Abstract
Schistosoma mansoni is a flatworm that causes schistosomiasis, a neglected tropical disease that affects more than 200 million people worldwide. There is only one drug indicated for treatment, praziquantel, which may lead to parasite resistance emergence. The ribonucleoside analogue 5-azacytidine (5-AzaC) is an epigenetic drug that inhibits S. mansoni oviposition and ovarian development through interference with parasite transcription, translation and stem cell activities. Therefore, studying the downstream pathways affected by 5-AzaC in S. mansoni may contribute to the discovery of new drug targets. Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with low or no protein coding potential that have been involved in reproduction, stem cell maintenance and drug resistance. We have recently published a catalog of lncRNAs expressed in S. mansoni life-cycle stages, tissues and single cells. However, it remains largely unknown if lncRNAs are responsive to epigenetic drugs in parasites. Here, we show by RNA-Seq re-analyses that hundreds of lncRNAs are differentially expressed after in vitro 5-AzaC treatment of S. mansoni females, including intergenic, antisense and sense lncRNAs. Many of these lncRNAs belong to co-expression network modules related to male metabolism and are also differentially expressed in unpaired compared with paired females and ovaries. Half of these lncRNAs possess histone marks at their genomic loci, indicating regulation by histone modification. Among a selected set of 8 lncRNAs, half of them were validated by RT-qPCR as differentially expressed in females, and some of them also in males. Interestingly, these lncRNAs are also expressed in other life-cycle stages. This study demonstrates that many lncRNAs potentially involved with S. mansoni reproductive biology are modulated by 5-AzaC and sheds light on the relevance of exploring lncRNAs in response to drug treatments in parasites.
Subject terms: Long non-coding RNAs, Parasite biology
Introduction
Schistosomiasis is a very debilitating disease, spread across three continents with a global burden estimated by the World Health Organization at 2,543,364 DALYs (Disease Adjusted Life Years)1. It is estimated that schistosomiasis affects more than 200 million people in 74 countries2,3. The disease is caused by parasitic trematodes of the genus Schistosoma, being the three main species Schistosoma mansoni, S. japonicum and S. haematobium4. S. mansoni is the prevalent species in Latin America, with 1 to 3 million people infected and over 25 million living in risk areas mainly in Brazil and Venezuela5.
Administration of praziquantel (PZQ) to infected individuals is the basis of current schistosomiasis therapy. PZQ is a safe, cheap and tolerable drug6, however, cure rates of less than 50% have been recorded7 and drug tolerance has already been reported8,9. This scenario reinforces the need of new and more efficient approaches in reducing morbidity or disease eradication, such as the development of a vaccine10 or alternative drugs11.
5-azacytidine (5-AzaC) is a ribonucleoside currently used to treat human myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML)12. 5-AzaC is considered an epigenetic drug as it can prevent DNA methylation by inhibition of DNA methyltransferases. It can also impede RNA methylation13 and decrease protein synthesis14. In S. mansoni, 5-AzaC has been shown to inhibit biological processes related to female metabolism, including egg production, egg maturation and normal ovarian development15. In addition, 5-AzaC also significantly alters S. mansoni adult female transcription, translation and stem cell activities16. Therefore, the study of the downstream pathways affected by 5-AzaC in S. mansoni may contribute to the understanding of the epigenetic control of gene expression and its physiological consequences in schistosomes and, in the future, to the possible development of new chemotherapeutic strategies against schistosomiasis.
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides with low or no protein coding potential17,18 that in humans are involved in a wide range of biological processes, including cell cycle regulation, reproduction, stem cell maintenance and drug resistance19. While the functions of lncRNAs have been explored20 and growing evidence suggests that they should be considered as drug targets in human diseases21, the mechanisms of regulation of lncRNA expression are much less understood17. In helminths other than S. mansoni, just a few works have reported the identification of lncRNAs using transcriptomic approaches22–24, however no further investigation of the mechanisms of lncRNA regulation or response to drug treatments were performed.
In S. mansoni, the expression of lncRNAs at different life-cycle stages was first detected by our group in 2011 using microarrays25 and then subsequently reported by many groups using RNA-Seq approaches26–31. However, it is largely unknown if S. mansoni lncRNA levels may be regulated by drugs. In the present work, we have evaluated the effect of 5-AzaC on lncRNA expression in S. mansoni adult worms by performing a re-analysis of the public RNA-Seq data from Geyer et al.16. We show, for the first time, that an epigenetic drug affects lncRNA levels in S. mansoni and that many of these lncRNAs are also differentially expressed in unpaired females and ovaries, indicating involvement in parasite reproductive biology. Understanding the mechanisms of control of lncRNAs expression will help the identification of potential new therapeutic targets and may contribute to the development of novel therapeutic strategies in the future.
Results
A set of lncRNAs is differentially expressed in S. mansoni females upon 5-AzaC in vitro treatment
We reanalyzed the RNA-Seq public data generated by Geyer et al.16 to search for long non-coding RNAs (lncRNAs) possibly affected by 5-AzaC treatment in S. mansoni females (Supplementary Table S1 shows the samples used and alignment statistics). In that study, Geyer et al.16 had treated S. mansoni adult worm pairs in vitro (Puerto Rican strain/NMRI, obtained from mice) with 5-AzaC at 491 μM for 48 h, extracted RNA from the females and then performed RNA-Seq. Thirty adult worm couples were used in each of three biological replicates16. Geyer et al.16 analyzed in that RNA-Seq dataset only the protein-coding genes differentially expressed in S. mansoni females after treatment with 5-AzaC. As lncRNA levels have been shown to be modulated by nucleoside analogs in other eukaryotes32,33, we hypothesized that lncRNA levels would also be modulated by 5-AzaC in S. mansoni.
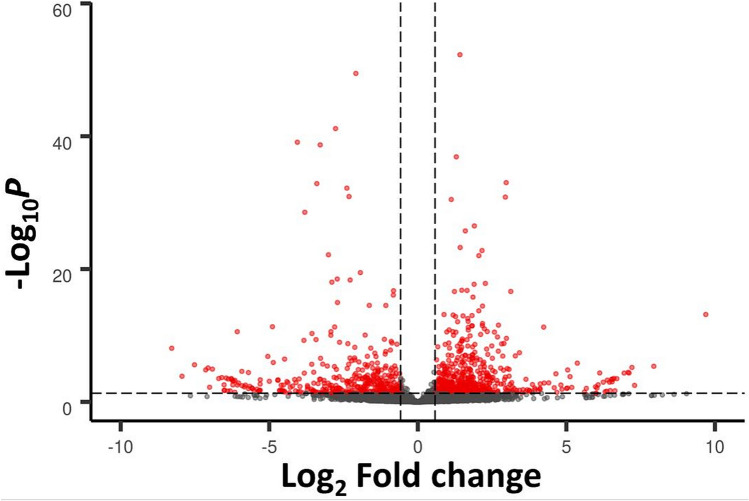
Indeed, the re-analysis of Geyer et al.16 RNA-Seq data with a reference transcriptome that is comprised of protein-coding genes as well as lncRNAs31 (see “Materials and methods”), found 912 lncRNAs differentially expressed in S. mansoni females upon 5-AzaC in vitro treatment. Among them, 522 were long intergenic non-coding RNAs (lincRNAs, being 353 upregulated and 169 downregulated), 358 were antisense non-coding RNAs (SmLNCAs, being 183 upregulated and 175 downregulated), and 32 were sense non-coding RNAs (SmLNCSs, being 16 upregulated and 16 downregulated) (Fig. 1). All differentially regulated protein-coding genes and lncRNAs are shown in Supplementary Table S2 with their transcript per million (TPM) values, and in Supplementary Table S3 with their raw counts.
Figure 1.
Differentially expressed long non-coding RNAs (lncRNAs) detected by RNA-Seq in adult S. mansoni females treated with 5-AzaC. These results were obtained by re-analysis of the RNA-Seq data from Geyer et al.16 using the S. mansoni lncRNA transcriptome published in Maciel et al.31 as reference. In Geyer et al.16, parasites were cultured either in the presence or absence of 491 μM 5-AzaC for 48 h. LncRNA gene expression levels identified in this RNA-Seq dataset are shown with a volcano plot, which displays the differentially expressed lncRNAs between 5-AzaC-treated and control S. mansoni females (red dots, showing FDR < 0.05 and log2FC > 0.59 or < -0.59, dotted lines). Grey dots represent non-differentially expressed lncRNAs. 912 lncRNAs were considered significantly differentially expressed, being 522 long intergenic ncRNAs, 358 antisense lncRNAs and 32 sense lncRNAs.
We also found in our analysis 3219 protein-coding genes (corresponding to 3693 transcripts, 1655 upregulated and 2038 downregulated) differentially expressed after 5-AzaC treatment, of which 1810 have also been found as differentially expressed by Geyer et al.16. Geyer et al.16 previously identified 4036 protein-coding genes differentially expressed after 5-AzaC treatment, and 3221 of these genes are contained in the S. mansoni genome v7 annotation; thus, we were able to retrieve 1810 out of 3221 (or 56%) protein-coding genes found as differentially expressed in that work, which is a reasonable proportion considering the difference in the genomes used for reads alignment (genome version 7 was used here versus genome version 5.2 in the previous work) and the different read-mapping and counting tools used for the analysis.
As expected, principle component analysis (PCA) resulted in transcriptomes of the 5-AzaC treated and control groups segregating broadly into two distinct regions with replicates from the same condition clustering together, both for control or 5-AzaC-treated groups (Supplementary Fig. S1).
