The transcription factor EIL1 has an additive role to SLIM1 in the regulation of sulfur-deficiency response.
Abstract
Sulfur, an indispensable constituent of many cellular components, is a growth-limiting macronutrient for plants. Thus, to successfully adapt to changing sulfur availability and environmental stress, a sulfur-deficiency response helps plants to cope with the limited supply. On the transcriptional level, this response is controlled by SULFUR LIMITATION1 (SLIM1), a member of the ETHYLENE-INSENSITIVE3-LIKE (EIL) transcription factor family. In this study, we identified EIL1 as a second transcriptional activator regulating the sulfur-deficiency response, subordinate to SLIM1/EIL3. Our comprehensive RNA sequencing analysis in Arabidopsis (Arabidopsis thaliana) allowed us to obtain a complete picture of the sulfur-deficiency response and quantify the contributions of these two transcription factors. We confirmed the key role of SLIM1/EIL3 in controlling the response, particularly in the roots, but showed that in leaves more than 50% of the response is independent of SLIM1/EIL3 and EIL1. RNA sequencing showed an additive contribution of EIL1 to the regulation of the sulfur-deficiency response but also identified genes specifically regulated through EIL1. SLIM1/EIL3 seems to have further functions (e.g. in the regulation of genes responsive to hypoxia or mediating defense at both low and normal sulfur supply). These results contribute to the dissection of mechanisms of the sulfur-deficiency response and provide additional possibilities to improve adaptation to sulfur-deficiency conditions.
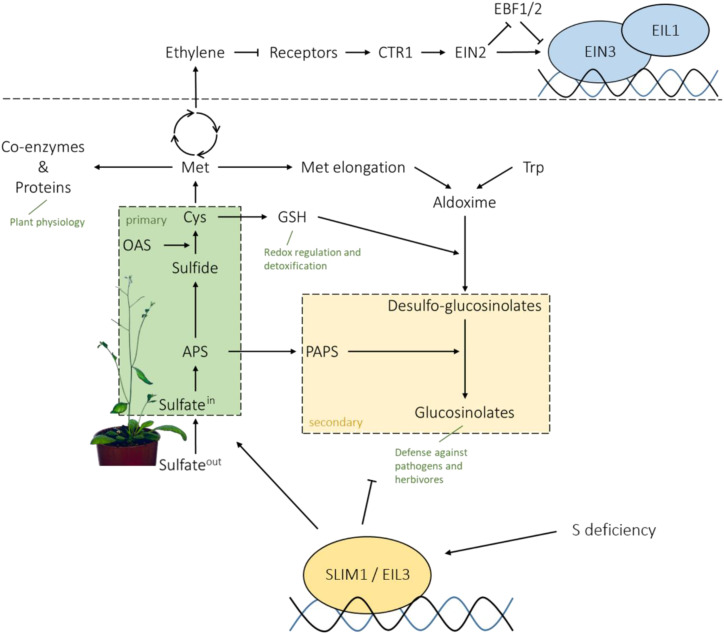
The assimilation of the macronutrient sulfur (Fig. 1) is a growth-limiting process, as sulfur is an indispensable active component of many essential metabolites. As an organic thiol, it is incorporated in the amino acids Cys and Met, which define protein translation start, activity, and structure. It is also present in several prosthetic groups and cofactors, such as CoA, iron-sulfur proteins, thiamine pyrophosphate, and biotin (Koprivova and Kopriva, 2014). In plants, sulfur for these metabolites is provided by the assimilation of sulfate, the oxidized, inorganic form of sulfur. The uptake of sulfate through the plasma membrane is achieved by transmembrane transport proteins against a negative membrane potential (Takahashi et al., 2012). Arabidopsis (Arabidopsis thaliana) possesses two high-affinity sulfate transporters in the root cortex, SULTR1;1 and SULTR1;2, which enable the plant to take up sulfate into the roots. Via xylem-based root-shoot translocation, sulfate moves to leaf cells, where it can be either transported to the plastids for assimilation or stored in the vacuole (Yoshimoto et al., 2002; Kataoka et al., 2004b; Cao et al., 2013). In plastids, the inert sulfate is activated to adenosine 5-phosphosulfate (APS) by ATP sulfurylase (ATPS; Takahashi et al., 2011). APS is a branching point of sulfur metabolism, as it can be further reduced for primary (reductive) assimilation or phosphorylated to 3′-phosphoadenosine 5′-phosphosulfate (PAPS), a sulfate donor for a number of secondary (oxidized) sulfur metabolites (Kopriva et al., 2012). In the primary sulfate assimilation pathway, APS reductase (APR) catalyzes the reduction of APS to sulfite, which subsequently is reduced by the ferredoxin-dependent sulfite reductase to sulfide. In addition to the SULTR uptake system, another crucial control point of the sulfate assimilation pathway is the first reduction step driven by three isoforms of APR (Vauclare et al., 2002; Koprivova and Kopriva, 2014). Sulfide is subsequently incorporated into the amino acid skeleton of O-acetyl-serine (OAS) by O-acetyl-serine (thiol) lyase (OAS-TL) to form Cys (Wirtz et al., 2004). Cys can be incorporated into proteins and peptides, such as glutathione (GSH), or provide sulfur for the synthesis of Met and coenzymes (Takahashi et al., 2011). In the secondary assimilation pathway, PAPS is needed, for example, for the sulfation of sulfated peptide hormones and glucosinolates (Sønderby et al., 2010; Koprivova and Kopriva, 2016b). This group of amino acid-derived secondary metabolites has a crucial function in plant defense against pathogens and herbivores, as they are toxic and deterrent upon breakdown (Halkier and Gershenzon, 2006; Bednarek et al., 2009). Sulfur metabolism, with a broad range of enzyme isoforms in different cellular compartments, is a highly complex process, regulated by sulfur availability and a diversity of environment-dependent signals (Takahashi et al., 2011).
Figure 1.
SLIM1/EIL3 in the regulation of sulfur-deficiency response and a link to EIN3/EIL1 function in ethylene signal transduction.
One of the environmental conditions strongly affecting sulfate assimilation is sulfur deficiency. During sulfur deficiency, the vast majority of enzymes mediating sulfate uptake and assimilation are coordinately regulated (Takahashi et al., 1997). Thereby, the acquisition and primary assimilation of sulfate is enhanced, whereas glucosinolate biosynthesis is repressed (Hirai et al., 2003, 2005; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003, 2005). This is predominantly achieved via transcriptional regulation (Maruyama-Nakashita et al., 2006). The transcription factor responsible for at least part of this sulfur-deficiency response is SULFUR LIMITATION1 (SLIM1), as it controls the induction of sulfate uptake and acquisition as well as the catabolic recycling of secondary sulfur compounds under low-sulfate conditions (Fig. 1; Maruyama-Nakashita et al., 2006).
SLIM1 is part of the six-member ETHYLENE-INSENSITIVE3-LIKE (EIL) family of transcription factors in Arabidopsis and, therefore, also referred to as EIL3 (Guo and Ecker, 2004; Maruyama-Nakashita et al., 2006). Other members of the EIL family have been associated with the control of ethylene signaling transduction. The eponym EIN3 and its functional homologs EIL1 and EIL2 regulate transcript levels of ethylene-responsive genes upon ethylene signaling transmitted through EIN2, resulting in a further transcriptional cascade through transcription factors such as ERF1 (Chao et al., 1997; Solano et al., 1998; Merchante et al., 2013). SLIM1/EIL3 was assumed to have a distinct function, as it did not complement ein3 phenotypes, in contrast to EIL1 and EIL2 (Chao et al., 1997), whereas the other EILs did not complement the loss of function of SLIM1/EIL3 (Maruyama-Nakashita et al., 2006). However, several recent studies indicated the participation of other EIL proteins in regulation of the sulfur-deficiency response. For example, heterodimerization of EIN3 and SLIM1, via the highly conserved N terminus of both transcription factors (EIN3, amino acids 1–299; SLIM1, amino acids 75–286), led to impaired DNA binding of SLIM1, suggesting that EIN3 acts as an inhibitor of SLIM1 (Wawrzyńska and Sirko, 2016). Also, EIL2 was shown to activate the expression of the sulfur deficiency-upregulated gene LSU in tobacco (Nicotiana tabacum) in a sulfur status-dependent manner (Wawrzyńska et al., 2010).
Despite the importance of SLIM1 for control of the sulfur-deficiency response, the mechanism of this regulation remains poorly understood. SLIM1 transcript is expressed preferentially in the vasculature of leaves, hypocotyl, and roots, particularly in parenchyma and xylem cells, but is not affected by the sulfur status (Maruyama-Nakashita et al., 2006; Aubry et al., 2014). Similarly, SLIM1 protein abundance and localization in nuclei are not altered upon sulfur deficiency (Maruyama-Nakashita et al., 2006). This prompted our attempt to understand the molecular mechanisms of the sulfur-deficiency response and the wider role of the EIL family in this regulation. We reveal here that EIL1 acts as an additional transcription factor regulating the sulfur-deficiency response. We compared the transcriptional responses of eil3, eil1, and a double eil1 eil3 mutant to sulfur deficiency using RNA sequencing (RNA-seq) and show that EIL1 has both overlapping and specific functions compared with SLIM1.
