Abstract
Resveratrol is a representative polyphenol of diet-derived putative cancer chemopreventive agents, which have attracted increasing interest in the cancer chemoprevention community. The inhibition of the action of human papillomavirus (HPV) E6 and E7 has been considered a key approach for cervical cancer therapy. Resveratrol has been shown to induce the apoptosis, and reduce both the viability and mitotic index of a number of cancer cell lines, including human cervical cancer cells. In the present study, it was confirmed that resveratrol inhibited the HPV E6 mRNA, HPV E6 protein and phosphorylated retinoblastoma protein (p-pRb1) levels, and increased the p53 protein levels in HeLa and Ca Ski cells, as well as in subcutaneous tumor tissue grown from HeLa cells. High-risk HPV uses a bicistronic RNA to control E6 and E7 genes simultaneously. On the whole, the present study demonstrates that resveratrol inhibits cervical cancer development by suppressing the transcription and translation of E6 and E7, and also by promoting the apoptosis and G1/S phase transition arrest. These findings may provide the basis for the development of resveratrol as a candidate for cervical cancer therapy.
Keywords: resveratrol, cervical cancer, human papillomavirus E6 and E7, cell apoptosis, cell cycle arrest
Introduction
Cervical cancer is a common malignant tumor affecting women worldwide. Its main risk factor is human papilloma-virus (HPV) infection. The HPV genome can be integrated into the host genome (1). The detection of the HPV DNA content has been used to screen cervical cancer (2). HPV types 16 and 18 were found in 22% of HPV-positive cases in Tampere, Finland (3). Among HPV-positive cervical cancers in Iranian women, 48.5% were HPV16 and 12.5% were HPV18 positive (4). HPV16 and HPV18 are highly prevalent in cervical cancer, and belong to a subset of high-risk HPVs.
HPV is a small non-enveloped virus containing a single double-stranded DNA molecule approximately 8 kb in length. The HPV genome contains 3 coding regions: The early region (E), late region (L) and upstream regulatory region. The E region comprising E1-E8 encodes proteins essential for replication, transcription and transformation (5). E6 and E7 play a critical role in the molecular pathogenesis of cervical cancer (6). The expression level of HPV E6/E7 is closely related to the malignant degree of cervical cancer and the poor prognosis of patients (7,8).
HPV E6 and E7 interact with regulatory proteins essential to cell fate and the cell cycle. E6 binds to a short leucine (L)-rich LXXLL consensus sequence within the cellular ubiquitin ligase E6AP. Subsequently, the E6/E6AP heterodimer recruits and degrades p53 (9-11) and inhibits the transcriptional activity of p53 (12,13). E6 promotes retinoblastoma protein (pRb) phosphorylation and cell cycle progression. The E6 protein markedly increases the activity of cyclin A/cyclin-dependent kinase 2, which is involved in Rb1 phosphorylation (14). E7 also inhibits p53 transcriptional activity and impairs the control of cell cycle checkpoints by p53 (15-17). The conserved N-terminal domain of E7 can bind and inactivate the Rb protein, promote the activation of the E2F transcription factor, and subsequently activates the transcription of S-phase DNA replication-related genes. The E7 protein can inhibit the effects of the cyclin-dependent kinase inhibitors, p27, p15, and p21, allowing cells to escape the G1/S phase check point and then triggering abnormal chromosome replication. Suppressing the action of E6 and E7 is a key approach for cervical cancer therapy (6,18).
Resveratrol is a natural polyphenol compound with a variety of biological activities, such as antioxidant, anti-inflammatory, immunoregulatory, chemopreventive and antitumor activities (19). Resveratrol treatment has been shown to trigger the apoptosis of the T-acute lymphoblastic leukemia (T-ALL) cell line, CCRF-CEM, via the upregulation of the apoptotic BAX gene and the downregulation of the anti-apoptotic BCL2 gene (20). Apoptosis has also been shown to be induced in TRAIL-resistant lung cancer cells following co-treatment with resveratrol and TRAIL via the suppression of Akt/NF-κB signaling (21). Resveratrol is a candidate for the treatment and prevention of various types of cancer via the inhibition of reactive oxygen species (ROS) (22). Resveratrol also plays a role in killing cervical cancer cells through multiple molecular mechanisms. Resveratrol has been shown to induce the autophagy and apoptosis (23,24), and suppress the migration and invasion of human cervical cancer cells (25). Resveratrol significantly inhibits the occurrence and development of cervical cancer by regulating phospholipid scramblase 1 (26). Resveratrol has also exhibited antitumor activity on an HPV E6-positive cervical cancer (27,28). The combination of resveratrol and curcumin, epicatechin gallate and tricurin has been shown to exert a unique synergistic suppressive effect on HPV E6, eliminating HPV+ cancer cells, and inhibiting tumor progression (29). However, whether or not the effects of resveratrol on HPV E7 are the same as those on HPV E6 remains unknown. The mechanisms through which resveratrol affects cervical cancer cells remains to be further elucidated.
In the present study, the effects of resveratrol on the expression of HPV E6 and E7, and the proliferation, apoptosis and cycle of HPV16/18 cervical cancer cells were investigated by using the HPV18-positive human cervical cancer cell line, HeLa (30), and the HPV16- and HPV18-positive Ca Ski cells (31,32). The present study demonstrates that resveratrol inhibits cervical cancer development by suppressing HPV E6 and E7 expression, and promoting p53, BAX and p16 expression, thereby promoting cell apoptosis and cell cycle arrest at the G1/S phase transition.
Materials and methods
Reagents and antibodies
High-glucose DMEM (11965-092) and 0.25% trypsin (25300054) were from Gibco; Thermo Fisher Scientific, Inc. Fetal Bovine Serum (FBS-HI-11A) was from Capricorn Scientific GmbH. Resveratrol (1602105-100MG) was from Sigma-Aldrich; Merck KGaA. The cell counting kit-8 (CCK-8; BS350B) was from Biosharp. The apoptosis test kit (abs50001) was from Absin Bioscience Inc. and the cell cycle detection kit (KGA512) was from KeyGEN BioTECH Corp., Ltd. TRIzol reagent (93289-100ML) was from Sigma-Aldrich; Merck KGaA. The reverse transcription kit (M1705) was from Promega Corporation and SYBR-Green PCR Mix (1725204) and the Bio-Rad DC Protein Assay kit (5000002) were from Bio-Rad Laboratories, Inc. Goat anti-mouse IgG (115-035-003) and goat anti-rabbit IgG (111-035-003) were from Jackson Immunoresearch Laboratories, Inc. ECL substrate (46640) was from Thermo Fisher Scientific, Inc. Mouse monoclonal anti-β-actin antibody (abs137975), mouse monoclonal anti-GAPDH antibody (abs137959), rabbit anti-β-tubulin polyclonal antibody (abs125209), mouse monoclonal anti-HPV18-E6 (abs128694), rabbit polyclonal anti-p53 (abs130605), rabbit polyclonal anti-phosphorylated pRb1 (p-pRb1; abs110536), rabbit anti-Rb1 (C-term) poly-clonal antibody (abs111662), mouse monoclonal anti-BAX (abs100264), and rabbit polyclonal anti-BCL-2 (abs115024) were from Absin Bioscience Co., Ltd. Mouse monoclonal anti-HPV18-E7 (ab100953) was from Abcam; rabbit polyclonal anti-p16-INK4A (10883-1-AP), rabbit polyclonal anti-Cyclin E1 (11554-1-AP), rabbit polyclonal anti-CDK4 (11026-1-AP) and rabbit polyclonal anti-E2F1 (12171-1-AP) were from Proteintech Group, Inc. DyLight 594 Orange goat anti-mouse (A23310) and DyLight 488 green goat anti-rabbit fluorescent antibody (A23240) were from Abbkine Scientific Co., Ltd.. DAPI dye (P0131) was obtained from the Beyotime Institute of Biotechnology, Inc.
