Abstract
Background/Aim:
Few studies have evaluated the role of miRNAs in pediatric acute lymphoblastic leukemia (ALL) relapse and a consensus of a clinically significant miRNA signature is yet to be identified. In this study, we evaluated miRNAs associated with pediatric B-ALL early relapse in two independent sample sets.
Materials and Methods:
We performed global miRNA profiling on diagnostic bone marrow specimens from six early relapse (≤3 years after diagnosis) and six age- and cytogenetics-matched prolonged remission (≥4 years) patients (first set) and an independent set of 14 early relapse and 14 matched prolonged remission specimens (second set).
Results:
Twelve and 39 top differentially expressed miRNAs were observed in the first and second sets, respectively; however, there was no overlap between the top candidates. In post-hoc analyses six miRNAs (miR-101–3p, miR-4774–5p, miR-1324, miR-631, miR-4699–5p and miR-922) among the top candidates in the second, but not the first set, were consistently upregulated in early relapse compared to remission specimens in both first (fold change=1.13–2.19, q<0.38) and second (fold change=1.48–4.78, all q<0.05) sets. Four (miR-631, mir-101–3p, miR-922 and miR-1324) of these miRNAs have been previously implicated in key functional oncogenic pathways in adult cancers.
Conclusion:
This study suggests that six candidate miRNAs, not previously implicated in pediatric ALL, are associated with early relapse in pediatric B-ALL. Validation and investigation of mechanistic roles of these miRNAs in a larger cohort are warranted, so that they may be used as prognostic markers for early relapse of pediatric B-ALL.
Keywords: Pediatric, miRNA, ALL, relapse, epigenetic
Acute lymphoblastic leukemia (ALL) is the most common cancer diagnosed among children, accounting for a quarter of all cancer diagnoses and three-quarters of childhood leukemia diagnoses (1, 2). The 5-year and overall survival rates of ALL have increased over the years to approximately 80% and 85%, respectively (1–3), largely due to risk-stratification and combination chemotherapy (4, 5). However, 10–15% of patients develop recurrence and, depending on the recurrence site, 5–50% will survive with only a third surviving long-term (6–9).
Knowledge on the mechanism of leukemia relapse is still limited (10–13). Few studies have evaluated the association between miRNA expression and pediatric ALL relapse (14). Although these studies have identified potential prognostic miRNAs, a consensus of a clinically significant miRNA signature is yet to be identified. In addition, several previous studies have investigated a limited number (n=1–723) of miRNAs (15–20) or have not included an independent replication dataset (17, 18). In this study, we provide additional evidence for the association between miRNAs and pediatric B-ALL early relapse by investigating a large number of miRNAs (~2,600 human miRNAs) in two independent sample sets.
Materials and Methods
Patient accrual and sample collection.
The study included two independent preselected sets of patients. The first set consisted of six patients with early relapse (≤3 years) and six cytogenetics (Lysine (K)- specific methyltransferase 2A gene, KMT2A, rearrangement and chromosomal translocation) matched prolonged remission (>3 years) patients with B-cell ALL diagnosed between July 2008 and October 2013 at the Johns Hopkins School of Medicine. The second set consisted 14 patients with early relapse and 14 age-, sex- and cytogenetics-matched remission patients with B-cell ALL obtained from the Children’s Oncology Group (21). Initial diagnostic bone marrow specimens were obtained for all patients in both sets. Clinical and pathological information was obtained from medical records (first set) or from the COG data center (second set). All samples and data were de-identified. The study, including sample acquisition, retrieval, processing and analysis, was approved by the Institutional review board of the Johns Hopkins All Children’s Hospital (IRB00056030).
miRNA microarray processing.
High quality total RNA (including miRNA) for the first and second set of samples were obtained from PAB’s lab at Johns Hopkins and COG’s biorepository, respectively. All samples were transferred to Moffitt Cancer Center for profiling using a standard Affymetrix protocol.
