Abstract
The past 30 years have seen the emergence and proliferation of isothermal amplification methods (IAMs) for rapid, sensitive detection and quantification of nucleic acids in a variety of sample types. These methods share dependence on primers and probes with quantitative PCR, but they differ in the specific enzymes and instruments employed, and are frequently conducted in a binary, rather than quantitative format. IAMs typically rely on simpler instruments than PCR analyses due to the maintenance of a single temperature throughout the amplification reaction, which could facilitate deployment of IAMs in a variety of environmental and field settings. This review summarizes the mechanisms of the most common IAM methods and their use in studies of pathogens, harmful algae and fecal indicators in environmental waters, feces, wastewater, reclaimed water, and tissues of aquatic animals. Performance metrics of sensitivity, specificity and limit of detection are highlighted, and the potential for use in monitoring and regulatory contexts is discussed.
Keywords: NASBA, LAMP, qPCR, water quality, harmful algal blooms, pathogens, fecal indicators
Introduction
Technological advances in the past few decades have provided rapid, quantitative methods for measuring specific microorganisms in the environment. Such methods can benefit human and ecosystem health by answering questions about environmental water quality in a precise and timely manner. For example, the sanitary (microbial) quality of recreational waters has historically been assessed by culturing fecal indicator bacteria (e.g. Escherichia coli, enterococci). Culture methods typically require incubation periods of 18-24 hours (U.S. Environmental Protection Agency 2002, 2006), therefore results are reported the day after sampling occurs. The one-day lag delays important decisions regarding beach closures or warnings, and can lead to exposure of beachgoers to waterborne pathogens (Korajkic et al. 2018). In order to mitigate this shortcoming, molecular methods have been adapted for recreational water quality monitoring, particularly quantitative real-time PCR (qPCR), which can produce results in as little as few hours (U.S. Environmental Protection Agency 2010). Similarly, qPCR has been a method of choice for microbial source tracking (MST) (Shanks et al. 2012), which enables identification of fecal pollution sources. MST is an important tool for water quality managers since not all fecal contamination poses equal risk to human health (Soller et al. 2010). However, qPCR is not without drawbacks, as it requires expensive instrumentation and highly skilled technicians, and it can also be subject to inhibition by various substances found in environmental samples (Casper et al. 2005).
Like many technologies, isothermal amplification methods (IAMs) originated in the clinical arena as simple and inexpensive diagnostic tools that also possessed the high target specificity inherent in methods based on nucleic acid sequences (Obande and Banga Singh 2020). IAMs were adopted by the food industry as a diagnostic test for pathogens (Shang et al. 2020), as well as a means to identify certain plant and animal species (Kitamura et al. 2019, Lee et al. 2017, Wang et al. 2019, Xiong et al. 2020). More recently, IAMs have been applied to environmental samples, including ambient water, wastewater, fecal material from humans and other animals, sediment/soil/vegetation as well as animal tissues (Chen et al. 2013, Heijnen and Medema 2009, Karanis et al. 2007, Koloren and Ayaz 2016, Kou et al. 2006, Martzy et al. 2017, Melville et al. 2014).
IAMs offer attractive alternatives to the more established PCR-based methods due to considerations of instrumentation and personnel requirements (training and labor), as well as potential for field use. Instrumentation can be less expensive because the assay temperature does not cycle (Qi et al. 2018), and training requirements for personnel may be less sophisticated.. As a contemporary example, an IAM device developed for use in the field (e.g., using the rechargeable, battery-operated AmpliFire®) currently cost ~$9,000.00, while colorimetric assays require no instrumentation and therefore instrumentation costs are minimal. By comparison, the majority of thermal-cyclers required for qPCR start at double that amount (~$20,000).
The use of IAMs in environmental testing is a relatively recent practice, emerging as an area of wide research interest in the last decade. Major knowledge gaps therefore remain about the performance of IAMs in environmental settings compared to the well-documented performance of qPCR methods (Harwood et al. 2014). Specificity metrics for a given target are frequently not available, limits of detection are not always documented and potential for inhibition of the amplification reaction is largely unexplored in the environment. Use of the technology in field settings is an even more recent development, and questions about sample preparation and potential for contamination in the field are under-explored. The goal of this review is to present an overview of IAMs used to analyze environmental samples for pathogenic microorganisms and fecal indicators, and to examine the performance metrics reported for the various methods.
Protocol for Inclusion and Exclusion of Articles
Articles included in this review were selected according to pre-defined eligibility criteria as recommended by the PRISMA-P 2015 statement for systematic reviews (Moher et al. 2015). We searched the databases PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), Web of Science (https://clarivate.com/webofsciencegroup/solutions/web-of-science/), Scopus (https://www.elsevier.com/solutions/scopus), ScienceDirect (https://www.sciencedirect.com/) and JSTOR (https://www.jstor.org/), using following keywords: isothermal amplification, nucleic acid sequence-based amplification (NASBA), loop mediated isothermal amplification (LAMP), strand displacement amplification (SDA), signal mediated amplification of RNA technology (SMART), circular helicase-dependent amplification (HDA), rolling circle amplification (RCA), multiple displacement amplification (MDA), and recombinase polymerase amplification (RPA). Search results were limited to articles published since 2005, and identified over 800 publications that included our search terms. Relevant articles, which totaled 97, were limited to those that analyzed microorganisms in ambient water, wastewater, sediment, vegetation, soil, shellfish or fecal samples. Sixty-seven of the 97 articles included at least one of the performance metrics for the IAM(s), i.e. sensitivity, specificity, limit of detection. Articles in which SDA, SMART, and MDA were utilized did not include any performance metric, and were therefore ruled out by these eligibility criteria.
Summary Statistics of Articles Reviewed
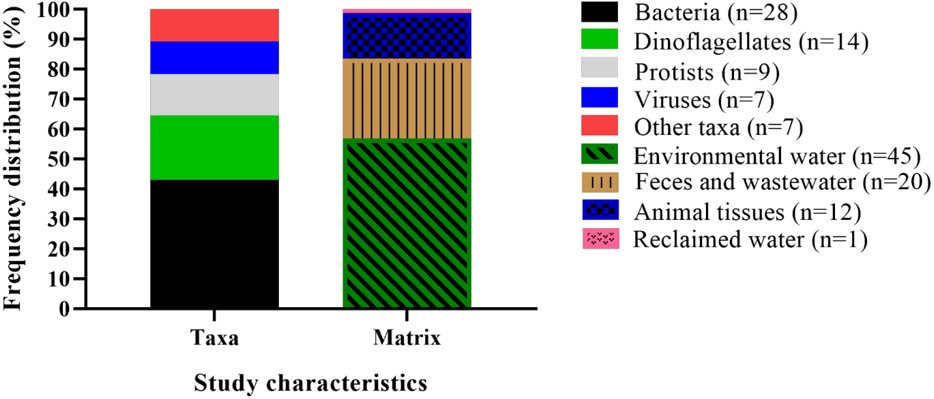
Assay targets varied widely among studies: bacteria were the most common assay targets (43%), followed by dinoflagellates (22%), protozoa (14%) and viruses (11%) (Figure 1; Table S1). Cyanobacteria, various phytoplankton, flukes, and marker genes are also represented but together comprise less than 11% of the articles included in this review (Figure 1; Table S1). Among the most frequently targeted organisms were Vibrio spp. (13), Cryptosporidium spp. (5), Karlodinium veneficum (4), Karenia spp. (3), noroviruses (3), Amphidinium carterae (2), Escherichia coli (2), Enterococci spp. (2), Giardia duodenalis (2), and Ostreopsis spp. (2), while remaining taxa represented by a single paper comprised of 37% of the total studies (Figure 1; Table S1). Matrices tested included freshwater, groundwater, marine or estuarine water (57.7%), sewage or feces (25.6%), animal tissues (15.4%) and reclaimed water (1.3%) (Figure 1; Table S1).
Figure 1.
Distribution of article subjects in terms of taxa (bacteria, dinoflagellates, protists, viruses, other organisms) and matrix (environmental water, animal feces, animal tissues, or other)
Overview of IAMs
Isothermal amplification methods (IAMs) share a basic premise with PCR: they require at least a primer and a nucleic acid template in order to amplify a specific nucleic acid sequence. All methods require at least one forward primer in the reaction mix (Table 1, Figure 2), and most methods also require a reverse primer. Each primer is typically ~20 - 35 nucleotides (Table 1). Reaction mixes also typically include the following: buffers (e.g., Tris-HCl, KCl, MgSO4), dNTPs, betaine to enhance amplification of GC-rich sequences, and Trixton X-100. A polymerase is required to synthesize nucleic acid, and most IAMS include at least one additional enzyme, which varies depending upon the method (Table 1) and whose functions are discussed below. Unlike the enzymes used in PCR methods, IAM polymerases do not have to be thermostable, which increases flexibility in method design. The template for amplification may be DNA, or cDNA derived from RNA (Table S1).
