Abstract
Oil palm is the most productive oilseed crop and its oil yield is seriously affected by frequent drought stress. However, little is known about the molecular responses of oil palm to drought stress. We studied the root transcriptomic responses of oil palm seedlings under 14-day drought stress. We identified 1293 differentially expressed genes (DEGs), involved in several molecular processes, including cell wall biogenesis and functions, phenylpropanoid biosynthesis and metabolisms, ion transport and homeostasis and cellular ketone metabolic process, as well as small molecule biosynthetic process. DEGs were significantly enriched into two categories: hormone regulation and metabolism, as well as ABC transporters. In addition, three protein–protein interaction networks: ion transport, reactive nitrogen species metabolic process and nitrate assimilation, were identified to be involved in drought stress responses. Finally, 96 differentially expressed transcription factors were detected to be associated with drought stress responses, which were classified into 28 families. These results provide not only novel insights into drought stress responses, but also valuable genomic resources to improve drought tolerance of oil palm by both genetic modification and selective breeding.
Subject terms: Genetics, Plant sciences
Introduction
Oil palm (Elaeis guineensis, Jacq.) is the most productive oilseed crop1. Palm oil extracted from palm fruits has been used as cooking oil and in making cosmetics, candles, soaps, biofuels, and lubricating greases, as well in processing tinplate and coating iron plates, and the demand for palm oil is increasing1. Oil palm is mainly cultivated in the tropical regions of Southeast Asia, Africa and South America2,3, where water is particularly important to the productivity of this crop. Oil palm needs ~ 2000 mm/year rain water to have a normal production and does not tolerate to drought for more than 90 days3. However, with unpredictable global climate changes, tropical regions are accidentally in danger of lacking of rainfall of up to months4. Continuous drought condition affects oil palm production frequently5. A previous study showed that deficit of 100 mm water reduced the yield by 20%6. Recent studies demonstrated that drought stress for 7–21 days induced physiological and growth changes in oil palm seedlings7–9. However, not much is known about transcriptomic response to drought stress. Therefore, it is essential to increase drought tolerance in oil palm to ensure sustainable development of palm oil industry. Understanding of more about the response of oil palm to drought stress will facilitate the breeding of drought tolerant oil palm varieties for sustainable oil production5,10,11.
An increasing number of studies have focused on understanding the molecular mechanisms of drought tolerance in agronomic plant species or model plant species (e.g. Arabidopsis) and have identified important genes and/or evolutionary conserved genetic elements that act to regulate drought tolerance12–14. Drought resistance is complex and determined by genetic and environmental factors, and their interactions. It takes very long time to increase drought tolerance by conventional breeding15. The mechanisms underlying drought tolerance are variable among different species16–18 and are still poorly understood for most non-model species19. Thus, identifying the genetic factors and understanding the underlying mechanisms are essential to improve drought tolerance by gene modifications20 and breeding5 in different species.
Plants may use diverse mechanisms to maintain water and ion homeostasis19. At the molecular levels, many drought-responsive genes and transcription factors have been identified21. Some gene classes playing crucial roles in defending against drought stress have been reported22. For example, some hormone-related genes can initiate drought resistance signalling23. Transporters, like ABC transport system, are essential for osmotic signal and osmosensing mechanism24. Endogenous abscisic acid (ABA), which works as an important phytohormone, has well-known functions in drought tolerance in plants, by inducing the expression of stress-related genes25. Another study also indicated that jasmonic acid (JA) was associated with stomatal closure to increase drought tolerance26. In addition, it was reported that transcription factors (TFs), including NAC, GmNAC, HD-STARTtype and NF-YB family members, played important roles in drought tolerance by regulation of hormone metabolisms27. Beyond individual genes, cascaded signalling pathways and regulatory networks were suggested to have played indispensable roles in drought tolerance28. ABA coupled with corresponding TFs and ABA responsive elements, could regulate the expressions of a wide range of genes under osmotic stress, via cis regulations28. A number of protein families in the calcium signalling pathways, mitogen-activated protein kinases (MAPKs) signalling pathways and phosphorylation cascades were also involved in drought stress responses29,30. Previous studies on the responses of plants to drought stress have shed new insights in understanding of the mechanisms of drought tolerance in different species. However, in the oil palm, transcriptomic responses to drought stress are still poorly understood.
The purpose of the current study was to identify genes, pathways, networks and transcription factors involved in drought response using RNA-seq and bioinformatics analysis to understand more about molecular responses of oil palm roots under drought stress. RNA-seq is a next-generation sequencing technology used to analyse the presence and quantity of RNA molecules in biological samples31. Herein, oil palm seedlings were firstly treated with drought stress. We studied the transcriptome responses of roots to drought in the primary tissue for stress signal perception and initiating of cascade gene regulation pathways in response to drought. A total of 1293 DEGs, including 96 transcription factors, were identified in drought stress responses. Besides individual genes, signalling pathways and protein–protein interaction networks, involving transcription factors, also likely have played crucial roles in drought tolerance. This study provides both novel insights of molecular response of oil palm to drought stress and genomic resources to improve and develop drought-tolerant oil palms for sustainable oil production.
Results and discussion
Morphological and physiological responses to drought
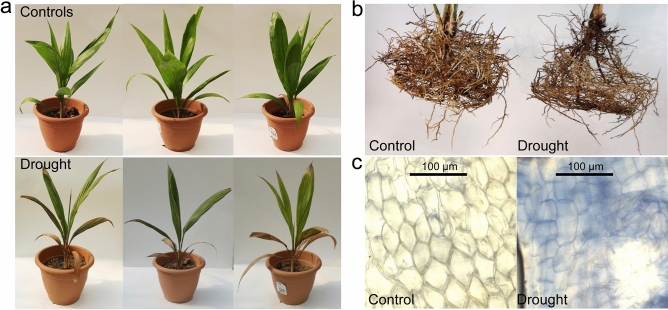
Obvious morphological changes in leaves and roots of oil palm seedlings under drought stress with a period of 14 days were observed. The effects of drought stress were first observed in leaf morphology, showing initial edge and tip necrosis and then wilting and yellowing for the drought treated samples (Fig. 1a). In comparison to the control, the drought-stressed palms showed significant decrease in the number of roots, root volume and overall biomass (Fig. 1b). Trypan blue staining showed that the drought treatment roots experienced not only obvious cell deformation but also more cell membrane injury than that of the well-watered controls (Fig. 1c). These observations are consistent with those of previous studies in oil palm7–9 and other plant species32–34 under drought stresses and undergoing water deprivation. These results indicate substantial physiological responses of the oil palm seedlings under drought stress35, and provided useful starting materials to study the genes, pathways and networks involved in drought responses using RNA-seq36 and bioinformatics analysis.
