SUMMARY
To grow and divide, cells must extract resources from dynamic and unpredictable environments. Many organisms use different metabolic strategies for distinct contexts. Budding yeast can produce ATP from carbon sources by mechanisms that prioritize either speed (fermentation) or yield (respiration). Withdrawing glucose from exponentially growing cells reveals variability in their ability to switch from fermentation to respiration. We observe two subpopulations of glucose-starved cells: recoverers, which rapidly adapt and resume growth, and arresters, which enter a shock state characterized by deformation of many cellular structures, including mitochondria. These states are heritable, and on high glucose, arresters grow and divide faster than recoverers. Recoverers have a fitness advantage during a carbon source shift but are less fit in a constant, high-glucose environment, and we observe natural variation in the frequency of the two states across wild yeast strains. These experiments suggest that bet hedging has evolved in budding yeast.
Keywords: Metabolism, mitochondria, bimodality, bet hedging, respiration, fermentation
Graphical Abstract
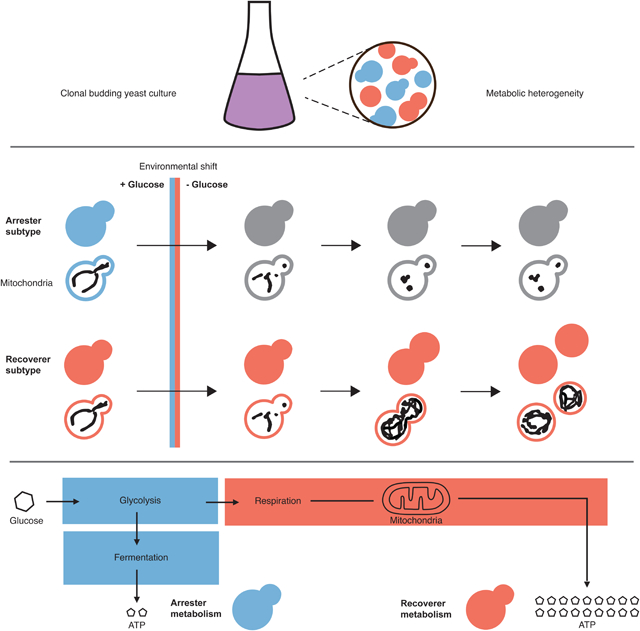
Bagamery et al. find that budding yeast display a bimodal response to acute glucose starvation. They identify two heritable states: recoverers are resistant to nutrient shifts, while arresters grow rapidly under constant, abundant glucose. Natural variation in the coexistence of the two types suggests that bet hedging has evolved in budding yeast.
INTRODUCTION
Metabolic flexibility--the ability to alternate between distinct metabolic pathways for performing functionally interchangeable anabolic and catabolic activities--allows cells to modulate their metabolism dynamically in response to external nutrient supplies, which often vary widely in quantity and quality. The budding yeast, Saccharomyces cerevisiae, can use multiple metabolic strategies to derive energy from carbon: in conditions permitting high consumption rates of their preferred carbon source, glucose, yeast rely on glycolysis and fermentation for the generation of ATP and biomass; cells perform respiration, a process with a higher ATP yield per unit sugar, when glucose is scarce, when they depend on a carbon source that can only be metabolized through oxidation [1,2].
How quickly cells can switch from fermentation to respiration varies between strains [3]. Budding yeast prefers to ferment, a bias that is enforced by catabolite repression, a complex and overlapping set of regulatory networks that repress the expression of proteins involved in respiration even in the presence of oxygen and carbon sources that can be oxidized [4]. The slow reversal of catabolite repression can delay cell proliferation. In a batch culture of yeast grown in typical synthetic medium, the initial reliance on heavy fermentation and the later induction of respiratory metabolism result in two separate exponential phases of growth with a variable period of slowed or stalled growth between the two, known as the diauxic lag (Figure 1A). One of the requirements for the transition is the activation of oxidative electron transport in mitochondria. In yeast, mitochondria exist as a network of branched tubules along the cell cortex. Mitochondrial network size and structure are flexible and responsive to environmental conditions: cells in the presence of high glucose possess minimal mitochondrial networks that can become more branched and increase in volume when forced to utilize a nonfermentable carbon source such as glycerol [5–8].
Figure 1. Heterogeneity in mitochondrial size scaling and structural integrity track with growth stalling during fermentative-to-respiratory transitions.
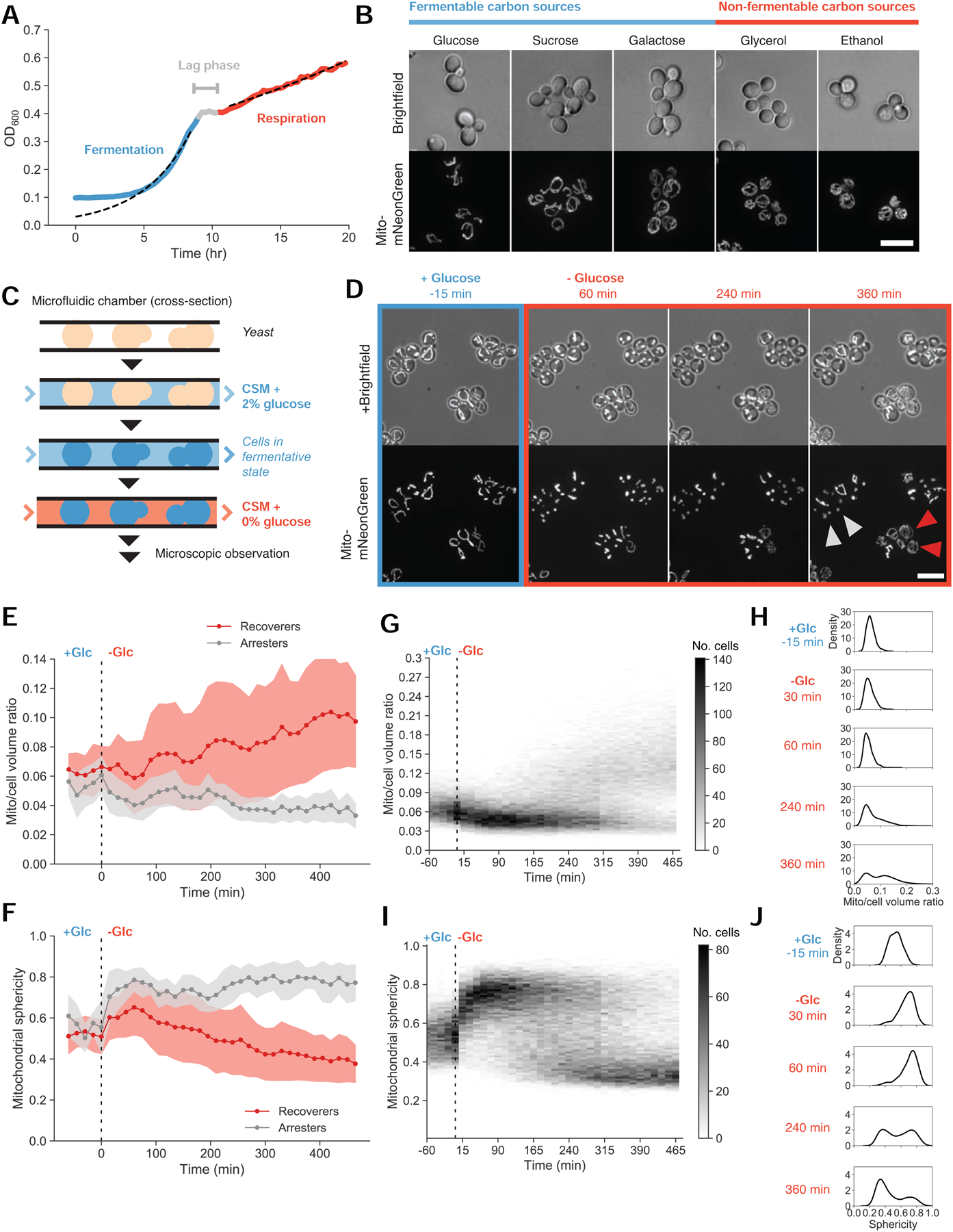
(A) Typical yeast fermentative and respiratory growth dynamics, expressed as the optical density of a batch yeast culture (yLB126) measured on a plate reader with continuous shaking. Dotted lines depict exponential fits to the indicated blue and red regions.
(B) Mitochondrial networks visualized by matrix-targeted mNeonGreen fluorescent protein (mito-mNeonGreen) (yLB126) in exponentially-growing yeast cells in synthetic media with indicated carbon sources. Scale bar, 10 μm.
(C) Experimental setup. Cells were immobilized in microfluidic plates and supplied with synthetic medium containing 2% glucose by continuous perfusion, promoting glucose repression and permitting fermentative growth. Cells were observed by microscopy during this pregrowth period and following abrupt replacement of the source medium with synthetic medium containing 0% glucose.
(D) Representative images of mito-mNeonGreen (yLB126) in high glucose and followed by abrupt switch to glucose-free medium. Labeled cells indicate examples of recoverer cells, which adapt and perform mitochondrial biogenesis (red arrows), and arrester cells, with extended mitochondrial collapse (gray arrows). Scale bar, 10 μm.
(E) Distributions of the ratio of mitochondrial to total cell volume for phenotypic classes above for all cells in the field of view in (D). Filled bands indicate one standard deviation from the mean. These sample trajectories are representative of 13 independent experiments as displayed in (G) and (H).
(F) Distributions of mitochondrial sphericity index (ratio of surface area of hypothetical sphere with the same volume as a mitochondrion to its actual surface area) for phenotypic classes in (D), in a given sample field of view. Filled bands indicate one standard deviation.
(G) and (H) Time-resolved heat maps of mitochondrial/cell volume ratio (G) and mitochondrial sphericity (H) in N = 1,329 cells, collected across 13 independent experiments, before and during acute glucose starvation beginning at 0 min. Intensity reflects absolute number of cells within the binning area.
(I) and (J) Histograms of mitochondrial/cell volume ratio (I) and sphericity (J) at the indicated time points from (G) and (H) displaying bimodality in mitochondrial morphology following glucose withdrawal.
Because different strains show different responses to shifts in their carbon sources, we asked if there was any heterogeneity in the response of individual cells, within a strain, to such shifts. We examined the dynamics of fermentative-to-respiratory shifts in single cells by using microfluidics to rapidly remove glucose from the medium, following mitochondrial network size and morphology as markers of cellular reorganization. In most cells, mitochondrial networks collapsed into spherical globules. Their later behavior divided them into two states: recoverers rapidly rebuild their mitochondrial networks and resume cell growth and proliferation, whereas arresters fail to reorganize their mitochondria or grow. This difference was heritable, and cells can be locked in either state by mutations that affect glucose sensing, signaling, or utilization. Removing glucose gives an advantage to recoverers, whereas arresters divide slightly faster than recoverers during exponential growth in high glucose, and the switching rate from recoverers to arresters is faster than the opposite transition. This environmental growth-rate tradeoff also occurs in natural yeast strains, and their different degrees of metabolic flexibility suggest that the extent of preparation against nutrient instability is under evolutionary selection and may constitute a form of bet hedging.
RESULTS
Bimodality in mitochondrial structural integrity and organelle size scaling during sudden glucose deprivation
We first sought an optical readout of the metabolic state of yeast cells. To assess whether mitochondrial network morphology reports on metabolic state, we examined mitochondria in cells growing in a variety of carbon sources. To visualize mitochondria, we fused a mitochondrial targeting sequence to the fluorescent protein mNeonGreen (mito-mNeonGreen), which we expressed constitutively. The gene product is transported to the mitochondrial matrix, and the targeting sequence is cleaved off, labeling the mitochondrial matrix with free fluorescent protein.
As described previously [5,7,8], microscopic images reveal divergent mitochondrial network architectures as a function of nutrient conditions. Cells possess minimal mitochondrial content in the presence of high levels of fermentable sugars (glucose, sucrose, galactose) and denser, more extensive mitochondrial networks when growing under respiratory conditions (glycerol, ethanol) (Figure 1B). We performed three-dimensional segmentation of mitochondrial networks and calculated the proportion of the total cell volume occupied by mitochondria. We found that distinct carbon sources produced mitochondrial volume ratios spanning a nearly two-fold range, from 0.06 under high glucose conditions to 0.11 in cells grown on ethanol (Figure S1A). To quantify the relationship between network size and metabolic output, we measured oxygen consumption rates as a proxy for respiratory activity. Cells with larger mitochondrial networks depleted oxygen from the medium faster, indicating that increased respiratory activity correlates with increased mitochondrial content (Figure S1B).
We then followed mitochondria during metabolic transitions by fluorescent microscopy. We used microfluidics to rapidly switch the media perfusing immobilized cells (Figure 1C). After an initial flow of synthetic medium containing 2% glucose, which permits fermentation, cells were shifted into glucose-free medium to prevent fermentation but allow the respiration of amino acids in the medium. We refer to this as glucose starvation.
