Advances in the complexity and functionality of in vitro models of the CNS mean that such models now have the potential to overcome many of the limitations of traditional model systems. Nikolakopoulou et al. provide an overview and user guide for in vitro CNS models, with a focus on ‘Organ-on-a-Chip’ technologies.
Keywords: organ-on-a-chip, in vitro model, CNS models, neurodegenerative disease, translational medicine
Abstract
The complexity of the human brain poses a substantial challenge for the development of models of the CNS. Current animal models lack many essential human characteristics (in addition to raising operational challenges and ethical concerns), and conventional in vitro models, in turn, are limited in their capacity to provide information regarding many functional and systemic responses. Indeed, these challenges may underlie the notoriously low success rates of CNS drug development efforts. During the past 5 years, there has been a leap in the complexity and functionality of in vitro systems of the CNS, which have the potential to overcome many of the limitations of traditional model systems. The availability of human-derived induced pluripotent stem cell technology has further increased the translational potential of these systems. Yet, the adoption of state-of-the-art in vitro platforms within the CNS research community is limited. This may be attributable to the high costs or the immaturity of the systems. Nevertheless, the costs of fabrication have decreased, and there are tremendous ongoing efforts to improve the quality of cell differentiation. Herein, we aim to raise awareness of the capabilities and accessibility of advanced in vitro CNS technologies. We provide an overview of some of the main recent developments (since 2015) in in vitro CNS models. In particular, we focus on engineered in vitro models based on cell culture systems combined with microfluidic platforms (e.g. ‘organ-on-a-chip’ systems). We delve into the fundamental principles underlying these systems and review several applications of these platforms for the study of the CNS in health and disease. Our discussion further addresses the challenges that hinder the implementation of advanced in vitro platforms in personalized medicine or in large-scale industrial settings, and outlines the existing differentiation protocols and industrial cell sources. We conclude by providing practical guidelines for laboratories that are considering adopting organ-on-a-chip technologies.
Introduction
The human brain has a complex physiology that distinguishes it from other animal brains. Features that are unique to the human brain include its large size, its highly folded cortex, and the unique pathophysiology of human neural diseases. The hierarchical complexity of the human brain (Supplementary Fig. 1A) poses a substantial challenge for the development of models for research of CNS pathologies: Animal models lack many essential human characteristics and are associated with many specific interspecies differences; in addition, they are expensive, their throughput is low, and they raise ethical concerns. Conventional in vitro models, in turn, cannot provide information regarding behavioural responses, many functional responses, or systemic responses (organ–organ interactions), and are therefore considered to be too simplistic for many practical applications. The lack of adequate CNS models is one explanation for the low success rate of CNS drug development (Kesselheim et al., 2015; Gribkoff and Kaczmarek, 2017); this low success rate has led many major pharmaceutical companies to limit their research and development investments in the neurological domain (Wegener and Rujescu, 2013).
The lack of adequate CNS models has spurred academic and industrial researchers to seek out new technologies for mimicking brain physiology and functionality in health and disease, by using tools such as induced pluripotent stem cells (iPSCs) (reviewed in Shi et al., 2017), organ-on-a-chip (OoC) systems, organoids (Pașca, 2018), 3D printed gels (Hopkins et al., 2015) and neuronal machine interfaces (Moxon and Foffani, 2015). Though none of these methods can fully capture the complex physiology, anatomy and functionality of the human brain (or of any other whole organ, for that matter), they are nonetheless showing very promising results in terms of their capacity to recapitulate certain human functions or pathological mechanisms, as well as to reveal new physiological interactions that could not have been identified with current standard tools in vitro or in vivo. To give the reader a quick overview of different models of the CNS we have compared the use of rodent models versus conventional cultures, organoids and OoCs, rating each model's usability for specific CNS studies (Table 1). Advanced in vitro platforms are rapidly becoming more accessible in terms of cost, ease of use and availability. So far, however, use of these tools has tended to be limited to the laboratories in which the technologies were developed, suggesting that their adoption by members of the wider CNS research community has lagged behind. This gap suggests a need to raise CNS researchers’ awareness of the variety of novel CNS models that are currently available and that might serve to complement traditional in vitro and in vivo models, thereby enhancing the overall accuracy of preclinical evaluations aimed at predicting clinical outcomes.
Table 1.
Overviewing comparison of rodent in vivo models
| Human relevance | Disease models | Systemic effects | Brain regions | Behaviour | Electrophysiology | Mechanistic studies | ADME/ TOX | HTS | Cost | |
|---|---|---|---|---|---|---|---|---|---|---|
| Standard 2D cultures | ||||||||||
| Human primary | +++ | ++ | – | ++ | – | ++ | ++ | + | + | ++ |
| Human iPSC | +++ | +++ | – | ++ | – | ++ | +++ | + | +++ | +++ |
| Rodent primary | – | ++ | – | +++ | – | +++ | +++ | + | + | ++ |
| Cell lines | + | + | – | – | – | + | +++ | + | +++ | ++ |
| Organoids | ||||||||||
| Human primary | +++ | +++ | – | ++ | – | – | ++ | – | – | ++ |
| Human iPSC | +++ | +++ | – | + | – | + | ++ | + | ++ | ++ |
| Rodent primary | – | + | – | +++ | – | + | ++ | – | – | ++ |
| Cell lines | + | + | – | – | – | + | ++ | – | ++ | + |
| OoC | ||||||||||
| Human primary | +++ | ++ | ++ | ++ | – | ++ | ++ | ++ | – | ++ |
| Human iPSC | +++ | +++ | ++ | ++ | – | ++ | +++ | ++ | – | ++ |
| Rodent primary | – | ++ | ++ | +++ | – | +++ | +++ | + | – | + |
| Cell lines | + | + | ++ | – | – | + | +++ | ++ | – | + |
| Rodent in vivo | – | ++ | +++ | +++ | +++ | ++ | + | ++ | – | +++ |
Table shows rodent in vivo models (the most commonly used mammal), standard 2D cell culture models, organoid cultures and OoC for their human specificity and their capacity to model human diseases, systemic effects, brain regionality, behaviour, drug absorption, distribution, metabolism and excretion, and toxicity (ADME/TOX). We also rate the possibility for electrophysiological studies, detailed mechanistic studies, high throughput studies (HTS), and the cost of the model. For the three in vitro models, we divided them into the accessible cell sources, human primary cells, rodent primary cell and hiPCS, and cell lines. Notably, we want to emphasize that human primary cells from the CNS are scarce. We further wish to highlight that this rating, the appropriateness of each model, varies for each specific study, and our rating should be used as a general guideline of what is possible to achieve with each model. – = poor/non-existent; + = OK; ++ = good; +++ = excellent.
Accordingly, in this review we will report on recent developments of in vitro CNS models, and will discuss how these models can address some of the challenges associated with current in vivo models. We focus our discussion on engineered in vitro models, i.e. models that involve a technical approach to control the organization of cells, and briefly mention self-assembled structures such as spheres and organoids. We begin by providing an overview of some of the main recent developments in in vitro CNS models. Next, given that a key goal of advanced in vitro platforms is to enhance the translatability of experimental results—which implies reliance on human tissue sources and, specifically, human-derived iPSCs (hiPSCs)—we discuss some of the challenges associated with integration of hiPSCs into these platforms. We then provide a detailed explanation of some of the fundamental principles underlying engineered in vitro models of the healthy CNS and discuss several applications of these models in practice. We subsequently discuss the most relevant models of CNS disease. We conclude by providing practical guidelines for biomedical labs that are considering adopting these technologies and give an outlook for future developments.
Overview of recent developments in in vitro CNS models
The CNS is an intricate cellular network, consisting of neurons, astrocytes, oligodendrocytes, pericytes, immune cells, and vascular endothelium embedded in a tissue-specific extracellular microenvironment (Rauti et al., 2019). Understanding the physiological cellular mechanisms of the CNS is essential for identifying potential drug targets, as well as for predicting drug side effects and the pathogenesis of neurological diseases.
In addition to the distinct cellular subtypes, organization and interconnectivity, it is imperative to recapitulate the extracellular milieu when generating translatable in vitro CNS models. Hence, all the physical, chemical, and mechanical cues of the extracellular matrix (ECM) should be considered (Frantz et al., 2010; Abdeen et al., 2016; Uwamori et al., 2017). The brain ECM has a distinct structure and composition that differentiates it from other organs and it is organized in three different compartments: the basement membrane, the perineuronal nets and the interstitial matrix (Novak and Kaye, 2000; Yamaguchi, 2000; Bonneh-Barkay and Wiley, 2009; Lau et al., 2013; Rauti et al., 2019). The basement membrane mainly consists of collagen IV, laminin–nidogen complexes, fibronectin, heparan sulphate proteoglycans such as perlecan and agrin (Baeten and Akassoglou, 2011; Xu et al., 2019) and a plethora of growth factors (Barcelona and Saragovi, 2015). The perineuronal nets are mainly composed of hyaluronan, proteoglycans, such as chondroitin sulphate proteoglycans (e.g. aggrecan, brevican, phosphacan and neurocan) and heparan sulphate proteoglycans (e.g. syndecan and glypican), while the interstitial matrix is composed of proteoglycans, hyaluronic acid, tenascins and fibrous proteins (e.g. collagen and elastin) as well as glycoproteins (Novak and Kaye, 2000; Rauti et al., 2019). The extracellular milieu of the brain serves as a physical support for cell migration as well relaying mechanical and biochemical stimuli that influences cell growth and differentiation (García-Parra et al., 2013; Levy et al., 2014; Potjewyd et al., 2018; Rauti et al., 2019).
Currently, the most extensively explored CNS-derived in vitro models consist of 2D and 3D cellular cultures of various species (Zhuang et al., 2018). The first 2D in vitro animal CNS models were established by Harrison and Hoadley (Hoadley, 1924; Waddington and Cohen, 1936), who were also the first to observe neurite extensions. Since then, improvements in the capacity to maintain healthy in vitro CNS cultures for long periods enabled additional primary models to be developed, based on cultures of hippocampal cells (Fig. 1A) (Dotti et al., 1988; Barrejón et al., 2019), cortical cells, midbrain cells (Brewer, 1995; Lingor et al., 1999; Pacitti et al., 2019), astrocytes and microglial cells (Giulian and Baker, 1986). In parallel, organotypic cultures, including cultures of hippocampal tissue (Fig. 1B), the substantia nigra, and many others also emerged (LaVail and Wolf, 1973; Knopfel et al., 1989; Robertson et al., 1997); these models were suggested to better resemble in vivo conditions compared with 2D monocultures, yet lack cytoarchitecture, physiological perfusion, and cannot be scaled to larger studies of human tissue (Humpel, 2015).
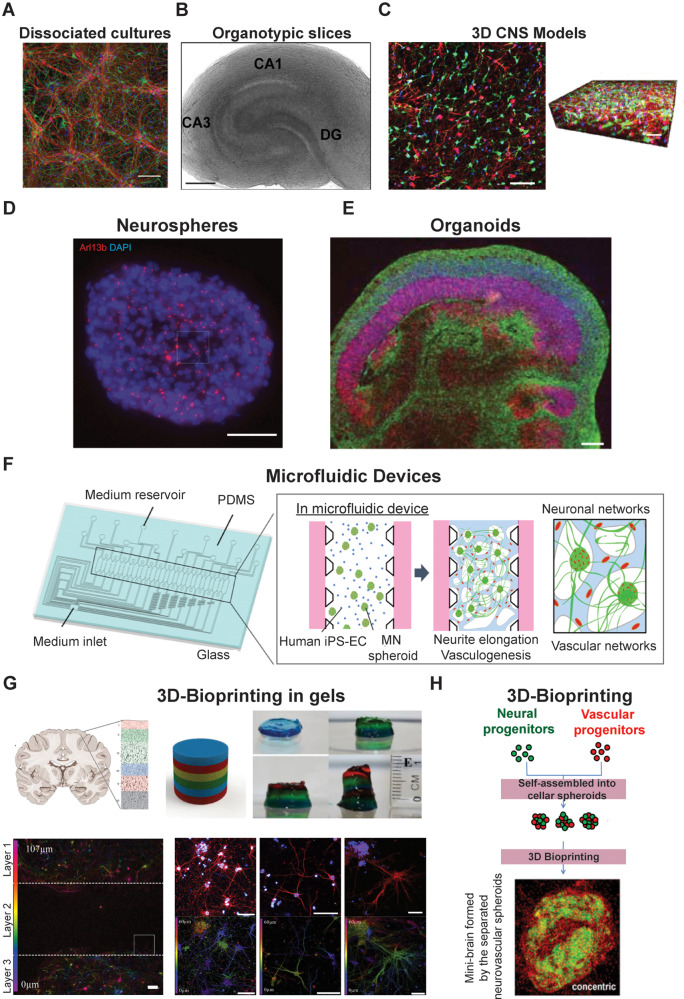
Figure 1.
In vitro CNS models. (A) Confocal micrographs showing 2D-hippocampal dissociated cultures, immunostained for the cytoskeletal component β-tubulin III (in red), the glial protein GFAP (in green) and DAPI to visualize neurons (in blue). Scale bar = 100 µm. Modified from Barrejón et al. (2019) with permission. (B) Light micrograph of a hippocampal slice [modified from Miller et al. (2015) with permission]. (C) Confocal section and 120-µm thick 3D stacks reconstruction showing a 3D hydrogel-encapsulated cortical neuronal network, immunostained for neurons (red, β-tubulin III), glia (green, S100) and nuclei (blue, DAPI). Scale bar = 50 µm [modified from Dana et al. (2014) with permission]. (D) Confocal image representing neurosphere processed for immunofluorescence against Arl13b (red) and DNA [modified from Shimada et al. (2017) with permission]. (E) A representative image of an organoid immunostained for neurons (TUJ1, green) and progenitors (SOX2, red) [modified from Lancaster and Knoblich (2014) with permission]. (F) Schematic image of a microfluidic device where vascular and neuronal networks were co-cultured [modified from Osaki et al. (2018b) with permission]. (G) Schematic representation of in vivo and in vitro cortical brain layer structures, in which each colour represents a different printed layer. In the bottom panel, confocal reconstructions of the neurons coloured for their z-axis distribution through the gel after 5 days of culture. Scale bar = 100 µm [modified from Lozano et al. (2015) with permission]. (H) Schematic sketch of a potential 3D-printing procedure to generate a mini-brain from cellular spheroids [modified from Han and Hsu (2017) with permission].
While both primary and organotypic CNS models can provide insights about cellular morphology and functionality, while retaining most of the cells’ in vivo properties (Balgude et al., 2001; Hopkins et al., 2013, 2015), they still come with many limitations, including the challenge in preserving their viability and sterility (Walsh et al., 2005), as well as the variability of cell maturation (Gähwiler, 1981). Importantly, these models are generally animal-derived, and thus do not resemble human physiology, including different degrees of circuit complexity and brain architecture (Herculano-Houzel, 2014; DeFelipe, 2015; Hopkins et al., 2015).
The isolation of human-derived stem cells and hiPSCs created exciting new opportunities for the development of scalable human models in neurobiology (Dubois-Dauphin et al., 2010; Hopkins et al., 2015; Pacitti et al., 2019; Silva and Haggarty, 2020). Moreover, the development of 3D in vitro culture systems (Fig. 1C) can recapitulate more complex cell–cell interactions, opening up the door to more closely resemble in vivo cell environments (Dana et al., 2014; Langhans, 2018; Pașca, 2018). Examples range from cell-self-assembly-based approaches (e.g. spheroids) to scaffold-based platforms (e.g. hydrogels).
Novel in vitro CNS 3D-models, such as neurospheres (Hogberg et al., 2013) and organoids (Lancaster et al., 2013), began to be developed based on stem cells. Neurospheres (Fig. 1D) are self-assembled, dense structures, mainly composed of neural stem cells, neuronal and glial-restricted progenitor cells, postmitotic neuronal cells and dead or dying cells. Neurospheres constitute valuable systems for studying neurogenesis and neural development, and also serve as an almost unlimited source of neural and progenitor cells. A major limitation of such systems is that, because of low access to oxygen and nutrients, cells growing in the centre of the neurospheres die (Bez et al., 2003; Jensen and Parmar, 2006). Other drawbacks that have limited their use include continuous loss in neurogenic potential after some rounds of subculturing, in addition to difficulties in merging findings between different laboratories. Lancaster and Knoblich (2014) introduced, for the first time, the concept of human brain organoids (Fig. 1E), which are self-assembled cells, derived from pluripotent stem cells, featuring lineages and structures of the early embryonic CNS. There has been an exponential surge in the use of this technology, especially as a tool for in vitro disease modelling (Lancaster et al., 2013; Mariani et al., 2015; Jo et al., 2016; Raja et al., 2016; Lee et al., 2017). Organoids are particularly useful for the identification and testing of novel therapeutic approaches, in light of their capacity to adapt to genome editing techniques or gene therapy (Yin et al., 2016; Gonzalez-Cordero et al., 2018). Pellegrini et al. (2020) recently published an in vitro organoid model, which shows key functions of the human choroid plexus: barrier formation and CSF secretion. Drawbacks of organoids include the fact that their spontaneous formation makes it difficult to create reproducible systems in terms of cell types and organization. Furthermore, they lack many features, such as vascular perfusion, mechanical cues and circulating immune cells, which are critical for the physiological functionality of organs (Ingber, 2016).
To combine the advantages of current in vitro and in vivo models (Table 1), a novel platform began to be developed: OoCs, microfluidic devices in which cells and tissues can be cultured in micro-scale volumes and in a controlled microenvironment, designed to mimic specific in vivo cues (Meyvantsson and Beebe, 2008; Meer and Berg, 2012; Halldorsson et al., 2015; MacKerron et al., 2017, Osaki et al., 2018c; Sosa-Hernández et al., 2018; Oddo et al., 2019). These microfluidic cell chambers are also collectively referred to as microphysiological systems. The simplest platform is a single, perfused microfluidic chamber in which one kind of cell type or cell mixture can be grown. In a more complex design, two or more cell-containing chambers in the same chip are separated by membranes, channels or gel regions to allow direct contact or secretome-mediated interactions (Figs 1F and 2), as further described the ‘Engineered in vitro models to mimic CNS physiology’ section. Yet another level of complexity is to connect two or more microdevices (Fig. 1F), containing different cell types, allowing the interaction of different tissues or tissue regions (e.g. blood–brain barrier) (Bhatia and Ingber, 2014; Phan et al., 2017; Oddo et al., 2019). Such systems might ultimately provide the possibility of studying multi-organ physiological systems (Esch et al., 2014; Maschmeyer et al., 2015; Ingber, 2016).
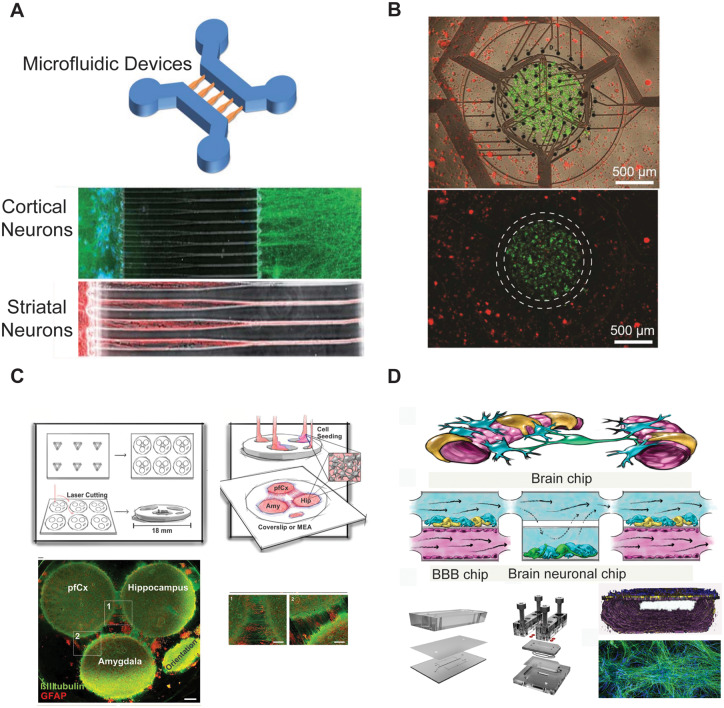
Figure 2.
In vitro culturing of multiple brain regions. (A) Representative image of a microfluidic device (top) and immunofluorescence micrographs (bottom) of cortical (in green) and striatal (in red) neurons growing inside the chips [modified from Peyrin et al. (2011) with permission from The Royal Society of Chemistry]. (B) Novel multielectrode array device used for co-culturing primary rodent hippocampal and cortical neurons [modified from Soscia et al. (2017) with permission]. (C) Schematic representation of a novel brain-on-a-chip model comprising the three different brain regions, prefrontal cortex, hippocampus and amygdala, shown via confocal images (bottom), stained for β-tubulin III (in green), and GFAP (in red), [modified from Dauth et al. (2017) with permission]. (D) Schematic representation of a microfluidic device allowing to metabolically couple neuronal and endothelial cells [modified from Maoz et al. (2018) with permission].
Microfluidic models offer several key advantages, including flexibility in design and low-cost fabrication compared to custom-made large-scale cell culture, fluidics, and robotic systems. Additional benefits over traditional cell culture formats can include lower risk of contamination, lower consumption of reagents, and efficient experimental throughput (Halldorsson et al., 2015). Importantly, OoC technology presents the possibility to apply mechanical forces to recreate physiological movements (Huh et al., 2010; Kim et al., 2016), as well as fluid flow and shear stresses (Bhatia and Ingber, 2014; Bischel et al., 2015; Benam et al., 2016; Ingber, 2016). Nevertheless, there are some challenges limiting the use of OoC systems, such as a fairly long prototyping time, the lack of standardized protocols, the requirement of specialized equipment and complex and time-consuming fabrication processes (Coluccio et al., 2019).
