Abstract
The Scripps Research Molecular Screening Center (SRMSC) was founded in 2004 and comprises over 22 million dollars of specialized automation. As part of the Translation Research Institute (TRI) it comprises early drug discovery labs and medicinal chemistry. Together with Scripps Research at the La Jolla CA campus, this represent one of the most competitive academic industrial screening centers worldwide. The SRMSC uses automated platforms; one a screening cell and the other a cherry-picking platform. Matched technologies are available throughout Scripps to allow scientists to develop assays and prepare them for automated screening. The library comprises over 1 million drug-like compounds including a proprietary collection of >665K molecules. Internal chemistry has included ∼40K unique compounds that are not found elsewhere. These collections are screened against a myriad of disease targets including cell based and biochemical assays that are provided by Scripps faculty or from global investigators. Scripps has proven competence in all detection formats including high content analysis, fluorescence, BRET, TR-FRET, FP, luminescence, absorbance, AlphaScreen and Ca++ signaling. These technologies are applied to NIH derived collaborations as well as biotech and pharma initiatives. The SRMSC and TRI are recognized for discovering multiple leads including Ozanimod.
Keywords: Drug Discovery, automation, MLPCN, probes, High Throughput Screening
Introduction
Scripps Florida was founded in 2004 as a division of Scripps Research, headquartered in La Jolla, California and is located in Jupiter, Florida (Palm Beach County). Using the latest cutting-edge technologies, researchers at Scripps Florida focus on basic biomedical research and drug discovery. More than 600 employees with 56 full-time faculty members work at the 350,000-squre-foot campus, which is comprised of three state-of-the-art research buildings. Much of the work at Scripps Florida is dedicated to basic biomedical research, a vital segment of medical research that seeks to decipher the most fundamental processes of life. Additionally, researchers at Scripps Florida are developing advanced technologies and applying these tools to the discovery of new therapeutic agents for a variety of devastating human diseases. Departments represented at Scripps are Immunology and Microbiology, Integrative Structural & Computational Biology, Neuroscience, Molecular Medicine and Chemistry. Scripps also has a graduate program offering Doctoral (PhD) training in chemical and biological sciences on the La Jolla and Florida campuses.
The Scripps Florida Campus is uniquely positioned to provide a multidisciplinary approach with ready access to advanced instrumentation for next-generation sequencing, proteomics, X-ray crystallography, structural biology and high-throughput drug screening; offering unique opportunities to discover new therapeutic targets and identify drug leads. Scripps Florida builds on its current strength in medicinal chemistry for biomedical research and focuses on the development of enabling chemical technologies to turn biological insights into small molecule probes, therapeutic leads and ultimately drugs to solve global health challenges.
Support for drug development efforts is also provided through the Drug Metabolism and Pharmacokinetics (DMPK) Core at Scripps Florida and is staffed by experienced professionals with backgrounds in the pharmaceutical and biotech industries. The Scripps Florida Animal Resources Center (ARC) is dedicated to providing a comprehensive-state-of-the-art program of animal care, comparative medicine and associated animal related resources in support of biomedical research performed by Scripps Florida Investigators. The Scripps Florida animal facility is an AAALAC accredited 25,000 square foot vivarium, which includes animal holding, procedure, behavioral rooms and Behavioral Core.
Scripps Research Molecular Screening Center
Currently under the directorship of Dr. Louis Scampavia and Timothy Spicer, the Scripps Research Molecular Screening Center (SRMSC), has been in nonstop operation since 2005 as a major academic HTS facility and formerly as one of the four comprehensive HTS providers during the NIH Molecular Libraries Programs Center Network era.
Annually, we conduct 20 to 30 large-scale HTS campaigns (+665,000 compounds) that require numerous smaller-scaled screening efforts to support these large-scale HTS campaigns. This includes pre-HTS campaign support with assay development, pilot screening, as well as post-HTS campaign support that includes confirmation, counter and dose titration screening; also providing medicinal SAR support with dose testing for potency and selectivity.
Since establishing operations in 2005, the SRMSC has completed over 300 primary campaigns against a large diversity of targets to generate more than 80 million data points for both industrial and academic collaborations. Numerous scientific papers (>110 from SRMSC alone) and presentations demonstrate our ability to produce excellent research-driven data. Our screening results have led to the development of over 72 bioactive molecular probes as novel drug targets for biomedical studies (https://hts.florida.scripps.edu/). Two probes have now been approved as investigational new drugs (IND) by the FDA as New Chemical Entities (NCEs) for clinical trials in the treatment of multiple sclerosis (MS) and cancer as licensed by Celgene and Cleave Biosciences1.
