Significance
Meiotic crossovers are essential for the production of gametes with balanced chromosome content. MutLγ (Mlh1–Mlh3) endonuclease, a mismatch repair heterodimer, also functions during meiosis to generate crossovers. Its activity requires Exo1 as well as the MutSγ heterodimer (Msh4–Msh5). Crossovers also require the polo kinase Cdc5 in a way that is poorly understood. We show that Exo1 directly interacts with Cdc5, and that this promotes the activation of MutLγ for crossovers. Since Cdc5 also controls other key meiotic processes, including kinetochores mono-orientation and disassembly of the synaptonemal complex, this direct interaction with the crossover proteins may be a way to coordinate meiotic chromosome segregation events to avoid untimely activation of the MutLγ endonuclease once recruited to future crossover sites.
Keywords: recombination, meiosis, crossovers, polo kinase, MutL
Abstract
Crossovers generated during the repair of programmed meiotic double-strand breaks must be tightly regulated to promote accurate homolog segregation without deleterious outcomes, such as aneuploidy. The Mlh1–Mlh3 (MutLγ) endonuclease complex is critical for crossover resolution, which involves mechanistically unclear interplay between MutLγ and Exo1 and polo kinase Cdc5. Using budding yeast to gain temporal and genetic traction on crossover regulation, we find that MutLγ constitutively interacts with Exo1. Upon commitment to crossover repair, MutLγ–Exo1 associate with recombination intermediates, followed by direct Cdc5 recruitment that triggers MutLγ crossover activity. We propose that Exo1 serves as a central coordinator in this molecular interplay, providing a defined order of interaction that prevents deleterious, premature activation of crossovers. MutLγ associates at a lower frequency near centromeres, indicating that spatial regulation across chromosomal regions reduces risky crossover events. Our data elucidate the temporal and spatial control surrounding a constitutive, potentially harmful, nuclease. We also reveal a critical, noncatalytic role for Exo1, through noncanonical interaction with polo kinase. These mechanisms regulating meiotic crossovers may be conserved across species.
Meiotic recombination provides crossovers that allow homologous chromosomes to be transiently physically attached together and then properly segregated (1). A lack or altered distribution of meiotic crossovers is a major source of aneuploidies, leading to sterility or disorders, such as Down syndrome, providing strong impetus for understanding how crossovers are controlled during meiosis. Meiotic recombination is triggered by the formation of programmed DNA double-strand breaks (DSBs), catalyzed by Spo11 together with several conserved protein partners (2). Following Spo11 removal, 5′ DSB ends are resected and the 3′ ends invade a homologous template, leading to formation of D-loop intermediates. After DNA synthesis, many of these intermediates are dismantled by helicases and repaired without a crossover (3). However, a subset is stabilized, and upon capture of the second DSB end, mature into a double Holliday junction (dHJ), which is almost exclusively resolved as a crossover (4, 5). Essential to this crossover pathway are a group of proteins, called ZMMs, which collectively stabilize and protect recombination intermediates from helicases (3, 6–8). The mismatch repair MutLγ (Mlh1–Mlh3) heterodimer is proposed to subsequently act on these ZMM-stabilized dHJs to resolve them into crossovers (9). Accordingly, MutLγ foci reflect the number and distribution of crossovers on both mammalian and plant meiotic chromosomes (reviewed in ref. 10).
In a distinct process, eukaryotic mismatch repair, a MutS-related complex (MutSα or MutSβ) recognizes a mismatch, and recruits a MutL-related complex to initiate repair together with proliferating cell nuclear antigen and Exo1 (reviewed in ref. 11). MutLα represents the major mismatch repair activity, which involves its endonuclease activity (12, 13). The related MutLγ is instead essential to meiotic crossover, a process that remains relatively ill-defined, although MutLγ endonuclease activity is required for crossover (9, 14–16). Despite its importance and cross-species relevance, the mechanism of dHJ resolution and crossover formation by MutLγ remains unknown. MutLγ alone does not resolve HJs in vitro (15–17), and may need additional partners, meiosis-specific posttranslational modifications, and a specific DNA substrate to promote specific nuclease activity and hence crossover formation.
Based on recent in vitro experiments and genome-wide analysis of meiotic recombination, it seems that rather than acting as a canonical resolvase, MutLγ may nick the DNA, which could result in dHJ resolution and crossover formation if two closely spaced nicks on opposite strands are made (17, 18). In vitro analyses with recombinant proteins also suggest that MutLγ may need to extensively polymerize along DNA in order to activate its DNA cleavage activity (17).
MutLγ partners with the ZMM MutSγ heterodimer (Msh4–Msh5), proposed to first stabilize recombination intermediates and then to work together with MutLγ for crossover resolution (reviewed in refs. 8 and 11). In addition, the exonuclease Exo1 interacts in vitro with Mlh1. Previous work showed that Exo1’s ability to interact with Mlh1 is important for MutLγ function in crossover formation (19). This leaves the mechanism and role of Exo1 in meiotic crossovers unclear, although broad genetic analysis suggests a noncatalytic role given that Exo1-null yeast show reduced crossovers and reduced viable gametes, but catalytic mutants of Exo1 do not (9, 20). Similarly, Exo1-null mice are sterile, but not Exo1 catalytic mutants, highlighting the conservation of this function (21, 22). How Exo1 is required for activating MutLγ is not known. Finally, the polo-like kinase Cdc5 (homolog of human PLK1) is required for meiotic crossovers (23). Cdc5 expression is induced by the Ndt80 transcription factor at the pachytene stage of meiotic prophase, once homolog synapsis and recombination intermediates formation are achieved (24–27). Cdc5 promotes multiple steps during exit from pachytene, including dHJ resolution, disassembly of the synaptonemal complex, and sister kinetochore mono-orientation (28–30). Among other targets, Cdc5 activates the Mus81–Mms4 structure-specific nuclease, by phosphorylating Mms4 (31), but this produces only a minority of crossovers. It is not known how Cdc5 promotes the remaining crossovers thought to come from the major, MutLγ-dependent, pathway.
In order to define how these factors come together to control crossovers, we undertook a comprehensive analysis of MutLγ activation and distribution genome-wide in budding yeast. We found that MutLγ and Exo1 form a constitutive complex that associates with recombination sites stabilized by ZMM proteins. Exo1 then interacts with the Cdc5 kinase through a direct, specific contact that is necessary for crossover formation by MutLγ. Finally, we found that MutLγ-dependent crossover formation is regulated at multiple levels to avoid regions around centromeres where deleterious outcomes could result.
Results
Mlh3 Foci on Yeast Meiotic Chromosomes Are Distinct from ZMM Foci.
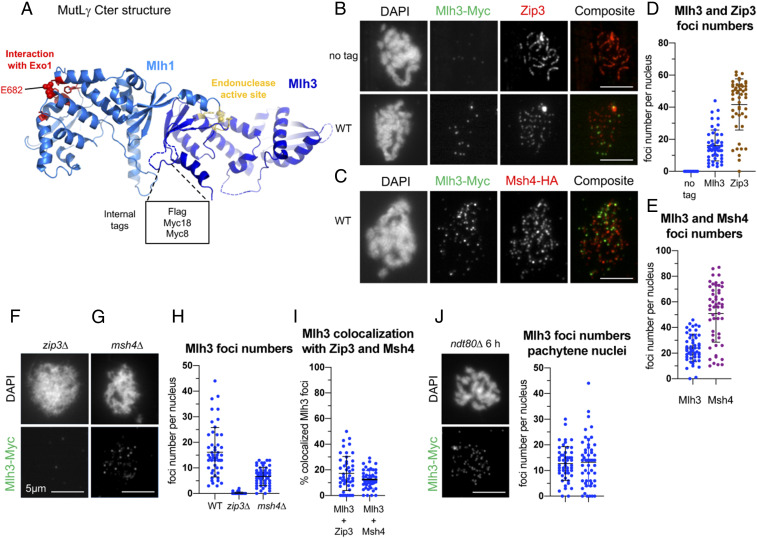
To follow how MutLγ is initially recruited, we examined its relationship to ZMM proteins, Zip3 and Msh4. A recent crystal structure of the C-terminal domain of Mlh1-3 shows that the last residues of Mlh1 are part of the Mlh3 endonuclease active site, rendering it impossible to C-terminally tag Mlh1 (Fig. 1A) (32). To preserve Mlh1-3 functionality in vivo, we introduced an internal tag at an unstructured loop in Mlh3 that is predicted not to affect Mlh1-3 function and catalytic activity (Fig. 1A). Tags inserted at this site had no major effect on mismatch repair and no effect on spore viability or meiotic crossover frequency, and are therefore suitable for further in vivo studies (SI Appendix, Fig. S1).
