser knock-in mice.
Figure 5.
Gait analysis of UQCRC1 p.Tyr314Ser knock-in mice. (A) Representative illuminated footprints recorded from (top) wild-type littermate controls and (bottom) UQCRC1 p.Tyr314Ser knock-in mice at the age of 12 months as they walked across the CatWalk platform. One stride length (white bracket) is illustrated in each track. (B–F) Different parameters of the CatWalk analyses of wild-type (black box-and-whiskers) and UQCRC1 p.Tyr314Ser knock-in (red box-and-whiskers) mice show that (B) stride length and (C) swing speed was significantly decreased, but (D) stances, (E) duty cycles, and (F) paw step cycles increased significantly in UQCRC1 p.Tyr314Ser knock-in mice compared to the littermate wild-type controls. (G) Serial analysis of swing speeds in littermate wild-type controls and UQCRC1 p.Tyr314Ser knock-in mice with, and without, l-DOPA treatment from 6 to 12 months of age. Wild-type littermate controls versus UQCRC1 p.Tyr314Ser knock-in mice were 62.8 ± 4.1 cm/s versus 61.4 ± 3.4 cm/s at 6 months, F(1,16) = 2.1, P = 0.76; 62.5 ± 5.1 cm/s versus 56.9 ± 3.7 cm/s at 9 months, F(1,16) = 6.2, P = 0.03; 63.2 ± 3.2 cm/s versus 52.4 ± 2.3 cm/s at 12 months; F(1,16) = 34.8, P = 0.01 by one-way ANOVA. UQCRC1 p.Tyr314Ser knock-in mice with (green line) versus without l-DOPA treatment was 60.9 ± 4.6 cm/s versus 61.4 ± 3.4 cm/s at 6 months, F(1,14) = 2.4, P = 0.89; 59.2 ± 4.3 cm/s versus 56.9 ± 3.7 cm/s at 9 months, F(1,14) = 5.3, P = 0.07; 60.1 ± 4.1 cm/s versus 52.4 ± 2.3 cm/s at 12 months; F(1,14) = 9.8, P = 0.04 by one-way ANOVA. n = 6–10 for each genotype in each experiment. LF = left front paw; LH = left hind paw; RF = right front paw; RH = right hind paw; *P < 0.05; **P < 0.01.
UQCRC1 p.Tyr314Ser knock-in mice show reduced striatal dopamine levels and loss of nigral TH-positive neurons
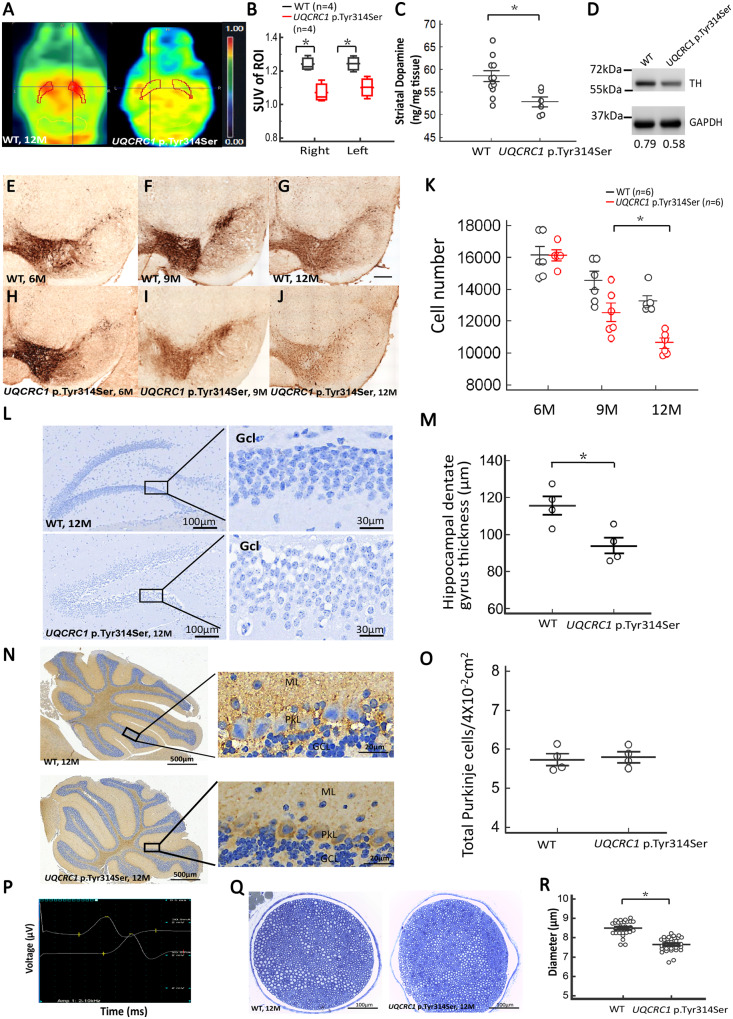
As the nigral-striatal dopaminergic system controls locomotor function (Smith and Villalba, 2008), we therefore performed 18F-DOPA PET scans to assess the integrity of the dopaminergic system in the mouse striatum. The uptake of 18F-DOPA in the striatum was significantly reduced in UQCRC1 p.Tyr314Ser knock-in mice compared to wild-type littermate controls at 12 months of age (Fig. 6A and B; P = 0.01). We next determined whether the observed decrease of 18F-DOPA uptake in the striatum of UQCRC1 p.Tyr314Ser knock-in mice was the result of a striatal dopamine deficiency by measuring the dopamine concentrations in striatal tissue lysates by HPLC. We found that striatal dopamine concentration was significantly lower in UQCRC1 p.Tyr314Ser knock-in mice than in age and gender-matched wild-type littermate controls at 12 months of age (Fig. 6C, P = 0.013). These changes were accompanied by a marked reduction of TH expression in nigral and striatal homogenates from UQCRC1 p.Tyr314Ser knock-in mice relative to wild-type littermates at 12 months of age, as assessed by western blot analysis (Fig. 6D, P = 0.02). Taken in concert, these data indicate a greater disruption of dopamine homeostasis in the striatum of UQCRC1 p.Tyr314Ser knock-in mice compared to age-matched littermate controls.