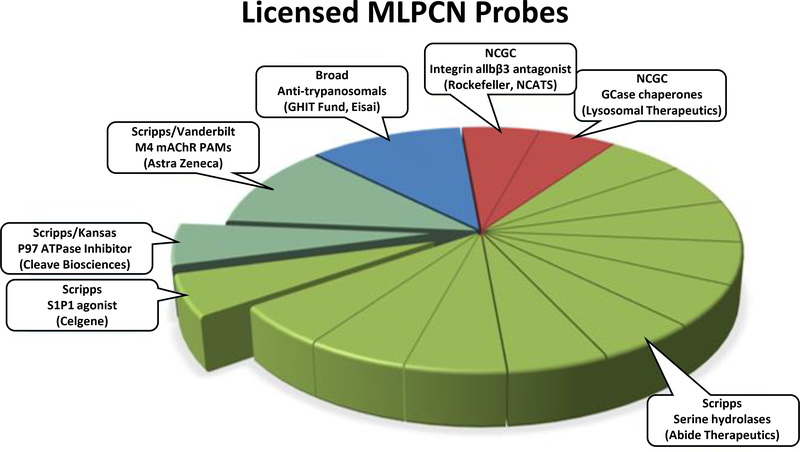
During the MLPCN era, the SRMSC was one of four comprehensive screening centers which supported screening, informatics and medicinal chemistry. During the MLPCN, SRMSC performed 235 HTS campaign from 2005 through 2013. Comparison to other MLPCN comprehensive centers are limited to data available from PubChem. This data includes total number of assays performed (multiple assays per HTS campaign), total number of substances tested and active number of substances tested. The SRMSC ranked first for total number of assays performed and second in both tested an active substances behind the NIH Chemical Genomics Center. Additional cross-center comparisons of more sophisticated descriptors such as assay reporters and assay types would be interesting to measure, however this is currently impossible to accomplish due to a lack of standardized assay descriptors, unique assays performed and terms in PubChem. Coupled to our HTS efforts is Scripps Florida's expertise in medicinal chemistry and pharmacology, Scripps Florida was responsible for 14 of the 18 licensed probes (Figure 1). Moreover, Scripps Florida was the only comprehensive HTS screening center to forward probes into IND for clinical trials.
Figure 1.
Fourteen of the eighteen probes licensed from the MLPCN initiative were discovered at Scripps Florida.
The SRMSC also supports research via fee-for-service collaborations with industrial partners. These collaborations allow for flexibility in screening the SRMSC 665,000 compound library, partner-provided libraries, or both. For partner provided libraries, SRMSC also offers compound management services for compound registration, reformatting, QC and cherrypicking. Typical turnaround times in fee-for-service collaborations entailing a full 665K library HTS campaign, subsequent confirmation and counterscreens, concentration response determination, LC-MS and final reports deliverables are in the four to six-month range. This is based on availability of reagents, timing of assay delivery and assay development requirements all dictated to a degree by the assay provider. Many of the collaborations established between the SRMSC and industrial partners have also resulted in publications, including research performed for Proteostasis Therapeutics2, Eli Lilly3 and Eutropics Pharmaceuticals4.
High Throughput Screening at SRMSC
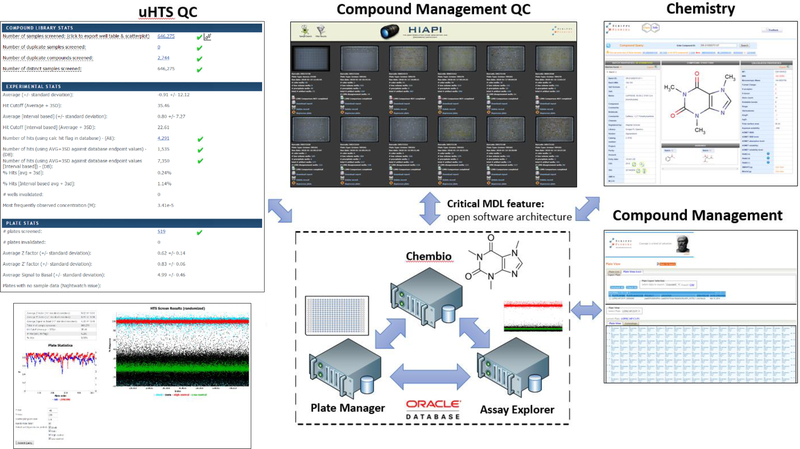
By combining robotics, biology, chemistry and informatics, the SRMSC is able to perform High-Throughput Screening, a fully automated and fully functional robotic process that tests hundreds of thousands of drug-like compounds for biological activity using biochemical interaction or cell-based assays. Currently, SRMSC employs a 1536 well plate format to screen its Scripps Drug Discovery Library (SDDL) of over 665,000 small molecule entities in a rapid and economical fashion. To this end, SRMSC has two industrial robotic platforms from GNF Systems dedicated to running uHTS campaigns and providing hit-picking compound management support (Figure 2). In addition to the deployment of commercial uHTS platforms, the SRMSC has also invested considerable engineering effort into further automating all aspects of the uHTS environment. This includes development of in-house software to fully integrate the GNF HTS and hitpicking platforms to the SRMSC LIMS and custom QC platforms used to ensure data fidelity.
Figure 2. Lead Identification high throughput screening and compound management automation.
(A) HTS Staubli arm, (B) GNF robotic incubator, (C) GNF pin tool transfer station, (D) Perkin Elmer EnVision and Viewlux plate readers, (E) GNF bottlevalve dispenser, (F) Molecular Devices FLIPR Tetra plate reader, (G) Thermo CellInsight high content reader, (H) Cherrypicker Staubli robotic arm, (I) GNF robotic compound plate carousel, (J) Tecan Freedom Evo span-8 cherrypicker, (K) Beckman Biomek FX, (L) REMP Small Sized Store, (M) Compound freezer farm.
Nearly all HTS-ready assay formats are supported including protein to protein interaction (PPI) assays, biochemical-based or cell-based assays. HTS campaigns are conducted either as target-or phenotypic-based readouts using all fluorescence modalities, absorbance, bio/chemo luminescence or high-content analysis.
