Abstract
Ciliopathies affect a variety of tissues during development including the heart, kidneys, respiratory tract, and retina. Though an increasing number of monogenic causes of ciliopathies have been described, many remain unexplained. Recently, recessive variants in NUP93 and NUP205 encoding two proteins of the inner ring of the nuclear pore complex were implicated as causes of steroid resistant nephrotic syndrome. In addition, we previously found that the inner ring nucleoporins NUP93 and NUP188 function in proper left-right patterning in developing embryos via a role at the cilium. Here, we describe the role of an additional inner ring nucleoporin NUP205 in cilia biology and establishment of normal organ situs. Using knockdown in Xenopus, we show that Nup205 depletion results in loss of cilia and abnormal cardiac morphology. Furthermore, by transmission electron microscopy, we observe a loss of cilia and mispositioning of intracellular ciliary structures such as basal bodies and rootlets upon depleting inner ring nucleoporins. We describe a model wherein NUP93 interacting with either NUP188 or NUP205 is necessary for cilia. We thus provide evidence that dysregulation of inner ring nucleoporin genes that have been identified in patients may contribute to pathogenesis through cilia dysfunction.
1. Introduction
Ciliopathies are a group of diseases characterized by genetic mutations that result in either abnormal formation or function of cilia (Braun and Hildebrandt, 2017). Since cilia are a component of nearly all cells in developing vertebrate organisms, cilia dysfunction results in pathologies that include hydrocephalus, polycystic kidney disease, retinal dystrophy, as well as congenital heart disease (CHD) (Bachmann-Gagescu and Neuhauss, 2019; Chaudhry and Henderson, 2019; Devlin and Sayer, 2019; Park et al., 2019). Advances in human genomics have led to the identification of numerous CHD candidate genes (Fakhro et al., 2011; Homsy et al., 2015; Jin et al., 2017). Given the relationship between cilia and heart development, the functional testing of these CHD candidate genes promises to uncover additional regulators of cilia structure and function.
Our recent efforts investigated one such candidate CHD gene NUP188, which is a key component of the nuclear pore complex (NPC). The NPC is a 100 MDa macro-molecular assembly that spans the nuclear envelope and mediates molecular exchange between the cytoplasm and nucleoplasm (Hampoelz et al., 2019). The NPC’s central transport channel is filled with intrinsically disordered Phe-Gly (FG) rich proteins. These FG-rich nucleoporins (nups) establish a size-selective diffusion barrier and provide binding sites for shuttling nuclear transport receptors (karyopherins, importins, and exportins) that ferry signal-bearing cargo across the NPC (Cautain et al., 2015). The FG-nups line a cylindrical scaffold consisting of concentric inner and outer ring complexes formed by repeating modular units of the NUP93 and NUP107-160 subcomplexes, respectively. The NUP93 subcomplex is the major subunit of the inner ring of the NPC and consists of NUP93, NUP35/53, NUP188 and NUP205 (Kosinski et al., 2016). NUP205 is a likely evolutionary paralog of NUP188 (Beck and Hurt, 2017) and both directly interact with NUP93, albeit mutually exclusively.
Recent work has underlined the importance of specific nup function in a developmental context. For example, both inner and outer-ring nups have distinct roles during renal development with dysfunction leading to nephrotic syndrome (Braun et al., 2018; Braun et al., 2016; Miyake et al., 2015). Nup variants may also contribute to a variety of congenital neurological diseases (Tullio-Pelet et al., 2000; Zanni et al., 2019). Further, it is emerging that, like its paralog NUP188, NUP205 may also impact CHD (Chen et al., 2019). Interestingly, in our search for a plausible mechanism for CHD pathogenesis, we discovered that Nup188 and Nup93 localize to the bases of cilia and are crucial for proper cilia development and organ situs (Del Viso et al., 2016). The extent to which other nups are required for cilia remains to be fully understood. Thus, given our finding that some inner ring nups localize to the cilium base and are essential for cilia, we sought to test the hypothesis that NUP205 may also play a role at cilia. Here, we describe a role for NUP205 in the cilia of Xenopus, which provides a plausible mechanism for CHD.
