Abstract
USP5 and USP8 (Deubiquitinating enzyme) are highly overexpressed and more recognized as poor prognosis marker in various cancers. Depleting USP5 or USP8 to assess the synergism with proteasome inhibitor (Bortezomib) were measured. Furthermore, in present finding USP5 cooperates hnRNPA1 & USP8 cooperate SF2/ASF1, therefore gain in expression of either hnRNPA1 or SF2/ASF1 is sufficient to promote cell survival. On the other side, apoptosis markers were more pronounced in U87 or T98G cells devoid of either USP5 or USP8. However, apparent increase in SF2/ASF1 in absence of USP5, providing resistant factor is new. Antiapoptotic activity due to rise in SF2/ASF1 was validated after co-knock down of SF2/ASF1 in addition to USP5 induces more apoptosis comparing to individual knock down of USP5 or SF2/ASF1. This reveals SF2/ASF1 (RNA binding protein) delayed the apoptotic effect due to loss of USP5, lends ubiquitination of hnRNPA1. In presence of USP5, PI3 kinase inhibition promotes even more interaction between USP5 and hnRNPA1, thereby stabilizes hnRNPA1 in U87MG. In that way hnRNPA1 and SF2/ASF1 impart oncogenic activity. In conclusion, siRNA based strategy against USP5 is not enough to inhibit glioma, moreover targeting additionally SF2/ASF1 by knocking down USP8 is suitably more effective to deal with glioma tumour reoccurrence by indirectly targeting both SF2/ASF1 and hnRNPA1 oncogene.
Keywords: USP5, USP8, hnRNPA1, SF2/ASF1, Apoptosis
Abbreviations: DUB, Deubiquitinating enzymes; USP5, Ubiquitin specific peptidase 5; USP8, Ubiquitin specific peptidase 8; hnRNPA1, Heterogeneous Nuclear Ribonucleoprotein A1; SF2/ASF1, Serine arginine rich alternative splice factor
Highlights
-
•
Deubiquitinating enzyme USP5 interact with hnRNPA1 and promotes hnRNPA1 ubiquitination is PI3 Kinase dependent.
-
•
USP5 knock down in glioma cell, stabilizes SF2/ASF1 expression act as resistance factor.
-
•
Depleting SF2/ASF1 and USP5 synergistically promotes apoptosis in glioma cell.
1. Introduction
The ubiquitin-proteasome system (UPS) collectively plays crucial role in maintaining the protein turn over vested to various cellular process such as cell differentiation, DNA repair, cell division, etc. [1]. Deubiquitinating (DUB's) family of enzymes are component of the Ubiquitin proteasome system (UPS), that cleaved out the ubiquitin from proteins and prevents its degradation thereby modulates the functionary circuit of proteins. Many Deubiquitinating enzymes are known to be highly expressed in the brain and reproductive organs [2]. A class of DUB's are described as Ubiquitin-specific protease [USP], where USP1, USP7, USP11, USP22, USP44 and USP49 are present in the nuclei, whereas as USP6 is found in Plasma membrane [3]. Ubiquitin-specific protease plays an essential role in cancer progression [[4], [5], [6]]. Study related with silencing of USP8 in Gefitinib resistant Non-small-cell lung carcinoma was shown to cause downregulation of receptor tyrosine kinases (RTK), including MET, EGFR, ERBB2, ERBB3 [7]. USP5 (Isopeptidase T), another USP family protein a member of the peptidase C19 family, cleaves multi-ubiquitin polymers with a marked preference for branched ubiquitin polymers [8]. Main function of USP5 is the recycling of dissemble polyubiquitin released at the proteasome entry site, thereby stabilizing cytosolic ubiquitin pool [9]. It is noteworthy that USP5 is highly expressed in Gliomas [2], where p53 stabilization effect caused due to the accumulation of unanchored polyubiquitin in the absence of USP5 causes cell cycle arrest [10]. It is reported that exopeptidase hydrolyses isopeptide bonds in between polyubiquitin from the free C-terminal end to produce monoubiquitin, which is reused in conjugating to substrate proteins [11]. Deletion of USP5 or its functional ortholog in yeast led to inhibition of the proteasome due to accumulation of free ubiquitin chains [12]. These studies provide evidence that cells strictly require to maintain the ubiquitin pool to sustain homeostasis.
USP5 expression promotes tumorigenesis in many cancers, like in non-small cell lung cancer overexpression of USP5 stabilizes the beta-catenin protein [13]. In Pancreatic cancer, USP5 was shown to encourage oncogenicity by modulating the cell cycle regulators, as inhibition of USP5 attenuated pancreatic cell growth [14]. In myeloma cells, USP5 stabilizes the c-Maf transcription factor, where inhibition of USP5 promotes c-Maf degradation and leads to apoptosis in myeloma cells [15]. Genome-wide array analysis has revealed a strong correlation between USP5 isoform 2 production and PTBP1 expression in GBM (Glioblastoma) tumor samples and cell lines. Moreover, USP isoform 2 production was also reported to be crucial for gliomagenesis, indicating that selective inhibition of USP5 isoform 2 is conducive to glioma therapy [16]. However long term effect in absence of USP5 in cancer cells were not demonstrated, to study tumor relapse effect because of very short glioma patient survival.
HnRNPA1, a member of the hnRNP A/B family, is aberrantly overexpressed in different cancers. These are nuclear proteins that bind to newly derived transcripts generated by RNA polymerase II [17,18]. They bind specifically to splicing silencer sequences on pre-mRNA and promote exon inclusion, thus acting as splicing repressors [19]. hnRNPA1 is known to play essential roles in key steps of mRNA metabolism involved in alternative splicing, mRNA export, translation, microRNA processing, and telomere maintenance [20]. Splice factor proteins are the key regulators of splicing, and their deregulation leads to the production of aberrantly mRNA spliced isoforms contributes to tumorigenesis [21]. Among the splice factor protein, TRAF6 an E3 ligase promotes hnRNPA1 ubiquitination and synthesizes lysine 63 Ub chains on its substrates [22]. Other way round overexpressed hnRNPA1 promotes the expression of antiapoptotic proteins like BCL-XL [23].
In the present study, our objective is to study in broad the secondary down-stream effect after depleting USP5 or USP8, which were initially showed to induce apoptosis in various cancers. Moreover, our study pointed out the SF2/ASF1 oncoprotein expression predicted to be resistant factor, which delayed the apoptosis effect after the loss of USP5, also promotes hnRNPA1 ubiquitination.
2. Results
2.1. Analysis of USP5 & hnRNPA1 expression in glioma cells
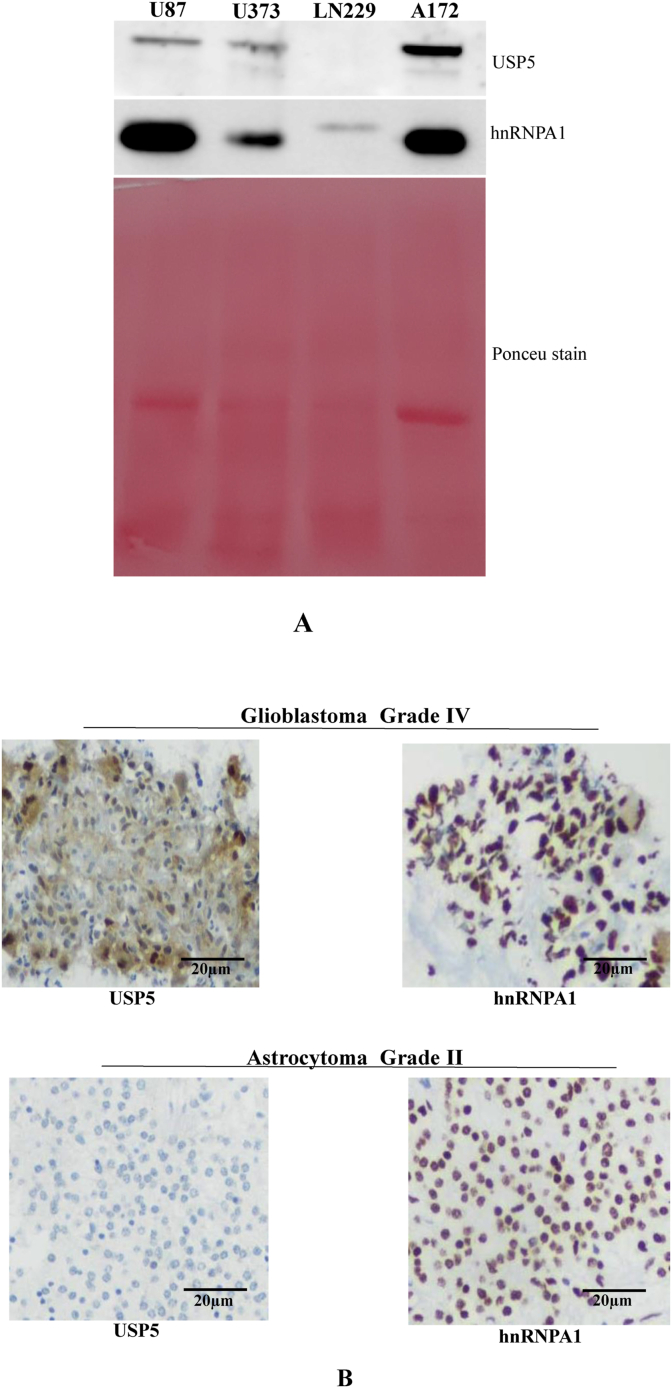
Expression of different Deubiquitinating enzymes (USP's) are up regulated in many cancers like myeloma, non-small cell lung cancer and pancreatic cancer. USP5 protein expressions were correlated with hnRNPA1 in different glioma cell lines, by western blotting technique. We observed that A172 glioma cell line has highest expression of USP5 and U87, U373 have almost moderate expression of USP5, whereas LN229 expresses weakly. Similarly, expression of hnRNPA1 was observed higher in U87 and A172 and less in LN229 (Fig. 1A), shows direct positive correlation.