HTS efforts typically begin with assay development and optimization. Successes in assay development/optimization often advance toward a small-scale pilot screen to validate HTS readiness and estimate hit rate. Pending the target type these pilot screen may use small collections of around ~1,000 compounds such as LOPAC (1280 compounds - Sigma Life Science's Library of Pharmacologically Active Compounds), TOCRIS SCREEN (1070 compounds), Prestwick (1,200 compounds), or larger libraries such as the Scripps FDA-drug approved library (~3,300 compounds) or the Maybridge HitFinder (14,400 compounds). Upon completion of a pilot screen, data is shared and discussed with the collaborating investigator to evaluate performance and make adjustments to the primary/secondary screening strategies to improve hit discovery and selection. Once an agreement is reached, the full deck uHTS campaign is scheduled and QA/QC testing of the robotic instrumentation is initiated to ensure the best results prior to any HTS campaign execution.
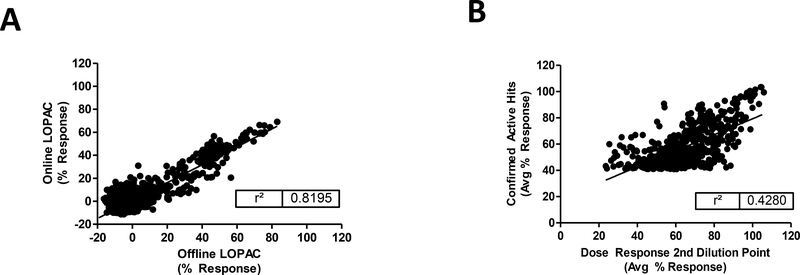
Once the assay implementation has been completed, a pilot screen is performed to validate the assay. The robotic assay validation is performed to confirm that the assay is performing well based assay statistics and a good reproducibility of the activity of the compounds between offline and online assays is been observed (Figure 3). Then the primary screen is performed in a singular (1X) testing mode against the SDDL collection of ~665K compounds; using good statistical separation for hit selection as determined by an acceptable Z'-factor of the controls. Typically, ~1% of the SDDL will be cherry-picked and advance to confirmation testing in triplicate (3x). In addition, secondary screenings will be used to rule out undesirable hits or false positives. Each HTS project will have a unique formulation of secondary screenings which typically include:
Figure 3.
(A) Representative graph of compound activity from a typical assay correlating offline vs. online LOPAC library assays during validation. (B) The results of the compounds tested at the dose response tertiary assays compared to the same activity of the confirmation active hits are shown.
Orthogonal Assay
An assay performed following the primary assay that targets the same biology but attempts to differentiate between compounds that generate false positives, typically as artifacts related to the primary assay reporter, from those compounds that are genuinely active against the target. A hit in this assay would be included for further pursuit.
Specificity Counterscreen
The assay used in the counter-screen is developed to identify compounds that are non-specific and or have the potential to interfere with the assay used in the primary screen. This is also used to eliminate compounds that possess undesirable properties; for example, cytotoxicity. This can also include an assay used to test the promiscuity of a compound against a related a homolog target (e.g. alternative GPCR, kinase, ion channel).
Hits triaged from confirmation and secondary assays are forwarded to 10-point dose titration profiling to determine the IC50, hillslope, dose profile and therapeutic window as compared to a secondary assay. Confirmation, secondary and dose titration assays are performed in triplicate, with averaged values reported along with standard deviation. Hits that demonstrate an acceptable potency and selectivity are then analyzed using in silico procedures to ascertain structural cluster profiles, Pan-Assay Interference Compounds (PAINS), Rule of Five compliance and historical promiscuity score5, 6. A comparison of the activity of the compounds at the point of secondary vs. the dose response data at the same concentration shows the selectivity of the most active compounds. (Figure 3). Data is compiled and typically reviewed with contract investigators and medicinal chemists to select tractable hit scaffolds for SAR development and hit to lead optimization.
Compound Management at SRMSC
The SRMSC Compound Management facility is comprised of software and hardware automation to enable rapid processing of samples in a quality control focused environment (Figure 2). Compound Management automation includes Beckman Coulter Biomek FX and NXp platforms along with a Tecan Freedom EVO which is integrated into a GNF hitpicking platform. All Compound Management automation is fully integrated with a corporate LIMS platform to eliminate the need for manual curation of files.
In addition to support for automated sample handling in formats up to 1536-well microtiter plates, the SRMSC Compound Management facility utilizes a custom in-house developed automated sample QC system known as the Plate Auditor7. Every compound plate which passes through the SRMSC Compound Management facility is processed through this instrument which uses machine vision to automatically identify artifacts in compound plates including empty wells, air bubbles, partially filled wells, precipitate and variations in color between samples. Plate Auditor results are automatically cross-correlated against the SRMSC LIMS to determine if any errors exist in a plate and if sample remediation is necessary prior to releasing plates for downstream processes. If errors are detected, Compound Management team members are notified in real time. Consistent usage of this technology, when combined with in-house LCMS capabilities ensures that sample fidelity is tracked over time and that potential errors in compound management are captured and corrected in real-time, prior to being released for downstream processes in HTS.