2. Results
2.1. Nup205 is Necessary for proper left-right patterning
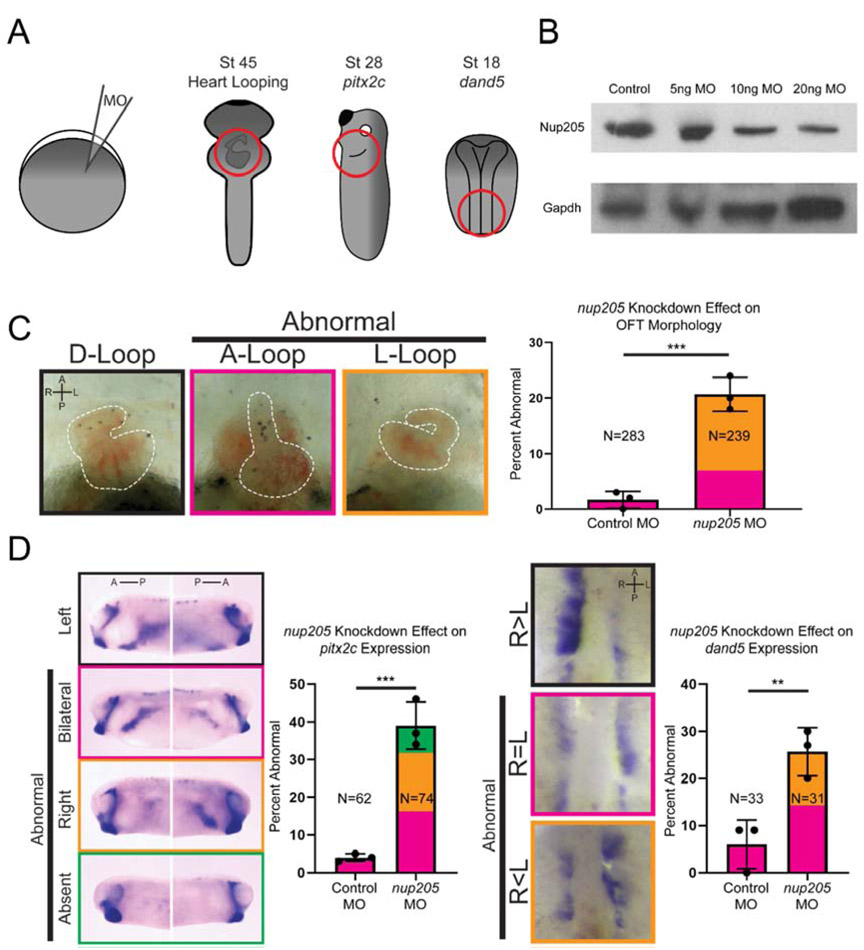
Xenopus is an advantageous model for studying situs and organ patterning (Fig. 1A). Gene dosage can be carefully controlled to test both gain and loss of function in a technically simple and cost-effective manner. Also, Xenopus has organ features such as a septated atria and trabeculations within the heart that resemble that of humans more closely than other aquatic organisms (Hwang et al., 2019; Sempou and Khokha, 2019). By injecting morpholino oligos (MOs) into one-cell embryos, we can obtain efficient knockdown of gene products. We tested the depletion of Nup205 in Xenopus and evaluated cardiac situs (Fig. 1B and C). During embryonic development, the heart initially forms as a midline heart tube that undergoes complex morphogenesis to loop to the right (a D loop). Remarkably, 21% of injected embryos displayed abnormal A (midline) or L (left) loop (Fig. 1C). Critically, we confirmed the specificity of our MOs by replicating our findings through the use of CRISPR based mosaic knockout of nup205 which displayed a similar phenotype (Fig. S1). In addition, expression of human NUP205 via electroporation could rescue the Nup205 MO depletion phenotype (see below).
Fig. 1. Knockdown of nup205 Disrupts LR Patterning.
(A) Schematic of one-cell injections and regions analyzed for LR patterning defects. (B) Representative blot of Nup205 protein levels in whole stage 45 Xenopus protein extracts. (C) Representative images of normal and abnormal cardiac OFT directionality (ventral views with anterior to the top) along with quantitation of frequency of abnormal morphology in control and nup205 morphant embryos. (D) Representative images of normal and abnormal pitx2c expression patterns (lateral views, dorsal to the top) along with quantitation of frequency of abnormal patterns in control and nup205 morphant embryos. (E) Representative images of normal and abnormal dand5 expression patterns (ventral views, anterior to the top) along with quantitation of frequency of abnormal patterns in control and nup205 morphant embryos. For all graphs, Ns indicate number of individual embryos assessed across a minimum of 3 replicates. Graph colors correspond to example border colors. Bar graph color coding depicts proportion of total MCCs assessed, while error bar is based on frequency of combined abnormal phenotypes. p<0.005 and p<0.0005 for ** and *** respectively by two-tailed T-Test. A – anterior, P – posterior, R – right, L-left.
To further ascertain the mechanism by which Nup205 contributes to organ patterning and situs, we examined the expression of two well characterized markers of left-right (LR) patterning at different stages of embryonic development: pitx2c and dand5. Pitx2c is a transcription factor expressed in the left lateral plate mesoderm and is essential for laterality of internal organs (Campione et al., 1999). Dand5 is a nodal antagonist which acts upstream of pitx2c and is initially expressed symmetrically at the flanks of the Left-Right Organizer (LRO), but later becomes asymmetrically expressed as the left-right axis is established (stage 19 in Xenopus) (Schweickert et al., 2010; Vonica and Brivanlou, 2007). In nup205 morphants the pattern of pitx2c expression was abnormal (Fig. 1D) in 39% of embryos. Depletion of Nup205 also led to abnormal dand5 expression (Fig. 1E) in 26% of embryos. Taken together, these data indicate a global defect in left-right patterning at the earliest steps in the left-right signaling cascade, likely due to abnormalities within the LRO.
As NUP205 dysfunction has previously been implicated in nephrotic syndrome (Braun et al., 2016), we additionally assessed kidney development in the context of nup205 depletion. In embryos injected at the two-cell stage, there was dysfunctional pronephric development on the side of the embryo injected with nup205 MO as compared to the contralateral side injected with control MO (Fig. S2A).