RESULTS
Loss of EIL1 Affects the Accumulation of Sulfur-Containing Metabolites during Sulfur Deficiency
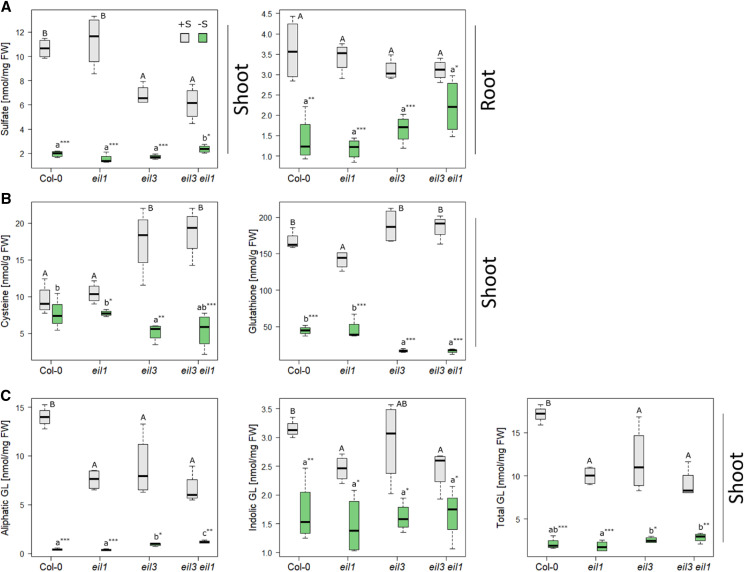
Previous studies showed that overexpression of other EIL proteins was not sufficient to rescue the fluorescence signal of pSULTR1;2PRO-GFP reporter in transgenic slim1-1 or slim1-2 seedlings (Maruyama-Nakashita et al., 2006). This, however, does not mean that they do not play potential roles in the sulfur-deficiency response, such as regulation of genes affected by sulfur deficiency independent of SLIM1. Alternatively, the EILs might play a subordinate role in the regulation of sulfur-deficiency response, and their function might be masked by the dominant activity of SLIM1. Therefore, as a first step to assess the importance of the EIL family for sulfur-deficiency response, we generated the double mutant eil3 eil1 and tested it and the single mutants for their response to sulfur-deficiency conditions. The eil3 T-DNA line was compared with the original slim1-1 and slim1-2 mutants and showed very similar defects in transcriptional response to sulfur deficiency (Supplemental Fig. S1). Seedlings of the Columbia-0 (Col-0) wild type, eil1, eil3, and eil3 eil1 were grown on modified Long Ashton Medium plates with either full sulfur (0.75 mm SO42− [+S]) or low sulfur (0.015 mm SO42− [−S]) supply for 18 d, and levels of anions, thiols, and glucosinolates were analyzed. In control plants, sulfate levels in shoots were reduced by approximately 50% in eil3 and eil3 eil1 mutants but remained the same in the roots (Fig. 2A). Sulfate levels strongly diminished in response to sulfate deficiency, except in shoots of eil3 eil1, where the reduction in sulfate accumulation was attenuated compared with the wild type or the single mutants. Foliar Cys levels in eil3 and eil3 eil1 were higher than in the wild type and eil1 in control conditions but were lower in low sulfur (Fig. 2B). Thus, no or only small differences in Cys concentration between control and –S conditions are found in the wild type and eil1, whereas the genotypes with disrupted SLIM1 exhibit a high difference in Cys. GSH in the leaves of sulfur-deficient plants shows a similar pattern as Cys: lower levels in eil3 and eil3 eil1 than in the wild type (Fig. 2B). In control conditions, however, it is the eil1 mutant that accumulates less GSH than the other genotypes. Again, the decrease in GSH is more pronounced in sulfur-deficient eil3 and eil3 eil1 than in the wild type and eil1. An attenuated –S response was also observed for shoot levels of aliphatic glucosinolates (Fig. 2C). Under normal sulfur supply, all three mutants had reduced glucosinolate levels compared with the wild type. Whereas in the wild type and eil1 the aliphatic glucosinolates were decreased to low levels by sulfur deficiency, this decrease was not as pronounced in eil3 and was even less in eil3 eil1. Indolic glucosinolates did not show substantial differences between the genotypes, except a slight reduction in control conditions in mutants with disrupted EIL1: eil1 and eil3 eil1 (Fig. 2C). These results suggest that EIL1 participates in the regulation of sulfur metabolism by sulfur deficiency.
Figure 2.
Metabolite analysis of eil1, eil3, and eil3 eil1. The mutants and wild-type Col-0 plants were grown for 18 d on either 0.75 mm SO42− (gray) or 0.015 mm SO42− (green). Sulfate content was determined via ion chromatography (A), whereas thiols (B) and glucosinolate (C) contents were measured by HPLC. Data from one representative example of at least two independent experiments are presented as box plots of four biological replicates, with lines marking medians. FW, Fresh weight. Different letters represent values significantly different using Student’s t test (P < 0.05). Asterisks indicate significant differences between values of one genotype at control or low sulfur using Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
EIL1 Is Required for the Maintenance of Sulfur Uptake and Allocation during Deficiency Conditions
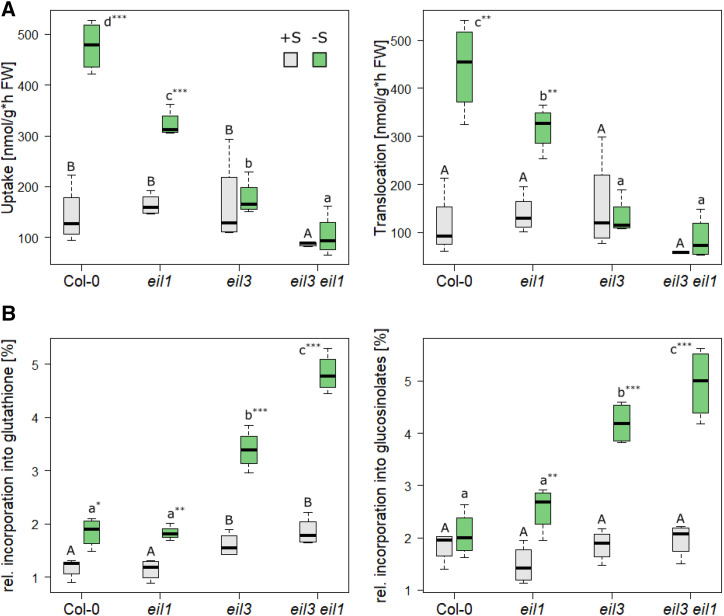
To evaluate whether the loss of EIL1 also affects the flux through sulfate assimilation, the different genotypes were grown under +S and −S supply for 18 d and subsequently fed with [35S]sulfate for 3 h. The incorporation of 35S into thiols, glucosinolates, and proteins, as well as the overall uptake and root-shoot translocation, were determined. Arabidopsis normally adapts to sulfur-deficiency conditions by increasing the uptake of sulfate into the root via the high-affinity sulfate transporters. This response was clearly visible in wild-type seedlings and was accompanied by enhanced sulfate translocation from root to shoot, the major site of assimilation. In eil1 seedlings, the increases in uptake and translocation were dramatically reduced, whereas in eil3, the sulfur-deficiency response was totally abolished (Fig. 3A). Interestingly, the double mutant seedlings were affected even stronger, since their sulfate uptake and translocation were not only the lowest under sulfur-deficiency conditions but were impaired already at normal sulfur supply.
Figure 3.
Flux through sulfate assimilation in eil1, eil3, and eil3 eil1. Plants were grown for 18 d on either 0.75 mm SO42− (gray) or 0.015 mm SO42− (green) and subsequently fed with [35S]sulfate. A, Uptake and root-shoot translocation were determined via scintillation counter. B, 35S incorporation to GSH and glucosinolates was measured via HPLC with a radiodetector or after elution from DEAE-Sephadex by scintillation counting, respectively. Data from one representative example of at least two independent experiments are presented as box plots of four biological replicates. FW, Fresh weight. Different letters represent values significantly different using Student’s t test (P < 0.05). Asterisks indicate significant differences between values of one genotype at control or low sulfur using Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
A similar pattern was observable for the incorporation of 35S into GSH (Fig. 3B), corresponding to previously observed reduced GSH levels (Fig. 2). Although sulfur deficiency leads to a decrease in glucosinolate accumulation, the flux into glucosinolates was unchanged in the wild type and was slightly increased in eil1 (Fig. 3B). This increase was more prominent in eil3 and eil3 eil1 (Fig. 3B). Indeed, in –S conditions, eil3 and eil3 eil1 seedlings incorporated twice as much 35S into glucosinolates than eil1 and the wild type, presumably because of the higher specific activity of GSH as the donor of sulfur for core glucosinolate synthesis and the diminished upregulation of SDI1 (see below). The incorporation of 35S into proteins did not show any alteration between the mutants and the wild type under both conditions. The reducing magnitude of the sulfur metabolism response to sulfur deficiency from the wild type, over eil1, to eil3 and to eil3 eil1 clearly indicates that EIL1 plays a role in the control of the sulfur-deficiency response, as shown for SLIM1 previously. Furthermore, it indicates that both transcription factors have an additive function for the regulation of acquisition, allocation, and assimilation of sulfate.
EIL1 Contributes to the Regulation of Marker Genes for Sulfur Deficiency
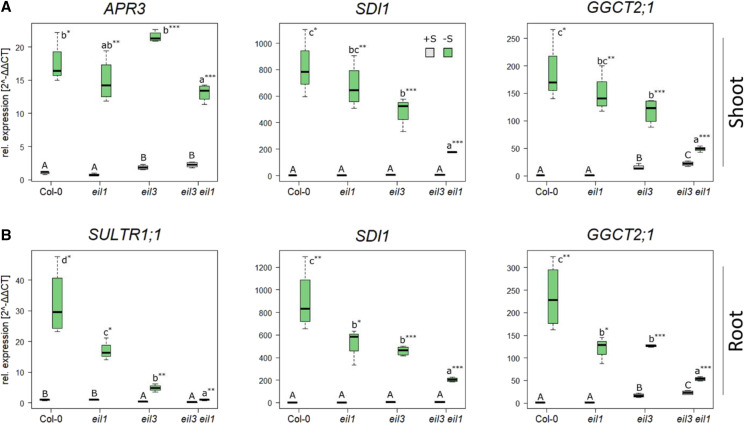
Sulfur deficiency leads to profound changes in gene expression in Arabidopsis. A great number of genes, such as SULFUR DEFICIENCY-INDUCED1 (SDI1) and GAMMA-GLUTAMYL CYCLOTRANSFERASE2;1 (GGCT2;1), are upregulated in a SLIM1-dependent manner, whereas others, like the APR isoforms, were shown to be SLIM1 independent (Maruyama-Nakashita et al., 2006). We, therefore, tested whether EIL1 contributes to the regulation of these marker genes by sulfur deficiency. Transcript levels of SDI1 and GGCT2;1, involved in the regulation of aliphatic glucosinolate biosynthesis and GSH catabolism, respectively, were highly upregulated in wild-type seedlings upon –S conditions both in shoots and roots (Fig. 4). As expected, this response was significantly decreased in eil3 mutants by approximately 50% in both organs. The same was true for SULTR1;1 in the roots (Fig. 4B), but APR3 in the leaves was upregulated to the same extent in eil3 and the wild type (Fig. 4A). Loss of EIL1 did not affect the sulfur-deficiency response in the leaves; however, in the roots, the upregulation of SDI1 and GGCT2;1 was attenuated to the same degree as in eil3, and for SULTR1;1 it was intermediate between the wild type and eil3 (Fig. 4). Loss of both SLIM1 and EIL1 simultaneously led to an additive effect on gene expression; in leaves, SDI1 and GGCT2;1 transcript levels were significantly less affected by sulfur deficiency in eil3 eil1 than in the wild type and the single mutants. The same was true for the roots, where particularly the upregulation of SULTR1;1 was very low. These findings clearly show that EIL1 is involved in the transcriptional regulation of the sulfur-deficiency response, where it seems to regulate gene transcription additively to SLIM1.