We observed a heterogeneous response to sudden glucose deprivation (Figure 1D and Videos S1–S2). All cells stopped growing--no longer increasing in size or dividing--when glucose was removed from the medium. In most cells, mitochondrial networks lost their tubular structure and condensed into spherical globules. In a subset of these cells, this collapse in mitochondrial structural integrity rapidly reversed itself at varying time intervals. The restoration of mitochondrial tubularity was followed by an increase in mitochondrial network volume and the eventual resumption of growth. The remaining cells remained arrested indefinitely, though they remained physiologically and genetically viable: they recovered their mitochondrial structure and resumed growth and division when glucose was returned to the environment (Video S2). Roughly 6 hr after starting glucose starvation, cells ceased to transition from the arrested to the recovering state, and these two states’ relative proportions remained stable for at least another 4 hr (Figure S1C). We thus divided the population into two phenotypes: recoverers, the cells that resumed growth within 6 hours of glucose depletion, and arresters, those that had not.
We quantified the dynamics of the morphological and volumetric changes in the mitochondrial network. Figure 1E displays mitochondrial trajectories for the cells shown in Figure 1D, classified as recoverers or arresters based on their possession of tubular or spherical mitochondria, respectively, 6 hr into glucose starvation. The fraction of a cell’s volume occupied by mitochondria approximately doubled in recoverers.
To assess the tubularity of the network, we calculated the sphericity index, a dimensionless metric for comparison to the proportions of a sphere (see Methods). We observed increases in mitochondrial sphericity beginning at the time glucose was removed and peaking approximately 1 hr into glucose starvation (Figure 1F). In arresters, this value remained constant over time. In recoverers, sphericity abruptly decreased to a steady-state lower than the initial pre-withdrawal value. This decline reflects both the reestablishment of mitochondrial tubularity and mitochondrial biogenesis, as a larger mitochondrial volume is arranged into longer, branched, rodlike structures within the same, limited cell volume. Mitochondrial sphericity thus allows us to distinguish the loss of structural integrity from remodeling activity associated with the establishment of new mitochondrial network sizes, detected as subtler decreases from the baseline.
We present the heterogeneity in the response glucose starvation responses on a population scale in Figure 1G. We observe initial mitochondrial-to-total-cell volume ratios near 0.06, with an asynchronous rise during glucose starvation to ratios centered about 0.13, as well as a subpopulation that does not increase its mitochondrial volume fraction (Figure 1G, with selected histograms in Figure 1H). Similarly, mitochondrial sphericity indices, initially concentrated between 0.4 and 0.6, rise to 0.8, then decrease in a subset of cells to values centered about 0.35 (Figures 1I and 1J). Histograms plotted from individual time points demonstrate the bimodality within the distributions of both mitochondrial volume ratio and sphericity: glucose-starved cells concentrated into two distinct peaks, reflecting mitochondrial biogenesis and the recovery of structural integrity in recoverers and the absence of these events in arresters (Figures 1H and 1I).
Bimodal effects on growth, pH homeostasis, and other cellular structures during environmental instability
The aim of metabolic adaptation is to support cell growth and proliferation in the altered environment. To determine the relationship between mitochondrial structural properties and proliferative capacity, we determined the time of first visible cell volume increase (or bud emergence) for a set of 357 cells from the recoverer subpopulation and measured their mitochondrial networks at these timepoints. Relative to the predominantly collapsed mitochondrial state observed 1 hr into glucose starvation, and the bimodal distribution of collapsed and recovered cells at 6 hr, the distribution of cells just beginning regrowth is a unimodal distribution of low sphericity, matching one of the two subpopulations within the larger data set (Figure 2A). Likewise, the mitochondrial volume ratios of growing cells in glucose-deprived conditions are uniformly high and overlap with the upper peak of the bimodal distribution (Figure 2B). We observed no growth in cells whose mitochondria did not recover from their initial collapse. Cells that did recover had restored mitochondrial tubularity and increased mitochondrial volume more than 90 min before resuming growth (Figures S2A and S2B). We conclude that cells in a state of mitochondrial collapse are nongrowing cells that have not begun preparatory functions for respiration, which would include mitochondrial network expansion.
Figure 2. Mitochondrial structural collapse is accompanied by internal pH drop and other intracellular aggregation.
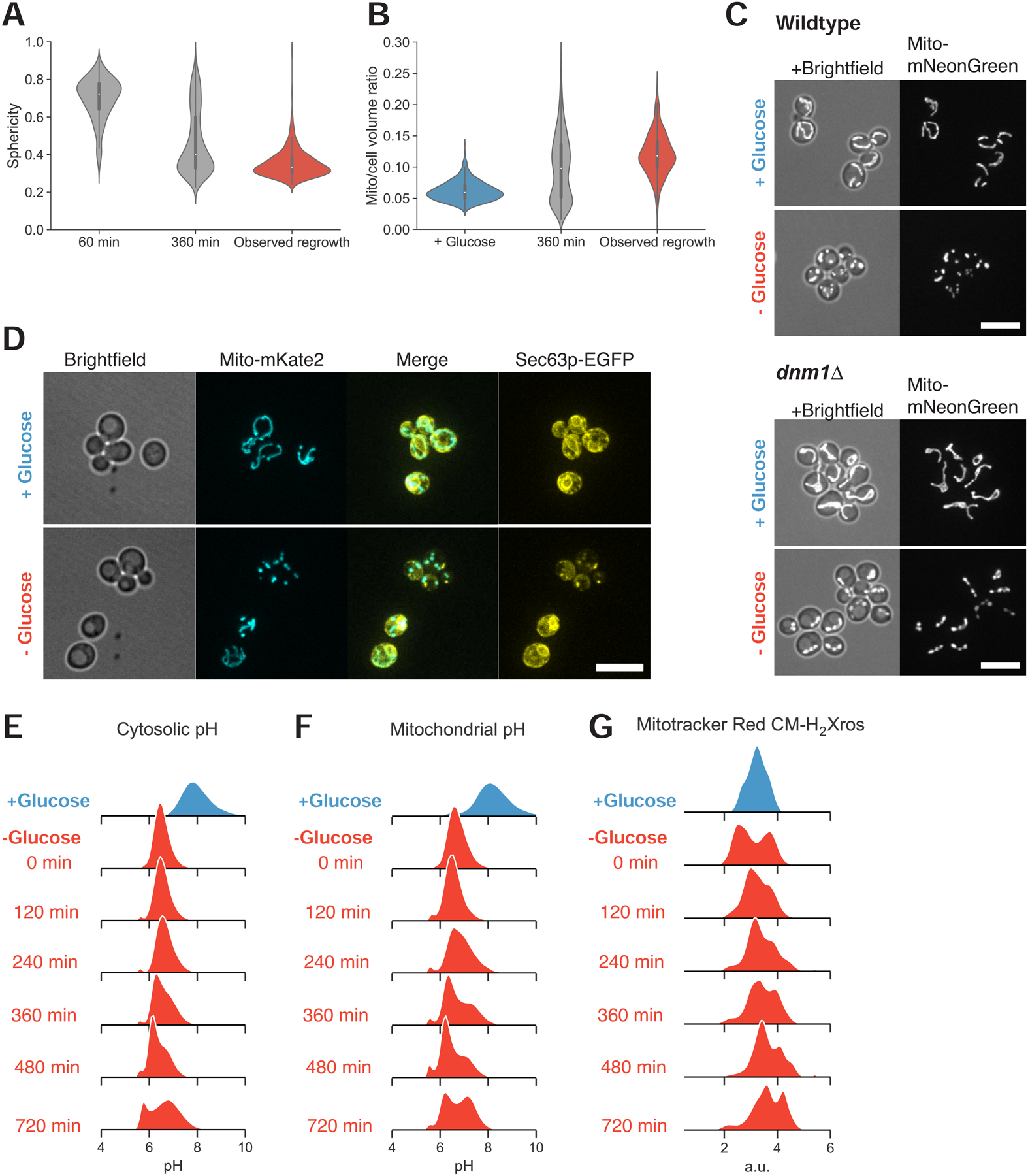
(A) Distributions of mitochondrial sphericity index following 1 and 6 hr acute glucose starvation and in a subpopulation of recovering cells at the time they resumed growth as determined by observable cell volume increase and/or budding. N ≥ 357 cells.
(B) Distributions of mitochondrial to total cell volume ratio in cells growing in synthetic media containing high glucose, cells starved of glucose for 6 hr, and starving cells at the first detectable resumption of growth.
(C) Mitochondrial matrix marker in wild-type (yLB126) and mitochondrial fission-defective dnm1Δ mutants (yLB134), in synthetic medium containing high glucose and following abrupt glucose withdrawal. Scale bars, 10 μm.
(D) Co-expression of mito-mNeptune and the endoplasmic reticulum marker Sec63pmNeonGreen (yLB41) during exponential growth in synthetic medium containing high glucose and following glucose washout. Scale bar, 10 μm.
(E)-(F) Dynamics of intracellular pH (E) and mitochondrial pH (F) in cells growing in high glucose and during starvation, detected by constitutively-expressed pHluorin2 localized to the cytosol or mitochondrial matrix, respectively (yLB397 and yLB219).
(G) Relative potential across the inner mitochondrial membrane before and following glucose starvation, measured in cells (yLB1) stained with potential-sensitive MitoTracker Red CM-H2-Xros. All distributions consist of three biological replicates of n = 40,000 cells.
See also Figure S2.
The disappearance of tubular mitochondria could be due to two mechanisms: an excess of mitochondrial fission over fusion events or the inhibition of the processes that maintain mitochondria’s rodlike structure. We examined the role of mitochondrial fission in these morphological changes by examining the dynamics of mitochondria as a fission-deficient dnm1Δ mutant was starved of glucose. When growing in the presence of high glucose, dnm1Δ cells possess a single mitochondrial network with extensive branching; once glucose is removed from the medium, the continuity of the network is maintained, but multiple structural collapses occur, resulting in a series of globules connected by small bridges (Figure 2C). We thus conclude that mitochondrial fission is not the mechanism of collapse.
To determine whether the collapse of mitochondria reflects broader changes in cellular properties, we examined other structures following glucose withdrawal. Because mitochondrial tubularity depends on contact with endoplasmic reticulum [9,10], we examined mitochondria in tandem with endoplasmic reticulum structure by fusing a fluorescent protein to the endoplasmic reticulum membrane protein Sec63p [11,12]. We found that arresters contained aggregates of Sec63p, which were absent in recoverer cells with tubular mitochondria (Figure 2D). Arresters also aberrantly localized the actin filament-binding protein Abp140p (Figure S2D) and the cortical actin patch protein Abp1p (Figure S2E).
Previous studies have observed the formation of stress granules and protein aggregates associated with nutrient starvation in budding yeast [13–16], postulated to be a consequence of cytosolic acidification concurrent with reduced ATP levels and an inability to maintain pH homeostasis [17–19]. We thus used pHluorin2, a fluorescent protein with a ratiometric signal that is pH-sensitive [20], to measure the cytosolic and mitochondrial pH of cells subjected to acute glucose starvation. We measured pH dynamics by flow cytometry, using samples arrested by centrifugation and resuspended in medium lacking glucose (see Methods).
Consistent with previous literature, we found that cytosolic pH rapidly dropped upon glucose depletion, from a mean of pH 7.6 to 6.5 (Figure 2E). Upon prolonged starvation, we observed a bimodal pH distribution centered about pH 6 and 6.7. One subpopulation recovered slightly from the initial acidification event, reminiscent of the heterogeneity in the recovery of mitochondrial morphology. We observed a subtle but consistent cytosolic pH decrease in cells containing collapsed mitochondria relative to cells retaining tubular structure when we examined glucose-starved populations by microscopy. This confirmed that the subset of cells with disrupted pH homeostasis are those with perturbed mitochondrial morphology (Figure S2E). We also observed a dramatic pH drop inside the mitochondrial matrix, followed by a bimodal distribution as a portion of cells managed to increase matrix pH thereafter (Figure 2F).
We applied a potential-sensitive, fluorescent mitochondrial dye, MitoTracker Red CM-H2Xros, to cells before and during glucose starvation to determine whether there is a disruption of the electrical potential across the inner mitochondrial membrane. This potential, in conjunction with the pH gradient created by the expulsion of protons from the mitochondrial matrix, provides the free energy gradient that drives protons through the ATP synthase complex and generates ATP [21,22]. We observed bimodality in mitochondrial potential immediately upon glucose withdrawal, with a subpopulation of cells experiencing a steep loss of potential. This initial response was followed by a continued bimodal distribution in which the worst-affected cells partially increased their membrane potential but remained consistently distinguishable from the second subpopulation (Figure 2G).
Sudden glucose starvation thus produces two populations, arresters and recoverers, that differ in mitochondrial structure and the integrity of a variety of cellular structures and processes.
Heterogeneity during nutrient transitions is a heritable trait specific to carbon source deprivation
To determine whether this unusual mitochondrial response and the variable capacity for recovery are features of a general starvation response or unique to carbon source starvation, we examined mitochondria during rapid starvation for another nutrient, nitrogen. We cultured prototrophic cells, capable of growing without external amino acid supplements, in minimal synthetic medium containing ammonium sulfate as the sole nitrogen source and glucose as the sole carbon source before switching the medium to deprive cells of either extracellular nitrogen or carbon.