Figure 1.
Phenotypic changes of oil palm seedlings to drought stress. The leaves (a) and roots (b) responses to severe drought stress in comparison to controls at 10 days post challenge, where both leaves and roots were significantly affected by drought, and (c) trypan blue staining of roots between control and drought stress groups, where damaged cells were stained.
Differentially expressed gene (DEGs) in roots of oil palm seedling under drought stress
An average of 70.2 million (M) cleaned reads out of 71.3 M raw reads, were obtained across all six samples. The controls had higher reads coverage than the drought-stressed palms (84.8 M vs 55.7 M) (Supplementary Table S1). Nevertheless, the sequence coverage of the drought-stressed palms (> 200 × of transcriptomes) was sufficient to construct transcripts and identify DEGs. Approximately 70% of cleaned reads were uniquely mapped to the reference genome of 31,640 annotated protein coding genes37. The drought stressed seedlings showed slightly higher uniquely mapping rates than the controls (72.1% vs 66.3%), indicating the duplicated genes also play important roles in response to drought stress38 in oil palm, a species of palaeotetraploid origin37 and future studies should also focus on paralogous genes and their potential functions in stress responses39.
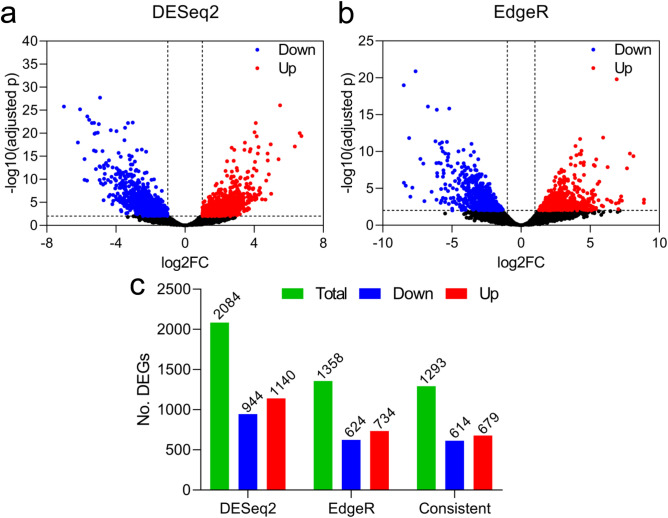
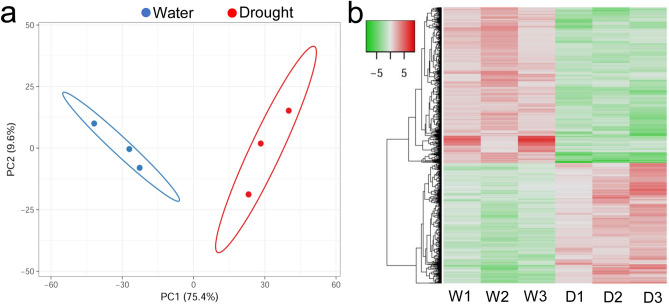
A total of 2084 and 1358 DEGs were identified using two approaches: DESeq2 and EdgeR, respectively, within which 1293 were shared by the two data sets (Fig. 2). DESeq2 identified 944 down-regulated and 1140 up-regulated DEGs, while EdgeR screened 624 down-regulated and 734 up-regulated DEGs. The number of common down- and up-regulated DEGs were 614 and 679, respectively, between the two approaches (Fig. 2; Supplementary Table S2). To obtain confident results, only the common DEGS were kept for further analysis. Based on the relative expression of DEGs across samples, the drought stressed and control samples were clearly differentiated by both PCA and hierarchical clustering analyses and showed substantial differences in expression profiles (Fig. 3). We further assessed the accuracy of the RNAseq data by comparing to the results of qPCR of randomly selected nine genes (Supplementary Table S3). We observed an overall high consistency of the expression patterns of these genes between RNA-seq and qPCR (Supplementary Fig. S1a), with a correlation coefficient of 0.978 (P < 0.0001), as examined using Pearson’s correlation test (Supplementary Fig. S1b). Taken together, these data indicate that the RNA-seq data is reliable.
Figure 2.
Comparison of differentially expressed genes (DEGs) in the roots of oil palm seedlings under drought stress, as shown by Volcano plot, identified by DESeq2 (a) and EdgeR (b). The numbers of DEGs that are down-regulated and up-regulated revealed by DESeq2 and EdgeR and the consistent DEGs between the two methods are shown in (c).
Figure 3.
Principal component analysis (PCA) among samples of the experimental (Drought) and control (Water) groups in oil palm (a) and hierarchical clustering (b) of the relationships of samples between experimental (Drought) and control (Water) groups based on randomly selected DEGs. W1, W2 and W3 are controls, while D1, D2 and D3 are experimental samples.
Interestingly, we observed that most of the DEGs in subcategories phenylalanine metabolism and tryptophan metabolism were down-regulated (Table 1). In plant species, phenylalanine and tryptophan metabolisms are more involved in pathogen related immune responses40. The down regulation of most DEGs within these categories implies the effects of metabolic compensation to drought stress responses by sacrificing the less important biological functions. In addition, we found two genes: two-component response regulator ORR9 and two-component response regulator ORR24 that were down- and up-regulated, respectively, were enriched into the subcategory: zeatin biosynthesis (Table 1), which plays important roles in drought stress response in Populus simonii33. Further studies on how the two genes are involved in the responses to drought stress in oil palm, are required.
Table 1.