Fig. 1.
Mlh3 forms foci on yeast pachytene meiotic chromosomes, distinct from ZMM foci. (A) Crystal structure of the C-terminal region of Saccharomyces cerevisiae Mlh1–Mlh3 heterodimer showing the position of the internal tags in Mlh3. The Mlh1 and Mlh3 regions are colored in light and dark blue, respectively. The Mlh1 binding motif for Exo1 and the endonuclease site of Mlh3 are colored in red and yellow, respectively. (B) Comparison of Mlh3–Myc18 and Zip3 foci. (C) Comparison of Mlh3–Myc18 and Msh4-HA foci. (D and E) Quantification of Mlh3, Zip3, and Msh4 foci from B and C. (F and G): Mlh3-myc foci in zip3∆ (F) and msh4∆ (G). (H) Quantification of Mlh3-Myc foci in zip3∆ and msh4∆ mutants from B, F, and G; 51 nuclei examined in each. (I) Colocalization quantification of Mlh3 with Zip3 or Msh4 foci. The percent of Mlh3 foci colocalizing is indicated. (J) Quantification of Mlh3 foci in pachytene-selected nuclei of wild-type (same as in B) and ndt80∆; 51 nuclei examined in each. (B–J) All experiments at 4 h in meiosis, except ndt80∆ (6 h in meiosis). Scale bars: 5 μm. See also SI Appendix, Fig. S1.
As in plants and mammals, Mlh3 formed foci on yeast meiotic chromosomes (Fig. 1 B–E). Importantly, Mlh3 foci were largely absent in zip3∆ and in substantially reduced numbers in msh4∆ mutants (Fig. 1 F–H), in agreement with genetic data showing that MutLγ acts downstream of ZMM proteins. Surprisingly, although Zip3 and to a lesser extent Msh4 are important for Mlh3 foci, Mlh3 showed very little colocalization with either Zip3 or Msh4 (Fig. 1 B, C, and I), suggesting that Mlh3 coexists only briefly with the other two components at the same intermediates. We conclude that Zip3 and Msh4 ZMM are required for normal Mlh3 foci formation, and propose that Zip3 and Msh4 form earlier foci on recombination intermediates, before Mlh3 binding, which may reflect their function for stabilization of intermediates.
Mlh3 Associates with Meiotic DSB Hotspots at a Late Step of Recombination.
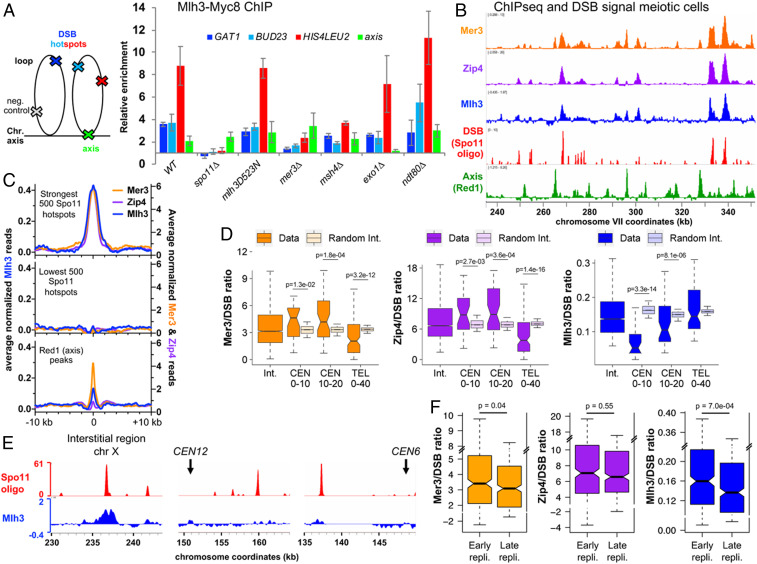
We next examined Mlh3 association with specific loci by chromatin immunoprecipitation (ChIP). Mlh3 associated with the three tested DSB hotspots during meiosis, reaching a maximum at the expected time of crossover formation (4 to 5 h) (Fig. 2A and SI Appendix, Fig. S2A) (5), with kinetics very similar to the ZMM proteins (33, 34). Mlh3 also weakly associated with the chromosome axis, where DSB sites transiently relocate during recombination (35, 36) (Fig. 2A). Mlh3 association with DSB hotspots required DSB formation, since it was not observed in a spo11∆ mutant (Fig. 2A and SI Appendix, Fig. S2B), but was independent of its nuclease activity (mlh3-D523N mutant) (Fig. 2A and SI Appendix, Fig. S2C). Mlh3 binding also required the ZMM protein, Mer3 (Fig. 2A and SI Appendix, Fig. S2D). In msh4∆ mutants, Mlh3 recruitment was strongly reduced (Fig. 2A and SI Appendix, Fig. S2E), consistent with the reduced number of Mlh3 foci in msh4∆ (Fig. 1 G and H). In contrast, Mlh3 bound at hotspots at nearly wild-type levels in exo1∆ mutants (Fig. 2A and SI Appendix, Fig. S2F). Finally, to understand how and when it functions, we assessed the role of Cdc5 polo kinase, required for meiotic crossover, in recruiting Mlh3 to recombination sites. We found that in a ndt80∆ mutant, where Cdc5 is not expressed, Mlh3 was still recruited at DSB sites (Fig. 2A and SI Appendix, Fig. S2G) and also formed foci on pachytene chromosomes (Fig. 1J). We conclude that the lack of dHJ resolution in the absence of Cdc5 is not due to a failure to recruit Mlh1-3 to recombination sites, but possibly a failure to activate Mlh1-3 once recruited.
Fig. 2.
Mlh3 associates with meiotic DSB hotspots and its distribution is influenced locally by specific chromosome features. (A) Mlh3–Myc levels at the three indicated meiotic DSB hotspots and one axis-associated site relative to a negative control site (NFT1) assessed by ChIP and qPCR during meiotic time courses. Values are the mean ± SEM from three (four in wild-type) independent experiments at the time-point of maximum enrichment. The full corresponding time courses are in SI Appendix, Fig. S2. The cartoon illustrates the position of sites analyzed by qPCR relative to the meiotic chromosome structure. (B) ChIP-seq binding of Mlh3 (at 5.5 h in meiosis) compared to the binding of the Mer3 and Zip4 (33) ZMMs, to DSBs (Spo11 oligos) (82), and to axis sites (Red1 ChIP-seq) (72). Normalized data are smoothed with a 200-bp window. (C) Average ChIP-seq signal at the indicated features. Same data as in B). The Mer3 and Zip4 ChIP-seq signals are aligned on the Spo11 hotspots midpoints from ref. 82, and Mlh3 ChIP-seq signal on the pCUP1-IME1 Spo11 hotspots midpoints (this study). (Bottom) ChIP-seq signal is aligned on the Red1 peaks summits from ref. 72. (D) ZMM and Mlh3 signals per DSB vary with the proximity to a centromere or a telomere. The ChIP-seq signal of each protein divided by their corresponding Spo11 oligo signal (Spo11 signal from ref. 82 for Mer3 and Zip4, and pCUP1-IME1 Spo11 signal for Mlh3) was computed on the width plus 1 kb on each side of the strongest corresponding 2,000 Spo11 hotspots at the indicated chromosome regions: Interstitial (1,805 and 1,829 hotspots); 0 to10 kb from a centromere (29 and 28 hotspots); 10 to 20 kb from a centromere (50 and 46 hotspots); 0 to 40 kb from a telomere (117 and 97 hotspots). For each region, the corresponding interstitial control was computed by randomly selecting groups of interstitial hotspot regions with the same median Spo11 oligo level; this step was repeated 10,000 times. Statistical differences (Mann–Whitney Wilcoxon test) between different regions and their interstitial control are indicated. (E) Examples of DSB (pCUP1-IME1 Spo11 oligo) and Mlh3 binding in an interstitial region and two pericentromeric regions. The normalized signal smoothed with a 200-bp window size is indicated. (F) The ZMM and Mlh3 signals per DSB vary with the timing of DNA replication. Same legend as in D for the indicated chromosomal regions, except that early and late regions were directly compared to each other. See also SI Appendix, Figs. S2–S5.
Determining that Mlh3 binds at a late step of recombination, depending on ZMM proteins, makes it a good candidate for being a marker for crossovers, as in other organisms.
