C-tail proteins are anchored in the inner membrane with a transmembrane segment at the C terminus in an N-in/C-out topology. Due to this topology, membrane insertion occurs only posttranslationally. Nevertheless, the C-tail-anchored protein SciP is targeted cotranslationally by SRP. We report here that two amino-terminal hydrophobic stretches in SciP are individually recognized by SRP and target the nascent protein to FtsY. The presence of two signal sequences may enable a retargeting mechanism, as already observed for multispanning membrane proteins, to make the posttranslational insertion of SciP by YidC more efficient.
KEYWORDS: C-tailed protein, membrane protein targeting, protein sorting, protein folding, signal recognition particle, signal sequences
ABSTRACT
During their synthesis, the C-tailed membrane proteins expose the membrane-spanning segment late from the ribosome and consequently can insert into the membrane only posttranslationally. However, the C-tailed type 6 secretion system (T6SS) component SciP uses the bacterial signal recognition particle (SRP) system for membrane targeting, which operates cotranslationally. Analysis of possible sequence regions in the amino-terminal part of the protein revealed two candidates that were then tested for whether they function as SRP signal peptides. Both sequences were tested positive as synthetic peptides for binding to SRP. In addition, purified ribosomes with stalled nascent chains exposing either sequence were capable of binding to SRP and SRP-FtsY complexes with high affinity. Together, the data suggest that both peptides can serve as an SRP signal sequence promoting an early membrane targeting of SciP during its synthesis. Like observed for multispanning membrane proteins, the two cytoplasmic SRP signal sequences of SciP may also facilitate a retargeting event, making the targeting more efficient.
IMPORTANCE C-tail proteins are anchored in the inner membrane with a transmembrane segment at the C terminus in an N-in/C-out topology. Due to this topology, membrane insertion occurs only posttranslationally. Nevertheless, the C-tail-anchored protein SciP is targeted cotranslationally by SRP. We report here that two amino-terminal hydrophobic stretches in SciP are individually recognized by SRP and target the nascent protein to FtsY. The presence of two signal sequences may enable a retargeting mechanism, as already observed for multispanning membrane proteins, to make the posttranslational insertion of SciP by YidC more efficient.
INTRODUCTION
Gram-negative bacteria are divided into four different cellular subcompartments, including the cytoplasm, the inner and outer membranes, and the periplasm. All of these compartments contain a distinct set of proteins fulfilling important biological functions. The challenge of compartmentalization is that all proteins are synthesized by ribosomes in the cytoplasm, and, subsequently, they have to be sorted and transported to their different target locations in the cell (1). Therefore, various protein sorting pathways exist. Proteins which are directed to the inner membrane of Escherichia coli are delivered either in a post- or cotranslational way, depending on their intrinsic signal elements (2, 3). Cotranslational delivery of inner membrane proteins involves the universally conserved signal recognition particle (SRP) pathway. The SRP pathway is comprised of the cytoplasmic ribonucleoprotein SRP, which consists of the protein component Ffh, the 4.5S RNA, and the membrane-associated SRP receptor FtsY (4–6). Both SRP and FtsY exhibit GTPase activity controlling the cotranslational targeting process (4, 7). SRP is associated with ribosomes by binding to the ribosomal proteins uL23, uL29, and bL32 and the ribosomal 23S RNA. It scans the growing nascent chain for the presence of an SRP signal sequence (8–11). As soon as an SRP signal sequence is exposed outside the ribosomal exit tunnel, it is bound with high affinity by SRP via its hydrophobic groove that is formed by the M domain (9, 12–14). Consequently, the ribosome-nascent chain complex (RNC) is targeted to the membrane-associated SRP receptor FtsY (15). The interaction of SRP with its receptor leads to the coordinated activation of the two GTPase domains in SRP and FtsY (16). After GTP hydrolysis, the nascent chain is forwarded to the Sec translocon or the YidC insertase to be integrated into the bilayer, and SRP enters a new targeting cycle (17, 18).
For SRP signal sequences, no consensus motif has been identified. However, an uninterrupted stretch of at least 8 hydrophobic amino acids was found to be crucial for SRP binding (19–22). In most cases, the N-terminal first transmembrane segment (TMS) located within the first 100 amino acids of an inner membrane protein serves as an SRP signal sequence (20). In addition to hydrophobicity, basic amino acids in the N-terminal part of an SRP signal sequence promote SRP binding, probably due to electrostatic interactions with the SRP RNA (23). In 2008, it was shown that SRP signal sequences are not limited to transmembrane domains of inner membrane proteins. The four-spanning potassium sensor protein KdpD has its first transmembrane domain 400 amino acids downstream from the N terminus (24). However, KdpD is still targeted by SRP early during translation; a short amphiphilic region, located between amino acid residues 22 to 48 in the cytoplasmic domain of KdpD, was identified to facilitate SRP-dependent targeting (25). Similar to KdpD, the C-tail-anchored protein SciP from enteroaggregative E. coli (EAEC) is targeted early during translation by SRP, although the insertion of the TMS occurs posttranslationally via YidC due to the special (C-out) topology (26). Interestingly, two short hydrophobic regions between amino acids 12 to 20 and 62 to 71 within the N-terminal cytoplasmic domain were found to be involved in membrane targeting of SciP. Probably, this early targeting mechanism ensures that the RNC is already located close to the YidC insertase to directly receive the C-terminal hydrophobic transmembrane domain and to protect it from the aqueous surrounding. However, no other SRP substrate with more than one SRP signal sequence in a cytoplasmic domain is described in the literature to date.
The question of whether both amino-terminal hydrophobic regions of SciP actually serve as SRP signal sequences, as suspected from in vivo experiments (26), is addressed in this study. In vitro interaction studies of purified SRP- and TnaC-stalled ribosomes (27) using microscale thermophoresis demonstrate that both hydrophobic regions of SciP are bound with similar affinity to SRP or an SRP-FtsY complex. We also show here, by in vitro cross-linking studies, that short peptides of both signal sequences are binding to the SRP M domain, similar to other known SRP substrates. The presence of two cytoplasmic SRP signal sequences may enable a retargeting event, as already observed for multispanning membrane proteins, to make the insertion of SciP by YidC more efficient.
RESULTS
Binding of SciP nascent chains to SRP in vitro.
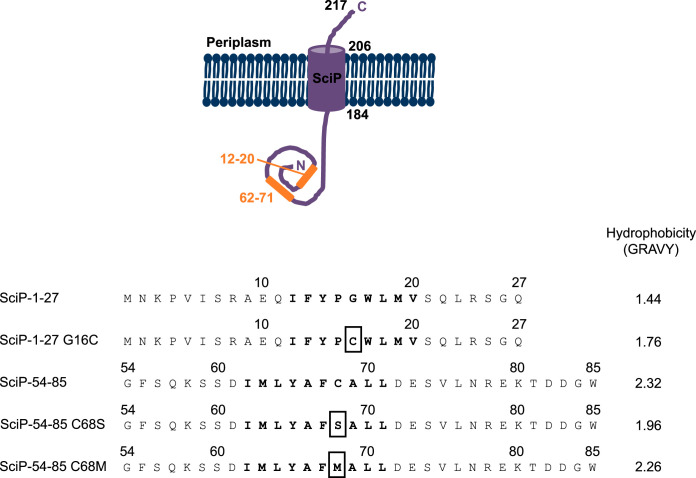
The C-tail-anchored protein SciP of EAEC consists of 217 amino acid residues and is anchored by one transmembrane segment located at the extreme C-terminal part of the protein between amino acid residues 184 and 206. This results in a short periplasmic C-tail of only 11 amino acids and a large N-terminal cytoplasmic domain of 183 amino acids (Fig. 1, top). Recently, we have shown that the insertion of SciP into the inner E. coli membrane depends on the SRP system and YidC (26). Two short hydrophobic stretches in the N-terminal cytoplasmic domain (amino acids [aa] 12 to 20 and 62 to 71) were identified as potential SRP signal sequences (Fig. 1), and both regions are able to target a superfolder green fluorescent protein (sfGFP) fusion construct to the membrane (26). In addition, deletion of both regions prevented targeting of SciP, as shown with fluorescence microscopy (26).
FIG 1.