The quality of the RNA in both sets was determined using NanoDrop (Thermo Scientific, Waltham, MA, USA) (all A260/A280 ≥1.8, except two samples that were 1.7) and run on a BioAnalyzer (Seattle, WA, USA). Global miRNA expression profiling was then performed using Affymetrix miRNA v 4.0 (Affymetrix, Santa Clara, CA, USA) on the two independent samples at different times. Labeling and hybridization were performed according to standard Affymetrix protocols using total RNA for each of the 40 samples.
Statistical analysis.
Demographic characteristics of study participants were summarized by relapse status for each sample set using descriptive statistics. Expression data were analyzed in the R environment (v3.4.2). We used the Affymetrix package and the pd.mirna.4.0 package for our specific array to load and process the data. Expression data were log transformed and normalized using the Robust Multi-array Average method. The Significance Analysis of Microarray (SAM) two-class unpaired method implemented in the samr Bioconductor package was used to identify differentially expressed miRNAs (23) in each set. The samr parameter delta=0.6 was selected to classify miRNAs as differentially expressed at a fold-change>1.0 and a q-value<0.05. In post-hoc analyses, we used a fold-change>1.0 with no q-value threshold to determine differentially expressed miRNAs in both datasets and then selected miRNAs differentially expressed in the same direction for both datasets. The analysis was limited to human miRNAs. We used the online tool, DNA Intelligent Analysis (DIANA) microT v5 (24), to predict targeted genes involved in leukemia for the identified miRNAs.
Results
Demographic and clinical characteristics of the study participants are shown in Table I. There were 8 females and 4 males in the first set compared to 14 females and 14 males in the second set. Participants in the second set had KMT2A wild type and were older with a longer median follow up compared to participants in the first set that had rearranged KMT2A and were infants with a shorter follow up.
Table I.
Patient characteristics.
| First set (n=12) | Second set (n=28) | |||
|---|---|---|---|---|
| Variable | Early relapse | Remission | Early relapse | Remission |
| Median age in years (range) | 0.3 (0.2–0.7) | 0.4 (0.0–0.6) | 4 (1–18) | 4 (1–16) |
| Gender | ||||
| Female | 3 | 5 | 5 | 8 |
| Male | 3 | 1 | 9 | 6 |
| KMT2A fusion partner | Positive | Positive | Negative | Negative |
| ENL | 3 | 3 | 0 | 0 |
| AF4 | 3 | 3 | 0 | 0 |
| Relapse-free survival time (months) | 8.5 (1.4–19.3) | 51.4 (27.3–77.0) | 17 (8.4–36.5) | 83.4 (47.3–125.6) |
KMT2A: Lysine (K)- specific methyltransferase 2A gene; ENL: Eleven nineteen leukemia; AF4: ALL-1 fuse gene from chromosome 4.
A two-class unpaired comparison of the global profiles in the first set revealed 12 upregulated miRNAs (fold change=1.48–4.45, all q<0.05) in early relapse samples compared to remission samples (Table II). A similar comparison of miRNA profiles in the second set revealed 39 up-regulated miRNAs (fold change=1.12–2.19, q<0.05) for relapse compared to remission samples. The top differentially expressed miRNAs in the second set were different from those of the first set (Table II). However, a post-hoc analysis that compared all the miRNA profiles in the two sets with no q-value threshold, revealed six miRNAs (miR-101–3p, miR-4774–5p, miR-1324, miR-631, miR-4699–5p and miR-922) consistently upregulated in early relapse compared to remission specimens in both the first (fold change=1.13–2.19, all q<0.38) and second (fold change=1.48–4.78, all q<0.05) sets (Figure 1); albeit all the miRNAs in the first set had q-values>0.05 (Table III). The six miRNAs were among the top most differentially expressed miRNAs in the second set, but not in the first set (Table II).
Table II.