Table 1.
Molecular underpinning of isothermal amplification methods
| Methods | Isothermal temperature1 |
Enzymes2 | Primers3 |
|---|---|---|---|
| Loop mediated isothermal amplification (LAMP) | 60-65°C | DNA polymerase with strand displacement activity, e.g. Bst polymerase | Two inner (forward and reverse), two outer (forward and reverse) primers; optional, two loop primers (forward and reverse) |
| Nucleic acid sequence-based amplification (NASBA) | 41°C | Avian myeloblastosis reverse transcriptase (AMV-RT), RNase H, T7 RNA polymerase | Forward and reverse primers |
| Rolling circle amplification (RCA) | 37-65°C | Ligase, DNA polymerase with strand displacement activity | Forward primer |
| Recombinase polymerase amplification (RPA) | 37-42°C | Recombinase, DNA polymerase | Forward and reverse primers |
| Helicase-dependent amplification (HDA) | 37-65°C | DNA helicase, DNA polymerase | Forward and reverse primers |
Assays using double-stranded DNA as a template require prior denaturation step at 94°C
DNA assays can be amended to amplify RNA template by inclusion of a reverse-transcriptase step utilizing RNA-dependent DNA polymerase and random primer sets
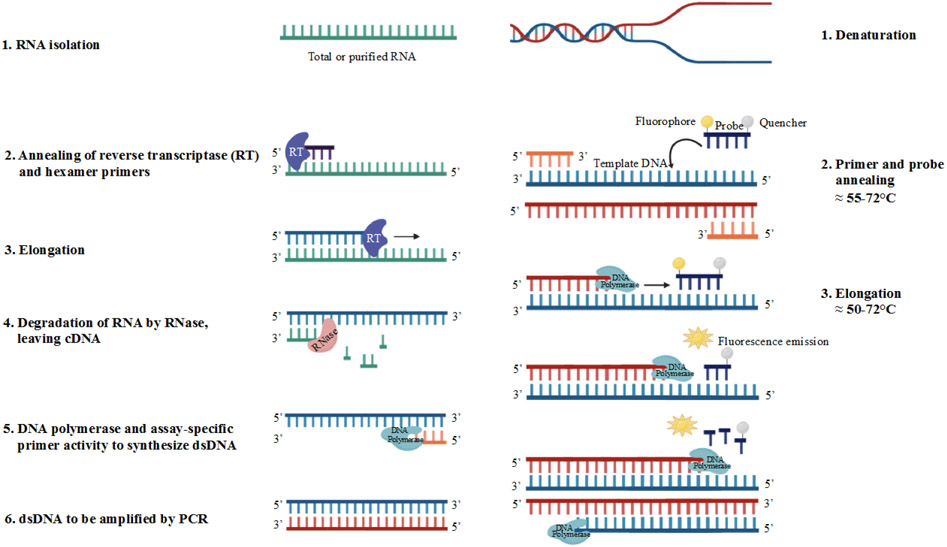
Figure 2.
Molecular underpinnings of quantitative PCR (qPCR) reactions using RNA (reverse-transcriptase qPCR; left column) or DNA (right column) as a template. [1] Purified template RNA or DNA in a reaction well (and denaturation if the template is double stranded DNA). [2] Hexamer primers and reverse transcriptase bind to target RNA or forward and reverse primers along with probe bind to template DNA. [3] Reverse transcriptase transcribes a complementary DNA strand to form a cDNA-RNA hybrid or DNA polymerase synthesizes dsDNA displacing the quencher from the probe; fluorescence emission is captured. [4] RNase hydrolyzes the RNA, leaving the cDNA strand free for [5] binding of DNA polymerase to synthesize dsDNA followed by steps [1], [2] and [3] from the qPCR reaction
In addition to common chemistry, IAMs share the basic sample preparation steps of cell lysis and purification of nucleic acids. Cell lysis methods vary among microorganisms, e.g., bead beating for Enterococcus spp. (Martzy et al. 2017), or lysis buffer containing β-mercaptoethanol for Karenia brevis (Casper et al. 2004). Some lysis protocols include the use of a weak alkali (Shiraho et al. 2016) or heat treatment (Hirayama et al. 2006). Post-lysis steps include purification and concentration of nucleic acids. These methods differ across IAMs, taxa and matrices because purification and concentration steps depend on the target template, sample type, and the potential for inhibitors.
All IAM reactions require the use of a polymerase to catalyze the synthesis of nucleic acids (Zhao et al. 2015). The polymerase used depends on the target template, the mechanism of amplification and the temperature of the IAM reaction (Table 1). For example, NASBA reactions employ T7 RNA polymerase to synthesize RNA from a RNA template at 41°C (Fakruddin et al. 2012), while a strand-displacing DNA polymerase (e.g., Bst DNA polymerase) is employed to synthesize DNA from a DNA template in LAMP reactions at 60°C – 65°C (Karanis et al. 2007). Isothermal temperatures for IAMs included in this review range from 37°C – 65°C (Table 1). For IAMs that can be used in a wider range of temperatures (e.g., RCA can be used from 37°C – 65°C), temperature selection depends on target nucleic acids (e.g., RNA or DNA), primer design, enzymes, buffer composition, pH, and instrumentation (i.e., water bath, test tube at ambient temperature, specialized proprietary instruments (Crannell et al. 2014, Notomi et al. 2015, Parida et al. 2008, Ulrich et al. 2015)).
Amplification products from IAMs can be visualized under UV light following gel electrophoresis (Tomita et al. 2008) or by a variety of other methods for binary and semi-quantitative applications. The most simplistic visualization strategy in terms of instrumentation is the colorimetric assay. For example, RCA and HDA reactions can be visualized in a presence/absence mode without additional instrumentation through the use of a colorimetric assay integrated into the reaction mix (Zhu et al. 2014). Reaction products can also be quantified with a microplate reader (e.g., a spectrophotometer) (Zhao et al. 2015), or by turbidity using the MgSO4 in the reaction mix to form magnesium pyrophosphate precipitates (Zhu et al. 2014). Turbidity can be assessed visually to determine presence/absence of amplification product, or measured by a microplate reader. RCA, HDA, and LAMP reactions can be visualized by a fluorescent immunoassay (Rames and Macdonald 2019) or a luminescence assay. The simplicity of these visualization methods affords end-users rapid screening of samples, particularly if the primary concern is presence/absence of a target taxa, such as bacteria (Lee et al. 2019, Martzy et al. 2017) or viruses (Farkas et al. 2020, Hamza et al. 2011) Figure 1 and Table S1 show taxa included in papers reviewed here.
The quantitative visualization method predominantly used by LAMP and NASBA is a fluorescent probe assay, in which a nucleic acid sequence containing a fluorophore is initially quenched by a second molecule attached to the probe. As the template is amplified, the fluorophore is released from its quencher, which will concurrently release a fluorescent or phosphorescent signal that is measured in relative fluorescence units by the instrument. These signals can be used in multiple ways. For example, in NASBA, a phosphorescent signal is released (Figure 3) and measurements are shown in real-time on the screen of a proprietary instrument. An internal control can be added to diagnose inhibition of the reaction (Casper et al. 2005). The rate of phosphorescence, or fluorescence, can be regressed against a standard curve to accurately quantify the concentration of unknown target in the sample.
