Abstract
The retinal degenerative disease, progressive retinal atrophy (PRA) is a major reason of vision impairment in canine population. Canine PRA signifies an inherently dissimilar category of retinal dystrophies which has solid resemblances to human retinis pigmentosa. Even though much is known about the biology of PRA, the knowledge about the intricate connection among genetic loci, genes and pathways associated to this disease in dogs are still remain unknown. Therefore, we have performed a genome wide association study (GWAS) to identify susceptibility single nucleotide polymorphisms (SNPs) of PRA. The GWAS was performed using a case–control based association analysis method on PRA dataset of 129 dogs and 135,553 markers. Further, the gene-set and pathway analysis were conducted in this study. A total of 1,114 markers associations with PRA trait at p < 0.01 were extracted and mapped to 640 unique genes, and then selected significant (p < 0.05) enriched 35 gene ontology (GO) terms and 5 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways contain these genes. In particular, apoptosis process, homophilic cell adhesion, calcium ion binding, and endoplasmic reticulum GO terms as well as pathways related to focal adhesion, cyclic guanosine monophosphate)-protein kinase G signaling, and axon guidance were more likely associated to the PRA disease in dogs. These data could provide new insight for further research on identification of potential genes and causative pathways for PRA in dogs.
Keywords: Canine, Genome wide association study (GWAS), Inherited disease, Gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Progressive retinal atrophy
INTRODUCTION
Progressive retinal atrophy (PRA) is a comprehensive term encompassing several inherited retinal diseases which all progress over time and eventually leads to vision loss in affected dogs. PRA in dogs has been reported to be genetically similar to the retinitis pigmentosa (RP) disease occurs in human[1]. Similar to RP, PRA shows genetically heterogeneous and phenotypic similarities [2]. The first significant clinical sign of this disease is night blindness, followed by total eyesight loss. [1]. In PRA disease, various ophthalmoscopic complication mostly takes place in eye such as retinal vascular degeneration, retinal detachment, loss of pigmentation, hyperreflectivity in tapetal area, and optic atrophy [3].
PRA disease has been first recognized during the early 20th century after an impulsive retinal deterioration due to PRA was reported in setter family dogs [4–6]. At present, several forms of PRA have been described in more than 100 different breeds of dogs [1], although many of those exhibit similar ophthalmologic signs; the difference between breeds is only in the rate of progression, aetiology, and age. The majority of hereditary PRA diseases in dog are autosomal recessive, while some are X-linked forms or autosomal dominant [1,5,7]. So far, researchers have identified as many as 31 mutations in 24 genes, as underlying cause for canine retinal disease [8]. The methods to identify susceptible genetic variants and genes discovery in canine have seen considerable improvement over the past 20 years [8,9]. Unfortunately, the real treatment for PRA disease in canine still does not exist. Hence, identification of PRA associated genetic variations using deferent approach like Single nucleotide polymorphism (SNP) microarrays and next generation sequencing, is essential for finding disease associated candidate genes.
Genome-wide association studies (GWAS) is considered worldwide as an influential tool for identifying causative loci and genes associated with genetic disease. Nevertheless, conducting robust GWAS analysis with the aim of detailed biological understanding is challenging due to certain factors like large number of samples, stringent statistical thresholds to account for multiple testing, small effect sizes of variants resulting in very few reaching the stringent threshold levels, and linkage disequilibrium [10–12]. GWAS cannot describe the fact that pathways and cluster of genes are connected with each other to implement a specific molecular function [13]. Additionally, GWAS possibly will not able to fully capture the effect of multi-allelic quantitative trait locus because of markers having two allelic variants [14]. Hence, the identification of genetic loci through GWAS only might offer inadequate insight into the polygenic disease like PRA.
Recommended resolution for addressing those issues mentioned earlier is to perform GWAS accompanied by gene-set and pathway analyses. The integration of GWAS, gene-set analysis, pathway and network analyses might help to overcome the limitations of GWAS and provide insights into the biological pathways affecting the particular trait. Such approaches have been used in humans [15], and dairy cattle [13,14]. Though several GWAS have been performed for PRA disease in canine [1,5,7,16], those studies have resulted in only few markers and have not exploited the use of pathway analyses for understanding the underlying biology of PRA. Therefore, aim of the present study was to carry out GWAS analysis for finding out the sensitive loci and genes associated to PRA disease. Additionally, we have identified regulatory gene ontology (GO) terms and pathways which are associated to canine PRA disease for understanding the biological functions regulating this disease.
MATERIALS AND METHODS
Animal and sample collection
Whole blood sample was collected from a total of 129 dogs (101 ophthalmoscopically normal dogs, 28 dogs diagnosed with PRA) of 18 different breeds (Table 1) and stabilized by ethylenediaminetetraacetic acid. We followed an appropriate instruction articulated by The Institutional Animal Care and Use Committee (IACUC) of National Institute of Animal Science (NIAS). These dogs were diagnosed individually by veterinary ophthalmologists from the division of animal disease and health, NIAS, Korea. Genomic DNA was isolated from blood sample followed by wizard® genomic DNA purification kit Protocol (Promega, Madison, WI, USA).
