Highlights
-
•
Fatty liver disease (hepatosteatosis) is a hallmark of ALD and NAFLD.
-
•
FABP4 is normally expressed in adipocytes and macrophages.
-
•
ALD leads to FABP4 synthesis/release from steatotic hepatocytes.
-
•
FABP4 stimulates hepatoma cell growth and migration.
Keywords: Alcohol, Liver, Hepatosteatosis, Fatty acid binding protein 4, Hepatocellular carcinoma
Abbreviations: HCC, hepatocellular carcinoma; EtOH, Ethanol; FABP, Fatty acid binding protein; ALD, Alcoholic liver disease; P450 2E1, Cytochrome P450 2E1; HFD, High-fat diet; CD, Control Diet; C-LD, Control liquid diet; E-LD, Ethanol-containing liquid diet; ERK, Extracellular regulated kinase, JNK, Janus kinase; p38MAPK, p38 Mitogen activated protein kinase; ELISA, enzyme linked immunosorbent assay; HBV, Viral hepatitis B
Abstract
Fatty liver disease (hepatosteatosis) is a common early pathology in alcohol-dependent and obese patients. Fatty acid binding protein-4 (FABP4) is normally expressed in adipocytes and macrophages and functions as a regulator of intracellular lipid movement/storage. This study sought to investigate hepatic FABP4 expression and function in alcoholic liver disease (ALD) and hepatocellular carcinoma (HCC). Using chronic ethanol fed mouse models and patient samples FABP4 expression was analyzed. Human HCC cells, and HCC cells transfected to express CYP2E1, were exposed to ethanol and analyzed for FABP4 expression, or exposed to rhFABP4 (in the absence/presence of ERK, p38-MAPK or JNK1/2 inhibitors) and cell proliferation and migration measured. Hepatosteatotic-ALD mouse models exhibited increased hepatic FABP4 mRNA and protein levels, with FABP4 expression confirmed in hepatocytes. In HCC cells, CYP2E1-dependent ethanol metabolism induced FABP4 expression in vitro and exogenous rhFABP4 stimulated proliferation and migration, effects abrogated by ERK and JNK1/2 inhibition. Increased FABP4 was also detected in ALD/ALD-HCC patients, but not patients with viral hepatitis/HCC. Collectively these data demonstrate ethanol metabolism induces hepatic FABP4 expression and FABP4 promotes hepatoma cell proliferation/migration. These data suggest liver-derived FABP4 may be an important paracrine-endocrine factor during hepatic foci expansion and/or hepatoma progression in the underlying setting of ALD.
Introduction
Liver tumors are the second leading cause of cancer-related mortality in the world and hepatocellular carcinoma (HCC) accounts for more than 80% of all primary hepatic neoplasms diagnosed [1]. The incidence of HCC is directly linked to exposure to known risk factors, the most common of which are viral hepatitis, chronic/heavy ethanol (EtOH) consumption, aflatoxin ingestion, and obesity [2], [3], [4]–5]. Hepatosteatosis (increased hepatic lipid uptake/storage) is a common pathological feature in alcohol-dependent and obese patients [2,6], and a significant factor in the progression of viral hepatitis-induced liver disease [7]. Sustained hepatic insult often leads to more severe liver diseases (alcoholic liver disease (ALD) to alcoholic steatohepatitis [8], non-alcoholic fatty liver disease to non-alcoholic steatohepatitis [2]) and increased risk of genetic damage, foci formation and HCC development/progression.
Fatty acid binding proteins (FABPs) are small (14–15 kDa), water-soluble proteins that bind long chain fatty acids to facilitate intracellular lipid transport [9]. To date, nine mammalian FABPs have been identified (FABP1–9) and FABP-subtype expression is [predominantly] tissue-specific [9]. For example, in the liver FABP1 is expressed in hepatocytes, while FABP4 is expressed in macrophages and adipocytes [9]. However, increasing evidence suggests FABP4 can act as a biologically active paracrine-endocrine signaling molecule in the setting of different disease pathologies [9], [10], [11]. In addition to metabolic diseases, FABP4 can also play a role in cancer development and progression [4]. For example, association of ovarian cancer omental cells with adipocytes stimulates FABP4 expression and metastasis in vitro, while [tumor cell] FABP4 depletion diminishes metastasis in vivo [12]. Similarly, in a prostate cancer model, bone marrow adipocytes stimulate FABP4 production and tumor metastasis [13].
With regard to HCC, increased FABP4 expression is reported in obesity-associated models of HCC in vivo, and exposure of hepatoma cells to free fatty acids increases FABP4 expression/secretion in vitro [14]. These data are consistent with other reports demonstrating 25-hydroxycholesterol promotes HCC metastasis via increased FABP4/matrix metallopeptidase 9 expression [15]. In contrast, Zhong et al., report increased hepatic FABP4 expression versus pair-matched HCC samples, a correlation between HCC-FABP4 expression and poorer recurrence-free survival, and FABP4-dependent inhibition of HCC proliferation [16].
The primary goals of this study were to examine changes in FABP4 expression in response to chronic ethanol consumption in experimental models and human subjects. The secondary goal was to use in vitro models to identify potential mechanisms whereby alcohol affects hepatic FABP expression and identify FABP-dependent signaling events in HCC progression.
Materials and methods
Human subjects
Informed written consent was obtained from human subjects taking part in this study according to a protocol governed by our Institutional Review Board.
Animal studies
Mice were purchased from Charles River Laboratories (Wilmington, MA). All studies were approved by our Institutional Animal Care and Use Committee and confirmed to the Guide for the Care and Use of Laboratory Animals.
Obesity-alcohol comorbidity model
Male C57BL/6 mice (35-days old) were randomized to either a 10% [kcal%] fat control diet (CD) or a 60% [kcal%] high fat diet (HFD) (N = 20 per group). At 35 weeks of age, mice from CD and HFD groups were randomized to either maintenance on drinking water (DW) alone, or a 10/20% (v/v) alternate day ethanol-drinking water (EtOH-DW) protocol (N = 10 per group). For mice randomized to EtOH-DW, mice were weaned onto EtOH-DW for 1 week (3 days 5% (v/v) EtOH-DW, 4 days 10% (v/v) EtOH-DW) prior to 10/20% (v/v) alternate day EtOH-DW. At 42 weeks, mice were anesthetized (isoflurane to surgical plane), euthanized by exsanguination, and serum and tissue collected and stored for analysis [17].
Lieber-DeCarli (LDC) model of chronic alcohol feeding and hepatocyte isolation
Male C57BL/6 mice (8–10 weeks) were randomized, weighed, and assigned to control (C-LD) or ethanol (E-LD) feeding groups. After weening onto liquid diets, mice were maintained on their respective diet for 4-weeks. At conclusion, mice were euthanized by exsanguination and serum and tissue collected for analysis, or hepatocytes isolated, purified, and collected for analysis [18].