Mlh3 Binding Levels Correlate Well with DSB Frequencies, Except in Centromeric and Late Replicating Regions.
Whether Mlh1-3–dependent crossover has regional specificity across chromosomal territories is an important question, knowing that crossover mislocalization can lead to aneuploidies (37). To address this, we mapped Mlh3 binding sites by ChIP-sequencing (ChIP-seq) from highly synchronized meiotic cultures thanks to a copper-inducible pCUP1-IME1 construct (38). We used the 5- and 5.5-h time points, when crossovers appear (SI Appendix, Fig. S3). We also mapped DSB sites by sequencing Spo11 oligonucleotides from pCUP1-IME1 synchronized cultures at 5 h when DSB levels are maximal (SI Appendix, Fig. S3). The Mlh3 binding map was highly similar to that of the Mer3 and Zip4 ZMMs: Mlh3 formed peaks around DSB hotspots, in addition to being weakly associated with axis binding sites (Fig. 2 B and C and SI Appendix, Fig. S4 A and B).
To get insight into the mode of action of Mlh1-3 once on recombination intermediates, we next asked which DNA region Mlh3 occupies on individual DSB hotspots. Mlh3 peaks at DSB hotspots had a width (2.2 kb) similar to that previously determined for the length of resection tracts at DSB hotspots (1.6 kb) (39) (SI Appendix, Fig. S4C), which is predicted to result in recombination intermediates of roughly similar size (18). Furthermore, in an exo1-D173A (exo1-nd) catalytic mutant that shows a 2.2-fold reduction in resection tracts lengths (19, 39), Mlh3 peaks at DSB hotspots were clearly less wide than in wild-type (SI Appendix, Fig. S4 C and D), consistent with the detectable Mlh1-3 distribution in vivo being mostly defined by the limits of recombination intermediates.
Among the 1,155 Mlh3 peaks identified (Dataset S1), 484 overlapped at least one of the strongest 1,000 DSB hotspots (Dataset S2); and among the strongest 200 DSB hotspots, 164 had a detected Mlh3 peak. In addition, the signal intensities of DSB and Mlh3 at the DSB hotspots were highly correlated (SI Appendix, Fig. S4E). Despite this high global correlation between DSB and Mlh3 binding frequencies, we observed a significant reduction of Mlh3 binding per DSB in pericentromeric regions (Fig. 2 D and E and SI Appendix, Fig. S5 A and B). In contrast, the ZMM Mer3 and Zip4 were not reduced at pericentromeric regions, but were reduced in subtelomeric regions (Fig. 2D), indicating a different regulation than Mlh3.
Finally, we asked if the binding of ZMM and Mlh3 on DSB hotspots is modulated by replication timing. Whereas ZMM binding was not affected, Mlh3 binding per DSB was reduced in late replicating regions (Fig. 2F); this was seen in samples from both 5 h (SI Appendix, Fig. S5C) and 5.5 h (Fig. 2F), suggesting that this effect is not due to temporal differences in Mlh3 loading between early and late replicating regions.
Altogether, our results indicate a regional control of Mlh3 binding to DSBs that operates negatively in centromeric and in late replicating regions.
MutLγ Forms a Complex with Exo1 and Interacts with MutSγ In Vivo.
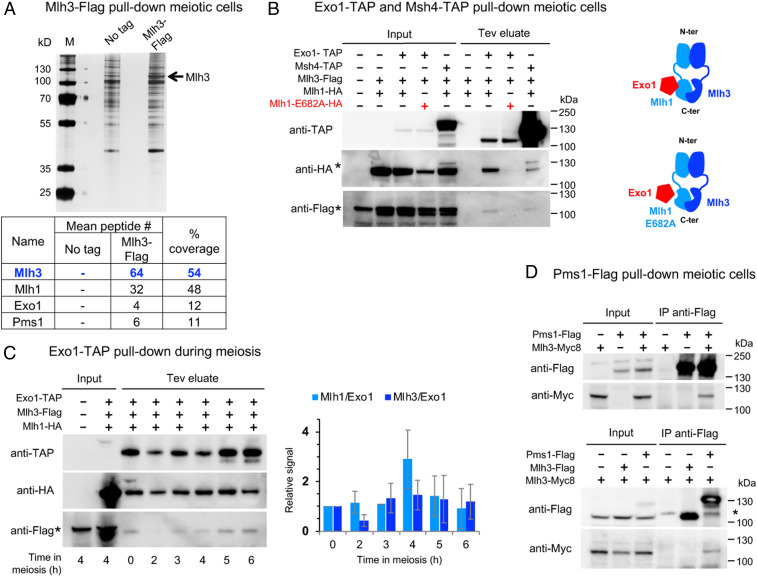
To further gain a mechanistic handle on the function of key players in meiotic crossover, we examined the in vivo interacting partners of MutLγ. We immunoprecipitated Mlh3 4 h after initiating meiosis, when crossovers appear (5), followed by proteomics mass spectrometry analysis (Fig. 3A). Mlh3 pulled down its MutLγ partner Mlh1, as expected, but also Exo1, providing in vivo support for previous proposals that it assists Mlh1-3 in crossover resolution (19) (Fig. 3A and Dataset S3). We confirmed this interaction by Western blot analysis of the reverse pull-down of Mlh3 by Exo1 (Fig. 3B). Interestingly, Exo1 pulled down Mlh1 and Mlh3 throughout meiosis, and is therefore a constitutive partner of MutLγ (Fig. 3C). This interaction required the Mlh1 E682 residue that mediates the direct interaction between Exo1 and Mlh1 (Fig. 3B), implying that the interaction between Exo1 and MutLγ is mediated by Mlh1. Consistent with these results, an mlh1E682A mutant displays meiotic crossover defects (19). We noticed that the Mlh1E682A protein is reproducibly present at lower levels than the wild-type Mlh1 protein, although previous studies showed that it is still proficient for mismatch repair. This may be due to its loss of interaction with several partners (Exo1, but also Sgs1, Ntg2) (40).
Fig. 3.
MutLγ forms a complex with Exo1 and transiently interacts with MutSγ in vivo. (A) Affinity pull-down of Mlh3-Flag from cells at 4 h in meiosis. Silver-stained gel of pulled-down proteins. Table: Mass-spectrometry analysis of selected proteins reproducibly identified in all replicates and not in the controls (no tag strain). The number of peptides of four independent experiments is shown (two with benzonase treatment, two without). Detail of pulled-down proteins in Dataset S3. (B) Coimmunoprecipitation by Exo1-TAP or Msh4-TAP from cells at 4 h in meiosis analyzed by Western blot. The asterisk indicates the uncleaved Msh4-TAP, of which the protein A region is weakly recognized by the anti-HA antibody. The tobacco etch virus (Tev)-cleaved Msh4-TAP is no longer recognized by the anti-HA. (C) Exo1-TAP pull-down throughout meiosis. The graph indicates the ratio of immunoprecipitated Mlh1 and Mlh3 relative to Exo1 in the Tev eluates at the indicated times in meiosis. Values are mean ± SD of two independent experiments. (D) Coimmunoprecipitation between Pms1-Flag and Mlh3-Myc (Upper) or between Mlh3-Flag and Mlh3–Myc (Lower) from cells at 4 h in meiosis analyzed by Western blot. Asterisks indicate a nonspecific cross-hybridizing bands. See also SI Appendix, Fig. S6.
Mlh3 immunoprecipitates also contained Pms1, the partner of Mlh1 in the MutLα heterodimer (Fig. 3A and Dataset S3), and we confirmed this interaction in reverse Pms1 pull-downs (Fig. 3D). This possible MutLα–MutLγ interaction prompted us to ask if MutLγ–MutLγ interactions also occur, possibly as a result of the proposed MutLγ polymerization needed to cleave DNA in vitro (17). However, we failed to detect any Mlh3–Mlh3 interaction in coimmunoprecipitation assays (Fig. 3 D, Lower, and SI Appendix, Fig. S6), suggesting either that MutLγ does not interact with itself in vivo or that this may be regulated and occur only at the time of crossover formation, making it difficult to detect.
Surprisingly, we did not recover any peptides from the Msh4–Msh5 (MutSγ) heterodimer or from other ZMM proteins in the Mlh3 immunoprecipitates (Dataset S3). A weak coimmunoprecipitation of Mlh1 and Mlh3 by Msh4 was detected, but the fraction of Mlh1 and Mlh3 was very low (Fig. 3B), consistent with the lack of visible colocalization of Mlh3 and Msh4 foci. This Mlh3–Msh4 interaction, even transient in vivo, supports recent in vitro data showing that the activity of the human MutLγ complex is strongly stimulated by both Exo1 and MutSγ (41, 42).
