Summary
Methylation of histone 3 at lysine 9 (H3K9) constitutes a roadblock for cellular reprogramming. Interference with methyltransferases or activation of demethylases by the cofactor ascorbic acid (AA) facilitates the derivation of induced pluripotent stem cells (iPSCs), but possible interactions between specific methyltransferases and AA treatment remain insufficiently explored. We show that chemical inhibition of the methyltransferases EHMT1 and EHMT2 counteracts iPSC formation in an enhanced reprogramming system in the presence of AA, an effect that is dependent on EHMT1. EHMT inhibition during enhanced reprogramming is associated with rapid loss of H3K9 dimethylation, inefficient downregulation of somatic genes, and failed mesenchymal-to-epithelial transition. Furthermore, transient EHMT inhibition during reprogramming yields iPSCs that fail to efficiently give rise to viable mice upon blastocyst injection. Our observations establish novel functions of H3K9 methyltransferases and suggest that a functional balance between AA-stimulated enzymes and EHMTs supports efficient and less error-prone iPSC reprogramming to pluripotency.
Keywords: H3K9 methylation, induced pluripotency, epigenetic remodeling, ascorbic acid, EHMT1, EHMT2 cellular reprogramming
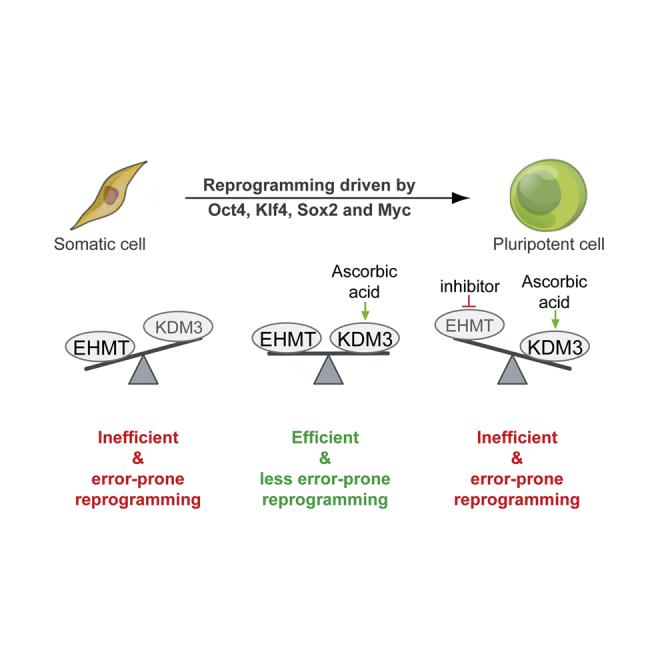
Graphical Abstract
Highlights
-
•
EHMT function during mouse cell reprogramming is modulated by ascorbic acid (AA)
-
•
EHMT inhibition counteracts reprogramming in the presence of AA
-
•
EHMT inhibition in the presence of AA results in global erasure of H3K9 dimethylation
-
•
Cells reprogrammed in the presence of EHMT inhibitor are functionally impaired
In this article, Stadtfeld and colleagues show that the activity of a specific repressive histone methyltransferase is important for efficient cellular reprogramming under conditions that favor global histone demethylation. Their observations suggest that a balance between antagonizing chromatin modifiers is important to achieve faithful resetting of the epigenome during the generation of induced pluripotent stem cells.
Introduction
Covalent chromatin modifications such as DNA and histone methylation modulate gene expression and stabilize epigenetic states in a wide variety of biological processes. The genome-wide and locus-specific abundance of chromatin marks is determined by counteracting enzymatic activities, such as histone methyltransferases and demethylases (Black et al., 2012). Histone 3 at lysine 9 (H3K9) methylation is an epigenetic mark predominantly associated with gene repression that is conserved during evolution and plays important regulatory functions during embryonic development, sex determination, neuronal plasticity, immune cell function, and tumorigenesis (Benevento et al., 2015; Casciello et al., 2015; Kuroki and Tachibana, 2018; Scheer and Zaph, 2017). H3K9 trimethylation in heterochromatic regions is established by SETDB1 and SUV39H1/2 (Kang, 2015; Peters et al., 2001; Rice et al., 2003), while the H3K9 mono- and dimethyltransferases EHMT1 and EHMT2 (also known as GLP and G9A, respectively) mediate gene silencing at euchromatic loci (Shinkai and Tachibana, 2011). Enzymes involved in the establishment of H3K9 methylation are repressors of core pluripotency-associated gene loci (Epsztejn-Litman et al., 2008) and, thus, have particular relevance for physiological and experimentally induced changes in cell identity (Becker et al., 2016; Feldman et al., 2006). Inefficient removal of H3K9 methylation is a frequent cause of incomplete transcriptional reprogramming after somatic cell nuclear transfer (Matoba et al., 2014) and has been reported to impede the binding of reprogramming factors to the genome during the derivation of induced pluripotent stem cells (iPSCs) (Soufi et al., 2012). Consequently, the formation of mouse (Chen et al., 2013b; Liang et al., 2012; Sridharan et al., 2013; Tran et al., 2015; Wang et al., 2011; Wei et al., 2017) and human (Onder et al., 2012; Soufi et al., 2012) iPSCs can be substantially facilitated by interference with H3K9 methyltransferases or by activation of the respective demethylases. Despite the importance of H3K9 methyltransferases for cellular reprogramming and different physiological and pathological processes (Shankar et al., 2013), our understanding of the regulatory interactions controlling the function of these enzymes in different cellular contexts remains incomplete. We reasoned that the systematic comparison of cells that reprogram at markedly different efficiencies could be utilized to discover unexplored aspects of the H3K9 methylation machinery. By taking this approach, we were able to assign specific and context-dependent functions to the EHMTs during the reprogramming of fibroblasts into iPSCs by the “Yamanaka transcription factors” OCT4, KLF4, SOX2, and MYC (OKSM). In particular, we report the unexpected finding that EHMT activity supports efficient and faithful establishment of pluripotency under conditions that favor histone demethylation.
Results
Context-Dependent Roles of EHMT Activity during Mouse Fibroblast Reprogramming
We used a well-established doxycycline (dox)-controllable transgenic system (Stadtfeld et al., 2010b) to compare the roles of H3K9 methylation during iPSC formation either driven solely by the OKSM factors (“basal reprogramming”) or further supported by chemical modulation of the transforming growth factor β and WNT signaling pathways (via iALK5 and CHIR99021, respectively) and of chromatin state (via ascorbic acid, AA) (“3c enhanced reprogramming”) (Vidal et al., 2014). These compounds are commonly used to facilitate reprogramming and pluripotent cell culture (Dakhore et al., 2018), and iPSCs generated via 3c enhanced reprogramming are developmentally highly competent (Amlani et al., 2018). 3c enhanced reprogramming yields approximately 50 times more stable iPSC colonies than basal reprogramming and does so in a shorter period of time (6 rather than 12 days of OKSM expression) (Penalosa-Ruiz et al., 2019; Saunders et al., 2017; Schwarz et al., 2018; Stelzer et al., 2015; Vidal et al., 2014). Since H3K9 methylation is a well-known roadblock for cellular reprogramming, we speculated that 3c might enhance iPSC formation by counteracting H3K9 methylation more efficiently than the OKSM factors alone. We focused our studies on EHMT1 and EHMT2, the major methyltransferases that catalyze the repressive H3K9me2 mark in mammalian cells.
