Abstract
Characterizing the allelic diversity within major histocompatibility complex (MHC) genes is an important way of determining the potential genetic resilience of a population to infectious and ecological pressures. For the koala (Phascolarctos cinereus), endemic diseases, anthropogenic factors and climate change are all placing increased pressure on this vulnerable marsupial. To increase the ability of researchers to study MHC genetics in koalas, this study developed and tested a high-throughput immunogenetic profiling methodology for targeting MHC class I UA and UC genes and MHC class II DAB, DBB, DCB and DMB genes in a population of 82 captive koalas. This approach was validated by comparing the determined allelic profiles from 36 koala family units (18 dam-sire-joey units and 18 parent-joey pairs), finding 96% overall congruence within family profiles. Cancers are a significant cause of morbidity in koalas and the risk factors remain undetermined. Our analysis of this captive population revealed several novel MHC alleles, including a potential link between the DBB*03 allele and a risk of developing cancer. This method offers a reliable, high-throughput protocol for expanded study into koala immunogenetics.
Keywords: Koala, Phascolarctos cinereus, MHC, Immunogenetic profiling
Major histocompatibility complex (MHC) genes play a critical role in the immune system. MHC molecules present antigens from either intracellular threats (such as viruses and cancerous proteins, via class I molecules) or phagocytosed antigens (such as bacteria and parasites, via class II molecules) to T lymphocytes to initiate an adaptive immune response (Punt et al. 2018). In vertebrates, MHC allelic variation in a population has been linked to biological traits from immune recognition and susceptibility to infectious and autoimmune diseases and to ecological success with mating preferences and pregnancy outcomes (Sommer 2005). For the last remaining member of the family Phascolarctidae, the koala (Phascolarctos cinereus), survival against both endemic disease (from Chlamydia pecorum and potentially koala retrovirus) and population fragmentation/genetic bottlenecking has reached a crisis point (Australia 2011; Hemming et al. 2018). This has recently led to an increased focus on studying of MHC genetic loci in koalas to understand their potential genetic resilience in the face of these ecological pressures.
There are 23 MHC class I and 23 MHC class II genes and pseudogenes annotated in the koala genome (Johnson et al. 2018). Detailed investigation into class I genes determined that 11 of these genes are actively transcribed in the koala, with three genes ubiquitously expressed as classical class Ia genes (Phci-UA, UB and UC) and eight genes with tissue restricted expressions as nonclassical class Ib genes (Phci-UD, UE, UF, UG, UH, UI, UJ and UK) (Cheng et al. 2018). All of the expressed MHC class I genes appear to be present as single copy genes in the genome (Cheng et al. 2018; Johnson et al. 2018). Within the MHC class II gene family, four class II subfamilies are recognized, consisting of alpha and beta subunits of DA, DB, DC and DM (Abts et al. 2018; Johnson et al. 2018; Lau et al. 2013). Studies investigating the allelic diversity of class II DA and DB genes have found that the beta subfamilies (DAB and DBB) contain more allelic diversity than the alpha subfamilies (DAA and DAA) (Lau et al. 2013). In addition, genome analysis and diversity studies indicate that DAB and DBB genes are present as three distinct loci in the genome, while DCB and DMB genes are present as single copy genes (Johnson et al. 2018; Lau et al. 2013).