Most of the lncRNAs differentially expressed upon 5-AzaC treatment in S. mansoni females belong to co-expression modules related to male metabolism
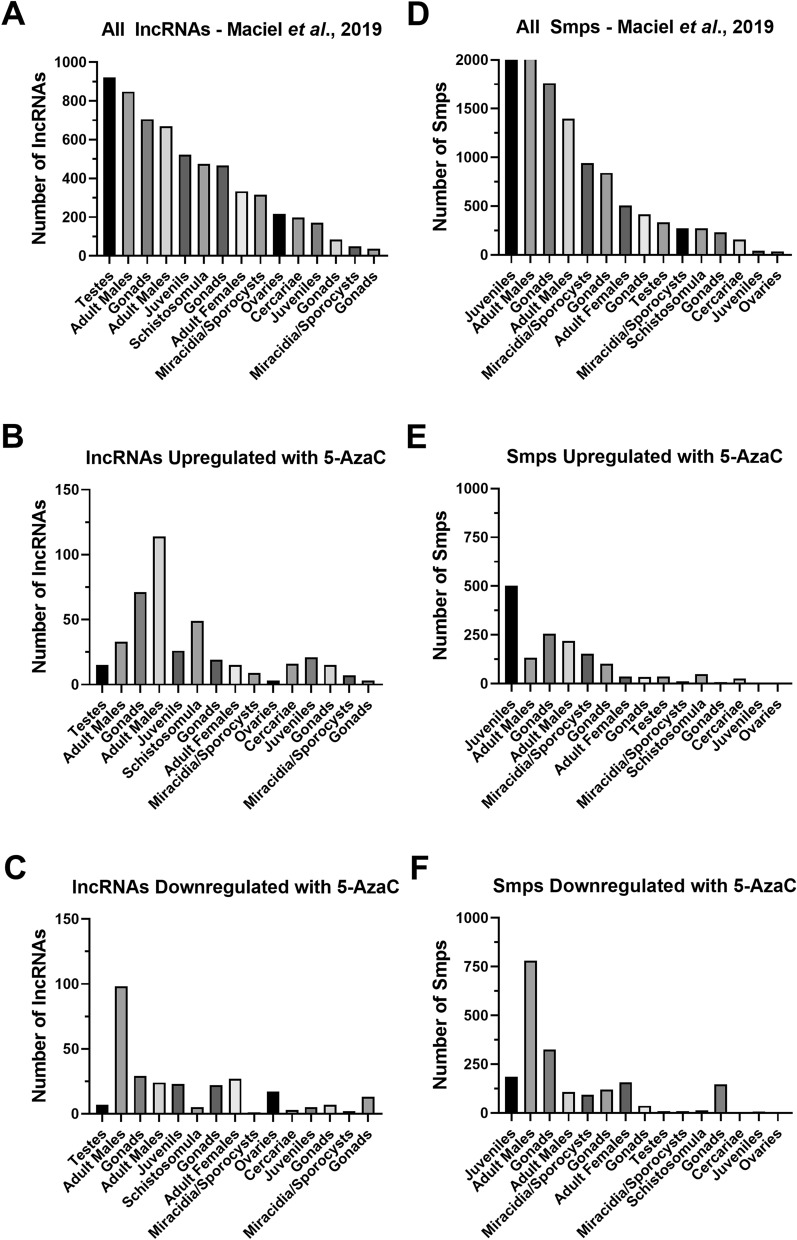
Previously, besides building a new S. mansoni transcriptome comprised of lncRNAs in addition to protein-coding genes31, we also showed by weighted gene co-expression network analyses (WGCNA) that 6016 out of 16,583 lncRNAs identified in different S. mansoni life-cycle stages and tissues belong to one of 15 different lncRNAs/mRNAs co-expression modules31 (Fig. 2A). Each of these 15 modules represents one cluster of highly interconnected lncRNA/mRNA genes that are more expressed in one given S. mansoni stage/tissue, including miracidia, sporocysts, cercariae, schistosomula, juveniles, adult males, adult females and gonads (testes and ovaries)31; also, some of the stage/tissues are represented by more than one module (Fig. 2A) (please refer to “Materials and methods” section for details).
Figure 2.
Distribution of 5-AzaC-affected long non-coding RNAs and protein-coding genes among the weighted gene co-expression network (WGCNA) modules (represented by life-cycle stages and tissues). (A) Number of lncRNAs detected in each of 15 different WGCNA modules, according to Maciel et al.31; note that different modules are associated to the same S. mansoni life-cycle stage/tissue. (B) Number of lncRNAs upregulated in 5-AzaC treated females in each of the 15 WGCNA modules. (C) Number of lncRNAs downregulated in 5-AzaC treated females in each of the 15 WGCNA modules. (D) Number of protein-coding genes detected in each of 15 different WGCNA modules, according to Maciel et al.31; note that different modules are associated to the same S. mansoni life-cycle stage/tissue. (E) Number of protein-coding genes upregulated in 5-AzaC treated females in each of the 15 WGCNA modules. (F) Number of protein-coding genes downregulated in 5-AzaC treated females in each of the 15 WGCNA modules.
Evaluating to which of the lncRNAs/mRNAs co-expression modules the lncRNAs differentially expressed after 5-AzaC treatment belong to, can help the understanding of the gene expression programs altered by this epigenetic drug on S. mansoni females. When we looked at the modules to which the 552 lncRNAs upregulated after 5-AzaC exposure in females belong to, we observed enrichment in a male-related module (Fig. 2B). Out of the 552 lncRNAs upregulated after 5-AzaC treatment, 450 were assigned to any module, being the top three most represented enriched modules: adult males (yellow module, 114 lncRNAs or 20% of the lncRNAs, p value < 0.0001, hypergeometric test), gonads (brown module, 71 lncRNAs or 13% of the lncRNAs, p value < 0.001, hypergeometric test) and schistosomula (magenta module, 49 lncRNAs or 9% of the lncRNAs, p value < 0.01, hypergeometric test) (Fig. 2B).
Out of the 360 lncRNAs downregulated after 5-AzaC treatment, 296 were assigned to any module, being the three top most represented modules: adult males (turquoise module, 98 lncRNAs or 27% of the lncRNAs, p value < 0.0001, hypergeometric test), gonads (brown module, 29 lncRNAs or 8% of the lncRNAs, p value = 0.057, hypergeometric test) and adult females (pink module, 27 lncRNAs or 7% of the lncRNAs, p value < 0.01, hypergeometric test) (Fig. 2C). It is possible that 5-AzaC treatment in females switches the lncRNA transcriptional program to a pattern more similar to that shown by males, as it was shown for protein-coding genes in females treated with GSK343, an histone methyltransferase EZH2 inhibitor34, or in unpaired females, in which the gonads are not developed35. This effect can impact on stem cell activity and egg production by females, as previously shown by Geyer et al.16.
Similar patterns of module distribution of protein-coding genes in the Maciel et al.31 dataset (Fig. 2D) and of protein-coding genes differentially expressed after 5-AzaC treatment (Fig. 2E,F) were obtained in our re-analysis of the Geyer et al.16 dataset.
The list of lncRNAs and protein-coding genes differentially expressed after 5-AzaC treatment, as well as the modules to which they belong are given in Supplementary Table S4.
Involvement of lncRNAs with the parasite reproductive biology
In order to evaluate if the lncRNAs differentially expressed after 5-AzaC treatment could be involved in S. mansoni reproductive biology, we checked if these lncRNAs are also differentially expressed in pairing-dependent conditions or in reproductive organs compared with whole worms. To do that, we cross compared the lncRNAs differentially expressed after 5-AzaC exposure in females with lncRNAs that we found to be differentially expressed in a re-analysis of the Lu et al. data35 (please refer to “Materials and methods” section for details) for lncRNAs differentially expressed between bisex (paired) females (bF) and single-sex females (sF), between bisex ovaries (bO) and single-sex ovaries (sO) and between bisex ovaries(bO) and bisex females (bF). We found that 60% of the lncRNAs downregulated after 5-AzaC exposure (216 out of 360 lncRNAs) are also present in at least one of these comparisons (Supplementary Fig. S2A,B, see overlap between the yellow oval and the other ovals). When the statistical significance of the overlaps between the lncRNAs downregulated after 5-AzaC exposure and each of the above comparisons was calculated, all of the overlaps were statistically significant, with all the p values obtained from the pairwise comparisons lower than 1.254e−10 (hypergeometric test).
In addition, 23% of the lncRNAs upregulated after 5-AzaC exposure (127 out of 552 lncRNAs) are also present in at least one of these comparisons (Supplementary Fig. S2C,D, see overlap between the yellow oval and the other ovals). When the statistical significance of the overlaps between the lncRNAs upregulated after 5-AzaC exposure and each of the comparisons was calculated, all of the overlaps were statistically significant (with all the p values obtained for the pairwise comparisons lower than 0.037, hypergeometric test), except for two pairwise comparisons: “lncRNAs upregulated after 5-AzaC versus bF > sF” (p value = 0.119) and “lncRNAs upregulated after 5-AzaC versus sF > bF” (p value = 0.181).
It has been shown that juvenile worms and schistosomula co-express transcripts that cluster into modules midnightblue and magenta, respectively31. These modules are among those with higher lncRNA/total transcripts ratio when compared with all modules: midnightblue is the second and magenta is the fourth module, out of 15 modules, with the highest lncRNAs/total transcript ratios. In midnightblue and magenta modules, lncRNAs correspond to 80% and 64% of all the transcripts, respectively31. As a high proportion of lncRNAs upregulated by 5-AzaC in S. mansoni females belongs to midnightblue and magenta modules (16.7% of all upregulated lncRNAs assigned to any module), we tested if 5-AzaC treatment would have any impact on schistosomula viability. We treated schistosomula with different concentrations of 5-AzaC and measured the viability at each day, along 5 days of treatment. No statistically significant reduction in schistosomula viability as measured by ATP levels was observed after 5-AzaC treatment at any of the concentrations and days tested (Supplementary Fig. S3A), with discrete phenotypic alterations observed only at day 5 post-treatment, at 245 µM, the highest concentration tested (Supplementary Fig. S3B). This is in agreement with observations that S. mansoni schistosomula possess lower detectable levels of 5-methylcytosine and of mRNAs encoding SmDnmt2 and SmMBD proteins involved with DNA methylation15, compared with other S. mansoni life-cycle stages, thus probably making schistosomula less susceptible to 5-AzaC treatment.