Figure 6.
Striatal dopamine, substantia nigra TH-positive cells, hippocampal dentate gyrus neurons, cerebellar Purkinje cells immunostaining and sciatic nerve fibre changes of wild-type and UQCRC1 p.Tyr314Ser knock-in mice. (A) Axial series of 6-18F-fluoro-l-DOPA PET images in 12-month-old (left) wild-type littermates (WT) and (right) UQCRC1 p.Tyr314Ser knock-in mice. The bilateral striatum was drawn manually to indicate the region of interest on the PET images. (B) The standard uptake values (SUVs) in the right and left regions of interest (shown in the PET images) were calculated and compared between wild-type littermates (black box-and-whiskers) and UQCRC1 p.Tyr314Ser knock-in (red box-and-whiskers) mice. The average SUV of right region of interest in wild-type littermates and UQCRC1 p.Tyr314Ser was 1.23 ± 0.06 and 1.06 ± 0.04, F(1,6) = 7.2, P = 0.02 by one-way ANOVA; the average SUV of the left region of interest in wild-type littermates and UQCRC1 p.Tyr314Ser was 1.27 ± 0.05 and 1.09 ± 0.03, F(1,6) = 6.9, P = 0.03 by one-way ANOVA. n = 4 for each genotype. (C) Scatterplot comparison of striatal dopamine concentration measured by HPLC in wild-type littermates and UQCRC1 p.Tyr314Ser knock-in mice at the age of 12 months. UQCRC1 p.Tyr314Ser knock-in mice versus control mice; 52.9 ± 2.6 µg/ml versus 58.5 ± 4.0 µg/ml; F(1,6) = 9.7, P = 0.013 by one-way ANOVA. (D) Schematic representations of western blot. At 12 months of age, UQCRC1 p.Tyr314Ser mice presented with lower nigral and striatal TH levels than littermate controls. The relative expression level of TH to the GAPDH in wild-type littermates versus UQCRC1 p.Tyr314Ser at the age of 12 months was 0.79 ± 0.04 versus 0.58 ± 0.07, F(1,4) = 10.2, P = 0.02 by one-way ANOVA. The experiments were repeated three times. *P < 0.05. (E–J) Representative mid-brain sections show anti-TH immunostaining of dopaminergic neurons in the SNc of (E–G) wild-type and (H–J) UQCRC1 p.Tyr314Ser knock-in mice at 6, 9, and 12 months of age, as indicated. ×100 magnification. (K) Average numbers of TH-positive neurons observed in the SNc of wild-type and UQCRC1 p.Tyr314Ser knock-in mice at 6, 9, and 12 months of age, determined with stereological counting. At 6 months, wild-type littermates versus UQCRC1 p.Tyr314Ser was 16 189 ± 621 versus 16 200 ± 735; P = 0.82; At 9 months, wild-type littermates versus UQCRC1 p.Tyr314Ser was 14 980 ± 683 versus 12 389 ± 702; P = 0.08; At 12 months, wild-type littermates versus UQCRC1 p.Tyr314Ser was 13 127 ± 591 versus 10 830 ± 782; P = 0.032 by one-way ANOVA. n = 6 mice in each genotype, for all analyses. Values are mean ± SEM. *P < 0.05. (L) Representative sections showed hippocampal dentate gyrus from wild-type (WT) and UQCRC1 p.Tyr314Ser knock-in mice at the age of 12 months using Nissl staining. Gcl = granule cell layer. (M) Quantitative evaluation of the hippocampal dentate gyrus thickness in wild-type and UQCRC1 p.Tyr314Ser knock-in mice at 12 months of age is shown. Wild-type littermates versus UQCRC1 p.Tyr314Ser was 115.57 ± 3.81 µm versus 86.30 ± 2.13 µm; F(1,7) = 0.65, P = 0.02 by one-way ANOVA. n = 4 mice in each genotype, for all analyses. Values are mean ± SEM. (N) Representative sections show posterior lobes of cerebellum from wild-type and UQCRC1 p.Tyr314Ser knock-in mice at the age of 12 months immunolabelled for the GABAergic enzyme, glutamate decarboxylase 67 (GAD67). GCL = granule cell layer; ML = molecular layer; PL = Purkinje cell layer. (O) The quantification of the mean number of GAD67-positive Purkinje cells in an area of 4 × 102 mm2 for wild-type littermate controls and UQCRC1 p.Tyr314Ser knock-in mice at 12 months of age is shown. Wild-type littermates versus UQCRC1 p.Tyr314Ser was 5.74 ± 0.31 versus 5.81 ± 0.26; F(1,7) = 0.11, P = 0.75 by one-way ANOVA. n = 4 mice in each genotype, for all analyses. Values are mean ± SEM. (P) Characteristic nerve conduction recordings of normal (top), distal, and (bottom) proximal motor responses. Sensitivity (y-axis): 2 µV/division; sweep speed (x-axis): 0.25 ms/division. (Q) Representative brightfield images of semithin sections of sciatic nerves derived from wild-type littermate controls and UQCRC1 p.Tyr314Ser knock-in mice at 12 months of age. Myelinated fibres were stained with toluidine blue. (R) Quantification of diameter of myelinated nerve fibres in sciatic nerve. Wild-type littermates versus UQCRC1 p.Tyr314Ser was 8.49 ± 0.23 µm versus 7.58 ± 0.31 µm; F(1,59) = 80.17, P = 0.01 by one-way ANOVA. n = 3 mice in each genotype. Data are presented as mean ± SEM. *P < 0.05, **P < 0.01.
