The NAC transcription factors OsNAC20 and OsNAC26 influence starch and storage protein accumulation by directly regulating the expression of starch and storage protein synthesis-related genes.
Abstract
Starch and storage proteins determine the weight and quality of cereal grains. Synthesis of these two grain components has been comprehensively investigated, but the transcription factors responsible for their regulation remain largely unknown. In this study, we investigated the roles of NAM, ATAF, and CUC (NAC) transcription factors, OsNAC20, and OsNAC26 in starch and storage protein synthesis in rice (Oryza sativa) endosperm. OsNAC20 and OsNAC26 showed high levels of amino acid sequence similarity. Both were localized in the aleurone layer, starchy endosperm, and embryo. Mutation of OsNAC20 or OsNAC26 alone had no effect on the grain, while the osnac20/26 double mutant had significantly decreased starch and storage protein content. OsNAC20 and OsNAC26 alone could directly transactivate the expression of starch synthaseI (SSI), pullulanase (Pul), glutelin A1 (GluA1), glutelin B4/5 (GluB4/5), α-globulin, and 16 kD prolamin and indirectly influenced plastidial disproportionating enzyme1 (DPE1) expression to regulate starch and storage protein synthesis. Although they could also bind to the promoters of ADP-Glc pyrophosphorylase small subunit 2b (AGPS2b), ADP-Glc pyrophosphorylase large subunit 2 (AGPL2), and starch branching enzymeI (SBEI), and the expression of the three genes was largely decreased in the osnac20/26 mutant, ADP-Glc pyrophosphorylase and starch branching enzyme activities were unchanged in this double mutant. In addition, OsNAC20 and OsNAC26 are main regulators of Pul, GluB4, α-globulin, and 16 kD prolamin. In conclusion, OsNAC20 and OsNAC26 play an essential and redundant role in the regulation of starch and storage protein synthesis.
Rice (Oryza sativa) is one of the most important staple foods worldwide. Its endosperm is composed of ∼80% starch and 10% storage protein. Starch is the main dietary source of energy, and protein plays an important role in the quality of rice grains. Therefore, revealing the basis of the regulation underlying starch and storage protein accumulation is essential for improvement of rice grain quality.
Although starch is a simple combination of Glc molecules, its biosynthesis in plant endosperm involves a series of complex and coordinated enzymatic actions. In the first step, ADP-Glc pyrophosphorylase (AGPase) converts Glc-1-phosphate into ADP-Glc, the substrate of starch synthesis (Beckles et al., 2001). Starch phosphorylase1 (Pho1) plays a critical role in starch initiation by extending the chain length of the initial primer (Satoh et al., 2008). Granule-bound starch synthaseI (GBSSI)/Waxy (Wx) produces amylose (Denyer et al., 2001). Amylopectin synthesis is mainly controlled by three types of biosynthetic enzymes, starch synthases (SSs), starch branching enzymes (SBEs), and debranching enzymes (DBEs; Nakamura, 2002; Davis et al., 2003; Tetlow, 2006). SSs and SBEs contribute to the extension of glucan chains and the formation of branch points, respectively. DBEs trim the irregular glucan structure created by SSs and SBEs to obtain ordered amylopectin branch chains. In addition, plastidial disproportionating enzyme1 (DPE1) has also been reported to take part in starch synthesis by transferring maltooligosyl groups from amylose and amylopectin to amylopectin (Dong et al., 2015).
Only a few regulators have been reported to participate in the regulation of starch synthesis, and most of these reports focus on rice and maize (Zea mays). Rice YB1 directly binds to the G-box of the Wx promoter and activates Wx transcription (Bello et al., 2019). Maize ZmEREB156 positively modulates ZmSSIIIa through the synergistic effect of Suc and abscisic acid (ABA; Huang et al., 2016; Li et al., 2018b). ZmbZIP91 is homologous to AtVIP1 and complements the altered starch phenotype of the vip1 mutant in Arabidopsis (Arabidopsis thaliana) leaves. Further analyses indicate that ZmZIP91 directly binds to the promoters of the starch synthesis-related genes AGPS1, SSI, SSIIIa, and ISA1 (Chen et al., 2016). In addition, OsbZIP33 has also been inferred to play a role in regulation of starch synthesis, as it is able to interact with the ACGT element buried in the promoter of SBEI and Wx (Cai et al., 2002). Recently, Song et al. (2020) reported that TubZIP28 in Triticum urartu and its homolog, TabZIP28 from common wheat (Triticum aestivum), play a role in the regulation of starch synthesis as TubZIP28 binds to the promoter of cytosolic AGPase and enhances its transcription and activity. Apart from the above-mentioned regulators that directly bind to the promoters of starch synthesis-related genes to regulate their expression, some other regulators such as ZmaNAC36, ZmNAC34, rice starch regulator1 (RSR1), disulfide isomerase1-2 protein (TaPDIL1-2), ZmMADS1a, and G protein γ-subunit DEP1/qPE9-1 also have an indirect or unknown role in the regulation of starch synthesis (Fu and Xue, 2010; Zhang et al., 2014, 2019a; Dong et al., 2019a, 2019b; Peng et al., 2019).
The major storage proteins in rice endosperm include acid- and alkaline-soluble glutelins, alcohol-soluble prolamins, and saline-soluble α-globulin. They comprise ∼70%, 20%, and 3% of the total storage proteins, respectively (He et al., 2013). Based on their sequence similarity, glutelin genes are classified into four subfamilies, designated as GluA, GluB, GluC, and GluD. Prolamin genes are divided into three families, designated as 10 kD, 13 kD, and 16 kD. The GluA, GluB, 10 kD, and 13 kD subfamilies are composed of several members (Saito et al., 2012; He et al., 2013).
Cis-regulatory elements, such as O2/O2-like and prolamin boxes, have been widely detected in the storage protein synthesis-related genes in cereal crops (Washida et al., 1999; Wu et al., 2000). Maize O2 directly binds to the O2 box to modulate almost all zein genes, except for genes encoding 16-kD γ- and 18-kD δ-zein, whereas O2 displays different activation activity for different zein genes (Cord Neto et al., 1995; Schmidt et al., 2011; Li et al., 2015). The maize endosperm-specific Dof (DNA binding one zinc finger) transcription factor (TF), prolamin box binding factor1 (PBF1), specifically recognizes the prolamin box to regulate 22-kD α-zein and 27-kD γ-zein genes (Vicente-Carbajosa et al., 1997; Wu and Messing, 2012). Likewise, rice O2-like protein, bZIP58 (also called RISBZ1) and the PBF subfamily member RPBF also play a role in the regulation of storage protein synthesis. Individual knockdown mutants of bZIP58 and RPBF show little change in most storage proteins, but a double knockdown mutant has a pronounced decrease of most storage protein, as bZIP58 and RPBF are able to directly activate the expression of GluA1, GluA2, GluA3, GluB1, GluD1, 10 kD Prolamin, 13 kD prolamin, and 16 kD prolamin (Yamamoto et al., 2006; Kawakatsu et al., 2009). In addition, maize O2 hetero-dimerizing protein1 (OHP1), OHP2, ZmbZIP22, and ZmMADS47 also can regulate zein gene transcription directly (Zhang et al., 2015; Qiao et al., 2016; Li et al., 2018a).
There are several TFs showing a regulation role in both protein and starch metabolism. Apart from the function in zein regulation, O2 and PBF1 also play a role in starch synthesis by directly binding to the promoters of PPDK1, PPDK2, and SSIII and by indirect regulation of SSIIa and SBEI (Zhang et al., 2016). Likewise, OsbZIP58 not only regulates storage protein accumulation, it can also bind directly to the promoters of the starch synthesis-related genes OsAGPL3, Wx, OsSSIIa, SBE1, OsBEIIb, and ISA2 to regulate their expression (Wang et al., 2013; Kim et al., 2017). OPAQUE11 (O11), a basic helix-loop-helix-type TF, directly modulates O2 and PBF1, which gives it a modulated function in both starch and storage protein accumulation (Feng et al., 2018). Recently, Zhang et al. (2019b) reported that deficiency of ZmNAC128 and ZmNAC130 decreases starch and protein accumulation, as ZmNAC128 and ZmNAC130 directly regulate 16-kD γ-zein and AGPS2 (Bt2) expression by binding to their promoters.
In general, starch and storage protein syntheses have been comprehensively investigated, but the plant regulators responsible for the syntheses remain largely unknown. NAM, ATAF1/2, and CUC2 (NAC) TFs are one of the largest plant-specific TF families (Souer et al., 1996; Aida et al., 1997; Zhao et al., 2016a). The most notable feature of the NAC TF family is that they contain a conserved NAC domain responsible for the binding properties in the N terminus. The C terminus of NAC TFs normally confers a diversified transcriptional regulatory region (TRR) that contributes to its activation or repression properties (Ooka et al., 2003; Olsen et al., 2005). NAC TFs have been reported to participate in various processes, such as biotic and abiotic stress responses, formation of a secondary cell wall, senescence, meristem regulation, and so on (Kubo et al., 2005; Fang et al., 2008; Yamaguchi et al., 2008; Balazadeh et al., 2011; Wu et al., 2012; Mao et al., 2017; Li et al., 2019; Sun et al., 2019, Sakuraba et al., 2020). However, their functions in the endosperm have not been fully elucidated.
A search of transcriptome data, a rice expression profile database (http://ricexpro.dna.affrc.go.jp/), and published papers (Fang et al., 2008; Nie et al., 2013; Mathew et al., 2016) yielded eight endosperm-specific NAC TFs: OsNAC20 (LOC_Os01g01470/Os01g0104500), OsNAC23 (LOC_Os02g12310/ Os02g0214500), OsNAC24 (LOC_Os05g34310/Os05g0415400), OsNAC25 (LOC_Os11g31330/Os11g0512000), OsNAC26 (LOC_Os01g29840/Os01g0393100), OsNAC127 (LOC_Os11g31340/Os11g0512100), OsNAC128 (LOC_Os11g31360/ Os11g0512200), and OsNAC129 (LOC_Os11g31380/Os11g0512600). Herein, the most highly expressed OsNAC26 (previously ONAC026) and its homologous gene, OsNAC20 (previously ONAC020), were selected for further analysis to detect whether they have a function in rice endosperm. We employed a reverse genetic approach to unveil the functions of OsNAC20 and OsNAC26 in synthesizing starch and storage protein in rice endosperm. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-generated osnac20 and osnac26 single mutants showed no alterations in their grain phenotype, but their double mutant, osnac20/26, displayed a floury grain caused by decreased starch and storage protein content. A series of assays were utilized to determine the target genes of OsNAC20 and OsNAC26. These results could enrich our knowledge of the regulatory network of starch and storage protein synthesis and accumulation.