Fig. 1.
Analysis of USP5 & hnRNPA1 (RNA Binding Protein) in glioma cells: A) Expression of USP5 & hnRNPA1 in glioma cells (U87, U373, LN229 and A172) were analysed using specific antibodies by western blotting. Ponceu stain was used to show the protein loading in every lane. B) Immunohistochemistry of USP5 and hnRNPA1 in Glioblastoma (Grade IV) and Astrocytoma (Grade II) tumour tissues. Protein expression was analysed using specific monoclonal antibodies following with secondary HRPO conjugated antibody, detection using DAB staining (brown colour). Blue colour marks nuclear staining. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also showed the expression analysis in the Glioblastoma grade IV and Low grade Astrocytoma tissue section by using IHC (Immunohistochemistry) technique. Here, we observed equally high expression of hnRNPA1 and USP5 in Grade IV Glioblastoma tissue in comparison to lowly expressing Grade II astrocytoma tissue with hnRNPA1 is more localized in nucleus (Fig. 1B).
2.2. Depletion of deubiquitin enzymes leads to glioma cell apoptosis
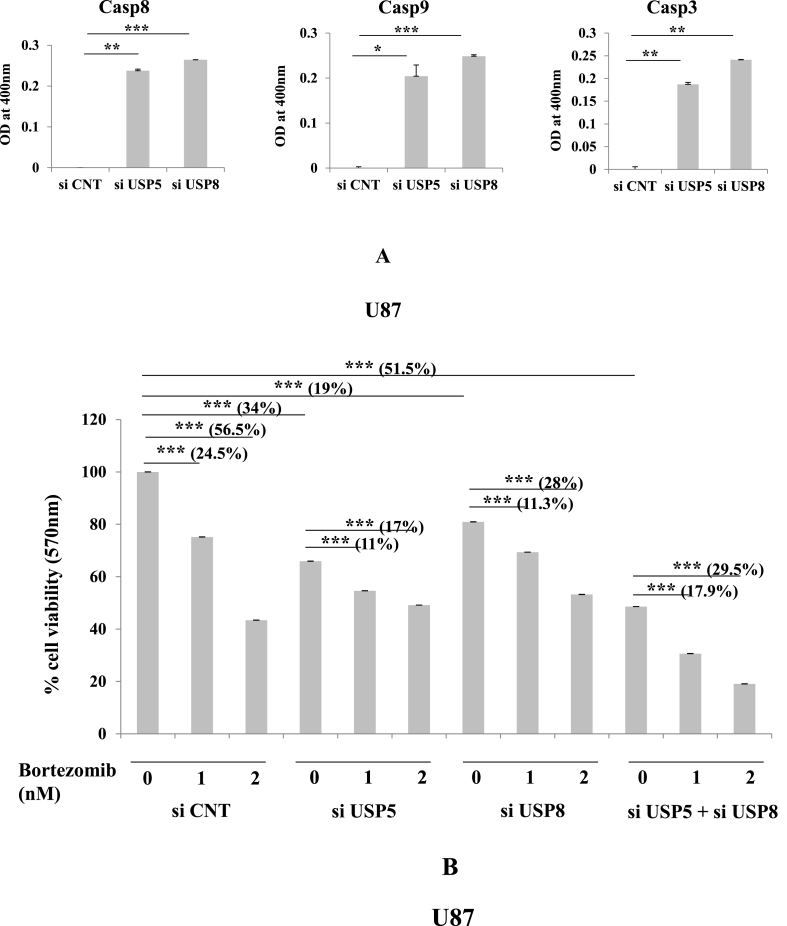
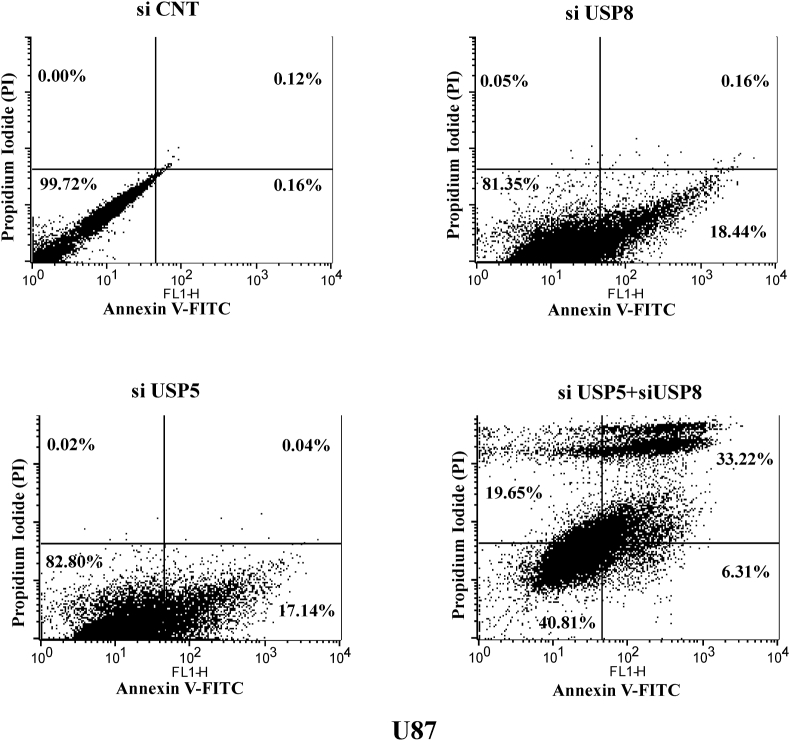
USP5 along with family protein USP8 was chosen to demonstrate apoptotic effect in glioma cells. Efficiency of knock down was more than 50%–60% and was standardized for subsequent experiment. The Caspase cascade was studied using colorimetric based cleavage assay (Methodology section) evaluated with significant increase in Casp8, Casp9 & Casp3 cleavage in both USP5 and USP8 knock down cells (Fig. 2A). In all the caspase colorimetric assays, experimental samples were normalized with control siRNA transfected samples to baseline level. We also measured cell viability using MTT assay in USP5 and USP8 si RNA transfected U87 glioma cells. siRNA treated samples were compared with control siRNA treated samples in presence or absence of Bortezomib (BR) (Proteosome inhibitor). Many literatures have suggested that Bortezomib promotes apoptosis in different types of cancer [24]. In our study also, we observed apoptosis but no further decrease in cell viability was observed after Bortizomib treatment in deubiquitinating enzyme knock down cell sets. Experimentally we followed USP5 and USP8 knock down approach individually as well as in combination. In control siRNA transfected cells Bortezomib treatment decreased the cell viability in dose dependent manner 0 nM, 1 nM (24.5% decrease), 2 nM (56.5% decrease) with IC50 dose at 2 nM effectively. However no further sensitization in USP5 or USP8 knock down cells was observed after Bortezomib treatment on same dose (0, 1, 2 nM). USP5 knock down shows only 11% and 17% decrease in cell viability with 1 nM & 2 nM bortezomib doses respectively & USP8 knock down shows only 11.3% & 28% decrease in cell viability after bortezomib treatment with 1 & 2 nM respectively, this suggest no additional cell death affect. Whereas in absence of Bortezomib, cell viability was decreased 34% in siUSP5 and 19% in siUSP8 glioma cells. Whereas, cells transfected in combination with siRNA against USP5 and USP8, observed synergistic reduction in cell viability around 51%, This suggest USP5 and USP8 regulates two independent pathways, enforced cancer cells to loss its viability was confirmed by detecting early apoptosis and late apoptosis by Annexin V FITC and Propidium iodide staining, using flow cytometry based assay (Fig. 5). Bortezomib (2 nM) treatment in siRNA co-knock down USP5 and USP8 shows loss of cell viability upto 29.5% (Fig. 2B). This shows loss of deubiquitin enzyme, specifically USP5 loss doesn't act synergistically with Bortezomib treatment because ubiquitinated hnRNPA1 or its degradation was not obstructed due loss of HSP27 in presence of Bortezomib (proteasome inhibitor), therefore additional apoptosis was not observed.