Topology of SciP and the amino acid sequences of the N-terminal cytoplasmic hydrophobic regions of SciP (aa 12 to 20 and 62 to 71; orange) involved in SRP targeting. The C-tail anchored protein SciP consists of one transmembrane domain located between amino acids 184 to 206, a short periplasmic domain of 11 amino acids, and a large cytoplasmic domain of 183 amino acids. The hydrophobic regions involved in SRP targeting are located between amino acids 1 to 27 and 54 to 85, and the hydrophobic stretch is displayed in bold letters. The mutations analyzed in this study are marked with a box. The grand average of hydropathy (GRAVY scale [42]) of each shown sequence is indicated.
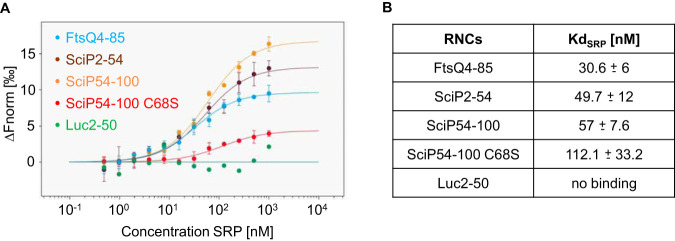
To explore whether the two hydrophobic stretches in the N-terminal cytoplasmic domain of SciP support the binding of SRP cotranslationally, ribosome-nascent chain complexes were generated. Hence, the two signal sequences of SciP with an extended sequence (amino acids 2 to 54 and 54 to 100) were fused to the TnaC-stalling sequence. This approach allows a tryptophan-dependent ribosomal stalling in vivo (27). Since about 30 amino acids are harbored in the ribosomal exit tunnel, the nascent chains with the signal sequences (aa 12 to 20 and 62 to 71) were extended to ensure that they are exposed outside the tunnel. The RNCs were produced in E. coli KC6 (28) and purified via an N-terminal His tag and a 10 to 40% sucrose gradient. The binding of the stalled ribosomes with purified SRP was investigated with microscale thermophoresis (MST) measurements (Fig. 2). The stalled ribosomes were labeled with the cysteine-reactive fluorescent dye NT-647. A fixed concentration of 5 nM labeled ribosomes was incubated with unlabeled SRP ranging from 1 μM to 0.5 nM. As a control, RNCs exposing the amino acids 4 to 85 of the SRP substrate FtsQ were used (29), resulting in a dissociation constant (Kd) of 30.6 ± 6 nM (Fig. 2A, blue dots). In contrast, the nascent chains of the cytoplasmic protein luciferase (residues 2 to 50) served as a negative control which showed no binding to SRP in this concentration range (green dots). First, ribosomes with nascent chains of the SciP residues 2 to 54 were incubated with SRP. The MST measurements (Fig. 2B) resulted in a binding event with an estimated Kd of 49.7 ± 12 nM (brown dots), which is in the same range as the positive-control FtsQ. Next, ribosomes with nascent chains of the SciP residues 54 to 100 were applied for the MST measurements with SRP. Also, in this case, a clear binding was observed with a Kd of 57 ± 7.6 nM (orange dots), comparable to the positive-control FtsQ. We conclude from the MST measurements that both hydrophobic regions of SciP can bind independently of each other to SRP when exposed from a ribosome.
FIG 2.
Microscale thermophoresis measurements of SRP with RNCs. Unlabeled SRP (1 μM to 0.49 nM) was incubated with 5 nM labeled RNCs for 5 min on ice and filled into premium capillaries (NanoTemper Technologies). (A) The change in normalized fluorescence (ΔFnorm) of FtsQ4-85, SciP2-54, SciP54-100, SciP54-100 C68S, and Luc2-50 is plotted against the concentration of unlabeled SRP. (B) Summary of the Kd values (three independently pipetted measurements are merged, and the error bars represent the standard deviation). The raw data of the MST measurements are shown in Fig. S1 and Table S1 in the supplemental material.
Mutations in the signal peptide affect SRP-dependent targeting.
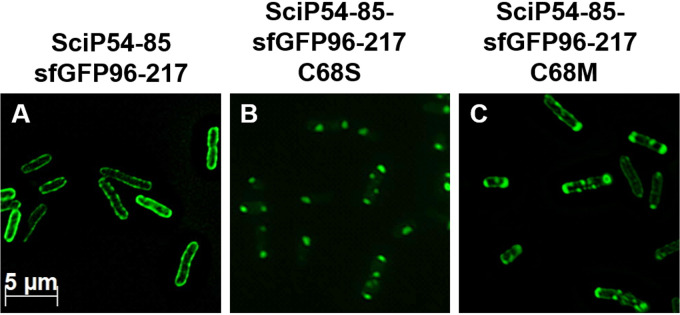
For SRP signal sequences, no consensus motif has been found. However, an uninterrupted stretch of at least 8 hydrophobic residues enables SRP binding (19–22). Also for the potassium sensor protein KdpD, it was shown that the hydrophobicity of its signal peptide is crucial for its SRP-dependent targeting (29). To monitor the membrane targeting of SciP, the second hydrophobic region (residues 54 to 85) had been fused to the N terminus of the fluorescent protein sfGFP (26). We used this construct to investigate whether mutations in this hydrophobic region of SciP affect the targeting to the membrane. The cysteine residue at position 68 in the hydrophobic stretch was substituted with methionine (C68M) or with a hydrophilic serine residue (C68S). The localization of the mutants was monitored in E. coli MC4100 cells using fluorescence microscopy (Fig. 3), and expression was analyzed by SDS-PAGE (see Fig. S2 in the supplemental material). The expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h at 30°C, and the samples were applied on poly-l-lysine-coated slides for fluorescence microscopy with a specific filter set for GFP. The fluorescent signal of the SciP54-85–sfGFP-96-217 construct was found at the membrane, demonstrating that the second hydrophobic region of SciP targets the fusion construct to the membrane (Fig. 3A). In contrast, the replacement of cysteine 68 with a serine residue resulted in a patchy fluorescent signal mainly found at the cell poles (Fig. 3B), indicating that membrane targeting is impaired. The replacement of the cysteine with a methionine residue partially restored the targeting defects of the C68S mutant (Fig. 3C). Taken together, the results obtained with the mutants of the targeting signals show that their hydrophobicity is a common feature for SRP-dependent targeting of SciP.
FIG 3.
Fluorescence microscopy of various SciP-sfGFP fusion proteins. E. coli MC4100 cells were transformed with the respective plasmid, and the expression was induced with 1 mM IPTG for 1 h at 30°C. (A) The fluorescent signal of SciP54-85 was clearly found at the membrane, indicating a targeting function of this region. (B) Replacement of the cysteine residue at position 68 with a serine residue led to distinct spots, probably due to the aggregated protein. (C) Replacement of the cysteine with a methionine residue resulted in a mixed phenotype with distinct spots, mainly at the cell poles and signals found at the membrane.
To test whether the targeting defects of the SciP C68S mutant that we had observed by fluorescence microscopy in vivo (Fig. 3B) correlate with the SRP binding ability, the affinity of the C68S mutant for SRP was measured with MST in vitro. Therefore, the C68S mutation was introduced in the SciP54-100 RNC construct (Fig. 2). The C68S mutation resulted in a 2-fold decrease in the binding affinity with a Kd of 112.1 ± 33.2 nM (Fig. 2A, red dots) compared to the SciP54-85 wild-type sequence (Kd of 57 ± 7.6 nM) (orange dots). We conclude that the less hydrophobic SciP signal sequence is also affected for its binding to SRP, explaining the impaired membrane targeting.
Binding of SciP-nascent chains to an SRP-FtsY complex.
Since in vivo, the ribosome nascent chain (RNC)-SRP complex is targeted to the SRP receptor at the membrane, we were interested to study the binding of the RNCs, SRP, and the SRP receptor FtsY in vitro. To test this, a preformed and stable SRP-FtsY complex was employed for the interaction studies with the RNCs as has been described earlier (13).
In vivo, the major amount of FtsY is located at the membrane; however, as shown in several other studies with Sec-dependent substrates, interaction and binding of SRP or an SRP-RNC complex with soluble FtsY may also occur (30–34). Based on this, it was concluded that in vitro binding of FtsY to an RNC-SRP complex in solution occurs, but to generate the ternary complex RNC-SRP-FtsY-SecYEG, the interaction of FtsY with lipids and the SecYEG translocon is required (35, 36).