We next dissected mouse brains at 6–12 months of age and prepared sections of the substantia nigra pars compacta (SNc) to investigate the number of TH-positive neurons using immunohistochemical methods and stereological counting. While results in wild-type littermates and UQCRC1 p.Tyr314Ser knock-in mice were comparable at 6 months of age (Fig. 6E and H, statistics in Fig. 6K), a modest deficit was observed at 9 months of age (Fig. 6F and I, statistics in Fig. 6K). Notably, TH staining was significantly reduced in UQCRC1 p.Tyr314Ser knock-in mice compared to wild-type littermates at 12 months of age (Fig. 6G and J, statistics in Fig. 6K).
We further examined whether the mutant UQCRC1 involved in the degeneration of other neuronal systems, including hippocampal and cerebellar neurons. We investigated the morphology of hippocampal neurons using Nissl staining. There was no significant difference in the thickness of the hippocampal dentate gyrus between wild-type littermates and UQCRC1 p.Tyr314Ser knock-in mice at the age of 9 months (data not shown). At the age of 12 months, we observed a significant reduction in the thickness of the hippocampal dentate gyrus in UQCRC1 p.Tyr314Ser knock-in mice compared to wild-type littermates (Fig. 6L, left; statistics in Fig. 6M). A decreased neuronal density with dispersed degenerated neurons in the granule cell layer of hippocampus was also noted (right panel in Fig. 6L). In addition, the number of cerebellar Purkinje cells immunolabelled for the GABAergic enzyme, glutamate decarboxylase 67 (GAD67), in the cerebellum at 12 months of age were comparable between wild-type littermates and UQCRC1 p.Tyr314Ser knock-in mice (Fig. 6N, statistics in Fig. 6O). These results suggest that the UQCRC1 p.Tyr314Ser mutation results in an age-dependent loss of TH-immunoreactive dopaminergic neurons in the SNc beginning at the age of 9 months, which may explain the reduction in 18F-DOPA uptake and dopamine levels observed in the striatum (Fig. 6A–D), and then involved the degeneration of hippocampus at an advanced age of 12 months. We then investigated whether the UQCRC1 p.Tyr314Ser mutation might lead to α-synucleinopathy at age of 12 months and even to the age of 15 months. However, we did not observe any abnormal phospho-α-synuclein immunostaining in nigra or hippocampal neurons (data not shown).
l-DOPA improves the locomotor deficits of aged UQCRC1 p.Tyr314Ser knock-in mice
As bradykinesia in patients with Parkinson’s disease is improved by l-DOPA treatment, we then examined the effects of l-DOPA on locomotor dysfunction of aged UQCRC1 p.Tyr314Ser knock-in mice to further confirm dopaminergic system dysfunction caused by the UQCRC1 mutation. We found that intraperitoneal injection of l-DOPA compared to vehicle (saline) in UQCRC1 p.Tyr314Ser knock-in mice improved motor phenotypes observed in open field testing, including total distance travelled, movement duration and velocity at the age of 12 months (Fig. 4F and G; statistics in Fig. 4H–J) and walking speed velocity by Catwalk analysis (P = 0.04 by one-way ANOVA, green line in Fig. 5G).
UQCRC1 p.Tyr314Ser knock-in mice exhibit peripheral neuropathy
In addition to features of parkinsonism, patients with heterozygous UQCRC1 p.Tyr314Ser mutation had mild signs of peripheral neuropathy with axonal degeneration (Table 1). We therefore examined whether heterozygous UQCRC1 p.Tyr314Ser knock-in mice had signs of peripheral neuropathy by performing nerve conduction studies on the sciatic nerves of UQCRC1 p.Tyr314Ser mice and wild-type littermates at 9 and 12 months of age (Fig. 6P). Compound muscle action potentials (CMAP) and nerve conduction velocities were comparable between wild-type and UQCRC1 p.Tyr314Ser knock-in mice at 9 months of age. However, at 12 months of age UQCRC1 p.Tyr314Ser knock-in mice exhibited significant reductions in distal CMAP amplitudes compared to wild-type littermates (littermate wild-type control versus UQCRC1 p.Tyr314Ser; 4.95 ± 0.38 µV versus 2.85 ± 0.31 µV; P = 0.028 by one-way ANOVA), as well as a slightly reduced conduction velocity but preserved distal motor latency. We further explored the alterations in peripheral nerve fibre morphology by obtaining semithin sections from sciatic nerves of wild-type and UQCRC1 p.Tyr314Ser knock-in mice at 12 months of age. We detected a marked decrease in the diameter of myelinated nerve fibres in UQCRC1 p.Tyr314Ser knock-in mice compared with littermate wild-type controls (Fig. 6Q, statistics in Fig. 6R), suggesting that large myelinated fibres are more susceptible to UQCRC1 p.Tyr314Ser mutation. Collectively, these results are suggestive of peripheral nerve degeneration, and support findings from nerve conduction and EMG studies in patients with UQCRC1 p.Tyr314Ser (Table 1).