RESULTS
OsNAC20 and OsNAC26 Are Specifically Expressed in Grain
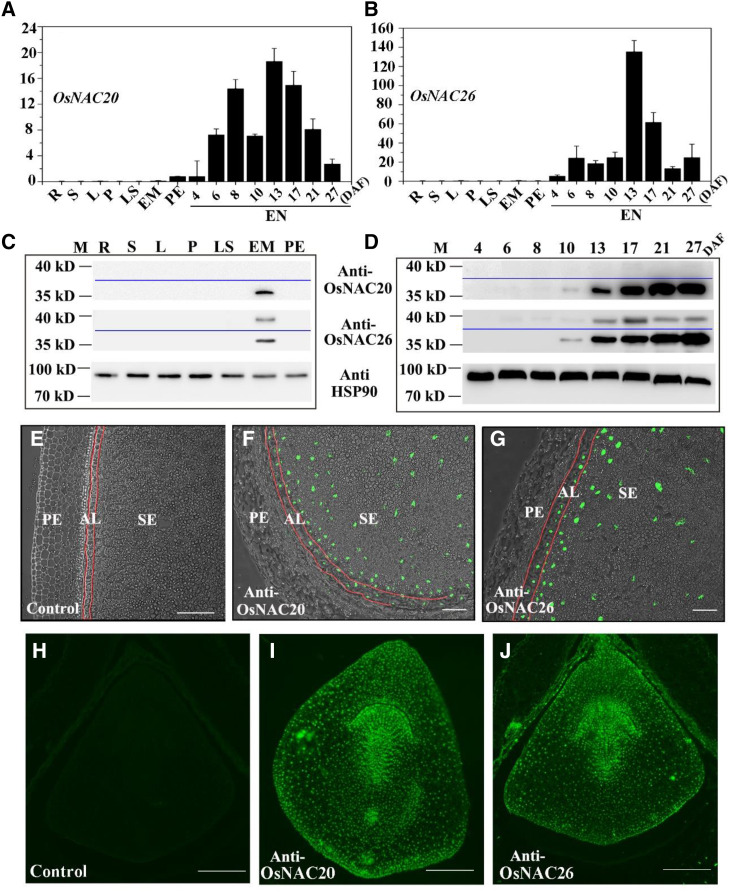
OsNAC20 and OsNAC26 share 89% sequence similarity in their coding sequence (CDS) and 85% sequence similarity in their amino acid sequence (Supplemental Fig. S1). Reverse transcription quantitative PCR (RT-qPCR) analysis showed that OsNAC20 and OsNAC26 were not detectable in a range of organs, including the root, stem, leaf, panicle, leaf sheath, embryo, and pericarp, but were expressed only in developing endosperm, with a peak at 13 d after flowering (DAF; Fig. 1, A and B; Supplemental Fig. S2A). The protein level was further analyzed by immunoblot. Anti-OsNAC20 and anti-OsNAC26 were produced against the synthetic peptide CRIFHKSSGIKKPVPVAPHQ and the expressed protein (160–333 amino acids), respectively (Supplemental Fig. S2B). The protein sizes for OsNAC20 and OsNAC26 are 35.74 and 37.39 kD, respectively. As shown in Figure 1, C and D, only one band was observed at ∼36 kD for anti-OsNAC20, but two bands were observed, at ∼36 kD and 38 kD, for anti-OsNAC26, which indicated that the anti-OsNAC20 is specific but anti-OsNAC26 can detect both OsNAC20 and OsNAC26. Interestingly, OsNAC20 and OsNAC26 both were detected not only in endosperm but also in the embryo at the protein level, although no mRNA in the embryo was detected by RT-qPCR assays (Fig. 1, A–D). In endosperm, OsNAC26 was expressed after 4 DAF, while OsNAC20 showed an obvious protein level at 10 DAF (Fig. 1D). Immunofluorescence assays with anti-OsNAC20 and anti-OsNAC26 showed that the fluorescence signals were situated in the nuclei of starchy endosperm and the aleurone layer (Fig. 1, E–G). The signals were also broadly distributed in the whole embryo nuclei (Fig. 1, H–J). In addition, subcellular localization of 35S:OsNAC20-GFP and 35S:OsNAC26-GFP expressed in Nicotiana benthamiana leaves was concentrated in the nuclei (Supplemental Fig. S3), which indicated that both OsNAC20 and OsNAC26 function in the nucleus. In conclusion, OsNAC20 and OsNAC26 are grain-specific instead of endosperm-specific genes, as both were located in nuclei of the aleurone layer, starchy endosperm, and embryo.
Figure 1.
Expression analysis of OsNAC20 and OsNAC26 in rice. A and B, RT-qPCR analysis of OsNAC20 and OsNAC26 expression in different organs and in developing endosperms (EN) of cv ZH11. Actin was used as an internal control. The y axis represents the expression level relative to that of the control. Samples of root (R), stem (S), leaf (L), and leaf sheath (LS) were derived from plants at the four-leaf stage. Panicles (P) were harvested at 3 d before heading. Both pericarp (PE) and embryo (EM) tissues were collected from developing caryopses at 8 DAF. Three biological experiments (three technical replicates for each biological experiment) were conducted in A and B. Values are means ± sd. C and D, Immunoblot analysis of OsNAC20 and OsNAC26 expression profiles in different organs and developing endosperms. HSP90 antibody was used as a sample loading control. Total protein samples were extracted from pooled tissues. M, Protein marker. E to J, Immunofluorescence assays of OsNAC20 and OsNAC26 in endosperm (E–G) and embryo (H–J) at 8 DAF. Both endosperm and embryo were transversely sectioned. Samples without anti-OsNAC20 and anti-OsNAC26 were used as control (E and H). AL, Aleurone layer; SE, starchy endosperm. Scale bars = 150 μm.
OsNAC20 and OsNAC26 Can Interact with Each Other
To determine whether OsNAC20 and OsNAC26 can form homo- or heterodimers, we performed bimolecular fluorescence complementation assays. Both OsNAC20 and OsNAC26 interacted with themselves (Supplemental Fig. S4A). More importantly, OsNAC20 and OsNAC26 could interact with each other in the nuclei (Supplemental Fig. S4A), which is consistent with the result for onion (Allium cepa) epidermal cells (Mathew et al., 2016). A His pull-down assay confirmed their heterodimerization (Supplemental Fig. S4B). Therefore, OsNAC20 can physically interact with OsNAC26 in plant cell nuclei.
Phylogenetic Analysis of OsNAC20 and OsNAC26
The most similar proteins to OsNAC20 and OsNAC26 are from wild rice species (Oryza spp.; Supplemental Fig. S5). Their whole protein sequences share 43% to 72% similarity with OsNAC20 and OsNAC26. The sequence similarity of other homologous proteins is limited to the NAC domain, which shows ∼70% similarity with OsNAC20 and OsNAC26. Phylogenetic analysis indicated that these homologous proteins with known functions are ZmNAC128 and ZmNAC130 in maize, and AtNAC87 in Arabidopsis. ZmNAC128 and ZmNAC130 have been reported to take part in the regulation of storage product accumulation (Zhang et al., 2019b). Expression of AtNAC87 is changed in response to Cabbage leaf curl virus in Arabidopsis (Ascencio-Ibáñez et al., 2008). These results indicated that OsNAC20 and OsNAC26 might play a role in storage product synthesis and stress response.
Simultaneous Deficiency of OsNAC20 and OsNAC26 Results in Floury Grains
To investigate whether deficiency of OsNAC20 and/or OsNAC26 affects grain development, we generated knockout mutants through the CRISPR/Cas9 technique. Each target locus was scanned for possible off-target matches through mapping of the whole genome (http://cbi.hzau.edu.cn/cgi-bin/CRISPR; Supplemental Table S1) to ensure the specificity of these target sequences.
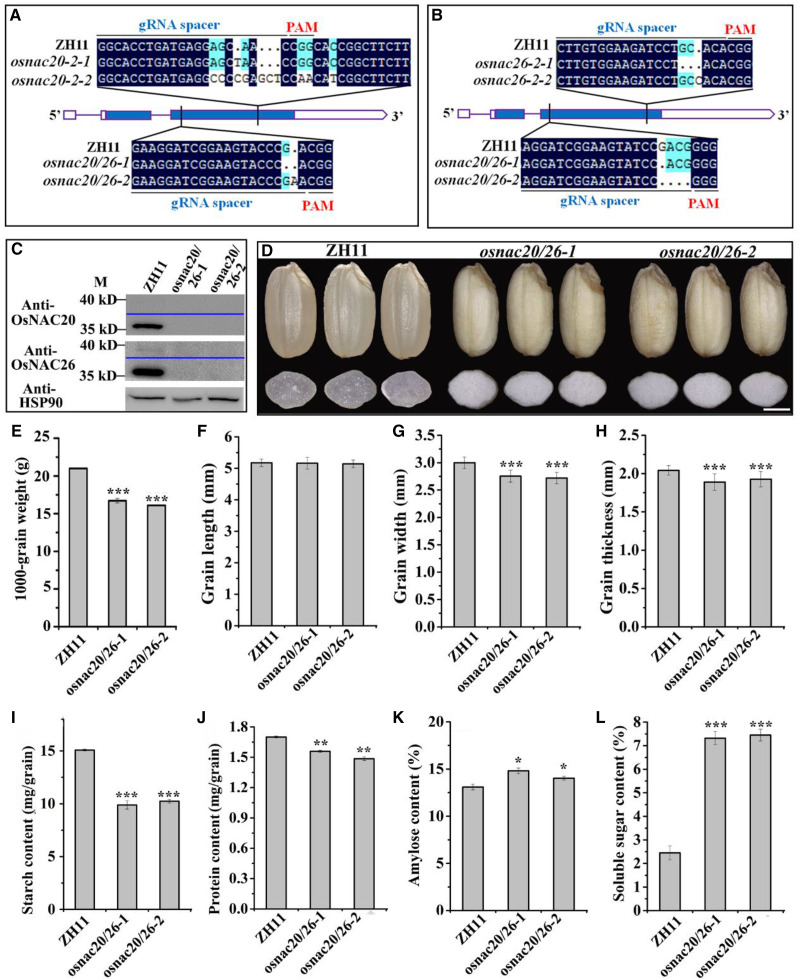
Two targets in the CDS regions were mutated to derive the single mutant for each gene, and the resulting mutants were designated as osnac20-1, osnac20-2, osnac26-1, and osnac26-2 (Supplemental Fig. S1A). For osnac20-1 and osnac26-1, unfortunately, we didn't genotype the corresponding single mutants, as the target sequences for these two mutants were highly similar and both genes were mutated in the two single mutants. The target for osnac20-2 is specific and no possible off-target sequence was detected (Supplemental Fig. S1A; Supplemental Table S1). Although the target site for osnac26-2 was also similar to the counterpart of OsNAC20, OsNAC20 was not mutated in the osnac26-2 mutant (Supplemental Figs. S1A and S6A; Supplemental Table S1). Therefore, the osnac20-2 and osnac26-2 mutants were used for further analysis. As shown in Supplemental Fig. S7, the single mutation of OsNAC20 and OsNAC26 had no effect on grain development.
The sites targeted for osnac20-1 and osnac26-1 were mutated to effectively derive the double mutant (Fig. 2, A and B; Supplemental Fig. S1A). Two homozygous independent double mutants were named osnac20/26-1 and osnac20/26-2 (Fig. 2, A and B). The possible off-target risk in OMTN1 (LOC_Os02g36880) in the double mutant was excluded (Supplemental Fig. S6B; Supplemental Table S1). Immunoblot analysis verified the loss of function of OsNAC20 and OsNAC26 in the two mutants (Fig. 2C). Phenotypic characterization showed no detectable change in such traits as plant height, panicle number, and flowering date was observed in the osnac20/26-1 and osnac20/26-2 mutants compared to cv Zhonghua11 (ZH11; Supplemental Fig. S8A), but the mutant grains showed a floury phenotype (Fig. 2D). The grain width, thickness, and 1,000-grain weight were significantly reduced (Fig. 2, E–H). Follow-up phenotyping of the two mutants revealed a consistent grain phenotype (Supplemental Fig. S8, B–F), which further indicated that this phenotype was caused by the deficiency of OsNAC20 and OsNAC26. The osnac20/26-1 line was used for further study in the following experiments.