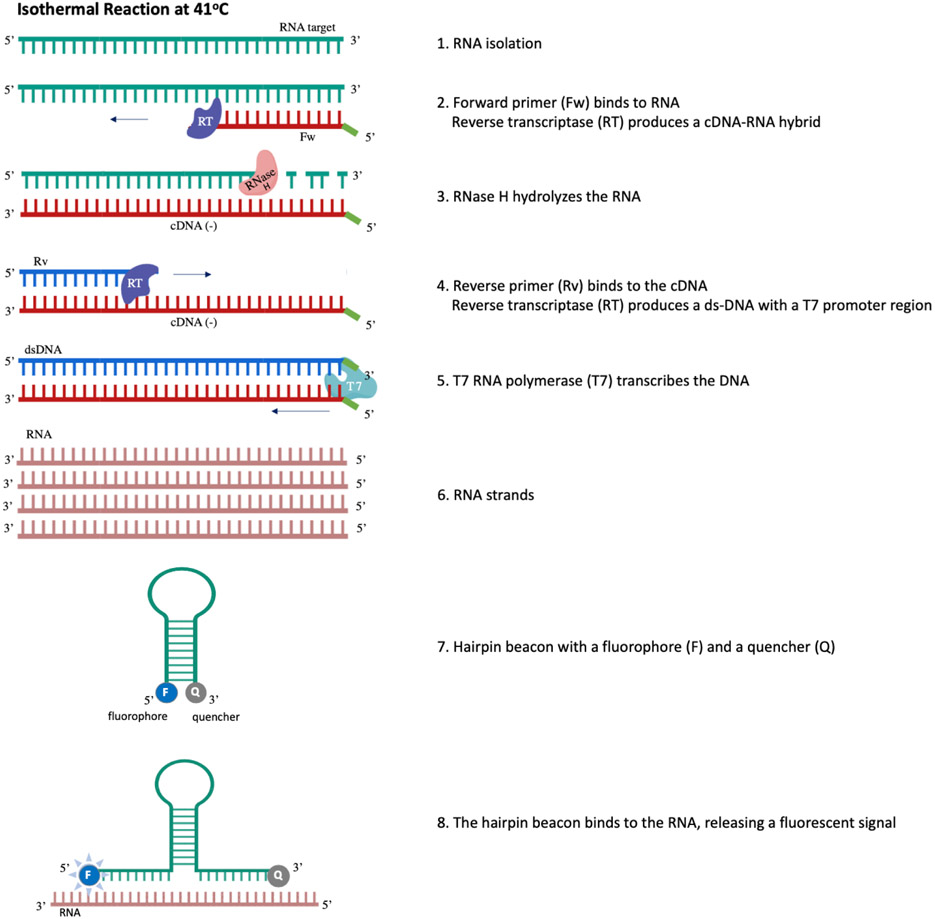
Figure 3.
Molecular underpinnings of a nucleic acid sequence-based amplification (NASBA) reaction. [1] Purified template RNA in a reaction well. [2] Forward primer binds to target RNA and reverse transcriptase transcribes a complementary DNA strand to form a cDNA-RNA hybrid. [3] RNase H hydrolyzes the RNA, leaving the cDNA strand free for [4] the reverse primer to bind to it; reverse transcriptase then produces a double-stranded DNA molecule with a complete T7 promoter region. [5] T7 RNA polymerase binds the T7 promoter region and transcribes the DNA to [6] anti-sense RNA strands. [7] A loop-structure hairpin beacon is self-hybridized to quench its fluorophore; however, it is attracted to the anti-sense RNA strands. [8] The beacon binds the anti-sense RNA, releasing a fluorescent signal that can be read on an instrument, either to quantify the concentration of the sample, or as a presence/absence metric.
Specific IAMs
Loop-mediated Isothermal Amplification (LAMP)
LAMP is carried out at temperatures between 60°C - 65°C (Table 1; Figure 4). LAMP employs a strand-displacing DNA polymerase (e.g., Bst DNA polymerase) to separate nucleic acid strands in lieu of the heat denaturation step that is characteristic of PCR. LAMP utilizes 4-6 primers specific to 6-8 targeted regions of a specific DNA or RNA sequence (Notomi et al. 2015). At least four primers are necessary to carry out the reaction: (1) a forward inner primer (FIP) that includes a reverse complementary sequence, (2) a forward outer primer, (3) a backward/reverse inner primer (RIP) that includes a reverse complementary sequence, (4) a backward/reverse outer primer. Two additional primers are optional: (5) a forward loop primer and (6) a backward/reverse loop primer. Though LAMP publications refer to “backward” primers, for the sake of consistency in this review, these primers will be referred to as reverse primers.
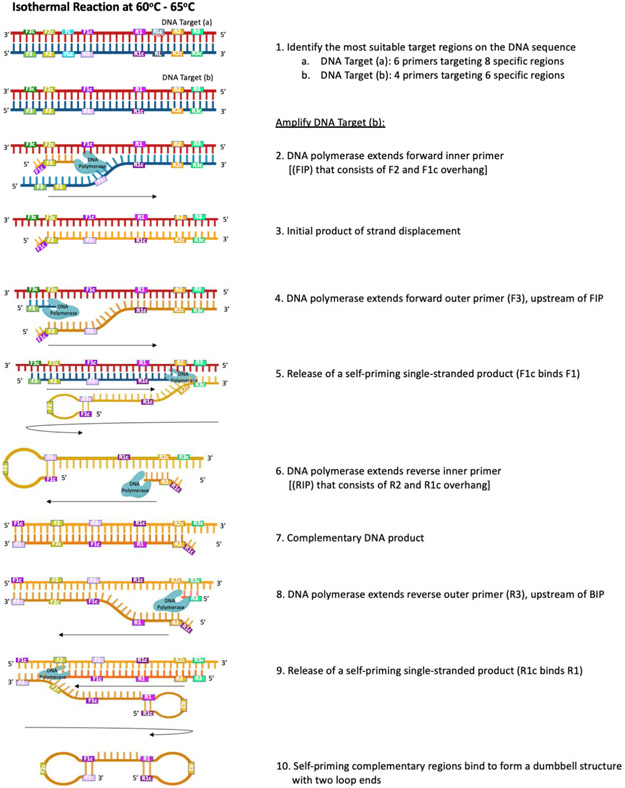
Figure 4.
Molecular underpinnings of a loop-mediated isothermal amplification (LAMP) reaction (shown with DNA template). [1] Six – eight target regions are included in a LAMP assay; [1a] 6 primers (including forward and reverse loop primers) target 8 regions to provide more sites for priming and increase the rate of amplification, as this affords an increase in concatemer formation, while [1b] 4 primers target 6 regions in a standard reaction. [2] The reaction begins as the forward inner primer (FIP), which contains a sequence for the F2 region of target DNA and a complementary sequence overhang (F1c), binds the F2c region on the 3’ end of the target. A strand-displacing DNA polymerase (e.g., Bst DNA polymerase) extends the FIP to [3] synthesize a complementary strand of DNA (the initial product). Next, [4] forward outer primer (F3) binds to a region upstream of the FIP binding site; it is extended by DNA polymerase to displace the initial product, which [5] self-primes into a single-end loop structure as F1c binds F1. [6] The reverse inner primer (RIP), which contains a sequence for the R2 region of the target DNA and a complementary sequence overhang (R1c), binds the R2 region on the 3’ end of the initial product; [7] DNA polymerase synthesizes complementary DNA in the 3’ to 5’ direction. [8] Reverse outer primer (R3) binds to a region upstream of the RIP binding site; it is extended by DNA polymerase to [9] displace the self-priming dumbbell product with two loop ends, which forms as [10] F1 binds F1c and R1c binds R1. This dumbbell structure forms the base for the cycling phase of the LAMP reaction, wherein multiple sites are available for binding by primers (FIP, BIP, and optional loop primers); these sites initiate the synthesis of concatemers (not pictured) that also contain initiation sites, which are subsequently used during the cycling phase, increasing overall amplification.
The LAMP reaction is initiated by FIP strand invasion; FIP binds at a secondary site on the 3’ end of the target DNA (Figure 4), and a strand-displacing DNA polymerase extends FIP to synthesize a complementary 5’ to 3’ product while separating the DNA duplex. The goal of the reaction is to create a dumbbell structure with two loops that will act as the basis for the cycling phase of LAMP (Tomita et al. 2008). The initial product is displaced by synthesis driven by DNA polymerase, when the forward outer primer anneals to a target region upstream of the FIP-bound region. The 5’ end of the displaced product forms a self-hybridizing loop structure, as the reverse complementary sequence included in the FIP binds its complementary region located on the single displaced strand (Figure 4). This annealing and displacement cycle repeats on the opposite end of the target DNA sequence, using RIP and reverse outer primer, to create a dumbbell structure that contains multiple sites for initiation of synthesis (Figure 4), and thereby for exponential LAMP amplification through the formation of concatemers.
Optional loop primers (forward and reverse) contain sequences complementary to the 5’ end of the loop regions on each resultant dumbbell structure, affording all single stranded loop structures usable regions to initiate concatemer synthesis; these additional sites reduce amplification time because concatemer formation is expedited (Tomita et al. 2008). Developing these optional loop primers can be challenging, as two additional unique sequence regions must be identified between the primary (e.g., F1 and R1) and secondary (e.g., F2 and R2) regions of the target DNA, thus a total of 8 distinct regions must be identified on the target DNA sequence in order to utilize loop primers (Figure 4).