To overcome these issues, researchers have been developing methodologies for 3D printing (Fig. 1G and H) in vitro brain models (Lozano et al., 2015; Han and Hsu, 2017; Hampson et al., 2018; Sivandzade and Cucullo, 2018). This approach enables CNS models to be fabricated along the z-axis using different materials (even living cells), thereby creating 3D structures that are biologically active (Xu et al., 2006; Gu et al., 2016, 2018; Bishop et al., 2017; Han and Hsu, 2017; Thomas and Willerth, 2017; Knowlton et al., 2018; Potjewyd et al., 2018; Oliveira et al., 2019). These in vitro models offer the opportunity to provide a more reliable representation of in vivo nervous tissue with customized design and precise fabrication, facilitating the generation of platforms with great consistency that can be used for drug testing and in clinical applications. Currently, in 3D printing, the most important limitations that need to be addressed are the development of 3D printing methods with minimal impact on cellular stress, reproducible cell technology, a better understanding of molecular gradients in the native nervous tissue (Zhuang et al., 2018), design of the ECM and validation of cellular function (Rauti et al., 2019).
It should be noted that, despite the benefits of 3D OoC setups, the 3D character of these systems makes imaging and data evaluation challenging (Booij et al., 2019). Nevertheless, today, most microfluidic devices are relatively thin (100–1000-µm height and thinner cell layers within these structures). As such, unlike conventional 3D cultures, these devices are often compatible with live imaging experiments, such as cell migration assays, and conventional immunohistochemistry assessment. For example, Deosarkar et al. (2015) used confocal imaging to image the independent vascular channels of their device with dimensions of 200 μm × 100 μm × 2762 μm (width × height × length) (Deosarkar et al., 2015). Recent technological developments in two-photon microscopy (Rakotoson et al., 2019) and advanced 3D imaging, which uses artificial intelligence and machine learning (Joshi et al., 2018; Masullo et al., 2018; Puls et al., 2018; Scheeder et al., 2018; Booij et al., 2019), integration of biosensors (Misun et al., 2016; Maoz et al., 2017), and hiPSC-derived cellular components are expected to advance 3D in vitro modelling. A recent Sciencemag Technical feature comment suggested that the incorporation of biosensors and microfluidics with tissue culture could soon reduce (if not replace in many cases), animal-based research (Dove et al., 2018).
In addition to advancements in cell-based in vitro platforms, it is important to acknowledge the development of non-cell-based in vitro models, which can serve as alternatives to cell-based strategies. Examples of such models include the immobilized artificial membrane assay (IAM), parallel artificial membrane permeability assay (PAMPA), and the solid supported lipid membrane assay (TRANSIL) (Vastag and Keserű, 2009; Sharma et al., 2019). Likewise, computer-based models and simulations, known as in silico models, are becoming increasingly sophisticated (e.g. integrating machine learning and deep learning methods; see Yuan et al., 2018), and can be used to supplement or even replace some experimental procedures. Computer-based models offer the possibility to synthetize, prescreen and virtually test novel drugs, limiting the need for intensive laboratory experiments and expensive clinical trials, and accelerating the drug development process (Naik and Cucullo, 2012; Alsarrani and Kaplita, 2019; Chlebek et al., 2019). Nevertheless, non-cell-based models are not yet sufficient on their own, and the results obtained with such studies must be validated by (cell-based) in vitro and in vivo studies (e.g. to determine the biological activity and the brain distribution of a specific compound) (Naik and Cucullo, 2012).
Indeed, validation, i.e. ensuring that a model faithfully recapitulates in vivo physiological and pathological processes, is essential for the translatability of any model. Such validation is highly challenging in CNS models, owing to the biological complexity of the system being reproduced. Accordingly, extensive efforts are continuously underway for determining the extent to which in vitro CNS responses are representative of their in vivo counterparts. For example, Belle et al. (2018) recently used electrophysiological measurements to detect quantifiable differences but also similarities between cortical neurons in vivo and in vitro. For engineered models, in vitro to in vivo comparisons are highly challenging-yet crucial nevertheless (Frazier, 1990; Belle et al., 2018; Jones et al., 2018). Currently, microfluidic devices are validated testing different annotated and well-known drug compounds.
Another challenge that hinders the translatability of in vitro CNS models relates to the cell populations they use. For example, though the primary purpose of OoCs is to mimic human microphysiological systems in vivo, many of the OoC studies cited in this review relied on (non-human) animal cells or on human primary cells, rather than on cells derived from hiPSCs, which are likely to offer greater translational value (Table 1). It should be noted that, though we believe that hiPSC-based OoCs hold great promise for the future of precision medicine, even hiPSC-based systems may not be perfectly translatable (see Doss and Sachinidis, 2019; Ortuño-Costela et al., 2019 for extensive reviews of the translational concerns raised by iPSCs). Reliance on non-hiPSC-based cell populations is largely driven by the substantial difficulties that scientists still face in their efforts to incorporate hiPSCs (and iPSCs in general) into in vitro systems. As these difficulties are a key hindrance to the establishment of hiPSC-based engineered in vitro models as a standard tool for brain research in both academic and industrial settings, we discuss them in detail before proceeding to describe the modelling platforms themselves.
In vitro modelling of the CNS using hiPSCs: benefits and challenges
Overview of hiPSC use in in vitro CNS models
Since Takahashi and Yamanaka reported iPSCs in 2006, iPSCs have revolutionized biomedical research and boosted hopes for personalized therapeutics (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). These pluripotent stem cells resemble embryonic stem cells yet can be generated from terminally differentiated adult somatic cells such as skin fibroblasts or peripheral blood; hence, the use of human-derived iPSCs does not trigger the ethical concerns associated with the use of foetal cells in clinical trials (Yu et al., 2007; Loh et al., 2009; Polo et al., 2010; Halevy and Urbach, 2014; King and Perrin, 2014; Choi et al., 2015; Shi et al., 2017; Volarevic et al., 2018). Note that in this review we use the term iPSCs to refer to the technology itself and how it can be used in in vitro CNS models; when we refer to human-derived cells, human-oriented strategies or challenges, we use the term hiPSCs.
Success in producing hiPSCs has led to an enormous boost for cell replacement strategies; adult, somatic and patient-specific cells may now be reprogrammed and converted into immature cells, which can undergo directed differentiation into specific cell types, and then be grafted into the patient (Sánchez Alvarado and Yamanaka, 2014). Nevertheless, substantial hurdles must still be overcome before hiPSCs can be integrated into CNS therapy, even though hiPSC-based clinical trials involving non-brain tissues are underway (Bragança et al., 2019; Ortuño-Costela et al., 2019). Such challenges stem from the fact that the immense cytoarchitectural complexity of the brain tissue, coupled with immune responses (even in the case of autologous sources), often results in unsuccessful implementation of CNS transplants (Zhao et al., 2011; Nikolakopoulou et al., 2016; Garreta et al., 2018). These concerns regarding in vivo trials highlight the need for effective personalized CNS in vitro models, which capture the tissue microenvironment and cellular interactions.
Nowadays, hiPSCs from both healthy donors and patients with CNS diseases are used worldwide to model the complexity of human brain tissue and shed light on the mechanisms that govern its function in health and disease. The use of hiPSCs in conjunction with advanced in vitro technologies, such as OoCs, has enabled researchers to recapitulate patient-specific complex aspects of the human CNS, such as the blood–brain barrier (Vatine et al., 2019). Indeed, hiPSC-based brain-on-chip devices are routinely used to study, among others, neurodevelopment and neurodegeneration, contributing substantially to the advancement of regenerative medicine, toxicology, and high-throughput investigations (Berg et al., 2019). Nevertheless, researchers still face multiple obstacles when attempting to integrate iPSCs into in vitro platforms. These obstacles, elaborated in detail in what follows, stem both from the innate characteristics of the brain tissue and from the biological characteristics of the cell source.
Current challenges for hiPSC-based advanced in vitro models
Donor variability and cell heterogeneity
Donor variability, a feature shared with primary cells, is a major hurdle in hiPSC-based in vitro modelling. Residual epigenetic memory, genetic background and specific characteristics acquired during the reprogramming and differentiation processes result in great diversity among hiPSC-derived cell lines (Kim et al., 2010, 2011; Polo et al., 2010; Bar-Nur et al., 2011; Boland et al., 2014). Moreover, several hiPSC lines show defective proliferation and differentiation potential due to incomplete reprogramming (Ohnuki et al., 2014). Advanced gene transduction and editing technologies [e.g. zinc-finger nucleases, transcription activator-like effector nuclease (TALEN), clustered regularly interspaced short palindromic repeats (CRISPR)/Cas-9 based gene or base editing] and next-generation sequencing are used widely to handle the variability issue in hiPSC lines (Komor et al., 2016; Brookhouser et al., 2017; Doss and Sachinidis, 2019).
Differentiation protocols, reproducibility, and maturity
Reliable translational models require both high-quality hiPSCs and effective differentiation protocols towards the desired cell fates. Thus, researchers in both academia and industry have been working continuously towards the development of CNS lineage-specific differentiation protocols. Nevertheless, the need for pure and mature cell types remains largely unmet. The cells frequently show immature functional characteristics with embryonic and foetal tissue attributes. Consequently, they can serve as good models for neurodevelopmental and early onset disease studies but cannot adequately mimic late onset disease and mature tissue (Miller et al., 2013). Interestingly, a recent study by Lu et al. (2019) highlights problems with iPSC-derived cells. The authors question the identity of iPSC-derived brain microvascular endothelial cells (BMECs) (Lu et al., 2019). According to the authors, the differentiated cells recapitulated properties from neuroectodermal epithelium rather than the blood–brain barrier. Moreover, several studies have reported biased differentiation potential of hiPSCs towards specific lineages due to residual epigenetic memory. It may be possible to reset these cells to pluripotential patterns by increasing the number of passages before differentiation (Polo et al., 2010; Bar-Nur et al., 2011; Kim et al., 2011; Boland et al., 2014; Kedziora and Purvis, 2017; Doss and Sachinidis, 2019). Still, overall, to increase the translatability of iPSC-based in vitro models, it remains a critical issue to produce high-quality hiPSCs and to develop current protocols further.
Immunogenicity of hiPSC-derived cells
Earlier studies performed in animals (de Almeida et al., 2014; Zhao et al., 2015) and in human co-culture systems (Huang et al., 2014) have shown that differentiated cells are less immunogenic than the corresponding iPSC populations. This observation suggests that hiPSC-derived tissues may replace autologous tissue transplants, since they can surpass the physiological immune responses and avoid rejection by the patients. This same feature suggests, however, that hiPSC-derived in vitro models may be lacking in predictivity in terms of physiological immune responses. In such a scenario, it is plausible that hiPSC-derived models may poorly describe immune mediated diseases of the brain tissue and thereby hinder successful drug development.
Using hiPSC-based disease models to model neurodegenerative diseases
Disease modelling using hiPSCs may be highly accurate for monogenic diseases, but the relevance for complex, polygenic and sporadic diseases is debatable. The reports so far compare cellular characteristics of one or a few patient cells to their respective family and gender-matched controls; any differences observed during phenotypic analysis are usually attributed to the mutation under investigation and thereby to the cause of the disease. This approach may prevent researchers from distinguishing disease-related variation from variation attributable to other factors such as epigenetic memory, genetic background and environmental cues. Consequently, and because most neurodegenerative diseases are sporadic in nature, it is necessary to use large numbers of patient-derived hiPSC lines to decrease the signal-to-noise ratio of the studies and increase the accuracy of the results (Doss and Sachinidis, 2019). Moreover, hiPSC derivatives are commonly used in 2D cultures, which lack the multi-faceted interactions in the human body in health and disease; engineered 3D models, on the other hand, provide in vivo-like platforms to investigate how, among others, genetics and environmental cues influence disease phenotypes (Sharma et al., 2020).
Regional identity of hiPSC-derived cells
The complexity of the human CNS arises from the active interactions among multiple neural cellular subtypes. Most neurological diseases, on the other hand, stem from defects in specific cellular subtypes, with the underlying mechanisms remaining largely elusive (Imaizumi et al., 2015). Accordingly, accurate disease modelling requires the use of differentiated cells with specified regionality, thereby providing relevance to the disease of interest. Indeed, researchers worldwide have used hiPSCs from patients to study several neurological diseases, including amyotrophic lateral sclerosis (ALS), Alzheimer’s disease, schizophrenia and Parkinson’s disease (Cooper et al., 2010; Zhang et al., 2014; Ahmad et al., 2018; Fujimori et al., 2018; Henstridge and Spires-Jones, 2018; Li et al., 2018; Mishima et al., 2018, Osaki et al., 2018c; Ishii et al., 2019; Penney et al., 2020). In these studies, hiPSCs were differentiated into the respective cell types, thereby providing cellular disease models with regional characteristics. The conclusions obtained in these studies, however, remain largely ambiguous, owing to the limited capacity of current differentiation protocols to provide highly pure cultures of the various neuronal subtypes. We believe that further development of the current protocols in terms of regional characteristics, combined with large cohorts to overcome patient-specific variability of derived cells, will be of great benefit for in vitro CNS modelling.
The need for a ‘universal medium’
An issue of great importance for iPSC-derived in vitro models based on OoCs is the need for a ‘universal medium’. To effectively mimic the multifaceted nature of brain tissue, it is necessary to co-culture several cell types. As discussed in a remark by CellPress editor Pavlovich, ‘there are biological factors: not every cell thrives in the same medium, responds to the same growth factors or differentiation cues, or adheres to the same matrix’ (Pavlovich, 2018). Therefore, research and industrial laboratories continue to invest enormous efforts into the development of xeno-free, defined protein substrates and media to support iPSC-derived cell expansion and differentiation. Alternative potential solutions include advanced microengineering, elaborate design and compartmentalized devices. It is important to note, however, that the latter approaches require extensive technical expertise.
Engineered in vitro models to mimic CNS physiology
Cellular organization in microfluidic platforms
A basic objective underlying the development of in vitro microfluidic platforms is the creation of an environment in which physiological compartments (e.g. vasculature and parenchymal tissue) are simplified and physically separated yet can still interact with one another-thereby facilitating observations that are effectively impossible to achieve in in vivo models. This simultaneous separation and interaction may be realized either by linking several OoCs or by using one compartmentalized device.
The concept of physically separating the neuronal body and its extending neurites was first proposed by Campenot in 1977 (Campenot, 1977), and since then was extensively applied in the so-called Campenot chambers. In 2003, Taylor et al. incorporated microgrooves in a cell culture chamber, taking advantage of microfluidic, micropatterning and microfabrication technology for the first time; the researchers were able to physically isolate neuronal soma and neurites, and to use the system to study local damage of neurites (Taylor et al., 2003; Neto et al., 2016). Since then, numerous engineered in vitro platforms have relied on single-chip CMDs to support physical isolation of the cellular populations and/or components on the microscale as a basis for mechanistic studies, including studies of axonal biology (Shin et al., 2010) or synapse formation and modulation (Taylor et al., 2010; Coquinco et al., 2014). More than a decade ago, in 2005, Taylor et al. pioneered a microfluidics-based in vitro platform to study axonal regeneration after injury (Taylor et al., 2005). The device was further advanced by Park and collaborators to investigate neuron-glia interactions and axon myelination (Taylor et al., 2005; Park et al., 2006, 2009a, 2012; Higashimori and Yang, 2012; Shi et al., 2013). As a further advancement, intra-system co-cultures of neuronal and glial populations (Park et al., 2009b; Yang et al., 2012; Shi et al., 2013), aiming to elucidate myelination pathways, have been developed with the aim of identifying novel treatment strategies for demyelinating diseases such as multiple sclerosis. Subsequent developments of such systems may include integration of immune cells and particularly hiPSC-derived microglia (Abud et al., 2017; Douvaras et al., 2017; Haenseler et al., 2017; Pandya et al., 2017; Garcia-Reitboeck et al., 2018; McQuade et al., 2018) and hiPSC-derived oligodendrocytes (Ehrlich et al., 2017), towards shedding light on the complex cellular processes occurring in the human demyelinating brain tissue.
In a multi-chip design, several OoCs are linked; thus, the complex cytoarchitecture of the nervous tissue may be largely replicated. For example, a linked three-chip arrangement revealed metabolic coupling between the endothelial and the neuronal cells in an engineered model of the human neurovascular unit (NVU) (Maoz et al., 2018).
Moreover, microfluidics have been used extensively to explore how biochemical cues affect axonal behaviour, outgrowth, pathfinding and synapse function (Wu et al., 2005; Cox et al., 2008; Taylor et al., 2009; Gumy et al., 2011; Park et al., 2014; Kung et al., 2015). Deglincerti et al. (2015) elegantly used the physical separation of axons from the neuronal soma to show that local protein synthesis and degradation are interconnected in growth cones. The study demonstrated that growth cones show elevated levels of ubiquitylation, and that the ubiquitin-proteasome system targets locally translated proteins. Thus, the authors suggested that axonal tuning responses towards guidance signals may incorporate local protein synthesis and degradation (Deglincerti et al., 2015; Neto et al., 2016). Local translation in axons and particularly in the growth cones is now widely accepted; this is a result of the capability to perform studies in isolated axonal fractions with the use of microfluidics.
In addition to providing a means of separating cell populations while enabling interactions to take place between them, OoCs offer the possibility to control a vast array of mechanical properties such as stiffness, geometric confinement, interstitial flow and shear stress, which affect cellular state and differentiation (Sundararaghavan et al., 2009; Peyrin et al., 2011; Song and Munn, 2011; Kim et al., 2013; Galie et al., 2014; Hattori et al., 2014; Asano et al., 2015; Osaki et al., 2018a). Moreover, biochemical gradients on the cellular components can be easily imposed on the devices to recapitulate early neurodevelopmental processes and perform mechanistic studies. This feature is useful, e.g. for manipulating concentration gradients of cytokines, such as BMP4, SHH, FGF, RA, and WNT3, which orchestrate cellular proliferation, differentiation and organogenesis in early brain tissue development (Park et al., 2009b; Demers et al., 2016; Uzel et al., 2016; Osaki et al., 2018a).
The above section briefly describes how OoCs may be used to investigate cellular organization in the brain tissue. Several of the studies mentioned here utilize cells of animal origin; nevertheless, we consider them of tremendous importance for further development of human in vitro models. We envision that incorporation of hiPSC-derived cellular entities in similar platforms may further advance our understanding of how cells organize themselves in the human brain.
The use of engineered microfluidic systems to recapitulate distinct brain regions and their connectivity
The human brain consists of more than 250 different brain regions (Ding et al., 2016), which have a highly specialized ECM (Dauth et al., 2016), architecture and functionality (Novak and Kaye, 2000; Lau et al., 2013; Dauth et al., 2016). Nevertheless, higher brain functions, as well as many neuropsychiatric disorders (Quadrato et al., 2016), are regulated by the interaction between multiple brain regions such as the hippocampus, cortex, thalamus, cerebellum, and amygdala (Kato-NegiShi et al., 2013). Therefore, it is of great importance to mimic the different physiologically connected brain regions in vitro. Currently, there are just a few reported in vitro models incorporating cells derived from two or more different brain regions (Kanagasabapathi et al., 2011; Peyrin et al., 2011; Kato-NegiShi et al., 2013; Dauth et al., 2017; Soscia et al., 2017). These include a model developed by Peyrin et al. (2011), who reported the ability to recreate a functional and synchronized cortico-striatal oriented network, using a microfluidic system. The two-chambered microfluidic device was used to culture primary murine cortical and striatal neurons and let them connect through controlled and highly ordered axons (Fig. 2A) (Peyrin et al., 2011); this enabled the researchers to observe that cortical neurons trigger the differentiation of spiny striatal neurons and the formation of dendritic spines. A similar neurofluidic device with microchannels was also used to provide insights into the interactions between the cortex and thalamus (Kanagasabapathi et al., 2011); the system provided easy accessibility and manipulation capabilities, which cannot be easily achieved in in vivo systems (e.g. the ability to investigate cortical-thalamic interactions in isolation, without the influence of other regions). The latter in vitro model allowed for neurite outgrowth, connections between the two different brain tissues, functional readouts (e.g. electrophysiology) and immunohistochemistry (Kanagasabapathi et al., 2012). By using this in vitro system, the authors demonstrated that burst events originate in the cortical region, triggering the cortical-thalamic network, confirming some previous data obtained from in vivo experiments.
The next advance was achieved by Kato-Negishi et al. (2013), who developed a 3D millimetre-sized neural building block, formed from rat hippocampal and cortical cells. This model allowed for Ca2+ imaging, gene transduction measurements, and immunohistochemistry, and it was able to show the formation of projections and synaptic contacts between cortical and hippocampal neurons, thereby enabling in vitro investigations of interactions between multiple brain regions. In a similar way, Soscia et al. (2017) developed a platform to co-culture hippocampal and cortical neurons (Fig. 2B), with the possibility of investigating how the different brain regions establish connections, integrate networks and increase their firing rate. Recently, Dauth et al. (2017) were the first to develop an in vitro multiregional CNS model implementing three brain regions, derived by culturing rat prefrontal cortex, hippocampus and amygdala-derived tissues, functionally connected through axons (Fig. 2C). Their model incorporated functional readouts by measuring extracellular field potentials with multielectrode arrays, together with biochemical readouts and immunohistochemistry. Their work demonstrated the significance of connecting different brain regions in vitro, showing that doing so changes the cellular composition, protein expression and electrophysiological properties of the co-cultured cells compared with those observed in monocultures. Moreover, the model was used to mimic the corticolimbic system and to examine the effects of phencyclidine hydrochloride (PCP) on one brain region and to identify how the other regions respond.