Table 1. Summary of phenotype (including breed, number of male and female dogs investigated and number of cases) used to perform GWAS in our analysis.
| No. | Breed name | No. of dogs investigated | PRA-affected dogs |
|---|---|---|---|
| 70 females 58 males | |||
| 1 | Beagle | 3 | 1 |
| 2 | Bichon Fris | 1 | 0 |
| 3 | Chihuahua | 6 | 1 |
| 4 | Cocker Spaniel | 8 | 3 |
| 5 | Dachshund | 3 | 1 |
| 6 | Doberman | 1 | 0 |
| 7 | German Shepherd | 1 | 0 |
| 8 | Golden Retriever | 1 | 0 |
| 9 | Maltese | 30 | 3 |
| 10 | Miniature Pinscher | 3 | 1 |
| 11 | Mixed | 19 | 2 |
| 12 | Parson Russell Terrier | 2 | 1 |
| 13 | Pomeranian | 3 | 1 |
| 14 | Poodle | 20 | 7 |
| 15 | Schnauzer | 4 | 1 |
| 16 | Shih Tzu | 10 | 2 |
| 17 | Spitz | 1 | 0 |
| 18 | Yorkshire Terrier | 12 | 4 |
GWAS, genome-wide association study; PRA, progressive retinal atrophy.
Genotyping and genome-wide association analysis
Genome-wide association analysis was performed in 28 cases and 101 controls using the Illumina Canine HD Bead-Chip, which genotypes 173,662 SNPs evenly spaced across the genome. The GWAS analysis was performed with PLINK software (version 1.9). Prior to the analysis, quality control (QC) filtering with a minor allele frequency < 5%, low genotyping call rate < 90%, missing genotype calls > 10%, Hardy–Weinberg equilibrium at p < 1E-06 were carried out. The final genotyping call rate was 98.5%. Following QC filtering, 135,553 SNPs and 128 animals remained for further analyses. The PRA phenotype used in this analysis was discrete. A case–control GWAS was performed to test the association between disease trait and allele in each SNP with the -- assoc option of PLINK software. We generated a total of 20 PC; the eigenvalues of all the PCs were fit as covariance to account for population stratification. The following statistical model equation was used in this GWAS study.
Where, y is experiential phenotype, x and z are known design matrices, b is a vector of fixed effects (sex, age, and breed), u is the additive genetic effect for each SNP and e is vector of residual terms. Manhattan and quantile-quantile (Q-Q) plots were generated with CMplot [17]. Bonferroni correction was deemed as too stringent; therefore, the significance level was set at nominal p< 0.01 to identify putative markers.
Gene ontology and pathway analysis
To identify the pathways and genes enriched for the PRA disease in dogs, we performed GO and pathway analysis following the process discussed by Dadousis et al. [13]. The SNPs (nominal p < 0.01) from GWAS analysis were used for the analysis. The markers were annotated to the nearby genes using the SnpEff software version 4.3 [18]. Then, the genes overrepresentation in every GO terms and pathway were tested using a Fisher’s exact test, which was conducted with the web-based functional enrichment analysis tool, the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.abcc.ncifcrf.gov/) [19]. The GO [20] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [21] databases were used to describe the GO terms and pathways related to the enriched genes. The GO terms/KEGG category pathways enriched by more than 10 and less than 1,000 genes were examined in order to narrow down the functional categories [22].
RESULTS
Genotyping and genome-wide association analysis
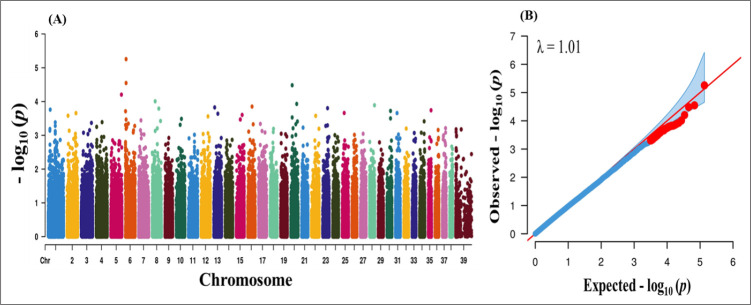
After QC, 4,108, 3,418 and 30,583 markers were removed due to missing genotype data, HWE (p < 0.000001) and MAF < 5%. A total of 135,553 markers were finally passed the QC and genotyped in the GWAS analysis. We performed case–control based association analysis with PRA. Manhattan plots were constructed based on the association p-values to find major loci associated with PRA disease. The physical location of SNPs in chromosome was plotted against −log10 p-values (Fig. 1A). Even though the number of cases was not sufficient for effective GWAS analysis, our findings provide some insight into major loci underlying PRA. However, no significant loci were found from the association analysis. A Q-Q plot of observed p-values was plotted against expected p-values for PRA is shown in Fig. 1B. There were no significant deviation (λ value = 1.01) which indicates absence of significant SNPs. The lack of highly significant loci might be due the inclusion of large number of breed (n = 18) and lower sample number.
Fig. 1. (A) Manhattan plot of SNP associations for PRA trait in dog. (B) The quantile-quantile (Q-Q) plot of 135,553 single nucleotide polymorphisms from 28 cases and 101 controls.
p-values of the Q-Q plot of the association test did not show any significant deviation (λ value = 1.01) which indicates absence of significant SNPs. SNP, single nucleotide polymorphism; PRA, progressive retinal atrophy.