Cells and cell culture
HeLa and Ca Ski cells were purchased from Hunan Feng Hui Biological Technology Co., Ltd. These cells were certified by an STR test, consistent with published reports (33-35). These cells were cultured in high-glucose DMEM containing 10% fetal bovine serum at 37°C in a 5% CO2 incubator.
Determination of cell proliferation by CCK-8 assay
HeLa and Ca Ski cells were seeded in 96-well plates at a density 5×103 cells/100 µl/well. Following culture in regular medium 37°C in a 5% CO2 incubator for 24 h, the cells were cultured in resveratrol-containing fresh medium. Following further incubation 37°C in a 5% CO2 incubator for 24, 48 and 72 h, 10 µl of CCK-8 buffer were added to each well and incubated at 37°C for 2 h. The plates were measured for absorbance (OD) at 450 nm on a plate reader (Spectra Max i3x, Molecular Devices, LLC). Cell viability and the half-maximal inhibitory concentration (IC50) values were calculated by using the analysis function of Graphpad Prism (GraphPad Software, Inc.) software, transforming the resveratrol concentrations to logarithms, then using non-linear regression to fit curve and select dose-response-inhibition. CCK-8 reagent contains WST-8, which is reduced to a highly water-soluble yellow formazan dye by dehydrogenase in cell mitochondria under the action of electron carrier (1-methoxy PMS). The amount of generated formazan is in direct proportion to the number of living cells.
Determination of cell apoptosis by flow cytometry
HeLa cells were treated with the vehicle (DMEM complete medium containing 1/2,500 volume of ethanol) and resveratrol (5, 10, 20 and 40 µM) for 24 h and then collected. After washing with pre-cooled PBS, the cells were collected by centrifugation at 300 × g for 5 min at 4°C, resuspended with 1X Binding Buffer of an apoptosis test kit, and mixed with 5 µl of Annexin V-FITC. The cells were incubated at room temperature for 15 min and then stained with 5 µl of PI for 5 min. Subsequently, 200 µl of 1X Binding Buffer were added, and the cells were analyzed on a BD FACSAria II flow cytometer (BD Biosciences) at 488 nm. The apoptotic ratio was analyzed by FlowJo™ v10.7 (version X; TreeStar, Inc.).
Determination of cell cycle by flow cytometry
HeLa and Ca Ski cells were treated with the vehicle (DMEM complete medium containing 1/2,500 volume of ethanol) and resveratrol (10, 20 and 40 µM) for 24 h and collected. After washing with PBS, the cells were collected by centrifugation at 300 × g, room temperature, for 5 min and then resuspended at a density 1×106/ml. The cells (1 ml) were aliquoted and collected by centrifugation at 300 × g, room temperature, for 5 min. After the supernatant was removed, 500 µl of cold 70% ethanol were added and stored at 4°C overnight. After washing with PBS, the cells were resuspended with 500 µl of the PI/RNase A working solution in the cell cycle detection kit in accordance with the manufacturer's instructions. Following incubation in the dark at room temperature for 45 min, the cells were analyzed on a BD FACSAria II flow cytometer (BD Biosciences) at 488 nm. Cell clusters meeting the analysis conditions were circled at the interface of FSC and SSC, and then circled single cells at the interface of PE-W and PE-A. Cell cycle curve fitting was completed by using the cell cycle function of FlowJo™ v10.7 (version X; TreeStar, Inc.).
Reverse transcription-quantitative PCR
Total RNA was extracted from the Ca Ski cells or tumor tissue grown from HeLa cells (as described below). A total of 2 µg total RNA was reverse-transcribed into cDNA in the reaction system of 25 µl. cDNA (0.5 µl) was used as a PCR template, and PCR analysis was performed using a SYBR-Green PCR Mix by following the program: Pre-denaturation at 95°C for 5 min, 40 cycles of 95°C for 15 sec, 56°C for 30 sec, and 72°C for 30 sec. The specificity of primers (Table I) was examined using a melting curve. GAPDH was used as the internal reference, and the relative expression of the target gene was calculated using the 2−∆∆Cq method (36). The primers were synthesized by Genecreate Biotechnology Co., Ltd.
Table I.
Primer pair sequences used for RT-qPCR.
| Gene | Orientation | Sequence (5′→3′) |
|---|---|---|
| HPV18-E6 | Sense | GCCAGAAACCGTTGAATCC |
| Antisense | AGTCTTTCCTGTCGTGCTCG | |
| HPV18-E7 | Sense | GCATGGACCTAAGGCAACA |
| Antisense C | TCGTCGGGCTGGTAAAT | |
| BCL-2 | Sense | TTCTTTGAGTTCGGTGGGG |
| Antisense | AGTCTTTCCTGTCGTGCTCG | |
| BAX | Sense | ACTGTGCGTGGAAAGCGTA |
| Antisense | TGAGACACTCGCTCAGCTTC | |
| BCL-xL | Sense | TTCTGCCCTCAACCGCAAAGAT |
| Antisense | ACCAGCGGTTGAAGCGTT | |
| p16 | Sense | TCCAGGTCATGATGATGGG |
| Antisense | ATGCGGGCATGGTTACTG | |
| p21 | Sense | GCCAGAAACCGTTGAATCC |
| Antisense | AGTCTTTCCTGTCGTGCTCG | |
| p27 | Sense | GCTAACTCTGAGGACACGCA |
| Antisense | GGGGAACCGTCTGAAACAT | |
| GAPDH | Sense | AGAAGGCTGGGGCTCATTTG |
| Antisense | AGGGGCCATCCACAGTTCTTC |
Western blot analysis
HeLa cells were treated with the vehicle (DMEM complete medium containing 1/2,500 volume of ethanol) and resveratrol (10, 20 and 40 µM) and Ca Ski cells were treated with the vehicle (1/2,500 ethanol) and resveratrol (50 µM) for 24 h, washed with cold PBS, collected, and then homogenized in lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM EDTA and 1 µg/ml leupeptin]. Tissue samples were homogenized in lysis buffer [10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EGTA, 50 mM NaF, 0.5 mM phenylmethylsul-fonylfluoride, 1 mM sodium vanadate, 1% Triton X-100, 0.5% Nonidet P-40 and and 1 µg/ml of aprotinin]. The homogenized samples were centrifuged at 12,000 × g for 15 min at 4°C, and the supernatants were collected. An aliquot of the super-natant was used to determine the protein concentration of each supernatant by using a Protein Assay kit. All samples containing 30 µg protein/sample were aliquoted, mixed with 5X loading buffer, and then loaded for electrophoresis on a 12% SDS-polyacrylamide gel. After electrophoresis, the resolved proteins in the gel were transferred onto PVDF (polyvinylidene difluoride) membranes. The membranes were blocked in 5% non-fat milk TBST buffer (20 mM Tris pH 7.4, 150 mM NaCl and 0.1% Tween-20) for 1 h at room temperature and then probed with primary antibodies over-night at 4°C. After washing with TBST buffer 3 times, the membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibody, goat anti-mouse IgG (1:1,000 dilution) or goat anti-rabbit IgG (1:1,000 dilution) for 1 h at room temperature and developed using the ECL substrate. The relative amount of protein on the blots was determined by densitometry using Lab Works software (UVP, LLC). Mouse monoclonal anti-β-actin (1:5,000 dilution) or mouse monoclonal anti-GAPDH antibody (1:5,000 dilution) was used as loading controls. Proteins were detected using the following primary antibodies: Mouse mono-clonal anti-HPV18-E6 (1:500 dilution), mouse monoclonal anti-HPV18-E7 (1:1,000 dilution), rabbit polyclonal anti-p53 (1:500 dilution), rabbit polyclonal anti-p-pRb1 (1:2,000 dilution), rabbit polyclonal anti-Rb1 (1:2,000 dilution), mouse monoclonal anti-BAX (1:500 dilution), rabbit polyclonal anti-BCL-2 (1:500 dilution), rabbit polyclonal anti-p16-INK4A (1:500 dilution), rabbit polyclonal anti-Cyclin E1 (1:1,000 dilution), rabbit polyclonal anti-CDK4 (1:500 dilution), and rabbit polyclonal anti-E2F1 (1:500 dilution).