Nucleic Acid Sequence-based Amplification (NASBA)
NASBA is an isothermal method that is carried out at 41°C and is used to amplify single-stranded nucleic acids (e.g., RNA, single-stranded DNA) (Cook 2003). NASBA requires two primers (forward and reverse; Table 1; Figure 3), both with T7 promoter region overhangs. Three enzymes work to complete the reaction: avian myeloblastosis virus reverse transcriptase (AMV-RT), T7 RNA polymerase, and RNase H. The reaction begins when the forward primer binds target nucleic acids (e.g., RNA; Figure 3). A cDNA strand is synthesized by AMV-RT, creating a cDNA-RNA hybrid. The RNA strand is then hydrolyzed by RNase H, allowing the reverse primer to bind the cDNA strand, and double-stranded DNA is synthesized by reverse transcriptase. These double strands, both with a T7 overhang, form a T7 promoter region that is bound by T7 polymerase to produce anti-sense RNA strands.
A key component of a NASBA reaction is its molecular beacon (Casper et al. 2004), which is designed as a hairpin loop-structure with a fluorophore and a quencher. The beacon is designed to be self-hybridizing in order to quench fluorescence in the absence of amplification. The beacon is also designed to be more attracted to the anti-sense RNA strands produced by the NASBA reaction than it is attracted to itself via self-hybridization; thus, the beacon will release a fluorescent signal when it breaks its internal bonds and anneals with the anti-sense RNA strands. This signal can be read in real-time on various instruments, some of which are handheld and battery-operated, facilitating field use.
Helicase-dependent Amplification (HDA)
HDA is an isothermal method carried out at temperatures ranging between 37°C – 65°C (Table 1) (Vincent et al. 2004). HDA employs two primers, DNA helicase, single-stranded DNA binding proteins (SSBs), and polymerase to produce short nucleic acid products (Jeong et al. 2009). HDA is used to amplify DNA targets, and reverse transcriptase can be added to amplify RNA targets (Goldmeyer et al. 2007, Yang et al. 2015). The temperature of the reaction primarily depends on which DNA helicase and polymerase are selected for the reaction, and thereby also depends on the nucleic acid (i.e., DNA or RNA) target of the enzymes.
DNA helicase in HDA reactions enzymatically separates two complementary strands of target DNA to produce single-stranded templates that are stabilized by SSBs and then bound by primers (forward primer at the 3’ end and reverse primer at the 5’ end), and extended by DNA polymerase (e.g., Bst) to synthesize double-stranded products that enter the next round of amplification and act as a template (Vincent et al. 2004).
Rolling Circle Amplification (RCA)
RCA is carried out at temperatures between 37°C - 65°C (Table 1). RCA employs a circular DNA template, a polymerase, and a forward primer, producing long, repeating single-stranded nucleic acid products (e.g., ssDNA) (Zhao et al. 2015). RCA can be used to amplify DNA templates (Chen 2015), and it has been modified to utilize additional probes (e.g., padlock probes) to amplify RNA (Takahashi et al. 2018) or other templates such as proteins (Shi et al. 2017).
RCA is initiated by a template-mediated enzymatic ligation, frequently catalyzed by T4 DNA ligase. The forward primer subsequently binds the circular DNA and a strand-displacing polymerase (most commonly T7 RNA polymerase for RNA amplification and Phi29 for DNA amplification (Zhao et al. 2015) synthesizes a long repeating product. The temperature of the reaction varies depending upon which polymerase is used; for example, the optimal temperature for Phi29 in these reactions is 37°C - 42°C.
Recombinase Polymerase Amplification (RPA)
The optimal temperature range for RPA is 37°C - 42°C (Table 1). The specific reaction temperature is dependent upon the specific strand-displacing polymerase and recombinase proteins utilized (Lillis 2016). RPA utilizes forward and reverse primers to sequence and amplify DNA. The 3’ ends of the primers are initially complexed in a helix with recombinase proteins via the action of a strand-displacing recombinase. As the primers enter the helix, they are stabilized as strands within the helix by single-strand binding proteins. Recombinase disassembles when the recombinase-primer complexes are stabilized by the single-strand binding proteins; this exposes the 3’ ends of the primers, and allows the forward primer to bind to the DNA template for synthesis by DNA polymerase.
Method Performance
Sensitivity and specificity.
A thorough understanding of method performance is an essential aspect of wide acceptance and use of any scientific methodology. Sensitivity and specificity are defined in this review by the conventions associated with clinical applications and microbial source tracking (Altman and Bland 1994, Stoeckel and Harwood 2007). Sensitivity is defined as: (TP/TP +FN) x 100 where TP= true positive and FN = false negative. Specificity is: (TN/TN +FP) x 100, where TN= true negative and FP= false positive. For example, if an assay targeting Karenia brevis was tested on 100 samples known to contain K. brevis and K. brevis was detected in 98 samples, the sensitivity of the given assay would be 98%. Conversely, if 100 non-target samples (e.g. pure cultures containing other algal species) were tested for K. brevis and 80 produced negative results (TN), but 20 produced false-positives (FP), the specificity of the assay would be 80%.
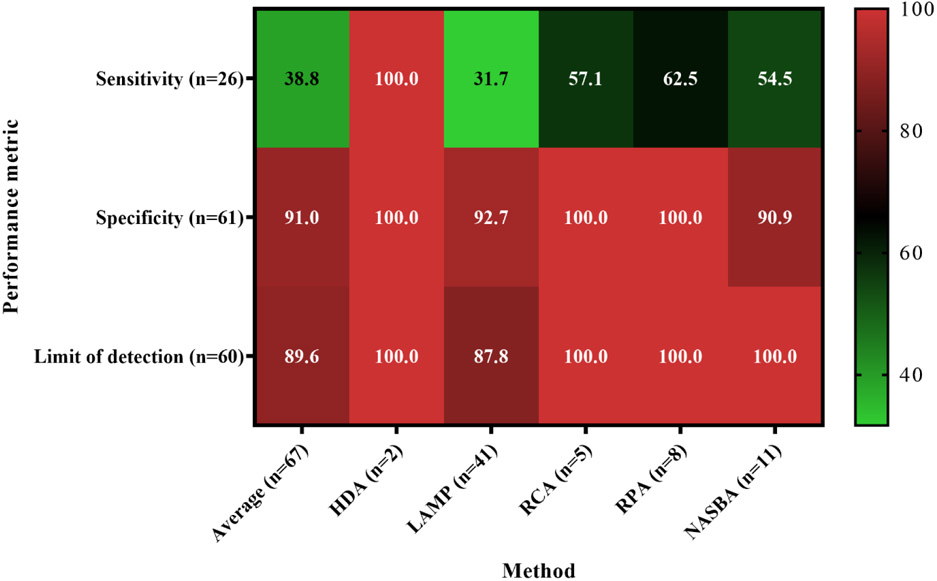
Within 67 papers that met our search criteria and reported performance metrics, LAMP was the most frequently employed method (61.2% of all studies included in this review) followed by NASBA (16.4%), RCA (12.2% each), RPA (11.9%), and finally HDA (1.5%). Irrespective of the method, the majority of studies reported data in a binary fashion (80.3%), while quantitative data reporting was rare (19.7%) (Table S1). All papers reported at least one performance metric for the method(s) tested. Specificity was the most commonly reported performance metric (91.0%), followed by limit of detection (89.6%) and finally sensitivity (38.8%) (Table 2; Figure 5). Of all the papers surveyed, 28.4% reported sensitivity, specificity, and limit of detection.
Table 2.
Summary of performance metrics for isothermal amplification methods. The mean value for each method/metric/unit combination was calculated. The number of studies in which the metric was measured is included parenthetically (). Where possible, units and values were converted to allow direct comparison, e.g. values originally reported as fg/μL were converted to ng/μL. All units representing nucleic acids (GC, ng) are normalized per μL of template volume. A detailed compilation of characteristics for each study is provided in supplemental Table S1.
| Method | Limit of Detection Unit |
Mean Limit of Detectionb |
Sensitivity | Specificity |
|---|---|---|---|---|
| HDA | GCa/μL | 6.46 x 100 (2) | 99% (2) | 100% (?) |
| LAMP | GC/μL | 8.78 x 101 (4) | 94.4% | 99.8% |
| GC/reactionc | 2.24 x 101 (4) | |||
| ng/μL | 2.19 x 10−3 (26) | |||
| cells/μL | 5.45 x 100 (7) | |||
| CFU/μLd | 1.15 x 100 (7) | |||
| CFU/μg fecese | 2.43 x 10−3 (2) | |||
| LF-RPA | GC/μL | 2.60 x 101 (1) | 93.3% | 100% |
| NASBA | GC/μL | 2.02 x 102 (2) | 100% | 95.5% |
| cells/μL | 1.97 x 100 (3) | |||
| CFU/μL | 8.34 x 101 (6) | |||
| PFU/μL | 2.00 x 100 (1) | |||
| ng/μg tissue | 6.67 x 10−8 (1) | |||
| RCA | GC/μL | 1.00 x 101 (1) | 100% | 99.8% |
| cells/μL | 1.00 x 100 (2) | |||
| ng/μL | 7.03 x 10−6 (4) | |||
| RPA | GC/μL | 2.43 x 101 (2) | 98.7% | 100% |
| cells/μL | 2.02 x 10−2 (5) | |||
| ng/μL | 2.51 x 100 (5) |
GC gene copies
Number of publications where units were comparable or could be readily converted for comparison.