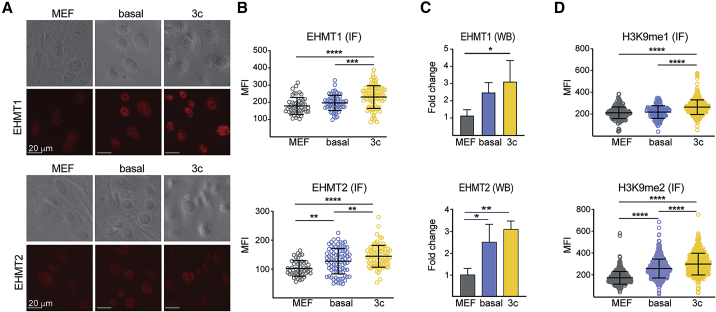
Analyses by quantitative PCR (Figure S1A) and immunofluorescence (IF) (Figures 1A and 1B) showed a surprising upregulation of EHMT1 and EHMT2 at both the mRNA and the protein level 24 h after initiation of OKSM expression in basal and 3c enhanced conditions compared with uninduced mouse embryonic fibroblasts (MEFs). The observed increase in EHMT1 and EHMT2 was further supported by western blot (WB) analysis (Figures 1C and S1B). Since we observed significant differences in the abundance of housekeeper proteins frequently used to normalize WB signals between fibroblasts and reprogramming intermediates (including increased levels of histone H3), we normalized signals based on total protein levels determined by sensitive fluorescence assessment (Kirshner and Gibbs, 2018) (Figure S1C). Total H3K9 mono- and dimethylation (H3K9me1/2) levels also increased early during reprogramming (Figures 1D and S1D). IF suggested opposite trends for H3K9me3 (Figures S1D and S1E) and the activating marks H3K4me2 and H3K4me3 (Figure S1E), supporting the notion that dynamic chromatin remodeling commences early during iPSC formation (Koche et al., 2011).
Figure 1.
Increased EHMT Activity during Early Stages of OKSM-Driven Reprogramming
(A) Representative bright-field and fluorescence images of MEFs and cells under the indicated reprogramming conditions 24 h after induction of OKSM and staining with antibodies against EHMT1 (top) or EHMT2 (bottom).
(B) Mean fluorescence intensities after IF with antibodies against EHMT1 and EHMT2 under the indicated conditions. At least 100 size-matched nuclei were analyzed. Similar results were obtained in three independent experiments.
(C) Representative mean fold change in EHMT1 and EHMT2 protein levels analyzed by WB during basal and enhanced reprogramming. n = 3 independent experiments.
(D) Same as (B) but after IF against indicated histone marks.
Significance in (B–D) with one-way ANOVA with Tukey post test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
We next evaluated the effect of treatment of MEFs undergoing reprogramming with UNC0638, an efficient and specific substrate inhibitor of EHMT activity (Vedadi et al., 2011). Standard doses (1 μM) of UNC0638 reduced levels of H3K9me2, particularly under 3c enhanced conditions, as detected by both IF and WB (Figures S2A–S2C). This suggests that EHMT-mediated accumulation of H3K9me2 is an early and previously unappreciated event during cellular reprogramming, which appears more pronounced during 3c enhanced than during basal iPSC reprogramming.
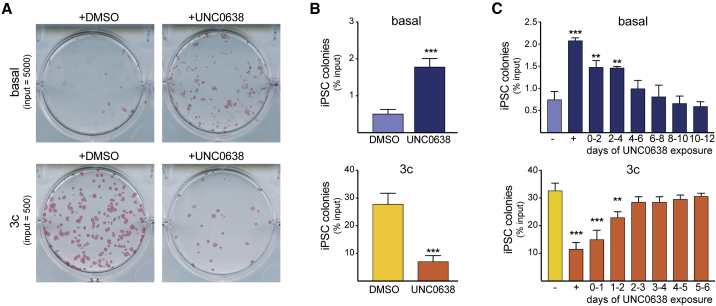
Chemical and genetic interference with EHMTs during OKSM-driven reprogramming of mouse neural progenitor cells (Shi et al., 2008) and fibroblasts (Sridharan et al., 2013) has been reported to facilitate iPSC formation, supporting the prevalent notion that H3K9 methyltransferase activity counteracts the induction of pluripotency. Accordingly, EHMT inhibition via UNC0638 in our system increased reprogramming efficiency under basal conditions (Figures 2A and 2B, top), generating significantly higher numbers of transgene-independent iPSC colonies—defined by their epithelial morphology and expression of an EGFP cassette inserted into the endogenous Pou5f1 locus, also known as Oct4 (Lengner et al., 2007)—after washout of dox (Figure S2D). To our surprise, iPSC formation was strongly impaired (3- to 5-fold) when UNC0638 was administered during 3c enhanced reprogramming (Figures 2A and 2B, bottom). We also obtained reduced numbers of iPSC colonies in the presence of UNC0638 during 3c enhanced reprogramming driven by OKS factors (no ectopic MYC) (Figures S2E and S2F) or when using lentiviral vectors expressing OKSM (Sommer et al., 2009) (Figure S2G), suggesting that our observations are not restricted to a single reprogramming approach. Together, these findings demonstrate that 3c enhanced reprogramming, in contrast to basal reprogramming, partially becomes dependent on EHMT activity.
Figure 2.
EHMT Activity Supports 3c Enhanced Reprogramming
(A) Alkaline phosphatase staining of iPSCs formed from the indicated input MEFs via basal and 3c enhanced reprogramming in the absence and presence of UNC0638, respectively. Images were taken 4 days after dox was removed to select for stably reprogrammed cells.
(B) Quantification of iPSC colony formation under the indicated conditions as percentage input MEFs. n = 3 independent experiments. Significance with unpaired t test: ∗∗∗p < 0.001.
(C) Percentage iPSC colonies formed from reprogrammable MEFs under basal or 3c enhanced conditions that were exposed to EHMT inhibitor during the indicated time windows (“+” indicates chronic exposure and “−” addition of DMSO instead of UNC0638). Input cells were 2,500 (basal) and 300 (3c enhanced) MEFs, respectively.
n = 3 independent experiments. Significance with one-way ANOVA with Dunnett post test: ∗∗p < 0.01 and ∗∗∗p < 0.001.