As in previous experiments, removing glucose led to the collapse of mitochondrial network structure and recovery--or retention of tubularity--followed by network size increase in only a subpopulation of cells (Figure 3A, left); unlike their behavior in media containing amino acids, recoverer cells with tubular mitochondria never resumed the cell cycle or produced new buds, consistent with the absence of an energy source. The comparison with our earlier results shows that cells adapting to the absence of glucose can utilize amino acids as an alternative carbon source to support growth and proliferation (Figure S3A). However, cells deprived of nitrogen displayed neither the mitochondrial collapse nor the subsequent mitochondrial biogenesis that occurs in cells recovering from carbon starvation (Figures 3A–B and S3B).
Figure 3. Glucose specificity and heritability of mitochondrial structural collapse.
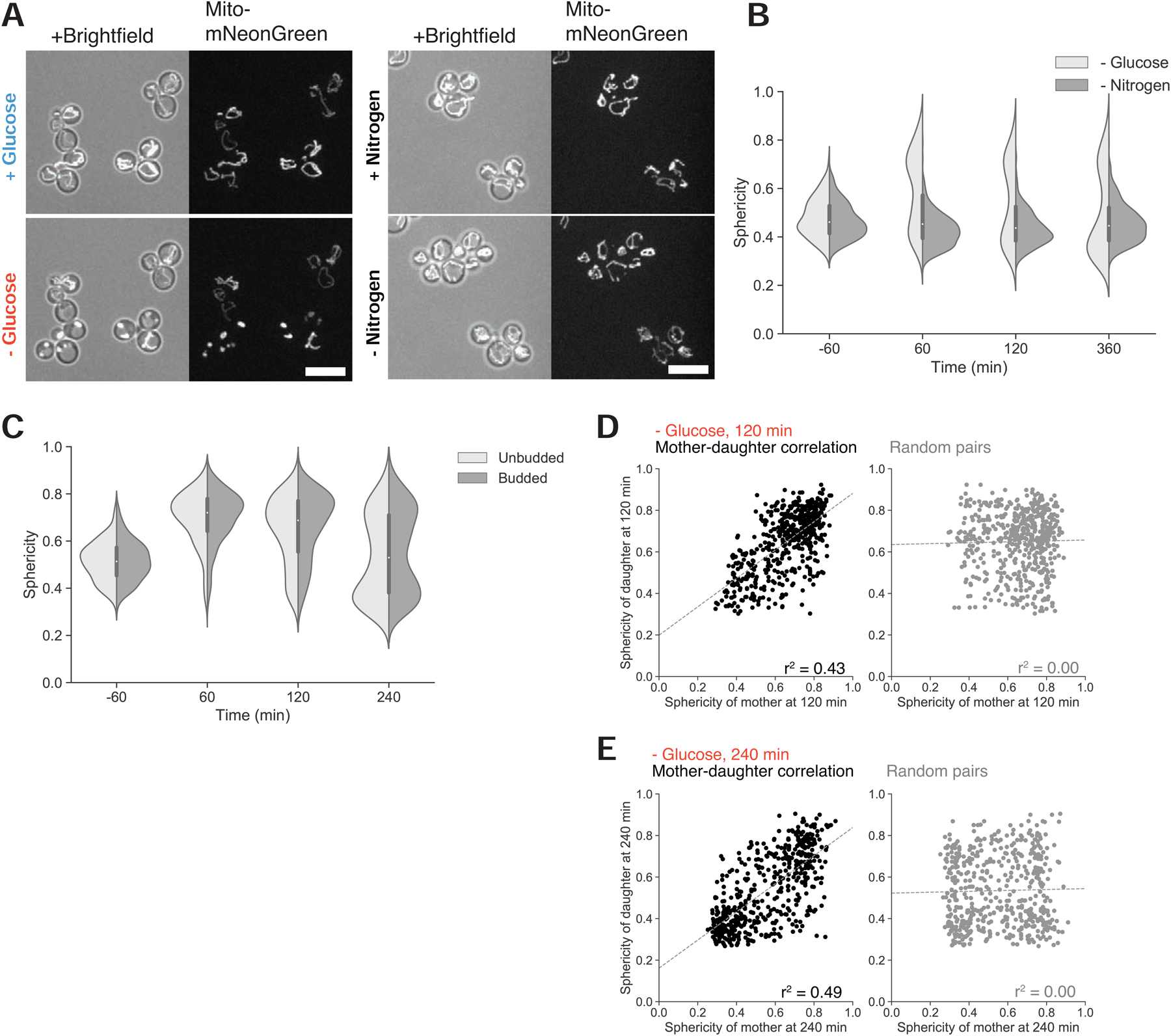
(A) Mitochondrial matrix fluorescent marker in prototrophic cells growing in synthetic medium lacking amino acids, in the presence of high glucose and 1 hr following glucose withdrawal (left), or in the same synthetic medium, before and 1 hr following nitrogen (ammonium sulfate) withdrawal (right panel). Scale bars, 10 μm.
(B) Distribution of mitochondrial sphericity in cells that were acutely deprived of glucose or nitrogen at time 0 min, N ≥ 360 cells.
(C) Mitochondrial sphericity in cells before and following glucose washout at time 0 min, partitioned by budded or unbudded state at the moment of glucose withdrawal. N ≥ 404 cells per type.
(D)-(E). Correlation between mitochondrial sphericity in mother-daughter pairs following starvation for 120 min (D) and 240 min (E), contrasted with correlation between mother and daughter cells when paired at random. Dotted lines depict regression fits calculated by least-squares method. N = 617 mother-daughter cell pairs.
See also Figure S3.
We tested whether mitochondrial collapse was due to glucose withdrawal rather than another perturbation, such as osmotic shock, that might accompany glucose disappearance. We simultaneously removed glucose and replaced it with galactose, a fermentable carbon source that requires additional enzymes, normally repressed in the presence of glucose, for its catabolism [23]. We again observed mitochondrial collapse and bimodality in mitochondrial structure (Figure S3C), suggesting that glucose starvation causes mitochondrial collapse by reducing glycolytic flux rather than by changing external osmolarity.
We wished to understand how a population of genetically identical cells could display two such divergent phenotypes upon sudden glucose starvation. The most obvious source of non-genetic heterogeneity is cell cycle state, as budding and division cycles typically proceed asynchronously within a yeast population. We examined whether differential sensitivity to glucose starvation and subsequent growth capacity could be explained by cell cycle heterogeneity by partitioning our data based on each individual cell’s budded or unbudded status at the moment of disappearance of glucose from the media. We found that each population displayed a dramatic initial increase in mitochondrial sphericity followed by the development of a bimodal distribution of cells, eliminating cell cycle stage as a major determinant of glucose starvation behavior (Figures 3C and S3D).
We next examined whether the response to abrupt glucose deprivation was instead driven by lineage-dependent heterogeneity. We identified pairs of mother and daughter cells and compared their responses to glucose withdrawal. We found that cells and their direct progenitors were likely to share the same response to starvation, with specific sphericity indices modestly correlated (r2 = 0.43) relative to randomly paired mothers and daughters (r2 = 0.00) following 2 hr glucose starvation (Figure 3D). This mother-daughter correlation was maintained as the adapting subpopulation began to alter its sphericity distribution at 4 hr post-glucose withdrawal (mother-daughter r2 = 0.49; random pairing r2 = 0.00) (Figure 3E). We conclude that the starvation response is heritable.
Glucose sensing, signaling, and utilization pathways modulate heterogeneity in response to sugar starvation
To elucidate the factors influencing cell response during acute glucose starvation, we searched for mutants with unimodal starvation behavior, focusing on the loss of gene products involved in glycolysis and associated regulatory activity. We identified four sets of single and double deletions that permit cells to recover uniformly and rapidly during sudden glucose loss by variously disrupting 1) external glucose sensing (rgt2Δ snf3Δ double mutant), 2) glucose phosphorylation (hxk2Δ), 3) glucose-dependent transcriptional repression (mig1Δ mig2Δ double mutant), or 4) the ability to inhibit Snf1p kinase, a crucial inducer of respiration (reg1Δ) (Figure 4A). The opposite phenotype, uniform and indefinite loss of mitochondrial structural integrity, is associated with loss of Snf1p (snf1Δ). Representative images of homogeneous mitochondrial collapse or recovery are shown in Figure 4B with their sphericity dynamics quantified in Figure 4C (see also Video S3). The mutants which prevent mitochondrial collapse do not display detectable increases in sphericity upon sudden glucose starvation. In contrast, snf1Δ mutants undergo similar structural collapse as wild-type cells but never recover (see also Figures S4A and S4B).
Figure 4. Glucose signaling and utilization pathways modulate starvation behavior.
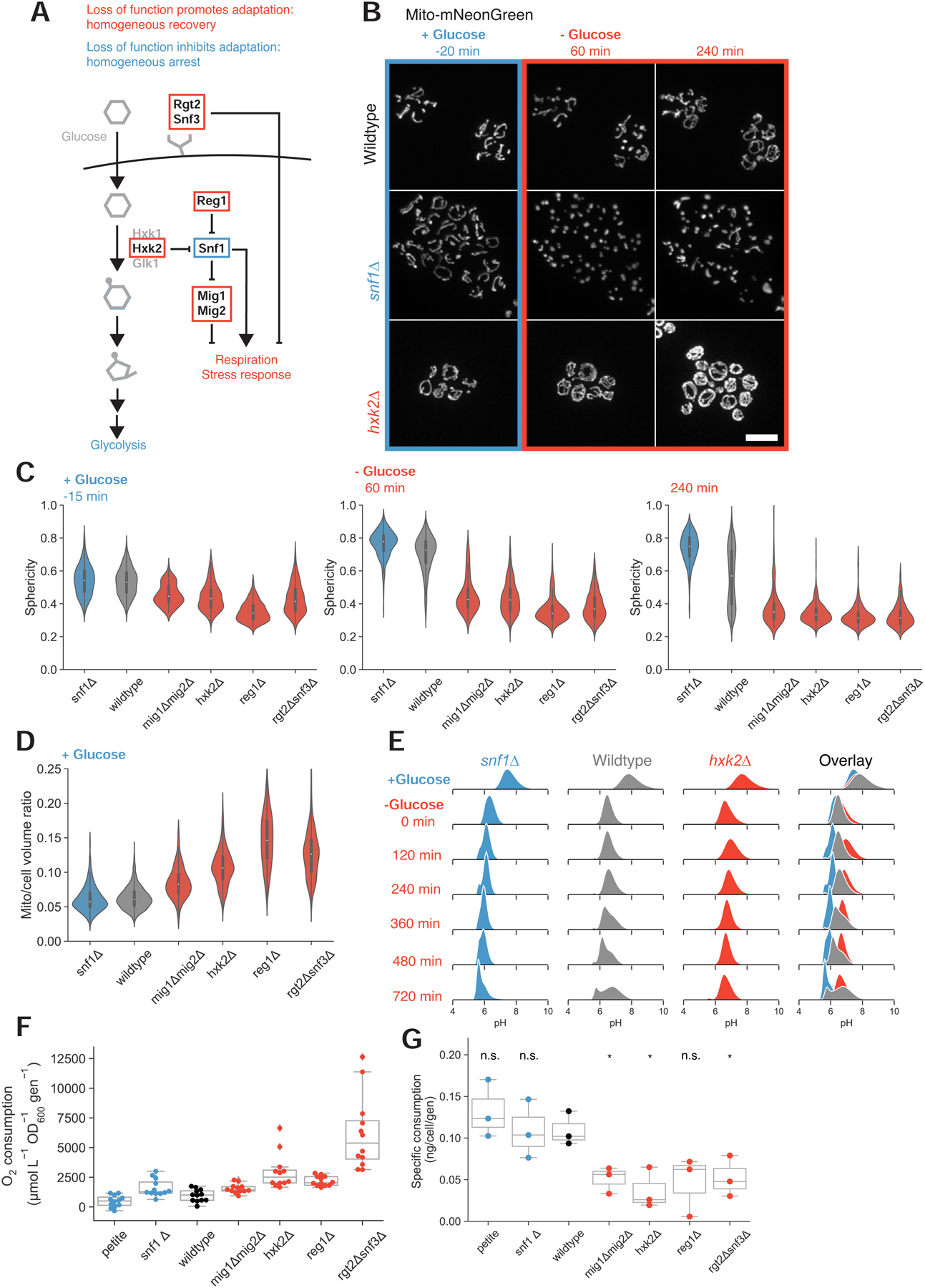
(A) Simplified depiction of selected glucose sensing and signaling pathways in budding yeast. Red boxes indicate gene products for which loss of function results in rapid, homogeneous adaptation during acute glucose starvation. The blue box identifies a gene product whose loss results in a homogeneous inability to adapt following glucose withdrawal.