Microarray analysis and gene expression
Total RNA was isolated from liver or hepatocytes (Quick-RNA Miniprep kit, Zymo Research, Irvine, CA) and reverse transcribed, amplified, and labelled using a GeneChipⓇ 3′ IVT Express Kit (Affymetrix Inc., Santa Clara, CA). Labelled complementary RNA (cRNA) was purified, fragmented and analyzed (2100 Bioanalyzer, Agilent, Santa Clara, CA) and cRNA samples/probe array controls hybridized to an Affymetrix GeneChipⓇ HT Mouse Genome-430 2.0 array. Expression analysis was performed in R (V.3.5.3) using Biocondutor's affy and limma packages. To analyze individual transcripts, total RNA from liver tissue was reverse-transcribed to cDNA (Improm-II kit, Promega; Madison, WI) and quantitative RT-PCR (qRT-PCR) performed (TaqManⓇ Universal Master Mix II, ThermoFisher Scientific, Grand Island, NY). FABP4 mRNA levels were calculated and normalized to 18s RNA.
Western blot analysis
Tissue and hepatocyte lysates were prepared and Western blot analysis performed as previously reported [14]. Successful protein transfer was determined using Ponceau S membrane staining, and images from Ponceau S membrane stain were used to correct for protein loading for signal detection using Image J software (National Institutes of Health, Bethesda, MD) [14]. An anti-FABP4 antibody was purchased from Cell Signaling Technology (Danvers, MA). Antibodies against total/phosphorylated extracellular regulated kinase(ERK1/2/pERK1/2),total/phosphorylatedjanuskinase1/2(JNK1/2/pJNK1/2),and total/phosphorylated p38 mitogen activated protein kinase (p38/pp38-MAPK) were purchased from Sant Cruz Biotechnology (Dallas, TX).
Serum FABP4 levels
Serum samples were prepared from whole blood and analyzed using an FABP4 enzyme linked immunosorbent assay (ELISA) kit (BosterBio, Pleasanton, CA) as per the manufacturer's instructions.
Cell culture
Human HepG2 HCC cells were purchased from ATCC (Manassas,VA). Human HuH7 HCC cells purchased from the JCRB Cell Bank (Sekisui XenoTech; Kansas City, KS). HepG2-E47 and HuH7-CYP (stably transfected with a plasmid expressing cytochrome P4502E1 (CYP 2E1)) were generous gifts from Dr. A. Cederbaum (Ichan School of Medicine, NY) and Dr. N. Osna (University of Nebraska, Omaha, NE), respectively. Cells were maintained in Dulbecco's Modified Eagle Medium (Gibco, Waltham, MA) as previously reported [19]. Recombinant human FABP4 (rhFABP4) was purchased from BioVision (Milipitas, CA).
Effect of ethanol on FABP4 production
HepG2/E47 and HuH7/HuH7-CYP cells were seeded in T-25 flasks and maintained overnight in growth medium (10% [v/v] FBS). Cells were serum-depleted (0.1% [v/v] FBS) for 24-hours prior to exposure to ethanol (0–100 mM). Cells and culture medium were collected after 48-hours and analyzed for FABP4 mRNA (qRT-pCR) and protein expression-secretion (ELISA).
Cell proliferation
HepG2 or HuH7 cells were seeded in 96-well plates and maintained overnight in growth medium. Cells were serum-depleted (0.1% [v/v] FBS) for 24-hours prior to exposure to SB203580 (20 µM; p38 MAPK inhibitor),U0126(10 µM; ERK inhibitor),orSP600125(50 µM; JNK1/2 inhibitor)(Cell Signaling Technologies, Danvers, MA) for 60-minutes followed by treatment with exogenous rhFABP4 (100 ng/mL). In parallel experiments, culture medium was replaced with 0.1 or 10% (v/v) FBS as negative/positive controls. A CyQuant NF cell proliferation assay (Invitrogen) was performed 48 h later as per the manufacturer's instructions.
Cell migration
Using Transwell Boyden chambers (8 µm pore size, Corning Inc., Lowell, MA) HepG2 or HuH7 cells were seeded in the upper chamber in 0.1% (v/v) FBS medium for 24 h. Cells were then pre-treated with pharmacological inhibitors SB203580 (20 µM), U0126 (10 µM), or SP600125 (50 µM) for 60-minutes prior to addition of rhFABP4 (200 ng/mL) to the lower chamber. After 48-hours cells remaining on the upper surface were removed using a wet cotton swab and cells on the lower surface fixed and stained with 0.2% (w/v) crystal violet solution in 100% EtOH (15 min), and the membrane removed and imaged on a fluorescence microscope. The crystal violet stain on the membrane was next dissolved (10% (w/v) sodium dodecyl sulfate) and absorbance measured at 592 nm.
Statistical analysis
Data are expressed as mean ± SEM. One- and two-way ANOVA were performed as appropriate using open source R statistical software (version 3.5.3). A P-value <0.05 was considered statistically significant.