LncRNAs differentially expressed upon 5-AzaC treatment have histone marks at their genomic loci
The presence of histone marks at the TSSs of lncRNAs adds another layer of functionality evidence for lncRNAs, indicating regulation by epigenetic mechanisms related to chromatin structure26,31. Therefore, to check if the lncRNAs differentially expressed after 5-AzaC treatment would have histone marks at their TSSs, we cross compared the lncRNAs affected by 5-AzaC treatment with lncRNAs expressed in S. mansoni and reported by Maciel et al.31 as having at least one histone mark obtained by ChIP-Seq (H3K4me3, that is generally associated with active transcription or H3K27me3, associated to transcription repression) in non-treated S. mansoni cercariae, schistosomula or adults. As reported in that work31, 8599 out of 16,583 lncRNAs identified in different S. mansoni life-cycle stages and tissues have at least one histone modification mark within 1 kb from their TSS31. In addition, gene expression control by DNA/RNA methylation, affected by 5-AzaC, has been linked to histone modifications in eukaryotes36,37.
A total of 461 out of 912 lncRNA transcripts differentially expressed after 5-AzaC treatment have at least one histone modification mark within 1 kb from their TSS, being 274 upregulated lncRNAs (Supplementary Table S5) and 187 downregulated lncRNAs (Supplementary Table S6). This represents 50% of all the 912 lncRNAs differentially expressed after 5-AzaC treatment, which is statistically significant (p value < 0.05, hypergeometric test).
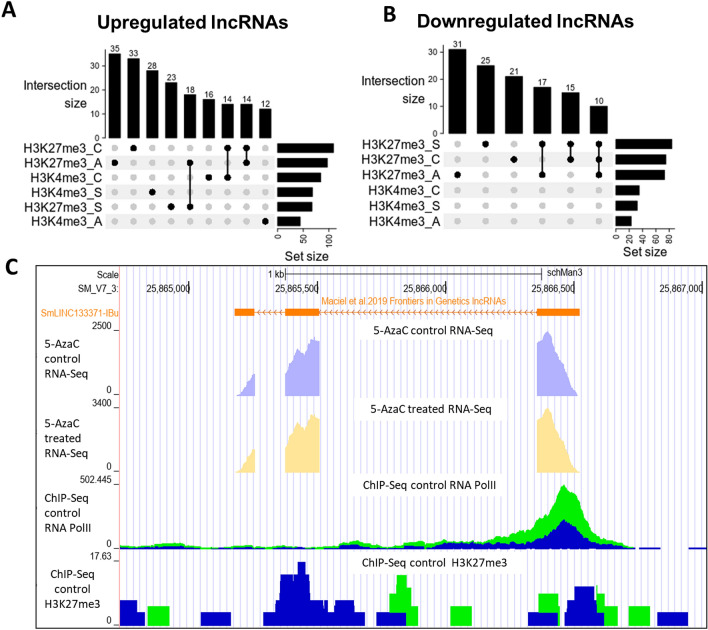
The most abundant mark found individually at the loci of the lncRNAs differentially expressed after 5-AzaC treatment was H3K27me3 in adults, for both upregulated lncRNAs (with 35 marks, Fig. 3A) and downregulated lncRNAs (with 31 marks, Fig. 3B). The second and third most present marks were, among the upregulated lncRNAs, H3K27me3 in cercariae and H3K4me3 in schistosomula, and among the downregulated lncRNAs H3K27me3 in schistosomula and H3K27me3 in cercariae.
Figure 3.
Hundreds of lncRNAs differentially expressed after 5-AzaC exposure in S. mansoni females have histone transcriptional activating or repressive marks at their TSSs. The UpSet intersection diagram shows the number of S. mansoni lncRNAs differentially expressed after 5-AzaC exposure (y-axis) that have been detected in each of the intersection sets, indicated by the connected points in the lower part of the plot, as having the H3K4me3 transcriptional activating marks and/or the H3K27me3 repressive marks within 1 kb (upstream or downstream) from their TSSs. Six histone mark datasets indicated at the bottom left were analyzed: H3K4me3_A in adults, H3K4me3_C in cercariae, H3K4me3_S in schistosomula, H3K27me3_A in adults, H3K27me3_S in schistosomula, and H3K27me3_C in cercariae, and each set size horizontal black bar represents the number of lncRNAs that contain the indicated histone mark at the indicated stage. The top enriched intersection sets are shown for the 5-AzaC upregulated (A) and downregulated (B) lncRNAs; all intersection sets and the lists of lncRNAs in each intersection set are shown in Supplementary Table S5, S6. (C) Snapshot of a S. mansoni genome browser image (www.schistosoma.usp.br), showing a region spanning 3 kb on chromosome 3, where the SmLINC133371-IBu is located. The orange track (top) represents lncRNAs from S. mansoni published by Maciel et al.31. Below the orange lncRNAs track, two other tracks show RNA-Seq data from control (light purple) or 5-AzaC treated S. mansoni females (light yellow). Below, two ChIP-Seq tracks are shown: RNA Polymerase II ChIP-Seq (ChIP-Seq Control RNA Pol II) and H3K27me3 histone mark ChIP-Seq (ChIP-Seq Control H3K27me3). The green and blue colours at the two ChIP-Seq tracks at the bottom represent each of two experimental biological replicates.
In addition, when computed together with other marks, the most abundant mark found in the upregulated lncRNAs upon 5-AzaC treatment (Fig. 3A) was the transcriptional repressive mark, H3K27me3, with 18 lncRNAs presenting this mark in adults and schistosomula and other 14 lncRNAs presenting this mark in adults and cercariae simultaneously. For the downregulated lncRNAs upon 5-AzaC treatment (Fig. 3B), H3K27me3 was also the most abundant mark found when the marks were computed together, with 17 lncRNAs presenting this mark in adults and schistosomula and other 10 lncRNAs presenting this mark in adults, schistosomula and cercariae simultaneously.
In Fig. 3C, we show the locus on chromosome 3 of SmLINC133371-IBu (orange track), a lincRNA that has H3K27me3 histone mark ChIP-Seq peaks (“ChIP-Seq Control H3K27me3” track at the bottom of the image) in adults (blue/green tracks). This lincRNA is upregulated 1.5 × in females after 5-AzaC treatment (yellow track, “5-AzaC treated RNA-Seq”), belongs to the greenyellow module and also has RNA Polymerase II peaks (“ChIP-Seq Control RNAPol II”) at its locus.
Validation of lncRNAs differential expression by RT-qPCR
We designed PCR primer pairs for a selected set of ten genes, including eight lincRNAs and two protein-coding genes, to validate their differential expression after 5-AzaC treatment. First, we treated adult worm couples with 5-AzaC at 491 µM for 48 h, extracted RNA from females and males separately and then performed RT-qPCR. As observed by Geyer et al.15, 5-AzaC was not lethal to adult worms even when they were treated with 5-AzaC at 491 µM, the limit of aqueous solubility. Here, we measured for the first time the amount of ATP in adult worms upon 5-AzaC exposure, as readout for worm viability. 5-AzaC exposure for 48 h did not alter significantly the ATP content of adult worms when compared with the controls (Supplementary Fig. S4A, p = 0.12). In addition, we observed a statistically significant 49% reduction in egg laying by adult worms treated with 5-AzaC at 491 µM (p = 0.02, Supplementary Fig. S4B). Eggs laid by adult worms treated with 5-AzaC show many phenotypic abnormalities, including lack of lateral spine on some eggs and eggs with smaller sizes (Supplementary Fig. S4C).
Quantitative real-time PCR (RT-qPCR) was then employed to validate results obtained by the RNA-Seq analysis. Two protein-coding genes were used as controls: Smp_151640 (Insulin-like growth factor I), which was 14.6 × upregulated in the RNA-Seq after 5-AzaC treatment (Supplementary Fig. S5A, left) and Smp_121390 (Genome polyprotein), which was downregulated 3.6 × in the RNA-Seq data (Supplementary Fig. S5B, left). In the RT-qPCR, both protein-coding genes were validated in females: Smp_151640 was upregulated 12.1 × (Supplementary Fig. S5A, right) and Smp_121390 was downregulated 2.5 × (Supplementary Fig. S5B, right) after 5-AzaC treatment. In addition, we also tested the expression of both Smp_151640 and Smp_121390 after 5-AzaC treatment in S. mansoni males. While Smp_151640 was found to be 51.4 × upregulated (Supplementary Fig. S6A), Smp_121390 was not differentially expressed after 5-AzaC in vitro treatment in males (Supplementary Fig. S6B).
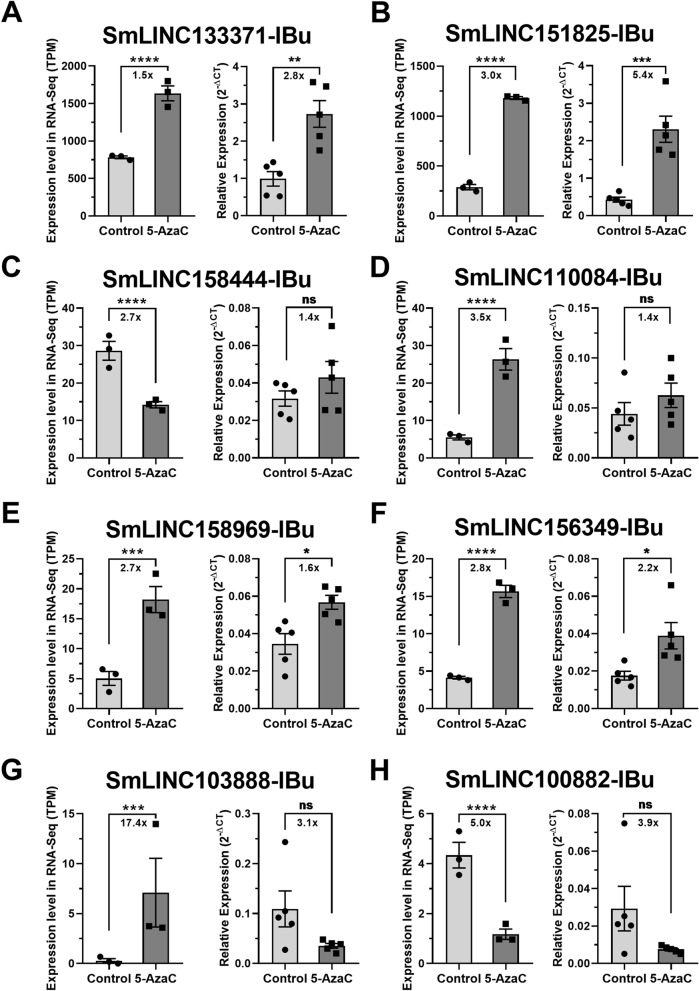
We then tested by RT-qPCR in S. mansoni females and males a selected set of eight lincRNAs found to be differentially expressed in the female RNA-Seq dataset: SmLINC133371-IBu, SmLINC151825-IBu, SmLINC158444-IBu, SmLINC110084-IBu, SmLINC158969-IBu, SmLINC156349-IBu, SmLINC103888-IBu and SmLINC100882-IBu. These lincRNAs were selected because they show a wide range of expression levels in the RNA-Seq (TPM from 4 to 1635 in at least one of the conditions, control or 5-AzaC treated), because they have fold-changes higher than 1.5 × in the RNA-Seq dataset and because they all show only one isoform at their loci, except for SmLINC151825-IBu.