Selected enriched KEGG pathways and DEGs in response to drought challenge in the roots of oil palm seedlings.
| Gene | Annotation | Subsignaling pathways | Expression |
|---|---|---|---|
| Plant hormone signal transduction | |||
| LOC105046997 | Jasmonic acid-amido synthetase JAR1 | a-Linolenic acid metabolism | Up |
| LOC105048226 | Jasmonic acid-amido synthetase JAR1 | a-Linolenic acid metabolism | Down |
| LOC105051062 | Protein TIFY 9 | a-Linolenic acid metabolism | Up |
| LOC105055031 | Transcription factor MYC2 | a-Linolenic acid metabolism | Up |
| LOC105049192 | Probable xyloglucan endotransglucosylase/hydrolase protein 23 | Brassinosteroid biosynthesis | Down |
| LOC105043050 | Probable protein phosphatase 2C 75 | Carotenoid biosynthesis | Up |
| LOC105053585 | Probable protein phosphatase 2C 75 | Carotenoid biosynthesis | Up |
| LOC105056896 | Probable protein phosphatase 2C 24 | Carotenoid biosynthesis | Up |
| LOC105059101 | Ethylene-responsive transcription factor 1B | Cysteine and methionine metabolism | Up |
| LOC105034229 | Transcription factor TGAL5 | Phenylalanine metabolism | Down |
| LOC105061166 | Pathogenesis-related protein 1B | Phenylalanine metabolism | Down |
| LOC105061545 | Pathogenesis-related protein PRB1-2 | Phenylalanine metabolism | Down |
| LOC105061554 | Pathogenesis-related protein PRB1-2 | Phenylalanine metabolism | Down |
| LOC105042180 | Auxin-responsive protein IAA30 | Tryptophan metabolism | Down |
| LOC105046556 | Auxin-responsive protein IAA30 | Tryptophan metabolism | Down |
| LOC105047880 | Auxin-responsive protein IAA1 | Tryptophan metabolism | Up |
| LOC105051115 | Indole-3-acetic acid-amido synthetase GH3.17 | Tryptophan metabolism | Down |
| LOC105051940 | Auxin transporter-like protein 3 | Tryptophan metabolism | Down |
| LOC105055859 | Auxin-responsive protein IAA30 | Tryptophan metabolism | Down |
| LOC105056155 | Probable indole-3-acetic acid-amido synthetase GH3.8 | Tryptophan metabolism | Up |
| LOC105049107 | Two-component response regulator ORR9 | Zeatin biosynthesis | Down |
| LOC105049543 | Two-component response regulator ORR24 | Zeatin biosynthesis | Up |
| MAPK signaling pathway | |||
| LOC105043050 | Probable protein phosphatase 2C 75 | Abscisic acid | Up |
| LOC105053585 | Probable protein phosphatase 2C 75 | Abscisic acid | Up |
| LOC105056896 | Probable protein phosphatase 2C 24 | Abscisic acid | Up |
| LOC105052054 | Protein MKS1-like | Pathogen infection | Up |
| LOC105061166 | Pathogenesis-related protein 1B | Pathogen infection | Down |
| LOC105061545 | Pathogenesis-related protein PRB1-2 | Pathogen infection | Down |
| LOC105061554 | Pathogenesis-related protein PRB1-2 | Pathogen infection | Down |
| LOC105055031 | Transcription factor MYC2 | Phytohomones | Up |
| LOC105059101 | Ethylene-responsive transcription factor 1B | Phytohomones | Up |
| LOC105059691 | Chitinase 1 | Phytohomones | Up |
| ABC transporters | |||
| LOC105043189 | ABC transporter C family member 5 | ABC1 subfamily | Up |
| LOC105032304 | ABC transporter B family member 11 | ABCB subfamily | Down |
| LOC105038824 | Putative multidrug resistance protein | ABCB subfamily | Down |
| LOC105041030 | ABC transporter B family member 9 | ABCB subfamily | Up |
| LOC105056548 | ABC transporter B family member 19 | ABCB subfamily | Down |
| LOC105060251 | Putative multidrug resistance protein | ABCB subfamily | Up |
Ontology enrichment analysis of genes responding to drought stress
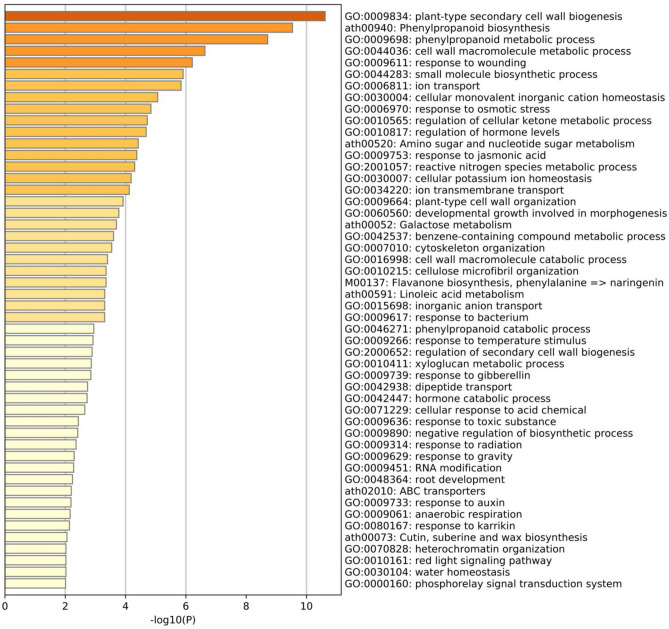
To understand the transcriptomic responses to the drought stress, we first carried out gene ontology enrichment analysis using the 1293 DEGs identified by both EdgeR and DESeq2. A total of 89 GO terms were significantly enriched, involving many categories of diverse functions (Supplementary Table S4). The most significant enrichment entities included GO terms related to cell wall biogenesis and functions (e.g., GO:0009834, GO:0044036, GO:0009664, GO:0016998 and GO:2000652), which was consistent with the our observation that the cell wall of the drought treatments has likely been damaged by severe drought stress and thus has triggered the mechanism of damage and repair (Fig. 4). Moreover, we also observed significant enrichments related to phenylpropanoid biosynthesis and metabolisms (e.g., ath00940 and GO:0009698 and GO: 0046271). It is known that phenylpropanoid pathway is activated by stress conditions, such as drought, salinity and extreme temperature, and leads to accumulation of phenolic compounds, which play critical physiological roles in regulation under abiotic stress to cope with environmental challenges41. Moreover, we found some GO terms classified into the groups related to ion transport and homeostasis (e.g., GO:0006811, GO:0030004 GO:0030007, GO:0015698 and GO:0034220) and response to osmotic stress and water homeostasis (e.g., GO:0006970 and GO:0030104). Differential expressions of genes in these functions likely result from the responses of plants to water deprivation by direct regulation of osmotic pressure42,43. In addition, a number of genes were enriched into the biological categories related to regulation of cellular ketone metabolic process (GO:0010565), suggesting that genes involved in ketone metabolic process play important roles in drought stress in oil palm44,45. Hormone regulations are also indispensable to stress responses of plant species. Here, we identified two enriched GO terms related to hormone regulation and metabolism (e.g., GO:0010817 and GO:0042447). Previous studies have shown that production of numerous secondary metabolites is essential for physiological processes to respond to abiotic stress46,47. Consistent with these results, we found several significant enrichments related to these terms: small molecule biosynthetic process (GO:0044283), amino sugar and nucleotide sugar metabolism (ath00520), galactose metabolism (ath00052), benzene-containing compound metabolic process (GO:0042537), linoleic acid metabolism (ath00591) and xyloglucan metabolic process (GO:0010411). Interestingly, we also identified significant enriched GO terms, like response to jasmonic acid (GO:0009753) and ABC transporters (ath02010), which play crucial roles in abiotic stress responses (Fig. 4).