Figure 2.
Mutation sequence and seed phenotypic analysis of the osnac20/26 mutant. A and B, Mutation of the CDS in OsNAC20 (A) and/or OsNAC26 (B) mutants. Two homozygous independent transgenic plants were derived for each mutation. All of these mutations resulted in loss of function of OsNAC20 and OsNAC26 due to a frame shift. Solid boxes represent the exons, solid lines the introns, and open boxes the untranslated region. C, Immunoblot analysis of anti-OsNAC20 and anti-OsNAC26 in the osnac20/26 double mutant. Anti-HSP90 antibody was used as the internal control. Total proteins were extracted from pooled endosperm samples at 13 DAF. M, Protein marker. D, Grain phenotypic features of cv ZH11, osnac20/26-1, and osnac20/26-2. Images shown were digitally extracted and scaled for comparison. Scale bar = 1.5 mm. E to H, Measurement of 1,000-grain weight (E), grain length (F), grain width (G), and grain thickness (H) of the wild type and the osnac20/26 mutant (n = 3 [E] and 50 [F–H]). I to L, Starch content (I), storage protein content (J), amylose content (K), and soluble sugar content (L) in mature grains of cv ZH11 and the osnac20/26 mutant. Data in E to L were measured from mature grains. Values are means ± sd from two (J and K) and three (I and L) biological replicates. The asterisk indicates a statistically significant difference between cv ZH11 and the osnac20/26 mutants, as calculated by Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Simultaneous Deficiency of OsNAC20 and OsNAC26 Results in a Significant Decrease in Starch Accumulation
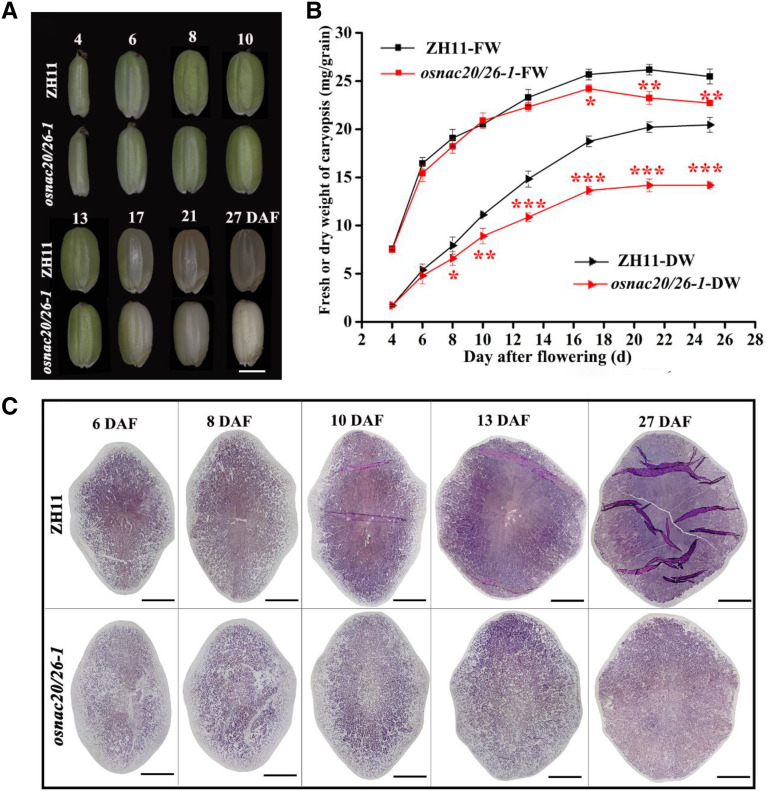
Shape observation of developing caryopsis showed no obvious differences between cv ZH11 and osnac20/26-1 caryopses before 4 DAF, but the osnac20/26-1 caryopsis became smaller after 4 DAF, and this difference was continuously detected with caryopsis development and was ascribed to defective dry matter accumulation (Fig. 3, A and B). In rice grains, most of the dry matter is starch. Two lines of evidence indicated that the significantly decreased dry matter/grain weight in the osnac20/26-1 mutant was mainly due to the significantly reduced starch content (Fig. 2I). First, the grain-filling rate was reduced in the osnac20/26-1 mutant after 4 DAF (Fig. 3B). Second, the starch granules in the osnac20/26-1 mutant were always packed less tightly than the corresponding region of cv ZH11 during the development stage (Fig. 3C).
Figure 3.
Phenotype, weight, and starch accumulation in developing caryopsis of the osnac20/26-1 mutant. A, Phenotype of the developing caryopsis. Scale bar = 2 mm. B, Fresh weight (FW) and dry weight (DW) of developing caryopsis. Thirty caryopses from the same part of three plants (10 for each plant) were considered as one replicate, and the weight of one caryopsis was calculated. Three replicates were conducted. Values are means ± sd. The asterisk indicates a statistically significant difference between cv ZH11 and the osnac20/26-1 mutant, as calculated by Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001). C, Microphotographs of iodine-stained transverse sections of developing caryopses at the midregion. Scale bars = 0.5 mm. Images shown in A and C were digitally extracted and scaled for comparison.
Interestingly, the morphology of starch granules did not show a significant difference between cv ZH11 and the osnac20/26-1 mutant, both of which displayed a polygonal shape in the central, dorsal, ventral, and lateral regions of the kernel. It was only that the size of starch granules in the mutant was less homogeneous than that of the control (Supplemental Fig. S9, A and B). The starch also showed a similar Mr distribution, except that the amylose content of the mutant was slightly higher than that of the control (Supplemental Fig. S9C; Supplemental Table S2), which was consistent with the results obtained from the iodine colorimetric method (Fig. 2K). In addition, starches from cv ZH11 and osnac20/26 had an A-type crystalline structure and similar gelatinization properties (Supplemental Fig. S9, D and E). Overall, the deficiency of OsNAC20 and OsNAC26 significantly changed starch accumulation but did not have a remarkable influence on starch morphology, molecular structure, and physicochemical properties.
Simultaneous Deficiency of OsNAC20 and OsNAC26 Results in a Pronounced Decrease in Storage Protein Content and a Defective Protein Body
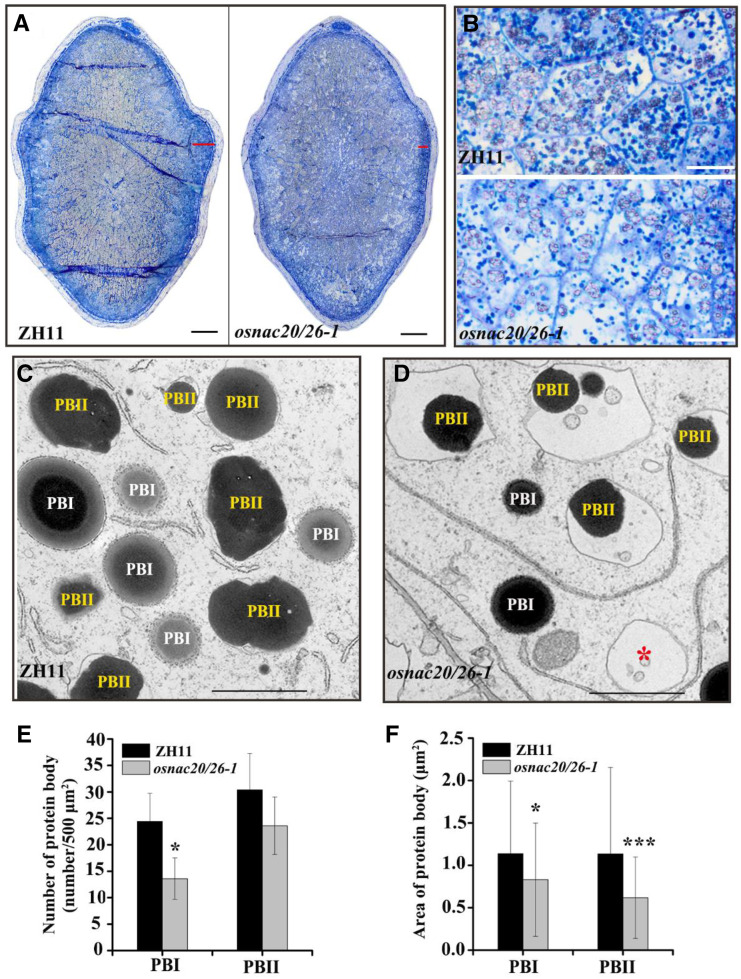
Apart from starch, storage protein is also an important component of dry matter in rice grains. The storage protein amount showed a pronounced decrease in the osnac20/26 mutant (Fig. 2J). The storage proteins accumulate in the protein body (PB), and therefore, the PB was further investigated using semithin sections stained with acid fuchsin and Coomassie brilliant blue, two dyes that are specific for protein staining (Fig. 4, A and B; Supplemental Fig. S10). The PBs were mainly distributed in the subaleurone region in cv ZH11 and osnac20/26-1 plants, but those in the double mutant were less densely packed than those in the control (Fig. 4, A and B; Supplemental Fig. S10). Furthermore, the number and area of PBs were investigated by transmission electron microscopy (TEM). PBI, which derives from the endoplasmic reticulum (ER) and consists of prolamin, showed spherical morphology and was surrounded by rough ER attached with ribosomes (Fig. 4C). Glutelin- and globulin-containing PBII accumulated in protein storage vacuoles (PSVs) and displayed an irregular shape (Fig. 4C). Although PBI morphology was similar in the osnac20/26-1 mutant, the density and area were decreased by ∼40% and 30%, respectively (Fig. 4, E and F). With regard to PBII, its filling was significantly affected. As shown in Figure 4D, PSVs were not completely occupied by storage proteins, and in some cases were empty, resulting in an extremely small PBII area in the osnac20/26-1 mutant compared to cv ZH11 (Fig. 4F). The above results indicated that storage protein synthesis is severely affected in this double mutant.
Figure 4.
Storage protein analysis in endosperm of the osnac20/26-1 mutant. A, Microphotographs of Coomassie brilliant blue-stained transverse sections of midregion caryopses from cv ZH11 (left) and osnac20/26 (right) at 10 DAF. Scale bar = 0.3 mm. B, Images are magnifications of the regions marked by a red line at the far right in the full-scale images of A (left and right, respectively). Scale bars = 10 μm. C and D, TEM photographs of PBI and PBII in endosperm of cv ZH11 (C) and osnac20/26 (D) at 10 DAF. The red asterisk in D indicates the empty PSV in the osnac20/26-1 mutant. Scale bars = 2 μm. Images shown in A to D were digitally extracted and scaled for comparison. E and F, Number and area of the PB in the same endosperm region of cv ZH11 and osnac20/26 at 10 DAF. Numbers of PBs were counted in the TEM photographs with the same magnification and area (500 μm2), and their means and sds were calculated from five TEM photographs. The areas of PBI and PBII were individually measured using Photoshop Software, and their means and sds were calculated from all PBs in the above-mentioned five TEM photographs. The asterisk indicates a statistically significant difference between cv ZH11 and the osnac20/26-1 mutant, as calculated by Student’s t test (*P < 0.05 and ***P < 0.001).