Figure 4.
Expression analysis of sulfur-deficiency marker genes in eil1, eil3, and eil3 eil1. Plants were grown for 18 d on either 0.75 mm SO42− (gray) or 0.015 mm SO42− (green). Relative gene expression (2−ΔΔCt) of –S marker genes APR3, SDI1, and GGCT2;1 in the leaves (A) and SULTR1;1, SDI1, and GGCT2;1 in the roots (B) was determined via reverse transcription quantitative PCR (RT-qPCR). A representative example of at least two independent experiments is presented. Box plots of four biological replicates are given, and lines represent medians. The values in Col-0 at control conditions were set to 1. Different letters represent values significantly different using Student’s t test (P < 0.05). Asterisks indicate significant differences between values of one genotype at control or low sulfur using Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
Global Transcriptional Network of EIL1 and EIL3 in Sulfur-Deficiency Response
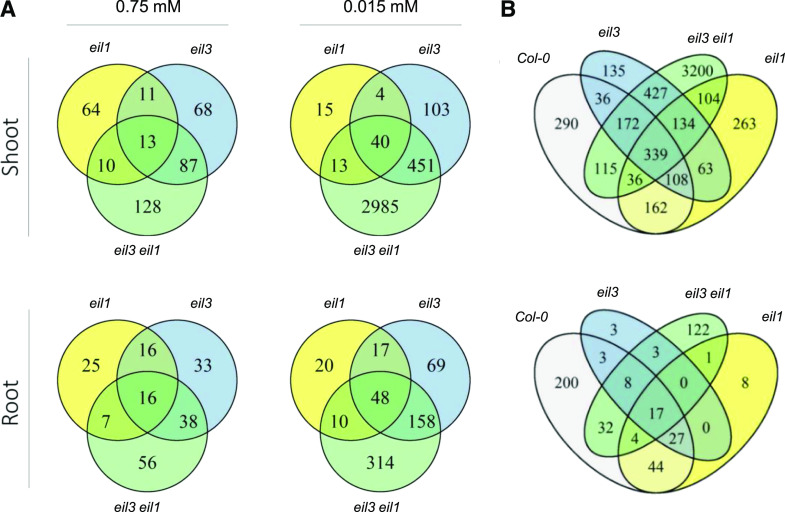
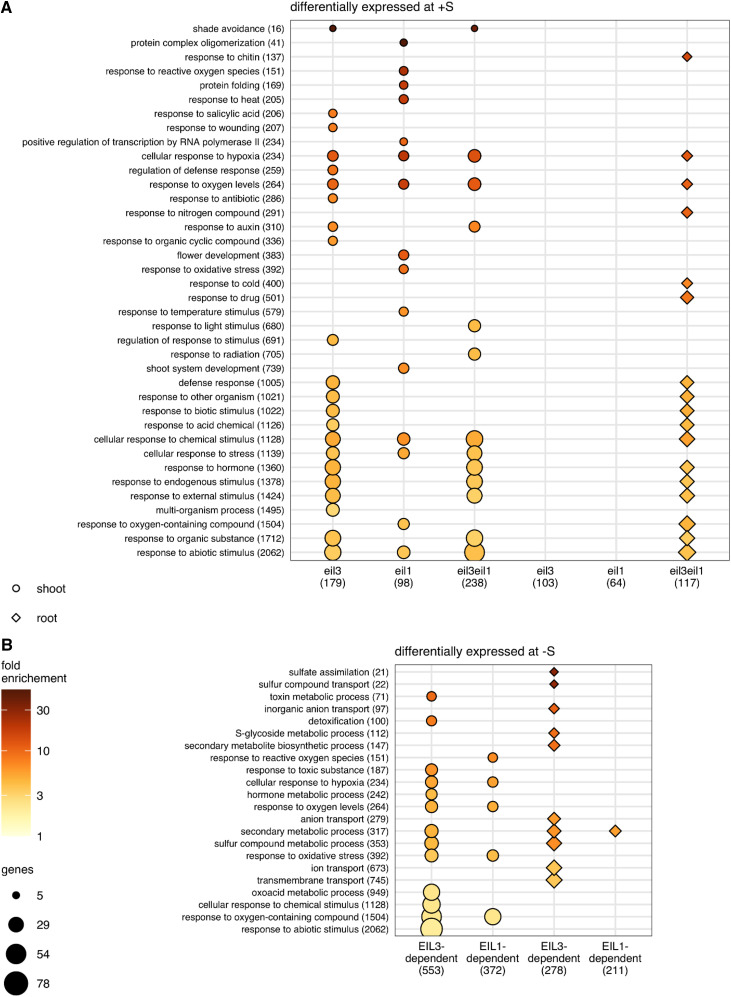
To obtain insights into the global transcription networks regulated by SLIM1 and EIL1, as well as their functional overlaps, we utilized an RNA-seq approach. RNA was isolated from shoots and roots of the four genotypes grown at normal and –S conditions. The RNA was subjected to RNA-seq using the Illumina platform at the Cologne Centre for Genomics (Supplemental Data Set S1). In order to shorten the process of transcript abundance quantification without losing accuracy, we performed kallisto pseudoalignment with subsequent sleuth analysis for the determination of differentially expressed genes (DEGs), with false discovery rate-adjusted P < 0.05 and fold change > 2. Already at control sulfur supply, 179 and 98 genes were differentially expressed in leaves of eil3 and eil1, respectively, compared with the wild type (Fig. 5A; Supplemental Table S1). In the double mutant, the number of DEGs was raised to 238. From these, 13 genes were differentially expressed in both parental mutants and the double mutant, but 68 and 64 DEGs were specific to eil3 and eil1, respectively. A total of 128 genes were differentially expressed only in the double mutant. Interestingly, several but not all of the typical sulfur-deficiency markers (LSU1, LSU2, LSU3, and SDI1) were slightly but significantly induced in the eil3 mutant, whereas in eil3 eil1 they were induced to a somewhat higher level, and additionally APR2 and APR3 were found among the induced DEGs. To identify processes differentially regulated in the mutants, Gene Ontology (GO) overrepresentation analysis was performed. Under normal sulfur supply, loss of SLIM1 affected genes annotated as being involved in response to hypoxia and oxygen levels, to other organisms, wounding, and salicylic acid, and, interestingly, to shade avoidance (Fig. 6A; Supplemental Table S2). Among the DEGs in eil1, genes for response to hypoxia and oxygen levels were also overrepresented, in addition to a number of genes involved in response to heat and reactive oxygen species (ROS) and in shoot and flower development, in line with the role of EIL1 in ethylene signaling. In roots, at normal sulfur supply, 103 and 64 DEGs were found in eil3 and eil1 and 117 DEGs were found in the double mutant, with 16 genes differentially expressed in all three mutants. Most of these genes were affected in an organ-specific way; only two and 12 genes were differentially expressed in both roots and shoots of eil1 and eil3, respectively, which increased to 16 genes (13.6% of root DEGs) in the eil3 eil1 double mutant. In the roots of single mutants eil1 and eil3, no functional category was overrepresented at a stringent P < 0.005. However, in eil3 eil1 roots, the genes for response to hypoxia and oxygen levels were overrepresented again, in addition to new categories, such as response to chitin, to nitrogen compounds, and to cold (Fig. 6A). Generally, a high number of the overrepresented GO terms in all three genotypes were related to stress response (Fig. 6A; Supplemental Table S2).
Figure 5.
Venn diagrams of DEGs. Plants were grown for 18 d on either 0.75 mm SO42− or 0.015 mm SO42−, and the RNAs from shoots and roots were subjected to transcriptome analysis by RNA-seq. DEGs (q > 0.05; –1 > log2 fold change [log2FC] > 1) were determined via kallisto pseudoalignment and sleuth analysis. A, DEGs between the mutants and Col-0 at control and low S. B, DEGs between control and low sulfur in individual genotypes.
Figure 6.
Functional enrichment. Overrepresentation analysis of GO terms annotated to lists of DEGs was performed with PANTHER using Fisher’s exact test. Biological process terms with Bonferroni-corrected P < 0.005, semantic similarity < 0.7 (as determined by REVIGO), and fewer than 10% of Arabidopsis genes annotated to them are visualized. GO terms are sorted on the y axis by size, with the most specific terms on top. The number of Arabidopsis genes annotated to each term is provided in parentheses after the term name. Fold enrichment of the GO term in the indicated gene list is represented by the point fill color. The number of DEGs in the gene list that are annotated to each term is represented by point size. The shape of the point specifies the tissue for the DEG list, and the size of the gene list is shown in parentheses. A, GO terms overrepresented in eil3, eil1, and eil3 eil1 at 0.75 mm SO42−. B, GO terms overrepresented among SLIM1- and EIL1-dependent genes responsive to sulfur deficiency.
At –S conditions, the number of DEGs between the wild type and the individual mutants in the leaves significantly increased for genotypes with disrupted SLIM1 expression, namely eil3 and eil3 eil1, to 598 and 3,489, respectively (Fig. 5A; Supplemental Table S1). On the other hand, in eil1, the number of DEGs decreased in –S compared with control conditions to 72. Forty genes were differentially expressed in all three mutants. In roots, 95 DEGs between eil1 and the wild type were detected under low S, 292 in eil3, and 530 in the double mutant, from which 48 were affected in all three genotypes. The overlap between roots and shoots was somewhat bigger in eil3 and eil3 eil1, where 70 genes (23.6% of root DEGs) and 188 genes (35.5% of root DEGs) were differentially expressed in both roots and shoots, whereas only four genes were affected in both organs in eil1.
