Abstract
Malignant abdominal fluid (ascites) frequently develops in women with advanced high-grade serous ovarian cancer (HGSOC) and is associated with drug resistance and a poor prognosis1. To comprehensively characterize the HGSOC ascites ecosystem, we used single-cell RNA-seq (scRNA-seq) to profile ~11,000 cells from 22 ascites specimens from 11 HGSOC patients. We found significant inter-patient variability in the composition and functional programs of ascites cells, including immunomodulatory fibroblast sub-populations and dichotomous macrophage populations. We find that the previously described “immunoreactive” and “mesenchymal” subtypes of HGSOC, which have prognostic implications, reflect the abundance of immune infiltrates and fibroblasts rather than distinct subsets of malignant cells2. Malignant cell variability was partly explained by heterogeneous copy number alterations (CNA) patterns or expression of a stemness program. Malignant cells shared expression of inflammatory programs that were largely recapitulated in scRNA-seq of ~35,000 cells from additionally collected samples, including three ascites, two primary HGSOC tumors and three patient-ascites-derived xenograft models. Inhibition of the JAK/STAT-pathway, which was expressed in both malignant cells and CAFs, had potent anti-tumor activity in primary short-term cultures and PDX models. Our work contributes to resolving the HSGOC landscape3–5 and provides a resource for the development of novel therapeutic approaches.
Despite recent therapeutic advances, recurrent ovarian cancer is incurable and portends a poor prognosis with a median survival of approximately one year6. Intra-tumor heterogeneity of ovarian cancer cells and associated non-malignant cells are important factors in driving treatment resistance, but remain poorly understood. Genomic analysis of high-grade serous ovarian cancer (HGSOC), the most common and aggressive histological subtype, revealed TP53 mutations, defects in homologous recombination DNA repair and extensive copy-number aberrations in most tumors2, and classified HGSOC into four transcriptional subtypes with distinct prognoses2,7. Ascites, comprised of a diverse collection of cell types, is present in one-third of ovarian cancer patients at the time of diagnosis, and frequently occurs in patients with chemotherapy-resistant disease8. Here, we study ascites samples from HGSOC patients, primary tumors and patient-derived xenograft (PDX) models by single-cell RNA-seq (scRNA-seq), to resolve the expression profiles of diverse cancer, immune and stromal cells, and their interactions, each of which may contribute to disease development and treatment resistance9–12.
scRNA-Seq of the ascites ecosystem of high-grade serous ovarian cancer
We collected scRNA-seq data from three high-grade serous ovarian cancer (HGSOC) cohorts totaling 35,957 high-quality cell profiles from a set of 22 ascites samples from 11 patients and a validation set (gathered separately) of three additional ascites samples and two primary tumors (Supplementary Table 1), collected through a translational workflow9,13. Together, this data compendium spans 4 treatment naïve, 2 on-treatment during initial chemotherapy, 18 on-treatment during disease recurrence, and 2 post-neo-adjuvant chemotherapy specimens that reflects the substantial, real-life diversity among HGSOC patients. We used three complementary profiling strategies (Fig. 1a, Extended Fig. 1a, Supplementary Table 1). First, to obtain a broad view of the different cell types in the ascites ecosystem, we analyzed 8 specimens (that were partly depleted of CD45+ immune cells, Methods) from 6 patients by massively parallel droplet scRNA-seq (Cohort 1) (Methods). Second, because even after CD45+ cell depletion, droplet-based profiling had a relatively low proportion of malignant cells (7.9%; see below), we complemented it by isolating 1,297 viable malignant EPCAM+CD24+ cells, which identify cancer cells with high sensitivity and specificity14, from 14 ascites specimens from 6 individuals by fluorophore-activated cell sorting (FACS) into 96-well plates, followed by full length scRNA-seq using a modified SMART-seq2 protocol15,16 (Cohort 2) (Methods). Third, as a validation set to examine the generalizability of our results, we assembled three additional ascites specimens (10,688 cells) and two primary tumors (14,505 cells) from the Human Tumor Atlas Pilot Project (HTAPP) (Cohort 3)17.
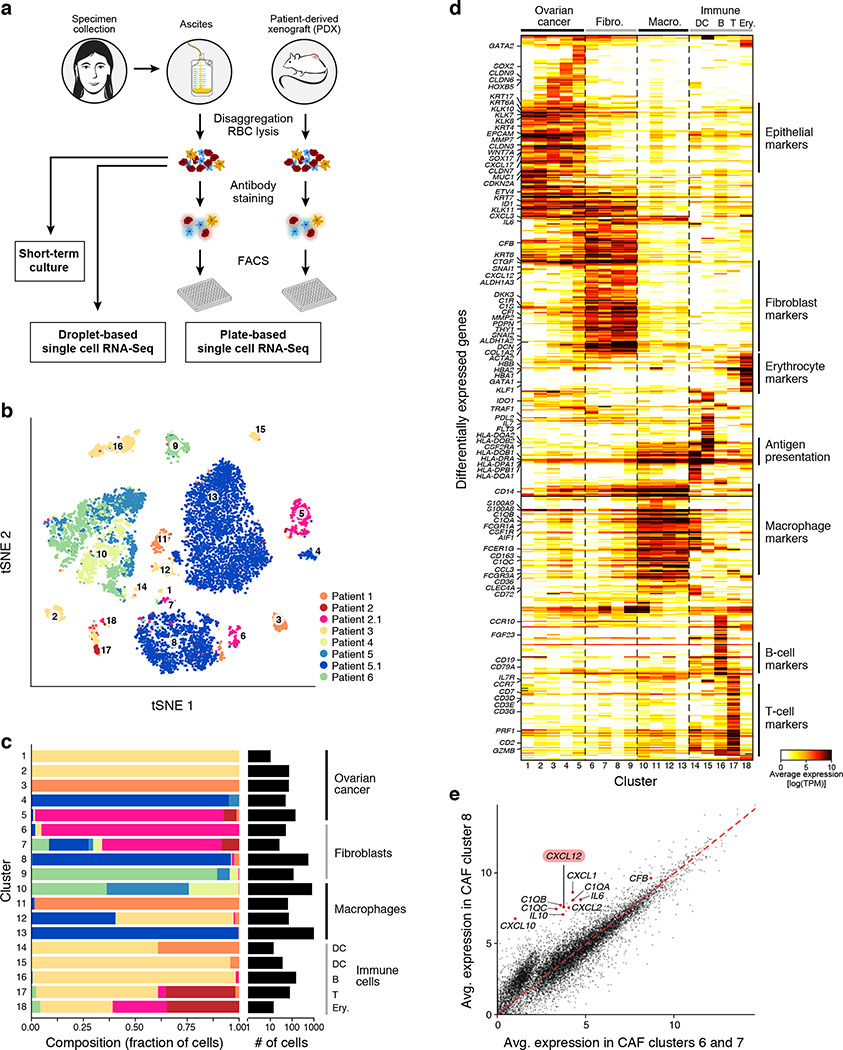
Figure 1. Charting the ovarian cancer ascites landscape by scRNA-seq.
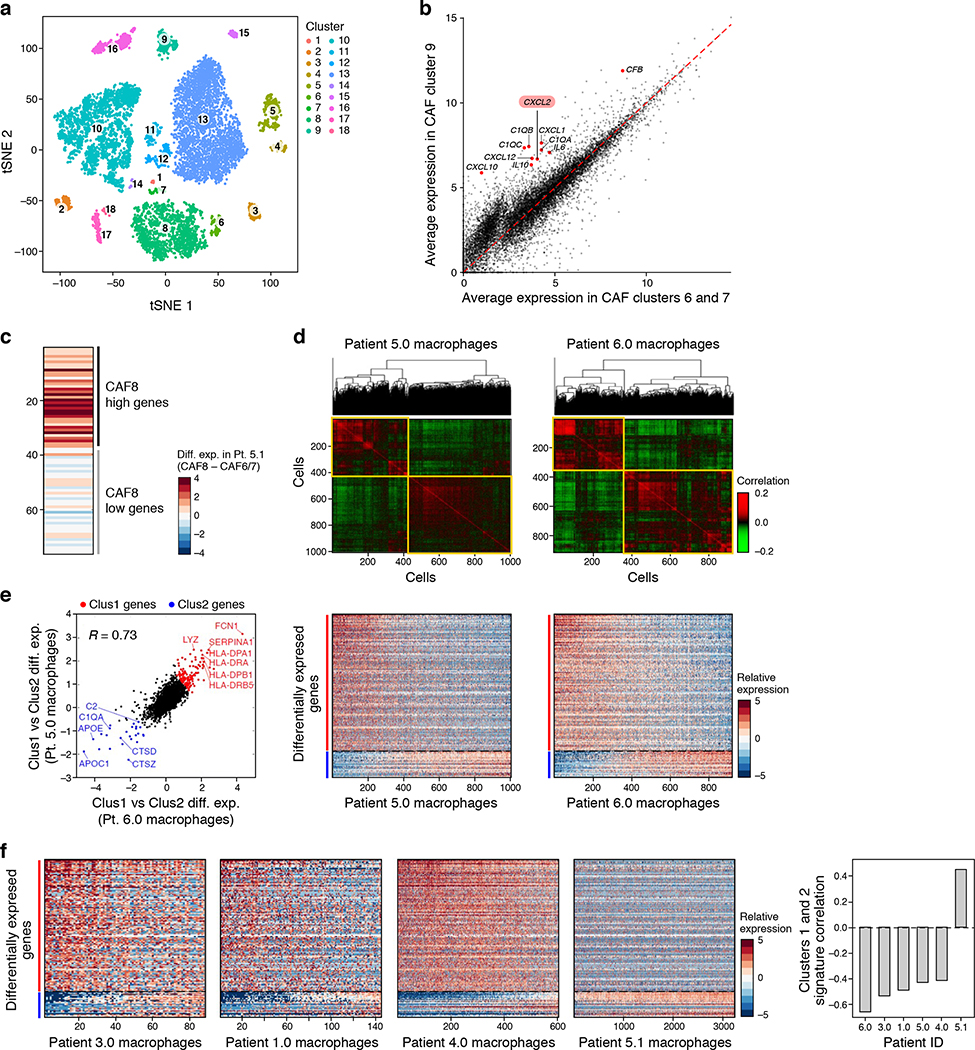
(a) Overview of sample collection and profiling strategy. (b,c) Malignant and non-malignant cell clusters in patient ascites by droplet-based scRNA-seq. (b) t-stochastic neighborhood embedding (tSNE) of 9,609 droplet-based scRNA-seq profiles from 8 samples, colored by sample-of-origin and numbered by unsupervised cluster assignment. (c) Fraction of cells (x axis) from each sample (color code, as in b) in each cluster (y axis). Clusters are labeled (right) by their post-hoc annotation based on differentially expressed genes (as in d). (d) Differentially expressed genes. Average expression (log2(TPM+1), color bar) of the top 30 genes (rows) that are differentially expressed in each cluster (columns). Genes are ordered by hierarchical clustering. (e) An inflammatory subset of CAFs. Comparison of the average expression (log2(TPM+1)) of each gene in CAF cluster 8 (y axis) vs. CAF clusters 6 and 7 (x axis). Red: immunomodulatory genes.
We first used the droplet based scRNA-seq (Cohort 1) to identify and annotate 18 distinct cell clusters spanning epithelial cells (5 clusters marked by EPCAM, cytokeratins, kallikreins), macrophages (4 clusters marked by CD14, AIF1, CSF1R, CD163), cancer-associated fibroblasts (CAFs) (4 clusters marked by PDPN, DCN and THY1), dendritic cells (2 clusters marked by CD1C, CD1E, CCR7, CD83), B cells (CD19, CD79A/B), T cells (CD2, CD3D/E/G) and erythrocytes (GATA1, hemoglobin) (Fig. 1b–d, Extended Fig. 2a, Supplementary Table 2, Methods). Although we depleted CD45+ cells, immune cells remained the most abundant component, comprising on average 67% of the cells in each sample (ranging 4–98%), highlighting a unique challenge of analyzing tumor cells from malignant effusions not usually encountered with solid tumor preparations17. The overall proportion of immune cells in each sample may reflect the differential efficiency of immune depletion (Methods), while other differences between samples may also be of biological origin (Fig. 1c). We examined whether different treatment histories explained differences in cellular ascites composition. Among cancer cells, there was significant inter-patient heterogeneity (Fig. 1b–d) which was not associated with prior treatment history (Extended Fig. 1b). Among CAFs and macrophages, expression profiles were similar across patients, indicating shared phenotypes, while the differences between patients were not clearly correlated with patient’s treatment histories (Extended Fig. 1c). Determining whether expression patterns are linked to prior treatment exposures will require larger patient cohorts.