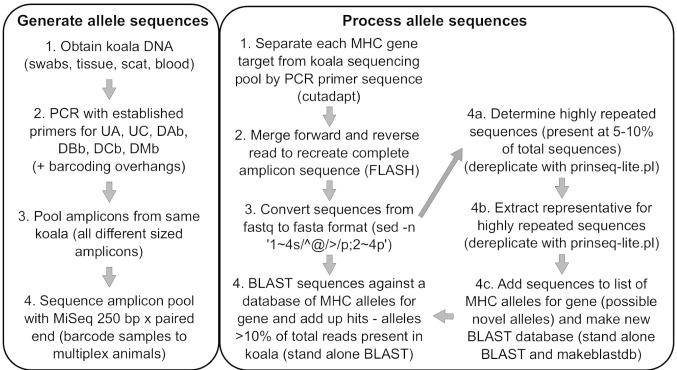
Several techniques have been used to identify the allelic diversity of MHC class I and II genes in wild and captive koala populations. Initial studies focused on class II DA and DB genes and utilized single-strand conformation polymorphism (OSCP) analysis (Lau et al. 2014a, 2014b, 2013). OSCP analysis involves PCR amplification of the target loci, lambda exonuclease digestion to remove the forward amplicon stand and acrylamide gel electrophoresis of the reverse amplicon strand to generate a banding pattern and excising of individual bands for direct sequencing or cloning and sequencing to determine allele sequences (Lau et al. 2013). While this approach has the advantage of ensuring all alleles within an individual are detected (via each allele’s unique banding position in the gel), this method is very labour intensive and low throughput. Later studies investigating class I UA, UB and UC genes or class II DA and DB genes opted for the directly cloning and sequencing of PCR amplicons from the target loci (Cheng et al. 2018; Quigley et al. 2018). While this approach improved throughput, it could not guarantee every allele for a tested gene was detected in the set of sequenced clones. Most recently, direct Illumina sequencing of PCR amplicons from the target loci was attempted for a range of class I and II gene loci (Abts et al. 2018). This approach allowed for higher throughput processing and greatly increased confidence that all the allelic diversity within a koala would be detected; however, sequence processing challenges related to amplicon size and multiple genes per loci limited the number of targets that generated reportable data. The field of koala MHC immunogenetics needs a comprehensive approach that combines the advantages of previous studies into a single, reliable, high-throughput technique. That is what this study achieved (Fig. 1).
Fig. 1.
Flowchart of high-throughput MHC allele determination method in koalas. The left panel summarizes the steps from sample acquisition to sequence generation while the right panel summarizes sequence processing to allele assignment (with example programs/commands necessary to complete each step given in parentheses)
In our current study, the immunogenetic profile of 82 captive koalas was generated in a high-throughput fashion. Blood samples were collected from koalas from Lone Pine Koala Sanctuary (Brisbane, Queensland, Australia) as part of routine health monitoring. DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen) as per manufacturer’s instructions. Established PCR primers that target the receptor binding grove (exon 2 region) of class I UA and UC genes and class II DAB, DBB, DCB and DMB genes (Table 1) (Abts et al. 2018; Cheng et al. 2018; Lau et al. 2013) were used to generate loci-specific amplicons between 200–397 bp. To increase throughput and reduce costs, adaptor sequences were added to the 5′ end of each primer to allow for multiplex barcoding of koala samples for sequencing. The six MHC gene target amplicons from each koala were pooled for barcoding (generating one barcoded sample per koala) and all 82 koala samples were pooled for sequencing on a single MiSeq 250 bp paired end Illumina run (Ramaciotti Centre for Genomics, Sydney) (Fig. 1).
Table 1.