EHMT Inhibition Counteracts Downregulation of a Subset of MEF-Associated Genes
To establish the temporal requirements of EMHT activity during reprogramming, we next exposed OKSM-expressing cells at specific intervals of time to UNC0638. We adjusted the intervals of exposure (1 day for 3c and 2 days for basal reprogramming) to reflect the substantially faster kinetics of 3c enhanced reprogramming (Vidal et al., 2014). This revealed that the reprogramming-promoting effect of EHMT inhibition on basal reprogramming, as well as the reprogramming-counteracting effect of EHMT inhibition on 3c enhanced reprogramming, was most pronounced when UNC0638 was administered during early reprogramming stages (days 0–4 during basal and days 0–2 during 3c enhanced reprogramming) (Figure 2C).
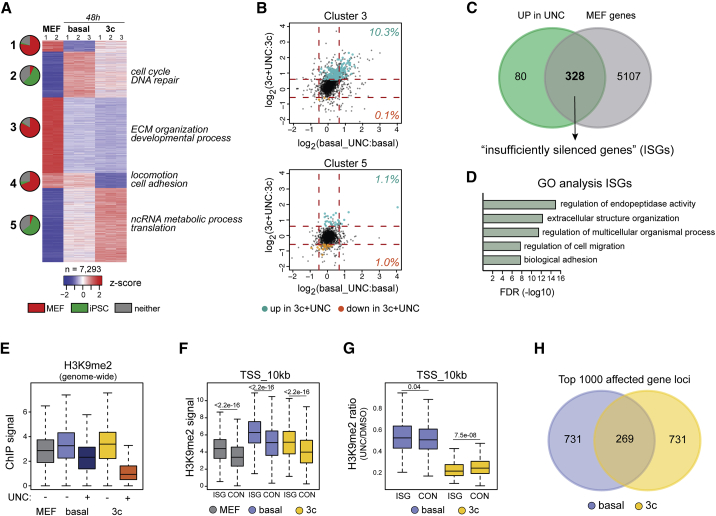
To gain insight into the genome-wide molecular consequences of EHMT inhibition during the early stages of iPSC formation we used RNA sequencing (RNA-seq), which clearly distinguished MEFs, basal intermediates, and 3c enhanced intermediates (Figure S3A). Unsupervised k-means clustering of genes differentially expressed between starting MEFs and cells 2 days after initiation of OKSM expression under either condition revealed five distinct groups (Figure 3A and Table S1). Three of these groups contained mostly genes expressed more highly in MEFs than in established iPSCs (fold change > 2; adjusted p < 0.05) that were downregulated early during reprogramming (clusters 1, 3, and 4), while the two other groups contained larger fractions of iPSC-associated genes that were already upregulated at this stage of iPSC formation (clusters 2 and 5) (Figure 3A). Gene ontology (GO) analysis revealed stronger activation of genes involved in RNA metabolism and translation (cluster 5) and more efficient silencing of genes required for cell adhesion (cluster 4) during 3c enhanced reprogramming (Figure 3A and Table S1). Direct comparison of differentially expressed genes between early intermediates of the two different reprogramming conditions confirmed more efficient downregulation of transcripts associated with cell adhesion and extracellular matrix organization during 3c enhanced reprogramming (Table S2). These observations demonstrate that basal and 3c enhanced reprogramming exhibit distinct molecular characteristics during early stages of iPSC formation.
Figure 3.
Molecular Consequences of EHMT Inhibition during Early Reprogramming Stages
(A) Unsupervised k-means clustering of genes differentially expressed between MEFs and cells expressing OKSM for 2 days under either basal or 3c conditions (adjusted p < 0.05; fold change [FC] > 2). Pie charts indicate the abundance of genes associated with either MEFs or iPSCs (adjusted p < 0.05; FC > 2) or neither cell type in the respective cluster. Select GO categories (false discovery rate [FDR] < 0.05) associated with identified clusters are highlighted.
(B) Effects of EHMT inhibition (UNC) on the expression levels of genes associated with cluster 3 (top) and cluster 5 (bottom) identified in (A) during both basal (x axis) and 3c enhanced (y axis) reprogramming. Transcripts with significantly changed abundance during 3c enhanced reprogramming (adjusted p < 0.05; FC > 1.5) are highlighted in green (failed downregulation) and orange (failed upregulation), respectively. Percentage of affected cluster-specific genes is shown.
(C) Venn diagram showing overlap between genes upregulated during 3c enhanced reprogramming in the presence of UNC0638 and genes significantly more highly expressed in MEFs than in iPSCs.
(D) GO terms associated with MEF-associated genes that are inefficiently silenced during 3c enhanced reprogramming in the presence of UNC0638 (ISGs).
(E) H3K9me2 ChIP signal across 2.5-kb tiling intervals across the mouse genome. Average values from two separate chromatin precipitations are shown for each of the indicated conditions.
(F) H3K9me2 ChIP signal over the 10 kb immediately upstream of the TSS in ISGs in the presence of UNC0638 during 3c reprogramming and 2,664 control MEF-associated genes that are efficiently silenced (CON). n = 2 independent experiments.
(G) Ratio of H3K9me2 signal in the presence and absence of UNC0638 at the TSS region of ISGs and control genes during basal and 3c reprogramming. Indicated p values in (F and G) were calculated with Wilcoxon rank-sum test. n = 2 independent experiments.
(H) Overlap of the 1,000 protein-coding gene loci with the highest degree of loss of H3K9me2 signal upon EHMT inhibition during basal and 3c reprogramming.
Next, we interrogated the consequences of EHMT inhibition on the transcriptional dynamics associated with the two different reprogramming regimens. Principal-component analysis (PCA) of early reprogramming intermediates confirmed close proximity of biological replicates and suggested a more pronounced effect of UNC0638 on 3c enhanced than on basal reprogramming (Figure S3B). Differential gene expression analysis confirmed this observation and identified a total of 456 genes associated with 3c enhanced reprogramming that were significantly altered (fold change > 1.5; adjusted p < 0.05) in the presence of UNC0638 (Table S3). In agreement with the established roles of EHMTs as transcriptional repressors, we observed more frequent gene activation upon inhibition of these enzymes (408 of 456 genes or 89.5%). Only 44 (or 9.7%) of the 456 genes affected during 3c enhanced reprogramming were also affected in their expression by UNC0638 treatment during basal reprogramming (Table S3). Analysis of affected genes in the aforementioned five clusters revealed that EHMT inhibition counteracted the downregulation of a subset of genes in the MEF-enriched clusters 1, 3, and 4, while minimally affecting the iPSC-related clusters 2 and 5 (Figures 3B and S3C). Overall, we found that the majority of genes upregulated in the presence of UNC0638 during 3c enhanced reprogramming (328 of 408 genes or 80.4%) were MEF associated. We will refer to this group of transcripts as “insufficiently silenced genes” (ISGs) (Figure 3C). GO analysis showed that ISGs represent biological processes such as biological adhesion and extracellular matrix remodeling (Figure 3D and Table S3). Together, these observations suggest that EHMTs during 3c enhanced reprogramming contribute to the silencing of somatic gene expression during iPSC formation.