Variation in CAFs and macrophage subsets within and across patients
Macrophages and CAFs each comprised four clusters (Fig. 1c,d), driven by both inter- and intra-patient variability. While some clusters were comprised almost entirely of cells from a single patient, most had cells from multiple patients (Fig. 1c), including fibroblast clusters composed of cells from three or more patients.
Among CAFs, we identified distinct cell states, including sub-populations with expression of immune-related genes, such as complement factors (C1QA/B/C, CFB), chemokines (CXCL1/2/10/12), and cytokines (IL6 and IL10) in clusters 8 and 9 compared to CAFs in clusters 6 and 7 (Fig. 1e, Extended Fig. 2b), suggesting a role as immunomodulatory CAFs18,19. This variation is also observed within a single patient: repeating the same analysis only with CAFs from Patient sample 5.1, recapitulates CAF sub-populations based on the differential expression of 80 genes (Extended Fig. 2c). Among other cytokines expressed by these fibroblast subsets, CXCL12 and IL6 activate JAK/STAT signaling across cancers20.
Among macrophages, Clusters 10 and 13 comprised the majority of cells. While most cells from Cluster 13 derived from one patient sample (5.1), Cluster 10 had cells from multiple patients. To examine intra-patient heterogeneity of macrophages, we focused on cells in cluster 10 from Patient samples 6 and 5.0, each separating into two sub-populations (Extended Fig. 2d, Groups 1 and 2), and characterized by consistent transcriptional programs. Group 1 cells co-expressed HLAs encoding for MHC Class II, IFNGR1 CD1D, CD36, and CD52, whereas Group 2 cells expressed complement factor components, cathepsins and APO genes (Extended Fig. 2e). Moreover, Group 1 cells expressed several genes identified as markers for M1 macrophages (IFNGR1, CD36, DDX5, MNDA) and as suppressors of M2 differentiation (C3AR1), while Group 2 cells expressed genes described in M2 macrophages, including those regulating M2 differentiation (e.g., AIF121, VISG422). While the M1/M2 dichotomy is currently being revised, this separation may indeed be associated with functional pro-/anti-tumor macrophage states. These findings generalized to samples with macrophages from all other patients (except Patient sample 5.1) (Extended Fig. 2f).
Inter- and intra-patient heterogeneity of malignant cells in HGSOC
To characterize the variation within the malignant compartment, we focused on the FACS-enriched, deeper-coverage full length scRNA-seq profiles (Cohort 2), and identified eight clusters (Fig. 2a,b), including six of epithelial cells, one of CAFs (cluster 7), and one of macrophages (cluster 8) (Fig. 2b,c, Extended Fig. 3). We confirmed that the epithelial cells were malignant by inferring chromosomal copy number alterations (CNAs)9 (Extended Fig. 4, Methods). In contrast to macrophages and CAFs, the malignant cells clustered by their patient of origin (Fig. 2a,b), highlighting inter-individual variation. Some of this variation reflects the distinct CNA profile of each tumor (Extended Fig. 4), but additional genetic and epigenetic effects likely contribute to inter-patient variability among malignant cells.
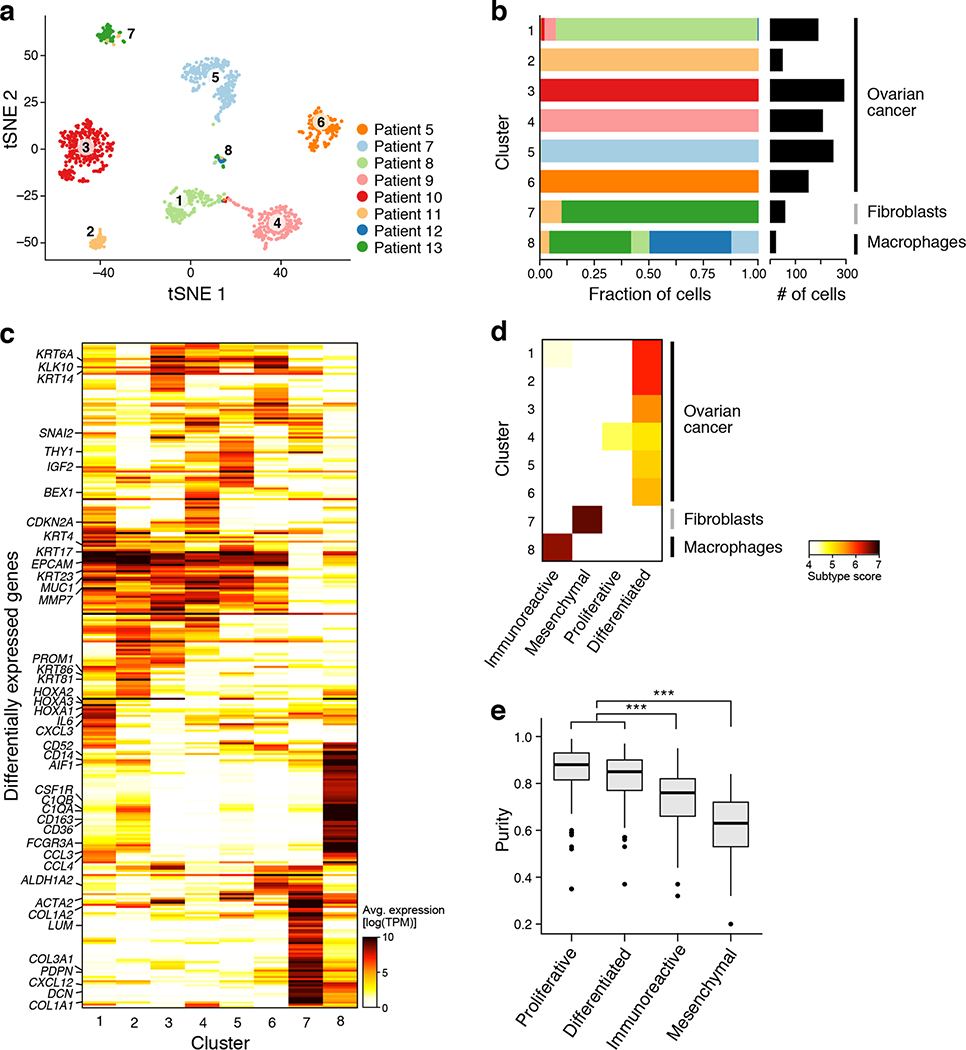
Figure 2. Malignant and non-malignant cell expression profiles help identify cellular basis of TCGA subtypes.
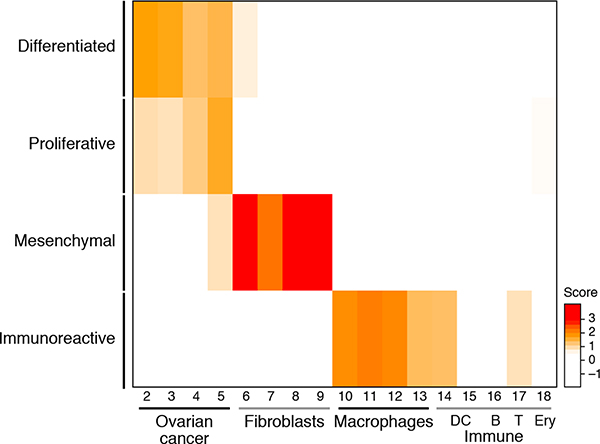
(a,b) Malignant cell clusters are enriched in patient ascites by FACS and plate-based scRNA-seq. (a) tSNE of 1,297 single cell profiles from 14 ascites samples profiled by plate-based scRNA-seq, colored and numbered by unsupervised cluster assignment. (b) Fraction of cells (x axis) from each sample (color code, as in a) in each cluster (y axis). Clusters are labeled (right) by their post-hoc annotation based on differentially expressed genes (as in c). (c) Differentially expressed genes. Average expression (log2(TPM+1), color bar) of the top 30 genes (rows) that are differentially expressed in each cluster (columns). Genes are ordered by hierarchical clustering. (d) The immunoreactive and mesenchymal subtypes reflect macrophages and fibroblast components. Subtype score (color bar), based on average expression of subtype-specific genes (Methods) of each clusters (rows) for each of four TCGA subtypes (column). (e) Immunoreactive and mesenchymal TCGA subtypes have lower overall purity than differentiated and proliferative. Distribution of a purity estimate value (y axis, ABSOLUTE41; Methods) for TCGA ovarian cancer tumors (n=282) in each subtype (x axis). Horizontal bar: mean; box: interquartile range, whiskers: minimum and maximum. Dots: outliers. ***p <10−10 (two-sided t test).
The Cancer Genome Atlas (TCGA) mesenchymal and immunoreactive subtypes reflect non-malignant contributions
We next asked whether inter-patient variability among malignant cells is consistent with the previously described TCGA subtypes - differentiated, proliferative, mesenchymal and immunoreactive – that have been derived from RNA expression profiling of bulk solid tumors from untreated HGSOC patients and have been associated with varying prognoses2. We thus scored each cluster for the expression of TCGA-derived subtype signatures (Methods).
All six malignant cell clusters highly expressed the “differentiated” signature and only one (cluster 4) strongly expressed the “proliferative” signature (Fig. 2d, Extended Fig. 5), whereas the “mesenchymal” and “immunoreactive” signatures showed weak or no expression in cancer cell clusters, but were highly expressed by the CAF and macrophage clusters, respectively. Thus, the mesenchymal and immunoreactive subtypes may represent the intra-tumoral abundance of CAFs and macrophages, respectively. This is consistent with previous reports that TCGA tumors with these classifications had a significantly lower purity23 (Fig. 2e), but the single-cell analysis shows that the immunoreactive and mesenchymal gene programs (such as those associated with epithelial-mesenchymal transition) are derived from the non-malignant immune cells and CAFs, respectively, as opposed to malignant cells. Thus, subtype classifications based on bulk RNA profiles likely predominantly reflect tumor ecosystem composition rather than cancer cell-derived HGSOC subtypes.