PCR primers used to identify MHC alleles
| MHC class | Gene target | Primer sequencea | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Class I | UA | MHCI_UA_F: ACCCCTGACCCTGCCGTGTC | 313 | Cheng et al. (2018) |
| MHCI_UA_R: CACCGCCTTCGCTCTGGTTGA | ||||
| UC | MHCI_UC_F: AAGGTCTCCAATGTTTCCGACTCA | 397 | Cheng et al. (2018) | |
| MHCI_UC_R: TCTCGCGCTAAGGCCATACC | ||||
| Class II | DAb | MHCII_DAb_F: ATGCCCCAAAGCACTTCAC | 271 | Lau et al. (2013) |
| MHCII_DAb_R: CGCACTRAGAAGGGCTCA | ||||
| DBb | MHCII_DBb_F: AGGGACATCCCAGAGGATTTCG | 282 | Lau et al. (2013) | |
| MHCII_DBb_R: TCTTCTGTCCACCGCGAAGG | ||||
| DCb | MHCII_DCb_F: GGTGAGGTCTGAGTGTCACA | 200 | Abts et al. (2018) | |
| MHCII_DCb_R: CATTCACTATGGACCTTGCAGT | ||||
| DMb | MHCII_DMb_F: CATGTGGAGAGTGGCTGTATG | 266 | Abts et al. (2018) | |
| MHCII_DMb_R: GTCCTTTGGGTCAACGCTC |
aFor Illumina MiSeq sequencing, adaptor sequences were added to the 5′ ends of each primer to allow for multiplex barcoding: forward adaptor: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG; reverse adaptor: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG
To deconvolute the raw sequencing results into MHC alleles present per koala, the sequences obtained from each koala were sorted, merged into complete amplicon sequences, processed to extract highly repeated sequences and identified against a database of known koala alleles (Fig. 1). Sequence files from each koala were first separated into individual target gene files based on the PCR primer sequence for each target gene, trimmed to remove the primers sequences and culled to remove any reads shorter than 150 bp using the program cutadapt (Martin 2011). Next, paired forward and reverse reads were merged to reassemble the complete amplicon sequence using the program FLASH (Magoc and Salzberg 2011). The sequence data was then converted from Fastq to Fasta format using the standard unix ‘sed’ command. Finally, sequences were BLAST searched against a list of known koala MHC alleles using stand-alone BLAST (Altschul et al. 1990; Camacho et al. 2009). For each gene target, reference alleles that represented more than 10% of the total sequence reads for that gene were considered present in the koala. To detect novel MHC alleles not represented in the reference list, sequence files were separately tested for highly repetitive sequences with the program prinseq (Schmieder and Edwards 2011) and novel sequences were added to the reference list (Fig. 1).
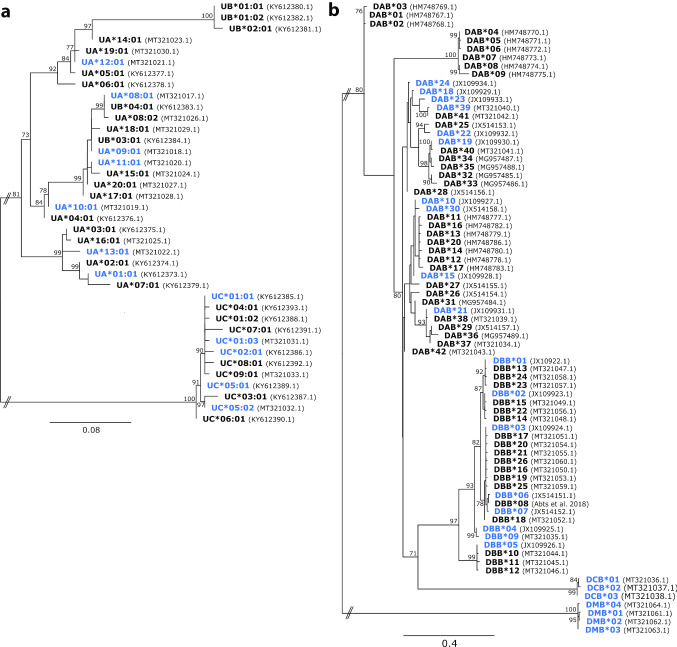
Using this high-throughput method, the allelic diversity of six MHC genes was determined for all 82 test koalas (Fig. 2). Overall, this population contained seven UA alleles (six novel), five UC alleles (three novel), 10 DAB alleles (one novel), eight DBB alleles, three DCB alleles (all novel) and four DMB alleles (all novel) (Fig. 2). Within these alleles, the expected range of 1 to 2 alleles per koala was retrieved from the single genome copy genes UC, DCB and DMC and 1 to 6 alleles per koala were retrieved from the three genome copy genes DAB and DBB. Interestingly, between 1 and 3 alleles per koala were retrieved from UA (a single copy gene). Sequence comparison revealed that the detected UA alleles designated UA*08:01 and UA*09:01 were identical to the previously published UB alleles UB*04:01 and UB*03:01, respectively (Fig. 2). This suggested that the UA PCR primer set was amplifying both UA and UB alleles, and both gene loci are represented in the UA allele results. Phylogenetic analysis of class I sequences supported the fact that UA and UB alleles are closely related, preventing segregation of alleles into UA or UB gene origin (Fig. 2a).