Immunocytochemical assay
HeLa cells were treated with resveratrol (40 µM) and the vehicle (DMEM complete medium containing 1/2,500 volume of ethanol) for 24 h and fixed in 4% paraformaldehyde for 15 min at room temperature. After washing with PBS 3 times, 0.5% Triton X-100 was added followed by incubation at room temperature for 15 min. After washing with PBS 3 times, the cells were blocked with horse serum for 1 h at room temperature and then probed with mouse monoclonal anti-HPV18-E6 (1:100 dilution), mouse monoclonal anti-HPV18-E7 (1:100 dilution), rabbit polyclonal anti-p53 (1:100 dilution), rabbit polyclonal anti-p-pRb1 (1:100 dilution) overnight at 4°C. After washing with PBS, the cells were probed with DyLight 594 Orange goat anti-mouse (1:2,000 dilution) and DyLight 488 green goat anti-rabbit fluorescent antibody (1:2,000 dilution) in the dark room at room temperature for 60 min. After washing with PBS 3 times, nuclei were labeled with DAPI at room temperature for 10 min and then washed with PBS 3 times. The cells were examined under a confocal laser microscope (Leica DMi8, Leica Microsystems GmbH) within 1 h.
Animal models and treatment
A total of 12 Balb/C female nude mice (4-6 weeks old, weighing 20.0±2.0 g) were purchased from Hunan SJA laboratory animal Co. Ltd. The mice were housed at a temperature of 20-22°C with 50-60% relative humidity and fed with standard laboratory chow and tap water ad libitum, and were allowed to adapt for 1 week. These nude mice were subcutaneously injected with HeLa cells (2×107/ml, 100 µl/mouse) into the right axillary midline. The volume of the tumors was measured using a caliper as follows: Tumor volume=(length) × (width)2 ×0.5 every 3 days. When length was ≥2 mm, the mice were randomly divided into 2 groups with 6 mice in each group: The vehicle group and the resveratrol treatment group. The resveratrol treatment group was administered intragastrically with 600 µl of 15 mg/kg resveratrol, whereas the vehicle group was administered the same volume of saline (containing 1/100 ethanol). Gastric administration was performed 3 times per week and lasted for approximately 5 weeks. The key time nodes are explained respective figure (see below). The maximum tumor volume in mice was not >1.5 cm3. During the administration, the mice in each group were observed in terms of viability, activity, diet and water intake. At the end of the treatment, all nude mice were euthanized by cervical dislocation. Tumors were isolated, weighed and aliquoted for RT-qPCR, western blot analysis, hematoxylin and eosin (H&E) staining, and immunohistochemical (IHC) staining assay. The present study was approved by the Ethics Committee for Animal Experimentation of Xiangyang No.1 People's Hospital (no. 2017DW038).
Immunohistochemical staining of tissues from HeLa-derived subcutaneous tumor xenografts in mice
Tumor tissues were fixed in 4% polyformaldehyde, paraffin-embedded and then sectioned into 5-µm-thick slices. The sections were then incubated with anti-HPV18-E6 (1:100 dilution) and mouse monoclonal anti-HPV18-E7 (1:100 dilution) antibody at 4°C overnight, and a biotinylated goat anti-rabbit antibody was used as a secondary antibody at room temperature for 50 min. The slides were then washed with PBS and incubated at room temperature with diaminobenzidine chromogen for 3-5 min. Brown-yellow staining was considered positive. The stained sections were examined under a microscope (IX73P2F, Olympus Corporation), and 4 fields were selected in areas with clear cell staining and good background. The view was recorded in images and analyzed using ImageJ (Fiji, National Institutes of Health) software. The relative expression of HPV18-E6 and HPV18-E7 was calculated based on the positively stained area of area and the total area in the views.
Statistical analysis
Data were analyzed using GraphPad Prism 7.0 (GraphPad Software, Inc.) software. The results are expressed as the means ± standard deviation for at least 3 independent experiments. Statistical comparisons between groups were performed using a Student's t-test or one-way ANOVA, followed by Dunnett's post hoc test. Data with P<0.05 were considered to indicate statistically significant differences.
Results
Resveratrol inhibits the proliferation of cervical cancer cells
Firstly, it was confirmed that resveratrol efficiently inhibited HeLa and Ca Ski cell growth. Under the present experimental conditions, the resveratrol IC50 value at 24 h was 155.8 µM, at 48 h was 71.66 µM and at 72 h it was 40.06 µM in the HeLa cells (Fig. S1A). Similar effects of resveratrol on Ca Ski cells were observed, and the IC50 value at 24 h was 172.4 µM, at 48 h was 100.8 µM and at 72 h it was 59.07 µM (Fig. S1B). This suggested that resveratrol inhibited the proliferation of cervical cancer cells in a dose- and time-dependent manner.