Top differentially expressed miRNAs in two independent sample sets.
| First set | Second set | ||||
|---|---|---|---|---|---|
| miRNA | Fold change | q-value | miRNA | Fold change | q-value |
| hsa-miR-548am-5p | 4.77 | <0.05 | hsa-miR-5586–5p | 1.67 | <0.05 |
| hsa-miR-4699–3p | 4.45 | <0.05 | hsa-miR-202–5p | 1.63 | <0.05 |
| hsa-miR-4498 | 4.33 | <0.05 | hsa-miR-20a-3p | 1.40 | <0.05 |
| hsa-miR-7161–3p | 4.17 | <0.05 | hsa-miR-649 | 1.34 | <0.05 |
| hsa-miR-6805–5p | 3.63 | <0.05 | hsa-miR-6777–5p | 1.26 | <0.05 |
| hsa-miR-4507 | 3.21 | <0.05 | hsa-miR-1307–5p | 1.23 | <0.05 |
| hsa-miR-1973 | 2.92 | <0.05 | hsa-miR-6887–5p | 1.21 | <0.05 |
| hsa-miR-103b | 2.64 | <0.05 | hsa-miR-4321 | 1.21 | <0.05 |
| hsa-miR-99a-3p | 2.18 | <0.05 | hsa-miR-890 | 1.2 | <0.05 |
| hsa-miR-7113–3p | 2.1 | <0.05 | hsa-miR-4699–5p | 1.18 | <0.05 |
| hsa-miR-4436b-3p | 1.73 | <0.05 | hsa-miR-1324 | 1.18 | <0.05 |
| hsa-miR-4260 | 1.48 | <0.05 | hsa-miR-922 | 1.15 | <0.05 |
Figure 1.
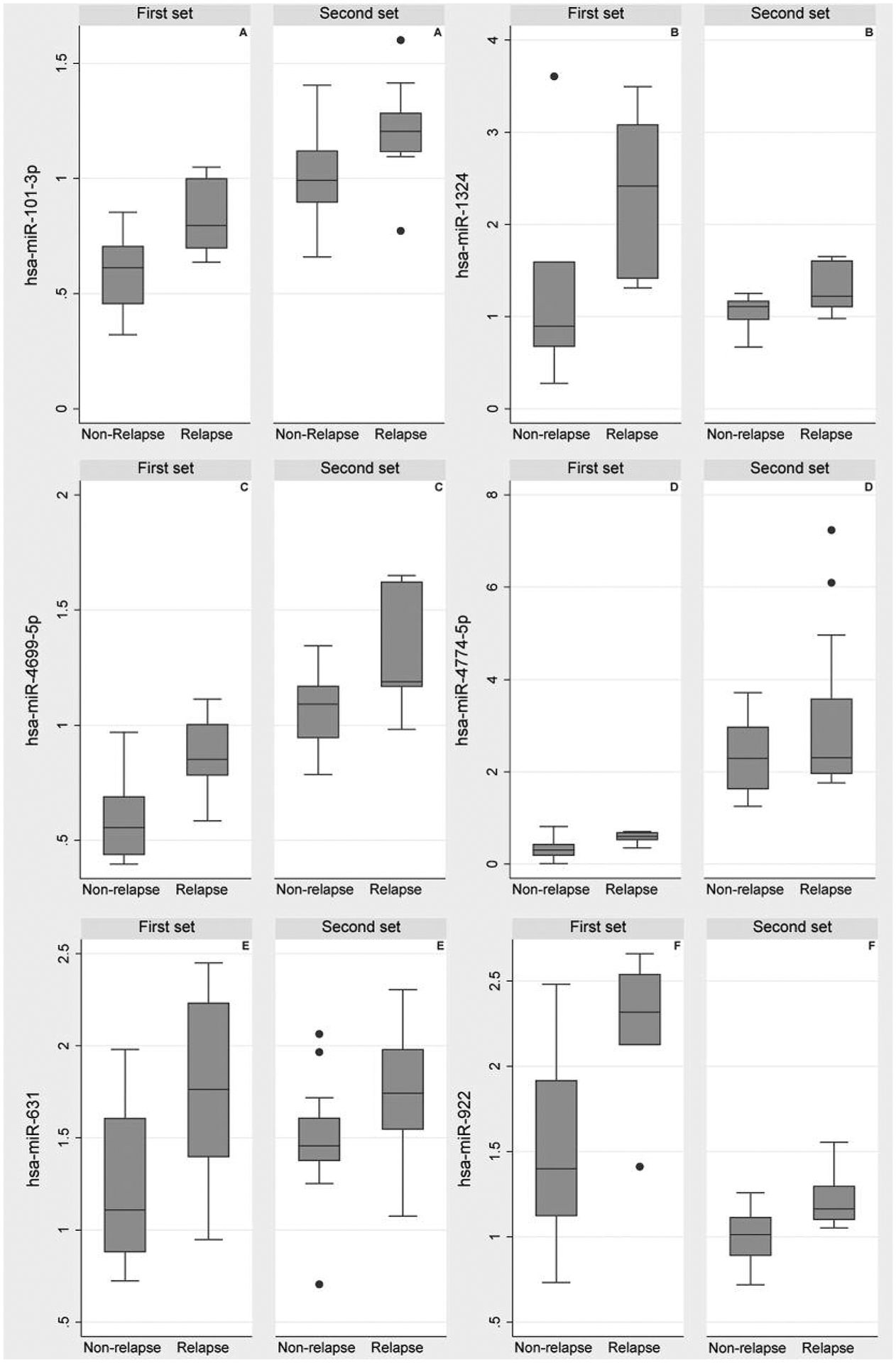
Relative fold change for miRNAs differentially expressed in two independent sample sets (first set, 12 samples from JHU; second set, 28 samples from COG) of pediatric B-ALL initial diagnostic samples.
Table III.
Differential expression of overlapping miRNAs in two independent sample sets.
| First set | Second set | |||
|---|---|---|---|---|
