Leaf angle across the sorghum canopy is genetically controlled by both common and layer-specific loci, which holds potential for the development of an optimized canopy associated with higher yields.
Abstract
A “smart canopy” ideotype has been proposed with leaves being upright at the top and more horizontal toward the bottom of the plant to maximize light interception and conversion efficiencies, and thus increasing yield. The genetic control of leaf angle has, to date, been studied on one or two leaves, or data have been merged from multiple leaves to generate average values. This approach has limited our understanding of the diversity of leaf angles across layers and their genetic control. Genome-wide association studies and quantitative trait loci mapping studies in sorghum (Sorghum bicolor) were performed using layer-specific angle data collected manually and via high-throughput phenotyping strategies. The observed distribution of angles in indoor and field settings is opposite to the ideotype. Several genomic regions were associated with leaf angle within layers or across the canopy. The expression of the brassinosteroid-related transcription factor BZR1/BES1 and the auxin-transporter Dwarf3 were found to be highly correlated with the distribution of angles at different layers. The application of a brassinosteroid biosynthesis inhibitor could not revert the undesirable overall angle distribution. These discoveries demonstrate that the exploitation of layer-specific quantitative trait loci/genes will be instrumental to reversing the natural angle distribution in sorghum according to the “smart canopy” ideotype.
Canopy architecture has been recognized as a major yield determinant in cereals (Long et al., 2006; Zhu et al., 2010; Ort et al., 2015). While erect canopies facilitate dense plantings, and thus generate higher yields per unit of land (Lambert and Johnson, 1978; Duvick et al., 2004; Duvick, 2005; Ma et al., 2014), a particular arrangement of leaves across the canopy has been proposed as the optimized structure for an increased photosynthetic conversion efficiency (Duncan, 1971; Long et al., 2006; Ku et al., 2010; Zhu et al., 2010; Ort et al., 2015). Increasingly horizontal leaves from the top to the bottom of the canopy is one of the fundamental components of this modeled plant architecture, recently called “smart canopy” (Ort et al., 2015). The other elements of a “smart canopy” include improved metabolic features such as a differential catalytic capacity of Rubisco, and contrasting antenna sizes and reaction center numbers across the canopy. While manipulating leaf angles might seem the most feasible alteration toward a “smart canopy” crop, the development of optimized architectures will require the investigation of the natural leaf angle distribution pattern, and knowledge about the genetic control of this trait at each canopy layer.
Numerous studies have been conducted to discover the genetic control of leaf angle in cereals (Moreno et al., 1997; Li et al., 1999; Hart et al., 2001; Mickelson et al., 2002; Liu et al., 2007; Ku et al., 2010, 2011; Tian et al., 2011; Zhao et al., 2016; Mantilla-Perez and Salas-Fernandez, 2017). A detailed review of those experimental designs revealed the underlying assumption of a common genetic control of angle throughout the plant (Mantilla-Perez and Salas-Fernandez, 2017). Therefore, either the flag or the preflag (PFL) leaf, the leaf above or below the ear in maize (Zea mays), or a randomly selected leaf, was the unit of investigation. In some cases, the “overall canopy” was qualitatively characterized, or angles of multiple leaves averaged to perform quantitative analyses. The well-known effects of auxins, gibberellins, and brassinosteroids (BRs) on leaf angle were discovered using similar experiments based on the observation of a limited number of leaves (Yamamuro et al., 2000; Morinaka et al., 2006; Sakamoto et al., 2006; Shimada et al., 2006; Divi and Krishna, 2009; Tong et al., 2014; Sun et al., 2015; Truong et al., 2015; Best et al., 2016; Feng et al., 2016; Mantilla-Perez and Salas-Fernandez, 2017). While these investigations unraveled the hormonal mechanisms affecting leaf angle, they do not provide evidence of the possible differential effects of hormone-related genes across the canopy.
Despite recent advances to exploit high-throughput phenotyping (HTP) technologies, the natural variation of leaf angle within and between canopy layers remains poorly described. Under field conditions, the estimation of angle of individual leaves presents challenges imposed by overlapping canopies, variable environmental conditions (e.g. wind), and the orientation of leaves relative to the phenotyping device. Therefore, most HTP attempts to characterize this trait were performed under controlled conditions on single potted plants (Cabrera-Bosquet et al., 2016; Duan et al., 2016; McCormick et al., 2016; Zhang et al., 2017). These investigations did not evaluate the entire canopy under field density conditions, or were conducted only at the seedling stage, or angles of individual leaves were averaged to conduct quantitative genetic analyses.
Here, first, we utilized sorghum (Sorghum bicolor) as both a model grass species and an economically important crop to first characterize the natural distribution of leaf angles throughout the canopy, and we discovered that the orientation of leaves does not follow the proposed “smart canopy” ideotype. Second, we utilized HTP methods to identify genomic regions controlling leaf angle at different canopy layers, which were validated using three biparental mapping populations. These quantitative genetic approaches facilitated the discovery of two types of quantitative trait loci (QTL)—those controlling leaf angles across the canopy, and those controlling the trait only at specific leaves or canopy layers. Third, we investigated the leaf-specific expression of two genes known to control angle in sorghum: the BR-related transcription factor BRASSINAZOLE RESISTANT 1/BRI1-EMS-SUPPRESSOR1 (BZR1/BES1; Mantilla-Perez et al., 2014; Mantilla-Perez and Salas-Fernandez, 2017) and the auxin transporter Dwarf3 (Dw3; Truong et al., 2015; Mantilla-Perez and Salas-Fernandez, 2017). The leaf-specific transcription levels of both genes were highly correlated with the distribution of angles across canopy layers. Finally, we applied the BR biosynthesis inhibitor propiconazole (pcz) to evaluate its potential effect across the canopy. Even though angles decreased, the treatment failed to revert the undesirable natural distribution of angles across the canopy. In combination, these discoveries fill important knowledge gaps about the genetic mechanisms underlying this complex trait at multiple levels of the canopy. While the differential manipulation of leaf angles across the plant might appear as the most achievable alteration of the “smart canopy” characteristics, this study reveals the importance of developing a breeding strategy, including phenotyping, focused on angle distribution across the canopy. The exploitation of layer-specific QTL/genes, such as those reported herein, will be instrumental to reverse the natural angle distribution in sorghum according to the proposed ideotype.
RESULTS
Natural Leaf Angle Distribution Throughout the Canopy Is Not Optimized in Sorghum
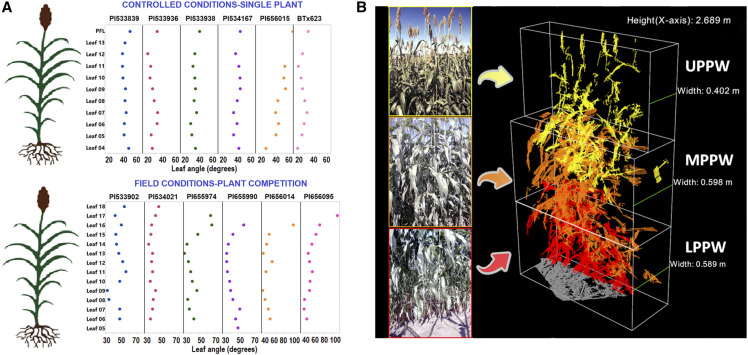
Six sorghum accessions with contrasting angles at the PFL leaf, as previously determined (Mantilla-Perez et al., 2014), were manually evaluated under controlled conditions to characterize this trait at all individual leaves. This investigation revealed consistent patterns that were contrary to the proposed ideotype. No linear trend was observed in leaf angle variation across the plant; the flag leaf was the most horizontal, and angles increased from the middle to the upper canopy (Fig. 1A). Under field conditions with plants grown at commercial planting densities and high competition, the analysis of another set of six accessions with variable plant architecture traits (Zhao et al., 2016; Salas-Fernandez et al., 2017) confirmed the previously observed pattern throughout the canopy (Fig. 1A). At larger scale, a natural angle distribution contrary to the proposed ideotype was also demonstrated by the phenotypic data collected on three individual leaves (PFL, leaf 4 [L4], and leaf 5 [L5] counting from the top after flowering) in three biparental recombinant inbred line (RIL) populations (Supplemental Table S1). The angle of leaves increased from middle (L5) to upper canopy layers (PFL), similarly to the detailed phenotypes obtained in the subset of lines (Supplemental Table S2). Therefore, there are multiple sources of evidence to conclude that the distribution of leaf angles across the sorghum canopy does not follow the proposed model of more erect leaves on the top, with gradually increasing angles toward the lower layers. The implementation of effective breeding or genetic engineering/editing strategies to redesign and manipulate this natural architecture will be dependent on a thorough understanding of the genetic control of this trait across the canopy.