Gene-set enrichment and pathway analysis
GWAS was supplemented by a functional enrichment analysis for detecting significant GOs and pathways associated to PRA. Among the total 135,553 SNPs used in this study, 91,779 SNPs were mapped to the genes. Based on reduced GWAS threshold of (p < 0.01) 1,114 SNPs were deemed to be associated with PRA (Supplementary Table S1). These SNPs were annotated to assign 640 unique genes, (Supplementary Table S1). Subsequently, the gene list was uploaded to DAVID for GO enrichment and pathway analysis. A total of 76 GO terms and 11 KEGG pathways were found to be enriched (Supplementary Table S2 and Table S3). Out of the total enriched GO terms, 35 significantly enriched (p < 0.05) GO terms with at least 4 genes are presented here. There were 26 biological processes related to cell adhesion, cell migration, protein kinase C, regulation of cell signaling, phosphorylation, ion transport, cell growth, cell proliferation, axon guidance, DNA replication, apoptosis and startle response; 4 cellular component GO terms including focal adhesion, endoplasmic reticulum, basal lamina, and early endosome membrane protein, and 4 molecular function GO including calcium ion binding, adenosine triphosphate (ATP) binding, protein kinase activity, potassium channel regulator activity, and heterodimerization activity (Table 2). ATP binding (GO:0005524) was the most significant with the highest number of genes (49) among all GO terms. Meanwhile, homophilic cell adhesion via plasma membrane adhesion molecules (GO:0007156) was the most significantly enriched biological process for PRA trait (p = 8.32E-06).
Table 2. Significant (p < 0.05) gene ontologies (GO) enriched for genes within 5-kb flanking region of associated SNP for PRA disease.
| GO ID | GO term | No. of genes | p-value | Genes |
|---|---|---|---|---|
| GO:0007156 | Homophilic cell adhesion via plasma membrane adhesion molecules | 13 | 8.32E-06 | CDH12, CDH8, CDH13, CRTAM, CDH9, PTPRM, FAT3, ROBO1, DSC3, CDHR3, ROBO2, PCDH15, CDH10 |
| GO:0090036 | Regulation of protein kinase C signaling | 4 | 4.86E-04 | AKAP12, SEZ6L, CAPZB, SEZ6 |
| GO:0006915 | Apoptotic process | 10 | 6.46E-04 | DLC1, DCC, ATCAY, PRUNE2, STK24, CLIC4, BNIP2, TFAP2B, VDAC1, HIP1 |
| GO:0035264 | Multicellular organism growth | 9 | 1.17E-03 | SLITRK1, TBL1XR1, STK40, CLIC4, APBA2, H3F3B, PCDH15, PKDCC, WWTR1 |
| GO:0046777 | Protein autophosphorylation | 11 | 1.59E-03 | EPHA4, TNIK, VRK2, STK24, NLK, PEAK1, LRRK2, EIF2AK3, DAPK3, EPHB1, KDR |
| GO:0005509 | Calcium ion binding | 28 | 3.24E-03 | SPOCK3, NELL1, NELL2, FAM20C, DLK1, PRKCSH, KCNIP4, CDH8, CDH9, FAT3, FSTL5, HPCAL1, CNTNAP4, C2CD5, CAPN9, LRP1B, CDHR3, PADI2, PCDH15, MAN1A1, ITPR2, CDH12, CDH13, CBLB, SULF2, DSC3, CACNA1E, CDH10 |
| GO:0042130 | Negative regulation of T cell proliferation | 5 | 3.67E-03 | MAD1L1, IL20RB, FOXJ1, PDE5A, DLG1 |
| GO:0005524 | ATP binding | 49 | 4.50E-03 | CDK19, RAD51C, ATP10B, PEAK1, EPHB1, IARS, GSS, MCM9, ADCK1, VRK2, STK40, MCCC1, CHD2, DHX35, CHD6, ROS1, CDK14, AKT2, ABCA12, ABCA 4, CDK1, TNIK, STK24, FLT3, NLK, MYLK4, CDK8, ATP1A4, RPS6KC1, PKDCC, ATP1A1, DGKI, OXSR1, PRKCE, DAPK3, KDR, MAP4K4, TARS, EPHA4, HUNK, ABCC9, ASCC3, NTRK2, RRM1, MAP3K10, CDC42BPA, LRRK2, EIF2AK3, PCCA |
| GO:0005783 | Endoplasmic reticulum | 24 | 6.31E-03 | MOGAT1, ATP10B, NDFIP2, MAN1A1, SEZ6L, PRKCE, CPED1, VASH1, FDFT1, KDR, VCAM1, NCSTN, PSMF1, TMEM117, BRINP1, SULF2, SRD5A3, SUMF1, SIL1, LRRK2, USP25, SEZ6, ZFYVE1, DLG1 |
| GO:0001701 | In utero embryonic development | 12 | 6.54E-03 | BMI1, NMT1, MSX1, HAND2, FOXF1, EDN1, DSC3, APBA2, SMAD2, PRKCSH, FOXP1, PITX2 |
| GO:0004672 | Protein kinase activity | 10 | 7.97E-03 | CDK19, MAP4K4, ADCK1, VRK2, STK40, STK24, NTRK2, PEAK1, PKDCC, FASTKD5 |
| GO:0010811 | Positive regulation of cell-substrate adhesion | 5 | 8.29E-03 | RSU1, FOXF1, DMP1, VIT, PRKCE |