Resveratrol promotes the apoptosis of cervical cancer cells
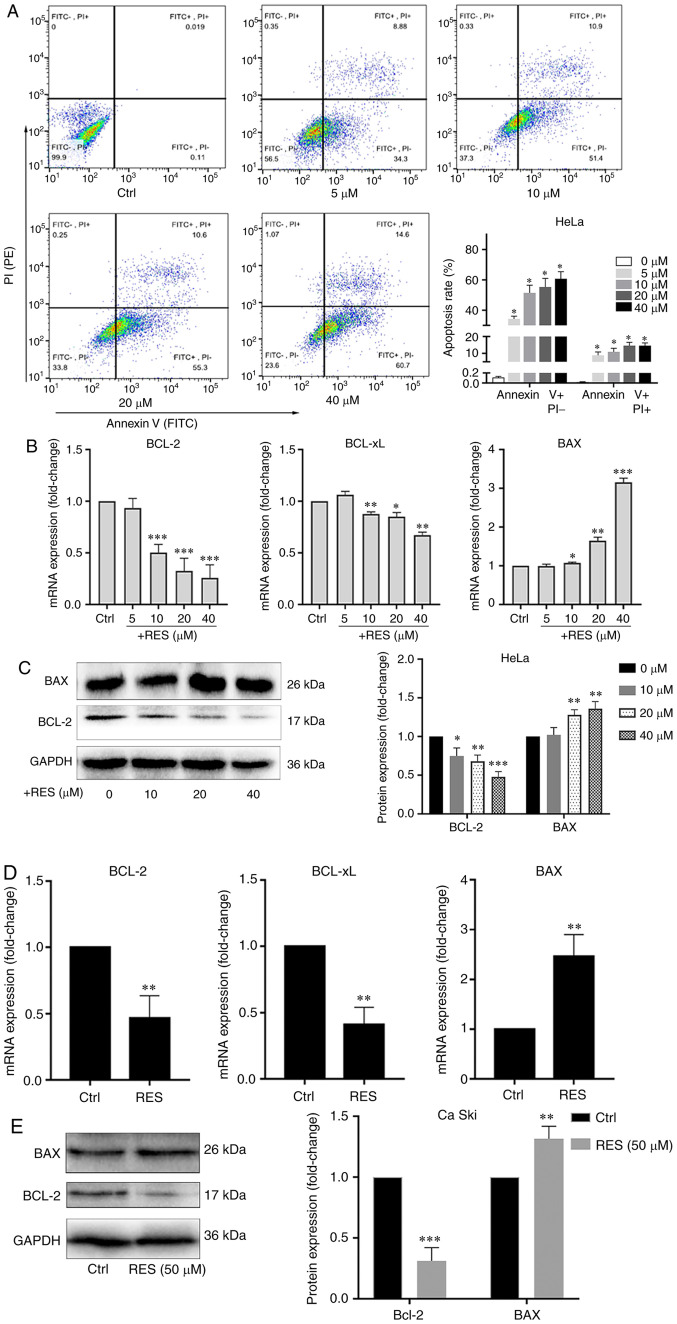
Using resveratrol, the apoptosis of HeLa and Ca Ski cells treated with resveratrol was examined. The apoptotic rate was determined by flow cytometry, and the BCL-2, BCL-xL and BAX mRNA and protein levels were measured by RT-qPCR and western blot analysis. The results of flow cytometry revealed that resveratrol increased the apoptotic rate of HeLa cells in a concentration-dependent manner, particularly in the late stage of apoptosis (Fig. 1A). The BCL-2 and BCL-xL mRNA levels (Fig. 1B) and the BCL-2 protein level (Fig. 1C) significantly decreased, and the BAX mRNA (Fig. 1B) and protein level (Fig. 1C) significantly increased in the HeLa cells treated with resveratrol compared with the cells treated with the vehicle control. Similarly, the BCL-2 and BCL-xL mRNA (Fig. 1D) and BCL-2 protein (Fig. 1E) levels decreased, and the BAX mRNA (Fig. 1D) and protein (Fig. 1E) levels increased in the Ca Ski cells treated with resveratrol compared with the cells treated with the vehicle. These results indicated that resveratrol promoted the apoptosis of cervical cancer cells.
Figure 1.
Resveratrol promotes the apoptosis of cervical cancer cells. (A) HeLa cells were treated with the vehicle (DMEM medium supplemented with 10% fetal bovine serum, and containing 1/2,500 volume of ethanol) or resveratrol at various concentrations (5, 10, 20 and 40 µM) for 24 h. Apoptotic cells were detected by flow cytometry. (B) BCL-2, BCL-xL and BAX mRNA levels in HeLa cells treated with resveratrol or the vehicle were determined by RT-qPCR. (C) BCL-2 and BAX protein levels in HeLa cells treated with resveratrol (10, 20 and 40 µM) or the vehicle for 24 h were determined by western blot analysis. (D) BCL-2, BCL-xL and BAX mRNA levels in Ca Ski cells treated with resveratrol (50 µM) or the vehicle (DMEM medium supplemented with 10% fetal bovine serum, and containing 1/2,500 volume of ethanol) for 24 h were determined by RT-qPCR. (E) BCL-2 and BAX protein levels in Ca Ski cells treated with resveratrol (50 µM) or the vehicle for 24 h were determined by western blot analysis. Data are presented as the means ± SD. *P<0.05, **P<0.01, ***P<0.001, compared with the vehicle control.
Resveratrol promotes G1/S arrest in cervical cancer cells
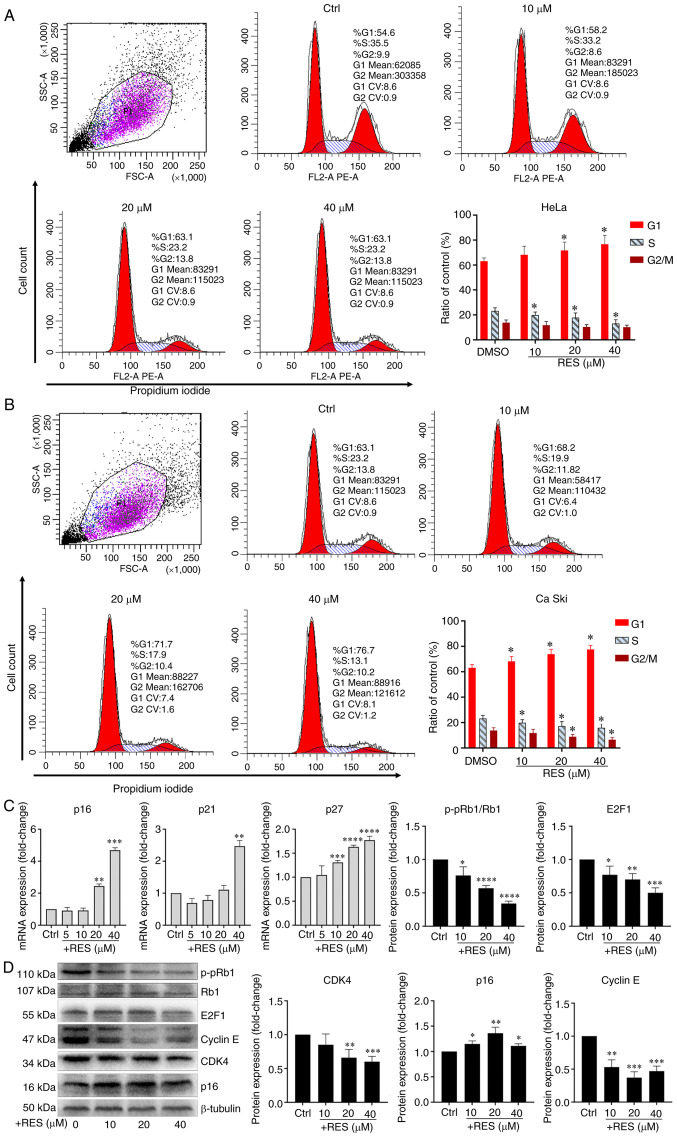
To examine the effects of resveratrol on the cell cycle of cervical cancer, HeLa and Ca Ski cells were treated with resveratrol or the vehicle. The results of flow cytometry revealed that treatment of the HeLa cells with resveratrol significantly increased the proportion of cells in the G1 phase and significantly decreased the proportion of cells in the S phase in a concentration-dependent manner (Fig. 2A). Similar effects of resveratrol on the cell cycle of Ca Ski cells were observed (Fig. 2B). It was also found that the p16, p21 and p27 mRNA levels significantly increased in the HeLa cells treated with resveratrol in a concentration-dependent manner (Fig. 2C). The CDK4, E2F1 and p-pRb1 protein levels significantly decreased in the HeLa cells treated with resveratrol in a concentration-dependent manner (Fig. 2D). Although the concentration of resveratrol used altered the protein levels of Cyclin E and p16, the level of Cyclin E decreased following treatment with resveratrol at up to 20 µM, but then increased at 40 µM, and the level of p16 increased following treatment with resveratrol at up to 20 µM, but then decreased at 40 µM. Thus, the levels of Cyclin E and p16 were not altered in a concentration-dependent manner (Fig. 2D). These results suggested that resveratrol promoted G1/S phase arrest in cervical cancer cells. Moreover, resveratrol reduced the expression of p-pRb1 and promoted the activity of hypo-phosphorylated Rb1 binding to the transcription factor, E2F1.