UQCRC1 p.Tyr314Ser knock-in mice have abnormal mitochondrial morphology and complex III activity in nigral neurons
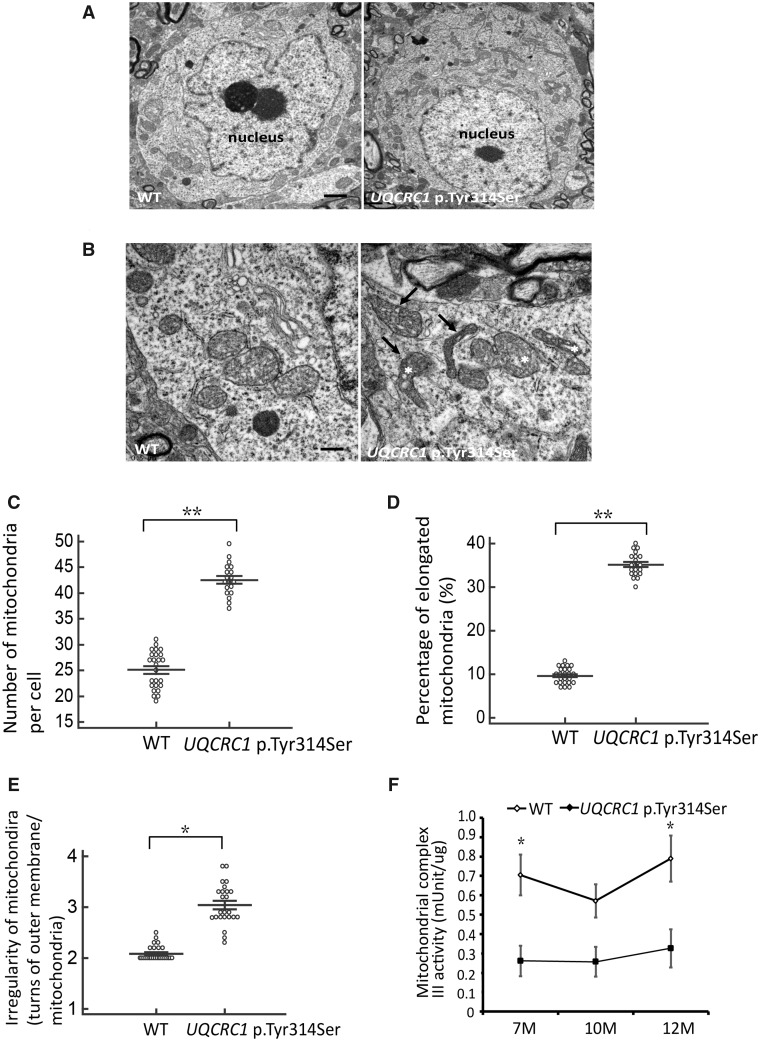
To confirm and extend our observations that the UQCRC1 mutation disrupts mitochondrial function, transmission electron microscopy (TEM) was used to visualize mitochondrial morphology in nigral neurons from UQCRC1 p.Tyr314Ser animals. Ultrastructural analysis revealed multiple morphological defects in neuronal mitochondria in UQCRC1 p.Tyr314Ser knock-in mice. At 9 months of age, compared to wild-type littermate controls, UQCRC1 p.Tyr314Ser knock-in mice had a significantly higher numbers of intra-neuronal mitochondria (Fig. 7A; statistics in Fig.7C). Notably, many mitochondria in UQCRC1 p.Tyr314Ser neurons were irregularly shaped and elongated (Fig. 7B) and appeared to be vacuolated (Fig. 7B). Specifically, 35% of neuronal mitochondria in UQCRC1 p.Tyr314Ser mutants were abnormally elongated (Fig. 7D, P = 0.007) and uncharacteristically irregular in shape with more turns in the outer membrane per mitochondrion compared to wild-type mice (Fig. 7E, P = 0.021).
Figure 7.
Altered mitochondrial ultra-structures and reduced complex III activity in nigral neurons. (A) Representative images of cell bodies of nigral neurons from (left) wild-type and (right) UQCRC1 p.Tyr314Ser knock-in mice at 9 months of age. Scale bar = 2 µm. (B) Close examination reveals that many mitochondria in (right) UQCRC1 p.Tyr314Ser neurons are irregularly shaped (arrows) and appear fragmented (asterisks) compared to (left) wild-type neurons. Scale bar = 0.5 µm. (C) The mean number of mitochondria counted per cell body. Wild-type versus UQCRC1 p.Tyr314Ser; 25.12 ± 0.24 versus 42.93 ± 0.51; F(1,42) = 285, P = 0.008 by one-way ANOVA. (D) The percentage of elongated mitochondria calculated by counting the elongated mitochondria and normalizing to the total number of mitochondria. Wild-type versus UQCRC1 p.Tyr314Ser; 9.8% ± 0.89% versus 35.12% ± 0.31%; F(1,42) = 1404, P = 0.007 by one-way ANOVA. (E) Irregularity of mitochondrial morphology was calculated as the number of turns in the outer membrane per mitochondria. Wild-type versus UQCRC1 p.Tyr314Ser; 2.12 ± 0.18 versus 3.05 ± 0.42; P = 0.021 by one-way ANOVA. n = 5 for each genotype; n = 20 images per group. (F) Mitochondrial fractions of tissue homogenates of substantia nigra and striatum from wild-type (n = 3) and UQCRC1 p.Tyr314Ser knock-in mice (n = 3) at variable ages were examined for complex III activity. There is a decrease of complex III activity in UQCRC1 p.Tyr314Ser knock-in mice compared to wild-type mice at the age of 7 months and 12 months. Wild-type versus UQCRC1 p.Tyr314Ser; 0.71 ± 0.01 versus 0.26 ± 0.04; F(1,4) = 15, P = 0.01 by one-way ANOVA for the age of 7 months; wild-type versus UQCRC1 p.Tyr314Ser; 0.57 ± 0.02 versus 0.25 ± 0.03; F(1,4) = 3.89, P = 0.08 by one-way ANOVA for the age of 10 months; wild-type versus UQCRC1 p.Tyr314Ser; 0.76 ± 0.08 versus 0.32 ± 0.04; F(1,4) = 11.13, P = 0.02 by one-way ANOVA for the age of 12 months. n = 3 for each genotype in individual ages. *P < 0.05, **P < 0.01.