Modelling the neurovascular unit (the blood–brain barrier)
The studies discussed above focused on modelling the brain’s complex neuronal architecture and functionality. Yet, recent studies have acknowledged the need for in vitro CNS models to take into account not only the brain’s parenchymal cells but also its unique vasculature, which differs from non-CNS vasculature in that it exhibits continuous tight junctions, is void of fenestrations and has a very low rate of transcytosis (Abbott et al., 2006). Perhaps most importantly, the brain’s vasculature includes a highly specialized endothelium—the blood–brain barrier—which tightly regulates the entry of compounds into the brain. In other words, in vitro models aimed at understanding the brain’s responses to various stimuli [e.g. mechanical perturbations such as traumatic brain injury (TBI), drug development, toxicology, etc.] must model the entirety of the NVU, which is composed of the blood–brain barrier and the perivasculature; brain pericytes, and closely interacting astrocytes and neurons. In general, ensuring that a given molecule can penetrate the blood–brain barrier and thus access the CNS is a major challenge in CNS drug development (Herland et al., 2020). In small molecular approaches, computational pharmaco-distribution and animal models have been fairly successful, but the increasing dominance of human-specific biological pharmaceuticals has led to a greater need for models that are highly predictive of human blood–brain barrier penetrance (Gribkoff and Kaczmarek, 2017).
Several engineered in vitro models have been developed to mimic the NVU (summarized in Table 2 and Supplementary Fig. 2). For example, a recent model by Maoz and colleagues connected three chips, one blood–brain barrier chip connected to a brain chip, which was connected to a second blood–brain barrier chip containing human BMECs, neuronal cells, glial cells and pericytes (Fig. 2D). They used this system to analyse the individual cell types comprising the NVU (Maoz et al., 2018), as well as to mimic the effect of intravascular administration of psychoactive drugs (e.g. methamphetamines) and to identify, for the first time, metabolic ‘crosstalk’ among the cellular components of the blood–brain barrier. Moreover, in silico models of the blood–brain barrier have also been extensively used to predict drug permeability in the blood–brain barrier (Cabrera et al., 2004; Suenderhauf et al., 2012; Miao et al., 2019; Roy et al., 2019).
Table 2.
Summary of in vitro models commonly used in blood–brain barrier research
| Model | Shear stress | Cell-cell interactions | High-throughput / cost | Similarity to human physiology |
|---|---|---|---|---|
| Transwella,b | No | Co-culturing possible, tri-culturing more challenging to evaluate cell populations | Yes / low | Minimal, ECM present only as anchoring points, 2D geometry |
| Porous-tube modelsc | Yes | Same as Transwell | Minimal / moderate | Improved similarity to human physiology (shear stress, 3D luminal geometry), but minimal ECM present |
| Microfluidic chips (membrane-based)d,e | Yes | Capability of compartmentalization and studying interactions between cell populations | Yes; however, more time consuming than Transwell / moderate | Same as porous-tube models |
| Microfluidic chips(ECM-based)f | Yes | Same as membrane-based microfluidic chips | Yes; however, more time consuming than Transwell / moderate | Utmost attempt at in vitro biomimicry (shear stress, 3D geometry, ECM present) |
NVC = neurovascular chip.
In this list, we consider studies that use Transwell in static cultures, there are, however, studies that implement flow in Transwell (Hinkel et al., 2019).
Zenker et al., 2003; Colgan et al., 2008; Helms et al., 2014; Labus et al., 2014; Canfield et al., 2017; Delsing et al., 2018.
In this list, microfluidic chips with a temporary membrane (i.e. a membrane that degrades over time) are not included, such as the work of Tibbe et al. (2018).
Below, we elaborate on the considerations that must be taken into account when modelling in vitro the properties of the NVU and blood–brain barrier, which is constantly under flow and is characterized by diverse types of cell–cell interactions.
Blood–brain barrier in vivo metrics
Evaluation of barrier properties is done by measuring transendothelial electrical resistance (TEER), passive permeability to small compounds (<1000 g/mol), and the activity of efflux and influx transporters (e.g. by the use of P-glycoprotein substrates and glucose, respectively) (Lippmann et al., 2012; Stebbins et al., 2016). A key component that affects these properties is the source of cells for the model. In particular, as noted above, the use of animal cells can raise concerns regarding inter-species differences. Primary human BMECs retain some blood–brain barrier phenotypes. However, TEER of human primary BMECs (does typically not exceed 200 Ω×cm2 (Mackic et al., 1999; Zenker et al., 2003), which accounts for only 10% of in vivo TEER measurements of the blood–brain barrier in rats and frogs (Crone and Olesen, 1982; Butt et al., 1990). In 2012, with the advent of hiPSCs, Lippmann was the first to report hiPSC-derived BMEC-like cells that exhibited TEER values >200 Ω×cm2 (Lippmann et al., 2012). The controversy of the identity of these cells were discussed in an earlier section. Since then, a plethora of studies on hiPSC-derived BMEC-like cells emerged, using similar (Lippmann et al., 2014; Hollmann et al., 2017) or conceptually different differentiation strategies (Orlova et al., 2014).
Real-time monitoring of salient features of the NVU-on-chip is a field of expanding interest. To date, researchers have integrated permeability evaluations and TEER measurements in 2D (Booth and Kim, 2012; Walter et al., 2016; Wang et al., 2017) and 3D cultures (Brown et al., 2015, Xu et al., 2016a; Partyka et al., 2017) on chips. However, real-time monitoring of metabolic processes with analytical microfluidic chips has so far been only implemented in conjunction with animal models (Lin et al., 2014).
Cell–cell interactions
Co-culturing cells of the NVU adds another level of complexity to the in vitro model, enabling the model to capture in vivo conditions more faithfully. In particular, perhaps unsurprisingly, co-culturing BMECs with CNS cells contributes to the blood–brain barrier-like properties of endothelial cells, e.g. through fortification of tight junctions and expression of polarized transporters (Kasa et al., 1991; Megard et al., 2002; Didier et al., 2002, 2003; Haseloff et al., 2005; Lippmann et al., 2012; Herland et al., 2016; Hollmann et al., 2017). Moreover, studies reveal that astrocytes and BMECs secrete factors that confer each other’s maturity (Janzer and Raff, 1987; Fukushima et al., 2009; Blanchette and Daneman, 2015).
The Transwell model (Table 2 and Supplementary Fig. 2) has been used extensively for co-culturing BMECs with CNS and non-CNS cells; this model allows for non-invasive TEER measurement, permeability assays, and evaluation of efflux pumps (Zenker et al., 2003; Colgan et al., 2008; Helms et al., 2014; Labus et al., 2014; Canfield et al., 2017; Delsing et al., 2018). However, this approach reflects a static environment with a non-continuous cell monolayer, and thus does not capture the in vivo blood–brain barrier setting.
Flow
Flow is a significant parameter that should be considered in the development of NVU models. Endothelial cells in the capillaries experience a force (shear stress) parallel to the endothelium, exerted from the blood flow. Jiang et al. (2019) recently discussed the impact of flow on cellular functionality in a comprehensive review of microfluidic models of the blood–brain barrier.
Siddharthan et al. (2007) showed a correlation between shear stress and the upregulation of the tight junction protein ZO-1 in BMECs. In 2011, Cucullo et al. reported a thorough evaluation of the impact of shear stress on the transcriptome of BMECs; shear stress upregulated tight and adherens junctions as well as multidrug resistance transporters (Cucullo et al., 2011), which is in accordance with the blood–brain barrier transcriptome footprint.
The emergence of OoCs has further enabled researchers to observe the functionality of vascular and other cells in vitro in the presence of flow stimulation. Huh et al. (2010) were the first to demonstrate flow-induced organ-level functions, in an OoC model, specifically a lung-on-a-chip model (Huh et al., 2010). Booth and Kim (2012) first reported an NVU-on-a-chip using brain endothelial and astrocytic cell lines, documenting that the NVU-on-a-chip resulted in higher TEER than the static conditions as well as permeability to tracers that resembled in vivo levels. However, another study using hiPSC-derived BMECs suggested that there was no difference in tight and adherens junction expression between static and dynamic conditions; this lack of difference may have been attributable to immaturity of the differentiated BMECs (DeStefano et al., 2017). Other studies have suggested that the permeability levels of tracers and compounds in NVU-on-chip systems are on par with in vivo data (Wang et al., 2017), nevertheless, these studies did not carry out direct comparisons between chips and Transwells. More thorough studies suggest that NVU-on-chip systems exhibit lower permeability than the Transwell model (Prabhakarpandian et al., 2013; Walter et al., 2016; Partyka et al., 2017). Recently, Vatine et al. (2019) demonstrated that flow significantly decreases the blood–brain barrier permeability in an isogenic NVU-on-chip. In addition, they perfused the isogenic NVU-on-chip with blood from the same donor and could detect inter-individual blood–brain barrier characteristics. The various discrepancies between different studies highlight the fact that variables such as cell sources, materials, media volumes as well as cell-to-medium ratios are important aspects when comparing various in vitro models.
Flow might also influence the alignment of BMECs; notably, however, in vitro studies investigating this alignment process have produced different results. A recent study by Moya et al. (2020), which used a brain endothelial cell line, suggested that BMECs align in the direction of the flow. Another study on primary BMECs also suggested that primary cells align in the direction of the flow (Garcia-Polite et al., 2017). However, several studies using primary BMECs (Ye et al., 2014; Reinitz et al., 2015) and iPSC-BMECs (DeStefano et al., 2017) have suggested that BMECs do not align in the direction of the flow. None of these in vitro studies addressed whether the BMECs (or human BMECs) have a venous or arterial phenotype. This omission is notable, given that, in vivo, the capacity of endothelial cells to align in the direction of the flow is dependent on variables such as the nature of the endothelium (arterial or venous) (dela Paz and D’Amore, 2009) as well as the level of shear stress (Masumura et al., 2009). These variables may have a direct implication on endothelial cells’ capacity to align in the direction of the flow.
Extracellular matrix
Mimicking the brain ECM in vitro is a great challenge (Rauti et al., 2019); the ECM components, discussed in the ‘Overview of recent developments in in vitro CNS models’ section, differ throughout the brain (i.e. different brain regions have unique ECM composition, and the brain vasculature ECM is different from the brain ECM) while that is also amenable to the developmental stage i.e. BMECs in the brain vasculature swift their signalling from fibronectin in development to laminin in adulthood (Herland et al., 2016; Adriani et al., 2017; Linville et al., 2019). To recapitulate the NVU in vitro, the ECM of both the brain vasculature and the rest of the brain should be considered to ensure the accurate replication and effectiveness of the NVU in vitro model (Rauti et al., 2019).
The majority of iPSC-derived BMECs protocols use a combination of collagen IV and fibronectin as a purification step during BMEC differentiation (Lippmann et al., 2012, 2014; Hollmann et al., 2017). Nevertheless, other types of ECM components have been used when replicating the NVU in vitro, such as type I collagen. Albeit type I collagen is not naturally present in the brain, the gelation properties of type I collagen have made it quite favourable in 3D in vitro rendering of the NVU (Herland et al., 2016; Partyka et al., 2017; Wevers et al., 2018; Grifno et al., 2019; Linville et al., 2019).
ECM-derived gels have been incorporated into microfluidic devices, adding yet another layer of complexity to the existing fluidic models (Herland et al., 2016; Adriani et al., 2017; Linville et al., 2019). Currently, microfluidic systems with incorporated ECM gels constitute the most comprehensive attempts at achieving biomimicry in NVU-on-chip models. Yet, the process of setting up systems that incorporate ECM gels is time-consuming, costly, complex, and the throughput of these systems is lower than that of traditional Transwells. Given the physiological importance, the recapitulation human relevant ECM in vitro is a growing research field, however with many challenges unsolved.
Engineered in vitro models mimicking CNS disease
Cell death or alterations in the CNS cellular microenvironment may lead to network disruption and pathologies, including neurodegenerative diseases, TBI and cancer (Osaki et al., 2018a).
The brain ECM is altered in some pathological conditions and ageing (Bonneh-Barkay and Wiley, 2009; Burnside and Bradbury, 2014; Caldeira et al., 2018). There are alterations that directly contribute to the progression of certain pathological conditions diseases (Baeten and Akassoglou, 2011) such as autism (Mercier et al., 2012), epilepsy (Dityatev, 2010; McRae and Porter, 2012), Alzheimer’s disease and schizophrenia (Lu et al., 2011; Berretta, 2012; Pantazopoulos et al., 2015; Reed et al., 2019). Moreover, recently it was shown that the brain ECM have a significant role in the development of neurodegenerative diseases such as Alzheimer’s disease, schizophrenia and bipolar disorders (Barcelona and Saragovi, 2015).
To elucidate the underlying mechanisms of brain tissue pathologies, it is necessary to emulate the occurring processes accurately in vitro. In CNS pathology, animal models have shown particularly low predictive capacity; Alzheimer’s is a striking example of how animal trials have shown promising results, whereas, one after the other, clinical trials have failed (De Felice and Munoz, 2016; Mofazzal Jahromi et al., 2019). Below, we discuss advanced engineered 2D and 3D in vitro models that have recently been developed to investigate brain pathologies.
Neurodegenerative disease
Neurodegenerative diseases are escalating in prevalence and have devastating effects on individual and societal well-being (Marras et al., 2018; Patterson, 2018; Fisher and Bannerman, 2019). Most neurodegenerative diseases are incurable, and the neurobiological mechanisms governing disease initiation, progression and therapy remain elusive (Centeno et al., 2018). Some of these diseases are monogenic, whereas for others, e.g. Alzheimer’s disease, Parkinson’s disease and ALS, the vast majority of cases (90% or more) have not been linked to a genetic cause, making it almost impossible to generate relevant animal models (Centeno et al., 2018). The use of hiPSC-based 3D engineered in vitro models offer a potential alternative to animal testing and provide human-specific mechanistic insights in both monogenic and sporadic disease pathology.
Alzheimer’s disease
Alzheimer’s is currently the most prevalent neurodegenerative disease (WHO, 2019), and extensive efforts have been devoted to the development of predictive in vitro models for this disease. Patients suffer from progressive cognitive dysfunction characterized by excessive amyloid-β accumulation and neurofibrillary tangles (Hardy and Selkoe, 2002; Tanzi and Bertram, 2005). Transgenic mouse models of Alzheimer’s disease mutations mimic aspects of the disease such as memory loss; however, they fail to recapitulate key characteristics of the condition such as the neurofibrillary tangle pathology (Chin, 2011).
In 2014, Tanzi and his team were the first to report a human 3D in vitro model that resembled the pathophysiology of the disease (Choi et al., 2014). Even though this was one of the greatest advancements in the field an important piece to the puzzle was still missing; the contribution of the blood–brain barrier breakdown to the disease phenotype could not be investigated. To fill the gap, in 2019 the team proposed an advanced, physiologically relevant 3D human microfluidic-based platform, which incorporated a tubular BMEC layer with barrier-like properties into their Alzheimer’s culture system (Shin et al., 2019). Scientists may now use this platform to model the progressive accumulation of amyloid-β peptides in the ECM and the sequential transport via the NVU microenvironment.
Incorporation of hiPSC-derived cells in the 3D culture platforms may bring us as a step closer to understanding the mechanics of this devastating disease and design effective human-oriented therapeutics (Choi and Tanzi, 2012). To this end, Zhang et al. (2014) generated a 3D culture system in which hiPSC-neurons were cultured in a commercially available soft hydrogel composed of laminin and a synthetic peptide (i.e. RADA-16), mimicking the 3D neural microenvironment, especially the tissue stiffness. Park et al. (2018) proposed a novel 3D organotypic tri-culture system, which incorporated neurons, astrocytes, and microglia in a microfluidic platform to study the pathogenesis of Alzheimer’s with respect to neuroinflammatory stimuli (Fig. 3A).
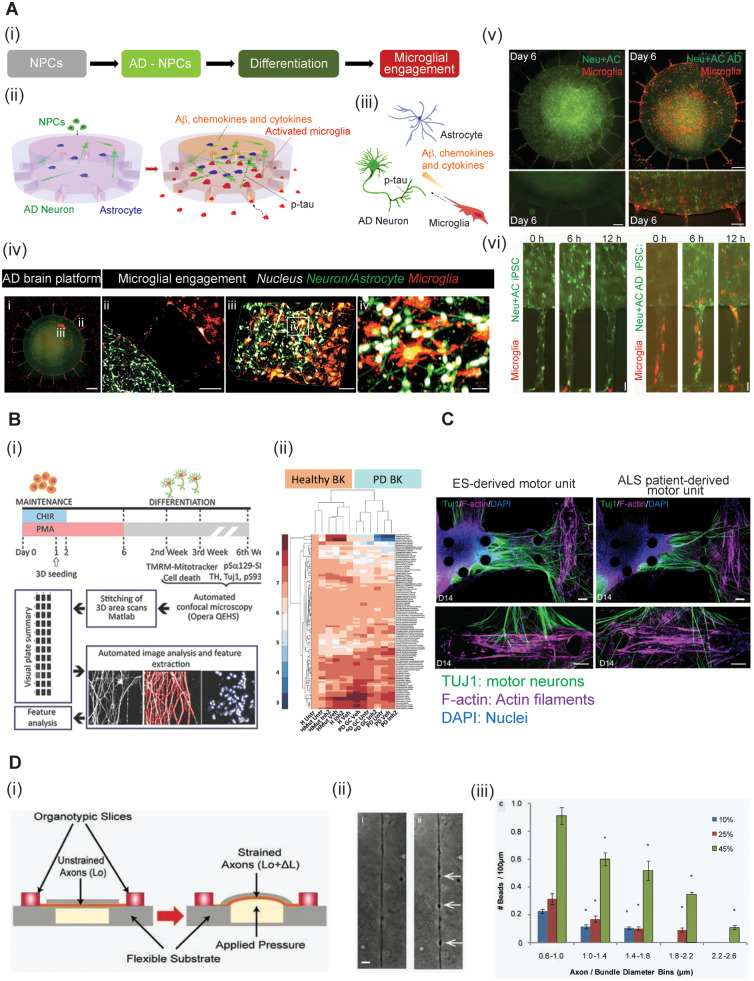
Figure 3.
3D engineered in vitro models for neurodegeneration and TBI. (A) A 3D organotypic human triculture model for Alzheimer’s disease (AD) (Park et al., 2014). (i) Neural progenitor cells (NPCs) were differentiated to Alzheimer’s disease neurons and astrocytes, while monitoring microglia recruitment. (ii–iii) Schematic of the multicellular interactions in the in vitro microfluidic AD model (ii) and in the AD brain (iii). (iv) Image i: Fluorescent image of the microfluidic platform. Alzheimer’s disease neurons (Neu)/astrocytes (AC) (green) are in the central chamber and microglia (red) are in the angular chambers. Scale bar = 250 μm. ii: Microglial recruitment across the angular microchannels. Scale bar = 250 μm. iii and iv: Confocal imaging confirms the 3D physiological intercellular communication among neurons (green), astrocytes (green) and microglia (red) in the central chamber. Nuclei are shown in white. Scale bars = 100 μm in iii; 40 μm in iv. (v) Comparison of microglial recruitment (red) by the control Neu + AC (green) and the AD Neu + AC (green). Scale bars = 250 μm (top) and 200 μm (bottom). (vi) Microglial recruitment by hiPSC AD Neu + AC. Scale bar = 10 μm. (B) 3D model of Parkinson’s disease (PD) dopaminergic (DA) neurons for high content phenotyping and drug screening (Bolognin et al., 2019). (i) Schematic illustration of the experimental procedure. The setup allows for automated image acquisition, segmentation, feature extraction and data analysis. (ii) A clear clustering of the lines according to genetic background is shown in the heat map. (C) A 3D ALS motor unit microfluidic model (Osaki et al., 2018c). The ALS motor unit (right) exhibits fewer thick neural fibres and decreased neuromuscular junction (NMJ) formation compared with the embryonic stem (ES) cell-derived motor unit (left). Motor neurons are stained with TUJ1 (green), actin filaments with F-actin (purple) and nuclei with DAPI (blue). Scale bars = 100 μm. (D) A brain-on-a-chip to model TBI. Schematic sketch of the uniaxial axonal strain device (i) an example of axonal beading observed before and after the strain injury (ii) and a bar plot representing the correlation between the diameter of the axon/bundle and the number of beads used to injure the cells (iii). Figure components are modified from Dollé et al. (2014), Osaki et al. (2018c), Park et al. (2018) and Bolognin et al. (2019) with permission.
Nevertheless, both in vivo and in vitro experimental platforms have this far failed to mimic tissue maturity. Recent studies have shown that epigenetic modifications underlie the link between ageing and disease progression (Fyfe, 2018; Nativio et al., 2018). Future incorporation of mature neurons and immune cells in in vitro platforms may substantially enhance predictivity and bring us closer to effective diagnostics and therapeutics for patients with Alzheimer’s disease.
Parkinson’s disease
Parkinson’s disease is characterized by selective loss of dopaminergic neurons in the substantia nigra (Antony et al., 2013). Since 2010, when hiPSC-derived dopaminergic neurons (Cooper et al., 2010; Hargus et al., 2010) were developed, researchers have used either organoids (Monzel et al., 2017) or OoC methods (Moreno et al., 2015; Bolognin et al., 2019) to mimic the parkinsonian brain in vitro. The G2019S mutation in the leucine-rich-repeat-kinase-2 (LRRK2) is frequently associated with the pathophysiology of both familial and sporadic forms of Parkinson’s disease (Paisán-Ruı´z et al., 2004; Zimprich et al., 2004; Healy et al., 2008; Simón-Sánchez et al., 2009; Abud et al., 2017; Islam and Moore, 2017). Animal models with core LRRK2 mutations are widely used in Parkinson’s research since LRRK2 has a druggable kinase domain and it is therefore considered a potential therapeutic target (Bolognin et al., 2019). Nevertheless, Bolognin et al. recently exploited microfluidics combined with high-content imaging technology to develop an advanced 3D in vitro model enabling pharmacogenomics in pathophysiological conditions (Fig. 3B). Intriguingly, the most penetrant disease phenotypes were a result of patients’ overall genetic background and were not solely dependent on the LRRK2-G2019S mutation (Bolognin et al., 2019).