Results
FABP4 is upregulated by obesity and chronic alcohol consumption in vivo
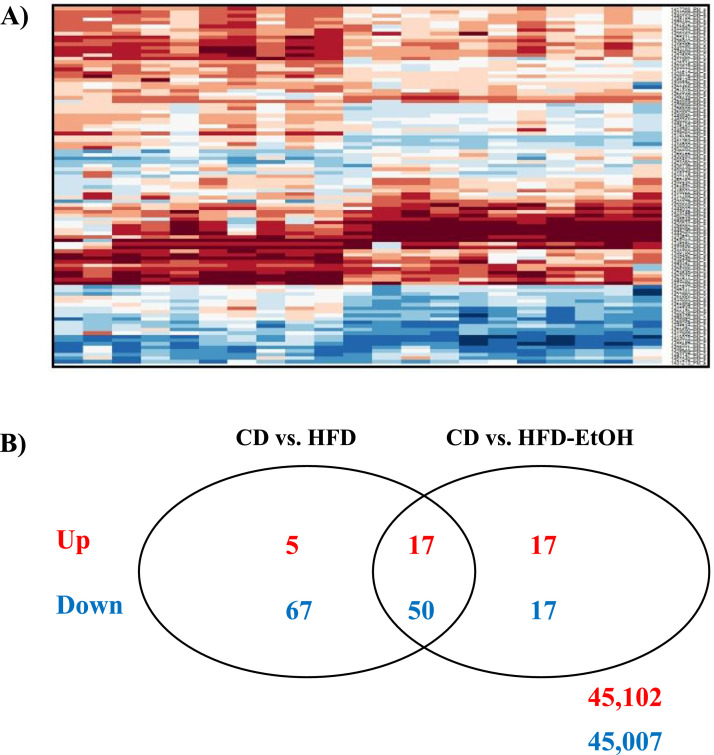
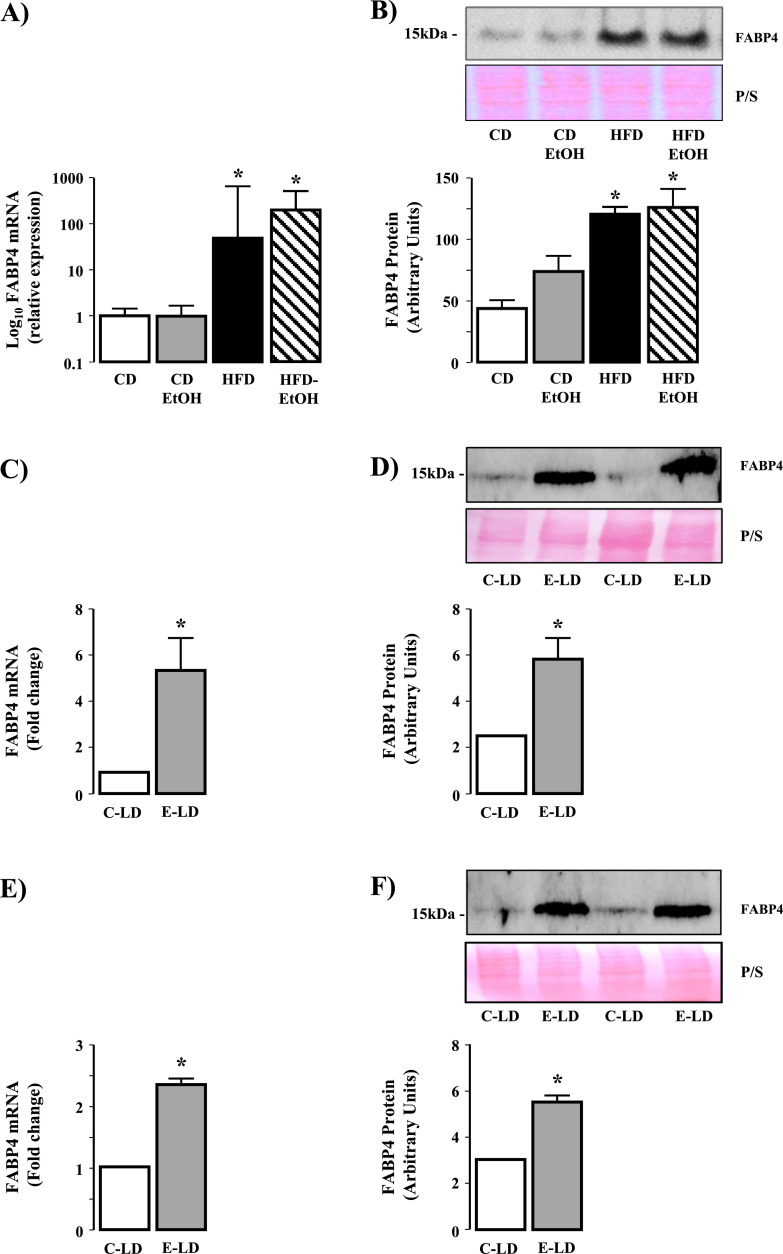
Microarray analysis revealed distinct expression patterns of the top 200 genes, with the most pronounced effects due to HFD (Fig. 1A). A subset analysis revealed 173 differentially expressed genes between CD, HFD and HFD-EtOH, with 67 genes (17 upregulated, 50 downregulated) commonly expressed in HFD and HFD-EtOH versus CD, and 34 genes (17 upregulated, 17 downregulated) unique to EtOH (Fig. 1B). Exploration of commonly dysregulated genes revealed genes associated with cellular and fatty acid metabolism were upregulated in HFD and HFD-EtOH versus CD (Table 1). qRT-PCR and Western blot revealed increased FABP4 mRNA and protein in HFD and HFD-EtOH versus CD, although no difference in FABP4 expression was detected between HFD and HFD-EtOH (Fig. 2A and B, P<0.05 HFD and HFD-EtOH versus CD).
Fig. 1.
High fat diet and chronic ethanol feeding dysregulate hepatic gene expression. Excised livers were processed for total RNA and gene array analyses performed. (A) Examination of a heat map demonstrating differentially expressed genes (top 200 in magnitude) between mice fed control diet with drinking water (CD) or ethanol in drinking water (CD-EtOH) and animals on high fat diet drinking water (HFD) or HFD and ethanol in drinking water (HFD-EtOH). (B) Venn Diagram identifying genes dysregulated between CD versus HFD and CD versus HFD-EtOH.
Table 1.
| Gene Symbol | Log FC | Biological function |
|---|---|---|
| Klf6 | 1.29 | Transcription |
| Foxq1 | −139 | Transcription |
| Hsd3b5 | −236 | Steroid biosynthetic process |
| Uap1l1 | 125 | Metabolic process |
| Sptlc3 | 2.63 | Metabolic process |
| Sdr9c7 | 1.12 | Metabolic process |
| Oxct1 | 1.19 | Metabolic process |
| Gstm3 | 133 | Metabolic process |
| Cbr3 | 134 | Metabolic process |
| Scd2 | 1.19 | Lipid metabolic process |
| Lp1 | 1.64 | Lipid metabolic process |
| Cyp39a1 | −120 | Lipid metabolic process |
| Cpt1b | 1.63 | Lipid metabolic process |
| Serpina12 | −131 | Glucose metabolic process |
| Fabp5 | −160 | Glucose metabolic process |
| Elovl1 | −1.63 | Fatty acid biosynthetic process |
| Fabp4 | 2.17 | Crokire production |
Fig. 2.
FABP4 is upregulated in models of obesity and ethanol consumption and expressed in hepatocytes from ethanol fed mice. (A) FABP4 mRNA expression in liver tissue isolated from mice fed control diet with drinking water (CD) or ethanol in drinking water (CD-EtOH), and mice on high fat diet drinking water (HFD) or HFD and ethanol in drinking water (HFD-EtOH). *P<0.05 versus CD and CD-EtOH. (B) Western blot analysis of FABP4 protein expression in liver tissue isolated from CD, CD-EtOH, HFD, HFD-EtOH (upper panel) and cumulative densitometric analysis of FABP4 protein expression (lower panel). *P<0.05 versus CD and CD-EtOH. (C) FABP4 mRNA expression in liver tissue from mice maintained on control Lieber-DeCarli liquid diet (C-LD) or ethanol containing Lieber-DeCarli liquid diet (E-LD). *P<0.05 versus C-LD. (D) Western blot analysis of FABP4 protein expression in liver tissue from mice maintained on C-LD or E-LD (upper panel) and cumulative densitometric analysis of FABP4 protein expression (lower panel). *P<0.05 versus C-LD. (E) FABP4 mRNA expression in isolated hepatocytes from mice maintained on C-LD or E-LD. *P<0.05 versus C-LD. (F) Western blot analysis of FABP4 protein expression in isolated hepatocytes from mice maintained on C-LD or E-LD (upper panel) and cumulative densitometric analysis of FABP4 protein expression (lower panel). *P<0.05 versus C-LD.