Identification of Genes Regulated by OsNAC20 and OsNAC26
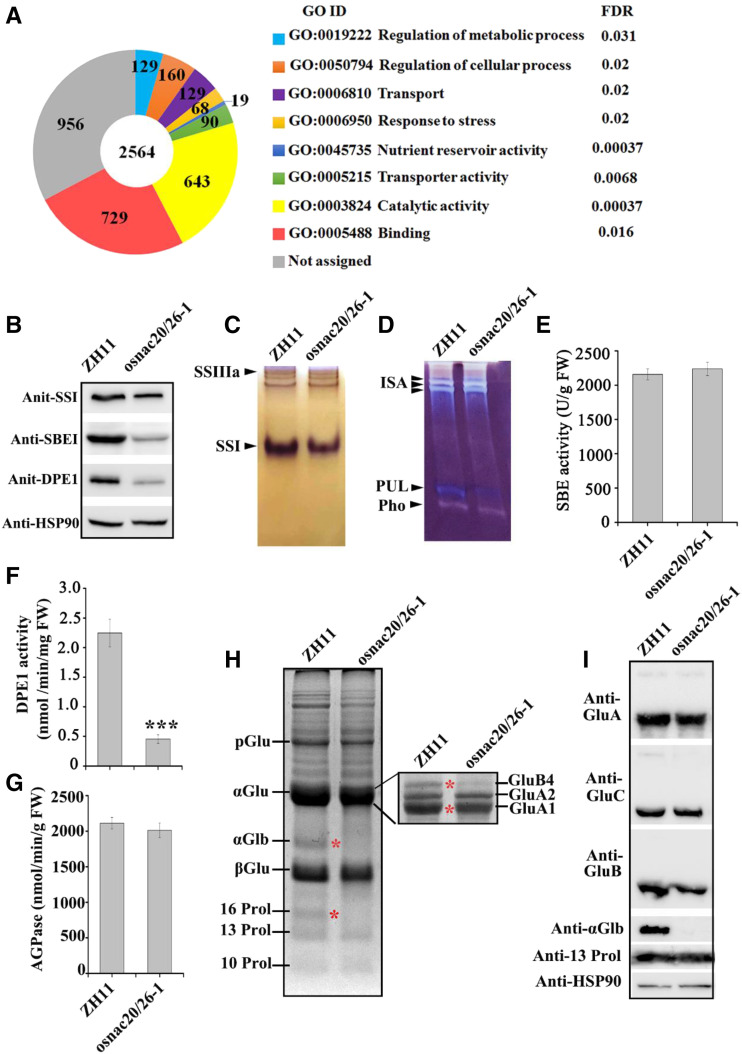
RNA-sequencing (RNA-seq) analysis was conducted to investigate the molecular mechanism of phenotypic defects in the osnac20/26 mutant (Supplemental Fig. S11). A total of 2,564 differentially expressed genes (DEGs) were characterized, including 1,631 downregulated genes and 933 upregulated genes (Supplemental Table S3). The Gene Ontology (GO) enrichment termed “response to stress” suggested again a potential function of OsNAC20 and OsNAC26 in stress, which is consistent with the result of phylogenetic analysis (Fig. 5A; Supplemental Fig. S5). Most importantly, the “nutrient reservoir” terms, mainly including glutelin, prolamin, and globulin synthesis-related genes, broadly changed expression profile in the osnac20/26-1 mutant; in which 11 DGEs were downregulated, and the reduction levels ranged from 68% to 93% (Table 1). Furthermore, we detected that several starch synthesis-related genes, including AGPS2b, AGPL2, SSI, SBEI, Pul, and DPEI, were significantly downregulated (Table 1). Among them, the transcription levels of AGPS2b and Pul were most markedly decreased, by 91.3% and 93.6%, respectively. RT-qPCR confirmed the decreased expression levels of these DGEs (Supplemental Fig. S12A).
Figure 5.
Genome-wide analyses of the OsNAC20- and OsNAC26-regulated network. A, GO classification of DEGs from the osnac20/26-1 RNA-seq analysis. The changes in gene expression level were calculated from three biological replicates (fold change >2.5 and Q < 0.001). Enrichment analysis of functional categories was performed at the agriGO Web site (http://bioinfo.cau.edu.cn/agriGO/analysis.php). The RNA sample for each replicate was extracted from independent pools of endosperm at 10 DAF. The numbers of genes classified within each GO term, as well as their Q values, are shown. Only GO terms with at least 10 annotated genes were kept. B, Immunoblot analysis of starch synthesis-related proteins in rice endosperm at 10 DAF. C and D, Native-PAGE/activity staining analysis of SS (C) and DBE (D). E to G, Activity of SBEI (E), DPE1 (F), and AGPase (G). Total protein samples were extracted from pooled endosperm at 10 DAF. Values are means ± sd from three biological replicates. The asterisk indicates a statistically significant difference between cv ZH11 and the osnac20/26-1 mutant as calculated by Student’s t test (***P < 0.001). H, SDS-PAGE analysis of storage proteins in mature endosperm from cv ZH11 and the osnac20/26 mutant. Red asterisks indicate the bands in the osnac20/26 mutant that are different from those of cv ZH11. The inset shows the αGlu further separated with a higher polyacrylamide concentration gradient gel and lower electric tension. I, Immunoblot analysis of storage protein synthesis-related proteins in mature endosperm. HSP90 was used as a loading control. pGlu, Glutelin precursors; αGlu, glutelin acidic subunits; βGlu, glutelin basic subunits; αGlb, α-globulin; 10 Prol, 13 Prol, and 16 Prol represent 10-kD, 13-kD, and 16-kD prolamin, respectively.
Table 1. Decreased expression of starch- and storage protein synthesis-related genes.
| Gene Identifier (RAP_Locus) | Gene Name | Fold Change | False Discovery Rate |
|---|---|---|---|
| Os08g0345800 | AGPS2b | 0.087 | 8.86E−41 |
| Os01g0633100 | AGPL2 | 0.279 | 3.17E−48 |
| Os06g0160700 | SSI | 0.358 | 3.66E−32 |
| Os06g0726400 | SBEI | 0.201 | 8.01E−09 |
| Os04g0164900 | Pul | 0.064 | 8.34E−14 |
| Os07g0627000 | DPE1 | 0.399 | 1.56E−17 |
| Os01g0762500 | GluA1 | 0.268 | 3.79E−07 |
| Os10g0400200 | GluA2 | 0.320 | 7.86E−07 |
| Os02g0249800 | GluB-1a | 0.222 | 2.63E−06 |
| Os02g0249900 | GluB-1b | 0.165 | 5.57E−14 |
| Os02g0249600 | GluB2 | 0.205 | 0.00012 |
| Os02g0268300 | GluB4 | 0.136 | 0.00064 |
| Os02g0268100 | GluB5 | 0.105 | 0.00091 |
| Os02g0453600 | GluC | 0.157 | 1.80E−05 |
| Os05g0499100 | α-globulin | 0.114 | 3.66E−12 |
| Os07g0206400 | 13 kD prolamin | 0.072 | 9.42E−06 |
| Os06g0507200 | 16 kD prolamin | 0.077 | 4.92E−38 |
Next, SDS-PAGE, immunoblot, and/or enzyme activity assays were performed in rice endosperm to examine whether the change in RNA expression of these DEGs affected the protein amount and/or enzyme activity. Similar to the reduced protein abundance, SSI and DPE1 activities were decreased by ∼34% and 80%, respectively. Pul activity was decreased ∼70% (Fig. 5, B–G). However, the activity levels of AGPase and SBE in the osnac20/26-1 mutant were not significantly different from those in cv ZH11, although AGPS2b, AGPL2, and SBEI were remarkably downregulated (Fig. 5, E and G; Table 1). In addition, the protein abundance of some other starch synthesis-related enzymes was similar between cv ZH11 and the double mutant, as was the RNA level (Supplemental Fig. S12B; Supplemental Table S3).
For storage proteins, decreased accumulation of glutelin acidic and basic subunits was detected in the mutant compared to cv ZH11 (Fig. 5H). When further separating glutelin acidic subunits, it was detected that the GluA2 band was not significantly different between cv ZH11 and osnac20/26-1, and the GluA1 abundance was slightly lower in the mutant than in cv ZH11, while GluB4 was severely decreased in the osnac20/26-1 mutant compared to cv ZH11 (Fig. 5H). Immunoblot analysis indicated that total GluA and GluB acidic subunits were decreased by ∼17% and 32%, respectively, in the osnac20/26-1 mutant compared to cv ZH11 due to the respective reductions in GluA1 and GluB4 (Fig. 5I). Most notably, α-globulin and 16 kD prolamin were almost completely absent from the osnac20/26-1 mutant (Fig. 5, H and I). These data indicated that decreased starch and storage protein synthesis are mainly attributed to the reduced expression of starch and storage protein synthesis-related genes, and they further suggested possible roles of OsNAC20 and OsNAC26 in the regulation of starch and storage protein synthesis in rice endosperm.
OsNAC20 and OsNAC26 Directly Target Endosperm Starch and Storage Protein Synthesis-Related Genes
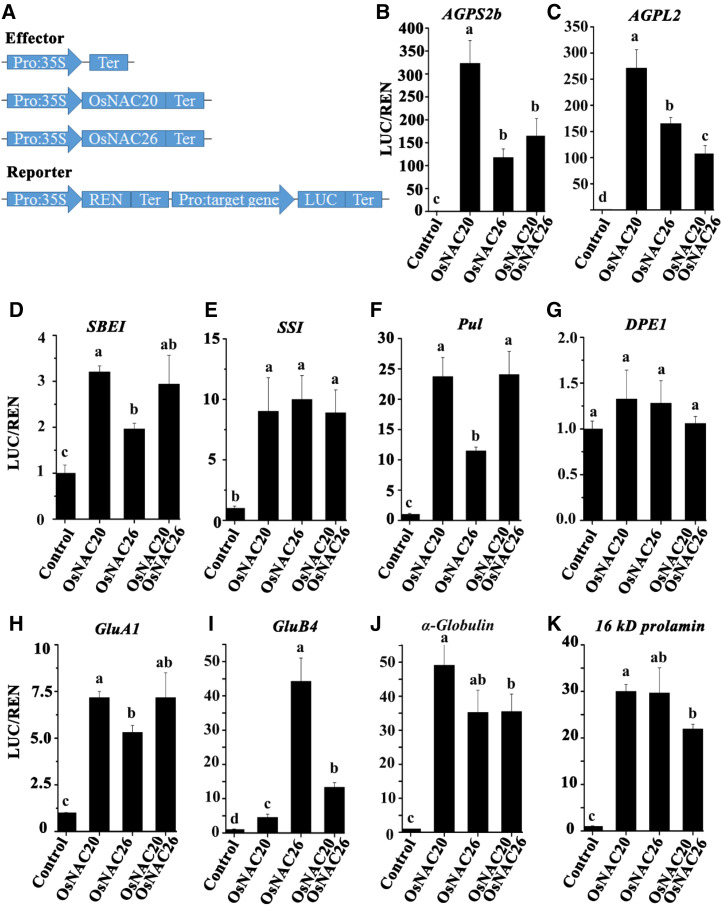
According to RNA-seq, RT-qPCR, SDS-PAGE, immunoblot, and enzyme activity assay results, starch and storage protein synthesis-related genes, including AGPS2b, AGPL2, SSI, SBEI, Pul, DPE1, α-glolulin, 16 kD prolamin, GluB4, and GluA1, were further investigated through a dual-luciferase (dual-luc) reporter assay to detect whether they can be directly transactivated by OsNAC20 and/or OsNAC26 (Figs. 5 and 6; Table 1). As shown in Figure 6, B to K, both OsNAC20 alone and OsNAC26 alone transactivated these genes well except in the case of DPE1; however, their activation level varied among different promoters. For example, OsNAC20 showed a 323-fold luciferase increase in activating the AGPS2b promoter, but OsNAC26 only produced a 117-fold increase; with regard to the 16 kD prolamin promoter, the two proteins showed a similar activation level, but coexpression did not enhance the LUC expression level compared to that of OsNAC20 or OsNAC26 alone. This result indicated that although OsNAC20 and OsNAC26 can interact with each other, they independently regulated the expression of starch and storage protein synthesis-related genes.