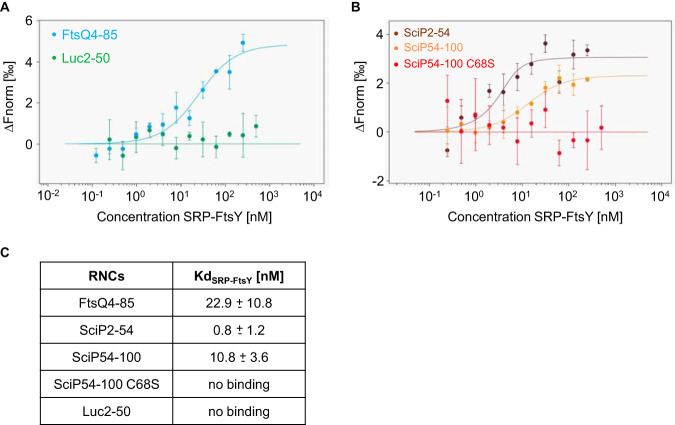
Purified SRP and the SRP receptor protein FtsY were preincubated in the presence of 0.2 mM GMP-PNP (guanosine 5′-imidotriphosphate trisodium salt hydrate) for the MST measurements (Fig. 4). A fixed concentration of 5 nM labeled RNCs was incubated with increasing SRP-FtsY complex concentrations ranging from 0.1 to 250 nM. First, RNCs with amino acids 2 to 54 of SciP as a nascent chain were analyzed with SRP-FtsY, resulting in a Kd of 0.8 ± 1.2 nM (Fig. 4B, brown dots). This binding affinity is even stronger than that for the positive-control FtsQ with an estimated Kd of 22.9 ± 10.8 nM (Fig. 4A, blue dots). RNCs with amino acids 54 to 100 of SciP were also bound by SRP-FtsY with a Kd of 10.8 ± 3.6 nM (Fig. 4B, orange dots). In contrast, RNCs exposing the nascent chain of luciferase did not show any binding to the SRP-FtsY complex (Fig. 4, green dots). From that, we conclude that both nascent chains exposing either one of the SciP hydrophobic regions bind to SRP-FtsY with high affinities and therefore represent true SRP substrates.
FIG 4.
Microscale thermophoresis measurements of a preincubated SRP-FtsY complex with RNCs. Unlabeled SRP-FtsY (500/250 nM to 0.24/0.12 nM) was mixed with 5 nM labeled RNCs, incubated for 5 min on ice, and filled into premium capillaries (NanoTemper Technologies). (A) The change in normalized fluorescence (ΔFnorm) of FtsQ4-85 and Luc2-50 is plotted against the concentration of unlabeled SRP-FtsY. (B) The change in normalized fluorescence (ΔFnorm) of SciP2-54, SciP54-100, and SciP54-100 C68S is plotted against the concentration of unlabeled SRP-FtsY. (C) Summary of the Kd values (three independently pipetted measurements are merged, and the error bars represent the standard deviation). The raw data of the MST measurements are shown in Fig. S3, Fig. S4, and Table S2.
The exchange of the cysteine residue at position 68 with a serine residue resulted in a 2-fold decreased affinity for SRP (Fig. 2). The MST measurements of the C68S mutant for SRP-FtsY complex binding revealed no affinity (Fig. 4B, red dots). This clearly shows that the C68S mutant is no functional SRP substrate.
Peptides composed of the two SRP signal sequences of SciP contact the SRP M domain.
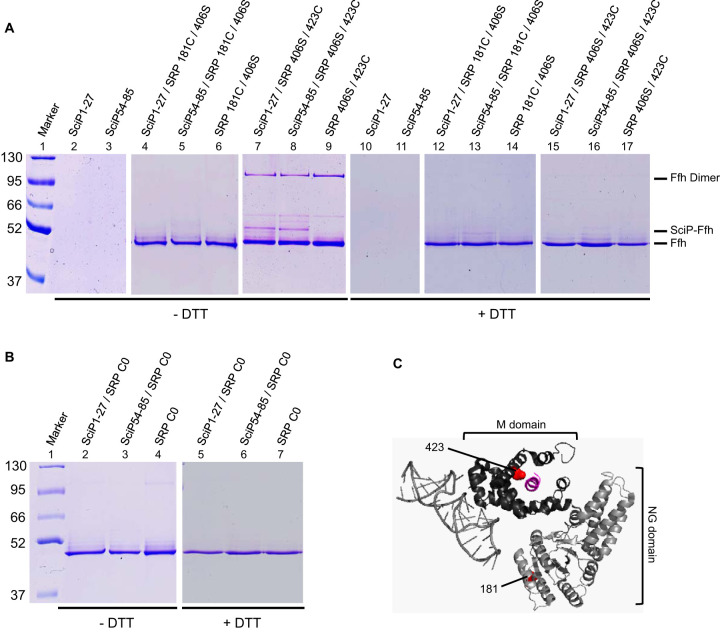
SRP signal sequences are bound after their exposure on the ribosome by a hydrophobic groove of the M domain within SRP (37). Whether the two SRP signal sequences of SciP both contact the SRP M domain was addressed with in vitro disulfide cross-linking studies using the oxidizing agent copper phenanthroline. Therefore, the two putative SRP signal sequences of SciP were synthesized in vitro as individual peptides. In a previous study, the short ΔEspP signal peptide was shown by fluorescence anisotropy to interact with SRP, suggesting that SRP is also able to bind to short peptides without the context of a ribosome (38). For disulfide-cross-linking studies, a single cysteine residue is required in each interaction partner. The SciP54-85 peptide naturally has a cysteine residue at position 68 (Fig. 1). However, for SciP1-27, a cysteine residue had to be incorporated since no endogenous cysteine residue was present. We decided to substitute the glycine residue at position 16 with a cysteine (Fig. 1). For the cross-linking studies, two mutant Ffh proteins (the protein component of SRP) with a cysteine residue at position 423 in the M domain or a cysteine residue at position 181 in the NG domain were used (Fig. 5C). In both Ffh mutants, the endogenous cysteine residue at position 406 had been substituted with a serine residue. Two micromolar of the respective reconstituted SRP were incubated with 20 μM of the respective SciP peptide and 1 mM copper phenanthroline for 1 h on ice. The samples were trichloroacetic acid (TCA) precipitated, resuspended in buffer with or without dithiothreitol (DTT), and analyzed by 10% SDS-PAGE. When the SciP peptides were incubated with SRP bearing a cysteine residue at position 423 in the M domain, an additional band at about 100 kDa appeared (Fig. 5A, lanes 7 and 8). This band corresponds to the Ffh dimer, formed by intermolecular cross-linking due to the accessible position of the incorporated cysteine residue at position 423 in the M domain. Although the cross-linked Ffh dimer probably leads to a competition event with the SciP peptides, an additional band was visible at about 52 kDa, corresponding to the SciP-Ffh cross-linking product (Fig. 5A, lanes 7 and 8). This band was also sensitive to the reducing agent DTT (lanes 15 and 16). In addition, no cross-linking band at 52 kDa appeared when the isolated peptides or SRP 406S/423C alone were incubated with copper phenanthroline (lanes 2, 3, and 9). Also, no additional band was visible when the two peptides were incubated with the SRP 181C/406S mutant (lanes 4 and 5) having a cysteine residue in the NG domain or no cysteine in the cysteine-less SRP C0 mutant (Fig. 5B, lanes 2 and 3).
FIG 5.
In vitro disulfide cross-linking of SRP and SciP1-27/SciP54-85 with copper phenanthroline. (A and B) We incubated 20 μM of the SciP1-27 or SciP54-85 peptides (single cysteines at positions 16 and 68, respectively) with 2 μM reconstituted SRP L181C/406S or SRP 406S/423C (A) or SRP C0 (B) and 1 mM copper phenanthroline for 1 h on ice. The samples were TCA precipitated, resuspended in buffer with or without DTT, and loaded on 10% SDS-PAGE. When the peptides were incubated with SRP 406S/423C, a 52-kDa cross-linking band (SciP-Ffh) appeared. The shifted bands were sensitive to the reducing agent DTT. No cross-linking band appeared with SRP 181C/406S and SRP C0. The SciP peptides and the SRP mutants alone were analyzed for control. (C) Crystal structure of the E. coli SRP bound to a signal sequence, displayed as a cartoon. The 4.5S RNA and the Ffh NG domain are colored in light gray, the Ffh M domain is in dark gray, and the signal sequence is in purple. The positions of the incorporated cysteine residues are displayed as red dots (image created with PyMOL 2.3.0; PDB ID 5GAD).
