SUMMARY
The gut microbiota influences development1–3 and homeostasis4–7 of the mammalian immune system, and is associated with human inflammatory-8 and immune diseases9,10 as well as patients’ responses to immunotherapy11–14. Still, our understanding of how gut bacteria modulate the immune system remains limited, particularly in humans where a lack of deliberate manipulations makes inference challenging. Here we study hundreds of hospitalized—and closely monitored—cancer patients receiving hematopoietic cell transplantation as they recover from chemotherapy and stem cell engraftment. This aggressive treatment causes large shifts in both circulatory immune cell and microbiota populations, allowing the relationships between the two to be studied simultaneously. Analysis of observed daily changes in circulating neutrophil, lymphocyte and monocyte counts and >10,000 longitudinal microbiota samples from patients revealed consistent associations between gut bacteria and immune cell dynamics. High-resolution clinical metadata and Bayesian inference allowed us to compare the effects of bacterial genera relative to those of immunomodulatory medications, revealing a considerable influence of the gut microbiota—in concert and over time—on systemic immune cell dynamics. Our analysis establishes and quantifies the link between the gut microbiota and the human immune system, with implications for microbiota-driven modulation of immunity.
The human gut microbiota is considered a major modulator of the immune system during development3, and in health and disease8,9. For example, children born preterm have distinct microbiome compositions alongside distinct developmental trajectories of peripheral immune cell populations3. In adults, the success of immunotherapies that rely on peripheral immune cells, such as checkpoint inhibitor treatments, was associated with the composition of the microbiome11–13,15. There is an increasing interest in using the microbiome to modulate the immune system and augment treatments7,16, including the burgeoning field of chimeric antigen receptor (CAR) T-cell therapy17. But our understanding of how the microbiota influences the dynamics of immune cells in humans, and how this compares to deliberate immunomodulatory interventions, remains limited due to a lack of feasible experiments.
To overcome this, we asked whether the gut microbiota could influence day-by-day changes in peripheral immune cell counts. We collected a vast dataset of immune reconstitution dynamics after allogeneic hematopoietic cell therapy (allo-HCT) from patients treated at Memorial Sloan Kettering (MSK) between 2003 and 2019 (Fig. 1a, Supplementary Table 1). The conditioning regimen of radiation and chemotherapy administered to HCT patients is the most severe perturbation to the immune system deliberately performed in humans: this offers a unique opportunity to investigate links between the gut microbiota and immune dynamics directly in humans.
Figure 1: Immune reconstitution and microbiome dynamics after HCT.
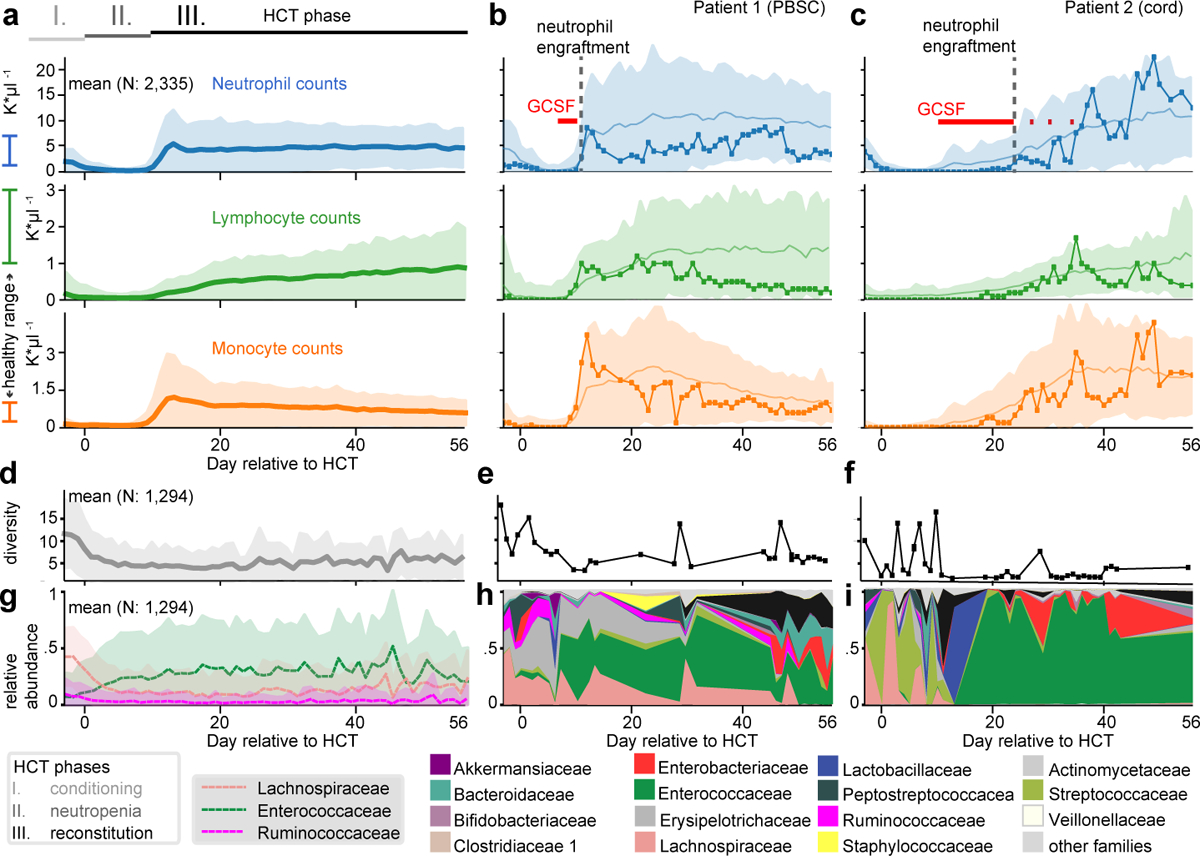
a Major periods of HCT: I) immunoablation during conditioning before HCT on day 0 followed by II) post-HCT neutropenia and III) reconstitution. Mean counts (shaded: ±1 standard deviation, σ) of neutrophils, lymphocytes and monocytes per day relative to HCT from patients transplanted between 2003 and 2019 (a), contrasted with individual patients (b,c) representative of the recovery trajectories for different stem cell graft sources; patient 1 received a PBSC graft and patient 2 received umbilical cord blood (line with circles: patient data, solid line and shaded region: mean±1σ of all PBSC/cord patients, red: GCSF administration). d-f) Loss of microbial diversity during HCT, measured by 16S rRNA gene sequencing of fecal samples, confirming previous smaller studies23,24 (d, line: mean per day, shaded: ±1σ; e,f: individual patients) coincides with loss of g-i commensal families (g, line: mean relative abundances of bacterial families, shaded: ±1σ); h,i: individual patients), often replaced by Enterococcaceae domination.
Conditioning depletes white blood cell (WBC) counts leading to neutropenia (<500 neutrophils/μl blood) until transplanted stem cells begin to release granulocytes from the bone marrow, initiating immune reconstitution (Fig. 1a–c). HCT therapies also damage the gut microbiota18 and reduce its biodiversity (Fig. 1d–i), a collateral effect associated with increased mortality in HCT patients19. Immune and microbiome reconstitution vary considerably between patients and treatment types (Fig. 1, Extended Data Fig. 1a) enabling analyses of associations between microbiome and immune system, and their comparison to immunomodulators such as granulocyte-colony stimulating factor (GCSF).
To detect a directional and causal link between the microbiota and circulatory WBCs, we first used data from a randomized trial of autologous fecal microbiota transplantation (auto-FMT)—a controlled microbiota manipulation experiment performed directly on our patients20 (Extended Data Fig. 2a). To investigate if auto-FMT affected WBC reconstitution, we compared the 24 enrolled patients’ neutrophil, lymphocyte and monocyte counts post-neutrophil engraftment (3 consecutive days >500 neutrophils per μl). FMTs were conducted at variable dates relative to neutrophil engraftment (Fig. 2a, Supplementary Table 2). Overall, we observed higher counts of each WBC type in patients who received an auto-FMT during the first 100 days post neutrophil engraftment (p<0.001, Fig. 2b,c, total WBCs Extended Data Fig. 2b–g).
Figure 2: Neutrophil, lymphocyte and monocyte counts increased in FMT-treated patients in weeks following treatment.
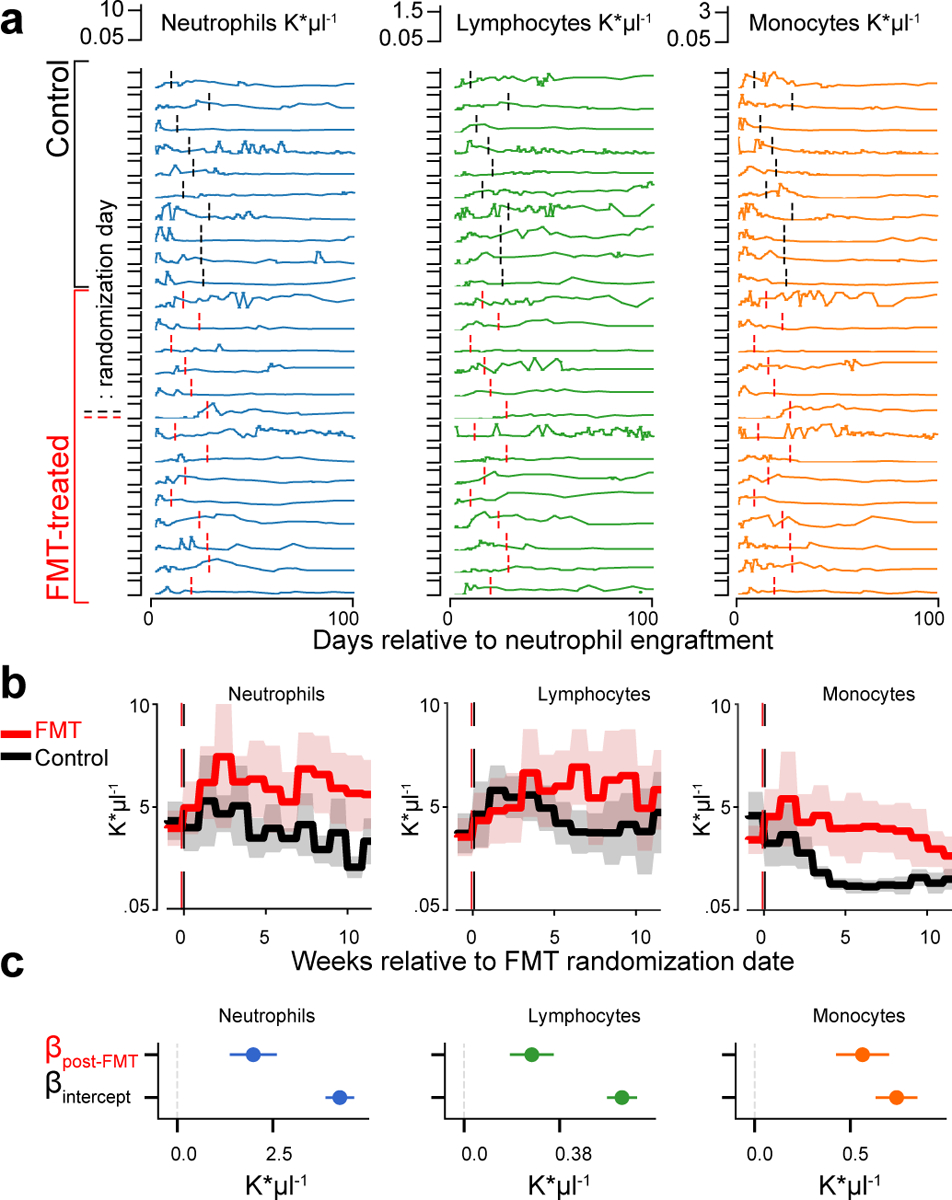
a Absolute counts of neutrophils (blue), lymphocytes (green) and monocytes (orange) in 10 control, 14 FMT-treated patients post-neutrophil engraftment; vertical lines indicate randomization dates. b Weekly mean cell counts aligned to the randomization date (FMT-treated: red, control: black). Line: mean per week, shaded region: 95%-CI. c Coefficient estimates from linear mixed effects models of neutrophils, lymphocytes and monocytes over time indicate an auto-FMT-induced increase of each white blood cell type (βFMT: p=4×10−11, p=2×10−10, p=2×10−16, respectively, full regression results in SI; b, c: N=24 subjects, n=921 blood samples).