We further assessed the effect of UQCRC1 p.Tyr314Ser on respiratory chain complex III activity in the mitochondrial fractions of SNc and striatum homogenates from wild-type littermates and UQCRC1 p.Tyr314Ser knock-in mice with age. The complex III activity in substantia nigra and striatum of UQCRC1 p.Tyr314Ser knock-in mice was significantly reduced compared to wild-type littermates, starting from 7 months of age (Fig. 7F). Collectively, these results suggest that UQCRC1 p.Tyr314Ser mutation causes altered ultrastructural morphology in mitochondria, suggesting a role for UQCRC1 in the maintenance of proper mitochondrial function in nigral dopaminergic neurons.
Discussion
We provide functional evidence to implicate mutations in UQCRC1, encoding a subunit of mitochondrial respiratory chain complex III, contribute to familial parkinsonism. As the identified genetic variants in UQCRC1 are very rare, functional analyses applying both in vitro and in vivo models are crucial to prove the pathogenicity of UQCRC1 mutations in Parkinson’s disease-related neurodegeneration. Human clinical phenotypes are observed in heterozygous animal models, whereas heterozygous UQCRC1 p.Tyr314Ser knock-in flies and mice successfully recapitulate aspects of the motor dysfunction, presynaptic nerve degeneration, and neurochemical dopamine insufficiency features of human disease.
We demonstrate that UQCRC1 p.Ile311Leu and p.Tyr314Ser coding substitutions segregate with parkinsonism in families with autosomal-dominant inheritance. Affected heterozygotes present with asymmetrical tremor-predominant parkinsonism and findings of Tc-99m TRODAT scan are compatible with idiopathic Parkinson’s disease. The age of onset ranges from the late forties to mid-sixties and all patients have a good response to levodopa. Neither mutation was observed among >1000 control subjects. Both substitutions perturb evolutionarily conserved amino acids within the same domain, and crystal structure analysis suggests that both are likely to be deleterious to protein function. The Japanese Family C with autosomal-dominant parkinsonism was also identified with a rare UQCRC1 variant and was proven to affect splicing, which may lead to pathogenicity. DNA samples for other family members of Family C were lacking and we were unable to assess segregation with disease. Additionally, a rare variant in CLCN6, p.D555N, was also shared by some affected members of Family A and was considered a potential candidate variant for disease. CLCN6 encodes a chloride channel exclusively expressed in the central and peripheral nervous systems, in late endosomes and proximal axons (Poët et al., 2006). CLCN6 disruption in mice leads to moderate behavioural abnormalities, reduced pain sensitivity and pathological enlargement of proximal axons with autofluorescence, contributing to a rare neurological disorder, neuronal ceroid lipofuscinosis (Poët et al., 2006). However, as the CLCN6 p.D555N variant is almost as rare as a mutation, further functional analysis for this variant is warranted to assess the independent and bigenic contribution with UQCRC1 in Parkinson’s disease. Overall, genetic evidence for pathogenicity of UQCRC1 is limited by the size of Families B and C, and the rarity of the changes observed, and hence functional analyses are critical to illustrate the pathogenicity of UQCRC1 mutations.
Mitochondrial respiratory chain complex III is composed of 11 subunits that are arranged as a homodimer, embedded in the inner mitochondrial membrane (Iwata et al., 1998). UQCRC1 is one of the core proteins of complex III, a subunit required for oxidative phosphorylation and ATP synthesis (Unni et al., 2019). There are crosswalks among the complex III subunits, which cooperate to integrate transfer of electrons from ubiquinone to cytochrome C and proton pumping from the mitochondrial matrix into the inter-membrane space. Therefore, although the mutations in UQCRC1 are in a heterozygous state, the genetic substitutions may destabilize the complex III homodimer protein structure, as predicted in our in silico structure analysis, and perturb mitochondria function. These effects may result in a dominant negative effect and an autosomal-dominant inheritance pattern (Veitia et al., 2018). Another typical example of dominant negative effect is provided by homodimeric membrane receptors. For example, piebaldism is an autosomal dominant pigment anomaly and is caused by heterozygous variants in the KIT gene that encodes a receptor tyrosine kinase (RTK), which is due to a dominant negative effect so that mutant receptors form non-functional dimers with wild-type receptors (Robertson et al., 2000). Complexes I and III are the main producers of superoxide and derived reactive oxygen species (Drose et al., 2012) and are essential for many physiological functions including cell signalling, immune responses and premature ageing (Kumar and Kumar, 2009). Loss of complex III activity results in elevated mitochondrial reactive oxygen species production and may lead to many neurodegenerative diseases, including Parkinson’s disease and neuropathy (Lee et al., 2011). Our findings show both coding substitutions and the aberrant splicing variant impair neurite growth, reduced mitochondrial respiration activity, decrease ATP and elevate reactive oxygen species production. UQCRC1 p.Tyr314Ser knock-in Drosophila exhibit age-dependent locomotor defects and preferential loss of TH-positive neurons. We complement and extend these observations by developing a UQCRC1 p.Tyr314Ser knock-in mouse model. Heterozygous animals show progressive age-associated impairments in motor function, beginning at 9 months, accompanied by functional deficits in striatal dopaminergic innervation and loss of TH-positive neurons in the SNc. Peripheral impairments in nerve conduction amplitudes and altered nerve fibre pathology in sciatic nerves from the UQCRC1 p.Tyr314Ser knock-in mice also mirror the observations made in mutant flies and may help explain the peripheral neuropathy described in heterozygous UQCRC1 p.Tyr314Ser patients. The changes seen in post-mitotic mitochondrial abundance, function, and ultrastructure in UQCRC1 p.Tyr314Ser knock-in mice were congruent with mitochondrial complex III dysfunction.