Rapid Loss of H3K9me2 upon EHMT Inhibition during 3c Enhanced Reprogramming
We next evaluated the global levels of H3K9me2 by chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) in starting fibroblasts and early (48 h) reprogramming intermediates. PCA revealed distinct clustering of all four reprogramming conditions (basal and 3c enhanced, with and without UNC0638) (Figure S3D). Integration of RNA-seq data revealed an inverse correlation between expression levels of gene loci and their H3K9me2 intensity in the respective samples (shown for MEFs in Figure S3E), in agreement with the reported repressive nature of this mark. Consistent with our IF analyses (Figures 1D and S1D), ChIP-seq showed a genome-wide increase in H3K9me2 levels in reprogramming intermediates and a marked decrease in the presence of UNC0638 that was significantly more pronounced during 3c enhanced reprogramming (Figure 3E). This increase in H3K9me2 was not restricted to downregulated genes. Similarly, the loss of this mark in the presence of UNC0638 extended beyond genes that were transcriptionally affected in early reprogramming intermediates by EHMT inhibition (Table S4). These observations argue against a simple correlation between the dynamics of global H3K9me2 changes and transcriptional output, at least during the early stages of iPSC formation we analyzed.
Next, we tested whether alterations in H3K9me2 levels could explain the insufficient silencing of specific MEF-associated genes during 3c reprogramming in the presence of UNC0638. We therefore focused on the 328 ISGs (Figure 3C and Table S3) and a control group of 2,664 MEF-associated genes that are equally efficiently downregulated in the absence and presence of EHMT inhibition (fold change between 0.9 and 1.1). Although both gene groups gained H3K9me2 compared with MEFs during reprogramming, ISGs on average showed significantly higher levels of this chromatin mark both immediately upstream of the transcription start site (TSS) (Figure 3F) and across the gene body (Figure S3F). Although both ISGs and control genes exhibited a drastic decrease in H3K9me2 levels upon UNC treatment (~50% in basal and ~80% in 3c-enhanced reprogramming), ISGs experienced a stronger loss of this mark, in particular under 3c conditions (Figures 3G and S3G). When we compared gene loci that were most strongly affected in their H3K9me2 levels by EHMT inhibition during either basal or 3c enhanced reprogramming, we observed limited overlap and distinct GO terms (Figures 3H and S3H and Table S4). Together, these experiments reveal widespread remodeling of H3K9me2 during early stages of iPSC formation and demonstrate both quantitative and qualitative differences with respect to target loci affected by EHMT inhibition during basal and 3c enhanced reprogramming. Figure S3I highlights H3K9me2 changes at select MEF-associated gene loci.
Interference with EHMT Activity Affects Intermediate and Late Markers of Reprogramming
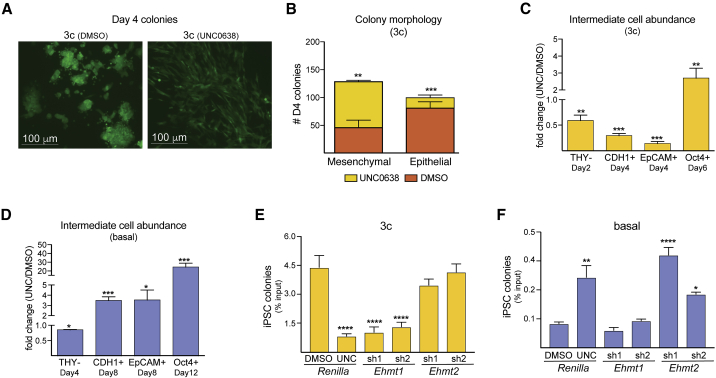
We turned to microscopy and flow cytometry to assess how EHMT inhibition affects later stages of iPSC formation. Visualization of 3c enhanced reprogramming intermediates labeled with a strong, dox-dependent EGFP viral transgene revealed a pronounced reduction in nascent colonies with epithelial features and a concomitant increase in colonies that consisted exclusively of cells retaining fibroblastic morphology in the presence of UNC0638 (Figures 4A and 4B). This correlated with reduced numbers of intermediate cells expressing the epithelial surface markers CDH1 (also known as E-cadherin) and EpCAM (also known as CD326) (Figure 4C). In addition, the intermediate cells that successfully upregulated CDH1 retained elevated levels of the fibroblast surface marker THY1 (Figures S4A and S4B). In contrast, EHMT inhibition during basal reprogramming led to a significant increase in intermediate cells expressing CDH1 and EpCAM (Figure 4D). The relative abundance of late reprogramming intermediates (measured at day 6 during 3c and at day 12 during basal reprogramming) that had reactivated expression of endogenous Oct4 was about 2-fold increased in 3c enhanced reprogramming and 20-fold increased in basal reprogramming (Figures 4C and 4D). These observations indicate that EHMT inhibition facilitates the reactivation of endogenous pluripotency loci, in particular under basal conditions, but under 3c enhanced conditions counteracts the efficient transition of reprogrammable fibroblasts to the intermediate, epithelialized stages of iPSC formation.
Figure 4.
Impact of EHMT Inhibition on Later Stages of iPSC Reprogramming
(A) Representative images of colonies formed upon 4 days of OKSM expression under 3c conditions in the absence (DMSO) or presence of UNC0638. Starting MEFs were labeled with a lentiviral vector expressing EGFP to visualize colony morphology.
(B) Quantification of day 4 colonies with mesenchymal or epithelial morphology under the indicated conditions. Significance with multiple two-tailed t tests: ∗∗p < 0.01 and ∗∗∗p < 0.001. At least 100 colonies were scored in three separate reprogramming experiments for each condition.
(C) Relative abundance in the presence versus absence of UNC0638 (UNC) of the indicated reprogramming intermediates during 3c enhanced reprogramming, as measured by flow cytometry. n = 3 independent experiments.
(D) Same as (C) for reprogramming intermediates during basal reprogramming. n = 3 independent experiments. Significance in (C and D) with multiple t tests using Holm-Sidak correction: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
(E) Percentage of stable iPSC colonies (per input MEFs) formed under 3c reprogramming conditions from cells transduced with indicated shRNAs.
(F) Like (E) but under basal reprogramming conditions.
Significance in (E and F) was calculated with one-way ANOVA with Tukey post test: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. n = 3 independent experiments.