Figure 6.
OsNAC20 and OsNAC26 stimulate the expression of starch and storage protein-related genes. A, Schematic diagram of the effector and reporter vectors for dual-luc assays. LUC, Firefly luciferase; REN, Renilla luciferase; Ter, Terminator. B to K, Relative luciferase activity was examined in N. benthamiana leaves cotransfected with the reporter and different effector constructs. A 1- to 3-kb region upstream of the start codon in these genes was selected as the gene promoter for firefly luciferase. Values are means ± sd from at least three biological replicates. Different lowercase letters indicate significant difference (P < 0.05) as determined by one-way ANOVA and Tukey’s honestly significant difference (HSD) mean-separation test.
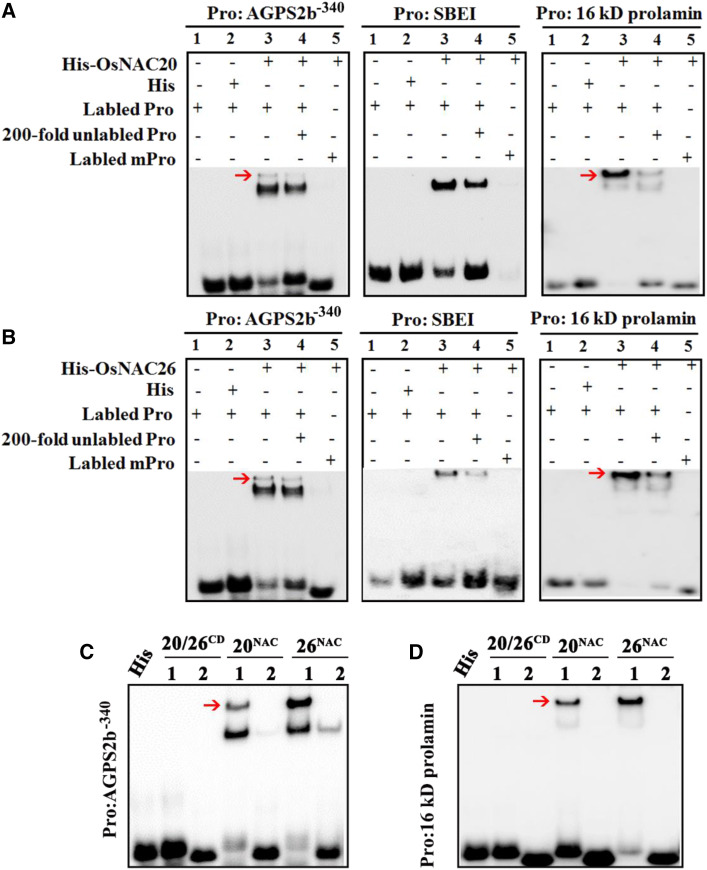
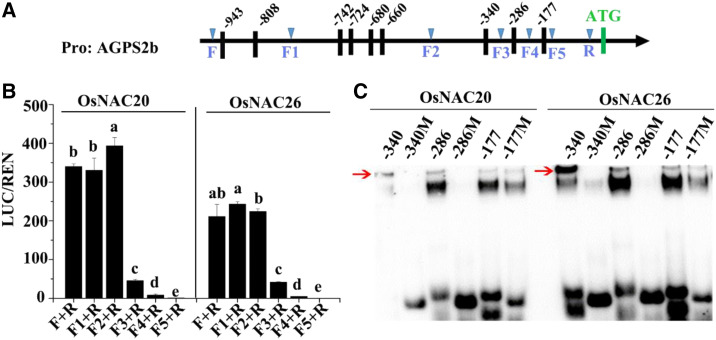
To further confirm the binding of OsNAC20 and OsNAC26 to these gene promoters, an electrophoretic mobility shift assay (EMSA) was conducted. As shown in Supplemental Table S4, the promoters of AGPL2, SSI, SBEI, Pul, GluB4/5, and 16 kD prolamin contain the ACGCAA motif, an element that has been reported to interact with maize NAC TFs to take part in storage product accumulation (Zhang et al., 2019b). Among these, SBEI and 16 kD prolamin promoters were further investigated to demonstrate their interaction with OsNAC20 and OsNAC26. The result showed that OsNAC20 and OsNAC26 were able to bind to the ACGCAA motif (Fig. 7, A and B; Supplemental Table S5). In the case of AGPS2b, there is no ACGCAA motif in its promoter. Therefore, the truncated AGPS2b promoters were produced to detect the cis-element targeted by OsNAC20 and OsNAC26. As shown in Figure 8, A and B, compared to the F+R, F1+R and F2+R fragments, which displayed strong transactivation activity, F3+R showed significantly decreased activity, F4+R demonstrated relatively weak activity, and F5+R had no activity. This result placed the cis-element at the −340, −286, and −177 sites in the AGPS2b promoter, especially the −340 site, which is mainly responsible for the binding activity. EMSA results showed that DNA sequences that contain the −340 and −286 sites were specifically bound by OsNAC20 and OsNAC26, respectively, but those containing the −177 fragment were not, which might be because some other NAC binding sites exist in it (Figs. 7A and 8C; Supplemental Table S5). Although there is also no ACGCAA element in the GluA1 and α-glolulin promoters, they contain the core binding sites of the −340 binding site (Supplemental Table S4).
Figure 7.
OsNAC20 and OsNAC26 directly bind to the promoters of AGPS2b, SBEI, and 16 kD prolamin. A and B, EMSA of OsNAC20 (A) and OsNAC26 (B) with the promoters of starch and protein synthesis-related genes, respectively. Pro, Promoter; mPro, mutated promoter. Both His-OsNAC20 and His-OsNAC26 were able to bind to the promoters of SBEI and 16 kD prolamin and induced a mobility shift (lane 3), while addition of an unlabeled probe competitively interacted with the recombinant proteins and resulted in a compromised band (lane 4). In addition, His and the mutated fragment alone did not cause this mobility shift (lanes 2 and 5, respectively). C, EMSA confirming the domain that interacts with the DNA sequence. 20/26CD indicates the same subdomain C and D in OsNAC20 and OsNAC26 (Supplemental Fig. S1B). 20NAC and 26NAC represent the NAC domains in OsNAC20 and OsNAC26, respectively. Numbers 1 and 2 represent the probe and mutated probe, respectively. Probe sequences are listed in Supplemental Table S5. The red arrow indicates the positive band.
Figure 8.
Identification of cis-elements targeted by OsNAC20 and OsNAC26 in the AGPS2b promoter. A, Schematic representation showing predicted NAC binding sites in the AGPS2b promoter. F to F5 and R represent the primers designed in the AGPS2b promoter. Black vertical bars represent the predicted NAC binding sites. Pro:AGPS2b, APGS2b promoter. B, Relative luciferase activity of different truncated AGPS2b promoters bound by OsNAC20 and OsNAC26. Values are means ± sd from at least three biological replicates. REN, Renilla luciferase; LUC, firefly luciferase. Different lowercase letters indicate significant difference (P < 0.05) as determined by one-way ANOVA and Tukey’s honestly significant difference (HSD) mean-separation test. C, EMSA identifying binding activity of the three DNA fragments containing the −340, −266, and −177 binding sites with OsNAC20 and OsNAC26. The red arrow indicates the positive band. M, Mutated probe. Probe sequences are listed in Supplemental Table S5. The core binding sites were deleted in the mutated probes.
Previous studies have reported that subdomains C and D in the NAC domain may interact with cis-acting elements (Wang and Culver, 2012). We generated two truncated versions of OsNAC20 and OsNAC26, OsNAC20NAC, OsNAC26NAC, and OsNAC20/26CD, and tested the binding of these three proteins to the target sequence to investigate whether the NAC domain or C and D subdomains are required for DNA binding. Results showed that the NAC domain, but not the C and D subdomains, was able to bind to the cis-elements (Fig. 7C), which might be due to the fact that except for the C and D subdomains, other subdomains are also required for assistance in binding to DNA sequences.
DISCUSSION
Expression Patterns of OsNAC20 and OsNAC26 in Seeds
The thinner caryopsis phenotype and decreased dry weight after 4 DAF in the osnac20/26 mutant indicated that OsNAC20 and OsNAC26 function after 4 DAF in the caryopsis. Starch and storage protein contents significantly increased between 4 and 10 DAF, and few of them are synthesized at the late developing stage (after 17 DAF; Fig. 3B; Yamagata et al., 1982). However, only faint bands of OsNAC20 and OsNAC26 were detected between 4 and 10 DAF, whereas higher content of these proteins was observed at the late stage (Fig. 1D). It is inferred that OsNAC20 and OsNAC26 might have a function in late seed development. Interestingly, we observed a preharvest sprouting phenotype (Supplemental Fig. S13), which is related to ABA-controlled seed dormancy. After 17 DAF, the seed prepares to enter into the dormancy stage. Combined with the downregulated OsRCAR1 (Os06g0527800; ABA receptor) expression in the double mutant (Supplemental Table S3), we reasonably speculated that OsNAC20 and OsNAC26 might have a function in controlling seed dormancy.
In previous studies, OsNAC20 and OsNAC26 are reported as endosperm-specific genes, as embryo tissue was not included in the investigated organs (Fang et al., 2008; Nie et al., 2013). In this study, OsNAC20 and OsNAC26 transcripts were not detected in the embryo at 8 DAF, but their proteins were detected by immunoblot and immunostaining at this stage (Fig. 1, A–D and H–J), which might be due to the fact that mRNA is transcribed first, followed by protein translation.
OsNAC20 and OsNAC26 Regulate Starch Synthesis
In the osnac20/26-1 mutant, the starch content was reduced by ∼30% (Figs. 2I and 3B), which was attributed to the reduced expression and activity of SSI, Pul, and DPEI instead of the unchanged AGPase and SBE activity, although OsNAC20 and OsNAC26 can bind to the promoters of AGPS2b, AGPL2, and SBEI to regulate their expression (Figs. 5–8; Table 1).