2.2. Nup205 is necessary for cilia in the LRO, epidermis, and pronephros
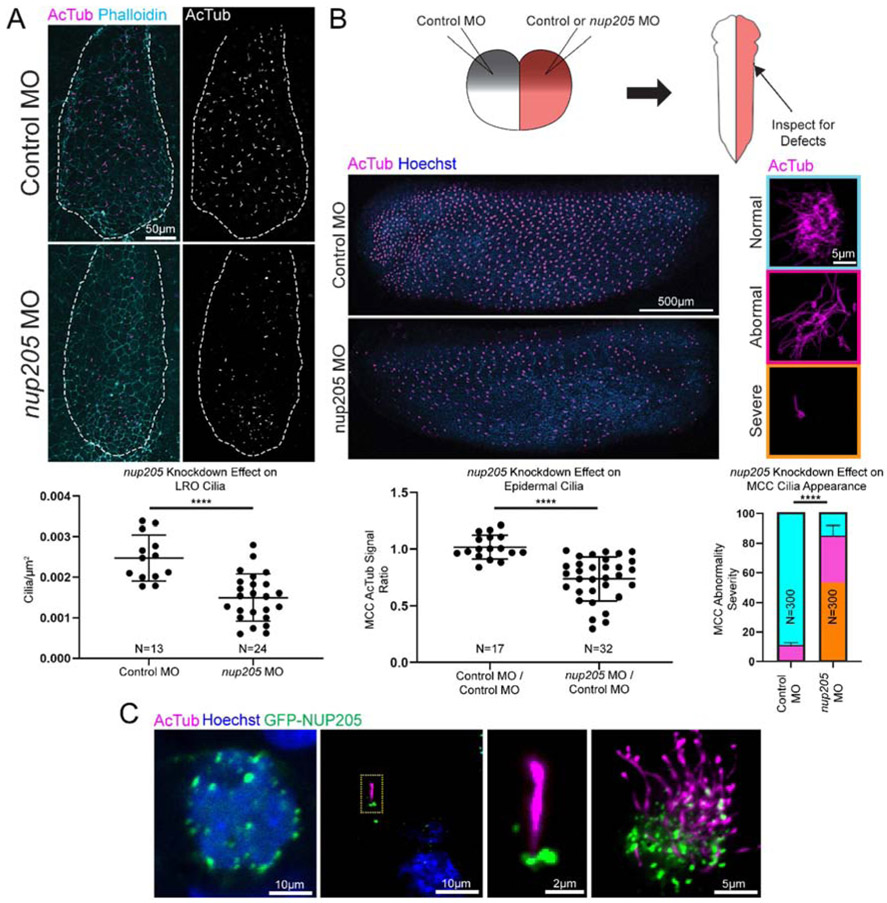
LR asymmetry is established by cilia within the LRO. Cilia beating generates a predominantly leftward extracellular fluid flow sensed by immotile primary cilia at the LRO periphery (Boskovski et al., 2013; McGrath et al., 2003; Nonaka et al., 1998; Okada et al., 1999; Schweickert et al., 2007). Since Nup205 morphants had abnormal dand5 and pitx2c expression, we speculated that cilia might be disrupted in these embryos. To address this hypothesis, we directly assayed cilia in the LRO by immunofluorescence staining of acetylated α-tubulin. In nup205 morphant LROs, there was a significant 40% reduction in cilia number compared to the LROs of control morphant embryos (Fig. 2A). We further assayed whether this cilia loss was generalizable to other tissues in the developing embryo. The embryonic epidermis of Xenopus has an array of multiciliated cells (MCCs) that utilize cilia to generate extracellular fluid flow reminiscent of the mammalian tracheal epithelium. In embryos injected at the two-cell stage, there was a loss of cilia in the MCCs on the side of the embryo injected with nup205 MO as compared to the contralateral side injected with control MO (Fig. 2B). Finally, given the kidney phenotype of patients, we examined the embryonic Xenopus pronephos, which is lined by cilia. In pronephroi depleted of nup205 there was also a loss of cilia (Fig. S2B).
Fig. 2. Knockdown of nup205 Disrupts Cilia in Multiple Tissues.
(A) Representative images and quantitation of cilia number for control and nup205 morphant embryo LROs (ventral view with anterior to the top). Ns indicate number of distinct LROs analyzed. (B) Schematic of two-cell injections and representative images of control and nup205 morphant sides of an embryo (lateral view with dorsal to the top, top panel left side, bottom panel right side) and severity of ciliation defects in individual MCCs along with quantitation of remaining multiciliated structures and severity of MCC defect frequency. Ns indicate number of distinct embryos for which both sides were analyzed (dot plot) and numer of MCCs analyzed on either control MO or nup205 MO injected sides of embryos with no more than 10 MCCs assessed per a single side of an embryo (bar graph). Graph colors correspond to example border colors. Bar graph color coding depicts proportion of total MCCs assessed, while error bar is based on frequency of combined severe and abnormal phenotypes. (C) Representative localization of overexpressed GFP-NUP205 at the nuclear periphery and cilium in the Xenopus LRO and MCCs (yellow dotted line designates magnified area). p<0.0001 for **** by two-tailed T-Test for dot plots or Chi Squared Test for bar graph. Graphed values were obtained across three replicates.
For the inner ring nups, Nup188 and Nup93, we previously demonstrated that they localized to the cilium base. Unfortunately, we did not have a suitable antibody for immunofluorescence detection of Nup205; therefore, we examined the localization of an N terminal tagged human GFP-NUP205 overexpressed in the LRO and MCCs. We observed localization both at the nuclear periphery as well as at the cilium base with GFP-NUP205 (Fig. 2C).
2.3. Analysis of inner ring nup morphants
Because cilia in MCCs are so easily accessible and plentiful, we next considered mechanisms that could explain the role of nup205 in cilia in this context. First, we considered the possibility that MCC cell fate specification could be disrupted. To test this, we used whole mount in situ hybridization to assess dnah9 and foxj1, markers of ciliated cell fate (Vick et al., 2009). These markers appeared unchanged in MO knockdown embryos of nup205 (Fig. S3). We then assayed whether depletion of nup205 could influence the generation of basal bodies required for ciliogenesis in MCCs. We assessed this through quantification of basal bodies in MCCs depleted of Nup205 as compared to contralateral control MO injected MCCs (Fig. S4). There was no significant difference in basal body number as observed through immunofluorescent labeling of γ-tubulin. We did however observe an abnormal distribution of basal bodies along the apicobasal axis. Normally, the basal bodies are docked at the apical surface of the cell, but in Nup205 morphants, multiple basal bodies could be seen sub-apically (Fig. S4).
We next turned to transmission electron microscopy (TEM) to examine the ultrastructural abnormalities in cilia in morphant embryos. We performed TEM on MCCs of stage 28 embryos at which point basal bodies have normally docked apically, and cilia are mature in control cells (Fig. 3A). Compared to control MO injected embryos, a subset of basal bodies and rootlets in Nup205 depleted embryos were mis-localized away from their stereotyped position near the apical cell surface. In fact, this phenotype is similar to that seen when other inner ring nups are depleted including Nup93 and Nup188 (Fig. 3B), suggesting a redundant role for these nups in apical localization of the basal bodies. Consistent with this idea, this phenotype was particularly severe in the context of depletion of Nup93 or depletion of both Nup188 and Nup205 (Fig. 3B,C).