The SciP mutant with the G16C mutation was then tested in vivo for membrane targeting in the SciP1-27–sfGFP fusion construct, its localization in the cell was analyzed with fluorescence microscopy (Fig. 6), and its expression was analyzed after induction with 1 mM IPTG in E. coli MC4100 cells for 1 h at 30°C (Fig. S2). As shown for both the wild-type SciP1-27 sequence (Fig. 6A) and the G16C mutant (Fig. 6B), the fluorescent signal was located at the membrane, indicating that SRP-dependent targeting is normal. Taking these results together, we conclude that both signal sequences of SciP bind to SRP in the hydrophobic groove of the M domain.
FIG 6.
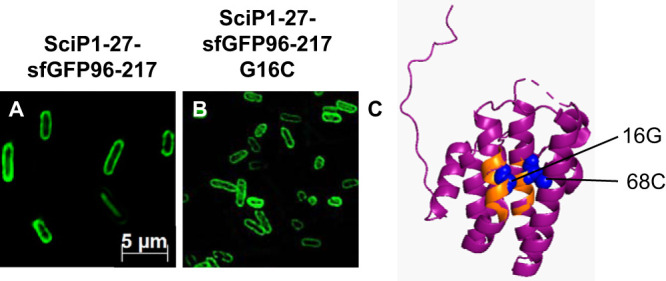
(A and B) Subcellular localization studies of SciP1-27-sfGFP96-217 and the mutant fusion protein. The fluorescent signals of SciP1-27-sfGFP96-217 (A) and SciP1-27-sfGFP96-217 G16C (B) were clearly found at the membrane, showing that the mutation G16C has no influence on SRP-dependent targeting. (C) In the structure of the cytoplasmic domain of SciP, the two hydrophobic regions (amino acids 12 to 20 and 62 to 71) are colored orange, and the glycine residue at position 16, as well as the cysteine residue at position 68, are highlighted as blue spheres, respectively (image created with PyMOL 2.3.0; PDB ID 3U66).
The membrane insertion of SciP requires only one of the two signal sequences.
In vitro, both hydrophobic regions of SciP are recognized and bound by the SRP M domain, which results in the recruitment of the SRP receptor FtsY. To explore whether the mutant proteins lacking one or both hydrophobic regions were also defective for the insertion of SciP in vivo, we deleted these regions individually or together. Membrane insertion was monitored with a periplasmic cysteine accessibility assay using 4-acetoamido-4′-maleimidylstilbene-2,2′disodium sulfonate (AMS). To do this, a cysteine residue was incorporated at position 218 of SciP (SciP-C). The insertion of SciP-C and the mutant proteins was tested in E. coli MC4100 cells. The expression was induced with 1 mM IPTG for 10 min, and 2.5 mM AMS was added 1 min prior to pulse-labeling with radioactive methionine/cysteine. After 2 min of pulse-labeling, nonradioactive methionine/cysteine was added for 10 min, and the AMS reaction was quenched with 10 mM DTT for an additional 10 min. The samples were TCA precipitated, immunoprecipitated with anti-His antibody, and analyzed qualitatively by 14% SDS-PAGE (Fig. S5). When both hydrophobic stretches were present, as is the case for SciP-C, the protein was modified with AMS, indicating an efficient translocation of the C-tail (Fig. S5, lane 1). The deletion of either one of the hydrophobic regions did not have an influence on the translocation efficiency of the C-tail since a comparable amount was modified with AMS, respectively (lanes 2 and 3). In contrast, the deletion of both regions led to a decreased translocation of the C-tail since a weaker modification with AMS was visible (lane 4). These results are the first indications that both regions are functional SRP-targeting signals, but it seems that the presence of only one is sufficient for the efficient insertion of SciP into the inner membrane. However, we cannot exclude the possibility that other factors compensate for the loss of SRP binding sites since a weak amount of SciP is still inserted in the absence of the two hydrophobic stretches.
DISCUSSION
In the N-terminal domain of the C-tailed protein SciP, two SRP signal sequences were found close to the N terminus at amino acid residues 12 to 20 and more distant at 62 to 71. We show here that both these sequences bind to SRP and the SRP-FtsY complex with a high affinity when they are exposed as RNCs. Deletion analysis further indicated that it is sufficient if one of the two signal sequences is present to efficiently insert SciP into the membrane of E. coli (see Fig. S5 in the supplemental material). However, when both hydrophobic stretches were deleted, structural effects and the fragility of the SciP mutant protein did not allow us to study the fate of the protein posttranslationally.
To our knowledge, this is the first case where two bona fide signal sequences have been found in a cytoplasmic domain of an SRP substrate. Presumably, the extra signal sequence may function as a backup system if one signal is skipped. Global profiling studies have recently suggested that RNCs can be bound multiple times by SRP, and the observed binding events correlate with the transmembrane domains in multispanning membrane proteins (39). It is suggested that RNCs exposing an only moderately hydrophobic TMS sometimes lose contact with the translocon and need to be retargeted by SRP by the binding of an exposed downstream TMS (39). Similarly, a retargeting mechanism could also occur in the cytoplasmic domain of a C-tail protein to support insertion into the inner membrane protein. The presence of the two SRP binding sites might prevent the SciP-RNC from losing contact with the membrane too early, before the TMS can contact the membrane insertase YidC, which is necessary for the insertion of SciP (26). Therefore, this mechanism may contribute to more efficient targeting and successful membrane insertion of SciP.
Our in vitro disulfide cross-linking studies show that both SRP signal sequences can bind to the SRP M domain, similar to the mechanism found with other SRP signal sequences. This is also in good agreement with the results obtained with the cytoplasmic SRP signal sequence of KdpD. As in this case, the SRP signal sequence is embedded in the hydrophobic groove formed by the SRP M domain (29). In another study, an SRP recognition sequence was detected for the cytoplasmic σ32 as it cross-linked to the hydrophobic groove of SRP (40). During the negative feedback regulation in heat shock response, σ32 is targeted to the membrane via the SRP system for degradation by FtsH (41). From these results, it was concluded that also cytoplasmic proteins can contain SRP signal sequences and that they are bound in a similar manner by SRP as SRP signal sequences of transmembrane proteins.
The ribosomal profiling studies showed that SRP can bind to nascent protein chains when the N terminus reaches a distance of 42 to 100 amino acid residues from the peptidyl transferase center (PTC), exposing an SRP signal sequence (39). In our arrested RNCs, the N termini of the two SRP signal sequences of SciP are located 73 and 66 amino acid residues from the PTC, respectively. These distances correlate well with the findings of the ribosomal profiling studies. In most cases, the N-terminal first TMS serves as an SRP signal sequence; however, SciP is targeted by SRP via two short hydrophobic sequences located in the N-terminal cytoplasmic region. The fact that SRP signal sequences are not limited to transmembrane domains was also shown for the potassium sensor protein KdpD of E. coli. A short amphiphilic region in the N-terminal cytoplasmic part between amino acid residues 22 to 48 serves as an SRP signal sequence and enables cotranslational targeting of KdpD (25). The two SRP signal sequences of SciP correlate well with the important characteristics for SRP binding identified in the KdpD SRP signal sequence. Both regions contain a hydrophobic core region with grand average of hydropathy (GRAVY) (42) scores of 1.44 and 2.54 compared to the KdpD signal sequence with a GRAVY score of 1.33. The first SRP region of SciP shows only moderate hydrophobicity compared to other known SRP signal sequences. Also, the KdpD signal sequence is only moderately hydrophobic. However, it is assumed that the lower hydrophobicity is compensated by the presence of three basic amino acid residues that contribute to the affinity for binding to SRP (29). The fact that positively charged amino acid residues promote SRP binding was also shown for the SecA-dependent proteins MBP and OmpA. Increasing the number of basic amino acid residues to the only moderately hydrophobic signal sequences of MBP and OmpA resulted in the rerouting of both proteins into the SRP pathway (23). The first moderately hydrophobic region of SciP is preceded by two positively charged amino acid residues. Presumably, the only moderately hydrophobic region (aa 12 to 20) is sufficient to be recognized by SRP due to the presence of these two basic amino acid residues, supporting a stable SRP binding by the generation of salt bridges to the SRP RNA. In contrast, the second signal sequence region of SciP is very hydrophobic and contains only one positively charged residue. Here, the higher hydrophobicity might be sufficient for high-affinity SRP binding, and additional basic amino acids are not needed for strengthening the binding.