Fig. 2.
Inhibition of Deubiquitinating enzymes leads to caspase activation and apoptosis: A) Caspase 8, Caspase9 & Caspase3 cleavage assay was performed in U87 glioma cells transfected si RNA (10 nM) against USP5 and USP8 were compared with control siRNAs. B) Cell viability assay using MTT reagent: U87 glioma cells transfected with specific siRNA in following set: USP5, USP8, USP5+USP8 were treated with or without Bortezomib (Proteasome inhibitor) in different dose concentrations (0, 1 & 2 nM). No significant p value > 0.05, if p value < 0.05 (∗), <0.01(∗∗), and <0.001(∗∗∗) are considered significant.
Fig. 5.
AnnexinV & PI staining to analysis apoptosis in glioma cells after inhibiting USP5 & USP8: U87 glioma cells were knock down by siRNA against USP5, USP8 and in co-knockdown USP5 + USP8, compared with control siRNA. Analysed the early and late apoptosis using Annexin V- FITC and PI staining kit by flow cytometer.
2.3. Deubiquitin enzyme regulates hnRNPA1 and SF2/ASF1 proteins in glioma cells
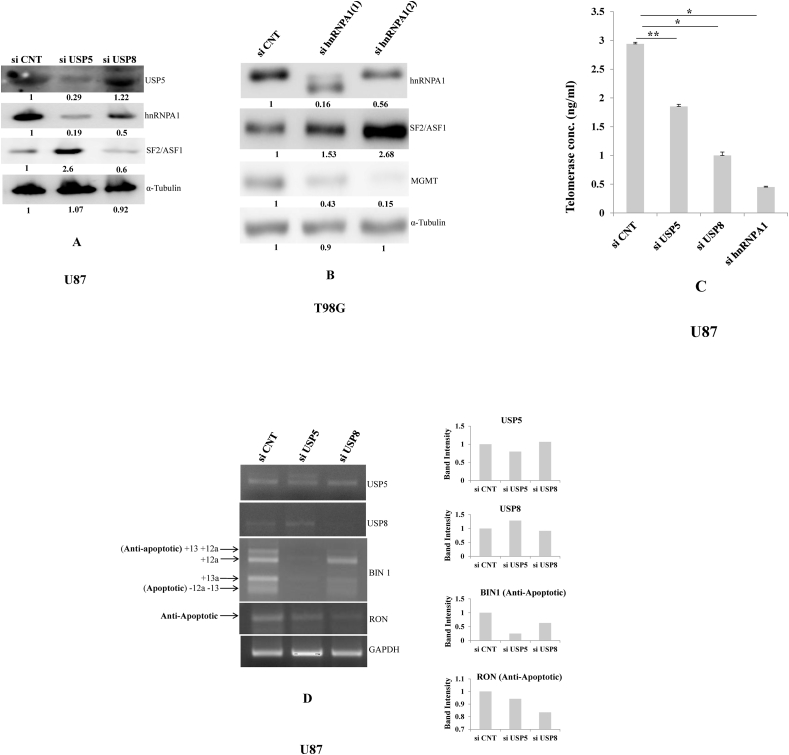
To determine the role of USP5 & USP8 in Gliomas, we knocked down USP5 & USP8 in gliomas cancer cell line (U87MG), using siRNAs approach. The protein expression of hnRNPA1 (Splicing repressor) was downregulated in both USP5 and USP8 knock down cells but SF2/ASF1 (Splicing enhancer) expression was found upregulated in USP5 knock down cells and downregulated in USP8 knock down cells (Fig. 3A). This important piece of result demonstrated that USP5 antagonistically regulates expression of splicing factor family proteins such as hnRNPA1 and SF2/ASF1. To confirm SF2/ASF1 is downstream factor after hnRNPA1 or USP5, validated by knocking down hnRNPA1 (splice factor) using two different siRNA's in T98G glioma cells. The protein expression of SF2/ASF1 was much elevated, but at the same time MGMT(O6)-Methylguanine-DNA methyltransferase) expression was down regulated (Fig. 3B). MGMT is a glioma poor prognosis marker against glioblastoma prescribed treatment with alkylating drug Temozolomide [25]. This means hnRNPA1 is a downstream target of USP5, thereafter affecting SF2/ASF1 expression level in absence of hnRNPA1. HnRNPA1 overexpression showed to replace the SF2/ASF1 binding site on pre-mRNA sequence suppressing the exon3b splicing in Rac1 gene [26].
Fig. 3.
Deubiquitinating enzyme regulates RNA binding family proteins: A) Western blotting of SF2/ASF1, hnRNPA1, USP5, α-Tubulin (loading control) was performed in U87 glioma cells transfected with si RNA against USP5 and USP8, were compared with control si RNA. B) T98G glioma cells transfected with two different hnRNPA1 siRNAs compared with control siRNA. Western blotting was performed to analyse the expression of MGMT and SF2/ASF1. C) Telomerase concentration (ng/ml) was measured in U87 glioma cells transfected with specific siRNAs (USP5, USP8, and hnRNPA1) were compared with control si RNA. Statistical analysis was performed. No significant p value > 0.05, if p value < 0.05 (∗), <0.01(∗∗), and <0.001(∗∗∗) are considered significant. D) Comparative mRNA alternative spliced gene variant of BIN1, & RON in USP5 (10 nM) and USP8 (10 nM) knock down U87 cells was determined, GAPDH housekeeping gene was used for loading control. Densitometry was analysed using Image j available (NIH) software. (n = 3, Mean ± SE).
Additionally, we validated the direct effect of hnRNPA1 in regulating telomerase concentration in U87 Glioma cells was confirmed by knocking down USP5, USP8 and hnRNPA1was positive control to quantitate telomerase. Observation leading to downregulation of telomerase concentration in USP5, USP8 and hnRNPA1 knock down cells shows to be hnRNPA1 dependent (Fig. 3C).
2.4. PCR based alternative splicing assay in USP5 and USP8 Knock down cells
To demonstrate further that hnRNPA1 and SF2/ASF1 are downstream factors of USP5 and USP8. Alternative splicing was performed using our standardized protocol to assess the apoptotic and antiapoptotic variant of BIN1 (Tumour suppressor) and RON (Receptor tyrosine kinase). We showed here a decrease in antiapoptotic variants of BIN1 in USP5 knock down cell, is hnRNPA1 dependent. Similarly, decrease in antiapoptotic band of RON receptor in USP5 and USP8 knock down was also observed (Fig. 3D), is SF2/ASF1 dependent.