Our findings corroborate and extend past literature by demonstrating UQCRC1 mutations in mitochondrial respiratory chain complex III may be directly involved in the pathogenesis of Parkinson’s disease. In contrast to PRKN (parkin) or PINK1 mutant and knock-out Drosophila, that display severe wing defects and male sterility (Clark et al., 2006; Park et al., 2006), UQCRC1 p.Tyr314Ser knock-in flies are comparable to wild-type flies in that regard. Parkin and PINK1 are critical regulators of mitophagy (Burman et al., 2012; Ashrafi et al., 2014). Lack of wing phenotypes of UQCRC1 p.Tyr314Ser knock-in flies suggests mutant UQCRC1 involves a different pathway way than PINK1-parkin, leading to functional mitochondrial deficits rather than mitophagy dysfunction. Nevertheless, our findings showed marked mitochondria morphology changes in the nigra neurons, which is similar with the findings in PINK1 knock-out mice (Wood-Kaczmar et al., 2008). One recent study revealed that fibroblasts from patients with mitochondrial respiratory chain complex I deficiency had altered mitochondria morphology and disruption of mitochondrial bioenergetics and dynamics (Leipnitz et al., 2018) supporting a link between respiratory chain function and mitochondria morphogenesis. As mitochondria morphology is maintained by a balanced mitochondrial dynamic and cellular bioenergetic demands, several studies have reported effects of mitochondrial dysfunction on mitochondrial morphology through regulating the interplay between mitochondria fusion and fission events (Wu et al., 2011). We hypothesize that increased reactive oxygen species and mitochondria dysfunction may affect the expression and function of fusion related (MFN1, MFN2, OPA1) or fission related proteins (DRP1), resulting in altered mitochondria ultrastructure changes. Future studies are needed to explore the potential molecular cross-talks between UQCRC1 and mitochondria dynamics-related proteins (Tilokani et al., 2018).
The UQCRC1 p.Tyr314Ser knock-in mice had age-dependent progressive dopaminergic neuronal loss with l-DOPA responsive locomotor defects but lack of intra-neuronal α-synuclein accumulations. As α-synuclein is a major component of Lewy bodies, which is pathognomonic in Parkinson’s disease, its aggregation is a leading candidate for the aetiology of the disease. However, consistent with our findings, numerous mouse models and cell culture models of monogenic Parkinson’s disease causative genetic mutations, except overexpression of SNCA, have failed to demonstrate intra-neuronal α-synuclein aggregations (Lim and Ng, 2009). For example, mutant PINK1 or PRKN have attempted to elucidate whether there is an interaction between mitochondria dysfunction and α-synuclein accumulations in neurons; and PINK1 was recently reported to regulate immune-mediated neuroinflammatory responses (Matheoud et al., 2019). However, to date, it is still unclear if this is the case (Stichel et al., 2007; Gautier et al., 2008). In addition, no mouse models of LRRK2 mutations, which are common risk factors for familial and sporadic Parkinson’s disease and associated with mitochondria dysfunction, develop Lewy body-like α-synuclein-positive inclusions (Yue and Lachenmayer, 2011). Thus, mitochondrial dysfunction and α-synuclein aggregations appear to be independent, albeit concomitant processes. How mitochondrial dysfunction impairs proteostasis and leads to α-synuclein accumulation is worthy of further investigation. Furthermore, in addition to age-dependent dopaminergic neuronal degeneration beginning at the age of 9 months of the UQCRC1 p.Tyr314Ser knock-in mutant mice, we also observed a significant loss of the hippocampal neurons in UQCRC1 p.Tyr314Ser knock-in mice at the age of 12 months. These observations were coherent with previous studies showing patients with Parkinson’s disease have three to six times higher risks of developing dementia than age-matched subjects who do not have Parkinson’s disease (Aarsland et al., 2001). Although patients of Family A did not reveal marked cognitive decline as shown by Mini-Mental State Examination (MMSE) scores, their brain MRI scans revealed mild diffuse cortical atrophy at advanced stage of the disease as shown in Table 1. However, the cognition function was evaluated using MMSE, a simple test for global cognitive impairment, in the current study. A future follow-up with detailed neuropsychological tests evaluating individual cognitive domains is needed to provide more information for assessing the impact of UQCRC1 mutations on cognitive function in affected members.
Although our findings demonstrate heterozygous UQCRC1 p.Tyr314Ser knock-in flies and mice successfully recapitulate human disease phenotypes and provide functional evidence of UQCRC1 variations disturbing mitochondrial function, our study has some limitations. First, the low frequency of mutations in UQCRC1 in familial Parkinson’s disease suggest variants in this gene are rare genetic cause for parkinsonism in Asians. Future studies screening UQCRC1 in autosomal-dominant parkinsonism with polyneuropathy in other ethnicities are needed to confirm our findings. Second, we did not have the chance to access the nerve conduction studies for affected patients in Families B and C, which may hamper us from having a complete landscape of clinical phenotypes of UQCRC1 mutations. Further screening of UQCRC1 in large cohorts of CMT families without common causative genetic mutations in different ethnicities is worth pursuing to have a better understanding of mitochondrial dysfunction and the role of UQCRC1 in peripheral nerve degeneration. Finally, post-mortem brain tissues from patients with UQCRC1 mutations are not available yet to confirm our findings from the in vivo models.
In conclusion, our study provides functional evidence demonstrating that rare genetic variants in the UQCRC1 gene coding for mitochondria complex III protein may lead to autosomal dominant inherited parkinsonism. Our findings provide an additional link between mitochondrial dysfunction and the pathogenesis of Parkinson’s disease.