Fig. 3. Knockdown of Inner-Ring NUPs Disrupts Cilia.
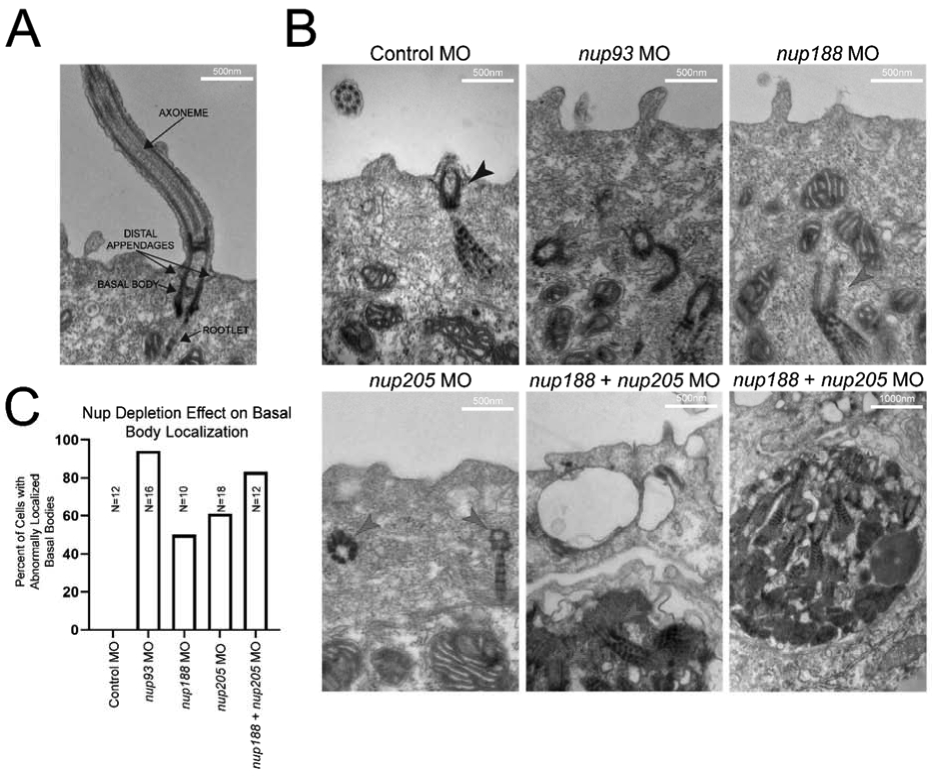
(A) Examples of cilia components. (B) Representative images of cilia within MCCs of control and inner-ring NUP knockdown morphant embryo epidermis. Black arrowheads indicate properly positioned ciliary components, red arrowheads indicate mispositioned ciliary components beneath the cell surface. Note the apparent inclusion body with multiple ciliary components in the cases of nup188 + nup205 knockdown. (C) Percentages of visualized cells with abnormally positioned basal bodies at cell surface for each condition (basal bodies within 100nm of surface were considered to be normally localized). Ns indicate number of cells assessed from a single embryo per condition.
2.4. Nup188 and Nup205 are functionally redundant for cilia in MCCs
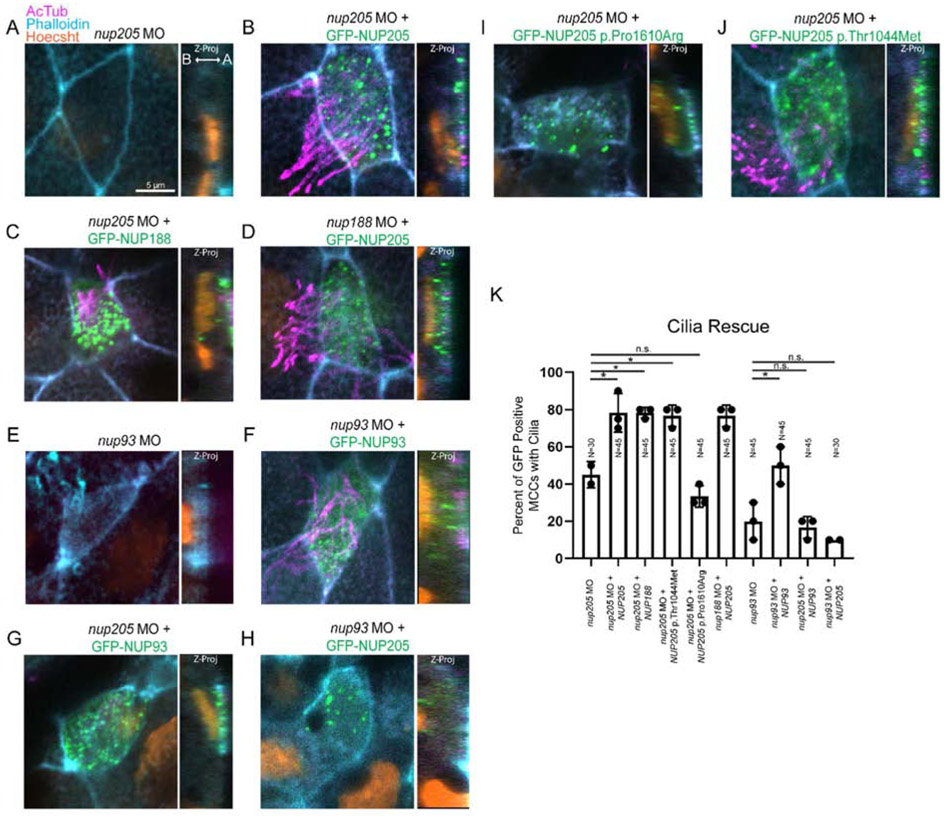
When we depleted nup93 or depleted nup188 and nup205 in combination, basal bodies failed to reach the apical cell surface, a prerequisite for cilia. Strikingly, in the context of Nup188 and Nup205 depletion, we observed abnormal electron dense bodies that included structures reminiscent of rootlets and basal bodies suggesting that these structures had formed an intracellular inclusion body (Fig. 3B). Since there appeared to be a synergistic effect of codepleting Nup188 and Nup205, and Nup188 and Nup205 are paralogous, we strongly suspected that they may carry out overlapping functions required for cilia in the MCCs.