(B) Mitochondrial matrix marker in wild-type (yLB126), hxk2Δ (yLB146), and snf1Δ (yLB168) cells, growing in synthetic medium containing high glucose and following abrupt removal of glucose by washout. Scale bar, 10 μm.
(C) Distribution of mitochondrial sphericity in snf1Δ (yLB168), wild-type (yLB126), mig1Δmig2Δ (yLB180), hxk2Δ (yLB146), reg1Δ (yLB196), and rgt2Δsnf3Δ (yLB233) strains growing in high glucose (left panel) and 60 min and 240 min post-glucose withdrawal. Mutants in red display relatively low sphericity and no increase during starvation. The snf1Δ mutant in blue initially resembles wild-type but uniformly fails to adapt during starvation. N ≥ 142 cells per genotype.
(D) Pre-starvation mitochondrial to cell volume ratios for the strains in (C). N ≥ 142 cells per genotype.
(E) Cytosolic pH in wild-type (yLB397), hxk2Δ (yLB416), and snf1Δ (yLB412) cells expressing ratiometric pHluorin2, prior to and following glucose starvation. All distributions consist of three biological replicates of N = 40,000 cells each.
(F) Oxygen consumption rates, normalized by optical density and growth rate, for petite (yLB73), snf1Δ (yLB167), wild-type (yLB1), mig1Δmig2Δ (yLB181), hxk2Δ (yLB145), reg1Δ (yLB194), and rgt2Δsnf3Δ (yLB232) strains growing in synthetic media containing high glucose. Data consist of three biological replicates, each comprised of four technical replicates. All Student’s two-tailed independent t test p-values against wild-type < 0.05.
(G) Glucose uptake rates, normalized by cell number and growth rate, for strains in (F). Distributions consist of three biological replicates. Student’s two-tailed independent t-test p-values ≥ 0.05 (marked “n.s.” for “not significant) against wild-type for petite (p = 0.38), snf1Δ (p = 0.99), and reg1Δ (p = 0.06). P-values < 0.05 for mig1Δmig2Δ (p = 0.02), hxk2Δ (p = 0.02), and rgt2Δsnf3Δ (p = 0.04) genotypes compared to wild-type. See also Figure S4 and Video S3.
The mitochondrial networks in our uniformly-recovering strains were larger than those of wild-type cells even before any nutrient shift, ranging from volume fractions of 0.092 (mig1Δmig2Δ) to 0.147 (reg1Δ), compared to fractions of 0.061 and 0.063 in wild-type and snf1Δ cells, respectively (Figure 4D).
The homogeneity in the response of these mutants extended to other intracellular responses, particularly the extreme pH drop observed immediately following glucose washout. The mean cytosolic pH of wild-type, hxk2Δ, and snf1Δ cells expressing pHluorin2 is initially similar, and all three strains experience cytosolic acidification upon loss of glucose (Figure 4E). However, although the heterogeneous adaptation of wild-type cells produces a bimodal distribution, the pH of each mutant is unimodal and overlaps with one of the two wild-type subpopulations: snf1Δ cells retain a low pH and hxk2Δ mutants reach a new steady-state of slightly increased pH several hours in advance of the recovering wild-type cells. Each of these mutant strains thus appears to recapitulate one of the two wild-type subpopulations, albeit with faster recovery dynamics in hxk2Δ cells.
Even in high-glucose environments, our fast-adapting, fully-recovering mutants are all closer in mitochondrial content to glucose-starved cells than they are to wild-type cells. We hypothesized that these mutations recover homogeneously because they were prepared for starvation: they had already begun to transition to a respiratory state before glucose suddenly disappeared from the medium. Prior work has demonstrated that a history of growth on a non-glucose carbon source can decrease lag time during subsequent shifts from glucose to other non-glucose sugars [24].
To test whether our recoverer strains were respiring in glucose, we measured oxygen consumption rates in the presence of high glucose for seven strains: wild-type, the five mutant strains discussed above, and a respiratory-deficient petite strain (Figure 4F). The oxygen consumption rates of all four mutants that lock cells in the recoverer state were significantly elevated above wild-type (p ≤ 0.02), consistent with the notion that prior respiratory activity is beneficial during sudden glucose starvation. Wild-type cells, in turn, consumed oxygen faster than petites (p = 0.03). Curiously, snf1Δ mutants, which cannot grow on non-fermentable carbon sources, consumed oxygen more rapidly than our wild-type strain (p = 0.02). This anomalous behavior has been observed previously and attributed to oxidation of amino acids [25].
Previous work has detected a correlation between glucose uptake rate and relative rates of fermentation and respiration, suggesting that metabolic activity may be regulated through the rate of sugar influx [26,27]. We therefore measured the rate of glucose uptake in wild-type and homogeneous arrester and recoverer strains. We found that wild-type, respiratory-deficient petite, and snf1Δ strains have similarly high levels of glucose uptake, while three of our four recoverer mutants displayed statistically significant decreases in glucose uptake (Figure 4G). The strain that does not, reg1Δ, grows very poorly and produced noisy data. We conclude that perturbations that reduce glycolytic flux force cells to respire more, locking them in the recoverer phenotype.
Preparation for future glucose starvation allows for faster adaptation at an immediate fitness cost
We wondered whether initial mitochondrial network size could explain the variability in the starvation response of wild-type populations. We ranked 1,329 cells by their mitochondrial volume fraction immediately before glucose starvation and tracked the least- and most-mitochondrially endowed 10% of cells as they responded to glucose removal. The high-mitochondrial subgroup had a higher fraction of recoverers than the general population and was enriched for cells that recovered within an hour of starvation (Figures 5A and 5C). In contrast, cells with the lowest mitochondrial volume fraction were less likely to decrease in sphericity after the initial mitochondrial collapse and cells that never increased in sphericity were absent from this subpopulation (Figures 5B and 5D). Thus the mitochondrial volume fraction partially predicts a cell’s future behavior. Because the prediction is imperfect, we believe that mitochondrial volume is not fully correlated with the parameter that determines whether cells arrest or recover.
Figure 5. Starvation adaptation is associated with preparatory activity that imposes a short-term fitness cost.
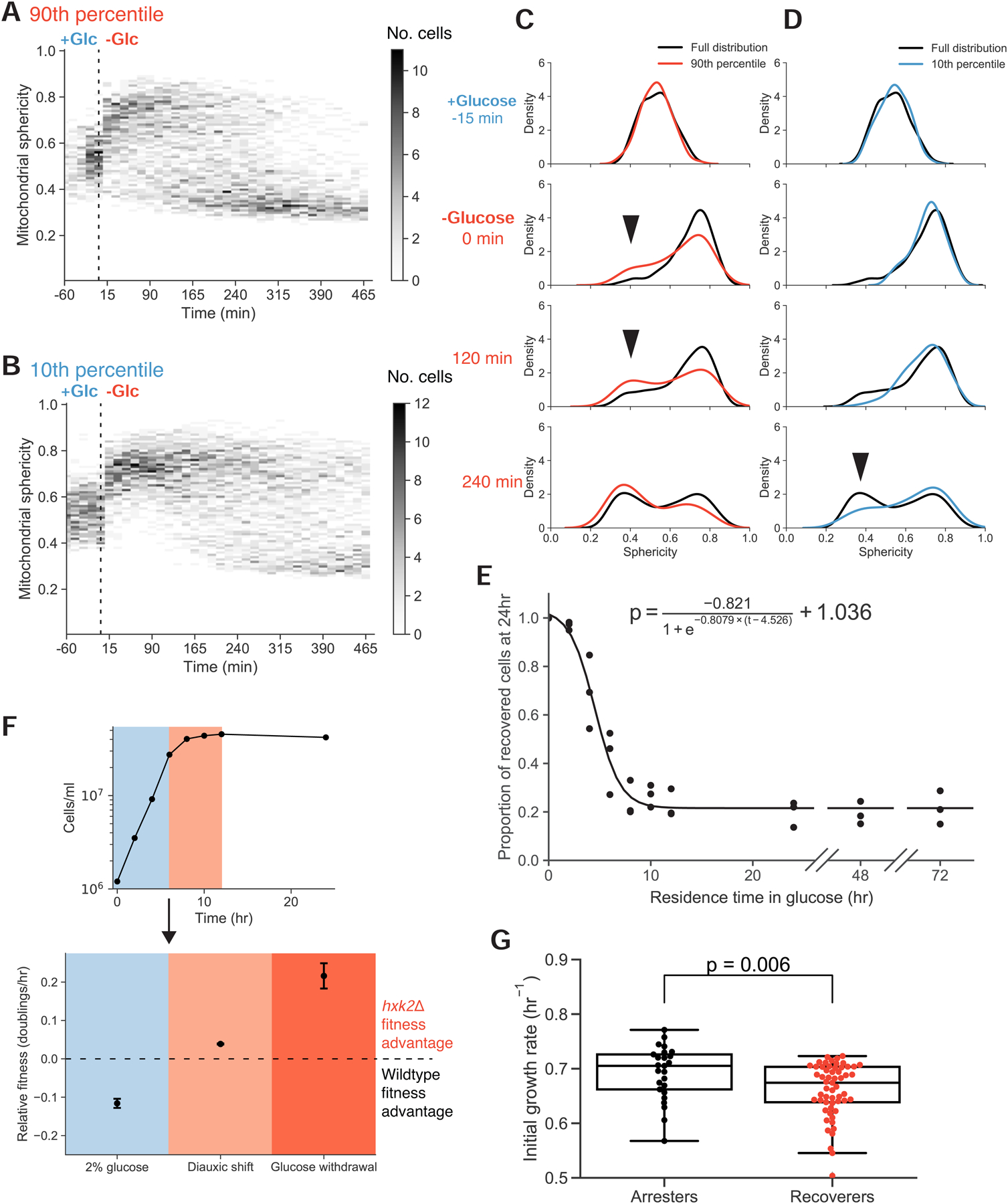
(A)-(B) Time-resolved heat maps of mitochondrial sphericity in the presence of high glucose and during acute glucose starvation for cells with the top 10% (A) and bottom 10% (B) mitochondrial to total cell volume ratios immediately prior to glucose washout (N = 133 and N = 132, respectively). Intensity reflects the absolute number of cells with a mitochondrial sphericity index within the given bin.
(C)-(D) Histograms of selected timepoints from (A) and (B), relative to the entire distribution. Arrows highlight discrepancies in relative subpopulation sizes.
(E) Adaptation probability as a function of residence time in media containing high glucose prior to glucose disappearance. Cells expressing Hxt3p-mNeonGreen (yLB432) were grown in synthetic medium lacking amino acids and containing non-fermentable potassium acetate as a carbon source, then switched into otherwise identical medium containing high glucose for the specified number of hours prior to sudden glucose starvation. Recovery was scored as the presence or absence of Hxt3p-mNeonGreen signal by flow cytometry. The solid line represents the logistic function, displayed above, calculated from the data by non-linear least squares fit. Three biological replicates, N = 40,000 cells for each time point.
(F) Upper panel: cell density of a culture initiated with equal proportions of wild-type (yLB365) and hxk2Δ (yLB373) in synthetic medium containing high glucose. Blue and red shaded regions indicate exponential and diauxic growth phases, respectively. Lower panel: relative fitness of wild-type and hxk2Δ mutants during exponential phase growth and after the diauxic shift (as in upper panel) calculated as the relative rates of cell doublings/hr and measured by the changes in the ratios of the two genotypes by flow cytometry. Equivalent fitness measurement, in rates of cell number increase in the first 8 hours following sudden glucose deprivation, is displayed in darker red. Error bars, one standard deviation. Three biological replicates, N = 40,000 cells analyzed by flow cytometry for all time points of all conditions.
(G) Distribution of growth rates of single-lineage microcolonies in the presence of high glucose, partitioned by the failure or success of lineages to complete one doubling in the 12 hr following glucose starvation (arresters and recoverers, respectively). Mann-Whitney U test p-value = 0.006.
See also Figure S5.
These data imply a history-dependent metabolic flexibility. Cells with larger mitochondrial networks, lower glycolytic flux, weaker glucose repression, and some respiration adapt faster to sudden glucose deprivation. We wished to test this directly. For higher-throughput batch experiments, we identified the abundance of the hexose transporter Hxt3p as a marker of presence or absence of metabolic adaptation to glucose starvation. Hxt3p is highly expressed during fermentative growth, but during growth on nonfermentable sugars, its transcription is repressed and existing protein is degraded [28–30]. Recovers degrade their Hxt3p, but in arresters, which fail to adapt, Hxt3p continues to reside on the plasma membrane. The behavior of Hxt3p during glucose starvation matched our expectations: it was lost from all hxk2Δ cells, retained on all snf1Δ cells, and lost on those wild-type cells that recovered and retained on those that did not (Figure S5A).Flow cytometry validated the bimodal response of Hxt3p to glucose starvation (Figure S5B).