Amyotrophic lateral sclerosis
ALS is a fatal neurodegenerative disease characterized by progressive degeneration of motor neurons, disturbed neuromuscular junction (NMJ) and muscle atrophy; in most cases death occurs within 3 years after ALS diagnosis due to respiratory failure (Centeno et al., 2018; Ionescu and Perlson, 2019). Osaki et al. (2018c) proposed a novel ALS-on-chip model, a motor unit, comprising 3D skeletal muscle bundles and optogenetic motor neurons from an ALS patient (Fig. 3C). The authors further enhanced their platform with an hiPSC-derived blood–brain barrier to study the CNS penetration of putative therapeutic agents (Osaki et al., 2018b). Future incorporation of mature neurons will mimic later stages of the disease; this may be accomplished either via genetic manipulation (Miller et al., 2013; Osaki et al., 2018c) or via direct differentiation of fibroblasts of ALS patients (Tang et al., 2017). Altman et al. (2019) developed a compartmentalized device to mimic the NMJ; their data depict the importance of mitochondrial accumulation for NMJ functionality (Altman et al., 2019). Replacement of the animal cells with hiPSC-derived populations might further enhance the translatability of the model and provide a platform to study the implication of mitochondria in the NMJ vulnerability in ALS.
We suggest that similar setups may be used to study neurodegenerative diseases that are associated with deterioration in muscle strength and motor skills, such as Alzheimer’s and Parkinson’s diseases (Boyle et al., 2009; Cano-de-la-Cuerda et al., 2010; Antony et al., 2013).
Traumatic brain injury
TBI is a severe health and socioeconomic problem (LaPlaca et al., 2005), and is also considered to be a risk factor for neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease and ALS (Sivanandam and Thakur, 2012). In vivo models of TBI have mainly focused on its behavioural and systemic effects, whereas in vitro studies provide a powerful tool to deeply investigate the cellular mechanisms. Currently, the ability to accurately model TBI in engineered in vitro models is limited because of the low physiological resemblance of standard neural in vitro models and incompletely understood pathophysiology manifested in mechanical shear, twist and compression forces as well as subsequent hypoxia (Kumaria, 2017). LaPlaca et al. (2005) pioneered a reproducible model of in vitro TBI. By using an electro-mechanical cell shearing device, they were able to mechanically perturb a 3D neuronal-glial model, with deformation rates and magnitudes comparable to those that occur in inertial human head injuries (LaPlaca et al., 2005). Dollé et al. (2013, 2014) developed a brain-on-a-chip microsystem investigating neuronal response to a mechanical injury (Fig. 3D). The device allowed easy manipulation of the dimensions of the microchannels, which influenced the strain on either individual axons or bundles of axons, thereby suggesting that axonal diameter plays a significant role in strain injury, and thus in TBI. While there has been a great development in new in vitro models for TBI (Morrison et al., 2011), the combination of the advantages of engineered platforms with in vitro capabilities of monitoring cellular mechanisms and functional response to mechanical injury is a powerful tool for studying TBI responses and developing potential therapeutics (Shrirao et al., 2018).
Cancer
Despite tremendous efforts to identify putative treatments for brain cancer, most drug candidates fail in human clinical trials (Huszthy et al., 2012; Caragher et al., 2019; Sontheimer-Phelps et al., 2019). The complex tumour microenvironment in neural tissue is difficult to recapitulate; thus, most proposed compounds are inadequate for treating brain tumours (Sontheimer-Phelps et al., 2019). To increase the predictivity of current experimental platforms, it is necessary to mimic tumour dissemination, reduction and metastasis, cancer stem cell proliferation and differentiation, drug penetration across the blood–brain barrier and immune responses.
Animal modelling of primary brain tumours has been the gold standard in cancer research; however, translatability of the results tends to be questionable (Denayer et al., 2014; Mak et al., 2014). Patient-derived xenograft animal models maintain most of the biological characteristics of the original tumour, and they hold promise for translation to humans (Choi et al., 2018; Yada et al., 2018). Nevertheless, the mice used in these models are usually immunocompromised to prevent possible rejection of the xenograft, and immunity is neglected (Choi et al., 2018). Two-dimensional in vitro models, on the other hand, fail to recapitulate the tumour microenvironment, intercellular communication and tumour cell metastasis. Recent developments of vascularized brains-on-chips that incorporate tumorigenic cells may offer the possibility of improved translatability and effective drug discovery (Saliba et al., 2018; Wang et al., 2018; Sontheimer-Phelps et al., 2019).
OoCs may also provide a means of investigating the cascade of events that contributes to tumor metastasis from other organs to the brain (Fig. 4A), towards identifying novel therapeutics (Caballero et al., 2017). Lei et al. (2016) developed an OoC to study interactions between cortical neurons and cancer cells. The authors showed that functional neurites promoted cancer migration to the neuronal compartment, while perturbed neurites inhibited neuronal signalling cascades, cancer progression and metastasis (Lei et al., 2016). As a future development, advanced 3D microscopy and machine learning (Kingston et al., 2019) may soon augment the predictivity of similar tumour extravasation models.
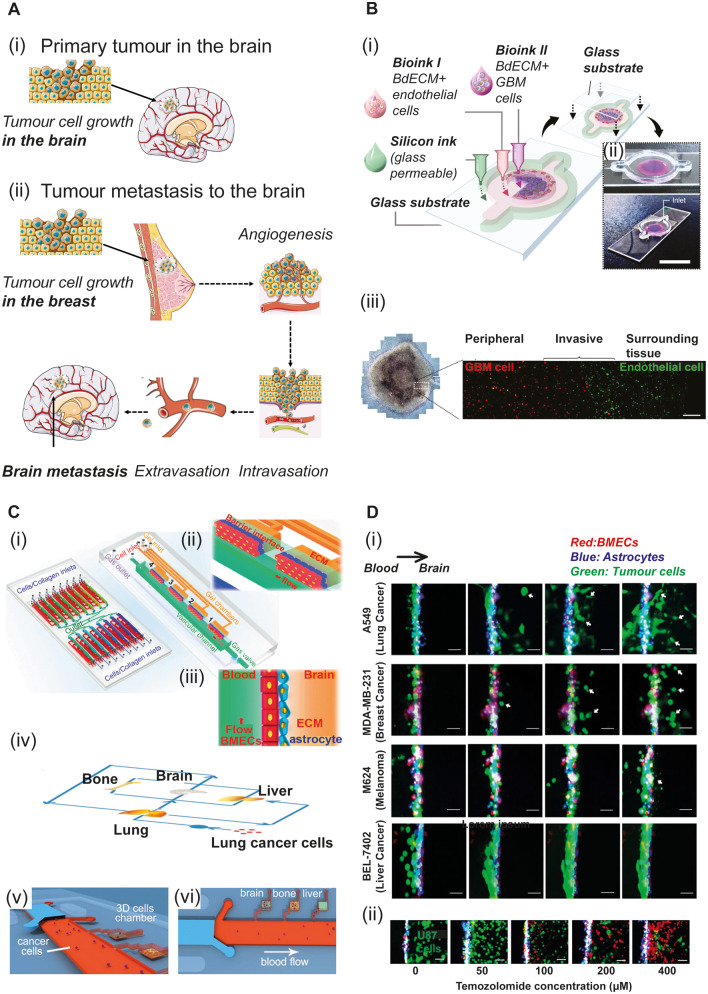
Figure 4.
3D engineered in vitro models for brain cancer. (A) Brain cancer development. Cancerous tumours are classified into two main categories: primary tumours, which begin within the brain tissue (i) and secondary tumours, which arise due to metastasis from other organs, such as the breast, following a series of events as illustrated in ii. Servier Medical Art (SMART) was used for the illustration. (B) Primary tumours: GBM. (i) Construction of a bioprinted GBM-on-a-chip (Yi et al., 2019); (ii) Photographs of the GBM-on-a-chip from above (top) and the corner (bottom). Scale bar = 2 cm. The brain decellularized extracellular matrix (BdECM) bioink includes human umbilical vein endothelial cells (HUVECs; magenta) or GBM cells (blue). (iii) Phase-contrast (left) and fluorescent image (right) of the GBM-on-a-chip. GBM cells are stained with DiI (red) and HUVECs with DiO (green). Scale bar = 200 μm. (C) OoCs to model cancer metastasis to the brain (i–iii) A physiologically relevant blood–brain barrier device (Xu et al., 2016a). The device consists of 16 independent functional units connected via microchannels (i, left). Detailed view of each functional unit (i, right). Magnified view (ii) and side view (iii) of the blood–brain barrier region composed of BMECs, astrocytes and ECM. The red arrow indicates the flow direction. (iv–vi) A multi-organ microfluidic chip to model lung cancer metastasis (Xu et al., 2016c). (iv) Schematic of lung cancer metastasis to distant organs including the brain. (v and vi) 3D cell cultures of different organs in distinct chambers. Lung cancer cells (A549) flow through the media in the microvascular channel (red) to mimic cancer metastasis via the blood vessels. (D) Blood–brain barrier-on-chip device to investigate metastatic brain tumours (Xu et al., 2016a). (i) Time-lapse imaging of different cancer cell types (green) across the blood–brain barrier via the vascular compartment. Cell extravasation to the brain was monitored for 72 h. Lung cancer cells (A549), breast cancer cells (MDA-MB-231) and melanoma cells (M624) disrupted the integrity of the blood–brain barrier and migrated to the brain, whereas liver cancer cells (BEL-7402) did not. (ii) Functional responses of the blood–brain barrier to therapeutic agents. The glioblastoma cells (U87) showed a dose-dependent response to the lipophilic and blood–barrier-permeable medication temozolomide, which was added to the vascular compartment of the chip. Green = live cells; red = dead cells. Scale bar = 25 μm. Figure components are modified from Xu et al. (2016a, c) and Yi et al. (2019) with permission.
Xu et al. (2016a) used a blood–brain barrier-on-a-chip to model tumour invasion of the brain via a disrupted barrier. Their study confirmed the synergic role of astrocytes and endothelial cells in maintaining barrier integrity, as well as the prohibitive role of astrocytes in cancer metastasis. The authors later extended their setup to incorporate a multi-chamber device accommodating organ-specific cell types. They used this system to study lung cancer metastasis to several organs, including the brain [Fig. 4C(iv–vi)] (Xu et al., 2016c). In a subsequent study, Xu and colleagues further addressed effects of inflammatory microvasculature on tumour extravasation, and they showed that TNFa-induced inflammation increases adhesion of adenocarcinoma cells to the inflammatory endothelium (Xu et al., 2017).
Additional in vitro models have sought to model solid brain tumours, which are characterized by high heterogeneity, obstruction of solute production, high waste accumulation, and a hypoxic inner microenvironment (Sleeboom et al., 2018; Wang et al., 2018). 3D scaffolds (Gomez-Roman et al., 2017), organoid-based 3D models (Lancaster et al., 2013; Hubert et al., 2016; Ogawa et al., 2018), bioprinting (Heinrich et al., 2019), glioblastoma multiforme (GBM)-on-chip (Ayuso et al., 2017), and combinatorial methods have been used for this purpose. A recent effort by Yi et al. (2019) describes a bioprinted human GBM-on-chip, which demonstrated patient-specific sensitivity to putative therapeutic agents (Fig. 4B). Shen et al. (2017) used a 3D in vitro platform for quantitative high-throughput screening and showed that several antiparasitic agents may have therapeutic potential for paediatric solid tumours. Incorporation of immune cells in these models might further elucidate the complex interplay among microglia and/or macrophages and tumour cells (Roesch et al., 2018; Sevenich, 2018).
High scalability and ease of use are crucial for effective and accurate cancer disease models in vitro. Phan et al., 2017 presented a novel and versatile OoC drug screening platform, requiring minimal equipment and no external pumps for flow generation. Custom fitted vascularized microtissues on a standard 96-well plate have been used for large-scale drug screenings (Fig. 4D), enabling anti-angiogenic and anti-cancer drugs to be detected (Phan et al., 2017). We envision that hiPSC-based, vascularized cancer-on-chips combined with advanced 3D imaging may pave the way for patient-specific therapeutics.
Host–pathogen interactions
Various pathogens such as bacteria, viruses, and fungi may cause life-threatening infections of the human CNS (Giovane and Lavender, 2018). The increased morbidity and mortality rate of these cases demand accurate disease models for infectious diseases. Upon CNS infection, multiple cells and tissues engage in a vivid interplay while innate and adaptive immune responses lead to cytokine overproduction widely known as ‘cytokine storm’ (Tisoncik et al., 2012; Koyuncu et al., 2013). This process exposes the neural tissue to a cascade of biological phenomena with tremendous long-term sequelae.
Scientists use animal models to mimic the complexity of infectious disease phenotypes (Swearengen, 2018). Even though these models have shed light on various aspects of disease initiation, progression, and manifestation, they often lack translatability to humans (Mestas and Hughes, 2004; Seok et al., 2013; Costamagna et al., 2019). In the case of neurotropic viruses, for example, animal models are challenged by the versatility and the complexity of the intruders. Their translatability is limited by (i) low reproducibility of subtle neurological clinical phenotypes; (ii) the use of transgenic mice to induce severe clinical phenotypes; (iii) inability to reproduce indirect disease mechanisms, which often enhance CNS virulence (e.g. inflammation); and (iv) differential viral infection kinetics due to animal-specific characteristics that result in faster virus clearance (Natoli et al., 2020). On the other hand, hiPSCs are susceptible to human pathogen infections (Costamagna et al., 2019); thus, hiPSC-based infectious disease models may have higher translational value.
In the past, hiPSC-derived brain organoids have been used extensively to model host-virus interactions in the human CNS. Zika virus (ZIKV) is associated with Guillain-Barré syndrome as well as congenital infection and microcephaly (Aragao et al., 2016; Qian et al., 2017; Costamagna et al., 2019). Several efforts have now established hiPSC-based brain organoids as an effective method to investigate ZIKV-induced microcephaly (Cugola et al., 2016; Garcez et al., 2016; Qian et al., 2016). ZIKV impaired the overall growth of the organoids; specifically, the infection resulted in neural precursor cell death and disruption in neurosphere formation. Moreover, Xu et al. (2016b) used forebrain-specific hiPSC-derived organoids to perform a big screen with the prospect of drug repurposing against ZIKV. This work led to the identification of compounds that inhibited either the ZIKV replication or the neural precursor cell death (Xu et al., 2016b). Herpesviruses are often responsible for CNS infections in humans, which mostly result in encephalitis, meningitis, or myelitis (Bulakbasi and Kocaoglu, 2008). Herpesvirus CNS infections exhibit dramatic sequelae especially in immunocompromised or elderly patients and newborns (Costamagna et al., 2019). Unfortunately, and despite early antiviral drug administration, infection of newborns results in encephalitis and high mortality (Kimberlin, 2004). Researchers have used hiPSC-derived cerebral organoids to mimic herpes simplex virus 1 (HSV-1) and cytomegalovirus (CMV) infection in vitro (D’Aiuto et al., 2018, 2019; Brown et al., 2019; Sun et al., 2020). These efforts have greatly advanced our understanding of disease development after infection and manifest the value of using human 3D organoid cultures to recapitulate virus-induced pathologies in the developing brain.
Meningococcal meningitis, caused by Neisseria meningitidis bacteria is a devastating CNS infection associated with increased mortality and frequent severe sequelae (WHO, 2018). N. meningitidis is a human-specific pathogen (Hodeib et al., 2020), thus animal models fail to replicate the pathophysiology of the disease. Martins Gomes et al. (2019) used hiPSC-derived BMECs as a cellular model of N. meningitidis infection. The authors showed barrier permeability, tight junction disruption, and bacterial transmigration into the CNS via the hiPSC-BMECs after N. meningitidis infection. We hypothesize that engineered in vitro models of the human NVU coupled with hiPSC technology might further elucidate the mechanisms underlying the pathophysiology of meningococcal meningitis.
Accumulating clinical evidence shows that respiratory viruses can escape the immune responses of the human body and not only cause severe respiratory issues but also migrate to other organs including the CNS (Vareille et al., 2011; Desforges et al., 2019). The infection of the resident neural populations may then lead to several pathologies including encephalitis (Bohmwald et al., 2018). Like other neuroinvasive viruses, human coronaviruses (hCoVs) can enter the CNS via the hematogenous route or the neuronal retrograde route (Desforges et al., 2019). After studies in both animals and microfluidic devices, Desforges et al. (2019) proposed a putative mechanism that the human coronavirus OC43 (HCoV-OC43) invades the CNS. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the coronavirus disease 2019 (COVID-19) and has led to the ongoing global pandemic (WHO, 2020). The COVID-19 outbreak is at a very early stage; nevertheless, clinical data support the neurotropic character of the virus as several patients suffer from neurological symptoms (Alvin et al., 2020; Mao et al., 2020; Natoli et al., 2020). Both neurons and endothelial cells express the angiotensin-converting enzyme 2 (ACE2) receptor, which hCoVs utilize to enter the cells (Li et al., 2003). SARS-CoV-2 binds the cell membrane via the CoV spike glycoprotein, which in turn binds ACE2 with high affinity. According to Natoli et al. (2020) this might explain the higher neuroinvasive potential of SARS-CoV2 compared to the previous SARS-CoV.
Effective mimicking of the complex CNS cytoarchitecture and cell responses due to SARS-CoV infection is pivotal to perform mechanistic studies, investigate the time course of infection, and disease progression. Beyond any doubt, patient-oriented disease models set the basis for the development of effective therapeutics. The existing mouse models fail to recapitulate human CNS infection and disease progression sufficiently; therefore, the development of humanized in vitro models is critical. Structural differences between ACE2 mouse and human proteins result in the poor tropism of SARS-CoV for mouse tissue; hence, the virus cannot infect mice efficiently (Cleary et al., 2020; Wan et al., 2020). The K18-hACE2 transgenic mice, first proposed by McCray et al. (2007), and now offered by Jackson laboratory, express the human ACE2 receptor and provide us with a valuable tool to study the course of SARS-CoV infection in humans. In these mice, however, the expression of ACE2, driven by the human keratin 18 (KRT18) promoter, is limited to the epithelia. Thus, K18-hACE2 mice are not useful to investigate the effects of the infection on the human brain endothelium. A recent study by Varga et al. (2020) highlights the implication of the endothelial cell layer in various tissues from COVID-19 patients. Moreover, Monteil et al. (2020) have recently shown with the use of human organoids as a model system that human recombinant soluble ACE2 (hrsACE2) but not mouse recombinant soluble ACE2 (mrsACE2) can significantly reduce SARS-CoV-2 infection. Certainly, SARS-CoV-2 research is at an immature state and the involvement of the CNS in disease progression is largely ambiguous (Li et al., 2020a, b; Turtle, 2020). Nevertheless, the severity of the disease and the detrimental impact on our society necessitates the development of accurate, patient-oriented disease models, which emulate the overall disease phenotype including the neurological symptoms (Zhou et al., 2020).
We believe that OoCs offer limitless opportunities for such studies. We envision that linked microdevices might serve as putative platforms to study how systemic inflammation due to respiratory infection contributes to neurological deficits and vice versa. Researchers can use OoCs to unravel how viruses use immune cells as ‘trojan horses’ to surpass the blood–brain barrier and transmigrate to the neural tissue. We foresee that the use of hiPSC-based OoCs may help the scientific society to surpass the translational limits of the animal disease models and pave the way towards improved diagnostics and patient-oriented therapeutics.
Neurotoxicity
For the pharma industry and regulatory authorities there is a growing need to increase the output and efficacy of current toxicity testing and to minimize the use of animals (Krewski et al., 2010; Crofton et al., 2011). In particular, the scientific community is striving to minimize animal use by using in silico studies, such as pharmacokinetic and pharmacodynamic models and quantitative structure activity relationship (QSAR) models (Raies and Bajic, 2016), coupled with advanced in vitro models.
Neurotoxicity testing is multifaceted, focusing not only on whether a potential drug can penetrate the blood–brain barrier but also the vulnerability of the CNS. The developing CNS is particularly sensitive and goes through several highly orchestrated processes such as cell migration, proliferation, and differentiation, and subsequent cell populations develop processes such as synaptogenesis, cell pruning, and myelination. Hence, the conventional dose-response battery of tests needs to factor in the developmental phase when evaluating a potentially harmful chemical (Rice and Barone, 2000; Giordano and Costa, 2012). Alarmingly, many drugs that are on the market have not undergone developmental neurotoxicity testing (Grandjean and Landrigan, 2014; Judson et al., 2014; Schwartz et al., 2015; Tohyama, 2016).
Specific OoC demonstrations of neurotoxicity have not been reported; instead, organoid approaches have so far been used. In 2012, Eiraku and colleagueswere the first group to consider 3D brain organoids (Eiraku and Sasai, 2012) as potential tools for investigating neurotoxicity, even if lacking vasculature and microglia. Schwartz et al. (2015) reported neural constructs encompassing vasculature, albeit not functional, and microglial cells that, in combination with machine learning approaches, resulted in a model that could identify 9 of 10 toxic chemicals in a blinded trial. Future advances for developing organoids for neurotoxicity testing steer towards generating organoids with a functional blood–brain barrier, possibly combined with OoC technology to increase bio-fidelity. Organoid culturing can indeed be a stepping stone towards recapitulating brain development in vitro that exhibits not only distinct developmental phases but also higher brain functions.