| GO:0008360 | Regulation of cell shape | 9 | 8.29E-03 | DLC1, VRK2, DIAPH1, ARHGAP18, LIMD1, DAPK3, FN1, KDR, DLG1 |
| GO:0060079 | Excitatory postsynaptic potential | 4 | 8.92E-03 | GRIK2, DGKI, LRRK2, SEZ6 |
| GO:0010575 | Positive regulation of vascular endothelial growth factor production | 4 | 1.15E-02 | CYP1B1, SULF2, RORA, EIF2AK3 |
| GO:0008283 | Cell proliferation | 9 | 1.22E-02 | CDK1, TYR, ASCC3, E2F8, TBX20, UCHL1, H3F3B, ROS1, OCA2 |
| GO:0005925 | Focal adhesion | 16 | 1.77E-02 | ACTB, DLC1, RSU1, PEAK1, AKAP12, ITGA11, ARF6, NCSTN, RND3, CDH13, YWHAQ, GIT2, SNTB1, PPP1R12A, HSPB1, LIMD1 |
| GO:0015459 | Potassium channel regulator activity | 4 | 2.63E-02 | SUMO1, KCNAB1, KCNIP4, DLG1 |
| GO:0001662 | Behavioral fear response | 4 | 2.66E-02 | BRINP1, GRIK2, GABRA5, VDAC1 |
| GO:0072659 | Protein localization to plasma membrane | 5 | 2.69E-02 | TNIK, C2CD5, AKT2, DLG1, ABCA12 |
| GO:0006260 | DNA replication | 5 | 2.70E-02 | POLD3, MCM9, GINS3, RRM1, REV3L |
| GO:0045944 | Positive regulation of transcription from RNA polymerase II promoter | 24 | 2.76E-02 | TBL1XR1, IKZF3, COPS5, EDN1, E2F8, EOMES, FHL5, RORA, WWTR1, FOXP1, CDH13, EPC1, NPAS2, PAX7, GALR2, THRAP3, EBF1, TFAP2B, PPP1R12A, POU3F3, NRG1, TCF4, BCAS3, PIK3R1 |
| GO:0007411 | Axon guidance | 7 | 3.32E-02 | DCC, KLF7, ROBO1, SEMA3C, CNTN4, ROBO2, RELN |
| GO:0048286 | Lung alveolus development | 4 | 3.33E-02 | STK40, FOXF1, PKDCC, ABCA12 |
| GO:0046982 | Protein heterodimerization activity | 4 | 3.38E-02 | PPARD, TFAP2B, ADRA1B, TCF4 |
| GO:0030336 | Negative regulation of cell migration | 6 | 3.53E-02 | DLC1, CYP1B1, STK24, ROBO1, CLIC4, KANK1 |
| GO:0031901 | Early endosome membrane | 4 | 3.53E-02 | MARCH1, EPHA4, PMEPA1, EPHB1 |
SNP, single nucleotide polymorphism; PRA, progressive retinal atrophy; ATP, adenosine triphosphate.
5 KEGG pathways were also significantly (p < 0.05) enriched for PRA trait including genes involved in focal adhesion, axon guidance, cyclic guanosine monophosphate (cGMP)-protein kinase G (PKG) signaling pathway, gastric acid secretion, and platelet activation (Table 3). Focal adhesion pathway was the most significantly enriched pathway with PRA associated genes. Besides, it was enriched with the highest number of genes (14) from the gene list uploaded for pathway analysis.
Table 3. Significant (p < 0.05) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for genes within 5-kb flanking region of associated SNP for PRA disease.
| KEGG | Pathway name | No. of genes | Genes | p-value |
|---|---|---|---|---|
| Cfa04510 | Focal adhesion | 14 | ACTB, DIAPH1, COL6A3, MYLK4, PPP1R12A, ITGA11, RELN, PDGFD, COL5A2, COL11A1, PIK3R1, FN1, AKT2, KDR | 2.56E-03 |
| Cfa04360 | Axon guidance | 10 | ABLIM1, DCC, EPHA4, CXCR4, ROBO1, NTN4, SEMA3C, ROBO2, EPHB1, RASA1 | 4.57E-03 |
| Cfa04022 | cGMP-PKG signaling pathway | 10 | MYLK4, PDE5A, PPP1R12A, ADRA1B, ATP1A4, ATP1A1, PRKCE, AKT2, ITPR2, VDAC1 | 1.98E-02 |
| Cfa04971 | Gastric acid secretion | 6 | SSTR2, SLC26A7, MYLK4, ATP1A4, ATP1A1, ITPR2 | 3.76E-02 |
| Cfa04611 | Platelet activation | 8 | ACTB, MYLK4, PPP1R12A, COL5A2, COL11A1, PIK3R1, AKT2, ITPR2 | 4.53E-02 |
SNP, single nucleotide polymorphism; PRA, progressive retinal atrophy; cGMP, cyclic guanosine monophosphate; PKG, protein kinase G.
DISCUSSION
The present study investigated functional enrichment based association analysis, a post-GWAS method to understand PRA disease biology in dogs. These approaches were not previously reported for revealing the functional associations among genes associated to PRA disease. The pathway-based association analyses effectively check the cluster of gene enriched in every pathway for providing the knowledge on the pathway-level divergence [10,23]. Moreover, it offers us different understandings about genes as well as their interaction with each other. Thus, these approaches have been quite successful in acquiring new information regarding a particular disease trait.