We asked how the fraction of recoverers within a population depended on the metabolic history of the culture. We induced cells expressing Hxt3p-mNeonGreen to respire by culturing them with a nonfermentable carbon source. We then transferred cells to media containing high concentrations of glucose for varying times before resuspending them in media lacking glucose. We performed these experiments in prototrophic strains, omitting amino acids from all media. Because neither arresters nor recoverers can divide in this context, this allowed us to score the probability with which cells can successfully switch metabolic states based on the proportion of cells that have lost Hxt3p-mNeonGreen signal. As the population spends more time in high glucose, the fraction of recoverers falls with the dynamics that fit a logistic decay curve (r2 = 0.95) (Figure 5E). Many generations after their ancestors last respired, the culture reaches a steady state in which roughly 20% of cells are recoverers.
If starvation recovery, or lack thereof, is a binary state as observed in both mitochondrial morphology and Hxt3p abundance, then all populations would consist of some combination of the two types. A culture grown from a small number of founding cells on a nonfermentable carbon source is, by definition, composed solely of the recoverer type, as this metabolic state is a prerequisite for growth and division in this context. These cells have already successfully assumed a respiratory metabolic state. As we observe a decrease in recovery probability with increasing exposure time to glucose, we thus see interconversion between the two states as a previously homogeneous population develops heterogeneity. The decrease in recovery probability as cells proliferate on glucose reflects the conversion rate between recoverer and arrester metabolic states. By fitting our data to a logistic curve, we estimate this rate to be 0.23 hr−1.
For a culture maintained in exponential phase in glucose-rich media for 24 hr or more, the steady state ratio of arrester cells to recoverer cells reveals the ratio of the switching rate from recoverer to arrester to that from arrester to recoverer. At steady state, we measured an arrester:recoverer ratio of 3.65, inferring that the rate at which arrester cells become recoverers must be 0.23 hr−1 / 3.65 = 0.06 hr−1. We also measured these rates directly by growing individual founder cells into extended lineages in our microfluidics device and detecting switching events through inconsistencies in cell pedigrees once these lineages were subjected to sudden starvation (see Methods). We calculated the recoverer-to-arrester switching rate to be 0.18 ± 0.02 hr−1 and the arrester-to-recoverer switching rate as 0.08 ± 0.02 hr−1, with both values consistent with our bulk assays.
Why do all cells not retain the metabolic flexibility to adapt to sudden environmental perturbations, given its obvious advantages? Our data suggest that recoverer cells are prepared for starvation due to partial glucose derepression, outright respiratory activity, or some intermediary status on the path to fully respiratory metabolism. If so, in environments featuring abundant glucose, recoverers would generate ATP at slower total rates or bear protein or transcriptional loads that other cells would not. These cells would grow and replicate more slowly to maintain their metabolic flexibility, sacrificing current growth potential for resistance to a future period of glucose starvation. Previous work has identified such a fitness tradeoff during shifts between glucose and maltose in hxk2Δ mutants [31].
We tested for context-dependent fitness differences between wild-type cells and uniformly-recovering hxk2Δ mutants. We inoculated cultures with equal numbers of cells of each genotype, each bearing a different fluorescent label. We tracked their relative changes in abundance in synthetic media containing glucose during exponential phase growth and through the diauxic shift. In separate cultures, we performed glucose withdrawal and measured the relative effect on each strain.
We found that hxk2Δ cells display a fitness defect of approximately 12% during growth in glucose-containing media but have a fitness advantage of 3.9% after glucose is depleted (Figures 5F, S5C, and S5D). During sudden glucose starvation, hxk2Δ cells out-proliferate wild-type cells by 21.6%. These reciprocal relative fitnesses in the presence or absence of glucose are consistent with our hypothesis.
We next examined whether a reciprocal fitness effect is detectable within wild-type populations. Single-cell growth rates are difficult to measure in S. cerevisiae as emerging buds are poorly resolved by microscopy. We instead pooled measurements across microlineages, tracking the total area encompassed by individual founding cells and their progeny across several cell divisions and assessing the collective growth of the pedigree before and following sudden glucose washout. We partitioned these microcolonies into two states, recoverers and arresters, and compared the pre-starvation growth rates across the two groups. The recoverer and arrester groups were enriched in slower-growing and faster-growing cells, respectively, in the original, high-glucose environment (p = 0.006, Figure 5G). These data indicate that cells best able to adapt during sudden glucose deprivation grow more slowly in high glucose.
Bimodal starvation behavior with natural variation in steady states in wild yeast isolates
If cells must balance their short-term competitive fitness against long-term stress resistance, then we might expect that strains dwelling in distinct niches would all obey a similar fitness tradeoff but would evolve to establish distinct arrester:recoverer ratios in response to their particular environmental challenges. To test this hypothesis, we integrated our Hxt3p-mNeonGreen marker for metabolic adaptation into ten S. cerevisiae strains isolated from a range of contexts [32]. We measured their pairwise fitnesses in the presence of high glucose and following sudden glucose starvation, using competition against one strain as a common reference.
We observed an inverse correlation between these strains’ relative fitness in the presence or sudden absence of glucose across all strains (slope = −0.71, r2 = 0.84) (Figure 6A). We examined mitochondria and Hxt3p, the diagnostic hexose transporter, in one of these natural isolates, BC187, during microfluidics-based glucose withdrawal, and saw similar bimodality between arresters and recoverers (Figures 6B and 6C). We conclude that the presence of these two physiological states extends beyond laboratory cultivation and that the balance between them is likely to have been shaped by local ecological forces.
Figure 6. Bimodal starvation behavior obeys a fitness tradeoff in natural yeast strains.
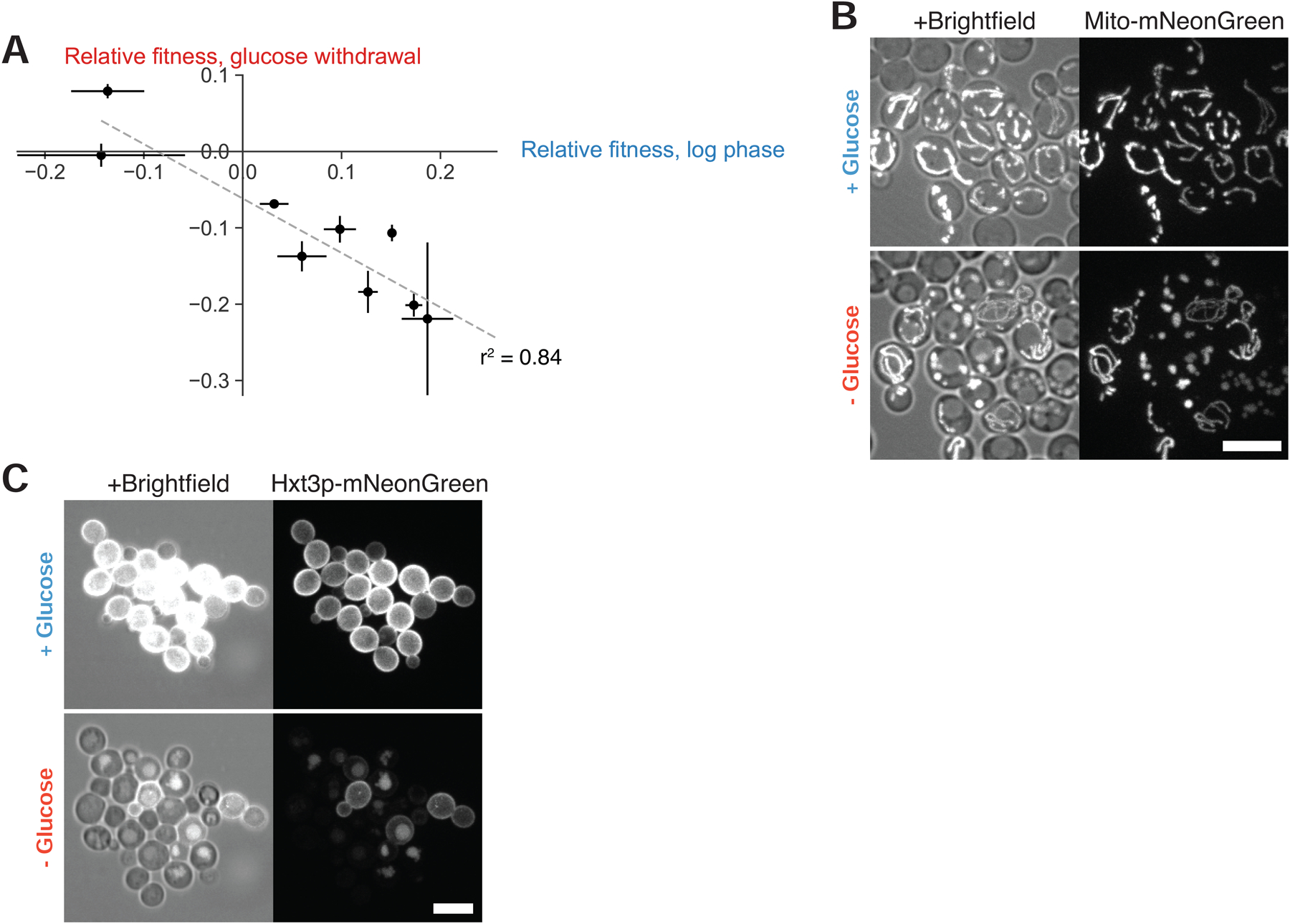
(A) Relative fitnesses of diploid derivatives of YJM978 (yLB463), CEN.PK (yLB467), DBVPG1373 (yLB470), L-1374 (yLB474), BC187 (yLB478), Y12 (yLB486), K11 (yLB492), YPS606 (yLB494), and UWOPS83–787.3 (yLB496) expressing mNeptune under the ACT1 promoter, measured against a common referance, YS2 (yLB480), when assayed in pairwise competitions, both in exponential phase growth in the presence of abundant glucose and following abrupt glucose deprivation. Three independent biological replicates measured for all competitions. Error bars, one standard deviation.
(B)-(C) Microscopic images revealing bimodality in mitochondrial matrix morphology (B) and Hxt3p-mNeonGreen (C) in diploid BC187 derivative (yLB453), in synthetic medium containing high glucose and 7 hr following abrupt glucose withdrawal. Scale bars, 10 μm.
DISCUSSION
We investigated the response of budding yeast cells to sudden glucose starvation using mitochondrial abundance and morphology as proxies for a cell’s metabolic state. Glucose starvation causes mitochondrial collapse in the majority of cells together with a fall in intracellular pH and electric potential across the mitochondria. As starvation continues, cells respond bimodally, reflecting an inherited state: recoverers return to a normal mitochondrial morphology, increase their mitochondrial content, raise intracellular pH and mitochondrial potential, and—when amino acids are present in the media--resume growth, while arresters remain arrested with collapsed mitochondria. Mutations that affect metabolic regulators can lock cells in either state. Recoverers grow more slowly in the presence of high glucose but have a strong fitness advantage during the switch from glucose to non-fermentable carbon sources. These two states are present in wild yeast strains, with variation in their relative proportions.
Based on the correlations between cell growth rates with mitochondrial morphology, intracellular pH, and mitochondrial potential, we propose a model of budding yeast metabolism illustrated in Figure 7. We argue that genetically homogeneous populations growing in the presence of abundant glucose are bimodal in rates of glycolysis and in broader metabolic state, with one subset of cells, arresters, fermenting heavily and another subpopulation, recoverers, displaying partial glucose derepression and respiration. We posit that when glucose is removed, arresters experience a profound drop in ATP production due to their reliance on glycolysis. This deficit cannot easily be remedied and leads to a prolonged arrest without functional mitochondria. Recoverers, already respiratory-competent, can easily respire amino acids in the medium (and, potentially, any storage carbohydrates) to restore intracellular ATP levels and resume growth.
Figure 7. A model of mitochondria and metabolic transitions.