Interestingly, the short hairpin RNA (shRNA)-mediated knockdown (KD) of Ehmt1 recapitulated the reprogramming-inhibiting effect of UNC0638 treatment during 3c enhanced reprogramming, while KD of Ehmt2 recapitulated the reprogramming-promoting effect of EHMT inhibition during basal reprogramming (Figures 4E and 4F), suggesting different and context-dependent functions of these enzymes during iPSC formation. Quantitative PCR confirmed a similar degree of specificity and efficiency of the shRNAs used to interfere with the EHMTs (Figure S4C). However, WB showed that loss of EHMT1 was associated with strongly reduced levels of EHMT2 protein but not vice versa (Figure S4D). This suggests that EHMT1 stabilizes EHMT2 in reprogramming intermediates, as has been previously reported for mouse embryonic stem cells (Tachibana et al., 2005), while EHMT1 is independent of EHMT2 levels. In addition to destabilization of EHMT2, KD of Ehmt1 but not Ehmt2 reduced the levels of H3K9me2 in early reprogramming intermediates, while H3K9me1 and H3K9me3 appeared unaffected (Figure S4E). These observations suggest that 3c enhanced reprogramming intermediates can compensate for a significant reduction in EHMT2 but not in EHMT1 levels, which might help explain the different effects that Ehmt1 and Ehmt2 KD has on the efficiency of iPSC formation (Figures 4E and 4F).
Evidence for a Balance between EHMT Activity and AA-Stimulated Enzymes during Reprogramming
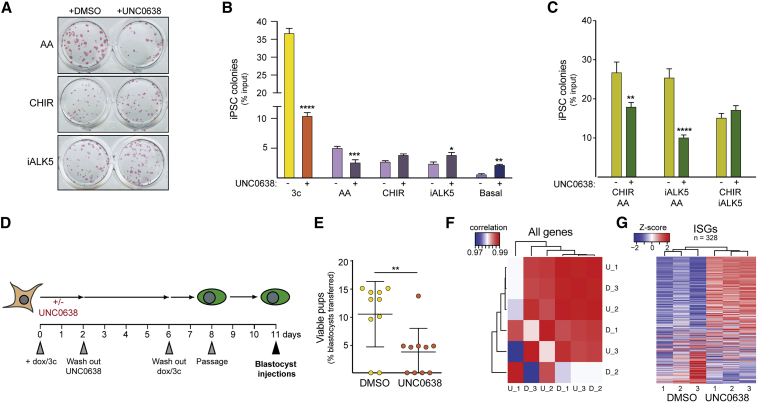
Next, we sought to investigate whether a specific component of the 3c mixture (AA, iALK5, and CHIR99021) is responsible for the reduction in iPSC formation observed upon EHMT inhibition. Reprogramming experiments conducted in the presence of single compounds revealed a significant reduction in iPSC colonies when UNC0638 was used together with AA (Figures 5A and 5B). In contrast, a slight increase in colony numbers was observed when EHMTs were inhibited in the presence of either CHIR99021 or iALK5, although this was not significant in all experiments conducted with CHIR99021 (Figure 5B). The fold-change reduction in colony numbers observed in the presence of AA alone was less dramatic than in the context of 3c conditions, raising the possibility that AA might synergize with one or both of the other two reprogramming enhancing compounds in establishing a requirement for EHMT activity. Indeed, iPSC colony numbers were strongly reduced when AA was combined with either iALK5 or CHIR99021 in the presence of UNC0638 (Figure 5C). Colony numbers remained unchanged when iALK5 was combined with CHIR99021 in the absence of AA (Figure 5C), suggesting that not all approaches to enhancing reprogramming efficiencies acquire dependence on EHMT activity.
Figure 5.
AA Establishes a Requirement for EHMT Activity during Enhanced iPSC Reprogramming
(A) Alkaline phosphatase staining of transgene-independent iPSC colonies obtained after reprogramming MEFs in the presence of the indicated compounds in the absence or presence of UNC0638.
(B) Quantification of iPSC colonies formed under the indicated conditions. n = 3 independent reprogramming experiments.
(C) Quantification of iPSC colonies formed in the presence of the indicated dual-compound conditions.
n = 3 independent reprogramming experiments. Significance in (B and C) was calculated by two-way ANOVA with Sidak post test with adjusted p values of ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
(D) Schematic of the approach taken to test for potential functional deficits of iPSCs caused by transient EHMT inhibition during reprogramming.
(E) Quantification of viable pups obtained after blastocyst injection with the indicated iPSCs. Each circle represents the offspring obtained from one recipient female (n = 10 for each condition) of 20 injected blastocysts. ∗∗p < 0.01 with Mann-Whitney test. Lines show mean with SD.
(F) Heatmap of Pearson's correlation coefficient for all expressed genes between three iPSC cultures derived in the presence of UNC0638 under 3c conditions and three iPSC cultures derived in the absence of UNC0638.
(G) Heatmap of the expression levels (RPKM) of the 328 ISGs (see Figure 3C) in established iPSCs derived in either the absence or the presence of UNC0638.
EHMT inhibition has been reported to counteract cellular proliferation in cancer cells (Casciello et al., 2015). In light of the importance of the proliferative potential for iPSC formation (Li et al., 2009; Utikal et al., 2009), we asked whether this might contribute to the observed reduction in colonies. Quantification of cells at early reprogramming stages revealed reduced cell numbers in the presence of UNC0638 under all conditions, including those where EHMT inhibition favors reprogramming (Figure S5A). This suggests that an effect on proliferation is unlikely to explain the reduction in iPSC reprogramming observed only under specific conditions.
AA has been recognized as a potent cofactor and stimulant of the enzymatic activity of chromatin-modifying enzymes (Cimmino et al., 2018; Monfort and Wutz, 2013), in particular of histone and DNA demethylases. Therefore, we attempted to restore efficient reprogramming in the presence of UNC0638 by shRNA-mediated KD of AA-stimulated H3K9 demethylases. Interference with KDM3B, which has recently been reported to be the main modulator of H3K9me2 downstream of AA in established pluripotent stem cells (Ebata et al., 2017), strongly impaired basal reprogramming but slightly increased colony numbers during 3c enhanced reprogramming in the absence of UNC0638 (Figure S5B). Interference with KDM3B also ameliorated the reduction in iPSC colony formation during 3c enhanced reprogramming in the presence of UNC0638 (Figure S5C). The incomplete rescue observed may suggest the involvement of additional enzymes or other regulators. KD of Kdm3a had no significant effect on any reprogramming condition assessed (Figures S5B and S5C), despite a similar reduction in mRNA and protein levels (Figures S5D and S5E). Together, these observations further support context-dependent functions of the H3K9 methylation and demethylation machinery during iPSC formation.