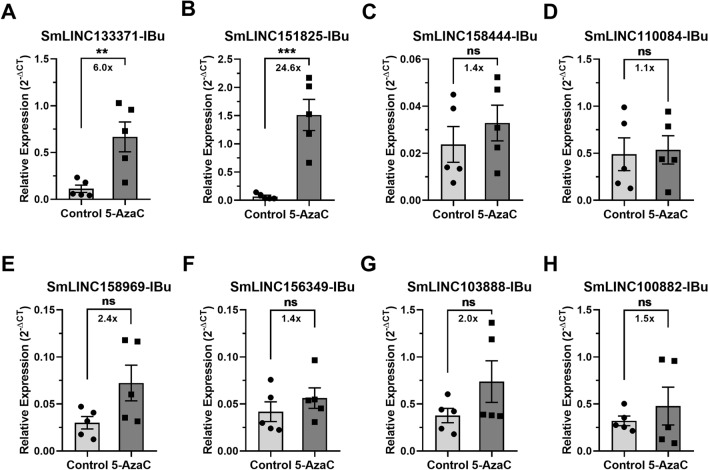
Four of these lincRNAs (SmLINC133371-IBu, SmLINC151825-IBu, SmLINC158969-IBu and SmLINC156349-IBu) were validated by RT-qPCR in females, confirming the RNA-Seq data; in our assays with S. mansoni females, they were upregulated 2.8 ×, 5.4 ×, 1.6 × and 2.2 ×, respectively (Fig. 4A,B,E,F). Additionally, four other lincRNAs tested were detected as expressed in the RT-qPCR assays; however, they were not differentially expressed after 5-AzaC treatment as predicted by the RNA-seq data (Fig. 4C,D,G,H). This indicates that there is variability of lncRNA expression and response to 5-AzaC exposure, probably related to the different parasite strains used in our RT-qPCR assays and in the RNA-Seq experiments from the literature16.
Figure 4.
Expression profiles in S. mansoni females of selected lincRNAs differentially expressed after 5-AzaC treatment (491 µM). Eight lincRNAs were selected after re-analysis of RNA-Seq public datasets of 5-AzaC treated S. mansoni females16 for validation by RT-qPCR in females. For each of the eight selected lincRNAs, the expression profiles obtained with RNA-Seq are shown on the left as TPM (transcripts per million), whereas the RT-qPCR results are shown on the right: (A) SmLINC133371-IBu; (B) SmLINC151825-IBu; (C) SmLINC158444-IBu; (D) SmLINC110084-IBu; (E) SmLINC158969-IBu; (F) SmLINC156349-IBu; (G) SmLINC103888-IBu; (H) SmLINC100882-IBu. For the RNA-Seq data, three biological replicates were analyzed; the fold-changes and p values represented by asterisks that are shown in the brackets were obtained using DESeq2. For the RT-qPCR data, mean ± SEM from five biological replicates are shown, and Student unpaired two-sided t test was applied. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns: not significant.
Considering the six genes in which the effect of 5-AzaC was validated in females by RT-qPCR (four lincRNAs and the two protein-coding genes), the extent of the effect measured by RT-qPCR mirrored the one obtained with RNA-Seq, as fold changes in expression were well correlated (Pearson correlation coefficient = 0.9334, p value = 0.0065, Supplementary Fig. S7).
Interestingly, two out of the eight lincRNAs that were tested (SmLINC133371-IBu and SmLINC151825-IBu) were also upregulated in males treated with 5-AzaC (6.0 × and 24.6 ×, respectively, Fig. 5A,B), indicating that these lincRNAs share similar regulatory mechanisms in both sexes. Expression of the other six tested lincRNAs was not significantly affected by 5-AzaC treatment of males (Fig. 5C–H).
Figure 5.
Expression profiles in S. mansoni males of selected lincRNAs differentially expressed after 5-AzaC treatment (491 µM). Eight lincRNAs were selected after re-analysis of RNA-Seq public datasets of 5-AzaC treated S. mansoni females16 for evaluation of differential expression by RT-qPCR in S. mansoni males. For each of the eight lincRNAs, the expression profiles in controls and in 5-AzaC treated S. mansoni males by RT-qPCR are shown: (A) SmLINC133371-IBu; (B) SmLINC151825-IBu; (C) SmLINC158444-IBu; (D) SmLINC110084-IBu; (E) SmLINC158969-IBu; (F) SmLINC156349-IBu; (G) SmLINC103888-IBu; (H) SmLINC100882-IBu. Mean ± SEM from five biological replicates are shown, and Student unpaired two-sided t test was applied; **p < 0.01, ***p < 0.001; ns not significant.
LncRNAs modulated by 5-AzaC are differentially expressed along S. mansoni life-cycle stages
To evaluate if the lncRNAs differentially expressed after 5-AzaC treatment tested by RT-qPCR here are also expressed in other S. mansoni life-cycle stages or tissues, we re-analyzed data from public RNA-Seq libraries from different S. mansoni life-cycle stages and tissues (Supplementary Table S7) to look for the expression patterns of the eight selected lincRNAs. First, we evaluated the stage-specificity of the different RNA-Seq datasets that we used for this re-analysis by confirming that five protein-coding genes previously described as stage markers38,39 were indeed more highly expressed at the predicted stages in our analysis (Supplementary Fig. S8). In addition, PCA analysis (Supplementary Fig. S9) shows that biological replicates of the same sample grouped according to the life-cycle stages and tissues, confirming the clustering of samples in expected segregating groups.
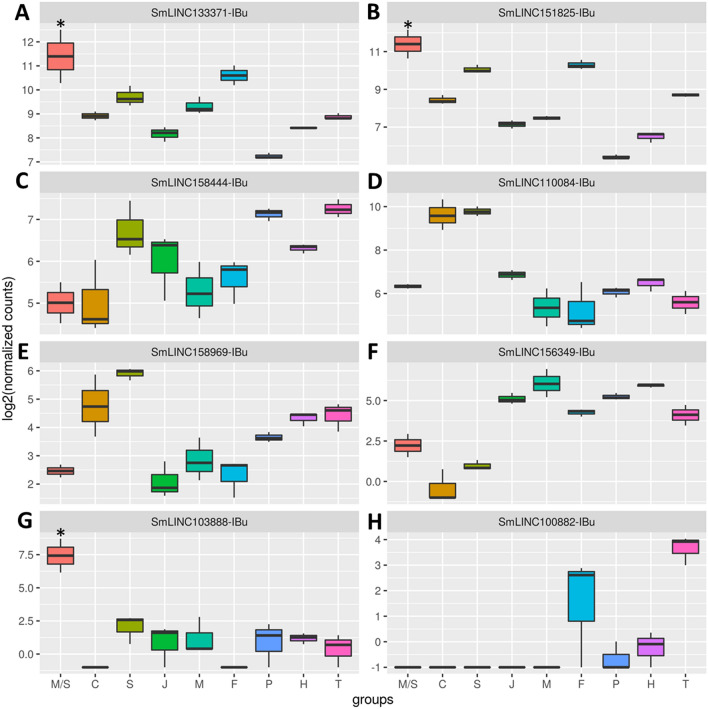
We then looked at the expression levels along S. mansoni life-cycle stages and tissues of the eight lincRNAs tested by RT-qPCR and observed a heterogeneous expression pattern distribution (Fig. 6). Expression of SmLINC133371-IBu (Fig. 6A), SmLINC151825-IBu (Fig. 6B) and SmLINC103888-IBu (Fig. 6G) is higher in miracidia and sporocysts stages, with SmLINC133371-IBu and SmLINC151825-IBu being also highly expressed in adult females and schistosomula (Fig. 6A,B). Whereas SmLIN158444-IBu shows higher expression in the posterior adult somatic tissues and tails (Fig. 6C), SmLINC110084-IBu has higher expression in schistosomula and cercariae (Fig. 6D). While SmLINC156349-IBu (Fig. 6E) and SmLINC158969-IBu (Fig. 6F) show broad expression in all the stages, SmLINC100882-IBu (Fig. 6H) is highly expressed in female adult worms and tails. These results show that most of the tested lncRNAs (except SmLINC103888-IBu and SmLINC100882-IBu) are not stage-specific and may play roles in other S. mansoni life-cycle stages.
Figure 6.
RNA-seq expression profiles at different S. mansoni stages of selected lincRNAs differentially expressed after 5-AzaC treatment (491 µM). The expression levels (shown as log2 of normalized counts) of the eight lincRNAs whose gene IDs are indicated at the top of each panel are shown. These lincRNAs were selected after re-analysis of RNA-Seq public datasets of 5-AzaC treated S. mansoni females16. The y-axis shows the expression level for each lincRNA in the RNA-seq assays (log2 of normalized counts) as determined at the stage indicated in the x-axis as follows: miracidia/sporocysts (M/S), cercariae (C), schistosomula (S), juveniles (J), adult males (M), adult females (F), posterior somatic tissues (P), heads (H) and tails (T). (A) SmLINC133371-IBu; (B) SmLINC151825-IBu; (C) SmLINC158444-IBu; (D) SmLINC110084-IBu; (E) SmLINC158969-IBu; (F) SmLINC156349-IBu; (G) SmLINC103888-IBu; (H) SmLINC100882-IBu. Only transcripts that were upregulated in one stage/tissue when compared with all others were considered as significantly more expressed in that stage/tissue and are marked with an asterisk. *p value < 0.05.