Figure 1.
Leaf angle distribution throughout the canopy and estimation of plot-based plant width (PPW) as a proxy for leaf angle. A, Natural leaf angle distribution throughout the canopy on diverse sorghum genotypes under field and controlled conditions. B, Original red-green-blue (RGB) images and derived 3D reconstruction of a sorghum plot with the independent estimation of PPW for each one-third of the canopy. Different colors indicate point clouds from each of the three sets of stereo cameras on the automated platform Phenobot 1.0 (Salas-Fernandez et al., 2017).
Leaf Angle Across Canopy Layers Is Controlled by a Common Set of Genomic Regions
The common genetic control of leaf angle across canopy layers was independently demonstrated by the results obtained from a linkage mapping analysis of three biparental populations, and a genome-wide association study (GWAS) using the sorghum association panel (SAP) of 342 accessions (Casa et al., 2008). Field image-based HTP was utilized to characterize the SAP for PPW at three canopy layers (Fig. 1B). This novel imaged-derived descriptor has been previously demonstrated to capture variation in leaf angle (Bao et al., 2018; Breitzman et al., 2019), and its value as a proxy for this trait has been confirmed herein (Fig. 1B; Supplemental Fig. S1). For linkage mapping analysis, manual measurements of leaf angle were performed after flowering at the PFL, L4, and L5 counting from the top, representing the upper (PFL) and middle canopy levels (L4 and L5). The three RIL populations were selected/developed based on the contrasting parental phenotypes for our target trait, and their segregating haplotypes on chromosome 7, previously associated with leaf angle variation (Supplemental Table S1; Zhao et al., 2016). The validation of the newly discovered common determinants of leaf inclination was accomplished by a comparative analysis between mapping studies in which coincident markers/genomic regions were identified.
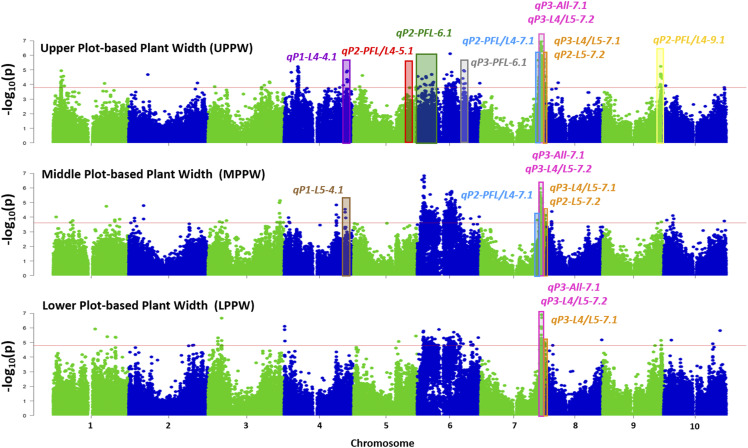
According to GWAS results, a common set of 31 single nucleotide polymorphisms (SNP) on chromosomes 6 and 7 were significantly associated with PPW at all canopy levels (upper plot-based plant width [UPPW], middle plot-based plant width [MPPW], and lower plot-based plant width [LPPW]; Fig. 2; Supplemental Table S3). These markers, which represent only 3.07% of the total number of significant polymorphisms, explained between 5.14% and 11.89% of the phenotypic variation. Additionally, the allelic effects had a consistent direction in the change of PPW across canopy layers. For example, allele G at S6_30802725 increased PPW at all levels (UPPW, MPPW, and LPPW) relative to allele C, but the magnitude of the effect varied depending on the canopy layer. In general, there was a larger effect on the upper and lower canopy levels (UPPW and LPPW) than the middle (MPPW).
Figure 2.
Genome-wide associations for UPPW, MPPW, LPPW, and validated regions by coincident QTL discovered using three independent biparental populations. Validated regions are indicated by color-shaded areas. Colors indicate the QTL identifiers with a physical position coincident with an associated region by GWAS.
When only two canopy layers were compared, 87 and 91 markers explained the phenotypic variation at adjacent levels MPPW/LPPW and UPPW/MPPW, respectively (Supplemental Tables S4 and S5). Common marker associations between the lower and upper levels were limited to only 0.29% of the significant SNPs (S7_57889967, S7_58540584, and S7_59428633). These results are consistent with the lower correlation observed between UPPW and LPPW (r = 0.16) relative to the other pairwise comparisons (rUPPW-MPPW = 0.61 and rMPPW-LPPW = 0.50).
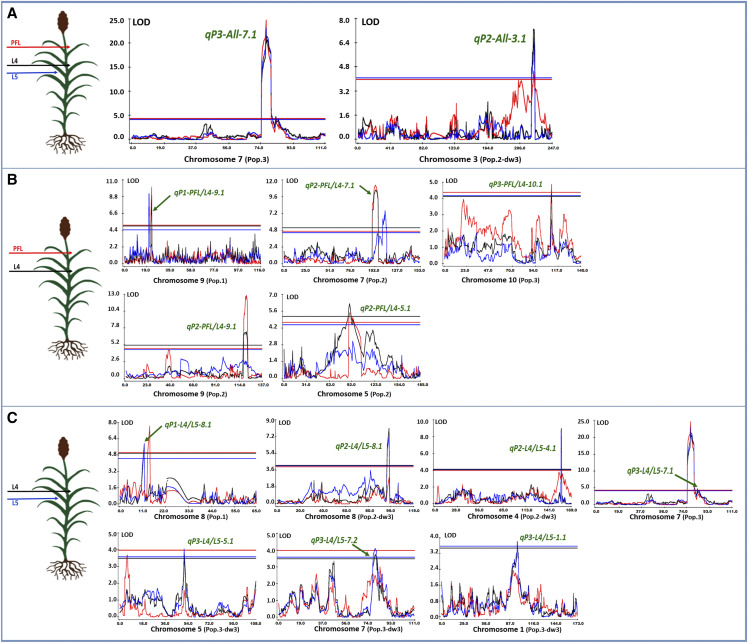
The control of angle by common genetic determinants across canopy layers was confirmed by the QTL results from two biparental populations (Pop. 2 and Pop. 3), in which the same genomic regions were identified for the top (PFL) and middle canopy levels (L4 and L5). In Pop. 3, derived from BTx623 and IS3620C, qP3-All-7.1 was consistently discovered for all leaves (PFL, L4, and L5), explaining a large percentage of angle variation (Fig. 3; Supplemental Table S6). This 205-Kb chromosomal interval (58573749–58779394) contains the auxin transporter gene Dw3 at position 58610248 to 58618253 (genome v1), which has been previously related to angle control of a single leaf (Truong et al., 2015). The second common QTL across the canopy (qP2-All-3.1) was identified on chromosome 3 using the Tx430 × P898012 RIL population (Pop. 2) and Dw3 as a covariate in the analysis (Fig. 3; Supplemental Table S6).
