Significance
In light of the genetic code, combinations of three nucleotides which are known as synonymous codons, can give rise to the same amino acid. Despite the homology at the protein level, these different codons are recognized distinctly by the translational machinery. The unequal use of synonymous codons influences protein expression. Surprisingly, we find that the coding sequences of KRAS and other frequently mutated cancer genes are adapted to be efficiently translated in proliferating cells in comparison to their family counterparts. Our work contributes to the unsolved question of why in tumors some members of cancer gene families show a higher mutation rate than their family counterparts. Thus, our results elucidate the relationship between tRNA expression, codon usage, and oncogenicity.
Keywords: codon usage, translation, tRNA, oncogene, KRAS
Abstract
It is well known that in cancer gene families some members are more frequently mutated in tumor samples than their family counterparts. A paradigmatic case of this phenomenon is KRAS from the RAS family. Different explanations have been proposed ranging from differential interaction with other proteins to preferential expression or localization. Interestingly, it has been described that despite the high amino acid identity between RAS family members, KRAS employs an intriguing differential codon usage. Here, we found that this phenomenon is not exclusive to the RAS family. Indeed, in the RAS family and other oncogene families with two or three members, the most prevalently mutated gene in tumor samples employs a differential codon usage that is characteristic of genes involved in proliferation. Prompted by these observations, we chose the RAS family to experimentally demonstrate that the translation efficiency of oncogenes that are preferentially mutated in tumor samples is increased in proliferative cells compared to quiescent cells. These results were further validated by assessing the translation efficiency of KRAS in cell lines that differ in their tRNA expression profile. These differences are related to the cell division rate of the studied cells and thus suggest an important role in context-specific oncogene expression regulation. Altogether, our study demonstrates that dynamic translation programs contribute to shaping the expression profiles of oncogenes. Therefore, we propose this codon bias as a regulatory layer to control cell context-specific expression and explain the differential prevalence of mutations in certain members of oncogene families.
Cancers arise due to mutations that confer a selective growth advantage on cells (1). These mutations can occur in oncogenes, which when activated by mutations contribute to the cancer proliferation phenotype. Interestingly, oncogenes often have closely related family members that are less frequently mutated in cancer. The RAS family is a striking example. Activating mutations in KRAS are among the most common mutations in human cancers (2). KRAS belongs to a family of three genes, with the other two members being HRAS and NRAS. The proteins encoded by these genes share a high sequence identity of 85% and therefore have similar structures and biochemical properties (3). However, the reasons behind the drastic variation in mutation incidence between the RAS genes remain enigmatic.
Several studies indicate that each RAS protein leads to different cellular responses and oncogenic phenotypes (4–9). These observations suggest that specificities at the level of function and expression might contribute to the RAS mutation patterns observed in human cancers (10). Part of the functional variation, for example, is mediated by the distinct amino acid sequences at the C-terminal hypervariable region that lead to distinct processing (11) and cellular localization (12). Also, the RAS proteins differ in their ability to activate downstream signaling pathways (13–15). Besides the biochemical differences between the RAS proteins, previous studies also suggest that the expression levels of HRAS, KRAS, and NRAS seem to play an important role in their biological response (8, 16). Compelling work in mouse embryogenesis showed that knockouts of KRAS, which are embryonic lethal (17), can be rescued by knocking in HRAS at the KRAS locus. This shows that the HRAS protein is capable of replacing the essential function of KRAS in mouse development (18). However, adult HRAS knockin mice showed a pathological cardiovascular phenotype (18), a direct consequence of the overexpression of the HRAS protein. Indeed, elimination of the wild-type (WT) copies of HRAS completely prevented these cardiovascular defects (9). With regards to RAS expression, differences have also been identified at the level of translation. Intriguingly, even though the RAS proteins are highly similar, they employ a different codon usage with only 15% codon identity (19). For instance, in comparison to the nucleotide sequence of HRAS, KRAS is enriched in rare codons, which are decoded by low-abundant tRNAs. This difference has been linked to poor translation efficiency for KRAS and high translation efficiency for HRAS (19).
Codon usage and tRNA abundance are important parameters for fine tuning protein synthesis. The functional influence of codon optimality and tRNA levels on the efficiency of protein production remains a topic of intense debate (20, 21). In recent years, studies have shown that tRNA levels are not static but dynamically regulated in different cellular contexts, leading to changes in the translation efficiency of transcripts depending on their codon composition (22–26). In mammalian cells, changes in tRNA abundance have been reported across different cell states, and specifically between healthy and cancer cells (22, 23). Interestingly, Gingold et al. (22) showed that a specific subset of tRNAs is up-regulated in proliferating cells, but down-regulated in differentiated or arrested cells. Additionally, the codon usage of genes that are necessary for cell division was found to be adapted to the tRNA repertoire of proliferative cells. Thus, changes in the expression of specific tRNAs could regulate an entire functional class of genes—for instance proliferative genes—to favor cell growth. Would a cancer cell take advantage of this translational program to modulate the expression of genes to its own growth advantage? Could a dynamic regulation of RAS translation efficiency determine the uneven mutation frequencies across RAS genes? Would this be a general phenomenon across other cancer gene families?
To answer the above questions, we first identified eight protein families of three members each (RAS, RAF, RAC, RHO, FOXA, FGFR, COL, and AKT) with high protein sequence similarity and at least one protein relevant to cancer. In seven of the eight families, we found that in contrast to its two homologous family members, the most frequently mutated gene had a codon usage signature characteristic of proliferation-related genes. The strongest association between mutation frequency and proliferation-related codon usage was observed for the RAS, RAF, and RAC families. Additionally, we found that this pattern holds true for gene pairs and even helps to identify candidate cancer gene families. Subsequently, we examined the RAS family in further detail. We measured how proliferation and quiescent cell states induce codon-dependent changes in KRAS protein levels. Finally, we found that different tRNA expression profiles between cell lines corresponded to differences in KRAS protein levels. This work suggests the existence of different translational programs such as the up-regulation of proliferative tRNAs that have the potential to boost the protein synthesis of oncogenes. Thus, our results suggest that dynamic changes in this fundamental cellular process may contribute to cancer and specifically to the prevalence of mutations in certain genes when compared to their closely related family members.
Results
Codon Usage of Cancer Genes.