Figure 4.
Enrichment of gene ontology (GO) of DEGs against drought challenge at the significance level of 0.01 in the roots of oil palm seedlings under drought stress.
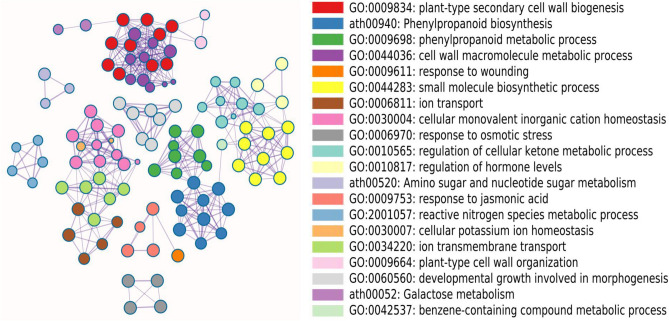
The interactions of these enriched GO terms were further investigated using network analysis. Eight enriched GO networks were identified, with each consisting of no less than 3 genes (Fig. 5). The major GO networks involved those related to cell wall related biogenesis and metabolism (GO:0009834 and GO:0044036), small molecule related biosynthetic and metabolic processes (ath00940, GO:0009698, GO:0044283 and GO:0010565) and ion transport and homeostasis related processes (GO:0006811, GO:0034220 and GO:0030004). These data imply that genes in these networks are more extensively induced to differentially express to respond to drought stress42,47,48. We further investigated the enriched KEGG pathways and found that the functions of the enriched pathways were generally consistent with those of the enriched GO terms as shown above (Supplementary Table S5). Above all, these enrichment analyses suggest that many genes, pathways and networks respond to the drought stress in the roots of oil palm seedlings. The DEGs, pathways and networks identified in this study provide valuable resources for future studies on their functions to improve drought tolerance of oil palm.
Figure 5.
Major gene networks among the top 20 enriched GO terms as shown in this figure, based on DEGs against drought challenge in the roots of oil palm seedlings. Each network and the corresponding GO term are indicated with the same color.
Plant hormone signal transduction in drought stress responses
Plant hormones not only play crucial roles in controlling growth and development, but also are indispensable in regulation of stress responses49. Herein, we first focused on the DEGs involved in plant hormone signal transduction pathway and found significant enrichments of DEGs within subcategories of KEGG pathways including a-Linolenic acid metabolism, carotenoid biosynthesis, phenylalanine metabolism, tryptophan metabolism and zeatin biosynthesis (Table 1). Previous studies revealed that genes related to a-Linolenic acid metabolism played important roles in drought stress responses50,51. We found that four DEGS were involved in a-Linolenic acid metabolism and three out of them were up-regulated. Interestingly, the down-regulated DEG, jasmonic acid-amido synthetase JAR1 (LOC105048226), was a duplicated copy of the up-regulated one, jasmonic acid-amido synthetase JAR1 (LOC105046997), suggesting functional divergence of paralogous genes since genome duplication events. Nevertheless, consistent with the expression patterns of most DEGs in this subcategory, the up-regulated jasmonic acid-amido synthetase JAR1 might be more important in regulation of drought stress response in oil palm. Moreover, we found that all of the DEGs, including probable protein phosphatase 2C 24 and two duplicated copies of probable protein phosphatase 2C 75, in the subcategory carotenoid biosynthesis, were up-regulated. These three DEGs were also enriched into the subcategory abscisic acid pathway (ABA) within MAPK signalling pathway (Table 1). Carotenoid biosynthesis signalling pathway is specifically induced by root and contributes to induce ABA production to regulate ion homeostasis, as studied in Arabidoposis52. ABA-independent signalling pathways are involved in the regulation of drought stress response in many plant species53. Therefore, our results suggest that ABA related genes also play important roles in drought stress responses of oil palm.