Several studies have reported that the cytosolic isoforms, including AGPS2b and AGPL2 in rice and AGPS1a (Bt2) and AGPL1 (Sh2) in maize, are mainly responsible for the AGPase activity in endosperm. Any mutation of these four genes can produce a significant reduction of AGPase activity, resulting in a shrunken seed phenotype due to the marked decrease of starch synthesis (Yano et al., 1984; Giroux and Hannah, 1994; Tuncel et al., 2014). One interesting result worth noting in the osnac20/26-1 mutant is that although the expression levels of AGPS2b and AGPL2 were markedly decreased, the AGPase activity in this double mutant paralleled that of cv ZH11 (Fig. 5G; Table 1). Therefore, the decreased starch content in the osnac20/26-1 mutant is unrelated to the reduced expression of AGPS2b and AGPL2. A similar case is detected in the mutant of ZmbZIP22, a gene that encodes a bZIP-type TF and regulates the 27-kD γ-Zein gene in maize (Li et al., 2018a). In the zmbzip22 mutant, the expression of AGPS1a and AGPL1 are also significantly reduced. Although AGPase activity was not examined in the study of Li et al. (2018a), we speculate that it is unchanged, as the total starch level is essentially unchanged in the zmbzip22 mutant. Furthermore, we investigated the gene expression of other plastid-localized AGPase subunits in osnac20/26-1 endosperm, and the results showed that the expression levels of AGPS1 (Os09g0298200) and AGPL3 (Os03g0735000) were not significantly different between cv ZH11 and the osnac20/26-1 mutant, and that of AGPL1 (Os05g0580000) was ∼58% reduced in the mutant (Supplemental Table S3). This indicated that AGPase complementation did not happen in this double mutant. A recent study has reported that AGPase is phosphorylated in wheat seeds, which indicated that AGPase can undergo posttranslational modification (Ferrero et al., 2020). Therefore, we inferred that the lack of change in AGPase activity in the osnac20/26 mutant might have been due to posttranslational modification of the residual transcripts of AGPS2b and AGPL2, which could maintain full AGPase activity.
OsNAC20 and OsNAC26 can bind to the SBEI promoter to regulate its expression, but the total SBE activity was not decreased in the double mutant. Two possibilities exist: either the SBEI activity was not changed, although its RNA and protein levels were decreased, or the reduced SBEI activity was complemented by SBEIIa and SBEIIb. We are inclined to favor the former hypothesis, as an extra SBEIIb activity has a remarkable effect on the molecular structure of starch (Tanaka et al., 2004), whereas the molecular structure and physicochemical properties of starch in the osnac20/26-1 mutant were not significantly changed (Supplemental Fig. S9, C–E). In addition, SBEI can be phosphorylated (Tetlow et al., 2004; Qiu et al., 2016), a posttranslational modification that might keep its full enzyme activity in the double mutant, even if its mRNA and protein level are decreased. With regard to DPE1, although OsNAC20 and OsNAC26 could not directly bind to its promoter to regulate its expression (Fig. 6G), its RNA level, protein amount, and enzyme activity were significantly decreased (Fig. 5, B and F). This might be attributed to a physical/functional interaction of DPE1 with other starch synthetic enzymes, and therefore, it was reduced along with other components in the isozyme complex. Hwang et al. (2016) reported that Pho1 assembles with DPE1 to form a protein complex that enhances the synthesis of malto-oligosaccharides. However, Pho1 was not significantly different between cv ZH11 and the osnac20/26-1 mutant. Therefore, this hypothesis might be impossible. More evidence is needed to demonstrate why DPE1 was affected in this double mutant.
Generally, a decrease in starch content is accompanied by a change in starch morphology. In the osnac20/26-1 mutant, starch showed a heterogeneous size, but it exhibited a polygonal shape (Supplemental Fig. S9, A and B). In addition, the amylose content in this double mutant was increased (Fig. 2K; Supplemental Fig. S9C; Supplemental Table S2). Previous reports have indicated that single mutations of SSI and Pul do not affect starch morphology and amylose content (Satoh et al., 2003; Fujita et al., 2006, 2009), whereas DPE1 deficiency results in changes in starch morphology and increased amylose content, similar to those observed in the osnac20/26-1 mutant (Dong et al., 2015), indicating that in the osnac20/26-1 mutant, the slight changes in starch morphology and amylose content were caused by decreases in DPE1 expression and activity. However, the starch content in DPE1-deficiency transgenic lines does not show a significant difference from that of the wild type (Dong et al., 2015). Therefore, it is concluded that the significantly decreased starch content in the osnac20/26-1 mutant is a combined result of the reduced expression of SSI, Pul, and DPEI. In addition, Pul was decreased largely at RNA and enzyme activity levels, which implies that OsNAC20 and OsNAC26 are the main regulators for Pul.
OsNAC20 and OsNAC26 Regulate Storage Protein Synthesis
OsNAC20 and OsNAC26 directly bound to the promoters of GluA1, GluB4, α-globulin, and 16 kD prolamin to regulate storage protein synthesis (Figs. 6 and 7). OsNAC20 and OsNAC26 deficiency caused an almost complete loss of α-globulin and 16 kD prolamin and severely decreased GluB4 (Fig. 5, H and I; Table 1); therefore, OsNAC20 and OsNAC26 are the main regulators for the three genes. The 16 kD prolamin protein is located in PBI, and its loss resulted in a pronounced decrease in PBI number and area (Fig. 4, C–F), indicating that 16 kD prolamin is required for PBI initiation and filling. A similar case in which prolamin is responsible for PB initiation has been reported in the maize 27-kD γ-zein protein, but 27-kD γ-zein is not required for the bulk of PB filling (Lopes and Larkins, 1995; Moro et al., 1995; Guo et al., 2013). GluA1, GluB4 and α-globulin are contained in PBII, and reduction of these proteins led to deficient filling of PBII.
We found that storage protein accumulation was not always mainly limited by its transcript amount. For example, GluA2, GluB1, GluB2, and GluC transcripts were reduced to ∼32%, 22%, 20%, and 16% of cv ZH11 levels, respectively, whereas their protein abundance was comparable to control levels (Fig. 5, H and I; Table 1; Supplemental Fig. S12C). Although GluA1 transcripts were dramatically decreased, the protein was not reduced to quite the same extent (Fig. 5H; Table 1). A similar phenomenon has also been reported in maize by other groups. Guo et al. (2013) observed that in 19- and 22-kD α-zein RNA interference transgenic plants, dramatic reduction of the transcripts of these two proteins does not result in a corresponding reduction of their protein abundance. Li et al. (2018b) found that in the zmbzip22 mutant, an obvious transcript increase in 15-kD β-zein did not cause a noticeable increase in this zein at the protein level. This finding suggests that an excessive transcript level of storage protein, available amino acids, and translational machinery may also be responsible for storage protein abundance (Guo et al., 2013).
The expression of genes encoding major prolamins and globulin was decreased (Table 1), but the expression of Os12g0269100, Os03g0766350, and Os05g0116000, which encode minor prolamins and globulin, was increased (Supplemental Table S3). The supplementary expression of the three genes is promoted to counterbalance the deficit. (Kusaba et al., 2003; Kawakatsu et al., 2010; Lee et al., 2015). A similar protein rebalancing phenomenon has also been reported in other species, including barley (Hordeum vulgare; Hansen et al., 2007), rapeseed (Brassica napus; Rolletschek et al., 2020), wheat (Barro et al., 2016), soybean (Glycine max; Schmidt et al., 2011), and maize (Wu and Messing, 2014). However, the compensatory mechanism has not been identified.
OsNAC20 and OsNAC26 Are Redundantly Required for Starch and Storage Protein Synthesis
Three lines of evidence indicate that OsNAC20 and OsNAC26 have a redundant function in the regulation of starch and storage protein syntheses. First, OsNAC20 and OsNAC26 proteins share high similarity and they activate the same targets. They have almost identical NAC domains, except for three amino acid deletions in the NAC domain of OsNAC20, and their TRR domains share 76% sequence similarity (Supplemental Fig. S1). The NAC domain is responsible for cis-element interaction and the TRR domain contributes to its activation or repression properties (Ooka et al., 2003; Olsen et al., 2005). In this study, the NAC domains of OsNAC20 and OsNAC26 bind to the same cis-elements and activate the same starch and protein synthase-related genes (Figs. 6–8). Second, their promoters share 91% sequence similarity in that they have highly similar cis-elements in their promoters (Supplemental Table S6), meaning that OsNAC20 and OsNAC26 might have similar expression patterns. This is further demonstrated by the RT-qPCR result, which showed that both OsNAC20 and OsNAC26 are only detected at developing endosperm and both have their lowest and highest expression at 4 and 13 DAF, respectively (Fig. 1, A and B). Third, single mutation of OsNAC20 and OsNAC26 did not affect the grain phenotype, but simultaneous mutation of these two genes resulted in a floury grain phenotype (Fig. 2; Supplemental Figs. S7 and S8). The normal phenotype in their respective single mutants is due to the increased expression level of the other OsNAC to compensate for the loss of function (Supplemental Fig. S14).
Several Other NAC TFs Might Take Part in Storage Product Accumulation
In RNA-seq data, we detected 8 other NAC TFs that changed their expression. OsNAC25 (Os11g0512000), OsNAC41 (Os01g0925400), OsNAC128 (Os11g0512200), and OsNAC129 (Os11g0512600) were downregulated in the endosperm of the osnac20/26 mutant. OsNAC25, OsNAC128, and OsNAC129 are endosperm-specific genes, and OsNAC25 has a similar expression pattern to that of OsNAC20 and OsNAC26. OsNAC41 (Os01g0925400) is a candidate for high-yield genes in rice (Huang et al., 2018). These four NAC TFs might play a role in storage product accumulation, but how they take part in this process and how they are regulated by OsNAC20 and OsNAC26 are worth further investigation. Four stress-associated genes, OsNAC2/OMTN2 (Os04g0460600), OsNAC11/OMTN4 (Os06g0675600), OsNAC39/OsCUC1 (Os03g0327100), and OsNAC45 (Os11g0127600), were upregulated in the endosperm of the osnac20/26 mutant (Fang et al., 2008, 2014; Zheng et al., 2009), which might contribute to the function of OsNAC20 and OsNAC26 in the stress response. In addition, their upregulation might be to compensate for the deficiency of OsNAC20 and OsNAC26 in the osnac20/26 mutant.
In conclusion, OsNAC20 and OsNAC26 redundantly and directly transactivate the expression of SSI, Pul, GluA1, GluB4/5, α-globulin, and 16 kD prolamin and indirectly influence DPE1 expression to regulate starch and storage protein synthesis. These findings have also expanded our understanding of the NAC family in the regulation of endosperm starch and storage protein synthesis in plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The CRISPR/Cas9 constructs were generated in a japonica rice (Oryza sativa) ‘Zhonghua11’ (ZH11) background using a CRISPR/Cas9 Kit (Biogle) that contains CRISPR plasmids BGK032 (for the single mutant) and BGK032-DSG (for the double mutant). Briefly, for the single mutant, the synthesized target single guide RNA (sgRNA) sequences were inserted between the OsU6 promoter and the sgRNA scaffold by recombination reaction. For the double mutant, one target was inserted between the OsU6 promoter and the sgRNA and another between OsU3 and the sgRNA. The transgenic T1 to T4 plants were planted in the transgenic closed experiment field of Yangzhou University (Yangzhou, Jiangsu Province, China) under natural field conditions from May to September during the period 2016 to 2019. T3 plants and T4 grains were used for analysis in this study. In addition, the T3 plants used for follow-up seed phenotype observation were planted in Sanya (Hainan Province, China) from December 2017 to April 2018.