Fig. 2.
Phylogenetic relationships of known koala MHC gene alleles from class I (a) and class II (b). These maximum likelihood phylogenetic trees were generated using DNA sequences aligned with mafft (Katoh et al. 2002) before ModelFinder determined the best fit model (HKY + F + G4 for (a); TIMe + G4 for (b) (Kalyaanamoorthy et al. 2017)) and IQ-TREE (Nguyen et al. 2015) and UFBoot2 (Hoang et al. 2018) constructed the tree with 1000 bootstrap replicates. Only bootstrap values above 70 are shown. Alleles highlighted in blue were detected in this study. The accession number for each allele is presented in parenthesises after the allele name
Within the captive koala study group, there were 18 family units (dam-sire-joey) and an additional 18 parent-offspring pairs (either dam-joey or sire-joey). This allowed for a detailed evaluation of the accuracy of this high-throughput koala immunogenetic approach (Tables 2 and 3). Knowing that MHC alleles must follow Mendel’s law of segregation of genes (offspring must inherit an allele from each parent) and that the genetics of an offspring should be composed of the genetics of their parents, the accuracy of allele assignment within the family groups was determined. Examining all 36 family units, there was 100% congruency between the parent(s)/joey genetic profiles for UA, DCB and DMB loci, 94% (34/36) congruency in DAB and DBB profiles and 86% (31/36) congruncy in UC profiles (Tables 2 and 3). Within these 216 comparisons, the nine inconsistent cases involved five cases where the joey was missing an allele from either their dam or sire (four UC, one DBB), three cases where the joey possessed an allele neither parent possessed (two DAB, one DBB) and one case where the joey did not have an allele from either parent (UC). Overall, these minor discrepancies resulted in this method having an overall congruency rate of 96%.
Table 2.
Immunogenetic profiles of dam-sire-joey family groups
| Family | Relationship | Name | UA | UC | DAB | DBB | DCB | DMB |
|---|---|---|---|---|---|---|---|---|
| 1 | Dam | Jazz | 08:01; 10:01; 11:01 | 01:01 | 10; 15; 19; 21 | 02; 05 | 01; 03 | 02 |
| Sire | Barney | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 | |
| Joey | Angelica | 08:01; 10:01; 11:01 | 01:01; 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01; 02 | |
| 2 | Dam | Rusa | 08:01; 09:01; 11:01 | 01:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 03 |
| Sire | Keeley | 08:01; 09:01 | 05:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 04 | |
| Joey | Myrtle | 08:01; 09:01; 11:01 | 02:01 | 15; 19; 21 | 04 | 01; 02 | 02; 03 | |
| 3 | Dam | Jubilee | 08:01; 09:01; 11:01 | 01:01; 05:01 | 10; 19; 21; 30 | 01; 03; 04 | 01; 03 | 02; 03 |
| Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| Joey | Kia | 08:01;10:01; 11:01 | 01:01 | 15; 19; 21; 30 | 02; 03 | 01 | 02; 03 | |