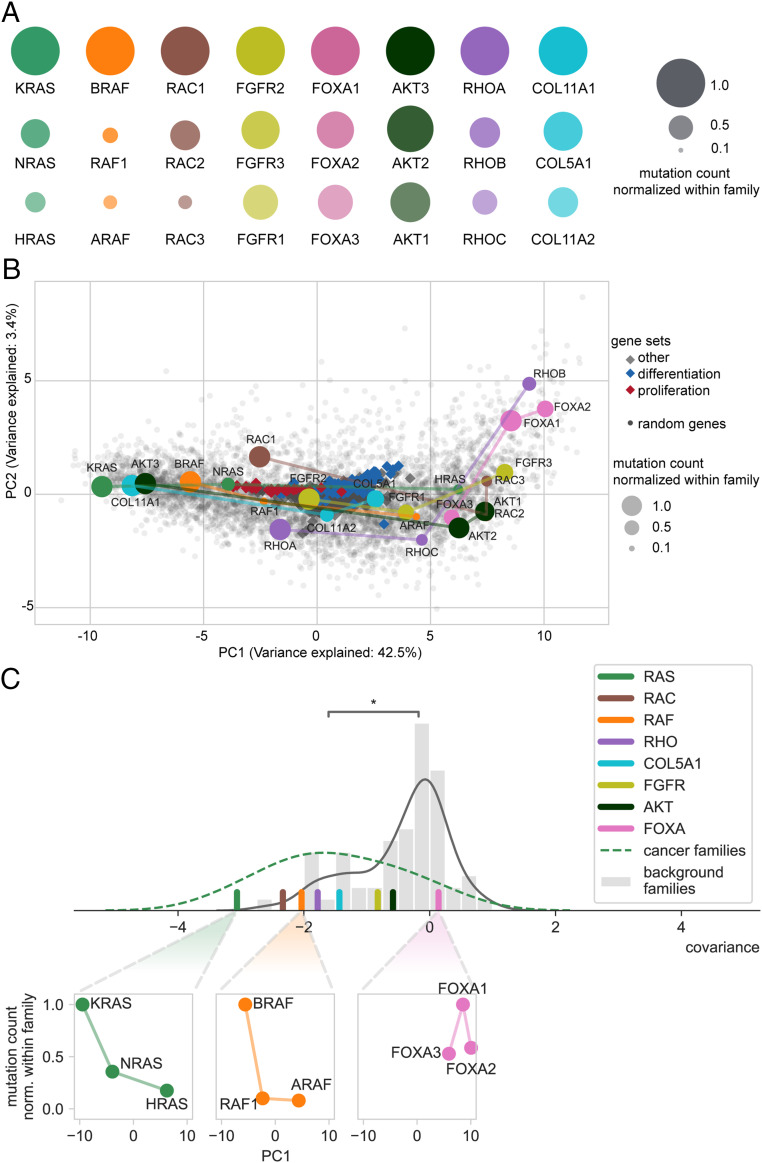
To explore whether the differences in mutation frequency observed for RAS genes also occur in other gene families, we performed a genome-wide survey of a pan-cancer dataset from The Cancer Genome Atlas (TCGA). To define the families, we clustered sets of proteins based on protein sequence similarity. We restricted the analysis to families containing at least one known cancer driver gene (2). We identified eight families, including the RAS family, that have a high degree of amino acid sequence similarity within the family (SI Appendix and Materials and Methods). We consistently observed one gene to be more frequently mutated (nonsynonymous mutation counts) than the other genes of the same family (Fig. 1A and SI Appendix, Table S1). This was especially true for the RAS, RAF, and RAC families, which showed at least a twofold variation in the mutation count number (i.e., fold change between the family members with the lowest and highest mutation count). For the RHO, FOXA, FGFR, and COL families, we observed a fold change between 1.30 and 1.95, and for the AKT family, this effect was milder, with a fold change of just 1.22.
Fig. 1.
Association between codon usage and mutation frequency in genes from eight different families. (A) Gene triplets with divergent mutation frequencies in cancer, mutation counts normalized within each family are represented. (B) PCA projection of the human codon usage. The location of each gene is determined by its codon usage. Distribution of GO gene sets along the main codon usage axis reveals the two functional poles: “proliferation” (negative PC1) and “differentiation” (positive PC1). The positions of each gene within the RAS, RAC, RAF, RHO, FGFR, AKT, COL, and FOXA families and their normalized mutation count are shown. (C) Distribution of the covariance of mutation count normalized within family and PC1 (lines are kernel density estimates as a guide for the eye). The covariance of cancer gene families is significantly more negative than that of background families (W.M.W. test, *P < 0.008). All families but one (FOXA) have a negative covariance.
As previously described, RAS genes have a high amino acid sequence identity (85%) but differ in their codon usage (15% codon identity) (19). The same observation applies to the other seven families we selected (SI Appendix, Fig. S1A). This raises the question of whether differences in mutation count could be related to variations in codon usage in addition to potential biochemical differences at the protein level.
Therefore, we investigated whether the codon usage of these genes is related to a specific translation program. Previously, Gingold et al. (22) described the average bias in codon usage for different gene functional groups and observed that genes in two cellular programs—differentiation and proliferation—preferentially use different synonymous codons. Additionally, they found that the tRNAs induced during proliferation corresponded to the codons that are enriched in the functional set of proliferation genes.
To test if functional adaptation to these cellular programs could have shaped the codon usage in the selected gene families, we examined how the codon usage of the selected genes correlates to the codon usage of proliferation- and differentiation-related genes. In order to visualize how the codon usage of the selected genes correlates with the codon usage of proproliferative genes, we used a similar approach to Gingold et al. (22) by applying principal component analysis (PCA) to the relative codon usage frequencies of all individual genes. By computing the projection of all major gene sets in the Gene Ontology (GO) classification, we reproduced the results of Gingold et al. (22) and revealed two distinct functional poles at the extremes of the codon usage main projected axis, the first principal component (PC1) (Fig. 1B). At one extreme (negative values of PC1), there is a strong enrichment of gene sets that are descendants of the “cell cycle” term (16 out of the top 30, Fisher’s exact test, two sided, P < 2.2e-17). At the other extreme (positive values of PC1), the majority of gene sets are descendants of the “multicellular organism development” or “cell differentiation” terms (14 out of the top 30, Fisher’s exact test, two sided, P < 5.8e-6). This observation, together with the previously described tRNA changes in proliferative versus nonproliferative cells (22), shows that the two poles of codon usage correspond to two cellular translation programs. We next calculated the average codon usage of each coding sequence of the selected cancer gene families and projected it in the PCA plane (Fig. 1B). In comparison to HRAS, we observed that the transcript of KRAS is composed of codons that are more frequently used by genes involved in proliferation. This seems to be a general phenomenon because the codon usage of proproliferative genes was more similar to the codon usage of the most frequently mutated family member than it was to the codon usage of the other cognate family members in all families studied except for the FOXA family. All FOXA genes have a codon usage that is at the opposite pole to proproliferation codon usage (Fig. 1B). In this family, the cancer driver gene FOXA1 can take the role of a tumor suppressor (27, 28); unlike the other families where the most frequently mutated gene is an oncogene with activating mutations, FOXA1 is typically inactivated by mutations (28) (binomial test, P < 0.007; SI Appendix, Fig. S1D). This suggests that the usage of proliferation-associated codons in cancer genes is a characteristic property of oncogenes.