Lowering the hydrophobicity of the second SRP signal sequence of SciP from a score of the GRAVY scale of 2.54 to 1.96 resulted in targeting defects, as shown by fluorescence microscopy (Fig. 3), and prevented SRP-FtsY binding, as shown with MST (Fig. 4). The binding of SRP-FtsY to an exposed SRP signal sequence is a crucial step in the SRP cycle and depends on several conformational changes of the two GTP binding domains in SRP and FtsY. FtsY recruitment represents another checkpoint for the discrimination between an SRP or a non-SRP substrate (43, 44). Hence, FtsY is only recruited, and a stable RNC-SRP-FtsY complex can only be formed if SRP is bound to a correct cargo (13). The C68S mutant can still be bound by SRP but with a weaker affinity than the wild type. However, the second checkpoint in the SRP pathway, the formation of a closed SRP-FtsY complex, was obviously inhibited. In addition, fluorescence microscopy of the C68S mutant cells showed a patchy fluorescent signal mainly found at the cell poles, which is probably due to the aggregation of the mutant protein in the cytoplasm since the interaction of SRP with FtsY is inhibited. From that, we concluded that the C68S mutant with a lower hydrophobicity is no longer recognized as an SRP substrate. These findings correlate with the results we had obtained with KdpD. Also, in this case, the decrease in hydrophobicity resulted in impaired SRP and SRP-FtsY binding to KdpD monitored with MST (29). Taking these results together, we conclude that the N-terminal cytoplasmic SRP signal sequences have to exceed a certain threshold level of hydrophobicity or have additional basic amino acids to ensure efficient recruitment of the SRP receptor FtsY and to complete the cotranslational SRP targeting cycle.
In the cotranslational pathway, SRP binding occurs early to an exposed nascent chain at the exit site of the ribosome. This shields a hydrophobic protein chain from the hydrophilic cytoplasm and also prevents premature folding of the protein. Therefore, the folding of the protein is possible only after the release from SRP, and this will occur when the RNC-SRP has contacted the SRP receptor at the membrane. In the case of SciP, the release of the nascent chain from SRP allows both signal sequences to fold into an α-helical bundle (Fig. 6C) that functions as a structural component of the type 6 secretion system (T6SS) necessary for the dimerization and assembly of a ring complex (45, 46).
The release of the nascent chain from SRP likely occurs after docking to the FtsY receptor at the membrane. In the SRP-YidC pathway, the handover of a substrate is probably facilitated by the direct interaction of both SRP and FtsY with YidC. Cross-linking experiments have shown that SRP and FtsY interact with YidC via binding to the first cytoplasmic (C1) loop (47). This would attribute the YidC-bound FtsY as the function of the membrane-docking site for SRP-YidC substrates. The C1 domain of YidC forms a helical hairpin structure at the entrance of the hydrophilic groove of YidC and is essential for YidC function. Cysteine accessibility assays with SciP and different YidC mutant proteins demonstrated that the second helix of the first cytoplasmic loop (CH2) of YidC is important for the insertion of SciP (48). In agreement with these results, we think that the C1 loop of YidC plays a crucial role in the recruitment of a cotranslationally targeted YidC substrate via the SRP system.
For RNCs exposing the first hydrophobic region of SciP, a 50-times-higher affinity for FtsY-SRP than for SRP alone was measured, while no increase in the affinity for the positive-control FtsQ was observed. The reason for this might be based on the amphiphilic α-helical structure of the SciP residues 6 to 27, which is not found for the FtsQ signal sequence, or just on the fact that the membrane insertion pathways are different for the two proteins. Whereas SciP requires YidC, FtsQ is a SecYEG-dependent protein (49).
YidC also interacts with translating ribosomes, exposing a YidC substrate. In contrast, nontranslating ribosomes only show a weak binding affinity to YidC (18, 50, 51). The interaction of YidC with RNCs was shown in a reconstituted system, even in the absence of SRP (50). This may be the explanation for the decreased insertion rate we had observed in the absence of both signal sequences (Fig. S5) or of SRP (26). In vivo, efficient targeting and insertion of SciP occur cotranslationally, and a nascent chain of SciP binds to the membrane. This is then probably received by FtsY that is bound to YidC and prepared for the interaction with the C-terminal transmembrane domain (TMD). After the insertion by YidC and release into the lipid bilayer, the SciP protein can dimerize via its TMDs (46, 52), enabling the structural function in the T6SS.
A previous study with two additional C-tail-anchored proteins, DjlC and Flk, had also suggested that SRP is involved in their membrane targeting. In contrast to SciP, the studies indicated that for DjlC and Flk, SRP binds to the C-terminal transmembrane domains and that these proteins are targeted by SRP in a posttranslational manner (53). This is different for SciP, where a membrane targeting of the C-terminal transmembrane domain fused to the fluorescent protein sfGFP did not occur (26). Therefore, different insertion pathways may exist in E. coli. Also for eukaryotic tail-anchored (TA) proteins, different targeting and insertion pathways exist. In mammalian cells and yeast, some of these can insert by a nonassisted pathway. In vitro studies with the TA proteins Cytb5 and tyrosine phosphatase 1B showed that they can insert into liposomes without the assistance of other proteins or nucleotides (54–56). However, it seems that the majority of the TA proteins use a chaperone-mediated pathway, which divides into an SRP, Hsc70/Hsp40, SND (SRP-independent targeting), or the GET (guided entry of tail-anchored proteins) pathway (57–59).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli BL21(DE3) (60), MC4100 (61), and KC6 (28) cells were grown overnight in LB medium with 100 μg ml−1 ampicillin. The next day, the cultures were back diluted in fresh LB medium supplemented with 100 μg ml−1 ampicillin and grown at 37°C.
Construction of the SciP-sfGFP mutants.
The replacement of the glycine with a cysteine residue at position 16 (SciP1-27 G16C) was done with site-directed mutagenesis on plasmid pMS-SciP1-27sfGFP96-217 (26) with the oligonucleotides 5′-CAG ATT TTT TAT CCC TGC TGG CTG ATG GTC-3′ and 5′-GAC CAT CAG CCA GCA GGG ATA AAA AAT CTG-3′. The substitutions in the second hydrophobic region of SciP were generated with site-directed mutagenesis on plasmid pMS-SciP54-85sfGFP96-217 (26). The oligonucleotides 5′-C ATG TTG TAT GCC TTC AGC GCC CTG CTG G-3′ and 5′-C CAG CAG GGC GCT GAA GGC ATA CAA CAT G-3′ were used to generate pMS-SciP54-85sfGFP96-217 C68S (named SciP54-85 C68S), whereas the oligonucleotides 5′-C ATG TTG TAT GCC TTC ATG GCC CTG CTG GAC G-3′ and 5′-C GTC CAG CAG GGC CAT GAA GGC ATA CAA CAT G-3′ resulted in the construction of pMS-SciP54-85sfGFP96-217 C68M (named SciP54-85 C68M).
Construction of plasmids for ribosome-nascent chain complexes.
The sequences coding for amino acids 2 to 54 or 54 to 100 of SciP were amplified from pMS-SciP-C using the oligonucleotides 5′-GGC CAA TTG AAT AAA CCT GTT ATC TCC CGG GC-3′ and 5′-CGG CCA TGG TCC TGC TTC GGC CAG CTC TTC ACG-3′ (SciP2-54) or 5′-GGC CAA TTG GGA TTC AGT CAG AAA AGC AGT GAC-3′ and 5′-CGG CCA TGG CGT ACC AAA AAA ATG AGC CTG CAG C-3′ (SciP54-100) flanking the PCR products with MfeI and NcoI restriction sites. The PCR products and the plasmid pMS-MscL115-TnaC (29) were digested with MfeI and NcoI and ligated to exchange the mscL sequence with the two sciP sequences, respectively. To generate pMS-SciP54-100-C68S-TnaC, site-directed mutagenesis was done on plasmid pMS-SciP54-100-TnaC using the oligonucleotides 5′-C ATG TTG TAT GCC TTC AGC GCC CTG CTG G-3′ and 5′-C CAG CAG GGC GCT GAA GGC ATA CAA CAT G-3′. All coding sequences were verified by sequence analysis.
As a negative control for the MST measurements, the plasmid pMS-Luc2-50 (29) was used. The plasmid pEM36-3C encoding amino acids 4 to 85 of FtsQ fused to the TnaC-stalling sequence (kindly provided by R. Beckmann, Munich, Germany) was used as a positive control.
Plasmid construction for cysteine accessibility assays.