ABC transporters in drought responses
Membrane transporters play vital roles in regulation of water and ion homeostasis of organisms, among which ATP-binding cassette (ABC) transporters constitute one of the largest protein families and act as both exporters and importers, driven by ATP hydrolysis54. ABC transporters play irreplaceable roles in transmembrane allocations of various molecules to adapt to rapidly changing environments, such as water scarcity, heavy metal stress and pathogen stress55. In order to survive in these changing abiotic conditions, it is necessary for cells to absorb nutritious chemical substances and discharge endogenous toxins, as well as exchange signalling molecules55. Thus, the ABC transporters occupy a diverse range of functions and hence the regulations upon stress responses are also complicated. Here, we found that six DEGs were enriched into the pathway of ABC transporters (Table 1). Five of them were ABCB subfamily members, among which three [ABC transporter B family member 11, ABC transporter B family member 19 and putative multidrug resistance protein (LOC105038824)] were down-regulated and two [ABC transporter B family member 9 and another putative multidrug resistance protein (LOC105060251)] were up-regulated. Such differential expression patterns of these ABCB subfamily transporters indicate the complicated functions in controlling of influx and efflux of chemical molecules56,57. In addition, we also identified an ABCC subfamily member, ABC transporter C family member 5, which was up-regulated. Interestingly, two putative multidrug resistance protein genes (LOC105038824 and LOC105060251) were differentially expressed against drought stress. As shown in previous studies, multidrug resistance‐associated proteins are widely involved in regulation of stress responses, such as salt stress, water deprivation, oxidative stress and fungal stress58. Taken together, these different types of ABC transporters likely play important roles in responses to drought stress in oil palm.
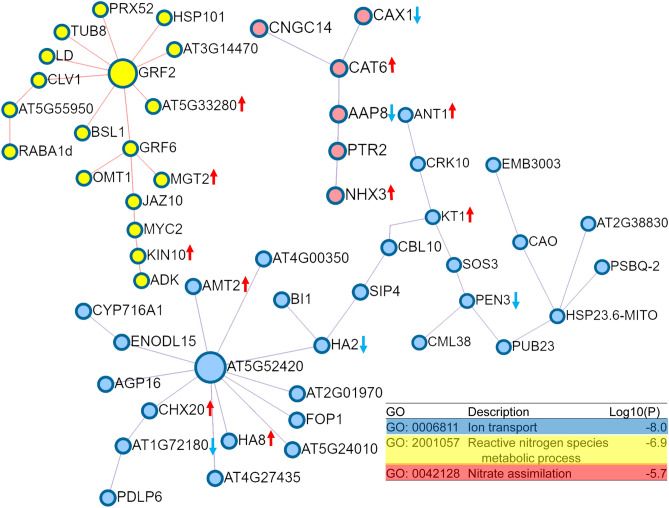
Protein–protein interaction networks in response to drought responses
Other than significantly enriched GO terms and KEGG pathways, we also identified three protein–protein interaction networks, focused on ion transport, reactive nitrogen species metabolic process and nitrate assimilation (Fig. 6; Table 2). Eight DEGs were involved in the ion transport network, among which five [ammonium transporter 2 member 1 (AMT2-1), amino acid transporter ANT1 (ANT1), cation/H(+) antiporter 20 (CHX20), plasma membrane ATPase 4 (PMA4) and potassium channel AKT1 (AKT1)] and three [receptor-like protein kinase HSL1 (HSL1), plasma membrane ATPase (PMA) and ABC transporter G family member 42 (ABCG42)] were up- and down-regulated, respectively (Table 2). Interestingly, most of the cation channel and transporter genes were up-regulated, including ammonium transporter 2 member 1 (AMT1), amino acid transporter ANT1 (ANT1), cation/H(+) antiporter 20 (CHX20) and potassium channel AKT1 (KT1), indicating their positive effects in regulating ion homeostasis in oil palm16. Nevertheless, we also observed that three DEGs were down-regulated in the same network, implying both positive and negative feedback regulations are acting on this network59. Reactive nitrogen species metabolic process is also suggested to have critical roles in stress responses, such as drought and salinity60. Consistently, we identified three up-regulated genes: magnesium transporter MRS2-1 (MGT2), putative chloride channel-like protein CLC-g (AT5G33280) and serine/threonine protein kinase OSK1 (KIN10), in this protein–protein interaction network. Nitrate assimilation is another biological process affecting salt and water stress tolerance in plants61. Here, we found four DEGs involved in this network: two were up-regulated [cationic amino acid transporter 6, chloroplastic (CAT6) and sodium/hydrogen exchanger 4 (NHX4)], while the other two were down-regulated [amino acid permease 8 (AAP8) and vacuolar cation/proton exchanger 1a (CAX1)]. As these protein–protein interaction networks play crucial roles in drought stress response, the DEGs involved in these networks provide important candidate genes to improve drought tolerance of oil palm by genetic engineering and/or selective breeding.
Figure 6.
Three significant enrichments of protein–protein interaction networks identified based on DEGs against drought challenge in the roots of oil palm seedlings. DEGs in the three networks are further enriched with GO terms, with each showing corresponding significance. The networks are shown in different colors. The DEGs within each network are indicated and the expression patterns are shown with colored up- and down-arrows.
Table 2.
DEGs in three enriched protein–protein interaction networks in response to drought challenge in the roots of oil palm seedlings.
| Gene | Annotation | Expression |
|---|---|---|
| Ion transport | ||
| LOC105045862 | Ammonium transporter 2 member 1 | Up |
| LOC105049578 | Amino acid transporter ANT1 | Up |
| LOC105037902 | Receptor-like protein kinase HSL1 | Down |
| LOC105033828 | Cation/H(+) antiporter 20 | Up |
| LOC105039277 | Plasma membrane ATPase | Down |
| LOC105052943 | Plasma membrane ATPase 4 | Up |
| LOC105047848 | Potassium channel AKT1 | Up |
| LOC105040536 | ABC transporter G family member 42 | Down |
| Reactive nitrogen species metabolic process | ||
| LOC105060165 | Magnesium transporter MRS2-1 | Up |
| LOC105050121 | Putative chloride channel-like protein CLC-g | Up |
| LOC105044259 | Serine/threonine protein kinase OSK1 | Up |
| Nitrate assimilation | ||
| LOC105037451 | Amino acid permease 8 | Down |
| LOC105052094 | Cationic amino acid transporter 6, chloroplastic | Up |
| LOC105032563 | Vacuolar cation/proton exchanger 1a | Down |
| LOC105038946 | Sodium/hydrogen exchanger 3 | Up |
Transcription factors in drought responses
To date, more and more studies focused on the biological functions of transcription factors as regulatory elements binding proteins21. Transcription factors are vital for development, response to intercellular and environmental signals and pathogenesis21. The expression changes are often associated with important cellular processes15. In this study, we identified 96 differentially expressed transcription factors that were classified into 28 families (Supplementary Table S6, Table 3). Previous studies have shown that transcription factors are broadly involved in drought/abiotic stress responses, such as the members of family MYB, WRKY, DREB, NAC and AP2/EREBP27,62–64. Here we also observed that genes in these transcription factor families were differentially expressed under drought stress in oil palm, further supporting their important roles in drought tolerance in plant species. Interestingly, we found several families of transcription factors that were rarely studied and involved in abiotic stress responses, such as the C2H2, LFY and TALE transcription families. Therefore, it is important to understand the mechanisms of regulatory functions of these genes, which might be useful to help improve drought tolerance of related plant species.