Protein Extraction, SDS-PAGE, and Immunoblot
The protein extraction for SDS-PAGE and immunoblot analysis of storage protein was conducted as described by Tian et al. (2013). All other protein samples were extracted as described previously (Wang et al., 2018). SDS-PAGE and immunoblot analyses were conducted according to the methods of Tian et al. (2013) and Wang et al. (2018). The starch synthesis- and storage protein synthesis-related antibodies were described previously (He et al., 2013; Liu et al., 2014).
Subcellular Localization Assays and Bimolecular Fluorescence Complementation
Both OsNAC20 and OsNAC26 were cloned into a 35S-GFP vector, 35S-1305, for subcellular localization analysis in Nicotiana benthamiana leaves. Both OsNAC20 and OsNAC26 were fused to pXY106 (nYFP-C) and pXY104 (N-cYFP) to observe their interactions in N. benthamiana leaves. The detailed transformation into the Agrobacterium tumefaciens strain and the infiltration into N. benthamiana leaves were performed as previously described (Cui et al., 2016). The primers used in this article are listed in Supplemental Table S7.
Immunofluorescence Analysis
The hulled grains at 8 DAF were fixed in phosphate buffer with paraformaldehyde and then embedded into paraffin. The samples were incubated with anti-OsNAC20 or anti-OsNAC26, followed by incubation with fluorescent secondary antibodies. The detailed immunofluorescence analysis was performed following the method of Wang et al. (2018).
Microscopic Observation
Semithin sections of developing and mature grains were investigated following the method of Zhao et al. (2016b). These sections were stained with iodine solution to observe starch granules, and stained with Coomassie brilliant blue and acid fuchsin to observe storage proteins. The stained sections were observed and photographed with a fluorescence microscope (BX51, Olympus). Transmission electron microscopy was conducted as described previously (Tian et al., 2013).
Pull-Down Assay
The CDSs of OsNAC20 and OsNAC26 were inserted into pET-32a and pGEX-6P-1 vectors, respectively. The empty and recombinant plasmids were transformed into Escherichia coli BL21 cells to express the fused protein, followed by protein purification using the corresponding beads. Glutahione S-transferase (GST)- or OsNAC26-GST coupled beads were used to capture His or OsNAC20-His and then their interaction was detected with anti-His antibodies.
Determination of Starch, Storage Protein, Amylose, and Soluble Sugar Contents and Starch Mr Distribution
The starch, storage protein, amylose, and soluble sugar contents were determined as described by Man et al. (2014) and Wang et al. (2018). The Mr distribution of starch was analyzed using gel-permeation chromatography described by Man et al. (2014).
Analyses of Thermal and Crystalline Properties
The thermal and crystalline properties of starch were investigated using differential scanning calorimetry and X-ray powder diffraction as described previously (Man et al., 2014).
Enzyme Activity Assay
Endosperm samples at 10 DAF were derived and homogenized on ice in extraction buffer (50 mm HEPES-NaOH [pH 7.4], 2 mm MgCl2, and 12.5% [v/v] glycerol). The resulting supernatant was used for further enzyme activity analysis. Native PAGE/activity staining assays for DBE and SS were conducted following the method of Nishi et al. (2001). AGPase activity was recorded based on the amount of Glc-1-P produced by coupling its formation to NADH production using phosphoglucomutase and Glc-6-P dehydrogenase (Nishi et al., 2001). The detailed procedure was conducted using the ADPG Pyrophosphorylase Assay Kit (Solarbio). DPE1 assessment was based on a published paper (Akdogan et al., 2011). Total SBE activity was measured using a SBE Assay Kit (Solarbio).
Transcriptome Analysis
Endosperm at 10 DAF was collected and used for RNA isolation with the Qiagen RNeasy mini kit. Paired-end libraries were synthesized through the TruSeq RNA Sample Preparation Kit (Illumina). Purified libraries were deep-sequenced using an Illumina HiSeq 2500. All sequencing raw data were preprocessed by filtering out other nonrelated reads and sequencing adapters and then mapped into the rice reference genome with two mismatches.
Dual-Luc Assay
Coding sequences of OsNAC20 and OsNAC26 were cloned into the 35S-1305 vector. The promoter sequences of starch synthesis- and storage protein synthesis-related genes were inserted into pGreenII-0080 vector. Then, all the recombinant vectors were transformed into Agrobacteria GV3101 or, in the case of the pGreenII-0080 constructs, cotransformed with pSoup-P19. The following procedures were conducted according to the method of Hellens et al. (2005). The activity of Renilla and LUC was determined using the Dual-Luciferase Reporter (DLR, Promega) and detected by BioTek Synergy2 (BioTek).
EMSA
The Web site http://plantpan2.itps.ncku.edu.tw/ was used to analyze the 1- to 3-kb upstream region of selected genes for the analysis of NAC protein binding sites. DNA fragments (30–50 bp) containing NAC protein biding sites were synthesized and labeled with the EMSA Probe Biotin Labeling Kit (Beyotime). The labeled probes were incubated with His-tagged OsNAC20 or OsNAC26 protein and then this mixture was run on an 8% (w/v) native polyacrylamide gel. After transferring to a nylon membrane, cross-linking, and blocking, the signal was detected with the Chemiluminescent EMSA Kit (Beyotime).
Statistical Analysis
The significance of intergroup differences was investigated by one-way ANOVA and Tukey’s honestly significant difference (HSD) mean-separation test. Significance of difference between two groups of data was assessed by Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsNAC20 (LOC_Os01g01470), OsNAC26 (LOC_Os01g29840), AGPS2b (LOC_Os08g25734), AGPL2 (LOC_Os01g44220), SBEI (LOC_Os06g51084), SBEIIa (LOC_Os04g33460), SBEIIb (LOC_Os02g32660), Pho1 (Os04g0164900; LOC_Os03g55090), Pul (LOC_Os04g08270), GBSSI (LOC_Os06g04200), ISA1 (LOC_Os08g40930), SSI (LOC_Os06g06560), SSIIa (LOC_Os06g12450), SSIIIa (LOC_Os08g09230), DPE1 (LOC_Os07g43390), GluA1 (LOC_Os01g55690), GluA2 (LOC_Os10g26060), GluB1 (LOC_Os02g15178), GluB2 (LOC_Os02g15150), GluC (LOC_Os02g25640), 13 kD prolamin (LOC_Os07g10570), and 16 kD prolamin (LOC_Os06g31070).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment analysis of coding and amino acid sequences of OsNAC20 and OsNAC26.
Supplemental Figure S2. Primers for RT-qPCR and antigen sequence for anti-OsNAC20 and anti-OsNAC26.
Supplemental Figure S3. Subcellular localization of OsNAC20-eGFP and OsNAC26-eGFP in N. benthamiana leaves.
Supplemental Figure S4. Interaction assay between OsNAC20 and OsNAC26.
Supplemental Figure S5. Phylogenetic analysis of OsNAC20 and OsNAC26.
Supplemental Figure S6. Sequencing results of possible off-target matches.
Supplemental Figure S7. Grain phenotypic analysis of the osnac20 and osnac26 single mutants.
Supplemental Figure S8. A follow up phenotype observation in the osnac20/26 mutant.
Supplemental Figure S9. Morphology, crystalline, and gelatinization properties of starch in cv ZH11 and osnac20/26.
Supplemental Figure S10. Storage protein phenotype analysis in endosperm of the osnac20/26-1 mutant.
Supplemental Figure S11. Repeatability between three independent biological replicates of RNA-seq.
Supplemental Figure S12. RT-qPCR and immunoblot analysis of starch synthesis- and storage protein synthesis-related genes and protein expressions.
Supplemental Figure S13. The preharvest sprouting and fast germination phenomenon in seeds from the osnac20/26 mutant.
Supplemental Figure S14. Relative expression levels of OsNAC20 and OsNAC26 in their respective single mutants.
Supplemental Table S1. Possible off-target matches.
Supplemental Table S2. Mr distribution of starch isolated from mature seeds.
Supplemental Table S3. Differentially expressed genes between cv ZH11 and the osnac20/26-1 mutant.
Supplemental Table S4. Predicted NAC binding sites in the promoters of target genes.
Supplemental Table S5. DNA sequences for the EMSA in this article.
Supplemental Table S6. Predicted cis-elements in the OsNAC20 and OsNAC26 promoters.
Supplemental Table S7. Primers used in this article.