Applicability of engineered in vitro models in neuroscience settings
The discussion above points to the vast potential of engineered in vitro systems to enhance our understanding of the CNS in health and in disease. Notably, adoption of such systems in commercial and academic labs may ultimately also be cost-effective: for example, continuing to develop OoC devices for drug screening is expected to reduce research and development costs for each new drug by 10–26%, within a timescale of 5 years (Franzen et al., 2019). Below, we elaborate on some of the considerations that labs should take into account when considering the adoption of an engineered in vitro model system.
There are several channels through which a commercial or academic neuroscience lab can adopt new engineered CNS in vitro models: (i) collaborating with model developers; (ii) fabricating models in-house; or (iii) purchasing commercial systems. When deciding which of these approaches to take, it is important to consider the complexity and portability of the system in relation to the biological question that is to be addressed.
Model fabrication in-house or with the aid of collaborators
Biological considerations
The constraints of biological entities such as cells, tissue slices and native structure of proteins should guide the design of a specific engineered device. Technically, geometry, porosity, mechanical parameters (such as stiffness of the surface), topography, transparency, thickness and material chemistry are just a few of the parameters to be considered in fabricating a microfluidic in vitro device (Kilic et al., 2016; Vernetti et al., 2017).
Microfabrication techniques and materials
Currently, microfabrication techniques that are suitable for fabricating engineered devices for a tissue-specific environment at the microscale, and that can easily be implemented in an academic lab, include soft lithography, photolithography, direct laser patterning and microcontact printing and 3D printing (Campisi et al., 2018). All these techniques allow for the development of devices at reasonable costs, and can also serve to reduce the amounts of chemicals and biological materials used compared to large-scale plate formats (Wang et al., 2018).
One of the most easily implemented materials in a non-engineering lab is poly(dimethylsiloxane) (PDMS). Templates or moulds defining the channels and shapes of the device can be generated to order by central foundries or by companies at low cost. PDMS is then simply cast in the mould and cured at ∼80°C. Additional treatments, such as oxygen plasma, may be needed for assembling separate parts into a complete device. An oxygen plasma chamber can be acquired by a non-engineering lab at relatively low cost. The reason to choose PDMS for small-scale (e.g. academic) chip fabrication is that it is the most widely used material to fabricate microfluidic devices, and it allows for fabrication easily adopted by non-experts, transparency, biocompatibility and gas permeability, essential requirements in developing engineered in vitro models. Specifically, gas permeability enables cells or tissues to be kept alive even for long-term experiments, while transparency allows cellular morphology to be monitored using various high-quality microscopy techniques (Park et al., 2015; Kilic et al., 2016). Yet, PDMS-based devices also suffer from several shortcomings. Primary limitations include absorption of small molecules and diffusion of water vapour, which affect concentrations of components in the cell culture, such as certain amino acids, vitamins, neurotransmitters and growth factors (Toepke and Beebe, 2006), as well as drugs, such as verapamil and nifedipine (Berthier et al., 2012; van Meer et al., 2017).
To overcome this limitation, Ingber and colleagues began to use polyurethane in fabricating polymer-based devices, which preserve PDMS properties, such as flexibility and transparency, without absorbing small hydrophobic molecules (Zhang et al., 2017). Another potential candidate is styrene-ethylene-butylene-styrene (SEBS) copolymers, whose use in constructing devices was patented by Emulate (Huh et al., 2011; Zhang et al., 2017). Polyetherimide (PEI), polycarbonate, silicon, glass, silk protein, and agarose are additional materials that might potentially be used. Importantly, many of these materials can be processed in non-engineering settings because of the relatively large dimensions (>100 mm) of OoCs. More detailed discussion about the production methods for engineered in vitro brain can be found in other relevant papers (Haring et al., 2017; Sosa-Hernández et al., 2018; Yu and Choudhury, 2019), and filmed chip fabrication protocols can be found in some recently published studies (van der Helm et al., 2017; Novak et al., 2018; Jagadeesan et al., 2020).
System setup
When an academic lab has chosen the chip design and material based on a specific biological question, the next step is to introduce a sterilization method, surface coatings, cells or tissue as well as a fluidic interface. For the latter, depending on budget and model design, there are a variety of solutions using external pumps and tubing or pumpless, gravity and diffusion driven solutions. Indeed, when developing and designing a highly accurate OoC system, fluid control (directing, monitoring, controlling) should be a key focus, particularly since the high surface-to-volume ratio necessitates frequent renewal of the cell culture medium. Different types of systems have been developed to accurately deliver and control flow in microfluidic devices (e.g. peristaltic and recirculating pumps (Skafte-Pedersen et al., 2009); pressure-control systems (Heo et al., 2016) or syringe pumps (Kuczenski et al., 2007; Chen et al., 2018). Furthermore, several recent studies have focused on reproducing physiological blood flow changes, implementing OoCs to generate laminar (Zheng et al., 2012), pulsatile (Shao et al., 2009) or interstitial flow (Kingsmore et al., 2016; Kaarj and Yoon, 2019).
Some of the parameters that need to be taken into consideration when developing new OoC devices are summarized in Box 1.
Box 1.
Overview of some important parameters when developing a new OoC device
| ADVANTAGES | LIMITATIONS |
|---|---|
| HUMAN TISSUE SOURCE | |
| Embryonic stem cells (ESCs) a , b | |
|
|
| Human induced pluripotent stem cells (hiPSCs) a , b , c , d , e , f | |
|
|
| Tissue biopsies b , f , g | |
|
|
| Cell lines a , b , f , h | |
|
|
| FLOW MANIPULATION | |
| Microfluidic systems f , h , I , j , k , l , m | |
|
|
| MATERIALS: BIOCOMPATIBLE POLYMERS | |
| Polydimethylsiloxane (PDMS) e , f , h | |
|
|
| Poly(methyl methacrylate) (PMMA) n , o , p | |
|
|
| Polycarbonate (PC) n , p | |
|
|
| FABRICATION TECHNIQUES | |
| Photolithography e , h , q | |
|
|
| 3D printing h , p , r | |
|
|
| Microcontact printing s | |
| Low cost and rapid prototyping | Difficulty in controlling the ink and the surface robustness |
| Laser-based patterning s | |
| Cells and any particles can be manipulated | Large instrumentation, complex setup |
| Injection moulding g | |
|
|
| Casting u | |
| Process, equipment setup and replication accuracy | Long process time (e.g. labour and lab costs) |
| CHIP DESIGN | |
| 2D system v , w | |
|
|
| 3D system v , w | |
| 3D architecture very close to in vivo model | Very complex and expensive to build and to control |
Jodat et al., 2018;
Importantly, a new OoC user should be informed of simple strategies to avoid air bubble formation, contamination and flow disturbances in their specific systems. We recommend visiting an OoC lab or participating in workshops to acquire the expertise needed to set up an in-house microfluidics system.
Commercial engineered tissue culture devices
Because of the promising results of novel in vitro models, several approaches to commercializing engineered tissue culture devices are ongoing (Zhang et al., 2017). Some of the leading developments in commercial microfluidic devices used for recapitulating brain tissues are summarized in Table 3 and Supplementary Fig. 3. GlaxoSmithKline, BASF, Sanofi and AbbVie funded MIMETAS B.V. (https://mimetas.com), an OoC company with a multiple-chip solution integrated on one plate, and which has recently begun to develop a microfluidic 3D cell culture system for neurotoxicity screening (Wevers et al., 2016; Kane et al., 2019). The Wyss Institute for Biologically Inspired Engineering at Harvard University is collaborating with the start-up Emulate, Inc. (Wyss Institute, 2014) for commercializing its OoC technology (Sances et al., 2018). TissUse GmbH is another company that, together with the HiPSTAR consortium, began to develop a new in vitro model of the human blood–brain barrier (https://www.tissuse.com/en/news/press-releases/); their work focuses on identifying new drugs and therapies targeting dementia. Hesperos (https://hesperosinc.com/technology/) commercialized a new microfluidic hiPSC-derived blood–brain barrier model capable of mimicking in vivo blood–brain barrier characteristics; their platform includes reliable in vitro transport mechanisms and accommodates rate measurements for drug permeability screening (Wang et al., 2018; Ramme et al., 2019). In 2019, the company AxoSim, already a cutting-edge facility for modelling human physiology in vitro, acquired Organome (https://www.axosim.com) and subsequently developed and patented two novel platforms (the nerve-on-a-chip and the mini-brain organoid), expanding the possibility to address the growing burden of neurodegenerative diseases, both at central and peripheral nervous level (Huval et al., 2015). Additional commercial options include a 3D-cell culture chip developed by AimBiotech (https://www.aimbiotech.com/about-us.html), which gave rise to the development of a new 3D blood–brain barrier model replicating the in vivo neurovascular organization (Campisi et al., 2018), offering a new platform for drug discovery. Additional manufacturers that warrant attention include Synvivo (https://www.synvivobio.com), Xona microfluidics (https://xonamicrofluidics.com/) and Ananda (https://anandadevices.com/application/extension-of-axons-isolated-using-anandas-neuro-device/); these companies have successfully created novel microfluidic devices to direct neuronal growth and axonal extension (Deosarkar et al., 2015; Magdesian et al., 2016; Paranjape et al., 2019).
Table 3.
Commercial OoC or chip providers
| Developer | Engineered devices | Strengths | Limitations |
|---|---|---|---|
| MIMETAS the organ-on-a-chip company | OrganoPlate®, a microfluidic 3D culture plate, made of 96 independent microfluidic chips Each culture cells contain a perfusion channel It can be used as culture system for different cell types, including neurons, endothelial cells and organoids |
|
Only operational with bi-directional flow |
| EMULATE | Dual-channel microfluidic chip able to recreate the body’s dynamic cellular microenvironment (e.g. tissue-tissue interaction, blood flow and mechanical forces) |
|
|
|
HUMIMIC Chip 4, microfluidic four-organ-chip devices, includes two separate microfluidic process, designed to host intestinal, liver, renal and brain cultures |
|
|
| AxoSim | Microengineered nerve-on-a-chip device enabling the growth of 3D neural fibres bundles for peripheral neurotoxicity and physiological testing |
|
|
| SynVivo | SynBBB, 3D OoC model, allowing real-time studies of cellular behaviour, drug delivery and drug discovery, closely mimicking in vivo cellular microenvironment |
|
|
| Xona® microfluidics | XonaChips®, multicompartment microfluidic chip, allowing neuron cell culture It offers the ability to isolate and grow axons, for specifically studying neuronal response to axonal damage, in an isolated fluidic environment |
|
|
| anandaTM | The Neuro Device enables to pattern neurons and direct axonal extension It enables the growth of 100 axons with more than 1 mm in length |
|
|
In this context, it is also important to mention Ibidi (https://ibidi.com/content/34-ibidi-at-a-glance) and the microfluidic ChipShop (https://www.microfluidic-chipshop.com/). These two companies have become some of the leading providers of innovative functional microfluidic chips currently utilized from various academic research and pharma. Many of these commercial models are interesting options for industrial researchers, and some are also affordable for academic researchers.
Concluding remarks and outlook
Advanced in vitro platforms comprising human cells, and that can recreate integrated human physiological functions, have vast potential to contribute to our understanding of cellular mechanisms and the pathogenesis of neurological disorders. Engineered platforms based on microfluidics, such as OoCs, have emerged as a particularly promising in vitro technology, one that is also versatile and flexible enough to be integrated in a wide range of experimental settings (Huang et al., 2012). The main advantage of OoC systems is the possibility to mimic human organ physiology while observing individual cell systems in isolation, and in a manner that is highly reproducible as well as cost effective. Though in vitro techniques are unlikely to replace in vivo models as the gold standard for cancer research (at least in the foreseeable future), engineered in vitro platforms such as those discussed herein hold substantial potential for overcoming many of the limitations of animal models (e.g. questionable translatability to humans). Importantly, as emphasized herein, OoC technologies have matured to the point where CNS researchers in both academic and commercial research labs should be able to easily adopt them—either alone or in conjunction with alternative methodologies—to study basic physiological mechanisms, disease, as well as drug pharmacokinetic and pharmacodynamic models.
Funding
This research was supported by the Wallenberg foundation, Grant 2015-0178, KTH Tenure Track and Forska Utan Djurforsok (A.H.); Azrieli Foundation (B.M.M), The Israel Science Foundation Grant 2248/19 (B.M.M.) and The Brainboost grant (B.M.M).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- ALS =
amyotrophic lateral sclerosis
- BMEC =
brain microvascular endothelial cells
- COVID-19 =
coronavirus disease 2019
- ECM =
extracellular matrix
- GBM =
glioblastoma multiforme
- (h)iPSC =
(human) induced pluripotent stem cell
- NVU =
neurovascular unit
- OoC =
organ-on-chip
- PDMS =
poly(dimethylsiloxane)
- SARS-CoV-2 =
severe acute respiratory syndrome coronavirus 2
- TBI =
traumatic brain injury
- TEER =
transendothelial electrical resistance
References
- Abbott NJ, Rönnbäck L, Hansson E.. Astrocyte–endothelial interactions at the blood–brain barrier. Nat Rev Neurosci 2006; 7: 41. [DOI] [PubMed] [Google Scholar]
- Abdeen AA, Lee J, Kilian KA.. Capturing extracellular matrix properties in vitro: microengineering materials to decipher cell and tissue level processes. Exp Biol Med (Maywood) 2016; 241: 930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-derived human microglia-like cells to study neurological diseases. Neuron 2017; 94: 278–93.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achyuta AK, Conway AJ, Crouse RB. et al. A modular approach to create a neurovascular unit-on-a-chip. Lab Chip 2013; 13: 542. doi: 10.1039/C2LC41033H. [DOI] [PubMed] [Google Scholar]
- Adriani G, Ma D, Pavesi A, Kamm RD, Goh ELK.. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood–brain barrier. Lab Chip 2017; 17: 448–59. [DOI] [PubMed] [Google Scholar]
- Ahadian S, Civitarese R, Bannerman D. et al. Organ-on-a-chip platforms: A convergence of advanced materials, cells, and microscale technologies. Adv Healthc Mater 2018; 7: 201700506. doi: 10.1002/adhm.201700506. [DOI] [PubMed] [Google Scholar]
- Ahmad R, Sportelli V, Ziller M, Spengler D, Hoffmann A.. Tracing early neurodevelopment in schizophrenia with induced pluripotent stem cells. Cells 2018; 7:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsarrani A, Kaplita PV. In silico and in vitro evaluation of brain penetration properties of selected nootropic agents. Future Drug Discov 2019; 1: 1. [Google Scholar]
- de Almeida PE, Meyer EH, Kooreman NG, Diecke S, Dey D, Sanchez-Freire V, et al. Transplanted terminally differentiated induced pluripotent stem cells are accepted by immune mechanisms similar to self-tolerance. Nat Commun 2014; 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman T, Geller D, Kleeblatt E, Gradus-Perry T, Perlson E.. An in vitro compartmental system underlines the contribution of mitochondrial immobility to the ATP supply in the NMJ. J Cell Sci 2019; 132: jcs234492. [DOI] [PubMed] [Google Scholar]
- Alvin MD, George E, Deng F, Warhadpande S, Lee SI.. The impact of COVID-19 on radiology trainees. Radiology 2020; 296: 246–8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H, Van D, Berg A. Microfabrication and microfluidics for tissue engineering: state of the art and future opportunities. Lab Chip 2004; 4: 98. doi: 10.1039/b314469k. [DOI] [PubMed] [Google Scholar]
- Antony PM, Diederich NJ, Kruger R, Balling R.. The hallmarks of Parkinson’s disease. FEBS J 2013; 280: 5981–93. [DOI] [PubMed] [Google Scholar]
- Aragao MDF, Linden VVD, Brainer-Lima AM, Coeli RR, Rocha MA, Silva PD, et al. Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: retrospective case series study. BMJ 2016; 353: i1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Ishizuka T, Morishima K, Yawo H.. Optogenetic induction of contractile ability in immature C2C12 myotubes. Sci Rep 2015; 5: 8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso JM, Monge R, Martínez-González A, Virumbrales-Muñoz M, Llamazares GA, Berganzo J, et al. Glioblastoma on a microfluidic chip: generating pseudopalisades and enhancing aggressiveness through blood vessel obstruction events. Neuro Oncol 2017; 19: 503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeten KM, Akassoglou K. Extracellular matrix and matrix receptors in blood-brain barrier formation and stroke. Dev Neurobiol 2011; 71: 1018. doi: 10.1002/dneu.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgude AP, Yu X, Szymanski A, Bellamkonda RV.. Agarose Gel Stiffness Determines Rate of DRG Neurite Extension in 3D Cultures. 2001; 8. [DOI] [PubMed] [Google Scholar]
- Barcelona PF, Saragovi HU.. A Pro-Nerve Growth Factor (proNGF) and NGF Binding Protein, α 2 -macroglobulin, differentially regulates p75 and TrkA receptors and is relevant to neurodegeneration Ex Vivo and In Vivo. Mol Cell Biol 2015; 35: 3396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N.. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 2011; 9: 17–23. [DOI] [PubMed] [Google Scholar]
- Barrejón M, Rauti R, Ballerini L, Prato M.. Chemically cross-linked carbon nanotube films engineered to control neuronal signaling. ACS Nano 2019; 13: 8879–89. [DOI] [PubMed] [Google Scholar]
- Becker H, Gärtner C. Polymer microfabrication technologies for microfluidic systems. Anal Bioanal Chem 2008; 390: 89. doi: 10.1007/s00216-007-1692-2. [DOI] [PubMed] [Google Scholar]
- Belle AM, Enright HA, Sales AP, Kulp K, Osburn J, Kuhn EA, et al. Evaluation of in vitro neuronal platforms as surrogates for in vivo whole brain systems. Sci Rep 2018; 8: 10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee H-H, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods 2016; 13: 151–7. [DOI] [PubMed] [Google Scholar]
- Berg AVD, Mummery CL, Passier R, Meer AVD.. Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 2019; 19: 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology 2012; 62: 1584–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier E, Young EWK, Beebe D.. Engineers are from PDMS-land, Biologists are from Polystyrenia. Lab Chip 2012; 12: 1224. [DOI] [PubMed] [Google Scholar]
- Bez A, Corsini E, Curti D, Biggiogera M, Colombo A, Nicosia RF, et al. Neurosphere and neurosphere-forming cells: morphological and ultrastructural characterization. Brain Res 2003; 993: 18–29. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE.. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32: 760–72. [DOI] [PubMed] [Google Scholar]
- Bischel LL, Beebe DJ, Sung KE.. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer 2015; 15: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop ES, Mostafa S, Pakvasa M, Luu HH, Lee MJ, Wolf JM, et al. 3-D bioprinting technologies in tissue engineering and regenerative medicine: current and future trends. Genes Dis 2017; 4: 185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Daneman R.. Formation and maintenance of the BBB. Mech Dev 2015; 138: 8–16. [DOI] [PubMed] [Google Scholar]
- Bohmwald K, Gálvez NMS, Ríos M, Kalergis AM.. Neurologic alterations due to respiratory virus infections. Front Cell Neurosci 2018; 12: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Nazor KL, Loring JF.. Epigenetic regulation of pluripotency and differentiation. Circ Res 2014; 115: 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognin S, Fossépré M, Qing X, Jarazo J, Ščančar J, Moreno EL, et al. 3D cultures of Parkinson’s disease-specific dopaminergic neurons for high content phenotyping and drug testing. Adv Sci 2019; 6: 1800927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneh-Barkay D, Wiley CA.. Brain extracellular matrix in neurodegeneration. Brain Pathol 2009; 19: 573–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij TH, Price LS, Danen EHJ.. 3D cell-based assays for drug screens: challenges in imaging, image analysis, and high-content analysis. SLAS Discov 2019; 24: 615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth R, Kim H.. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 2012; 12: 1784–92. [DOI] [PubMed] [Google Scholar]
- Boyle PA, Buchman AS, Wilson RS, Leurgans SE, Bennett DA.. Association of muscle strength with the risk of alzheimer disease and the rate of cognitive decline in community-dwelling older persons. Arch Neurol 2009; 66: 1339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragança J, Lopes JA, Mendes-Silva L, Almeida Santos JM.. Induced pluripotent stem cells, a giant leap for mankind therapeutic applications. World J Stem Cells 2019; 11: 421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ. Serum-free B27/neurobasal medium supports differentiated growth of neurons from the striatum, substantia nigra, septum, cerebral cortex, cerebellum, and dentate gyrus. J Neurosci Res 1995; 42: 674–83. [DOI] [PubMed] [Google Scholar]
- Brookhouser N, Raman S, Potts C, Brafman DA.. May I Cut in? Gene editing approaches in human induced pluripotent stem cells. Cells 2017; 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, et al. Recreating blood-brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics 2015; 9: 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM, Rana PSJB, Jaeger HK, O’Dowd JM, Balemba OB, Fortunato EA.. Human Cytomegalovirus Compromises Development of Cerebral Organoids. J Virol 2019; 93: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulakbasi N, Kocaoglu M.. Central nervous system infections of herpesvirus family. Neuroimaging Clin N Am 2008; 18: 53–84. [DOI] [PubMed] [Google Scholar]
- Burnside ER, Bradbury EJ.. Review: manipulating the extracellular matrix and its role in brain and spinal cord plasticity and repair: manipulating the matrix for CNS repair. Neuropathol Appl Neurobiol 2014; 40: 26–59. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Li YF, Matsa E. et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016; 22: 547. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Jones HC, Abbott NJ.. Electrical resistance across the blood-brain barrier in anaesthetized rats: a developmental study. J Physiol 1990; 429: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero D, Kaushik S, Correlo VM, Oliveira JM, Reis RL, Kundu SC.. Organ-on-chip models of cancer metastasis for future personalized medicine: from chip to the patient. Biomaterials 2017; 149: 98–115. [DOI] [PubMed] [Google Scholar]
- Cabrera MA, Bermejo M, Pérez M, Ramos R.. TOPS-MODE approach for the prediction of blood–brain barrier permeation. J Pharmaceut Sci 2004; 93: 1701–17. [DOI] [PubMed] [Google Scholar]