Chronic ethanol consumption increases hepatocyte FABP4 expression in vivo
Analysis of liver tissue and isolated hepatocytes revealed increased hepatic FABP4 mRNA and protein in E-LD versus C-LD liver tissue (Fig. 2C and D, P<0.05 E-LD versus C-LD, n = 4 per group), changes that were maintained in isolated hepatocytes (Fig. 2E & F, P<0.05 E-LD versus C-LD, n = 4 per group).
Ethanol metabolism enhances FABP4 expression in vitro
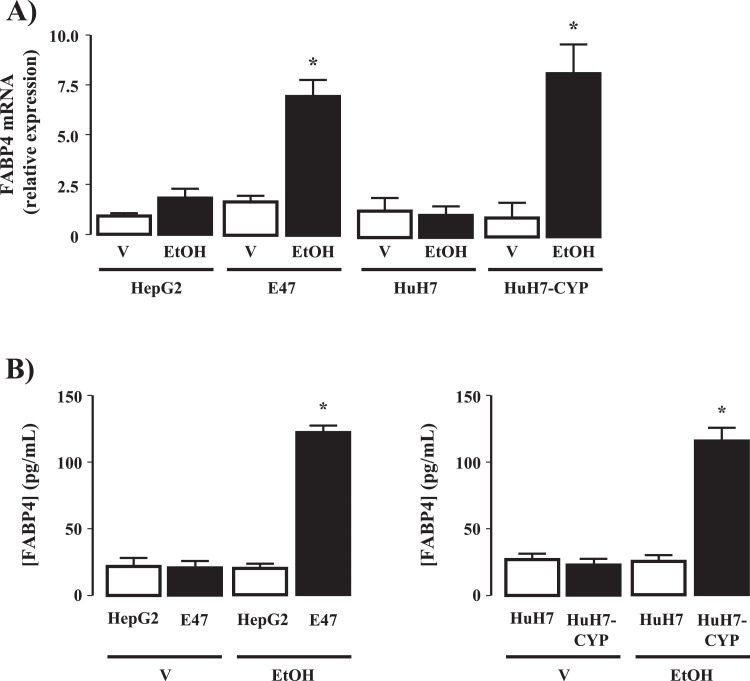
Exposure of HepG2 and HuH7 cells to EtOH did not significantly alter FABP4 mRNA expression (48 h-) or protein expression-secretion (48 h-) at any of the concentrations of EtOH employed (0–100 mM; Supplemental Fig. 1, Fig. 3A [100 mM EtOH]). In contrast, significant EtOH-concentration dependent increases in FABP4 mRNA (48-hours) and protein expression (48 h-) was detected in CYP2E1-expressing HepG2-E47 and HuH7-CYP hepatoma cells (Supplemental Fig. 1 and Fig. 3A and B; P<0.05 HepG2-E47 versus HepG2, and HuH7-CYP versus Huh7, n = 4 separate experiments).
Fig. 3.
Cytochrome P4502E1 (CYP2E1) dependent ethanol metabolism induces FABP4 expression in vitro. (A) FABP4 mRNA expression in HepG2 and HuH7 cells and HepG2 and HuH7 stably transfected to express CYP2E1(E47 and HuH7-CYP) following exposure to vehicle (V; 0.1mL phosphate buffered saline) or 100 mM ethanol (EtOH; 48h). *P<0.05 EtOH versus V, transfected versus non-transfected, N = 4 independent experiments. FABP4 secretion into culture medium from (B) HepG2/E47 and (C) HuH7/HuH7-CYP cells following exposure to V or 50 mM EtOH (48h). *P<0.05 EtOH versus V transfected versus non-transfected, N = 4 independent experiments.
Exogenous rhFABP4 stimulates HCC cell proliferation and migration in vitro
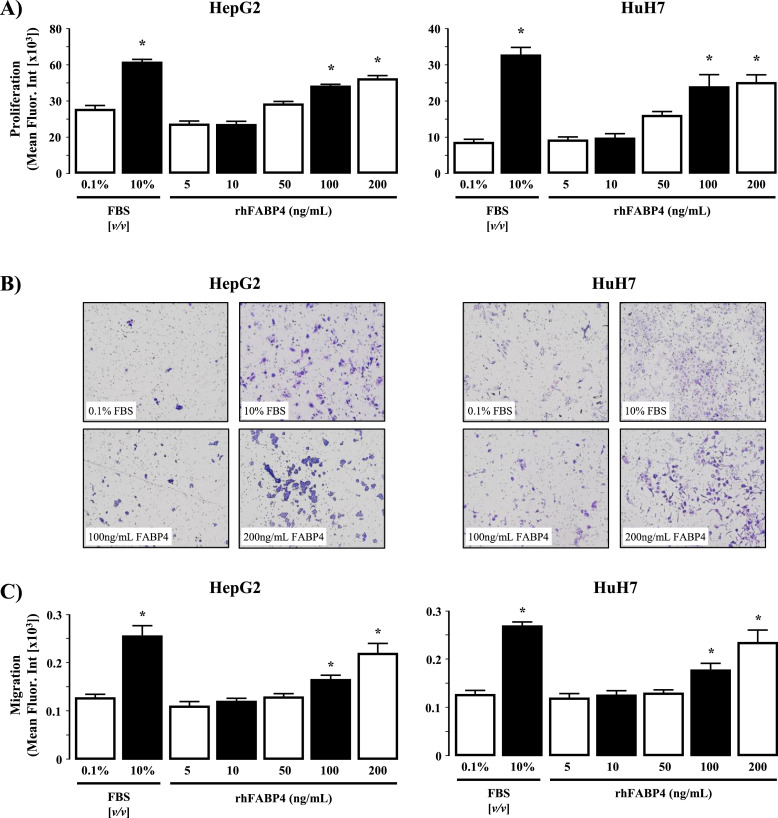
Addition of rhFABP4 to cell culture medium stimulated both cell proliferation and migration in HepG2 and HuH7 cells in a dose-dependent manner (Fig. 4A-C, P<0.05 rhFABP4 versus 0.1% [v/v] FBS).