Figure 3.
Common QTL across leaves controlling leaf angle in three biparental populations (Pop.1, Pop.2, and Pop.3). A to C, Sections correspond to the QTL-controlling angle of PFL, L4, and L5 (A), PFL and L4 (B), and L4 and L5 (C).
Five loci on chromosomes 9, 5, 7, and 10 were simultaneously associated with angles of the PFL and L4 (Fig. 3; Supplemental Table S7). Even though these genomic regions were significant across canopy layers, there was no evidence to suggest they also control L5, physically close to L4. In addition to the across-layer QTL (either for all leaves or PFL and L4), there were seven genomic intervals affecting angle variation in both L4 and L5, representing only the middle canopy level (Fig. 3; Supplemental Table S8).
From these common genomic regions, it was evident that, as observed in the GWAS, any particular QTL exerted its effect with a consistent direction for all leaves. In general, while the magnitude of the effect decreased from the top (PFL) to lower layers (L5), the percentage of the phenotypic variation explained for each leaf was similar (Supplemental Tables S6–S8).
The comparison between biparental QTL and GWAS experiments provided an independent validation of these common chromosomal intervals determining leaf angle across canopy layers. Seven out of the 16 SNPs on chromosome 7 significantly associated with all PPW layers (UPPW, MPPW, and LPPW) were encompassed within qP3-All-7.1 (Fig. 2; Supplemental Table S6). Additionally, GWAS markers associated with UPPW and MPPW colocalized with qP2-PFL/L4-5.1, qP2-PFL/L4-7.1, and qP2-PFL/L4-9.1, which control our target trait across PFL and L4 (Fig. 2; Supplemental Table S7), providing another independent validation of the existence of an across-canopy genetic control of leaf angle.
Layer-Specific Leaf Angle Is Determined by Distinct Genes/QTL
The GWAS facilitated the discovery of distinct regions controlling PPW at different levels of the canopy, suggesting that an independent control of angle at each layer also exists (Fig. 2). A total of 269, 362, and 164 SNPs were exclusively associated with variation in either UPPW, MPPW, or LPPW, respectively (Supplemental Table S9). Interestingly, the proportions of the phenotypic variance explained by these markers (maximum 13.7%), localized in all chromosomes, were within the ranges observed for markers associated across canopy layers (Supplemental Table S9).
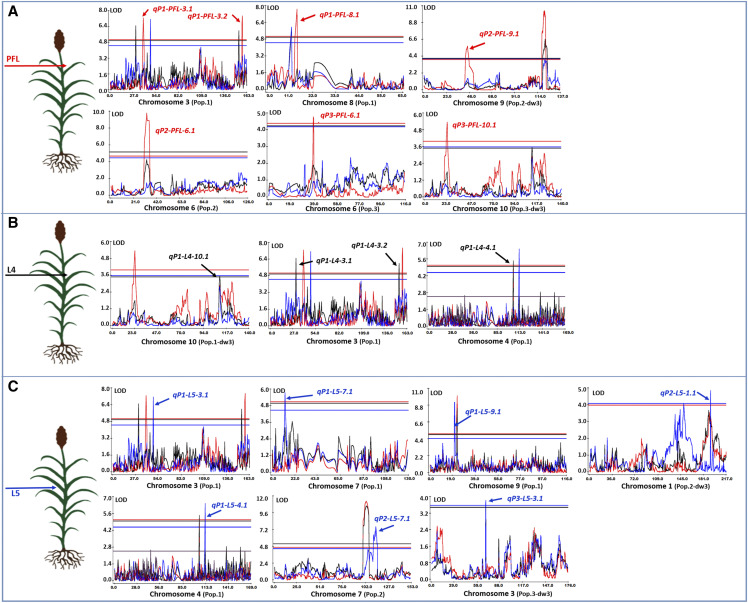
Leaf-specific QTL were also identified in the three biparental populations tested under field conditions. The smallest number of loci (four) were associated with angle variation in L4, while seven chromosomal intervals were exclusively controlling the PFL and L5 (Fig. 4; Supplemental Tables S10–S12). The percentage of angle variation explained by leaf-specific QTL ranged from 1.5% to 11.1%, and there was at least one locus for each leaf accounting for >10% of the variation. Acknowledging that effects are population-specific and might change when introduced into novel backgrounds, it is encouraging to discover that these leaf-specific QTL are not restricted to small effect loci with limited chances to impose a change in the overall canopy structure.
Figure 4.
Leaf-specific QTL controlling leaf angle in three biparental populations (Pop.1, Pop.2, and Pop.3). A to C, Sections correspond to QTL specific for PFL (A), L4 (B), and L5 (C). “Pop.X-dw3” indicates QTL discovered using the Dw3 gene as a covariate in the analysis.
A comparative analysis, similar to the one conducted to validate common regions across canopy layers, was performed between results from HTP-characterized specific levels and those of individual leaves. Five QTL from biparental populations (qP2-PFL-6.1, qP3-PFL-6.1, qP1-L4-4.1, qP1-L5-4.1, and qP2-L5-7.2) validated markers associated with UPPW or MPPW (Fig. 2; Supplemental Tables S10–S12). This validation provides strong evidence of the leaf or layer-specific genetic control of leaf angle that could be exploited to manipulate angle distribution to achieve the desirable arrangement according to the “smart canopy” ideotype.
BZR1/BES1 and Dw3 Genes Control Leaf Angle Across the Canopy with Variable Effects at Each Layer
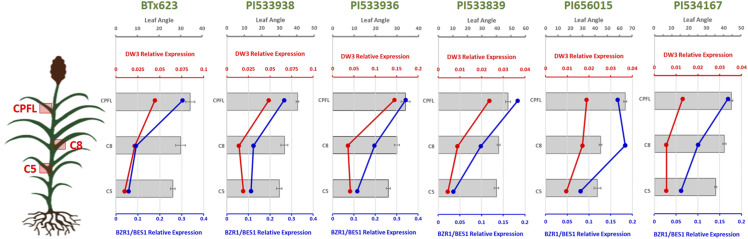
Dw3, a known leaf angle gene in sorghum (Truong et al., 2015), has been associated herein with trait variation at all canopy levels. Therefore, its expression profile in collar tissues from different canopy layers was investigated to determine the potential relationship between the leaf-specific transcription level and its corresponding angle. Similarly, BZR1/BES1 expression was monitored in the same tissues because previous studies in cereals have demonstrated that BR genes control overall leaf angle (Mantilla-Perez and Salas-Fernandez, 2017), and that, in sorghum, this gene is associated with angle variation in the PFL leaf (Mantilla-Perez et al., 2014). Using six diverse sorghum accessions (PI533839, PI534167, PI656015, PI533938, PI533936, and BTx623) with different across-canopy angle profiles (Fig. 1), the reverse transcription quantitative PCR (RT-qPCR) data obtained from collars of L5 (C5), L8 (C8), and collar of the PFL (CPFL; counting from the bottom) demonstrated that the expression of BZR1/BES1 was highly and positively correlated (r > 0.95) with leaf angle across the canopy, except for PI656015 (r = 0.45). As the expression of BZR1/BES1 increased from the C5, to the C8 and CPFL, the angle of the corresponding leaves also increased in all accessions (Fig. 5). The expression of Dw3 varied across canopy layers similarly to BZR1/BES1 in all genetic backgrounds (Fig. 5), as demonstrated by the high correlation (r > 0.73) observed in all accessions. Interestingly, the expression of Dw3 was higher in all collars of BTx623, PI533936, and PI533938, which harbor the dysfunctional dw3/dw3 haplotype that produces a mal-functioning auxin transporter (Multani et al., 2003; Truong et al., 2015). In line with the functional Dw3 haplotype (PI533839, PI656015, and PI534167), the across-canopy expression profile was similar to the one observed for the dysfunctional haplotype, although within a generally lower transcription level (Fig. 5). Therefore, these results clearly demonstrate that, while a single gene can control the target trait across the canopy, its increasing expression toward upper layers could explain, at least partly, the distribution of angles opposite to the proposed ideotype.