Figure 2.
Resveratrol promotes G1/S phase arrest of cervical cancer cells. (A) HeLa cells and (B) Ca Ski cells were treated with resveratrol at various concentrations (0, 10, 20 and 40 µM) and the vehicle (DMEM medium supplemented with 10% fetal bovine serum, and containing 1/2,500 volume of ethanol) for 24 h. Cell cycle was determined by flow cytometry. Cell cycle curve fitting was completed by using the cell cycle function of FlowJo™ v10.7 (Version X). (C) p16, p21 and p27 mRNA levels in HeLa cells treated with resveratrol or vehicle for 24 h were determined by RT-qPCR. (D) p16, p21, p27, CDK4, Cyclin E, E2F1 and p-pRb1 protein levels in HeLa cells treated with resveratrol or the vehicle for 24 h were determined by western blot analysis and quantitative analysis of western blot analysis results. Data are presented as the means ± SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, compared with the vehicle control.
Resveratrol inhibits the expression of HPV E6 and HPV E7 in cervical cancer cells
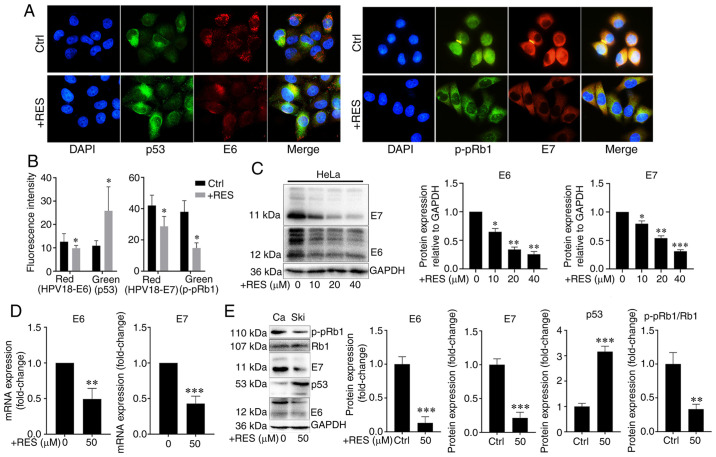
To examine the effects of resveratrol on the expression of HPV E6 and HPV E7 in cervical cancer cells, HeLa and Ca Ski cells were treated with resveratrol or the vehicle. The expression and localization of HPV E6/E7, p53 and p-pRb1 in HeLa cells were determined using immunocyto-chemistry and western blot analysis, and the HPV E6/E7 mRNA levels were measured by RT-qPCR. Immunofluorescence assay revealed that the amounts of HPV E7 and p-pRb1 were reduced in the nuclei, whereas the nuclear localization signal of HPV E6 and p53 exhibited no marked changes in the HeLa cells treated with resveratrol (Fig. 3A). The HPV E6/E7 and p-pRb1 protein levels decreased and the p53 protein levels increased in the HeLa cells treated with resveratrol compared with those treated with the vehicle control (Fig. 3A and B). Western blot analysis revealed that the HPV E6 and E7 protein levels in the HeLa cells were significantly inhibited by resveratrol in a concentration-dependent manner (Fig. 3C). Similarly, the HPV E6/E7 mRNA levels significantly decreased (Fig. 3D), the HPV E6 and E7 and p-pRb1 protein levels decreased, and the p53 protein levels increased in the Ca Ski cells treated with resveratrol compared with the cells treated with the vehicle control (Fig. 3E). These results suggested that resveratrol significantly inhibited the expression of HPV E6 and HPV E7 in cervical cancer cells. Therefore, it was hypothesized that the effects of resveratrol on apoptosis and the cell cycle of cervical cancer cells were due to its inhibition of HPVE6 and E7 (Fig. 6).
Figure 3.
Resveratrol inhibits the expression of HPV18-E6 and HPV18-E7 in cervical cancer cells. (A) HPV18 E6/E7, p53 and p-pRb1 levels in HeLa cells treated with resveratrol (40 µM) or the vehicle (DMEM medium supplemented with 10% fetal bovine serum, and containing 1/2,500 volume of ethanol) for 24 h were determined using immunocytochemistry. (B) Quantitative results of (A). (C) HPV18 E6/E7 protein levels in HeLa cells treated with resveratrol (40 µM) or the vehicle for 24 h were determined by western blot analysis. (D) HPV18 E6/E7 mRNA levels in Ca Ski cells treated with resveratrol (50 µM) or the vehiclefor 24 h were determined by RT-qPCR. (E) HPV18-E6/E7, p53 and p-pRb1 protein levels in Ca Ski cells treated with resveratrol (50 µM) or the vehicle for 24 h were determined by western blot analysis. Data are presented as the means ± SD. *P<0.05, **P<0.01, ***P<0.001, compared with the vehicle control.
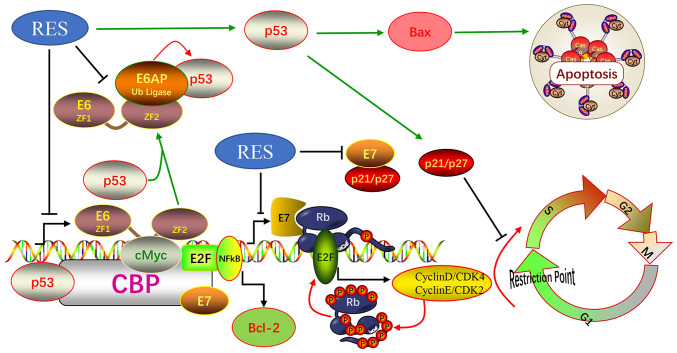
Figure 6.
Graphical representation illustrating the mechanisms through which resveratrol inhibits HPV18 E6/E7 expression to induce cervical cancer cell apoptosis and cycle arrest.
Resveratrol inhibits the growth of HeLa-derived subcutaneous tumor xenografts in mice
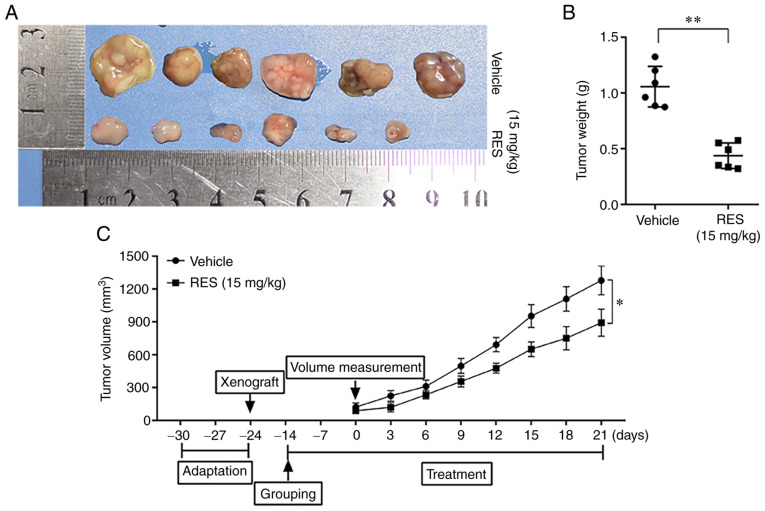
To examine the effects of resveratrol on the growth of cervical cancer cells in vivo, subcutaneous tumor xenograft derived from HeLa cells were established in Balb/C female nude mice and the mice were then treated with resveratrol or saline. The tumor tissues were examined, and the weight of tumor mass and the volumes of tumors were measured. The results revealed that the weight and volumes of tumors in the resveratrol treatment group were significantly lower than those in the saline treatment group (Fig. 4). The results suggested that resveratrol significantly inhibited the growth of cervical cancer cells in nude mice.