Cdc5 Kinase Interacts with Both MutLγ and Exo1 Bound to Recombination Sites.
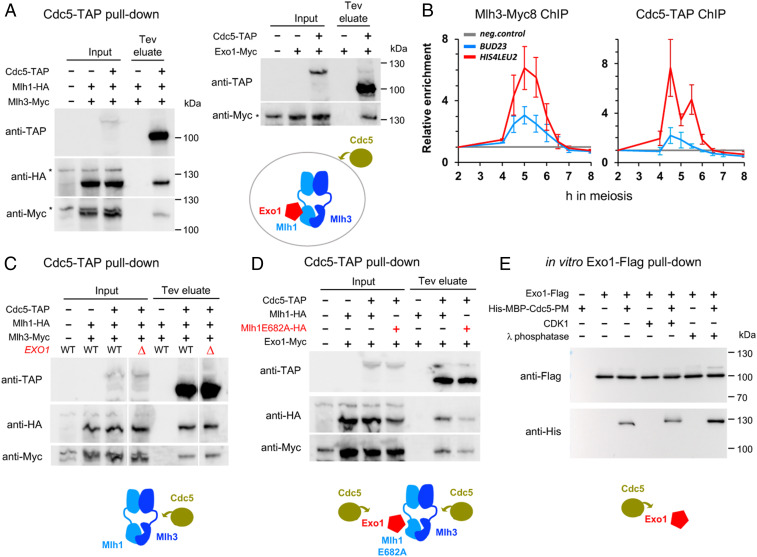
We have shown that Cdc5 is not required for Mlh3 binding to DSB hotspots, and moreover its targets relevant for crossover formation by the Mlh1–Mlh3 pathway are still unknown (23). We did not detect Cdc5 in our meiotic Mlh3 pull-downs (Dataset S3), but to address this issue, we examined if Cdc5 interacts with MutLγ and Exo1 in highly synchronous meiotic cells. At the time when crossovers start to appear (5.5 h) (SI Appendix, Fig. S3), Cdc5 robustly immunoprecipitated Mlh1, Mlh3, and Exo1 (Fig. 4A), raising the possibility that it may directly activate MutLγ. Indeed, Cdc5 associated with recombination sites during meiosis, at the same time as Mlh3 (Fig. 4B). Since MutLγ associates with recombination sites in ndt80∆ mutants, where Cdc5 is absent (Fig. 2A), we propose that Cdc5 is recruited to a preexisting Exo1–MutLγ complex bound at crossover sites.
Fig. 4.
The Cdc5 kinase interacts with both MutLγ and Exo1 bound on recombination sites. (A) Coimmunoprecipitation by Cdc5-TAP of Mlh1-HA, Mlh3-Myc, and Exo1-Myc from pCUP1-IME1 synchronized cells at 5.5 h in meiosis analyzed by Western blot. (B) Mlh3-Myc and Cdc5-TAP association with the indicated DSB hotspots as revealed by ChIP and qPCR at the indicated times of a pCUP1-IME1 synchronized meiotic time course. Same normalization as in Fig. 2. Values are the average of three (Mlh3-Myc) or four (Cdc5-TAP) independent experiments ± SEM (C) Coimmunoprecipitation by Cdc5-TAP of Mlh1-HA and Mlh3-Myc is independent of Exo1. Same conditions as in A. (D) Coimmunoprecipitation by Cdc5-TAP of Exo1 is independent of Exo1 interaction with MutLγ. Same conditions as in A. (E) Direct, phosphorylation-independent interaction between recombinant Exo1 and Cdc5 proteins. Western blot showing the pull-down of purified Cdc5-PM (phosphomimetic; see Materials and Methods) by Exo1-Flag, in the presence or absence of CDK1 or λ-phosphatase. See also SI Appendix, Fig. S7.
To know which of the MutLγ–Exo1 components is a direct target of Cdc5, we assessed their interdependency for in vivo coimmunoprecipitation with Cdc5. Strikingly, Cdc5 still associated at normal levels with Mlh1 and Mlh3 in the absence of Exo1 (Fig. 4C), suggesting that MutLγ itself or another of its interacting partners may be a phosphorylation target of Cdc5. Moreover, Exo1 still coimmunoprecipitated with Cdc5 in the absence of interaction with MutLγ (Fig. 4D) (mlh1E682A mutant, where Exo1 interaction with Mlh1-3 is abrogated), demonstrating that Cdc5 also interacts with Exo1.
Cdc5 Directly Interacts with Exo1 to Promote Crossover Formation.
We tested whether Cdc5 interacts directly with the above factors. Using a yeast two-hybrid assay, we found that the Cdc5 polo-box domain (PBD), known to be involved in interaction with Cdc5 targets, did interact with Exo1, whereas it did not with either Mlh1 or Mlh3 (SI Appendix, Fig. S7A). Cdc5 PBD usually binds a priming phosphate, often formed by CDK1, in the context of a consensus motif, S-S(Phos)/T(Phos)-P (43, 44). However, recombinant purified Exo1 strongly interacted with purified Cdc5 (Fig. 4E and SI Appendix, Fig. S7B), even after pretreating Exo1 with phosphatase, ruling out that this direct interaction is mediated through Exo1 phosphorylation.
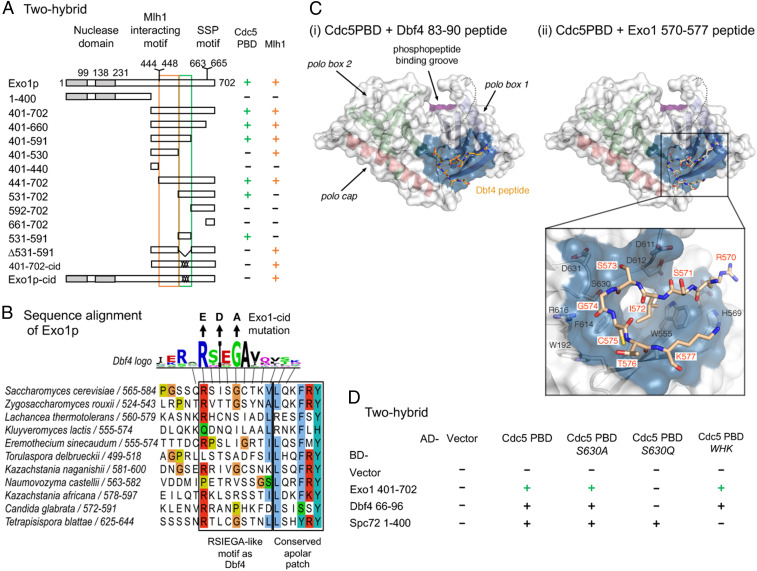
A few other cases of phosphorylation-independent Cdc5 interactions have been described, including that between Cdc5 and Dbf4 (45–47). Domain mapping defined a small region of Exo1 (531 to 591) necessary and sufficient for interaction with Cdc5 PBD using two-hybrid assays (Fig. 5A and SI Appendix, Fig. S7C). This region contains a motif (570 to 575) conserved among yeast species, resembling the RSIEGA motif found in Dbf4 (Fig. 5B) (47). The Cdc5 PBD structure has recently been solved, and shows a specific binding surface for the Dbf4 motif, opposite from the canonical phosphopeptide binding site (Fig. 5 C, i) (48). Interestingly, the Exo1 RSIEGA-like motif could be modeled interacting exactly with the same Cdc5 surface as Dbf4 (Fig. 5 C, ii). Importantly, mutation of the key residue of this surface (S630Q mutation) abolished interaction with Exo1, as it did for Dbf4, but kept intact the interaction with Spc72, a canonical interacting substrate of Cdc5 (Fig. 5D and SI Appendix, Fig. S7D) (48). Conversely, mutation of the WHK motif of Cdc5 involved in phosphopeptide recognition (44) did not alter interaction with Exo1 or Dbf4, but reduced interaction with Spc72 (Fig. 5D and SI Appendix, Fig. S7D) (47, 48). Furthermore, mutating the equivalent of the key residues of the Dbf4 RSIEGA motif in Exo1 (R570E I572D G574A, hereafter called exo1-cid, Cdc5 interaction-deficient) abolished the two-hybrid interaction between Exo1 and Cdc5 (Fig. 5A and SI Appendix, Fig. S7C), confirming that Exo1 and Dbf4 bind in a similar way to Cdc5.
Fig. 5.