| miRNA | Fold change | q-value | Fold change | q-value |
| hsa-miR-101–3p | 1.18 | 0.29 | 1.15 | <0.05 |
| hsa-miR-1324 | 2.04 | 0.20 | 1.18 | <0.05 |
| hsa-miR-4699–5p | 1.20 | 0.29 | 1.18 | <0.05 |
| hsa-miR-4774–5p | 1.18 | 0.38 | 1.71 | <0.05 |
| hsa-miR-631 | 1.44 | 0.26 | 1.19 | <0.05 |
| hsa-miR-922 | 1.65 | 0.11 | 1.15 | <0.05 |
In silico prediction of targeted genes of the six candidate miRNAs using DIANA microT revealed that all of them (miR-101–3p, miR-4774–5p, miR-1324, miR-631, miR-4699–5p and miR-922) targeted genes implicated in leukemia (24) (Table IV). In addition, four (miR-631, mir-101–3p, miR-922 and miR-1324) out of the six overlapping miRNAs have been previously implicated in key functional oncogenic pathways in adult cancers, including multiple myeloma, prostate, lung, liver, breast and head and neck cancers (25–31) (Table V).
Table IV.
Predicted target genes of miRNAs associated with relapse in pediatric B-ALL samples.
| MicroRNA | DIANA microT predicted target genes in leukemia (24) |
|---|---|
| hsa-miR-101–3p | RUNX1T1, TGFB3 |
| hsa-miR-1324 | ZBTB16, NRAS, CBL, STAT5 |
| hsa-miR-4699–5p | E2F3, SOS1 |
| hsa-miR-4774–5p | CBL, NRAS, CCND1, PTPN11, SMAD4 |
| hsa-miR-631 | RUNX1T1, TGFB3 |
| hsa-miR-922 | PIMI, CDK6, BRAF, BCL2L1, IKBKB, PIK3R3, MECOM, NRAS, RUNX1, SMAD3, CRKL, SOS1, RAF1, PIK3R1, MAPK1, TGFBR1, PIK3R2 |
Table V.