When not in use, all samples are stored in secure freezers. When sample access is required, labware containing samples of interest can be placed on the GNF hitpicking platform or in the REMP Small Sized Store, both of which provide fully automated retrieval of sample materials. All sample records are maintained in a secure Oracle database with access restricted to necessary personnel. Full sample and plate genealogies are tracked from the entry of the sample into the lab through use in HTS campaigns, follow-up work and delivery to external collaborators. This tracking includes full automated imaging of compound plates via the Plate Auditor which provides a visual record of plate and sample status at each checkpoint throughout the life of a sample.
Informatics at SRMSC
The SRMSC institutional informatics infrastructure is built around a commercial LIMS platform originally developed by Symyx/MDL. This infrastructure consists of three core components: Chembio for compound registration, Plate Manager for plate registration and Assay Explorer for assay registration. These three core components are built upon an Oracle database and are tightly integrated with one another, resulting in straightforward exchange of data without any need for intermediate manual curation or manipulation of data by end users. Compounds registered in Chembio are automatically associated with plates in Plate Manager when new plates are registered. Similarly, assays created in Assay Explorer automatically aggregate plate metadata with compound metadata and combine both with assay results to provide a single point of access for reviewing and performing QC of assay results.
Access to the institutional LIMS database and all associated software products is tightly restricted not only to relevant users, but with specific roles assigned to individual users to ensure access is project and function specific. Databases are also further segregated by funding source and isolated from each other by distinct login parameters, requiring specific user access to be granted for each instance.
In addition to the software platforms being fully integrated with one another, a Symyx/MDL software API is available which allows SRMSC to further extend the functionality of the existing software by developing in-house solutions which directly interact with the core Symyx/MDL platform (Figure 4). Over the past 13 years, the SRMSC has developed software to provide web-based interfaces for each of the three core components and to further extend and automate many routine compound management and HTS tasks. The web-based interfaces developed by SRMSC have been linked to one another to enable approved users to seamlessly browse datasets across various stages of the drug discovery effort without having to load many different interfaces to access the information of interest.
Figure 4.
In-house software has been developed to extend the core Symyx/MDL LIMS platform functionality and provide user access via web interfaces.
Extended software automation
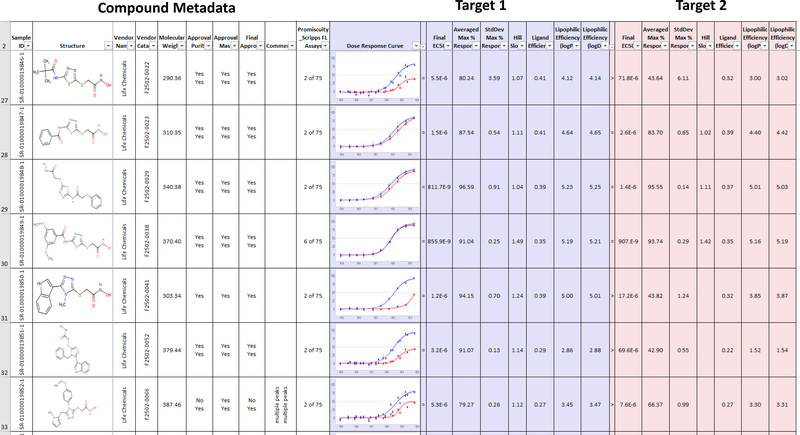
Dose Response Reporter aggregates dose response curve results with compound metadata in an Excel format that is easily sharable with collaborators without requiring proprietary software to review. This report can be run by assay scientists via a web interface which allows for user selection of data to be brought into the report. Curves from multiple dose response assays can be plotted alongside data from primary screen, confirmation and counterscreen assays to provide a report of overall compound activity over the course of a project. SRMSC also maintains a list of approved drugs and associated metadata which is added to all reports as appropriate. Historical activity of compounds in the form of a promiscuity index is also included, providing insight into compound selectivity across assay type and target. All report generation is fully automated via the core Symyx/MDL databases and requires no manual manipulation of data by the user (Figure 5).
Figure 5.
Example of standard automated report generated without human intervention at end of HTS campaign including compound metadata, dose response curves with associated data and LCMS results.
Plato is a web-based interface to Plate Manager which allows users to register new plates, query for existing plates and review plate data in a variety of formats. In addition to interfacing with Plate Manager, Plato is integrated with the GNF screening & hitpicking platforms, allowing robot inventories to be monitored in real-time. Assay daughter plates from screening campaigns are automatically associated with parent compound plates, including transfer volumes, assay diluent volume and full genealogical plate information. Additionally, Plato has been developed to support data uptake from various compound management automation including Beckman Biomek FX and NXp units. Biomek liquid handling platforms are capable of reformatting millions of compounds with full barcode tracking and data export for registration in Plato. SRMSC routinely processes large compound collections for HTS efforts with all data tracking and management performed in an automated fashion.
Cheminfo is a web-based interface to Chembio which allows users to review compound metadata. Assay results are also available for review, providing a full history of compound activity across all associated drug discovery activities including HTS campaigns and DMPK studies. The ChemInfo interface also allows users to quickly browse out to related PubChem compound and substance summaries along with PubChem bioactivity analysis. SRMSC has also utilized ChemInfo to provide compound metadata beyond what is available in Chembio, by including calculated properties such as PAINS descriptors, AlogP, polar surface area, H acceptors/donors, rotatable bonds and solubility.