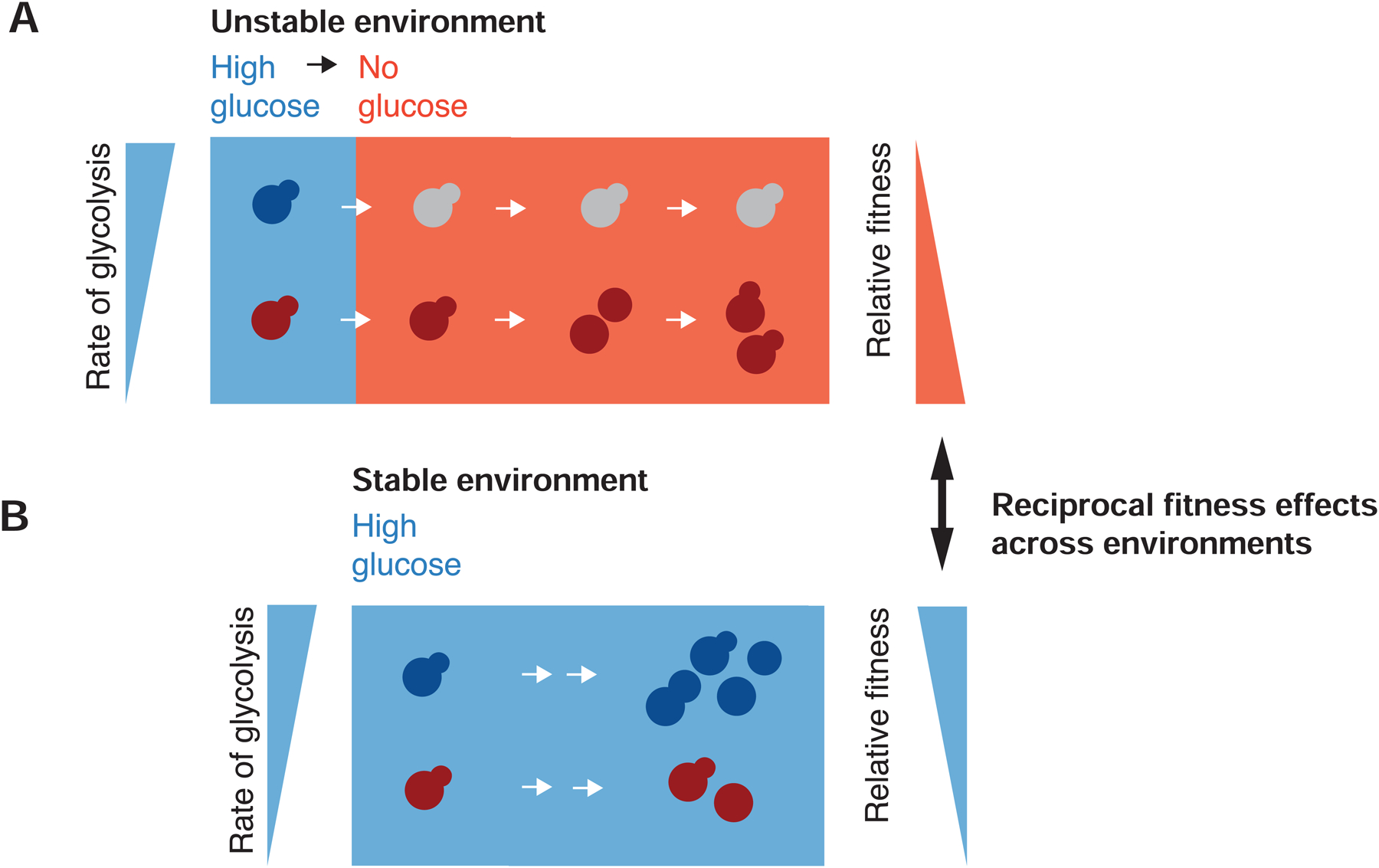
(A) A clonal population of yeast growing in the presence of abundant glucose (blue background) is composed of cells in two distinct metabolic states in which the relative rate of glycolysis is either high or low (blue and red cells, respectively). Upon sudden glucose starvation (red background), cells with high glycolytic activity arrest indefinitely, while those with low activity adapt and ultimately resume growth. Lower initial rates of glycolysis result in a fitness advantage during starvation.
(B) In stable, high-glucose conditions, maximizing the rate of glycolysis (blue cells) can sustain a higher growth rate and thus produce a fitness advantage.
Although recoverers have an advantage upon glucose starvation, arresters grow faster at high glucose concentrations, most likely by investing less in producing mitochondria and more rapidly generating ATP from glycolysis (Figure 5G). The arrester strategy emphasizes carbon source specialization, utilizing high levels of glucose as efficiently as possible and is thus vulnerable to glucose starvation. The recoverer strategy is generalist: cells grow more conservatively in rich environments to retain growth potential during environmental instability. A genetically homogeneous population contains cells of each type: those who are more competitive in the current context, and those who will be more resistant to unexpected stresses. The combination helps the population to balance its current fitness concerns against future environmental uncertainty.
This metabolic generalist-specialist tradeoff bears a resemblance to the growth behavior of E. coli during the transition from glucose, a preferred carbon source, to gluconeogenic sources. Experimental evolution produces two genetically distinct, coexisting subpopulations of cells: one which grows rapidly before the diauxic shift but slowly afterwards, and another which grows more slowly before diauxy but shows a smaller lag at the transition [33]. These experiments indicate that the dynamics of nutrient depletion offer multiple niches for distinct metabolic phenotypes. Similar bistability, in which one subpopulation adapts while another arrests, has also been observed within genetically homogenous E. coli populations [34]. Our findings indicate that such diversification is present within the carbon metabolic strategies of eukaryotes as well.
What determines the state of an individual cell? We have demonstrated that daughter cells are likely to display the same phenotype as their mothers, indicating that metabolic state is heritable. Our collective experiments have ruled out, in addition to long metabolic memory, age dependence (given the heritability of our phenotype, as yeast daughters are born young at the expense of their mothers), cell cycle status, and cell size in the determination of these states. We observe these state ratios in well-mixed liquid cultures as well as in microfluidic chambers, precluding spatial effects in cell state establishment. We therefore propose that metabolic state switching occurs through stochastic changes in gene expression. Genome sequencing and expression analysis of E. coli metabolic types indicates that state-switching capacity is governed by multiple metabolic enzymes and substrate uptake transporters [33–35]; these precedents, together with the diversity in glucose uptake rates observed in our mutant strains, offer a wealth of candidate mechanisms by which metabolic activity could fluctuate.
The adaptive value of recoverer cells (or “persisters”), which have prepared for starvation by reducing their current growth rate, depends on the probability of a sudden environmental challenge. If no such event arrives, recoverers reduce the population’s overall fitness. Evolutionary forces should shape the switching rates between the two states as a function of the relative frequency with which a population with access to abundant glucose experiences sudden starvation: more frequent starvation should be associated with a higher steady-state frequency of recoverers. Accordingly, we observe variation in carbon utilization strategies across natural yeast isolates.
In its simplest form, bet hedging is defined as a stochastic variation between two phenotypes, each of which has superior fitness on one of two environments. The fitness of a bet hedging population in a temporally varying environment is higher than that of populations that uniformly adopt either of the two phenotypes. Is the behavior we observed bet hedging? Our experiments identify phenotypic heterogeneity within clonal populations that produces two cell states, each of which displays a fitness advantage in one of two environments. In addition, strains from the wild obey similar tradeoffs between growth in abundant glucose compared to growth during glucose starvation, suggesting that this tradeoff is under natural selection. As it stands, this makes our study of heterogeneity in the response to glucose starvation a strong case for the existence of bet hedging in budding yeast.
A conclusive demonstration of bet hedging must satisfy multiple criteria: it must establish that there are two phenotypic states, that these two states have reciprocally different fitness values in two environments, that a mixture of both phenotypes is adaptive for an individual population within a fluctuating environment, and that the ratio of the two states (determined by the switching rates between them) is optimized for the particular frequency of environmental fluctuation under which the population has evolved [36,37]. The latter two criteria directly link the nature and degree of phenotypic heterogeneity to the underlying environmental instability, which distinguishes bet hedging from a cellular response to a cue from the environment [36,37]. While previous claims of bet hedging in yeast (e.g. [38,39]) have failed to meet several of these criteria, our experiments fulfill all but the last two. We cannot show that the wild-type populations, which switch phenotypes, are superior to populations locked in either state, because the mutations that force cells into either state have additional fitness costs. The final criterion, the ability of selection to alter the dynamics of non-genetic individuality, has only rigorously been shown for bacterial persistence [40]. The observation that wild yeast isolates have different tradeoffs between the two environments and show the same arrester and recoverer phenotype leads us to speculate that selection has optimized this mechanism to the dynamics of the environments that these strains occupy. Moreover, while previous work identified natural variation in the duration of the diauxic lag phase and a reciprocal fitness effect in the presence of high glucose and another carbon source [3,31,41], our combination of following the behavior of individual cells, lineage analysis, and fitness measurements in microlineages allows us to build upon these prior advances, establishing the cell biological nature and reproductive consequences of this heterogeneity.
When the dynamics of natural environmental fluctuations are unknown, as is the case for our natural yeast strains, we cannot verify the correspondence between the statistics of bet hedging and those of the environment. However, our nutrient-withdrawal paradigm, applied in the context of experimental evolution, would permit direct testing of both the existence of bet hedging and, if applicable, the genetic mechanisms by which the frequencies and properties of recoverers and arresters are controlled. By evolving yeast under conditions that fluctuate between glucose-rich media and sudden glucose starvation, with nutrient shifts occurring at varying average frequencies, it would be possible to determine whether the rate of metabolic state switching between cell types optimizes the geometric mean fitness determined by the rate of environmental instability. If successful, this could offer insight into the mechanisms that control this bimodality, the rates of interconversion between states, and the nature of bet hedging in budding yeast.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andrew Murray (awm@mcb.harvard.edu).
Materials availability
Yeast strains and plasmids created in this study are available upon request from the Lead Contact.
Data and code availability
Mitochondrial morphological data and code used in the analysis and production of figures are available at https://github.com/bagitmery/yeast_mitochondria_and_metabolism.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains and media
All experiments were performed using strains constructed in a modified W303 background in which the bud4 and rad5-535 alleles were replaced with their respective, functional loci from the S288C background (MATa leu2-3,112 can1-100 ura3-1 his3-11,15 BUD4-S288C RAD5). The mito-mNeonGreen construct was derived from pVT100U-mtGFP, a gift from Benedikt Westermann (Addgene plasmid 45054) [42] and consists of a pRS403 integrating vector [43] harboring an ADH1 promoter, the first 69 amino acids of subunit 9 of the Neurospora crassa F0 ATPase (the mitochondrial presequence preSu9), a short linker sequence [44], mNeonGreen [45], and an ADH1 terminator. All sequences were codon-optimized for expression in yeast.
Integrating plasmids carrying fluorescent pH reporters were constructed by introducing a yeast-optimized sequence encoding ratiometric pHluorin2 [20] and an ADH1 terminator into the pRS403 integrating vector. In the mitochondrial pH reporter, pHluorin2 was placed under the control of the ADH1 promoter, directly downstream of preSu9; in the cytosolic pH reporter, expression was controlled by the ACT1 promoter.
All other fluorescent fusion constructs consist of a common linker and a fluorescent protein joined to the C terminus of the labeled protein, expressed from its native locus and under its native promoter. Fusion sequences were joined by isothermal assembly [46]. All constructs and markers for both fluorescent labeling and gene disruptions were introduced by standard yeast genetic methods [47] and confirmed by PCR and, where applicable, DNA sequencing.
Respiratory-deficient petite strains were produced by incubating cells in 25 μg/ml ethidium bromide for 24 hr. Loss of mitochondrial DNA was verified by DAPI staining and the absence of growth on non-fermentable carbon sources.
Experiments were performed in synthetic complete (SC) media prepared from 10x yeast nitrogen base (YNB); 100x stocks of adenine, tryptophan, and uracil; 2 g/L complete amino acids lacking the three amino acids above; 10x stock of the specified carbon source, if any; and sterilized water. For bulk assays, YNB was prepared according to the Wickerham specifications [48]; for microscopy assays, this recipe was modified to omit riboflavin and folic acid, the two primary contributors to autofluorescence [44]. Where noted, select bulk assays and microfluidics-based rate-switching assays were performed in minimal media lacking all amino acids but otherwise prepared as described for SC media. Ingredients for the preparation of these media were purchased from Sigma-Aldrich.
For growth and starvation behavior assays, strains were streaked from stocks stored in 15% glycerol and sterilized water at −70°C onto agar plates of the appropriate selective dropout media. After two days of growth at 30°C, single colonies were used to inoculate cultures that were continuously maintained in early exponential phase (a density of less than 5 × 106 cells/ml as measured on a Beckman Coulter Counter), also at 30°C, for a minimum of 24 hr and no more than 36 hr (unless otherwise noted), prior to the initiation of any experiment. All experiments consist of a minimum of three biological replicates performed on separate days.
METHOD DETAILS
Imaging sample preparation and microfluidics
Single-timepoint images of cells grown in varying carbon sources were imaged in liquid media in 96-well plates with optical glass bottoms (Grenier Bio-One), precoated with 0.5 mg/ml concanavalin A (MP Biomedicals) in sterile water.
Cells examined during and following abrupt nutrient shifts were immobilized in silicone CellASIC Y04 plates for haploid yeast cells, and the flow of media was controlled with the CellASIC ONIX microfluidic system (EMD Millipore). Plates were pretreated through perfusion of 0.5 mg/ml concanavalin A, freshly dissolved in a buffer of 10 mM Na2HPO4 and 0.5 mM CaCl2, pH 6.5, immediately prior to use. Priming with this solution was performed at an initial flow rate of 5 psi for 5 min, followed by a 90 min perfusion at 2 psi, then washout with yeast media for 5 min at 5 psi. Cell cultures, prepared as described above, were loaded into the chamber at 4 psi for 10 sec. Cells were continually supplied with growth or starvation media at a rate of 2 psi thereafter; based on the manufacturer’s specified flow properties, we estimate this to result in roughly 4.5 complete media exchanges per minute within the cell chamber. Cells were allowed to equilibrate within the chamber for a minimum of 1 hr before the primary carbon source was removed.
For measuring mitochondrial potential, cells were incubated with 0.5 μM MitoTracker Red CM-H2Xros in the dark for 15 min prior to imaging or flow cytometry.