Values normalized per reaction in the publication were converted to per μL template volume unless template volume was not provided.
Converted to CFU/μL from CFU/mL for comparison
Converted to CFU/μg feces from CFU/g feces for ease of comparison
Figure 5.
Percentage of studies reporting performance metrics (sensitivity, specificity, limit of detection) of IAM methods used for environmental sample analyses
Both performance metrics, sensitivity and specificity, were close to 100% for most studies (Table S1). The mean value of all methods was >90% for both metrics, although a few exceptions were noted (Table 2, Table S1). The reference materials for most studies were pure cultures of target or non-target organisms, although a few studies targeting fecal microorganisms used fecal samples or known positive environmental samples (e.g., (Kolm et al. 2019, Plutzer and Karanis 2009)). The lowest reported sensitivity was 68% for a LAMP method targeting the translation elongation factor of Giardia duodenalis (Plutzer and Karanis 2009), in which fecal and water samples known to contain the parasite (tested by immunofluorescence microscopy) provided the reference material. The authors attributed the false-negative results to the primers, whose sequence may not have captured single nucleotide polymorphisms.
The lowest specificity in the articles reviewed here was 86% in a NASBA assay targeting the rbcS gene of Pseudo-nitzschia multiseries (Delaney et al. 2011). The assay produced false-positive results against other Pseudo-nitzschia species but did not amplify RNA from algal species belonging to other genera. A qPCR assay targeting the same gene was 100% specific for P. multiseries. The number of samples used for sensitivity and specificity calculations was not clearly stated in every publication. Among the studies where sample number could be determined, only ten studies used 20 or more target organisms for sensitivity testing, and only 12 used 20 or more non-target organisms for specificity testing. While there are no formal guidelines establishing the optimal number of samples for sensitivity and specificity testing, the greater number of samples is likely to result in a more robust testing and more accurate assessment of method performance metrics (Harwood et al. 2014).
Limit of detection.
The assay limit of detection (LOD) is a measurement used to describe analytical sensitivity. LOD and analytical sensitivity have an inverse relationship; lower LODs are indicative of increased analytical sensitivity. LOD was reported in 91.2% of papers, but the units utilized varied widely, complicating direct comparisons across the studies or among the methods. For example, some studies reported the LOD normalized to concentration of nucleic acid (e.g., ng/μL), while others expressed it as number of cells/colony forming units/plaque forming units per unit volume or as gene copies per reaction or per μL of reaction volume. Where possible, we normalized units to allow direct comparison (e.g., fg or pg/μL were recalculated as ng/μL) (Table 2). The studies that measured LOD in ng/μL utilized purified nucleic acid from target microorganisms normalized to template volume, which produced very low LODs that are not intuitively relevant to concentrations in the environment (Table 2). Units such as CFU/mL water or /g feces are more readily interpreted in an environmental context, but LODs are higher than those calculated in units such as ng/μL due to the nature of the unit. For example, a RCA study on the dinoflagellate Karlodinium veneficum found a LOD of 8.00 x 10−8 ng/μL for the D1/D2 domain of the large-subunit ribosomal RNA, which corresponded to 1.00 x 10−5 cells/μL (Table S1) (Liu et al. 2019).
LAMP assay LODs were frequently measured in ng/μL (27 of 50 measurements in the papers reviewed here, or 54%). The mean LOD of these assays calculated from the studies reviewed was 2.19 x 10−3 ng/μL (Table 2), with values ranging from a low of 1.00 x 10−7 ng/μL for the S-adenosyl-L-methionine synthetase gene of Cryptosporidium species (Koleren et al 2016) to a high of 2.00 x 10−2 ng/μL for the Vibrio parahaemolyticus thr gene (Malcolm et al 2015) (Table S1). The higher LOD may well have been due to the shellfish tissue matrix investigated in Malcolm et al 2015. LAMP assay LODs measured by gene copies (GC) provided higher values, which is expected since many copies of a given gene would be present in one ng nucleic acid. The mean LOD for LAMP assays expressed in GC/μL was 8.78 x 101 (Table 2).
It is difficult to compare the LODs of LAMP and NASBA assays, as NASBA LODs were most frequently expressed in terms of CFU/volume or cells/volume (Table 2; Table S1), as opposed to gene copies or ng/volume. Mean NASBA LODs calculated from multiple studies were 8.34 x 101 CFU/μL and 1.97 x 10° cells /μL (Table 2), which were higher than those observed for LAMP (Table 2; Table S1). LOD values for LAMP assays in individual studies which were expressed in terms of CFU ranged from 1.0 x 10−−1 CFU/μl for E. coli (Heijnen and Medema 2009) to 5.0 x 102 CFU/μl for V. cholerae (Fykse et al. 2012).
Other IAMs displayed similar trends in LODs (Table 2; Table S1). An exception was the markedly higher mean LOD of RPA expressed in ng/μL of 2.51 x 100; however, the mean LOD of measurements calculated in cells/μL was 2.02 x 10−2, which compared very favorably with other methods. The LOD mean for RPA in ng/μL was skewed by one observation of a relatively high LOD of 10 ng/μL for a dipstick method to detect Karlodinium veneficum (Fu et al. 2019).
Comparison with PCR and inhibition.
Direct comparisons between IAMs and PCR were generally conducted with conventional PCR, rather than QPCR, and produced binary results (presence/absence). Those studies that compared IAMs with end-point PCR generally found the LOD of IAMs to be 10 – 100-fold lower than PCR (Chen et al. 2013, Li et al. 2019). However, outcomes with qPCR comparisons are more ambiguous. Some studies found IAMs to be more sensitive than qPCR (Pang et al. 2019, Rutjes et al. 2006) and this finding was attributed to IAMs resilience to inhibition by humic acids and similar compounds (Rutjes et al. 2006, Stedtfeld et al. 2016). It was suggested that the Bst polymerase of Bacillus stearothermophilus used in LAMP assays is less susceptible to inhibitors than the Taq polymerase frequently used for qPCR (Francois et al. 2011, Martzy et al. 2017, Martzy et al. 2019); however, direct comparisons between PCR and IAMs susceptibility to inhibition were rare, and environmental samples were generally not tested for inhibition. The strategy commonly used in PCR of dilution of samples to reduce the effect of inhibitors can also be used with IAMs (Kolm et al. 2019), although diluting samples also raises the LOD, decreasing the likelihood that rare gene targets can be detected.
Research that supports the hypothesis that LAMP assays are less susceptible to inhibition than PCR assays include the recent report in which humic acid, a known inhibitor of PCR, was added to qPCR and LAMP assays (Stedtfeld et al. 2016). While the PCR and qPCR assays were able to amplify the Dehalobacter target with up to 1 mg/L concentrations of humic acid, LAMP amplified target DNA on unextracted samples with up to 100 mg/L of added inhibitor (Stedtfeld et al. 2016). A study focusing on the fecal indicator bacteria E. coli and Enterococcus faecalis also demonstrated that PCR assays were more susceptible to inhibition than LAMP assays in surface water (Lee et al. 2019). Conversely, others reported that lower LOD and greater sensitivity and specificity were achieved by (RT)qPCR methods compared to IAMs (Delaney et al. 2011, Fykse et al. 2012).
Use of IAM Methods in the Field
An advantage of isothermal amplification methods, as compared to more conventional amplification methods like qPCR, is its potential for field use. While qPCR assays generally require complex specialized thermocyclers to rapidly switch between programmed temperatures, isothermal amplification methods only require a single constant temperature for amplification and can more readily be carried out in the field or in low-technology laboratory settings, although quantification may be sacrificed.
The potential for use of IAMs in the field to detect specific microorganisms has been explored in a number of studies, including modifications designed to adapt methods to field conditions (Fu et al. 2019, Hamburger et al. 2013, Wang et al. 2019, Wu et al. 2019). A desirable quality for field assays is that detection of amplification is accomplished by eye or by a specialized portable device that is simple and sturdy enough to perform in less than ideal conditions. Also desirable is that the assay requires minimal sample preparation, is not highly susceptible to inhibition, and that minimal cold storage is required for reagents. Although no current device or system has allowed for autonomous, self-contained field-deployable detection of nucleic acids using IAM, recent developments in sample preparation and signal detection have been progressing toward an all-in-one device (Kolm et al. 2019, Tang et al. 2019).