We have previously shown that 3c reprogramming yields iPSCs that pass the most stringent functional assays (Amlani et al., 2018), raising the question whether EHMT activity is required to generate developmentally highly competent iPSCs by this method. We observed no differences in the morphology or expression of endogenous Oct4 between iPSCs that were derived using 3c enhanced reprogramming in either the absence or the presence of UNC0638 (Figures S5F and S5G). However, when conducting tetraploid blastocyst injections with iPSCs derived from cells exposed to UNC0638 during the first 48 h of 3c enhanced reprogramming, we obtained significantly reduced numbers of viable pups compared with control iPSCs (Figures 5D and 5E). To further investigate the reasons for this observation, we initially analyzed the imprinting status of Dlk1-Dio3, since loss of imprinting (LOI) at this gene cluster frequently occurs during basal reprogramming and results in developmentally impaired iPSCs (Carey et al., 2011; Stadtfeld et al., 2010a), but is normally not observed after 3c enhanced reprogramming (Swanzey and Stadtfeld, 2016). Analysis of iPSCs carrying a sensitive fluorescent reporter system for imprint stability at Dlk1 revealed a very low degree of LOI in 3c iPSCs derived in either the absence or the presence of UNC0638 (Figures S5H and S5I). This argues against a causal role of dysregulation at Dlk1-Dio3 for the observed reduced survival of mice obtained from 3c UNC iPSCs. Next, we subjected the iPSC lines used for blastocyst injections to RNA-seq analysis, which was performed at P3 (injections had been performed at P1). PCA confirmed an overall very high degree of similarity between iPSCs independent of derivation regimen (Figure 5F). However, when we focused on the 328 MEF-associated genes that failed to be efficiently downregulated in early reprogramming intermediates upon UNC0638 treatment (ISGs) (Figure 3C), the iPSCs clustered by treatment group, revealing elevated expression levels of a substantial subset of these genes in cells derived in the presence of compound (Figure 5G). Together, these observations suggest that even transient treatment with UNC0638 during reprogramming can have lasting molecular consequences in iPSCs that interfere with the developmental potential of these cells or the proper function of derivative tissues.
Discussion
A plethora of experimental evidence has demonstrated that H3K9 methyltransferases can counteract the induction of pluripotency ex vivo. By studying the consequences of EHMT inhibition in two well-defined reprogramming conditions, our work provides several lines of insight into how interference with the H3K9 methylation machinery can affect cellular and molecular changes during iPSC formation.
First, our work confirms that counteracting H3K9 methylation facilitates reactivation of the Pou5f1 locus (Chen et al., 2013b; Sridharan et al., 2013), which is consistent with the developmental role of EHMT2 (Feldman et al., 2006). Second, EHMTs appear to be involved in the early downregulation of fibroblast-associated genes, indicating that H3K9 methyltransferases can contribute to the early silencing of the somatic program (Li et al., 2017). Why this effect is restricted to specific loci remains to be determined, but the observation that gene loci transcriptionally affected by UNC0638 administration exhibit higher levels of H3K9me2 methylation already in fibroblasts suggests that this chromatin modification might be of special importance for the regulation of these genes. The dramatic, genome-wide drop in H3K9me2 upon EHMT inhibition during 3c enhanced reprogramming might lower the levels of this chromatin mark at specific gene loci below a threshold required for appropriate regulation. The more pronounced loss of H3K9me2 during enhanced reprogramming might be due to the enzymatic stimulation of histone demethylases by AA or the faster cell cycle under these conditions. The observation that H3K9me2 levels at different gene loci are most strongly affected in basal and 3c reprogramming could be due to context-dependent upregulation or stabilization of a protein cofactor that recruits EHMTs to specific gene loci, as has been reported for MYC in cancer cells (Tu et al., 2018). Third, EHMTs can exert context-dependent functions during the mesenchymal-to-epithelial transition necessary for reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010). In basal reprogramming, chemical inhibition of EHMT facilitates the upregulation of epithelial markers such as CDH1 and EpCAM, suggesting that H3K9 methylation contributes to the stable silencing of additional pluripotency-associated gene loci in somatic cells beyond Pou5f1. In striking contrast, EHMT inhibition counteracts epithelization during our enhanced reprogramming approach. This might be a consequence of the impaired silencing of fibroblast-associated gene loci discussed above or reflective of interference with other aspects of the molecular change that distinguish enhanced from basal reprogramming, such as the much more dramatic alterations in H3K9me2 patterns.
The requirement for EHMT activity during enhanced reprogramming appears to be caused by AA. To our knowledge, none of the prior studies reporting facilitated reprogramming upon interference with the EHMTs utilized AA, explaining why this interaction has been missed. In addition, most efforts to improve iPSC formation by interfering with H3K9 methylation were done at late stages during reprogramming or by using partially reprogrammed cells (Chen et al., 2013b; Sridharan et al., 2013; Tran et al., 2015). Of note, a genetic screen for epigenetic regulators conducted during human iPSC formation (which are routinely derived and cultured in the presence of AA) reported reduced reprogramming efficiencies upon KD of EHMT1 (Onder et al., 2012). To explain the context-dependent consequences of EHMT inhibition, we propose that a balance between these histone methyltransferases and AA-stimulated enzymes such as KDM3B is important for iPSC reprogramming. The concept of a functional balance between H3K9 methyltransferases and AA-stimulated enzymes might help develop new strategies targeting diseases driven by the dysregulation of chromatin-modifying enzymes. Of note, the impact of TET1 on the success of iPSC formation is also modulated by the presence of AA (Chen et al., 2013a), raising the possibility that DNA demethylases might functionally interact with H3K9 methyltransferases and demethylases during enhanced reprogramming. Of note, Tet1 expression was linked to H3K9 demethylase activity during the conversion of partially into fully reprogrammed iPSCs (Tran et al., 2019), supporting the interplay of DNA and histone methylation during reprogramming. It is noteworthy that our results strongly suggest that EHMT1 and EHMT2 have at least partially different roles during fibroblast reprogramming. These enzymes predominantly function as a heterodimer in embryonic stem cells (Tachibana et al., 2005), while distinct functions of these methyltransferases have been reported in other cell types (Battisti et al., 2016).
Our data also suggest that even transient inhibition of H3K9me2 methyltransferase activity during reprogramming can result in impaired iPSC function, suggesting the introduction of persistent epigenetic aberrations. The precise molecular abnormalities caused by EHMT inhibition remain to be determined, but this observation represents a cautionary note for potential undesired consequences when targeting chromatin modifiers to facilitate iPSC formation. In conclusion, our results show that H3K9 methyltransferases can function as flexible regulators of cellular reprogramming that influence not only molecular change during the process, but also the properties of resultant iPSCs.