Microscopy
Images were acquired using a Nikon TI inverted microscope equipped with a Yokogawa CSU-10 dual spinning disk confocal unit, 16.0 μm-pixel Hamamatsu ImagEM EM-CCD camera, Nikon 100x NA 1.45 TIRF objective, and MetaMorph software. 488 and 594 nm lasers were used with 525/45 nm (GFP) and 609/57 nm (RFP) bandpass filters for imaging of green (mNeonGreen) and red (mNeptune) fluorophores, respectively, with exposure times of 122.12 ms. Z-stacks were collected in 0.2 μm slices with a minimum of 8 μm total depth. The two-dimensional images shown were flattened by maximum intensity z-projection. Microscopy was performed at room temperature.
Bulk glucose withdrawal
Cells grown in batch culture conditions were starved of glucose after allowing a clonal culture, derived from a single, freshly streaked colony, to grow exponentially for a minimum of 24 hr in the presence of high glucose as described above. Cells in 5–10 ml culture volumes were collected by centrifugation at 600 rpm for 5 min, washed twice with 50 ml media lacking glucose, and resuspended in 5–10 ml of glucose-free media. At regular intervals, samples were analyzed by flow cytometry as described below.
Flow cytometry
Samples were prepared for flow cytometry by treating 1 ml aliquots of cell culture with cycloheximide (Sigma-Aldrich) added to a final concentration of 100 μg/ml. Samples were stored at 4°C in the dark until analysis, performed within one day. Cycloheximide stocks were prepared at 100x in ethanol and stored at −70°C for no more than one year. Analysis was performed on an LSR II flow cytometer (BD Biosciences) using the 488 nm laser and a 505 nm long pass filter and 530/30 nm bandpass filter to measure mNeonGreen intensity. For pH assays, samples were analyzed live, in the original media environment and without additional treatment, using the 405 nm and 488 nm lasers, each coupled with a 505 nm long pass and 530/30 nm bandpass filter. For all experiments, 40,000 cells were analyzed. Data were imported into the Python 2.7 environment using the FlowCytometryTools package and custom Python scripts were used to isolate the single-cell population on the basis of forward and side scatter profiles and to examine the distributions of fluorescent signals.
Competition assays
Strains labeled with either mNeonGreen of mNeptune, expressed under the ACT1 promoter from a construct integrated at the HIS3 locus, were grown separately in log phase in complete synthetic medium containing 2% glucose for a minimum of 24 hr prior to fitness testing. Cells were mixed at a 1:1 ratio in similar complete synthetic medium at a density of 4 × 106 cells/ml and either maintained in the presence of high glucose or subjected to bulk glucose withdrawal as described above. Samples were extracted every 1–2 hr, treated with 100 μg/ml cycloheximide and analyzed promptly by flow cytometry, with the ratio of each cell type calculated from clustering based on a Gaussian mixture model with full covariance matrix, implemented via the scikit-learn library in Python 2.7. Relative growth dynamics were quantified as the ratio of doublings per hour.
Calibration and measurement of pH
In situ calibration of the pHluorin2 ratiometric reporter was performed by collecting cells expressing the pHluorin2 construct and growing them in exponential phase in synthetic complete medium, resuspending them in PBS, and treating with 100 μl/ml digitonin, to permeabilize the plasma membrane, for 10 min. Cells were then separated into nine aliquots and resuspended in a series of citric acid-Na2HPO4 buffers spanning pH 5.0–9.0 in half-pH-unit intervals. Cells were analyzed immediately by flow cytometry as described above. The relative signals obtained from the 405 nm and 488 nm laser settings in three biological replicates were plotted against pH and fit to an exponential function. In subsequent live-cell experiments, this standard curve was used to compute pH from relative 405 nm / 488 nm signal intensity.
Oxygen depletion measurements
Oxygen depletion was measured in 96-well microplates containing embedded oxygen optical sensors (OxoPlate OP96C, PreSens). Cells were grown for 24–48 hr in log phase in synthetic medium containing 2% glucose (or other carbon sugars as noted) and added to microplate wells at a density of 4 × 106 cells/ml. Sensor output was measured using a Synergy Neo2 plate reader (BioTek) with a monochromator set for excitation at 540 ± 2 nm and emission at 509/20 nm (reference signal) and 650/20 nm (indicator signal). Fluorescent signal and absorption at 600 nm were measured every 15 min for 8 hr at 30°C with plate shaking between time points.
Glucose uptake rate measurements
Glucose uptake rates were inferred from the depletion rates of glucose from culture medium. Cell density (by Beckman Coulter Counter) and glucose concentration (Glucose HK Assay Kit, Sigma) were measured at regular intervals in cells growing exponentially in synthetic medium containing 2% glucose. Absorbance at 340 nm, directly proportional to glucose concentration in the medium, was measured on a NanoDrop 1000 Spectrophotometer (Thermo Fisher). Cultures were incubated at 30°C with continuous shaking between measurements.
Starvation phenotype state-switching measurements
The rates of interconversion between starvation phenotypes were calculated from microfluidics experiments in which cells expressing Hxt3p-mNeonGreen (yLB432) were pregrown in synthetic medium lacking amino acids and containing 2% glucose for 6 hr, after which the flow was replaced with identical medium containing 0% glucose. Cells were classified as arrested or recovering on the basis of their retention or turnover, respectively, of Hxt3p-mNeonGreen 6 hr post-glucose withdrawal. See analysis below.
QUANTIFICATION AND STATISTICAL ANALYSIS
General reporting of statistical parameters
The exact sample numbers of cells N for all experiments are included within their respective figure legends and were collected over a minimum of three biological replicates. Figure panels variously display data from individual cells, population distributions, and/or population means and standard deviations as described in their associated legends. Other parameters, including r2 values of best-fit lines and p-values, are described below and in each figure’s accompanying text.
Image analysis
Brightfield images of cells were segmented with the CellStar algorithm run via MATLAB plugin [49]. The resulting cell masks were manually inspected and corrected in the event of over- or under-segmentation. Cell volume was calculated by fitting the generated cell contour to an ellipse and projecting into three dimensions based on the geometry of a prolate ellipsoid. Buds smaller than 20 μm3 were excluded from analysis; this was determined to be the threshold for the generation of reliable masks by the manual examination of > 200 cells. Tracking, also performed by the CellStar algorithm, was used to assign unique identification numbers to all masks and allow for analysis of single-cell trajectories. Prior to mitochondrial analysis, cell masks were used to crop images and generate single-cell z-stacks. Background noise as calculated within a five-pixel band about the cell contour was used to populate all pixels outside of the cell mask. Cell volumetric calculations and preprocessing for mitochondrial analysis were performed by custom MATLAB scripts available at https://github.com/bagitmery/yeast_mitochondria_and_metabolism.
Mitochondrial segmentation was performed using MitoGraph software [50]. Three-dimensional reconstructions of mitochondrial content in Visualization ToolKit (VTK) format were imported into a Python 2.7 environment via the Python VTK wrapper library. Mitochondrial and whole-cell volumetric data were matched by cell identification numbers and statistical analysis and plotting were performed in Python. Mitochondrial/cell volume ratios were calculated as the total mitochondrial volume divided by the total cell volume. For sphericity index, the volume of each mitochondrion was used to calculate the surface area of a sphere of equivalent volume, which was divided by the actual surface area of that mitochondrion. This value was calculated for all topologically distinct mitochondrial units individually, and an overall sphericity index was calculated as the average of all distinct mitochondrial sphericity values within a cell, weighted by the volume of each mitochondrion. Individual mitochondrial units of less than 45 nm3 total volume were flagged as potential artifacts; this was determined to be the threshold of reliable segmentation by the manual examination of > 200 cells. Cells containing five or more such fragments were excluded from analysis.
Cells’ budded or unbudded state, mother-daughter relationships, and time of visible growth resumption during glucose starvation were annotated manually and integrated with mitochondrial data and analyzed in Python.
Composite images, maximum intensity z-projections, and Videos were assembled for presentation using the Fiji distribution of ImageJ [51].
Oxygen consumption analysis
In oxygen depletion assays, oxygen saturation was computed from the ratiometric signal according to the manufacturer’s instructions, using air-saturated and oxygen-depleted water (0.01 g/ml Na2SO3) as calibration standards, measured in parallel with the biological samples. Oxygen depletion rates were calculated by normalizing oxygen readings to optical density, performing a linear regression on the linear portion of the depletion trace by least squares fitting, and reporting the obtained slope. Slopes were measured for three biological replicates, each consisting of four technical replicates. P-values were calculated using a Student’s two-tailed independent t-test.
Glucose uptake rate analysis
Glucose concentrations in extracellular media were calculated from absorbance at 340 nm according to the assay kit manufacturer’s instructions, using serial dilutions of glucose at 0.0625 – 1.0 mg/ml as standards. Glucose uptake rate for cell cultures was calculated as change in extracellular glucose concentration over the observed time interval, normalized by cell density and the culture’s growth rate. Rates were obtained for three biological replicates. P-values were calculated using a Student’s two-tailed independent t-test.
Starvation phenotype state-switching analysis
Cells imaged in state-switching experiments described above were classified as arrested or recovering on the basis of their retention or turnover, respectively, of Hxt3p-mNeonGreen 6 hr post-glucose withdrawal. The birth state of each cell was inferred by comparing its phenotype to those of its ancestors and descendants and assuming the fewest state switches necessary to produce the observed phenotypic patterns within a lineage. These data, along observation time between each cell’s birth and its sudden starvation, were collected for a total of 2,702 cells across five independent experiments. These data were partitioned based on the initial phenotype of each cell at birth, and the proportion of cells not experiencing a phenotypic switch was calculated for all cells with a shared observed lifetime (binned into 15-min increments, the frame rate at which fluorescent images were collected). These probabilities were fit to the k = 0 case of the Poisson distribution in which the average number of events per interval is a product of the switching rate and the length of the observation window. We calculated a recovererto-arrester switching rate of 0.18 ± 0.02 hr−1, with our data fitting a Poisson probability mass function with r2 = 0.77. For arrester-to-recoverer switching, we restricted our analysis to cells with observed lifetimes of 90 min or less, as the probability of switching decreased after 90 min, likely due to the relatively high probability of cells switching back to the arrester phenotype during longer observation times. We calculated an arrester-to-recoverer switching rate of 0.08 ± 0.02 hr−1, with r2 = 0.70.
Single-lineage growth measurements
The MATLAB implementation of the Canny edge detection algorithm was used to determine the boundaries of microcolonies within brightfield microscopic images. Edges were subsequently connected by dilation. The two-dimensional area circumscribed by these boundaries was measured during growth in the presence of high glucose and following glucose deprivation, performed by microfluidics as described above. Pre-starvation growth rate was calculated by performing a linear regression on the increase in base two logarithm of observed area over time in the presence of high glucose. The ability of cells to switch metabolic states at any point within the lineage, though rare, results in microlineages of mixed states and confounds the resulting measurements. We therefore made comparisons between two classes of lineages: those in which all members collectively completed one round of division following 12 hr of glucose starvation, and those which collectively failed to achieve one doubling (Figure S5E). Distributions of pre-starvation growth rates were compared by Mann-Whitney U test.
Supplementary Material
Video S1. Heterogeneity in mitochondrial morphology during sudden glucose starvation. Related to Figure 1. Time lapse imaging of mito-EGFP (yLB113) (right panel), with brightfield overlay (left panel) during growth in synthetic medium containing glucose (blue background) and during abrupt glucose washout (red background). Cells display heterogeneity in their ability to retain mitochondrial tubular structure and perform mitochondrial biogenesis. The presence of multiple clonal clusters showing no collapse is unusual, and this field is shown here for the purposes of clearly displaying the differences between divergent phenotypes. Time stamp indicates hours:minutes relative to glucose withdrawal occurring at time 0 hr. Time interval between frames, 45 min. Scale bar, 10 μm.
Video S2. Diversity in mitochondrial structural state during acute glucose starvation, then uniform morphology during refeeding. Related to Figure 1. Time lapse of cells shown in Figure 1D expressing mito-mNeonGreen (yLB126) (right panel) with brightfield overlay (left panel), imaged during growth in synthetic medium containing glucose (blue background), for 8 hr glucose starvation (red background), and for 4 hr during the refeeding on high-glucose media (blue background). Time stamp indicates hours:minutes relative to glucose withdrawal occurring at time 0 hr. Glucose is returned to the media at 8 hr. Time interval between frames, 15 min. Scale bar, 10 μm.