Fig. 4.
rhFABP4 stimulates hepatoma cell proliferation and migration in vitro. (A) HepG2 and HuH7 cells were exposed to rhFABP4 (0–200 ng/mL, 48h) and assayed for cell proliferation. Negative and positive controls were 0.1% [v/v] and 10% [v/v] fetal bovine serum (FBS). *P<0.05 versus 0.1% [v/v] FBS, N = 4 independent experiments. (B) Representative images of HepG2 and HuH7 cell migration following 48h in the presence of 0.1% [v/v] FBS (control), 10% [v/v] FBS, 100 ng/mL or 200 ng/mL rhFABP4. (C) Cumulative analysis of HepG2 and HuH7 cell migration (crystal violet mean fluorescent intensity) following 48h in the presence of 0.1% [v/v] FBS (control), 10% [v/v] FBS, or rhFABP4 (5–200 ng/mL). *P<0.05 versus 0.1% [v/v] FBS and 5 ng/mL rhFABP4, N = 4 independent experiments.
FABP4 stimulates HCC cell proliferation and migration via activation of ERK and/or JNK signaling pathways
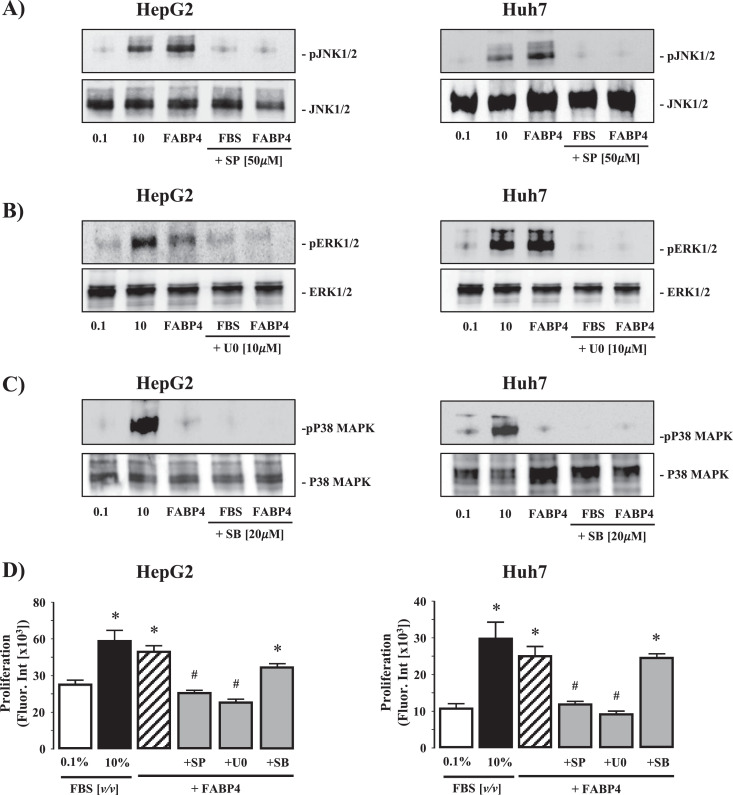
Exposure of quiescent (0.1% FBS (v/v), 24h) HepG2 and HuH7 cells to 10% (v/v) FBS led to robust detection of phosphorylated (activated) JNK 1/2, ERK 1/2, and p38MAPK by Western blot (Fig. 5A-C). Conversely, while exogenous rhFABP4 (200 ng/mL) stimulated JNK 1/2 and ERK 1/2 activity, rhFABP4 failed to stimulate p38MAPK activation (Fig. 5A-C). In parallel studies, pretreatment of HepG2 and HuH7 cells with SP600125 (20μM; JNK 1/2 inhibitor) or U0126 (10μM; ERK 1/2 inhibitor) abrogated the effects of both 10% (v/v) FBS and rhFABP4 (200 ng/mL) on phosphorylated JNK 1/2, ERK 1/2 expression, while pretreatment with SB203580 (50μM; p38-MAPK inhibitor) abolished the effect of 10% (v/v) FBS on phosphorylated p38MAPK expression (Fig. 5A-C).
Fig. 5.
Intracellular signaling pathways involved in rhFABP4-stimulated hepatoma cell proliferation and migration in vitro. (A) Representative Western blot analysis of phosphorylated/total JNK1/2 (pJNK1/2 - JNK1/2) in HepG2 and HuH7 HCC cells following exposure to 0.1 or 10% (v/v) fetal bovine serum (FBS), rhFABP4 (200 ng/mL), or 10% (v/v) FBS/rhFABP4 in the presence of SP 600,125 [SP; JNK inhibitor] (50 μM). (B) Phosphorylated/total ERK1/2 (pERK 1/2 - ERK 1/2) in HepG2 and HuH7 HCC cells following exposure to FBS, rhFABP4 (200 ng/mL), or FBS/rhFABP4 in the presence of U0126 [U0; ERK1/2 inhibitor] (10 μM). (C) Phosphorylated/total p38 mitogen-activate protein kinase (pp38MAPK - p38MAPK) in HepG2 and HuH7 HCC cells following exposure to FBS, rhFABP4 (100 ng/mL), or FBS/rhFABP4 in the presence of SB 203,589 [SB; p38MAPK inhibitor] (20 μM). D) HepG2 and HuH7 cells were pretreated with SB, U0, or SP for 60 min prior to exposure to exogenous rhFABP4 (200 ng/mL; 24 h) and assayed for proliferation. Negative and positive controls were 0.1% [v/v] and 10% [v/v] FBS respectively. *P<0.05 versus 0.1% [v/v] FBS, #P<0.05 versus rhFABP4, N = 4.
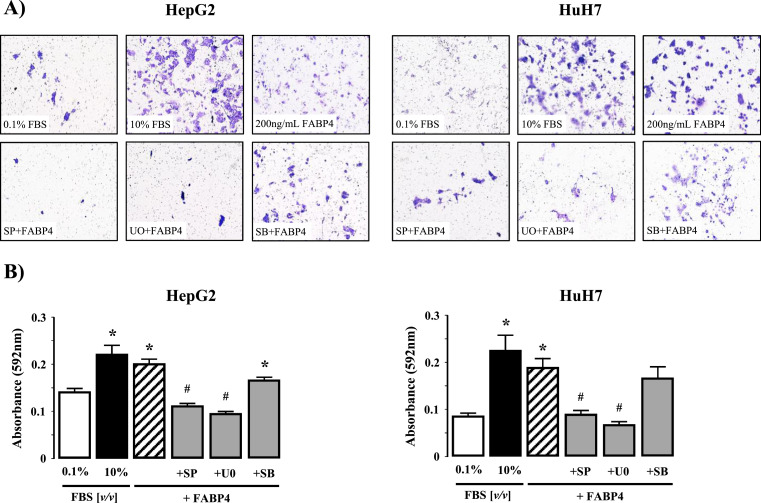
Using concentrations of rhFABP4 demonstrated to stimulate proliferation (100 ng/mL) and migration (200 ng/mL) we next examined the effect of inhibition of JNK 1/2, ERK 1/2, or p38MAPK on proliferation and migration. Inhibition of JNK1/2 (SP600125) or ERK (U0126) abolished the effect of exogenous rhFABP4 on both proliferation (Fig. 5D; P<0.05 200 ng/mL rhFABP4 versus 0.1% [v/v] FBS, n = 4) and migration (Fig. 6A and B; P<0.05 200 ng/mL rhFABP4 versus 0.1% [v/v] FBS, n = 4 independent experiments). In contrast, pre-treatment with SB203580 (p38-MAPK inhibitor failed to prevent a significant increase in proliferation or migration compared to control (0.1% (v/v) FBS) (Fig. 5A–C and Fig. 6A & B; n = 4 independent experiments).