Discussion
Here, we have shown that long non-coding RNAs levels can be modulated in S. mansoni by 5-AzaC, a DNA methyltransferase inhibitor that is currently used to treat myelodysplastic syndrome and acute myeloid leukemia in humans12,40. Hundreds of the lncRNAs differentially expressed after 5-AzaC exposure in S. mansoni females belong to co-expression modules related to male metabolism, have histone marks at their genomic loci and are also differentially expressed in unpaired compared with paired S. mansoni females and ovaries. While short RNAs (especially miRNAs) have been more explored in various helminths41–44, lncRNAs have received little attention, being identified by transcriptomic approaches only in a few helminths other than S. mansoni22–24 or studied in a limited number of free-living nematodes45,46. In addition, unlike miRNAs47–52, the mechanisms of regulation of lncRNAs are largely unknown in parasites and, to our knowledge, this is the first report of modulation of lncRNAs levels by an epigenetic drug in any helminth.
In the past few years, human lncRNAs have been proposed as drug targets in many diseases, especially in cancer and neurological syndromes21,53–55. In parasitic diseases, there is a clear need to develop new and inexpensive drugs, especially with the emerging reports of drug resistance6,56–58. We believe that it is time to consider lncRNAs as possible drug targets also in parasitic diseases, especially because they show lower conservation in their primary sequences between species than protein-coding genes17,19,59,60, which in principle would reduce side effects in therapeutic strategies.
The choice of lncRNAs to be further validated as drug targets will rest on the appropriate selection of lncRNA candidates. This selection should be guided by functional characterization of the lncRNA as well as by the demonstration of the lncRNA relevance to the parasite biology. Here, we found that 38% of the lncRNAs differentially expressed after 5-AzaC treatment in S. mansoni females (343 out of 912 lncRNAs) are also differentially expressed between paired and unpaired females or ovaries35, whilst 24% of them (221 out of 912 lncRNAs) belong to co-expression modules related to “gonads”31, indicating an important involvement of lncRNAs on parasite sexual maturation and reproductive biology. In addition, 50% of the lncRNAs differentially expressed after 5-AzaC treatment in S. mansoni females (461 out of 912 lncRNAs) have at least one histone mark at their TSSs previously detected at S. mansoni life-cycle stages31. These lncRNAs with evidence of chromatin marks at their genomic loci could be prioritized in further functional assays to elucidate their relevance, roles and mechanisms of action in S. mansoni biology. Technologies for lncRNAs targeting should be considered in these studies, including cell and tissue localization, silencing by CRISPR or antisense oligonucleotides methods in vitro and in vivo, and discovery of lncRNA partners (DNA, RNA or proteins)61–63.
The expression patterns along life-cycle stages may also be criteria for the selection of lncRNAs to be tested in functional assays. Some of the lncRNAs tested by RT-qPCR here in S. mansoni female and male adult worms are also expressed at high levels (TPM > 100) in other life-cycle stages, including SmLINC133371-IBu and SmLINC151825-IBu with high expression levels in miracidia, sporocysts and schistosomula. In addition, SmLINC158444-IBu and SmLINC110084-IBu are highly expressed in posterior somatic tissues and schistosomula, respectively. All these lncRNAs, except SmLINC100882-IBu are expressed in schistosomula, another life-cycle stage of interest regarding drug targeting, as praziquantel has no efficacy against schistosomula64.
Additionally, many lncRNAs have been associated with drug resistance in human cancers65–67. Here, by measuring ATP levels, we confirm that 5-AzaC treatment has no effect on the viability of S. mansoni adult worms, as previously shown15,16. Moreover, we show that S. mansoni schistosomula viability is also not affect by 5-AzaC. It is unclear why the parasites’ viability is not affected by 5-AzaC, but since schistosomes show nucleoside auxotrophy68, precise regulation of nucleoside analogs uptake may control their toxicity. It is also possible that the lncRNAs differentially expressed after 5-AzaC exposure may be involved in a 5-AzaC drug resistance mechanism, as shown for human cancer-related lncRNAs such as HOTAIR and XIST69–71.
Understanding the mechanisms of lncRNA expression regulation may help the selection of lncRNAs for the development of new therapeutic strategies in the future. These mechanisms, which include epigenetic regulation by histone modification72–74 at lncRNA genomic loci and DNA/RNA methylation already described in human lncRNAs75–78 are, however, less understood than those of protein-coding genes79,80. It is now clear that epigenetic processes play important roles on schistosomes81–83. In fact, epigenetic mechanisms participate in schistosome phenotypic plasticity84,85, in egg production and adult worm viability34,86,87 as well as in schistosomula survival88–92. DNA methylation, one of the most studied epigenetic mechanisms, has been detected in S. mansoni15, although the significance of DNA cytosine methylation (5mC) in this parasite has been somewhat controversial93–95. Here, we observed that 912 lncRNAs are differentially expressed after 5-AzaC exposure in S. mansoni females, all of them expressed at an average TPM > 0.1 in control or 5-AzaC treated samples. As previous analysis identified 9229 lncRNAs expressed in females (TPM > 0.1) out of all 16,583 detected at any S. mansoni life-cycle stage31, we estimate that 10% of all lncRNAs expressed in females are differentially expressed upon 5-AzaC exposure.
Although the presence of DNA methylation in many invertebrates has been already reported96,97, previous work was unable to detect functional roles of DNA methylation in invertebrates98. 5-AzaC is an inhibitor of DNA methyltransferase that has been shown in S. mansoni to inhibit female specific biological processes including egg production, egg maturation and normal ovarian development15,16, phenotypic effects confirmed in our treatments. These effects are achieved by modifications of adult female transcription and translation, with 81% inhibition in de novo protein synthesis in female schistosomes16. As 5-AzaC incorporates preferentially into RNAs, with only 20% being incorporated into DNA99, it is more likely that 5-AzaC interferes preferentially in lncRNA stability through lncRNA methylation impediment than through promoter DNA methylation. In fact, many lncRNAs were shown to be regulated by RNA methylation in humans and in Arabidopsis100–102, although some human lncRNAs have been also identified as regulated by DNA methylation at their promoter regions32,33,103.
5-AzaC also affects S. mansoni females’ stem cells, leading to a 95% reduction in the number of proliferating stem cells16. Remarkably, lncRNAs actively participate in human stem cell pluripotency, maintenance and differentiation104. Thus, it is possible that some of the lncRNAs found here as modulated by 5-AzaC play important roles in parasite stem cells. Further analyses of lncRNA expression in spatially-distinct S. mansoni female stem cell populations under 5-AzaC exposure, including vitelline S1 stem cells which are vital for egg production, may uncover lncRNA functional roles on stem cell maintenance. Alternatively, 5-AzaC may modulate lncRNA expression levels by exerting pleiotropic effects similar to those reported in human cell lines such as suppressing lipid metabolism105, inhibition of pathways that regulate DNA synthesis/repair106 or de-repression of retroviral expression107. Future studies aiming to elucidate the precise mechanism of action of 5-AzaC in lncRNA regulation108,109 could offer starting points for lncRNA targeting and manipulation in S. mansoni.
In summary, this study adds another layer on the understanding of the effects of 5-AzaC in S. mansoni and sheds light on the relevance of looking at lncRNA regulation in response to drug treatment in parasites. Although the use of 5-AzaC against schistosomiasis is unlikely as its effects are not parasite selective, the lncRNAs affected by 5-AzaC identified here, together with downstream pathways already described as affected by 5-AzaC, could represent new targets for the development of alternative chemotherapeutic strategies against schistosomiasis.
Material and methods
Analysis of 5-AzaC RNA-Seq data
Public RNA-Seq data from Geyer et al.16 for S. mansoni females were downloaded from the SRA-NCBI database (project number PRJNA428470; controls #SRR6490481, #SRR6490482 and #SRR6490483; treated with 491 µM 5-AzaC #SRR6490480, #SRR6490484 and #SRR6490485). Adapters and bad quality reads were filtered out using fastp v. 0.19.5 with default parameters110. For transcripts expression quantitation the genome sequence v.7, and a GTF file containing the protein-coding transcriptome v 7.1 were downloaded from the WormBase ParaSite resource (version WBPS14)111. The latter was merged with the lncRNA transcriptome sequences identified by Maciel et al.31 and the resulting GTF, which is available at http://schistosoma.usp.br/, was used as the reference. The filtered RNA-Seq reads were aligned with STAR v 2.7112 and quantified with RSEM v 1.3.1113, both using default parameters, and with the RSEM “estimate-rspd parameter on” option. Transcripts with counts lower than 10 were removed and differential expression analysis was performed using DESeq2 package114 v. 1.24.0 with an FDR threshold of 0.05. The Volcano plot shows the − log10(p value) vs log2(fold-change) for the lncRNAs obtained in the DESeq2 analysis, using EnhancedVolcano (R package version 1.6.0), available at https://github.com/kevinblighe/EnhancedVolcano. To look for the expression patterns of lincRNAs at different S. mansoni life-cycle stages and tissues we re-analyzed data from the public RNA-Seq libraries indicated in Supplementary Table S7, using the same pipeline described above. Pairwise differential expression analysis was performed between each two stages and/or tissues using DESeq2 with an FDR threshold of 0.05. Only transcripts that were upregulated in one stage/tissue when compared with all others, were considered as significantly more expressed in that stage/tissue.
PCA plot was obtained after normalization using the vst function followed by the plotPCA function from DESeq2.
Analysis of the features of lncRNAs differentially expressed after 5-AzaC treatment
The lncRNAs differentially expressed in S. mansoni females after 5-AzaC treatment were compared with the lists of lncRNAs identified in four different datasets, namely: (1) lncRNAs belonging to one of the 15 weighted gene co-expression network analyses (WGCNA) modules previously published by Maciel et al.31 to check to which modules the lncRNAs differentially expressed after 5-AzaC treatment belong to. In Maciel et al.31, 90 libraries from S. mansoni miracidia, sporocysts, schistosomula, cercariae, and gonads (testes and ovaries) were analyzed using the unsupervised WGCNA115 co-expression analysis approach and 15 different WGCNA modules were obtained31, each of them representing one cluster of highly interconnected genes that are more expressed in a given stage/tissue. That analysis31 resulted in the identification of two modules representing miracidia/sporocysts (black and purple), two modules representing juveniles (blue and midnight blue), two modules representing adult males (turquoise and yellow) and four modules representing gonads (brown, green, greenyellow and salmon). Regarding the other five modules, each of them represents only one stage/tissue: cyan (ovaries), magenta (schistosomula), pink (adult females), red (testes) and tan (cercariae); (2) lncRNAs differentially expressed between bisex females (paired, bF) and single-sex females (unpaired, sF) and between bisex ovaries (paired, bO) and single-sex ovaries (unpaired, sO) and whole worms, which we determined by a re-analysis of the transcriptomes previously obtained by Lu et al.35 (see below); (3) lncRNAs previously published by Maciel et al.31 as having at least one histone mark (H3K4me3 or H3K27me3) at their TSSs, to check for the presence of histone marks at the TSS of lncRNAs differentially expressed after 5-AzaC treatment; (4) the expression patterns of lncRNAs along S. mansoni life-cycle stages, previously published by Maciel et al.31.