To delete the sequence coding for amino acids 12 to 20 and 62 to 71 of SciP, site-directed mutagenesis was done on plasmid pMS-SciP-C (26) using the oligonucleotide pairs 5′-CT GTT ATC TCC CGG GCT GAA CAG AGC CAG CTG CGC AGC-3′ with 5′-GCT GCG CAG CTG GCT CTG TTC AGC CCG GGA GAT AAC AG-3′ (Δ12 to Δ20) and 5′-GCA GGA TTC AGT CAG AAA AGC AGT GAC GAC GAG AGT GTA CTG AAC CGC G-3′ with 5′-C GCG GTT CAG TAC ACT CTC GTC GTC ACT GCT TTT CTG ACT GAA TCC TGC-3′ (Δ62 to Δ71), resulting in plasmids pMS-SciP-CΔ12-20, pMS-SciP-CΔ62-71, and pMS-SciP-CΔ12-20 and Δ62-71, respectively. The coding regions of the constructs were verified by sequence analysis.
Fluorescence microscopy.
The plasmids pMS-SciP1-27sfGFP96-217 (26), pMS-SciP1-27sfGFP96-217 G16C, pMS-SciP54-85sfGFP96-217 (26), pMS-SciP54-85sfGFP96-217 C68S, and pMS-SciP54-85sfGFP96-217 C68M were transformed in E. coli MC4100 cells, respectively. The cells were grown to an optical density at 600 nm (OD600) of 0.5, induced with 1 mM IPTG for 1 h at 30°C, and washed twice with fresh LB medium. The cells were resuspended in 2 mM EDTA and 50 mM Tris-HCl (pH 8) and were applied to poly-l-lysine-coated cover slides for fluorescence microscopy with the Axio Imager M1 and a filter set for GFP.
Purification and labeling of ribosome-nascent chain complexes.
E. coli KC6 cells were transformed with plasmids pMS-SciP2-54-TnaC, pMS-SciP54-100-TnaC, pMS-SciP54-100-C68S-TnaC, pMS-Luc2-50-TnaC, and pEM36-3C. The purification and labeling were done as described by Pross and Kuhn (29). The cells were grown to an OD600 of 0.5 in 2 liters LB with 100 μg ml−1 ampicillin and induced with 1 mM IPTG for 1 h at 37°C. The cells were harvested and resuspended in 20 ml buffer ARNC {20 mM HEPES (pH 7.2), 250 mM potassium acetate (KOAc), 25 mM magnesium acetate [Mg(OAc)2], 2 mM l-tryptophan, and 0.1% dodecyl-β-d-maltoside (DDM)}. We added 0.2 mM phenylmethylsulfonyl fluoride (PMSF), the cells were lysed at 1.23 kbar using the OneShot, and the lysate was centrifuged for 20 min at 31,000 × g. The supernatant (10 ml) was loaded onto 45 ml of 750 mM sucrose in buffer ARNC and centrifuged for 20 h at 49,000 × g. The ribosomal pellet was resuspended in 20 ml buffer ARNC and loaded into a column with 3 ml Ni-nitrilotriacetic acid (Ni-NTA) (GE Healthcare) (blocked by incubation with 10 μg ml−1 E. coli tRNA for 1 h at 4°C). After 1 h at 4°C on a rotary wheel, the flowthrough was collected, and the matrix was washed with 30 ml buffer BRNC (50 mM HEPES [pH 7.2], 500 mM KOAc, 25 mM MgCl2, 2 mM l-tryptophan, and 0.1% DDM). The His-tagged ribosomes were eluted with 3 ml buffer BRNC supplemented with 150 mM imidazole and 3 ml buffer BRNC with 300 mM imidazole in 1-ml fractions. The elution fractions were pooled, loaded on a linear sucrose gradient from 10 to 40% sucrose in buffer BRNC, and centrifuged for 3.5 h at 114,000 × g. The sucrose gradient was collected in 1-ml fractions, and the fractions containing the ribosomes (determined via the absorbance at 260 nm) were pooled and centrifuged at 114,000 × g for 4 h at 4°C. The pellet was resuspended in buffer CRNC [20 mM HEPES (pH 7.2), 50 mM KOAc, 5 mM Mg(OAc)2, and 2 mM l-tryptophan] and stored at −80°C until labeling.
The purified RNCs were adjusted to a concentration of 2 μM in 100 μl buffer CRNC and labeled with the cysteine-reactive dye NT-647 and the red-maleimide labeling kit (NanoTemper Technologies). Therefore, 6 μM of the dye in 100 μl labeling buffer (provided in the kit) was mixed with the RNC solution. The labeling reaction mixture was incubated for 1 h at room temperature in the dark and purified using the column of the kit to remove the free dye. The concentration of labeled RNCs was calculated by measuring the absorbance at 260 nm, and they were stored at −80°C until the measurement.
Purification of Ffh, Ffh L181C/406S, Ffh 406S/M423C, and FtsY.
The purification of Ffh, the Ffh mutants, and FtsY was done as described by Pross and Kuhn (29). The plasmids pMS-Ffh-C-Strep, pMS-Ffh-L181C-C-Strep, and pMS-Ffh-M423C-C-Strep (29) were transformed in E. coli BL21(DE3) for purification. The overnight culture was back diluted 1:100 in 2 liters LB medium with 100 μg ml−1 ampicillin and grown to an OD600 of 0.5. The expression was induced with 1 mM IPTG for 3 h at 37°C. The cells were harvested and resuspended in buffer AFfh (20 mM HEPES [pH 8], 350 mM NaCl, 10 mM MgCl2, 10 mM KCl, and 10% glycerol), and 0.2 mM PMSF was added. After cell disruption with the OneShot at 1.23 kbar, the lysate was centrifuged 2 times for 30 min at 20,000 × g, and the supernatant was loaded onto a 3-ml Strep-Tactin matrix (IBA Lifesciences). The flowthrough was collected, and the matrix was washed with 50 ml buffer WFfh (20 mM HEPES [pH 8], 500 mM NaCl, 10 mM MgCl2, and 100 mM KCl). Twenty milliliters of buffer EFfh (20 mM HEPES [pH 8], 350 mM NaCl, 10 mM MgCl2, 10 mM KCl, 10% glycerol, and 2.5 mM desthiobiotin) were used for the elution of the protein in 2-ml fractions. The elution fractions were pooled and purified using an Äkta system and the Superdex 75 16/60 column in buffer GFFfh (20 mM HEPES [pH 8], 200 mM NaCl, 10 mM MgCl2, 10 mM KCl, and 10% glycerol).
The plasmid pTrc99-FtsY-His (provided by H.G. Koch, Freiburg, Germany) was used for the expression of E. coli FtsY in E. coli BL21(DE) cells. Two liters of LB medium supplemented with 100 μg ml−1 ampicillin was inoculated 1:100 with an overnight culture, and the culture was grown to an OD600 of 0.5 at 37°C. After induction with 1 mM IPTG for 4 h at 37°C, the cells were harvested and resuspended in buffer AFtsY [50 mM HEPES (pH 7.6), 1 M NH4Ac, 10 mM Mg(OAc)2, 10% glycerol, and 1 mM DTT]. We added 0.2 mM PMSF, and the cells were lysed using the OneShot at 1.23 kbar and centrifuged for 30 min at 4,300 × g. The supernatant was further centrifuged in a Beckman Ti60 rotor for 1 h at 38,000 rpm, 30 mM imidazole was added, the supernatant was mixed with 2 ml Ni-NTA and incubated for 1 h at 4°C on a rotary wheel. The resin was washed with 20 ml buffer WFtsY [50 mM HEPES (pH 7.6), 1 M NH4Ac, 10 mM Mg(OAc)2, 1 mM DTT, and 30 mM imidazole], and the protein was eluted in 2-ml fractions using 20 ml buffer EFtsY [50 mM HEPES (pH 7.6), 1 M NH4Ac, 10 mM Mg(OAc)2, 400 mM imidazole, and 10% glycerol]. The elution fractions were pooled and purified using the Äkta system and the Superdex 200 16/60 column (GE Healthcare) in buffer GFFtsY [100 mM HEPES (pH 7.6), 200 mM KOAc, 20 mM Mg(OAc)2, 2 mM DTT, and 10% glycerol].
In vitro synthesis of 4.5S RNA and SRP reconstitution.