Supplementary Material
Acknowledgements
We thank all the patients that participated in this study. We are also grateful for the technical assistance provided by the First and Second Core Labs in the National Taiwan University College of Medicine, Taipei, Taiwan. We thank the staff of the Human Disease Modeling Center and Gene Knockout/in Cell Line Modeling Core at the First Core Labs, National Taiwan University College of Medicine, for technical assistance. We are also grateful for technical assistance of Electron Microscope Laboratory of Tzong Jwo Jang, College of Medicine, Fu Jen Catholic University and Dr. Chung-Yin Lin from Chang Gung University for her assistance in high performance liquid chromatography (HPLC) assay.
Funding
We are grateful to the following organizations for their support of this work: Ministry of Science and Technology (MOST106-2314-B-002-072-MY3 and MOST 108-2321-B-002-060-MY2), Excellent Translational Medicine Research Projects of National Taiwan University College of Medicine, and National Taiwan University Hospital (97-N-979, UN106-010, 107C101-82 and 108C101-22). This project was also supported in part through funds from the Canada Excellence Research Chair, British Columbia Leadership Endowment funds and the Canadian Foundation for Innovation (M.J.F.).
Competing interests
The authors report no competing interests
Supplementary material
Supplementary material is available at Brain online.
Glossary
- NMJ =
neuromuscular junction
References
- Aarsland D, Andersen K, Larsen JP, Lolk A, Nielsen H, Kragh-Sorensen P.. Risk of dementia in Parkinson's disease: a community-based, prospective study. Neurology 2001; 56: 730–6. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeby-Moller K, Berge OG, Hamers FP.. Using the CatWalk method to assess weight-bearing and pain behaviour in walking rats with ankle joint monoarthritis induced by carrageenan: effects of morphine and rofecoxib. J Neurosci Methods 2008; 174: 1–9. [DOI] [PubMed] [Google Scholar]
- Ashrafi G, Schlehe JS, LaVoie MJ, Schwarz TL.. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol 2014; 206: 655–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL.. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 2013; 4: 220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT.. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci 2000; 3: 1301–6. [DOI] [PubMed] [Google Scholar]
- Burman JL, Yu S, Poole AC, Decal RB, Pallanck L.. Analysis of neural subtypes reveals selective mitochondrial dysfunction in dopaminergic neurons from parkin mutants. Proc Natl Acad Sci USA 2012; 109: 10438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charan J, Kantharia ND.. How to calculate sample size in animal studies? J Pharmacol Pharmacother 2013; 4: 303–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006; 441: 1162–6. [DOI] [PubMed] [Google Scholar]
- Drose S, Brandt U.. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol 2012; 748: 145–69. [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A.. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007; 39: 175–91. [DOI] [PubMed] [Google Scholar]
- Ferreira M, Massano J.. An updated review of Parkinson's disease genetics and clinicopathological correlations. Acta Neurol Scand 2017; 135: 273–84. [DOI] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J.. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci USA 2008; 105: 11364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb DJ, Oliver E, Gilman S.. Diagnostic criteria for Parkinson disease. Arch Neurol 1999; 56: 33–9. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP.. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 2004; 166: 1775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen HJ, Jensen EB, Kiêu K, Nielsen J.. The efficiency of systematic sampling in stereology–reconsidered. J Microsc 1999; 193: 199–211. [DOI] [PubMed] [Google Scholar]
- Guo R, Zong S, Wu M, Gu J, Yang M.. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 2017; 170: 1247–57. [DOI] [PubMed] [Google Scholar]
- Hardy J, Lewis P, Revesz T, Lees A, Paisan-Ruiz C.. The genetics of Parkinson's syndromes: a critical review. Curr Opin Genet Dev 2009; 19: 254–65. [DOI] [PubMed] [Google Scholar]
- Hoffman GG, Lee S, Christiano AM, Chung-Honet LC, Cheng W, Katchman S, et al. Complete coding sequence, intron/exon organization, and chromosomal location of the gene for the core I protein of human ubiquinol-cytochrome c reductase. J Biol Chem 1993; 268: 21113–9. [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 1998; 281: 64–71. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW.. Biochemical assays of respiratory chain complex activity. Methods Cell Biol 2007; 80: 93–119. [DOI] [PubMed] [Google Scholar]
- Kumar P, Kumar A.. Possible role of sertraline against 3-nitropropionic acid induced behavioral, oxidative stress and mitochondrial dysfunctions in rat brain. Prog Neuropsychopharmacol Biol Psychiatry 2009; 33: 100–8. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015; 524: 309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DW, Selamoglu N, Lanciano P, Cooley JW, Forquer I, Kramer DM, et al. Loss of a conserved tyrosine residue of cytochrome b induces reactive oxygen species production by cytochrome bc1. J Biol Chem 2011; 286: 18139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NC, Shieh YD, Chien YH, Tzen KY, Yu IS, Chen PW, et al. Regulation of the dopaminergic system in a murine model of aromatic L-amino acid decarboxylase deficiency. Neurobiol Dis 2013; 52: 177–90. [DOI] [PubMed] [Google Scholar]