Intra-patient variation in expression programs of malignant cells
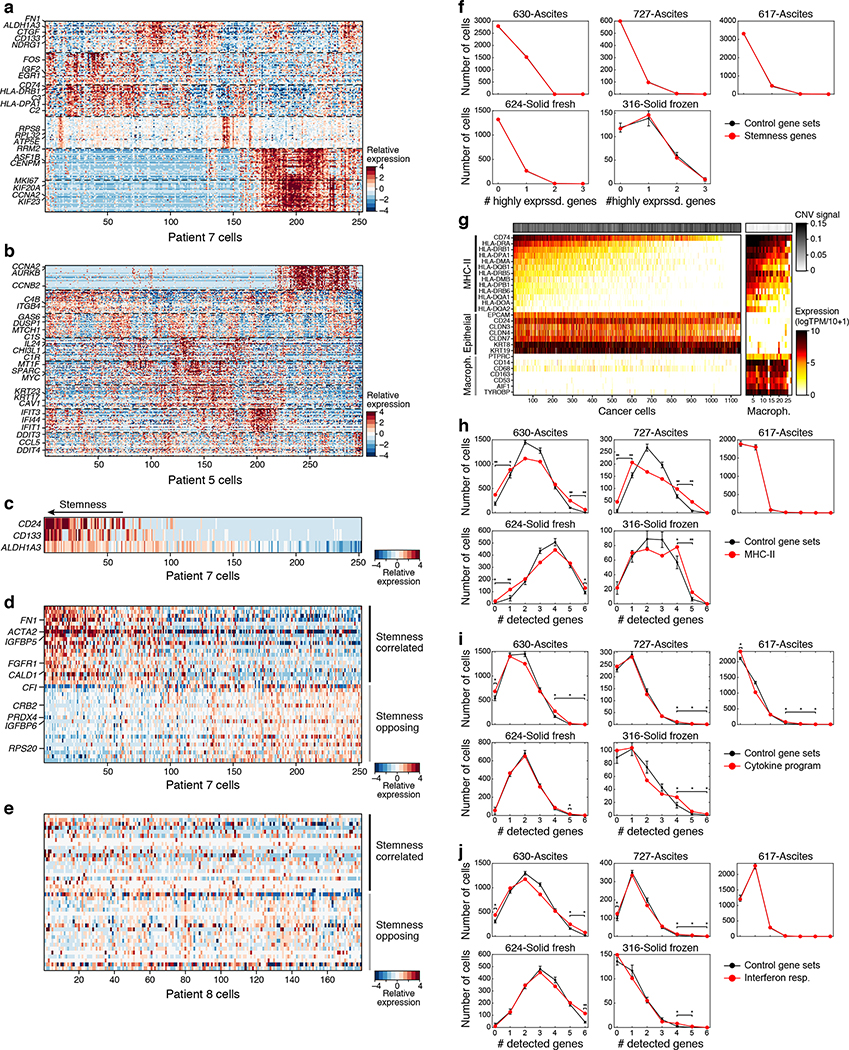
We next identified programs that vary among each patient’s malignant cells. We used non-negative matrix factorization (NMF) and identified a total of 35 modules across malignant cells with coherently co-varying gene expression (Fig. 3a–c, Extended Fig. 6a,b and Supplementary Table 3). The modules spanned diverse functions, reflected by their top-scoring genes, including cell cycle (e.g., CCNA2, CCNB2, AURKB), inflammation (e.g., IL6, IL32, TNF, IFI6), and stress or activation (e.g., HSPA5–7, ATF4, JUN, DDIT3). One intriguing module consisted of prominent stemness24 (ALDH1A3 and CD133/PROM1) and mesenchymal (FN1, ACTA2 and MYL9) markers, and AXL and its only known ligand GAS6, which is implicated in drug resistance25 (Extended Fig. 6c–e); however, this program was unique to patient 7 and was not detected in cancer cells across cohorts examined here (Extended Fig. 6c–f). Profiling of larger patient cohorts will be required to examine whether this is a recurrent stemness signature in HGSOC. Together, and in line with previous work9–11,4,3, we find inter-patient variation across cancer cells and intra- and inter-individual heterogeneity of non-malignant cells, suggesting different functional subpopulations that contribute to shaping the HGSOC ecosystem.
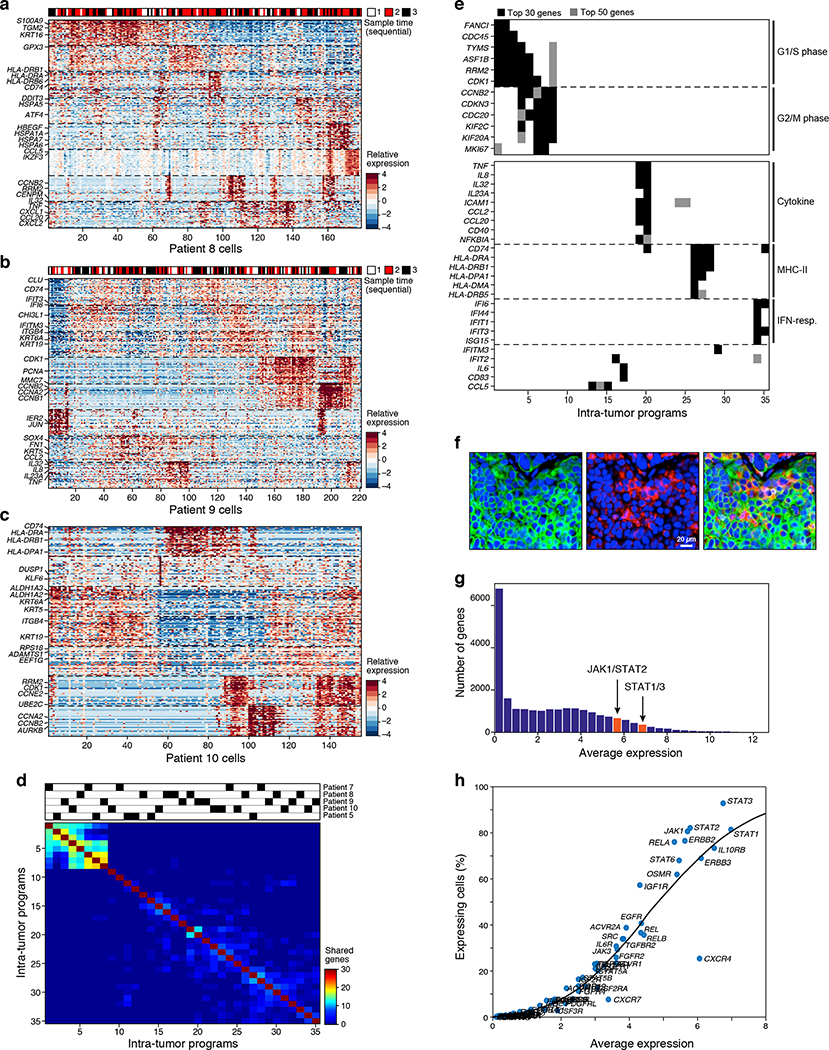
Figure 3. Inflammatory programs in malignant cells from patient ascites predict a role for JAK-STAT signaling.
(a-c) Intra-tumoral expression modules. Relative expression (color bar, Methods) of the top 30 module-specific genes (rows) in each module (ordered by module, dashed horizontal lines), as defined by NMF (Methods) across all cancer cells (columns; ordered by hierarchical clustering) from patients 8 (a), 9 (b), or 10 (c). Selected genes are annotated. Top bar (a and b): sample time in 3 sequential samples from the same patient. (d,e) Cell cycle and inflammatory/immune programs recur across patients. (d) Number of top genes (color bar) shared between each pair of modules (rows and columns, ordered by hierarchical clustering). Top: module’s patient-of-origin. (e) Module membership in the top 30 (black) or 50 (grey) of selected cell cycle (top) and immune-related (bottom) genes (rows) across all modules (columns), ordered as in (d). (f) MHC-II expressing cancer cells in situ. Representative immunofluorescence staining of HGSOC primary tissue staining for nucleus (blue), pan-keratin (green) and MHC Class II (red). Size bar: 20μm. Overlay (right): co-expression of pan-keratin and MHC Class II, indicating cancer cell-autonomous expression of MHC Class II in a subset of cancer cells. (g,h) Broad and high expression of JAK-STAT pathway components across malignant cells. (g) Distribution of average expression of genes (x axis, log2(TPM+1)) for all detected genes (y axis). Red: STAT1/3 expression bin. (h) Mean expression (x axis, log2(TPM+1)) and percentage of expressing cells (y axis) of signaling genes. Key nodes of the JAK/STAT-pathway are labeled. Line: LOWESS regression curve.
Shared expression of cancer cell-autonomous inflammatory programs across patients and PDX models
To identify cancer cell programs shared across patients, we next compared the modules across different patients (Fig. 3d). As expected, there was strong overlap of cell cycle modules, indicating the presence of proliferating subpopulations of cells in all specimens. In addition, three programs dominated by immune- or inflammation-associated genes were shared among patients (Fig. 3d,e): an inflammatory cytokines module (e.g., IL6, TNF, IL8, IL32), an MHC-class II antigen presentation module (e.g., CD74, HLA-DRA), and an interferon-response module (e.g., IFI6, IFIT1, ISG15). These immune-related programs were also detected in cells from Cohort 3, including three additional ascites samples and two primary tumors (Extended Fig. 6h–j). Cancer cell-intrinsic expression of MHC-Class II protein has recently been described in other epithelial cells26,27, tumor-initiating cells28, and cancers, such as melanoma29, and can be associated with response to immunotherapies even in the absence of MHC Class I expression29. We confirmed the expression of the MHC Class II module in CNV-bearing malignant cells (Extended Fig. 6g) and showed by immunofluorescence staining of independent primary HGSOC tumors the presence of a subpopulation of MHC Class II expressing cancer cells (among other cells that express MHC Class II) (Fig. 3f).
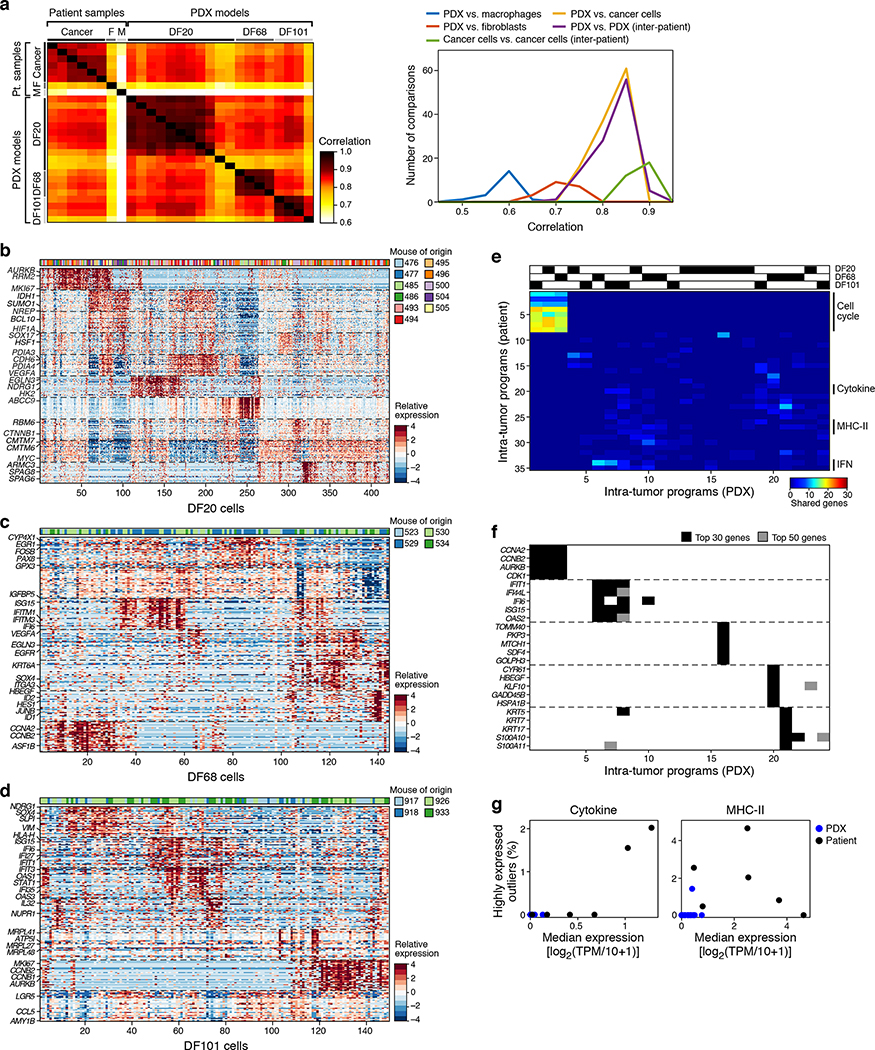
Inflammatory cancer-cell programs may be induced by the ascites microenvironment or reflect an endogenous property of the cancer cells. To evaluate these possibilities, we profiled 795 cells by scRNA-seq of three previously established PDX models (DF20, DF68 and DF101) (Methods) grown as subcutaneous (s.c.) tumors in immunocompromised animals30 collected pre-, on- and post-platinum therapy (Methods). Globally, PDX and patient cancer cells were as strongly correlated with each other (Pearson r=0.819 on average) as cancer cells of different PDX models (r=0.822 on average) or cancer cells across patients (“inter-patient” comparison; r=0.882 on average) (Extended Fig. 7a). We next identified 24 programs in the PDX models by NMF (Extended Fig. 7b–d, Supplementary Table 4), and compared them to those identified in patient ascites (Extended Fig. 7e,f).