Video S3. Mutants in glucose signaling and utilization adopt homogeneous cell fates during sudden nutrient shifts. Related to Figure 4. Time lapse imaging of wild-type (yLB126, top row), hxk2Δ (yLB146, middle row), and snf1Δ (yLB168, bottom row) cells expressing mito-mNeonGreen, with and without brightfield overlay (left and right columns, respectively), during growth in synthetic medium containing high glucose and following abrupt washout of glucose from the media (blue and red backgrounds, respectively). Wild-type cells display heterogeneous arrest and recovery, while hxk2Δ mutants recover homogeneously and snf1Δ mutants arrest homogeneously. Time stamp indicates hours:minutes relative to glucose washout at time 0 hr. Time interval between frames, 15 min. Scale bar, 10 μm.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Concanavalin A | MP Biomedicals | Cat#0219528301 |
| Cycloheximide | MilliporeSigma | Cat#C7698 |
| MitoTracker Red CM-H2Xros | ThermoFisher | Cat#M7513 |
| Critical Commercial Assays | ||
| Glucose (HK) Assay Kit | MilliporeSigma | Cat#GAHK20 |
| Deposited Data | ||
| Raw mitochondrial data and analysis scripts | This study, GitHub | https://github.com/bagitmery/yeast_mitochondria_and_metabolism |
| Experimental Models: Organisms/Strains | ||
| Yeast strains | See Table S1 | N/A |
| Recombinant DNA | ||
| Plasmids | See Table S2 | N/A |
| pVT100U-mtGFP | [42] | Addgene plasmid #45054 |
| Software and Algorithms | ||
| Analysis scripts | This study, GitHub | https://github.com/bagitmery/yeast_mitochondria_and_metabolism |
| CellStar | [49] | https://www.cellstar-algorithm.org |
| MitoGraph v 2.0 | [50] | https://github.com/vianamp/MitoGraph |
| Fiji | [51] | https://imagej.net/Fiji |
| Other | ||
| CellASIC ONIX Microfluidic Platform | MilliporeSigma | Cat#C117908 |
| CellASIC ONIX Microfluidic Yeast Plates | MilliporeSigma | Cat#Y04C-02 |
| OxoPlate | PreSens | Cat#OP96C |
Highlights:
Non-genetic heterogeneity is present in the budding yeast glucose starvation response
Starvation capacity is bimodal and heritable
Ability to adapt is inversely correlated with high-glucose growth rate
Natural variation in the bimodal distribution suggests the existence of bet hedging
ACKNOWLEDGMENTS
This work was supported by NIH grants DP2AI117923-01 to E.C.G., R01GM043987 to A.W.M. and the NSF/Simons Center for Mathematical & Statistical Analysis of Biology at Harvard (#1764269 (NSF) and #594596 (Simons Foundation)). L.E.B. was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1144152. The authors thank the Harvard Bauer Core Facility for technical assistance. We thank P. Stoddard, S. Srikant, Y. Sun, and other members of the Murray and Garner labs for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Johnston M, and Carlson M (1992). Carbon regulation in Saccharomyces. Molecular and Cellular Biology of the Yeast Saccharomyces, Broach JR, Pringle JR, Jones EW (eds) Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 2.Zaman S, Lippman SI, Zhao X, and Broach JR (2008). How Saccharomyces responds to nutrients. Annu. Rev. Genet 42, 27–81. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Atolia E, Hua B, Savir Y, Escalante-Chong R, and Springer M (2015). Natural variation in preparation for nutrient depletion reveals a cost-benefit tradeoff. PLoS Biol. 13, e1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kayikci Ö, and Nielsen J (2015). Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 15 Available at: 10.1093/femsyr/fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yotsuyanagi Y (1962). [Study of yeast mitochondria. I. Variations in mitochondrial ultrastructure during the aerobic growth cycle]. J. Ultrastruct. Res 7, 121–140. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann HP, and Avers CJ (1973). Mitochondrion of yeast: ultrastructural evidence for one giant, branched organelle per cell. Science 181, 749–751. [DOI] [PubMed] [Google Scholar]
- 7.Stevens BJ (1977). Variation in number and volume of the mitochondria in yeast according to growth conditions. A study based on serial sectioning and computer graphics reconstitution. J. Microsc. Biol. Cell Available at: http://agris.fao.org/agris-search/search.do?recordID=US201302523462.
- 8.Egner A, Jakobs S, and Hell SW (2002). Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochondria in live yeast. Proc. Natl. Acad. Sci. U. S. A 99, 3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess SM, Delannoy M, and Jensen RE (1994). MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J. Cell Biol 126, 1375–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sogo LF, and Yaffe MP (1994). Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol 126, 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyn J, Hibbs AR, Sanz P, Crowe J, and Meyer DI (1988). In vivo and in vitro analysis of ptl1, a yeast ts mutant with a membrane-associated defect in protein translocation. EMBO J. 7, 4347–4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Sanders SL, Feldheim DA, and Schekman R (1991). Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349, 806–808. [DOI] [PubMed] [Google Scholar]
- 13.Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, and Marcotte EM (2009). Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. U. S. A 106, 10147–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrovska I, Nüske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz J-M, et al. (2014). Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife. Available at: 10.7554/eLife.02409. [DOI] [PMC free article] [PubMed]
- 15.Zacharogianni M, Aguilera-Gomez A, Veenendaal T, Smout J, and Rabouille C (2014). A stress assembly that confers cell viability by preserving ERES components during amino-acid starvation. Elife 3 Available at: 10.7554/eLife.04132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suresh HG, da Silveira Dos Santos AX, Kukulski W, Tyedmers J, Riezman H, Bukau B, and Mogk A (2015). Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae. Mol. Biol. Cell 26, 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orij R, Postmus J, Ter Beek A, Brul S, and Smits GJ (2009). In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155, 268–278. [DOI] [PubMed] [Google Scholar]
- 18.Dechant R, Binda M, Lee SS, Pelet S, Winderickx J, and Peter M (2010). Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munder MC, Midtvedt D, Franzmann T, Nüske E, Otto O, Herbig M, Ulbricht E, Müller P, Taubenberger A, Maharana S, et al. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife 5 Available at: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahon MJ (2011). pHluorin2: an enhanced, ratiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol 2, 132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholls DG (2002). Mitochondrial function and dysfunction in the cell: its relevance to aging and aging-related disease. Int. J. Biochem. Cell Biol 34, 1372–1381. [DOI] [PubMed] [Google Scholar]
- 22.Nicholls DG (2004). Mitochondrial membrane potential and aging. Aging Cell 3, 35–40. [DOI] [PubMed] [Google Scholar]
- 23.Timson DJ (2007). Galactose metabolism in Saccharomyces cerevisiae. Dyn. Biochem. Process Biotechnol. Mol. Biol 1, 63–73. [Google Scholar]
- 24.Cerulus B, Jariani A, Perez-Samper G, Vermeersch L, Pietsch JM, Crane MM, New AM, Gallone B, Roncoroni M, Dzialo MC, et al. (2018). Transition between fermentation and respiration determines history-dependent behavior in fluctuating carbon sources. Elife 7 Available at: 10.7554/eLife.39234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicastro R, Tripodi F, Guzzi C, Reghellin V, Khoomrung S, Capusoni C, Compagno C, Airoldi C, Nielsen J, Alberghina L, et al. (2015). Enhanced amino acid utilization sustains growth of cells lacking Snf1/AMPK. Biochim. Biophys. Acta 1853, 1615–1625. [DOI] [PubMed] [Google Scholar]
- 26.Elbing K, Larsson C, Bill RM, Albers E, Snoep JL, Boles E, Hohmann S, and Gustafsson L (2004). Role of hexose transport in control of glycolytic flux in Saccharomyces cerevisiae. Appl. Environ. Microbiol 70, 5323–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huberts DHEW, Niebel B, and Heinemann M (2012). A flux-sensing mechanism could regulate the switch between respiration and fermentation. FEMS Yeast Res. 12, 118–128. [DOI] [PubMed] [Google Scholar]
- 28.Perez M, Luyten K, Michel R, Riou C, and Blondin B (2005). Analysis of Saccharomyces cerevisiae hexose carrier expression during wine fermentation: both low- and high-affinity Hxt transporters are expressed. FEMS Yeast Res. 5, 351–361. [DOI] [PubMed] [Google Scholar]
- 29.Roberts GG, and Hudson AP (2006). Transcriptome profiling of Saccharomyces cerevisiae during a transition from fermentative to glycerol-based respiratory growth reveals extensive metabolic and structural remodeling. Mol. Genet. Genomics 276, 170–186. [DOI] [PubMed] [Google Scholar]
- 30.Snowdon C, Schierholtz R, Poliszczuk P, Hughes S, and van der Merwe G (2009). ETP1/YHL010c is a novel gene needed for the adaptation of Saccharomyces cerevisiae to ethanol. FEMS Yeast Res. 9, 372–380. [DOI] [PubMed] [Google Scholar]
- 31.New AM, Cerulus B, Govers SK, and Perez-Samper G (2014). Different levels of catabolite repression optimize growth in stable and variable environments. PLoS. Available at: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed]
- 32.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spencer CC, Bertrand M, Travisano M, and Doebeli M (2007). Adaptive diversification in genes that regulate resource use in Escherichia coli. PLoS Genet. 3, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotte O, Volkmer B, Radzikowski JL, and Heinemann M (2014). Phenotypic bistability in Escherichia coli’s central carbon metabolism. Mol. Syst. Biol 10, 736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Gac M, Brazas MD, Bertrand M, Tyerman JG, Spencer CC, Hancock REW, and Doebeli M (2008). Metabolic changes associated with adaptive diversification in Escherichia coli. Genetics 178, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Jong IG, Haccou P, and Kuipers OP (2011). Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays 33, 215–223. [DOI] [PubMed] [Google Scholar]
- 37.Simons AM (2011). Modes of response to environmental change and the elusive empirical evidence for bet hedging. Proc. Biol. Sci 278, 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levy SF, Ziv N, and Siegal ML (2012). Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 10, e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Giardina DM, and Siegal ML (2018). Control of nongenetic heterogeneity in growth rate and stress tolerance of Saccharomyces cerevisiae by cyclic AMP-regulated transcription factors. PLoS Genet. 14, e1007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fridman O, Goldberg A, Ronin I, Shoresh N, and Balaban NQ (2014). Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421. [DOI] [PubMed] [Google Scholar]
- 41.Venturelli OS, Zuleta I, Murray RM, and El-Samad H (2015). Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 13, e1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westermann B, and Neupert W (2000). Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427. [DOI] [PubMed] [Google Scholar]
- 43.Sikorski RS, and Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheff MA, and Thorn KS (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670. [DOI] [PubMed] [Google Scholar]
- 45.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- 47.Amberg DC, and Strathern JN (2005). Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual (CSHL press; ). [Google Scholar]
- 48.Wickerham LJ (1951). Taxonomy of yeasts (U.S. Dept. of Agriculture).
- 49.Versari C, Stoma S, Batmanov K, Llamosi A, Mroz F, Kaczmarek A, Deyell M, Lhoussaine C, Hersen P, and Batt G (2017). Long-term tracking of budding yeast cells in brightfield microscopy: CellStar and the Evaluation Platform. J. R. Soc. Interface 14 Available at: 10.1098/rsif.2016.0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viana MP, Lim S, and Rafelski SM (2015). Quantifying mitochondrial content in living cells. Methods Cell Biol. 125, 77–93. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Heterogeneity in mitochondrial morphology during sudden glucose starvation. Related to Figure 1. Time lapse imaging of mito-EGFP (yLB113) (right panel), with brightfield overlay (left panel) during growth in synthetic medium containing glucose (blue background) and during abrupt glucose washout (red background). Cells display heterogeneity in their ability to retain mitochondrial tubular structure and perform mitochondrial biogenesis. The presence of multiple clonal clusters showing no collapse is unusual, and this field is shown here for the purposes of clearly displaying the differences between divergent phenotypes. Time stamp indicates hours:minutes relative to glucose withdrawal occurring at time 0 hr. Time interval between frames, 45 min. Scale bar, 10 μm.
Video S2. Diversity in mitochondrial structural state during acute glucose starvation, then uniform morphology during refeeding. Related to Figure 1. Time lapse of cells shown in Figure 1D expressing mito-mNeonGreen (yLB126) (right panel) with brightfield overlay (left panel), imaged during growth in synthetic medium containing glucose (blue background), for 8 hr glucose starvation (red background), and for 4 hr during the refeeding on high-glucose media (blue background). Time stamp indicates hours:minutes relative to glucose withdrawal occurring at time 0 hr. Glucose is returned to the media at 8 hr. Time interval between frames, 15 min. Scale bar, 10 μm.
Video S3. Mutants in glucose signaling and utilization adopt homogeneous cell fates during sudden nutrient shifts. Related to Figure 4. Time lapse imaging of wild-type (yLB126, top row), hxk2Δ (yLB146, middle row), and snf1Δ (yLB168, bottom row) cells expressing mito-mNeonGreen, with and without brightfield overlay (left and right columns, respectively), during growth in synthetic medium containing high glucose and following abrupt washout of glucose from the media (blue and red backgrounds, respectively). Wild-type cells display heterogeneous arrest and recovery, while hxk2Δ mutants recover homogeneously and snf1Δ mutants arrest homogeneously. Time stamp indicates hours:minutes relative to glucose washout at time 0 hr. Time interval between frames, 15 min. Scale bar, 10 μm.
Data Availability Statement
Mitochondrial morphological data and code used in the analysis and production of figures are available at https://github.com/bagitmery/yeast_mitochondria_and_metabolism.