Experimental Procedures
Mice
Derivation, handling, and genotyping of reprogrammable mice (JAX011001) with the Oct4-GFP allele were described previously (Stadtfeld et al., 2010b). All animal experiments were in accordance with the guidelines of the NYU School of Medicine Institutional Animal Care and Use Committee.
Basic Cell Culture and Cell Culture-Based Assays
Reprogrammable MEFs were heterozygous for Rosa26-rtTA and for Oct4-GFP and either heterozygous for an inducible OKSM allele (Stadtfeld et al., 2010b) or homozygous for an inducible OKS allele (Borkent et al., 2016). Culture of MEFs and iPSCs and reprogramming experiments were conducted as previously described (Vidal et al., 2014). If applicable, UNC0638 (1 μM) was added.
Immunofluorescence
The following primary antibodies were used: H3K9me1 (ab9045; 1:200), H3K9me2 (ab1220; 1:200), H3K9me3 (ab8898; 1:200), H3K4me2 (ab7766; 1:200), H3K4me3 (ab8580; 1:200), EHMT1 (ab41969; 1:200), and EHMT2 (C6H3, 1:50).
Tetraploid Blastocyst Injections
Embryo injections were conducted with 5–10 iPSCs per blastocyst as previously described (Stadtfeld et al., 2012).
shRNA-Mediated Knockdown
Oligonucleotides against specific target genes were designed using the splashRNA algorithm (Pelossof et al., 2017) and are listed in Table S5.
Lentiviral Reprogramming
Reprogramming was carried out with dox-inducible mouse OKSM (Sommer et al., 2009) and rtTA (FUdeltaGW-rtTA, Addgene, 19780) (Maherali et al., 2008) lentiviruses.
Flow Cytometry
Single-cell suspensions were incubated with eFluor 450-conjugated anti-THY1 (53-2.1), biotin-conjugated anti-CDH1 (DECMA-1), and PE/Cy7-conjugated anti-EpCAM (G8.8), followed by incubation with streptavidin-APC (all eBiosciences).
Western Blot Analysis
The following antibodies were used: EHMT1 (Ab41969) 1:1,000, EHMT2 (C6H3) 1:800, H3K9me2 (Ab8896) 1:800, H3K9me2 (Ab1220) 1:800, H3K9me3 (Ab8898) 1:800, H3 (Ab1791) 1:1,000, KDM3A (12835-1-AP) 1:500, and KDM3B (19915-1-AP) 1:500.
RNA-Seq Library Preparations and Analysis
Total RNA was subjected to Automated TruSeq stranded total RNA with RiboZero Gold library preparation (Illumina) and sequencing with a HiSeq 2500. Reads were aligned to differentially expressed genes identified with DESeq (Anders and Huber, 2010). GO analysis was conducted using Gorilla (Eden et al., 2009) and REVIGO (Supek et al., 2011).
ChIP-Seq
Native ChIP-seq was performed with 10 million cells as previously described (Chen et al., 2018), using H3K9me2 antibody (Abcam, ab1220) and Protein G Dynabeads. Libraries were prepared using the KAPA HyperPrep Kit (Roche) and sequenced (paired-end 50) on an Illumina HiSeq 4000.
ChIP-Seq Analysis
Alignment of sequenced reads (Langmead et al., 2009) and enrichment analysis of ChIP signal (Quinlan and Hall, 2010) were performed as described.
Data and Code Availability
Raw RNA-seq and ChIP-seq data were submitted to Gene Expression Omnibus under accession number GEO: GSE130490.
Author Contributions
S.V. and M.S. conceived the study and supervised data analysis. S.V. performed the initial characterization of the effect of EHMT inhibition on reprogramming, conducted reprogramming experiments, and optimized and conducted the isolation of reprogramming intermediates for RNA-seq. A.P. conducted bioinformatic analysis with the support of Y.G. and A.T. K.C. conducted WB analysis and reprogramming experiments, L.E. isolated material for ChIP-seq, E.S. quantified the effect of EHMT inhibition of Dlk1 imprinting, J.M.V. conducted reprogramming experiments with individual compounds, H.W. generated and validated knockdown constructs, C.P. and B.A. provided cell lines and other reagents, V.S. and J.A.S. assisted with RNA-seq, S.T. advised on the biology of histone methyltransferases, S.K. conducted blastocyst injections, E.A. was involved in experimental planning and supervised data analysis, and M.S. conducted reprogramming, flow cytometry, and IF experiments and wrote the manuscript with input from all other authors.
Acknowledgments
We thank Konrad Hochedlinger and current and past members of the Stadtfeld and Apostolou labs for helpful suggestions on the manuscript and during the course of this project. We are grateful to the Cytometry and Cell Sorting Laboratory and the Genomics Core at NYU Langone for expert help with our experiments and the Sfeir lab for help with cell counting and for being excellent neighbors. We thank Carol Chen and Matthew Lorincz for expert advice on H3K9me2 ChIP-seq. M.S. was supported by the National Institutes of Health (1R01GM111852-01). L.E. is a New York Stem Cell Foundation–Druckenmiller Fellow. This research was supported by the New York Stem Cell Foundation.
Published: September 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.08.011.