Figure 4.
Resveratrol inhibits the growth of HeLa-derived subcutaneous tumor xenografts in nude mice. Balb/C female nude mice were subcutaneously injected with HeLa cells (2×107/ml, 100 µl/mouse) into the right axillary and treated with resveratrol 15 mg/kg, 600 µl intragastric administration or saline (contain 1/100 ethanol) 3 times a week for 5 consecutive weeks. (A) Tumor tissues in each treatment group. (B) Weight of the tumor mass in the resveratrol group was significantly smaller than that in the saline group. (C) Volume of the tumor mass in the resveratrol group was significantly smaller than that in the saline group. Data are presented as the means ± SD. *P<0.05, **P<0.01, compared with the vehicle control.
Resveratrol inhibits the expression of HPV E6 and E7 in subcutaneous tumor xenograft tissues from nude mice
To examine the effects of resveratrol on the expression of HPV E6 and E7 in subcutaneous tumor xenograft tissues in vivo, the HPV E6, E7, p53 and p-pRb1 mRNA and protein levels in tumor tissues from subcutaneous xenograft were determined by RT-qPCR, western blot analysis and immunohistochemistry as appropriate. The results revealed that the tumor tissue HPV E6 and E7 mRNA levels were lower in the resveratrol treatment group than in the vehicle treatment group (Fig. 5A). The results of western blot analysis revealed that the tumor tissue HPV E6, E7, and p-pRb1 protein levels significantly decreased, and the p53 protein level significantly increased in the resveratrol treatment group compared with the vehicle treatment group (Fig. 5B and C). Consistently, immunohistochemical assay revealed that HPV E6 and E7 expression in the cancer tissues significantly decreased in the resveratrol treatment group compared with the vehicle treatment group (Fig. 5D and E). These results indicated that resveratrol significantly inhibited the expression of HPV E6 and E7, promoted the expression of p53 protein, and inhibited the phosphorylation of Rb1 protein in cervical cancer tissues in nude mice.
Figure 5.
Resveratrol inhibits HPV18 E6/E7 expression in cancer tissues of HeLa subcutaneous xenograft tumor in nude mice. (A) HPV18 E6/E7 mRNA levels in cancer tissues were determined by RT-qPCR. (B) HPV18-E6/E7, p53 and p-pRb1 protein levels in cancer tissues were determined by western blot analysis. (C) Quantitative analysis of the results in (B). (D) HPV18 E6/E7 expression is cancer tissues was determined by immunohistochemistry. (E) Quantitative analysis of the results in (D) Data are presented as the means ± SD. *P<0.05, **P<0.01, ***P<0.001, compared with the vehicle control.
Discussion
In the present study, it was found that resveratrol inhibited the proliferation of cervical cancer cells in a concentration- and time-dependent manner. Resveratrol treatment also decreased the weight and volumes of tumors derived from HeLa cells in Balb/C female nude mice. These data were consistent with those of previous studies (23,24,26-28). In addition, a previous study demonstrated that resveratrol suppressed the migration and invasion of human cervical cancer cells (25). Therefore, resveratrol inhibits the development and progression of cervical cancer and is a candidate for cervical cancer therapy.
E6 and E7 are highly expressed in HPV18-positive HeLa cells (30) and HPV16- and HPV18-positive Ca Ski cells (31,32). E6 and E7, both encoded by HPV, play a critical role in the molecular pathogenesis of cervical cancer (6). High-risk HPV E6 and E7 are two viral oncoproteins that respectively, induce protein degradation of tumor suppressors p53 and pRb1 (37). A previous study found that resveratrol downregulated E6 and induces and upregulated p53 protein levels (28). Consistently, the present study found that resveratrol inhibited the HPV E6 mRNA and protein and phosphorylated pRb1 protein levels, and also increased the p53 protein levels in HeLa and Ca Ski cells and cervical cancer tissues grown from HeLa cells. In addition, resveratrol inhibited the HPV18 E7 mRNA and protein levels in HeLa and Ca Ski cells and cervical cancer tissues grown from HeLa cells. These results suggest that resveratrol inhibits the development of cervical cancer by suppressing the E6 and E7 mRNA and protein levels in HPV16- and HPV18-positive cervical cancers.
In high-risk HPV, the splicing of the E6 intron from a bicistronic E6E7 pre-mRNA is a crucial step to control expression of E6 and E7 oncogenes (38,39). while the unspliced E6/E7 mRNA encodes for the full length E6 protein, the E6*I and E6*II mRNAs may favor the E7 expression by a termination-re-initiation process or leaky scanning mechanisms and the production of shorter E6 peptides (38,40). The HPV16 E6*I isoform have oncogenic activities, including disruption of mitochondrial functions and promotion of ROS production (41). Resveratrol has mitochondrial protection and ROS exert scavenging effects. It is hypothesized that resveratrol may affect the splicing of E6 introns, thereby inhibiting the transcription and translation of E6 and E7. Further studies are required to understand the precise role of resveratrol on splicing processes of E6/E7 mRNA.
p53 promotes apoptosis by boosting the expression of the pro-apoptotic protein, BAX. A recent study associated the chemopreventive activity of resveratrol with its ability to block the NF-κB pathway through IκB kinase inhibition (28). In the present study, the results of flow cytometry revealed that resveratrol increased the apoptotic rate of HeLa cells in a concentration-dependent manner, particularly in the late stage of apoptosis. In addition, resveratrol decreased BCL-2 expression and increased BAX expression in HeLa and Ca Ski cells. BCL-2 is an anti-apoptotic factor, and BAX is a pro-apoptotic factor. The data of the present study are consistent with those of previous studies (24,27,42). These results suggest that the induction of apoptosis is a mechanism through which resveratrol inhibits the development of cervical cancer. Of note, the published literature indicate that resveratrol uses different mechanisms to induce cell death in cervical cancer cell line with or without expression of viral oncoproteins (24).
The expression of HPV E6 and E7 in human cervical tissue destroys cell cycle G1/S checkpoints and promotes cells to proliferate indefinitely (14-16). Resveratrol regulates the cancer cell cycle. It impairs G1/S phase transition in MDA-MB-231 and MCF-7 cells (43) and induces G1/S arrest in human prostate cancer cell lines (44,45). The E2F1 complex binds specifically hypophosphorylated Rb1. During the cell cycle, Rb1 becomes phosphorylated in mid-to-late G1 phase, detaches from the DRTF1/E2F complex, rendering E2F transcriptionally active. Viral oncoprotein HPV E7 is capable of sequestering Rb1, thus releasing the active complex (46). In the present study, the treatment of HeLa and Ca Ski cells with resveratrol significantly increased the proportion of G1 phase cells and significantly decreased the proportion of S phase cells in a dose-dependent manner. Furthermore, resveratrol significantly increased the expression levels of p53, p16, p21 and p27, which inhibited G1/S phase progression, and significantly decreased the expression levels of CDK4, Cyclin E, E2F1 and p-pRb1, which promote G1/S phase progression. These data suggest that resveratrol promotes the G1/S arrest of HeLa and Ca Ski cells. The induction of G1/S arrest is also a possible mechanism through which resveratrol inhibits the development of cervical cancer. Therefore, resveratrol can also reduce the inhibition of p53 transcriptional activity by inhibiting the expression of HPV E6 and E7 and restore the role of P53 at the G1/S detection point of cells, and the expression of pro-apoptotic protein BAX, as shown in Fig. 6.