Detecting amplification from isothermal methods is one of the challenges to creating a field-adapted test. One strategy developed to simplify field detection is the use of lateral flow strips for rapid detection (Kolm et al. 2019, Wang et al. 2019, Wu et al. 2017). This method, which has been paired with HDA (Kolm et al. 2017), RPA (Wu et al. 2017), and RCA (Liu et al. 2019), works by attaching antibodies that complex with probes targeted to specific nucleic acid sequences onto a test strip of paper. When the isothermal amplification product is added, any amplicons present will attach to the antibody probe, which then binds to a biotin ligand on the test line on the strip, producing a visual result. These tests are often very fast, producing qualitative results in 70 minutes (Wang et al. 2019). Another detection method for RCA involves multiplexed, hyperbranched RCA with dot blot hybridization as the detection mechanism, although this may not be ideal for field use (Zhang et al. 2018). Another, simpler visualization method is the addition of a fluorescent DNA dye, such as SYBR Green I; this method allows for fast qualitative results of an isothermal method using UV light to detect amplified nucleic acid (Chen 2015, Siddique et al. 2017). These fluorescent methods can also be paired with portable fluorescence scanners, such as the Gene-Z system (Stedtfeld et al. 2016, Stedtfeld et al. 2017, Stedtfeld et al. 2012). This device allows for extracted DNA to be added to disposable chips with pre-loaded LAMP reagents, resulting in quick assays with real-time fluorescent detection data which can upload to an attached iPod or smartphone (Stedtfeld et al. 2012).
Development of simple, effective methods for nucleic acid purification in field settings are among the greatest challenges in creating field deployable IAMs. Although crude lysis or minimally purified nucleic acids can be amplified in samples where target microorganisms are concentrated (Stedtfeld et al. 2016), a simple one-step nucleic acid extraction method would be preferable to accomplish rapid sample processing without the use of a sterile lab (Wu et al. 2019). One possible solution is the use of FTA Elute cards that contain reagents to lyse cells and solubilize DNA, which can then be extracted using water and heat (Nishiuchi et al. 2014).
Reagents that require minimal refrigeration or freezing, or, ideally, are stable at ambient temperature provide a great advantage in field studies. This approach has been a particular focus in developing isothermal amplification protocols for detection of Vibrio cholerae in field environments in undeveloped countries, where a laboratory is unlikely to be found. A technique that uses thermostabilized NASBA reagents paired with an ELIZA assay showed promising results for field detection of V. cholerae (Lee et al. 2008). Thermostabilized LAMP reagents have also been designed for use in V. cholerae detection (Syafirah et al. 2018). These thermostabilized reagents greatly simplify the field use of isothermal amplification by eliminating the cold-chain requirements that complicate these assays in field settings. However, direct comparisons between field-adaptable nucleic acid purification procedures and those performed in a laboratory environment are still needed to evaluate the feasibility of this approach, including purity of the resulting extracts and the potential for contamination in field environment.
It is also advantageous for field-based assays to be easily transferable so that minimally-trained students or workers can accurately and reliably set up and run the assay. For example, Hamburger et al. (2013) tested the performance of pre-mixed reaction chemicals, enzymes and primers held at ambient temperature in a LAMP assay conducted in a field laboratory. The assay was designed to amplify Schistosoma haematobium and S. mansoni DNA extracted from snail tissue. Sucrose was used to stabilize reaction components other than primers, which were packaged separately in a lyophilized form. The method used SYBR Green to detect amplification in a binary (+/−) mode. Of 103 field tests conducted, 54 were positive by PCR and qPCR, while 50 were positive in field LAMP assays. The authors concluded that this low-technology approach could be useful for molecular monitoring of endemic infections of snails by Schistosoma spp. in areas where infection prevalence in humans is persistent (Hamburger et al. 2013). Stedtfeld et al (2017) described a field experiment involving minimally trained technicians, where a field-adapted LAMP assay kit was sent to a community college. These students were able to accurately estimate the amount of antibiotic resistance genes present in bacterial communities of environmental waters, with results that correlated strongly with qPCR assays run in a lab (Stedtfeld et al. 2017).
Conclusions and Future Directions
IAMs are emerging methodologies for detection or quantification of microorganisms in environmental samples. While IAMs have a decades-long history of use in clinical settings (e.g. van der Vliet et al. 1993) and food microbiology (e.g. Uyttendaele et al. 1995), their use in environmental samples began about a decade later (Abd el-Galil et al. 2005) and has increased steadily since that time. The reported advantages of IAMs include ease of performance, requirement of less sophisticated instrumentation than PCR/qPCR, cost-effectiveness, and less susceptibility to inhibition than PCR/qPCR; however, such assertions are often made without empirical evidence.
Recommendation 1:
Perform more head-to head studies of qPCR and IAM methods that include performance metrics and assess susceptibility to inhibition in environmental samples.
The benefits of IAMs come with certain trade-offs, mostly in the form of accurate quantification as IAM results are largely reported in a binary fashion. The majority of IAM observations made in the studies reviewed here reported binary data (80.3%), with only 19.7% of sample results reported quantitively (Table S1). The remaining 17.1% of observations were reported as binary, but standard curves were created for these studies in order to assess LODs, therefore quantification for these assays was possible (Table S1). In some cases a certain threshold concentration of microorganisms is the benchmark for safe versus unsafe conditions, e.g., the threshold for potential bloom forming conditions for K. brevis is 5 x 103 cells/L (Pierce and Henry 2008), while 70 CFU/100 mL enterococci is the beach action value criterion for poor water quality at marine and freshwater recreational waters (U.S. Environmental Protection Agency 2012). IAMs could be used for either application in a binary detection mode (above or below threshold) in conjunction with a pre-calculated standard curve. However, if detection of trends is needed (e.g., increase or decrease in microbial concentrations), then binary reporting is not sufficient and IAMs would need to be capable of fully quantitative systems.
Recommendation 2:
Determine which IAMs are best suited for threshold detection, or binary modes, and fully quantitative modes in environmental studies.
Based on the publications reviewed here, IAMs represent a promising screening tool for potential regulatory environmental applications (e.g., early detection of harmful algal blooms and monitoring of recreational waters for fecal indicator bacteria) that may eventually be field-deployable. Additional research and refinement is needed for reliable target quantification. Ultimately, IAMs may be superior tools to inform spatiotemporal distributions of undesirable microorganisms in the environment such as harmful algal blooms and sewage spill plumes.
Supplementary Material
Acknowledgements
The authors would like to thank James Conrad (USF) for proof-reading the method diagrams.