Cdc5 directly interacts with Exo1 through a noncanonical site. (A) Delineation of the Exo1 motif responsible for interaction with Cdc5 PBD by two-hybrid assays. The GAL4-BD fusions with indicated Exo1 fragments were tested in combination with a GAL4–AD–Cdc5–PBD fusion; “+” indicates an interaction. Exo1-cid: Cdc5 interaction-deficient. (B) Conservation of the Exo1 region interacting with Cdc5 PBD and illustration of the Exo1-cid mutation. The Dbf4 motif interacting with Cdc5 PBD (47) is indicated (consensus from 17 Saccharomycetaceae family species; see Materials and Methods). (C) Modeling of Dbf4 and Exo1 motifs on the crystal structure of Cdc5 PBD. (i) Crystal structure of the Cdc5 PBD bound to a Dbf4-derived peptide encompassing the RSIEGA motif (PDB ID code 6MF6) (48). The structural elements of the polo box domain are color coded. The region of the domain where phosphorylated substrates bind is labeled for reference. (ii) Model of the Cdc5–Exo1 interaction based on the crystal structure of the Cdc5–Dbf4 complex with Cdc5 shown in the same orientation and color scheme as in (i). The inset shows the residues mediating the interaction between the RSIEGA motif of Exo1 (red labels) and Cdc5 (black labels). (D) The same surface of Cdc5 used for interaction with Dbf4 is used for interaction with Exo1. The GAL4–BD fusions with indicated fragments were tested in combination with a GAL4–AD–Cdc5–PBD fusion with the indicated mutations (WHK stands for W517F H641A K643M); “+” indicates an interaction. See also SI Appendix, Figs. S7 and S8.
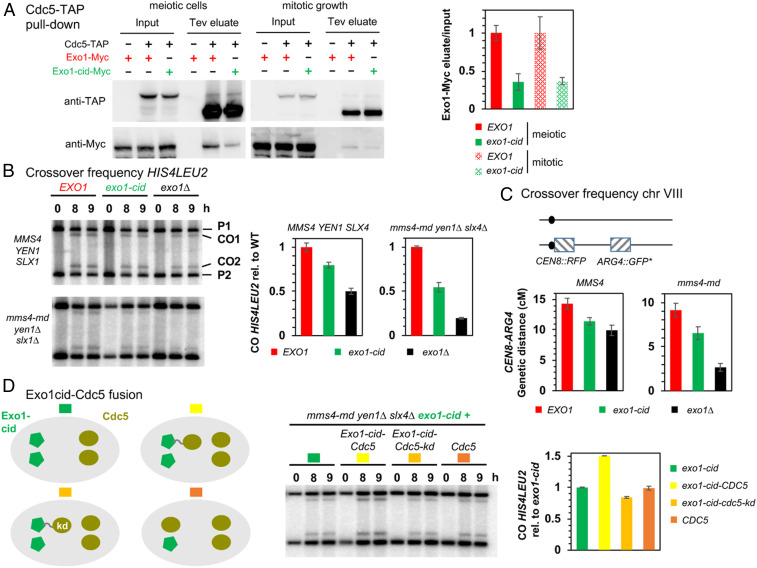
Interaction between Exo1-cid and Cdc5 was also strongly reduced in in vivo coimmunoprecipitation assays, to around 35% of wild-type levels (Fig. 6A). In addition, exo1-cid mutants showed decreased crossovers, on two different tested chromosomes, at the HIS4LEU2 hotspot on chromosome III (Fig. 6B), and in the CEN8-ARG4 interval on chromosome VIII (Fig. 6C). Moreover, crossovers were especially reduced by the exo1-cid mutation combined with a triple nuclease mutant where only MutLγ-dependent crossovers remain (Fig. 6B) (mms4-md yen1∆ slx4∆) (9), or with the single mms4-md mutant (Fig. 6C). In all cases examined, crossover levels in exo1-cid were greater than in the exo1∆ mutant, but decreased to the same extent as in mutants that impair Exo1 interaction with MutLγ [exo1-FF447AA and mlh1E682A mutants (19)], highlighting that the in vivo crossover function of Exo1 is lost in the exo1-cid mutant. To ensure that the crossover defects of Exo1-cid are due to its loss of interaction with Cdc5, we asked if forced recruitment of Cdc5 to this mutant protein restores crossovers. We therefore built strains where one copy of exo1-cid is fused to CDC5, behind the endogenous EXO1 promoter. To avoid confounding effects of expressing CDC5 at the same time as EXO1, and therefore throughout meiosis, we made a control where CDC5 was expressed from the EXO1 promoter without fusion to EXO1 (Fig. 6D). Furthermore, to avoid premature activation of the structure-specific nucleases due to early expression of CDC5 (31, 49), we conducted the experiment in the triple nuclease background. Remarkably, only the fusion of Cdc5, but not its kinase-dead version, to Exo1-cid increased crossovers to levels approaching wild-type Exo1 (Fig. 6D). Our results therefore demonstrate the direct function of Exo1 in recruiting Cdc5 for downstream phosphorylation of targets relevant for crossover.
Fig. 6.
Cdc5 direct interaction with Exo1 promotes crossover formation. (A) Coimmunoprecipitation by Cdc5-TAP of Exo1-Myc in meiotic cells or in cells growing mitotically. Same conditions as in Fig. 4A. (Right) Quantification of Exo1-Myc levels in the Tev eluate relative to the input. Error bars represent SD of two independent experiments. (B) Meiotic crossover frequencies at the HIS4LEU2 hotspot in the exo1-cid mutant. (Left) Representative Southern blot analysis of crossovers in the indicated exo1 mutants, in an otherwise wild-type (MMS4 YEN1 SLX4) or triple nuclease mutant (mms4-md yen1∆ slx4∆) background. mms4-md stands for pCLB2-mms4. (Right) Quantification of crossovers. Values are the mean ± SEM of four independent experiments (wild-type background) or mean ± SD of two independent experiments (triple nuclease mutant), normalized to the corresponding EXO1 value. (C) Meiotic crossovers on chromosome VIII. (Left) Illustration of the fluorescent spore set-up (78). (Right) Genetic distances measured in the CEN8-ARG4 genetic interval for each indicated genotype. (D) Meiotic crossover frequencies at the HIS4LEU2 hotspot with an Exo1-cid–Cdc5 fusion protein. (Left) Scheme illustrating the different experimental setups. In each cell, proteins are expressed from the two allelic endogenous EXO1 promoters (Left) or CDC5 promoters (Right). Same legend as in B. Values are the mean ± SD of two independent experiments, normalized to the homozygous exo1-cid-Myc value.
Since Exo1 directly interacts with Cdc5, we assessed if Exo1 phosphorylation is important for crossovers. We mapped the in vivo Exo1 phosphorylation sites by mass spectrometry of Exo1-TAP purified from meiotic cells after induction of Cdc5, and identified S664 (within the CDK consensus SSP motif of Exo1) as being phosphorylated, along with eight other sites, including three previously described as resulting from DNA damage checkpoint activation (50) (SI Appendix, Fig. S8 A and B). However, mutation of all nine phosphorylated residues of the Exo1 (mutant exo1-9A) did not affect crossovers (SI Appendix, Fig. S8C). These results converge to suggest that Cdc5 does not phosphorylate Exo1 itself, or if it does, it is not important for crossover formation. Instead, our data suggest that Exo1 interacts with Cdc5, to activate in situ the kinase activity of Cdc5 toward its targets, MutLγ or its partners.