Data from Lu et al.35 were obtained from SRA (Project number PRJEB14695). In that work, Lu et al.35 performed RNA-Seq in bisex (paired) females (bF), single-sex females (sF), bisex (paired) males (bM), single-sex males (sM), bisex ovaries (bO), single-sex ovaries (sO), bisex testes (bT) and single-sex testes (sT), but only the protein-coding genes were analyzed. Here, a re-analysis of Lu et al.35 raw data to detect lncRNAs was performed using the same genome, annotation files and bioinformatics tools and parameters that were used to analyze the data from Geyer et al.16, as described above. The Venn diagram tool at http://jvenn.toulouse.inra.fr/app/index.html was used to compare the lists of lncRNAs detected as differentially expressed in the present study and in the gonad-specific and pairing-dependent study35.
Parasite materials
All parasite materials were from a BH isolate of S. mansoni maintained by passage through golden hamster (Mesocricetus auratus) and Biomphalaria glabrata snails. Cercariae were collected from snails infected with 10 miracidia each. Thirty-five days after infection, the snails were placed in the dark in water and then illuminated for 2 h to induce shedding. Schistosomula were obtained by mechanical transformation of cercariae and separation of their bodies as previously described116, with some modifications. Briefly, cercariae were collected as described above and then suspended in 15 ml of M169 medium (Vitrocell, cat number 00464) containing penicillin/streptomycin, amphotericin (Vitrocell, cat number 00148). Mechanical transformation was performed by passing the cercariae 10 times through a 23G needle. To separate schistosomula from the tails, the tail-rich supernatant was decanted and the sedimented bodies resuspended in a further 7 ml of M169 medium. The procedure was repeated until less than 1% of the tails remained. The newly transformed schistosomula were maintained for 72 h in M169 medium (Vitrocell, cat number 00464) supplemented with penicillin/streptomycin, amphotericin, gentamicin (Vitrocell, cat number 00148), 2% fetal bovine serum, 1 μM serotonin, 0.5 μM hypoxanthine, 1 μM hydrocortisone, and 0.2 μM triiodothyronine at 37 °C and 5% CO2. Schistosomula cultivated for 72 h were used for 5-AzaC exposure. Adult S. mansoni worms were recovered by perfusion of golden hamsters that had been infected 7 weeks before with 250 cercariae, as previously described116,117. After perfusion, the adult worm pairs were kept for 3 h at 37 °C and 5% CO2 in DMEM (Gibco, catalogue number 11995-065-500) supplemented with 10% fetal bovine serum (FBS) (Vitrocell) and 100 mg/ml penicillin/streptomycin (Vitrocell). After 3 h of incubation, the adult worm pairs were used for 5-AzaC treatment.
Parasite treatment with 5-AzaC
Schistosoma mansoni schistosomula and adult worms were treated with different final concentrations of 5-AzaC (Sigma, A2385) in culture medium specific to each stage: adult worms were treated with 5-AzaC at 491 μM (same treatment as in Geyer et al.16) and schistosomula were treated with 5-AzaC from 245 to 7.7 μM, as indicated in the Results. Adult male and female schistosome couples were cultivated in the presence (or absence) of 5-AzaC according to the methodology described in Geyer et al.15. 5-AzaC was added to 30 worm pairs for each of five biological replicates, while additional five replicates, lacking 5-AzaC, were included as controls. The schistosome cultures were incubated at 37 °C for 48 h in a humidified atmosphere with a 70% media exchange performed after 24 h. After 48 h, eggs were counted and schistosome worms were collected, washed three times with PBS and stored in RNAlater (Ambion) until RNA extraction. Before the extraction of RNA from males or females, adult worm pairs were manually separated in RNAlater (Ambion) using tweezers. Adult worm couple viability was evaluated 2 days post culture initiation using 9 worm pairs that were cultivated in 5 ml medium in 6 well tissue culture plates (n = 5 biological replicates; 5-AzaC at 491 μM treated or control worms).
Newly transformed schistosomula (NTS) were maintained in culture34 for 72 h and then treatment with 5-AzaC was initiated. Schistosomula viability was measured after 24, 48, 72, 96 and 120 h of treatment (n = 2 biological replicates).
Viability assay
The viability of S. mansoni schistosomula and adult worms after treatment with 5-AzaC was determined by a cytotoxicity assay based on the CellTiter-Glo Luminescent Cell Viability Assay (G7570, Promega)34,118. The assay determines the amount of ATP present in freshly lysed adults or in intact schistosomula; the assay signals the presence of metabolically active cells.
RNA extraction, quantification, and quality assessment
RNA extraction, quantification, and quality assessment were performed according to Maciel et al.31. Male or female adult worms were first disrupted in Qiagen RLT buffer using glass potters and pestles. RNAs from males or females were then extracted and purified using the Qiagen RNeasy Mini Kit (Cat number 74104), according to the manufacturer’s instructions, except for the DNase I treatment: the amount of DNase I was doubled, and the time of treatment was increased to 45 min.
The integrity of all RNAs was verified using the Agilent RNA 6000 Pico Kit (5067-1513 Agilent Technologies) in a 2100 Bioanalyzer Instrument (Agilent Technologies) and quantified using the Qubit RNA HS Assay Kit (Q32852, Thermo Fisher Scientifc). Purity was assessed by 260/280 nm and 260/230 nm ratios using Nanodrop (Thermo Fisher Scientific). Five biological replicates were assessed for 5-AzaC treated or control males or females.
Reverse transcription and quantitative PCR (RT-qPCR) assays
The reverse transcription (RT) reactions were performed with 200 ng total RNA of each control and 5-AzaC treated female samples and with 30 ng total RNA of each control and 5-AzaC treated male samples. For the RT reactions, the SuperScript IV FirstStrand Synthesis System (18091050; Life Technologies) and random hexamer primers were used in a 20 μl final volume. The obtained complementary DNAs (cDNAs) were diluted four times in DEPC water, and quantitative PCR was performed using 2.5 μl of each diluted cDNA in a total volume of 10 μl containing 1 × LightCycler 480 SYBR Green I Master Mix (04707516001, Roche Diagnostics) and 800 nM of each primer in a LightCycler 480 System (Roche Diagnostics). Primers for selected transcripts (Supplementary Table S8) were designed using the Primer 3 online tool, and each RT-qPCR was run in three technical replicates. The results were analyzed by comparative Ct method119. Ct values are shown in Supplementary Table S9. Real-time qPCR data were normalized in relation to the level of expression of two reference genes previously used in the literature, namely Smp_900000120–122 and Smp_123610117.
Statistical analyses
Two-tailed unpaired t test was used for pairwise comparisons, and GraphPad Prism software was used to perform the analyses (version 8.0). Hypergeometric test was used for enrichment calculations, using the online https://stattrek.com/online-calculator/hypergeometric.aspx tool. Quantification of data are represented as mean ± SEM and p value thresholds were * < 0.05, ** < 0.01, *** < 0.001 and **** < 0.0001.
Ethics statement
The experimental protocols were in accordance with the Ethical Principles in Animal Research adopted by the Brazilian College of Animal Experimentation (COBEA) and the protocol/experiments have been approved by the Ethics Committee for Animal Experimentation of Instituto Butantan (CEUAIB n˚ 1777050816).
Supplementary Information
Acknowledgements
We acknowledge Dr. David da Silva Pires and Dr. Ana Carolina Tahira for help in the generation of high-throughput data genome tracks.
Author contributions
M.S.A. and S.V.-A. conceived the project, analyzed the data and wrote the manuscript. M.S.A. carried out all experiments. M.S.A. and L.F.M. performed bioinformatics analysis. M.S.A., G.O.S., G.G.O.O. and J.V.P.L. designed and carried out RT-qPCR experiments. M.S.A. and L.K.I. performed schistosomula in vitro treatment experiments. A.S.A.P. gave support on hamster perfusion, adult worm recovery and separation of adult male from female worms after treatments. P.A.M. and E.N. provided biological material. All authors reviewed the manuscript and approved the submitted version.
Funding
This work was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (Thematic grant number 2018/23693-5 to SV-A). G.O.S., LFM and A.S.A.P. received fellowships from FAPESP (18/24015-0, 18/19591-2 and 2016/10046-6, respectively); G.G.O.O. received a fellowship from CNPq (116733/2019-5); S.V-A laboratory was also supported by institutional funds from Fundação Butantan and S.V-A received an established investigator fellowship award from CNPq, Brasil.
Data availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-78669-5.
References
- 1.WHO. Global Health Estimates 2016: Disease Burden by Cause, Age, Sex, by Country and by Region, 2000–2016. Geneva, World Health Organization.2016. https://www.who.int/healthinfo/global_burden_disease/estimates/en/index1.html Accessed Jan 2020. (2016).