Next, we wanted to assess the significance of the correlation between codon usage and mutation frequency. Our main observation was that the most frequently mutated gene member is the one that has a codon usage most adapted to the proliferation codon usage pole (see the negative pole of PC1 in the PCA projection). Thus, we expected PC1 and mutation frequency to be negatively correlated. For the 63 gene families that do not contain any cancer driver gene (noncancer gene families) but are characterized by a high degree of amino acid sequence similarity (as is the case within the cancer gene families; see Materials and Methods), we assumed that there was no specific relationship between codon usage and mutation frequency, such that the correlation should be randomly distributed around zero (SI Appendix, Fig. S1B). We also assumed that the pattern is more significant when, within a cancer gene family, both a large variation in codon usage and mutation frequency are observed. Thus, we compared the distribution of the covariance of PC1 and mutation frequency for cancer gene families to the distribution for the background gene families (Fig. 1C and Dataset S1). As the covariance tends to be large, more weight is given to the families that present a large variation in codon usage and mutation frequency. Families with little variation in either codon usage or mutation frequency, on the other hand, present a smaller covariance. We observed that the covariance of cancer gene families is significantly more negative than the covariance of background families (Wilcoxon–Mann–Whitney (W.M.W.) test, P < 0.008; SI Appendix, Fig. S1B). In particular, seven out of the eight families (RAS, RAF, COL, RAC, RHO, AKT, and FGFR) presented a negative correlation, with RAS, RAF, and RAC being the families with the highest negative covariance (Fig. 1C and SI Appendix, Fig. S1D).
Next, we examined whether the observed association between codon usage and mutation frequency was driven by positive selection acting upon mutations in proliferation-associated genes rather than by a higher background probability of mutations occurring at the loci of the cancer-associated family members. To this end, we repeated the correlation analysis using synonymous mutations, assuming that these mutations do not undergo any, or at least less selection than the nonsynonymous mutations. We observed no significant differences between background families and cancer families (W.M.W. test, P < 0.76), thereby supporting the notion that mutations in the family members with proliferation-related codon usage confer a selective advantage (SI Appendix, Fig. S1C).
We then asked whether we could use covariance to identify additional cancer genes that have not been classified as cancer drivers in Lawrence et al. (2). The covariance of two background families, LINGO and CSNK1G, was as negative as the covariance of the RAS, RAC, and RAF families (SI Appendix, Fig. S1E). Even though none of the members of these two background families have been categorized as cancer driver genes based on the mutation frequency approach we used (2), as well as in cancer driver database IntOGen (29), we found evidence that the representatives from each family with the strongest proliferation-associated codon usage (LINGO2 and CSNK1G3; SI Appendix, Fig. S1E) indeed are linked to cancer. In fact, it has been experimentally shown that an increase in the levels of LINGO2 (most frequently mutated member of the family with a proliferation-related codon usage) leads to cellular proliferation and the development of other features common to cancer cells (30). On the other hand, CSNK1G3 has been linked to hepatocellular carcinoma (31), prostate cancer (32), and renal cell carcinoma (33, 34). However, the functional role of CSNK1G3 in cancer remains to be mechanistically characterized.
In the previous analyses, we examined gene family triplets that have a comparable degree of sequence similarity to the sequence similarity found within the RAS family. Remarkably, when we extended the analysis of covariance between codon usage and mutation frequency to gene pairs (of which at least one is an oncogene), we observed a covariance of 0 or lower for all tested pairs (SI Appendix, Fig. S2). For example, the most negative covariance was found for PTPN11/PTPN6. In contrast, when examining a different set of families in which pairs had at least one tumor suppressor, three out of four pairs showed a positive covariance. This result confirms that negative covariance between mutation counts and proliferation-related codon usage is a specific feature of oncogenes (binomial test, P < 0.019).
Codon Usage-Specific Changes in KRAS Protein Abundance.
The above analyses suggest that oncogenes with a codon usage signature characteristic of proliferation-related genes are expressed at higher levels under a proliferative cell state. To test this hypothesis, we decided to examine the RAS family in further detail.
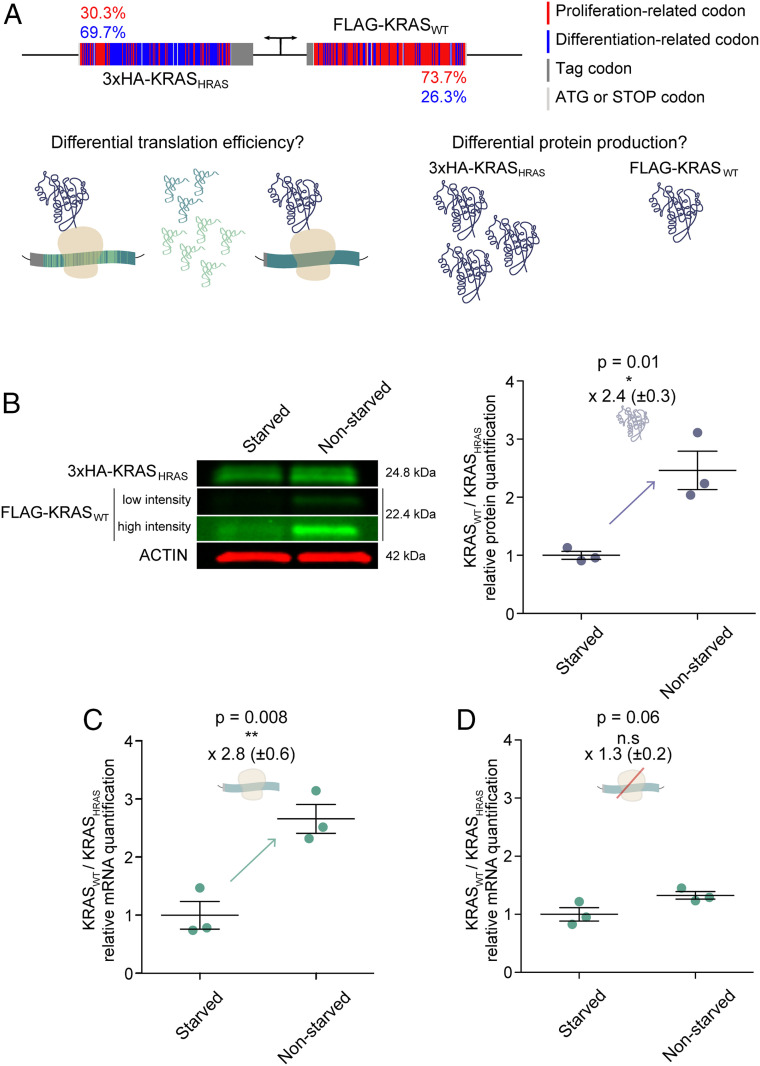
To determine if the changes in KRAS protein abundance seen in different cell states occur in a codon-dependent manner, we established a series of modified cells that coexpress KRAS wild type (KRASWT) together with a protein that is identical to KRAS in sequence but has the codon composition of HRAS (KRASHRAS) (19). The two coding sequences were cloned with different N-terminal tags (FLAG and 3×HA) so that they could be distinguished by their size. A bidirectional symmetrical promoter controls the simultaneous expression of the two genes. This design, which permits controlled expression, enabled us to exclusively assess codon-dependent changes in protein abundance while minimizing the impact of other factors (e.g., transcriptional efficiency and biochemical properties of the protein). Moreover, both genes are cloned in the same plasmid and therefore are integrated into the genome with an equal stoichiometry (Fig. 2A and Dataset S6). As a control, the experiments described below were also performed with: 1) identical constructs but with the tags swapped, and 2) identical constructs but their position switched in relation to the promoter. The latter control was designed to verify that no expression bias occurs as a result of position (SI Appendix, Fig. S3 A and B).