Discussion
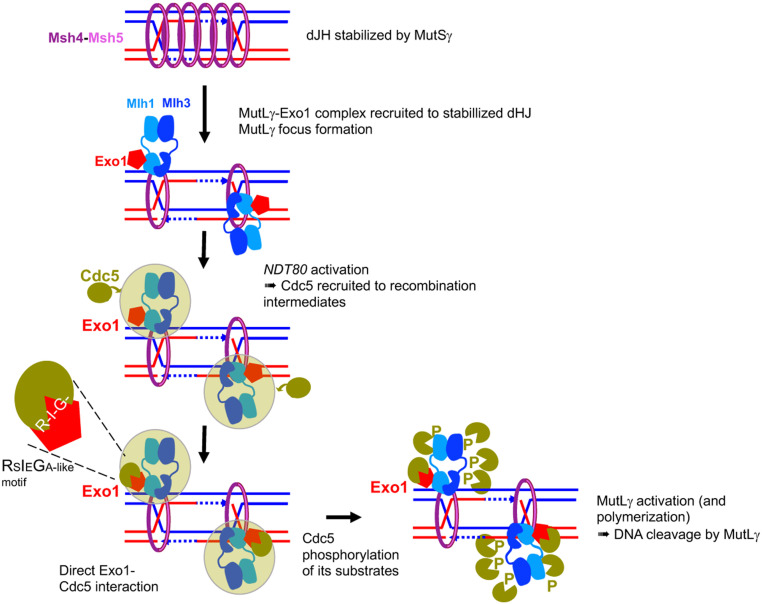
In this study, we provide thorough in vivo characterization of MutLγ complex function in meiotic crossovers. We show that MutLγ forms foci on meiotic chromosomes and that globally, its binding to recombination sites is repressed close to centromeres and in late replicating regions. This predicts that the outcome of meiotic recombination is specifically modulated at specific chromosomal regions. Relevant to MutLγ activation, we show a weak, possibly very transient physical interaction with MutSγ during meiosis and a robust, constitutive interaction with Exo1. We reveal a noncatalytic function of Exo1 in interacting with Cdc5 to activate crossover formation. Our data highlight a noncanonical, direct interaction, which does not seem to be accompanied by Cdc5-mediated Exo1 phosphorylation. We propose that Exo1 serves as a matchmaker and that direct binding of Cdc5 to Exo1 allows Cdc5 to be active and locally phosphorylate MutLγ (Fig. 7).
Fig. 7.
Model of MutLγ binding to sites of recombination and activation for crossover formation. MutSγ stabilizes dHJ intermediates and forms foci on chromosomes. MutLγ, in complex with Exo1, binds dJH intermediates that have been stabilized by MutSγ and other ZMM proteins. For simplicity, MutSγ is shown embracing both DNA duplexes of the intermediates, but recent data also suggest it may embrace separately each DNA duplex (74). Once bound on the dHJ, MutLγ may form a focus when interacting with MutSγ (model shown here), or later in the process of its activation (not shown). Upon NDT80 activation, the Cdc5 kinase is induced, and interacts with the MutLγ–Exo1 complex through multiple interactions (green cloud). Among these interactions, the direct interaction of Cdc5 with Exo1 is important for MutLγ-driven crossover formation. We propose that this interaction allows Cdc5 to phosphorylate MutLγ, which activates its nuclease function and produces crossover formation. This may occur through transient MutLγ polymerization.
The Map of Mlh3 Binding Sites Is Highly Correlated with That of DSBs except in Specific Chromosome Regions.
The choice to make a crossover during meiotic DSB repair may be modulated along chromosomes, but a precise and high-resolution comparison of DSB and crossover levels is missing (reviewed in ref. 51). In mice, studies of a few hotspots suggest that crossover/noncrossover ratios can vary from one hotspot to another (52–55). Recent data comparing a genome-wide DSB map with existing genetic maps have shown that in female mice, crossover formation is repressed at subtelomeric regions (56). Improper placement of crossovers in the vicinity of centromeres is infrequent and negatively influences meiotic chromosome segregation (37, 57, 58). Although recombination close to centromeres is already reduced at the level of DSB formation, residual DSBs increase the risk of aneuploidy (59). One way to prevent crossover formation by pericentromeric DSBs is to repair them from the sister chromatid or as noncrossovers (60, 61). We found that Mlh3/DSB ratios are lower in pericentromeric regions than along chromosome arms, which may also contribute to the lower crossover levels per DSB observed in pericentromeres. Since ZMM binding is not significantly reduced, we propose that there is a deselection of ZMM-bound at pericentromeric recombination sites, which are then preferentially repaired without MutLγ binding, as noncrossovers. In this regard, the phosphatase PP2A associated to Shugoshin has been shown to counteract cohesin kinases, including Cdc5, in pericentromeric regions (62). It may also prevent Cdc5-driven MutLγ activation in these regions.
Prior to our study, it was not clear if replication timing also influenced the choice to repair DSBs as a crossover (63, 64). The fact that Mlh3 binding per DSB is less frequent in late replicating regions suggests there may be a preferred time window for loading MutLγ onto recombination intermediates, such that DSBs occurring late as a consequence of late replication would be less likely to load MutLγ. It remains to be determined if these DSBs are repaired by the other nucleases able to generate crossovers (such as Mus81–Mms4) or if they are repaired through the noncrossover pathway. Recent cytological data in tomato suggested that the different crossover promoting nucleases (MutLγ and the structure-specific nucleases) are differently distributed along chromosomes (65).
Mlh3 Associates with DSB Sites, at a Late Step of Recombination, and Forms Foci Distinct from ZMM-Bound Foci.
MutLγ is predicted to act on recombination intermediates that have been stabilized by ZMM proteins, consistent with our finding that, in their absence, Mlh3 binding is absent (zip3∆ and mer3∆) or reduced (msh4∆). The fact that we did not see colocalization between ZMM and Mlh3 foci suggests that they are two temporally distinct species that form on recombination intermediates, at least as visible entities. Indeed, we detect much fewer (16 on average) Mlh3 foci than the estimated Mlh1–Mlh3-dependent crossovers (around 80). Zip3 and Msh4 foci would precede Mlh3 foci, to preserve the dHJs and hand them over to MutLγ, thereby positioning and activating MutLγ on the recombination intermediates, without staying on chromatin as visible foci when the Mlh3 focus forms (Fig. 7). Our results are consistent with those in mice, where early colocalization between Msh4 and Mlh1 foci disappears at midpachytene (66, 67).
In budding yeast, ZMM foci (around 45 per nucleus) are proposed to all give rise to crossovers (around 75 ZMM-dependent crossovers per meiosis) (51, 68), in contrast to plants, mammals, and Sordaria where only a subset of ZMM foci result in crossovers (69–72). However, the fact that we detect fewer Mlh3 foci (16 per nucleus) than ZMM suggests that a similar selection of a subset of ZMM-bound sites may operate in budding yeast, to become bound by MutLγ, the remaining being dismantled by helicases into noncrossover products. In Sordaria, Msh4 foci numbers diminish between early and midpachytene, a time frame in which Mlh1–Mlh3 is believed to be recruited (73). An alternative hypothesis is that Mlh3 residence time on late crossover intermediates may be limited and therefore Mlh3 is not detected as a focus within a single nucleus on all crossover sites. Finally, we cannot exclude that the foci we detect, which are much fewer (16 on average) than predicted MutLγ-dependent crossovers (75), represent only a subset (unproductive, or at a particular recombination stage) of all of the Mlh3 bound sites.
MutLγ Forms a Constitutive Complex with Exo1 in Meiotic Cells but Interacts Only Transiently with MutSγ.
In many organisms (budding yeast, mice, and plants), msh4 or msh5 mutants have an earlier meiotic defect than MutLγ mutants, but it is attractive to propose, by analogy to mismatch repair, that MutSγ also works directly with MutLγ–Exo1 for crossover formation. Consistent with an early role for MutSγ, we show that most Msh4 foci do not colocalize with Mlh3, but that Msh4 is required for the association of Mlh3 with recombination sites. In addition, the detected interaction of a small fraction of MutSγ with MutLγ in yeast meiotic cells fits also with a late function for MutSγ through direct activation of MutLγ. A similar interaction was also described for the cognate mouse proteins (67). A recent study reports that human MutSγ strongly stimulates human MutLγ endonuclease in vitro (41). Together with our cytological and interaction data, we can then propose that in vivo, MutSγ contributes by creating the proper substrate where the MutLγ–Exo1 ensemble loads, and that a subset of MutSγ may subsequently activate MutLγ catalysis by direct interaction with MutLγ (Fig. 7). However, it is worth noting that, in Caenorhabditis elegans, MutSγ promotes crossovers and remains bound to crossover sites, although crossover resolution is not achieved by MutLγ, suggesting that, in this organism, MutSγ cooperates, directly or indirectly, with a nuclease other than MutLγ (74, 75). Further work aiming at genetically separating these two functions of MutSγ will clarify these issues.
Another Example of Noncanonical Cdc5 Binding to Its Substrates, Involving the RSIEGA Motif.
We found that Exo1 interacts with Cdc5 through the same mode as the only example described so far, Dbf4. Besides the importance for the meiotic recombination context, our study therefore adds another case where the recently described interaction surface on Cdc5, distinct from the phosphopeptide recognition surface, promotes interaction with a protein relevant for a biological process (48). This surface is conserved in human PLK, and our work should open new avenues to search for other partners of Cdc5 or human PLKs with similar interacting motifs, and to inhibitors that may specifically target this new interacting surface.