- 2.Mutapi F, Maizels R, Fenwick A, Woolhouse M. Human schistosomiasis in the post mass drug administration era. Lancet Infect. Dis. 2017;17:e42–e48. doi: 10.1016/S1473-3099(16)30475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RA. Schistosomiasis then and now: What has changed in the last 100 years? Parasitology. 2020;147:507–515. doi: 10.1017/S0031182020000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383:2253–2264. doi: 10.1016/S0140-6736(13)61949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoni AC, Catala L, Ault SK. Schistosomiasis prevalence and intensity of infection in Latin America and the Caribbean Countries, 1942–2014: A systematic review in the context of a regional elimination goal. PLoS Negl. Trop. Dis. 2016;10:e0004493. doi: 10.1371/journal.pntd.0004493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergquist R, Utzinger J, Keiser J. Controlling schistosomiasis with praziquantel: How much longer without a viable alternative? Infect. Dis. Poverty. 2017;6:74. doi: 10.1186/s40249-017-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gryseels B, et al. Epidemiology, immunology and chemotherapy of Schistosoma mansoni infections in a recently exposed community in Senegal. Trop. Geogr. Med. 1994;46:209–219. [PubMed] [Google Scholar]
- 8.Melman SD, et al. Reduced susceptibility to praziquantel among naturally occurring Kenyan isolates of Schistosoma mansoni. PLoS Negl. Trop. Dis. 2009;3:e504. doi: 10.1371/journal.pntd.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vale N, et al. Praziquantel for schistosomiasis: Single-drug metabolism revisited, mode of action, and resistance. Antimicrob. Agents Chemother. 2017;61:e02582-16. doi: 10.1128/AAC.02582-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewitson JP, Maizels RM. Vaccination against helminth parasite infections. Expert Rev. Vaccines. 2014;13:473–487. doi: 10.1586/14760584.2014.893195. [DOI] [PubMed] [Google Scholar]
- 11.Gouveia MJ, Brindley PJ, Gartner F, Costa J, Vale N. Drug repurposing for schistosomiasis: Combinations of drugs or biomolecules. Pharmaceuticals (Basel) 2018;11:15. doi: 10.3390/ph11010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cataldo VD, Cortes J, Quintas-Cardama A. Azacitidine for the treatment of myelodysplastic syndrome. Expert Rev. Anticancer Ther. 2009;9:875–884. doi: 10.1586/era.09.61. [DOI] [PubMed] [Google Scholar]
- 13.Lu LJ, Randerath K. Mechanism of 5-azacytidine-induced transfer RNA cytosine-5-methyltransferase deficiency. Cancer Res. 1980;40:2701–2705. [PubMed] [Google Scholar]
- 14.Reichman M, Penman S. The mechanism of inhibition of protein synthesis by 5-azacytidine in HeLa cells. Biochim. Biophys. Acta. 1973;324:282–289. doi: 10.1016/0005-2787(73)90145-7. [DOI] [PubMed] [Google Scholar]
- 15.Geyer KK, et al. Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat. Commun. 2011;2:424. doi: 10.1038/ncomms1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyer KK, et al. The anti-fecundity effect of 5-azacytidine (5-AzaC) on Schistosoma mansoni is linked to dis-regulated transcription, translation and stem cell activities. Int. J. Parasitol .Drugs Drug Resist. 2018;8:213–222. doi: 10.1016/j.ijpddr.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 2020;21:102–117. doi: 10.1038/s41576-019-0184-5. [DOI] [PubMed] [Google Scholar]
- 21.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos LN, et al. De novo assembly and characterization of the Trichuris trichiura adult worm transcriptome using Ion Torrent sequencing. Acta Trop. 2016;159:132–141. doi: 10.1016/j.actatropica.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Azlan A, Halim MA, Azzam G. Genome-wide identification and characterization of long intergenic noncoding RNAs in the regenerative flatworm Macrostomum lignano. Genomics. 2020;112:1273–1281. doi: 10.1016/j.ygeno.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Ross E, Blair D, Guerrero-Hernandez C, SanchezAlvarado A. Comparative and transcriptome analyses uncover key aspects of coding- and long noncoding RNAs in flatworm mitochondrial genomes. G3 (Bethesda) 2016;6:1191–1200. doi: 10.1534/g3.116.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira KC, Carvalho ML, Maracaja-Coutinho V, Kitajima JP, Verjovski-Almeida S. Non-coding RNAs in schistosomes: An unexplored world. An. Acad. Bras. Cie. 2011;83:673–694. doi: 10.1590/s0001-37652011000200026. [DOI] [PubMed] [Google Scholar]
- 26.Vasconcelos EJR, et al. The Schistosoma mansoni genome encodes thousands of long non-coding RNAs predicted to be functional at different parasite life-cycle stages. Sci. Rep. 2017;7:10508. doi: 10.1038/s41598-017-10853-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Q, et al. Identification of long noncoding RNAs in Schistosoma mansoni and Schistosoma japonicum. Exp. Parasitol. 2018;191:82–87. doi: 10.1016/j.exppara.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira VF, et al. Identification of 170 new long noncoding RNAs in Schistosoma mansoni. Biomed. Res. Int. 2018;2018:1264697. doi: 10.1155/2018/1264697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasconcelos EJR, et al. Atlas of Schistosoma mansoni long non-coding RNAs and their expression correlation to protein-coding genes. Database (Oxford) 2018;2018:bay068. doi: 10.1093/database/bay068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HC, Khalil AM, Jolly ER. LncRNAs in molluscan and mammalian stages of parasitic schistosomes are developmentally-regulated and coordinately expressed with protein-coding genes. RNA Biol. 2020;17:805–815. doi: 10.1080/15476286.2020.1729594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maciel LF, et al. Weighted gene co-expression analyses point to long non-coding RNA hub genes at different Schistosoma mansoni life-cycle stages. Front Genet. 2019;10:823. doi: 10.3389/fgene.2019.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumegawa K, et al. A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci. Rep. 2016;6:26699. doi: 10.1038/srep26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diaz-Lagares A, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA. 2016;113:E7535–E7544. doi: 10.1073/pnas.1608585113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira ASA, et al. Inhibition of histone methyltransferase EZH2 in Schistosoma mansoni in vitro by GSK343 reduces egg laying and decreases the expression of genes implicated in DNA replication and noncoding RNA metabolism. PLoS. Negl. Trop. Dis. 2018;12:e0006873. doi: 10.1371/journal.pntd.0006873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Z, et al. Schistosome sex matters: A deep view into gonad-specific and pairing-dependent transcriptomes reveals a complex gender interplay. Sci. Rep. 2016;6:31150. doi: 10.1038/srep31150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeltsch A, Broche J, Bashtrykov P. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes (Basel) 2018;9:566. doi: 10.3390/genes9110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker-Manuel SJ, Ivens AC, Dillon GP, Wilson RA. Gene expression patterns in larval Schistosoma mansoni associated with infection of the mammalian host. PLoS Negl. Trop. Dis. 2011;5:e1274. doi: 10.1371/journal.pntd.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson L, et al. Schistosoma mansoni egg, adult male and female comparative gene expression analysis and identification of novel genes by RNA-Seq. PLoS Negl. Trop. Dis. 2015;9:e0004334. doi: 10.1371/journal.pntd.0004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan D, Rampal R, Mascarenhas J. Clinical developments in epigenetic-directed therapies in acute myeloid leukemia. Blood Adv. 2020;4:970–982. doi: 10.1182/bloodadvances.2019001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao CC, et al. Comparative analysis of microRNA profiles between adult Ascaris lumbricoides and Ascaris suum. BMC Vet. Res. 2014;10:99. doi: 10.1186/1746-6148-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fontenla S, Rinaldi G, Smircich P, Tort JF. Conservation and diversification of small RNA pathways within flatworms. BMC Evol. Biol. 2017;17:215. doi: 10.1186/s12862-017-1061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macchiaroli N, et al. Identification and expression profiling of microRNAs in Hymenolepis. Int. J. Parasitol. 2019;49:211–223. doi: 10.1016/j.ijpara.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Holz A, Streit A. Gain and loss of small RNA classes-characterization of small RNAs in the parasitic nematode family strongyloididae. Genome Biol. Evol. 2017;9:2826–2843. doi: 10.1093/gbe/evx197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodelsperger C, Menden K, Serobyan V, Witte H, Baskaran P. First insights into the nature and evolution of antisense transcription in nematodes. BMC Evol. Biol. 2016;16:165. doi: 10.1186/s12862-016-0740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei S, et al. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol. 2019;20:7. doi: 10.1186/s13059-018-1619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y, Cai X, Bradley JE. microRNAs in parasites and parasite infection. RNA Biol. 2013;10:371–379. doi: 10.4161/rna.23716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Britton C, Winter AD, Gillan V, Devaney E. microRNAs of parasitic helminths—identification, characterization and potential as drug targets. Int. J. Parasitol. Drugs Drug Resist. 2014;4:85–94. doi: 10.1016/j.ijpddr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tritten L, et al. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl. Trop. Dis. 2014;8:e2971. doi: 10.1371/journal.pntd.0002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks ND, et al. Profiling microRNAs through development of the parasitic nematode Haemonchus identifies nematode-specific miRNAs that suppress larval development. Sci. Rep. 2019;9:17594. doi: 10.1038/s41598-019-54154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meningher T, et al. Schistosomal extracellular vesicle-enclosed miRNAs modulate host T helper cell differentiation. EMBO Rep. 2020;21:e47882. doi: 10.15252/embr.201947882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, et al. Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PLoS Pathog. 2019;15:e1007817. doi: 10.1371/journal.ppat.1007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, et al. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat. Commun. 2018;9:3192. doi: 10.1038/s41467-018-05495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prabhakar B, Zhong XB, Rasmussen TP. Exploiting long noncoding RNAs as pharmacological targets to modulate epigenetic diseases. Yale J. Biol. Med. 2017;90:73–86. [PMC free article] [PubMed] [Google Scholar]
- 55.Blokhin I, Khorkova O, Hsiao J, Wahlestedt C. Developments in lncRNA drug discovery: Where are we heading? Expert Opin. Drug Discov. 2018;13:837–849. doi: 10.1080/17460441.2018.1501024. [DOI] [PubMed] [Google Scholar]
- 56.Geary TG, Sakanari JA, Caffrey CR. Anthelmintic drug discovery: Into the future. J. Parasitol. 2015;101:125–133. doi: 10.1645/14-703.1. [DOI] [PubMed] [Google Scholar]