Role of candidate miRNAs (associated with relapse in pediatric B-ALL pediatric) in other cancers.
| miRNA | Cancer | Role | Reference |
|---|---|---|---|
| hsa-miR-101–3p | TNBC | Over-expression inhibits proliferation | Liu P et al., 2016 (27) |
| NSCLC | Over-expression inhibits proliferation, migration and invasion | Zhang X et al., 2017 (30) | |
| ACC | Over-expression inhibits proliferation, migration and invasion | Liu X et al., 2015 (28) | |
| hsa-miR-631 | MM | Decreased expression associated with bortezomib resistance in multiple myeloma | Xi H et al., 2017 (29) |
| PCa | Down-regulated | Fu D et al., 2016 (25) | |
| hsa-miR-1324 | HCC | Inhibition enhances proliferation, migration and invasion | Zhu Q et al., 2017 (31) |
| hsa-miR-922 | HCC | Up-regulation promoting proliferation | Liu J et al., 2017 (26) |
TNBC: Triple negative breast cancer; NSCLC: non-small cell lung cancer; ACC: adenoid cystic carcinoma; MM: multiple myeloma; PCa: prostate cancer; HCC: hepatocellular carcinoma.
Discussion
In this study, we evaluated the association between miRNA expressions and pediatric ALL early relapse in two independent samples. The top differentially expressed miRNAs in the two sample sets did not overlap. However, in a post-hoc analysis we observed six miRNAs (miR-101–3p, miR-4774–5p, miR-1324, miR-631, miR-4699–5p and miR-922) among the top candidates of the second set that were consistently upregulated in the first set, but were not among the top candidates in this set. Exploration of the potential function of these miRNAs using in silico analyses revealed that all six miRNAs target genes previously implicated in leukemia (24).
Previous studies have suggested the association between pediatric ALL relapse and miRNAs, such as miR-24, miR-128b, miR-223, miR-210, miR-335, miR-130b, miR-126, miR-222, miR-345, miR-708 and miR-23a (14). However, none of the six candidate miRNAs in this study have been previously implicated in pediatric B-ALL relapse. Four out of six, though, have been previously implicated in other cancers. miR-631, located on chromosome 15q24.2, is implicated in multiple myeloma, where a decreased expression is associated with bortezomib resistance (29). Expression of miR-631 is also downregulated in prostate cancer tissue and prostate cancer cell lines (25). Genetic variants of this miRNA are associated with risk of esophageal cancer (32) and survival and recurrence in renal cell carcinoma (33).
Mir-101–3p is located on chromosome 1p31.3 and is involved in negative regulation of protein ubiquitination and positive regulation of blood vessel endothelial cell proliferation. It is implicated in triple negative breast cancer (TNBC) and ectopic over-expression of mir-101–3p in TNBC cells in vitro inhibits cell proliferation (27). In addition, miR-101–3p expression is decreased in non-small cell lung cancer cells and overexpression inhibits proliferation, migration and invasion of these cells in vitro (30). Similarly, adenoid cystic carcinoma (ACC) samples and corresponding cell lines that have a high potential for metastasis have decreased miR-101–3p expression; and ectopic expression of miR-101–3p in ACC cell lines enhances the induction of apoptosis and inhibits invasion, proliferation, colony formation, and establishment of xenografts in nude mice (28).
MiR-922 is located on 3q29. Its expression is upregulated in hepatocellular carcinoma (HCC) samples and cell lines and plays a key role in the promotion of HCC cell proliferation (26). miR-1324 is located on chromosome 3p13. A recent study showed that inhibition of miR-1324 enhances proliferation, migration and invasion of HCC cells (31). The other two candidate miRNAs (miR-4774–5p and miR-1324) observed in this study are located on 2q24.3 and 3p12.3, respectively, but have not been previously implicated in other cancers.