- Caldeira J, Sousa A, Sousa DM, Barros D, Extracellular matrix constitution and function for tissue regeneration and repair In: Barbosa MA, Martins MCL, editors. Peptides and proteins as biomaterials for tissue regeneration and repair. Elsevier; Amsterdam, Netherlands: Woodhead Publishing; 2018. p. 29–72. [Google Scholar]
- Campenot RB. Local control of neurite development by nerve growth factor. Proc Natl Acad Sci USA 1977; 74: 4516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi M, Shin Y, Osaki T, Hajal C, Chiono V, Kamm RD.. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 2018; 180: 117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield SG, Stebbins MJ, Morales BS, Asai SW, Vatine GD, Svendsen CN, et al. An isogenic blood-brain barrier model comprising brain endothelial cells, astrocytes and neurons derived from human induced pluripotent stem cells. J Neurochem 2017; 140: 874–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-de-la-Cuerda R, Pérez-de-Heredia M, Miangolarra-Page J, Muñoz-Hellín E, Fernández-de-las-Peñas C.. Is There muscular weakness in Parkinson’s disease? Am J Phys Med Rehabil 2010; 89: 70–6. [DOI] [PubMed] [Google Scholar]
- Caragher S, Chalmers AJ, Gomez-Roman N.. Glioblastoma’s next top model: novel culture systems for brain cancer radiotherapy research. Cancers (Basel) 2019; 11: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavero I, Guillon JM, Holzgrefe HH. Human organotypic bioconstructs from organ-on-chip devices for human-predictive biological insights on drug candidates. Expert Opin Drug Saf 2019; 18: 651. doi: 10.1080/14740338.2019.1634689. [DOI] [PubMed] [Google Scholar]
- Centeno EGZ, Cimarosti H, Bithell A.. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol Neurodegener 2018; 13: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapanian R, Amsden BG. . Combined and sequential delivery of bioactive VEGF165 and HGF from poly(trimethylene carbonate) based photo-cross-linked elastomers. J Control Release 2010; 143: 53. doi: 10.1016/j.jconrel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- Chen M-C, Lake JR, Heyde KC, Ruder WC.. Three-dimensional printing of thermoplastic materials to create automated syringe pumps with feedback control for microfluidic applications. J Vis Exp 2018; 138: 57532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. Selecting a mouse model of Alzheimer’s disease In: Roberson ED, editor. Alzheimer’s disease and frontotemporal dementia: methods and protocols. Totowa, NJ: Humana Press; 2011. p. 169–89. [DOI] [PubMed] [Google Scholar]
- Chlebek J Korábečný J Doležal R Štěpánková Š Pérez DI Hošťálková A, . et al. In Vitro and In Silico acetylcholinesterase inhibitory activity of thalictricavine and canadine and their predicted penetration across the blood-brain barrier. Molecules 2019; 24: 30959739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 2015; 33: 1173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014; 515: 274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Tanzi RE.. iPSCs to the rescue in Alzheimer’s research. Cell Stem Cell 2012; 10: 235–6. [DOI] [PubMed] [Google Scholar]
- Choi Y, Lee S, Kim K, Kim S-H, Chung Y-J, Lee C.. Studying cancer immunotherapy using patient-derived xenografts (PDXs) in humanized mice. Exp Mol Med 2018; 50: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SJ, Pitchford SC, Amison RT, Carrington R, Robaina Cabrera CL, Magnen M, et al. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br J Pharmacol 2020. doi: 1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan OC, Collins NT, Ferguson G, Murphy RP, Birney YA, Cahill PA, et al. Influence of basolateral condition on the regulation of brain microvascular endothelial tight junction properties and barrier function. Brain Res 2008; 1193: 84–92. [DOI] [PubMed] [Google Scholar]
- Coluccio ML, Perozziello G, Malara N, Parrotta E, Zhang P, Gentile F, et al. Microfluidic platforms for cell cultures and investigations. Microelectron Eng 2019; 208: 14–28. [Google Scholar]
- Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, et al. Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci 2010; 45: 258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquinco A, Kojic L, Wen W, Wang YT, Jeon NL, Milnerwood AJ, et al. A microfluidic based in vitro model of synaptic competition. Mol Cell Neurosci 2014; 60: 43–52. [DOI] [PubMed] [Google Scholar]
- Costamagna G, Andreoli L, Corti S, Faravelli I.. iPSCs-based neural 3D systems: a multidimensional approach for disease modeling and drug discovery. Cells 2019; 8: 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR.. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol 2008; 10: 149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofton KM, Mundy WR, Lein PJ, Bal-Price A, Coecke S, Seiler AEM, et al. Developmental neurotoxicity testing: recommendations for developing alternative methods for the screening and prioritization of chemicals. ALTEX 2011; 28: 9–15. [PubMed] [Google Scholar]
- Crone C, Olesen SP.. Electrical resistance of brain microvascular endothelium. Brain Res 1982; 241: 49–55. [DOI] [PubMed] [Google Scholar]
- Cucullo L, Hossain M, Puvenna V, Marchi N, Janigro D.. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neurosci 2011; 12: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016; 534: 267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aiuto L, Bloom DC, Naciri JN, Smith A, Edwards TG, McClain L, et al. Modeling Herpes Simplex Virus 1 infections in human central nervous system neuronal cells using two- and three-dimensional cultures derived from induced pluripotent stem cells. J Virol 2019; 93: e00111–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aiuto L, Naciri J, Radio N, Tekur S, Clayton D, Apodaca G, et al. Generation of three-dimensional human neuronal cultures: application to modeling CNS viral infections. Stem Cell Res Ther 2018; 9: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Marom A, Paluch S, Dvorkin R, Brosh I, Shoham S.. Hybrid multiphoton volumetric functional imaging of large-scale bioengineered neuronal networks. Nat Commun 2014; 5: 3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauth S, Grevesse T, Pantazopoulos H, Campbell PH, Maoz BM, Berretta S, et al. Extracellular matrix protein expression is brain region dependent: brain Extracellular Matrix Distribution Analysis. J Comp Neurol 2016; 524: 1309–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauth S, Maoz BM, Sheehy SP, Hemphill MA, Murty T, Macedonia MK, et al. Neurons derived from different brain regions are inherently different in vitro: a novel multiregional brain-on-a-chip. J Neurophysiol 2017; 117: 1320–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice FG, Munoz DP.. Opportunities and challenges in developing relevant animal models for Alzheimer’s disease. Ageing Res Rev 2016; 26: 112–4. [DOI] [PubMed] [Google Scholar]
- DeFelipe J. The anatomical problem posed by brain complexity and size: a potential solution. Front Neuroanat 2015; 9: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A, Liu Y, Colak D, Hengst U, Xu G, Jaffrey SR.. Coupled local translation and degradation regulate growth cone collapse. Nat Commun 2015; 6: 6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsing L, Donnes P, Sanchez J, Clausen M, Voulgaris D, Falk A, et al. Barrier properties and transcriptome expression in human iPSC-derived models of the blood-brain barrier. Stem Cells 2018; 36: 1816–27. [DOI] [PubMed] [Google Scholar]
- Demers CJ, Soundararajan P, Chennampally P, Cox GA, Briscoe J, Collins SD, et al. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development 2016; 143: 1884–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer T, Stöhr T, Van Roy M.. Animal models in translational medicine: validation and prediction. New Horizons Transl Med 2014; 2: 5–11. [Google Scholar]
- Deosarkar SP, Prabhakarpandian B, Wang B, Sheffield JB, Krynska B, Kiani MF.. A novel dynamic neonatal blood-brain barrier on a chip. PLoS One 2015; 10: e0142725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges M, Le Coupanec A, Dubeau P, Bourgouin A, Lajoie L, Dubé M, et al. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses 2019; 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano JG, Xu ZS, Williams AJ, Yimam N, Searson PC.. Effect of shear stress on iPSC-derived human brain microvascular endothelial cells (dhBMECs). Fluids Barriers CNS 2017; 14: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier N, Banks WA, Créminon C, Dereuddre-Bosquet N, Mabondzo A.. HIV-1-induced production of endothelin-1 in an in vitro model of the human blood-brain barrier. Neuroreport 2002; 13: 1179–83. [DOI] [PubMed] [Google Scholar]
- Didier N, Romero IA, Créminon C, Wijkhuisen A, Grassi J, Mabondzo A.. Secretion of interleukin-1beta by astrocytes mediates endothelin-1 and tumour necrosis factor-alpha effects on human brain microvascular endothelial cell permeability. J Neurochem 2003; 86: 246–54. [DOI] [PubMed] [Google Scholar]
- Ding S-L, Royall JJ, Sunkin SM, Ng L, Facer BAC, Lesnar P, et al. Comprehensive cellular-resolution atlas of the adult human brain: adult human brain atlas. J Comp Neurol 2016; 524: 3127–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov 2006; 5: 210. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- Dityatev A. Remodeling of extracellular matrix and epileptogenesis: extracellular matrix and epileptogenesis. Epilepsia 2010; 51: 61–5. [DOI] [PubMed] [Google Scholar]
- Dollé J-P, Morrison B, Schloss RS, Yarmush ML.. Brain-on-a-chip microsystem for investigating traumatic brain injury: axon diameter and mitochondrial membrane changes play a significant role in axonal response to strain injuries. Technology 2014; 02: 106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollé J-P, Morrison B, Schloss RS III, Yarmush ML. An organotypic uniaxial strain model using microfluidics. Lab Chip 2013; 13: 432–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss MX, Sachinidis A.. Current challenges of iPSC-based disease modeling and therapeutic implications. Cells 2019; 8: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti C, Sullivan C, Banker G.. The establishment of polarity by hippocampal neurons in culture. J Neurosci 1988; 8: 1454–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep 2017; 8: 1516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove A. In vitro veritas: Biosensors and microarrays come to life. Science | AAAS 2018[ 2018.
- Dubois-Dauphin ML, Toni N, Julien SD, Charvet I, Sundstrom LE, Stoppini L.. The long-term survival of in vitro engineered nervous tissue derived from the specific neural differentiation of mouse embryonic stem cells. Biomaterials 2010; 31: 7032–42. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Mozafari S, Glatza M, Starost L, Velychko S, Hallmann A-L, et al. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc Natl Acad Sci USA 2017; 114: E2243–E2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Sasai Y.. Self-formation of layered neural structures in three-dimensional culture of ES cells. Curr Opin Neurobiol 2012; 22: 768–77. [DOI] [PubMed] [Google Scholar]
- Esch MB, Mahler GJ, Stokol T, Shuler ML.. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014; 14: 3081–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini GS, Chiu DT. Disposable microfluidic devices: fabrication, function, and application. Biotechniques 2005; 38: 429. doi: 10.2144/05383RV02. [DOI] [PubMed] [Google Scholar]
- Fisher EMC, Bannerman DM.. Mouse models of neurodegeneration: know your question, know your mouse. Sci Transl Med 2019; 11: eaaq1818. [DOI] [PubMed] [Google Scholar]
- Frantz C, Stewart KM, Weaver VM.. The extracellular matrix at a glance. J Cell Sci 2010; 123: 4195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzen N, van Harten WH, Retèl VP, Loskill P, van den Eijnden-van Raaij J, IJzerman M.. Impact of organ-on-a-chip technology on pharmaceutical R&D costs. Drug Discov Today 2019; 24: 1720–4. [DOI] [PubMed] [Google Scholar]
- Frazier JM. Validation of In Vitro Models. J Am Coll Toxicol 1990; 9: 355–9. [Google Scholar]
- Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat Med 2018; 24: 1579–89. [DOI] [PubMed] [Google Scholar]
- Fukushima M, Setoguchi T, Komiya S, Tanihara H, Taga T.. Retinal astrocyte differentiation mediated by leukemia inhibitory factor in cooperation with bone morphogenetic protein 2. Int J Dev Neurosci 2009; 27: 685–90. [DOI] [PubMed] [Google Scholar]
- Fyfe I. Alzheimer disease: epigenetics links ageing with Alzheimer disease. Nat Rev Neurol 2018; 14: 254. [DOI] [PubMed] [Google Scholar]
- Gähwiler BH. Organotypic monolayer cultures of nervous tissue. J Neurosci Methods 1981; 4: 329–42. [DOI] [PubMed] [Google Scholar]
- Galie PA, Nguyen D-HT, Choi CK, Cohen DM, Janmey PA, Chen CS.. Fluid shear stress threshold regulates angiogenic sprouting. Proc Natl Acad Sci USA 2014; 111: 7968–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Costa R. D, Higa LM, Trindade P, Delvecchio R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016; 352: 816–8. [DOI] [PubMed] [Google Scholar]
- García-Parra P, Maroto M, Cavaliere F, Naldaiz-Gastesi N, Álava JI, García AG, et al. A neural extracellular matrix-based method for in vitro hippocampal neuron culture and dopaminergic differentiation of neural stem cells. BMC Neurosci 2013; 14: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Polite F, Martorell J, Del Rey-Puech P, Melgar-Lesmes P, O’Brien CC, Roquer J, et al. Pulsatility and high shear stress deteriorate barrier phenotype in brain microvascular endothelium. J Cereb Blood Flow Metab 2017; 37: 2614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reitboeck P, Phillips A, Piers TM, Villegas-Llerena C, Butler M, Mallach A, et al. Human induced pluripotent stem cell-derived microglia-like cells harboring TREM2 missense mutations show specific deficits in phagocytosis. Cell Rep 2018; 24: 2300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta E, Sanchez S, Lajara J, Montserrat N, Belmonte JCI.. Roadblocks in the path of iPSC to the clinic. Curr Transpl Rep 2018; 5: 14–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gencturk E, Mutlu S, Ulgen KO. Advances in microfluidic devices made from thermoplastics used in cell biology and analyses. Biomicrofluidics 2017; 11: 051502. doi: 10.1063/1.4998604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano G, Costa LG.. Developmental neurotoxicity: some old and new issues. ISRN Toxicol 2012; 2012: 814795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovane RA, Lavender PD.. Central nervous system infections. Prim Care Clin Office Pract 2018; 45: 505–18. [DOI] [PubMed] [Google Scholar]
- Giulian D, Baker T.. Characterization of ameboid microglia isolated from developing mammalian brain. J Neurosci 1986; 6: 2163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Stevenson K, Gilmour L, Hamilton G, Chalmers AJ.. A novel 3D human glioblastoma cell culture system for modeling drug and radiation responses. Neuro Oncol 2017; 19: 229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Cordero A, Goh D, Kruczek K, Naeem A, Fernando M, Kleine Holthaus S-M, et al. Assessment of AAV vector tropisms for mouse and human pluripotent stem cell–derived RPE and photoreceptor cells. Hum Gene Ther 2018; 29: 1124–39. [DOI] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ.. Neurobehavioural effects of developmental toxicity. Lancet Neurol 2014; 13: 330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribkoff VK, Kaczmarek LK.. The need for new approaches in CNS drug discovery: why drugs have failed, and what can be done to improve outcomes. Neuropharmacology 2017; 120: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifno GN, Farrell AM, Linville RM, Arevalo D, Kim JH, Gu L, et al. Tissue-engineered blood-brain barrier models via directed differentiation of human induced pluripotent stem cells. Sci Rep 2019; 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Tomaskovic-Crook E, Lozano R, Chen Y, Kapsa RM, Zhou Q, et al. Functional 3D neural mini-tissues from printed gel-based bioink and human neural stem cells. Adv Healthcare Mater 2016; 5: 1429–38. [DOI] [PubMed] [Google Scholar]
- Gu Q, Tomaskovic-Crook E, Wallace GG, Crook JM, Engineering human neural tissue by 3D bioprinting In: Chawla K, editor. Biomaterials for tissue engineering. New York, NY: Springer New York; 2018. p. 129–138. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Yeo GSH, Tung Y-CL, Zivraj KH, Willis D, Coppola G, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA 2011; 17: 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, et al. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Rep 2017; 8: 1727–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy T, Urbach A.. Comparing ESC and iPSC—based models for human genetic disorders. J Clin Med 2014; 3: 1146–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldorsson S, Lucumi E, Gómez-Sjöberg R, Fleming RMT.. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron 2015; 63: 218–31. [DOI] [PubMed] [Google Scholar]
- Hampson SM, Rowe W, Christie SDR, Platt M.. 3D printed microfluidic device with integrated optical sensing for particle analysis. Sens Actuat B Chem 2018; 256: 1030–7. [Google Scholar]
- Han H-W, Hsu S.. Using 3D bioprinting to produce mini-brain. Neural Regen Res 2017; 12: 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ.. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 2002; 297: 353–6. [DOI] [PubMed] [Google Scholar]
- Hargus G, Cooper O, Deleidi M, Levy A, Lee K, Marlow E, et al. Differentiated Parkinson patient-derived induced pluripotent stem cells grow in the adult rodent brain and reduce motor asymmetry in Parkinsonian rats. Proc Natl Acad Sci USA 2010; 107: 15921–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring AP, Sontheimer H, Johnson BN.. Microphysiological human brain and neural systems-on-a-chip: potential alternatives to small animal models and emerging platforms for drug discovery and personalized medicine. Stem Cell Rev Rep 2017; 13: 381–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff RF, Blasig IE, Bauer HC, Bauer H.. In search of the astrocytic factor(s) modulating blood-brain barrier functions in brain capillary endothelial cells in vitro. Cell Mol Neurobiol 2005; 25: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori K, Munehira Y, Kobayashi H, Satoh T, Sugiura S, Kanamori T.. Microfluidic perfusion culture chip providing different strengths of shear stress for analysis of vascular endothelial function. J Biosci Bioeng 2014; 118: 327–32. [DOI] [PubMed] [Google Scholar]
- Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol 2008; 7: 583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J.. 3D-bioprinted mini-brain: a glioblastoma model to study cellular interactions and therapeutics. Adv Mater 2019; 31: 1806590. [DOI] [PubMed] [Google Scholar]
- van der Helm MW, Odijk M, Frimat J-P, van der Meer AD, Eijkel JCT, van den Berg A, et al. Fabrication and validation of an organ-on-chip system with integrated electrodes to directly quantify transendothelial electrical resistance. J Vis Exp 2017; 127: 56334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms HC, Hersom M, Kuhlmann LB, Badolo L, Nielsen CU, Brodin B.. An electrically tight in vitro blood-brain barrier model displays net brain-to-blood efflux of substrates for the ABC transporters, P-gp, Bcrp and Mrp-1. AAPS J 2014; 16: 1046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henstridge CM, Spires-Jones TL.. Modeling Alzheimer’s disease brains in vitro. Nat Neurosci 2018; 21: 899–900. [DOI] [PubMed] [Google Scholar]
- Heo YJ, Kang J, Kim MJ, Chung WK.. Tuning-free controller to accurately regulate flow rates in a microfluidic network. Sci Rep 2016; 6: 23273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. The glia/neuron ratio: how it varies uniformly across brain structures and species and what that means for brain physiology and evolution: the Glia/Neuron Ratio. Glia 2014; 62: 1377–91. [DOI] [PubMed] [Google Scholar]
- Herland A, Maoz BM, Fitzgerald EA. et al. Proteomic and metabolomic characterization of human neurovascular unit cells in response to methamphetamine. Adv Biosyst 2020; e1900230. [DOI] [PubMed] [Google Scholar]
- Herland A, van der Meer AD, FitzGerald EA, Park T-E, Sleeboom JJF, Ingber DE.. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS One 2016; 11: e0150360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimori H, Yang Y.. Imaging Analysis of Neuron to Glia Interaction in Microfluidic Culture Platform (MCP)-based Neuronal Axon and Glia Co-culture System. J Vis Exp 2012; 68: e4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkel S, Mattern K, Dietzel A, Reichl S, Müller-Goymann CC. Parametric investigation of static and dynamic cell culture conditions and their impact on hCMEC/D3 barrier properties. Int J Pharm 2019; 566: 434. [DOI] [PubMed] [Google Scholar]
- Hoadley L. The independent differentiation of isolated chick Primordia in chorioallantoic grafts. I. The eye, nasal region, otic region, and mesencephalon. Biol Bull 1924; 46: 281–315. [Google Scholar]
- Hodeib S, Herberg JA, Levin M, Sancho-Shimizu V.. Human genetics of meningococcal infections. Hum Genet 2020; 139: 961–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogberg HT, Bressler J, Christian KM, Harris G, Makri G, O’Driscoll C, et al. Toward a 3D model of human brain development for studying gene/environment interactions. Stem Cell Res Ther 2013; 4: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann EK, Bailey AK, Potharazu AV, Neely MD, Bowman AB, Lippmann ES.. Accelerated differentiation of human induced pluripotent stem cells to blood-brain barrier endothelial cells. Fluids Barriers CNS 2017; 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AM, De Laporte L, Tortelli F, Spedden E, Staii C, Atherton TJ, et al. Silk hydrogels as soft substrates for neural tissue engineering. Adv Funct Mater 2013; 23: 5140–9. [Google Scholar]
- Hopkins AM, DeSimone E, Chwalek K, Kaplan DL.. 3D in vitro modeling of the central nervous system. Prog Neurobiol 2015; 125: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Liu P, Li X, Chen S, Wang L, Qin L, et al. Neural progenitor cells from human induced pluripotent stem cells generated less autogenous immune response. Sci China Life Sci 2014; 57: 162–70. [DOI] [PubMed] [Google Scholar]
- Huang Y, Williams JC, Johnson SM.. Brain slice on a chip: opportunities and challenges of applying microfluidic technology to intact tissues. Lab Chip 2012; 12: 2103. [DOI] [PubMed] [Google Scholar]
- Hubert CG, Rivera M, Spangler LC, Wu Q, Mack SC, Prager BC, et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res 2016; 76: 2465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Hamilton GA, Ingber DE.. From 3D cell culture to organs-on-chips. Trends Cell Biol 2011; 21: 745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE.. Reconstituting organ-level lung functions on a chip. Science 2010; 328: 1662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpel C. Organotypic brain slice cultures: a review. Neuroscience 2015; 305: 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PØ, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol 2012; 14: 979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huval RM, Miller OH, Curley JL, Fan Y, Hall BJ, Moore MJ.. Microengineered peripheral nerve-on-a-chip for preclinical physiological testing. Lab Chip 2015; 15: 2221–32. [DOI] [PubMed] [Google Scholar]
- Imaizumi K, Sone T, Ibata K, Fujimori K, Yuzaki M, Akamatsu W, et al. Controlling the regional identity of hPSC-derived neurons to uncover neuronal subtype specificity of neurological disease phenotypes. Stem Cell Rep 2015; 5: 1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell 2016; 164: 1105–9. [DOI] [PubMed] [Google Scholar]
- Ionescu A, Perlson E.. Patient-derived co-cultures for studying ALS. Nat Biomed Eng 2019; 3: 13–4. [DOI] [PubMed] [Google Scholar]