We wished to directly test whether these inner ring nups were interchangeable in their function via rescue experiments. Another important consideration is whether these nups regulate cilia via a role at NPCs or at the base of the cilium. Recently, we demonstrated that turnover of Nup188 within the pericentriolar material is much faster than at the NPCs (Vishnoi et al., 2020). Thus, we speculated that introducing these inner ring nups as the MCCs form could preferentially target them to the basal bodies rather than the NPCs. We depleted Nup205, Nup188, and Nup93 through microinjection of a MO into the 1 cell stage embryo. We then electroporated plasmids encoding either human NUP205, NUP188 or NUP93 each tagged with GFP at stage 16. The use of electroporation allowed us to temporally control our rescue experiments. With electroporation, we could introduce exogenous nups after the terminal differentiation of the MCCs. We speculated that at this late stage exogenous nups would preferentially localize to the bases of cilia rather than at the NPC, because inner ring nups are extremely stable and do not turnover from NPCs (D’Angelo et al., 2009; Savas et al., 2012; Toyama et al., 2013). And indeed, we did not observe marked accumulation of GFP-nups at the nuclear envelope, but instead they clearly decorated the bases of cilia in MCCs. Remarkably, despite not localizing to NPCs, we observed a clear rescue of cilia in the nup205 morphants (Fig. 4A and B). Thus, we favor an interpretation that this rescue is due to a direct function of Nup205 at cilia bases. Interestingly, and consistent with our hypothesis, the expression of GFP-NUP188 was also able to rescue cilia in nup205 morphants to levels equal to that of NUP205 (Fig. 4C). And likewise, the expression of NUP205 could complement cilia loss in nup188 morphants (Fig. 4D). These data reinforce the concept that Nup188 and Nup205 are functionally interchangeable in this context. In contrast, only NUP93 could complement cilia loss in nup93 morphants (Fig. 4E-H). Overall, these data support a model where both Nup93 and either Nup205 or Nup188 are required for cilia through a specific function at cilia bases, not NPCs.
Fig. 4. Inner-Ring Nucleoporin Roles Overlap.
(A-D) Knockdown of nup205 loss of MCC cilia can be rescued with GFP-NUP188 or GFP-NUP205 respectively. Similarly, knockdown of nup188 loss of MCC cilia can be rescued with GFP-NUP205 (E-H) Knockdown of nup93 and loss of MCC cilia can be rescued with GFP-NUP93, but knockdown of nup205 and loss of MCC cilia cannot be rescued not be rescued with GFP-NUP93 nor can knockdown of nup93 and loss of MCC cilia be rescued with GFP-NUP205 (I-J) Knockdown of nup205 and loss of MCC cilia can be rescued with GFP-NUP205(3131C>T) but not GFP-NUP205(4829C>G). (K) Quantitation of rescue efficiencies amongst morphant and reintroduced NUPs/NUP variants with pairwise comparisons assessed via two-tailed T-test with * designating p < 0.05. Ns indicate number of MCCs assessed with 3 MCCs assessed per embryo across 3 replicates per condition. A: Apical B: Basal.
2.5. Patient Nup205 variants are dysfunctional at cilia
As expression of human NUP205 could functionally complement the loss of epidermal cilia in Xenopus morphant MCCs, it provided the opportunity to test whether two recently identified CHD patient variants in NUP205, NUP205 p.Thr1044Met and NUP205 p.Pro1610Arg (Chen et al., 2019), may impact cilia function. Indeed, NUP205 p.Pro1610Arg failed to rescue cilia loss in nup205 morphants suggesting that it may be a loss of function allele (Fig. 4I). However, NUP205 p.Thr1044Met was able to complement cilia loss to levels similar to wildtype NUP205 (Fig. 4J). By assessing the available structural information for the interaction between NUP205 and NUP93 in the context of the NPC (Kosinski et al., 2016), the NUP205 p.Pro1610Arg variant would introduce a change that may increase steric hindrance and alter the local polarity of the protein structure of NUP205 at a close proximity to NUP93, potentially impairing their interaction (Fig. S5).
3. Discussion
Here, we provide additional evidence that disruption of specific nups that comprise the inner ring of the NPC contribute to LR patterning in the developing embryo and provide a rationale for why NUP188 and NUP205 could contribute to CHD. Based on our findings, we suggest that Nup205, like Nup188, acts to ensure the formation of both primary cilia (e.g. in the LRO) and motile cilia in the MCCs of the Xenopus epidermis. Indeed, our data are most consistent with the interpretation that Nup188 and Nup205 act redundantly as simultaneous depletion of both transcripts results in the severe loss of cilia in MCCs (Fig. 3). Further, it is clear from our complementation analysis that NUP188 and NUP205 are functionally interchangeable; these data are consistent with the idea that NUP188 and NUP205 function as paralogs.
The conclusion that NUP188 and NUP205 can functionally substitute for one another is also borne out by their established physical interactions with NUP93. In vitro reconstitution studies have demonstrated that NUP93 either binds to NUP205 or NUP188 but is unable to bind them both simultaneously (Amlacher et al., 2011; Theerthagiri et al., 2010). Our genetic data nicely mirror this biochemistry as loss of Nup93 would be predicted to phenocopy the co-depletion of Nup205 and Nup188, which it does (Fig. 3). Further, it would predict that neither NUP205 nor NUP188 would be able to complement the cilia loss in nup93 morphants, as is seen in our studies (Fig. 4). While a subset of Nup205 proteins appears to localize apically even upon depletion of Nup93 (Fig. 4H) this pool of Nup205 is smaller than when we deplete Nup188 or Nup205. As we are overexpressing Nup205, the apical localization of the protein in the context of Nup93 depletion may simply be a result of overexpression which is consistent with the fact that cilia are not recovered. Thus, these data suggest that Nup93 may be required for proper function of either Nup188 or Nup205 at the cilium base and suggests that the biochemical interactions that help build the NPC may also be ported to the cilium as well. Such a hypothesis is further supported by the inability of the NUP205 p.Pro1610Arg allele to complement the cilia loss in the nup205 morphant, as this mutation potentially disrupts the biochemical interaction between NUP205 and NUP93 as observed from patient cells (Chen et al., 2019).