| 4 | Dam | Elata | 01:01 | 01:01 | 10; 19; 21 | 02; 04; 05; 09 | 03 | 01; 02 |
| Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| Joey | Sargent | 01:01; 13:01 | 05:02 | 10; 15; 19; 21 | 01; 04 | 01; 03 | 02; 03 | |
| 5 | Dam | Sinammon | 08:01; 10:01; 11:01 | 05:02 | 10; 15; 19; 21 | 01 | 01 | 03 |
| Sire | Fingal | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03; 05 | 01 | 01; 04 | |
| Joey | Milton | 08:01; 10:01; 13:01 | 01:01; 05:02 | 10; 19; 21 | 01; 03 | 01 | 01; 03 | |
| 6 | Dam | Minx | 08:01; 11:01 | 05:02 | 15; 19; 21; 22; 24; 39 | 04; 06; 07 | 01 | 01; 02 |
| Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| Joey | Tully | 08:01: 10:01; 11:01 | 5:02 | 15; 19; 21 | 01; 06; 07 | 01 | 02; 03 | |
| 7 | Dam | Halle | 01:01; 10:01 | 01:01 | 15; 19; 21 | 02; 05 | 01; 03 | 02 |
| Sire | Keeley | 08:01; 09:01 | 05:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 04 | |
| Joey | Jester | 01:01; 08:01; 09:01 | 01:01; 05:01 | 15; 19; 21 | 02; 05 | 02; 03 | 02; 04 | |
| 8 | Dam | Kirra | 08:01; 09:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Sire | Strudel | 01:01; 12:01 | 01:01; 01:03 | 10; 19; 21; 23 | 02; 03 | 01 | 01 | |
| Joey | Esmeralda | 12:01; 13:01 | 01:01 | 15; 19; 21; 23 | 02; 03; 04 | 01 | 01; 02 | |
| 9 | Dam | Dama | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 |
| Sire | Strudel | 01:01; 12:01 | 01:01; 01:03 | 10; 19; 21; 23 | 02; 03 | 01 | 01 | |
| Joey | Feenie | 08:01; 11:01; 12:01 | 01:03 | 15; 19; 21; 23 | 02; 05 | 01 | 01 | |
| 10 | Dam | Victory | 08:01; 11:01; 12:01 | 01:01; 05:01 | 15; 19; 21; 30 | 02; 06; 07 | 01 | 02; 04 |
| Sire | Aster | 08:01; 10:01; 11:01 | 01:01 | 15; 19; 21; 23 | 02; 04 | 01; 02 | 03; 04 | |
| Joey | Nat | 08:01; 11:01 | 01:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 03 | |
| 11 | Dam | Rookie | 08:01; 09:01; 12:01 | 01:01 | 10; 15; 19; 21 | 02; 04; 05 | 02; 03 | 01; 03 |
| Sire | Strudel | 01:01; 12:01 | 01:01; 01:03 | 10; 19; 21; 23 | 02; 03 | 01 | 01 | |
| Joey | Davis | 01:01; 08:01; 09:01 | 01:01; 01:03 | 10; 15; 19; 21 | 02; 03; 04; 05 | 01; 02 | 01; 03 | |
| 12 | Dam | Rookie | 08:01; 09:01; 12:01 | 01:01 | 10; 15; 19; 21 | 02; 04; 05 | 02; 03 | 01; 03 |
| Sire | Barney | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 | |
| Joey | Drew | 08:01; 11:01; 12:01 | 01:01 | 10; 19; 21; 22; 24; 39 | 02; 04; 05 | 01; 03 | 01 | |
| 13 | Dam | Guppy | 08:01; 09:01; 10:01 | 01:01 | 10; 19; 21; 22; 24; 39 | 02; 04 | 01; 03 | 01; 02 |
| Sire | Yeti | 01:01; 08:01; 11:01 | 01:01 | 15; 19; 21; 23 | 01; 03 | 01 | 01; 02 | |
| Joey | Urchin | 01:01; 08:01; 09:01 | 01:01 | 10; 19; 21; 23 | 03; 04 | 01 | 01 | |
| 14 | Dam | Zap | 01:01; 10:01 | 01:01 | 10; 15; 19; 21 | 02; 05 | 01; 03 | 02; 03 |
| Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| Joey | Hamlet | 10:01; 13:01 | 01:01 | 15; 19; 21 | 02; 05 | 01; 03 | 02 | |