Fig. 6.
(A) Representative images of HepG2 and HuH7 cell migration following exposure to 0.1 or 10% (v/v) fetal bovine serum (FBS), rhFABP4 (200 ng/mL), or 10% (v/v) FBS/rhFABP4 in the presence of SP 600,125 [SP; JNK inhibitor] (50 μM). U0126 [U0; ERK1/2 inhibitor] (10 μM), or SB 203,589 [SB; p38MAPK inhibitor] (20 μM). (B) Cumulative analysis of HepG2 and HuH7 cell migration (mean crystal violet fluorescent intensity). *P<0.05 versus 0.1% [v/v] FBS, #P<0.05 versus rhFABP4, N = 4.
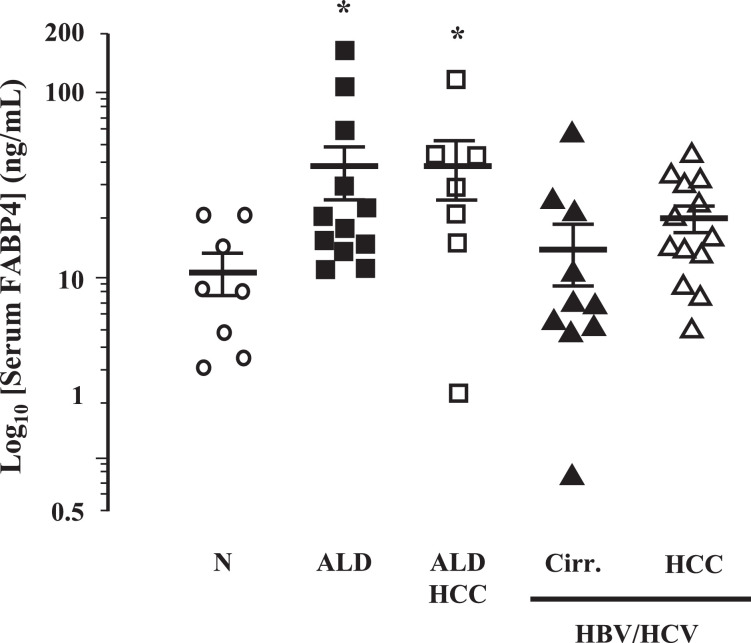
Patients with alcoholic liver disease have elevated serum FABP4
Serum FABP4 levels were significantly increased in ALD patients (n = 12) and ALD patients with HCC (n = 7) compared to individuals without underlying liver disease (n = 8) (Fig. 7; P<0.05 ALD and ALD+HCC versus Normal). Conversely, patients diagnosed with hepatic cirrhosis arising as a result of viral hepatitis infection (n = 10), and patients diagnosed with HCC arising as a result of viral hepatitis infection (n = 13), exhibited serum FABP4 levels that were not different to individuals without underlying liver disease (n = 8) (Fig. 7).
Fig. 7.
Serum FABP4 is elevated in ALD patients. Serum isolated from whole blood was collected from human subjects with normal livers (Norm, N = 8), patients with alcoholic liver disease (ALD; N = 12) or HCC caused by ALD (ALD-HCC; N = 7), or patients with cirrhosis caused by viral hepatitis (HBV/HCV-Cirr; N = 10) and viral hepatitis associated HCC (HBV/HCV-HCC; N = 13) and assayed for FABP4 levels. *P< 0.05 versus Norm.
Discussion
Hepatic steatosis is the most frequent hepatic pathology observed in obese and early stage alcohol-dependent patients, and plays a significant role in liver disease progression in patients with viral hepatitis. Data presented herein demonstrate FABP4, an FABP subtype usually expressed in macrophages and adipocytes, is elevated in hepatocytes from animal models of chronic alcohol feeding and in serum from patients with ALD/ALD-HCC. Using human hepatoma cells in vitro, CYP2E1-dependent alcohol metabolism induced FABP4 expression and secretion, and exogenous rhFABP4 stimulated proliferation and migration in HCC cells via ERK and JNK-dependent signaling pathways.
Previous studies using FABP4−/− mice [20], or a methionine-choline deficient mouse model of nonalcoholic steatohepatitis [11], indicate a role for FABP4 in metabolic disease progression. Additionally, FABP4 secreted from omental or bone marrow adipocytes induces tumor cell metastasis in ovarian and prostate cancer [12,13,21]. While these data support our hypothesis that FABP4 can promote HCC expansion, other studies report contradictory findings. For example, Zhong et al., report relative FABP4 expression (between HCC and non-HCC liver tissue) in HCC patients correlates with disease-free survival, while exogenous FABP4 inhibits HCC proliferation in vitro and over-expression of FABP4 inhibits tumor growth in an orthotopic tumor model in vivo [16]. However, it should be noted that in this study the [Chinese] patient population was identified as HBV-positive and the cell lines utilized in vitro were derived from HBV-positive patients. Relative to our findings, we report increased serum FABP4 in ALD and ALD-HCC patients but not in HBV/HCV-cirrhotic/HCC patients. Similarly, the cell lines we employed are HBV-negative, suggesting the impact of changes in FABP4 expression/signaling may depend (at least in part) on underlying viral status. Similarly, analysis of FABP4 expression in BEL-7404 hepatoma cells and non-tumorigenic L-02 hepatic cell line reports lower FABP4 expression in BEL-7404 cells. [22] However, when comparing these findings to our data, we suggest they should be taken into context within the respective study settings. That is, our data indicate both hepatocytes and HCC cell lines express FABP4 when challenged with stimuli that promote intracellular steatosis (alcohol). In contrast, this phenotype was not present in the cell culture or animal models utilizing FABP4 overexpressing cells further suggesting the underlying pathology in which changes in FABP4 expression occur may be of equal significance as the expression of FABP4 per se.