The plasmid pUC18-4.5S RNA (provided by Irmgard Sinning, Heidelberg, Germany), which consists of the 4.5S RNA gene downstream of the T7 promoter, was used for in vitro synthesis with the HiScribe T7 high yield RNA synthesis kit (New England BioLabs). Prior to the in vitro synthesis, the plasmid was linearized with BamHI and gel purified. One microgram of the linearized plasmid was used as a template. The sample was incubated for 16 h at 37°C, purified with the RNA Clean & Concentrator-25 kit (Zymo Research), and analyzed on a 2% agarose gel in 1× Tris-borate-EDTA buffer. For reconstitution, the 4.5S RNA was incubated for 2 min at 75°C and chilled on ice for 1 min. The purified Ffh was mixed with a 1.5-fold molar excess of 4.5S RNA in MST buffer [20 mM HEPES (pH 7.2), 50 mM KOAc, 5 mM Mg(OAc)2, 2 mM l-tryptophan, and 0.05% Tween 20] and incubated for 10 min at 20°C. The reconstituted SRP was stored on ice.
SciP peptide synthesis.
The two peptides SciP1-27 G16C (MNKPVISRAEQIFYPCWLMVSQLRSGQ) and SciP54-85 (GFSQKSSDIMLYAFCALLDESVLNREKTDDGW) were synthesized in vitro by the custom peptide synthesis services from Genosphere Biotechnologies (France) with N-terminal acetylation and C-terminal amidation and purity of >95%. The SciP1-27 G16C peptide was dissolved in H2O-CH3CN (3:1), whereas the SciP54-85 peptide was dissolved in dimethyl sulfoxide (DMSO).
In vitro disulfide cross-linking with copper phenanthroline.
For cross-linking studies, the SciP1-27 G16C peptide was synthesized with a cysteine residue at position 16 (G16C).
For in vitro cross-linking, 2 μM purified Ffh L181C/406S or Ffh 406S/423C were reconstituted with 3 μM 4.5S RNA in bufferSRP [20 mM HEPES (pH 7.2), 50 mM KOAc, and 5 mM Mg(OAc)2]. The reconstituted SRP was mixed with 20 μM of the synthesized peptide in buffercrosslink (50 mM Tris [pH 7.4], 150 mM NaCl, and 10 mM MgCl2), 1 mM copper phenanthroline was added, and the mixture was incubated for 1 h on ice. After the incubation, the mixture was precipitated with 10% TCA, resuspended in sample buffer without or with DTT (1 mM), and loaded on 10% SDS-PAGE.
Microscale thermophoresis.
For the interaction studies of RNCs with SRP, the labeled RNCs were diluted to 10 nM in MST buffer. With the reconstituted SRP, a series of 1:1 dilutions in MST buffer was prepared, resulting in a concentration from 1 μM to 0.49 nM. One volume of labeled RNCs was added to the dilutions, resulting in an RNC concentration of 5 nM. The samples were incubated for 5 min on ice, filled in Monolith NT premium treated capillaries (Nano Temper Technologies), and measured at 22°C with the Monolith NT.115 device. Thermophoresis was carried out with 5 s laser off, 20 to 30 s laser on, and 5 s laser off; LED power was 40 to 50%, and the MST power was low. Three independently pipetted measurements were merged and analyzed with MO.Affinity Analysis v2.3 (NanoTemper Technologies) using the manual evaluation (cold region start/end, −1 s/0 s; hot region start/end, 5.00 s/10.01 s).
The interaction studies of RNCs with SRP-FtsY were done using a preincubated closed SRP-FtsY complex. The reconstituted SRP was mixed with a 4-fold molar excess of purified FtsY in MST buffer supplemented with 200 μM of the nonhydrolyzable GTP analogue GppNHp. The sample was incubated for 10 min at 25°C and chilled on ice until the measurement. A series of 1:1 dilutions was prepared in MST buffer with 200 μM GppNHp (Sigma-Aldrich), resulting in a complex concentration of 500/250 nM to 0.25/0.12 nM, and one volume of labeled RNCs was added (final concentration, 5 nM). The samples were incubated for 5 min on ice, filled in Monolith NT premium treated capillaries, and measured at 22°C with the Monolith NT.115 device. Thermophoresis was carried out with 5 s laser off, 20 to 30 s laser on, and 5 s laser off; LED power of 60%; and the MST power low. Three independently pipetted measurements were merged and analyzed with the MO.Affinity Analysis v2.3 using the manual evaluation (cold region start/end, −1 s/0 s; hot region start/end, 5.01 s/10.05 s).
Periplasmic cysteine accessibility assay with AMS.
The E. coli strain MC4100 was transformed with the plasmids pMS-SciP-C, pMS-SciP-CΔ12-20 (named Δ12-20), pMS-SciP-CΔ62-71 (named Δ62-71), and pMS-SciP-CΔ12-20, Δ62-71 (named Δ12-20 Δ62-71), respectively. The cells were grown to an OD600 of 0.5, washed twice, and resuspended in M9 minimal media lacking methionine and cysteine. After shaking for 1 h at 37°C, the expression was induced with 1 mM IPTG. After 10 min, 2.5 mM AMS (Molecular Probes) was added, and after 1 min, the cells were pulsed-labeled with 15 to 30 μCi [35S]methionine-cysteine for 2 min. The pulse-labeling was chased with nonradioactive methionine-cysteine for 10 min, and the AMS reaction was quenched with 10 mM DTT for another 10 min. The samples were TCA precipitated and resuspended in 10 mM Tris–2% SDS, followed by the immunoprecipitation with anti-His antibody and the separation on a 14% SDS-PAGE. The visualization was done by phosphorimaging.
Data availability.
All data generated or analyzed during this study are included in this article (and the supplemental material) or are available from the corresponding author on reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank I. Seitl for providing pMS-Ffh-C-Strep and pMS-Ffh-M423C-C-Strep and for support and discussions, D. Lupo for discussions, B. Schmidt for assistance in the lab, S. Kraus for plasmid pMS-Ffh-L181C, and R. Beckmann, S. O. Shan, I. Sinning, and H. G. Koch for strains and plasmids.
This work was supported by a stipend of the Landesgraduiertenförderung Baden-Württemberg and by DFG grant KU749/5.
The experiments were designed by A.K. and performed by E.P. The results were discussed, and the manuscript was written by A.K. and E.P.