| 15 | Dam | Kirra | 08:01; 09:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Sire | Yeti | 01:01; 08:01; 11:01 | 01:01 | 15; 19; 21; 23 | 01; 03 | 01 | 01; 02 | |
| Joey | Merlin | 08:01; 09:01; 11:01 | 01:01 | 10; 15; 19; 21 | 01; 03 | 01 | 01; 02 | |
| 16 | Dam | Crumble | 10:01; 12:01 | 05:01 | 10; 19; 21 | 04; 06; 07 | 01; 03 | 01; 04 |
| Sire | Wisely | 01:01; 12:01 | 01:01; 01:03 | 15; 19; 21 | 02; 05 | 01 | 02 | |
| Joey | Waffle | 01:01; 10:01 | 01:01; 05:01 | 10; 15; 18; 19; 21 | 02; 06; 07 | 01 | 02; 04 | |
| 17 | Dam | Zap | 01:01; 10:01 | 01:01 | 10; 15; 19; 21 | 02; 05 | 01; 03 | 02; 03 |
| Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| Joey | Cordelia | 10:01 | 01:01 | 15; 19; 21 | 02 | 01; 03 | 02 | |
| 18 | Dam | Minx | 08:01; 11:01 | 05:02 | 15; 19; 21; 22; 24; 39 | 04; 06; 07 | 01 | 01; 02 |
| Sire | O'Malley | 08:01; 10:01; 11:01 | 01:01; 05:02 | 15; 19; 21; 30 | 01; 03 | 01 | 03 | |
| Joey | Archer | 08:01; 10:01; 11:01 | 01:01; 05:02 | 15; 19; 21; 30 | 03; 06; 07 | 01 | 02; 03 |
Italicized entries are alleles incongruent with Mendelian laws of genetics
Table 3.
Immunogenetic profiles of parent-joey family pairs
| Family | Relationship | Name | UA | UC | DAB | DBB | DCB | DMB |
|---|---|---|---|---|---|---|---|---|
| 19 | Dam | Elata | 01:01 | 01:01 | 10; 19; 21 | 02; 04; 05; 09 | 03 | 01; 02 |
| Joey | Major | 01:01; 08:01; 11:01 | 01:01 | 10; 19; 21; 30 | 03; 04 | 01; 03 | 02; 03 | |
| 20 | Dam | Minx | 08:01; 11:01 | 05:02 | 15; 19; 21; 22; 24; 39 | 04; 06; 07 | 01 | 01; 02 |
| Joey | Barney | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 | |
| 21 | Dam | Dama | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 |
| Joey | Sprocket | 08:01; 09:01; 11:01 | 01:01; 01:03 | 10; 15; 19; 21 | 02; 03; 05 | 01 | 01; 02 | |
| 22 | Dam | Mooloolah | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Joey | Clifton | 08:01; 09:01; 13:01 | 01:01; 05:02 | 10; 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| 23 | Dam | Mooloolah | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Joey | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| 24 | Dam | Liana | 10:01; 12:01 | 01:01; 01:03 | 21; 23 | 02; 03; 05 | 01 | 01; 02 |
| Joey | Daiquiri | 08:01; 09:01; 10:01 | 01:01 | 15; 19; 21; 23 | 03 | 01; 03 | 01; 02 | |
| 25 | Dam | Elata | 01:01 | 01:01 | 10; 19; 21 | 02; 04; 05; 09 | 03 | 01; 02 |
| Joey | Virginea | 01:01; 10:01 | 02:01 | 10; 15; 19; 21 | 04 | 02; 03 | 02; 03 | |
| 26 | Dam | Mooloolah | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Joey | Jervis | 08:01; 11:01; 13:01 | 01:01 | 10; 19; 21; 30 | 03; 04 | 01 | 01 | |
| 27 | Dam | Rusa | 08:01; 09:01; 11:01 | 01:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 03 |
| Joey | Aster | 08:01; 10:01; 11:01 | 01:01 | 15; 19; 21; 23 | 02; 04 | 01; 02 | 03; 04 | |
| 28 | Dam | Dama | 08:01; 11:01 | 01:03 | 10; 15; 19; 21 | 02; 05 | 01 | 01 |
| Joey | Fraggle | 01:01; 08:01; 11:01 | 01:01; 01:03 | 10; 15; 19; 21 | 01; 02; 05 | 01 | 01; 02 | |