The HTS Dashboard automates a number of routine HTS QC and reporting tasks. From the HTS Dashboard interface, users can quickly validate all relevant HTS parameters prior to releasing data. The HTS Dashboard also automatically generates standardized visualizations for ease of reporting and QC. In addition to QC and reporting, the HTS Dashboard provides users with tools to explore compound promiscuity across multiple assays, to query for LCMS results and to perform compound enrichment searches when specific compound properties are desired for validation or small-scale screens.
SRMSC informatics capabilities are further extended past the Symyx/MDL platform and in-house tools with other commercial drug discovery informatics tools including Spotfire, Pipeline Pilot, GraphPad Prism and ChemAxon.
Innovation in HTS biology and laboratory automation at SRMSC
Efforts to advance screening technologies at the SRMSC include exploration of 3D cell culture in 1536 well format, the introduction of sophisticated informatics support for high content screens, neuron-based probe discovery platforms capable of HTS and development of custom software and hardware automation.
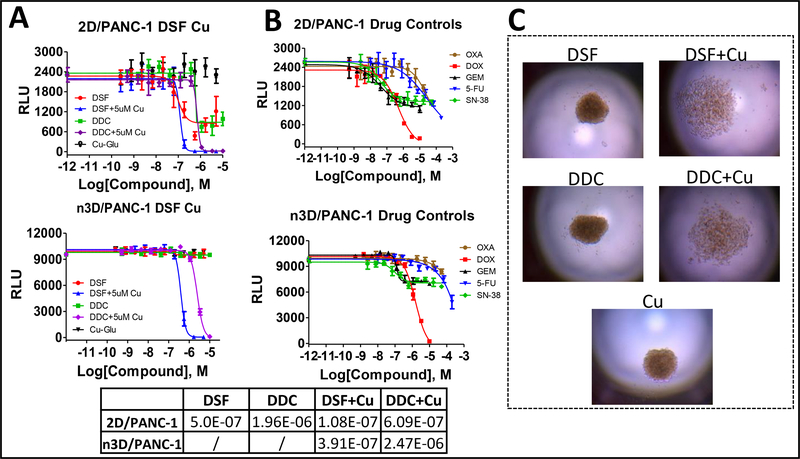
Traditional high-throughput drug screening in oncology routinely have been performed on two-dimensional (2D) cell culture monolayers, which inadequately recapitulate the physiology context of cancer. Three-dimensional (3D) cell culture models have recently gained popularity thanks to their ability to better mimic the complexity of in vivo systems. Three-dimensional (3D) cell models have been described as more physiologically relevant for a variety of applications, especially with regard to cancer research and viability/cytotoxicity assessment in vivo systems. Therefore, physiologically relevant 3D cell culture has been recognized as a potential bridge between traditional in vitro 2D culture and in vivo animal studies.8, 9 It is thus critical to advance the development of scalable and affordable methods of producing 3D spheroids and/or organoids suitable for high throughput cancer drug discovery. Creating three-dimensional (3D) culture systems utilizing patient-derived tumors for rapid testing in high-density format would constitute an important achievement toward precision medicine and regenerative therapies. 3D cellular models have been developed in our screening center that are cost effective and HTS amenable by combining the use of a cell-repellent surface with a bioprinting technology incorporating magnetic force (Figure 6). We have validated this homogenous process by evaluating the effects of well-characterized anticancer agents against four patient-derived pancreatic cancer KRAS mutant-associated primary cells, including cancer-associated fibroblasts10. In testing this scenario, we identified multiple compounds in the 3D assay format that had significantly different results from the traditional 2D monolayer assay format. In a direct comparison between the 2D and 3D assays, most of the tested drugs were less active in 3D, but a few drugs showed preferential cytotoxicity against 3D models over 2D culture, which proved to be both cell and drug-dependent. One such example was Disulfiram, which was more active in the 2D than the 3D format models and its addition to the clinical studies in metastatic pancreatic cancer11, prostate cancer12 and glioblastoma13, 14, seem to confirm that we are using a more phenotypically-relevant strategy which could translate into the development of precision medication initiatives for oncology research. Disulfiram (DSF) and its metabolite Diethyldithiocarbamate (DDC) and Cu-gluconate were tested in primary pancreatic cancer cells and PANC-1 in 2D and 3D format. Results suggested that the addition of DSF or DDC alone has the expected activity in 2D but not in 3D format. The addition of Cu sensitize DSF or DDC, enhancing their cytotoxic effects15 in 2D and 3D formats. (Figure 7)
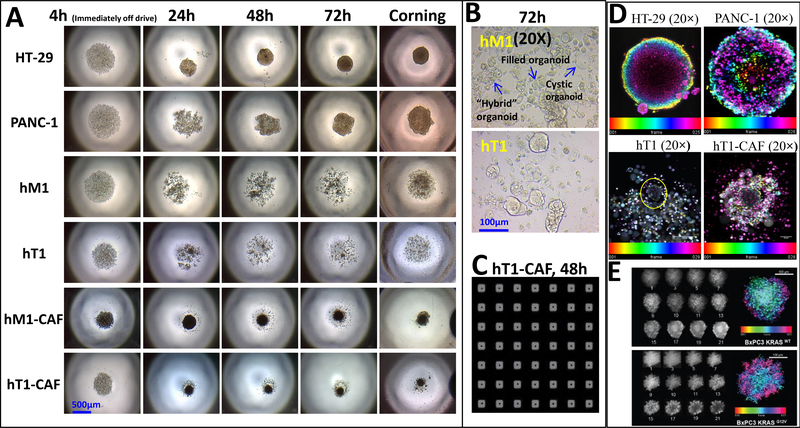
Figure 6. A panel of pancreatic cancer-derived cells was evaluated for their ability to form 3D structure using n3D bioprinting technology.