The higher WBCs in patients receiving auto-FMT could result from the successful reconstitution of a complex microbiota20 and associated metabolic capabilities21, or it could be a systemic response to a severe therapy which introduced billions of intestinal organisms at once via an enema (no enema was administered to control patients20). Moreover, chance differences in extrinsic factors such as different immunomodulator medications may have affected this result due to the small cohort size. Nonetheless, it supported the notion that the microbiota can modulate the peripheral immune system. High counts of lymphocytes during immune reconstitution had been associated with improved clinical outcomes22, and 3-year survival was positively associated with higher mean levels of WBCs during 100 days after neutrophil engraftment in our HCT patients (hazard ratio: 0.91, p=0.04). Determining which taxa modulate immune dynamics could therefore open new ways to improve immune reconstitution—critical for clinical outcomes.
To investigate the links between the gut microbiota and the dynamics of WBC recovery we turned to our large observational cohort of HCT patients. Homeostasis of circulatory WBC counts is a complex, dynamic process: WBCs are formed and released into the blood de novo by differentiation of hematopoietic progenitor cells from the bone marrow, and can be mobilized from thymus and lymph nodes (lymphocytes), spleen, liver and lungs (neutrophils); WBCs can also migrate from the blood to other tissues when needed23. To identify modulators of these dynamics processes, we developed a two-stage approach analyzing the changes of WBC counts between two days (Fig. 3a). Stage 1 served as a clinical- and metadata feature selection stage using blood and medication data of 1,096 patients without available microbiome information (Extended Data Fig. 1b for data inclusion). Stage 2 was performed on data from an independent cohort of 841 different patients from whom concurrent microbiome samples were available to detect associations between microbiome and peripheral immune cell dynamics.
Figure 3: Bayesian inference reveals associations between the microbiota and dynamics of circulatory white blood cell counts.
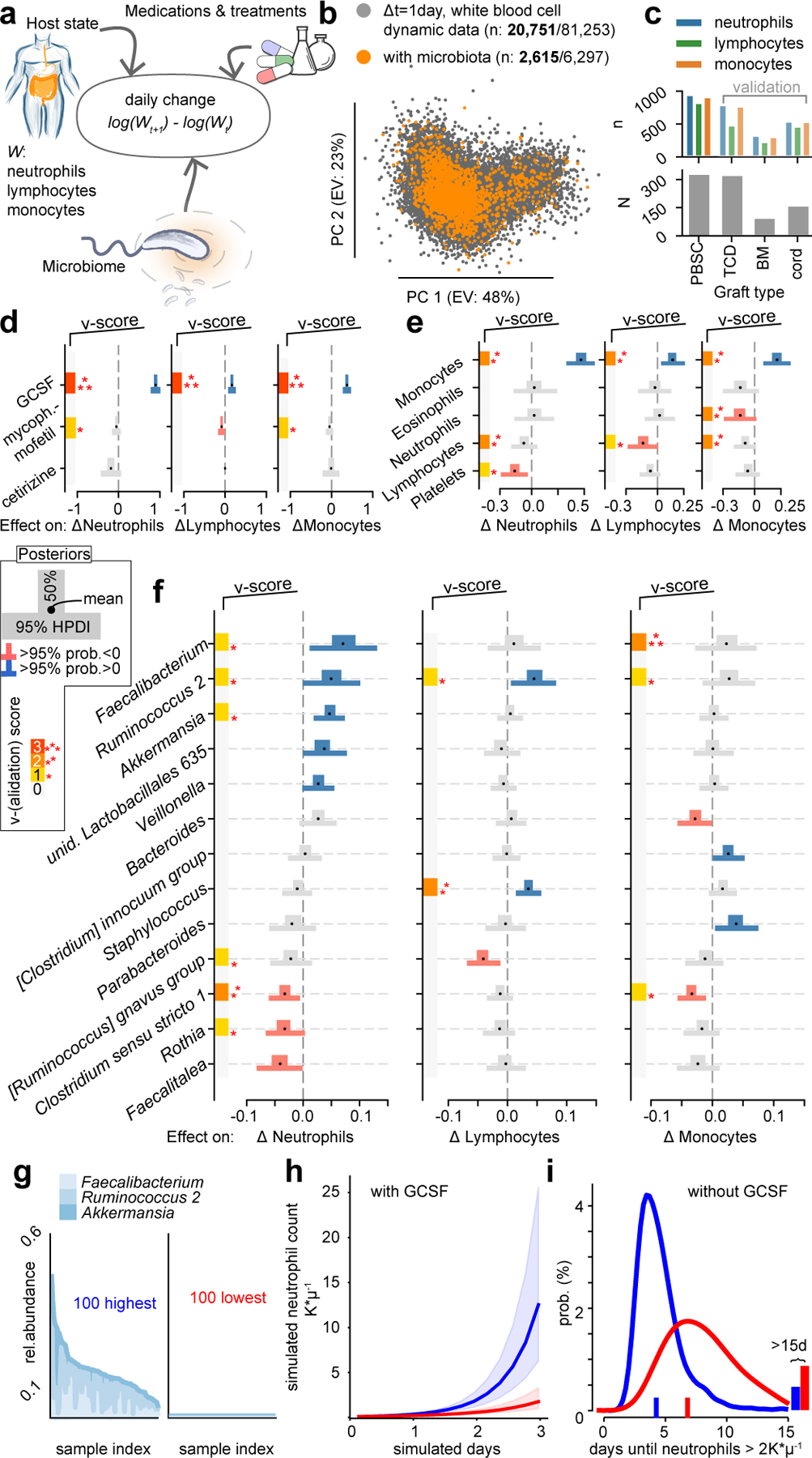
a Cartoon of the model: observed changes in WBC counts between two consecutive days are associated with the current state of the host in the form of blood cell counts in circulation, immunomodulatory medications, clinical metadata, and the state of the microbiome. b Visualization of the data of WBC dynamics; scatter plot of the principal components (PC) of observed daily changes of neutrophils, lymphocytes and monocytes without (grey) and with (orange) available concurrent microbiota samples (bold: post neutrophil-engraftment samples). c PBSC patients (N=312) provided most paired blood dynamics and microbiota samples (n=995); TCD, BM and cord patient data sets were used for validation. d-f Bayesian inference results from PBSC patient data; posterior coefficient distributions of associations between treatments (d), WBC counts (e), and log10-relative abundances of microbial genera (f) and daily changes in neutrophils, lymphocytes and monocytes. v-score: number of validation cohorts confirming associations, set to zero if invalidated by validation cohorts (additional coefficients in Extended Data Fig. 4a–c). g 100 microbiota samples with highest (left) or lowest (right) abundances of Faecalibacterium, Ruminococcus 2 and Akkermansia. h Simulation of neutrophil dynamics in presence of GCSF and microbiota compositions sampled either from those high (blue) or low (red) in Faecalibacterium, Ruminococcus 2 and Akkermansia relative abundance as shown in g); line: median of 1,000 simulations, shaded regions: interquartile range of simulated trajectories. i Time until neutrophil counts first reach >2K*μl−1 in equivalent simulations without GCSF.
In stage 1 we analyzed the changes in neutrophils, lymphocytes and monocytes during patients’ recovery from >20,000 pairs of post-engraftment blood samples separated by a single day (Fig. 3b). A cross-validated feature selection approach detected medications and HCT parameters associated with WBC dynamics (Extended Data Fig. 3a–c, Supplementary Table 3). In stage 2 we sought to identify the additional contribution of the gut microbiome. We performed Bayesian inferences using data from different sets of patients with available microbiome samples (Supplementary Table 4). Stage 1 had identified—as expected—that stem cell graft sources are associated with immune reconstitution kinetics (e.g. umbilical cord blood slower than peripheral blood, PBSC24), and we therefore stratified our patients by graft source in stage 2. The dynamic systems model of stage 2 now included bacterial genera as predictors of daily changes in WBC counts, in addition to the medications selected in stage 1, clinical features (conditioning intensity, age, sex), and the current state of the blood in the form of counts of neutrophils, lymphocytes, monocytes, eosinophils, and platelets (Fig. 3a). The data comprised 841 patients, but approximately 60% of the stool samples paired with a daily change in WBC counts were taken before neutrophil engraftment, i.e. when WBC counts were zero. Nevertheless, >2,000 post-engraftment observations of WBC changes during immune reconstitution provided a large sample to analyze dynamics (Fig. 3b). Stage 2 focused on data from the largest (Fig. 3c) cohort: PBSC graft recipients. We withheld the other cohorts (bone marrow, BM; T-cell depleted graft (ex-vivo) by CD34+selection, TCD; and cord) for validation scoring, and included data from patients treated at Duke University for additional validation (Supplementary Table 5).
Notably, as a verification of our approach, we detected associations between immunomodulator administrations and consequent immune cell dynamics that were consistent with known biological mechanisms (Fig. 3c, Extended Data Fig. 4a–f). The strongest association across all predictors is the well-known neutrophil-increasing effect of GCSF25; GCSF administration—used to accelerate recovery from chemotherapy-induced neutropenia25—was associated with a +140% increase in the rate of neutrophil changes from one day to the next ([+114%, +170%], 95 percent probability density interval [HPDI95]). This finding was observed in all MSK (v-score=3, Fig. 3d) and Duke validation data sets (Extended Data Fig. 4g–j). We furthermore found a GCSF-associated increase of +43% ([+30%, +58%]HPDI95, v-score=3) in monocyte rates, and, although smaller, in lymphocyte rates (+16%, [+5%, +27%]HPDI95, v-score=3). Neutrophil and lymphocyte rates decreased following antihistamine or immunosuppressive medication (cetirizine −18%, [−35%, +5%]HPDI95, mycophenolate mofetil −8% [−15%,+1%]HPDI95, respectively). Finally, less intensive chemotherapeutic conditioning regimens (non-ablative and reduced intensity) were associated with increased lymphocyte and monocyte rates (Extended Data Fig. 4c).
Beyond the mechanistically plausible associations of medications, our analysis detected associations between the current count of WBCs and their rates of change: negative associations of neutrophil and lymphocyte counts with the rates of monocytes, and negative associations of platelet and lymphocyte counts with the rates of lymphocytes and neutrophils (Fig. 3e). Conversely, we found positive associations between monocytes and the rates of each of the investigated WBC subsets. These associations, derived from daily counts of WBCs, could reflect a complex network underlying the regulation of blood immune cell composition23. More importantly, the associations quantified for these potential homeostatic feedbacks and medications provided a benchmark against which we could compare gut microbial taxa.
We identified bacterial genera that consistently associated with WBC dynamics (Fig. 3f). Higher relative abundances of Faecalibacterium (+8%, [+1%, +14%]HPDI95 per log10), Ruminococcus 2 (+5%, [0%, +10%]HPDI95) and Akkermansia (+4%, [+1%, +7%]HPDI95) were associated with increased neutrophil rates, whereas Rothia (−3%, [−7%, 0%]HPDI95), and Clostridium sensu stricto 1 (−3%, [−6%, 0%]HPDI95) associated with reduced neutrophil rates. These results were validated in univariate analyses of the Duke cohort (Extended Data Fig. 4g–i). We also used total bacterial abundances as predictors instead of relative abundances; this confirmed Faecalibacterium as most strongly associated with neutrophil dynamics (Extended Data Fig. 5). Ruminococcus 2 (+5%, [+1%, +9%]HPDI95) and Staphylococcus (+4%, [+1%, +6%]HPDI95) were positively associated with lymphocyte rates. Faecalibacterium and Ruminococcus 2 also associated with increases in monocyte rates; this association was validated in other cohorts (v-score 3 and 1, respectively), but there was higher uncertainty of the association estimate (HPDI50>0). Clostridium sensu strictu 1 (−3% [−5%, −1%]HPDI95) associated with decreased rates of monocytes. The associations we identified—and validated in other cohorts—between gut microbial taxa and daily changes in WBCs support the idea that hematopoiesis and mobilization respond to the composition of the gut microbiome, influencing systemic immunity26.