In this study, no locus was identified at the genome wide and suggestive significant levels in our GWAS data set. Therefore, nominal p < 0.01 from GWAS analysis was used to subset for SNPs for further functional enrichment analysis. In our gene-set enrichment analysis, the most significant biological process (GO:0007156) enriched for the PRA trait was “Homophilic cell adhesion via plasma membrane adhesion molecules”. This biological process is enriched with 13 significant genes (CDH12, CDH8, CDH13, CRTAM, CDH9, PTPRM, FAT3, ROBO1, DSC3, CDHR3, ROBO2, PCDH15, and CDH10) and all of them except two (ROBO2 and PTPRM) encode proteins that belong to cadherin superfamily. Interestingly, some of these genes, specially belong to cadherin family are also enriched in calcium ion binding molecular function (GO:0005509). It is widely known that cadherins, a class of transmermbrane protein depends on calcium ion binding to function, cell-cell adhesion [24]. Notably, these cadherin proteins have been significantly associated with PRA because of their involvement in retinal detachment and proliferative vitreoretinopathy [24,25]. The retinal separation was reported to influence the neural retinal atrophy by cutting choroidal blood supply to the outer segments of retinal pigment epithelium [26,27]. Moreover, cadherin proteins are found to be an important adhesion molecule in retinal pigmental epithelial cells [28]. In addition, another adhesion related biological process for PRA disease was GO:0005925 term “focal adhesion” shared one gene (CDH13) with other GO term “homophilic cell adhesion via plasma membrane adhesion molecules”. The gene (CDH13) encodes cadherin13 protein, which was reported to be extremely associated with cancer disease including retina cancer, lung cancer and breast cancer [29]. The disease relevance of cadherin13 studies found that the expression of this protein was invariable in retinoblastoma eye specimen of human than healthy eye [29]. Moreover, the association of CDH13 with Ca (2+)-dependent cell-cell adhesion molecule suggested distinctive connection in retinal atrophy [30].
Apoptosis or programmed cell death process is implicated in several neurodegenerative diseases, including the inherited retinal degenerations knows as RP diseases [31,32]. Marigo [33] identified two apoptotic pathways in undergoing cell death photoreceptors of autosomal recessive RP mouse model . They even suggested the use of anti-apoptotic strategies against such apoptosis may open new perception about innovative therapeutic strategy for treatment of retinal deterioration [33]. In this study we found the GO term apoptotic process (GO:0006915) was to be significantly enriched with PRA associated genes. Among the genes included in this specific GO, deleted in colorectal carcinoma (DCC) gene was significantly enriched in another biological process, axon guidance (GO:0007411). The DCC gene has a key role in the dual functionality of netrin-1 in axon guidance biological process [34]. Moreover, this axon guidance was detected to be associated with axonal degradation of retinal ganglion cells, which is directly linked to PRA disease [35].
The maximum number of genes was found to be enriched in GO term “ATP binding” (GO:000524). Specially, retina-specific gene ABCA4 and Na+/K+ transporting gene ATP1A4 is of particular interest. The ABCA4 gene encodes ATP-binding cassette, subfamily A, member 4, protein which was described to be an essential protein for human retinal degeneration disease. Moreover, this gene was found as an causal gene associated to generalized PRA disease in different breeds of dogs [36]. Besides, ATP1A, gene comes under Na+/K+ transporting ATPase alpha 1 family, has been a causative gene for autosomal recessive cone-rod dystrophy (CORD8) disease [37]. Moreover, this Na+/K+ ATPase was exhibited recently to be important in photoreceptor cell [38]. These above descriptions suggest an indirect link to the ATP binding term and PRA disease.
Another important GO related to “endoplasmic reticulum” was significantly enriched for PRA. Endoplasmic reticulum (ER) has vital contribution in membrane protein folding [39]. In cells, ER generally goes under stress upon failing to match the protein folding capacity demand [39,40]. Interestingly, this ER stress was found to be elevated in multiple animal model of RP disease [41]. The key role of ER in retinal neurodegeneration has already been described [40–43]. ER leads the degradation of pigment in photoreceptor cells which ultimately causes blindness [39]. Now, it can be hypothesized that GO term “endoplasmic reticulum” is strongly associated with the retinal disease including PRA.
Among the 5 significant KEGG pathway enriched for PRA, Focal adhesion and Axon guidance were found to be common in GO and KEGG categories. The gene sets enriched for Focal adhesion and Axon guidance varies between GO and pathway analysis. Focal adhesion pathway (Cfa04510) was the most significantly enriched pathway for the PRA trait. Focal adhesion kinase, member of the focal adhesion pathway, has been associated in retinal angiogenesis, which is a major feature of eye diseases that can lead to blindness [44–46]. Hence, a causal relationship among focal adhesion and retinal degeneration can be hypothesized (part of the PRA disease). In addition, among the included genes in focal adhesion pathway, AKT2 is of particular interest. This gene was common in another two pathways in our result such as cGMP-PKG signaling pathway and platelet activation pathway. This gene is closely related with serine/threonine protein kinases (AKT1, AKT2, and AKT3) and was reported to have contribution in light stress-induced retinal photoreceptor degeneration [47]. Furthermore, axon guidance pathway was significantly enriched with 10 genes including ABLIM1, DCC, EPHA4, CXCR4, ROBO1, NTN4, SEMA3C, ROBO2, EPHB1, and RASA1. The relationship between axon guidance pathway and degeneration of retina (causing progressive vision loss), was described above.