Fig. 2.
The codon usage of KRASWT is adapted for efficient translation during proliferation. (A) Experimental design: the construct coexpresses two genes coding for the same KRAS protein, but uses different codons. KRASWT is composed of its WT codons, whereas KRASHRAS is primarily enriched in HRAS codons. We depict how the codon composition is associated with proliferation- or differentiation-related codons. While KRASWT has a proliferation-related codon composition of 73.7%, that of KRASHRAS is 30.3%. The two genes are differentiated by size using different tags (3×HA and FLAG). The KRAS protein is represented in dark purple. Distinct tRNAs pools appear in green and blue. (B) Western blot analysis of the levels of KRASWT and KRASHRAS in starved and nonstarved BJ/hTERT cells. Low- and high-scan intensities are shown, whereby the high-scan intensity enables the visualization of the FLAG-KRASWT signal in the starved condition. The KRASWT/KRASHRAS protein ratio significantly increases from the quiescent to the proliferative state. (C) Relative quantification of KRASWT/KRASHRAS by qPCR. The KRASWT/KRASHRAS transcript ratio significantly increases between the two cell states. (D) Translation is inhibited by removing the translation initiation site and ATG site. We observe that translation inhibition reduces the effect on transcript levels. Results in B–D are representative of three independent experiments with three technical replicates each. Values are relative to the starved condition (KRASWT/KRASHRAS = 1). Error bars represent SEM. n.s, not significant; *P < 0.05; **P < 0.01 (unpaired Student t test).
Gingold et al. (22) previously reported changes in tRNA profiles of BJ/hTERT fibroblasts under different cell states: a quiescent state in which the cells are starved and a proliferative state in which they are not starved. Therefore, we first coexpressed KRASWT and KRASHRAS in BJ/hTERT fibroblasts and quantified the ratio of KRASWT/KRASHRAS protein in these two different cell states. We observed that this ratio increases by more than twofold when the cells are proliferating (Fig. 2B). This suggests that KRAS codons are more adapted for efficient translation during proliferation than HRAS codons. Similar results were found in HEK293 cells (SI Appendix, Fig. S4A). However, we did not see any significant difference in HeLa cells. The analysis of the cell cycle by flow cytometry revealed that in contrast to HeLa cells, BJ/hTERT and HEK293 cells enter the G1 phase when cells are starved (SI Appendix, Fig. S3C). As such, HeLa cells are not sensitive to serum starvation and therefore are not an adequate model to test expression changes among proliferative and nonproliferative cell states.
We also measured the KRASWT/KRASHRAS ratio at the transcript level and, interestingly, found the same effect as observed at the protein level: the ratio increases during proliferation (versus starvation) in both BJ/hTERT and HEK293 cells (Fig. 2C and SI Appendix, Fig. S4B). Previous studies have shown that in different species codon optimality has a high impact on transcript stability (35–37). An interesting hypothesis is that the dynamics of ribosomal elongation influences mRNA decay. Ribosome translocation is slower through nonoptimal transcripts and promotes mRNA decay; in Saccharomyces cerevisiae, this is mediated by the DEAD-box protein Dhh1p (38). Thus, codon content directly modulates both translation efficiency and mRNA stability. Our study suggests that KRASWT is composed of codons that are optimal for its expression in proliferative cells but nonoptimal for expression in starved cells. Therefore, to determine if changes in KRAS transcript abundance were due to differences in translation efficiency and not transcriptional regulation, we prevented translation by deleting the translation initiation site and the ATG start codon. We first confirmed that cells established with the nonproductive expression cassette had no KRAS expression (SI Appendix, Fig. S3C). After blocking the translation of the two genes, we observed that the difference in the KRASWT/KRASHRAS ratio at the transcript level was not significant between the nonstarved and starved states (Fig. 2D). Therefore, changes in the KRASWT/KRASHRAS ratio are mainly due to a differential translation efficiency (that also increases the corresponding mRNA level) between the quiescent state and the proliferative state.
Our results provide evidence supporting dynamic translational efficiency by cell state-specific codon usage of transcripts.
Effect of Differences in tRNA Levels on KRAS Abundance.