Sulfur deficiency had a large effect on gene expression. The analysis revealed 1,258 and 335 sulfur deficiency-regulated genes in shoots and roots of the wild type, respectively (Fig. 5B; Supplemental Table S3). The DEGs with the highest fold change in expression included genes identified by previous microarray analyses (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003), such as MIR395, LSU1, LSU2, SULTR1;1, SULTR1;2, SULTR2;1, SHM7, APS3, APS4, APR2, APR3, SERAT3;2, SDI1, CYP83A1, BCAT4, MAM1, BGLU28, and GGCT2;1 (Supplemental Table S3). These transcripts encode proteins involved in sulfate uptake and allocation, activation, and reduction, Cys biosynthesis, glucosinolate biosynthesis, as well as catabolism of secondary sulfur compounds, and thus cover a large part of the sulfate assimilation pathway. Whereas in leaves of eil1 and eil3 a similar number of genes were differentially expressed as in the wild type, specifically 1,210 and 1,414, respectively, almost four times as many DEGs (4,527) were found in eil3 eil1 (Fig. 5B). In the roots, the number of DEGs was reduced in all mutants compared with the wild type, with only 61 DEGs in eil3 (Fig. 5B; Supplemental Table S3).
Contribution of SLIM1 and EIL1 to Sulfur-Deficiency Response
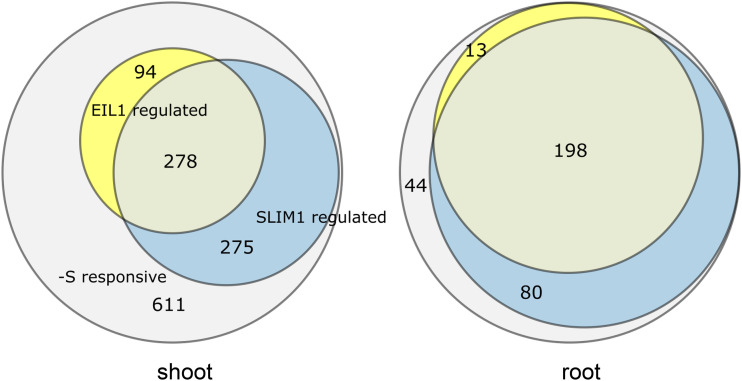
To identify the contribution of SLIM1 and EIL1 to the sulfur-deficiency response, we defined SLIM1- and EIL1-regulated genes as genes that were either differentially expressed in the wild type and not in the mutants eil3 and eil1 or in which the fold change was at least twofold lower in the mutants than in the wild type. In the roots, from the 335 DEGs in the wild type, 211 were regulated by EIL1 and 278 by SLIM1 (Fig. 7; Supplemental Table S4). From these, 198 genes were regulated by both transcription factors, 13 by EIL1 but not SLIM1, and 80 by SLIM1 and not EIL1. Thus, SLIM1 is responsible for the regulation of 83% of the sulfur-deficiency response in roots, with GO terms related to sulfate assimilation, sulfur compound transport, and glucosinolate synthesis all highly overrepresented (Fig. 6B; Supplemental Table S2). Interestingly, none of these GO biological process terms were overrepresented among EIL1-dependent sulfur-deficiency response genes. Among the SLIM1-independent genes were two APR isoforms, APR2 and APR3, as shown before (Maruyama-Nakashita et al., 2006) as well as several genes for enzymes of glucosinolate synthesis, CYP79B2, FMO_GS-OX3, AOP2, or SOT17 (Supplemental Table S4). Glucosinolate synthesis was also the most highly enriched GO biological process term among the SLIM1-independent genes in roots (Supplemental Table S2). Other SLIM1-independent genes included those for amino acid and organic acid metabolism. Several glucosinolate-related genes (CYP79B2, FMO_GS-OX3, and IPMI1), which were regulated by sulfur limitation in the same way in eil3 and the wild type, however, were among DEGs specific for the double mutant.
Figure 7.
Contributions of SLIM1 and EIL1 to control of the sulfur-deficiency response. DEGs of the sulfur-deficiency response (gray), which are regulated by SLIM1 (blue) and EIL1 (yellow), were defined as either being differentially expressed in the wild type and not in the mutants eil3 and eil1 or with the fold change at least twofold lower in the mutants than in the wild type.
From 1,258 DEGs in leaves, 372 were under the control of EIL1, 553 genes were regulated by SLIM1, and 278 were regulated by both (Fig. 7). Interestingly, from these 278 genes, 59 were regulated normally in the double eil3 eil1 mutant, pointing to opposite regulation of these genes by the two factors (Supplemental Table S4). The loss of both transcription factors in the double mutant affected 667 genes regulated by sulfur deficiency in leaves (i.e. 53%, with a stronger [44%] contribution of SLIM1 than of EIL1 [22%]), so they play a weaker role in leaves than in the roots. These genes include all those previously shown to be regulated by SLIM1 via microarray analysis (Supplemental Table S4; Maruyama-Nakashita et al., 2006). The 667 genes include 191 genes that only differed from the wild type in the double mutant but not in the single mutants. Among these 191 genes were again many glucosinolate-related genes and also APR1 and APR3 but not APR2 (Supplemental Table S4). The DEG set of eil3 eil1 was enriched with genes involved in the metabolism of sulfur compounds, including GSH and glucosinolates, and also in response to abiotic and biotic stress (Supplemental Table S2).
SLIM1 is considered the key regulator of the sulfate assimilation pathway, but the complete extent of its involvement is unknown. To quantify the contribution of SLIM1 and EIL1 to regulation of the pathway by sulfur deficiency, a heat map of all genes related to sulfate assimilation and connected pathways was constructed (Supplemental Fig. S2). This indicated that both SLIM1 and EIL1 are largely responsible for the regulation of sulfate uptake and the primary assimilation pathway (except APR) and additionally regulate the biosynthesis and catabolism of glucosinolates. On the other hand, both transcription factors seemed to have only a minor function in the biosynthesis of Met, GSH, and ethylene (Supplemental Fig. S2). Comparing gene expression in all four genotypes revealed that EIL1 often acts as an additive to SLIM1 (Supplemental Fig. S2). In addition, this comparison indicated that many genes in sulfur metabolism are not transcriptionally regulated by SLIM1 or EIL1.
Analysis of SLIM1-responsive genes revealed that SLIM1 possesses additional functions besides regulation of the sulfur-deficiency response. In shoot and root, 399 and 154 genes, respectively, were differentially expressed under sulfur deficiency in eil3 seedlings compared with the wild type. The GO enrichment analysis revealed SLIM1 regulation of gene transcripts encoding for proteins involved in response to stress, wounding, and hormones, such as JAZ13, JAZ5, and an acyl-CoA N-acyltransferase superfamily protein (Supplemental Table S2). Additionally, several genes related to the abscisic acid (ABA) response, glucosinolate biosynthesis, and nitrate transport were regulated by SLIM1 but not sulfur deficiency. By contrast, the –S response seemed not to be exclusively regulated by SLIM1 and EIL1. A total of 420 genes in shoots (33%) and 23 in roots (6.9%) were regulated by sulfur deficiency to the same extent in all four genotypes (Supplemental Table S4). Among the root transcripts were transcripts involved in glucosinolate biosynthesis, like MAM3, SOT17, several GSTs, and CYPs, in addition to isoforms of genes directly associated with the primary assimilation pathway of sulfate, such as APR3, APK2, ATPS1, SERAT1;1, OAS-TL C, and GSH1. Interestingly, transcripts of several redox-related enzymes, like glutaredoxin and thioredoxins, were found to be –S responsive but not under SLIM1 and EIL1 control in shoots, and correspondingly, the GO category oxidation-reduction process was enriched, together with GO terms on stress response and metabolism of Trp and indolic compounds (Supplemental Table S2).
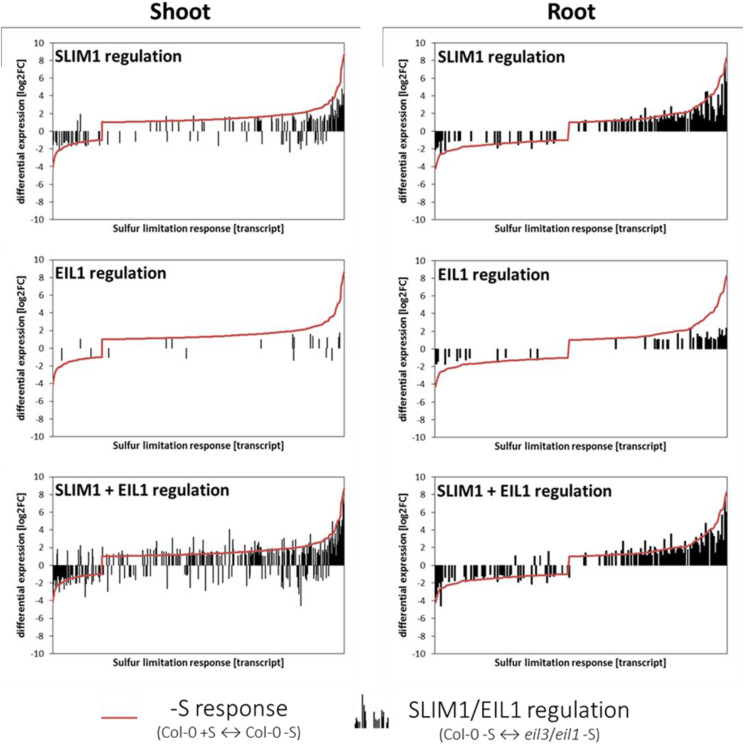
The overall contribution of SLIM1 and EIL1 to the global –S response showed that whereas the numbers of upregulated and downregulated transcripts were balanced in the roots, the majority of transcripts were upregulated in shoots (Fig. 8). DEGs that exceeded a log2FC of 2 or were below –2 formed only a small portion of the DEGs. SLIM1 and also EIL1 seemed to play major functions affecting this fraction of transcripts. By contrast, our data showed that the broad range of genes altered by a log2FC between 1 and 2, as well as –1 and –2, are often controlled independently of these transcription factors. The extent of the regulation (black bars) furthermore showed again that SLIM1 is the more important regulator and that EIL1 has mainly a supportive function (Fig. 8).
Figure 8.