Acknowledgments
We thank Xiuling Cai (Yangzhou University) and Ying He (Huazhong Agricultural University) for providing the antibodies for starch biosynthesis-related enzymes and storage protein synthesis-related protein, respectively. We thank Jianxiang Liu (Zhejiang University) for providing the dual-luc-related plasmids. We thank Dongping Zhang, Chen Chen, Baixiao Niu, Jian Wu, and Yijun Wang (Yangzhou University) for providing the advice about this research.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31800269), the Talent Project of Yangzhou University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Articles can be viewed without a subscription.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M(1997) Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdogan G, Kubota J, Kubo A, Takaha T, Kitamura S(2011) Expression and characterization of rice disproportionating enzymes. J Appl Glycosci 58: 99–105 [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L(2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B(2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4: 346–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barro F, Iehisa JCM, Giménez MJ, García-Molina MD, Ozuna CV, Comino I, Sousa C, Gil-Humanes J(2016) Targeting of prolamins by RNAi in bread wheat: Effectiveness of seven silencing-fragment combinations for obtaining lines devoid of coeliac disease epitopes from highly immunogenic gliadins. Plant Biotechnol J 14: 986–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T(2001) A cytosolic ADP-glucose pyrophosphorylase is a feature of graminaceous endosperms, but not of other starch-storing organs. Plant Physiol 125: 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BK, Hou Y, Zhao J, Jiao G, Wu Y, Li Z, Wang Y, Tong X, Wang W, Yuan W, et al. (2019) NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol J 17: 1222–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XL, Xie DL, Wang ZY, Hong MM(2002) Interaction of rice bZIP protein REB with the 5′-upstream region of both rice sbe1 gene and waxy gene. Chin Sci Bull 47: 310–314 [Google Scholar]
- Chen J, Yi Q, Cao Y, Wei B, Zheng L, Xiao Q, Xie Y, Gu Y, Li Y, Huang H, et al. (2016) ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J Exp Bot 67: 1327–1338 [DOI] [PubMed] [Google Scholar]
- Cord Neto G, Yunes JA, da Silva MJ, Vettore AL, Arruda P, Leite A(1995) The involvement of Opaque 2 on β-prolamin gene regulation in maize and Coix suggests a more general role for this transcriptional activator. Plant Mol Biol 27: 1015–1029 [DOI] [PubMed] [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F(2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JP, Supatcharee N, Khandelwal RL, Chibbar RN(2003) Synthesis of novel starches in planta: Opportunities and challenges. Starch 55: 107–120 [Google Scholar]
- Denyer KAY, Johnson P, Zeeman S, Smith AM(2001) The control of amylose synthesis. J Plant Physiol 158: 479–487 [Google Scholar]
- Dong J, Zheng Y, Fu Y, Wang J, Yuan S, Wang Y, Zhu Q, Ou X, Li G, Kang G(2019b) PDIL1-2 can indirectly and negatively regulate expression of the AGPL1 gene in bread wheat. Biol Res 52: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Wang F, Kong J, Xu Q, Li T, Chen L, Chen H, Jiang H, Li C, Cheng B(2019a) Functional analysis of ZmMADS1a reveals its role in regulating starch biosynthesis in maize endosperm. Sci Rep 9: 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhang D, Liu J, Liu QQ, Liu H, Tian L, Jiang L, Qu Q(2015) Plastidial disproportionating enzyme participates in starch synthesis in rice endosperm by transferring maltooligosyl groups from amylose and amylopectin to amylopectin. Plant Physiol 169: 2496–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L(2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65: 2119–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L(2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280: 547–563 [DOI] [PubMed] [Google Scholar]
- Feng F, Qi W, Lv Y, Yan S, Xu L, Yang W, Yuan Y, Chen Y, Zhao H, Song R(2018) OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30: 375–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero DML, Piattoni CV, Asencion Diez MD, Rojas BE, Hartman MD, Ballicora MA, Iglesias AA(2020) Phosphorylation of ADP-glucose pyrophosphorylase during wheat seeds development. Front Plant Sci 11: 1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F-F, Xue H-W(2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Toyosawa Y, Utsumi Y, Higuchi T, Hanashiro I, Ikegami A, Akuzawa S, Yoshida M, Mori A, Inomata K, et al. (2009) Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J Exp Bot 60: 1009–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y(2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux MJ, Hannah LC(1994) ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Mol Gen Genet 243: 400–408 [DOI] [PubMed] [Google Scholar]
- Guo X, Yuan L, Chen H, Sato SJ, Clemente TE, Holding DR(2013) Nonredundant function of zeins and their correct stoichiometric ratio drive protein body formation in maize endosperm. Plant Physiol 162: 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Lange M, Friis C, Dionisio G, Holm PB, Vincze E(2007) Antisense-mediated suppression of C-hordein biosynthesis in the barley grain results in correlated changes in the transcriptome, protein profile, and amino acid composition. J Exp Bot 58: 3987–3995 [DOI] [PubMed] [Google Scholar]
- He Y, Wang S, Ding Y(2013) Identification of novel glutelin subunits and a comparison of glutelin composition between japonica and indica rice (Oryza sativa L.). J Cereal Sci 57: 362–371 [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Xie S, Xiao Q, Wei B, Zheng L, Wang Y, Cao Y, Zhang X, Long T, Li Y, et al. (2016) Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci Rep 6: 27590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li J, Zhou J, Wang L, Yang S, Hurst LD, Li WH, Tian D(2018) Identifying a large number of high-yield genes in rice by pedigree analysis, whole-genome sequencing, and CRISPR-Cas9 gene knockout. Proc Natl Acad Sci USA 115: E7559–E7567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SK, Koper K, Satoh H, Okita TW(2016) Rice endosperm starch phosphorylase (Pho1) assembles with disproportionating enzyme (Dpe1) to form a protein complex that enhances synthesis of malto-oligosaccharides. J Biol Chem 291: 19994–20007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Hirose S, Yasuda H, Takaiwa F(2010) Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol 154: 1842–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F(2009) Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J 59: 908–920 [DOI] [PubMed] [Google Scholar]
- Kim JS, Chae S, Jun KM, Pahk YM, Lee TH, Chung PJ, Kim YK, Nahm BH(2017) Genome-wide identification of grain filling genes regulated by the OsSMF1 transcription factor in rice. Rice (N Y) 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T(2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T(2003) Low glutelin content1: A dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15: 1455–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Jo YM, Lee JY, Lim SH, Kim YM(2015) Lack of globulin synthesis during seed development alters accumulation of seed storage proteins in rice. Int J Mol Sci 16: 14717–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, et al. (2015) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of Opaque2 in maize. Plant Cell 27: 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yue Y, Chen H, Qi W, Song R(2018a) The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 30: 2402–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chang Y, Ma S, Shen J, Hu H, Xiong L(2019) Genome-wideidentification of SNAC1-targeted genes involved in drought response in rice. Front Plant Sci 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yu G, Lv Y, Long T, Li P, Hu Y, Liu H, Zhang J, Liu Y, Li WC, et al. (2018b) Combinatorial interaction of two adjacent cis-active promoter regions mediates the synergistic induction of Bt2 gene by sucrose and ABA in maize endosperm. Plant Sci 274: 332–340 [DOI] [PubMed] [Google Scholar]
- Liu D, Wang W, Cai X(2014) Modulation of amylose content by structure-based modification of OsGBSS1 activity in rice (Oryza sativa L.). Plant Biotechnol J 12: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA(1995) Genetic analysis of opaque2 modifier gene activity in maize endosperm. Theor Appl Genet 91: 274–281 [DOI] [PubMed] [Google Scholar]
- Man J, Lin L, Wang Z, Wang Y, Liu Q, Wei C(2014) Different structures of heterogeneous starch granules from high-amylose rice. J Agric Food Chem 62: 11254–11263 [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F(2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew IE, Das S, Mahto A, Agarwal P(2016) Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Sci 7: 1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro GL, Lopes MA, Habben JE, Hamaker BR, Larkins BA(1995) Phenotypic effects of opaque2 modifier genes in normal maize endosperm. Cereal Chem 72: 94–99 [Google Scholar]
- Nakamura Y.(2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol 43: 718–725 [DOI] [PubMed] [Google Scholar]
- Nie DM, Ouyang YD, Wang X, Zhou W, Hu CG, Yao J(2013) Genome-wide analysis of endosperm-specific genes in rice. Gene 530: 236–247 [DOI] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H(2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127: 459–472 [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K(2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Peng X, Wang Q, Wang Y, Cheng B, Zhao Y, Zhu S(2019) A maize NAC transcription factor, ZmNAC34, negatively regulates starch synthesis in rice. Plant Cell Rep 38: 1473–1484 [DOI] [PubMed] [Google Scholar]
- Qiao Z, Qi W, Wang Q, Feng Y, Yang Q, Zhang N, Wang S, Tang Y, Song R(2016) ZmMADS47 regulates zein genetranscription through interaction with Opaque2. PLoS Genet 12: e1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Hou Y, Tong X, Wang Y, Lin H, Liu Q, Zhang W, Li Z, Nallamilli BR, Zhang J(2016) Quantitative phosphoproteomic analysis of early seed development in rice (Oryza sativa L.). Plant Mol Biol 90: 249–265 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Schwender J, König C, Chapman KD, Romsdahl T, Lorenz C, Braun HP, Denolf P, Van Audenhove K, Munz E, et al. (2020) Cellular plasticity in response to suppression of storage proteins in the Brassica napus embryo. Plant Cell 32: 2383–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S, et al. (2012) Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant J 70: 1043–1055 [DOI] [PubMed] [Google Scholar]
- Sakuraba Y, Kim D, Han SH, Kim SH, Piao W, Yanagisawa S, An G, Paek NC(2020) Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 32: 630–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y(2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang S-K, Okita TW, Kaneko N, Fujita N, Yoshida M, et al. (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MA, Barbazuk WB, Sandford M, May G, Song Z, Zhou W, Nikolau BJ, Herman EM(2011) Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol 156: 330–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Luo G, Shen L, Yu K, Yang W, Li X, Sun J, Zhan K, Cui D, Liu D, et al. (2020) TubZIP28, a novel bZIP family transcription factor from Triticum urartu, and TabZIP28, its homologue from Triticum aestivum, enhance starch synthesis in wheat. New Phytol 226: 1384–1398 [DOI] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R(1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170 [DOI] [PubMed] [Google Scholar]
- Sun D, Zhang X, Zhang Q, Ji X, Jia Y, Wang H, Niu L, Zhang Y(2019) Comparative transcriptome profiling uncovers a Lilium regale NAC transcription factor, LrNAC35, contributing to defence response against cucumber mosaic virus and tobacco mosaic virus. Mol Plant Pathol 20: 1662–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y(2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol J 2: 507–516 [DOI] [PubMed] [Google Scholar]
- Tetlow IJ.(2006) Understanding storage starch biosynthesis in plants: A means to quality improvement. Can J Bot 84: 1167–1185 [Google Scholar]
- Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, Esposito S, Kosar-Hashemi B, Morell MK, Emes MJ(2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16: 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dai LL, Yin ZJ, Fukuda M, Kumamaru T, Dong XB, Xu XP, Qu Q(2013) Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J Exp Bot 64: 2831–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuncel A, Kawaguchi J, Ihara Y, Matsusaka H, Nishi A, Nakamura T, Kuhara S, Hirakawa H, Nakamura Y, Cakir B, et al. (2014) The rice endosperm ADP-glucose pyrophosphorylase large subunit is essential for optimal catalysis and allosteric regulation of the heterotetrameric enzyme. Plant Cell Physiol 55: 1169–1183 [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ(1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu P, Lin L, Chen Z, Liu Q, Wei C(2018) Gradually decreasing starch branching enzyme expression is responsible for the formation of heterogeneous starch granules. Plant Physiol 176: 582–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL(2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Culver JN(2012) DNA binding specificity of ATAF2, a NAC domain transcription factor targeted for degradation by Tobacco mosaic virus. BMC Plant Biol 12: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F(1999) Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol Biol 40: 1–12 [DOI] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor M-I, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, et al. (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Washida H, Onodera Y, Harada K, Takaiwa F(2000) Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: Minimal cis-element requirements for endosperm-specific gene expression. Plant J 23: 415–421 [DOI] [PubMed] [Google Scholar]
- Wu Y, Messing J(2014) Proteome balancing of the maize seed for higher nutritional value. Front Plant Sci 5: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Messing J(2012) Rapid divergence of prolamin gene promoters of maize after gene amplification and dispersal. Genetics 192: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z(1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T(2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55: 652–664 [DOI] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F(2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Isono Y, Satoh H, Omura T(1984) Gene analysis of sugary and shrunken mutants of rice, Oryza sativa L. Jpn J Breeding 34: 43–49 [Google Scholar]
- Zhang D, Zhang M, Zhou Y, Wang Y, Shen J, Chen H, Zhang L, Lü B, Liang G, Liang J(2019a) The rice G protein γ subunit DEP1/qPE9–1 positively regulates grain-filling process by increasing auxin and cytokinin content in rice grains. Rice (N Y) 12: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen J, Yi Q, Hu Y, Liu H, Liu Y, Huang Y(2014) Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol Biol 84: 359–369 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J(2019b) NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci USA 116: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang J, Wu Y(2015) Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of Opaque2, Prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng X, Yang J, Messing J, Wu Y(2016) Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc Natl Acad Sci USA 113: 10842–10847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Ma J, Li L, Fan S, Guo Y, Song M, Wei H, Pang C, Yu S(2016a) GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L). Gene 576: 268–274 [DOI] [PubMed] [Google Scholar]
- Zhao L, Pan T, Cai C, Wang J, Wei C(2016b) Application of whole sections of mature cereal seeds to visualize the morphology of endosperm cell and starch and the distribution of storage protein. J Cereal Sci 71: 19–27 [Google Scholar]
- Zheng X, Chen B, Lu G, Han B(2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379: 985–989 [DOI] [PubMed] [Google Scholar]