(A) The 3D structure formation of primary pancreatic cancer cells (hT1 and hM1), their associated fibroblasts (hT1-CAF and hM1-CAF) and standard cell lines (HT-29 and PANC-1) was monitored using standard microscopy (4× objective) in a Greiner Bio-One 384-well cell-repellent flat-bottom plate. These cells were also cultured in 384 Corning U-bottom spheroid plate as a point of comparison. (B) Enlarged images of hM1 and hT1 3D culture using a 20× objective representing small organoid-like structures in primary pancreatic cancer culture. (C) A portion of the full 384-well plate image obtained using the Scripps HIAPI instrument is shown to demonstrate homogeneous hT1-CAF spheroids were formed in each well of a 384-well plate using bioprinting technology. (D) 3D morphology of primary pancreatic cancer hT1 cells, associated fibroblasts, hT1-CAF and standard cell lines (HT29 and PANC1) cultured using n3D bioprinting technology were imaged using INCell 6000 confocal microscopy. Z-stack images were obtained by nuclei staining with Hoechst, then 2 or 5 micron image slices with confocal microscopy and stacked using ImageJ software. HT-29 and PANC-1 spheroids were used as standards. Z-stack images confirmed 3D structure formation of primary pancreatic cancer cells and their associated fibroblasts. (E) Confirmation of spheroidicity of BxPC-3-KRASWT and BxPC-3-KRASG12V stable cell lines was performed by confocal imaging. Z-stack images were taken at 10um increments from the equator of Hoechst-stained spheroids of both cell lines on a GE IN Cell 6000 Analyzer (10 x objective). Maximum intensity projection along the z axis of the 12 individual planes aligned in Image J to generate an intensity projection biased by color scale are shown in both panels.
Figure 7.
(A) CRCs of raw data (RLU) studying the effects of Disulfiram (DSF), its metabolite Diethyldithiocarbamate (DDC) and copper (Cu) gluconate on PANC-1 in both 2D and 3D formats. (B) CRCs for the 5 reference cancer drug control compounds (oxaliplatin, doxorubicin, gemcitabine, 5-fluorouracil and SN-38) versus PANC1 cells in 2D and 3D formats in 384 well. (C). The 3D spheroid formation of the pancreatic cancer cells after 96hrs of drug treatment.
3D cell culture developments also included the execution of a novel 3D HTS campaign to find inducers of a mutant KRAS16 oncogenic gene. The 3D spheroid approach facilitated the identification of a selective inhibitor of cells, Proscillaridin A. SRMSC has collaborations with industrial partners such as n3D Biosciences and Greiner Bio-One for development of a HTS compatible magnetic cell culture platform10 and Corning Incorporated for the development of cell repellent surface micro-cavity microtiter plates for spheroid culture17.
Additionally, High Content Screening (HCS) has gained significant attention as another development in complex screening technologies. HTS provides a single read out of activity for individual assays, whereas HCS can provide data and measurements for multiple features of individual cells in assays all at once. More recently, research with HCS has even been enhanced with the introduction of 3D culture cell lines as opposed to working with the traditional 2D monolayer cell cultures. HCS has the potential to both identify lead compounds and predict compound toxicity which has specific prominence in drug discovery18. Predicting toxicity and efficiency is extremely powerful and part of what makes HCS so complex. In collaboration with Michael Conn from University of Texas, we performed a drug screening in a cell-based high content assay into 1536-well plate format studying pharmacoperones associated with oxalosis kidney disease, which is a rare metabolic disorder.
Another collaboration using cell based high content screening was performed with Scripps Professor Gavin Rumbaugh. There is an unmet need to develop cell-based assays that involves the use of induced neuron based probe discovery platforms assays using primary human cells to study neuropsychiatric disorders19. These methodological improvements simplify the creation of highly scalable neuron-based phenotypic assays designed to improve drug discovery in CNS disorders.