Table 3.
Categorization of differentially expressed transcription factors and the patterns of their expressions after drought stress in the roots of oil palm seedlings.
| Family | Number | Down | Up |
|---|---|---|---|
| AP2 | 2 | 1 | 1 |
| ARR-B | 2 | 0 | 2 |
| B3 | 3 | 0 | 3 |
| bHLH | 10 | 4 | 6 |
| bZIP | 5 | 2 | 3 |
| C2H2 | 5 | 1 | 4 |
| C3H | 1 | 0 | 1 |
| Dof | 1 | 0 | 1 |
| E2F/DP | 1 | 0 | 1 |
| ERF | 9 | 1 | 8 |
| FAR1 | 1 | 0 | 1 |
| G2-like | 2 | 1 | 1 |
| GATA | 1 | 0 | 1 |
| GRAS | 2 | 0 | 2 |
| GRF | 1 | 1 | 0 |
| HB-other | 1 | 0 | 1 |
| HD-ZIP | 6 | 3 | 3 |
| HSF | 3 | 0 | 3 |
| LBD | 1 | 0 | 1 |
| LFY | 1 | 0 | 1 |
| MYB | 15 | 9 | 6 |
| NAC | 17 | 9 | 8 |
| RAV | 1 | 0 | 1 |
| TALE | 1 | 0 | 1 |
| TCP | 1 | 0 | 1 |
| Trihelix | 1 | 0 | 1 |
| WRKY | 1 | 0 | 1 |
| YABBY | 1 | 0 | 1 |
| Total | 96 | 32 | 64 |
Conclusions
We investigated transcriptomic response of root against drought stress in oil palm seedlings. We identified over 1000 DEGs responding to the drought stress, including the genes mainly involved in cell wall biogenesis and functions, phenylpropanoid biosynthesis and metabolisms and ion transport and homeostasis. We functionally enriched the genes in plant hormone signal transduction and ABC transporters pathways, which likely have played crucial roles in regulation of water deprivation. Three protein–protein interaction networks were identified that were related to ion transport, reactive nitrogen species metabolic process and nitrate assimilation. We also detected 96 transcription factors that were differentially expressed upon drought stress. The identified DEGs, pathways and protein–protein interaction networks, and transcription factors likely play important roles in drought tolerance of oil palm. This study helps understand more about the mechanism of drought stress response and provides valuable resources for future genetic improvement of drought tolerance in oil palm. Future studies should analyse the functions of DEGs identified, in combination with metabolomics and morpho-physiological approaches to obtain a comprehensive overview of drought stress impact on oil palm.
Materials and methods
Plant materials and drought treatment
Seeds of Tenera palms (Elaeis guineensis, Jacq.) were geminated with a standard protocol3,53. The seedlings were grown in a nursery for 120 days before drought stress treatment. Eight oil palm Tenera seedlings were planted in pots with diameter of 20 cm containing natural soil with water content of 23% (2.3 g water per 10 g soil), and placed in a greenhouse with a natural tropical temperature ranging from 28 to 34 °C, 30–50% relative humidity and natural photoperiod. Four seedlings were watered twice a week to maintain water content of > 23% while the other four seedlings were used as drought treatment without watering for two weeks. After drought stress challenge of 14 days, the mortality rate of experimental group was estimated at ~ 50%. The root tissues of both the control and experimental groups were harvested and measured, respectively. The samples were then preserved at − 80 °C for RNA isolation.
RNA extraction and sequencing
Total RNA was isolated from roots using RNeasy Plant Mini Kit (Qiagen, Germany), according to the manufacturer’s instructions. RNA quality was assessed by agarose gels and concentration was measured by NanoDrop (Thermo Fisher Scientific, USA). One µg total RNA from each sample was firstly treated with RNase-free DNase I (Sigma-Aldrich, Singapore) and then used for mRNA library construction with Illumina TruSeq RNA Library Prep Kit v2 (Illumina, USA), according to the manufacturer’s instructions. The libraries were paired-end sequenced (2 × 75 bp) using an Illumina NextSeq500 (Illumina, USA). Three biological replicates were sequenced for both control and drought treated samples. For validation of RNA sequencing data using real-time quantitative PCR (qPCR), two µg total RNA was treated with RNase-free DNase I (Sigma-Aldrich, Singapore) and was then used for synthesizing cDNA with the MMLV reverse transcriptase (Promega, USA).
Identification of differentially expressed genes (DEGs)
Raw sequencing reads were processed using the program process_shortreads in Stacks package65, to demultiplex samples, filter adaptors and clean up low quality reads. The program STAR66 was employed to align and map the cleaned reads to the reference genome of oil palm37, with default parameters. Only uniquely mapped reads were used to analyse the expression patterns of annotated genes. The program HTSeq-count67 was then used to count the expression level of each annotated gene, based on the information of gene features in the genome annotation file. We used both DESeq268 and EdgeR69 to normalize the relative expression of transcripts across samples. Only transcripts with the number of counts per million (CPM) mapped reads of > 1 were retained for further analysis. Transcripts with a fold change (FC) value of > 2 or < − 2 and with a significance value of 0.01 after application of Benjamini–Hochberg false discovery rate (FDR)70 were considered as differentially expressed genes, between drought treatment and control groups. Only DEGs that were consistently identified by both DESeq2 and EdgeR, were used for further analysis.