A major limitation of the study is that the candidate miRNAs were identified in a post-hoc analysis in two datasets each with a small sample size. Therefore, a larger study with a prespecified hypothesis and approach to evaluate the identified candidates is warranted. Although we observed an overlap of miRNAs between the two datasets in a post-hoc analysis, the overlapping miRNAs in the first dataset were not among the top differentially expressed in that set in terms of fold change and q-value. It is plausible that the lack of common statistically significant overexpressed miRNAs could be attributed to the different genetic characteristics and age in the two independent sets. The lack of overlap could also suggest that miRNAs that play an important role in relapse might be different for the two populations, therefore, miRNAs for each set might reflect the biology of early ALL relapse in each group (infant ALL vs. ALL at an older age). However, the six overlapping miRNAs were consistently over-expressed in both datasets and in silico analyses predicted target genes in leukemia. These miRNAs may reflect a shared biology for early relapse in the two groups. Again, these promising preliminary findings are based on a post-hoc analysis and further evaluation of the suggested candidate miRNAs is warranted. Another limitation is the use of global miRNA expression in both datasets. Ideally, candidate miRNAs identified in the first set would be validated using a different assay, for example real-time quantitative polymerase chain reaction, on a second independent sample set. Additionally, target genes of the candidate miRNAs were determined in silico and need to be validated in the laboratory to determine their functional role in pediatric ALL relapse.
Despite these limitations, this study identified candidate miRNAs that may be evaluated further in pediatric B-ALL relapse as prognostic markers for pediatric B-ALL relapse. However, these candidates need to be evaluated and validated in a larger study and their biological role in pediatric B-ALL relapse should also be determined.
Acknowledgements
This work was supported by a generous grant from the All Children’s Hospital Foundation, St Petersburg, Florida. Part of the specimen and data were obtained from the Children’s Oncology Group through NIH/NCI NCTN Operations Center Grant 180886 and COG Biospecimen Bank Grant U24CA196173.
Footnotes
Conflicts of Interest
The Authors declare no potential conflicts of interest.
References
- 1.Inaba H, Greaves M and Mullighan CG: Acute lymphoblastic leukaemia. Lancet 381(9881): 1943–1955, 2013. DOI: 10.1016/S0140-6736(12)62187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Robison LL and Look AT: Acute lymphoblastic leukaemia. Lancet 371(9617): 1030–1043, 2008. DOI: 10.1016/S0140-6736(08)60457-2 [DOI] [PubMed] [Google Scholar]
- 3.Demidowicz E, Pogorzala M, Lecka M, Zolnowska H, Marjanska A, Kubicka M, Kurylo-Rafinska B, Czyzewski K, Debski R, Koltan A, Richert-Przygonska M and Styczynski J: Outcome of pediatric acute lymphoblastic leukemia: Sixty years of progress. Anticancer Res 39(9): 5203–5207, 2019. DOI: 10.21873/anticanres.13717 [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Campana D, Pei D, Bowman WP, Sandlund JT, Kaste SC, Ribeiro RC, Rubnitz JE, Raimondi SC, Onciu M, Coustan-Smith E, Kun LE, Jeha S, Cheng C, Howard SC, Simmons V, Bayles A, Metzger ML, Boyett JM, Leung W, Handgretinger R, Downing JR, Evans WE and Relling MV: Treating childhood acute lymphoblastic leukemia without cranial irradiation. NEJM 360(26): 2730–2741, 2009. DOI: 10.1056/NEJMoa0900386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pui CH, Mullighan CG, Evans WE and Relling MV: Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood 120(6): 1165–1174, 2012. DOI: 10.1182/blood-2012-05-378943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaynon PS: Childhood acute lymphoblastic leukaemia and relapse. BJH 131(5): 579–587, 2005. DOI: 10.1111/j.1365-2141.2005.05773.x [DOI] [PubMed] [Google Scholar]