Figure 5.
Leaf angle and relative normalized expression of BZR1/BES1 and Dw3 by RT-qPCR in different genetic backgrounds under control conditions. Angles (gray bars) increase from L5 to the PFL leaf. The expression of BZR1/BES1 (blue) and Dw3 (red) also increase, as the plant develops, from the lower to the upper canopy. Error bars indicate se (n = 3).
Inhibiting BR Biosynthesis Caused Erect Canopies but Did Not Reverse the Undesirable Angle Distribution
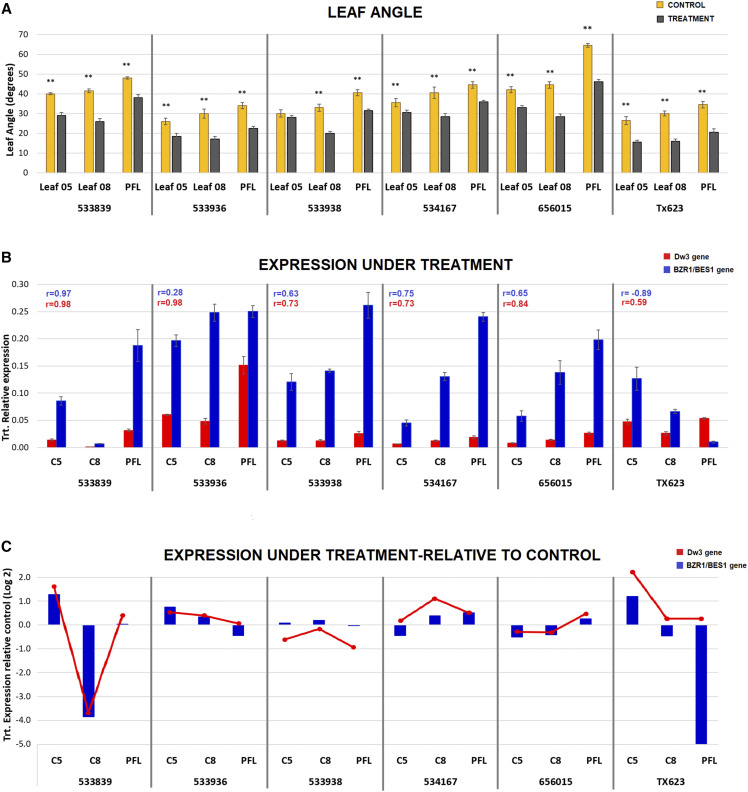
Even though there is abundant evidence of the overall reduction in leaf angle exerted by the decline in BR concentrations imposed by biosynthesis inhibitors (Mantilla-Perez and Salas-Fernandez, 2017), the potential use of this strategy to alter the angle distribution across the canopy is unknown. Therefore, the BR biosynthesis inhibitor pcz was applied to investigate the layer-specific effects of manipulating BR concentrations. After evaluating seven concentrations of pcz (0, 10, 20, 50, 100, 200, and 400 μm), 50 μm was selected as the optimal level to generate a maximum change in leaf angle throughout the canopy (Supplemental Fig. S2A), without severely affecting plant height (Supplemental Fig. S2B). At this concentration of pcz applied throughout development, plants of the same six diverse accessions produced more erect leaves but changed the distribution pattern of angle across the canopy (Fig. 6A; Supplemental Table S13). For instance, in four out of six accessions (PIs 533839, 533936, 656015, and 534167), L8 was among the two leaves with a maximum decrease in angle, while L9 to L12 were the most significantly affected leaves in BTx623 and PI 533938 (Supplemental Table S13). Therefore, the trend of increasing angle toward the upper canopy from L5 to L8 to PFL was altered to an arrangement in which the inclination of L8 was smaller than L5 in most cases (Fig. 6A). However, upon pcz application, the PFL remained the most horizontal leaf of the entire plant, as observed under control conditions. This experiment provided evidence of the leaf-specific sensitivity to the treatment, suggesting independent physiological and genetic determinants of angle across the canopy and at different developmental stages. Additionally, it was demonstrated that the undesirable horizontal orientation of leaves on the upper canopy could not be reverted by the general inhibition of BR biosynthesis.
Figure 6.
Phenotypic and gene expression response to the BR inhibitor pcz. A, Leaf angle under control and pcz treatment. Asterisks indicate significance by ANOVA (**P < 0.01). B, RT-qPCR expression profiles of BZR1/BES1 and Dw3 under treatment calculated as 2-ΔCT. Pearson correlation values were calculated between expression levels of each gene (indicated by colors) and the corresponding leaf angle after pcz treatment. C, RT-qPCR expression profiles of BZR1/BES1 and Dw3 under treatment relative to control calculated as 2−ΔΔCT. Negative values indicate lower expression than under control conditions. C5, C8, and CPFL (leaves counted from bottom to top). Trt., pcz treatment at 50 μm. Error bars indicate se (n = 3).
When BR biosynthesis was inhibited by pcz, the patterns of BZR1/BES1 and Dw3 expression across the canopy were modified, and their correlations with the leaf angle distribution were reduced in four out of the six accessions (Tx623, PI533936, PI533938, and PI534167; Fig. 6B). BZR1/BES1 expression was more substantially affected in an accession-dependent manner. For example, in Tx623 the expression level decreased from the C5 to the PFL, which rendered a negative correlation with leaf angle under treatment (r = −0.89). For PI533839, the observed associations between expression levels and leaf angle were still high, despite a clear decline in the angle of C8, because the transcript levels of both BZR1/BES1 and Dw3 also suffered a drastic decline in C8 (Fig. 6, A and B). Remarkably, when the expressions of both genes were plotted simultaneously and relative to control levels, their coincident profiles across tissues and development was evident (Fig. 6C). Even for Tx623 and PI533839, with a distinctive expression pattern, the crosstalk between BZR1/BES1 and Dw3 was demonstrated across the canopy (Fig. 6C). Despite some common overall trends in transcript levels across the canopy, treatment-induced changes in phenotypes and expression profiles demonstrated that accessions had different sensitivities to pcz at specific canopy layers/developmental stages.
DISCUSSION
Leaf angle is a trait of historical importance for yield enhancement in cereal crops. A better arrangement of leaves across the canopy directly affects yield by: improving light interception and conversion efficiency that translates into higher yields per plant (Lambert and Johnson, 1978; Murchie et al., 1999; Sinclair and Sheehy, 1999; Long et al., 2006; Zhu et al., 2008; Murchie and Niyogi, 2011; Truong et al., 2015); and increasing plant density for enhanced yield per unit of land (Lambert and Johnson, 1978; Duvick et al., 2004; Morinaka et al., 2006; Sakamoto et al., 2006; Fischer and Edmeades, 2010). Numerous studies have documented the impactful changes in leaf angle of maize germplasm over time (Pendleton et al., 1968; Duvick et al., 2004; Duvick, 2005; Lee and Tollenaar, 2007; Lauer et al., 2012). Breeding for increased plant densities from the 1930s to the 2000s was based on an indirect selection on leaf angle, generating germplasm with overall erect canopies, improved light interception efficiency and thus, grain yield (Duvick et al., 2004). The additional benefit of a differential distribution of leaf angle across the canopy has been experimentally confirmed in maize. Lines with high leaf area index (>3.0) had higher yields when the upper canopy was erect and the lower canopy had more horizontal leaves (Duncan, 1971; Tollenaar and Wu, 1999; Ku et al., 2010).