That key interactions that help build the inner ring of the NPC may also be relevant at cilia bases underlines the persistent, and still unmet, challenge of defining nup alleles that specifically disrupt a cilia function while conclusively leaving a nuclear transport function intact. A clear demonstration that NUP205 is recruited to the cilium base by NUP93 would certainly help solidify these assumptions. In our complementation studies, exogenous nup-GFP that rescues cilia appears localized predominately outside the nucleus. However, we cannot rule out that some incorporation into the NPCs may be relevant for this rescue of cilia. Of note, recent studies suggest that NUP205 may in fact physically interact with Nephrocystin-4 and Inversin, components of the ciliary transition zone (Blasius et al., 2019; Sang et al., 2011). While this certainly supports the concept that NUP205 may function at cilia, it is inconsistent with our genetic analysis and the idea that NUP205 is interchangeable with NUP188 and may thus share a similar localization and function at cilia bases. Specifically, as our prior work suggests that NUP188 is a component of pericentriolar material (PCM) (Del Viso et al., 2016), it is localized below the transition zone where it directly interacts with the PCM component CEP152 (Vishnoi et al., 2020). We recognize, however, that a higher resolution view of Nup205 localization at cilia bases remains to be presented and is the focus of our future work.
While a definitive demonstration of Nup205’s localization at cilia bases remains to be established, we argue that the cilia defects observed at the ultrastructural level suggest a key role for these proteins in ciliogenesis rather than the establishment of a ciliary diffusion barrier, as has been previously suggested (Kee et al., 2012). Specifically, we observe the striking accumulation of basal bodies and rootlets throughout the cytosol suggesting a stalling of the ciliogenesis pathway possibly before plasma membrane docking. This could conceivably stem from a role related to NPHP proteins that they have been proposed to interact with inner ring nups at cilia (Blasius et al., 2019; Sang et al., 2011). Nephrocystin-4 in particular has been shown to function in the regulation of cortical F-actin that forms a framework for ciliation in MCCs (Mollet et al., 2005; Yasanuga et al., 2015). Meanwhile additional regulators of cortical actin such as WDR5 have emerged as crucial components of the ciliation process in MCCs (Kulkarni et al. 2018). Thus, exploring this potential crosstalk between nups and actin networks in ciliated cells could provide further insight into the role of nups in cilia function. This concept awaits direct testing but could provide a unifying model to explain why these proteins function in two different subcellular locations in ways that are directly relevant to human health.
Finally, a driving force to understand the role of nups at the bases of cilia is the accumulating evidence that several nups play a role in developmental disease states (Braun et al., 2018; Braun et al., 2016; Chen et al., 2019; Miyake et al., 2015; Tullio-Pelet et al., 2000; Zanni et al., 2019). Our data recapitulate the laterality defects seen in patients with NUP205 variants and support that some of these NUP205 alleles are indeed loss of function supporting their role in disease causality. Establishing disease causality of these variants is an important step in further understanding both the disease and the biology that underlies it. Overall, this work serves to further support the role for nups in the context of cilia but also the importance of these functions for human disease.
4. Materials and methods
4.1. Xenopus Embryonic Manipulations
Xenopus tropicalis were housed and cared for according to established protocols approved by Yale Institutional Animal Care and Use Committee.
Using standard protocols, we injected MOs with fluorescent tracers or guide RNAs with Cas9 protein into one or two-cell Xenopus embryos, and assayed gene expression through WISH and cardiac looping by visual inspection through the translucent ventral aspect of the embryos. The following translation blocking MOs were injected: nup93 (5 ng 5’-TCCAAACCCTTCTCCATCCATTGTC-3’), nup188 (5 ng 5’-CCATCTTCACGCC CCCTTCACGGCC-3’), nup205 (10 ng 5’-ATTTAGCGCCAACTGCGCCGCCATC-3’). The guide RNA used for CRISPR/Cas9 editing targeted GGAAATATGGCAGACCGTGG in the first exon of nup205. Editing was assessed via fragment analysis (Forward Primer: 5’-TCCCAGTCACGACGTGACGTTAAACTGAGCAACG ACC-3’, Reverse Primer: 5’-GCACCATGTTCTGTGTTCCAC-3’) as previously described (Bhattacharya et al., 2015). Stage 16 embryos were electroporated with plasmids at a concentration of 200 ng/μl. Electroporation was carried out in 1/9 modified Ringer’s solution in chambers containing a platinum plate below the embryo while holding a platinum electrode above, with five pulses of 5 V (50 ms on, 100 ms off). Electroporated plasmids were composed of the pCDNA6.2-N-emGFP backbone along with either NUP93, NUP188, or NUP205. NUP205 variant sequence containing plasmids were generated using the Q5 site directed mutagenesis kit (NEB). Prior to processing, embryos were fixed in 4% paraformaldehyde for 1-2 hours at room temperature except for embryos where γ-tubulin antibody staining was carried out in which case fixation was carried out in Dent’s fixative overnight at 4 degrees Celsius.