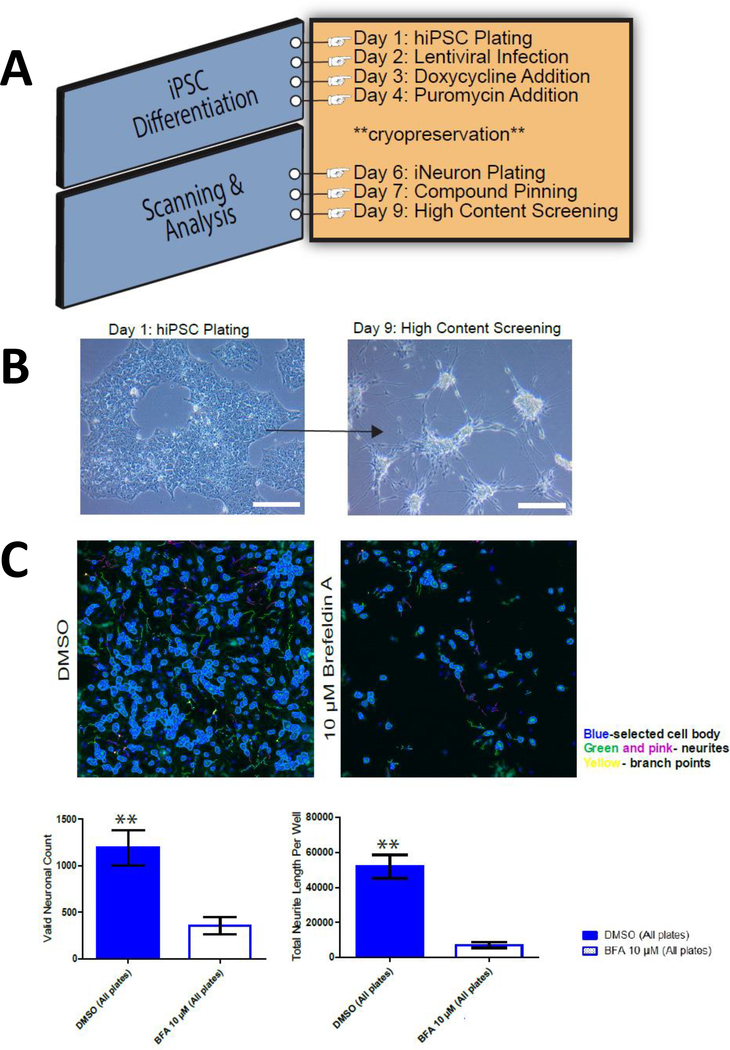
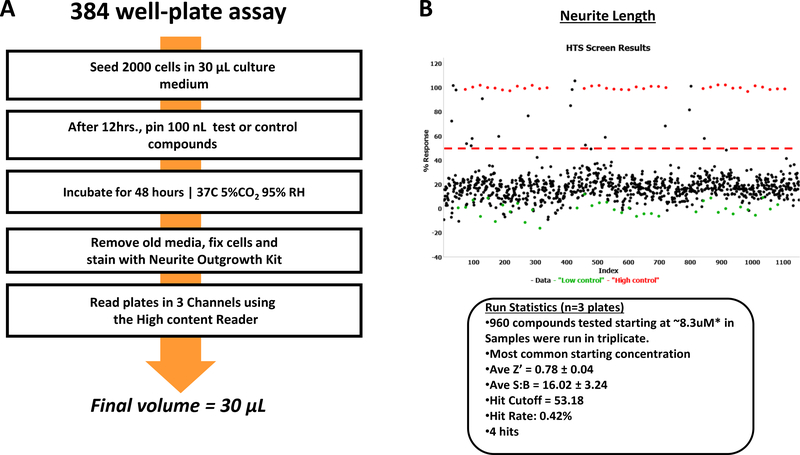
Thus a new direction is being investigated with exploiting the use of HCS and primary human cells to create induced human neurons for use in this research (Figure 8). The lack of robust phenotypic high content screening assays using induced neurons (iNs) is still a challenge due to the multistage and long differentiation protocols. We have been developing a simple method for scaling the production of human pluripotent stem cells (iPSC)-derived induced neurons. As an example, induced pluripotent stem cells, were used as human derived neurons in functional screening assays to measure mitochondrial activity and formation20. Using both HTS and HCS, these induced neurons could be screened for toxicity and against multiple compounds for cell activity in a miniaturized, cost, and time efficient, throughput method. We have been successfully used iNs in 384 well plate format to screen LOPAC pilot library (Figure 9) The end goal is to apply the iPSC derived lines from patients with genetic diseases to evaluate neurotoxicity in early drug discovery.
Figure 8.
(A) Schematic representation of NGN2 lentiviral-based neuronal induction starting from hiPSC to make glutamatergic neurons for use with HCS (B) Representative bright-field images of hiPSC differentiation to iNs at relevant time points. Day 1 was hiPSC plating and Day 9 denotes the end of screening time point. Scale bar = 100 μM. (C) Representative high content image processing of Day 9 iNs where neurites were traced according to predefined optimized parameters under the neuronal profiling module of Thermo's Cellomics ® software.
Figure 9.
(A) Overview of automated quantification protocol of neurite outgrowth from iNs (B) Scatter plot data analysis of the LOPAC pilot screen run in triplicate, including high (red) and low (green) controls, 24 wells each are displayed. High controls are wells containing cells and Brefeldin A and low controls wells containing DMSO. Results are shown as the % response calculated based on compound activity on each plate. Compounds above the red dashed line are considered hits.
Finally, the SRMSC also features a modern engineering lab space for repair and maintenance of existing laboratory instrumentation and development of novel automation. The engineering lab space houses a Tormach 770 CNC mill, an Ultimaker 2+ 3D printer and electronics workbench. These fabrication capabilities allow SRMSC engineers to provide rapid support for novel HTS workflows and to develop custom tools which improve overall lab efficiency. In addition to the previously described Plate Auditor, this also includes development of a real-time HTS liquid dispenser QC monitoring system21, creation of custom robotic incubator shelving for 3D spheroid HTS workflows22, electronic pipetting LED light guides and a wide variety of one-off automation modifications and repairs.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contributions of Dr. Peter Hodder, Mark Southern, Dr. Franck Madoux, Peter Chase, Dr. Shurong Hou, Dr. Dmitriy Minond and Dr. Sanjay Saldanha for their help in the initiation of the screening center and for their efforts during the MLPCN.