- 57.Fairweather I, Brennan GP, Hanna REB, Robinson MW, Skuce PJ. Drug resistance in liver flukes. Int. J. Parasitol. Drugs Drug Resist. 2020;12:39–59. doi: 10.1016/j.ijpddr.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Partridge FA, et al. Anthelmintic drug discovery: Target identification, screening methods and the role of open science. Beilstein J. Org. Chem. 2020;16:1203–1224. doi: 10.3762/bjoc.16.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warner KD, Hajdin CE, Weeks KM. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018;17:547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu SJ, Lim DA. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018;19:e46955. doi: 10.15252/embr.201846955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 2018;24:257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greenberg RM. New approaches for understanding mechanisms of drug resistance in schistosomes. Parasitology. 2013;140:1534–1546. doi: 10.1017/S0031182013000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai E, et al. ncDR: A comprehensive resource of non-coding RNAs involved in drug resistance. Bioinformatics. 2017;33:4010–4011. doi: 10.1093/bioinformatics/btx523. [DOI] [PubMed] [Google Scholar]
- 66.Merry CR, et al. Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer. Oncotarget. 2016;7:53230–53244. doi: 10.18632/oncotarget.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang WT, Han C, Sun YM, Chen TQ, Chen YQ. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019;12:55. doi: 10.1186/s13045-019-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levy MG, Read CP. Purine and pyrimidine transport in Schistosoma mansoni. J. Parasitol. 1975;61:627–632. doi: 10.2307/3279455. [DOI] [PubMed] [Google Scholar]
- 69.Xue X, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, et al. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology. 2017;22:208–216. doi: 10.1080/10245332.2016.1258152. [DOI] [PubMed] [Google Scholar]
- 71.Zhu J, et al. Knockdown of long non-coding RNA XIST inhibited doxorubicin resistance in colorectal cancer by upregulation of miR-124 and downregulation of SGK1. Cell Physiol. Biochem. 2018;51:113–128. doi: 10.1159/000495168. [DOI] [PubMed] [Google Scholar]
- 72.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu SC, Kallin EM, Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010;20:1109–1116. doi: 10.1038/cr.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amin V, et al. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat. Commun. 2015;6:6370. doi: 10.1038/ncomms7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Z, et al. DNA methylation and gene expression profiles characterize epigenetic regulation of lncRNAs in colon adenocarcinoma. J. Cell Biochem. 2020;121:2406–2415. doi: 10.1002/jcb.29463. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S, et al. Cancer-associated methylated lncRNAs in patients with bladder cancer. Am. J. Transl. Res. 2019;11:3790–3800. [PMC free article] [PubMed] [Google Scholar]
- 77.Jacob R, Zander S, Gutschner T. The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 2017;18:2387. doi: 10.3390/ijms18112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA. 2019;10:e1510. doi: 10.1002/wrna.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Z, et al. Regulation of lncRNA expression. Cell Mol. Biol. Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bunch H. Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genom. 2018;293:1–15. doi: 10.1007/s00438-017-1370-9. [DOI] [PubMed] [Google Scholar]
- 81.Geyer KK, Hoffmann KF. Epigenetics: A key regulator of platyhelminth developmental biology? Int. J Parasitol. 2012;42:221–224. doi: 10.1016/j.ijpara.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 82.Cosseau C, et al. (Epi)genetic inheritance in Schistosoma mansoni: A systems approach. Trends Parasitol. 2017;33:285–294. doi: 10.1016/j.pt.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fneich S, et al. Epigenetic origin of adaptive phenotypic variants in the human blood fluke Schistosoma mansoni. Epigenet. Chromatin. 2016;9:27. doi: 10.1186/s13072-016-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roquis D, et al. Histone methylation changes are required for life cycle progression in the human parasite Schistosoma mansoni. PLoS Pathog. 2018;14:e1007066. doi: 10.1371/journal.ppat.1007066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Augusto RC, Duval D, Grunau C. Effects of the environment on developmental plasticity and infection success of Schistosoma parasites—an epigenetic perspective. Front Microbiol. 2019;10:1475. doi: 10.3389/fmicb.2019.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carneiro VC, et al. Epigenetic changes modulate schistosome egg formation and are a novel target for reducing transmission of schistosomiasis. PLoS Pathog. 2014;10:e1004116. doi: 10.1371/journal.ppat.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Padalino G, Ferla S, Brancale A, Chalmers IW, Hoffmann KF. Combining bioinformatics, cheminformatics, functional genomics and whole organism approaches for identifying epigenetic drug targets in Schistosoma mansoni. Int. J. Parasitol. Drugs Drug Resist. 2018;8:559–570. doi: 10.1016/j.ijpddr.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson L, et al. Histone deacetylase inhibition modulates histone acetylation at gene promoter regions and affects genome-wide gene transcription in Schistosoma mansoni. PLoS Negl. Trop. Dis. 2017;11:e0005539. doi: 10.1371/journal.pntd.0005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dubois F, et al. Histone deacetylase inhibitors induce apoptosis, histone hyperacetylation and up-regulation of gene transcription in Schistosoma mansoni. Mol. Biochem. Parasitol. 2009;168:7–15. doi: 10.1016/j.molbiopara.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 90.Heimburg T, et al. Structure-based design and synthesis of novel inhibitors targeting HDAC8 from Schistosoma mansoni for the treatment of schistosomiasis. J. Med. Chem. 2016;59:2423–2435. doi: 10.1021/acs.jmedchem.5b01478. [DOI] [PubMed] [Google Scholar]
- 91.Lobo-Silva J, et al. The antischistosomal potential of GSK-J4, an H3K27 demethylase inhibitor: Insights from molecular modeling, transcriptomics and in vitro assays. Parasit. Vectors. 2020;13:140. doi: 10.1186/s13071-020-4000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whatley KCL, et al. The repositioning of epigenetic probes/inhibitors identifies new anti-schistosomal lead compounds and chemotherapeutic targets. PLoS Negl. Trop. Dis. 2019;13:e0007693. doi: 10.1371/journal.pntd.0007693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raddatz G, et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc. Natl. Acad. Sci. USA. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aliaga B, Bulla I, Mouahid G, Duval D, Grunau C. Universality of the DNA methylation codes in Eucaryotes. Sci. Rep. 2019;9:173. doi: 10.1038/s41598-018-37407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fantappie MR, Gimba ER, Rumjanek FD. Lack of DNA methylation in Schistosoma mansoni. Exp. Parasitol. 2001;98:162–166. doi: 10.1006/expr.2001.4630. [DOI] [PubMed] [Google Scholar]
- 96.Salzberg A, Fisher O, Siman-Tov R, Ankri S. Identification of methylated sequences in genomic DNA of adult Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2004;322:465–469. doi: 10.1016/j.bbrc.2004.07.134. [DOI] [PubMed] [Google Scholar]
- 97.del Gaudio R, Di Giaimo R, Geraci G. Genome methylation of the marine annelid worm Chaetopterus variopedatus: Methylation of a CpG in an expressed H1 histone gene. FEBS Lett. 1997;417:48–52. doi: 10.1016/s0014-5793(97)01262-3. [DOI] [PubMed] [Google Scholar]
- 98.Regev A, Lamb MJ, Jablonka E. The role of DNA methylation in invertebrates: Developmental regulation or genome defense? Mol. Biol. Evol. 1998;15:880–891. doi: 10.1093/oxfordjournals.molbev.a025992. [DOI] [Google Scholar]
- 99.Li LH, Olin EJ, Buskirk HH, Reineke LM. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer Res. 1970;30:2760–2769. [PubMed] [Google Scholar]
- 100.Amort T, et al. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013;10:1003–1008. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.David R, et al. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell. 2017;29:445–460. doi: 10.1105/tpc.16.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 104.Fico A, Fiorenzano A, Pascale E, Patriarca EJ, Minchiotti G. Long non-coding RNA in stem cell pluripotency and lineage commitment: Functions and evolutionary conservation. Cell Mol. Life. Sci. 2019;76:1459–1471. doi: 10.1007/s00018-018-3000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poirier S, et al. The epigenetic drug 5-azacytidine interferes with cholesterol and lipid metabolism. J. Biol. Chem. 2014;289:18736–18751. doi: 10.1074/jbc.M114.563650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aimiuwu J, et al. RNA-dependent inhibition of ribonucleotide reductase is a major pathway for 5-azacytidine activity in acute myeloid leukemia. Blood. 2012;119:5229–5238. doi: 10.1182/blood-2011-11-382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Strick R, Strissel PL, Baylin SB, Chiappinelli KB. Unraveling the molecular pathways of DNA-methylation inhibitors: Human endogenous retroviruses induce the innate immune response in tumors. Oncoimmunology. 2016;5:e1122160. doi: 10.1080/2162402X.2015.1122160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Romano G, Veneziano D, Nigita G, Nana-Sinkam SP. RNA methylation in ncRNA: Classes, detection, and molecular associations. Front Genet. 2018;9:243. doi: 10.3389/fgene.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. WormBase ParaSite—a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017;215:2–10. doi: 10.1016/j.molbiopara.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Basch PF. Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J. Parasitol. 1981;67:179–185. doi: 10.2307/3280632. [DOI] [PubMed] [Google Scholar]
- 117.Pereira ASA, et al. In vitro activity of aryl-thiazole derivatives against Schistosoma mansoni schistosomula and adult worms. PLoS One. 2019;14:e0225425. doi: 10.1371/journal.pone.0225425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Panic G, Flores D, Ingram-Sieber K, Keiser J. Fluorescence/luminescence-based markers for the assessment of Schistosoma mansoni schistosomula drug assays. Parasit. Vectors. 2015;8:624. doi: 10.1186/s13071-015-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 120.Avelar L, et al. Smp38 MAP kinase regulation in Schistosoma mansoni: Roles in survival, oviposition, and protection against oxidative stress. Front Immunol. 2019;10:21. doi: 10.3389/fimmu.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jeremias WJ, et al. Comparative sequence analysis reveals regulation of genes in developing schistosomula of Schistosoma mansoni exposed to host portal serum. PLoS One. 2017;12:e0178829. doi: 10.1371/journal.pone.0178829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marek M, Kannan S, Hauser A-T, Mourão MM, Caby S, Cura V, Stolfa DA, Schmidtkunz K, Lancelot J, Andrade L, Renaud J-P, Oliveira G, Sippl W, Jung M, Cavarelli J, Pierce RJ, Romier C. Structural basis for the inhibition of histone deacetylase 8 (HDAC8), a key epigenetic player in the blood fluke Schistosoma mansoni. PLoS Pathog. 2013;9:e1003645. doi: 10.1371/journal.ppat.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files).