- 7.Gaynon PS, Trigg ME, Heerema NA, Sensel MG, Sather HN, Hammond GD and Bleyer WA: Children’s Cancer Group trials in childhood acute lymphoblastic leukemia: 1983–1995. Leukemia 14(12): 2223–2233, 2000. DOI: 10.1038/sj.leu.2401939 [DOI] [PubMed] [Google Scholar]
- 8.Pui CH, Campana D and Evans WE: Childhood acute lymphoblastic leukaemia--current status and future perspectives. Lancet Oncol 2(10): 597–607, 2001. DOI: 10.1016/S1470-2045(01)00516-2 [DOI] [PubMed] [Google Scholar]
- 9.Pogorzala M, Kubicka M, Rafinska B, Wysocki M and Styczynski J: Drug-resistance profile in multiple-relapsed childhood acute lymphoblastic leukemia. Anticancer Res 35(10): 5667–5670, 2015. [PubMed] [Google Scholar]
- 10.Bhojwani D and Pui CH: Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol 14(6): e205–217, 2013; DOI: 10.1016/S1470-2045(12)70580-6 [DOI] [PubMed] [Google Scholar]
- 11.Staal FJ, de Ridder D, Szczepanski T, Schonewille T, van der Linden EC, van Wering ER, van der Velden VH and van Dongen JJ: Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia 24(3): 491–499, 2010. DOI: 10.1038/leu.2009.286 [DOI] [PubMed] [Google Scholar]
- 12.Sood R, Hansen NF, Donovan FX, Carrington B, Bucci D, Maskeri B, Young A, Trivedi NS, Kohlschmidt J, Stone RM, Caligiuri MA, Chandrasekharappa SC, Marcucci G, Mullikin JC, Bloomfield CD and Liu P: Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia 30(2): 501–504, 2016. DOI: 10.1038/leu.2015.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, Lapillonne H, Renneville A, Ragu C, Figeac M, Celli-Lebras K, Lacombe C, Micol JB, Abdel-Wahab O, Cornillet P, Ifrah N, Dombret H, Leverger G, Jourdan E and Preudhomme C: Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood 127(20): 2451–2459, 2016. DOI: 10.1182/blood-2015-12-688705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale V, Hale GA, Brown PA and Amankwah EK: A review of DNA methylation and microRNA expression in recurrent pediatric acute leukemia. Oncology 92(2): 61–67, 2017. DOI: 10.1159/000452091 [DOI] [PubMed] [Google Scholar]
- 15.Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, Zhang P, Xu L and Chen YQ: A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Gen 20(24): 4903–4915, 2011. DOI: 10.1093/hmg/ddr428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mei Y, Gao C, Wang K, Cui L, Li W, Zhao X, Liu F, Wu M, Deng G, Ding W, Jia H and Li Z: Effect of microRNA-210 on prognosis and response to chemotherapeutic drugs in pediatric acute lymphoblastic leukemia. Can Sci 105(4): 463–472, 2014. DOI: 10.1111/cas.12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nemes K, Csoka M, Nagy N, Mark A, Varadi Z, Danko T, Kovacs G, Kopper L and Sebestyen A: Expression of certain leukemia/lymphoma related microRNAs and its correlation with prognosis in childhood acute lymphoblastic leukemia. Path Oncol Res 21(3): 597–604, 2015. DOI: 10.1007/s12253-014-9861-z [DOI] [PubMed] [Google Scholar]