Functional annotation of DEGs
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) accessions 71 were retrieved for each DEG, according to the PalmXplore database of oil palm72. We first clustered all the samples using both principal component analysis (PCA) and heatmap approaches with the program ClustVis73, based on the relative expression of DEGs, to investigate the overall expression patterns between drought treatment and control groups. Gene ontology enrichment analysis was carried out using the program Metascape 74. The Metascape program74 was further employed to study the protein–protein interactions using network analysis by referencing to Arabidopsis. The candidate signalling pathways associated DEGs were classified and enriched by annotating against the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database75 of oil palm.
Validation of RNA-seq data using qPCR
DEGs were randomly selected and the relative expression patterns revealed by RNA-seq were examined by qPCR, to assess the effectiveness and accuracy of the whole DEGs dataset. Primers of randomly selected DEGs were designed according to the coding sequences, obtained from the annotated reference genome, using the program Primer376. Both β-actin gene and glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) were used as housekeeping genes to normalize the relative expression of genes, according to our previous study77. The 2−ΔΔCT method was used to quantify the expression level according to our previous method78. The experiment was carried out with three biological replications, each with three technical replicates.
Ethics declarations
All authors have reviewed the final version of the manuscript and agree to publish the data.
Supplementary Information
Acknowledgements
We thank Dr Zi Yi Wan and the intern ZY Sam for technical assisting in the experiments.
Author contributions
G.H.Y. designed the study. M.L. and B.Y. challenged samples and constructed libraries. M.L. sequenced the libraries. L.W. carried out qPCR. L.W. analysed the data. L.W. and G.H.Y. drafted the manuscript. All authors read and approved the final manuscript. This research was supported by the Internal Funds of the Temasek Life Sciences Lab (9200).
Data availability
Raw sequencing reads used in this study have been deposited to the NCBI SRA database with an accession no. PRJDB9517.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-78297-z.
References
- 1.EPOA. Palm Oil Production. https://www.palmoilandfood.eu/en/palm-oil-production. (2019).
- 2.Zhang C, et al. Transcriptional and physiological data reveal the dehydration memory behavior in switchgrass (Panicum virgatum L.) Biotechnol. Biofuels. 2018;11:91. doi: 10.1186/s13068-018-1088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corley RHV, Tinker PB. The Oil Palm. New York: Wiley; 2008. [Google Scholar]
- 4.Ummenhofer CC, D’Arrigo RD, Anchukaitis KJ, Buckley BM, Cook ER. Links between Indo-Pacific climate variability and drought in the Monsoon Asia Drought Atlas. Clim. Dyn. 2013;40:1319–1334. doi: 10.1007/s00382-012-1458-1. [DOI] [Google Scholar]
- 5.Corley R, Rao V, Palat T, Praiwan T. Breeding for drought tolerance in oil palm. J. Oil Palm Res. 2017;30:26–35. [Google Scholar]
- 6.Olivin J. Étude pour la localisation d'un bloc industriel de palmiers à huile. Oleagineux. 1968;23:499–504. [Google Scholar]
- 7.Cao H-X, Sun C-X, Shao H-B, Lei X-T. Effects of low temperature and drought on the physiological and growth changes in oil palm seedlings. Afr. J. Biotech. 2011;10:2630–2637. doi: 10.5897/AJB10.1272. [DOI] [Google Scholar]
- 8.Cha-um S, Yamada N, Takabe T, Kirdmanee C. Physiological features and growth characters of oil palm (Elaeis guineensis Jacq.) in response to reduced water-deficit and rewatering. Aust. J. Crop Sci. 2013;7:432. [Google Scholar]
- 9.Silva PA, et al. Drought tolerance in two oil palm hybrids as related to adjustments in carbon metabolism and vegetative growth. Acta Physiol. Plant. 2017;39:58. doi: 10.1007/s11738-017-2354-4. [DOI] [Google Scholar]
- 10.Bai B, et al. Genome-wide identification of markers for selecting higher oil content in oil palm. BMC Plant Biol. 2017;17:93. doi: 10.1186/s12870-017-1045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M, et al. A consensus linkage map of oil palm and a major QTL for stem height. Sci. Rep. 2015;5:1–7. doi: 10.1038/srep08232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo PJ, et al. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganguly DR, Crisp PA, Eichten SR, Pogson BJ. The Arabidopsis DNA methylome is stable under transgenerational drought stress. Plant Physiol. 2017;175:1893–1912. doi: 10.1104/pp.17.00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, et al. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019;98:697–713. doi: 10.1111/tpj.14267. [DOI] [PubMed] [Google Scholar]
- 15.Golldack D, Lüking I, Yang O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30:1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- 16.Osakabe Y, et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell. 2013;25:609–624. doi: 10.1105/tpc.112.105700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, et al. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson DE, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinclair TR. Challenges in breeding for yield increase for drought. Trends Plant Sci. 2011;16:289–293. doi: 10.1016/j.tplants.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Mittler R, Blumwald E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010;61:443–462. doi: 10.1146/annurev-arplant-042809-112116. [DOI] [PubMed] [Google Scholar]
- 21.Joshi R, et al. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016;7:1029. doi: 10.3389/fpls.2016.01029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: Discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 2006;17:113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Ko JH, Yang SH, Han KH. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuromori T, Sugimoto E, Shinozaki K. Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 2011;67:885–894. doi: 10.1111/j.1365-313X.2011.04641.x. [DOI] [PubMed] [Google Scholar]
- 25.Iuchi S, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 26.Alam MM, Nahar K, Hasanuzzaman M, Fujita M. Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different Brassica species. Plant Biotechnol. Rep. 2014;8:279–293. doi: 10.1007/s11816-014-0321-8. [DOI] [Google Scholar]