Summary of SLIM1/EIL1 contributions to the –S response. Plants were grown for 18 d on either 0.75 mm SO42− or 0.015 mm SO42−. DEGs (q > 0.05; –1 > log2FC > 1) of RNA-seq (Illumina sequencing) were determined via kallisto pseudoalignment and sleuth analysis. DEGs regulated by –S response (Col-0 +S versus Col-0 –S; red line), SLIM1 (Col-0 –S versus eil3 –S; inverted black bars), EIL1 (Col-0 –S versus eil1 –S; inverted black bars), and SLIM1 + EIL1 (Col-0 –S versus eil3 eil1 –S; inverted black bars) in shoot and root tissue are compared.
New Genes Regulated by Sulfur Deficiency
Since previous investigations of the sulfur-deficiency response were performed using microarrays, another advantage of our RNA-seq approach was an opportunity to identify further genes, those not included in the ATH-1 Array, which are responsive to sulfur deficiency and/or regulated by SLIM1 and EIL1. In shoots and roots, 238 and 67 such genes, respectively, were differentially expressed upon sulfur deficiency (Supplemental Fig. S3; Supplemental Table S5). Not surprisingly, these genes included other isoforms of known –S-responsive genes but also many undescribed genes. The latter included, for example, SULFATE UTILIZATION EFFICIENCY4 (SUE4; AT3G55880), a gene already associated with sulfur deficiency and heavy metal and oxidative stress tolerance (Wu et al., 2010); AT5G24920 encoding GLN DUMPER5, one of seven transmembrane proteins involved in amino acid export into the apoplast (Pratelli et al., 2010); AT1G49640, encoding an α/β-hydrolase superfamily protein with methyl indole-3-acetate esterase activity; AT3G21351, encoding a putative transmembrane protein expressed in guard cells; and AT2G18193, encoding a protein with possible ATPase activity and metal-ion binding (Supplemental Fig. S4). In addition, several unknown genes were highly induced by sulfur deficiency, such as AT2G34655, AT1G22065, and AT2G32487 (Supplemental Fig. S4), the latter being coregulated with genes of the OAS cluster (Hubberten et al., 2012). Gene expression analysis via RT-qPCR was used to confirm the regulation of these genes and to validate the RNA-seq results (Supplemental Fig. S5). The two methods showed an excellent correlation (Supplemental Fig. S5) and revealed that transcript levels of AT1G22065 and AT1G49640 were upregulated in response to deficiency conditions in an EIL1- and SLIM1-dependent manner. On the other hand, AT2G18193 was not responsive to sulfur deficiency in the wild type and was repressed by EIL1 and SLIM1 under both conditions. Interestingly, transcript levels of AT2G18193 in shoots became –S responsive in all mutants, the highest in eil3 eil1. These genes may represent additional components of the sulfur regulatory network and await further characterization.
DISCUSSION
EIL1 Contributes to Regulation of the Sulfur-Deficiency Response
Several lines of evidence led us to hypothesize that, besides SLIM1, other members of the EIL family may be involved in regulation of the sulfur-deficiency response. EIL proteins share 50% to 80% identity in their amino acid sequences, and EIN3, EIL1, and EIL2 share redundant functions in the regulation of gene expression (Chao et al., 1997; Solano et al., 1998; Guo and Ecker, 2004; Merchante et al., 2013). EIL proteins are able to bind the same motif, the tobacco EIN3-binding site [A(C/T)G(A/T)A(C/T)CT], which is present in promoters of ethylene-regulated genes (Kosugi and Ohashi, 2000). Tobacco EIN3-binding sites are also found in promoters of some sulfur deficiency-induced genes arranged in tandem and forming a UPE box, which is bound by SLIM1 homodimers (Wawrzyńska et al., 2010; Wawrzyńska and Sirko, 2016). In addition, EIN3 forms heterodimers with SLIM1 and modulates its binding to promoters in vitro (Wawrzyńska and Sirko, 2016). Overexpression of EILs in transgenic slim1-1 and slim1-2 mutants was not sufficient to recover the fluorescence signal of the pSULTR1;2-GFP reporter under sulfur-deficient conditions. However, eil mutants were not tested for response to sulfur deficiency (Maruyama-Nakashita et al., 2006). Thus, it is possible to hypothesize that other EILs contribute to the sulfur-deficiency transcriptional response independently of SLIM1, for example, in regulating the expression of APR. Furthermore, analyses of eil mutants under sulfur deficiency might unravel additional mechanisms of this regulatory process. This would be reminiscent of members of the same transcription factor families sharing control of other nutrient-related processes, such as PHR1 and PHL1/PHL2 in the phosphate-deficiency response (Bustos et al., 2010; Sun et al., 2016) and NLP7 and NLP6 in nitrate signaling (Konishi and Yanagisawa, 2013). Since we presumed that the contribution of EIL1 in an eil1 single mutant might be masked by the strong activity of SLIM1, we also generated and analyzed an eil3 eil1 double mutant.
Analyses of metabolite levels indicated a contribution of EIL1 to the maintenance of GSH and glucosinolate pools already at +S supply (Fig. 2, B and C). The sulfur-deficiency response was not dramatically affected by the loss of EIL1 in terms of steady-state metabolite levels. However, the sulfate uptake and flux measurements showed the importance of EIL1 in regulation of the assimilatory pathway much more clearly, both at normal and −S supply (Fig. 3). In particular, the analysis of the double mutant eil3 eil1 revealed a dramatic effect of the loss of EIL1 in the eil3 background, pointing clearly to a role of EIL1 in the regulation of sulfate assimilation and the sulfur-deficiency response. The expression analyses confirmed this assumption. In the shoot, the contribution of EIL1 to the control of sulfur-deficiency marker genes was visible only in the double mutant, but in the root, the genes were less upregulated already in the single eil1 mutants (Fig. 4). This was particularly dramatic for SULTR1;1, which in eil3 eil1 was upregulated by sulfur deficiency to a much lesser degree than in eil3. This agrees well with the very low sulfate uptake capacity of the eil3 eil1 plants. The expression analysis, however, also refuted our initial hypothesis that EIL1 might control the upregulation of APR. The discovery of EIL1 function in the regulation of the sulfur-deficiency response begs the question of whether this response is dependent on the canonical ethylene signaling; however, this is beyond the scope of this study.
Global Effect of the Loss of EIL1 on the Arabidopsis Transcriptome
The finding that EIL1 is involved in regulation of the sulfur-deficiency response triggered the question of the global extent of its contribution. Whereas the Arabidopsis transcriptomic response to sulfur deficiency was analyzed numerous times (Hirai et al., 2003, 2005; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003; Higashi et al., 2006; Bielecka et al., 2015), each of these analyses was based on microarray technology. In addition, the information on SLIM1-dependent expression is limited to roots, based on ATH1 microarrays and therefore missing analysis of ∼13,000 genes, and was determined with only two biological replicates (Maruyama-Nakashita et al., 2006). Therefore, a new comprehensive analysis using RNA-seq from roots and shoots separately was expected to yield substantial further understanding regarding both the global contribution of EIL1 to the sulfur-deficiency response and the function of SLIM1 beyond this response.
Indeed, the RNA-seq analysis confirmed an important role for EIL1 in the regulation of the sulfur-deficiency response. Although transcript levels of many genes were affected by the loss of EIL1 alone, the loss of EIL1 was particularly large in the eil3 background, as almost fourfold more DEGs were found in leaves of eil3 eil1 mutants than in leaves of the individual ones (Fig. 5). Most of the EIL1-regulated transcripts are also regulated by SLIM1, particularly in the roots (Fig. 6); however, presumably because of the subordinate function in the sulfur-deficiency response, fewer GO functional terms are overrepresented among the EIL1-regulated genes at the stringent significance threshold (Fig. 7). Thus, when SLIM1 is impaired, EIL1 might partially fulfill the role of SLIM1 under sulfate limitation to prevent a complete collapse of the response. Nonetheless, this function of EIL1 seems to be only supportive, since EIL1 is not sufficient for the full induction of the sulfur-deficiency response and the eil3 mutant still shows a strongly deficient response to sulfate limitation. It can be hypothesized that this insufficiency is due to the different expression patterns of EIL1 and SLIM1. However, this does not seem to be the case because (1) based on the transcripts per kilobase million values in our RNA-seq data set, EIL1 transcript levels are 4- to 5-fold more abundant than those of SLIM1 (Supplemental Data Set S1); (2) the tissue-specific expression patterns of EIL1 and SLIM1 in eFP Browser (Winter et al., 2007) are very similar; and (3) both genes did not show any enrichment in bundle sheath cells (Aubry et al., 2014).
Yet, a small number of prominent sulfur-deficiency marker genes are strongly regulated by EIL1 in roots, affecting essential parts of the sulfate assimilation pathway. For example, EIL1 regulates the ability to enhance the catabolism of various sulfur-containing compounds via the regulation of BGLU28/30, TGG1, and NSP5 for glucosinolates as well as GGCT2;1 for the degradation of GSH (Maruyama-Nakashita, 2017). EIL1 also contributes to enhanced sulfate uptake and root-shoot translocation upon sulfate demand (Fig. 3; Supplemental Fig. S2). EIL1 has minor but distinct effects on several sulfate transporters (SULTR1;1, SULTR1;2, SULTR2;2, SULTR4;1, and SULTR4;2), particularly when SLIM1 is impaired (Supplemental Fig. S2). In particular, the well-described upregulation of SULTR1;1 and SULTR2;1 transcripts is also partially controlled by EIL1. Additionally, EIL1 contributes to the control of xylem-based root-shoot transport via regulation of the low-affinity SULTR2;1 but without altering mRNA levels of its facilitator SULTR3;5 or SULTR1;3 responsible for phloem-based source-to-sink sulfate translocation (Yoshimoto et al., 2002; Kataoka et al., 2004a). This might be due to the EIL1 contribution to the upregulation of miR395 (Supplemental Fig. S2). Previous studies showed that an increase in miR395 is strongly dependent on SLIM1 (Kawashima et al., 2009, 2011). Phloem-specific expression of miR395 confines its target mRNA SULTR2;1 to xylem parenchyma cells and results in enhanced sulfate translocation to the shoot (Kawashima et al., 2009). All isoforms of miR395 transcripts are highly upregulated by sulfur deficiency in shoot and root tissue of wild-type plants, with miR395d and miR395f showing slightly weaker regulation. In shoots, miR395f is the only isoform affected by the loss of EIL1, whereas in roots, miR395c, miR395d, miR395e, and miR395f are all induced to a lesser extent in eil1.