- Ishii T, Ishikawa M, Fujimori K, Maeda T, Kushima I, Arioka Y, et al. In vitro modeling of the bipolar disorder and schizophrenia using patient-derived induced pluripotent stem cells with copy number variations of PCDH15 and RELN. eNeuro 2019; 6: ENEURO.0403-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MS, Moore DJ.. Mechanisms of LRRK2-dependent neurodegeneration: role of enzymatic activity and protein aggregation. Biochem Soc Trans 2017; 45: 163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesan S, Workman MJ, Herland A, Svendsen CN, Vatine GD.. Generation of a human iPSC-based blood-brain barrier chip. J Vis Exp 2020; 157: 60925. [DOI] [PubMed] [Google Scholar]
- Janzer RC, Raff MC.. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature 1987; 325: 253–7. [DOI] [PubMed] [Google Scholar]
- Jensen JB, Parmar M.. Strengths and limitations of the neurosphere culture system. Mol Neurobiol 2006; 34: 153–62. [DOI] [PubMed] [Google Scholar]
- Jiang L, Li S, Zheng J, Li Y, Huang H.. Recent progress in microfluidic models of the blood-brain barrier. Micromachines (Basel) 2019; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran H-D, Göke J, et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 2016; 19: 248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodat YA, Kang MG, Kiaee K. et al. Human-derived organ-on-a-chip for personalized drug development. Curr Pharm Des 2018 24: 5471–86.doi: 10.2174/1381612825666190308150055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones I, Yelhekar TD, Wiberg R, Kingham PJ, Johansson S, Wiberg M, et al. Development and validation of an in vitro model system to study peripheral sensory neuron development and injury. Sci Rep 2018; 8: 15961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Datar A, Yu K-N, Kang S-Y, Lee M-Y.. High-content imaging assays on a miniaturized 3D cell culture platform. Toxicolo In Vitro 2018; 50: 147–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Knudsen T, Thomas RS, Sipes N, et al. In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast Programme. Basic Clin Pharmacol Toxicol 2014; 115: 69–76. [DOI] [PubMed] [Google Scholar]
- Kaarj . Methods of delivering mechanical stimuli to organ-on-a-chip. Micromachines 2019; 10: 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagasabapathi TT, Massobrio P, Barone RA, Tedesco M, Martinoia S, Wadman WJ, et al. Functional connectivity and dynamics of cortical–thalamic networks co-cultured in a dual compartment device. J Neural Eng 2012; 9: 036010. [DOI] [PubMed] [Google Scholar]
- Kanagasabapathi TT, Massobrio P, Tedesco M, Martinoia S, Wadman WJ, Decre MMJ, An experimental approach towards the development of an in vitro cortical-thalamic co-culture model In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Boston, MA: IEEE; 2011. p. 648–651. [DOI] [PubMed] [Google Scholar]
- Kane KIW, Moreno EL, Hachi S, Walter M, Jarazo J, Oliveira MAP, et al. Automated microfluidic cell culture of stem cell derived dopaminergic neurons. Sci Rep 2019; 9: 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Chung BG, Langer R, Khademhosseini A. Microfluidics for drug discovery and development: from target selection to product lifecycle management. Drug Discov Today 2008; 13: 1–13. doi: 10.1016/j.drudis.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasa P, Pakaski M, Joó F, Lajtha A.. Endothelial cells from human fetal brain microvessels may be cholinoceptive, but do not synthesize acetylcholine. J Neurochem 1991; 56: 2143–6. [DOI] [PubMed] [Google Scholar]
- Kato-Negishi M, Morimoto Y, Onoe H, Takeuchi S.. Millimeter-sized neural building blocks for 3D heterogeneous neural network assembly. Adv Healthcare Mater 2013; 2: 1564–70. [DOI] [PubMed] [Google Scholar]
- Kedziora KM, Purvis JE.. The persistence of memory. Nature 2017; 549: 343–4. [DOI] [PubMed] [Google Scholar]
- Kesselheim AS, Hwang TJ, Franklin JM.. Two decades of new drug development for central nervous system disorders. Nat Rev Drug Discov 2015; 14: 815–6. [DOI] [PubMed] [Google Scholar]
- Kilic O, Pamies D, Lavell E, Schiapparelli P, Feng Y, Hartung T, et al. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 2016; 16: 4152–62. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Li H, Collins JJ, Ingber DE.. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA 2016; 113: E7–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467: 285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 2011; 29: 1117–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee H, Chung M, Jeon NL.. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip 2013; 13: 1489–500. [DOI] [PubMed] [Google Scholar]
- Kimberlin DW. Neonatal Herpes Simplex Infection. Clin Microbiol Rev 2004; 17: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NM, Perrin J.. Ethical issues in stem cell research and therapy. Stem Cell Res Ther 2014; 5: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore KM, Logsdon DK, Floyd DH, Peirce SM, Purow BW, Munson JM.. Interstitial flow differentially increases patient-derived glioblastoma stem cell invasion via CXCR4, CXCL12, and CD44-mediated mechanisms. Integr Biol 2016; 8: 1246–60. [DOI] [PubMed] [Google Scholar]
- Kingston BR, Syed AM, Ngai J, Sindhwani S, Chan WCW.. Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc Natl Acad Sci USA 2019; 116: 14937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopfel T, Rietschin L, Gahwiler BH.. Organotypic co-cultures of rat locus coeruleus and hippocampus. Eur J Neurosci 1989; 1: 678–89. [DOI] [PubMed] [Google Scholar]
- Knowlton S, Anand S, Shah T, Tasoglu S.. Bioprinting for neural tissue engineering. Trends Neurosci 2018; 41: 31–46. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016; 533: 420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu OO, Hogue IB, Enquist LW.. Virus infections in the nervous system. Cell Host Microbe 2013; 13: 379–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krewski D, Acosta D, Andersen M, Anderson H, Bailar JC, Boekelheide K, et al. Toxicity testing in the 21st century: a vision and a strategy. J Toxicol Environ Health B Crit Rev 2010; 13: 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczenski B, LeDuc PR, Messner WC.. Pressure-driven spatiotemporal control of the laminar flow interface in a microfluidic network. Lab Chip 2007; 7: 647. [DOI] [PubMed] [Google Scholar]
- Kumaria A. In vitro models as a platform to investigate traumatic brain injury. Altern Lab Anim 2017; 45: 201–11. [DOI] [PubMed] [Google Scholar]
- Kung F, Wang J, Perez-Castillejos R, Townes-Anderson E.. Position along the nasal/temporal plane affects synaptic development by adult photoreceptors, revealed by micropatterning. Integr Biol 2015; 7: 313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labus J, Häckel S, Lucka L, Danker K.. Interleukin-1β induces an inflammatory response and the breakdown of the endothelial cell layer in an improved human THBMEC-based in vitro blood-brain barrier model. J Neurosci Methods 2014; 228: 35–45. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA.. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014; 9: 2329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013; 501: 373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans SA. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol 2018; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlaca MC, Cullen DK, McLoughlin JJ, Cargill RS.. High rate shear strain of three-dimensional neural cell cultures: a new in vitro traumatic brain injury model. J Biomech 2005; 38: 1093–105. [DOI] [PubMed] [Google Scholar]
- Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW.. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat Rev Neurosci 2013; 14: 722–9. [DOI] [PubMed] [Google Scholar]
- LaVail JH, Wolf MK.. Postnatal development of the mouse dentate gyrus in organotypic cultures of the hippocampal formation. Am J Anat 1973; 137: 47–65. [DOI] [PubMed] [Google Scholar]
- Lee C-T, Bendriem RM, Wu WW, Shen R-F.. 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders. J Biomed Sci 2017; 24: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Li J, Wang N, Yang X, Hamada Y, Li Q, et al. An on-chip model for investigating the interaction between neurons and cancer cells. Integr Biol (Camb) 2016; 8: 359–67. [DOI] [PubMed] [Google Scholar]
- Levy AF, Zayats M, Guerrero-Cazares H, Quiñones-Hinojosa A, Searson PC.. Influence of basement membrane proteins and endothelial cell-derived factors on the morphology of human fetal-derived astrocytes in 2D. PLoS One 2014; 9: e92165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Jiang H, Zhang B, Feng J.. Modeling Parkinson’s Disease Using Patient-specific Induced Pluripotent Stem Cells. J Parkinsons Dis 2018; 8: 479–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426: 450–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-C, Bai W-Z, Hashikawa T.. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol 2020. a; 92: 552–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu T, Yang N, Han D, Mi X, Li Y, et al. Neurological manifestations of patients with COVID-19: potential routes of SARS-CoV-2 neuroinvasion from the periphery to the brain. Front Med 2020. b; 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yu P, Hao J, Wang Y, Ohsaka T, Mao L.. Continuous and simultaneous electrochemical measurements of glucose, lactate, and ascorbate in rat brain following brain ischemia. Anal Chem 2014; 86: 3895–901. [DOI] [PubMed] [Google Scholar]
- Lingor P, Unsicker K, Krieglstein K.. Midbrain dopaminergic neurons are protected from radical induced damage by GDF-5 application. J Neural Transm 1999; 106: 139–44. [DOI] [PubMed] [Google Scholar]
- Linville RM, DeStefano JG, Sklar MB, Xu Z, Farrell AM, Bogorad MI, et al. Human iPSC-derived blood-brain barrier microvessels: validation of barrier function and endothelial cell behavior. Biomaterials 2019; 190–191: 24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Al-Ahmad A, Azarin SM, Palecek SP, Shusta EV.. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci Rep 2014; 4: 4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Azarin SM, Kay JE, Nessler RA, Wilson HK, Al-Ahmad A, et al. Human blood-brain barrier endothelial cells derived from pluripotent stem cells. Nat Biotechnol 2012; 30: 783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-H, Agarwal S, Park I-H, Urbach A, Huo H, Heffner GC, et al. Generation of induced pluripotent stem cells from human blood. Blood 2009; 113: 5476–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R, Stewart EM, et al. 3D printing of layered brain-like structures using peptide modified gellan gum substrates. Biomaterials 2015; 67: 264–73. [DOI] [PubMed] [Google Scholar]
- Lu P, Takai K, Weaver VM, Werb Z.. Extracellular Matrix Degradation and Remodeling in Development and Disease. Cold Spring Harb Perspect Biol 2011; 3: a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TM, Redmond D, Magdeldin T, Nguyen D-HT, Snead A, Sproul A, et al. Human induced pluripotent stem cell-derived neuroectodermal epithelial cells mistaken for blood-brain barrier-forming endothelial cells. bioRxiv 2019; 699173. [Google Scholar]
- Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol 2014; 25: 45. doi: 10.1016/j.copbio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- MacKerron C, Robertson G, Zagnoni M, Bushell TJ.. A microfluidic platform for the characterisation of CNS active compounds. Sci Rep 2017; 7: 15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackic JB, Stins M, Jovanovic S, Kim KS, Bartus RT, Zlokovic BV.. Cereport (RMP-7) increases the permeability of human brain microvascular endothelial cell monolayers. Pharm Res 1999; 16: 1360–5. [DOI] [PubMed] [Google Scholar]
- Magdesian MH, Lopez-Ayon GM, Mori M, Boudreau D, Goulet-Hanssens A, Sanz R, et al. Rapid mechanically controlled rewiring of neuronal circuits. J Neurosci 2016; 36: 979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M.. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res 2014; 6: 114–8. [PMC free article] [PubMed] [Google Scholar]
- Mao L, Wang M, Chen S, He Q, Chang J, Hong C, et al. Neurological manifestations of hospitalized patients with COVID-19 in Wuhan, China: a retrospective case series study. medRxiv 2020; 2020.02.22.20026500.
- Maoz BM, Herland A, FitzGerald EA, Grevesse T, Vidoudez C, Pacheco AR, et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol 2018; 36: 865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maoz BM, Herland A, Henry OYF, Leineweber WD, Yadid M, Doyle J, et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017; 17: 2294–302. [DOI] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell 2015; 162: 375–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino A, Tricinci O, Battaglini M. et al. A 3D real-scale, biomimetic, and biohybrid model of the blood-brain barrier fabricated through two-photon lithography. Small 2018; 14: doi: 10.1002/smll.201702959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marras C, Beck JC, Bower JH, Roberts E, Ritz B, Ross GW, et al. Prevalence of Parkinson’s disease across North America. Npj Parkinsons Dis 2018; 4: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rivas A, González-Quijano GK, Proa-Coronado S, Séverac C, Dague E. Methods of micropatterning and manipulation of cells for biomedical applications. Micromachines (Basel) 2017; 8: 347. doi: 10.3390/mi8120347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins Gomes SF, Westermann AJ, Sauerwein T, Hertlein T, Förstner KU, Ohlsen K, et al. Induced pluripotent stem cell-derived brain endothelial cells as a cellular model to study Neisseria Meningitidis infection. Front Microbiol 2019; 10: 1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015; 15: 2688–99. [DOI] [PubMed] [Google Scholar]
- Masullo LA, Bodén A, Pennacchietti F, Coceano G, Ratz M, Testa I.. Enhanced photon collection enables four dimensional fluorescence nanoscopy of living systems. Nat Commun 2018; 9: 3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumura T, Yamamoto K, Shimizu N, Obi S, Ando J.. Shear stress increases expression of the arterial endothelial marker EphrinB2 in murine ES cells via the VEGF-notch signaling pathways. Arterioscler Thromb Vasc Biol 2009; 29: 2125–31. [DOI] [PubMed] [Google Scholar]
- McCray PB, Pewe L, Wohlford-Lenane C, Hickey M, Manzel L, Shi L, et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 2007; 81: 813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, Blurton-Jones M.. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener 2018; 13: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae PA, Porter BE.. The perineuronal net component of the extracellular matrix in plasticity and epilepsy. Neurochem Int 2012; 61: 963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer AVD, Berg AVD.. Organs-on-chips: breaking the in vitro impasse. Integr Biol 2012; 4: 461. [DOI] [PubMed] [Google Scholar]
- van Meer BJ, de Vries H, Firth KSA, van Weerd J, Tertoolen LGJ, Karperien HBJ, et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun 2017; 482: 323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megard I, Garrigues A, Orlowski S, Jorajuria S, Clayette P, Ezan E, et al. A co-culture-based model of human blood-brain barrier: application to active transport of indinavir and in vivo-in vitro correlation. Brain Res 2002; 927: 153–67. [DOI] [PubMed] [Google Scholar]
- Mercier F, Kwon YC, Douet V.. Hippocampus/amygdala alterations, loss of heparan sulfates, fractones and ventricle wall reduction in adult BTBR T+ tf/J mice, animal model for autism. Neurosci Lett 2012; 506: 208–13. [DOI] [PubMed] [Google Scholar]
- Mestas J, Hughes CCW.. Of Mice and Not Men: differences between Mouse and Human Immunology. J Immunol 2004; 172: 2731–8. [DOI] [PubMed] [Google Scholar]
- Meyvantsson I, Beebe DJ.. Cell culture models in microfluidic systems. Annu Rev Anal Chem 2008; 1: 423–49. [DOI] [PubMed] [Google Scholar]
- Miao R, Xia L-Y, Chen H-H, Huang H-H, Liang Y.. Improved classification of blood-brain-barrier drugs using deep learning. Sci Rep 2019; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AP, Shah AS, Aperi BV, Budde MD, Pintar FA, Tarima S, et al. Effects of blast overpressure on neurons and glial cells in rat organotypic hippocampal slice cultures. Front Neurol 2015; 6: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 2013; 13: 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima T, Fujioka S, Fukae J, Yuasa-Kawada J, Tsuboi Y.. Modeling Parkinson’s disease and atypical parkinsonian syndromes using induced pluripotent stem cells. Int J Mol Sci 2018; 19: 3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misun PM, Rothe J, Schmid YRF, Hierlemann A, Frey O.. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst Nanoeng 2016; 2: 16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofazzal Jahromi MA, Abdoli A, Rahmanian M, Bardania H, Bayandori M, Moosavi Basri SM, et al. Microfluidic brain-on-a-chip: perspectives for mimicking neural system disorders. Mol Neurobiol 2019; 56: 8489–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 2020; 181: 905–13.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep 2017; 8: 1144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno EL, Hachi S, Hemmer K, Trietsch SJ, Baumuratov AS, Hankemeier T, et al. Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab Chip 2015; 15: 2419–28. [DOI] [PubMed] [Google Scholar]
- Morrison B, Elkin BS, Dollé J-P, Yarmush ML.. In vitro models of traumatic brain injury. Annu Rev Biomed Eng 2011; 13: 91–126. [DOI] [PubMed] [Google Scholar]
- Moxon KA, Foffani G.. Brain-machine interfaces beyond neuroprosthetics. Neuron 2015; 86: 55–67. [DOI] [PubMed] [Google Scholar]
- Moya ML Triplett M Simon M Alvarado J Booth R Osburn J, . et al. A Reconfigurable In Vitro Model for Studying the Blood-Brain Barrier. Ann Biomed Eng 2020; 48: 780–93. doi: 10.1007/s10439-019-02405-y. [DOI] [PubMed] [Google Scholar]
- Nahmias Y, Schwartz RE, Verfaillie CM, Odde DJ. Laser-guided direct writing for three-dimensional tissue engineering. Biotechnol Bioeng 2005; 92: 129. doi: 10.1002/bit.20585. [DOI] [PubMed] [Google Scholar]
- Naik P, Cucullo L.. In vitro blood–brain barrier models: current and Perspective Technologies. J Pharm Sci 2012; 101: 1337–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Donahue G, Berson A, Lan Y, Amlie-Wolf A, Tuzer F, et al. Dysregulation of the epigenetic landscape of normal aging in Alzheimer’s disease. Nat Neurosci 2018; 21: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli S, Oliveira V, Calabresi P, Maia LF, Pisani A.. Does SARS-Cov-2 invade the brain? Translational lessons from animal models. Eur J Neurol 2020; 10.1111/ene.14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto E, Leitão L, Sousa DM, Alves CJ, Alencastre IS, Aguiar P, et al. Compartmentalized microfluidic platforms: the unrivaled breakthrough of in vitro tools for neurobiological research. J Neurosci 2016; 36: 11573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus W, Lauer R, Oelzant S, Fringeli UP, Ecker GF. , Noe CR. A novel flow based hollow-fiber blood-brain barrier in vitro model with immortalised cell line PBMEC/C1. J Biotechnol 2006; 125: 127–41. 10.1016/j.jbiotec.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Nikolakopoulou P, Poser SW, Masjkur JR, Celis M. D, Toutouna L, Andoniadou CL, et al. STAT3-Ser/Hes3 signaling: a new molecular component of the neuroendocrine system? Horm Metab Res 2016; 48: 77–82. [DOI] [PubMed] [Google Scholar]
- Novak R, Didier M, Calamari E, Ng CF, Choe Y, Clauson SL, et al. Scalable fabrication of stretchable, dual channel, microfluidic organ chips. J Vis Exp 2018; 140: 58151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U, Kaye AH.. Extracellular matrix and the brain: components and function. J Clin Neurosci 2000; 7: 280–90. [DOI] [PubMed] [Google Scholar]
- Oddo A, Peng B, Tong Z, Wei Y, Tong WY, Thissen H, et al. Advances in microfluidic blood-brain barrier (BBB) Models. Trends Biotechnol 2019; 37: 1295–314. [DOI] [PubMed] [Google Scholar]
- Ogawa J, Pao GM, Shokhirev MN, Verma IM.. Glioblastoma model using human cerebral organoids. Cell Rep 2018; 23: 1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci USA 2014; 111: 12426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira EP, Malysz-Cymborska I, Golubczyk D, Kalkowski L, Kwiatkowska J, Reis RL, et al. Advances in bioinks and in vivo imaging of biomaterials for CNS applications. Acta Biomater 2019; 95: 60–72. [DOI] [PubMed] [Google Scholar]
- Orlova VV, van den Hil FE, Petrus-Reurer S, Drabsch Y, Ten Dijke P, Mummery CL.. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc 2014; 9: 1514–31. [DOI] [PubMed] [Google Scholar]
- Ortuño-Costela MDC, Cerrada V, García-López M, Gallardo ME.. The challenge of bringing iPSCs to the patient. Int J Mol Sci 2019; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki T, Shin Y, Sivathanu V, Campisi M, Kamm RD.. In vitro microfluidic models for neurodegenerative disorders. Adv Healthc Mater 2018. a; 7. [DOI] [PubMed] [Google Scholar]
- Osaki T, Sivathanu V, Kamm RD.. Engineered 3D vascular and neuronal networks in a microfluidic platform. Sci Rep 2018. b; 8: 5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki T, Uzel SGM, Kamm RD.. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci Adv 2018. c; 4: east5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitti D, Privolizzi R, Bax BE.. Organs to cells and cells to organoids: the evolution of in vitro central nervous system modelling. Front Cell Neurosci 2019; 13: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisán-Ruı´z C, Jain S, Evans EW, Gilks WP, Simón J, Brug M. V D, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s Disease. Neuron 2004; 44: 595–600. [DOI] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci 2017; 20: 753–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Markota M, Jaquet F, Ghosh D, Wallin A, Santos A, et al. Aggrecan and chondroitin-6-sulfate abnormalities in schizophrenia and bipolar disorder: a postmortem study on the amygdala. Transl Psychiatry 2015; 5: e496–e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape SR, Nagendran T, Poole V, Harris J, Taylor AM.. Compartmentalization of human stem cell-derived neurons within pre-assembled plastic microfluidic chips. J Vis Exp 2019; 3: doi: 10.3791/59250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim S, Park SI, Choe Y, Li J, Han A.. A microchip for quantitative analysis of CNS axon growth under localized biomolecular treatments. J Neurosci Methods 2014; 221: 166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Koito H, Li J, Han A.. Microfluidic compartmentalized co-culture platform for CNS axon myelination research. Biomed Microdevices 2009. a; 11: 1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Koito H, Li J, Han A.. Multi-compartment neuron-glia co-culture platform for localized CNS axon-glia interaction study. Lab Chip 2012; 12: 3296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee S-H.. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 2015; 15: 141–50. [DOI] [PubMed] [Google Scholar]
- Park J, Wetzel I, Marriott I, Dréau D, D’Avanzo C, Kim DY, et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci 2018; 21: 941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL.. Microfluidic culture platform for neuroscience research. Nat Protoc 2006; 1: 2128. [DOI] [PubMed] [Google Scholar]
- Park JY, Kim S-K, Woo D-H, Lee E-J, Kim J-H, Lee S-H.. Differentiation of neural progenitor cells in a microfluidic chip-generated cytokine gradient. Stem Cells 2009. b; 27: 2646–54. [DOI] [PubMed] [Google Scholar]
- Partyka PP, Godsey GA, Galie JR, Kosciuk MC, Acharya NK, Nagele RG, et al. Mechanical stress regulates transport in a compliant 3D model of the blood-brain barrier. Biomaterials 2017; 115: 30–9. [DOI] [PubMed] [Google Scholar]
- Pașca SP. The rise of three-dimensional human brain cultures. Nature 2018; 553: 437–45. [DOI] [PubMed] [Google Scholar]
- Patterson C. World Alzheimer Report 2018 - The state of the art of dementia research: New frontiers. 2018.