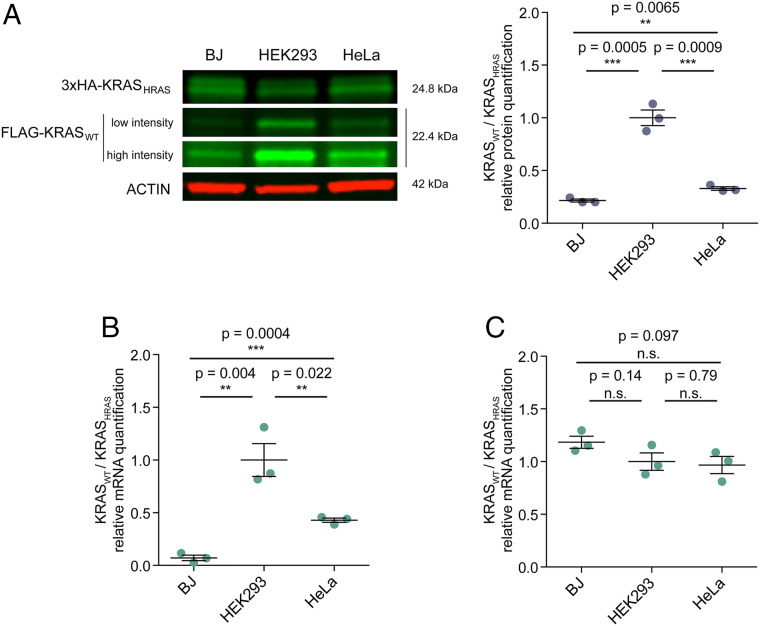
To investigate whether condition-specific translation efficiency is mediated by differential tRNA expression, we explored the effect of cell line-specific tRNA abundances on KRAS expression. One previous study (39) already reported a cell line-specific expression of KRASWT and KRASHRAS. We therefore hypothesized that the tRNA content of different cell lines varies as a function of the proliferation rate and that this in turn may influence translation efficiency in a codon-dependent manner. To test our hypothesis, we compared changes in the KRASWT/KRASHRAS ratio among BJ/hTERT, HEK293, and HeLa cells. We observed that the expression results in changes of both protein and mRNA (Fig. 3 A and B). Of these three cell lines, HEK293 exhibits the highest proliferation rate (SI Appendix, Fig. S5A) and the highest abundance of KRASWT. We also observed the same effect when switching the position of the FLAG and 3×HA tags (SI Appendix and Fig. 3A), thereby confirming that the tags had no influence. As before, the removal of the translation initiation site and start codon led to similar transcript levels in all three cell lines (Fig. 3C), indicating that translation is an important determinant of mRNA stability. Similar results were found when performing the same experiment in the RAC family after replacing the codons of RAC1 with those of RAC3 (least mutated family member); HEK293 cells presented a higher RAC1WT/RAC1RAC3 ratio than HeLa cells (SI Appendix, Fig. S5B and Dataset S6).
Fig. 3.
Codon-related changes in KRAS levels in different cell lines. (A) Western blot analysis of the levels of KRASWT and KRASHRAS in BJ/hTERT, HEK293, and HeLa cells. Low- and high-scan intensities are shown, whereby the high-scan intensity enables better visualization of the FLAG-KRASWT signal in BJ/hTERT and HeLa cells. The KRASWT/KRASHRAS protein ratio varies between the different cell lines. (B) Relative quantification of KRASWT/KRASHRAS by qPCR. The KRASWT/KRASHRAS transcript ratio also varies between the cell lines. (C) Translation inhibition reduces the differential effect on transcript level observed in the cell lines. Results in A–C are representative of three independent experiments with three technical replicates each. Values are relative to HEK293 cells (KRASWT/KRASHRAS = 1). Error bars represent SEM. n.s, not significant; **P < 0.01; ***P < 0.001 (unpaired Student t test).
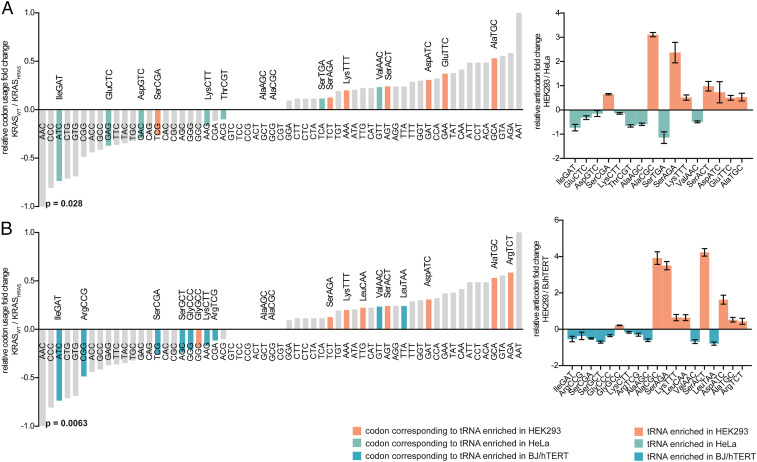
The above observations suggested that the effect of codon bias may be differentially regulated in different cell types. We therefore hypothesized that if translation efficiency is different in each cell type, it should match the cell type’s tRNA anticodon abundance. More specifically, we expected that the relative synonymous codon frequencies (relative to the amino acid) of KRASWT would more accurately match the relative abundances of cognate tRNAs in HEK293 than in HeLa or BJ/hTERT. Therefore, to associate the amount of tRNAs with codon usage, we performed hydro-tRNA sequencing (40) and quantified the relative abundance of tRNAs in BJ/hTERT, HEK293, and HeLa cells (Datasets S2 and S3). We found 16 tRNAs showing significant differences (q < 0.05, t test) between HEK293 and HeLa cells (SI Appendix, Fig. S6A and Dataset S3). Six of these tRNAs are more highly expressed in HEK293 cells than in HeLa cells and match codons enriched in the coding sequence of KRASWT (TCT, AAA, AGT, GAT, GAA, and GCA). One tRNA, tRNASerCGA, is more highly expressed in HEK293 cells but its associated codon TCG is not enriched in KRASWT. On the other hand, five of the tRNAs whose expression levels were significantly higher in HeLa cells corresponded to codons enriched in HRAS and therefore in KRASHRAS (ATC, GAG, GAC, ACG, and AAG). Only two tRNAs with a higher expression in HeLa cells (tRNASerTGA and tRNAValAAC) did not match codons enriched in KRASHRAS (Fig. 4A). Overall, we found 11 codons matching the expected tRNAs (P < 0.028, binomial test). Similarly, we found seven of the tRNAs that were more highly expressed in HEK293 cells than in BJ/hTERT cells (SI Appendix, Fig. S6B and Dataset S3) to match the codons enriched in KRASWT (TCT, AAA, AGT, TTG, GAT, GCA, and AGA). One exception was tRNAGlyCCC, which is more highly expressed in HEK293 cells but the associated codon GGG is not enriched in KRASWT. With regards to those tRNAs whose expression levels were significantly higher in BJ/hTERT cells, seven corresponded to enriched codons in KRASHRAS (ATC, CGG, TCG, AGC, CGA, GGG, and AAG). Overall, we found 14 out of 16 codons matching the expected tRNAs when comparing HEK293 and BJ/hTERT (P < 0.0063, binomial test; Fig. 4B). Therefore, the difference in relative tRNA supply between cell lines could explain the observed variation in protein levels of KRASWT and KRASHRAS. In a previous study where different codons of KRAS were changed, it was observed that certain replacements resulted in significant increases in KRAS expression, cumulatively reaching the levels of HRAS (19). These codon changes included GCA to GCC, AAA to AAG, and ATT to ATC, which correspond to the anticodons of tRNAs that are differentially expressed between HEK293, BJ/hTERT, and HeLa cells. In contrast, changing GAA to GAG and CCT to CCC did not result in any protein abundance changes (SI Appendix, Fig. S6C).