Nine PDX modules were analogous to those from patient samples, including three reflecting cell cycle (PDX modules 1–3) and three interferon-response programs (PDX modules 6–8) (Extended Fig. 7f), highlighting this as the most significant similarity (apart from the cell cycle) between patterns of heterogeneity in patients and PDX models. The other two inflammation-related programs found in patients (cytokine and antigen presentation modules) were not detected in PDX models (Extended Fig. 7g), suggesting that their expression in cancer cells may depend on an intact immune system (mostly absent in nod scid gamma mouse (NSG) mice) or other microenvironmental cues.
JAK/STAT activation may represent a shared and targetable vulnerability in HGSOC
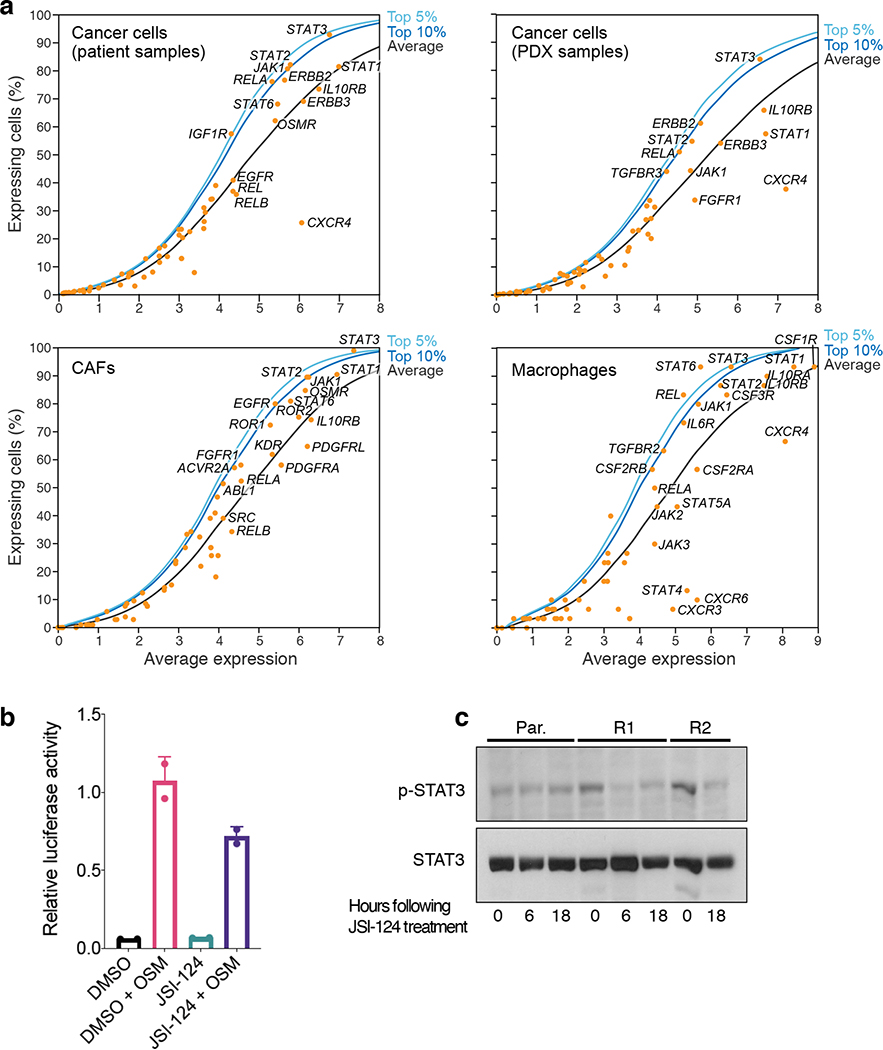
Multiple observations led us to consider the JAK/STAT-pathway as a potential vulnerability. First, as described above, subpopulations of cancer cells highly expressed three immune-related programs that may be downstream to the JAK/STAT pathway. Second, cells in the ascites microenvironment, such as CAFs, highly expressed genes of secreted ligands (e.g. IL6, CXCL12) that activate the JAK/STAT-pathway. Third, analysis of a large set of signaling genes highlighted a particularly high and ubiquitous expression of JAK/STAT-pathway components both in malignant and in non-malignant cells (Fig. 3g,h, Extended Fig. 8a).
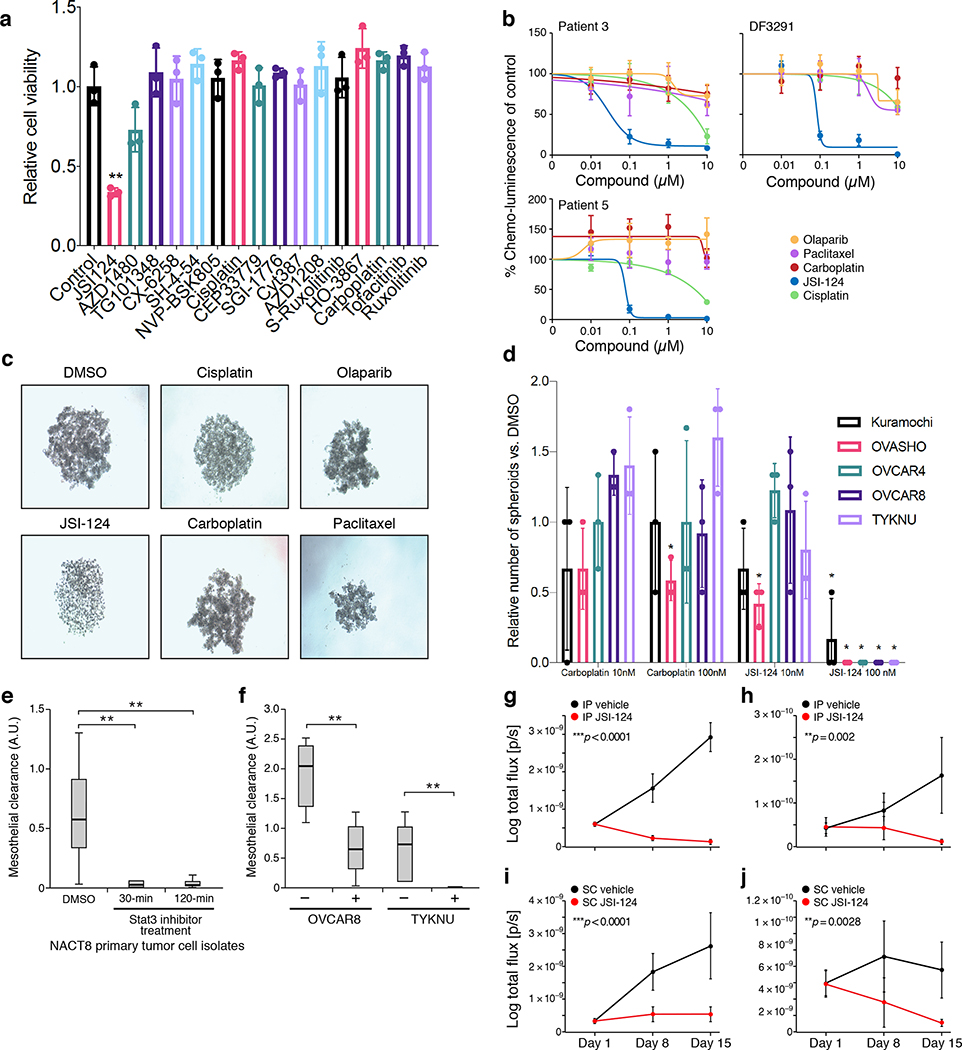
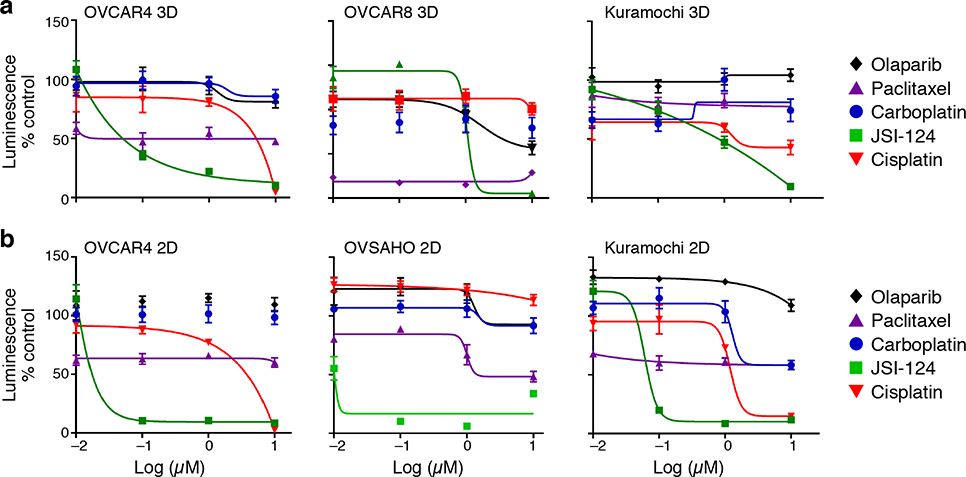
To determine the impact of JAK/STAT inhibition, we performed a drug screen using 15 compounds targeting different nodes of the pathway or its effectors and platinum-chemotherapies (Supplementary Table 5) in HGSOC cell line OVCAR4, and identified JSI-12431 as a potent inhibitor of cell viability (Fig. 4a; Extended Fig. 8b,c). JSI-124 showed anti-tumor activity at nanomolar doses in three different patient-derived cell culture models and additional HGSOC cell lines 32, while other drugs frequently used for the treatment of HGSOC patients had little to no activity (Fig. 4b,c; Extended Fig. 9). JSI-124 also reduced the formation of three-dimensional clusters (spheroids) and their invasion through a mesothelial monolayer (modeling the abdominal peritoneum, which represents an important barrier for metastatic disease) (Fig. 4d–f, Online Videos 1–4). In the PDX model DF20, which has substantial transcriptional similarity to patient ascites cancer cells (Extended Fig. 7a), early initiation of JSI-124 treatment (7 days after intraperitoneal [I.P.] or subcutaneous [S.C.] injection of tumor cells) abrogated the development of malignant ascites and tumor growth, respectively (Fig. 4g,i). Furthermore, JSI-124 significantly reduced disease burden in models where I.P. ascites and S.C. tumors were grown for 21 days prior to treatment initiation (Fig. 4h,j). Together, these results suggest that JAK/STAT inhibition may be a potent therapeutic option for patients with HGSOC, through action on malignant cells, non-malignant cells or both.
Figure 4. JAK/STAT-inhibition reduces viability, spheroid formation, and invasion of HGSOC models ex vivo and in vitro.