Exo1 Serves as a Cdc5 Recruiting/Activating Platform for MutLγ Crossover Formation.
Our study establishes a functional molecular link between Cdc5 and the MutLγ–Exo1 complex, providing some explanation for why this kinase is important for meiotic crossovers (23). We show that Exo1 is constitutively associated with MutLγ, and therefore the binding of Exo1 to MutLγ is not sufficient in vivo to activate resolution. Rather, our data, in particular the observation that mutants blocking Exo1–Cdc5 interaction have crossover defects, suggest that resolution is activated by Cdc5 through its direct interaction with Exo1. What could be the downstream event triggered by Cdc5 binding to Exo1?
Cdc5 function in MutLγ crossovers involves phosphorylation of at least a substrate, because induction of a kinase-dead Cdc5 in meiosis is not sufficient to promote crossover formation (23) and because the forced recruitment of Cdc5 to Exo1 only stimulates crossover when Cdc5 kinase activity is intact (our results). It is unlikely that Exo1 is the relevant phosphotarget, since mutation of Exo1’s meiotic phosphorylated residues that we have detected had no effect on crossovers. Another obvious candidate for phosphorylation, once Cdc5 binds Exo1, is the MutLγ complex itself. However, attempts failed to detect Cdc5 activation-associated change in the mobility of Mlh1 or Mlh3 (Fig. 3C) (this study and ref. 76), and published meiotic phosphoproteome datasets do not contain meiotic phosphorylation sites on Mlh1 or Mlh3 that would appear upon Cdc5 induction (77). Another possible target is the Chd1 protein, recently described as a binding partner of Exo1 and important for MutLγ crossovers (76). A proposed function of this chromatin remodeler is to allow MutLγ polymerization along DNA, an event suggested important for in vitro nuclease activity on naked DNA (17, 76) and that would require chromatin remodeling in vivo. Although we failed at detecting indications that MutLγ polymerizes (no MutLγ–MutLγ interaction, no large spreading of Mlh3 peaks around DSB hotspots), we cannot exclude that once loaded, MutLγ may polymerize transiently when becoming active on a region broader than recombination intermediates (Fig. 7). Further experiments examining Mlh3 dynamics at the time of dHJ resolution and the influence of Cdc5 and Chd1 will be required to examine this possibility.
It is noteworthy that despite the reduced crossovers seen when Cdc5 no longer directly interacts with Exo1, Cdc5 is able to coprecipitate with MutLγ from meiotic cells even in the absence of Exo1, and that the Exo1-cid mutant, although totally deficient for two-hybrid interaction with Cdc5, also shows residual coprecipitation with Cdc5 in vivo, likely via its interaction with MutLγ. Therefore, rather than being essential to bring Cdc5 to the MutLγ–Exo1 complex, direct Exo1 binding may serve as the molecular switch to allow Cdc5 phosphorylate its targets (Fig. 7).
As a whole, our study reveals that MutLγ activation is tightly controlled in vivo, both locally through the direct coupling of crossover formation with key meiotic progression steps by the Cdc5 kinase, and globally by the underlying chromosome structure. Much of this regulation is likely to operate in mammals, where the key proteins are conserved. Our study provides an illustration of how nucleases at risk for impairing genome integrity in the germline are kept in check through multiple control levels.
Materials and Methods
Yeast Manipulations.
All yeast strains are derivatives of the SK1 background except those used for two-hybrid and mutation analyses (SI Appendix, Table S1), and the strains used in specific figures are listed in SI Appendix, Table S2. All experiments were performed at 30 °C. Growth conditions and strain constructions are described in SI Appendix, Supplemental Materials and Methods.
Analysis of Crossover Frequencies.
For crossover analysis at HIS4LEU2, cells were harvested from meiotic time courses at the indicated time point. Two micrograms of genomic DNA was digested with XhoI and analyzed by Southern blot using a labeled DNA probe A, as described previously (5). The radioactive signal was detected by a PhosphorImager (Typhoon, GE Healthcare) and quantified using the Image Quant software, as described previously (32). For genetic distances on chromosome VIII, diploids were sporulated in liquid medium, and recombination between fluorescent markers was scored after 24 h in sporulation, by microscopy analysis, as described previously (78). Two independent sets of each strain were combined and at least 600 tetrads were counted. Genetic distances in the CEN8-ARG4 interval were calculated from the distribution of parental ditype (PD), nonparental ditype (NPD), and tetratype (T) tetrads and genetic distances (cM) were calculated using the Perkins equation: cM = (100 (6NPD + T))/(2(PD + NPD + T)). SEs of genetic distances were calculated using Stahl Lab Online Tools (https://elizabethhousworth.com/StahlLabOnlineTools/).
Flag-Affinity Pull-Down, Proteomics Mass Spectrometry, and Coimmunoprecipitation Analyses.
For Flag pull-down and mass spectrometry analysis, 2 × 1010 cells or 1.2 × 109 cells were harvested from synchronous meiotic cultures at the indicated time after meiosis induction. Details are provided in the SI Appendix, Supplemental Materials and Methods.
Recombinant Proteins and Interaction Assays.
Recombinant yeast Exo1-Flag and Cdc5 were purified from insect cells. Details of protein purification and Exo1-Flag pull-downs are in the SI Appendix, Supplemental Materials and Methods.
Yeast Two-Hybrid Assays.
Yeast two-hybrid assays were performed exactly as described previously (32). Cloning details are in the SI Appendix, Supplemental Materials and Methods.
Modeling of Dbf4 and Exo1 Peptides on Cdc5 PBD Structure.
The RSIEGA motif of Exo1 was modeled based the structure of Cdc5 bound to a Dbf4-derived peptide encompassing the RSIEGA motif of Dbf4 (PDB ID code 6MF6) (48). The sequence was edited in Coot (79) and the rotamers of each residue chosen to prevent clashes between Cdc5 and the modeled peptide.
Cytology.
For cytology, 4 × 107 cells were harvested at the indicated time-point and yeast chromosome spreads were prepared as described previously (80). Mlh3-myc18 was stained with primary anti-myc rabbit antibody (Abcam, 1:600) and secondary FITC-conjugated anti-rabbit (Thermo Fischer; 1:600) for all cytological experiments except for the colocalization with Zip3, for which primary 9E11 anti-myc mouse antibody was used together with a secondary Cy3-conjugated anti-mouse (Jackson ImmunoResearch; 1:600). Msh4-HA3 was stained with primary anti-HA mouse antibody (Anopoli; 1:1,000) and secondary Cy3-conjugated anti-mouse antibody (Thermo Fischer; 1:600) when used in parallel with rabbit based anti-myc. Zip3 was stained with a primary anti-Zip3 rabbit antibody (1:2,000), a gift from Akira Shinohara (Osaka University, Osaka, Japan). The secondary antibody was Cy5-conjugated anti-rabbit (Amersham; 1:500). Fluorescent microscopy was carried out on a ZEISS AXIO Imager M2 with a ZEISS Plan-Neofluar 100×, and a 2× additional magnification by a Zeiss optovar. Images were taken at an exposure of 1 s for DAPI (BFP channel), Cy3 (Cy3 channel), FITC (FITC channel), and 2 s for Cy5 (AF660 channel). The Light source: Sola SM II (Llumencor); camera: CoolSNAP HQ2 (Visitron Systems GmbH); acquisition software: Visiview (Visitron Systems). Nuclei acquired with this setup were analyzed by Fiji software and R-scripts.
ChIP, Real-Time Quantitative PCR, and ChIP-Seq.
For each meiotic time point, 2 × 108 cells were processed as described previously (32). Details are in the SI Appendix, Supplemental Materials and Methods.
Spo11 Oligonucleotide Mapping.
Spo11-Flag oligonucleotides were purified and processed for sequencing library preparation and analyzed as previously described (81), from synchronous pCUP1-IME1 strains (SI Appendix, Supplemental Materials and Methods).
Sequencing Data Processing and Bioinformatic Analyses.