- Leipnitz G, Mohsen AW, Karunanidhi A, Seminotti B, Roginskaya VY, Markantone DM, et al. Evaluation of mitochondrial bioenergetics, dynamics, endoplasmic reticulum-mitochondria crosstalk, and reactive oxygen species in fibroblasts from patients with complex I deficiency. Sci Rep 2018; 8: 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Ng CH.. Genetic models of Parkinson disease. Biochim Biophys Acta 2009; 1792: 604–15. [DOI] [PubMed] [Google Scholar]
- Lin CH, Chen PL, Tai CH, Lin HI, Chen CS, Chen ML, et al. A clinical and genetic study of early-onset and familial parkinsonism in Taiwan: an integrated approach combining gene dosage analysis and next-generation sequencing. Mov Disord 2019; 34: 506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Lin HI, Chen ML, Lai TT, Cao LP, Farrer MJ, et al. Lovastatin protects neurite degeneration in LRRK2-G2019S parkinsonism through activating the Akt/Nrf pathway and inhibiting GSK3beta activity. Hum Mol Genet 2016; 25: 1965–78. [DOI] [PubMed] [Google Scholar]
- Lin CH, Tsai PI, Wu RM, Chien CT.. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ss. J Neurosci 2010; 30: 13138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N.. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 2008; 40: 476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheoud D, Cannon T, Voisin A, Penttinen AM, Ramet L, Fahmy AM, et al. Intestinal infection triggers Parkinson's disease-like symptoms in Pink1-/- mice. Nature 2019; 571: 565–9. [DOI] [PubMed] [Google Scholar]
- Niu J, Yu M, Wang C, Xu Z.. Leucine-rich repeat kinase 2 disturbs mitochondrial dynamics via Dynamin-like protein. J Neurochem 2012; 122: 650–8. [DOI] [PubMed] [Google Scholar]
- Oruganty-Das A, Ng T, Udagawa T, Goh EL, Richter JD.. Translational control of mitochondrial energy production mediates neuron morphogenesis. Cell Metab 2012; 16: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006; 441: 1157–61. [DOI] [PubMed] [Google Scholar]
- Poët M, Kornak U, Schweizer M, Zdebik AA, Scheel O, Hoelter S, et al. Lysosomal storage disease upon disruption of the neuronal chloride transport protein ClC-6. Proc Natl Acad Sci USA 2006; 103: 13854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringsheim T, Jette N, Frolkis A, Steeves TD.. The prevalence of Parkinson's disease: a systematic review and meta-analysis. Mov Disord 2014; 29: 1583–90. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SC, Tynan JA, Donoghue DJ.. RTK mutations and human syndromes: when good receptors turn bad. Trends Genet 2000; 16: 368. [DOI] [PubMed] [Google Scholar]
- Rossor AM, Polke JM, Houlden H, Reilly MM.. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat Rev Neurol 2013; 9: 562–71. [DOI] [PubMed] [Google Scholar]
- Salabei JK, Gibb AA, Hill BG.. Comprehensive measurement of respiratory activity in permeabilized cells using extracellular flux analysis. Nat Protoc 2014; 9: 421–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Gegg M.. Mitochondrial contribution to Parkinson's disease pathogenesis. Parkinsons Dis 2011; 2011: 159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Y, Villalba R.. Striatal and extrastriatal dopamine in the basal ganglia: an overview of its anatomical organization in normal and Parkinsonian brains. Mov Disord 2008; 23: S534–547. [DOI] [PubMed] [Google Scholar]
- Stichel CC, Zhu XR, Bader V, Linnartz B, Schmidt S, Lübbert H.. Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum Mol Genet 2007; 16: 2377–93. [DOI] [PubMed] [Google Scholar]
- Su YC, Qi X.. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum Mol Genet 2013; 22: 4545–61. [DOI] [PubMed] [Google Scholar]
- Tilokani L, Nagashima S, Paupe V, Prudent J.. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem 2018; 62: 341–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh J, Farrer M.. Advances in the genetics of Parkinson disease. Nat Rev Neurol 2013; 9: 445–54. [DOI] [PubMed] [Google Scholar]
- Tsai PI, Lin CH, Hsieh CH, Papakyrikos AM, Kim MJ, Napolioni V, et al. PINK1 phosphorylates MIC60/Mitofilin to control structural plasticity of mitochondrial crista junctions. Mol Cell 2018; 69: 744–56. [DOI] [PubMed] [Google Scholar]
- Tzoulis C, Tran GT, Schwarzlmüller T, Specht K, Haugarvoll K, Balafkan N, et al. Severe nigrostriatal degeneration without clinical parkinsonism in patients with polymerase gamma mutations. Brain 2013; 136: 2393–404. [DOI] [PubMed] [Google Scholar]
- Unni S, Thiyagarajan S, Srinivas Bharath MM, Padmanabhan B.. Tryptophan oxidation in the UQCRC1 subunit of mitochondrial complex III (ubiquinol-cytochrome C reductase) in a mouse model of myodegeneration causes large structural changes in the complex: a molecular dynamics simulation study. Sci Rep 2019; 9: 10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA, Caburet S, Birchler JA.. Mechanisms of Mendelian dominance. Clin Genet 2018; 93: 419–28. [DOI] [PubMed] [Google Scholar]
- Wood-Kaczmar A, Gandhi S, Yao Z, Abramov AY, Miljan EA, Keen G, et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PloS One 2008; 3: e2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhou F, Zhang Z, Xing D.. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J 2011; 278: 941–54. [DOI] [PubMed] [Google Scholar]
- Xia RH, Yosef N, Ubogu EE.. Dorsal caudal tail and sciatic motor nerve conduction studies in adult mice: technical aspects and normative data. Muscle Nerve 2010; 41: 850–6. [DOI] [PubMed] [Google Scholar]
- Yue M, Hinkle KM, Davies P, Trushina E, Fiesel FC, Christenson TA, et al. Progressive dopaminergic alterations and mitochondrial abnormalities in LRRK2 G2019S knock-in mice. Neurobiol Dis 2015; 78: 172–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Lachenmayer ML.. Genetic LRRK2 models of Parkinson's disease: dissecting the pathogenic pathway and exploring clinical applications. Mov Disord 2011; 26: 1386–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med 2010; 2: 52–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author.