Exposing hepatoma cells to exogenous rhFABP4 in vitro in the absence/presence of pharmacological inhibitors revealed FABP4-dependent proliferation and migration were regulated, at least in part, by JNK and/or ERK signaling. Given the extensive literature identifying the roles of both JNK and ERK signaling during migration and proliferation, including that of hepatoma cells [23,24], these findings raise interesting questions regarding the mechanisms by which FABP4 signals. Previous studies report the importance of FABP4 binding to FFAs and other ligands intracellularly to undergo conformational change and localization to the nucleus as a means to regulate cell function, including gene transcription [25]. Indeed, Laouirem et al. report FABP4-dependent increases in proliferation in vitro and in vivo were abrogated by BMS309403 [26] (a biphenyl azole inhibitor that binds with high affinity to the FABP4-binding cavity [27]). However, data from our study raise the possibility that secreted/exogenous FABP4 may bind to (an as yet unidentified) cell membrane-bound receptor as a means to regulate intracellular signaling activity. This possibility is supported by reports in which exogenous FABP4 stimulates intracellular signaling [28,29], and the significance of adjacent, adipose-derived FABP4 during tumor progression [4,30,31]. In addition, the potential existence of an FABP4-receptor provides further explanation as to why over-expressing FABP4 in cell inoculation-tumor models may exert different responses when compared to endocrine/paracrine FABP4 release.
When performing in vitro analyses of FABP4 expression and function, it is of interest to note the relative differences between the amount of FABP4 secreted in response to EtOH and the amount of exogenous rhFABP4 required to stimulate proliferation and migration. Such differences may be attributable to the culture conditions used (cell were made quiescent prior to EtOH/rhFABP4 addition), or the absence of physical and physiological structures that exist in vivo when compared to in vitro culture conditions (3-dimensional/multi-cell, cell-cell contact-interaction, vascularization, nutrient/oxygen availability). However, these differences may also relate to potential differences in protein structure/folding between exogenous rhFABP4 and endogenously synthesized/secreted FABP4, mechanism of FABP4 secretion, and/or the paracrine-autocrine signaling mechanisms by which FABP4 exerts its effect[s] following release. Addressing these mechanisms at the cellular level, in addition to utilizing gene silencing and/or knock-out animal models will provide important insight into the mechanisms by which FABP4 is both synthesized/released, and the means by which it affects target cell function.
When considering our data, it is important to highlight potential limitations associated with our study. Firstly, our focus was on the role of FABP4 in tumor progression within the setting of a specific disease state (ALD), rather than the role of hepatic FABP4 during initial hepatocyte transformation per se. Significant advances have been made in understanding the role of alcohol metabolism, oxidative stress and DNA, protein, and lipid adduct formation in alcohol-dependent-HCC, and it will be of interest to determine which (if any) of these factors may affect FABP4 expression during transformation. Secondly, while potential roles for JNK and ERK were identified in FABP4-dependent signaling, significant work remains to identify potential downstream effectors, especially given the widespread involvement or JNK and ERK in proliferation and migration via interaction with different downstream mediators [32]. Finally, our preliminary analysis of serum from ALD and viral hepatitis patients indicated differences in FABP4 were confined to those patients with ALD. However, increased non-parenchymal cell FABP4 expression is reported in HCC patients, but less frequently observed is elevated expression of FABP4 in hepatic parenchymal cells [16]. These findings are not necessarily surprising considering cirrhosis and HCC development in patients with viral hepatitis is independent of obesity/hepatic steatosis. In addition, advanced liver fibrosis in patients with non-alcoholic steatohepatitis and ALD is often accompanied by a reduction in hepatic fat, a phenomenon termed ‘burn-out NASH’. [33] Thus, the source of FABP4 during HCC initiation may vary during tumor progression, either due to changes in underlying hepatic pathology and/or the influence of comorbid factors such as elevated body-mass index and cardiovascular disease.
Conclusion
Chronic ethanol ingestion/hepatic ethanol metabolism induce hepatic FABP4 mRNA and protein expression in vivo and in vitro, and exposure of hepatoma cells to rhFAB4 stimulates proliferation and migration. These data suggest liver-derived FABP4 may play a role in hepatoma expansion and/or progression.
CRediT authorship contribution statement
Neha Attal: Data curation, Formal analysis, Methodology, Supervision, Writing - original draft, Writing - review & editing. Mariel T. Sullivan: Data curation, Formal analysis, Investigation, Writing - review & editing. Cara A. Girardi: Data curation, Formal analysis, Investigation, Writing - review & editing. Kyle J. Thompson: Conceptualization, Data curation, Formal analysis, Methodology, Supervision, Validation, Writing - original draft, Writing - review & editing. Iain H. McKillop: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
None of the authors have any conflicts of interest to declare.
Funding
Studies were supported from institutional funds (Atrium Health)
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100975.
Contributor Information
Neha Attal, Email: Neha.Attal@atriumhealth.org.
Mariel T. Sullivan, Email: Mariel.Sullivan@atriumhealth.org.
Cara A. Girardi, Email: Cara.Girardi@atriumhealth.org.
Kyle J. Thompson, Email: Kyle.Thompson@atriumhealth.org.
Iain H. McKillop, Email: Iain.McKillop@atriumhealth.org.
Appendix. Supplementary materials
References
- 1.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R., Priyadarshi R.N., Anand U. Non-alcoholic fatty liver disease: growing burden, adverse outcomes and associations. J. Clin. Transl. Hepatol. 2020;8:76–86. doi: 10.14218/JCTH.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCullough A.K., Lloyd R.S. Mechanisms underlying aflatoxin-associated mutagenesis - Implications in carcinogenesis. DNA Repair (Amst.) 2019;77:76–86. doi: 10.1016/j.dnarep.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKillop I.H., Girardi C.A., Thompson K.J. Role of fatty acid binding proteins (FABPs) in cancer development and progression. Cell Signal. 2019;62 doi: 10.1016/j.cellsig.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Sagnelli E., Macera M., Russo A., Coppola N., Sagnelli C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection. 2020;48:7–17. doi: 10.1007/s15010-019-01345-y. [DOI] [PubMed] [Google Scholar]