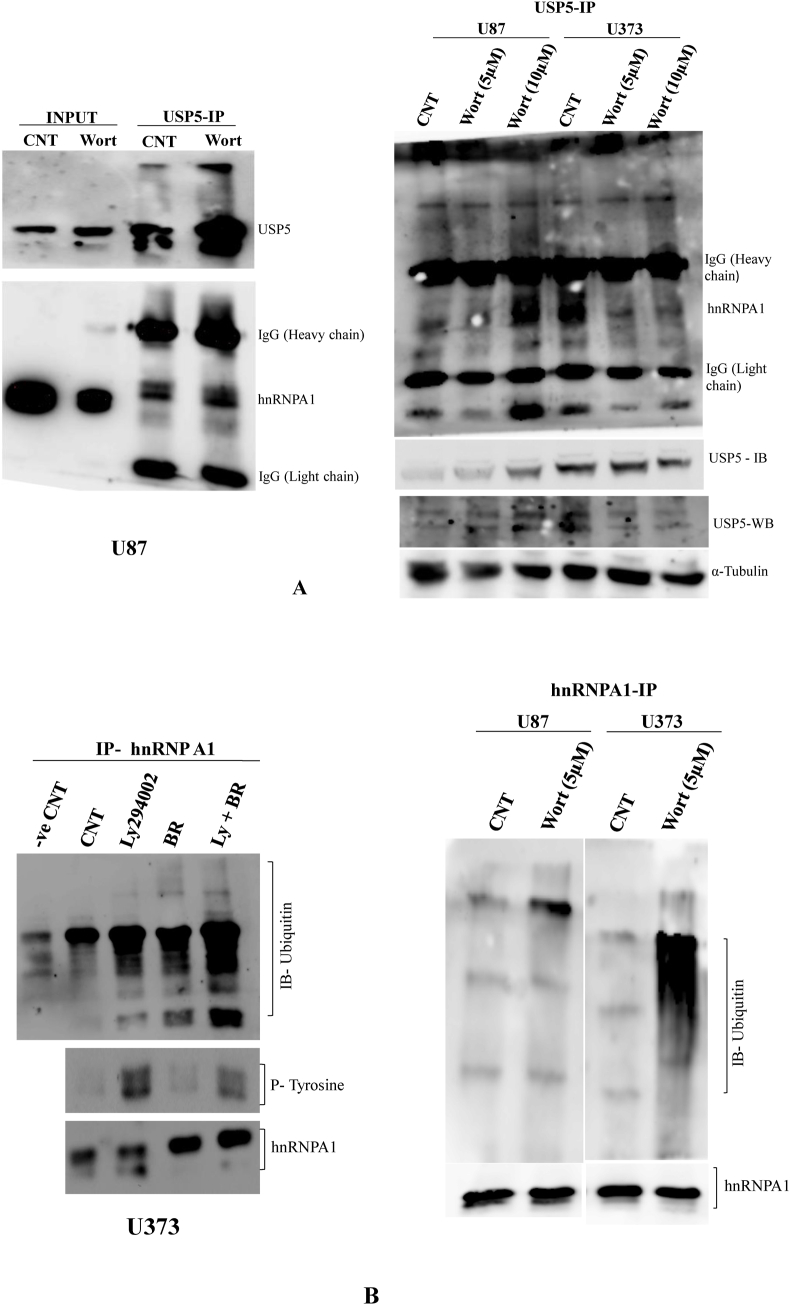
2.5. Interaction between USP5 and hnRNPA1 & hnRNPA1 ubiquitination in U87 & U373 glioma cells
As showed positive cooperative expression between USP5 and hnRNPA1 in glioma cell lines. Immunoprecipitation technique were carried out to demonstrate the interaction between USP5 and hnRNPA1 in U87 & U373 glioma cell line. Both USP5 and hnRNPA1 were showed to interact with each other and found USP5 dependent hnRNPA1 ubiquitination, however differently regulated in U373 in comparison to U87MG. It may be because of the differentially expression two splice variants of USP5 in two different glioma cell lines U87 & U373, accordingly drive ubiquitin modification of hnRNPA1. Interestingly, using PI3 Kinase selective inhibitor wortmannin, promotes interaction between USP5 and hnRNPA1 in U87 cells, and loss of interaction between USP5 and hnRNPA1. The experiment was demonstrated after immunoprecipitating with USP5 antibody showed pull down of high molecular weight hnRNPA1, comparing to the molecular weight of hnRNA1 in input sample (10%). This interaction effect probably due to PI3 kinase dependent inactivation of GSK3 kinase [27], where AKT kinase phosphorylates hnRNPA1 at serine 199 [28], showed lesser interaction between hnRNPA1 and USP5 in U373 cells in two different wortmannin dose treated (5 μM, 10 μM) (Fig. 4A).
Fig. 4.
Interaction between USP5 & hnRNPA1 in presence of wortmannin (PI3kinase) and hnRNPA1 ubiquitination in U87 & U373 Glioma Cells: A) USP5 interacts with hnRNPA1 in U87 cells treated with PI3 kinase inhibitor (Wortmannin) (10 μM) for 24 h. USP5 (Santa Cruz) antibody was used for the immunoprecipitation following with immunoblot with USP5 and hnRNPA1. 10% total protein lysates were used as an input for positive control. Second picture: The interaction between USP5 and hnRNPA1 was compared between U87 and U373. Both the cell lines were treated with PI3 kinase inhibitor (Wortmannin) at two different doses 5 μM & 10 μM for 24 h. USP5 (Santa Cruz) antibody was used for the immunoprecipitation, followed with immunoblotting with USP5 and hnRNPA1 (Santa Cruz). Non-immunoprecipitated western blotting of USP5 in U87 & U373 was determined in wortmannin treated samples and Alpha tubulin (Millipore) antibody was used, as loading control. B) U373 cells were treated with or without PI3 kinase inhibitor (LY294002) (10 μM), in presence or absence of Proteosome inhibitor (Bortezomib) (5 nM) for 24 h. After treatment total protein lysate was prepared in Triton cell lysis buffer. After protein quantification, hnRNPA1 protein was immunoprecipitated with specific monoclonal antibody from 1 mg total protein lysate from all the experimental samples followed with SDS-PAGE and western blotting. Membrane was immunoblotted with Ubiquitin antibody (Santa Cruz), Phosphotyrosine (Santa Cruz), and hnRNPA1 (Santa Cruz) antibodies to compare the hnRNPA1 ubiquitination, phosphorylation and total hnRNPA1 in all the samples. Second picture: U87 & U373 cells were treated with PI3 kinase inhibitor (Wortmannin) (5 μM) for 24 h, following treatment total protein lysate was prepared in Triton cell lysis buffer. After protein quantification, hnRNPA1 immunoprecipitation with its specific antibody from 1 mg total protein lysate from each of the samples followed with SDS-PAGE and western blotting. Membrane was immunoblotted with Ubiquitin (Santa Cruz), and Total hnRNPA1 (Santa Cruz) antibodies to compare the hnRNPA1 ubiquitination, and pull down of hnRNPA1 in all the samples.
Furthermore, demonstrated hnRNPA1 ubiquitination in both U87 & U373 glioma cells after inhibiting PI3 kinase using Ly294002 or wortmannin, showed high hnRNPA1 ubiquitination in U373 drug treated cells, and hnRNPA1 was found more accumulated in proteasome inhibitor treated in comparison to hnRNPA1 ubiquitination in U87 MG (Fig. 4B).
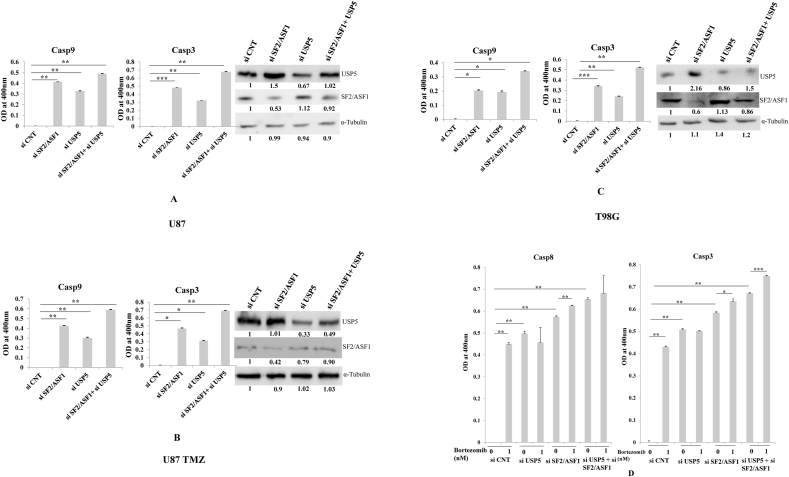
2.6. Caspase9 and Caspase3 cleavage after in SF2/ASF1 & USP5 knock down cell
As previously shown, USP5 & USP8 combination knock down led to more glioma cell apoptosis in comparison to individual USP5 or USP8 knock down. In USP5 knock down hnRNPA1 was down regulated & SF2/ASF1 was upregulated but in USP8 knock down both hnRNPA1 & SF2/ASF1 were downregulated. Moreover, functionally hnRNPA1 is a splicing suppressor and SF2/ASF is a splicing enhancer but both are highly expressed in many cancers and antagonistically act primarily to bring splicing diversity hallmark in cancer progression [29]. Preferred experiments related with endogenous SF2/ASF1 overexpression in USP5 knock down cell were performed: USP5 and SF2/ASF1 knock down individually or in combination using specific siRNAs against SF2/ASF1+USP5 in different glioma cell lines (U87, U87 TMZ, and T98G). U87 TMZ cells are Temozolomide (alkylating drug) resistant cells made in our laboratory. Knock down efficiency of mentioned genes are around ≥50%, using these experimental cell sets, we analysed Casp9 & Casp3 cleavage assay. The prominent Casp9 and Casp3 cleavage in SF2/ASF1 knockdown cells in comparison to USP5 knock down cells were observed, whereas in combination knock down SF2/ASF1 + USP5 was found more effective in promoting Casp9 and Cas3 cleavage. Additionally, knockdown of SF2/ASF1 leads to more USP5 expression is new observation, where with siUSP5 in combination it sensitizes cells to undergo more Caspase cascade activation (Fig. 6 A, B, C). Therefore, targeting directly SF2/ASF1 using selective siRNA or through USP8 knockdown is a good approach to indirectly down regulate SF2/ASF1 (Fig. 3 A) to overcome therapeutic resistance challenge.