| 29 | Dam | Mooloolah | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Joey | Fingal | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03; 05 | 01 | 01; 04 | |
| 30 | Dam | Elata | 01:01 | 01:01 | 10; 19; 21 | 02; 04; 05; 09 | 03 | 01; 02 |
| Joey | Pellita | 01:01; 12:01 | 01:01 | 10; 19; 21; 23 | 03; 04 | 01; 03 | 01; 02 | |
| 31 | Dam | Mooloolah | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 |
| Joey | Kirra | 08:01; 09:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03 | 01 | 01; 02 | |
| 32 | Dam | Rusa | 08:01; 09:01; 11:01 | 01:01 | 15; 19; 21 | 02; 04 | 01; 02 | 02; 03 |
| Joey | Bkley | 08:01; 09:01; 11:01 | 01:01 | 15; 19; 21 | 03; 04 | 01; 02 | 01; 03 | |
| 33 | Sire | Byron | 10:01; 13:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 |
| Joey | Tango | 08:01; 10:01; 11:01 | 01:01 | 10; 15; 19; 21 | 02 | 01 | 02 | |
| 34 | Sire | Rory | 10:01 | 02:01 | 10; 19; 21; 22; 24; 39 | 04 | 01; 03 | 01; 02 |
| Joey | Hermit | 08:01; 10:01; 11:01 | 02:01 | 10; 19; 21 | 04 | 03 | 01; 02 | |
| 35 | Sire | Orinoco | 08:01; 10:01; 11:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 03 |
| Joey | Ficus | 08:01; 10:01; 11:01 | 01:01; 05:02 | 15; 19; 21 | 01; 02 | 01 | 02; 03 | |
| 36 | Sire | Fingal | 08:01; 11:01; 13:01 | 01:01 | 10; 15; 19; 21 | 02; 03; 05 | 01 | 01; 04 |
| Joey | Claret | 08:01; 11:01; 13:01 | 01:01 | 15; 19; 21; 23 | 02; 04; 05 | 01; 03 | 02; 04 |
Italicized entries are alleles incongruent with Mendelian laws of genetics
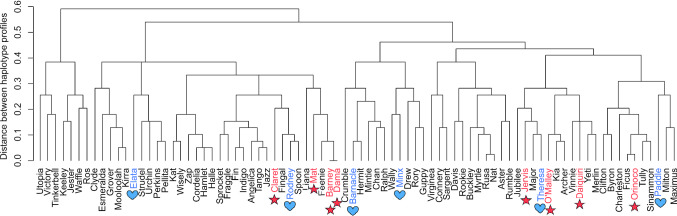
To examine the immunogenetic diversity within this koala captive population, MHC haplotypes were clustered in R (R_Core_Team 2014) based Gower’s coefficient of similarity (Gower 1971) in daisy and with complete linkage in hclust (Fig. 3). After sampling was done for this study, eight koalas developed cancer (primarily lymphoma or leukemia) and another six koalas died of natural causes (age-related). To determine if there were any associations between developing cancer and MHC alleles, both combined haplotype and individual allele prevalence were examined in this subset of deceased koalas. While there was no significant difference detected in the overall MHC haplotypes of these koalas (χ2 = 23.946, df = 29, p = 0.7316) (graphically seen in the lack of clustering by causes of death in Fig. 3), allele DBB*03 was significantly more prevalent in koalas that developed cancer (5/8; 63%) than koalas that died of natural causes (0/6; 0%) (Fisher exact p = 0.031). It should be acknowledged that association of an MHC allele with neoplasia provides no evidence of causation, as this outcome could be related to another linked genetic or retroviral trait. As the sample size in this analysis was relatively small, monitoring will continue in these koalas and reanalysis will be undertaken when sample sizes are larger.
Fig. 3.