(a) JSI-124 reduces viability of in OVCAR4 ovarian cancer cell line. Relative (mean) viability compared to control in GILA (y axis) following 2 days of treatment of the OVCAR4 cell line with each of 14 inhibitors of the JAK/STAT-pathway, carboplatin and cisplatin (x axis). (**adjusted p=0.0032 for JSI-124, one-way ANOVA with Holm-Bonferroni correction with Holm-Šídák-extension). Error bars: SD, n=3. (b) JSI-124, but not other compounds routinely used for the treatment of ovarian cancer reduces mean viability of patient-derived ex vivo cultures. Percent viability relative to DMSO treated cells (y axis) in ex vivo cultures derived from patients 3, 5, and DF3291, each treated for 48 hours with increasing doses (x axis, μM) of JSI-124, carboplatin, cisplatin, paclitaxel, or olaparib. Error bars: SD, n=4. Representative of biological duplicates. (c,d) JSI-124 leads to spheroid disintegration. (c) Examples of light microscopy images of spheroids treated with indicated compounds (representative of biological triplicates). (d) Average number of spheroids (relative to DMSO treated control, y axis) formed with five established ovarian cancer cell lines (x axis) treated with two doses of JSI-124 (blue bars) or carboplatin (red bars) (*adjusted p<0.05, one-way ANOVA with Holm-Bonferroni correction with Holm-Šídák-extension). Error bars: SEM, n=3. (e,f) JSI-124 treatment reduces mesothelial clearance by patient-derived spheroids from patient-derived cultures and established cell lines. Mesothelial clearance (y axis) by patient derived cells (NACT8, e) treated with either JSI-124 (for 30 or 120 min) vs. DMSO, or by ovarian cancer cell lines OVCAR8 and TYKNU treated for 30 min. 20 spheroids clusters assessed per iteration. ** p<0.01 (one-way ANOVA and post hoc Tukey-Kramer test), two independent experiments with n=20 spheroids/condition. Horizontal bar: mean; box: interquartile range, whiskers: minimum and maximum. (g-j) JSI-124 prevents tumor growth and eliminates established tumors in PDX models. Mean log BLI signal (y axis, log total flux in p/s) from PDX mice injected with DF20 tumor cells and treated with either vehicle (black) or JSI-124 (red) and monitored over time (x axis, days). Error bars: SEM. N=5 mice per group. All statistical tests are two-sided t test comparing mean±SD at Day 15 of treatment. (g) Mice injected intraperitoneally (IP) and started treatment one week later for 14 days. ***p<0.0001. (h) Mice were injected IP, malignant ascites were allowed to form, and treatment started at 21 days, for a total of another 14 days. **p=0.002. (i) Mice injected subcutaneously (SC) and started treatment one week later for 14 days. ***p<0.0001. (j) Mice were injected SC, tumors were allowed to form, and treatment started at 21 days, for a total of another 14 days. **p=0.0028.
DISCUSSION
Our scRNA-seq study of malignant ascites from patients with advanced HGSOC reveals significant variability in cellular states and programs among malignant and non-malignant cells. Among non-malignant cells we observed diversity among CAFs, with a subset expressing immunomodulatory programs, as recently proposed in other cancer types, such as pancreatic ductal adenocarcinoma19, where “inflammatory” CAFs strongly express IL6 and other cytokines and may promote tumor growth and drug resistance. Shared activation of the JAK/STAT-pathway in cancer cells and CAFs suggests that paracrine (and/or autocrine) signaling via this pathway may contribute to the pathogenesis of malignant ascites and drug resistance, and provide one example of how cell-to-cell interactions shape the ascites ecosystem. Macrophage diversity primarily involved one major axis of variation driven by two gene programs: one including MHC Class II, IFNGR1 and M1-associated genes, and the other by complement factors, and M2-related genes, such as AIF121 and VSIG422, suggesting that a balance of these phenotypes exists within the ascites ecosystem. Previous reports indicate that platinum-therapy may further push this balance towards M2 macrophages by altering monocyte differentiation33. Such changes may occur rapidly, as indicated in a shift from M1-like to M2-like macrophage programs in one patient pair (5.0 and 5.1), where we examined pre-treatment and on-treatment with platinum chemotherapy.
Variation across cancer cells was driven primarily by inter-patient variation, including CNA patterns, but additional subtler intra-patient variation is also present, such as a putative stemness program unique to a subpopulation of cells in patient 7. Some of the intra-patient patterns of heterogeneity were consistent across multiple patients. For example, subsets of malignant cells expressing the MHC Class II program were present in multiple patients and may be associated with increased abundance of tumor-infiltrating lymphocytes (TILs), improved prognosis and response to immunotherapies34. MHC Class II expressing subsets were not identified in PDXs, suggesting that they may depend on immune cell interactions. An interferon-response and a cytokine program also co-varied across malignant cells in multiple patients. Thus, significant cellular and transcriptional forces within the ascites ecosystem, pro-tumorigenic or pro-immunogenic, may balance disease progression and responses to therapies. Therapeutically shifting this balance may be one avenue for reshaping the drug resistant milieu.
Previous work focused on intra-patient and intra-lesion genetic variability showed that development and response to therapy of primary tumors or metastatic lesions emerge as a result of co-evolution of malignant and non-malignant compartments35,36. Similarly, we hypothesize that interactions between CAFs and macrophages in the ascites ecosystem regulate or enhance cancer cell-autonomous programs. One example is the putative interaction between CAFs secreting IL6 to stimulate JAK/STAT signaling in cancer cells, which is associated with poor prognosis and resistance to chemotherapies37. Consistently, JAK/STAT inhibition promoted anti-tumor activity in several pre-clinical models. Clinical trials, such as phase I/II study using combination therapies with JAK/STAT inhibitor ruxolitinib (NCT02713386) will help clarifying the role of such therapies in HGSOC.
HGSOC subtypes defined by TCGA have been associated with prognosis and drug response2,38.
In our study, the vast majority of cancer cells across patients strongly expressed the “differentiated” subtype program, and a minority of cells from one patient also expressed the “proliferative” subtype program. In contrast, the “mesenchymal” and “immunoreactive” subtype programs were not expressed by cancer cells, but reflected programs expressed by CAFs and macrophages, respectively, and therefore represent tumor composition rather than salient cancer cell programs. While previous work could not evaluate the relative contribution of CAFs and cancer cells to the mesenchymal subtype (which also includes EMT genes), our results suggest that most, if not all, of this subtype can be explained by CAFs. This is consistent with findings in colorectal39 and head and neck cancer12. Increased CAF infiltration may contribute to the low response rates to certain therapies, such as immune checkpoint inhibitors, whose efficacy is impacted by the tumor microenvironment40.
Future studies should enhance our work in two main ways. First, profiling a larger number of patient samples would allow us to test for the generality of programs identified only in one patient (e.g., stemness program in patient 7) in our study. Second, single cell profiling of well-stratified clinical cohorts – rather than the diverse broad ranging and heterogenous patient population in our analysis (included to recapitulate true-to-life clinical heterogeneity) – should enhance inter-patient comparisons and identify converging aspects of tumor biology and drug resistance, to improve our understanding of HGSOC.
Materials and Methods
Collection of patient specimens
Specimens were collected from patients with ovarian cancer at Brigham and Women’s Hospital and Dana-Farber Cancer Institute under IRB approved protocols 02–051 and 11–104. Ascites fluid was drained by an interventional radiologist and transferred for further processing in closed vacuum bottles. De-identified patient information, including their ovarian cancer histology, stage, treatment history, and BRCA mutation status were collected.
Sample handling, flow cytometry and single cell isolation
Immediately following drainage, malignant ascites was transported on ice, aliquoted into 50 mL conical tubes (BD Falcon) and spun for 5 min at 580 × g at 4°C. The supernatant was aspirated and the remaining pellet was resuspended with 5 mL of hypotonic lysis buffer ACK (Life technologies) and incubated on ice for 5 min. 20 ml of PBS was used to quench the lysis buffer. Cell suspensions were pooled and pipetted into a new 50 mL conical tube through a 100 μm mesh and spun for 5 min at 580 × g at 4°C. Hypotonic lysis was repeated until no visible red blood cell component was present, usually for a total of 2–3 times (for the plate-based approach). Following red blood cell lysis, the cell pellet was resuspended in PBS with 2% (v/v) FBS. These cells were then used for both plate-based and droplet-based scRNA-seq. For plate-based scRNA-seq, cells were labeled with the following fluorophore-conjugated flow-cytometry antibodies: live/dead stain with Calcein-AM (LifeTechnologies), 7-AAD (7-Aminoactinomycin D) (LifeTechnologies), CD45-FITC (VWR, 304006, Clone HI30), EPCAM-PE (Miltenyi Biotec, 130–111-116, Clone REA764), and CD24-PE/Cy7 (BioLegend, 311119, Clone ML5). Cells were incubated for 30 min on ice in the dark. Cells were washed twice by resuspending with PBS with 2% (v/v) FBS and spun 5 min at 580 × g at 4°C. Flow-cytometry and sorting was performed on a BD Biosciences cell sorter. Following doublet exclusion, Calceinhigh7AAD-CD45-EPCAM+CD24+ single cells were sorted into 96-well microtiter plates (on a plate chiller) that were prepared with 10 μL cell lysis buffer (TCL + 1% β-mercaptoethanol). Following completion of cell sorting, plates were covered with aluminum lids, vortexed for 10 seconds, centrifuged for 2 min at 580 × g at 4°C and immediately placed on dry ice prior to storage at −80°C.
Plate-based single-cell RNA-seq
Plate-based scRNA-seq following FACS enrichment was used to strongly enrich for malignant cells. This approach complements efforts using droplet-based method (below) where the non-malignant compartment makes up the vast majority of cells, but also includes a relatively small portion of cancer cells but with more shallow data. To balance both approaches, we used both to complement different types of analyses. For plate-based sc-RNA-seq, we performed Whole Transcriptome Amplification (WTA) with a modified Smart-seq2 protocol, as described previously 15,16 with Maxima Reverse Transcriptase (Life Technologies) instead of Superscript II. Next, WTA products were cleaned with Agencourt XP DNA beads and 70% ethanol (Beckman Coulter, Brea, CA) and Illumina sequencing libraries were prepared using Nextera XT (Illumina, San Diego, CA), as previously described16. The 96 samples of a multiwell plate were pooled, and cleaned with two 0.8X DNA SPRIs (Beckman Coulter). Library quality was assessed with a high sensitivity DNA chip (Agilent) and quantified with a high sensitivity dsDNA Quant Kit (Life Technologies). Barcoded single cell transcriptome libraries were sequenced with 38bp paired end reads on an Illumina NextSeq 500 Instrument.
Droplet-based single-cell RNA-seq
Single cells were isolated from patient-derived ascites as described above for all but one solid tumor, which was prepared for single nucleus isolation as recently described17. Upon drainage of ascites, we immediately processed fresh specimens by removal of red blood cells using ACK lysis buffer, filtration and isolation of a single cell suspension. Next, CD45+ cells were depleted using the MACS beads and columns per manufacturer’s instructions (Miltenyi Biotec). While this approach led to only partial depletion of CD45+ immune cells (the main component of ascites), we avoided repeated bead-based depletion, because it results in RNA degradation and compromises subsequent scRNA-seq17. Next, cells were counted and resuspended in PBS supplemented with 0.04% BSA for loading for single-cell library construction on the 10X Genomics platform. Experiments were performed with the Chromium Single Cell 3′ Library & Gel Bead Kit v2 and Chromium Single Cell 3′ Chip kit v2 according to the manufacturer’s instructions in the Chromium Single Cell 3′ Reagents Kits V2 User Guide. Briefly, ~6,000 cells were loaded to each channel, then partitioned into Gel Beads in Emulsion in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Barcoded single cell transcriptome libraries were sequenced with 38bp paired end reads on an Illumina NextSeq 500 Instrument. The HTAPP cohort was processed and analyzed as recently described17.
Cell culture
High-grade serous ovarian cancer cell lines Kuramochi, Ovsaho, Ovcar4, Ovcar8, and Tyknu were provided by the CCLE project at the Broad Institute. All cell lines were cultured in RPMI1640 Medium (Gibco), supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin (Invitrogen), and were maintained in an incubator at 37°C and 5% CO2. Sub-culturing of cell lines was done by detaching cells using 0.05% trypsin EDTA, quenching, washing and re-suspending the cell pellet in fresh media.