Fig. 6.
Caspase 9 and 3 cleavage were estimated in si RNA transfected experimental set (SF2/ASF1, USP5, and co-knock down SF2/ASF1+USP5) were performed and compared with control. Westorn blotting of USP5 and SF2/ASF1 was analysed: A) U87 cells.; B) U87TMZ; C) T98G; D) T98G as mentioned in section of siRNA transfected cells were treated or non-treated with Bortezomib (1 nM), colorimetric estimation of Casp8 and Casp3 was performed. Statistical analysis was performed. No significant p value > 0.05, if p value < 0.05 (∗), <0.01(∗∗), and <0.001(∗∗∗) are considered significant.
In separate experiment we did knock down of SF2/ASF1, USP5 individually or in combination using specific siRNAs against SF2/ASF1+USP5 in T98G glioma cell line with or without 1 nM bortezomib (proteasome inhibitor). Followed with analysis of Casp8 and Casp3 cleavage. Bortezomib (Proteosome inhibitor) treatment causes glioma cells to get more sensitized in control siRNA transfected, whereas in USP5+SF2/ASF1 or SF2/ASF1 or USP5 knockdown, presence of bortezomib dose (1 nM) was not that effective (Fig. 6D). Moreover, knock down of SF2/ASF1 showed USP5 overexpression as showed in previous figures, act as positive feed back loop deciphered in co knock down (SF2/ASF1 + USP5) showed more apoptosis effect. In all the caspase colorimetric assays we have normalized the control samples to baseline level for experimental evaluation.
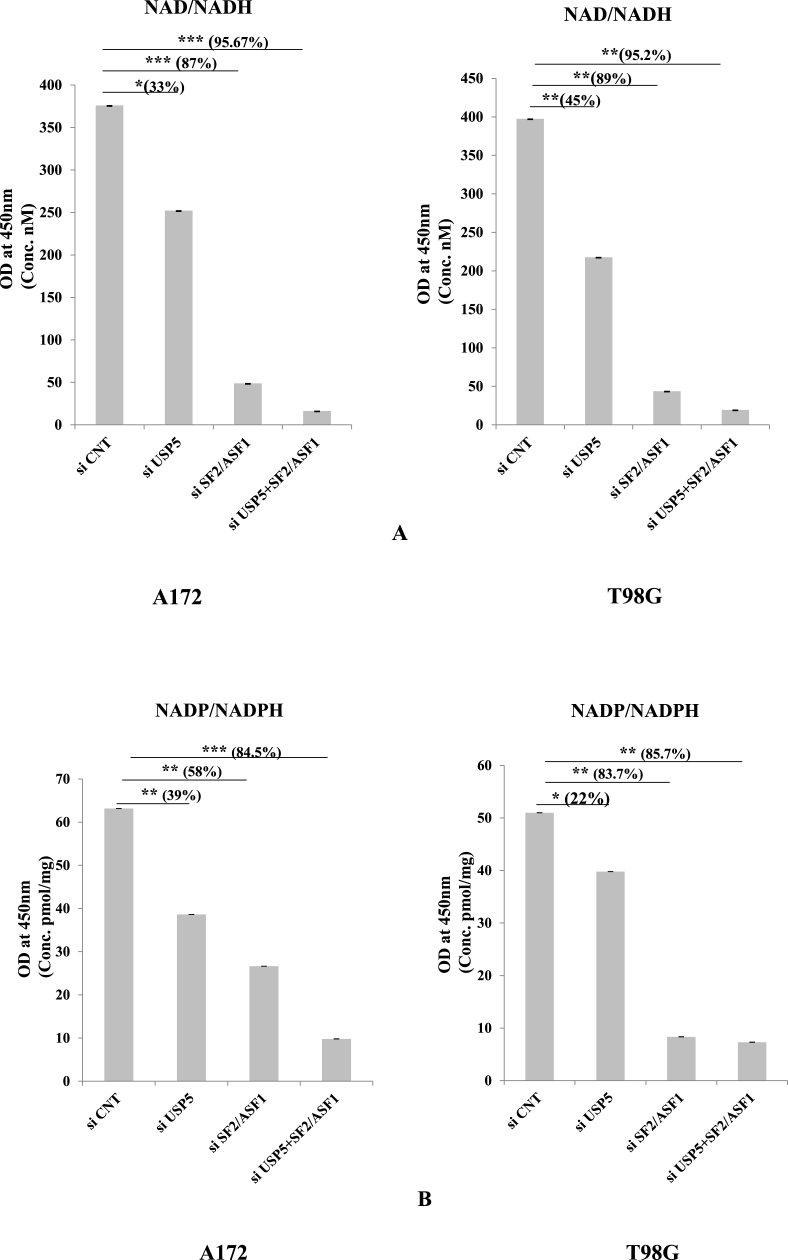
2.7. NAD/NADH & NADP/NADPH pathway in glioma cells
The involvement of the nicotinamide adenine dinucleotide (NAD+) pathway in cancer cell survival is poorly understood. NAD+ salvage pathway modulates cancer cell survival through the rarely mutated tumour suppressor p73 [30]. So after analysing the apoptosis pattern via caspases we demonstrated the expression of NAD/NADH & NADP/NADPH in glioma cells. Using similar strategy knock down cells were used as above (SF2/ASF1, USP5 and co-knockdown USP5+SF2/ASF1) compared with control siRNAs: After 72 h of transfection we calculated the total amount of NAD/NADH present in the cancer cells with the help of estimation kit (Cayman). Here we observed the decreased concentration of NAD/NADH in all the knock down treatment but in combination of both USP5 & SF2/ASF1, the concentration was decreased almost 95% in comparison to control siRNA cells in both A172 & T98G glioma cells. We observed 87% decrease in SF2/ASF1 knock down and 33% decrease in the USP5 knockdown in A172 glioma cells and in T98G cells; 89% decrease in SF2/ASF1 and 45% decrease in USP5 knock down cells (Fig. 7A). We also measured the NADP/NADPH concentration. NADP/NADPH are involved in the ROS (Reactive oxygen species), produced by tumor cells [31]. Same as NAD/NADH concentration, NADP/NADPH concentration was also measured in A172 & T98G glioma cells after knocking down USP5, SF2/ASF1 and USP5+SF2/ASF1 compared with si RNA control cells. Here we found 39% in USP5 knock down, 58% in SF2/ASF1 and 84.5% decrease in SF2/ASf1 & USP5 co-knock down cells. NADP/NADPH concentration in T98G is 22% in USP5 knock down and 83.7% decrease in SF2/ASF1 knock down decrease, and 85.7% in USP5+ SF2/ASF1 knockdown respectively in T98G cells. Interestingly, knock down of both USP5 and SF2/ASF1 in combination in both A172 & T98G cells, showed significant more then 85% was observed. (Fig. 7B).
Fig. 7.
Knock down SF2/ASF1, USP5, and co-knockdown SF2/ASF1+USP5 with specific siRNAs were performed and were compared with control siRNA in A172 & T98G glioma cells. Cells were also treated with Bortezomib (1 nM) (Proteosome inhibitor) and analysed the NAD and NADP concentration in nM, using colorimetric kit. Absorbance was taken at 450 nm.
A) NAD/NADH estimation
B) NADP/NADPH assay
Statistical analysis was performed. No significant p value > 0.05, if p value < 0.05 (∗), <0.01(∗∗), and <0.001(∗∗∗) are considered significant.
2.8. hnRNPA1 and SF2/ASF1 stability in USP5 knockdown cells
Stability of hnRNPA1 were assessed in T98G glioma cell line after transfection of control siRNA and USP5 siRNA for, following cycloheximide treatment (100 μg/ml) in absence or presence of MG132 (proteasome pathway inhibitor). hnRNPA1 expression as mentioned earlier was down regulated in USP5 knockdown cells in comparison to control, similarly we measure the stability of SF2/ASF1 in USP5 knock down cells in comparison to control and here found slow decrease in SF2/ASF1 stability was detected (Supplementary Fig. 1). Moreover, stability of hnRNPA1 is restored after inhibiting proteasome pathway using MG132, in USP5 si RNA as well as in control siRNA (Supplementary Fig. 2). This suggests loss of USP5 down regulate hnRNPA1 stability. Very interestingly, leading E3 Ligase TRAF6 known to be involved in ubiquitination dependent degradation of hnRNPA1 [32]. This prompted us to further evaluate TRAF6 stability in USP5 knock down, where TRAF6 was stable up to 2 h in cycloheximide treated cell.