ChIP-seq and Spo11 oligo data were analyzed and normalized mostly as described previously (33, 81). Details are provided in the SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Wolf D. Heyer for helpful suggestions; Michael Lichten and Arnaud De Muyt for critical comments on the manuscript; Hajime Murakami for the meiotic replication data; Angelika Amon and Folkert van Werven for the pCUP1-IME1 construct; Hardeep Kaur and Michael Lichten for the pGAL1-CDC5 strain; Rajeev Kumar for constructing the Mlh3-Myc8 allele; and Akira Shinohara for the anti-Zip3. We thank the Institut Curie Cell and Tissue Imaging Platform (PICT-IBISA) Pasteur Imaging facility, member of the France Bioimaging National Infrastructure (ANR-10-INBS-04), and the next-generation sequencing platform, supported by Grants ANR-10-EQPX-03 and ANR10-INBS-09-08 and by the Cancéropôle Ile-de-France. This work was supported by the Institut Curie and the CNRS; by Agence Nationale de la Recherche ANR-15-CE11-0011 (to V.B., J.-B.C., and P.C.); by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05 (to J.-B.C.); and by “Région Ile-de-France” and Fondation pour la Recherche Médicale medical grants (to D.L.). A.S. was supported by a postdoctoral fellowship from the Fondation ARC and by the Labex DEEP (ANR-11-LBX-0044).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013012117/-/DCSupplemental.
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession nos. GSE133108 for Spo11 oligos in pCUP1-IME1 and GSE132850 for Mer3 and Mlh3 ChIP-seq). Proteomic data were deposited on the Proteome Xchange database (identifiers PXD014180 for Mlh3 pull-downs and PXD014185 for Exo1 phosphosites). All data are included in the main text and Dataset S4.
References
- 1.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam I., Keeney S., Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7, a016634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Muyt A., et al. , BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Jessop L., Rockmill B., Roeder G. S., Lichten M., Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2, e155 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh S. D., et al. , BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pyatnitskaya A., Borde V., De Muyt A., Crossing and zipping: Molecular duties of the ZMM proteins in meiosis. Chromosoma 128, 181–198 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Zakharyevich K., Tang S., Ma Y., Hunter N., Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manhart C. M., Alani E., Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair (Amst.) 38, 84–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gueneau E., et al. , Structure of the MutLα C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site. Nat. Struct. Mol. Biol. 20, 461–468 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Kadyrov F. A., et al. , Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J. Biol. Chem. 282, 37181–37190 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishant K. T., Plys A. J., Alani E., A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179, 747–755 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjha L., Anand R., Cejka P., The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J. Biol. Chem. 289, 5674–5686 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogacheva M. V., et al. , Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J. Biol. Chem. 289, 5664–5673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manhart C. M., et al. , The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLoS Biol. 15, e2001164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsolier-Kergoat M. C., Khan M. M., Schott J., Zhu X., Llorente B., Mechanistic view and genetic control of DNA recombination during meiosis. Mol. Cell 70, 9–20.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Zakharyevich K., et al. , Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double holliday junctions. Mol. Cell 40, 1001–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsubouchi H., Ogawa H., Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 2221–2233 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaetzlein S., et al. , Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc. Natl. Acad. Sci. U.S.A. 110, E2470–E2479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei K., et al. , Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 17, 603–614 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sourirajan A., Lichten M., Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 22, 2627–2632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu S., et al. , The transcriptional program of sporulation in budding yeast. Science 282, 699–705 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Chu S., Herskowitz I., Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1, 685–696 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Tung K.-S., Roeder G. S., Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics 149, 817–832 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu L., Ajimura M., Padmore R., Klein C., Kleckner N., NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 6572–6581 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clyne R. K., et al. , Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5, 480–485 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Lee B. H., Amon A., Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300, 482–486 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Lee B. H., Amon A., Polo kinase—Meiotic cell cycle coordinator. Cell Cycle 2, 400–402 (2003). [PubMed] [Google Scholar]
- 31.Matos J., Blanco M. G., Maslen S., Skehel J. M., West S. C., Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147, 158–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duroc Y., et al. , Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. eLife 6, e21900 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Muyt A., et al. , A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serrentino M. E., Chaplais E., Sommermeyer V., Borde V., Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 9, e1003416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panizza S., et al. , Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Zickler D., Kleckner N., Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Lamb N. E., et al. , Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum. Mol. Genet. 6, 1391–1399 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Chia M., van Werven F. J., Temporal expression of a master regulator drives synchronous sporulation in budding yeast. G3 (Bethesda) 6, 3553–3560 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mimitou E. P., Yamada S., Keeney S., A global view of meiotic double-strand break end resection. Science 355, 40–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dherin C., et al. , Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 29, 907–918 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cannavo E., et al. , Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni D. S., et al. , PCNA activates the MutLγ endonuclease to promote meiotic crossing over. Nature 586, 623–627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng K. Y., Lowe E. D., Sinclair J., Nigg E. A., Johnson L. N., The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 22, 5757–5768 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elia A. E., et al. , The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Archambault V., D’Avino P. P., Deery M. J., Lilley K. S., Glover D. M., Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev. 22, 2707–2720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonner A. M., et al. , Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc. Natl. Acad. Sci. U.S.A. 110, E1222–E1231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y. C., Weinreich M., Dbf4 regulates the Cdc5 Polo-like kinase through a distinct non-canonical binding interaction. J. Biol. Chem. 285, 41244–41254 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almawi A. W., et al. , Distinct surfaces on Cdc5/PLK Polo-box domain orchestrate combinatorial substrate recognition during cell division. Sci. Rep. 10, 3379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arter M., et al. , Regulated crossing-over requires inactivation of Yen1/GEN1 resolvase during meiotic prophase I. Dev. Cell 45, 785–800.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morin I., et al. , Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 27, 2400–2410 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrentino M. E., Borde V., The spatial regulation of meiotic recombination hotspots: Are all DSB hotspots crossover hotspots? Exp. Cell Res. 318, 1347–1352 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Cole F., Keeney S., Jasin M., Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. Cell 39, 700–710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Boer E., Heyting C., The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115, 220–234 (2006). [DOI] [PubMed] [Google Scholar]
- 54.de Boer E., Jasin M., Keeney S., Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes Dev. 29, 1721–1733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarbajna S., et al. , A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum. Mol. Genet. 21, 2029–2038 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Brick K., et al. , Extensive sex differences at the initiation of genetic recombination. Nature 561, 338–342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuhl L. M., Vader G., Kinetochores, cohesin, and DNA breaks: Controlling meiotic recombination within pericentromeres. Yeast 36, 121–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rockmill B., Voelkel-Meiman K., Roeder G. S., Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics 174, 1745–1754 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan J., et al. , A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S. Y., et al. , Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincenten N., et al. , The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife 4, e10850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galander S., Barton R. E., Kelly D. A., Marston A. L., Spo13 prevents premature cohesin cleavage during meiosis. Wellcome Open Res. 4, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borde V., Goldman A. S., Lichten M., Direct coupling between meiotic DNA replication and recombination initiation. Science 290, 806–809 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Buhler C., Borde V., Lichten M., Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 5, e324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson L. K., et al. , Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. U.S.A. 111, 13415–13420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kneitz B., et al. , MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000). [PMC free article] [PubMed] [Google Scholar]
- 67.Santucci-Darmanin S., et al. , The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum. Mol. Genet. 11, 1697–1706 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chelysheva L., et al. , The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Muyt A., et al. , E3 ligase Hei10: A multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 28, 1111–1123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reynolds A., et al. , RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun X., et al. , Transcription dynamically patterns the meiotic chromosome-axis interface. eLife 4, e07424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Storlazzi A., et al. , Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141, 94–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woglar A., Villeneuve A. M., Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173, 1678–1691.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zalevsky J., MacQueen A. J., Duffy J. B., Kemphues K. J., Villeneuve A. M., Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153, 1271–1283 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wild P., et al. , Network rewiring of homologous recombination enzymes during mitotic proliferation and meiosis. Mol. Cell 75, 859–874.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suhandynata R. T., Wan L., Zhou H., Hollingsworth N. M., Identification of putative Mek1 substrates during meiosis in Saccharomyces cerevisiae using quantitative phosphoproteomics. PLoS One 11, e0155931 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thacker D., Lam I., Knop M., Keeney S., Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 189, 423–439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Xaver M., Huang L., Chen D., Klein F., Smc5/6-Mms21 prevents and eliminates inappropriate recombination intermediates in meiosis. PLoS Genet. 9, e1004067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lam I., Mohibullah N., Keeney S., Sequencing Spo11 oligonucleotides for mapping meiotic DNA double-strand breaks in yeast. Methods Mol. Biol. 1471, 51–98 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Zhu X., Keeney S., High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201, 525–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession nos. GSE133108 for Spo11 oligos in pCUP1-IME1 and GSE132850 for Mer3 and Mlh3 ChIP-seq). Proteomic data were deposited on the Proteome Xchange database (identifiers PXD014180 for Mlh3 pull-downs and PXD014185 for Exo1 phosphosites). All data are included in the main text and Dataset S4.