The Sulfur-Deficiency Response Is Not Entirely Controlled by SLIM1 and EIL1
The RNA-seq analysis allowed us to obtain the first complete picture of the transcriptional response to sulfur deficiency, including the full gene families (Supplemental Fig. S2). Although sulfate assimilation is the major pathway regulated by sulfur deficiency, some transcripts in the primary and secondary sulfate assimilation pathways were not regulated under these conditions. This is somewhat unexpected, since the pathway was previously shown to be coordinately regulated, for example by jasmonate (Jost et al., 2005), or in its spatial expression in bundle sheath cells of Arabidopsis (Aubry et al., 2014). Some of the genes found not to be regulated by sulfur deficiency are key for the sulfate assimilation pathway. SULTR3;1 encodes a sulfate transporter localized to the plastid envelope, providing sulfate for the reduction in plastids (Cao et al., 2013). ATPS1 is the major isoform of ATP sulfurylase responsible for approximately 50% of the activity in leaves (Koprivova et al., 2013). However, ATPS1 is a target of miR395, and two processes maintain its transcript levels, cleavage by the microRNA and increased transcription, which are well coordinated, so that the steady-state transcript levels do not change (Kawashima et al., 2011). Likewise, although SLIM1 is considered the key regulator of the sulfur-deficiency response, many genes associated with the sulfate assimilation pathway (Supplemental Fig. S2) are regulated independently of SLIM1 (and EIL1). Some of these genes have been known for a long time, such as the three isoforms of APR (Maruyama-Nakashita et al., 2006; Hubberten et al., 2012), but some have been added in this study, such as SULTR2;2, the 12-oxophytodienoic acid reductase OPR2, or BAT5 and some glucosinolate synthesis genes.
RNA-seq allowed us to quantify the contribution of the two transcription factors to the control of the sulfur-deficiency response. Quantifying this contribution is important, as SLIM1 has been considered a key regulator of this response (Maruyama-Nakashita et al., 2006; Koprivova and Kopriva, 2014). Whereas this is clearly true for the roots, where 83% of sulfur deficiency-regulated genes are under the control of SLIM1, in leaves, the contribution of SLIM1 is much lower and reaches only 44% (Fig. 7). EIL1 modulates the expression of 63% of the sulfur deficiency-responsive genes in roots and 30% in shoots. However, very few genes with a fold change > 5 are independent of SLIM1 and EIL1, among which are APR and some glucosinolate synthesis genes. Most of the SLIM1-independent genes are only moderately affected by sulfur deficiency, and their contribution to the physiological response needs to be established. Similarly, it needs to be shown whether these genes are controlled by one or a few transcription factors or whether their regulation depends on a large number of regulators.
Pathways Regulated by Sulfur Deficiency beyond Sulfur Metabolism
Obviously, cellular processes other than sulfate uptake and metabolism are affected by sulfur deficiency, as evidenced in the RNA-seq analysis. For example, several DEGs belong to other nutrient and phytohormone pathways. Transcripts of phosphate, nitrate, and molybdenum transporters were upregulated under sulfur-deficiency conditions in shoots and roots (Supplemental Table S3). This regulation was largely independent of SLIM1 and EIL1. Another big group of genes regulated by sulfur deficiency consists of genes involved in redox signaling (Supplemental Table S3). This is not surprising, since the connection between ROS/redox and hormone signaling seems to optimize plant physiology under normal and stress conditions and sulfur deficiency results in a lower concentration of the redox buffer GSH (Rouhier et al., 2015). Changes in the transcript levels of redox genes indicate alterations of the redox state during sulfur deficiency. Several peroxidases and oxidases are upregulated during sulfur-deficiency conditions in a SLIM1-independent manner, such as PEROXIDASE2, PEROXIDASE CB, RESPIRATORY BURST OXIDASE HOMOLOG B (RBOHB), RBOHC, and RBOHD. Indeed, NADP(H) oxidases and peroxidases were shown to produce superoxide and hydrogen peroxide, which subsequently are second messengers in many processes associated with plant growth and development (Mhamdi and Van Breusegem, 2018). Interestingly, these oxidases were also upregulated upon phosphorus, nitrogen, and potassium deficiency (Shin et al., 2005). Thus, (transient) ROS pulses might be the initial response to general nutrient imbalance, as these genes are controlled independently of SLIM1. Altered expression of genes encoding for ROS-scavenging enzymes, like COPPER/ZINC SUPEROXIDE DISMUTASE1, FE SUPEROXIDE DISMUTASE1, ACONITASE2 (ACO2), ACO3, and GLUTATHIONE PEROXIDASE6, might indicate the contribution of cellular ROS homeostasis to the initiation of the sulfur-deficiency response (Mittler, 2002). In addition, several members of CC-type glutaredoxins (ROXY4, ROXY12, ROXY13, ROXY15, ROXY16, and ROXY17) were downregulated during deficiency conditions in a SLIM1-dependent manner. Whether their repression might act as a secondary response of SLIM1 regulation, either as a negative feedback loop or an all-or-nothing principle, needs further elucidation. Both are possible, since ROS plays a dual function in abiotic stress by elevating the damage but also mediating the initial signal for the activation of defense responses (for review, see Dat et al., 2000).
It was shown that persistent increase of hydrogen peroxide levels leads to transcriptional expression profiles similar to the response to biotic and abiotic stress, including upregulation of marker genes associated with ethylene signaling (Vandenabeele et al., 2003). Moreover, a transient ROS burst is involved in plant hormone signaling (Noctor et al., 2018). Correspondingly, response to stress and response to ROS were among the functional categories affected by sulfur deficiency (Fig. 6; Supplemental Table S2), and a number of jasmonic acid- and ABA-responsive genes, such as ALLENE OXIDE CYCLASE2, CORONATINE INDUCED1, VEGETATIVE STORAGE PROTEIN1 (VSP1) and VSP2, COPPER AMINE OXIDASE, OPEN STOMATA1, and HIGHLY ABA-INDUCED PP2C GENE2, were upregulated under sulfur-deficiency conditions, which indicates the involvement of ROS-mediated hormone signaling (Jones et al., 2003; Stenzel et al., 2012; Kim et al., 2013; Mittler and Blumwald, 2015). In fact, sulfur deficiency seems to mimic or induce jasmonic acid and ABA signaling.
GO enrichment analysis of DEGs regulated by SLIM1 also indicated its function in the control of defense responses to pathogens, primarily through jasmonate signaling (Fig. 6). This is not surprising, as links between jasmonate signaling and sulfate assimilation have been long known. Jasmonate treatment induces genes involved in sulfate reduction, GSH biosynthesis, and glucosinolate metabolism, which leads to glucosinolate accumulation (Xiang and Oliver, 1998; Harada et al., 2000; Jost et al., 2005; Frerigmann and Gigolashvili, 2014). The effect of sulfur deficiency on jasmonate-related processes, however, might occur at different levels of regulation; the SLIM1/EIL1-dependent regulation of JAZ proteins in response to sulfur deficiency is similar to the strong upregulation of SDI1 protein, which binds MYB28 and inhibits its function (Aarabi et al., 2016). Interestingly, JAZ1, JAZ5, JAZ7, and JAZ8, which were differentially regulated in RNA-seq, are target genes of two other transcription factors, WRKY40 and WRKY18 (Pandey et al., 2010). Several other WRKY-associated jasmonate-responsive genes, like LIPOXYGENASE2 and PLANT DEFENSIN1.2, are regulated in a SLIM1- and/or EIL1-dependent manner. Induction of jasmonate biosynthesis in response to sulfur deficiency might serve the enhancement of defense capabilities of the plants when sulfur-containing defense compounds are less available (Koprivova and Kopriva, 2016a).
SLIM1 and EIL1 Function Independently of Sulfur Deficiency
Although EIL1 and particularly SLIM1 have been mainly discussed to control the sulfur-deficiency response, they affect sulfate metabolism and other processes also during normal sulfur supply. Indeed, the contents of glucosinolates are decreased in leaves of both eil3 and eil1 mutants under normal conditions, as is sulfate in eil3 and GSH in eil1 (Fig. 2). RNA-seq data confirmed their regulatory function toward several genes associated with glucosinolate biosynthesis in a mild manner (–0.6 > log2FC > 0.6) at +S supply. Breakdown of glucosinolates into Glc and an unstable aglycone intermediate that can be further rearranged into highly bioactive isothiocyanate, nitrile, thiocyanate, or cyclic compounds is mediated and catalyzed by myrosinases (Halkier and Gershenzon, 2006). EIL1 has a distinct function in the control of the thioglucosidases TGG1/2 and BGLU30 under normal conditions. Also, under adequate sulfur supply, transcript levels of SULTR2;1, SULTR4;2, GGCT2;1, and surprisingly APR1 and APR3 are higher in eil3 and to a lesser extent in eil1. This suggests a repressor function of SLIM1, as has been discussed previously (Wawrzyńska and Sirko, 2014). Upregulation of mRNA levels for GGCT2;1 and APR3 in eil3 and eil3 eil1 seedlings might also explain the decrease in sulfate and increase of Cys levels (Figs. 2 and 4). Interestingly, several LSU transcripts are also upregulated in eil3 in roots and shoots under normal conditions.
GO enrichment analysis of eil3 and eil1 under normal sulfur conditions, moreover, revealed the function of SLIM1 and EIL1 in various other processes, such as hormone signaling, stress response, and ROS and redox signaling (Supplemental Table S2). The upregulation of INDOLE-3-ACETIC ACID INDUCIBLE gene transcripts (IAA6, IAA19, IAA20, IAA29, and IAA30), SMALL AUXIN REGULATED RNAs (SAUR10, SAUR46, SAUR68, and SAUR76), as well as 1-AMINO-CYCLOPROPANE-1-CARBOXYLATE SYNTHASE8 (ACS8) in leaves points to a repressive function of SLIM1 in auxin signaling. Interestingly, we could also identify SLIM1 regulation of several ERFs, among them ERF11, ERF13, ERF71, and ERF98. However, as ERFs can also be activated by jasmonate or ABA signaling, it needs to be shown whether SLIM1 is able to directly transduce ethylene perception (Chao et al., 1997; Lorenzo et al., 2003).