Fig. 4.
Association of differentially expressed tRNAs and relative codon usage of KRASWT and KRASHRAS. (A) Log2 fold change of the relative codon usage (pseudocount +1) between KRASWT and KRASHRAS. Codons corresponding to tRNAs that are differentially expressed between HEK293 and HeLa cells are highlighted. The Right represents the log2 fold change of relative tRNA abundance of the tRNAs that are differentially expressed between HEK293 and HeLa cells. (B) Log2 fold change of the relative codon usage (pseudocount +1) between KRASWT and KRASHRAS. Codons corresponding to tRNAs that are differentially expressed between HEK293 and BJ/hTERT are highlighted. The Right represents the log2 fold change of relative tRNA abundance of the tRNAs that are differentially expressed between HEK293 and BJ/hTERT cells. Error bars represent SEM of three independent experiments. Binomial tests were performed in A and B (one-sided, P values shown below plots) by calculating the probability of the correct number of associations between relative tRNA expression and codon usage.
Additionally, we investigated whether the codons corresponding to the tRNAs that significantly change are also enriched in the most prevalent oncogenes of the RAS, RAF, and RAC families. Overall, the AAA, GCA, GAA, AGT, and GAT codons were enriched in KRAS, BRAF, and RAC1, and the matching tRNAs had significantly higher levels in HEK293 cells (SI Appendix, Fig. S7A). Conversely, the codons enriched in HRAS, RAF, and RAC3 (ACG, AAG, GAG, and GAC) matched the tRNAs in HeLa (SI Appendix, Fig. S7A). This leads to a significant association between relative tRNA abundance and relative codon usage (P < 0.0018, binomial test; SI Appendix, Fig. S7B). Similarly, we found a significant relationship between the relative tRNA abundance of HEK293 and BJ/hTERT cells and the codon usage of the three families (P < 0.0035, binomial test; SI Appendix, Fig. S8 A and B). Next, we repeated the analysis considering wobble base pairs, which are known to play an important role in codon–anticodon interactions. We calculated normalized weights within each amino acid family (Materials and Methods). Again, we found a significant association to the codon usage of the studied genes (SI Appendix, Figs. S9–S11 and Dataset S4). Finally, we associated proliferation-specific codons to the tRNAs that are differentially expressed in the studied cell lines. For example, we found that AspATC and LysTTT from HEK293 cells match the codons that are at the extreme of the proliferation pole (SI Appendix, Fig. S12).
Finally, as our analyses hinted toward a link between an adaptation of KRAS translation to proliferation and positive selection for mutations in cancer, we tested if the RAS genes differ in their mutation frequencies between low- and high-proliferating cancer types. The expression of KI-67 is associated with cell proliferation and growth (41). Therefore, we estimated cancer type-specific proliferation rates by averaging the expression values of KI-67 in tumor samples from the TCGA and divided cancer types into those with low and those with high proliferation. We observed that compared to NRAS and HRAS, KRAS has significantly higher mutation rates in high proliferation tumors (P < 2.2e-16, chi-squared test; SI Appendix, Fig. S13 A and B). We next asked whether the cancer type-specific association between KI-67 expression and KRAS mutation frequency holds true at the patient level. To this end, we stratified samples by the mutation status of RAS isoforms in six cancer types (Materials and Methods) and compared KI-67 expression between the groups (SI Appendix, Fig. S13C). In all cancer types, KRAS-mutated samples were associated with a higher KI-67 expression than NRAS-mutated samples, which in turn showed a higher KI-67 expression than HRAS-mutated samples. The groupwise expression differences for each cancer type were not or only marginally significant due to the high variation within the groups and the low number of HRAS-mutated samples. However, all comparisons followed the expected trend of median KI-67 expression in KRAS-mutated samples > median KI-67 expression in NRAS-mutated samples > median KI-67 expression in HRAS-mutated samples (P = 0.008, binomial test).
Altogether, our results provide evidence to support a dynamic translational program in which specific changes in tRNA abundance can shape the expression of proliferation-related transcripts.
Discussion
Codon usage and tRNA abundance are crucial for efficient and accurate translation of mRNA into protein. Previous studies have found that tRNA repertoires are dynamic in a manner that facilitates selective translation of specific transcripts (22–24, 26). Here, we investigate whether an oncogenic translation program shapes the abundance of cancer driver genes. We describe protein families that have strong differences in both codon usage and mutation frequencies within the family. The observed codon bias reveals a proliferation-specific codon usage by the family members that are more prevalent in cancer. Specifically, the RAS, RAF, and RAC families exhibit the largest negative covariance between mutation frequency and proliferation-associated codon usage. Additionally, we observed the same pattern in cancer families with two members (e.g., PTPN) and in families that have been shown to be involved in cancer (e.g., LINGO) despite not being classified as cancer drivers. This raises the question of whether these transcripts are more effectively translated in proliferative cells than their closely related family members.
We focused on the RAS family and experimentally showed that the translation efficiency of KRASWT is up-regulated in proliferative cells in comparison to the translation efficiency of KRASHRAS. This same tendency was also observed with RAC1WT and RAC1RAC3. Additionally, we found that translation efficiency is a determinant of transcript abundance. This observation has been described previously in Homo sapiens (37), S. cerevisiae (35) and Escherichia coli (36). Here, we consistently show that the changes in KRASWT/KRASHRAS transcript abundance between cell types and cell states decrease when translation is suppressed.
Our observations are in agreement with recently identified alterations in transcript-specific translation that emerge as drivers of cellular transformation. For example, it has been shown that up-regulation of specific tRNAs (tRNAGluTTC and tRNAArgCCG) in metastatic cells leads to an increase in the amount of certain proteins, specifically EXOSC2 and GRIPAP1, that play an important role in metastasis (23). Indeed, we observed similar results here for KRAS. Consistent with previous reports (42, 43), we also observed that specific tRNAs vary between different cell lines, which could explain the differences in KRAS expression between BJ/hTERT, HEK293, and HeLa cells. One of these tRNAs, tRNAGluTTC, is also up-regulated in metastatic breast cancer cells as mentioned above. Moreover, we found differentially expressed tRNAs corresponding to codons that were previously reported to change KRAS protein levels when synonymously mutated (19). Taken together, our results suggest that the expression of only a few specific tRNAs, rather than the expression of multiple tRNAs, is required to increase KRAS translation efficiency. Particularly, we observed that codons corresponding to the tRNAs that mediate these changes are also enriched in the BRAF and RAC1 oncogenes. Therefore, our results suggest that certain tRNAs could be used as markers of oncogene-specific translation. Determining the tRNA abundance of different cell types may reveal previously unseen connections between translation and oncogene prevalence in cancer. It would also be interesting to investigate how tRNA modifications could influence oncogene translation. Furthermore, Supek et al. (44) showed that selection acts on somatic synonymous mutations of oncogenes in tumor evolution. In many cases these synonymous mutations are associated with changes in oncogene splicing in tumors. Importantly, synonymous mutations in KRAS have been shown to impact its expression (45) and transforming potential (46). It would be interesting to further investigate whether some of the recurrent synonymous mutations in oncogenes correspond to changes that enrich their coding sequence with proliferation-related codons, thereby yielding a greater translation efficiency. Importantly, it should be considered that the translational output from any given mRNA is also influenced by its secondary structure aside from codon usage. Disentangling the individual roles played by each of these two factors in translation remains difficult.