3. Discussion
Splicing factors play a pivotal role in stabilizing the splicing machinery by the maintaining dynamic control between splice factor family proteins, localization and activity [33]. Splicing enhancer SF2/ASF1 and suppressor hnRNPA1, distinctly modulate the capacity of cancer cells to bypass apoptotic pathway [34]. Evidently, HnRNPA1 is a downstream transcription regulator of c-Myc (proto-oncogene) whereas USP22 positively regulates c-Myc stability and promotes tumorigenesis [35,36]. Antagonistic action of hnRNPA1 against SF2/ASF1 manifest EMT/MET transition by switching the alternatively spliced variant isoforms of important proteins such as RON and Rac1b [26]. Heterogeneity in gliomas tumor also attributed to unregulated expression of SF2/ASF1 and hnRNPA1. Our results suggest that deubiquitinating enzyme USP5 is a major regulatory factor of splice factor proteins hnRNPA1. Deubiquitinating enzyme USP5 modulates hnRNPA1 at post-translational step via interaction dependent subsequent hnRNPA1 stabilization.
PI3Kinase are intracellular signal transducer enzymes, involved in various cellular functions such as cell growth and proliferation [27]. Inhibiting PI3 kinase pathway by Wortmannin inhibitor raisies more interaction between hnRNPA1 and USP5, caused inhibition of ubiquitination-dependent degradation of hnRNPA1 in U87MG cells. Whereas, in U373 cells Wortmannin induces ubiquitination of hnRNPA1 due to loss of hnRNPA1-USP5 interaction in presence of PI3 Kinase inhibitor. Since these two glioma cells U87 and U37 response differently to PI3 Kinase inhibitor, therefore interested to screen expression of hnRNPA1 and SF2/ASF1 in USP5 knockdown cells. SF2/ASF1 prominently expresses in USP5 knock down glioma cell lines, this is probably a hidden cancer escape survival pathway.
HnRNPA1 is the best candidate target where knockdown of hnRNPA1 downregulate ABCC4, ABCC6 transporters and CD44 v6 & v10 isoforms, also down regulate telomerase concentration and glycolysis pathway [[37], [38], [39], [40]]. Recently, another Splice factor family protein SF2/ASF1 been showed as a prominent oncodriver via MYO1B splice switching [41], progresses cancer. This gives a lead that hnRNPA1 down regulation is not enough to sensitize glioma apoptosis. We have demonstrated here that the expression of splice factor family protein hnRNPA1 and SF2/ASF1 are under the regulatory effect of the same enzyme USP5. So instead of directly targeting hnRNPA1, it's modulator USP5 was hypothesized as the candidate target.
It was noted that rise in SF2/ASF1 expression with the decrease in hnRNPA1 in USP5 depleted cells, therefore we propose that co-knockdown of SF2/ASF1 along with USP5 should able to enhances apoptosis to a high rate, through apoptotic pathways.
Additionally, T98G and U87 TMZ (Temozolomide resistant cells) along with U87MG cells were taken for our study having endogenous expression of MGMT is poor prognosis and Temozolomide resistance marker in gliomas [25]. Knocking down of USP5 along with SF2/ASF1 in these glioma cells sensitized cells to apoptosis through Caspase 9, 3 cleavage dependent pathway. Knock down of USP5 & SF2/ASF1 in combination also decreased the NAD and NADP concentration in A172 & T98G glioma cells. The nicotinamide adenine dinucleotide (NAD+)/reduced NAD+ (NADH) and NADP+/reduced NADP+ (NADPH) redox couples are important for maintaining cellular redox homeostasis maintain numerous biological events, including cellular metabolism. Deficiency or imbalance of these two redox couples has been associated with many pathological disorders and cancer [42], were showed downregulated.
In conclusion, hnRNPA1 and SF2/ASF1 splice factor proteins were found to be highly expressed in different cancers in a number of studies. Targeting heterogeneous populations of cancer cells with a varying proportion of hnRNPA1 or SF2/ASF1 expression patterns promotes cancer relapse cases. Therefore, to overcome cancer recurrence, an approach to target USP5 along with USP8 showed to be optimum from our experiments, though in vivo experiments in mouse models can potentially validate this hypothesis, with valuable therapeutic insights to get cure from cancer.
4. Material & methods
4.1. Cell culture material
U87, U373, T98G, A172 & LN229 cells (Generous support from Prof. Subrata Sinha, N.B.R.C, Manesar, Gurgaon (India)) and U87 TMZ cells (Resistant cell developed against Temozolomide till 150 μM) were propagated in Dulbecco's modified Eagle's medium (DMEM) (PAN Biotech) composition: 10% fetal bovine serum (PAN Biotech), 100 units/ml penicillin-streptomycin (PAN Biotech). Cells were grown under 37 °C and humidified chamber under 5% CO2.
4.2. Antibodies
Western blotting of all essential proteins was probed using primary monoclonal antibodies anti-hnRNPA1, anti SF2/ASF1, anti-USP5 (Santa Cruz biotech. Inc.), anti-Mgmt (Cell signalling Technology), anti-TRAF6 (Biospes), anti-Phospho Tyrosin (Millipore) and anti-α-Tubulin (Abcam) followed with HRP conjugated mouse or Rabbit secondary antibodies (GeNei company).
4.3. si RNA transfection
U87 cells were transfected next day after plating, with specific USP5 (10 nM), USP8 (10 nM), siRNA and scrambled siRNA (Santa Cruz Biotechnology, Inc.) as per protocol. In another experiment T98G cells were transfected next day after plating with two different specific siRNAs (Eurogentec) against hnRNPA1 (10 nM). Protein lysates were prepared after 72 h of transfection in cell lysis buffer (Cell Signaling Technology) containing protease inhibitor (Abcam) and phosphatase inhibitor (Santa Cruz Biotechnology, Inc.). In all experiment siRNA mediated knock down efficiency was achieved ≥50% to 60% using Interferrin transfection reagent from Polyplus Company. All experiment were performed at least three times, representative pictures were reported here.
4.4. Immunoblotting & immunoprecipitation
Protein lysate were prepared in cell lysis buffer and proteins were quantitated using Bradford reagent (Biorad) as per manufactures protocol. After quantification protein was run on the SDS PAGE and transfer into the PVDF membrane (MDI) overnight at 15 V in transfer buffer. Next day membrane was blocked with 5% milk for 4hrs and processed with specific primary and secondary antibodies followed develop the membrane using ECL (Biorad) (chemiluminescent reagent).
To study the protein-protein interaction and ubiquitination of hnRNPA1 we treated U87 and U373 cells with different PI3 kinase inhibitors (LY294002 & Wortmannin). In one experiment we treated U87 cells with wortmannin (10 μM) as compared with control. In another experiment we treated U87 and U373 cells with two different doses of Wortmannin (5 μM & 10 μM) as compared with control. In last experiment we treated U87 cells with LY294002 (PI3 kinase inhibitor), Bortezomib (Proteosome inhibitor) and combination of both LY and BR. Protein was isolated after 24 h of drug treatment followed with protein quantification with the kit (BioRad). Quantitated protein lysates were taken and add triton buffer for the volume makeup of each tube, add 1 μg of specific antibody which we want to pull down or immunoprecipitate. Incubate overnight at 4 °C. Next day add protein A/G agarose beads (Santa cruz biotechnology, INC.) in each tube and incubate at 4 °C for 4 h. Washing was done with triton buffer (Lysis buffer) 5 times at 4000 rpm for 5 min. Add 1X protein loading dye and boil for 10 min. Load the samples in SDS- PAGE and processed same as western blotting to check ubiquitination and protein interaction.
4.5. IHC (immunohistochemistry)
Immunohistochemical staining was performed. We got the paraffin embedded sections from the Rajiv Gandhi Cancer Institute and Research Center (RGCIRC), New Delhi, Delhi. Additionally, all samples were human ethic approved for use based on informed consent from both RGCIRC & from our institution. Protocol is as follows: 6-m paraffin-embedded sections were routinely deparaffinized with xylene, rehydrated through a graded alcohol series, and incubated in 0.5% (v/v) hydrogen peroxide to block endogenous peroxidase activity followed 3 times washing with 1X PBS. After this, sections were incubated with 5% (v/v) Bovine Serum Albumin (BSA) for 45 min to block nonspecific staining. Sections were incubated with a primary antibody (USP5 & hnRNPA1, 1:50) (Santa Cruz Biotechnology) at 4 °C in a humidified chamber overnight, followed by 4 times washing with 1X PBS. After that sections were incubated with HRP-conjugated Mouse secondary antibody (1:100) (GeNei) for 1 h at 4 °C. Washing was done 4 times with 1X PBS. The DAB method was used to detect USP5 & hnRNPA1 protein expression. The absence/presence of the USP5 & hnRNPA1 in the tissues was confirmed by Phase contrast microscopy. In addition, any nuclear staining was confirmed with the DAPI staining under flouroscence microscope (24).