4.2. Whole mount in situ hybridization
WISH was carried out as previously described (Henrique et al., 1995). Briefly, Xenopus embryos were fixed in 4% paraformaldehyde with 0.1% glutaraldehyde and dehydrated through washes in methanol. Embryos were rehydrated in PBS with 0.1% tween-20. Embryos were then hybridized with digoxigenin-labeled antisense RNA probes complementary to pitx2c, dand5, dnah9, or foxj1 generated using the T7 High Yield RNA Synthesis kit (NEB, E2040S) and DIGdUTP (Sigma). Embryos were then washed and blocked prior to incubation with anti-DIG-Fab fragments (Roche) overnight at 4 degrees Celsius. BM purple (Sigma) was used to visualize expression prior to post-fixation in 4% paraformaldehyde with 0.1% glutaraldehyde.
4.3. Transmission electron microscopy
Stage 26 Xenopus embryos were fixed with Karnovsky fixative for 1 hour at 4 degrees Celsius, washed with 0.1 M sodium cacodylate (pH 7.4), then post-fixed with Palade’s osmium for 1 h at 4 degrees Celsius, shielded from light. Following a second wash, embryos were stained with Kellenburger’s solution for 1 hour at room temperature, washed in double distilled water, then dehydrated in a series of ethanol, propylene oxide, 50/50 propylene oxide/epon, and incubated in 100% epon. Embedded embryos were sectioned at 400nm before staining with 2% uranyl acetate. Images were obtained using a FEI Tecnai G2 Spirit BioTWIN electron microscope.
4.4. Immunofluorescence microscopy
Fixed samples were first briefly permeabilized in 0.1% tween-20 in phosphate buffered saline (PBS). Samples were then cleared in Sca/eS4 for 5 days (Hama et al., 2015). Samples were then blocked overnight at 4 degrees Celsius in 10% goat serum in PBS, incubated in primary antibody overnight at 4 degrees Celsius, and incubated in secondary antibody overnight at 4 degrees Celsius. Antibodies are listed in Supplemental Table 1. Immunofluorescence images were obtained with either a Zeiss Observer outfitted with optical interference (Apotome) microscopy (Fig. 2B and C) or a Zeiss LSM880 confocal microscope (Fig. 2A, Fig.4A-J, and Fig. S2B). Fluorescence images were processed and analyzed with Fiji (Schindelin et al., 2012). For cilia and basal body counting the “analyze particles” function was employed (Fig. S6).
Statistical analysis
All experiments were performed a minimum of three times and numbers stated in graphs are the composite of multiple experiments. Statistical significance for each experiment was tested using two-tailed T-tests in GraphPad Prism version 8 (GraphPad Software; www.graphpad.com). For all experiments, statistical significance was defined as p < 0.05.
Supplementary Material
Nup205 is required for cilia and development of the left/right axis in Xenopus embryos.
Depletion of inner-ring nups causes defects in basal body distribution.
Nup205 and Nup188 have overlapping roles at cilia.
The Nup205 p.Pro1610Arg variant uncovered in a patient with congenital heart disease and heterotaxy is dysfunctional at cilia.
Acknowledgements:
We wish to thank Michael Slocum and Sarah Kubek for animal husbandry. We are indebted to the Yale Center for Cellular and Molecular Imaging core for fluorescence microscopy and electron microscopy resources. We thank Friedhelm Hildebrandt for kindly providing the NUP expression plasmids used in this study.
Funding
J.M. was supported by the Yale MSTP NIH T32GM007205 Training Grant, the Yale Predoctoral Program in Cellular and Molecular Biology T32GM007223 Training Grant, and the Paul and Daisy Soros Fellowship for New Americans. This work was supported by the NIH R01HL124402 Grant to C.P.L. and M.K.K.
Footnotes
Data and materials availability
All data is available in the main text or the supplementary materials.
Declaration of competing interests
Authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E, 2011. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146, 277–289. [DOI] [PubMed] [Google Scholar]
- Bachmann-Gagescu R, Neuhauss SC, 2019. The photoreceptor cilium and its diseases. Curr Opin Genet Dev 56, 22–33. [DOI] [PubMed] [Google Scholar]
- Beck M, Hurt E, 2017. The nuclear pore complex: understanding its function through structural insight. Nature reviews. Molecular cell biology 18, 73–89. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK, 2015. CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev Biol 408, 196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius TL, Takao D, Verhey KJ, 2019. NPHP proteins are binding partners of nucleoporins at the base of the primary cilium. PLoS One 14, e0222924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK, 2013. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature 504, 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Hildebrandt F, 2017. Ciliopathies. Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Lovric S, Schapiro D, Schneider R, Marquez J, Asif M, Hussain MS, Daga A, Widmeier E, Rao J, Ashraf S, Tan W, Lusk CP, Kolb A, Jobst-Schwan T, Schmidt JM, Hoogstraten CA, Eddy K, Kitzler TM, Shril S, Moawia A, Schrage K, Khayyat AIA, Lawson JA, Gee HY, Warejko JK, Hermle T, Majmundar AJ, Hugo H, Budde B, Motameny S, Altmuller J, Noegel AA, Fathy HM, Gale DP, Waseem SS, Khan A, Kerecuk L, Hashmi S, Mohebbi N, Ettenger R, Serdaroglu E, Alhasan KA, Hashem M, Goncalves S, Ariceta G, Ubetagoyena M, Antonin W, Baig SM, Alkuraya FS, Shen Q, Xu H, Antignac C, Lifton RP, Mane S, Nurnberg P, Khokha MK, Hildebrandt F, 2018. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest 128, 4313–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, Gee HY, Ashraf S, Lawson JA, Shril S, Airik M, Tan W, Schapiro D, Rao J, Choi WI, Hermle T, Kemper MJ, Pohl M, Ozaltin F, Konrad M, Bogdanovic R, Buscher R, Helmchen U, Serdaroglu E, Lifton RP, Antonin W, Hildebrandt F, 2016. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nature genetics 48, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M, 1999. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development (Cambridge, England) 126, 1225–1234. [DOI] [PubMed] [Google Scholar]