Additional Genes Regulated by Sulfur Deficiency
Our RNA-seq data further detail the sulfur-deficiency response with the identification of up to 20% of additional sulfur deficiency-responsive DEGs: 238 in shoots and 67 in roots. Whereas this gene set includes several new isoforms of known sulfur deficiency-responsive genes, 50% to 60% of the newly identified genes have not been previously described in this context. It can be assumed that a large proportion of these genes are involved in sulfur metabolism, oxidative stress response, and redox or phytohormone signaling. This is supported, for example, by confirming the upregulation of SUE4, a gene previously associated with sulfur deficiency, heavy metal, and oxidative stress tolerance (Wu et al., 2010). A gain-of-function mutant showed improved tolerance to –S conditions and improved root establishment. SUE4, a small protein with four transmembrane domains, might act in GSH homeostasis-related processes, since GSH levels were enhanced under both conditions compared with the wild type (Wu et al., 2010). Whereas the function of SUE4 has not been shown, it is similar to several α/β-hydrolases, with AT2G40095 as the closest homolog. Interestingly, we identified further nine genes encoding α/β-hydrolase superfamily proteins as upregulated under sulfur-deficiency conditions (AT1G68620, AT4G39955, AT4G10955, AT3G23570, AT1G49640, AT1G72620, AT1G56630, AT1G73480, and AT3G02410). As many proteins of this superfamily function as peroxidases, these proteins might be involved in redox signaling. Therefore, they might constitute interesting proteins that contribute to signaling within the sulfur-deficiency response (Nardini and Dijkstra, 1999; Lenfant et al., 2013).
Another gene of interest is GLN DUMPER5 (GDU5), a member of a family of seven transmembrane proteins involved in amino acid export into the apoplast (Pratelli et al., 2010). Upon sulfur deficiency, GDU5 was downregulated in leaves and roots, whereas another member of the family, GDU6, was upregulated. Both genes were found to be controlled by SLIM1 and EIL1. Their relative GDU1 is the best-studied Gln dumper and regulates amino acid export from plant cells (Pratelli et al., 2012). As the GDU proteins act in amino acid transport, they might play a role in GSH biosynthesis and homeostasis (Pilot et al., 2004; Pratelli et al., 2010). In the plasma membrane, GDU1 interacts with the RING E3 ubiquitin ligase LOG2 (Pratelli et al., 2012), which was shown to be a positive regulator of ABA-mediated drought- and salt-stress tolerance mechanisms (Kim and Kim, 2013). Interestingly, LOG2 transcript levels were upregulated upon –S conditions as well. Further experiments are needed to identify the specific functions of these identified genes.
Altogether, with EIL1 we identified a second transcriptional activator regulating the sulfur-deficiency response in Arabidopsis. EIL1 possesses a subordinate and additive function to SLIM1 in the control of the sulfur-deficiency response, particularly in the roots, but also in fine-tuning sulfur homeostasis at normal sulfur supply. We performed a comprehensive RNA-seq analysis to obtain a complete picture of the transcriptional response to sulfur deficiency and the role of SLIM1 and EIL1 within and beyond this response. We confirmed that SLIM1 is the main regulator of the sulfate assimilation pathway and showed that it has additional functions in the regulation of phytohormone-, defense-, and redox-related processes at both –S and normal sulfur supply. However, we also showed that more than 50% of the transcriptional response to sulfur deficiency in leaves is independent of SLIM1 and EIL1 and that further components of the regulatory circuit await identification.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants of the wild-type ecotype Col-0 and the T-DNA insertion lines (for schemes of T-DNA insertion, see Supplemental Fig. S1) eil1 (AT2G27050; SALK_042113), eil3 (AT1G73730; SALK_089129), and the double mutant eil3 eil1 were used for experiments.
Seeds were surface sterilized with chlorine gas for 4 h. Under sterile conditions, seeds were placed on modified Long Ashton Medium agarose plates in either +S (0.75 mm MgSO4) or −S (0.75 mm MgCl2 + 0.015 mm MgSO4) conditions and stratified for 3 d at 4°C in the dark. Afterward, the plates were incubated in Sanyo light chambers at 22°C in long-day conditions with a 16/8-h light cycle and 100 µmol photons m−2 s−1. After 18 d, shoot and root samples were collected and immediately frozen in liquid nitrogen. Each biological replicate was collected from seedlings grown on different plates. Results were obtained from at least two independent experiments.
Sulfate Analysis
For sulfate measurement, plant material was homogenized in 200 μL of deionized water and transferred to a screw-cap tube with another 800 µL of deionized water. The whole extract was shaken for 1 h at 4°C and afterward heated at 95°C for 15 min. After 15 min of centrifugation at 4°C and 16,000g, 100 μL of supernatant was transferred into an ion chromatography vial with 900 µL of water. Inorganic anions were measured with the Dionex ICS-1100 chromatography system and separated on a Dionex IonPac AS22 RFIC 4× 250-mm analytic column (Thermo Scientific). Solution of 4.5 mm Na2CO3 and 1.4 mm NaHCO3 was used as running buffer. A standard curve of sulfate was generated by using the external standards of 0.5, 1, and 2 mm K2SO4.
Isolation and Quantification of Low-Molecular-Weight Thiols
For thiols measurement, ∼20 mg of plant material was extracted in exactly a 10-fold volume of 0.1 m HCl and afterward centrifuged for 10 min at 4°C and 16,000g. In a new tube, 25 µL of supernatant was incubated with 25 µL of 0.1 m NaOH and 1 µL of freshly prepared 100 mm dithiothreitol for 15 min at 37°C in the dark. Subsequently, 10 µL of 1 m Tris-HCl, pH 8, 35 µL of water, and 5 µL of 100 mm monobromobimane (Thiolyte MB; Calbiochem) in acetonitrile were added to conjugate sulfhydryl groups of Cys and GSH for 15 min at 37°C in the dark. A total of 100 µL of 9% (v/v) acetic acid was used to stabilize bimane conjugates. A total of 180 µL of supernatant was transferred into HPLC vials. The thiol conjugates were separated and measured via HPLC (Spherisorb ODS2, 250 × 4.6 mm, 5 µm; Waters) and detected fluorimetrically with a 474 detector (excitation, 390 nm; emission, 480 nm). Two solvents were used in a linear gradient of 95% to 82% A (10% [v/v] methanol and 0.25% [v/v] acetic acid, pH 3.9) in B (90% [v/v] methanol and 0.25% [v/v] acetic acid, pH 3.9) with a constant flow rate of 1 mL min−1.
Isolation and Quantification of Glucosinolates
Glucosinolate content was determined by reverse-phase HPLC via UV detection (Burow et al., 2006) after isolation as described previously (Aghajanzadeh et al., 2015). Right before isolation, specific columns were prepared using 1-mL tips plugged with nonabsorbent cotton wool and a layer of 0.5 mL of DEAE-Sephadex A-25. The samples were extracted twice with 250 µL of hot 70% (v/v) methanol, with the addition of 10 µL of sinigrin as internal standard, and incubated at 70°C for 45 min. The samples were cooled and spun down at 16,000g. The supernatants were transferred onto the prepared columns, washed twice with 0.5 mL of deionized water, and subsequently twice with 0.5 mL of 0.02 m sodium acetate buffer. With a new tube placed underneath each column, 75 µL of sulfatase solution was added directly on the surface of the column. After incubation at room temperature overnight, the produced desulfoglucosinolates were eluted twice with 0.5 mL of water and finally with 0.25 mL of deionized water, vortexed, and centrifuged. A total of 350 µL of the supernatant was added into an HPLC vial. Desulfoglucosinolates were measured via HPLC (Spherisorb ODS2, 250 × 4.6 mm, 5 µm; Waters) by UV absorption at 229 nm, identified by retention time of the peaks, and quantified with the help of the internal standard sinigrin. For separation, a gradient of acetonitrile in water (5%–30% [v/v] in 8 min, 30%–50% [v/v] in 7 min) was used.
Sulfate Uptake and Flux Analyses
Sulfate uptake and flux through the assimilation pathway were measured in seedlings grown for 18 d under sulfur-sufficient or -deficient conditions. After short incubation of two seedlings in 2 mL of nonradioactive Long Ashton nutrient solution containing 0.2 mm sulfate on 24-well plates, the medium was exchanged with 1 mL of the same solution supplemented with 12 µCi of [35S]sulfuric acid and incubated for 3 h in the light. Whole seedlings were washed thoroughly, blotted dry, and shoot and root samples were stored separately in liquid nitrogen until further processing on the same day. Samples were extracted in a 10-fold volume of 0.1 m HCl. Ten microliters of extract was used to determine sulfate uptake, and 50-µL aliquots of each extract were collected for quantification of 35S incorporation into thiols, proteins, and glucosinolates exactly as previously described (Mugford et al., 2011).
Gene Expression Analyses
Total RNA was isolated by phenol-chloroform-isoamyl alcohol extraction and LiCl precipitation. DNase treatment and first-strand cDNA synthesis were performed with QuantiTect Reverse Transcription with 800 ng of RNA in a 6-µL reaction according to the manufacturer’s instructions. RT-qPCR was performed using SYBR Green in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Transcript levels were normalized to TIP41 using the 2−ΔΔCT method. RT-qPCR primers (Supplemental Table S6) were designed via the Universal ProbeLibrary Assay Design Center (Roche Diagnostics).
RNA-Seq Experiment
RNA-seq was performed by the Cologne Centre for Genomics (portal.ccg.uni-koeln.de) using Illumina Polymerase-based sequencing-by-synthesis, obtaining a read length of 150 bp (paired end with 75 bp each) and coverage of approximately 40 million reads per sample. Data from three biological replicates were processed with the Trimmomatic v.0.36 trimming tool to remove adapters and low-quality reads (http://www.usadellab.org/cms/?page=trimmomatic; Bolger et al., 2014). Trimmed, paired files underwent a second quality control via FastQC. FastQ files were further processed with kallisto v.0.43.1 (Bray et al., 2016) to quantify transcript abundances in pseudoalignment to the Arabidopsis reference transcriptome (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/TAIR10_gene_lists/) as transcripts per kilobase million. Ninety-six percent to 98% of reads reached processability and could be pseudoaligned. DEGs (q < 0.05) were determined from transcript abundance files using the package sleuth and compared with DESeq2 in the R environment (Love et al., 2014; Pimentel et al., 2017). Highmeans and lowmeans from the sleuth table were used to manually calculate fold changes and log2FC with Windows Excel.