In sorghum, historical analyses of improvements in grain yield and its component traits are limited. Current breeding efforts seeking high-yielding grain sorghum lines have focused on reducing height and adjusting flowering to temperate regions (Thurber et al., 2013), with no emphasis on improving leaf angle distribution. Evaluation of historical hybrids demonstrated that, contrary to maize (Duvick et al., 2004), there have been no changes in leaf angle of sorghum germplasm generated by public breeding programs over time, while those released by the private sector slightly increased angles from 1961 to 2015 at a rate of 0.076° annually (Pfeiffer et al., 2019). Interestingly, the rate of genetic gain for grain yield was the same for both public and private programs (0.008 t ha−1), irrespective of the direction of change in leaf angle. This apparent disconnection between grain yield improvements and leaf angle changes suggests that planting density and/or row spacing, together with canopy architecture, could be further optimized to maximize light interception efficiency and yield per unit of land. This optimization should be environment-specific because sorghum is produced under diverse conditions, ranging from rich soils with nonlimited rainfall, to irrigated production systems, or dry environments with marginal soils. Despite the known yield compensation mechanisms, such as varying seed number per panicle and tillering (Berenguer and Faci, 2001; Lafarge and Hammer, 2002; Tolk and Schwartz, 2017), numerous studies have concluded that narrow rows and/or higher densities are associated with higher average yields (Blum, 1970; Myers and Foale, 1981; Gerik and Neely, 1987; Jones and Johnson, 1991; Whish et al., 2005). Therefore, we have a valuable opportunity to strategically design canopy ideotypes and set breeding goals to optimize light interception and photosynthetic conversion efficiency in sorghum, to improve yield per unit of land.
In addition to the historical analysis demonstrating that more erect sorghum canopies have not been developed (Russell, 1991; Pfeiffer et al., 2019), our research proved that the distribution of angles across the canopy is opposite to the proposed ideotype, i.e. the angle decreases from the flag leaf downward (Fig. 1; Supplemental Tables S2 and S13). In all three biparental populations, the PFL leaf exhibited a larger angle than L4 and L5 in the middle canopy. These findings clearly indicate that there is a need to reverse the leaf angle distribution throughout the sorghum canopy, if the proposed ideotype is considered optimal for the target environmental conditions. An initial step to accomplish that goal is to utilize quantitative genetics to identify genes/regions that independently control angle at different leaves/layers (Mantilla-Perez and Salas-Fernandez, 2017). We have implemented that strategy and discovered QTL that could be exploited in breeding programs to realize the ideotype. Loci controlling angles both throughout the canopy and only for specific leaves were discovered (Figs. 2 and 3; Supplemental Tables S3–S12), and this knowledge opens the opportunity to design and genetically manipulate individual layers for an optimized canopy with higher conversion efficiency.
HTP technologies are essential to characterize canopy structure and could be further exploited to segment and dissect the canopy at microscales. In this study, multiview stereo 3D imaging enabled 3D reconstruction of canopy architecture under field conditions. Plot-based canopy architecture traits were extracted at a finite number of height levels. For future studies, it may be possible to estimate light interception from 3D plant surface models (Cabrera‐Bosquet et al., 2016; Gaillard et al., 2020; Zhu et al., 2020). In addition to 3D imaging, integrating fluorescence imaging and thermal imaging could provide spatial estimations of photosynthesis and stomatal conductance, respectively (Vialet-Chabrand and Lawson, 2019; Herritt et al., 2020). Combining these different imaging systems into a compact camera module is expected to facilitate multimodal image registration. Furthermore, deep learning-based visual perception would be essential for segmenting individual structural components of plants (e.g. leaves, stems, and tassels) from the stereo imagery (Baweja et al., 2018; Xiang et al., 2020). In summary, multimodal imaging in conjunction with deep learning-based image analysis has great potential for mapping the spatial variation of physiological processes at the microscale canopy level.
Undoubtedly, the Dw3 containing region on chromosome 7 (qP3-All-7.1) is an important determinant of leaf angle in sorghum (Multani et al., 2003; Brown et al., 2008; Truong et al., 2015; Zhao et al., 2016) and, based on expression analysis, partly responsible for the undesirable angle distribution throughout the canopy. However, previous evidence of the existence of other angle-associated genes in this interval (Truong et al., 2015; Zhao et al., 2016) was corroborated in this study. The presence of the nearby loci qP3-L4/L5-7.1, qP2-PFL/L4-7.1, and pP3-L4/L5-7.2, even when Dw3 is used as a covariate (Figs. 2 and 3), indicates that dissecting this region and cloning the corresponding gene/s will be challenging but necessary for an effective manipulation of angle throughout the canopy.
Under control conditions, a gradient of BZR1/BES1 and Dw3 transcripts increased in collar tissues over development from the bottom to the top of the canopy. A similar gradient has been proposed for auxin concentration across sorghum plants based on functional analyses of Dw3 (Brown et al., 2008) that would explain its known effect on plant height. The model assumes that plants with a dysfunctional dw3/dw3 allele would not efficiently transport auxins from the shoot apical meristem to the base of the plant, affecting internode elongation of low and middle sections of the stem (Brown et al., 2008). We have generated knowledge about the functional characterization of Dw3 as a leaf angle determinant, because we discovered an increasing expression gradient from C5 to CPFL (Fig. 5), regardless of the Dw3 haplotype. Even though all lines had similar patterns, the relative expression was higher for those with the dysfunctional haplotype dw3/dw3 (0.01–0.15 versus 0.005–0.018). In model species, BZR1/BES1 have been characterized as transcription factors that activate numerous genes involved in cell elongation and other plant growth processes (Wang et al., 2002; Yin et al., 2002; Sun et al., 2010). However, there are no previous reports of BZR1/BES1 expression in collar tissue at different canopy layers or specific developmental stages. Our results demonstrate that, together with Dw3, the BZR1/BES1 expression gradient through the sorghum canopy (Fig. 5) explains, at least partly, the observed leaf angle distribution across the plant in all genetic backgrounds.
Considering the known effects of BR-related genes on overall leaf angle determination in cereals, we evaluated the potential effectiveness of a BR-inhibitor to manipulate angles at specific canopy layers. As expected, a reduction in angles was observed in all leaves but the final distribution did not correspond to the proposed ideotype, i.e. the PFL was still the most horizontal across the canopy (Fig. 6A; Supplemental Table S13). While BR-related genes have been successfully manipulated to reduce overall angles, increase planting density, and boost final yields per unit of land, altering BRs did not contribute to achieve an optimized arrangement of leaves throughout the canopy. Similarly, alternative alleles of Dw3, known to generate very contrasting height phenotypes, did not correlate with changes in the angle distribution across the canopy. Therefore, the leaf-specific markers/QTL discovered in this study are a valuable tool to engineer sorghum germplasm according to the proposed ideotype.