Footnotes
Disclaimer
The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. The views expressed in this article are those of the author(s) and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Abd el-Galil KH, el-Sokkary MA, Kheira SM, Salazar AM, Yates MV, Chen W and Mulchandani A (2005) Real-time nucleic acid sequence-based amplification assay for detection of hepatitis A virus. Appl Environ Microbiol 71(11), 7113–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman DG and Bland JM (1994) Diagnostic tests. 1: Sensitivity and specificity. BMJ: British Medical Journal 308(6943), 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper ET, Patterson SS, Smith MC and Paul JH (2005) Development and evaluation of a method to detect and quantify enteroviruses using NASBA and internal control RNA (IC-NASBA). J Virol Methods 124(1-2), 149–155. [DOI] [PubMed] [Google Scholar]
- Casper ET, Paul JH, Smith MC and Gray M (2004) Detection and quantification of the red tide dinoflagellate Karenia brevis by real-time nucleic acid sequence-based amplification. Appl Environ Microbiol 70(8), 4727–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cai P, Zhang C, Wang Y, Zhang S, Guo C, Lu DD (2015) Hyperbranched rolling circle amplification as a novel method for rapid and sensitive detection of Amphidinium carterae. Harmful Algae 47, 66–74. [Google Scholar]
- Chen Y, Wen T, Lai DH, Wen YZ, Wu ZD, Yang TB, Yu XB, Hide G and Lun ZR (2013) Development and evaluation of loop-mediated isothermal amplification (LAMP) for rapid detection of Clonorchis sinensis from its first intermediate hosts, freshwater snails. Parasitology 140(11), 1377–1383. [DOI] [PubMed] [Google Scholar]
- Cook N (2003) The use of NASBA for the detection of microbial pathogens in food and environmental samples. Journal of Microbiological Methods 53(2), 165–174. [DOI] [PubMed] [Google Scholar]
- Crannell ZA, Rohrman B and Richards-Kortum R (2014) Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PloS one 9(11), e112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JA, Ulrich RM and Paul JH (2011) Detection of the toxic marine diatom Pseudo-nitzschia multiseries using the RuBisCO small subunit (rbcS) gene in two real-time RNA amplification formats. Harmful Algae 11, 54–64. [Google Scholar]
- Fakruddin M, Mazumdar RM, Chowdhury A and Mannan K (2012) Nucleic acid sequence based amplification (NASBA)-prospects and applications. Int J Life Sci Pharma Res 2(1), 106–121. [Google Scholar]
- Farkas K, Mannion F, Hillary LS, Malham SK and Walker DI (2020) Emerging technologies for the rapid detection of enteric viruses in the aquatic environment. Current Opinion in Environmental Science & Health 16, 1–6. [Google Scholar]
- Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD and Schrenzel J (2011) Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 62(1), 41–48. [DOI] [PubMed] [Google Scholar]
- Fu M, Chen G, Zhang C, Wang Y, Sun R and Zhou J (2019) Rapid and sensitive detection method for Karlodinium veneficum by recombinase polymerase amplification coupled with lateral flow dipstick. Harmful Algae 84, 1–9. [DOI] [PubMed] [Google Scholar]
- Fykse EM, Nilsen T, Nielsen AD, Tryland I, Delacroix S and Blatny JM (2012) Real-time PCR and NASBA for rapid and sensitive detection of Vibrio cholerae in ballast water. Marine pollution bulletin 64(2), 200–206. [DOI] [PubMed] [Google Scholar]
- Goldmeyer J, Kong H and Tang W (2007) Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. The Journal of Molecular Diagnostics 9(5), 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger J, Abbasi I, Kariuki C, Wanjala A, Mzungu E, Mungai P, Muchiri E and King CH (2013) Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. The American journal of tropical medicine and hygiene 88(2), 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza IA, Jurzik L, Überla K and Wilhelm M (2011) Methods to detect infectious human enteric viruses in environmental water samples. International journal of hygiene and environmental health 214(6), 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood VJ, Staley C, Badgley BD, Borges K and Korajkic A (2014) Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38(1), 1–40. [DOI] [PubMed] [Google Scholar]
- Heijnen L and Medema G (2009) Method for rapid detection of viable Escherichia coli in water using real-time NASBA. Water Res 43(12), 3124–3132. [DOI] [PubMed] [Google Scholar]
- Hirayama H, Kageyama S, Takahashi Y, Moriyasu S, Sawai K, Onoe S, Watanabe K, Kojiya S, Notomi T and Minamihashi A (2006) Rapid sexing of water buffalo (Bubalus bubalis) embryos using loop-mediated isothermal amplification. Theriogenology 66(5), 1249–1256. [DOI] [PubMed] [Google Scholar]
- Jeong Y-J, Park K and Kim D-E (2009) Isothermal DNA amplification in vitro: the helicase-dependent amplification system. Cellular and molecular life sciences 66(20), 3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanis P, Thekisoe O, Kiouptsi K, Ongerth J, Igarashi I and Inoue N (2007) Development and preliminary evaluation of a loop-mediated isothermal amplification procedure for sensitive detection of cryptosporidium oocysts in fecal and water samples. Appl Environ Microbiol 73(17), 5660–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura M, Kazato A, Yamamuro T, Ando H, Sasaki Y, Suzuki R and Shirataki Y (2019) Rapid identification of Aconitum plants based on loop-mediated isothermal amplification assay. BMC Res Notes 12(1), 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm C, Martzy R, Brunner K, Mach RL, Krska R, Heinze G, Sommer R, Reischer GH and Farnleitner AH (2017) A complementary isothermal amplification method to the US EPA quantitative polymerase chain reaction approach for the detection of enterococci in environmental waters. Environ Sci Technol 51(12), 7028–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolm C, Martzy R, Fuhrer M, Mach RL, Krska R, Baumgartner S, Farnleitner AH and Reischer GH (2019) Detection of a microbial source tracking marker by isothermal helicase-dependent amplification and a nucleic acid lateral-flow strip test. Sci Rep 9(1), 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koloren Z and Ayaz E (2016) Genotyping of Cryptosporidium spp. in environmental water in Turkey. Acta Parasitol 61(4), 671–679. [DOI] [PubMed] [Google Scholar]
- Korajkic A, McMinn BR and Harwood VJ (2018) Relationships between Microbial Indicators and Pathogens in Recreational Water Settings. Int J Environ Res Public Health 15(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X, Wu Q, Zhang J and Fan H (2006) Rapid detection of noroviruses in fecal samples and shellfish by nucleic acid sequence-based amplification. J Microbiol 44(4), 403–408. [PubMed] [Google Scholar]
- Lee MS, Su TY, Lien YY and Sheu SC (2017) The development of loop-mediated isothermal amplification (LAMP) assays for the rapid authentication of five forbidden vegetables in strict vegetarian diets. Sci Rep 7, 44238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chan Y, Elina H, Ravichandran M, Zainul F and Lalitha P (2008) Development of a Thermostabilized Cholera-NASBA-ELISA Assay for the Specific Detection of Vibrio cholerae. International Journal of Infectious Diseases 12, e60. [Google Scholar]
- Lee S, Khoo VSL, Medriano CAD, Lee T, Park SY and Bae S (2019) Rapid and in-situ detection of fecal indicator bacteria in water using simple DNA extraction and portable loop-mediated isothermal amplification (LAMP) PCR methods. Water Res 160, 371–379. [DOI] [PubMed] [Google Scholar]
- Li J, Wang C, Yu X, Lin H, Hui C, Shuai L and Zhang S (2019) Rapid detection of Cyanobacteria by recombinase polymerase amplification combined with lateral flow strips. Water Supply 19(4), 1181–1186. [Google Scholar]
- Lillis L, Siverson J, Lee A, Cantera J, Parker M, Piepenburg O, Lehman DA, Boyle DS (2016) Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol Cell Probes 30(2), 74–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FG, Chen GF, Zhang CY, Wang YY and Zhou J (2019) Exponential rolling circle amplification coupled with lateral flow dipstick strips as a rapid and sensitive method for the field detection of Karlodinium veneficum. Journal of Applied Phycology 31(4), 2423–2436. [Google Scholar]
- Martzy R, Kolm C, Brunner K, Mach RL, Krska R, Sinkovec H, Sommer R, Farnleitner AH and Reischer GH (2017) A loop-mediated isothermal amplification (LAMP) assay for the rapid detection of Enterococcus spp. in water. Water Res 122, 62–69. [DOI] [PubMed] [Google Scholar]
- Martzy R, Kolm C, Krska R, Mach RL, Farnleitner AH and Reischer GH (2019) Challenges and perspectives in the application of isothermal DNA amplification methods for food and water analysis. Anal Bioanal Chem 411(9), 1695–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville L, Kenyon F, Javed S, McElarney I, Demeler J and Skuce P (2014) Development of a loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Haemonchus contortus eggs in ovine faecal samples. Vet Parasitol 206(3-4), 308–312. [DOI] [PubMed] [Google Scholar]
- Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, Shekelle P, Stewart LA and Group P-P (2015) Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi Y, Tamaru A, Suzuki Y, Kitada S, Maekura R, Tateishi Y, Niki M, Ogura H and Matsumoto S (2014) Direct detection of Mycobacterium avium in environmental water and scale samples by loop-mediated isothermal amplification. Journal of water and health 12(2), 211–219. [DOI] [PubMed] [Google Scholar]