FUNDING
This SRMSC and TRI was supported by the Molecular Library Probe Production Centers Network (MLPCN) Grant Proposal Number #5U54 MH084512 and now currently in part by the National Cancer Institute of the National Institutes of Health under Award Number R33CA206949. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Schreiber S; Kotz J; Li M; et al. Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes. Cell 2015, 161, 1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith E; Giuliano KA; Shumate J; et al. A Homogeneous Cell-Based Halide-Sensitive Yellow Fluorescence Protein Assay to Identify Modulators of the Cystic Fibrosis Transmembrane Conductance Regulator Ion Channel. Assay Drug Dev Technol 2017, 15, 395–406. [DOI] [PubMed] [Google Scholar]
- 3.Jambrina E; Cerne R; Smith E; et al. An Integrated Approach for Screening and Identification of Positive Allosteric Modulators of N-Methyl-D-Aspartate Receptors. Journal of Biomolecular Screening 2016, 21, 468–479. [DOI] [PubMed] [Google Scholar]
- 4.Richard DJ; Lena R; Bannister T; et al. Hydroxyquinoline-derived compounds and analoguing of selective Mcl-1 inhibitors using a functional biomarker. Bioorganic & Medicinal Chemistry 2013, 21, 6642–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baell JB Observations on screening-based research and some concerning trends in the literature. Future Med Chem 2010, 2, 1529–46. [DOI] [PubMed] [Google Scholar]
- 6.Baell JB; Holloway GA New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. Journal of Medicinal Chemistry 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- 7.Baillargeon P; Scampavia L; Einsteder R; et al. Monitoring of HTS Compound Library Quality via a High-Resolution Image Acquisition and Processing Instrument. Journal of Laboratory Automation 2011, 16, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hruban RH; Goggins M; Parsons J; et al. Progression Model for Pancreatic Cancer. Clinical Cancer Research 2000, 6, 2969–2972. [PubMed] [Google Scholar]
- 9.Boehnke K; Iversen PW; Schumacher D; et al. Assay Establishment and Validation of a High-Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures. J Biomol Screen 2016, 21, 931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou S; Tiriac H; Sridharan BP; et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS DISCOVERY: Advancing Life Sciences R&D 2018, 23, 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ClinicalTrials.gov Disulfiram and Gemcitabine Hydrochloride in Treating Patients With Unresectable Solid Tumors or Metastatic Pancreatic Cancer. U.S. National Library of Medicine; February 2016, NCT02671890, https://clinicaltrials.gov/show/NCT02671890. [Google Scholar]
- 12.ClinicalTrials.gov A Phase Ib Study of Intravenous Copper Loading With Oral Disulfiram in Metastatic, Castration Resistant Prostate Cancer. U.S. National Library of Medicine; November 2016, NCT02963051, https://clinicaltrials.gov/show/NCT02963051. [Google Scholar]
- 13.ClinicalTrials.gov Disulfiram/Copper With Concurrent Radiation Therapy and Temozolomide in Patients With Newly Diagnosed Glioblastoma. U.S. National Library of Medicine; March 2016, NCT02715609, https://clinicaltrials.gov/show/NCT02715609. [Google Scholar]
- 14.ClinicalTrials.gov Safety, Tolerability and Efficacy of Disulfiram and Copper Gluconate in Recurrent Glioblastoma. U.S. National Library of Medicine; January 2017, NCT03034135, https://clinicaltrials.gov/show/NCT03034135. [Google Scholar]
- 15.Skrott Z; Mistrik M; Andersen KK; et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 2017, 552, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kota S; Hou S; Guerrant W; et al. A novel three-dimensional high-throughput screening approach identifies inducers of a mutant KRAS selective lethal phenotype. Oncogene 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madoux F; Tanner A; Vessels M; et al. A 1536-Well 3D Vaibility Assay to Assess the Cytotoxic Effect of Drugs on Spheroids. SLAS Discovery 2017, 22, 516–524. [DOI] [PubMed] [Google Scholar]
- 18.Hou S; Madoux F; Scampavia L; et al. Drug Library Screening for the Identification of Ionophores That Correct the Mistrafficking Disorder Associated with Oxalosis Kidney Disease. SLAS DISCOVERY: Advancing Life Sciences R&D 2017, 22, 887–896. [DOI] [PubMed] [Google Scholar]
- 19.Spicer TP; Hubbs C; Vaissiere T; et al. Improved Scalability of Neuron-Based Phenotypic Screening Assays for Therapeutic Discovery in Neuropsychiatric Disorders. Molecular Neuropsychiatry 2017, 3, 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rana P; Luerman G; Hess D; et al. Utilization of iPSC-derived human neurons for high-throughput drug-induced peripheral neuropathy screening. Toxicol In Vitro 2017, 45, 111–118. [DOI] [PubMed] [Google Scholar]
- 21.Shumate J; Baillargeon P; Spicer TP; et al. IoT for Real-Time Measurement of High-Throughput Liquid Dispensing in Laboratory Environments. SLAS TECHNOLOGY: Translating Life Sciences Innovation 0, 2472630318769454. [DOI] [PubMed] [Google Scholar]
- 22.Baillargeon P; Shumate J; Hou S; et al. Automating a 3D Bioprinting Technology for High Throughput Screening. SLAS Technology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]