Intestinal bacteria may affect circulatory WBC counts by influencing either their sources in the bone marrow or their cytokine profiles27 and proliferation rates in the blood, their sinks in different organs, or both. The immune system in turn can interact with the microbiota and modulate its composition, for example via immunoglobulin-A as studied in mice28–30. To investigate a reverse effect of the immune system onto bacterial populations, we employed an analogous approach to the stage 1 analysis. Dynamics of WBCs could be estimated from changes in absolute cell counts, and to obtain necessary absolute bacterial counts, we measured total bacterial 16S rRNA gene copies per gram of stool for a subset of our samples (3,995 samples from 481 patients) to jointly infer the bidirectional association network between microbiota and the peripheral immune system dynamics. All of our patients received antibiotics on some days during their treatment18 and their strong impact on microbiota dynamics were the dominant effects that survived feature selection (Extended Data Fig. 6). Relaxing the regularization strength (methods), however, revealed several bi-directional relationships between WBCs and gut bacterial dynamics (Extended Data Fig. 7). Of note, we detected a negative association of absolute [Ruminococcus] gnavus group abundance with lymphocytes rates, confirming our main result based on relative bacterial abundances (Fig. 3f). In the reverse direction, we saw a positive association of lymphocyte counts with [Ruminococcus] gnavus group growth rates. Ruminococcus gnavus is associated with inflammatory bowel diseases (IBD)31 and auto-immune disorders10; our analysis suggests it may drive high neutrophil to lymphocyte ratios that are broadly characteristic for poor disease outcomes in IBD32 and beyond33,34.
Several of the bacterial taxa positively associated with WBC rates were obligate anaerobes, some of which produce cell-wall molecules1,35 and short-chain fatty acids (SCFAs)36 that modulate immune responses and granulopoiesis37. Ruminococcus 2, for example, contains keystone species that release nutrients from complex dietary starch38 and such nutritional support from the microbiota improved hematopoietic reconstitution in mice21. To identify a similar association in our patients, we estimated the microbiota reconstitution potency per sample (methods). Shotgun metagenomic sequences from 124 of our samples revealed that positive microbiota potency samples were enriched in cholate degradation, vitamin-B1 synthesis and butanoate formation pathways (Extended Data Fig. 8). In line with evolutionary theory39, our results suggest that such broadly available microbial traits may be co-opted by the host as part of the homeostatic interplay between immune system and microbiota. Reassuringly, the genera Faecalibacterium, Ruminococcus 2 and Akkermansia that we associated with increased WBCs rates were among those best reconstituted by auto-FMT20, potentially explaining the higher counts of neutrophils, monocytes and lymphocytes in auto-FMT treated patients.
Our analyses show that the gut microbiome is associated with immune cell dynamics in humans. The inferred associations should be interpreted as net effects since they do not distinguish the microbiota’s impact on de novo hematopoiesis, for example, from its impact on other sources and sinks. Unlike the plausible role of obligate anaerobe fermenters in augmenting hematopoiesis via nutritional support21, the positive association detected between Staphylococcus and lymphocyte dynamics could instead result from reduced extravasation of T cells from circulation into the gut epithelium40, especially since high abundances of Staphylococcus are associated with low gut microbiota diversity (p<0.001, Extended Data Fig. 9a), which indicates a depleted microbiota.
Nevertheless, our approach allows us to leverage the chronology of events and assess “mathematical causality”41. Due to the observational nature of these data there are risks of confounding, e.g. from undetected infections or dietary components, that could explain some of the associations found, but the close temporal correspondence41 between microbiota and WBC dynamics reduces the number of plausible confounders. Notwithstanding potential confounders, our results suggest candidate bacterial taxa that might improve immune reconstitution, and focused follow-up studies are required to evaluate their immunomodulatory efficacy. Intriguingly, members of Faecalibacterium and Ruminococcus in one study12, and Akkermansia in another11, were associated with better responses to anti–PD-1 immunotherapy, which suggested a disagreement42. Our results, however, revealed Faecalibacterium, Ruminococcus 2, and Akkermansia as the taxa with the strongest immune cell dynamics associations, therefore agreeing with the finding of both studies that these taxa are associated with human immune modulation. Furthermore, our work enables us to directly compare their inferred effect sizes to the effects of immunomodulatory drugs. These genera are common in healthy people43, but they can drop below detection in patients after HCT18. Realistic ranges of 3–5 orders of magnitude in bacterial relative abundances (Figure 3g, Extended Data Fig. 9b,c) could yield effect sizes similar to the homeostatic feedbacks inferred between WBCs and even immunomodulatory medications (e.g. a change in Ruminococcus 2 from below detection to 1% relative abundance associated with a +67% and +63% increase in neutrophil and lymphocyte rates, respectively). The effect sizes of gut bacteria at first may appear small relative to those of immunomodulatory drugs, yet their effect could be considerable as they refer to changes in exponential rates of WBCs and would accumulate each day while those bacteria remain abundant. To demonstrate this accumulation over time, we simulated WBC dynamics using our posterior coefficient distributions (methods). We simulated 1,000 time series for microbiota compositions either chosen from the 100 samples highest or lowest in Faecalibacterium, Ruminococcus 2 and Akkermansia (Fig. 3g), in presence (Fig. 3h) or absence (Fig. 3i) of GCSF administration. Simulations predict that microbiota enriched in these genera accelerate immune reconstitution, and reduce the time until neutrophils reach >2K*μl−1 in absence of GCSF by 2.4 days, from predicted 6.8 (CI: [6.5, 7]) to 4.4 days (CI: [4.3,4.5]) days. Gut bacteria, in concert and over time, could therefore influence steady-state immune homeostasis considerably, even in individuals with less severely injured microbiomes.
In sum, our work links the gut microbiota to the dynamics of the human immune system via peripheral white blood cell dynamics. Our analysis uses WBCs counted directly from patients, which are coarse-grained clinical analyses conducted at large scale but lack details such as lymphocyte and other immune cell subsets. Nonetheless, our work demonstrates how analysis of massive clinical datasets can reveal novel biological insights. Because our study is in humans, it fills an important gap at a critical time for microbiome research when the clinical relevance of animal models of microbiome-immune interaction has been questioned44. By studying a large number of patients over time, we were able to infer and quantify the association between gut bacteria and systemic immune cell dynamics, and our results help to consolidate previous findings12,11 that seemed in conflict with each other42. Our demonstration that the microbiota influences systemic immunity in humans opens the door towards an exploration of potential microbiota-targeted interventions to improve immunotherapy and treatments for immune-mediated and inflammatory diseases8,10–12.
Methods
Ethics approval and informed consent
The participants in the auto-FMT trial (NCT02269150) provided written informed consent to participate in the trial (#14–025). Participants in the observational cohorts at both MSK and at Duke provided written informed consent for the use of their fecal specimens and clinical data. The use and analysis of these specimens for the work herein was approved by IRBs at both institutions: MSK (#16–834) and Duke (PRO0006268 and Pro00050975).
Complete blood count collection and characterization
Absolute white blood cells count data were obtained from routine complete blood counts ordered by clinicians during normal clinical practice, used to obtain informative diagnostic and monitoring information. Blood samples received in the clinical hematology laboratory were analyzed using Sysmex XN automated hematology analyzers (Sysmex, Lincolnshire, IL) and, when needed based on specific flags and parameters as per MSKCC standard operating procedures, were validated manually using the Sysmex DI-60 Slide Processing System or CellaVision DM9600 Automated Digital Morphology System (Sysmex, Lincolnshire, IL).
16S rRNA gene amplification and multiparallel sequencing
For each sample, duplicate 50-μl PCRs were performed, each containing 50 ng of purified DNA, 0.2 mM deoxynucleotide triphosphates, 1.5 mM MgCl2, 2.5 U Platinum Taq DNA polymerase, 2.5 μl of 10× PCR buffer, and 0.5 μM of each primer designed to amplify the V4-V5: 563F (5′-nnnnnnnn-NNNNNNNNNNNN-AYTGGGYDTAAAGNG-3′) and 926R (5′-nnnnnnnn-NNNNNNNNNNNN-CCGTCAATTYHTTTRAGT-3′). A unique 12-base Golay barcode (Ns) precedes the primers for sample identification45, and one to eight additional nucleotides were placed in front of the barcode to offset the sequencing of the primers. Cycling conditions were 94°C for 3 min, followed by 27 cycles of 94°C for 50 s, 51°C for 30 s, and 72°C for 1 min. For the final elongation step, 72°C for 5 min was used. Replicate PCRs were pooled, and amplicons were purified using the QIAquick PCR Purification Kit (Qiagen). PCR products were quantified and pooled at equimolar amounts before Illumina barcodes and adaptors were ligated, using the Illumina TruSeq Sample Preparation protocol. The completed library was sequenced on an Illumina MiSeq platform following the Illumina recommended procedures with a paired-end 250 × 250 bp kit
Sequence analysis
The 16S (V4-V5) paired-end reads were merged and demultiplexed. Amplicon sequence variants (ASVs) were identified using the Divisive Amplicon Denoising Algorithm (DADA2) pipeline including filtering and trimming of the reads46. Reads were trimmed to the first 180 bp or the first point with a quality score Q<2. Reads were removed if they contained ambiguous nucleotides (N) or if two or more errors were expected based on the quality of the trimmed read. We assigned taxonomy to ASVs using a 8-mer based classifier trained by IDTaxa47 using the SILVA database48. We determined the copy number of 16S rRNA genes per gram of stool for 4,158 of our samples as reported previously18, by quantitative PCR on total DNA extracted from fecal samples.
Quantification of total microbiota density per gram of stool and estimation of total genus abundances.
Quantitative PCR (qPCR) was performed on DNA extracted from the 1g wet weight of a stool sample using DyNAmo SYBR Green qPCR kit (Finnzymes) and 0.2 μM of the universal bacterial primer 8F (5′-AGAGTTTGATCCTGGCTCAG) and the broad-range bacterial primer 338R (5′-TGCTGCCTCCCGTAGGAGT-3′). Standard curves were prepared by serial dilution of the PCR blunt vector (Invitrogen) containing 1 copy of the 16s rRNA gene. Cycling conditions were 95°C for 10 minutes followed by 40 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 1 minute. We used the measurements of total 16S rRNA gene counts per gram of stool to multiply the relative abundances of taxa obtained from 16S amplicon sequencing to obtain the estimate of their total abundance per gram of stool (supplementary information). Importantly, this does not account for 16S copy number variation between taxa, but the observed dynamic ranges in total abundances of taxa in our data set span up to 9 orders of magnitude, exceeding the potential inaccuracies due to copy number variation.
Diversity calculations
Microbiome alpha-diversity was measured by the inverse Simpson (IS) index of a sample. It was calculated by , where p is the relative abundance of the jth ASV out of N total ASVs in sample i.
Linear mixed-effects model of white blood cell counts
To study the effect of auto-FMT on white blood cells, we investigated the white blood cell counts of 24 enrolled patients of this trial from the day of neutrophil engraftment until 100 days after. FMT occurred on different days relative to neutrophil engraftment. Thus, we performed an analogous analysis to that conducted in the original publication that demonstrated how FMT re-established a diverse microbiome in the post-FMT period20. To answer if white blood cell counts differed post-FMT, we used a linear mixed effects model of white blood cell counts, y, modeled as a function of the FMT treatment as well as patient and timepoint specific random effects. We included random intercept terms for each day i and each patient j, and a fixed effects term for the post-FMT period with associated coefficient “armpost”, using the indicator variable “FMT”, that is 1 when a patient was from the FMT treated arm of the trial and day was greater than or equal to the day of the FMT procedure. We conducted independent analyses for neutrophil, lymphocyte and monocyte counts. This resulted in the following model of a cell count, y, for patient j on day i:
with prior distributions , and , independent error and fixed intercept β0, for the D days post neutrophils engraftment and P patients, (D=100, P=24). For convenience of those interested in reanalyzing our data, the part of our data concerning the auto-FMT analysis is available in tidy format (supplementary information), and the analysis code conducted in the R programming language is available as an exported notebook (fmt_effect_on_wbc.pdf) on github: https://github.com/jsevo/wbcdynamics_microbiome/.49 We conducted an additional analysis with “day” as a continuous predictor which did not change our conclusions (supplementary information).