MHC haplotype clustering of captive koalas in this study. Koalas that developed cancer (primarily lymphomas and leukemias) are indicated in red with red stars, while koalas that died from natural causes (related to old age) are indicated in blue with blue hearts
In conclusion, this study designed and tested a high-throughput protocol to determine the MHC allelic profile of koala classic class I and class II beta subfamily genes. Using established PCR primer sets, standard Illumina paired end sequencing and freely available software, this method resulted in 96% congruence of allele assignment within 36 koala family units over six MHC loci. Alleles detected in this study expanded the list of known koala MHC alleles, and an association between the presence of DBB*03 and koalas developing cancer was detected. This protocol offers a reliable method for expanded study in the important area of koala immunogenetics.
Funding
This work was funded by Lone Pine Koala Sanctuary, Queensland, Australia.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This work received University of the Sunshine Coast ethics approval ANE1942.
References
- Abts KC, Ivy JA, DeWoody JA. Demographic, environmental and genetic determinants of mating success in captive koalas (Phascolarctos cinereus) Zoo Biol. 2018;37:416–433. doi: 10.1002/zoo.21457. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Australia Co (2011) ‘The koala - saving our national icon’. Report of the Senate Standing Committees on Environment and Communications on the inquiry into the status, health and sustainability of Australia’s koala population. https://www.aph.gov.au/Parliamentary_Business/Committees/Senate/Environment_and_Communications/Completed_inquiries/2010-13/koalas/report/inde
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Polkinghorne A, Gillett A, Jones EA, O'Meally D, Timms P, Belov K. Characterisation of MHC class I genes in the koala. Immunogenetics. 2018;70:125–133. doi: 10.1007/s00251-017-1018-2. [DOI] [PubMed] [Google Scholar]
- Gower J. A general coefficient of similarity and some of its properties. Biometrics. 1971;27:857–874. doi: 10.2307/2528823. [DOI] [Google Scholar]
- Hemming V, Hoffmann M, Jarrad F, Rumpff L (2018) NSW Koala reserach plan: expert elicitation of knowledge gaps. In Research CoEaE (ed.). Office of Environment and Heritage, The University of Melbourne, Australia
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RN, O'Meally D, Chen Z, Etherington GJ, Ho SYW, Nash WJ, Grueber CE, Cheng Y, Whittington CM, Dennison S, Peel E, Haerty W, O'Neill RJ, Colgan D, Russell TL, Alquezar-Planas DE, Attenbrow V, Bragg JG, Brandies PA, Chong AY, Deakin JE, Di Palma F, Duda Z, Eldridge MDB, Ewart KM, Hogg CJ, Frankham GJ, Georges A, Gillett AK, Govendir M, Greenwood AD, Hayakawa T, Helgen KM, Hobbs M, Holleley CE, Heider TN, Jones EA, King A, Madden D, Graves JAM, Morris KM, Neaves LE, Patel HR, Polkinghorne A, Renfree MB, Robin C, Salinas R, Tsangaras K, Waters PD, Waters SA, Wright B, Wilkins MR, Timms P, Belov K. Adaptation and conservation insights from the koala genome. Nat Genet. 2018;50:1102–1111. doi: 10.1038/s41588-018-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Q, Griffith JE, Higgins DP. Identification of MHCII variants associated with chlamydial disease in the koala (Phascolarctos cinereus) PeerJ. 2014;2:e443. doi: 10.7717/peerj.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Q, Jaratlerdsiri W, Griffith JE, Gongora J, Higgins DP. MHC class II diversity of koala (Phascolarctos cinereus) populations across their range. Heredity (Edinb) 2014;113:287–296. doi: 10.1038/hdy.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Q, Jobbins SE, Belov K, Higgins DP. Characterisation of four major histocompatibility complex class II genes of the koala (Phascolarctos cinereus) Immunogenetics. 2013;65:37–46. doi: 10.1007/s00251-012-0658-5. [DOI] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011;2011(17):3. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt J, Stranford SA, Jones PP, Owen JA. Kuby immunology. New York: Mcmillian Education; 2018. [Google Scholar]
- Quigley BL, Carver S, Hanger J, Vidgen ME, Timms P. The relative contribution of causal factors in the transition from infection to clinical chlamydial disease. Sci Rep. 2018;8:8893. doi: 10.1038/s41598-018-27253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Core_Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]