- 18.Organista-Nava J, Gomez-Gomez Y, Illades-Aguiar B, Del Carmen Alarcon-Romero L, Saavedra-Herrera MV, Rivera-Ramirez AB, Garzon-Barrientos VH and Leyva-Vazquez MA: High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol Rep 33(4): 1639–1649, 2015. DOI: 10.3892/or.2015.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Jiang N, Huang G, Tay JL, Lin B, Bi C, Koh GS, Li Z, Tan J, Chung TH, Lu Y, Ariffin H, Kham SK, Yeoh AE and Chng WJ: Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. BJH 163(1): 93–103, 2013. DOI: 10.1111/bjh.12489 [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L and Chen YQ: MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PloS One 4(11): e7826, 2009. DOI: 10.1371/journal.pone.0007826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duyu M, Durmaz B, Gunduz C, Vergin C, Yilmaz Karapinar D, Aksoylar S, Kavakli K, Cetingul N, Irken G, Yaman Y, Ozkinay F and Cogulu O: Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. Biomed Res Int 2014: 967585, 2014. DOI: 10.1155/2014/967585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen EC, Devidas M, Chen S, Salzer WL, Raetz EA, Loh ML, Mattano LA, Jr., Cole C, Eicher A, Haugan M, Sorenson M, Heerema NA, Carroll AA, Gastier-Foster JM, Borowitz MJ, Wood BL, Willman CL, Winick NJ, Hunger SP and Carroll WL: Dexamethasone and high-dose methotrexate improve outcome for children and young adults with high-risk B-acute lymphoblastic leukemia: A report from Children’s Oncology Group Study AALL0232. JCO 34(20): 2380–2388, 2016. DOI: 10.1200/JCO.2015.62.4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tusher VG, Tibshirani R and Chu G: Significance analysis of microarrays applied to the ionizing radiation response. PNAS 98(9): 5116–5121, 2001. DOI: 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41: W169–173, 2013. DOI: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu D, Liu B, Zang LE and Jiang H: MiR-631/ZAP70: A novel axis in the migration and invasion of prostate cancer cells. Biochem and Biophys Res Comm 469(3): 345–351, 2016. DOI: 10.1016/j.bbrc.2015.11.093. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Su Z, Zeng Y, Zhang H, Yang S and Liu G: miR-922 regulates CYLD expression and promotes the cell proliferation of human hepatocellular carcinoma. Oncol Rep 37(3): 1445–1450, 2017. DOI: 10.3892/or.2017.5431 [DOI] [PubMed] [Google Scholar]
- 27.Liu P, Ye F, Xie X, Li X, Tang H, Li S, Huang X, Song C, Wei W and Xie X: mir-101–3p is a key regulator of tumor metabolism in triple negative breast cancer targeting AMPK. Oncotarget 7(23): 35188–35198, 2016. DOI: 10.18632/oncotarget.9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu XY, Liu ZJ, He H, Zhang C and Wang YL: MicroRNA-101–3p suppresses cell proliferation, invasion and enhances chemotherapeutic sensitivity in salivary gland adenoid cystic carcinoma by targeting Pim-1. Am J Cancer Res 5(10): 3015–3029, 2015. [PMC free article] [PubMed] [Google Scholar]
- 29.Xi H, Li L, Du J, An R, Fan R, Lu J, Wu YX, Wu SX, Hou J and Zhao LM: hsa-miR-631 resensitizes bortezomib-resistant multiple myeloma cell lines by inhibiting UbcH10. Oncol Rep 37(2): 961–968, 2017. DOI: 10.3892/or.2016.5318 [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, He X, Liu Y, Zhang H, Chen H, Guo S and Liang Y: MiR-101–3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed Pharmacother 93: 1065–1073, 2017. DOI: 10.1016/j.biopha.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 31.Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D and Ni Y: CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem and Biophys Res Comm 497(2): 626–632, 2018. DOI: 10.1016/j.bbrc.2018.02.119 [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA and Wu X: Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 1(6): 460–469, 2008. DOI: 10.1158/1940-6207.CAPR-08-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG and Wu X: Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 31(10): 1805–1812, 2010. DOI: 10.1093/carcin/bgq168 [DOI] [PMC free article] [PubMed] [Google Scholar]