In vitro and ex vivo drug sensitivity testing
The Growth In Ultra-Low Attachment (GILA) Assay42 was used to assess drug sensitivity of patient-derived cell lines and established cell lines. Five ovarian cancer cell lines (Kuramochi, Ovsaho, Ovcar4, Ovcar8, Tyknu) were each plated at 5,000 cells/100 μL AR-5 medium (ACL4 Media with 5% FBS)43 per well in both flat-bottom high attachment (HA) (Corning Inc., 353072) and round-bottom ultra-low attachment (ULA) (Sigma-Aldrich Co. LLC CLS7007–24EA) 96-well plates. For the initial screen with JAK/STAT-pathway inhibitors, cells were treated with AZD1480 (AstraZeneca), NVP-BSK805 (Selleck Chemicals), TG101348 (STEMCELL Technologies Inc), CX-6258 (Selleck Chemicals), CEP33779 (Selleck Chemicals), Ruxolitinib (Novartis), Tofacitinib (Pfizer), SGI-1776 (Sigma-Aldrich, Inc), Cyt387 (Selleck Chemicals), S-Ruxolitinib (Selleck Chemicals), AZD1208 (AstraZeneca), HO-3867 (Cayman Chemical), SH-4–54 (Selleck Chemicals), trametinib (GlaxoSmithKline), JSI-124 (Sigma Aldrich), cisplatin (APP Pharmaceuticals, Inc) or carboplatin (Hospira, Inc) using 1 μM in both ULA and HA plates. Cells were collected on Day 0 (control) or after 48 hours of treatment, covered with an aluminum lid, and flash frozen at −80°C. Within 24 hours of freezing, cell viability of the samples was determined using CellTiter-Glo® Luminescent Cell Viability (CTG) Assay (Promega Corporation, G7572). The CTG reagent was thawed and diluted with 1 × PBS in a 1:1 ratio prior to use. Plates were thawed and resuspended in equal volumes of CTG, shaken on an orbital shaker at 100 rpm for 2 min at room temperature to mix contents and to induce cell lysis. The plates were incubated at room temperature for 10 min to stabilize luminescent signal. The cell culture and CTG reagent mixture was transferred to a white 96-well plates (Thermo Fisher Scientific, 07200722) and the luminescence signal was read on a 2103 EnVision™ Multilabel Plate Reader (PerkinElmer). Data analysis was performed using Excel and Prism.
Primary cell cultures were generated by removing RBCs and depleting CD45-positive cells using the MACS beads and columns per manufacturer’s instructions (Miltenyi Biotec), and 5,000 cells/100 μL AR-5 medium per well were plated in both HA and ULA 96-well plates. For the GILA assay, 5,000 cells/well were seeded, maintained for 24 hours and treated with either JSI-124 (Sigma Aldrich), cisplatin (APP Pharmaceuticals, Inc), carboplatin (Hospira, Inc), olaparib (AstraZeneca), or paclitaxel (Life Technologies) at indicated doses of 10 μM 1 μM, 100 μM, and 10 μM in both ULA and HA plates. After three or five days of treatment, plates were covered with aluminum lids and flash frozen at −80°C followed by CTG assay as described above. Data analysis was performed using Excel and Prism.
Spheroid formation inhibition assay
Five ovarian cancer cell lines (Kuramochi, Ovsaho, Ovcar4, Ovcar8, Tyknu) were plated in flat bottomed ULA plates with either 0.1% DMSO, JSI-124 or carboplatin, at 10 nM, 100 nM, and 1 μM dosing. After 48 and 72 hours, spheroids with a diameter over 200 μm were counted using a CK40 Culture Microscope (OLYMPUS America, Inc). The relative spheroid count was determined as ratio of spheroids in the treatment conditions compared to DMSO treated cells.
Culturing of patient-derived spheroids
Ovarian primary cells were frozen with 90% FBS and 10% DMSO at −80°C and transferred to a liquid nitrogen tank for long-term storage. For spheroid assays, cells were thawed, washed in PBS, re-suspended in AR-5 media and divided into flat bottom ultra-low attachment 6-well plates (Sigma-Aldrich Co. LLC, CLS3471–24EA) and maintained in an incubator at 37°C and 5% CO2 for four days. Media was replaced every 48 hours. After 96 hours, the cell suspension was passed through a 20 μm filter (Miltenyi μm filter (Miltenyi Biotec, 130–101-812) in order to capture the spheroids larger than 20 μm in diameter. Reverse filtration was performed with AR-5 media to capture the spheroids that remain on the filter. The spheroids were collected and plated in ultra-low attachment 6-well plates for short-term culture.
Mesothelial clearance assay
Primary ovarian cancer cells (NACT8) were cultured in a low-attachment 6-well plate for 96 hours. The culture was treated with 10 μM JSI-124 or DMSO for 30 or 120 min. Spheroids were isolated by passing the suspension through a 20 μm filter. Reverse filtration was performed to collect spheroids larger than 20 μm into a 6-well plate. Cells were washed and spheroids were collected for the mesothelial clearance assay. A mesothelial cells monolayer was prepared by plating mesothelial cells on glass-bottom dishes (Mat-TEK Corporation) coated with 5 μg/mL of fibronectin (Sigma, USA). Cells were maintained in culture until confluent (~48 hours after plating). Suspended NACT8 cell spheroids were collected and added to a confluent mesothelial monolayer expressing green fluorescent protein (GFP), allowed to attach for 30–60 min, and imaged for up to 16 hours using a Nikon Ti-E Inverted Motorized Widefield Fluorescence Microscope equipped with incubation chamber. Only spheres that remained attached during the experiment were used for quantification. Mesothelial clearance was quantified as previously described44,45.
Protein extraction and Western blot analysis
Cells were lysed in RIPA lysis buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1 SDS, 50 mM Tris pH 8.0, with protease and phosphatase inhibitors) on ice for 30 min. Phosphatase and protease inhibitors added to the RIPA buffer (Sigma-Aldrich) were purchased as phosSTOP and complete EDTA free mini tablets respectively (Roche). Westerns were performed as described46 using the indicated antibodies: Phospho- Tyr705 STAT3 (9131; Cell Signaling Technology), STAT3 antibody (sc-482, Santa Cruz Biotechnology), and Tubulin (T5168, Sigma).
Luciferase assay
Heya8 cells were transfected with the STAT3 responsive luciferase reporter M67-luc (kindly provided by Jacqueline Bromberg, Memorial Sloan Kettering) and renilla luciferase (Promega) using lipofectamine 200046. Cells were pretreated with 1 μM JSI-124 for 1 hour and then stimulated with 10 ng/mL oncostatin M (OSM; Peprotech) for 6 hours. Luciferase activity was measured using a dual luciferase kit (Promega) on a luminoskan luminometer. Firefly luciferase activity is normalized to renilla and expressed relative to media controls.
Immunofluorescence
Formalin-fixed, paraffin-embedded (FFPE) tissues were cut at a thickness of 5 μm and mounted on glass slides. Direct immunofluorescence was performed as previously described47 using the following antibodies (manufacturer, clone, dilution): anti-HLA-DPB1-Alexa 647 (Abcam, Clone EPR11226, 1:50) and anti-Pan-Cytokeratin-eFluor 570 (ThermoFisher Science, AE1/AE3, 1:100). Images were acquired on CyteFinder slide scanning fluorescence microscope (RareCyte Inc. Seattle WA) using a 10X objective.
Patient-derived xenograft model experiments
HGSOC PDX models derived from patients with different treatment histories were selected for implantation: DF20 (BRCAWT treatment-naïve, clinically platinum sensitive); DF101 (BRCA1 mutant, 2 lines of prior therapy, clinically platinum resistant); DF68 (BRCA1 mutant, 6 lines of prior therapy, clinically platinum resistant)30. For all experiments, we used NOD-SCID IL2Rγnull mice (NSG, Jackson Laboratory). For the carboplatin treatment study, to facilitate detecting minimal residual disease, we used a subcutaneous (SC) model instead of an intraperitoneal (IP) model.
Frozen stocks of tumor cells were briefly thawed and 5×106 cells were injected subcutaneously into the flanks of NOD-SCID IL2Rγnull mice (NSG, Jackson Laboratory). After tumors were established (150–300 mm3), animals were divided into two groups per model: vehicle (n=5) and carboplatin (n=30). Animals were treated with DMSO or with IP carboplatin at 70 mg/kg weekly for 3 total doses. Mice were monitored by weight, clinical appearance, and tumor burden by measurements by calipers and BLI. For BLI imaging, the mice were anesthetized and imaged every 1–2 weeks. Vehicle mice were harvested at endpoint using predefined criteria, 40% weight gain, 20% weight loss, ascites, or humane reasons. Carboplatin-treated mice for the minimal residual disease (MRD) group were harvested at the nadir of BLI signal and size. The remaining carboplatin-treated mice continued weekly to biweekly BLI monitoring and were harvested at endpoint using the same criteria as the vehicle mice. For the MRD cohort, the entirety of the tumor was harvested for scRNA-seq. For the remaining carboplatin cohort, at final harvest the majority of the tumor was harvested for scRNA-seq with one fragment to 10% neutral buffered formalin and 1 fragment snap-frozen. Solid organs were also placed in formalin. Tumors/tissues were disaggregated as previously described9. Tumor cells expressing mCherry were isolated and flow-sorted into 96-well plates as described above and subjected to plate-based scRNA-seq and analyzed in a pooled fashion.
For the JSI-124 treatment, PDX model DF20 was selected and 5×106 tumor cells were injected into two cohorts of mice, one cohort SC to induce tumors and one cohort IP to induce ascites (n=5 per group). For the first experiment (tumor formation) at 7 days post-injection of the tumor cells, the animals were treated with DMSO vs. IP JSI-124 at 1mg/kg daily for 14 days. For the second experiment (established tumors), tumors were allowed to grow for 3 weeks after cell injection, and then DMSO vs. IP JSI-124 dosing proceeded daily for 14 days. In each experiment animals were monitored by BLI weekly and sacrificed at study endpoint at day 16.
Plate-based scRNA-seq data processing
Expression levels were quantified as Ei,j = log2(TPMi,j/10+1) where TPMi,j refers to transcripts per million for gene i in cell j, as calculated by RSEM for Smart-seq2 samples48. TPM values were divided by 10 since we estimate the complexity of single-cell libraries to be about 100,000 transcripts and would like to avoid counting each transcript ~10 times. This would be the case with TPM, which may inflate the difference between the expression level of a gene in cells in which the gene is detected and those in which it is not detected. This modification has a minimal influence on the expression values, but decreases the difference between the expression values of undetected genes (i.e., zero) and that of detected genes (data not shown), thereby reducing the impact of dropouts on downstream analysis.
For each cell, we quantified two quality measures:
The number of genes for which at least one read was mapped, which is indicative of library complexity;
The average expression level (E) of a curated list of housekeeping genes, which is meant to verify that genes which are expected to be expressed highly, regardless of cell type, are indeed detected as highly expressed.
Scatterplot analyses of all profiled cells separated low and high quality cells based on the these two measures (data not shown), and we therefore conservatively excluded all cells with either fewer than 2,000 detected genes or an average housekeeping expression level (E) below 2.5, as done in previous studies9,49. In each sample, we further excluded cells with ad-hoc thresholds in case there was a subset of cells with fewer detected genes that appeared to be of low quality by manual inspection. For cells passing these quality controls, the median number of detected genes was 7892.
We used the remaining cells (k = 1297 for human samples and k = 795 for mouse samples) to identify genes that are expressed at sufficient levels by calculating the aggregate expression of each gene i across the k cells, as Ea(i) = log2(Average(TPMi)1,…,k+1), and excluded genes with Ea < 4. For the remaining cells and genes, we defined relative expression by centering the expression levels, Eri,j = Ei,j − Average(Ei)1,…,k. The relative expression levels, across the remaining subset of cells and genes, were used for downstream analysis.
Droplet based scRNA-seq data processing
The droplet-based data processing followed similar lines to that of the plate-based data, with the necessary changes to accommodate the change in platform:
TPM values were obtained from CellRanger.
Modified threshold values for the number of detected genes in order to accommodate for the lower detection rate of the droplet-based platform. The minimal number of detected genes was set to 1,000.
In addition, the droplet-based platform also enabled quantification of the number of transcripts (i.e. UMI), and therefore we used a second filtering parameter of at least 4,000 transcripts. We did not use a threshold for housekeeping genes.
Genes were chosen for downstream analyses if they were detected with more than 5 transcripts by more than 5 cells.
Dimensionality reduction and clustering
Following the initial processing steps described above, we clustered the cells using tSNE (with perplexity of 30 and default parameters of the Matlab’s tsne function) followed by density clustering using DBscan (with parameters epsilon=5 and min-points=10). In the case of smartseq2 clusters, cluster 1, which was dominated by cells of patient Patient 8, was also assigned several outlier cells from patients Patient 9 and Patient 10, which were manually excluded from the downstream analysis. Clusters were annotated based on expression of marker genes (as described above) and based on the top 30 most upregulated genes in each cluster (defined by fold change between the average expression in the corresponding cluster compared to average expression in all other clusters) (Fig. 1D, 2C). The malignant cell clusters were further supported by CNAs which were estimated as described previously9,49 (code is available at https://github.com/broadinstitute/inferCNV).