We declare that we do not have any competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Danese PN, Silhavy TJ. 1998. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet 32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 2.Cross BCS, Sinning I, Luirink J, High S. 2009. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol 10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 3.Driessen AJM, Nouwen N. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 5.Lütcke H. 1995. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur J Biochem 228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- 6.Kuhn A, Koch H-G, Dalbey RE. 7 March 2017, posting date. Targeting and insertion of membrane proteins. EcoSal Plus 2017 doi: 10.1128/ecosalplus.ESP-0012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang M-J, Zhang X. 2011. Molecular dynamics simulations reveal structural coordination of Ffh-FtsY heterodimer toward GTPase activation. Proteins 79:1774–1785. doi: 10.1002/prot.23000. [DOI] [PubMed] [Google Scholar]
- 8.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. 2006. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 9.Jomaa A, Boehringer D, Leibundgut M, Ban N. 2016. Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat Commun 7:10471. doi: 10.1038/ncomms10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtkamp W, Lee S, Bornemann T, Senyushkina T, Rodnina MV, Wintermeyer W. 2012. Dynamic switch of the signal recognition particle from scanning to targeting. Nat Struct Mol Biol 19:1332–1337. doi: 10.1038/nsmb.2421. [DOI] [PubMed] [Google Scholar]
- 11.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. 2006. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 12.Noriega TR, Chen J, Walter P, Puglisi JD. 2014. Real-time observation of signal recognition particle binding to actively translating ribosomes. Elife 3:e04418. doi: 10.7554/eLife.04418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saraogi I, Akopian D, Shan S-O. 2014. Regulation of cargo recognition, commitment, and unloading drives cotranslational protein targeting. J Cell Biol 205:693–706. doi: 10.1083/jcb.201311028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hainzl T, Huang S, Meriläinen G, Brännström K, Sauer-Eriksson AE. 2011. Structural basis of signal-sequence recognition by the signal recognition particle. Nat Struct Mol Biol 18:389–391. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- 15.Seluanov A, Bibi E. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem 272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 16.Egea PF, Shan S-O, Napetschnig J, Savage DF, Walter P, Stroud RM. 2004. Substrate twinning activates the signal recognition particle and its receptor. Nature 427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 17.Connolly T, Rapiejko PJ, Gilmore R. 1991. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science 252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 18.Welte T, Kudva R, Kuhn P, Sturm L, Braig D, Müller M, Warscheid B, Drepper F, Koch H-G. 2012. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol Biol Cell 23:464–479. doi: 10.1091/mbc.e11-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martoglio B, Dobberstein B. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol 8:410–415. doi: 10.1016/S0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 20.Lee HC, Bernstein HD. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc Natl Acad Sci U S A 98:3471–3476. doi: 10.1073/pnas.051484198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde RS, Bernstein HD. 2006. The surprising complexity of signal sequences. Trends Biochem Sci 31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Hainzl T, Sauer-Eriksson AE. 2015. Signal-sequence induced conformational changes in the signal recognition particle. Nat Commun 6:7163. doi: 10.1038/ncomms8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson JH, Woolhead CA, Bernstein HD. 2003. Basic amino acids in a distinct subset of signal peptides promote interaction with the signal recognition particle. J Biol Chem 278:46155–46162. doi: 10.1074/jbc.M309082200. [DOI] [PubMed] [Google Scholar]
- 24.Zimmann P, Puppe W, Altendorf K. 1995. Membrane topology analysis of the sensor kinase KdpD of Escherichia coli. J Biol Chem 270:28282–28288. doi: 10.1074/jbc.270.47.28282. [DOI] [PubMed] [Google Scholar]
- 25.Maier KS, Hubich S, Liebhart H, Krauss S, Kuhn A, Facey SJ. 2008. An amphiphilic region in the cytoplasmic domain of KdpD is recognized by the signal recognition particle and targeted to the Escherichia coli membrane. Mol Microbiol 68:1471–1484. doi: 10.1111/j.1365-2958.2008.06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pross E, Soussoula L, Seitl I, Lupo D, Kuhn A. 2016. Membrane targeting and insertion of the C-tail protein SciP. J Mol Biol 428:4218–4227. doi: 10.1016/j.jmb.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Bischoff L, Wickles S, Berninghausen O, van der Sluis EO, Beckmann R. 2014. Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nat Commun 5:4103. doi: 10.1038/ncomms5103. [DOI] [PubMed] [Google Scholar]
- 28.Calhoun KA, Swartz JR. 2006. Total amino acid stabilization during cell-free protein synthesis reactions. J Biotechnol 123:193–203. doi: 10.1016/j.jbiotec.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Pross E, Kuhn A. 2019. The SRP signal sequence of KdpD. Sci Rep 9:8717. doi: 10.1038/s41598-019-45233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jagath JR, Rodnina MV, Wintermeyer W. 2000. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J Mol Biol 295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- 31.Ataide SF, Schmitz N, Shen K, Ke A, Shan S-O, Doudna JA, Ban N. 2011. The crystal structure of the signal recognition particle in complex with its receptor. Science 331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Estrozi LF, Boehringer D, Shan S-O, Ban N, Schaffitzel C. 2011. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol 18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Loeffelholz O, Knoops K, Ariosa A, Zhang X, Karuppasamy M, Huard K, Schoehn G, Berger I, Shan S-O, Schaffitzel C. 2013. Structural basis of signal sequence surveillance and selection by the SRP-FtsY complex. Nat Struct Mol Biol 20:604–610. doi: 10.1038/nsmb.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loeffelholz O. v, Jiang Q, Ariosa A, Karuppasamy M, Huard K, Berger I, Shan S-O, Schaffitzel C. 2015. Ribosome-SRP-FtsY cotranslational targeting complex in the closed state. Proc Natl Acad Sci U S A 112:3943–3948. doi: 10.1073/pnas.1424453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam VQ, Akopian D, Rome M, Henningsen D, Shan S-O. 2010. Lipid activation of the signal recognition particle receptor provides spatial coordination of protein targeting. J Cell Biol 190:623–635. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draycheva A, Bornemann T, Ryazanov S, Lakomek N-A, Wintermeyer W. 2016. The bacterial SRP receptor, FtsY, is activated on binding to the translocon. Mol Microbiol 102:152–167. doi: 10.1111/mmi.13452. [DOI] [PubMed] [Google Scholar]
- 37.Keenan RJ, Freymann DM, Walter P, Stroud RM. 1998. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94:181–191. doi: 10.1016/S0092-8674(00)81418-X. [DOI] [PubMed] [Google Scholar]
- 38.Bradshaw N, Neher SB, Booth DS, Walter P. 2009. Signal sequences activate the catalytic switch of SRP RNA. Science 323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schibich D, Gloge F, Pöhner I, Björkholm P, Wade RC, von Heijne G, Bukau B, Kramer G. 2016. Global profiling of SRP interaction with nascent polypeptides. Nature 536:219–223. doi: 10.1038/nature19070. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki R, Yura T, Suzuki T, Dohmae N, Mori H, Akiyama Y. 2016. A novel SRP recognition sequence in the homeostatic control region of heat shock transcription factor σ32. Sci Rep 6:24147. doi: 10.1038/srep24147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim B, Miyazaki R, Neher S, Siegele DA, Ito K, Walter P, Akiyama Y, Yura T, Gross CA. 2013. Heat shock transcription factor σ32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol 11:e1001735. doi: 10.1371/journal.pbio.1001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 43.Shan S-O, Walter P. 2003. Induced nucleotide specificity in a GTPase. Proc Natl Acad Sci U S A 100:4480–4485. doi: 10.1073/pnas.0737693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shan S-O, Stroud RM, Walter P. 2004. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol 2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R, Durand E. 2018. Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3:1404–1416. doi: 10.1038/s41564-018-0260-1. [DOI] [PubMed] [Google Scholar]
- 46.Zoued A, Duneau J-P, Durand E, España AP, Journet L, Guerlesquin F, Cascales E. 2018. Tryptophan-mediated dimerization of the TssL transmembrane anchor is required for type VI secretion system activity. J Mol Biol 430:987–1003. doi: 10.1016/j.jmb.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Petriman N-A, Jauß B, Hufnagel A, Franz L, Sachelaru I, Drepper F, Warscheid B, Koch H-G. 2018. The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci Rep 8:578. doi: 10.1038/s41598-017-19019-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spann D, Pross E, Chen Y, Dalbey RE, Kuhn A. 2018. Each protomer of a dimeric YidC functions as a single membrane insertase. Sci Rep 8:589. doi: 10.1038/s41598-017-18830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urbanus ML, Scotti PA, Froderberg L, Saaf A, de Gier J, Brunner J, Samuelson JC, Dalbey RE, Oudega B, Luirink J. 2001. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep 2:524–529. doi: 10.1093/embo-reports/kve108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kedrov A, Wickles S, Crevenna AH, van der Sluis EO, Buschauer R, Berninghausen O, Lamb DC, Beckmann R. 2016. Structural dynamics of the YidC:ribosome complex during membrane protein biogenesis. Cell Rep 17:2943–2954. doi: 10.1016/j.celrep.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kedrov A, Sustarsic M, Keyzer J. d, Caumanns JJ, Wu ZC, Driessen AJM. 2013. Elucidating the native architecture of the YidC:ribosome complex. J Mol Biol 425:4112–4124. doi: 10.1016/j.jmb.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 52.Durand E, Zoued A, Spinelli S, Watson PJH, Aschtgen M-S, Journet L, Cambillau C, Cascales E. 2012. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J Biol Chem 287:14157–14168. doi: 10.1074/jbc.M111.338731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peschke M, Le Goff M, Koningstein GM, Karyolaimos A, de Gier J-W, van Ulsen P, Luirink J. 2018. SRP, FtsY, DnaK and YidC are required for the biogenesis of the E. coli tail-anchored membrane proteins DjlC and Flk. J Mol Biol 430:389–403. doi: 10.1016/j.jmb.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Brambillasca S, Yabal M, Makarow M, Borgese N. 2006. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J Cell Biol 175:767–777. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, Hegde RS, Borgese N. 2005. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J 24:2533–2542. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Colombo SF, Longhi R, Borgese N. 2009. The role of cytosolic proteins in the insertion of tail-anchored proteins into phospholipid bilayers. J Cell Sci 122:2383–2392. doi: 10.1242/jcs.049460. [DOI] [PubMed] [Google Scholar]
- 57.Borgese N, Righi M. 2010. Remote origins of tail-anchored proteins. Traffic 11:877–885. doi: 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 58.Johnson N, Powis K, High S. 2013. Post-translational translocation into the endoplasmic reticulum. Biochim Biophys Acta 1833:2403–2409. doi: 10.1016/j.bbamcr.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Rabu C, Schmid V, Schwappach B, High S. 2009. Biogenesis of tail-anchored proteins: the beginning for the end? J Cell Sci 122:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 61.Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article (and the supplemental material) or are available from the corresponding author on reasonable request.