The numerous QTL, genes, or genomic regions associated with variation in leaf angle by forward or reverse genetic approaches in different species, have established a solid foundation for the genetic improvement of this complex trait (Mantilla-Perez and Salas-Fernandez, 2017). However, because most studies characterized one or two leaves but not at multiple levels of the canopy, the discoveries are not applicable for the differential manipulation of leaf inclination throughout the plant. This study aimed to perform a comprehensive characterization of leaf angle control in sorghum, shifting the research approach from a single leaf to the whole canopy. Considering that leaf angle has not been a target trait for improvements in sorghum, our results can be leveraged in multiple ways. Overall erect canopies can be generated by utilizing the genomic regions reported herein that consistently control angle across layers, and by manipulating the BR pathway. Additionally, we have generated knowledge (layer-specific QTL) to design efficient breeding/engineering/editing strategies to optimize canopy architecture according to the proposed “smart canopy” ideotype. Finally, the optimization of planting densities according to environments (marginal versus irrigated/nonlimiting) deserves further attention in sorghum, and the utilization of treatments like pcz offers an opportunity to experimentally test the impact of overall erect versus horizontal canopies on grain yield under contrasting environmental conditions and densities.
A better leaf angle distribution throughout the canopy of cereals is, with no doubt, one of the key tactics for increasing food, feed, fiber, and fuel production to an ever-growing population that relies on the progress of agriculture to provide these resources. In this sense, the knowledge generated in this study has established a foundation for the development of superior sorghum lines with an optimized inclination of leaves throughout the canopy, capable of maximizing light interception, conversion efficiency, photosynthetic capacity, and productivity per plant and unit of land.
MATERIALS AND METHODS
Populations
The SAP, consisting of 342 diverse sorghum (Sorghum bicolor) accessions, was grown in a randomized complete block design in two environments as described by Salas-Fernandez et al. (2017).
For QTL mapping, three biparental populations were selected based on haplotypes of parental lines for the region on chromosome 7 reported by Zhao et al. (2016; Supplemental Table S1). Population 1 consisted of 339 F2:5 lines generated using a modified single seed descent method from the cross of SC603 (PI533936) and SC558 (PI533938), and Population 2 included 242 RILs derived from Tx430 and P898012 (Li et al., 2015). Population 3 was a subset of 146 RILs derived from BTx623 and IS3620C, characterized for multiple plant architecture traits, including leaf angle (Hart et al., 2001; Truong et al., 2015; McCormick et al., 2016). The three biparental populations were grown in two environments (Boone, and Greenfield, Iowa) in RT-qPCR with two replications per location.
Phenotypes
Considering that leaf angle measurements are time-consuming and labor-intensive, the use of images obtained from the HTP platform Phenobot 1.0 (Salas-Fernandez et al., 2017; Bao et al., 2018) was one of the approaches utilized in this study to generate knowledge about this phenotype. Phenobot 1.0 is an autonomous navigation field-based platform equipped with stereo cameras (Salas-Fernandez et al., 2017). PPW was generated by reconstructing two-view stereo images into 3D point clouds using a 3D minimum spanning tree algorithm on Middlebury Stereo Evaluation 3.0 (Bao et al., 2018; Breitzman et al., 2019). An axis-aligned bounding box was extracted from the point cloud of each plot and was partitioned into 20 volume slices (Nslice) along the plot row direction. PPW was estimated as a weighted median of the Nslice considering the Z axis or width coordinate (Bao et al., 2018). This descriptor was first validated with ground-truth measurements using a set of six diverse sorghum accessions that were imaged using Phenobot 1.0 and manually measured to obtain angle data on the same day (Breitzman et al., 2019). The high correlation observed between average leaf angle across the canopy and PPW (r = 0.65; Supplemental Fig. S1) confirmed that this image-derived descriptor can be used as a proxy for leaf inclination in a quantitative genetic study. PPW was independently estimated for each third of the canopy height, generating the descriptors UPPW, MPPW, and LPPW (Fig. 1B). This set of image-derived layer-specific descriptors was extracted for the SAP (342 diverse accessions; Casa et al., 2008) planted in two locations in 2014 (Breitzman et al., 2019).
In biparental populations, leaf angle was measured as the inclination between the midrib of the leaf blade and the stem. The PFL, L4, and L5, counting from the flag leaf, were measured from three randomly-selected plants per plot at flowering time. These leaves are representative of the upper and middle canopy levels.
Genotyping
The public genotypic datasets for SAP, Pop 2, and Pop 3 were obtained using genotyping by sequencing (GBS Technologies), and included 265,000 SNPs (Morris et al., 2013), 8,961 SNPs (Li et al., 2015), and 10,389 polymorphisms (SNPs and Indels; Truong et al., 2015), respectively. Pop 1 was genotyped using sequence-based genotyping technology at the University of Minnesota Genomics Center. After imputation of missing calls, and filtering for <30% missing data, a total of 11,817 SNPs was retained for QTL analysis. The 882-bp tandem duplication of the dysfunctional allele dw3 was genotyped in individuals of Pop 3 and SAP, because this gene has been previously associated with leaf angle variation (Truong et al., 2015).
Statistical Analysis
The following linear model was used to obtain the variance components and test the effect of location, replication within location, genotype, and genotype by location interaction using PROC MIX (SAS Institute):
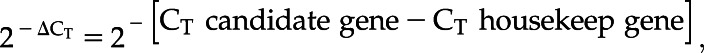 |
where Yijk is the response variable, μ is the overall mean, Li is the location effect, R(i)j is the replication nested within the location, Gk is the genotype (accession or RIL) effect, LGik is the effect of the interaction between the location and the genotype, and ε(i)jk is the residual. All factors in the model were treated as random variables. For genotypes, best linear unbiased predictions were estimated and used as the phenotype of individuals.
Genetic Map and QTL Analysis
Genetic maps were created using ICIMapping (Meng et al., 2015) and the Kosambi map function with no more than 30% of missing data (Supplemental Table S14). The QTL analysis was performed using composite interval mapping in QTL Cartographer 2.5 (Wang et al., 2012), with a window size of 2 m, and a walk speed of 1 m. In cases of QTL in close proximity, the window size was reduced and/or the number of background markers was increased. The tandem duplication of Dw3 was scored in individuals of Pop.2 and Pop.3 and used as covariate in the analysis. The significance threshold was obtained by 1,000 permutations.
GWAS
GWAS analysis was performed using TASSEL 5.2.12 (Bradbury et al., 2007) and the Mixed Linear model that accounts for population structure (Q, fixed effect) and kinship (K, random effect) to minimize spurious associations (Zhang et al., 2010). Q and K were calculated using STRUCTURE 2.2.3 (Pritchard et al., 2000) and SPAGeDi 1.4 (Hardey and Vekemans, 2002) respectively, as reported by Mantilla-Perez et al. (2014). Markers with >40% missing calls and a minimum allele frequency <5% were discarded before analysis (Zhao et al., 2016). SNPs developed for a priori BRs, gibberellic acid, photosynthesis, and photo-protection candidate genes (Mantilla-Perez et al., 2014; Zhao et al., 2016; Ortiz et al., 2017) were added to the public dataset (http://www.morrislab.org/data), for a final total of 134,600 markers. False discovery rate was used to account for multiple comparisons using the R package “qvalue” (Storey, 2002).
Gene Expression Analysis
Seven concentrations of pcz (0, 10, 20, 50, 100, 200, and 400 μm) were tested on BTx623 to determine the best treatment to induce changes in leaf angle (Supplemental Fig.S2). Once the pcz concentration was defined, six lines from the SAP (BTx623, PI 533839, PI 533936, PI 533938, PI 656015, and PI 534167) were selected based on two criteria: lines with different haplotypes for the associated BR-candidate genes (Mantilla-Perez et al., 2014); and lines with different haplotypes for Dw3 (Supplemental Table S13). Twelve plants per genotype were tested: six received the weekly pcz treatment (50 μm), and six were used as controls (distilled water as mock treatment). Each plant received 500 mL of distilled water with the corresponding treatment (pcz) once per week during the entire growth cycle, and additional 500 mL of distilled water 3 d later. Leaf angle was measured weekly, 24 h after treatment, from the three-leaf stage to grain filling.