Expression programs of intra-tumoral heterogeneity
For each of five patients and each of the 3 PDX models for which we had profiles of >100 malignant cells, we used non-negative matrix factorization (NMF) to identify 6–9 expression modules of genes coherently co-varying across the cells within each tumor separately. For this purpose, we used non-negative matrix factorization (as implemented by the MATLAB nnmf function, with the number of factors set to 10) to identify variable expression programs. NNMF was applied to the relative expression values (Er), by transforming all negative values to zero. Notably, undetected genes include many drop-out events (genes that are expressed but are not detected in particular cells due to the incomplete transcriptome coverage), which introduce challenges for normalization of single-cell RNA-seq; since NNMF avoids the exact normalized values of undetected genes (as they are all zero), it may be beneficial in analysis of single-cell RNA-seq (data not shown). We retained only programs for which the standard deviation in cell scores within the respective tumor was larger than 0.8, which resulted in a total of 35 programs across the 5 human samples and 24 across the mouse models. The programs were compared by hierarchical clustering, using the number of overlapping genes (among the 50 top-scoring genes of each program) as a similarity metric. Five clusters of programs (two cell cycle and three inflammatory programs) were identified in the human samples based on a minimal overlap of 10 genes between programs and used to define meta-signatures. For each cluster, NNMF gene scores were log2-transformed and then averaged across the programs in the cluster, genes were ranked by their average scores, and the top 30 genes were defined as the meta-signature.
To evaluate if similar programs of intra-tumor heterogeneity recur in the test dataset, we defined a small set of n core genes for each meta-signature, consisting of those genes that were identified in multiple tumors and/or are established as related to the programs inferred function. We then examined if there is an enrichment of cells in which a large number of those genes are detected as expressed. We counted the number of cells in which expression of X of those genes is detected, for X=[1...n]. To assess the significance of the observed counts, we repeated the analysis 10,000 times with other sets of n genes referred to as control gene-sets. Each control gene-set was chosen such that it has a similar distribution of expression levels to that of the signature’s core genes. To that end, we first partitioned all analyzed genes into 50 bins based on their average expression across all cancer cells. Next, we defined each control gene-set by randomly sampling from each bin the same number of genes that are in that bin among the signature’s core genes. The fraction of simulations (out of 10,000) in which an equal number of the core signature genes were detected as expressed, was used to define the p-value of observed counts.
TCGA subtype scores and purity estimate
Bulk RNA-seq data of samples, as well as NNMF clustering and differential expression analysis were downloaded from the Broad Firehose website (https://gdac.broadinstitute.org/), along with additional tumor and clinical annotations. Classification of tumors to predefined molecular subtypes was done based on the NNMF clustering with four factors, and the average expression of top 100 differentially expressed genes for each cluster was defined as the subtype signatures, for which single cell clusters were scored. Purity data for defined by including ABSOLUTE41.
Data Availability Statement
Processed data is be available at the Gene Expression Omnibus (GSE146026) and raw data will be available via the Broad Institute Data Use Oversight System (DUOS, https://duos.broadinstitute.org/#/home). Detailed instructions on establishing a DUOS account can be found on https://duos.broadinstitute.org/#/home.
Code Availability Statement:
Specific code will be made available upon request (without restrictions) to itay.tirosh@weizmann.ac.il.
Extended Data
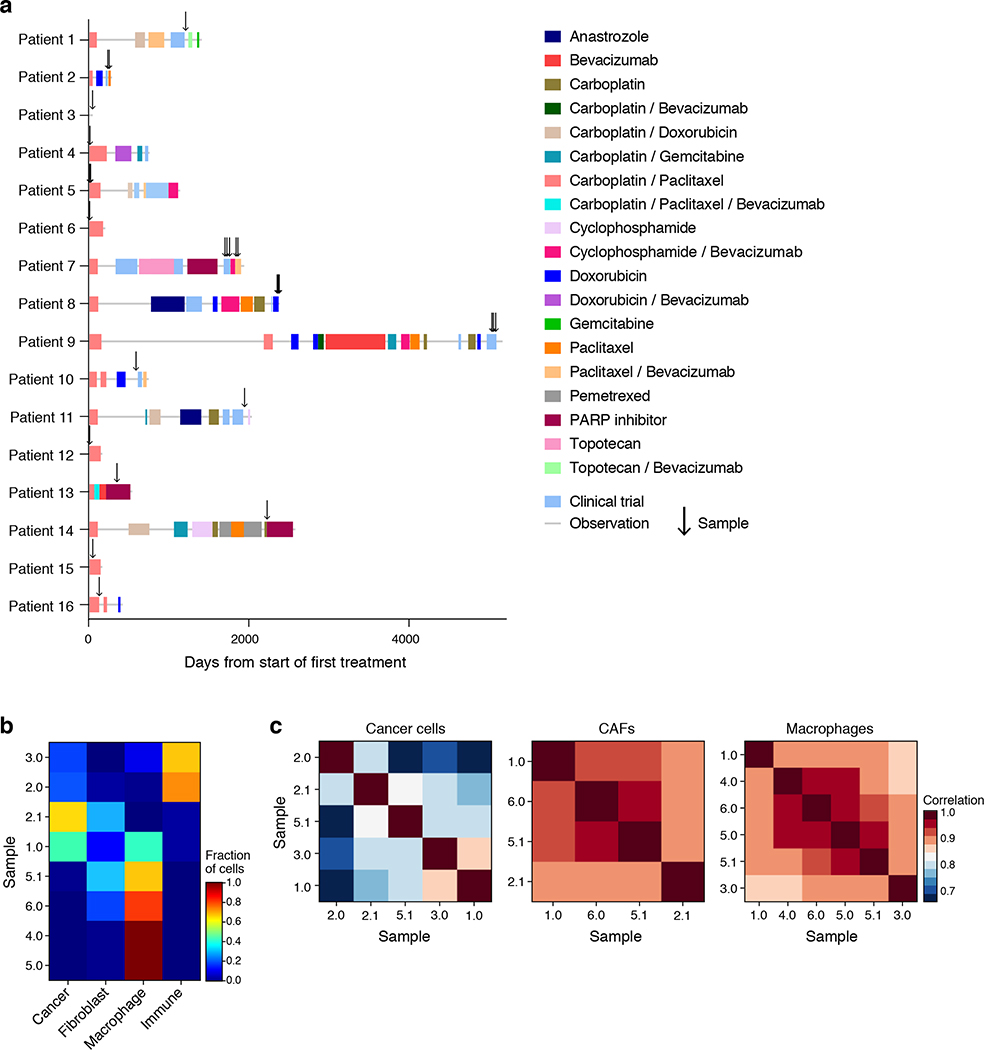
Extended Figure 1. Patient and sample characteristics.
(a) Timing (x axis, days) of therapies (color blocks) and sample collection (arrows) in each patient (y axis). (b) Cell type composition does not group samples by treatment history. Proportion (color bar) of the four major cell types (columns) in each of the ascites samples (rows) profiled by droplet-based scRNA-seq. (c) Cell intrinsic profiles do not group samples by treatment history. Pearson correlation coefficient (color bar) between the mean profiles of cancer cells (left), CAF (middle) or macrophages (right) of each pair of samples (rows, columns) profiled by droplet-based scRNA-seq and having at least 20 cells in each type.
Extended Figure 2. Clustering and characterization of malignant and non-malignant cell clusters in patient ascites by droplet scRNA-seq.
(a) t-stochastic neighborhood embedding (tSNE) of 9,609 droplet-based scRNA-seq profiles from 8 samples (as in Fig. 1b), colored by unsupervised cluster assignment. (b) Cluster 9 is an inflammatory subset of CAFs. Comparison of the average expression (log2(TPM+1)) of each gene in CAF cluster 9 (y axis) vs. CAF clusters 6 and 7 (x axis). Red: immunomodulatory genes. (c) CAF diversity observed within a single sample. Differential expression (log2(TPM+1)) between CAF8 and CAF6/7 cells in patient 5.1 only of the top up- and down- regulated genes from (b). (d-f) Two distinct macrophage programs. (d) Hierarchical clustering of macrophages (rows, columns) from cluster 10 from either Patient 5.0 (left) or Patient 6 (right). Shown are the Pearson correlation coefficients (color bar) between expression profiles of macrophages, ordered by the clustering. Yellow lines highlight the separation into two main clusters. (e) Left: Differential expression (log2(fold change)) for each gene (dot) between the two clusters identified in (d) for Patient 6 (x axis) or patient 5 (y axis), demonstrating high consistency. Top left corner: Pearson’s r. Genes significantly differentially up or down regulated in both patients are marked in red and blue, respectively. Middle and Right: Expression levels (color bar, log2(fold change)) of the highlighted differentially expressed genes from the left panel (rows) across macrophages from Patient 5 (middle) and Patient 6 (right) sorted by the hierarchical clustering of (d). (f) As in (e) for each other samples tested.
Extended Figure 3. Consistent clusters among droplet and plate based scRNA-seq.
(a) Pearson correlation coefficient (color bar) between the average expression profiles of 302 cluster marker genes in cells in clusters defined from either droplet-based or plate-based scRNA-seq (rows, columns; ordered by hierarchical clustering). (b) Pearson correlation coefficient of the mean profile of cell type specific clusters comparing droplet based and plate-based scRNA-seq.
Extended Figure 4. Inferred CNA of single cells from plate based scRNA-seq profiles.
Average relative copy number (color bar) in each chromosomal position (y axis) based on the average expression of the 100 genes surrounding that position9 in each cell in the malignant cell clusters 1–6 (x axis), compared to non-cancer clusters used as a reference, when using the original data (left) or when randomly ordering the genes across the genome and repeating the analysis (right), as control.
Extended Figure 5. Mesenchymal and immunoreactive TCGA subtypes reflect CAFs and macrophages by comparison to droplet based scRNA-seq profiles.
Subtype score (color bar), based on average expression of subtype-specific genes (Methods) of each cluster from the droplet-based scRNA-seq dataset (rows) for each of four TCGA subtypes (column). Only clusters with > 10 cells are represented in this figure.
Extended Figure 6. A putative stemness program in Patient 7 modules.
(a,b) Intra-tumoral expression modules in patients 7 and 5. Relative expression (color bar, Methods) of the top 30 module-specific genes (rows) in each module (ordered by module, dashed vertical lines), as defined by NMF (Methods) across all cancer cells (columns; ordered by hierarchical clustering) from patients 7 (a), or 5 (b, same as Fig. 4c). Selected genes are annotated. (c) Co-variation of stemness related genes in patient 7. Relative expression of three putative stemness markers (rows) in cells from patient 7, rank ordered by the cell’s average expression of the three markers. (d,e) Stemness related co-varying module present in patient 7 but not patient 8. Relative expression of the stemness score of patient 7 (top 20 genes (row) positively (top) or negatively (bottom)) correlated with the average expression of the three stemness genes in (c) in either cells from patient 7 (d) or patient 8 cells (e), with cells ordered by their average expression of the putative stemness score. (f) Stemness program is not detected in other ascites and primary tumor samples from our test cohort. Number of cells (y axis) expressing increasing numbers (x axis) of genes defining the stemness program from Patient 7 (CD24, CD133 (PROM1) and ALDH1A3) in patient cohort 3 (red) or expressing control genes with similar expression pattern in 10,000 simulations (g) Identification of cells expressing MHC Class II as cancer cells. Expression (color bar, log(TP100K+1)) of MHC Class II program, epithelial (cancer cell) markers, and macrophage markers (rows) in cancer cells (defined by marker expression and CNA) and macrophages (columns). Top panel: CNA signal, defined as the square of the inferred copy-number log-ratios, averaged across all genes. (h-j) MHC-II, cytokine and interferon programs are detected in other ascites and primary tumor samples from our test cohort. As in (f) for the three major immune programs defined as (h) MHC Class II (core genes (CD74, HLA-DRA, HLA-DRB1, HLA-DRB5, HLA-DMA, HLA-DPA1), (i) cytokines (core genes TNF, CXCL8, IL32, ICAM1, CCL2, CCL20, NFKBIA); and (j) interferon (IFN) program (core genes IFI6, IFI44, IFIT1, IFIT3, ISG15, MX1). Error bars: SD, *=p<0.05, **=p<0.001; empirical p-value is the fraction of simulations in which an equal number of stemness-program genes are detected as expressed.