Dynamic systems analyses
We analyzed factors associated with the observed changes of absolute counts of neutrophils, lymphocytes and monocytes between two days. In the following we describe how chronology of events and biological samples were encoded, and the models used to infer a role of medications, clinical parameters and the microbiome on dynamics of white blood cells.
To reveal factors that associate with day-to-day changes in white blood cell counts, we started from a first-order differential equation of white blood cell (W) dynamics:
Where gr represents the intercept, i.e. the base line rate of change during immune reconstitution, and βj are the to-be-estimated coefficients of the P predictors Xj, j ∈ P, of the white blood cell dynamics. This equation was then linearized to
And we parameterized the corresponding discrete difference equation:
where Δln(W) is the log-difference between single days of neutrophils, lymphocytes or monocytes counts, and Δt=1 for all intervals. Predictors include the counts of neutrophils, lymphocytes, monocytes, eosinophils and platelets during an interval (homeostatic feedbacks), immunomodulatory medication and clinical observations such as a blood stream infection and the onset of graft versus host disease, HCT parameters such as graft types and conditioning regimens, and, additionally, the microbiota composition in “stage 2” of our analysis (supplementary information for data exclusion and additional details on interval definitions). Importantly, by parameterizing a dynamic equation and analyzing rates of change, our coefficient estimates have an immediate causal interpretation within our modeling framework (i.e. a βj>0 implies that higher levels of the corresponding Xj increases the rate of change of white blood cell type, W). To differentiate such results from other associations, they have been described by the term “mathematical causality”41.
Stage 1 analysis: Feature selection. Identifying medications and clinical observations associated with white blood cell dynamics from patients without microbiome data
Stage 1 uses data of patients without any available microbiome samples and the following model of white blood cell changes, y:
with intercept, gr.The predictors, X, include dummy variables for the HCT graft type, patients’ age on the date of HCT, sex, 13 most frequently observed positive blood cultures with remaining other blood stream infections grouped into a separate category “other infections”, an indicator for the onset of graft versus host disease, administrations of 55 different, most common immunomodulatory medications and platelet transfusion events, and HCT conditioning intensity regimens as well as the log-transformed geometric mean counts of neutrophils, lymphocytes, monocytes, eosinophils and platelets during the respective interval. We used elastic net regression50 for feature selection using the sklearn package for the Python programming language51. For elastic net regression with 50% L1-penalty, predictors were scaled between zero and 1, and we used 10-fold cross validation (i.e. leaving out 10% of patients at each cross-validation step) to choose the regularization strength, λ, solving for
Stage 1 yielded a sparse coefficient matrix of predictors used to design the model in stage 2.
Expanded analysis on patients with microbiome data – stage 2
To identify associations between microbiota and white blood cell dynamics, we conducted an analogous, Bayesian regression using the package PyMC3 for the Python programming language52. Stage 1 identified important differences between transplant types, and we therefore stratified our data into 4 cohorts according to their stem cell graft source. Using data independently from each cohort, we applied “no U-turn” sampling53 to produce 10,000 posterior samples from 5 independent MCMC chains that parameterized the model:
with uninformative prior distributions
where y is the observed daily change of a focal white blood cell type as in stage 1 with normal distributed mean, μ, and σ, the model uncertainty with a thick-tailed half Cauchy prior (importantly, our posterior estimates do not depend on this choice as we obtain the same results with an inverse Gamma prior, Extended Data Fig. 10b). μ was a function of the baseline growth rate, gr, and predictors, : medications with non-zero coefficients in stage 1, the white blood cell counts, patient age and sex, and HCT conditioning intensities; additionally, now included the log-abundances of microbial genera as measured by 16S sequencing from DNA in the stool collected on the second day of a daily interval (see supplementary information for details). We considered taxa that were among the 100 most abundant, or had reached maximum relative abundances of at least 10%, and selected those who were non-zero in more than 75% of our samples. White blood cell counts and microbiota data present during a daily interval were log-transformed, and zeros were filled with half of the minimum observed non zero counts (i.e. 0.5e3 and 2e-6, respectively). We focused on the largest cohort (PBSC) and used the independent inference results from TCD, BM, and cord cohorts for validation.
Validation score
Coefficients learnt from the PBSC patient cohort were assigned a “validation score” based on the results obtained from the other three MSK patient cohorts. Our requirements for validation were conservative; we required evidence from our validation data sets as well as absence of counter evidence. For regression results from each of the validation graft type cohorts, i.e. TCD, BM, and cord, we checked if a coefficient had more than 75% probability (50%HPDI) to have the same sign as the mean of the PBSC coefficient posterior for a given predictor. If so, this was considered evidence of validation, and we summed the evidence over the three validation sets (i.e. maximum score of 3, 1 from each of TCD, BM, and cord cohorts). Conversely, if we found more than 75% probability among any of the validation data sets that a given predictor had the opposite sign as the posterior mean calculated from PBSC data, this was considered counter evidence and the validation score was always set to zero.
Analysis of white blood cell dynamics with absolute bacterial abundances as predictors instead of relative abundances
We conducted an ordinary least squares regression using the statsmodels package in the Python programming language of the same model as in the main Bayesian analysis using total bacterial abundances as predictors. This was only possible on a subset of 389 neutrophil, 331 lymphocyte and 376 monocyte rate observations from PBSC patients.
Forwards simulation of predicted immune system reconstitution kinetics
To assess the impact of the estimated microbiota coefficients on immune system dynamics, we conducted 1,000 simulations of the system of 3 differential equations describing the dynamics of neutrophils, lymphocytes and monocytes. We ran 1,000 simulations four times: in presence and absence of GCSF, each with microbiota compositions enriched or depleted in Faecalibacterium, Ruminococcus 2 and Akkermansia. To identify these compositions, we ranked the observed microbiota compositions by these taxa, and chose randomly either from the top or bottom 100. The coefficients for white blood cell interactions, interactions with the microbiota and the effect of GCSF were sampled from our posterior coefficient distributions. Using these coefficients sampled at the start of the simulation, and using 50 cells*μl−1 of neutrophils, lymphocytes and monocytes as initial values, we simulated these differential equations forwards in time using the odeint function of the scipy package for the Python programming language.
Validation on data from Duke University
We analyzed 9,603 blood samples with 25,581 associated administrations of immunomodulatory medications, and 741 microbiota samples from Duke as an orthogonal data set to validate our findings. The temporal resolution of this data was much lower, and after filtering for samples from the relevant post neutrophil engraftment period, and by requiring daily intervals, 83 valid, complete data points were available. Using these data, we correlated daily blood cell changes individually in univariate, or jointly in a partial least squares regression, with those predictors that achieved more than 95% probability density in the positive or negative domain in the PBSC data regression. For each of these predictors, we present the sign of slopes and Bonferroni corrected p-values from individual linear regressions.
Joint analysis of the effect of antibiotics and white blood cell counts on the microbiota and the microbiota and immunomodulatory medications on white blood cell counts
Analogous to stage 1, we performed cross-validated, regularized linear regressions (ElasticNet) using the scikit-learn package for the Python programming language to jointly estimate the association network between microbiota and circulatory white blood cells. For this, we constructed a block matrix X of predictor matrices Xi that include the absolute bacterial abundances, drug data (antibiotics for bacterial dynamics and immune modulators for white blood cell dynamics), as well as the counts of white blood cells and a separate intercept term per block. Each block , with nl observations and pl predictors (l=0…k), on the diagonal of X corresponds to the indices of the observed daily log-changes of one of the 41 bacterial genera considered in our main analysis or the log changes in neutrophil, lymphocyte and monocyte counts from PBSC patients contained in Y (in total we calculated 15,833 rates from 256 patients). Our regression problem can thus be written as:
with k=44, i.e. 41 bacterial genera and 3 white blood cell types, the to-be estimated coefficient vector β and 0 the zero matrix. This system is underdetermined and we therefore chose the same approach as in stage 1, elastic net regression, for feature selection. Predictors were scaled between zero and 1, and we used 3-fold cross validation, leaving out 1/3rd of the patients at each iteration to identify a global regularization strength, λ, solving for
where η is the total number of observed daily log changes in genera and white blood cells, and ρ the total number of predictors. This yielded a strongly regularizing λs, and thus few predictors. To characterize potential bidirectional relationships between white blood cell counts and the gut microbiota, we iteratively reduced the regularization strength until the strongest interaction between microbiota and white blood cell dynamics, i.e. Faecalibacterium with neutrophil dynamics, was detected. We than re-ran the regression with this reduced regularization strength, λr.
Shotgun sequencing
Sequencing of 124 post-neutrophil engraftment was conducted on the Illumina HiSeq platform. For details and the processing of the FASTQ files, see supplementary information. We used the HUMAnN2 pipeline54 with default settings for functional profiling of our samples, with the UniRef90 data base and ChocoPhlAn for alignment, and we renormalized our samples by library depth to copies per million. We used MetaCyc to obtain stratified and unstratified pathway abundances.
Statistical analysis of shotgun data
We calculated the predicted microbiota potency score for each sample and separately for neutrophils, lymphocytes and monocytes, by multiplying the abundances of taxa in each of the 124 samples with the corresponding posterior coefficients obtained from the PBSC inference. To distinguish the sets of metabolic functions that separate samples with positive and negative predicted potencies, we converted the pathway abundances into presence and absences profiles. We performed a linear discriminant analysis between positive and negative potency samples with a least squares solver and automatic shrinkage using the Ledoit-Wolf lemma using the sklearn package for the Python programming language51. To assess differences in the presence or absence of pathways between samples with positive and negative potency, we used Fisher’s exact test for each pathway.
Extended Data
Extended Data Fig. 1: Blood cell counts over time.
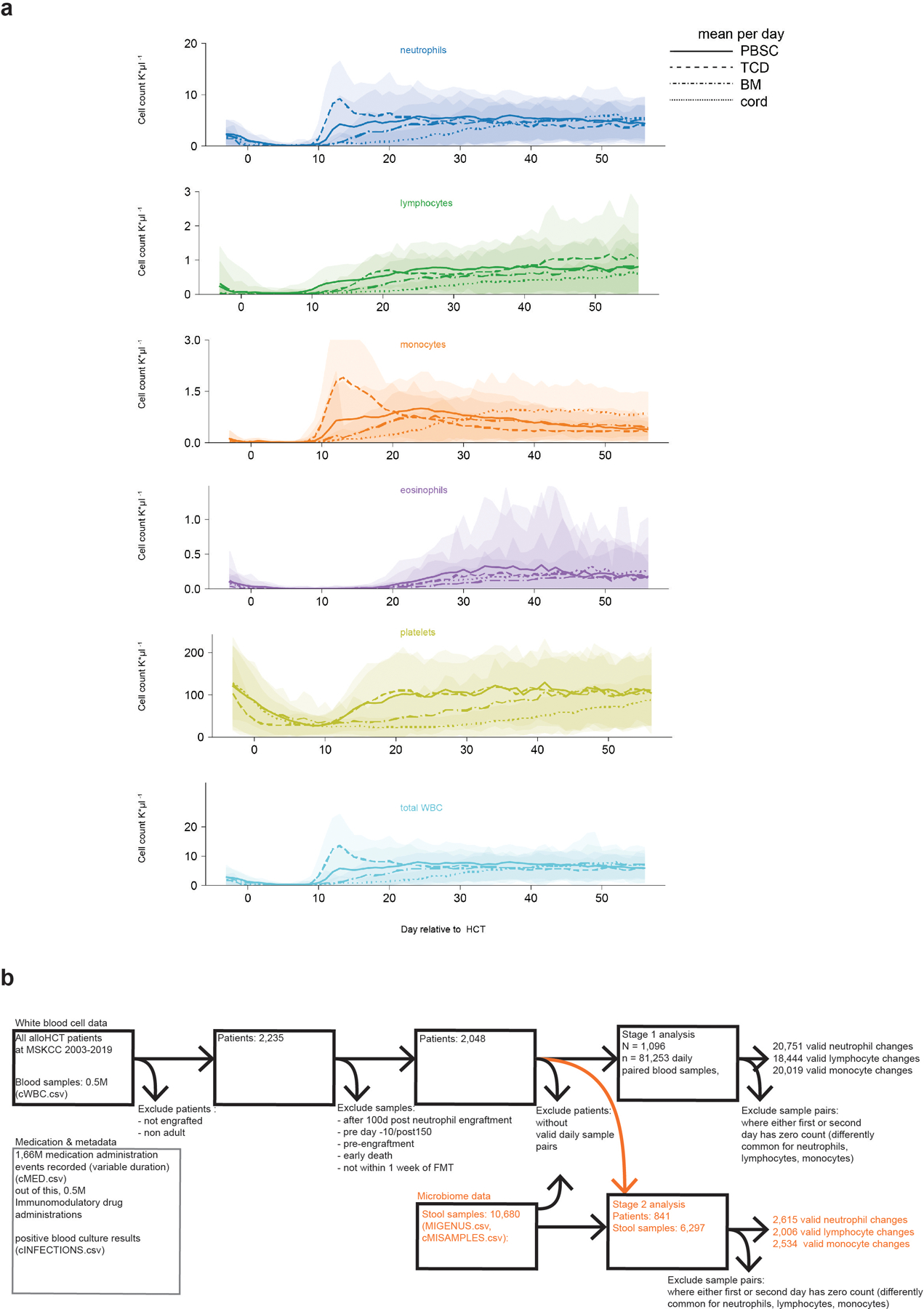
a, White blood cell counts and platelet counts per graft source over the first 100 days post HCT per day relative to HCT from N=2,235 adult patients (detailed demographics in supplementary table 1); lines: mean, shaded: ± standard deviations. b, data exclusion diagram.
Extended Data Fig. 2: FMT increases WBC counts.
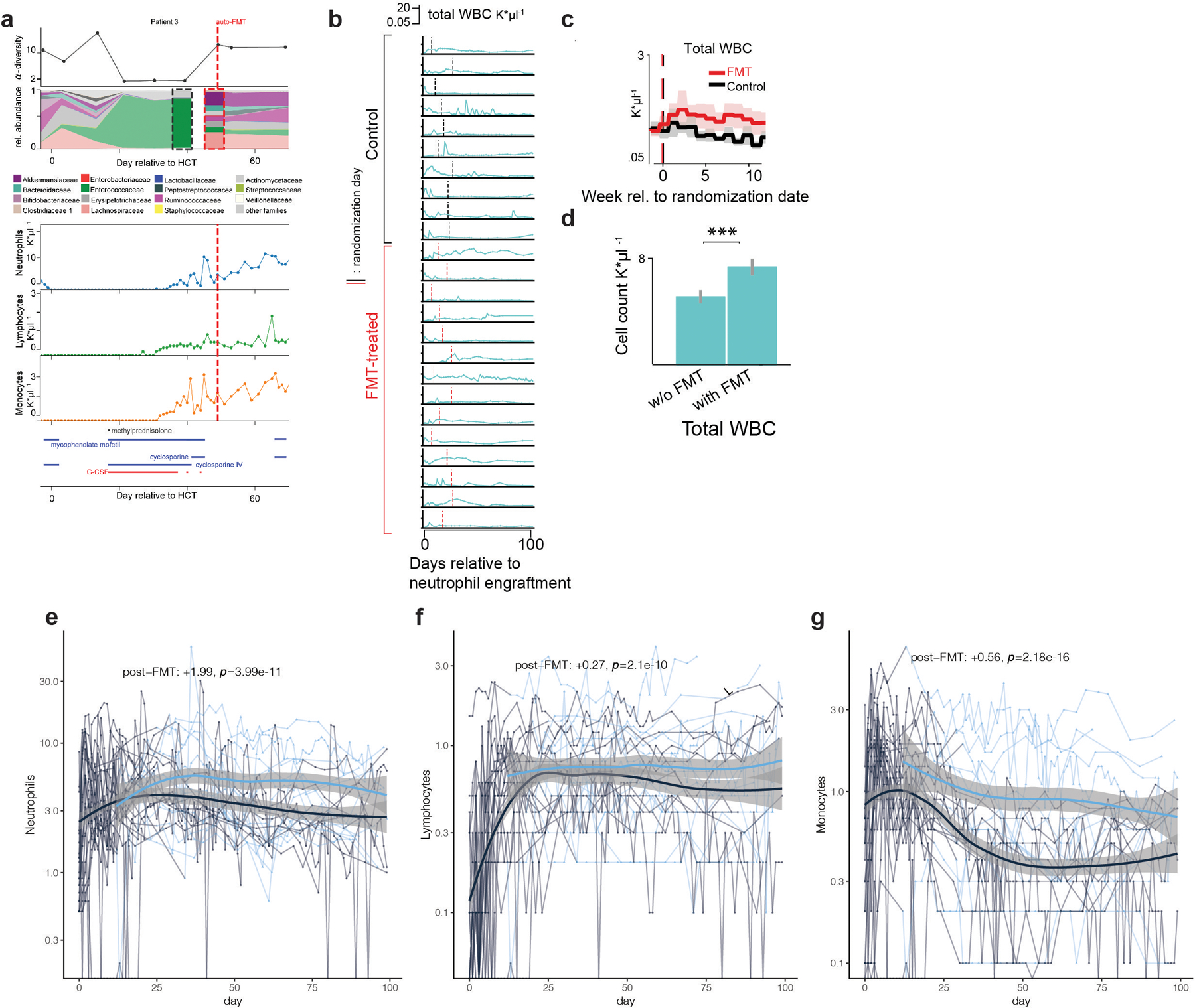
a, HCT patient who received an autologous fecal microbiota transplant (auto-FMT, dashed red line) that restored commensal microbial families and ecological diversity in the gut microbiota, with concurrent cell counts of peripheral neutrophils, lymphocytes and monocytes and immunomodulatory drug administrations. b, total white blood cell counts in 24 enrolled patients (10 control, 14 treated) post-neutrophil engraftment; vertical lines indicate randomization dates. c, weekly mean WBC counts aligned to the randomization date (FMT-treated: red, control: black). Line: mean per week, shaded region: 95%-CI. d, coefficient estimates (mean vs. mean + FMT effect) from linear mixed effects models of total WBC counts over time indicate an auto-FMT-induced increase of WBCs (βFMT: p=7×10–14). e, f, g, respectively: neutrophil, lymphocyte and monocyte count trajectories of 24 FMT trial patients. Thin lines: raw data (blue: post-FMT); thick black: mean per day, thick blue: mean+post-FMT coefficient. Means and confidence intervals (shaded region) without (black) and after FMT (blue), as well as the coefficient estimate for FMT treatment and its p-value from a linear mixed effects model relating cell counts over time to the FMT treatment (methods).
Extended Data Fig. 3: Results of the feature selection stage 1 regression.
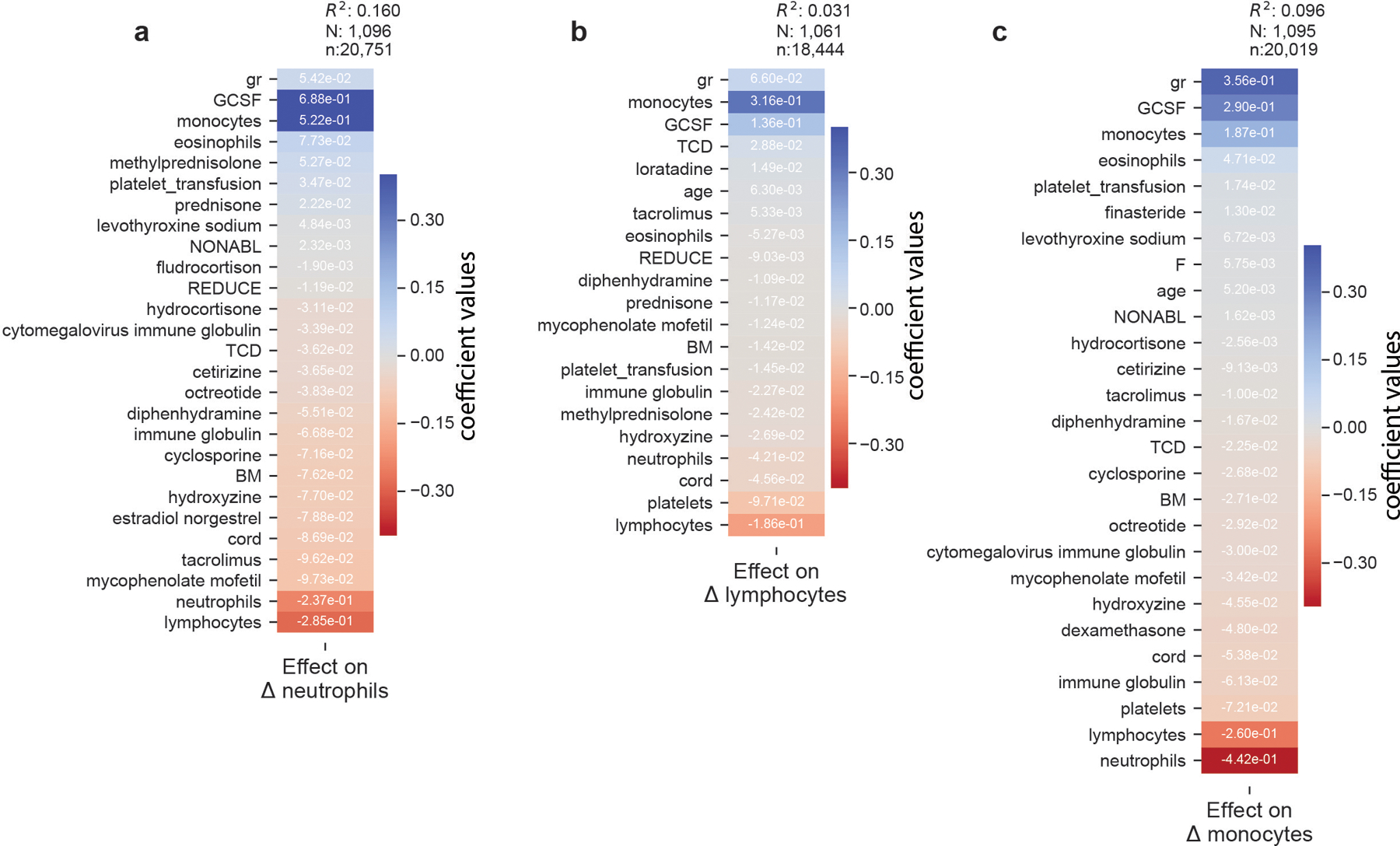
a-c, “Stage 1” regression on neutrophil, lymphocyte, and monocyte dynamics, respectively, on patients without microbiome data. Coefficients from 10-fold cross-validated elastic net regression daily changes in neutrophils. gr: intercept; TCD: T-cell depleted graft (ex-vivo) by CD34+selection; PBSC: peripheral blood stem cells; BM: bone marrow; cord: umbilical cord blood; NONABL: Nonmyeloablative; REDUCE: reduced-intensity conditioning regimen; F: female; N: patients, n: samples (daily changes in neutrophils).
Extended Data Fig. 4: Additional coefficients, posterior convergence evaluation and validation.
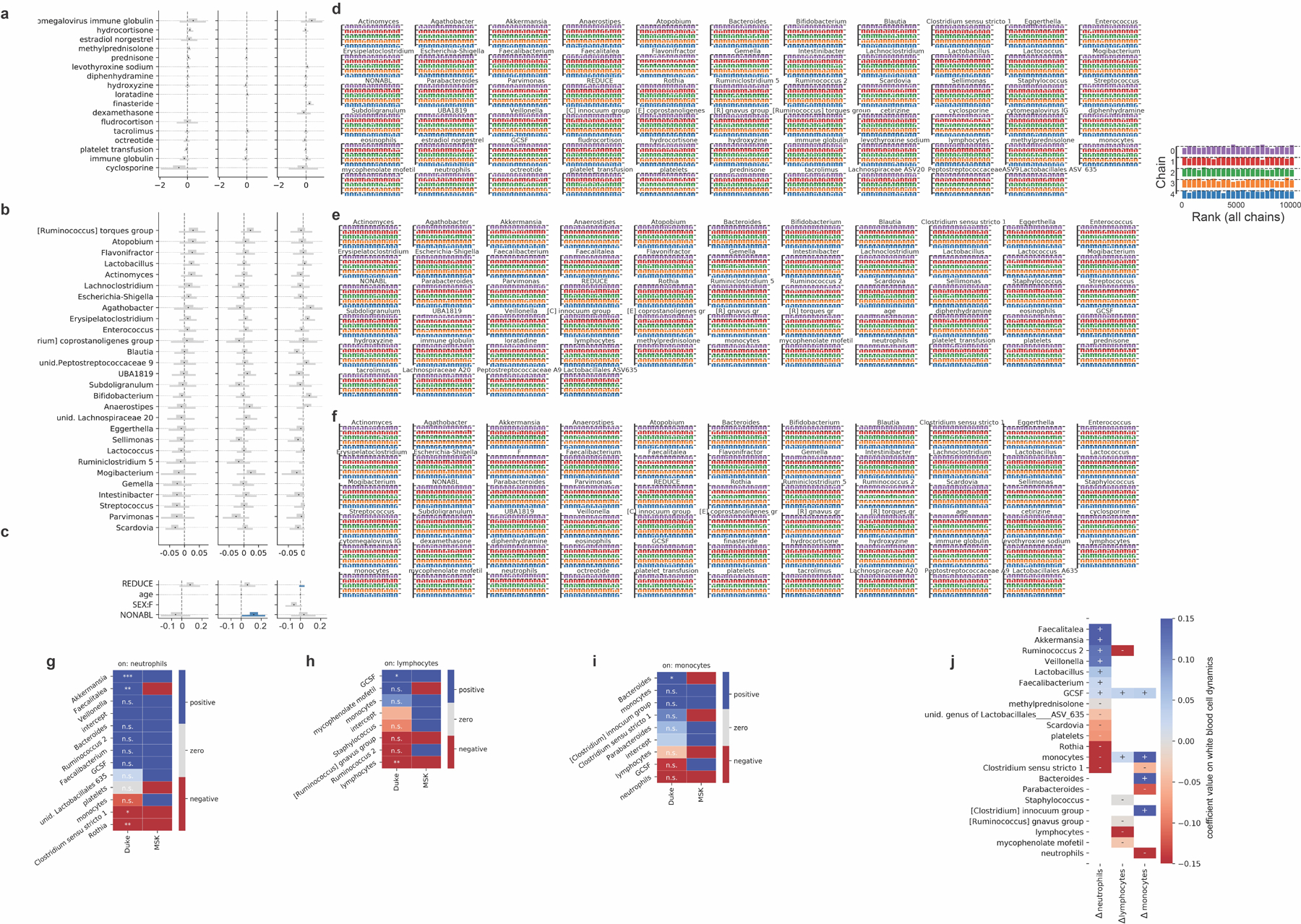
a-c, Additional posterior coefficient estimates of medications, additional genera and HCT metadata from the Bayesian stage 2 regression, see also Fig. 3. REDUCE: reduced-intensity conditioning regimen; NONABL: non-myeloablative conditioning regimen. F: female. d-f, posterior sampling convergence. Histograms of the ranked posterior draws from the model of neutrophil, lymphocyte and monocyte dynamics, respectively, in PBSC patients (ranked over all chains), plotted separately for each chain show no substantial differences between chains. g-i, Predictors of white blood cell dynamics using data from patients treated at Duke. Heatmaps indicate the slope coefficients from individual univariate regressions of microbiome and clinical predictors with changes in neutrophils, lymphocytes and monocyte, and for comparison the corresponding coefficients signs from the Bayesian multiple linear regressions in stage 2 of the analysis of white blood cell dynamics in MSK patients (Fig. 3). P-values were adjusted for multiple hypothesis testing using Bonferroni correction: ***<0.001, **<0.01, *<0.05; p>0.05: n.s. Sign of coefficients from MSK PBSC patients for comparison. j, equivalent validation analysis from patients treated at Duke using partial least squares regression of microbiome and clinical predictors identified in stage 2 of our analysis on daily changes in neutrophils, lymphocytes and monocyte.
Extended Data Fig. 5: Validation using absolute instead of relative abundance bacterial genus data.
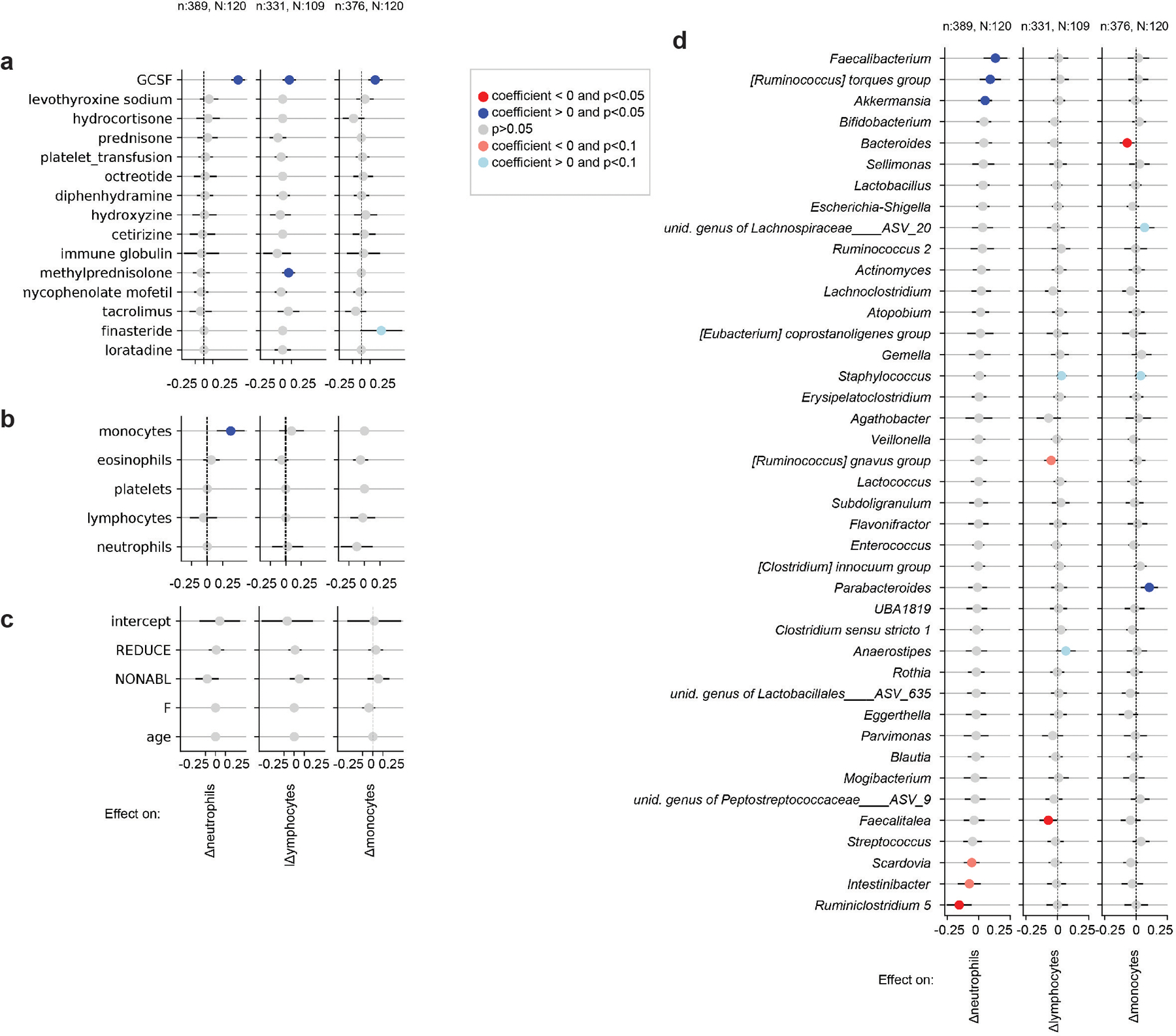
a-d Validation analysis of the main model using absolute bacterial abundances as predictors instead of relative abundances in Fig. 3. Results show inferred coefficients and p-values from multiple linear regressions. One regression per analyzed white blood cell type dynamics, i.e. neutrophil, lymphocyte and monocyte daily log-changes, was conducted, and coefficients for medications (A), white blood cell feedbacks (B) metadata (C) and total genus abundances (D) are shown. This was only possible for only a subset of the data used in the main analysis for which we obtained absolute bacterial abundance estimates (methods), n: samples, N: patients.
Extended Data Fig. 6: Jointly inferred association network between white blood cell and bacterial genus dynamics (methods).
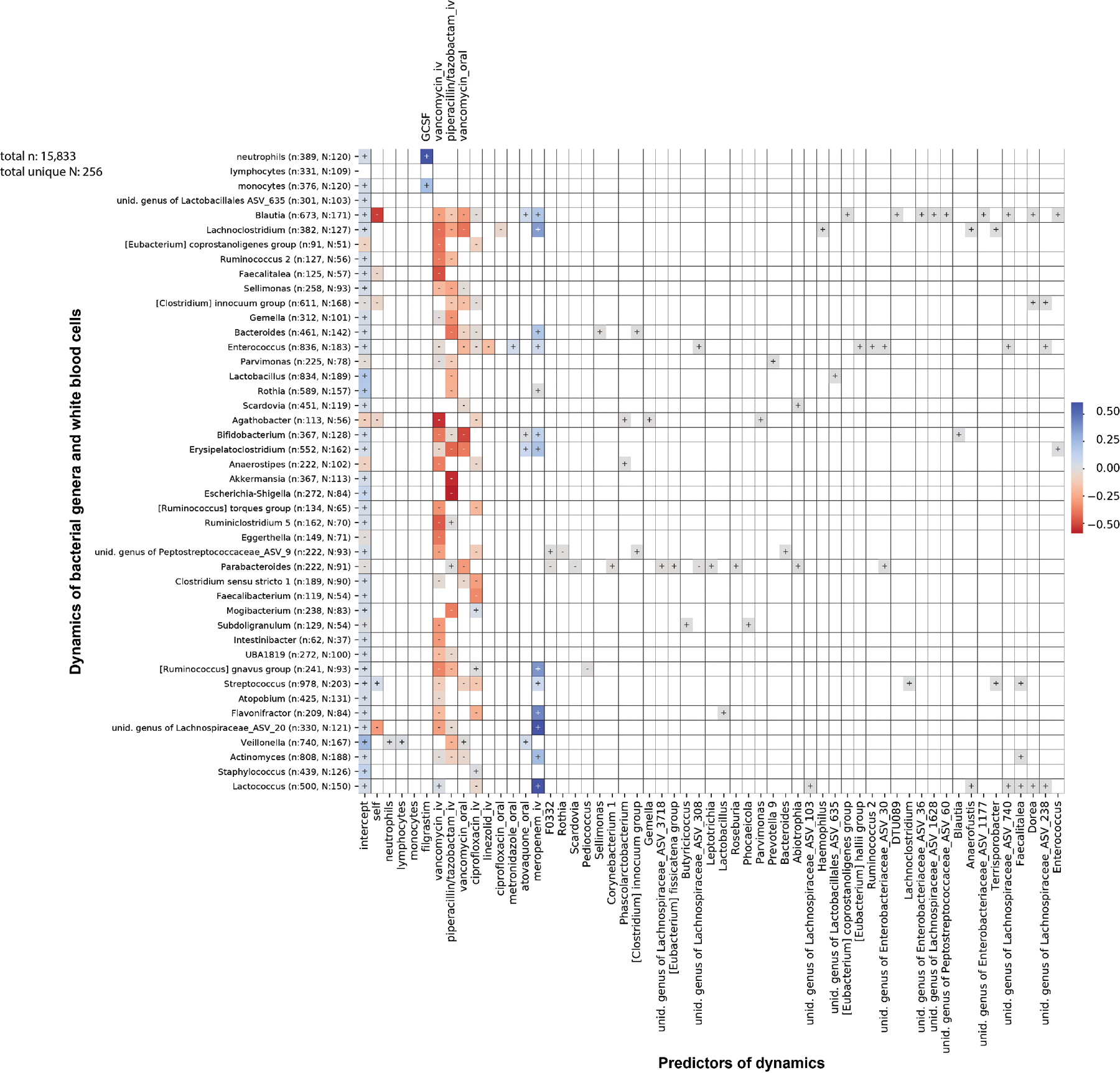
Strong regularization yields few non-zero coefficients and antibiotics dominate the dynamics.
Extended Data Fig. 7: Jointly inferred association network between white blood cell and bacterial genus dynamics with reduced regularization.
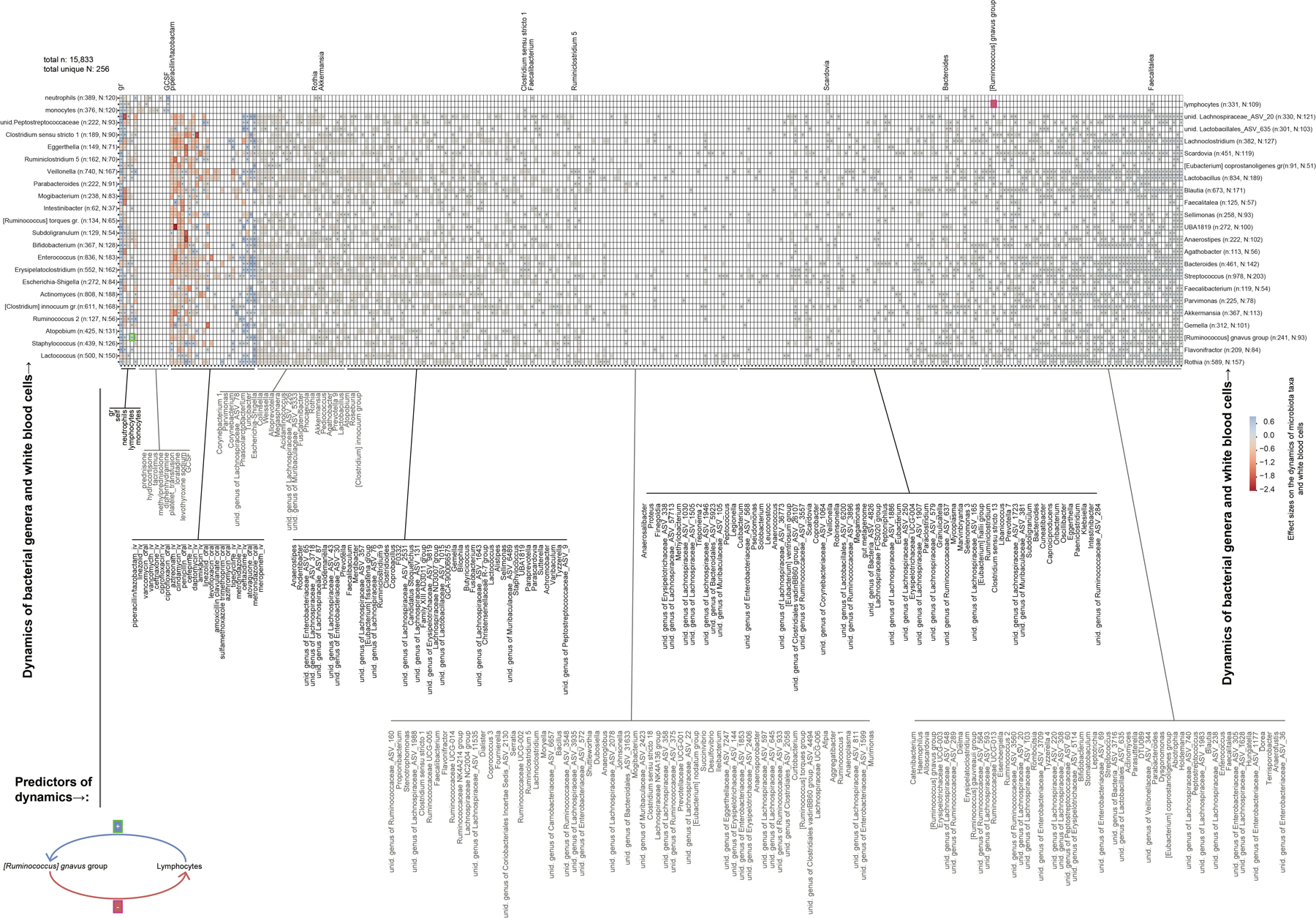
Reducing regularization strength (methods) indicates potential bidirectional feedbacks, e.g. between lymphocytes and [Ruminococcus] gnavus group (highlighter green boxes, and cartoon).
Extended Data Fig. 8: Functional analysis of microbiota samples.
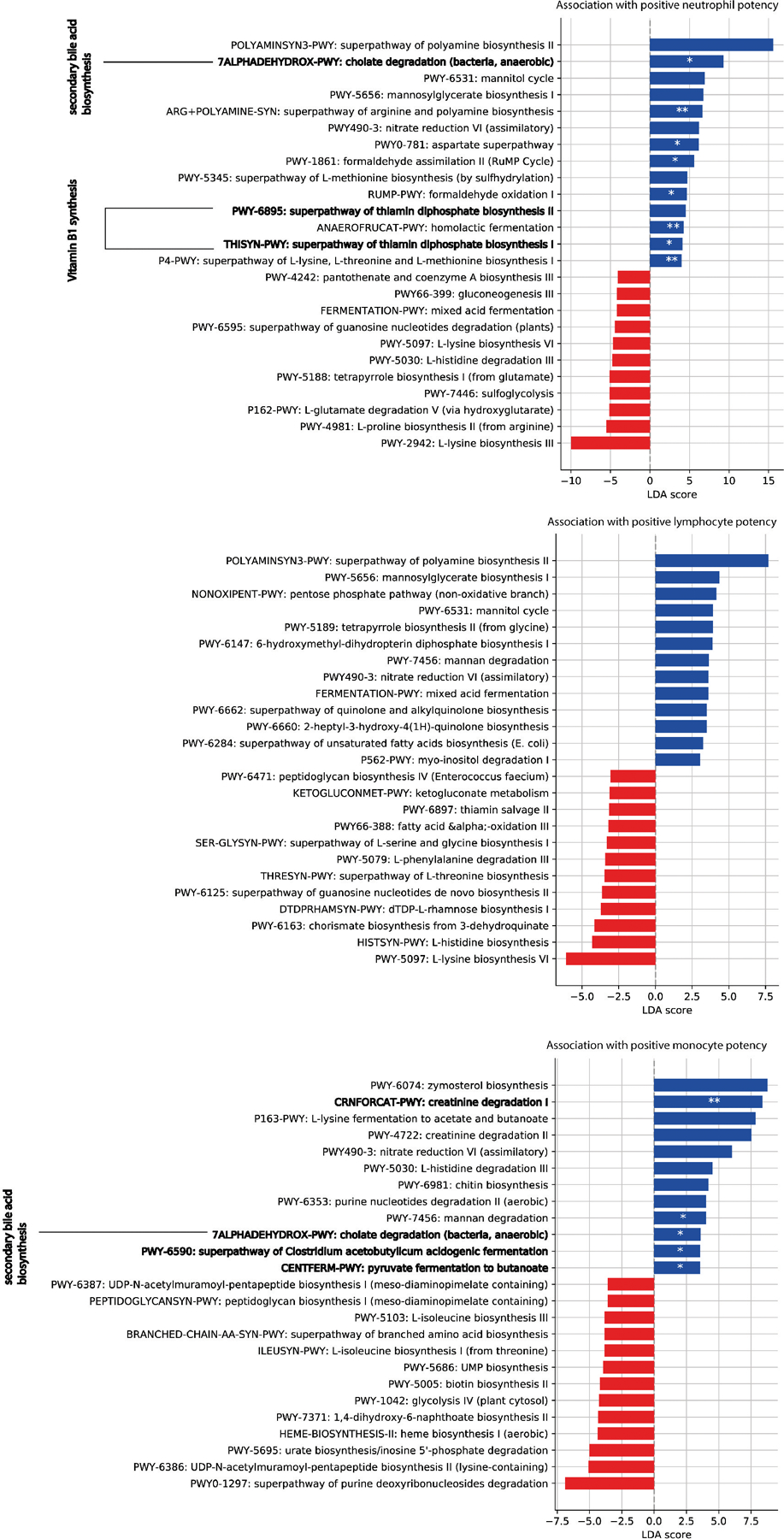
To distinguish samples predicted to increase rates of white blood cells, a microbiota potency score was calculated from posterior coefficients (Fig. 3, methods) and the relative abundance of taxa in samples. Bars show linear discriminant analysis (LDA) scores of MetaCyc pathway profiles from 124 shotgun sequenced samples that distinguished positive and negative potency samples the most (LDA-score magnitude in the 95th percentile). Highlighted pathways are discussed in the main text. For each pathway, we tested if pathway presence was enriched (depleted) in positive (negative) potency samples using one-sided Fisher’s exact test; p-value <0.001: ***, <0.01:**, <0.05:*.
Extended Data Fig. 9: Abundance profiles of bacterial genera across analyzed samples.
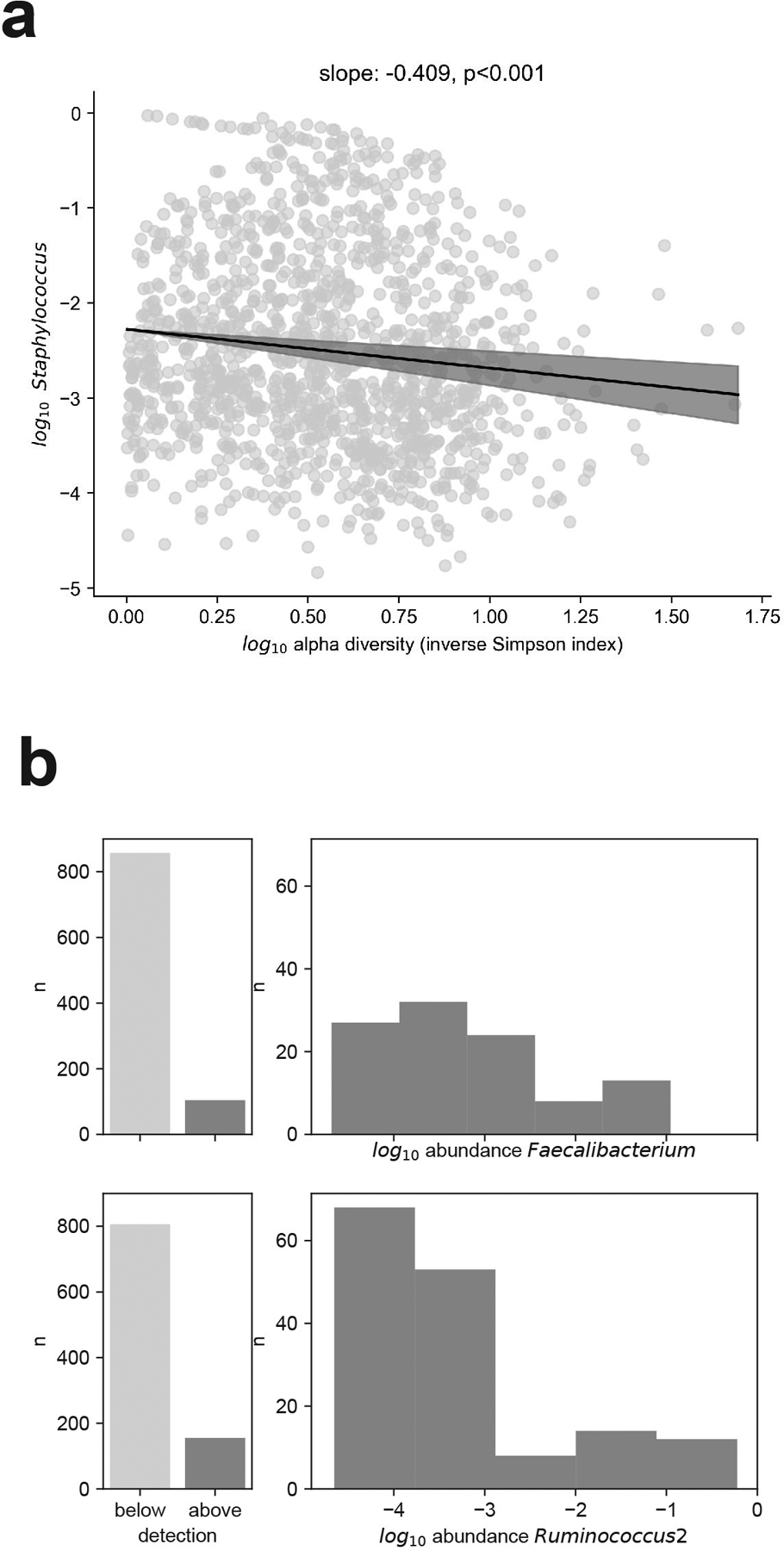
a, The relative non-zero abundance of Staphylococcus is inversely related to microbiome alpha diversity, bold line: regression line from a linear model of the mean of the log10 Staphylococcus relative abundance, shaded: 95% confidence intervals (n=1,381 samples with non-zero Staphylococcus abundances). b, c, Abundance profiles of the two genera, Faecalibacterium (b) and Ruminococcus 2 (c), most strongly associated with white blood cell increase; number of times detected (left) and log10 abundance distribution when above detection (right).
Extended Data Fig. 10: Survival analysis and confirmation of model results with different priors.
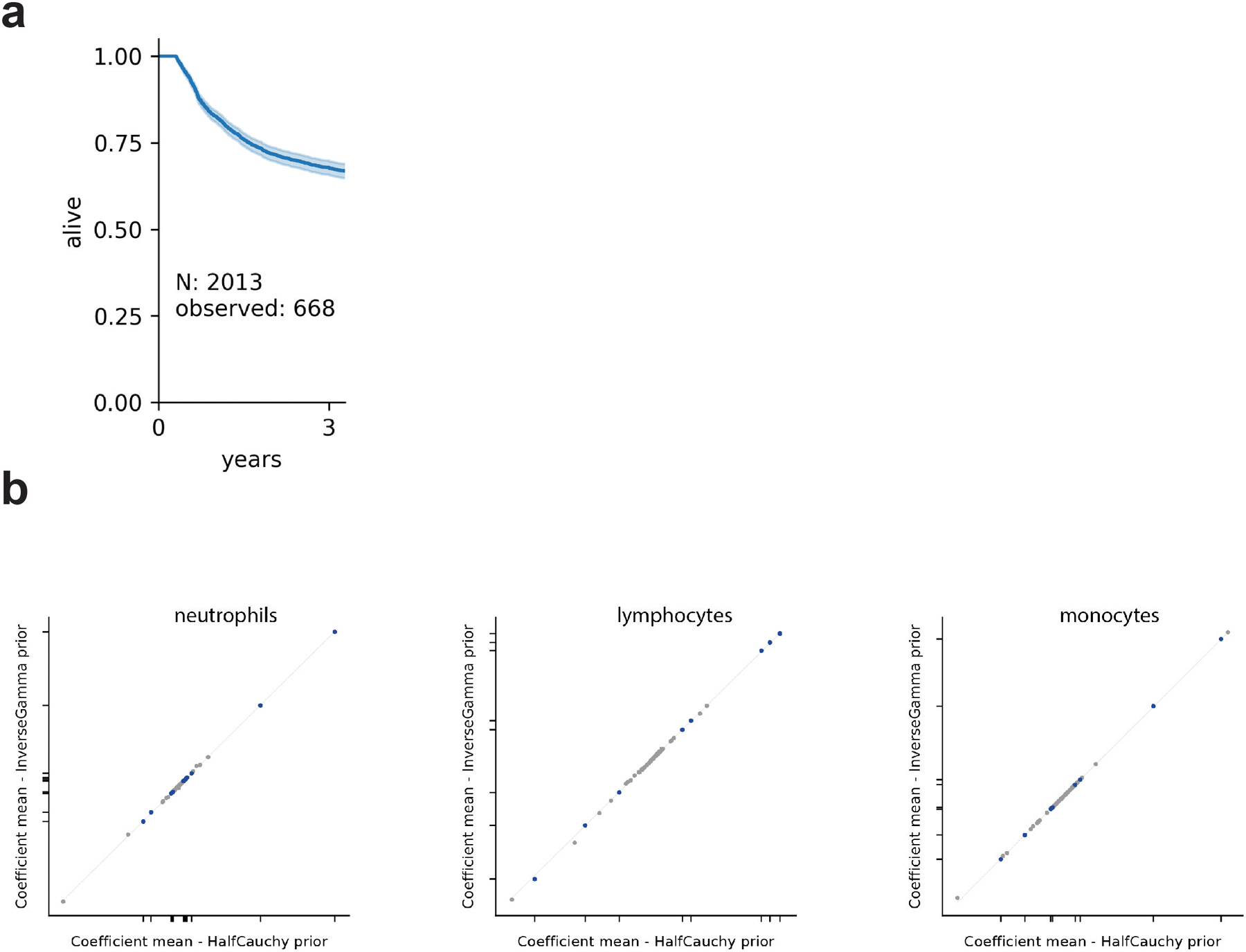
a, Kaplan-Meier plot of patient 3-year survival with sufficient available blood data (Supplementary Information, Extended Data Fig. 1). b, posterior association coefficients do not depend on the choice of prior for σ in the main Bayesian model. Plotted are the posterior means from our main analysis against the equivalent inference with an inverse Gamma prior (alpha=1, beta=1).
Supplementary Material
Acknowledgments
We thank Marc Lipsitch, Sandra B. Andersen, Kevin R. Foster, Jonathan Kevin Sia, Eric G. Pamer, Kat Coyte, Sibylle Mitschka and the members of the Xavier lab for helpful discussion and comments on the manuscript. This work was supported by the National Institutes of Health (NIH) grants U01 AI124275, R01 AI137269 and U54 CA209975 to JBX, by the MSKCC Cancer Center Core Grant P30 CA008748, the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center, the Sawiris Foundation, the Society of Memorial Sloan Kettering Cancer Center, MSKCC Cancer Systems Immunology Pilot Grant and Empire Clinical Research Investigator Program. MS received funding from the Burroughs Wellcome Fund Postdoctoral Enrichment Program, the Damon Runyon Physician-Scientist Award, and the Robert Wood Johnson Foundation. TMH is investigator in the Pathogenesis of Infectious Diseases from the Burroughs Wellcome Fund, and funded via an award from Geoffrey Beene Foundation, and NIH RO1 AI093808. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing Interests
MRMvdB and JUP received financial support from Seres Therapeutics. M-AP has received honoraria from AbbVie, Bellicum, Bristol-Myers Squibb, Incyte, Merck, Novartis, Nektar Therapeutics, and Takeda; has received research support for clinical trials from Incyte, Kite (Gilead) and Miltenyi Biotec; and serves on data and safety monitoring boards for Servier and Medigene and scientific advisory boards for MolMed and NexImmune.
Data Accessibility
- cGENUS.csv: relative taxon abundances in fecal microbiota samples from 12,633 stool samples
- cHCTMETA.csv: HCT characteristics
- cINFECTIONS.csv: positive blood culture results
- cMISAMPLES.csv: NCBI SRA accession number, diversity (inverse Simpson index), total 16S (where available), stool consistency for each fecal microbiota sample
- cMED.csv: medication data
- cPIDMETA.csv: anonymized patient demographics
- cWBC.csv: absolute counts of neutrophils, lymphocytes, monocytes, eosinophils, and platelets with indication if included in analyses
- cDUKE__GENUS.csv: relative taxon abundances in fecal microbiota samples from 12,633 stool samples
- cDUKE__WBC.csv: absolute counts of neutrophils, lymphocytes, monocytes, eosinophils, and platelets with indication if included in analyses
- cDUKE__MED.csv: medication data
- cFMT_analysis.csv: convenience table for Figure 2
- meta data: doi.org/10.6084/m9.figshare.12016986.v4
- 16S counts: doi.org/10.6084/m9.figshare.12016989.v3
- 16S taxonomy: doi.org/10.6084/m9.figshare.12016992.v1
Code Availability
All of the steps of the analyses that were performed in this study are described in detail to allow reproduction of the results. Relevant analysis code available publicly (https://github.com/jsevo/wbcdynamics_microbiome).
Supplementary Information
SI available online.
References
- 1.Mazmanian SK, Liu CH, Tzianabos AO & Kasper DL An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Gomez de Agüero M et al. The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Olin A et al. Stereotypic immune system development in newborn children. Cell 174, 1277–1292.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan TG et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 113, E8141–E8150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshmukh HS et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med 20, 524–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov II et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geva-Zatorsky N et al. Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd-Price J et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markey KA et al. The microbe-derived short-chain fatty acids butyrate and propionate are associated with protection from chronic GVHD. Blood 136, 130–136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azzouz D et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis 78, 947–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Routy B et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan V et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vétizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matson V et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanoue T et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Brandi G & Frega G Microbiota: Overview and Implication in Immunotherapy-Based Cancer Treatments. Int. J. Mol. Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin Yu J, Hubbard-Lucey VM & Tang J The global pipeline of cell therapies for cancer. Nat. Rev. Drug Discov 18, 821–822 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Morjaria S et al. Antibiotic-Induced Shifts in Fecal Microbiota Density and Composition during Hematopoietic Stem Cell Transplantation. Infect. Immun 87, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peled JU et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med 382, 822–834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taur Y et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staffas A et al. Nutritional Support from the Intestinal Microbiota Improves Hematopoietic Reconstitution after Bone Marrow Transplantation in Mice. Cell Host Microbe 23, 447–457.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savani BN et al. Absolute lymphocyte count on day 30 is a surrogate for robust hematopoietic recovery and strongly predicts outcome after T cell-depleted allogeneic stem cell transplantation. Biol. Blood Marrow Transplant 13, 1216–1223 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheiermann C, Frenette PS & Hidalgo A Regulation of leucocyte homeostasis in the circulation. Cardiovasc. Res 107, 340–351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson PA et al. Umbilical cord blood graft engineering: challenges and opportunities. Bone Marrow Transplant. 50 Suppl 2, S55–62 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Gabrilove JL et al. Effect of granulocyte colony-stimulating factor on neutropenia and associated morbidity due to chemotherapy for transitional-cell carcinoma of the urothelium. N. Engl. J. Med 318, 1414–1422 (1988). [DOI] [PubMed] [Google Scholar]
- 26.Belkaid Y & Hand TW Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schirmer M et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell 167, 1125–1136.e8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL & Foster KR Host selection of microbiota via differential adhesion. Cell Host Microbe 19, 550–559 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Hooper LV, Littman DR & Macpherson AJ Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palm NW et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henke MT et al. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 116, 12672–12677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okba AM et al. Neutrophil/lymphocyte ratio and lymphocyte/monocyte ratio in ulcerative colitis as non-invasive biomarkers of disease activity and severity. Auto Immun. Highlights 10, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi S-J et al. High neutrophil-to-lymphocyte ratio predicts short survival duration in amyotrophic lateral sclerosis. Sci. Rep 10, 428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y et al. Neutrophil/lymphocyte ratio is a more sensitive systemic inflammatory response biomarker than platelet/lymphocyte ratio in the prognosis evaluation of unresectable pancreatic cancer. Oncotarget 8, 88835–88844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hergott CB et al. Peptidoglycan from the gut microbiota governs the lifespan of circulating phagocytes at homeostasis. Blood 127, 2460–2471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith PM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balmer ML et al. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol 193, 5273–5283 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Ze X, Duncan SH, Louis P & Flint HJ Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster KR, Schluter J, Coyte KZ & Rakoff-Nahoum S The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y-Y et al. T Cell Recruitment to the Intestinal Stem Cell Compartment Drives Immune-Mediated Intestinal Damage after Allogeneic Transplantation. Immunity 51, 90–103.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerber GK The dynamic microbiome. FEBS Lett. 588, 4131–4139 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Jobin C Precision medicine using microbiota. Science 359, 32–34 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project. Nature 569, 641–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter J, Armet AM, Finlay BB & Shanahan F Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell 180, 221–232 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Caporaso JG et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murali A, Bhargava A & Wright ES IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6, 140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quast C et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinheiro JC, Bates DM, DebRoy SS & Sarkar D Nlme: Linear and Nonlinear Mixed Effects Models. (2013).
- 50.Tibshirani R Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society: Series B (Methodological) 58, 267–288 (1996). [Google Scholar]
- 51.Pedregosa F et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research (2011). [Google Scholar]
- 52.Salvatier J, Wiecki TV & Fonnesbeck C Probabilistic programming in Python using PyMC3. PeerJ Computer Science 2, e55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffman MD & Gelman A The No-U-Turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. Journal of Machine Learning Research (2014). [Google Scholar]
- 54.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