Activating mutations of oncogenes are a product of the selection that occurs during tumor initiation to produce the ideal level of signaling. It is plausible that selection acting upon a gene depends on the level of expression of that gene as well as on the function of the gene product itself. However, it is difficult to disentangle how differences in protein sequences and expression levels of RAS family members contribute to RAS protein-specific roles and selection advantage. The physiological role of the RAS proteins during development is intriguing because KRAS for example is the only family member that is embryonically lethal in homozygous null mice (17). This feature is also observed within the RAC family (47, 48). Interestingly, when the KRAS locus is substituted to encode the HRAS protein after exon 2 (i.e., KRAS codons are conserved in exon 1 and exon 2), mice are viable, yet display cardiomyopathies in adulthood (18). Relatedly, Pershing et al. (49) observed that upon replacing KRAS codons with HRAS codons in one exon, mice became more resistant to lung tumors and the number of KRAS mutations decreased. It is therefore likely that in addition to subtle differences in particular functions, the changes in the levels of the RAS proteins are important determinants of selection during tumor evolution. Specifically, our results suggest that translation efficiency might contribute to mutation frequency differences between genes. It is therefore tempting to speculate that cancer cells could take advantage of the translation program of dividing cells to boost the translation of transcripts to their own growth advantage.
Our results indicate that in oncogene families the gene most frequently mutated in cancer is the one that has a codon usage adapted to cell proliferation. There are several possible explanations for this. The most obvious is that oncogenes with a codon usage adapted to cell proliferation will be more expressed in dividing cells like cancer than their counterparts and therefore undergoing stronger selection to be mutated in cancer. An alternative explanation is that in oncogene families some members have different biochemical properties that favor cell division compared to the others. Overexpression of these genes in resting cells could promote tumorigenesis and therefore they evolved to have a codon usage that favors translation only in cells which are dividing (i.e., in embryogenesis or fast dividing cells like colon epithelial cells). These different possible explanations for our observations should be tested experimentally to elucidate which one is correct.
It has already been suggested that differences in codon usage might be related to the imbalance of mutation frequencies within the RAS family; the constitutively activated form of highly translated HRAS could lead to an overactivation of the MAPK pathway, ultimately leading to oncogene-induced senescence and therefore a possible favoring of KRAS mutations (50). On the other hand, an overactivation of RAS signaling can be beneficial for many tumors. Maintaining the balance between growth advantage and senescence evasion creates a sweet spot of optimal RAS signaling activation (51) (SI Appendix, Fig. S16). Our observation that genes adapted for optimized translation in proliferating cancer cells are under stronger positive selection for activating mutations suggests that in addition to the type of activating mutation, protein abundance is also important to place KRAS in the sweet spot for optimal signaling (51, 52).
Taken together, our work not only addresses a fundamental aspect of RAS biology but also provides insight into the controversial issue of how codon bias can influence protein expression. Collectively, our findings demonstrate that codon-driven translation efficiency can modulate protein expression of oncogenes in different cell contexts.
Materials and Methods
Data sources, computational analysis, sample preparation, and experimental procedures are fully described in SI Appendix, Materials and Methods.
Paralogs Ensembl.
To define gene families, we retrieved information regarding protein sequence similarity and family membership from Ensembl. Further details are provided in SI Appendix.
TCGA.
Mutation data were obtained from TCGA. We retrieved somatic mutations in coding regions for 20 types of cancer. Further details are provided in SI Appendix.
Cancer Gene Catalog.
We considered cancer driver genes to be those genes that had a significant (q < 0.01) number of nonsilent mutations in at least 1 out of 21 cancer types in 4,742 patients as defined by Lawrence et al. (2).
Coding Sequences.
The coding sequences of H. sapiens were downloaded from the Consensus Coding Sequence (CCDS) project (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/), release 2016/09/08. In the case of noncancer genes, one unique canonical coding sequence was arbitrarily chosen for each protein based on Uniprot mapping to the CCDS. For those genes belonging to the selected cancer gene families, the canonical coding sequence was chosen according to the corresponding canonical protein as defined in Uniprot.
GO Gene Sets.
Gene ontology was downloaded as a MySQL dump of the amiGO database, release 2017/01, and human gene annotations were downloaded from the amiGO database, release 2018/01/04. Further details are provided in SI Appendix.
Codon Usage PCA.
We applied PCA to the relative synonymous codon frequencies (52) of all individual human coding sequences. Further details are provided in SI Appendix.
Quantification of tRNA Expression.
tRNAseq mapping was performed using a specific pipeline for tRNAs (53). The basic pipeline was adapted to paired-end sequencing data. Further details are provided in SI Appendix.
Statistical Analyses.
For hypothesis testing, we performed two-sided Student t tests, two-sided Wilcoxon–Mann–Whitney tests, and one-sided binomial tests. In the differential expression analyses, a false discovery rate (FDR) correction was used to account for multiple testing.
Hydro-tRNA Sequencing.
Total RNA from HEK293, BJ/hTERT, and HeLa cells were extracted using the miRNeasy Mini kit (Qiagen). For each sample, 20 µg of total RNA was treated following the hydro-tRNAseq protocol (40). Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Eva Maria Novoa and Manuel Irimia for stimulating and critical discussions. We thank Disa Tehler for providing BJ/hTERT cells. We would also like to thank Tony Ferrar for scientific and language editing. The work of X.H.-A. has been supported by a PhD fellowship from the Fundación Ramón Areces. We acknowledge the support of the Spanish Ministry of Science and Innovation to the European Molecular Biology Laboratory partnership, the Centro de Excelencia Severo Ochoa and the Centres de Recerca de Catalunya Programme/Generalitat de Catalunya.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016119117/-/DCSupplemental.
Data Availability.