In conclusion, the data of the present study indicate that resveratrol inhibits cervical cancer development by suppressing E6 and E7 expression to promote apoptosis and cell cycle arrest at the G1/S phase transition. The findings of the present study may serve as a reference for the future development of resveratrol as a candidate for cervical cancer therapy.
Supplementary Data
Acknowledgments
The authors wish to thank Professor Xinsheng Gu of the Department of Pharmacology (Hubei University of Medicine) for his careful modification of the manuscript. The authors would also like to thank the Director of the Key Laboratory of Wudang Local Chinese Medicine Research (Hubei University of Medicine), Professor Xuanbin Wang, for providing the facilities to carry out the experiments.
Funding
The present study was supported by grants from the National Natural Science Foundation (no. 81903005) and the Natural Science Foundation of Hubei Province of China (nos. 2020DFE025, 2019AHB068 and 2018CFB701). The present study was also supported by the Open Project of Hubei Key Laboratory of Wudang Local Chinese Medicine Research (grant no. WDCM2018009), the Innovative Team Project (grant no. 2017YHKT02) from the Institute of Medicine and Nursing at Hubei University of Medicine. The funding bodies were not involved in the design of this study, in the collection, analysis, and interpretation of the data, or in writing of the manuscript
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The original data are available upon request to the corresponding author.
Authors' contributions
MS, XS and PF conceived and designed the experiments. XS, PF, LX, QX and LZ performed the experiments. SC, NJ and XW analyzed the data. XS and MS contributed to the reagents, materials and analysis tools, MS, XS and PF wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Committee for Animal Experimentation of Xiangyang No.1 People's Hospital (no. 2017DW038). All animal experiments in this study were undertaken in accordance with the provisions of the Declaration of Helsinki (as revised in Edinburgh 2000).
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
References
- 1.Xu B, Chotewutmontri S, Wolf S, Klos U, Schmitz M, Dürst M, Schwarz E. Multiplex identification of human papillomavirus 16 DNA integration sites in cervical carcinomas. PLoS One. 2013;8:e66693. doi: 10.1371/journal.pone.0066693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duvlis S, Popovska-Jankovic K, Arsova ZS, Memeti S, Popeska Z, Plaseska-Karanfilska D. HPV E6/E7 mRNA versus HPV DNA biomarker in cervical cancer screening of a group of Macedonian women. J Med Virol. 2015;87:1578–1586. doi: 10.1002/jmv.24199. [DOI] [PubMed] [Google Scholar]
- 3.Kares S, Veijalainen O, Kholova I, Tirkkonen M, Vuento R, Huhtala H, Tuimala V, Mäenpää J, Kujala P. HIGH-RISK HPV testing as the primary screening method in an organized regional screening program for cervical cancer: The value of HPV16 and HPV18 genotyping? APMIS. 2019;127:710–716. doi: 10.1111/apm.12990. [DOI] [PubMed] [Google Scholar]
- 4.Fani M, Mahmoodi P, Emadzadeh M, Avan A, Karimi E, Ferns GA, Rezayi M, Amiri IS. Correlation of human papil-lomavirus 16 and 18 with cervical cancer and their diagnosis methods in Iranian women: A systematic review and meta-analysis. Curr Probl Cancer. 2020;44:100493. doi: 10.1016/j.currproblcancer.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 5.DiGiuseppe S, Bienkowska-Haba M, Sapp M. Human papillomavirus entry: Hiding in a bubble. J Virol. 2016;90:8032–8035. doi: 10.1128/JVI.01065-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taghizadeh E, Jahangiri S, Rostami D, Taheri F, Renani PG, Taghizadeh H, Gheibi Hayat SM. Roles of E6 and E7 human papillomavirus proteins in molecular pathogenesis of cervical cancer. Curr Protein Pept Sci. 2019;20:926–934. doi: 10.2174/1389203720666190618101441. [DOI] [PubMed] [Google Scholar]
- 7.Wang HY, Park S, Lee D, Kim S, Kim G, Park KH, Lee H. Prevalence of type-specific oncogenic human papillomavirus infection assessed by HPV E6/E7 mRNA among women with high-grade cervical lesions. Int J Infect Dis. 2015;37:135–142. doi: 10.1016/j.ijid.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Zhang JJ, Cao XC, Zheng XY, Wang HY, Li YW. Feasibility study of a human papillomavirus E6 and E7 oncoprotein test for the diagnosis of cervical precancer and cancer. J Int Med Res. 2018;46:1033–1042. doi: 10.1177/0300060517736913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/MCB.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papilloma-virus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Zapien D, Ruiz FX, Poirson J, Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande Pol S, Podjarny A, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crook T, Fisher C, Masterson PJ, Vousden KH. Modulation of transcriptional regulatory properties of p53 by HPV E6. Oncogene. 1994;9:1225–1230. [PubMed] [Google Scholar]
- 13.Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malanchi I, Accardi R, Diehl F, Smet A, Androphy E, Hoheisel J, Tommasino M. Human papillomavirus type 16 E6 promotes retinoblastoma protein phosphorylation and cell cycle progression. J Virol. 2004;78:13769–13778. doi: 10.1128/JVI.78.24.13769-13778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer M, Uxa S, Stanko C, Magin TM, Engeland K. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci Rep. 2017;7:2603. doi: 10.1038/s41598-017-02831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slebos RJ, Lee MH, Plunkett BS, Kessis TD, Williams BO, Jacks T, Hedrick L, Kastan MB, Cho KR. P53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massimi P, Banks L. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology. 1997;227:255–259. doi: 10.1006/viro.1996.8315. [DOI] [PubMed] [Google Scholar]
- 18.Almeida AM, Queiroz JA, Sousa F, Sousa A. Cervical cancer and HPV infection: Ongoing therapeutic research to counteract the action of E6 and E7 oncoproteins. Drug Discov Today. 2019;24:2044–2057. doi: 10.1016/j.drudis.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Z, Chen K, Cheng L, Yan B, Qian W, Cao J, Li J, Wu E, Ma Q, Yang W. Resveratrol and cancer treatment: Updates. Ann NY Acad Sci. 2017;1403:59–69. doi: 10.1111/nyas.13466. [DOI] [PubMed] [Google Scholar]
- 20.Zadi Heydarabad M, Vatanmakanian M, Abdolalizadeh J, Mohammadi H, Azimi A, Mousavi Ardehaie R, Movasaghpour A, Farshdousti Hagh M. Apoptotic effect of resveratrol on human T-ALL cell line CCRF-CEM is unlikely exerted through alteration of BAX and BCL2 promoter methylation. J Cell Biochem. 2018;119:10033–10040. doi: 10.1002/jcb.27333. [DOI] [PubMed] [Google Scholar]