- Notomi T, Mori Y, Tomita N and Kanda H (2015) Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53(1), 1–5. [DOI] [PubMed] [Google Scholar]
- Obande GA and Banga Singh KK (2020) Current and Future Perspectives on Isothermal Nucleic Acid Amplification Technologies for Diagnosing Infections. Infect Drug Resist 13, 455–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Wang Q, Fei Y, Zhu P, Qiao L, Huang H, Dang C and Gao W (2019) A real-time recombinase polymerase amplification assay for the rapid detection of Vibrio harveyi. Mol Cell Probes 44, 8–13. [DOI] [PubMed] [Google Scholar]
- Parida M, Sannarangaiah S, Dash PK, Rao P and Morita K (2008) Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Reviews in medical virology 18(6), 407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce RH and Henry MS (2008) Harmful algal toxins of the Florida red tide (Karenia brevis): natural chemical stressors in South Florida coastal ecosystems. Ecotoxicology 17(7), 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutzer J and Karanis P (2009) Rapid identification of Giardia duodenalis by loop-mediated isothermal amplification (LAMP) from faecal and environmental samples and comparative findings by PCR and real-time PCR methods. Parasitol Res 104(6), 1527–1533. [DOI] [PubMed] [Google Scholar]
- Qi H, Yue S, Bi S, Ding C and Song W (2018) Isothermal exponential amplification techniques: From basic principles to applications in electrochemical biosensors. Biosens Bioelectron 110, 207–217. [DOI] [PubMed] [Google Scholar]
- Rames EK and Macdonald J (2019) Rapid assessment of viral water quality using a novel recombinase polymerase amplification test for human adenovirus. Applied microbiology and biotechnology 103(19), 8115–8125. [DOI] [PubMed] [Google Scholar]
- Rutjes SA, van den Berg HH, Lodder WJ and de Roda Husman AM (2006) Real-time detection of noroviruses in surface water by use of a broadly reactive nucleic acid sequence-based amplification assay. Appl Environ Microbiol 72(8), 5349–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Sun J, Ye Y, Zhang J, Zhang Y and Sun X (2020) Loop-mediated isothermal amplification-based microfluidic chip for pathogen detection. Crit Rev Food Sci Nutr 60(2), 201–224. [DOI] [PubMed] [Google Scholar]
- Shanks OC, Sivaganesan M, Peed L, Kelty CA, Blackwood AD, Greene MR, Noble RT, Bushon RN, Stelzer EA, Kinzelman J, Anan'eva T, Sinigalliano C, Wanless D, Griffith J, Cao Y, Weisberg S, Harwood VJ, Staley C, Oshima KH, Varma M and Haugland RA (2012) Interlaboratory comparison of real-time PCR protocols for quantification of general fecal indicator bacteria. Environ Sci Technol 46(2), 945–953. [DOI] [PubMed] [Google Scholar]
- Shi H, Mao X, Chen X, Wang Z, Wang K and Zhu X (2017) The analysis of proteins and small molecules based on sterically tunable nucleic acid hyperbranched rolling circle amplification. Biosens Bioelectron 91, 136–142. [DOI] [PubMed] [Google Scholar]
- Shiraho EA, Eric AL, Mwangi IN, Maina GM, Kinuthia JM, Mutuku MW, Mugambi RM, Mwandi JM and Mkoji GM (2016) Development of a loop mediated isothermal amplification for diagnosis of Ascaris lumbricoides in fecal samples. Journal of parasitology research 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, Jang W, Lee J, Ahn S, Suraiya S, Kim C and Kong I (2017) gro EL is a suitable genetic marker for detecting Vibrio parahaemolyticus by loop-mediated isothermal amplification assay. Letters in applied microbiology 65(2), 106–113. [DOI] [PubMed] [Google Scholar]
- Soller JA, Schoen ME, Bartrand T, Ravenscroft JE and Ashbolt NJ (2010) Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44(16), 4674–4691. [DOI] [PubMed] [Google Scholar]
- Stedtfeld RD, Stedtfeld TM, Samhan F, Kanitkar YH, Hatzinger PB, Cupples AM and Hashsham SA (2016) Direct loop mediated isothermal amplification on filters for quantification of Dehalobacter in groundwater. J Microbiol Methods 131, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Stedtfeld TM, Waseem H, Fitschen-Brown M, Guo X, Chai B, Williams MR, Shook T, Logan A and Graham A (2017) Isothermal assay targeting class 1 integrase gene for environmental surveillance of antibiotic resistance markers. Journal of environmental management 198, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld RD, Tourlousse DM, Seyrig G, Stedtfeld TM, Kronlein M, Price S, Ahmad F, Gulari E, Tiedje JM and Hashsham SA (2012) Gene-Z: a device for point of care genetic testing using a smartphone. Lab on a Chip 12(8), 1454–1462. [DOI] [PubMed] [Google Scholar]
- Stoeckel DM and Harwood VJ (2007) Performance, design, and analysis in microbial source tracking studies. Appl Environ Microbiol 73(8), 2405–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syafirah EEN, Najian AN, Foo PC, Ali MRM, Mohamed M and Yean CY (2018) An ambient temperature stable and ready-to-use loop-mediated isothermal amplification assay for detection of toxigenic Vibrio cholerae in outbreak settings. Acta tropica 182, 223–231. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Ohkawachi M, Horio K, Kobori T, Aki T, Matsumura Y, Nakashimada Y and Okamura Y (2018) RNase H-assisted RNA-primed rolling circle amplification for targeted RNA sequence detection. Sci Rep 8(1), 7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Choi G, Nouri R and Guan W (2019) Loop-Mediated Isothermal Amplification-Coupled Glass Nanopore Counting Toward Sensitive and Specific Nucleic Acid Testing. Nano Lett 19(11), 7927–7934. [DOI] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H and Notomi T (2008) Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3(5), 877–882. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (2002) Method 1603: Escherichia coli (E. coli) in Water by Membrane Filtration Using Modified membrane-Thermotolerant Escherichia coli Agar (Modified mTEC) Washington, D.C. [Google Scholar]
- U.S. Environmental Protection Agency (2006) Method 1600: Enterococci in Water by Membrane Filtration Using membraneEnterococcus Indoxyl-Beta-D-Glucoside Agar (mEI) Washington, D.C. [Google Scholar]
- U.S. Environmental Protection Agency (2010) Method A: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR) Assay Washington, D.C. [Google Scholar]
- U.S. Environmental Protection Agency (2012) Recreational Water Quality Criteria, U.S. Environmental Protection Agency. [Google Scholar]
- Ulrich RM, John DE, Barton GW, Hendrick GS, Fries DP and Paul JH (2015) A handheld sensor assay for the identification of grouper as a safeguard against seafood mislabeling fraud. Food Control 53, 81–90. [Google Scholar]
- Uyttendaele M, Schukkink R, Van Gemen B and Debevere J (1995) Development of NASBA®, a nucleic acid amplification system, for identification of Listeria monocytogenes and comparison to ELISA and a modified FDA method. International journal of food microbiology 27(1), 77–89. [DOI] [PubMed] [Google Scholar]
- van der Vliet GM, Schukkink RA, van Gemen B, Schepers P and Klatser PR (1993) Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J Gen Microbiol 139(10), 2423–2429. [DOI] [PubMed] [Google Scholar]
- Vincent M, Xu Y and Kong H (2004) Helicase-dependent isothermal DNA amplification. EMBO reports 5(8), 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Feng J and Tian X (2019) Application of loop-mediated isothermal amplification (LAMP) for rapid detection of Atlantic cod (Gadus morhua), Pacific cod (Gadus macrocephalus) and haddock (Melanogrammus aeglefinus). Mol Cell Probes 47, 101420. [DOI] [PubMed] [Google Scholar]
- Wu L, Ye L, Wang Z, Cui Y and Wang J (2019) Utilization of recombinase polymerase amplification combined with a lateral flow strip for detection of Perkinsus beihaiensis in the oyster Crassostrea hongkongensis. Parasit Vectors 12(1), 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xu M, Wang Q, Zhou C, Wang M, Zhu X and Zhou D (2017) Recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for detection of Toxoplasma gondii in the environment. Vet Parasitol 243, 199–203. [DOI] [PubMed] [Google Scholar]
- Xiong X, Huang M, Xu W, Li Y, Cao M and Xiong X (2020) Rainbow Trout (Oncorhynchus Mykiss) Identification in Processed Fish Products Using Loop-Mediated Isothermal Amplification (LAMP) and PCR Assays. J Sci Food Agric In press. [DOI] [PubMed] [Google Scholar]
- Yang Z, McLendon C, Hutter D, Bradley KM, Hoshika S, Frye C and Benner SA (2015) Helicase dependent isothermal amplification of DNA and RNA using self-avoiding molecular recognition systems. Chembiochem: a European journal of chemical biology 16(9), 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen G, Wang Y, Sun R, Nie X and Zhou J (2018) MHBMDAA: membrane-based DNA array with high resolution and sensitivity for toxic microalgae monitoring. Harmful Algae 80, 107–116. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chen F, Li Q, Wang L and Fan C (2015) Isothermal Amplification of Nucleic Acids. Chem Rev 115(22), 12491–12545. [DOI] [PubMed] [Google Scholar]
- Zhu P, Zhang B-F, Wu J-H, Dang C-Y, Lv Y-T, Fan J-Z and Yan X-J (2014) Sensitive and rapid detection of microcystin synthetase E Gene (mcyE) by loop-mediated isothermal amplification: a new assay for detecting the potential microcystin-producing Microcystis in the aquatic ecosystem. Harmful Algae 37, 8–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.