C5, C8, and PFL leaves were utilized to extract RNA, conduct complementary DNA synthesis, and RT-qPCR. Primers (Supplemental Table S15) for the sorghum genes BZR1/BES1, Dw3, and the housekeeping gene Ser/Thr protein phosphatase 2A (Sudhakar Reddy et al., 2016) were utilized to perform RT-qPCR on two biological replications and three experimental replications for each treatment/control and sorghum accession (Supplemental Figs. S3 to S5). Normalized individual data points were calculated as
because the samples of each sorghum accession were different and no accession could be used as a calibrator (Schmittgen and Livak, 2008). Under treatment conditions, relative gene expression was conducted to understand changes in expression due to treatment and the control conditions were used as calibrator. The methodology used was 2−ΔΔCT (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008), according to the following calculation:
“Candidate gene” refers to either BZR1/BES1 or Dw3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Ground-truth validation of image-derived PPW relative to leaf angle.
Supplemental Figure S2. Effects of six pcz concentrations on leaf angle and plant height.
Supplemental Figure S3. RT-qPCR of five 10-fold dilution series, melting curves, and reaction efficiency calculation for Ser/Thr protein phosphatase 2A, used as the housekeeping gene.
Supplemental Figure S4. RT-qPCR of five 10-fold dilution series, melting curves, and reaction efficiency calculation for Dw3.
Supplemental Figure S5. RT-qPCR of five 10-fold dilution series, melting curves, and reaction efficiency calculation for BZR1/BES1.
Supplemental Table S1. Description of biparental populations used in QTL mapping studies.
Supplemental Table S2. Descriptive statistics of the three leaf angle measurements in biparental populations, showing a clear trend of increasing angles from middle to upper canopy.
Supplemental Table S3. Genome-wide association results in which the same markers were significant for upper, middle, and lower canopy layers described in millimeters as UPPW, MPPW, and LPPW.
Supplemental Table S4. Genome-wide association results in which the same markers were significant for middle and lower canopy layers described in millimeters as MPPW and LPPW.
Supplemental Table S5. Genome-wide association results in which the same markers were significant for upper and middle canopy layers described in millimeters as UPPW and MPPW.
Supplemental Table S6. Common QTL across all leaves (PFL, L4, and L5) and coincident GWAS markers for PPW at more than one layer.
Supplemental Table S7. Common QTL across PFL and L4, and coincident GWAS markers for PPW.
Supplemental Table S8. Common QTL across L4 and L5, and coincident GWAS markers for PPW.
Supplemental Table S9. GWAS results in which markers were significant only for a single canopy layer described in millimeters as UPPW, MPPW, and LPPW.
Supplemental Table S10. Unique QTL controlling only PFL, and coincident GWAS markers only associated with UPPW.
Supplemental Table S11. Unique QTL controlling only L4, and coincident GWAS markers for PPW.
Supplemental Table S12. Unique QTL controlling L5 only, and coincident GWAS markers for PPW.
Supplemental Table S13. Changes in leaf angle in response to 50 μM of pcz treatment.
Supplemental Table S14. Total number of markers, BINs, and genetic distance for each biparental population.
Supplemental Table S15. Candidate genes used for expression analysis and their correlation with leaf angle across the canopy.
Acknowledgments
We thank Lisa Coffey (Schnable Lab) and Nicole Lindsey (Salas-Fernandez Lab) for their assistance designing and conducting the sorghum field experiments with Phenobot 1.0. We also acknowledge Virginia Arruti, Juan Panelo, Facundo Curin, Joshua Kemp, and Ezequiel Delfino (Iowa State University) for their help with the manual phenotypic data collection. Matthew Breitzman (Iowa State University) contributed to the ground-truth data collection of leaf angle on a subset of the SAP. Finally, we acknowledge Dr. Jianming Yu (Iowa State University) for contributing Population 2, its genotypic data, and access to the corresponding field plots.
Footnotes
This work was supported by the U.S. Department of Agriculture’s National Institute of Food and Agriculture (grant nos. IOW04314 to M.G.S.-F., and 2012–67009–19713), the Plant Sciences Institute at Iowa State University, and the R.F. Baker Center for Plant Breeding at Iowa State University.
Articles can be viewed without a subscription.
References
- Bao Y, Tang L, Breitzman MW, Salas-Fernandez MG, Schnable PS(2018) Field-based robotics phenotyping of sorghum plant architecture using stereo vision. J Field Robot 36: 397–415 [Google Scholar]
- Baweja HS, Parhar T, Mirbod O, Nuske S (2018) Stalknet: A deep learning pipeline for high-throughput measurement of plant stalk count and stalk width. In Hutter M, Siegwart R, eds., Field and Service Robotics. Springer Proceedings in Advanced Robotics, Vol. 5. Springer, Cham, Switzerland, pp 271–284 [Google Scholar]
- Berenguer MJ, Faci JM(2001) Sorghum (Sorghum bicolor L. Moench) yield compensation processes under different plant densities and variable water supply. Eur J Agron 15: 43–55 [Google Scholar]
- Best NB, Hartwig T, Budka J, Fujioka S, Johal G, Schulz B, Dilkes BP(2016) Nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol 171: 2633–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A.(1970) Effect of plant density and growth duration on grain sorghum yield under limited water supply. Agron J 62: 333–336 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES(2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Breitzman MW, Bao Y, Tang L, Schnable PS, Salas-Fernandez MG(2019) Linkage disequilibrium mapping of high-throughput image-derived descriptors of plant architecture traits under field conditions. Field Crops Res 244: 107619 [Google Scholar]
- Brown PJ, Rooney WL, Franks C, Kresovich S(2008) Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180: 629–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Bosquet L, Fournier C, Brichet N, Welcker C, Suard B, Tardieu F(2016) High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol 212: 269–281 [DOI] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S(2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48: 30–40 [Google Scholar]
- Divi UK, Krishna P(2009) Brassinosteroid: A biotechnological target for enhancing crop yield and stress tolerance. N Biotechnol 26: 131–136 [DOI] [PubMed] [Google Scholar]
- Duan T, Chapman SC, Holland E, Rebetzke GJ, Guo Y, Zheng B(2016) Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J Exp Bot 67: 4523–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan WG.(1971) Leaf angles, leaf area, and canopy photosynthesis. Crop Sci 11: 482–485 [Google Scholar]
- Duvick DN.(2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50: 193–202 [Google Scholar]
- Duvick DN, Smith JSC, Cooper M(2004) Long-term selection in a commercial hybrid corn breeding program: past, present, and future. Plant Breed Rev 25: 109–151 [Google Scholar]
- Feng Z, Wu C, Wang C, Roh J, Zhang L, Chen J, Zhang S, Zhang H, Yang C, Hu J, et al. (2016) SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J Exp Bot 67: 4241–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RAT, Edmeades GO(2010) Breeding and cereal yield progress. Crop Sci 50: 85–98 [Google Scholar]
- Gaillard M, Miao C, Schnable J, Benes B (2020) Voxel carving based 3D reconstruction of sorghum identifies genetic determinants of radiation interception efficiency. bioRxiv, doi:10.1101/2020.04.06.028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik TJ, Neely CL(1987) Plant density effects on main culm and tiller development of grain sorghum. Crop Sci 27: 1225–1230 [Google Scholar]
- Hardey OJ, Vekemans X(2002) SPAGeDI: A versatile computer program to analyze spatial genetic structure at the individual of population levels. Mol Ecol Notes 2: 618–620 [Google Scholar]
- Hart GE, Schertz KF, Peng Y, Syed NH(2001) Genetic mapping of Sorghum bicolor (L.) Moench QTLs that control variation in tillering and other morphological characters. Theor Appl Genet 103: 1232–1242 [Google Scholar]