Our pathway analysis revealed that a vital signaling pathway such as cGMP-PKG signalling pathway (Cfa04022) was amid the enriched pathways in our study. The cGMP- dependent (PKG) signaling pathway is known to be intricately linked to the hereditary retinal degeneration such as RP [48]. The importance of cGMP signaling pathway for neurodegenerative disease retinal degeneration was recently reviewed [49]. A previous study showed that cGMP-dependent PKG activation is a hallmark in photoreceptor degeneration in human homologus mouse [48]. Moreover, the same authors suggested to target PKG in cGMP dependent PKG pathway as a unique therapy for retinal degeneration disease [48].
Additionally, the gastric acid secretion pathway was significantly enriched for PRA trait. The ATP1A1 (ATPase Na+/K+ Transporting Subunit Alpha 1) gene encodes the protein which was reported to be connected to photoreceptor cell, which is a part of retina [38]. There is reported relation between Gastric acid secretion pathway and PRA trait. Last of all, the platelet activation (Cfa04611) was significantly enriched pathway for PRA trait. Among the most of the genes in this pathway were shared with focal adhesion pathway and cGMP-PKG signaling pathway. Moreover, there is an evidence of association between platelet activating factor and pathogenesis of age-related eye disease [50]. Hence, it suggests an unintended link between platelet activation pathway and PRA trait.
Seemingly, aside from ontologies and pathway that could be directly allied with PRA disease (e.g., GO term apoptotic process and cGMP-PKG signaling pathway); ontologies and pathway not strictly associated to the traits of our interest were also identified (negative regulation of T cell proliferation, platelet activation). Moreover, it is obvious that some of our result may need further validation. It is not noteworthy to mention here that the GO and KEGG pathways of canine which are accessible for public are remain scarce and not adequately elucidated. Therefore, an RNA-Seq analysis may be more useful for identifying critical pathways and genes influencing PRA.
CONCLUSION
The present study is the first to report about GWAS analysis together pathway and gene-set enrichment analysis for PRA in dogs. Our results showed new insight into gene sets and pathways associated to PRA suggesting that this disease is the influence of combined consequence of a variety of genes work together in a pathway. Out of many GO term for PRA, in particular calcium ion binding, endoplasmic reticulum, homophilic cell adhesion via plasma membrane adhesion molecules, apoptotic process were found to be strongly related to PRA trait. Among the pathways, two pathways (focal adhesion and cGMP-PKG signaling) were observed to be closely related to PRA. Thus, the genes associated in highlighted GOs and pathways identified may be valuable candidate for further studies regarding the underlying cause of PRA disease. However, additional gene expression studies are needed to identify rare genes and regulatory SNPs which play significant role behind the PRA disease.
SUPPLEMENTARY MATERIALS
Supplementary Tables
Acknowledgements
This study was supported by 2020 the RDA Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.
Competing interests
No conflict of interest exists among authors associated to current article.
Funding sources
The present research was funded by AGENDA project (Grant no. PJ012833022019) of the National Institute of Animal Science, RDA, Korea.
Availability of data and material
The data and materials of present study can be accessible from the corresponding author after reasonable appeal.
Authors’ contributions
Conceptualization: Sheet S, Krishnamoorthy S, Choi BH.
Data curation: Krishnamoorthy S, Park W, Lim D
Formal analysis: Ko M.
Methodology: Sheet S, Krishnamoorthy S, Choi BH.
Software: Krishnamoorthy S, Ko M.
Validation: Park W, Park JE, Choi BH.
Investigation: Lim D, Choi BH.
Writing - original draft: Sheet S.
Writing - review &l editing: Krishnamoorthy S, Park W, Lim D, Park JE, Choi BH.
Ethics approval and consent to participate
An appropriate instruction articulated by The Institutional Animal Care and Use Committee (IACUC) of National Institute of Animal Science (NIAS).