- Pavlovich M. How to build a better organ-on-a-chip. 2018.
- dela Paz NG, D’Amore PA.. Arterial versus venous endothelial cells. Cell Tissue Res 2009; 335: 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, Lancaster MA.. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science 2020; 369: eaaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney J, Ralvenius WT, Tsai L-H.. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol Psychiatry 2020; 25: 148–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin J-M, Deleglise B, Saias L, Vignes M, Gougis P, Magnifico S, et al. Axon diodes for the reconstruction of oriented neuronal networks in microfluidic chambers. Lab Chip 2011; 11: 3663–73. [DOI] [PubMed] [Google Scholar]
- Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, et al. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 2017; 17: 511–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol 2010; 28: 848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potjewyd G, Moxon S, Wang T, Domingos M, Hooper NM.. Tissue engineering 3D neurovascular units: a biomaterials and bioprinting perspective. Trends Biotechnol 2018; 36: 457–72. [DOI] [PubMed] [Google Scholar]
- Prabhakarpandian B, Shen M-C, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, et al. SyM-BBB: a microfluidic blood brain barrier model. Lab Chip 2013; 13: 1093–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls TJ, Tan X, Husain M, Whittington CF, Fishel ML, Voytik-Harbin SL.. Development of a novel 3D tumor-tissue invasion model for high-throughput, high-content phenotypic drug screening. Sci Rep 2018; 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Jacob F, Song H, Ming G-L.. Using brain organoids to understand Zika virus-induced microcephaly. Development 2017; 144: 952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 2016; 165: 1238–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Brown J, Arlotta P.. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med 2016; 22: 1220–8. [DOI] [PubMed] [Google Scholar]
- Raies AB, Bajic VB.. In silico toxicology: computational methods for the prediction of chemical toxicity. Wires Comput Mol Sci 2016; 6: 147–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja WK, Mungenast AE, Lin Y-T, Ko T, Abdurrob F, Seo J, et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS One 2016; 11: e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotoson I, Delhomme B, Djian P, Deeg A, Brunstein M, Seebacher C, et al. Fast 3-D imaging of brain organoids with a new single-objective planar-illumination two-photon microscope. Front Neuroanat 2019; 13: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramme AP, Koenig L, Hasenberg T, Schwenk C, Magauer C, Faust D, et al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Science OA 2019; 5: FSO413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauti R, Renous N, Maoz BM.. Mimicking the brain extracellular matrix in Vitro: a review of current methodologies and challenges. Isr J Chem 2019; [Google Scholar]
- Reed MJ, Damodarasamy M, Banks WA.. The extracellular matrix of the blood–brain barrier: structural and functional roles in health, aging, and Alzheimer’s disease. Tissue Barriers 2019; 7: 1651157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinitz A, DeStefano J, Ye M, Wong AD, Searson PC.. Human brain microvascular endothelial cells resist elongation due to shear stress. Microvasc Res 2015; 99: 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Zhou J, Wu H. Materials for microfluidic chip fabrication. Acc Chem Res 2013; 46: 2396. doi: 10.1021/ar300314s. [DOI] [PubMed] [Google Scholar]
- Rice D, Barone S.. Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ Health Perspect 2000; 108: 511–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson RT, Baratta J, Kageyama GH, Ha DH, Yu J.. Specificity of attachment and neurite outgrowth of dissociated basal forebrain cholinergic neurons seeded on to organotypic slice cultures of forebrain. Neuroscience 1997; 80: 741–52. [DOI] [PubMed] [Google Scholar]
- Rodrigues OR, Limahelder RA, Gomes T, Silva A. Polymer microfluidic devices: an overview of fabrication methods. J Eng 2017; 1: 67. doi: 10.24840/2183-6493_001.001_0007. [DOI] [Google Scholar]
- Roesch S, Rapp C, Dettling S, Herold-Mende C.. When immune cells turn bad-tumor-associated microglia/macrophages in glioma. Int J Mol Sci 2018; 19: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-on-a-chip: A fast track for engineered human tissues in drug development. Cell Stem Cell 2018; 22: 310. doi: 10.1016/j.stem.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Hinge VK, Kovalenko A.. To pass or not to pass: predicting the blood–brain barrier permeability with the 3D-RISM-KH molecular solvation theory. ACS Omega 2019; 4: 16774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba J, Daou A, Damiati S, Saliba J, El-Sabban M, Mhanna R.. Development of microplatforms to mimic the in vivo architecture of CNS and PNS physiology and their diseases. Genes (Basel) 2018; 9: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S, Ho R, Vatine G, West D, Laperle A, Meyer A, et al. Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep 2018; 10: 1222–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Yamanaka S.. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 2014; 157: 110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeder C, Heigwer F, Boutros M.. Machine learning and image-based profiling in drug discovery. Curr Opin Syst Biol 2018; 10: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Costa VS, et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc Natl Acad Sci USA 2015; 112: 12516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA 2013; 110: 3507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenich L. Brain-resident microglia and blood-borne macrophages orchestrate central nervous system inflammation in neurodegenerative disorders and brain cancer. Front Immunol 2018; 9: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Wu L, Wu J, Zheng Y, Zhao H, Jin Q, et al. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip 2009; 9: 3118. [DOI] [PubMed] [Google Scholar]
- Sharma A, Sances S, Workman MJ, Svendsen CN.. Multi-lineage Human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 2020; 26: 309–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Luhach K, Kulkarni GT, In vitro and in vivo models of BBB to evaluate brain targeting drug delivery In: Gao H, Gao X editors, Brain targeted drug delivery system. Amsterdam, Netherlands: Elsevier; 2019. p.53–101. [Google Scholar]
- Shen M, Asawa R, Zhang Y-Q, Cunningham E, Sun H, Tropsha A, et al. Quantitative high-throughput phenotypic screening of pediatric cancer cell lines identifies multiple opportunities for drug repurposing. Oncotarget 2017; 9: 4758–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Majumdar D, Gao Y, Brewer BM, Goodwin CR, McLean JA, et al. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab Chip 2013; 13: 3008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, Yamanaka S.. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 2017; 16: 115–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada IS, Badgandi H, Somatilaka BN, Mukhopadhyay S.. Using primary neurosphere cultures to study primary cilia. J Vis Exp 2017; 122: 55315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HS, Kim HJ, Min SK, Kim SH, Lee BM, Jeon NL.. Compartmental culture of embryonic stem cell-derived neurons in microfluidic devices for use in axonal biology. Biotechnol Lett 2010; 32: 1063–70. [DOI] [PubMed] [Google Scholar]
- Shin Y, Choi SH, Kim E, Bylykbashi E, Kim JA, Chung S, et al. Blood–brain barrier dysfunction in a 3D in vitro model of Alzheimer’s disease. Adv Sci 2019; 6: 1900962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrirao AB, Kung FH, Omelchenko A, Schloss RS, Boustany NN, Zahn JD, et al. Microfluidic platforms for the study of neuronal injury in vitro. Biotechnol Bioeng 2018; 115: 815–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddharthan V, V. Kim Y, Liu S, Kim KS.. Human astrocytes/astrocyte conditioned medium and shear stress enhance the barrier properties of human brain microvascular endothelial cells. Brain Res 2007; 1147: 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MC, Haggarty SJ.. Human pluripotent stem cell-derived models and drug screening in CNS precision medicine. Ann N Y Acad Sci 2020; 1471: 18–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 2009; 41: 1308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam V, Gijs MA. Exploring living multicellular organisms, organs, and tissues using microfluidic systems. Chem Rev 2013; 113: 3214. doi: 10.1021/cr200432q. [DOI] [PubMed] [Google Scholar]
- Sivanandam TM, Thakur MK.. Traumatic brain injury: a risk factor for Alzheimer’s disease. Neurosci Biobehav Rev 2012; 36: 1376–81. [DOI] [PubMed] [Google Scholar]
- Sivandzade F, Cucullo L.. In-vitro blood–brain barrier modeling: A review of modern and fast-advancing technologies. J Cereb Blood Flow Metab 2018; 38: 1667–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skafte-Pedersen P, Sabourin D, Dufva M, Snakenborg D.. Multi-channel peristaltic pump for microfluidic applications featuring monolithic PDMS inlay. Lab Chip 2009; 9: 3003. [DOI] [PubMed] [Google Scholar]
- Sleeboom JJF, Eslami Amirabadi H, Nair P, Sahlgren CM, den Toonder JMJ.. Metastasis in context: modeling the tumor microenvironment with cancer-on-a-chip approaches. Dis Model Mech 2018; 11: dmm033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Munn LL.. Fluid forces control endothelial sprouting. Proc Natl Acad Sci USA 2011; 108: 15342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer-Phelps A, Hassell BA, Ingber DE.. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer 2019; 19: 65. [DOI] [PubMed] [Google Scholar]
- Sosa-Hernández JE, Villalba-Rodríguez AM, Romero-Castillo KD, Aguilar-Aguila-Isaías MA, García-Reyes IE, Hernández-Antonio A, et al. Organs-on-a-chip module: a review from the development and applications perspective. Micromachines 2018; 9: 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soscia D, Belle A, Fischer N, Enright H, Sales A, Osburn J, et al. Controlled placement of multiple CNS cell populations to create complex neuronal cultures. PLoS One 2017; 12: e0188146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins MJ, Wilson HK, Canfield SG, Qian T, Palecek SP, Shusta EV.. Differentiation and characterization of human pluripotent stem cell-derived brain microvascular endothelial cells. Methods 2016; 101: 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenderhauf C, Hammann F, Huwyler J.. Computational prediction of blood-brain barrier permeability using decision tree induction. Molecules 2012; 17: 10429–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Chiuppesi F, Chen X, Wang C, Tian E, Nguyen J, et al. Modeling human cytomegalovirus-induced microcephaly in human iPSC-derived brain organoids. Cell Rep Med 2020; 1: 100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaghavan HG, Monteiro GA, Firestein BL, Shreiber DI.. Neurite growth in 3D collagen gels with gradients of mechanical properties. Biotechnol Bioeng 2009; 102: 632–43. [DOI] [PubMed] [Google Scholar]
- Swearengen JR. Choosing the right animal model for infectious disease research. Anim Model Exp Med 2018; 1: 100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861–72. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–76. [DOI] [PubMed] [Google Scholar]
- Tang Y, Liu M-L, Zang T, Zhang C-L.. Direct reprogramming rather than iPSC-based reprogramming maintains aging hallmarks in human motor neurons. Front Mol Neurosci 2017; 10: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Bertram L.. Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 2005; 120: 545–55. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon NL, Cotman CW.. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci 2009; 29: 4697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL.. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods 2005; 2: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, Ito HT, Kim SA, Schuman EM.. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron 2010; 66: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Rhee SW, Tu CH, Cribbs DH, Cotman CW, Jeon NL.. Microfluidic multicompartment device for neuroscience research. Langmuir 2003; 19: 1551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Willerth SM.. 3-D Bioprinting of neural tissue for applications in cell therapy and drug screening. Front Bioeng Biotechnol 2017; 5: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbe MP, Leferink AM, Van Den Berg A, Eijkel JCT, Segerink LI. Microfluidic gel patterning method by use of a temporary membrane for organ-on-chip applications. Adv Mater Technol 2018; 3: 1700200. doi: 10.1002/admt.201700200. [DOI] [Google Scholar]
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG.. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 2012; 76: 16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepke MW, Beebe DJ.. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006; 6: 1484. [DOI] [PubMed] [Google Scholar]
- Tohyama C. Developmental neurotoxicity test guidelines: problems and perspectives. J Toxicol Sci 2016; 41: SP69–SP79. [DOI] [PubMed] [Google Scholar]
- Turtle L. Respiratory failure alone does not suggest central nervous system invasion by SARS-CoV-2. J Med Virol 2020; 92: 705–6. [DOI] [PubMed] [Google Scholar]
- Uwamori H, Higuchi T, Arai K, Sudo R.. Integration of neurogenesis and angiogenesis models for constructing a neurovascular tissue. Sci Rep 2017; 7: 17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzel SGM, Amadi OC, Pearl TM, Lee RT, So PTC, Kamm RD.. Simultaneous or sequential orthogonal gradient formation in a 3d cell culture microfluidic platform. Small 2016; 12: 612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vareille M, Kieninger E, Edwards MR, Regamey N.. The airway epithelium: soldier in the fight against respiratory viruses. Clin Microbiol Rev 2011; 24: 210–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020; 395: 1417–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastag M, Keserű GM.. Current in vitro and in silico models of blood-brain barrier penetration: a practical view. Curr Opin Drug Discov Devel 2009; 12: 115–24. [PubMed] [Google Scholar]
- Vatine GD, Barrile R, Workman MJ, Sances S, Barriga BK, Rahnama M, et al. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 2019; 24: 995–1005.e6. [DOI] [PubMed] [Google Scholar]
- Velve-Casquillas G, Le Berre M, Piel M, Tran PT. Microfluidic tools for cell biological research. Nano Today 2010; 5: 28. doi: 10.1016/j.nantod.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, et al. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep 2017; 7: 42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volarevic V, Markovic BS, Gazdic M, Volarevic A, Jovicic N, Arsenijevic N, et al. Ethical and safety issues of stem cell-based therapy. Int J Med Sci 2018; 15: 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH, Cohen A.. Experiments on the development of the head of the chick embryo. J Exp Biol 1936; 13: 219–36. [Google Scholar]
- Walsh K, Megyesi J, Hammond R.. Human central nervous system tissue culture: a historical review and examination of recent advances. Neurobiol Dis 2005; 18: 2–18. [DOI] [PubMed] [Google Scholar]
- Walter FR, Valkai S, Kincses A, Petneházi A, Czeller T, Veszelka S, et al. A versatile lab-on-a-chip tool for modeling biological barriers. Sens Actuat B Chem 2016; 222: 1209–19. [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F.. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020; 94: e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Q, Pei J.. Microfluidic-based 3D engineered microvascular networks and their applications in vascularized microtumor models. Micromachines (Basel) 2018; 9: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YI, Abaci HE, Shuler ML.. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng 2017; 114: 184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener G, Rujescu D.. The current development of CNS drug research. Int JNeuropsychopharmacol 2013; 16: 1687–93. [DOI] [PubMed] [Google Scholar]
- Wevers NR, Kasi DG, Gray T, Wilschut KJ, Smith B, van Vught R, et al. A perfused human blood-brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS 2018; 15: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers NR, van Vught R, Wilschut KJ, Nicolas A, Chiang C, Lanz HL, et al. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci Rep 2016; 6: 38856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Meningococcal meningitis [Internet]. 2018. https://www.who.int/news-room/fact-sheets/detail/meningococcal-meningitis (3 May 2020, date last accessed).
- WHO. Mental Health. Towards a dementia plan: a WHO guide [Internet]. 2019. https://www.who.int/mental_health/neurology/dementia/en/
- WHO. Coronavirus Disease (COVID-19) - events as they happen [Internet]. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen (6 May 2020, date last accessed).
- Wnorowski A, Yang H, Wu JC. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv Drug Deliv Rev 2019; 140: 3. doi: 10.1016/j.addr.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, et al. Local translation of RhoA regulates growth cone collapse. Nature 2005; 436: 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss Institute. Wyss Institute’s technology translation engine launches ‘Organs-on-Chips’ company. [Press release] 2014. https://wyss.harvard.edu/wyss-institutes-technology-translation-engine-launches-organs-on-chips-company/.
- Xu H, Li Z, Guo Y, Peng X, Qin J.. Probing the response of lung tumor cells to inflammatory microvascular endothelial cells on fluidic microdevice. Electrophoresis 2017; 38: 311–9. [DOI] [PubMed] [Google Scholar]
- Xu H, Li Z, Yu Y, Sizdahkhani S, Ho WS, Yin F, et al. A dynamic in vivo-like organotypic blood-brain barrier model to probe metastatic brain tumors. Sci Rep 2016. a; 6: 36670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Nirwane A, Yao Y.. Basement membrane and blood-brain barrier. Stroke Vasc Neurol 2019; 4: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang W-K, Qian X, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016. b; 22: 1101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Gregory C, Molnar P, Cui X, Jalota S, Bhaduri S, et al. Viability and electrophysiology of neural cell structures generated by the inkjet printing method. Biomaterials 2006; 27: 3580–8. [DOI] [PubMed] [Google Scholar]
- Xu Z, Li E, Guo Z, Yu R, Hao H, Xu Y, et al. Design and construction of a multi-organ microfluidic chip mimicking the in vivo microenvironment of lung cancer metastasis. ACS Appl Mater Interfaces 2016. c; 8: 25840–7. [DOI] [PubMed] [Google Scholar]
- Yada E, Wada S, Yoshida S, Sasada T.. Use of patient-derived xenograft mouse models in cancer research and treatment. Future Sci OA 2018; 4: FSO271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix: CMLS. Cell Mol Life Sci 2000; 57: 276–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IH, Gary D, Malone M, Dria S, Houdayer T, Belegu V, et al. Axon myelination and electrical stimulation in a microfluidic, compartmentalized cell culture platform. Neuromol Med 2012; 14: 112–8. [DOI] [PubMed] [Google Scholar]
- Ye M, Sanchez HM, Hultz M, Yang Z, Bogorad M, Wong AD, et al. Brain microvascular endothelial cells resist elongation due to curvature and shear stress. Sci Rep 2014; 4: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H-G, Jeong YH, Kim Y, Choi Y-J, Moon HE, Park SH, et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng 2019; 3: 509. [DOI] [PubMed] [Google Scholar]
- Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O.. Engineering stem cell organoids. Cell Stem Cell 2016; 18: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Choudhury D.. Microfluidic bioprinting for organ-on-a-chip models. Drug Discov Today 2019; 24: 1248–57. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318: 1917–20. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zheng F, Zhan C-G.. Improved prediction of blood–brain barrier permeability through machine learning with combined use of molecular property-based descriptors and fingerprints. AAPS J 2018; 20: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker D, Begley D, Bratzke H, Rübsamen-Waigmann H, von Briesen H.. Human blood-derived macrophages enhance barrier function of cultured primary bovine and human brain capillary endothelial cells. J Physiol 2003; 551: 1023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A.. A 3D Alzheimer’s disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials 2014; 35: 1420–8. [DOI] [PubMed] [Google Scholar]
- Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci USA 2017; 114: E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang Z, Westenskow PD, Todorova D, Hu Z, Lin T, et al. Humanized mice reveal differential immunogenicity of cells derived from autologous induced pluripotent stem cells. Cell Stem Cell 2015; 17: 353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zhang Z-N, Rong Z, Xu Y.. Immunogenicity of induced pluripotent stem cells. Nature 2011; 474: 212–5. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, et al. In vitro microvessels for the study of angiogenesis and thrombosis. Proc Natl Acad Sci USA 2012; 109: 9342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhang M, Wang J, Gao J.. Sars-Cov-2: underestimated damage to nervous system. Travel Med Infect Dis 2020; 101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang P, Sun AX, An J, Chua CK, Chew SY.. 3D neural tissue models: from spheroids to bioprinting. Biomaterials 2018; 154: 113–33. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant Parkinsonism with pleomorphic pathology. Neuron 2004; 44: 601–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.