- 27.Huang Q, et al. TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 2015;15:268. doi: 10.1186/s12870-015-0644-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016;21:677–685. doi: 10.1016/j.tplants.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Haider MS, et al. Grapevine immune signaling network in response to drought stress as revealed by transcriptomic analysis. Plant Physiol. Biochem. 2017;121:187–195. doi: 10.1016/j.plaphy.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Abdullah HM, et al. Comparative transcriptome and metabolome analysis suggests bottlenecks that limit seed and oil yields in transgenic Camelina sativa expressing diacylglycerol acyltransferase 1 and glycerol-3-phosphate dehydrogenase. Biotechnol. Biofuels. 2018;11:335. doi: 10.1186/s13068-018-1326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magalhães AP, et al. RNA-Seq and gene network analysis uncover activation of an ABA-dependent signalosome during the cork oak root response to drought. Front. Plant Sci. 2016;6:1195. doi: 10.3389/fpls.2015.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Song Y, Zhang H, Zhang D. Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol. Biol. Rep. 2013;31:946–962. doi: 10.1007/s11105-013-0563-6. [DOI] [Google Scholar]
- 34.Huang L, et al. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genom. 2014;15:1026. doi: 10.1186/1471-2164-15-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao H-B, Chu L-Y, Jaleel CA, Zhao C-X. Water-deficit stress-induced anatomical changes in higher plants. C.R. Biol. 2008;331:215–225. doi: 10.1016/j.crvi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Ozsolak F, Milos PM. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature. 2013;500:335–339. doi: 10.1038/nature12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou C, Lehti-Shiu MD, Thomashow M, Shiu S-H. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 2009;5:e1000581. doi: 10.1371/journal.pgen.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teotia S, Lamb RS. The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol. 2009;151:180–198. doi: 10.1104/pp.109.142786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tonnessen BW, et al. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015;87:273–286. doi: 10.1007/s11103-014-0275-9. [DOI] [PubMed] [Google Scholar]
- 41.Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.2307/3870059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu JK. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
- 43.Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ranjan A, et al. Comparative transcriptomic analysis of roots of contrasting Gossypium herbaceum genotypes revealing adaptation to drought. BMC Genom. 2012;13:680. doi: 10.1186/1471-2164-13-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng X, et al. Transcriptome analysis revealed the drought-responsive genes in Tibetan hulless barley. BMC Genom. 2016;17:386. doi: 10.1186/s12864-016-2685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niinemets Ü. Uncovering the hidden facets of drought stress: Secondary metabolites make the difference. Tree Physiol. 2016;36:129–132. doi: 10.1093/treephys/tpw007. [DOI] [PubMed] [Google Scholar]
- 47.Akula R, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011;6:1720–1731. doi: 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Safronov O, et al. Detecting early signs of heat and drought stress in Phoenix dactylifera (date palm) PLoS One. 2017;12:e0177883. doi: 10.1371/journal.pone.0177883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daszkowska-Golec A, Szarejko I. Open or close the gate–stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 2013;4:138. doi: 10.3389/fpls.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upchurch RG. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotech. Lett. 2008;30:967–977. doi: 10.1007/s10529-008-9639-z. [DOI] [PubMed] [Google Scholar]
- 51.León J, Sánchez-Serrano JJ. Molecular biology of jasmonic acid biosynthesis in plants. Plant Physiol. Biochem. 1999;37:373–380. doi: 10.1016/S0981-9428(99)80043-6. [DOI] [Google Scholar]
- 52.Ruiz-Sola MÁ, Arbona V, Gómez-Cadenas A, Rodríguez-Concepción M, Rodríguez-Villalón A. A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS One. 2014;9:e90765. doi: 10.1371/journal.pone.0090765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida T, Mogami J, Yamaguchi-Shinozaki K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014;21:133–139. doi: 10.1016/j.pbi.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 54.Broehan G, Kroeger T, Lorenzen M, Merzendorfer H. Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genom. 2013;14:6. doi: 10.1186/1471-2164-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinoia E, et al. Multifunctionality of plant ABC transporters–more than just detoxifiers. Planta. 2002;214:345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- 56.Yazaki K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006;580:1183–1191. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 57.Cho M, Cho H. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013;8:642–654. doi: 10.4161/psb.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolfger H, Mamnun YM, Kuchler K. Fungal ABC proteins: Pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 2001;152:375–389. doi: 10.1016/S0923-2508(01)01209-8. [DOI] [PubMed] [Google Scholar]
- 59.Valliyodan B, Nguyen HT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006;9:189–195. doi: 10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Filippou P, Bouchagier P, Skotti E, Fotopoulos V. Proline and reactive oxygen/nitrogen species metabolism is involved in the tolerant response of the invasive plant species Ailanthus altissima to drought and salinity. Environ. Exp. Bot. 2014;97:1–10. doi: 10.1016/j.envexpbot.2013.09.010. [DOI] [Google Scholar]
- 61.Azcón R, Tobar RM. Activity of nitrate reductase and glutamine synthetase in shoot and root of mycorrhizal Allium cepa: Effect of drought stress. Plant Sci. 1998;133:1–8. doi: 10.1016/S0168-9452(96)04533-5. [DOI] [Google Scholar]
- 62.Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015;16:15811–15851. doi: 10.3390/ijms160715811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta. 2014;239:255–266. doi: 10.1007/s00425-013-1985-y. [DOI] [PubMed] [Google Scholar]
- 64.Liu Q, Zhao N, Yamaguch-Shinozaki K, Shinozaki K. Regulatory role of DREB transcription factors in plant drought, salt and cold tolerance. Chin. Sci. Bull. 2000;45:970–975. doi: 10.1007/BF02884972. [DOI] [Google Scholar]
- 65.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thissen D, Steinberg L, Kuang D. Quick and easy implementation of the Benjamini–Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ. Behav. Stat. 2002;27:77–83. doi: 10.3102/10769986027001077. [DOI] [Google Scholar]
- 71.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;20:gkaa970. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanusi NSNM, et al. PalmXplore: Oil palm gene database. Database. 2018;2018:bay095. doi: 10.1093/database/bay095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, et al. RNA-Seq revealed the impairment of immune defence of tilapia against the infection of Streptococcus agalactiae with simulated climate warming. Fish Shellfish Immunol. 2016;55:679–689. doi: 10.1016/j.fsi.2016.06.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads used in this study have been deposited to the NCBI SRA database with an accession no. PRJDB9517.