- Cautain B, Hill R, de Pedro N, Link W, 2015. Components and regulation of nuclear transport processes. FEBS J 282, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry B, Henderson DJ, 2019. Cilia, mitochondria, and cardiac development. J Clin Invest 129, 2666–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Y, Yang S, Shi Z, Zeng W, Lu Z, Zhou X, 2019. Bi-Allelic Mutations in NUP205 and NUP210 Are Associated With Abnormal Cardiac Left-Right Patterning. Circ Genom Precis Med 12, e002492. [DOI] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW, 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Viso F, Huang F, Myers J, Chalfant M, Zhang Y, Reza N, Bewersdorf J, Lusk CP, Khokha MK, 2016. Congenital Heart Disease Genetics Uncovers Context-Dependent Organization and Function of Nucleoporins at Cilia. Dev Cell 38, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin LA, Sayer JA, 2019. Renal ciliopathies. Curr Opin Genet Dev 56, 49–60. [DOI] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M, 2011. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America 108, 2915–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A, 2015. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529. [DOI] [PubMed] [Google Scholar]
- Hampoelz B, Andres-Pons A, Kastritis P, Beck M, 2019. Structure and Assembly of the Nuclear Pore Complex. Annu Rev Biophys 48, 515–536. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D, 1995. Expression of a Delta homologue in prospective neurons in the chick. Nature 375, 787–790. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA Jr., Kim R, Bilguvar K, Lopez-Giraldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, Russell MW, Mital S, Newburger JW, Gaynor JW, Breitbart RE, Iossifov I, Ronemus M, Sanders SJ, Kaltman JR, Seidman JG, Brueckner M, Gelb BD, Goldmuntz E, Lifton RP, Seidman CE, Chung WK, 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350, 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Marquez J, Khokha MK, 2019. Xenopus: Driving the Discovery of Novel Genes in Patient Disease and Their Underlying Pathological Mechanisms Relevant for Organogenesis. Front Physiol 10, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, Hung WC, Haider S, Zhang J, Knight J, Bjornson RD, Castaldi C, Tikhonoa IR, Bilguvar K, Mane SM, Sanders SJ, Mital S, Russell MW, Gaynor JW, Deanfield J, Giardini A, Porter GA Jr., Srivastava D, Lo CW, Shen Y, Watkins WS, Yandell M, Yost HJ, Tristani-Firouzi M, Newburger JW, Roberts AE, Kim R, Zhao H, Kaltman JR, Goldmuntz E, Chung WK, Seidman JG, Gelb BD, Seidman CE, Lifton RP, Brueckner M, 2017. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature genetics 49, 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ, 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol 14, 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, Hurt E, Beck M, 2016. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science (New York, N.Y.) 352, 363–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M, 2003. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61–73. [DOI] [PubMed] [Google Scholar]
- Miyake N, Tsukaguchi H, Koshimizu E, Shono A, Matsunaga S, Shiina M, Mimura Y, Imamura S, Hirose T, Okudela K, Nozu K, Akioka Y, Hattori M, Yoshikawa N, Kitamura A, Cheong HI, Kagami S, Yamashita M, Fujita A, Miyatake S, Tsurusaki Y, Nakashima M, Saitsu H, Ohashi K, Imamoto N, Ryo A, Ogata K, Iijima K, Matsumoto N, 2015. Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. American journal of human genetics 97, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N, 1998. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. [DOI] [PubMed] [Google Scholar]
- Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N, 1999. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell 4, 459–468. [DOI] [PubMed] [Google Scholar]
- Park SM, Jang HJ, Lee JH, 2019. Roles of Primary Cilia in the Developing Brain. Front Cell Neurosci 13, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, Huntzicker EG, Sfakianos MK, Sandoval W, Bazan JF, Kulkarni P, Garcia-Gonzalo FR, Seol AD, O’Toole JF, Held S, Reutter HM, Lane WS, Rafiq MA, Noor A, Ansar M, Devi AR, Sheffield VC, Slusarski DC, Vincent JB, Doherty DA, Hildebrandt F, Reiter JF, Jackson PK, 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR 3rd, Hetzer MW, 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M, 2010. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr Biol 20, 738–743. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K, Blum M, 2007. Cilia-driven leftward flow determines laterality in Xenopus. Curr Biol 17, 60–66. [DOI] [PubMed] [Google Scholar]
- Sempou E, Khokha MK, 2019. Genes and mechanisms of heterotaxy: patients drive the search. Curr Opin Genet Dev 56, 34–40. [DOI] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W, 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. The Journal of cell biology 189, 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR 3rd, Hetzer MW, 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio-Pelet A, Salomon R, Hadj-Rabia S, Mugnier C, de Laet MH, Chaouachi B, Bakiri F, Brottier P, Cattolico L, Penet C, Begeot M, Naville D, Nicolino M, Chaussain JL, Weissenbach J, Munnich A, Lyonnet S, 2000. Mutant WD-repeat protein in triple-A syndrome. Nature genetics 26, 332–335. [DOI] [PubMed] [Google Scholar]
- Vick P, Schweickert A, Weber T, Eberhardt M, Mencl S, Shcherbakov D, Beyer T, Blum M, 2009. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Dev Biol 331, 281–291. [DOI] [PubMed] [Google Scholar]
- Vishnoi N, Dhanasekeran K, Chalfant M, Surovstev I, Khokha MK, Lusk CP, 2020. Differential turnover of Nup188 controls its levels at centrosomes and role in centriole duplication. The Journal of cell biology 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH, 2007. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev Biol 303, 281–294. [DOI] [PubMed] [Google Scholar]
- Zanni G, De Magistris P, Nardella M, Bellacchio E, Barresi S, Sferra A, Ciolfi A, Motta M, Lue H, Moreno-Andres D, Tartaglia M, Bertini E, Antonin W, 2019. Biallelic Variants in the Nuclear Pore Complex Protein NUP93 Are Associated with Non-progressive Congenital Ataxia. Cerebellum 18, 422–432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.