REFERENCES
- 1.Wiik A, Ropstad E, Ekesten B, Karlstam L, Wade C, Lingaas F. Progressive retinal atrophy in Shetland sheepdog is associated with a mutation in the CNGA1 gene. Anim Genet. 2015;46:515–21. doi: 10.1111/age.12323. [DOI] [PubMed] [Google Scholar]
- 2.Parry H. Degenerations of the dog retina: II. generalized progressive atrophy of hereditary origin. Br J Ophthalmol. 1953;37:487–502. doi: 10.1136/bjo.37.8.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen-Jones SM. Animal models of human retinal dystrophies. Eye. 1998;12:566–70. doi: 10.1038/eye.1998.146. [DOI] [PubMed] [Google Scholar]
- 4.Magnusson H. Om nattblindhet hos hund sasom foljd av slaktskapsafvel. Sven Vet Tidskr. 1909;14:462–6. [Google Scholar]
- 5.Karlskov-Mortensen P, Proschowsky H, Gao F, Fredholm M. Identification of the mutation causing progressive retinal atrophy in Old Danish Pointing dog. Anim Genet. 2018;49:237–41. doi: 10.1111/age.12659. [DOI] [PubMed] [Google Scholar]
- 6.Parry H. Recent advances in canine medicine, with particular regard to blindness, viral hepatitis and post-infective neurological disorders. Vet Rec. 1951;63:323–6. doi: 10.1136/vr.63.18.323. [DOI] [PubMed] [Google Scholar]
- 7.Kelawala DN, Patil DB, Parikh PV, Sheth MJ, Joshi CG, Reddy B. Clinical studies on progressive retinal atrophy in 31 dogs. Iran J Vet Res. 2017;18:119–23. [PMC free article] [PubMed] [Google Scholar]
- 8.Miyadera K. Inherited retinal diseases in dogs: advances in gene/mutation discovery. J Anim Genet. 2014;42:79–89. doi: 10.5924/abgri.42.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasparini SJ, Llonch S, Borsch O, Ader M. Transplantation of photoreceptors into the degenerative retina: current state and future perspectives. Prog Retin Eye Res. 2019;69:1–37. doi: 10.1016/j.preteyeres.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Yoon S, Nguyen HCT, Yoo YJ, Kim J, Baik B, Kim S, et al. Efficient pathway enrichment and network analysis of GWAS summary data using GSA-SNP2. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao PY, Leung KH, Chan LW, Yip SP, Yap MK. Pathway analysis of complex diseases for GWAS extending to consider rare variants multi-omics and interactions. Biochim Biophys Acta Gen Subj. 2017;1861:335–53. doi: 10.1016/j.bbagen.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 12.Lee CM, Song DW, Ro WB, Kang MH, Park HM. Genome-wide association study of degenerative mitral valve disease in Maltese dogs. J Vet Sci. 2019;20:63–71. doi: 10.4142/jvs.2019.20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dadousis C, Pegolo S, Rosa GJM, Gianola D, Bittante G, Cecchinato A. Pathway-based genome-wide association analysis of milk coagulation properties, curd firmness, cheese yield, and curd nutrient recovery in dairy cattle. J Dairy Sci. 2017;100:1223–31. doi: 10.3168/jds.2016-11587. [DOI] [PubMed] [Google Scholar]
- 14.Dadousis C, Pegolo S, Rosa GJ, Bittante G, Cecchinato A. Genome-wide association and pathway-based analysis using latent variables related to milk protein composition and cheesemaking traits in dairy cattle. J Dairy Sci. 2017;100:9085–102. doi: 10.3168/jds.2017-13219. [DOI] [PubMed] [Google Scholar]
- 15.Fanous AH, Zhou B, Aggen SH, Bergen SE, Amdur RL, Duan J, et al. Genome-wide association study of clinical dimensions of schizophrenia: polygenic effect on disorganized symptoms. Am J Psychiatry. 2012;169:1309–17. doi: 10.1176/appi.ajp.2012.12020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downs LM, Wallin-Håkansson B, Bergström T, Mellersh CS. A novel mutation in TTC8 is associated with progressive retinal atrophy in the golden retriever. Canine Genet Epidemiol. 2014:1–4. doi: 10.1186/2052-6687-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li N, Zheng H, Cui J, Wang J, Liu H, Sun J, et al. Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice. 2019:12–24. doi: 10.1186/s12284-019-0285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pegolo S, Mach N, Ramayo-Caldas Y, Schiavon S, Bittante G, Cecchinato A. Integration of GWAS pathway and network analyses reveals novel mechanistic insights into the synthesis of milk proteins in dairy cows. Sci Rep. 2018:8–566. doi: 10.1038/s41598-017-18916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–97. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 24.Bolz H, Reiners J, Wolfrum U, Gal A. The role of cadherins in Ca2+-mediated cell adhesion and inherited photoreceptor degeneration. In: Baehr W, Palczewski K, editors. Photoreceptors and calcium. Boston: Springer; 2002. pp. 399–410. p. [DOI] [PubMed] [Google Scholar]
- 25.Hiscott P, Sheridan C, Magee RM, Grierson I. Matrix and the retinal pigment epithelium in proliferative retinal disease. Prog Retin Eye Res. 1999;18:167–90. doi: 10.1016/S1350-9462(98)00024-X. [DOI] [PubMed] [Google Scholar]
- 26.Herzlich AA, Patel M, Sauer TC, Chan CC. Retinal anatomy and pathology. In: Nguyen QD, Rodrigues EB, Farah M, Mieler WF, editors. Retinal pharmacotherapy e-book. Saunders Elsevier; 2010. pp. 5–14. p. [DOI] [Google Scholar]