All data generated during this study are included in this published article and its SI Appendix. Sequencing data generated during this study have been deposited in ArrayExpress database (54) under accession number E-MTAB-8144. BJ/hTERT sequencing data are available from Gene Expression Omnibus (55) under the accession code GSE137834 (56). All TCGA data (57) are available for download through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Codon usage of oncogenes and tRNA mapping are reported in the GitHub repository: https://github.com/webermarcolivier/codon_usage_oncogenes; https://github.com/hexavier/tRNA_mapping.
References
- 1.Nowell P. C., The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Lawrence M. S., et al. , Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hobbs G. A., Der C. J., Rossman K. L., RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129, 1287–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ninomiya Y., et al. , K-Ras and H-Ras activation promote distinct consequences on endometrial cell survival. Cancer Res. 64, 2759–2765 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Fotiadou P. P., Takahashi C., Rajabi H. N., Ewen M. E., Wild-type NRas and KRas perform distinct functions during transformation. Mol. Cell. Biol. 27, 6742–6755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parikh C., Subrahmanyam R., Ren R., Oncogenic NRAS, KRAS, and HRAS exhibit different leukemogenic potentials in mice. Cancer Res. 67, 7139–7146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haigis K. M., et al. , Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600–608 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q., et al. , Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood 117, 2022–2032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten M., et al. , H-Ras and K-Ras oncoproteins induce different tumor spectra when driven by the same regulatory sequences. Cancer Res. 77, 707–718 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahearn I., Zhou M., Philips M. R., Posttranslational modifications of RAS proteins. Cold Spring Harb. Perspect. Med. 8, a031484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apolloni A., Prior I. A., Lindsay M., Parton R. G., Hancock J. F., H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20, 2475–2487 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J., Roy S., Apolloni A., Lane A., Hancock J. F., Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 273, 24052–24056 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Voice J. K., Klemke R. L., Le A., Jackson J. H., Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 274, 17164–17170 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Wang M.-T., et al. , K-ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell 163, 1237–1251 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Omerovic J., Hammond D. E., Clague M. J., Prior I. A., Ras isoform abundance and signalling in human cancer cell lines. Oncogene 27, 2754–2762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koera K., et al. , K-ras is essential for the development of the mouse embryo. Oncogene 15, 1151–1159 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Potenza N., et al. , Replacement of K-ras with H-ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 6, 432–437 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lampson B. L., et al. , Rare codons regulate KRas oncogenesis. Curr. Biol. 23, 70–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson G., Coller J., Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rak R., Dahan O., Pilpel Y., Repertoires of tRNAs: The couplers of genomics and proteomics. Annu. Rev. Cell Dev. Biol. 34, 239–264 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Gingold H., et al. , A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Goodarzi H., et al. , Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165, 1416–1427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torrent M., Chalancon G., de Groot N. S., Wuster A., Madan Babu M., Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 11, eaat6409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bornelöv S., Selmi T., Flad S., Dietmann S., Frye M., Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 20, 119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guimaraes J. C., et al. , A rare codon-based translational program of cell proliferation. Genome Biol. 21, 44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri C. E., et al. , Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder M. P., Rubio-Perez C., Tamborero D., Gonzalez-Perez A., Lopez-Bigas N., OncodriveROLE classifies cancer driver genes in loss of function and activating mode of action. Bioinformatics 30, i549–i555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Perez A., et al. , IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods 10, 1081–1082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo J. H., et al. , Novel gastric cancer stem cell-related marker LINGO2 is associated with cancer cell phenotype and patient outcome. Int. J. Mol. Sci. 20, 555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bard-Chapeau E. A., et al. , Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat. Genet. 46, 24–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S., et al. , Widespread and functional RNA circularization in localized prostate cancer. Cell 176, 831–843.e22 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Masuda K., et al. , Downregulation of Cap43 gene by von Hippel-Lindau tumor suppressor protein in human renal cancer cells. Int. J. Cancer 105, 803–810 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Nishie A., et al. , High expression of the Cap43 gene in infiltrating macrophages of human renal cell carcinomas. Clin. Cancer Res. 7, 2145–2151 (2001). [PubMed] [Google Scholar]
- 35.Presnyak V., et al. , Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boël G., et al. , Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529, 358–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Q., et al. , Translation affects mRNA stability in a codon-dependent manner in human cells. elife 8, e45396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radhakrishnan A., et al. , The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122–132.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu J., Dang Y., Counter C., Liu Y., Codon usage regulates human KRAS expression at both transcriptional and translational levels. J. Biol. Chem. 293, 17929–17940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gogakos T., et al. , Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholzen T., Gerdes J., The Ki-67 protein: From the known and the unknown. J. Cell. Physiol. 182, 311–322 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Dittmar K. A., Goodenbour J. M., Pan T., Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavon-Eternod M., et al. , tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37, 7268–7280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Supek F., Miñana B., Valcárcel J., Gabaldón T., Lehner B., Synonymous mutations frequently act as driver mutations in human cancers. Cell 156, 1324–1335 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Sharma Y., et al. , A pan-cancer analysis of synonymous mutations. Nat. Commun. 10, 2569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters A. M., Bagni R., Portugal F., Hartley J. L., Single synonymous mutations in KRAS cause transformed phenotypes in NIH3T3 cells. PLoS One 11, e0163272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugihara K., et al. , Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 17, 3427–3433 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Corbetta S., et al. , Generation and characterization of Rac3 knockout mice. Mol. Cell. Biol. 25, 5763–5776 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pershing N. L. K., et al. , Rare codons capacitate Kras-driven de novo tumorigenesis. J. Clin. Invest. 125, 222–233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bodemann B. O., White M. A., Ras GTPases: Codon bias holds KRas down but not out. Curr. Biol. 23, R17–R20 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Li S., Balmain A., Counter C. M., A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 18, 767–777 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Sharp P. M., Li W. H., The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann A., et al. , Accurate mapping of tRNA reads. Bioinformatics 34, 2339 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Kolesnikov N., et al. , ArrayExpress update–simplifying data submissions. Nucleic. Acids Res. 43, D1113–D1116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic. Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hernandez-Alias X., Benisty H., Schaefer M. H., Serrano L., Translational efficiency across healthy and tumor tissues is proliferation-related. Mol. Syst. Biol. 16, e9275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinstein J. N. et al.,Cancer Genome Atlas Research Network , The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article and its SI Appendix. Sequencing data generated during this study have been deposited in ArrayExpress database (54) under accession number E-MTAB-8144. BJ/hTERT sequencing data are available from Gene Expression Omnibus (55) under the accession code GSE137834 (56). All TCGA data (57) are available for download through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Codon usage of oncogenes and tRNA mapping are reported in the GitHub repository: https://github.com/webermarcolivier/codon_usage_oncogenes; https://github.com/hexavier/tRNA_mapping.