Supplemental Information
References
- Amlani B., Liu Y., Chen T., Ee L.S., Lopez P., Heguy A., Apostolou E., Kim S.Y., Stadtfeld M. Nascent induced pluripotent stem cells efficiently generate entirely iPSC-derived mice while expressing differentiation-associated genes. Cell Rep. 2018;22:876–884. doi: 10.1016/j.celrep.2017.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti V., Pontis J., Boyarchuk E., Fritsch L., Robin P., Ait-Si-Ali S., Joliot V. Unexpected distinct roles of the related histone H3 lysine 9 methyltransferases G9a and G9a-like protein in myoblasts. J. Mol. Biol. 2016;428:2329–2343. doi: 10.1016/j.jmb.2016.03.029. [DOI] [PubMed] [Google Scholar]
- Becker J.S., Nicetto D., Zaret K.S. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 2016;32:29–41. doi: 10.1016/j.tig.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento M., van de Molengraft M., van Westen R., van Bokhoven H., Kasri N.N. The role of chromatin repressive marks in cognition and disease: a focus on the repressive complex GLP/G9a. Neurobiol. Learn Mem. 2015;124:88–96. doi: 10.1016/j.nlm.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkent M., Bennett B.D., Lackford B., Bar-Nur O., Brumbaugh J., Wang L., Du Y., Fargo D.C., Apostolou E., Cheloufi S. A serial shRNA screen for roadblocks to reprogramming identifies the protein modifier SUMO2. Stem Cell Rep. 2016;6:704–716. doi: 10.1016/j.stemcr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Casciello F., Windloch K., Gannon F., Lee J.S. Functional role of G9a histone methyltransferase in cancer. Front. Immunol. 2015;6:487. doi: 10.3389/fimmu.2015.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C.L., Goyal P., Karimi M.M., Abildgaard M.H., Kimura H., Lorincz M.C. H3S10ph broadly marks early-replicating domains in interphase ESCs and shows reciprocal antagonism with H3K9me2. Genome Res. 2018;28:37–51. doi: 10.1101/gr.224717.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Cimmino L., Neel B.G., Aifantis I. Vitamin C in stem cell reprogramming and cancer. Trends Cell Biol. 2018;28:698–708. doi: 10.1016/j.tcb.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhore S., Nayer B., Hasegawa K. Human pluripotent stem cell culture: current status, challenges, and advancement. Stem Cells Int. 2018;2018:7396905. doi: 10.1155/2018/7396905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebata K.T., Mesh K., Liu S., Bilenky M., Fekete A., Acker M.G., Hirst M., Garcia B.A., Ramalho-Santos M. Vitamin C induces specific demethylation of H3K9me2 in mouse embryonic stem cells via Kdm3a/b. Epigenetics Chromatin. 2017;10:36. doi: 10.1186/s13072-017-0143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S., Feldman N., Abu-Remaileh M., Shufaro Y., Gerson A., Ueda J., Deplus R., Fuks F., Shinkai Y., Cedar H. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Kang Y.K. SETDB1 in early embryos and embryonic stem cells. Curr. Issues Mol. Biol. 2015;17:1–10. [PubMed] [Google Scholar]
- Kirshner Z.Z., Gibbs R.B. Use of the REVERT((R)) total protein stain as a loading control demonstrates significant benefits over the use of housekeeping proteins when analyzing brain homogenates by Western blot: an analysis of samples representing different gonadal hormone states. Mol. Cell Endocrinol. 2018;473:156–165. doi: 10.1016/j.mce.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koche R.P., Smith Z.D., Adli M., Gu H., Ku M., Gnirke A., Bernstein B.E., Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki S., Tachibana M. Epigenetic regulation of mammalian sex determination. Mol. Cell Endocrinol. 2018;468:31–38. doi: 10.1016/j.mce.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C.J., Camargo F.D., Hochedlinger K., Welstead G.G., Zaidi S., Gokhale S., Scholer H.R., Tomilin A., Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu J., Yang X., Zhou C., Guo J., Wu C., Qin Y., Guo L., He J., Yu S. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–833 e6. doi: 10.1016/j.stem.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Canamero M., Blasco M.A., Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Liang G., He J., Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat. Cell Biol. 2012;14:457–466. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S., Liu Y., Lu F., Iwabuchi K.A., Shen L., Inoue A., Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A., Wutz A. Breathing-in epigenetic change with vitamin C. EMBO Rep. 2013;14:337–346. doi: 10.1038/embor.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelossof R., Fairchild L., Huang C.H., Widmer C., Sreedharan V.T., Sinha N., Lai D.Y., Guan Y., Premsrirut P.K., Tschaharganeh D.F. Prediction of potent shRNAs with a sequential classification algorithm. Nat. Biotechnol. 2017;35:350–353. doi: 10.1038/nbt.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalosa-Ruiz G., Bousgouni V., Gerlach J.P., Waarlo S., van de Ven J.V., Veenstra T.E., Silva J.C.R., van Heeringen S.J., Bakal C., Mulder K.W. WDR5, BRCA1, and BARD1 Co-regulate the DNA damage response and modulate the mesenchymal-to-epithelial transition during early reprogramming. Stem Cell Rep. 2019;12:743–756. doi: 10.1016/j.stemcr.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Saunders A., Li D., Faiola F., Huang X., Fidalgo M., Guallar D., Ding J., Yang F., Xu Y., Zhou H. Context-dependent functions of NANOG phosphorylation in pluripotency and reprogramming. Stem Cell Rep. 2017;8:1115–1123. doi: 10.1016/j.stemcr.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer S., Zaph C. The lysine methyltransferase G9a in immune cell differentiation and function. Front. Immunol. 2017;8:429. doi: 10.3389/fimmu.2017.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz B.A., Cetinbas M., Clement K., Walsh R.M., Cheloufi S., Gu H., Langkabel J., Kamiya A., Schorle H., Meissner A. Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell. 2018;23:289–305 e285. doi: 10.1016/j.stem.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S.R., Bahirvani A.G., Rao V.K., Bharathy N., Ow J.R., Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8:16–22. doi: 10.4161/epi.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Do J.T., Desponts C., Hahm H.S., Scholer H.R., Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors' initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Gonzales-Cope M., Chronis C., Bonora G., McKee R., Huang C., Patel S., Lopez D., Mishra N., Pellegrini M. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat. Cell Biol. 2013;15:872–882. doi: 10.1038/ncb2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., Shioda T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Borkent M., Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y., Shivalila C.S., Soldner F., Markoulaki S., Jaenisch R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015;163:218–229. doi: 10.1016/j.cell.2015.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanzey E., Stadtfeld M. A reporter model to visualize imprinting stability at the Dlk1 locus during mouse development and in pluripotent cells. Development. 2016;143:4161–4166. doi: 10.1242/dev.138255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K.A., Dillingham C.M., Sridharan R. Coordinated removal of repressive epigenetic modifications during induced reversal of cell identity. EMBO J. 2019;38:e101681. doi: 10.15252/embj.2019101681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran K.A., Jackson S.A., Olufs Z.P., Zaidan N.Z., Leng N., Kendziorski C., Roy S., Sridharan R. Collaborative rewiring of the pluripotency network by chromatin and signalling modulating pathways. Nat. Commun. 2015;6:6188. doi: 10.1038/ncomms7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu W.B., Shiah Y.J., Lourenco C., Mullen P.J., Dingar D., Redel C., Tamachi A., Ba-Alawi W., Aman A., Al-Awar R. MYC interacts with the G9a histone methyltransferase to drive transcriptional repression and tumorigenesis. Cancer Cell. 2018;34:579–595.e8. doi: 10.1016/j.ccell.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M., Barsyte-Lovejoy D., Liu F., Rival-Gervier S., Allali-Hassani A., Labrie V., Wigle T.J., Dimaggio P.A., Wasney G.A., Siarheyeva A. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S.E., Amlani B., Chen T., Tsirigos A., Stadtfeld M. Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Rep. 2014;3:574–584. doi: 10.1016/j.stemcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M.A., Pan G. The histone demethylases jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wei J., Antony J., Meng F., MacLean P., Rhind R., Laible G., Oback B. KDM4B-mediated reduction of H3K9me3 and H3K36me3 levels improves somatic cell reprogramming into pluripotency. Sci. Rep. 2017;7:7514. doi: 10.1038/s41598-017-06569-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq and ChIP-seq data were submitted to Gene Expression Omnibus under accession number GEO: GSE130490.