4.6. Semiquantitative PCR (polymerase chain reaction)
U87 cells were grown and after attaining 50–60% of confluency, transfected with specific USP5 (10 nM), USP8 (10 nM), siRNA and scrambled siRNA (Santa Cruz Biotechnology, Inc.) as per protocol. After 72 h of transfection total mRNA was extracted using TRIsoln (GeNei). 1 μg of total RNA was reverse transcribed using RT PCR kit (BIO RAD) following the manufacturer's protocol. Gene was amplified using specific forward and reverse primer (Table S1) with the help of Taq DNA Polymerase in thermocycler (BIO RAD). PCR cycling conditions were as follows: Initial denaturation 95 °C for 2 min. 35 cycles of Denaturation 95 °C for 30 s, different Annealing temp for different genes for 30 s, and Extension 72 °C for 45 s followed with final extension of 72 °C for 10 min and final hold the PCR at 4 °C.
4.7. Cell viability & telomerase assay in glioma cells
U87 cells was transfected after 17 h of freshly plated cells with specific siRNA's (CNT (10 nM), Customized si RNA's against USP5 (10 nM), USP8 (10 nM) and in combination of USP5&USP8 siRNA. After 48 h of si RNA transfection, Bortezomib (Proteosome inhibitor) was added in different concentration (0, 1 & 2 nM), after 24 h of incubation in CO2 incubator at 37 °C, cell viability was assessed using MTT colorimetric assay. Based on the ability of viable cells to reduce yellow MTT (3-(4-5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide tetrazole) (Amresco) to purple formazan product, absorbance measures at 570 nm. Experiment was performed in triplicate.
In another experiment U87 cells was transfected after 17 h of freshly plated cells with specific siRNA's (CNT (10 nM), USP5 (10 nM), USP8 (10 nM) & hnRNPA1 (10 nM). After 72 h of transfection cell lysate were prepared. The quantitation of telomerase was performed as per protocol mentioned in Telomerase estimation kit (MYBioSource.com).
4.8. Caspase (8, 9, & 3) colorimetric assay
U87 cells was transfected after 17 h of freshly plated cells with specific siRNA's (CNT (10 nM), USP5 (10 nM), USP8 (10 nM)). In another experiment U87, T98G, & U87 TMZ cells were transfected with specific siRNA (CNT (10 nM), USP5 (10 nM), SF2/ASF1 (10 nM), USP5+SF2/ASF1 (10 nM)). In one more experiment T98G cells were transfected with specific siRNA (CNT (10 nM), USP5 (10 nM), SF2/ASF1 (10 nM), USP5+SF2/ASF1 (10 nM)) with treatment of Botezomib (Proteosome inhibitor) (1 nM). After 72 h of transfection protein lysate were prepared in cell lysis buffer without SDS. Proteins were quantitated with the help of Bradford reagent (BIO RAD) as per manufactures protocol. 50 μg of protein was taken for calculating the Caspase 8, Caspase 9, and Caspase 3 cleavage using caspase colorimetric kit (Biovision) as per the manufacture's protocol. Absorbance was taken at 400 or 405 nm.
4.9. Flow cytometric analysis of apoptosis by FITC-Annexin V/PI
FITC-labelled Annexin V was used as a probe to detect exposed translocated membrane phospholipid on the early onset of apoptosis for flow cytometric analysis. Propidium iodide (PI) is used in conjunction with Annexin V staining to differentiate the probable apoptotic stages of the U87 cells. U87 cells were transfected with specific siRNA (CNT (10 nM), USP5 (10 nM), USP8 (10 nM), USP5+USP8 (10 nM)) for 72 h. Cells were harvested, washed twice with cold PBS and then re-suspended in 100 μL of binding buffer. Then 100 μL of the solution were transferred to a culture tube, followed by the addition of 5 μL of FITC-conjugated Annexin V and 5 μL of Propidium iodide PI (Abgenex Pvt. Ltd). The cells were gently vortexes and then incubated for 20 min at room temperature, in the dark. Immediately, 400 μL of binding buffer was added to each tube and the samples were analysed by flow cytometry FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA) to examine the early and late stages of apoptosis. Each sample was analysed using Cell Quest Pro software (BD Biosciences, San Jose, CA, USA) within 1 h.
4.10. NAD/NADH & NADP/NADPH colorimetric assay
T98G & A172 cells were transfected after 17 h of freshly plated cells with specific siRNA's (CNT (10 nM), USP5 (10 nM), SF2/ASF1 (10 nM), USP5+SF2/ASF1 (10 nM)). Cells were also treated or not treated with bortezomib 1 nM (Proteosome inhibitor). After 72 h of transfection protein lysate were prepared in cell lysis buffer without SDS. Proteins were quantified with the help of Bradford reagent (BIO RAD) as per manufactures protocol. 50 μg of protein was taken for calculating the NAD/NADH (Cayman) and NADP/NADPH (Biovision) concentration present in glioma cells as per the manufacture's protocol. Absorbance was taken at 450 nm.
4.11. Cycloheximide and MG132 treatment: westorn blotting
Glioma cell line T98G cells were plated day before, next day after 17 h cells were transfected with scrambled siRNA (10 nM) and USP5 si RNA (10 nM), 72hrs after transfection lysates were made after Cycloheximide (100 μg/ml) treatment at different time points 0hr, 2hrs, 4 h, 6 h. Protein lysates were made in a cell lysis buffer (cell signaling technology) containing protease inhibitor and phosphatase inhibitor.
Similarly in another experiment T98G glioma cell line, 72hrs after transfection of Scramble si RNA and USP5 si RNA, MG132 (Proteosome inhibitor) 10 μM concentration was added for 6 h in all transfected siRNA cells, following with Cycloheximide (100 μg/ml) treatment at different time point 0hrs, 2hrs, 4hrs, 6hrs. Protein lysates were made in a cell lysis buffer (cell signaling technology) containing protease inhibitor and phosphatase inhibitor. Westorn blotting was performed for analyzing USP5, hnRNPA1, SF2/ASF1, TRAF6 protein expression using mouse monoclonal antibodies as per protocol (Supplementary figures).
4.12. Statistical analysis
Numerical values obtained from individual experiments were expressed here, as mean ± standard error of the mean, student T-test, unpaired and p value < 0.05.
CRediT authorship contribution statement
Vidhi Vashistha: Writing - original draft, performed the experiments and drafted the manuscript. Sachin Bhardwaj: Writing - original draft, performed the experiments and drafted the manuscript. Birendra K. Yadav: Formal analysis, Tumor tissue slide section related experiment and analysis were performed. Ajay K. Yadav: Writing - original draft, Conceived & designed all experiments and wrote the manuscript.
Declaration of competing interest
None of the authors has no conflict of interest.