- 27.Chen HJ, Ma ZZ. N-cadherin expression in a rat model of retinal detachment and reattachment. Invest Ophthalmol Vis Sci. 2007;48:1832–8. doi: 10.1167/iovs.06-0928. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Chung JY, Rai U, Esumi N, Lewin AS. Cadherins in the retinal pigment epithelium (RPE) revisited: P-cadherin is the highly dominant cadherin expressed in human and mouse RPE in vivo. PLOS ONE. 2018;13:e0191279. doi: 10.1371/journal.pone.0191279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchong MN, Chen D, Corson TW, Lee C, Harmandayan M, Bowles E, et al. Minimal 16q genomic loss implicates cadherin-11 in retinoblastoma. Mol Cancer Res. 2004;2:495–503. [PubMed] [Google Scholar]
- 30.Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang GQ, Hao Y, Wong F. Apoptosis: final common pathway of photoreceptor death in rd rds, and mutant mice. Neuron. 1993;11:595–605. doi: 10.1016/0896-6273(93)90072-Y. [DOI] [PubMed] [Google Scholar]
- 32.Doonan F, Donovan M, Cotter TG. Caspase-independent photoreceptor apoptosis in mouse models of retinal degeneration. J Neurosci. 2003;23:5723–31. doi: 10.1523/JNEUROSCI.23-13-05723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marigo V. Programmed cell death in retinal degeneration: targeting apoptosis in photoreceptors as potential therapy for retinal degeneration. Cell Cycle. 2007;6:652–5. doi: 10.4161/cc.6.6.4029. [DOI] [PubMed] [Google Scholar]
- 34.Finci L, Zhang Y, Meijers R, Wang JH. Signaling mechanism of the netrin-1 receptor DCC in axon guidance. Prog Biophys Mol Biol. 2015;118:153–60. doi: 10.1016/j.pbiomolbio.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teotia P, Van Hook MJ, Fischer D, Ahmad I. Human retinal ganglion cell axon regeneration by recapitulating developmental mechanisms: effects of recruitment of the mTOR pathway. Development. 2019;146:dev178012. doi: 10.1242/dev.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lippmann T, Pasternack SM, Kraczyk B, Dudek SE, Dekomien G. Indirect exclusion of four candidate genes for generalized progressive retinal atrophy in several breeds of dogs. J Negat Results Biomed. 2006:5–19. doi: 10.1186/1477-5751-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naureen K, Naqsh-e-Zahra S, Awan W. Molecular modeling of discoidin domain receptor tyrosine kinase 2 (DDR2) and ATPase Na+/K+transporting, alpha 4 polypeptide (ATP1A4): candidate genes for autosomal recessive cone-rod dystrophy (CORD8) Int J Rehabil Sci. 2012;1:53–7. [Google Scholar]
- 38.Landfried B, Samardzija M, Barben M, Schori C, Klee K, Storti F, et al. Digoxin-induced retinal degeneration depends on rhodopsin. Cell Death Dis. 2017;8:e2670. doi: 10.1038/cddis.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang WC, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT, et al. Robust endoplasmic reticulum-associated degradation of rhodopsin precedes retinal degeneration. Mol Neurobiol. 2015;52:679–95. doi: 10.1007/s12035-014-8881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Wang L, Huang K, Zheng L. Endoplasmic reticulum stress in retinal vascular degeneration: protective role of resveratrol. Invest Ophthalmol Visual Sci. 2012;53:3241–9. doi: 10.1167/iovs.11-8406. [DOI] [PubMed] [Google Scholar]
- 41.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Wang JJ, Yu Q, Wang M, Zhang SX. Endoplasmic reticulum stress is implicated in retinal inflammation and diabetic retinopathy. FEBS Lett. 2009;583:1521–7. doi: 10.1016/j.febslet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ha Y, Dun Y, Thangaraju M, Duplantier J, Dong Z, Liu K, et al. Sigma receptor 1 modulates endoplasmic reticulum stress in retinal neurons. Invest Ophthalmol Visual Sci. 2011;52:527–40. doi: 10.1167/iovs.10-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos ARC, Corredor RG, Obeso BA, Trakhtenberg EF, Wang Y, Ponmattam J, et al. β1 integrin-focal adhesion kinase (FAK) signaling modulates retinal ganglion cell (RGC) survival. PLOS ONE. 2012;7:e48332. doi: 10.1371/journal.pone.0048332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gariano RF, Gardner TW. Retinal angiogenesis in development and disease. Nature. 2005;438:960–6. doi: 10.1038/nature04482. [DOI] [PubMed] [Google Scholar]
- 46.Kornberg LJ, Shaw LC, Spoerri PE, Caballero S, Grant MB. Focal adhesion kinase overexpression induces enhanced pathological retinal angiogenesis. Invest Ophthalmol Visual Sci. 2004;45:4463–9. doi: 10.1167/iovs.03-1201. [DOI] [PubMed] [Google Scholar]
- 47.Li G, Anderson RE, Tomita H, Adler R, Liu X, Zack DJ, et al. Nonredundant role of Akt2 for neuroprotection of rod photoreceptor cells from light-induced cell death. J Neurosci. 2007;27:203–11. doi: 10.1523/JNEUROSCI.0445-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paquet-Durand F, Hauck SM, Van Veen T, Ueffing M, Ekström P. PKG activity causes photoreceptor cell death in two retinitis pigmentosa models. J Neurochem. 2009;108:796–810. doi: 10.1111/j.1471-4159.2008.05822.x. [DOI] [PubMed] [Google Scholar]
- 49.Tolone A, Belhadj S, Rentsch A, Schwede F, Paquet-Durand F. The cGMP pathway and inherited photoreceptor degeneration: targets, compounds, and biomarkers. Genes. 2019:10–453. doi: 10.3390/genes10060453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He YG, Liao D, Zhao B, Owens J, Johnston JM. Evidence for a role of platelet-activating factor (PAF) in the pathogenesis of age-related macular degeneration (AMD) Invest Ophthalmol Visual Sci. 2007:48–2200. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables