Abstract
The rhesus macaque is an important animal model for AIDS and other infectious diseases. However, the investigation of Fc-mediated antibody responses in macaques is complicated by species-specific differences in Fcγ receptors (FcγRs) and IgG subclasses relative to humans. To assess the effects of these differences on FcγR-IgG interactions, reporter cell lines expressing common allotypes of human and rhesus macaque FcγR2A and FcγR3A were established. FcγR-mediated responses to B cells were measured in the presence of serial dilutions of anti-CD20 antibodies with Fc domains corresponding to each of the four subclasses of human and rhesus IgG and with Fc variants of IgG1 that enhance binding to FcγR2A or FcγR3A. All of the FcγRs were functional and preferentially recognized either IgG1 or IgG2. Whereas allotypes of rhesus FcγR2A were identified with responses similar to variants of human FcγR2A with higher (H131) and lower (R131) affinity for IgG, all of the rhesus FcγR3A allotypes exhibited responses most similar to the higher affinity V158 variant of human FcγR3A. Unlike responses to human IgGs, there was little variation in FcγR-mediated responses to different subclasses of rhesus IgG. Phylogenetic comparisons suggest that this reflects limited sequence variation of macaque IgGs as a result of their relatively recent diversification from a common IGHG gene since humans and macaques last shared a common ancestor. These findings reveal species-specific differences in FcγR-IgG interactions with important implications for investigating antibody effector functions in macaques.
Introduction
The Fcγ receptors (FcγRs) are a functionally diverse group of cell-surface glycoproteins that bind to the Fc domain of IgG antibodies. They include activating and inhibitory receptors with a range of affinities for IgG that are expressed by different hematopoietic cell types. Whereas inhibitory FcγRs have regulatory functions, the activating FcγRs mediate effector functions important for defense against infectious diseases and tumors. Upon recognition of antibody-antigen complexes, these receptors trigger cellular immune responses, including phagocytosis, cell-mediated cytotoxicity and cytokine release (1, 2).
Humans express up to six different FcγRs that belong to three different classes: FcγR1, FcγR2 (FcγR2A, FcγR2B and FcγR2C) and FcγR3 (FcγR3A and FcγR3B). Of these, the low affinity activating receptors FcγR2A and FcγR3A are responsible for cell-mediated antibody effector functions. FcγR2A (CD32a) is expressed by phagocytic cells, such as macrophages and neutrophils, and mediates the uptake of antibody-opsonized antigens or cells by antibody-dependent cellular phagocytosis (ADCP) (1). FcγR3A (CD16) is expressed on the surface of natural killer (NK) cells and some macrophages, and serves as the principal receptor for antibody-dependent cellular cytotoxicity (ADCC) (1). FcγR2C (CD32c) may also be expressed on NK cells and can contribute to ADCC, but is not present in most individuals due to a premature stop codon in the most common allele for this receptor (3, 4).
FcγR2A and FcγR3A differentially recognize four subclasses of human IgG (IgG1–4). Both of these receptors preferentially bind to IgG1 and IgG3, but generally exhibit weak or negligible interactions with IgG2 and IgG4 (5, 6). However, polymorphisms in FcγR2A and FcγR3A can affect IgG binding. There are two common variants of FcγR2A with either arginine (R) or histidine (H) at position 131 (7). Whereas the H131 polymorphism enhances binding to IgG2, the R131 variant binds poorly to this IgG subclass (5, 8). Interestingly, homozygosity for FcγR2A R131 has been associated with greater susceptibility to bacterial diseases and more rapid CD4+ T cell decline during HIV-1 infection (9–11). There are also two common variants of FcγR3A with either valine (V) or phenylalanine (F) at position 158. Compared to FcγR3A F158, the V158 variant exhibits higher affinity for IgG1 and IgG3 and is associated with greater efficacy of certain tumor immunotherapies (12, 13).
Although macaques express orthologs of FcγR2A and FcγR3A, sequence comparisons have identified polymorphisms that are not found in human FcγRs (14–19). Macaques also express four different subclasses of IgG with species-specific differences relative to human IgGs (20, 21). Similar to human IgGs, the macaque IgGs are numbered according to their relative abundance in serum (IgG1>IgG2>IgG3>IgG4); however, they are more similar to one another than to human IgGs and exhibit more limited sequence and structural diversity (20, 22, 23). As a result of these FcγR and IgG differences, it is not possible to predict FcγR-IgG interactions in macaques based on the interactions of these molecules in humans. Thus, one cannot assume that human antibodies will have the same FcγR-mediated functions in macaques as they do in humans or that antibody responses elicited in macaques will accurately reflect antibody effector functions in humans.
As macaques have become increasingly important models for the pre-clinical evaluation of antibody-based vaccines and therapies for HIV, dengue virus, Zika virus, SARS-CoV-2, and other infectious diseases (24–33), a better understanding of the molecular interactions of macaque FcγRs with human and macaque antibodies is needed for investigating antibody-mediated effector functions in these species. While biophysical studies with recombinant proteins corresponding to the extracellular domains of rhesus macaque FcγRs have revealed differences in affinity for human and macaque IgGs (34, 35), the functional interactions of antibodies with these receptors have not been assessed. This is especially important as the stimulation of responses through low affinity FcγRs is dependent on the formation of immune complexes with multiple FcγRs and cellular studies with human FcγRs and IgG subclass variants have revealed functional interactions that are not detectable with soluble, monomeric FcγRs by biophysical methods such as surface plasmon resonance (36). In the present study, we therefore establish reporter cell lines transduced with selected allotypes of human and macaque FcγR2A and FcγR3A that represent the most common variants of these receptors and quantitatively compare responses to different subclasses and Fc variants of human and rhesus macaque IgG.
Materials and Methods
Ethics Statement
Indian origin rhesus macaques (Macaca mulatta) were used in this study. The animals were housed at the Wisconsin National Primate Research Center (WNPRC) according to the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) and the University of Wisconsin Research Animal Resources Center (UWRARC) standards. Animal experiments were approved by the UWRARC (protocol number G005141) and conducted in compliance with the principles described in the Guide for the Care and Use of Laboratory Animals (37).
Cell lines
CEM.NKR-CCR5 cells (NIH AIDS Reagent Program) (38, 39) and Raji cells (ATCC) were cultured in RPMI medium supplemented with 10% FBS, L-glutamine and penicillin/streptomycin (R10 medium). Jurkat NFAT-Luciferase (JNL) cells (Signosis) were cultured in R10 medium plus 0.2 mg/ml hygromycin B (Dot Scientific). GP2–293 cells (Clontech Laboratories) were cultured in DMEM supplemented with 10% FBS, L-glutamine and penicillin/streptomycin (D10 medium). KHYG-1 cells transduced with rhesus macaque CD16 were maintained in R10 medium with 1 μg/ml cyclosporine A (Sigma Aldrich), 10 U/ml recombinant human IL-2 (R&D systems) and primocin (Invitrogen) as described previously (38, 40, 41).
JNL cells expressing human and rhesus macaque (Macaca mulatta) Fcγ receptors (FcγRs) were established by retroviral transduction. Full-length cDNA sequences for FCGR2A and FCGR3A alleles were either synthesized (ThemoFisher GeneArt or IDT) or amplified from mRNA isolated from PBMCs by RT-PCR using the Protoscript AMV First Strand cDNA synthesis Kit (New England Biolabs) and cloned into the retroviral vector pQCXIP. These constructs were packaged into VSV-G- pseudotyped murine leukemia virus (MLV) particles by co-transfecting GP2–293 cells with pQCXIP-FCGR2A or -FCGR3A clones and pVSV-G using GenJet In Vitro DNA Transfection Reagent (Ver. II) (Signagen) according to the manufacturer’s instructions. The cell culture medium was replaced after overnight incubation and virus-containing supernatant was collected the following day, centrifuged at 1,200 g to remove cell debris and concentrated using Amicon Ultra-15 Centrifugal Units with a 50 kDa cutoff (Millipore Sigma). JNL cells (5×105) were incubated overnight with 300 μl concentrated viral supernatant and brought to a volume of 1 ml with 40 μg/ml polybrene (Millipore Sigma). The following day, the cells were placed under selection in R10 medium containing 0.8 μg/ml puromycin (Invitrogen). After selection for puromycin resistance, FcγR expression was confirmed by flow cytometry. The GenBank accession numbers for human FCGR sequences are as follows: FCGR2A H131 (NP_001129691), FCGR2A R131 (AAA35932.1), FCGR3A V158 (HQ447137), and FCGR3A F158 (NM_000569). Accession numbers for rhesus FCGR sequences are as follows: FCGR2A:01:01 (MT304962), FCGR2A:02:01 (MT304964), FCGR2A:08:01 (MT304968), FCGR2A:10:01 (MT304970), FCGR2A:11:01 (MT304972), FCGR2A:12:01 (MT304974), FCGR2A:13:01 (MT304976), FCGR3A:01:01 (MT305008), FCGR3A:01:02 (MT305010), FCGR3A:02:01 (MT305014) and FCGR3A:03:01 (MT305018).
Monoclonal Antibodies
For anti-CD20 antibodies, cDNA sequences for the variable heavy (VH) and variable light (VL) domains of rituximab were cloned into separate pCEP4 (Invitrogen) constructs in-frame with expression-optimized sequences for the constant domains of human and rhesus macaque (IgGH1–4) and the corresponding kappa light chains (IgGκ) of each species, respectively. Human IgHG1–4 and rhesus macaque IgHG3–4 cDNAs were synthesized (IDT). Rhesus macaque IgHG1 and IgHG2 cDNAs were provided by Dr. Michael Farzan (The Scripps Research Institute, Jupiter, FL). The DEL substitutions were introduced into IgGH1 during cDNA synthesis and the G236A substitution was added to IgGH1 constructs by oligonucleotide-directed PCR mutagenesis. The GenBank accession numbers for antibody sequences are as follows: rituximab VH (AX556949), rituximab VL (AX556951), human IgG1H (AX556949), human IgG2H (AAG00910.2), human IgG3H (P01860.2), human IgG4H (BC025985.1), human IgGκ (AX556951), rhesus macaque IgG1H (AAQ57550.1), rhesus IgG2H (AAQ57565.1), rhesus IgG3H (ATV90898.1), rhesus IgG4H (MT310723) and rhesus IgGκ (ACN96964.1).
Monoclonal antibodies were produced by co-transfecting Expi293 cells (9×107 cells) with pCEP4 constructs expressing IgG heavy and light chains in small-scale (30 ml) cultures using serum-free medium and the Expifectamine transfection kit according to the manufacturer’s instructions (Gibco). Cell culture supernatants were harvested eight days later, clarified by centrifugation and filtered with 0.45 μM PVDF syringe filters (Millipore Sigma). The mAbs were purified using protein-A Gravitrap columns, or in the case of IgG3 protein-G Gravitrap columns (GE Healthcare Life Sciences). The columns were washed with tris-buffered saline (TBS) and bound antibodies were eluted with Gentle Elution Buffer (ThermoFisher). The antibodies were concentrated, and the buffer was exchanged with 50 mM sodium citrate using Amicon Ultra-15 Centrifugal Units with a 50 kDa cutoff (Millipore Sigma) before examination by SDS-PAGE with colloidal Coomassie stain. All antibody solutions were treated with 0.02% NaN3 for storage and centrifuged prior to use.
FCGR genotyping
Rhesus macaques were genotyped for Mamu-FCGR2A and Mamu-FCGR3A transcripts as described previously (18). Briefly, RNA was isolated from fresh whole blood or cryopreserved PMBC using Maxwell 16 LEV simplyRNA Blood or Cells Kits (ProMega). The SuperScript First-Strand Synthesis System (Thermo Fisher Scientific) was used to generate templates for cDNA/PCR with Phusion High-Fidelity PCR Master Mix (New England BioLabs) and barcoded primers that flank complete coding sequences (18). After normalization and pooling, SMRTbell libraries were prepared and sequenced on a PacBio Sequel instrument at the University of Wisconsin-Madison Biotechnology Center according to manufacturer’s protocols (Pacific BioSciences). Raw PacBio sequence datasets were processed using SMRT Link v6.0.0 tools (https://www.pacbio.com/support/software-downloads) as described recently by Shortreed et al (42). The resulting circular consensus sequences from each animal were mapped against a reference database of all known rhesus macaque FCGR2A and FCGR3A sequences using Geneious Prime 2020.0.4 software tools (Biomatters).
Flow cytometry
i. FcγR staining.
JNL cells were stained with Live/Dead NearIR or Aqua Live/Dead amine-reactive compound (ARC) (Invitrogen), washed in PBS with 1% FBS (FACS buffer) to quench the ARC, and stained with PE-conjugated anti-CD32 antibody (clone 2E1, Miltenyi Biotech) to detect human FcγR2A or goat anti-CD32 polyclonal antibody (R&D Systems) followed by AF647-conjugated rabbit anti-goat antibody (Jackson Immunoresearch) in the presence of 100 μg/ml rabbit IgG (Sigma) to detect rhesus macaque FcγR2A. Human and rhesus macaque FcγR3A were detected by staining with PE-conjugated anti-CD16 antibody (clone 3G8, Becton-Dickinson). CD20 was detected by staining CD20-transduced CEM.NKR-CCR5 cells with an A700-conjugated anti-CD20 antibody (clone 2H7, BD Pharmingen). Samples were washed with FACS buffer and fixed in 2% paraformaldehyde (PFA) before flow cytometry. The mean fluorescence intensity (MFI) of FcγR staining was measured after gating on the live singlet cell population. Data was collected using a BD LSRII SORP instrument and analyses were performed with FlowJo 9.9 software (TreeStar Inc.).
ii. CD20 staining.
Raji cells were washed with PBS and stained with Aqua Live/Dead ARC. After 20 min, cells were washed in FACS buffer and stained for 30 mins with anti-CD20 IgG (5 μg/ml) followed by an AF647-conjugated goat anti-human IgG polyclonal antibody. As a control for non-specific staining, cells were also stained with the goat anti-human IgG secondary antibody without the anti-CD20 primary antibody. Samples were washed with FACS buffer and fixed in 2% PFA. The MFI of CD20 staining was measured after gating on the live singlet cell population and divided by the secondary-only control to measure the fold-change in MFI. Data was collected using a BD LSRII SORP instrument and analyzed with FlowJo 9.9 software.
JNL reporter cell assay
FcγR interactions with IgGs were measured by the dose-dependent upregulation of luciferase by Jurkat NFAT-luciferase (JNL) cells expressing FcγRs in response to antibody-opsonized target cells over a range of IgG concentrations. FcγR-transduced JNL cells (2×105) were co-incubated with Raji cells (1×105) in the presence of serial dilutions of anti-CD20 IgG antibodies. JNL cells were incubated overnight or for 4 hours with Raji cells in triplicate wells at each antibody concentration (200 μl/well final volume) in opaque white, flat-bottom, 96-well plates (Corning). At the end of the incubation, 50 μl BriteLite Plus luciferase substrate (PerkinElmer) was added to each well and luminescence was measured using a Victor X4 multiplate reader (PerkinElmer). Ramos cells (ATCC), which do not express FcγR2B, were also tested for antibody-mediated recognition by FcγR-transduced JNL cells. The different IgG subclasses and Fc domain variants of human and rhesus macaque IgG were run in parallel for each experiment.
ADCC assays
Rhesus macaque PBMCs were isolated from peripheral blood by centrifugation over Ficoll Paque Plus (GE Healthcare) and incubated overnight in plastic T75 flasks laid on their side to deplete monocytes and other adherent cell types. Raji cells or CD20-transduced CEM-NKR.CCR5 cells were labelled with 10 μM calcein acetoxymethyl ester (CAM) (ThermoFisher) for 1 hour at 37 °C, washed with PBS, and resuspended in phenol red-free R10 medium containing 10 U/ml recombinant IL-2. KHYG-1 cells transduced with rhesus macaque FcγR3A:02 (5×104) (38) or macaque PBMCs (2×105) were co-incubated with CAM-labelled Raji or CD20+ CEM-NKR.CCR5 cells (1×104) in the presence of serial dilutions of anti-CD20 IgG. The cells were incubated for four hours in triplicate wells at each antibody concentration (200 μl/well final volume) in round-bottom, 96-well plates. Wells with only CAM-labelled target cells served as controls for background CAM release and wells treated with 1% NP-40 (Sigma) indicated maximum CAM release. After four hours, the plates were centrifuged (500 g) and 100 ul of supernatant was transferred from each well to corresponding wells of black, clear-bottom, 96-well plates. The release of CAM into the supernatant was measured using a Victor X4 multiplate reader (excitation 485 nm and absorbance 535 nm). Percent-specific lysis was calculated as (experimental release – spontaneous release)/(maximum release – spontaneous release) × 100.
Phylogenetic analysis of IGHG sequences
Chimpanzee IGHG sequences were located using the NCBI Genome Data Viewer and exons with high similarity to human IGHG sequences were extracted. The LOCI identifiers for the chimpanzee IGHG sequences are as follows: IgHG1 (LOC749366), IgHG2 (LOC453229), IgHG3 (LOC749390), IgHG4 (LOC749354). The accession numbers for baboon, pigtail macaque, and mouse IGHG sequences are as follows: baboon IgHG1 (AY125048), baboon IgHG2 (AY125049), baboon IgHG3 (AY125050), baboon IgHG4 (AY125051), pigtail macaque IgHG1 (JQ868732), pigtail macaque IgHG2 (JQ868733), pigtail macaque IgHG3 (JQ868734), pigtail macaque IgHG4 (JQ868735), mouse IgG1 (LT160966), mouse IgG2a (AB097847). Human and rhesus macaque IGHG accession numbers are provided above. The accession numbers for baboon, pigtail macaque, chimpanzee, and mouse FcγR sequences are as follows: baboon FcγR1 (XP_009178311.1), baboon FcγR2A (XM_021926122.2), baboon FcγR3A (NM_001112647.1), pigtail macaque FcγR1 (XP_011735031.1), pigtail macaque FcγR2A (KF234400), pigtail macaque FcγR3A (XM_011770191.2), chimpanzee FcγR1 (XP_009428355.1), chimpanzee FcγR2A (NM_001009077.1), chimpanzee FcγR3A (XM_024349652.1), mouse FcγR2B (NM_001077189.1). Human and rhesus macaque FcγR1 accession numbers are (P12314) and (AFD32558.1) respectively. Human and rhesus FcγR2A and FcγR3A accession numbers are provided above using the most frequent rhesus alleles. Amino acid sequences were aligned using Geneious and subjected to a substitution model test using MEGAX. A phylogenetic tree was constructed with MEGAX using the maximum likelihood method and Jones-Taylor-Thorton substitution model with 1,000 bootstrap replicates. Nodes with bootstrap values less than 0.5 were collapsed using TreeGraph (version 2.15.0–887 beta).
Statistical analysis
Graphing and statistical analysis were performed using GraphPad Prism (Version 8.2). Antibody concentrations for half-maximal responses (EC50) were calculated from sigmoidal curves fit to mean RLU data from triplicate wells at each antibody dilution. Mean EC50 values were calculated from at least three independent experiments and were compared using a two-tailed, unpaired student’s t-test.
Results
Rhesus macaque FcγR2A and FcγR3A polymorphisms
Analysis of full-length FCGR cDNA sequences from 17 Indian-origin rhesus macaques identified seven FCGR2A and four FCGR3A allelic variants. Table I provides the relative frequencies of these FCGR sequences in an expanded cohort of 155 rhesus macaques from the Wisconsin National Primate Research Center. The predicted amino acid sequences of these rhesus macaque alleles are aligned with human FcγR2A and FcγR3A in Figure 1. In addition to species-specific differences relative to human FcγR2A (huFcγR2A), polymorphisms in rhesus macaque FcγR2A (rhFcγR2A) were apparent at multiple positions, including residues predicted to contact IgG (Fig. 1A). Whereas five of the seven allotypes of rhFcγR2A have a histidine at position 131, which corresponds to the H131 polymorphism in huFcγR2A that enhances binding to IgG2, two of the rhFcγR2A variants (rhFcγR2A:11 and rhFcγR2A:12) have a proline at this position (P131) (Fig. 1A). Along with these species-specific polymorphisms, all allotypes of rhFcγR2A have a potential N-linked glycosylation site (PNG) at position 135. Two allotypes, rhFcγR2A:01 and rhFcγR2A:13, have an additional PNG at position 128 (Fig. 1A).
Table I. Rhesus macaque FcγR allele frequencies.
Frequencies of rhesus macaque FCGR2A and FCGR3A alleles. The frequencies of the FCGR2A and FCGR3A alleles identified in 17 Indian-origin rhesus macaques were determined relative to an internal database of FCGR sequences from 155 animals.
| FCGR2A Allele | Haplotypes | Frequency (%) | Accession # |
|---|---|---|---|
| 2A:01:01 | 94/302 | 31.1 | MT304962 |
| 2A:02:01 | 46/302 | 15.2 | MT304964 |
| 2A:08:01 | 51/302 | 16.9 | MT304968 |
| 2A:10:01 | 71/302 | 23.5 | MT304970 |
| 2A:11:01 | 8/302 | 2.6 | MT304972 |
| 2A:12:01 | 10/302 | 3.3 | MT304974 |
| 2A:13:01 | 4/302 | 1.3 | MT304976 |
| other FCGR2A | 18/302 | 6.0 | - |
| FCGR3A Allele | Haplotypes | Frequency (%) | Accession # |
| 3A:01:01 | 87/250 | 34.8 | MT305008 |
| 3A:01:02 | 23/250 | 9.2 | MT305010 |
| 3A:02:01 | 125/250 | 50.0 | MT305014 |
| 3A:03:01 | 11/250 | 4.4 | MT305018 |
| other FCGR3A | 4/250 | 1.6 | - |
FIGURE 1.

Amino acid sequence alignments of human and rhesus macaque FcγR2A (A) and FcγR3A (B) allotypes. Predicted IgG contact sites based on crystal structures of IgG in complex with human FcγR2A and FcγR3A are shaded gray (57, 58). Amino acid positions are indicated with dots for identity, dashes for deletions and single-letter amino acid codes for differences.
Rhesus FcγR3A (rhFcγR3A) allotypes have fewer amino acid differences relative to human FcγR3A (huFcγR3A) and exhibit considerably less polymorphism than rhFcγR2A. The three most common allotypes of rhFcγR3A differ from huFcγR3A by 20–21 amino acids, including three positions predicted to contact IgG (Fig. 1B). The only extracellular domain polymorphism among these variants is a conservative isoleucine versus valine difference at position 158, which is at the same position as the V158F polymorphism in huFcγR3A. This residue is an isoleucine (I158) in rhFcγR3A:01 and rhFcγR3A:02 and a valine (V158) in rhFcγR3A:03 (Fig. 1B). Additional polymorphisms previously implicated in the efficiency of B cell depletion with an anti-CD20 antibody were observed in the transmembrane and cytoplasmic domains of rhFcγR3A:01 (M211 and I215) (Fig. 1B) (43).
FcγR2A responses to human and rhesus macaque IgG subclasses and Fc variants
To assess FcγR2A interactions with IgG, reporter cell lines were established by transducing Jurkat cells that contain a NFAT-inducible luciferase reporter gene (JNL cells) with different alleles of human and rhesus macaque FCGR2A. These alleles encoded the H131 and R131 variants of huFcγR2A and five of the most common allotypes of rhFcγR2A (rhFcγR2A:01, rhFcγR2A:02, rhFcγR2A:08, rhFcγR2A:10 and rhFcγR2A:11), which represent 89% of the haplotypes of FCGR2A genotyped animals (Table I & Supplemental Fig. 1A). Recombinant IgGs were also produced with identical variable regions from the anti-CD20 antibody rituximab, but different heavy chain constant domains corresponding to each of the four subclasses of human and rhesus macaque IgG (huIgG1–4 and rhuIgG1–4) (Supplemental Fig. 2A–D). Additional variants of IgG1 were produced with Fc domain substitutions that selectively enhance binding to FcγR2A (G236A) or FcγR3A (DEL: S239D, I332E and A330L) (44–46). The purity of these antibodies was verified by SDS-PAGE and similar binding to CD20 on the surface of Raji cells was confirmed by flow cytometry (Supplemental Fig. 2E & 2F). FcγR2A-transduced JNL cells were incubated overnight with Raji cells in the presence of serial dilutions of anti-CD20 antibodies and FcγR ligation was measured by the dose-dependent upregulation of luciferase over a 6-log range of antibody concentrations (Fig. 2A). Antibody concentrations for half-maximal responses, or 50% effective concentrations (EC50), were compared to determine the relative strength of FcγR-IgG interactions (Table II). Raji cells maintained consistently high levels of CD20 expression during passage in culture as reflected by similar CD20 staining on three different occasions separated by at least three-week intervals (Supplemental Fig. 1C).
FIGURE 2.
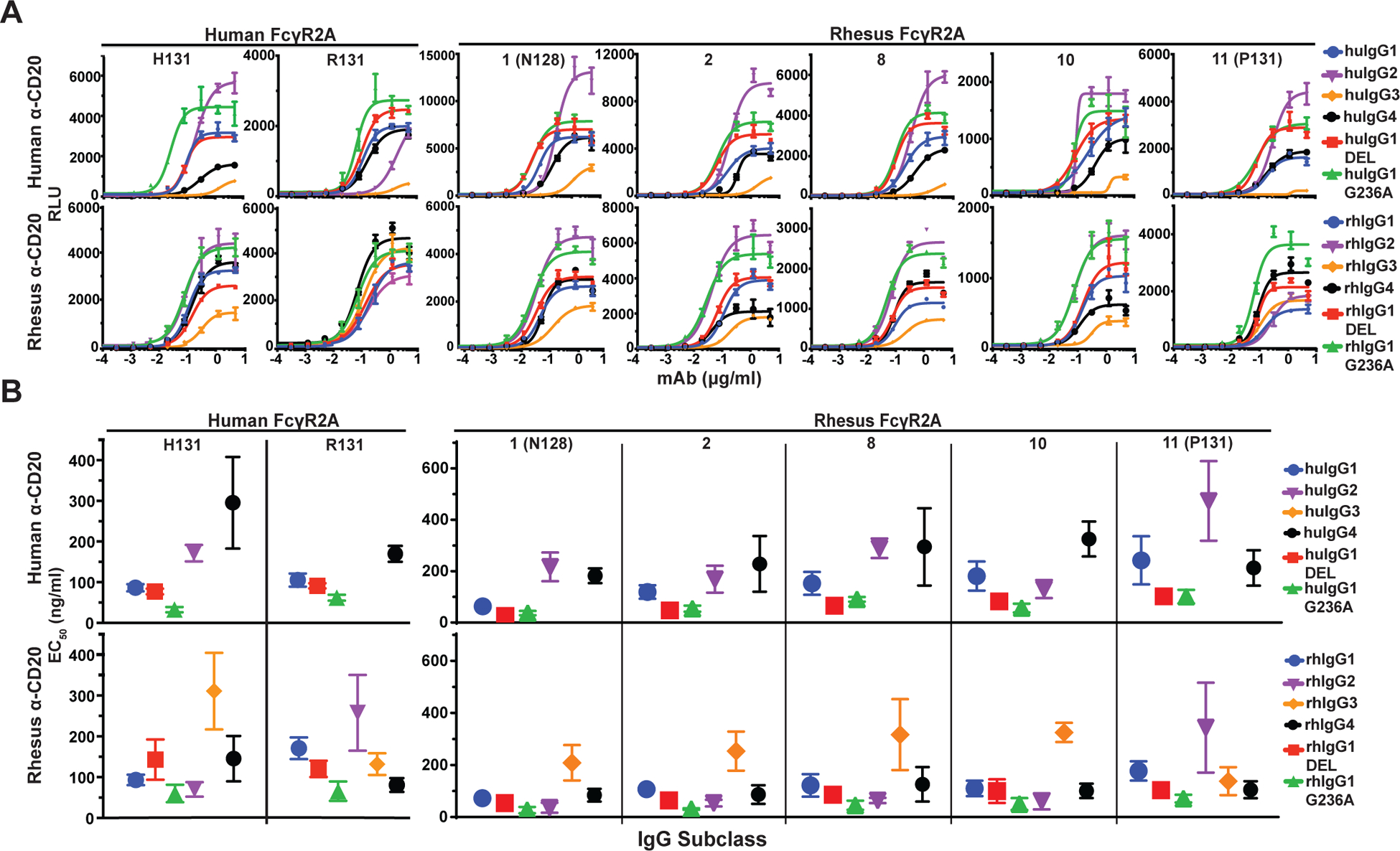
FcγR2A-mediated responses. (A) Jurkat NFAT-luciferase cells expressing individual allotypes of human and rhesus macaque FcγR2A were incubated overnight with Raji cells at a 2:1 E:T ratio in the presence of serial dilutions of anti-CD20 antibodies with Fc domains corresponding to each of the four subclasses of human and rhesus macaque IgG and variants of IgG1 that enhance binding to FcγR2A (G236A) or FcγR3A (DEL). Curves were fit to the data using GraphPad Prism software. Error bars indicate SD of the mean for triplicate measurements at each antibody concentration. (B) Antibody concentrations for half-maximal responses (EC50) were calculated from dose response curves. EC50 values that could not be determined within the range of antibody concentrations tested are not shown. Error bars indicate SD of the mean for at least three independent experiments.
Table II. Average EC50 values from JNL dose-response curves.
EC50 values for FcγR2A- and FcγR3A-mediated responses. Average antibody concentrations for half-maximal responses (EC50) and standard deviations of the mean (±) were calculated from at least three independent experiments with each allotype of FcγR2A and FcγR3A in combination with each subclass of human and rhesus IgG (top panels) and Fc domain variants of IgG1 with G236A and DEL substitutions (bottom panels).

|
As expected, JNL cells expressing huFcγR2A exhibited the strongest responses to huIgG1 and these interactions were significantly enhanced by the G236A substitution (Fig. 2B). P-values for statistical comparisons are shown in Table III. Moreover, consistent with previous observations (5, 8, 47–50), JNL cells expressing huFcγR2A H131 responded significantly better to huIgG2 than the R131 variant of this receptor (Fig. 2B & Table III). Weaker huFcγR2A H131-mediated responses to huIgG4 are also consistent with previous observations (5). However, the responses to huIgG3 were surprisingly weak (Fig. 2A) (5, 35). The reason for this difference is presently unclear, since the purity and integrity of huIgG3 was verified by SDS-PAGE (Supplemental Fig. 2F) and this antibody stimulated FcγR3A responses in subsequent experiments (Fig. 3A). These results suggest context-dependent differences in the activity of huIgG3 that may be related to the greater flexibility afforded by the extended hinge region of this antibody.
Table III. Statistical comparisons of EC50 values.
Statistical comparison of EC50 values for FcγR2A-mediated responses. The p-values are shown for statistical comparisons of mean antibody concentrations (EC50) for half-maximal responses and were determined using an unpaired, two-tailed Student’s t test. Dashes indicate comparisons with undefined EC50 values.

|
FIGURE 3.
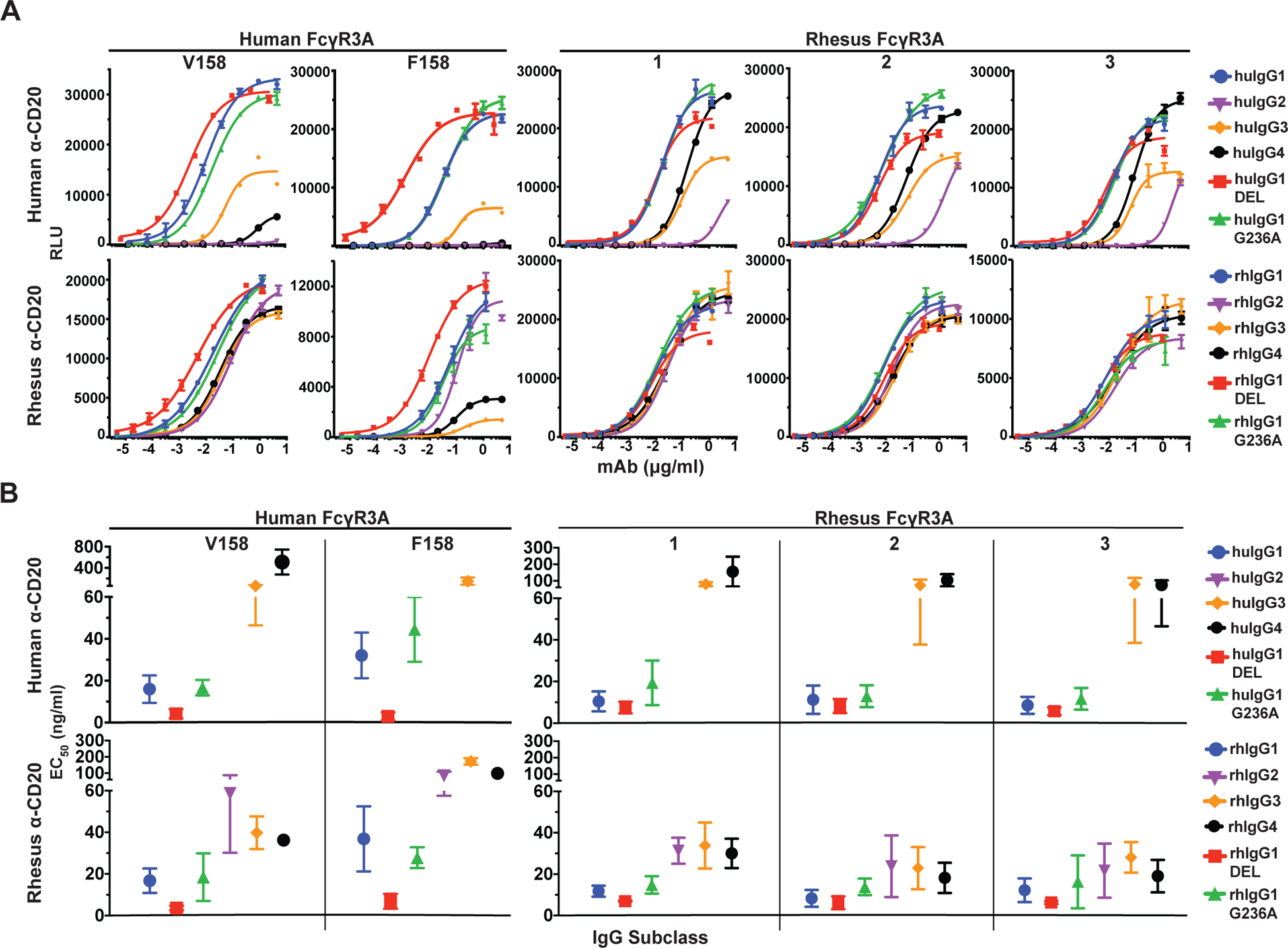
FcγR3A-mediated responses. (A) Jurkat NFAT-luciferase cells expressing individual allotypes of human and rhesus macaque FcγR3A were incubated overnight with Raji cells at a 2:1 E:T ratio in the presence of serial dilutions of anti-CD20 antibodies with Fc domains corresponding to each of the four subclasses of human and rhesus macaque IgG and variants of IgG1 that enhance binding to FcγR2A (G236A) or FcγR3A (DEL). Curves were fit to the data using GraphPad Prism software. Error bars indicate SD of the mean for triplicate measurements at each antibody concentration. (B) Antibody concentrations for half-maximal responses (EC50) were calculated from dose response curves. EC50 values that could not be determined within the range of antibody concentrations tested are not shown. Error bars indicate SD of the mean for at least three independent experiments.
Consistent with the presence of histidine at position 131 in four of the five allotypes of rhFcγR2A (rhFcγR2A:01, rhFcγR2A:02, rhFcγR2A:08 and rhFcγR2A:10), these receptors exhibited similar responses to huIgG1 and huIgG2 as huFcγR2A H131 (Fig. 2A). With the exception of rhFcγR2A:10, which preferentially recognized huIgG2, these rhFcγR2A variants responded more efficiently to huIgG1 than huIgG2 (Fig. 2B & Table II). All of the rhFcγR2A variants also recognized huIgG4 (Fig. 2B). The G236A substitution had a more modest effect on JNL cells bearing rhFcγR2A than huFcγR2A. Although the G236A substitution reduced the antibody concentration for half-maximal responses for all five allotypes of rhFcγR2A compared to wild-type IgG1 (Fig. 2B & Table II), this difference was only significant for rhFcγR2A:02 (Table III). These observations are in accordance with a recent study showing that the G236A substitution does not increase huIgG1 binding to rhFcγR2A to the same extent as its human counterpart (35). However, contrary to binding studies with recombinant FcγRs, which suggest that the N128 and P131 variants of rhFcγR2A are functionally impaired (34), JNL cells expressing receptors with these polymorphisms respond to all four subclasses of huIgG. As reflected by their EC50 values, rhFcγR2A:01 (N128) and rhFcγR2A:11 (P131) exhibited the strongest and the weakest responses to huIgG1, respectively (Fig. 2B & Table II).
JNL cells expressing rhFcγR2A responded better to rhIgGs than huIgGs and exhibited less variation among subclasses. The pattern of rhIgG recognition by different allotypes of rhFcγR2A also bore a striking resemblance to the pattern of rhIgG recognition by each of the huFcγR2A variants. Similar to huFcγR2A H131, all four allotypes of rhFcγR2A with the H131 polymorphism (rhFcγR2A:01, rhFcγR2A:02, rhFcγR2A:08 and rhFcγR2A:10) responded most efficiently to rhIgG2 (Fig. 2B). These rhFcγR2A variants recognized rhIgG1 and rhIgG4 at similarly low EC50 values (Fig. 2B & Table II). Conversely, the P131 variant of rhFcγR2A (rhFcγR2A:11) responded poorly to rhIgG2, but recognized rhIgG3 as efficiently as rhIgG1 and rhIgG4, which mirrors the recognition of rhIgGs by huFcγR2A R131 (Fig. 2B). Whereas the G236A substitution had a modest effect on rhFcγR2A interactions with human IgG1, the introduction of this substitution into rhIgG1 significantly enhanced interactions with rhFcγRIIa:11 (P131), rhFcγRIIa:02, and huFcγR2A R131 (Table III). This substitution may therefore be used in the context of rhIgG1 to increase interactions with rhFcγR2A. Collectively, these results reveal functional parallels between allotypes of rhFcγR2A and huFcγR2A and indicate that these receptors are less sensitive to differences among rhIgG subclasses than huIgG subclasses.
FcγR3A responses to human and rhesus macaque IgG subclasses and Fc variants
The responses of JNL cells expressing different allotypes of human and rhesus macaque FcγR3A were also tested. These included the V158 and F158 variants of huFcγR3A and the three most common variants of rhFcγR3A (rhFcγR3A:01, rhFcγR3A:02 and rhFcγR3A:03), which represent 98% of the haplotypes of FCGR3A genotyped animals. Stable JNL cell lines expressing similar levels of these receptors were established (Supplemental Fig. 1B) and incubated with Raji cells in the presence of serial dilutions of anti-CD20 antibodies corresponding to each of the human and rhesus macaque IgG subclasses and Fc domain variants (Fig. 3A & Supplemental Fig. 2). As expected, both huFcγR3A variants preferentially responded to huIgG1 and these interactions were significantly enhanced by the DEL substitutions designed to increase binding to huFcγR3A (Fig. 3B). P-values for statistical comparisons are shown in Table IV. The EC50 of the huIgG1 response was also lower for huFcγR3A V158 than for huFcγR3A F158, consistent with the higher IgG1 binding affinity of the V158 variant. Weaker responses to huIgG3 were observed for both receptors, which differs from studies reporting better interactions for IgG3 than IgG1 (5, 6), but is consistent with cytolysis assays done with primary human PBMCs (51–53). As expected, huFcγR3A interactions with huIgG2 and huIgG4 were negligible (Fig. 3B).
Table IV. Statistical comparisons of EC50 values.
Statistical comparison of EC50 values for FcγR3A-mediated responses. The p-values are shown for statistical comparisons of mean antibody concentrations (EC50) for half-maximal responses and were determined using an unpaired, two-tailed Student’s t test. Dashes indicate comparisons with undefined EC50 values.
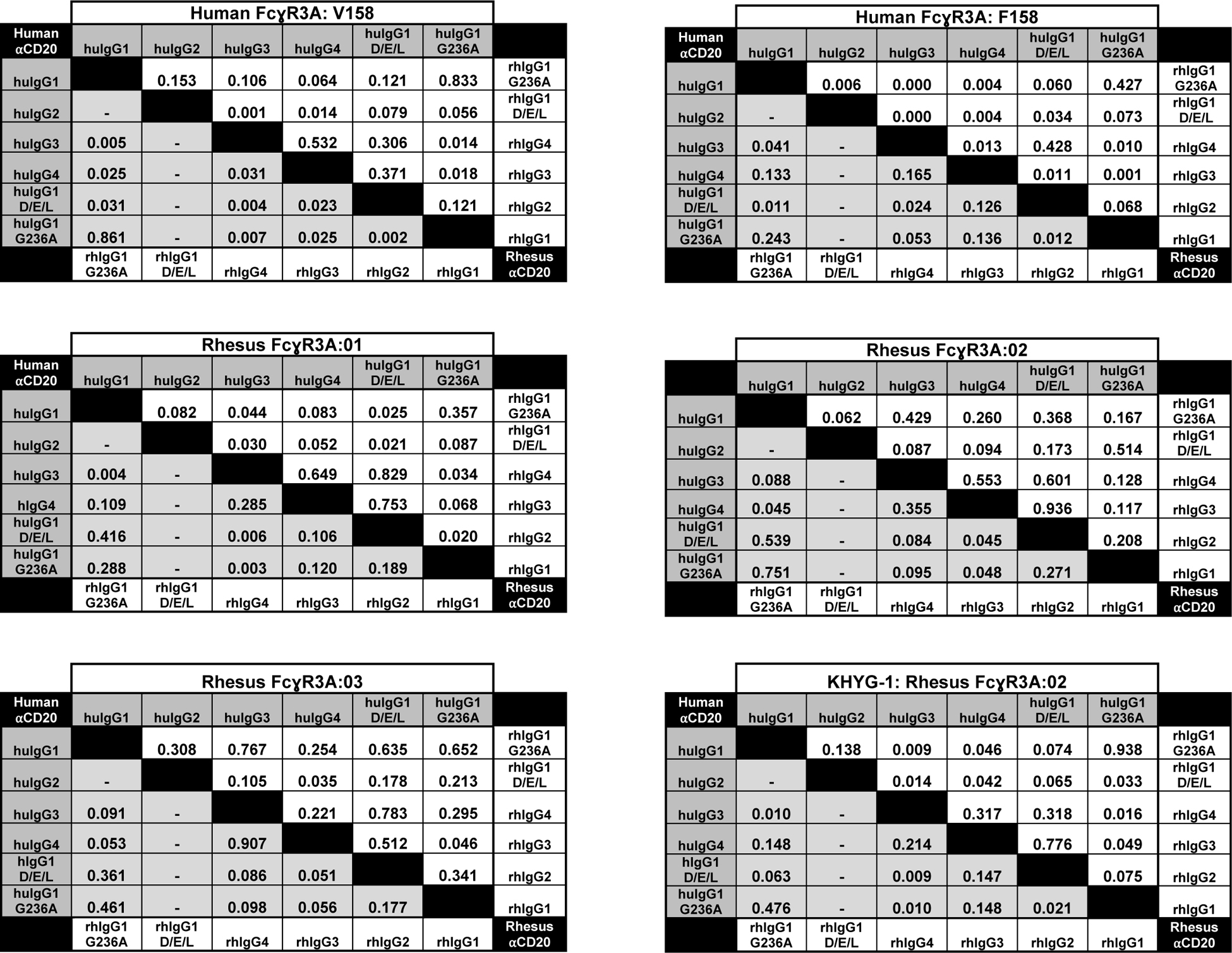
|
JNL cells expressing rhesus macaque FcγR3A variants exhibited a similar pattern of responses to the human IgGs as human FcγR3A V158. All three allotypes efficiently recognized huIgG1 and mediated measurable responses to huIgG3 and huIgG4, but did not respond to huIgG2 (Fig. 3). However, the DEL substitutions did not significantly enhance the already strong interactions of huIgG1 with these receptors (Fig. 3B & Table IV). Each of the rhFcγR3A variants also responded to huIgG4 better than either of the huFcγR3A variants, suggesting that stronger cross-species recognition of human antibodies of this subclass may trigger FcγR-mediated responses in macaques not observed in humans.
JNL cells bearing rhesus macaque FcγR3A exhibited a similar pattern of responses to the rhesus IgGs as human FcγR3A V158, but with surprisingly little variation among subclasses. Similar to their responses to huIgGs, all three rhFcγR3A variants responded most efficiently to rhIgG1 (Fig. 3B). Although the DEL substitutions did not improve these interactions to the same extent as they did for huIgG1 (Table II), these substitutions appeared to have a greater effect after a shorter 4 hour incubation than after an overnight incubation (Supplemental Fig. 3). Unlike responses to huIgGs, the rhFcγR3A variants also mediated strong responses to rhIgG2, rhIgG3 and rhIgG4 (Fig. 3). The EC50 values of the responses to rhIgG2–4 were in a similar range and approximately 2- to 3-fold higher than the EC50 values for rhIgG1 (Table II). The similarity of responses for rhFcγR3A:02 (I158) and rhFcγR3A:03 (V158), which only differ by a single amino acid at position 158, are consistent with data indicating that this polymorphism does not significantly impact IgG binding (34). The similarity of responses for rhFcγR3A:01 and rhFcγR3A:02, which differ by one residue in the transmembrane domain and one residue in the cytoplasmic domain, also suggests that these polymorphisms do not have a significant effect on signaling. Hence, these results reveal an overall lack of variability in rhFcγR3A-mediated responses to human or rhesus IgG subclasses, with each allotype exhibiting patterns of IgG recognition most similar to the high affinity V158 variant of huFcγR3A. Moreover, in accordance with the limited structural diversity among rhIgGs (16, 23), there was little variation in rhFcγR3A-mediated responses to different subclasses of rhIgG.
FcγR3A-mediated responses correlate with ADCC
To further corroborate FcγR3A-mediated responses, we took advantage of a previously established human NK cell line expressing rhesus macaque FcγR3A (38). KHYG-1 cells transduced with rhFcγR3A:02 were incubated with CAM-labelled Raji cells in the presence of serial dilutions of anti-CD20 antibodies and ADCC was measured as the dose-dependent release of CAM into the culture supernatant (Fig. 4A). The ADCC responses of this NK cell line mirrored luciferase induction from rhFcγR3A:02+ JNL cells. As observed for rhFcγR3A+ JNL cells, ADCC responses were strongest for human and rhesus IgG1 and were further enhanced (significantly so in the case of rhIgG1) by the DEL substitutions (Fig. 4A & Table IV). ADCC responses were also in a similar range for rhIgG2, rhIgG3 and rhIgG4, and weaker responses were detectable for huIgG3 and huIgG4 (Table II). Comparison of the antibody concentrations for half-maximal ADCC and JNL cell activation confirmed a strong correlation between these measures of rhFcγR3A-mediated responses (Fig. 4B).
FIGURE 4.
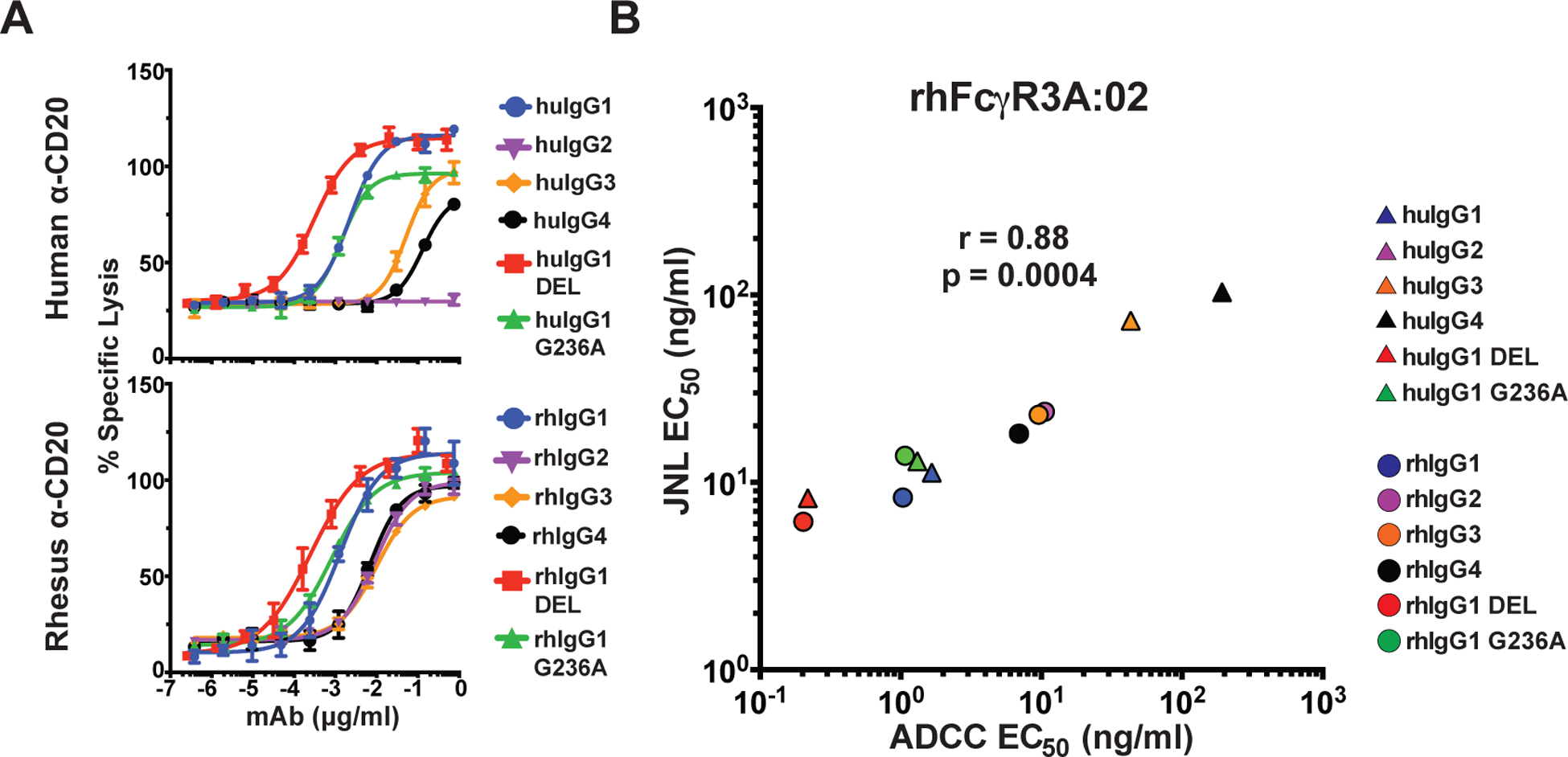
Correlation of rhFcγR3-mediated ADCC and JNL cell responses. (A) An NK cell line expressing rhFcγR3A:02 was incubated with CAM-labeled Raji cells for four hours at a 5:1 E:T ratio in the presence of serial dilutions of anti-CD20 antibodies. Percent-specific lysis was calculated from the release of CAM into the supernatant as measured using a Victor X4 multiplate reader (excitation 485 nm and absorbance 535 nm). Error bars indicate SD of the mean for triplicate measurements at each antibody concentration. (B) Mean EC50 values for ADCC responses were calculated from three independent experiments (Table II) and were compared to mean EC50 values for rhFcγR3A:02+ JNL cell responses by two-tailed Pearson correlation.
ADCC assays with primary rhesus macaque PBMC yielded similar results. For these experiments, the activation of rhFcγR3A:02+ JNL cells and the ADCC activity of unstimulated PBMCs from two different macaques were measured against a CD20-transduced CEM.NKR-CCR5 cell line (Fig. 5A & 5B). PBMCs were incubated overnight in plastic flasks to deplete monocytes and other adherent cells and CD20+ CEM.NKR-CCR5 cells were used as targets, since Raji cells are highly susceptible non-specific lysis by macaque NK cells in the absence of antibodies (54). Area under the curve values were compared, since antibody concentrations for 50% ADCC could not be determined in all cases. Although more variable than rhFcγR3A:02+ KHYG-1 cells, ADCC responses measured with PBMCs from two different animals correlated well with rhFcγR3A-mediated JNL cell responses, as indicated by the inverse relationship between ADCC and EC50 values for luciferase induction (Fig. 5C). Among the IgG subclasses, huIgG1 and rhIgG1 consistently resulted in the higher ADCC responses, which were further enhanced by the DEL substitutions (Fig. 5B). Lower ADCC responses were also measurable for rhIgG2, rhIgG3 and rhIgG4, but were negligible for huIgG2 and huIgG4 (Fig. 5B). These results demonstrate a good correspondence between rhFcγR3A-mediated responses of JNL cells and primary macaque NK cells.
FIGURE 5.
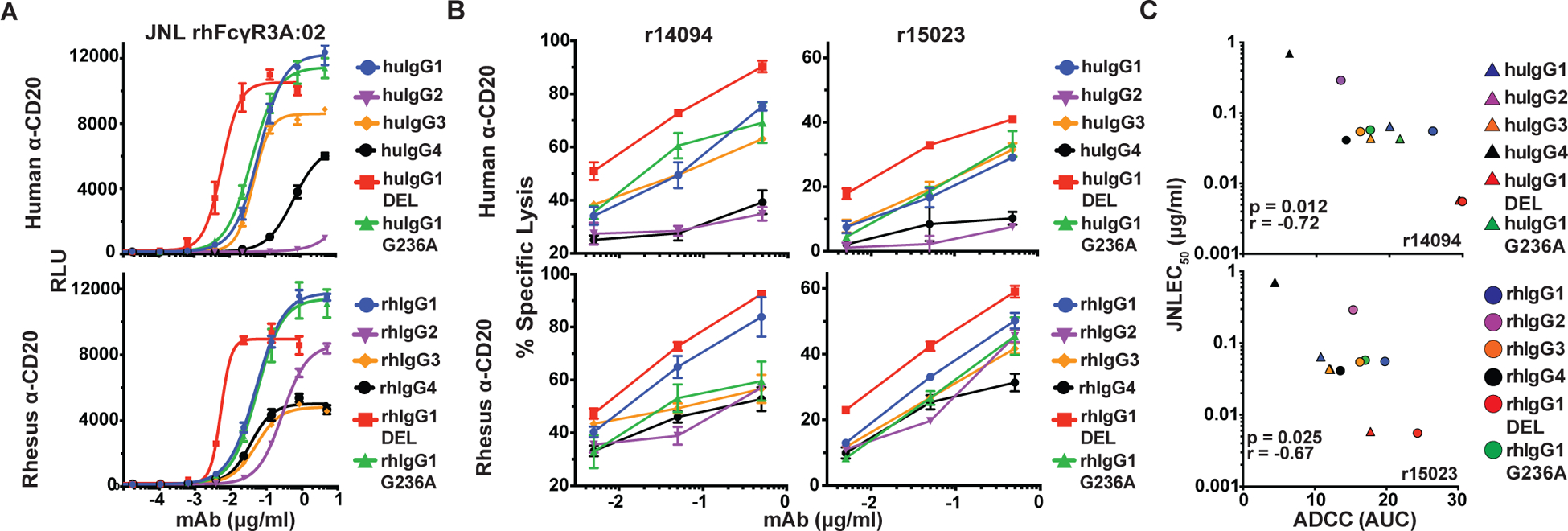
Correlation of ADCC by rhesus macaque PBMC and rhFcγR3-mediated JNL cell responses. (A) JNL cells expressing rhFcγR3A:02 were incubated for six hours with CD20-transduced CEM.NKR-CCR5 at a 1:1 E:T ratio with serial dilutions of anti-CD20 antibodies and luciferase activity was measured using a Victor X4 multiplate reader. (B) Unstimulated PBMCs from two different rhesus macaques (r14094 and r15023) were incubated for four hours with CAM-labeled CD20+ CEM.NKR-CCR5 cells at a 20:1 E:T ratio in the presence of anti-CD20 antibodies. Percent-specific lysis was calculated from the amount of CAM released in the supernatant and error bars indicate SD of the mean for triplicate measurements at each antibody concentration. (C) The relationship between the mean rhFcγR3A:02+ JNL cell responses (EC50) from two independent experiments and the mean area under the curve (AUC) values for ADCC responses was determined by two-tailed Pearson correlation.
Responses to a FcγR2B-negative B cell line
Raji cells express FcγR2B, which could interfere with IgG ligation of FcγRs on JNL cells by binding to anti-CD20 antibodies in cis (55). To determine if the presence of FcγR2B on Raji cells interferes with the availability of antibodies for the stimulation of FcγR2A and FcγR3A on JNL cells, we compared FcγR-mediated JNL cell responses to Raji cells with responses to Ramos cells, which are another CD20+ B cell line that does not express FcγR2B (55, 56). These experiments were performed with representative alleles of human and macaque FcγR2A and FcγR3A and each of the IgG subclasses and Fc variants. Although Ramos cells and Raji cells express similar levels of CD20 (Supplemental Fig. 4A), the Ramos cells resulted in a lower magnitude of luciferase induction, possibly reflecting their smaller size and lower surface area for FcγR ligation. Nevertheless, with the exception of slightly higher responses for huIgG3 using Ramos cells, the overall hierarchy and pattern of responses to Raji cells and Ramos cells were very similar (Supplemental Fig. 4B & 4C). This suggests that the presence of FcγR2B on Raji cells did not have a significant impact on the availability of antibodies for the stimulation of FcγRs on JNL cells.
Phylogenetic comparisons of the IgG subclasses of apes and Old World monkeys
The capacity of each of the allotypes of rhesus macaque FcγR2A and FcγR3A to recognize all four subclasses of rhesus IgG, albeit with variable efficiency, contrasts with preferential FcγR recognition of human IgG subclasses. To investigate the evolutionary relationships between human and macaque IgG subclasses, we performed a phylogenetic analysis that included sequences corresponding to the IgG1–4 heavy chain constant regions of two hominid species (humans and chimpanzees) and three species of Old World monkeys (baboons, pig-tailed macaques and rhesus macaques) (Fig. 6A). For comparison, we performed a similar analysis of the FcγR1, FcγR2A and FcγR3A sequences of these species (Fig. 6B).
FIGURE 6.
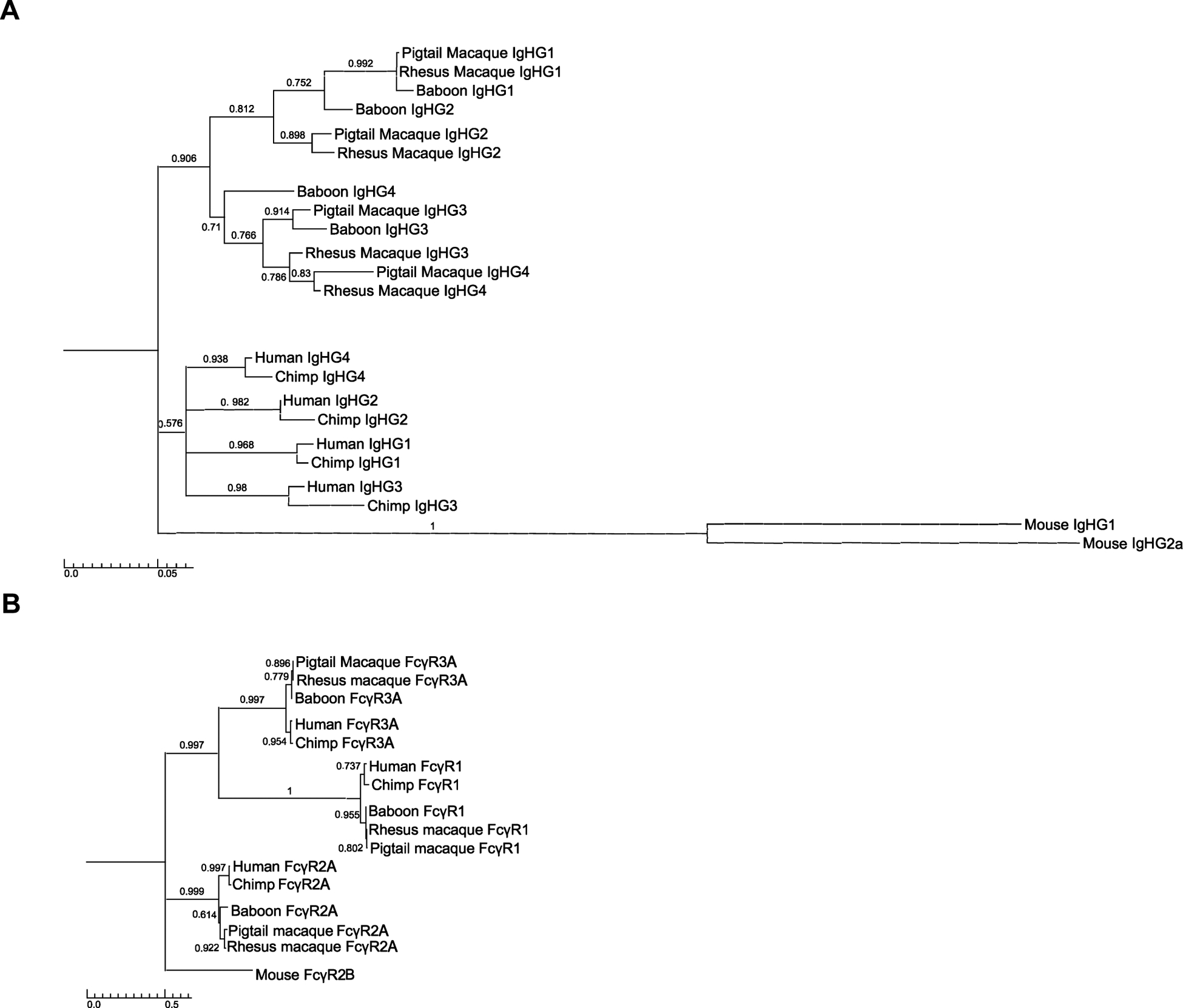
Phylogenetic analysis of hominid and Old World monkey IgGs and FcγRs. Phylogenetic trees were generated from the amino acid sequences of each of the IgG subclasses (IgG1–4) (A) and three FcγRs (FcγR1, FcγR2A and FcγR3A) (B) of humans, chimpanzees, baboons, pig-tailed macaques and rhesus macaques. Trees were constructed with using the maximum likelihood method and Jones-Taylor-Thornton substitution model with 1,000 bootstrap replicates. Trees were rooted on murine IgHG1, IgGh2a and FcγR2B sequences and nodes with bootstrap values less than 0.5 were collapsed using TreeGraph software. The scale bars indicate amino acid substitutions per site.
In contrast to the FcγRs, which cluster according to their respective gene products across all five primate species (Fig. 6B), the IgG subclasses of hominids and Old World monkeys segregate into distinct lineages (Fig. 6A). Whereas the human and chimpanzee IgG sequences cluster together according to subclass, reflecting an orthologous relationship among hominid species, the IgG sequences of macaques and baboons form separate branches (Fig. 6A). On one branch, the macaque and baboon IgG1 sequences cluster together with the IgG2 sequences of these species, and on another, the IgG3 and IgG4 sequences of these species intermingle (Fig. 6A). This suggests that unlike the FcγRs, the IgGs of macaques and baboons are products of gene duplication events that occurred after the divergence of apes and Old World monkeys. Thus, the relatively recent origin of the Old World monkey IgGs explains their limited sequence diversity and lack of orthology with human IgG subclasses.
Discussion
Macaques have become increasingly important animal models for the pre-clinical evaluation of antibody-based vaccines and therapies for HIV/AIDS and for other infectious diseases. However, the investigation of Fc-mediated antibody responses in macaques is complicated by species-specific differences and polymorphisms in macaque Fcγ receptors and IgG subclasses with respect to their human counterparts. To investigate the effects of these differences on FcγR-IgG interactions, we established Jurkat NFAT-Luciferase reporter cell lines expressing common allotypes of human and rhesus macaque FcγR2A and FcγR3A. We focused on FcγR2A and FcγR3A, since these are the receptors responsible for ADCP and ADCC effector functions. Five allotypes of rhesus FcγR2A and three allotypes of rhesus FcγR3A were selected, which represent approximately 89% and 98% of the alleles for these receptors among FCGR genotyped animals. FcγR-mediated responses were measured by incubating the JNL reporter cells with Raji cells in the presence of serial dilutions of anti-CD20 antibodies with Fc domains corresponding to each of the four subclasses of human and rhesus macaque IgG, or with Fc variants of IgG1 that selectively enhance FcγR binding. Our results show that all the allotypes of rhFcγR2A and rhFcγR3A tested are functional and preferentially recognize either IgG1 or IgG2. However, in contrast to the selective recognition of human IgG subclasses, which is a hallmark of FcγR-mediated effector functions in humans, the variability of responses to different rhesus IgG subclasses was more limited.
Of the rhesus macaque FcγRs, FcγR2A is the most polymorphic and exhibited the greatest diversity of responses. Consistent with the presence of histidine at position 131, four allotypes of rhFcγR2A (rhFcγR2A:01, rhFcγR2A:02, rhFcγR2A:08 and rhFcγR2A:11) exhibited patterns of IgG recognition similar to huFcγR2A H131. Among the human IgGs, these responses were characterized by efficient recognition of huIgG1 followed by huIgG2, and among the rhesus IgGs, preferential recognition of rhIgG2, efficient recognition of rhIgG1 and rhIgG4, and weaker interactions with rhIgG3. Conversely, rhFcγR2A:11, which has a proline at position 131, exhibited interactions that were more similar to huFcγR2A R131. Although the responses of rhFcγR2A:11 to human and rhesus IgG2 were weaker than the other rhFcγR2A allotypes, responses to rhIgG3 were comparable to rhIgG1. These results suggest that macaques have high and low affinity allotypes of FcγR2A that are functionally analogous to human FcγR2A H131 and FcγR2A R131. Whereas the majority of rhFcγR2A allotypes have the H131 polymorphism and exhibit a pattern of IgG recognition similar to huFcγR2A H131, rhFcγR2A:11 has a P131 polymorphism and exhibits responses more similar to huFcγR2A R131.
These results differ from a previous biophysical characterization of these receptors, which did not detect consistent IgG binding to the N128 and P131 variants of rhFcγR2A (34). In contrast to these experiments, responses to all four subclasses of rhIgG were detectable for rhFcγR2A:01 (N128) and rhFcγR2A:11 (P131). The responses of rhFcγR2A:01 to rhIgG1, rhIgG2 and rhIgG4 were somewhat stronger than the other H131 variants of rhFcγR2A, indicating that the additional potential N-linked glycosylation site of this receptor does not functionally impair interactions with IgG. In the case of rhFcγR2A:11, responses to rhIgG1 and rhIgG2 were weaker than the other allotypes. However, this receptor still mediated measurable responses to all four rhIgGs, including stronger responses to rhIgG3. Hence, our results complement previous binding studies to show that N128 and P131 variants of rhFcγR2A are functional, but differ in their recognition of rhIgG subclasses.
Comparatively little variation in IgG recognition was observed among different allotypes of rhesus macaque FcγR3A. Despite a number of species-specific differences with respect to human FcγR3A, the majority of rhFcγR3A allotypes only differ by a single amino acid in their extracellular domains (I158V) or a pair of residues in their transmembrane and cytoplasmic domains (V211M & I215V). Consistent with biophysical studies (34, 35), I158 and V158 variants of rhFcγR3A (rhFcγR3A:02 and rhFcγR3A:03) exhibited nearly identical responses to each of the subclasses of human and rhesus IgG. Likewise, the same pattern of responses was observed for rhFcγR3A:01, which only differs from rhFcγR3A:02 at positions 211 and 215. The responses mediated by each of the rhFcγR3A allotypes were most similar to the high affinity V158 variant of human FcγR3, as reflected by preferential recognition of huIgG1, weaker interactions with huIgG3 and huIgG4, and negligible responses to huIgG2. These observations support studies indicating that all allotypes of rhFcγR3A characterized thus far bind IgG1 with high affinity comparable to huFcγR3A V158, and that rhesus macaques lack a low affinity variant of this receptor analogous to huFcγR3A F158 (16, 35). With the caveat that our assays were performed with human cell lines, the similarity of responses for rhFcγR3A:01 and rhFcγR3A:02 also suggests that polymorphisms in the transmembrane and cytoplasmic domains of these receptors previously implicated in the efficiency of B cell depletion with rituximab (43) do not have a significant impact on rhFcγR3A-mediated responses.
Compared to FcγR recognition of human IgGs, there was remarkably little variation in FcγR-mediated responses to different subclasses of rhesus macaque IgG. This was particularly evident for allotypes of rhFcγR3A, which efficiently recognized all four subclasses of rhIgG within a narrow three-fold range of EC50 values. These observations are consistent with sequence comparisons indicating that different subclasses of rhIgG are more similar to one another than to any of the huIgGs and with recent crystal structures revealing less structural diversity among the Fc domains of rhIgGs (22, 23). To better understand the evolutionary relationships between human and macaque IgGs underlying these differences in Fc diversity, we performed a phylogenetic comparison of the IgG1–4 heavy chain constant regions of hominids and Old World monkeys. This analysis suggests that macaque IgGs are products of gene duplication events that occurred after the divergence of apes and Old World monkeys. Thus, macaque and human IgG subclasses are not orthologous and the limited structural and functional diversity of rhesus macaque IgGs can be explained by their recent divergence from a common IGHG gene since humans and macaques last shared a common ancestor.
The effects of Fc domain substitutions designed to selectively enhance IgG1 binding to human FcγR2A and FcγR3A were context dependent. The G236A substitution significantly enhanced huFcγR2A-mediated responses to huIgG1 as expected (44, 45). This substitution also enhanced rhFcγR2A-mediated responses to both huIgG1 and rhIgG1. However, in accordance with biophysical measurements (35), G236A consistently resulted in greater increases in rhFcγR2A responses in the context of rhIgG1 than huIgG1. Similar differences were observed in the effects of the DEL substitutions on FcγR3A recognition of human versus rhesus IgG1. Whereas these substitutions significantly enhanced huFcγR3A-mediated responses to huIgG1, the effects of these changes on rhFcγR3A-mediated responses in the context of either huIgG1 or rhIgG1 were more modest. In this case, the smaller effects of the DEL changes on rhFcγR3A recognition may reflect incremental enhancement of already strong IgG interactions with these receptors.
Our findings are generally consistent with studies characterizing IgG binding to macaque FcγRs and have important implications regarding the use of rhesus macaques to model Fc-mediated antibody responses. Although rhesus macaques express orthologs of FcγR2A with high and low affinity for IgG, similar to the H131 and R131 variants of huFcγR2A, and have orthologs of FcγR3A with high affinity for IgG similar to the huFcγR3A V158, the IgG subclasses of macaques are not orthologous to human IgGs and exhibit considerably less diversity in FcγR recognition. Whereas human and rhesus IgG1, which are the most abundant subclasses in their respective species, are efficiently recognized by human and rhesus macaque FcγRs, these interactions differ for other IgG subclasses. Unlike huIgG2–4, rhIgG2–4 are generally well recognized by rhFcγR2A and rhFcγR3A. Although there were some differences in responses to rhIgG2 and rhIgG3 for different allotypes of rhFcγR2A, the responses of the rhFcγR3A allotypes to rhIgGs were remarkably similar. This implies that rhIgG2–4 may contribute to FcγR-mediated effector functions in macaques to a greater extent than huIgG2–4 do in humans.
These differences in FcγR recognition of human versus macaque IgGs are fundamental to the interpretation of studies in macaques involving passive antibody transfer or vaccine-induced antibody responses. Since all allotypes of rhFcγR2A and rhFcγR3A recognize huIgG1 and rhIgG1 with similar efficiency as their corresponding human receptors, human and rhesus IgG1 are expected to have similar effector functions. The passive transfer of huIgG1 or rhIgG1 to rhesus macaques may therefore be useful for assessing FcγR-mediated functions of certain antibodies in humans. Likewise, vaccine-induced IgG1 responses in macaques may have similar effector functions as IgG1 responses in humans.
However, the other subclasses of rhesus IgG do not follow the same patterns of FcγR recognition as human IgGs. In accordance with the close phylogenetic relationship between rhIgG1 and rhIgG2, rhIgG2 was recognized at least as efficiently as rhIgG1 by four of the five allotypes of rhFcγR2A, and nearly as efficiently as rhIgG1 by all three allotypes of rhFcγR3A. Rhesus IgG4 was also recognized as efficiently as rhIgG1 by all allotypes of rhFcγR2A and with similar efficiency by each of the allotypes of rhFcγR3A. In the case of rhIgG3, interactions with rhFcγR2A were variable, but detectable for all allotypes, and interactions with rhFcγR3A were similar to the other rhIgG subclasses. In some cases, huIgGs were recognized better by rhFcγRs than huFcγRs. Thus, passive transfer experiments in which human or rhesus IgG2, IgG3 or IgG4 are administered to macaques should be interpreted with caution, since rhIgG2–4 may direct rhFcγR2A- and rhFcγR3A-mediated responses with similar potency as rhIgG1, and huIgG2–4 may trigger stronger responses than would normally occur in humans. Likewise, rhIgG2–4 responses elicited by vaccination are not expected to follow the same paradigms of FcγR recognition as huIgG2–4 and may contribute more broadly to antibody effector functions.
The incubation of FcγR-transduced JNL cells together with anti-CD20 antibodies and CD20+ target cells provides a simple, robust, and reproducible system to compare human and macaque FcγR-IgG interactions under the same conditions. Nevertheless, this approach has some limitations. Because of variation in signaling as result of species-specific differences and polymorphisms in the cytoplasmic domains of the FcγRs and/or epigenetic differences in luciferase expression among independently isolated cell lines, it is difficult to make quantitative comparisons between JNL cells expressing different FcγRs. For this reason, we have avoided quantitative comparisons between cell lines, focusing instead on comparisons of EC50 values for different antibodies measured with the same cells and qualitative comparisons of the overall hierarchies and patterns of responses among different cell lines. Additionally, certain features of Rituximab antibody binding to CD20 on Raji cells may not translate to other antibody-antigen combinations. For instance, the orientation of antibody binding may affect the accessibility of the Fc domain for FcγR ligation. Smaller targets such as viral particles may also stimulate FcγRs differently than antigens expressed on the cell surface. Indeed, the size of immune complexes has been shown to differentially affect functional interactions between human FcγRs and IgG subclass and Fc variants in cellular assays (36).
In conclusion, this study provides a quantitative comparison of the responses of the most common allotypes of rhesus macaque FcγR2A and rhFcγR3A to each of the four subclasses of human and rhesus IgG. To our knowledge, this is the first functional characterization of individual allotypes of rhesus macaque FcγRs. Our findings reveal parallels and key differences between the FcγR-IgG interactions of humans and macaques important for investigating FcγR-mediated effector functions of antibodies in the rhesus macaque.
Supplementary Material
Key Points.
Rhesus FcγR2A allotypes differ in their interactions with different IgG subclasses.
Rhesus FcγR3A allotypes show little variation in IgG subclass interactions.
Human and macaque IgG1, but not IgG2–4, exhibit similar IgG-FcγR interactions.
Acknowledgments
This work was supported by Public Health Service Grants AI121135, AI095098, AI148379 and AI098485 (to DTE), RR021745 (to DHO) and OD011106 (to the WNPRC). DTE is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
Abbreviations used in this article:
- FcγR
Fc gamma receptor
- huFcγR
human FcγR
- rhFcγR
rhesus FcγR
- IgG
immunoglobulin G
- huIgG
human IgG
- rhIgG
rhesus IgG
- JNL
Jurkat NFAT luciferase
- ADCC
antibody-dependent cellular cytotoxicity
- ADCP
antibody-dependent cellular phagocytosis
References
- 1.Nimmerjahn F, and Ravetch JV. 2008. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8: 34–47. [DOI] [PubMed] [Google Scholar]
- 2.Mellor JD, Brown MP, Irving HR, Zalcberg JR, and Dobrovic A. 2013. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metes D, Ernst LK, Chambers WH, Sulica A, Herberman RB, and Morel PA. 1998. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood 91: 2369–2380. [PubMed] [Google Scholar]
- 4.van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, and Kuijpers TW. 2012. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J. Immunol 188: 1318–1324. [DOI] [PubMed] [Google Scholar]
- 5.Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, and Daëron M. 2009. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 113: 3716–3725. [DOI] [PubMed] [Google Scholar]
- 6.Hogarth PM, and Pietersz GA. 2012. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov 11: 311–331. [DOI] [PubMed] [Google Scholar]
- 7.Clark MR, Clarkson SB, Ory PA, Stollman N, and Goldstein IM. 1989. Molecular basis for a polymorphism involving Fc receptor II on human monocytes. J Immunol 143: 1731–1734. [PubMed] [Google Scholar]
- 8.Warmerdam PA, van de Winkel JG, Vlug A, Westerdaal NA, and Capel PJ. 1991. A single amino acid in the second Ig-like domain of the human Fc gamma receptor II is critical for human IgG2 binding. The Journal of Immunology 147: 1338–1343. [PubMed] [Google Scholar]
- 9.Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB, and Montoya B. 2007. FcgammaRIIa genotype predicts progression of HIV infection. J Immunol 179: 7916–7923. [DOI] [PubMed] [Google Scholar]
- 10.Platonov AE, Kuijper EJ, Vershinina IV, Shipulin GA, Westerdaal N, Fijen CA, and van de Winkel JG. 1998. Meningococcal disease and polymorphism of FcgammaRIIa (CD32) in late complement component-deficient individuals. Clin Exp Immunol 111: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yee AM, Phan HM, Zuniga R, Salmon JE, and Musher DM. 2000. Association between FcgammaRIIa-R131 allotype and bacteremic pneumococcal pneumonia. Clin Infect Dis 30: 25–28. [DOI] [PubMed] [Google Scholar]
- 12.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, Lamy A, Penault-Llorca F, Frébourg T, Michel P, Sabourin JC, and Boissière-Michot F. 2009. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol 27: 1122–1129. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang Y, Xu W, Shen Y, and Li J. 2010. Fcγ receptor polymorphisms and clinical efficacy of rituximab in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk 10: 347–352. [DOI] [PubMed] [Google Scholar]
- 14.Rogers KA, Scinicariello F, and Attanasio R. 2006. IgG Fc receptor III homologues in nonhuman primate species: genetic characterization and ligand interactions. J Immunol 177: 3848–3856. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen DC, Scinicariello F, and Attanasio R. 2011. Characterization and allelic polymorphisms of rhesus macaque (Macaca mulatta) IgG Fc receptor genes. Immunogenetics 63: 351–362. [DOI] [PubMed] [Google Scholar]
- 16.Crowley AR, and Ackerman ME. 2019. Mind the Gap: How Interspecies Variability in IgG and Its Receptors May Complicate Comparisons of Human and Non-human Primate Effector Function. Front Immunol 10: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogarth PM 2015. Fc Receptors: Introduction. Immunol Rev 268: 1–5. [DOI] [PubMed] [Google Scholar]
- 18.Haj AK, Arbanas JM, Yamniuk AP, Karl JA, Bussan HE, Drinkwater KY, Graham ME, Ericsen AJ, Prall TM, Moore K, Cheng L, Gao M, Graziano RF, Loffredo JT, Wiseman RW, and O’Connor DH. 2019. Characterization of Mauritian Cynomolgus Macaque FcγR Alleles Using Long-Read Sequencing. J Immunol 202: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trist HM, Tan PS, Wines BD, Ramsland PA, Orlowski E, Stubbs J, Gardiner EE, Pietersz GA, Kent SJ, Stratov I, Burton DR, and Hogarth PM. 2014. Polymorphisms and interspecies differences of the activating and inhibitory FcγRII of Macaca nemestrina influence the binding of human IgG subclasses. J Immunol 192: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scinicariello F, Engleman CN, Jayashankar L, McClure HM, and Attanasio R. 2004. Rhesus macaque antibody molecules: sequences and heterogeneity of alpha and gamma constant regions. Immunology 111: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen DC, Sanghvi R, Scinicariello F, Pulit-Penaloza J, Hill N, and Attanasio R. 2014. Cynomolgus and pigtail macaque IgG subclasses: characterization of IGHG genes and computational analysis of IgG/Fc receptor binding affinity. Immunogenetics 66: 361–377. [DOI] [PubMed] [Google Scholar]
- 22.Boesch AW, Osei-Owusu NY, Crowley AR, Chu TH, Chan YN, Weiner JA, Bharadwaj P, Hards R, Adamo ME, Gerber SA, Cocklin SL, Schmitz JE, Miles AR, Eckman JW, Belli AJ, Reimann KA, and Ackerman ME. 2016. Biophysical and Functional Characterization of Rhesus Macaque IgG Subclasses. Front Immunol 7: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolbert WD, Subedi GP, Gohain N, Lewis GK, Patel KR, Barb AW, and Pazgier M. 2019. From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses. MAbs 11: 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, and Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esquivel RN, Patel A, Kudchodkar SB, Park DH, Stettler K, Beltramello M, Allen JW, Mendoza J, Ramos S, Choi H, Borole P, Asija K, Bah M, Shaheen S, Chen J, Yan J, Durham AC, Smith TRF, Broderick K, Guibinga G, Muthumani K, Corti D, Humeau L, and Weiner DB. 2019. In Vivo Delivery of a DNA-Encoded Monoclonal Antibody Protects Non-human Primates against Zika Virus. Mol Ther 27: 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, Marx PA, and Burton DR. 2007. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449: 101–104. [DOI] [PubMed] [Google Scholar]
- 27.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, and Burton DR. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 15: 951–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magnani DM, Rogers TF, Maness NJ, Grubaugh ND, Beutler N, Bailey VK, Gonzalez-Nieto L, Gutman MJ, Pedreño-Lopez N, Kwal JM, Ricciardi MJ, Myers TA, Julander JG, Bohm RP, Gilbert MH, Schiro F, Aye PP, Blair RV, Martins MA, Falkenstein KP, Kaur A, Curry CL, Kallas EG, Desrosiers RC, Goldschmidt-Clermont PJ, Whitehead SS, Andersen KG, Bonaldo MC, Lackner AA, Panganiban AT, Burton DR, and Watkins DI. 2018. Fetal demise and failed antibody therapy during Zika virus infection of pregnant macaques. Nat Commun 9: 1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Navio JM, Fuchs SP, Pantry SN, Lauer WA, Duggan NN, Keele BF, Rakasz EG, Gao G, Lifson JD, and Desrosiers RC. 2019. Adeno-Associated Virus Delivery of Anti-HIV Monoclonal Antibodies Can Drive Long-Term Virologic Suppression. Immunity 50: 567–575.e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prévost J, Nedellec R, von Bredow B, Abbink P, Cottrell CA, Kulp DW, Tokatlian T, Nogal B, Bianchi M, Li H, Lee JH, Butera ST, Evans DT, Hangartner L, Finzi A, Wilson IA, Wyatt RT, Irvine DJ, Schief WR, Ward AB, Sanders RW, Crotty S, Shaw GM, Barouch DH, and Burton DR. 2019. Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity 50: 241–252.e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, Diskin R, Bjorkman PJ, Eckhaus MA, Klein F, Mouquet H, Cetrulo Lorenzi JC, Gazumyan A, Burton DR, Nussenzweig MC, Martin MA, and Nishimura Y. 2014. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med 211: 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welles HC, Jennewein MF, Mason RD, Narpala S, Wang L, Cheng C, Zhang Y, Todd JP, Lifson JD, Balazs AB, Alter G, McDermott AB, Mascola JR, and Roederer M. 2018. Vectored delivery of anti-SIV envelope targeting mAb via AAV8 protects rhesus macaques from repeated limiting dose intrarectal swarm SIVsmE660 challenge. PLoS Pathog 14: e1007395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, Kuang D, Yang M, Yang J, Ma C, Xu J, Qian X, Li H, Zhao S, Li J, Wang H, Long H, Zhou J, Luo F, Ding K, Wu D, Zhang Y, Dong Y, Liu Y, Zheng Y, Lin X, Jiao L, Zheng H, Dai Q, Sun Q, Hu Y, Ke C, Liu H, and Peng X. 2020. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv: 2020.2004.2008.031807. [Google Scholar]
- 34.Chan YN, Boesch AW, Osei-Owusu NY, Emileh A, Crowley AR, Cocklin SL, Finstad SL, Linde CH, Howell RA, Zentner I, Cocklin S, Miles AR, Eckman JW, Alter G, Schmitz JE, and Ackerman ME. 2016. IgG Binding Characteristics of Rhesus Macaque FcγR. J Immunol 197: 2936–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boesch AW, Miles AR, Chan YN, Osei-Owusu NY, and Ackerman ME. 2017. IgG Fc variant cross-reactivity between human and rhesus macaque FcγRs. MAbs 9: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lux A, Yu X, Scanlan CN, and Nimmerjahn F. 2013. Impact of immune complex size and glycosylation on IgG binding to human FcγRs. J Immunol 190: 4315–4323. [DOI] [PubMed] [Google Scholar]
- 37.National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals., and Institute for Laboratory Animal Research (U.S.) 2011. Guide for the care and use of laboratory animals. National Academies Press, Washington, D.C. [Google Scholar]
- 38.Alpert MD, Heyer LN, Williams DE, Harvey JD, Greenough T, Allhorn M, and Evans DT. 2012. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86: 12039–12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trkola A, Matthews J, Gordon C, Ketas T, and Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol 73: 8966–8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Bredow B, Arias JF, Heyer LN, Moldt B, Le K, Robinson JE, Zolla-Pazner S, Burton DR, and Evans DT. 2016. Comparison of Antibody-Dependent Cell-Mediated Cytotoxicity and Virus Neutralization by HIV-1 Env-Specific Monoclonal Antibodies. J Virol 90: 6127–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Bredow B, Arias JF, Heyer LN, Gardner MR, Farzan M, Rakasz EG, and Evans DT. 2015. Envelope Glycoprotein Internalization Protects Human and Simian Immunodeficiency Virus-Infected Cells from Antibody-Dependent Cell-Mediated Cytotoxicity. J Virol 89: 10648–10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shortreed CG, Wiseman RW, Karl JA, Bussan HE, Baker DA, Prall TM, Haj AK, Moreno GK, Penedo MCT, and O’Connor DH. 2020. Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques. Immunogenetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller CJ, Genescà M, Abel K, Montefiori D, Forthal D, Bost K, Li J, Favre D, and McCune JM. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J Virol 81: 5024–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moldt B, Schultz N, Dunlop DC, Alpert MD, Harvey JD, Evans DT, Poignard P, Hessell AJ, and Burton DR. 2011. A panel of IgG1 b12 variants with selectively diminished or enhanced affinity for Fcγ receptors to define the role of effector functions in protection against HIV. J Virol 85: 10572–10581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards JO, Karki S, Lazar GA, Chen H, Dang W, and Desjarlais JR. 2008. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Mol Cancer Ther 7: 2517–2527. [DOI] [PubMed] [Google Scholar]
- 46.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, Chan C, Chung HS, Eivazi A, Yoder SC, Vielmetter J, Carmichael DF, Hayes RJ, and Dahiyat BI. 2006. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci U S A 103: 4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warmerdam PA, van de Winkel JG, Vlug A, Westerdaal NA, and Capel PJ. 1991. A single amino acid in the second Ig-like domain of the human Fc gamma receptor II is critical for human IgG2 binding. J Immunol 147: 1338–1343. [PubMed] [Google Scholar]
- 48.Salmon JE, Edberg JC, Brogle NL, and Kimberly RP. 1992. Allelic polymorphisms of human Fc gamma receptor IIA and Fc gamma receptor IIIB. Independent mechanisms for differences in human phagocyte function. J Clin Invest 89: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shashidharamurthy R, Zhang F, Amano A, Kamat A, Panchanathan R, Ezekwudo D, Zhu C, and Selvaraj P. 2009. Dynamics of the interaction of human IgG subtype immune complexes with cells expressing R and H allelic forms of a low-affinity Fc gamma receptor CD32A. J Immunol 183: 8216–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders LA, Feldman RG, Voorhorst-Ogink MM, de Haas M, Rijkers GT, Capel PJ, Zegers BJ, and van de Winkel JG. 1995. Human immunoglobulin G (IgG) Fc receptor IIA (CD32) polymorphism and IgG2-mediated bacterial phagocytosis by neutrophils. Infect Immun 63: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brüggemann M, Williams GT, Bindon CI, Clark MR, Walker MR, Jefferis R, Waldmann H, and Neuberger MS. 1987. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J Exp Med 166: 1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niwa R, Natsume A, Uehara A, Wakitani M, Iida S, Uchida K, Satoh M, and Shitara K. 2005. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J Immunol Methods 306: 151–160. [DOI] [PubMed] [Google Scholar]
- 53.Redpath S, Michaelsen TE, Sandlie I, and Clark MR. 1998. The influence of the hinge region length in binding of human IgG to human Fcgamma receptors. Hum Immunol 59: 720–727. [DOI] [PubMed] [Google Scholar]
- 54.Carver FM, and Thomas JM. 1988. Natural killer cells in rhesus monkeys: properties of effector cells which lyse Raji targets. Cell Immunol 117: 56–69. [DOI] [PubMed] [Google Scholar]
- 55.Lim SH, Vaughan AT, Ashton-Key M, Williams EL, Dixon SV, Chan HT, Beers SA, French RR, Cox KL, Davies AJ, Potter KN, Mockridge CI, Oscier DG, Johnson PW, Cragg MS, and Glennie MJ. 2011. Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood 118: 2530–2540. [DOI] [PubMed] [Google Scholar]
- 56.Vaughan AT, Iriyama C, Beers SA, Chan CH, Lim SH, Williams EL, Shah V, Roghanian A, Frendéus B, Glennie MJ, and Cragg MS. 2014. Inhibitory FcγRIIb (CD32b) becomes activated by therapeutic mAb in both cis and trans and drives internalization according to antibody specificity. Blood 123: 669–677. [DOI] [PubMed] [Google Scholar]
- 57.Ramsland PA, Farrugia W, Bradford TM, Sardjono CT, Esparon S, Trist HM, Powell MS, Tan PS, Cendron AC, Wines BD, Scott AM, and Hogarth PM. 2011. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J Immunol 187: 3208–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sondermann P, Huber R, Oosthuizen V, and Jacob U. 2000. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 406: 267–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