- 21.Rasheduzzaman M, Jeong JK, Park SY. Resveratrol sensitizes lung cancer cell to TRAIL by p53 independent and suppression of Akt/NF-KB signaling. Life Sci. 2018;208:208–220. doi: 10.1016/j.lfs.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 22.Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, Bilsland AE, Boosani CS, Chen S, Ciriolo MR, et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol. 2015;35(Suppl 1):S199–S223. doi: 10.1016/j.semcancer.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu KF, Wu CL, Huang SC, Wu CM, Hsiao JR, Yo YT, Chen YH, Shiau AL, Chou CY. Cathepsin L mediates resveratrol-induced autophagy and apoptotic cell death in cervical cancer cells. Autophagy. 2009;5:451–460. doi: 10.4161/auto.5.4.7666. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Zepeda SP, Garcia-Villa E, Diaz-Chavez J, Hernandez-Pando R, Gariglio P. Resveratrol induces cell death in cervical cancer cells through apoptosis and autophagy. Eur J Cancer Prev. 2013;22:577–584. doi: 10.1097/CEJ.0b013e328360345f. [DOI] [PubMed] [Google Scholar]
- 25.Kim YS, Sull JW, Sung HJ. Suppressing effect of resveratrol on the migration and invasion of human metastatic lung and cervical cancer cells. Mol Biol Rep. 2012;39:8709–8716. doi: 10.1007/s11033-012-1728-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Yuan X, Li X, Zhang Y. Resveratrol significantly inhibits the occurrence and development of cervical cancer by regulating phospholipid scramblase 1. J Cell Biochem. 2018 2018 Oct 22; doi: 10.1002/jcb.27335. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Chatterjee K, Mukherjee S, Vanmanen J, Banerjee P, Fata JE. Dietary polyphenols, resveratrol and pterostilbene exhibit anti-tumor activity on an HPV E6-Positive cervical cancer model: An in vitro and in vivo Analysis. Front Oncol. 2019;9:352. doi: 10.3389/fonc.2019.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatterjee K, AlSharif D, Mazza C, Syar P, Al Sharif M, Fata JE. Resveratrol and pterostilbene exhibit anticancer properties involving the downregulation of HPV Oncoprotein E6 in cervical cancer cells. Nutrients. 2018;10:243. doi: 10.3390/nu10020243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee S, Debata PR, Hussaini R, Chatterjee K, Baidoo JNE, Sampat S, Szerszen A, Navarra JP, Fata J, Severinova E, et al. Unique synergistic formulation of curcumin, epicatechin gallate and resveratrol, tricurin, suppresses HPV E6, eliminates HPV+ cancer cells, and inhibits tumor progression. Oncotarget. 2017;8:60904–60916. doi: 10.18632/oncotarget.16648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popescu NC, DiPaolo JA, Amsbaugh SC. Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet Cell Genet. 1987;44:58–62. doi: 10.1159/000132342. [DOI] [PubMed] [Google Scholar]
- 31.Choo CK, Rorke EA, Eckert RL. Differentiation-independent constitutive expression of the human papillomavirus type 16 E6 and E7 oncogenes in the CaSki cervical tumour cell line. J Gen Virol. 1994;75:1139–1147. doi: 10.1099/0022-1317-75-5-1139. [DOI] [PubMed] [Google Scholar]
- 32.Pattillo RA, Hussa RO, Story MT, Ruckert AC, Shalaby MR, Mattingly RF. Tumor antigen and human chorionic gonadotropin in CaSki cells: A new epidermoid cervical cancer cell line. Science. 1977;196:1456–1458. doi: 10.1126/science.867042. [DOI] [PubMed] [Google Scholar]
- 33.Saha SK, Khuda-Bukhsh AR. Berberine alters epigenetic modifications, disrupts microtubule network, and modulates HPV-18 E6-E7 oncoproteins by targeting p53 in cervical cancer cell HeLa: A mechanistic study including molecular docking. Eur J Pharmacol. 2014;744:132–146. doi: 10.1016/j.ejphar.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 34.Souza RP, Bonfim-Mendonca PS, Gimenes F, Ratti BA, Kaplum V, Bruschi ML, Nakamura CV, Silva SO, Maria-Engler SS, Consolaro ME. Oxidative stress triggered by apigenin induces apoptosis in a comprehensive panel of human cervical cancer-derived cell lines. Oxid Med Cell Longev. 2017;2017:1512745. doi: 10.1155/2017/1512745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner JD. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol. 1999;80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Tornesello ML, Annunziata C, Tornesello AL, Buonaguro L, Buonaguro FM. Human Oncoviruses and p53 tumor suppressor pathway deregulation at the origin of human cancers. Cancers (Basel) 2018;10:213. doi: 10.3390/cancers10070213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippova M, Evans W, Aragon R, Filippov V, Williams VM, Hong L, Reeves ME, Duerksen-Hughes P. The small splice variant of HPV16 E6, E6, reduces tumor formation in cervical carcinoma xenografts. Virology. 2014;450-451:153–164. doi: 10.1016/j.virol.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerasuolo A, Buonaguro L, Buonaguro FM, Tornesello ML. The role of RNA splicing factors in cancer: Regulation of viral and human gene expression in human papillomavirus-related cervical cancer. Front Cell Dev Biol. 2020;8:474. doi: 10.3389/fcell.2020.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ajiro M, Jia R, Zhang L, Liu X, Zheng ZM. Intron definition and a branch site adenosine at nt 385 control RNA splicing of HPV16 E6*I and E7 expression. PLoS One. 2012;7:e46412. doi: 10.1371/journal.pone.0046412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans W, Filippova M, Filippov V, Bashkirova S, Zhang G, Reeves ME, Duerksen-Hughes P. Overexpression of HPV16 E6* Alters β-Integrin and mitochondrial dysfunction pathways in cervical cancer cells. Cancer Genomics Proteomics. 2016;13:259–273. [PubMed] [Google Scholar]
- 42.Zhang P, Li H, Yang B, Yang F, Zhang LL, Kong QY, Chen XY, Wu ML, Liu J. Biological significance and therapeutic implication of resveratrol-inhibited Wnt, Notch and STAT3 signaling in cervical cancer cells. Genes Cancer. 2014;5:154–164. doi: 10.18632/genesandcancer.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medina-Aguilar R, Marchat LA, Arechaga Ocampo E, Gariglio P, García Mena J, Villegas Sepúlveda N, Martínez Castillo M, López-Camarillo C. Resveratrol inhibits cell cycle progression by targeting Aurora kinase A and Polo-like kinase 1 in breast cancer cells. Oncol Rep. 2016;35:3696–3704. doi: 10.3892/or.2016.4728. [DOI] [PubMed] [Google Scholar]
- 44.Wang TT, Schoene NW, Kim YS, Mizuno CS, Rimando AM. Differential effects of resveratrol and its naturally occurring methylether analogs on cell cycle and apoptosis in human androgen-responsive LNCaP cancer cells. Mol Nutr Food Res. 2010;54:335–344. doi: 10.1002/mnfr.200900143. [DOI] [PubMed] [Google Scholar]
- 45.Hsieh TC, Wu JM. Differential effects on growth, cell cycle arrest, and induction of apoptosis by resveratrol in human prostate cancer cell lines. Exp Cell Res. 1999;249:109–115. doi: 10.1006/excr.1999.4471. [DOI] [PubMed] [Google Scholar]
- 46.Fischer M, Muller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol. 2017;52:638–662. doi: 10.1080/10409238.2017.1360836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The original data are available upon request to the corresponding author.