Extended Figure 7. Some programs in malignant cells recur between patient ascites and PDX.
(a) Congruent cancer cell profiles between patient and PDX cells. Left: Pearson correlation coefficient (color bar) between mean profiles (rows, columns) among major cell types discovered by plate-based scRNA-seq (cancer cells, macrophages and CAFs) in patient samples and three patient-derived xenograft models (DF20, DF68 and DF101). Right: Distribution of Pearson correlation coefficient (x axis) between different subsets. n=27 (8 patient samples and 19 PDX samples). (b-d) Intra-tumoral expression modules. Relative expression (color bar, Methods) of the top 30 module-specific genes (rows) in each module (ordered by module, dashed horizontal lines), as defined by NMF (Methods) across all cancer cells (columns; ordered by hierarchical clustering) from PDX models DF20 (b), DF68 (c), and DF101 (d). Selected genes are annotated. Top bar (b and c): cell of origin from individual mice. (e,f) Cell cycle and inflammatory/immune programs recur across PDX models. (e) Number of top genes (color bar) shared between pairs of patients (rows, ordered as in Fig. 3e) and PDX (columns; ordered by hierarchical clustering) modules. Top: origin of each PDX module. (f) Module membership in the top 30 (black) or 50 (grey) of selected genes (rows) from cell cycle (top), immune-related (middle), and other (bottom) modules across all modules (columns), ordered as in (e). All genes included were shared between a corresponding PDX module and patient ascites module. (g) Cytokine and MHC-II programs are only identified in patient samples. Median expression (x axis) and % of outlier highly expressing cells (y axis; average log2(TPM+1)>5 and more than 2 SD larger than the mean of all cells) of the cytokine (left) and MHC-II (right) programs in each patient (black) and PDX (blue) samples. N=25 (6 patient samples and 19 PDX samples).
Extended Figure 8. On-target activity of JSI-124.
(a) Prominent expression of JAK-STAT pathway genes. Mean gene expression (x axis, log2(TPM+1)) and percentage of expressing cells (y axis) across the entire cell’s transcriptomes with highlighted signaling genes in patient cancer cells (top left), PDX models (top right), patient-derived CAFs (bottom left) and macrophages (bottom right). Black curve: LOWESS regression curve. Dark and light blue: top 5 and 10 percentiles calculated in a moving average of 200 genes. STAT3 activity induced by Oncostatin M. Relative (mean) luciferase activity (y axis) in Heya8 ovarian cancer cells transfected with a STAT3 responsive reporter that were stimulated with OSM to activate STAT3 for 6 hours (dark blue) or untreated (light blue) with either 1h pre-treatment with JSI-124 (1 μM) or vehicle (x axis) for 1 hour. p=0.09, t test. Error bars: SD. (c) JSI-124 treatment reduced pSTAT3. Cropped immunoblot (representative of duplicates; uncropped available in Source Data) of STAT3 and phosphorylated (p-)STAT3 from cells treated with 1μM JSI-124 for the indicated hours (bottom). Par=parental cell line, and R1 and R2 refer to two independently generated platinum-resistant cell lines.
Extended Figure 9. Dose response of JSI-124 in 2D cultures or 3D spheroids.
Relative (mean) viability (y axis, relative luminescence signal compared to DMSO control) of three ovarian cancer cell lines (labels, top) grown for 4 days in either ultra-low attachment conditions eliciting formation of spheroids (a) or in 2D cultures in regular plastic culture surfaces (b), and treated with JSI-124, carboplatin, paclitaxel, cisplatin or olaparib at indicated doses (x axis, log μM). Error bars: SD. n=4. Representative of biological duplicates.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their families. We thank Lauren MacDonald, Alexandra McKelvey, Julia Gelissen, Jeeva Jacob, Laura Frackiewicz, and Laura Dellostritto, Karla Helvie, Nichole Straub from the Center for Cancer Genomics at the Dana Farber Cancer Institute for coordinating clinical samples; members of the Belfer Center for Applied Cancer Science at Dana-Farber Cancer Institute, particularly Sangeetha Palakurthi and Paul Kirschmeier, for assistance with PDX experiments; and Leslie Gaffney for help with artwork. This work was supported by a Marsha Rivkin Scientific Scholar Award (B.I. and E.S.), Susan F. Smith Center for Women’s Cancers Cancer Research Award (B.I. and E.S.), the Wong Family Award for Translational Cancer Research (B.I.), the Ludwig Center for Cancer Research at Harvard (B.I.) and at MIT (Av.R.), the Burroughs Wellcome Fund Career Award for Medical Scientists (B.I.), DFCI Claudia Adams Barr Program for Innovative Cancer Research (B.I.), K08CA222663 from the National Cancer Institute (B.I.), the Israel Cancer Research Fund (I.T.), the STARR cancer consortium (I.T. and Av.R.), the Human Frontiers Science Program (I.T.), the Mexican Friends New Generation (I.T.), the Benoziyo Endowment Fund for the Advancement of Science (I.T.), grant U54CA225088 from the National Cancer Institute (P.K.S. and B.I.), the Koch Institute Support (core) grant P30CA14051 from the National Cancer Institute (Av.R.), grant R33-CA202820 (Av.R. and As. R.), and the Klarman Cell Observatory (Av.R.). Av.R. is an Investigator of the Howard Hughes Medical Institute.
CONFLICT OF INTEREST
B.I. is a consultant for Merck. L.A.G. is an employee of Genentech. L.A.G. was previously an employee of Eli Lilly and Co. L.A.G. was a paid consultant for Novartis, Foundation Medicine, and Boehringer-Ingelheim; he held equity in Foundation Medicine and was a recipient of a grant from Novartis. P.K.S is a member of the SAB or Board of Directors and holds equity in Applied Biomath, Glencoe Software and RareCyte Inc. As.R is a consultant to eGenesis, a member of the SAB in NucleAI and an holds equity in Celsius. A.R. is a SAB member of ThermoFisher Scientific, Neogene Therapeutics, Asimov and Syros Pharmaceuticals, a cofounder of and equity holder in Celsius Therapeutics, and an equity holder in Immunitas Therapeutics. B.I., I.T., E.H.S., L.A.G., O.R.-R, As.R. and Av.R. have filed a provisional patent for the use of JSI-124 for the treatment of ovarian cancer. All other authors do not disclose any conflicts of interest.
REFERENCES
- 1.Matulonis UA et al. Ovarian cancer. Nat. Rev. Dis. Primer 2, 16061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Network, T. C. G. A. R. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winterhoff BJ et al. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol. Oncol 144, 598–606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih AJ et al. Identification of grade and origin specific cell populations in serous epithelial ovarian cancer by single cell RNA-seq. PloS One 13, e0206785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z et al. The Repertoire of Serous Ovarian Cancer Non-genetic Heterogeneity Revealed by Single-Cell Sequencing of Normal Fallopian Tube Epithelial Cells. Cancer Cell 37, 226–242.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA. Cancer J. Clin 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Patch A-M et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ahmed N & Stenvers KL Getting to Know Ovarian Cancer Ascites: Opportunities for Targeted Therapy-Based Translational Research. Front. Oncol 3, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tirosh I et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venteicher AS et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puram SV et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 171, 1611–1624.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izar B & Rotem A GILA, a Replacement for the Soft-Agar Assay that Permits High-Throughput Drug and Genetic Screens for Cellular Transformation. Curr. Protoc. Mol. Biol 116, 28.8.1–28.8.12 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Peterson VM et al. Ascites analysis by a microfluidic chip allows tumor-cell profiling. Proc. Natl. Acad. Sci. U. S. A 110, E4978–4986 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picelli S et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Trombetta JJ et al. Preparation of Single-Cell RNA-Seq Libraries for Next Generation Sequencing. Curr. Protoc. Mol. Biol. Ed. Frederick M Ausubel Al 107, 4.22.1–4.22.17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slyper M et al. A single-cell and single-nucleus RNA-seq toolbox for fresh and frozen human tumors. bioRxiv 761429 (2019) 10.1101/761429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirri P & Chiarugi P Cancer associated fibroblasts: the dark side of the coin. Am. J. Cancer Res 1, 482–497 (2011). [PMC free article] [PubMed] [Google Scholar]
- 19.Öhlund D et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med 214, 579–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iliopoulos D, Hirsch HA & Struhl K An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai H et al. Colony-stimulating factor-1-induced AIF1 expression in tumor-associated macrophages enhances the progression of hepatocellular carcinoma. Oncoimmunology 6, e1333213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J et al. VSIG4 inhibits proinflammatory macrophage activation by reprogramming mitochondrial pyruvate metabolism. Nat. Commun 8, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aran D, Sirota M & Butte AJ Systematic pan-cancer analysis of tumour purity. Nat. Commun 6, 8971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva IA et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 71, 3991–4001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X et al. AXL kinase as a novel target for cancer therapy. Oncotarget 5, 9546–9563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biton M et al. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 175, 1307–1320.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smillie CS et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao Y et al. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell 177, 1172–1186.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodig SJ et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med 10, (2018). [DOI] [PubMed] [Google Scholar]
- 30.Liu JF et al. Establishment of Patient-Derived Tumor Xenograft Models of Epithelial Ovarian Cancer for Preclinical Evaluation of Novel Therapeutics. Clin. Cancer Res (2016) 10.1158/1078-0432.CCR-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaskovich MA et al. Discovery of JSI-124 (Cucurbitacin I), a Selective Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling Pathway Inhibitor with Potent Antitumor Activity against Human and Murine Cancer Cells in Mice. Cancer Res. 63, 1270–1279 (2003). [PubMed] [Google Scholar]
- 32.Barretina J et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dijkgraaf EM et al. Chemotherapy Alters Monocyte Differentiation to Favor Generation of Cancer-Supporting M2 Macrophages in the Tumor Microenvironment. Cancer Res. 73, 2480–2492 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Axelrod ML, Cook RS, Johnson DB & Balko JM Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 25, 2392–2402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiménez-Sánchez A et al. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 170, 927–938.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang AW et al. Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell 173, 1755–1769.e22 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Duan Z et al. Signal transducers and activators of transcription 3 pathway activation in drug-resistant ovarian cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 12, 5055–5063 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Kommoss S et al. Bevacizumab May Differentially Improve Ovarian Cancer Outcome in Patients with Proliferative and Mesenchymal Molecular Subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 23, 3794–3801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calon A et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet 47, 320–329 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Pitt JM et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol 27, 1482–1492 (2016). [DOI] [PubMed] [Google Scholar]
References
- 41.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rotem A et al. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. U. S. A 112, 5708–5713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gazdar AF & Oie HK Re: Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 46, 6011–6012 (1986). [PubMed] [Google Scholar]
- 44.Davidowitz RA et al. Mesenchymal gene program-expressing ovarian cancer spheroids exhibit enhanced mesothelial clearance. J. Clin. Invest 124, 2611–2625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwanicki MP et al. Mutant p53 regulates ovarian cancer transformed phenotypes through autocrine matrix deposition. JCI Insight 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson EA et al. Nifuroxazide inhibits survival of multiple myeloma cells by directly inhibiting STAT3. Blood 112, 5095–5102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin J-R et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel AP et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Processed data is be available at the Gene Expression Omnibus (GSE146026) and raw data will be available via the Broad Institute Data Use Oversight System (DUOS, https://duos.broadinstitute.org/#/home). Detailed instructions on establishing a DUOS account can be found on https://duos.broadinstitute.org/#/home.