Acknowledgements
Thanks to Funding agencies Science Engineering Research Board, Govt India (Grant number: EMR/2015/001927)., UGC-SAP, New Delhi (INDIA), ICMR Senior Research Fellowship (V.V). DST-Purse funding facility from University of Delhi (INDIA). In house funding from Dr. B. R Ambedkar Centre for Biomedical Research, University Of Delhi.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2020.100846.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Buckley S.M., Aranda-Orgilles B., Strikoudis A., Apostolou E., Loizou E., Moran-Crusio K., Farnsworth C.L., Koller A.A., Dasgupta R., Silva J.C., Stadtfeld M., Hochedlinger K., Chen E.I., Aifantis I. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11(6):783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin W.L., Mao X.Y., Qiu G.Z. Targeting deubiquitinating enzymes in glioblastoma multiforme: expectations and challenges. Med. Res. Rev. 2016 doi: 10.1002/med.21421. [DOI] [PubMed] [Google Scholar]
- 3.Clague M.J., Barsukov I., Coulson J.M., Liu H., Rigden D.J., Urbe S. Deubiquitylases from genes to organism. Physiol. Rev. 2013;93(3):1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 4.Komander D., Clague M., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 5.Oh Young Mi. USP8 modulates ubiquitination of LRIG1 for Met degradation. Sci. Rep. 2014;4:4980. doi: 10.1038/srep04980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeong M. USP8 suppresses death receptor-mediated apoptosis by enhancing FLIP L stability. Oncogene. 2017;36(4):458–470. doi: 10.1038/onc.2016.215. [DOI] [PubMed] [Google Scholar]
- 7.Byun S., Lee S.Y., Lee J., Jeong C.H., Farrand L., Lim S., Reddy K., Kim J.Y., Lee M.H., Lee H.J., Bode A.M., Won Lee K., Dong Z. USP8 is a novel target for overcoming gefitinib resistance in lung cancer. Clin. Canc. Res. 2013;19(14):3894–3904. doi: 10.1158/1078-0432.CCR-12-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson Keith D. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34(44):14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 9.Hadari T. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem. 1992;267(2):719–727. [PubMed] [Google Scholar]
- 10.Dayal S., Sparks A., Jacob J., Allende-Vega N., Lane D.P., Saville M.K. Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 2009;284(8):5030–5041. doi: 10.1074/jbc.M805871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y., Wang W.M., Zou L.Y., Li L., Feng L., Pan M.Z., Lv M.Y., Cao Y., Wang H., Kung H.F., Pang J.X., Fu W.M., Zhang J.F. Ubiquitin specific peptidase 5 mediates Histidine-rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14-P53 signaling. Proteomics. 2017;17(12) doi: 10.1002/pmic.201600350. doi: 10. [DOI] [PubMed] [Google Scholar]
- 12.AYu Amerik, Swaminathan S., Krantz B.A., Wilkinson K.D., Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16(16):4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X., Qi W., Pan H., Yang F., Deng J. Overexpression of USP5 contributes to tumorigenesis in non-small cell lung cancer via the stabilization of β-catenin protein. Am. J. Canc. Res. 2018;8(11):2284–2295. [PMC free article] [PubMed] [Google Scholar]
- 14.Kaistha B.P., Krattenmacher A., Fredebohm J., Schmidt H., Behrens D., Widder M., Hackert T., Strobel O., Hoheisel J.D., Gress T.M., Buchholz M. The deubiquitinating enzyme USP5 promotes pancreatic cancer via modulating cell cycle regulators. Oncotarget. 2017;8(39):66215–66225. doi: 10.18632/oncotarget.19882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Juan J., Zhang Z., Du Y., Xu Y., Tong J., Cao B., Moran M.F., Zeng Y., Mao X. Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis. 2017;8(9) doi: 10.1038/cddis.2017.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izaguirre D.I., Zhu W., Hai T., Cheung H.C., Krahe R., Cote G.J. PTBP1-dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol. Carcinog. 2012;1(11):895–906. doi: 10.1002/mc.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Zheng-Jun. Overexpression of HnRNP A1 promotes tumor invasion through regulating CD44v6 and indicates poor prognosis for hepatocellular carcinoma. Int. J. Canc. 2013;132(5):1080–1089. doi: 10.1002/ijc.27742. [DOI] [PubMed] [Google Scholar]
- 18.Ushigome Mitsunori. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int. J. Oncol. 2005;26(3):635–640. [PubMed] [Google Scholar]
- 19.Del Gatto-Konczak Fabienne. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell Biol. 1999;19(1):251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jean-Philippe, Jacques, Paz Sean, Caputi Massimo. hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci. 2013;14(9):18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oltean S., Bates D. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 22.Fang J., Bolanos L.C., Choi K., Liu X., Christie S., Akunuru S., Kumar R., Wang D., Chen X., Greis K.D., Stoilov P., Filippi M.D., Maciejewski J.P., Garcia-Manero G., Weirauch M.T., Salomonis N., Geiger H., Zheng Y., Starczynowski D.T. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 2017 Feb;18(2):236–245. doi: 10.1038/ni.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy Rajat. Emerging roles of hnRNPA1 in modulating malignant transformation. Wiley Interdiscipl. Rev.: RNA. 2017;8(6):e1431. doi: 10.1002/wrna.1431. [DOI] [PubMed] [Google Scholar]
- 24.Sun J., Hu Q., Peng H., Peng C., Zhou L., Lu J., Huang C. The ubiquitin-specific protease USP8 deubiquitinates and stabilizes Cx43. J. Biol. Chem. 2018;293:8275–8284. doi: 10.1074/jbc.RA117.001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansouri Alireza, Hachem Laureen D., Mansouri Sheila, Nassiri Farshad, Laperriere Normand J., Xia Daniel, Neal I Lindeman, Wen Patrick Y., Chakravarti Arnab, Mehta Minesh P., Hegi Monika E., Stupp Roger, Aldape Kenneth D., Zadeh Gelareh. MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019;21(2):167–178. doi: 10.1093/neuonc/noy132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonomi S., di Matteo A., Buratti E., Cabianca D.S., Baralle F.E., Ghigna C., Biamonti G. HnRNP A1 controls a splicing regulatory circuit promoting mesenchymal-to-epithelial transition. Nucleic Acids Res. 2013 Oct;41(18):8665–8679. doi: 10.1093/nar/gkt579. Epub 2013 Jul 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalhoub Nader, Baker Suzanne J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin Jheralyn. Phosphomimetic substitution of heterogeneous nuclear ribonucleoprotein A1 at serine 199 abolishes AKT-dependent internal ribosome entry site-transacting factor (ITAF) function via effects on strand annealing and results in mammalian target of rapamycin complex 1 (mTORC1) inhibitor sensitivity. J. Biol. Chem. 2011;286(18):16402–16413. doi: 10.1074/jbc.M110.205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zerbe Laura K. vol. 41. 2004. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: implications for pre-mRNA processing; pp. 187–196. (Molecular Carcinogenesis: Published in Cooperation with the University of Texas MD Anderson Cancer Center). 4. [DOI] [PubMed] [Google Scholar]
- 30.Sharif T., Ahn D.G., Liu R.Z., Pringle E., Martell E., Dai C., Nunokawa A., Kwak M., Clements D., Murphy J.P., Dean C., Marcato P., McCormick C., Godbout R., Gujar S.A., Lee P.W. The NAD (+) salvage pathway modulates cancer cell viability via p73. Cell Death Differ. 2016 Apr;23(4):669–680. doi: 10.1038/cdd.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy K., Wu Y., Meitzler J.L., Juhasz A., Liu H., Jiang G., Lu J., Antony S., Doroshow J.H. NADPH oxidases and cancer. Clin. Sci. (Lond.) 2015 Jun;128(12):863–875. doi: 10.1042/CS20140542. [DOI] [PubMed] [Google Scholar]
- 32.Fang J., Bolanos L.C., Choi K., Liu X., Christie S., Akunuru S., Kumar R., Wang D., Chen X., Greis K.D., Stoilov P., Filippi M.D., Maciejewski J.P., Garcia-Manero G., Weirauch M.T., Salomonis N., Geiger H., Zheng Y., Starczynowski D.T. Ubiquitination of hnRNPA1 by TRAF6 links chronic innate immune signaling with myelodysplasia. Nat. Immunol. 2017 Feb;18(2):236–245. doi: 10.1038/ni.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyd Florian, Lynch Kristen W. Degrade, move, regroup: signaling control of splicing proteins. Trends Biochem. Sci. 2011;36(8):397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbanski L.M., Leclair N., Anczuków O. vol. 9. Wiley Interdiscip Rev RNA; 2018. p. e1476. (Alternative-splicing Defects in Cancer: Splicing Regulators and Their Downstream Targets, Guiding the Way to Novel Cancer Therapeutics). 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David Charles J., Chen Mo, Assanah Marcela, Canoll Peter, James L., Manley HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010 Jan 21;463(7279):364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D., Hong A., Park H.I., Shin W.H., Yoo L., Jeon S.J., Chung K.C. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J. Cell. Physiol. 2017 Dec;232(12):3664–3676. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 37.Namgail T., Kumar D., Vashistha V., Aquib A., Yadav A. 2020. HnRNPA1 and Its Effect In The Expression of ABCC (ABCC4 and ABCC6) Transporter In Glioma Cell Lines. Preprints. 2020070193. [DOI] [Google Scholar]
- 38.Aquib A., Yadav A.K. 2020. CD44 Alternative Splice Variants v6 and v10. Preprints. 2020060255. [DOI] [Google Scholar]
- 39.LaBranche H., Dupuis S., Ben-David Y., Bani M.R., Wellinger R.J., Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 1998;19(2):199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 40.Luan Wenkang, Wang Yingyi, Chen Xincheng, Shi Yan, Wang Jiajia, Zhang Junxia, Qian Jin, Li Ri, Tao Tao, Wei Wenjin, Qi Hu, Liu Ning, You Yongping. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Xuexia. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J. Clin. Invest. 2019;129(2):676–693. doi: 10.1172/JCI120279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciccarese Francesco, Ciminale Vincenzo. Escaping death: mitochondrial redox homeostasis in cancer cells. Front. Oncol. 2017;7 –:117. doi: 10.3389/fonc.2017.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.