- 6.Neuman M.G., Malnick S., Maor Y., Nanau R.M., Melzer E., Ferenci P., Seitz H.K., Mueller S., Mell H., Samuel D., Cohen L.B., Kharbanda K.K., Osna N.A., Ganesan M., Thompson K.J., McKillop I.H., Bautista A., Bataller R., French S.W. Alcoholic liver disease: clinical and translational research. Exp. Mol. Pathol. 2015;99:596–610. doi: 10.1016/j.yexmp.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathurin P., Bataller R. Trends in the management and burden of alcoholic liver disease. J. Hepatol. 2015;62:S38–S46. doi: 10.1016/j.jhep.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thumser A.E., Moore J.B., Plant N.J. Fatty acid binding proteins: tissue-specific functions in health and disease. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:124–129. doi: 10.1097/MCO.0000000000000031. [DOI] [PubMed] [Google Scholar]
- 10.Hotamisligil G.S., Bernlohr D.A. Metabolic functions of FABPs–mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015;11:592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larter C.Z., Yeh M.M., Williams J., Bell-Anderson K.S., Farrell G.C. MCD-induced steatohepatitis is associated with hepatic adiponectin resistance and adipogenic transformation of hepatocytes. J. Hepatol. 2008;49:407–416. doi: 10.1016/j.jhep.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Nieman K.M., Kenny H.A., Penicka C.V., Ladanyi A., Buell-Gutbrod R., Zillhardt M.R., Romero I.L., Carey M.S., Mills G.B., Hotamisligil G.S., Yamada S.D., Peter M.E., Gwin K., Lengyel E. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herroon M.K., Rajagurubandara E., Hardaway A.L., Powell K., Turchick A., Feldmann D., Podgorski I. Bone marrow adipocytes promote tumor growth in bone via FABP4-dependent mechanisms. Oncotarget. 2013;4:2108–2123. doi: 10.18632/oncotarget.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson K.J., Austin R.G., Nazari S.S., Gersin K.S., Iannitti D.A., McKillop I.H. Altered fatty acid-binding protein 4 (FABP4) expression and function in human and animal models of hepatocellular carcinoma. Liver Int. 2018;38:1074–1083. doi: 10.1111/liv.13639. [DOI] [PubMed] [Google Scholar]
- 15.Wang S., Yao Y., Wang X., Zheng G., Ouyang W., Chen W. 25-HC promotes hepatocellular carcinoma metastasis through up-regulation of TLR4 dependent FABP4. Am. J. Cancer Res. 2019;9:2140–2155. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong C.Q., Zhang X.P., Ma N., Zhang E.B., Li J.J., Jiang Y.B., Gao Y.Z., Yuan Y.M., Lan S.Q., Xie D., Cheng S.Q. FABP4 suppresses proliferation and invasion of hepatocellular carcinoma cells and predicts a poor prognosis for hepatocellular carcinoma. Cancer Med. 2018;7:2629–2640. doi: 10.1002/cam4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson K.J., Swan R.Z., Walling T.L., Iannitti D.A., McKillop I.H., Sindram D. Obesity, but not ethanol, promotes tumor incidence and progression in a mouse model of hepatocellular carcinoma in vivo. Surg. Endosc. 2013;27:2782–2791. doi: 10.1007/s00464-013-2808-8. [DOI] [PubMed] [Google Scholar]
- 18.Powell R.D., Swet J.H., Kennedy K.L., Huynh T.T., McKillop I.H., Evans S.L. Resveratrol attenuates hypoxic injury in a primary hepatocyte model of hemorrhagic shock and resuscitation. J. Trauma Acute Care Surg. 2014;76:409–417. doi: 10.1097/TA.0000000000000096. [DOI] [PubMed] [Google Scholar]
- 19.Thompson K.J., Humphries J.R., Niemeyer D.J., Sindram D., McKillop I.H. The effect of alcohol on Sirt1 expression and function in animal and human models of hepatocellular carcinoma (HCC) Adv. Exp. Med. Biol. 2015;815:361–373. doi: 10.1007/978-3-319-09614-8_21. [DOI] [PubMed] [Google Scholar]
- 20.Hotamisligil G.S., Johnson R.S., Distel R.J., Ellis R., Papaioannou V.E., Spiegelman B.M. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 21.Cho K.R., Shih Ie M. Ovarian cancer. Annu. Rev. Pathol. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L.R., Zeng R., Shao X.X., Wang N., Xu Y.H., Xia Q.C. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis. 2000;21:3058–3068. doi: 10.1002/1522-2683(20000801)21:14<3058::AID-ELPS3058>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Min L., He B., Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 2011;21:10–20. doi: 10.1016/j.semcancer.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Wang J., Tai G. Role of C-Jun N-terminal kinase in hepatocellular carcinoma development. Target Oncol. 2016;11:723–738. doi: 10.1007/s11523-016-0446-5. [DOI] [PubMed] [Google Scholar]
- 25.Gillilan R.E., Ayers S.D., Noy N. Structural basis for activation of fatty acid-binding protein 4. J. Mol. Biol. 2007;372:1246–1260. doi: 10.1016/j.jmb.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laouirem S., Sannier A., Norkowski E., Cauchy F., Doblas S., Rautou P.E., Albuquerque M., Garteiser P., Sognigbe L., Raffenne J., van Beers B.E., Soubrane O., Bedossa P., Cros J., Paradis V. Endothelial fatty liver binding protein 4: a new targetable mediator in hepatocellular carcinoma related to metabolic syndrome. Oncogene. 2019;38:3033–3046. doi: 10.1038/s41388-018-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulsky R., Magnin D.R., Huang Y., Simpkins L., Taunk P., Patel M., Zhu Y., Stouch T.R., Bassolino-Klimas D., Parker R., Harrity T., Stoffel R., Taylor D.S., Lavoie T.B., Kish K., Jacobson B.L., Sheriff S., Adam L.P., Ewing W.R., Robl J.A. Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP) Bioorg. Med. Chem. Lett. 2007;17:3511–3515. doi: 10.1016/j.bmcl.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 28.Guaita-Esteruelas S., Bosquet A., Saavedra P., Guma J., Girona J., Lam E.W., Amillano K., Borras J., Masana L. Exogenous FABP4 increases breast cancer cell proliferation and activates the expression of fatty acid transport proteins. Mol. Carcinog. 2017;56:208–217. doi: 10.1002/mc.22485. [DOI] [PubMed] [Google Scholar]
- 29.Girona J., Rosales R., Plana N., Saavedra P., Masana L., Vallve J.C. FABP4 induces vascular smooth muscle cell proliferation and migration through a MAPK-dependent pathway. PLoS ONE. 2013;8:e81914. doi: 10.1371/journal.pone.0081914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabia B., Andrade S., Carreira M.C., Casanueva F.F., Crujeiras A.B. A role for novel adipose tissue-secreted factors in obesity-related carcinogenesis. Obes. Rev. 2016;17:361–376. doi: 10.1111/obr.12377. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Calvo R., Girona J., Alegret J.M., Bosquet A., Ibarretxe D., Masana L. Role of the fatty acid-binding protein 4 in heart failure and cardiovascular disease. J. Endocrinol. 2017;233:R173–R184. doi: 10.1530/JOE-17-0031. [DOI] [PubMed] [Google Scholar]
- 32.Harjes U., Bridges E., McIntyre A., Fielding B.A., Harris A.L. Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. J. Biol. Chem. 2014;289:23168–23176. doi: 10.1074/jbc.M114.576512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Poorten D., Samer C.F., Ramezani-Moghadam M., Coulter S., Kacevska M., Schrijnders D., Wu L.E., McLeod D., Bugianesi E., Komuta M., Roskams T., Liddle C., Hebbard L., George J. Hepatic fat loss in advanced nonalcoholic steatohepatitis: are alterations in serum adiponectin the cause? Hepatology. 2013;57:2180–2188. doi: 10.1002/hep.26072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.