Enrichment Analysis
Enrichment analyses of GO biological process terms (GO database release 2019-12-09) was performed with PANTHER (http://www.pantherdb.org; Mi et al., 2019) using Fisher’s exact test and Bonferroni correction for multiple testing. Significantly overrepresented GO terms (P < 0.005) in each list of DEGs were scored and clustered based on their semantic similarity (simRel calculation) using REVIGO (http://revigo.irb.hr; Supek et al., 2011).
Statistical Analysis
The statistical difference of two populations was tested by two-tailed, unpaired Student’s t test. When results were tested for hypotheses based on knowledge about SLIM1 function and physiological changes under sulfur-deficiency conditions, samples of two populations were statistically compared by one-tailed, unpaired Student’s t test in Excel (Microsoft Office 365). To test equality of variances, Levene’s test was performed in advance. With accepted null hypothesis (P < 0.05, Levene’s test), homoscedastic Student’s t test was performed. Different letters represent significant differences between values of two genotypes within one treatment (P < 0.05, Student’s t test). Capital letters are assigned to normal and small letters to sulfur-deficiency conditions. Asterisks indicate significant differences between values of one genotype at control or low sulfur (*, P < 0.05; **, P < 0.01; and ***, P < 0.001, Student’s t test). All experiments, except the RNA-seq, were independently repeated at least twice.
Accession Numbers
The RNA-seq data from this article can be found in the National Center for Biotechnology Information Gene Expression Omnibus data repository under accession number GSE157765.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of the eil3 mutant with slim1-1 and slim1-2.
Supplemental Figure S2. DEGs, regulated by –S response, SLIM1, and EIL1.
Supplemental Figure S3. Newly identified DEGs, regulated by –S response, in a SLIM1- and EIL1-dependent manner.
Supplemental Figure S4. Expression analyses of new genes regulated by sulfur deficiency.
Supplemental Figure S5. Comparison of RNA-seq and RT-qPCR results.
Supplemental Table S1. DEGs between wild-type Col-0 and eil1, eil3, and eil3 eil1 mutants at control and low sulfur.
Supplemental Table S2. Enrichment analyses.
Supplemental Table S3. DEGs in the four genotypes and two organs between control and low sulfur.
Supplemental Table S4. Dependence of DEGs in wild-type Col-0 on SLIM1 and EIL1.
Supplemental Table S5. Newly identified sulfur deficiency-responsive genes.
Supplemental Table S6. Primers used in the study.
Supplemental Data Set S1. RNA-seq data.
Acknowledgments
We thank the Cologne Centre for Genomics for performing the RNA-seq.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft under Germany’s Excellence Strategy (grant no. EXC 2048/1, project 390686111) and by the Max Planck Society (to S.J.W. and R.H.).
Articles can be viewed without a subscription.
References
- Aarabi F, Kusajima M, Tohge T, Konishi T, Gigolashvili T, Takamune M, Sasazaki Y, Watanabe M, Nakashita H, Fernie AR, et al. (2016) Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Sci Adv 2: e1601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanzadeh T, Kopriva S, Hawkesford MJ, Koprivova A, De Kok LJ(2015) Atmospheric H2S and SO2 as sulfur source for Brassica juncea and Brassica rapa: Impact on the glucosinolate composition. Front Plant Sci 6: 924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Smith-Unna RD, Boursnell CM, Kopriva S, Hibberd JM(2014) Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J 78: 659–673 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bielecka M, Watanabe M, Morcuende R, Scheible WR, Hawkesford MJ, Hesse H, Hoefgen R(2015) Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Front Plant Sci 5: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B(2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L(2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527 [DOI] [PubMed] [Google Scholar]
- Burow M, Müller R, Gershenzon J, Wittstock U(2006) Altered glucosinolate hydrolysis in genetically engineered Arabidopsis thaliana and its influence on the larval development of Spodoptera littoralis. J Chem Ecol 32: 2333–2349 [DOI] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J(2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Wirtz M, Hell R, Oliver DJ, Xiang CB(2013) SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J 73: 607–616 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR(1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F(2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T(2014) Update on the role of R2R3-MYBs in the regulation of glucosinolates upon sulfur deficiency. Front Plant Sci 5: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR(2004) The ethylene signaling pathway: New insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J(2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57: 303–333 [DOI] [PubMed] [Google Scholar]
- Harada E, Kusano T, Sano H(2000) Differential expression of genes encoding enzymes involved in sulfur assimilation pathways in response to wounding and jasmonate in Arabidopsis thaliana. J Plant Physiol 156: 272–276 [Google Scholar]
- Higashi Y, Hirai MY, Fujiwara T, Naito S, Noji M, Saito K(2006) Proteomic and transcriptomic analysis of Arabidopsis seeds: Molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J 48: 557–571 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K(2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Klie S, Caldana C, Degenkolbe T, Willmitzer L, Hoefgen R(2012) Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. Plant J 70: 666–677 [DOI] [PubMed] [Google Scholar]
- Jones PR, Manabe T, Awazuhara M, Saito K(2003) A new member of plant CS-lyases: A cystine lyase from Arabidopsis thaliana. J Biol Chem 278: 10291–10296 [DOI] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, Bogs J, Gershenzon J, Hähnel U, Hänsch R, Hartmann T, Kopriva S, Kruse C, et al. (2005) Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res 86: 491–508 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H(2004a) Root-to-shoot transport of sulfate in Arabidopsis: Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol 136: 4198–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H(2004b) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Matthewman CA, Huang S, Lee BR, Yoshimoto N, Koprivova A, Rubio-Somoza I, Todesco M, Rathjen T, Saito K, et al. (2011) Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J 66: 863–876 [DOI] [PubMed] [Google Scholar]
- Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T(2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57: 313–321 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim WT(2013) The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol 162: 1733–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lee Y, Park J, Lee N, Choi G(2013) HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol 54: 555–572 [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S(2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Kopriva S, Mugford SG, Baraniecka P, Lee BR, Matthewman CA, Koprivova A(2012) Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Front Plant Sci 3: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Giovannetti M, Baraniecka P, Lee BR, Grondin C, Loudet O, Kopriva S(2013) Natural variation in the ATPS1 isoform of ATP sulfurylase contributes to the control of sulfate levels in Arabidopsis. Plant Physiol 163: 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S(2014) Molecular mechanisms of regulation of sulfate assimilation: First steps on a long road. Front Plant Sci 5: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S(2016a) Hormonal control of sulfate uptake and assimilation. Plant Mol Biol 91: 617–627 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S(2016b) Sulfation pathways in plants. Chem Biol Interact 259: 23–30 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y(2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenfant N, Hotelier T, Bourne Y, Marchot P, Chatonnet A(2013) Proteins with an alpha/beta hydrolase fold: Relationships between subfamilies in an ever-growing superfamily. Chem Biol Interact 203: 266–268 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R(2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A.(2017) Metabolic changes sustain the plant life in low-sulfur environments. Curr Opin Plant Biol 39: 144–151 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H(2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H(2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Alonso JM, Stepanova AN(2013) Ethylene signaling: Simple ligand, complex regulation. Curr Opin Plant Biol 16: 554–560 [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Van Breusegem F(2018) Reactive oxygen species in plant development. Development 145: dev164376. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD(2019) PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R.(2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E(2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Lee BR, Koprivova A, Matthewman C, Kopriva S(2011) Control of sulfur partitioning between primary and secondary metabolism. Plant J 65: 96–105 [DOI] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW(1999) Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr Opin Struct Biol 9: 732–737 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R(2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: Interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R(2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Reichheld JP, Foyer CH(2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE(2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 64: 912–923 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB(2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L(2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Pratelli R, Guerra DD, Yu S, Wogulis M, Kraft E, Frommer WB, Callis J, Pilot G(2012) The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol 158: 1628–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Voll LM, Horst RJ, Frommer WB, Pilot G(2010) Stimulation of nonselective amino acid export by glutamine dumper proteins. Plant Physiol 152: 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Cerveau D, Couturier J, Reichheld JP, Rey P(2015) Involvement of thiol-based mechanisms in plant development. Biochim Biophys Acta 1850: 1479–1496 [DOI] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP(2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR(1998) Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA(2010) Biosynthesis of glucosinolates: Gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Otto M, Delker C, Kirmse N, Schmidt D, Miersch O, Hause B, Wasternack C(2012) ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: Tissue- and organ-specific promoter activities and in vivo heteromerization. J Exp Bot 63: 6125–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D(2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T(2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Buchner P, Yoshimoto N, Hawkesford MJ, Shiu SH(2012) Evolutionary relationships and functional diversity of plant sulfate transporters. Front Plant Sci 2: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R(2011) Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Yamazaki M, Sasakura N, Watanabe A, Leustek T, Engler JA, Engler G, Van Montagu M, Saito K(1997) Regulation of sulfur assimilation in higher plants: A sulfate transporter induced in sulfate-starved roots plays a central role in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 11102–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al. (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos P, Krähenbühl U, den Camp RO, Brunold C(2002) Flux control of sulphate assimilation in Arabidopsis thaliana: Adenosine 5′-phosphosulphate reductase is more susceptible than ATP sulphurylase to negative control by thiols. Plant J 31: 729–740 [DOI] [PubMed] [Google Scholar]
- Wawrzyńska A, Lewandowska M, Sirko A(2010) Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. J Exp Bot 61: 889–900 [DOI] [PubMed] [Google Scholar]
- Wawrzyńska A, Sirko A(2014) To control and to be controlled: Understanding the Arabidopsis SLIM1 function in sulfur deficiency through comprehensive investigation of the EIL protein family. Front Plant Sci 5: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyńska A, Sirko A(2016) EIN3 interferes with the sulfur deficiency signaling in Arabidopsis thaliana through direct interaction with the SLIM1 transcription factor. Plant Sci 253: 50–57 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ(2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M, Hell R(2004) O-Acetylserine (thiol) lyase: An enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. J Exp Bot 55: 1785–1798 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, Xiang CB(2010) Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis. J Exp Bot 61: 3407–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ(1998) Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10: 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Takahashi H, Smith FW, Yamaya T, Saito K(2002) Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J 29: 465–473 [DOI] [PubMed] [Google Scholar]