- Herritt MT, Pauli D, Mockler TC, Thompson AL(2020) Chlorophyll fluorescence imaging captures photochemical efficiency of grain sorghum (Sorghum bicolor) in a field setting. Plant Methods 16: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones OR, Johnson GL(1991) Row width and plant density effects on Texas high plains sorghum. J Prod Agric 4: 613–621 [Google Scholar]
- Ku L, Wei X, Zhang S, Zhang J, Guo S, Chen Y(2011) Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS One 6: e20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku LX, Zhao WM, Zhang J, Wu LC, Wang CL, Wang PA, Zhang WQ, Chen YH(2010) Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays L.). Theor Appl Genet 121: 951–959 [DOI] [PubMed] [Google Scholar]
- Lafarge TA, Hammer GL(2002) Tillering in grain sorghum over a wide range of population densities: Modelling dynamics of tiller fertility. Ann Bot 90: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R, Johnson R(1978) Leaf angle, tassel morphology, and the performance of maize hybrids. Crop Sci 18: 499–502 [Google Scholar]
- Lauer S, Hall BD, Mulaosmanovic E, Anderson SR, Nelson B, Smith S(2012) Morphological changes in parental lines of Pioneer brand maize hybrids in the U.S. central corn belt. Crop Sci 52: 1033–1043 [Google Scholar]
- Lee EA, Tollenaar M(2007) Physiological basis of successful breeding strategies for maize grain yield. Crop Sci 47: S202–S215 [Google Scholar]
- Li X, Li X, Fridman E, Tesso TT, Yu J(2015) Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc Natl Acad Sci USA 112: 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Paterson AH, Pinson SRM, Stansel JW(1999) RFLP facilitated analysis of tiller and leaf angles in rice (Oryza sativa L.). Euphytica 109: 79–84 [Google Scholar]
- Liu T, Zhang J, Wang M, Wang Z, Li G, Qu L, Wang G(2007) Expression and functional analysis of ZmDWF4, an ortholog of Arabidopsis DWF4 from maize (Zea mays L.). Plant Cell Rep 26: 2091–2099 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR(2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- Ma DL, Xie RZ, Niu XK, Li SK, Long HL, Liu YE(2014) Changes in the morphological traits of maize genotypes in China between the 1950s and 2000s. Eur J Agron 58: 1–10 [Google Scholar]
- Mantilla-Perez MB, Salas-Fernandez MG(2017) Differential manipulation of leaf angle throughout the canopy: Current status and prospects. J Exp Bot 68: 5699–5717 [DOI] [PubMed] [Google Scholar]
- Mantilla-Perez MB, Zhao J, Yin Y, Hu J, Salas-Fernandez MG(2014) Association mapping of brassinosteroid candidate genes and plant architecture in a diverse panel of Sorghum bicolor. Theor Appl Genet 127: 2645–2662 [DOI] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Mullet JE(2016) 3D Sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Li H, Zhang L, Wang J(2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3: 269–283 [Google Scholar]
- Mickelson SM, Stuber CS, Senior L, Kaeppler SM(2002) Quantitative trait loci controlling leaf and tassel traits in a B73xMO17 population of maize. Crop Sci 42: 1902–1909 [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M(1997) Liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Morinaka Y, Sakamoto T, Inukai Y, Agetsuma M, Kitano H, Ashikari M, Matsuoka M(2006) Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol 141: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS(2003) Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84 [DOI] [PubMed] [Google Scholar]
- Murchie EH, Chen Yz, Hubbart S, Peng S, Horton P(1999) Interactions between senescence and leaf orientation determine in situ patterns of photosynthesis and photoinhibition in field-grown rice. Plant Physiol 119: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Niyogi KK(2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RJK, Foale MA(1981) Row spacing and population density in grain sorghum-a simple analysis. Field Crops Res 4: 147–154 [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, Hu J, Salas-Fernandez MG(2017) Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J Exp Bot 68: 4545–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ(1968) Field investigations of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J 60: 422–424 [Google Scholar]
- Pfeiffer BK, Pietsch D, Schnell RW, Rooney WL(2019) Long-term selection in hybrid sorghum breeding programs. Crop Sci 59: 150–164 [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P(2000) Association mapping in structured populations. Am J Hum Genet 67: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WA.(1991) Genetic improvement of maize yields. Adv Agron 46: 245–298 [Google Scholar]
- Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al. (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24: 105–109 [DOI] [PubMed] [Google Scholar]
- Salas-Fernandez MG, Bao Y, Tang L, Schnable PS(2017) A high-throughput, field-based phenotyping technology for tall biomass crops. Plant Physiol 174: 2008–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ(2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M(2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE(1999) Erect leaves and photosynthesis in rice. Science 283: 1455 [Google Scholar]
- Storey JD.(2002) A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol 64: 479–498 [Google Scholar]
- Sudhakar Reddy P, Srinivas Reddy D, Sivasakthi K, Bhatnagar-Mathur P, Vadez V, Sharma KK(2016) Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front Plant Sci 7: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen D, Li X, Qiao S, Shi C, Li C, Shen H, Wang X(2015) Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev Cell 34: 220–228 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber CS, Ma JM, Higgins RH, Brown PJ(2013) Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Genome Biol 14: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES(2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Tolk JA, Schwartz RC(2017) Do more seeds per panicle improve grain sorghum yield? Crop Sci 57: 490–496 [Google Scholar]
- Tollenaar M, Wu J(1999) Yield improvement in temperate maize is attributed to greater stress tolerance. Crop Sci 39: 1597–1604 [Google Scholar]
- Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C(2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong SK, McCormick RF, Rooney WL, Mullet JE(2015) Harnessing genetic variation in leaf angle to increase productivity of Sorghum bicolor. Genetics 201: 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialet-Chabrand S, Lawson T(2019) Dynamic leaf energy balance: Deriving stomatal conductance from thermal imaging in a dynamic environment. J Exp Bot 70: 2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Basten J, Zeng ZB(2012) Windows QTL Cartographer 25. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Whish J, Butler G, Castor M, Cawthray S, Broad I, Carberry P, Hammer G, McLean G, Routley R, Yeates S(2005) Modelling the effects of row configuration on sorghum yield reliability in north-eastern Australia. Aust J Agric Res 56: 11–23 [Google Scholar]
- Xiang L, Tang L, Gai J, Wang L (2020) PhenoStereo: A high-throughput stereo vision system for field-based plant phenotyping-with an application in sorghum stem diameter estimation. Paper no. 2001190. 2020 ASABE Annual International Virtual Meeting. July 13-15, 2020. doi:10.13031/aim.202001190
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M(2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J(2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang C, Wu D, Qiao F, Li W, Duan L, Wang K, Xiao Y, Chen G, Liu Q, et al. (2017) High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol 173: 1554–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Mantilla Perez MB, Hu J, Salas-Fernandez MG(2016) Genome-wide association study for nine plant architecture traits in sorghum. Plant Genome 9: 1–14 [DOI] [PubMed] [Google Scholar]
- Zhu B, Liu F, Xie Z, Guo Y, Li B, Ma Y(2020) Quantification of light interception within image-based 3-D reconstruction of sole and intercropped canopies over the entire growth season. Ann Bot 126: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR(2010) Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol 61: 235–261 [DOI] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR(2008) What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr Opin Biotechnol 19: 153–159 [DOI] [PubMed] [Google Scholar]