Abstract
Paired amphipathic helix protein (SIN3B) is a transcription corepressor for many genes. Here we show a different regulation mechanism of integrin αV gene expression by SIN3B in human hepatocellular carcinoma (HCC). We first observed a close relationship between Integrin αV and SIN3B expressions in HCC patients and tumor cell lines with different metastatic potentials. Overexpression of SIN3B significantly accelerated the cell migration rate of SMMC-7721, but failed when integrin αV expression was silenced. Interestingly, SIN3B stimulated integrin αV subunit promoter activity only in the presence of sulfatide. Importantly, SIN3B was identified in the complex with sulfatide by mass spectrometry. Fat blot assay indicated that SIN3B specifically interacted with sulfatide. Molecular modeling suggested that sulfatide induced the conformational change of SIN3B from compacted α-helices to a relaxed β-sheet in PAH2 domain. The data of immunoprecipitation and ChIP assay indicated that altered SIN3B lost the binding affinity with MAD1 and HDAC2, which reduced the recruitment of HDAC2 on integrin αV gene promoter and prevented the deacetylation of the histone 3. In conclusion, this study demonstrated that SIN3B promoted the transcriptional activation of the integrin αV subunit gene promoter by reducing interaction with HDAC2.
Keywords: paired amphipathic helix protein, ITGAV, sulfatide, HCC, histone deacetylation
Introduction
Paired amphipathic helix protein SIN3B is generally considered as an inhibitory transcriptional factor, which lacks any known motif binding to DNA or intrinsic enzymatic activity, but forms a multiple complex through diverse protein–protein interaction (Grzenda et al., 2009). The protein structure of SIN3 is highly conserved and its basic structure is a plurality of continuous paired amphoteric helix domains. There are three PAH domains (paired amphipathic α-helix) in mammalian SIN3B, one histone deacetylase interact domain (HID) and a highly conserved C-terminal tail (HCR) (Grzenda, et al., 2009). SIN3B forms a SIN3/HDAC polyprotein corepressor complex with histone deacetylase (HDACs) through its HID domain (Laherty et al., 1997; David et al., 2008; Weng et al., 2014). While SIN3B can bind to the N-terminal SID (SIN3 interact domain) of Max dimerization protein (MXD) by the PAH2 domain (Spronk et al., 2000). SIN3B is often considered as a tumor suppressor since it recruits MXD which interacts with MYC associated factor X (Max), and competitively bind to E-Box (CANNTG) of the target gene promoter with c-MYC (Laherty et al., 1997; Hurlin and Huang, 2006). SIN3B interaction with c-MYC results in deacetylation and protein degradation of c-MYC (Garcia-Sanz et al., 2014).
SIN3B was found to be promoted by oncogene Ras in mouse embryonic fibroblasts and recruited to the promoters of E2F genes, which leads to the heterochromatinization and gene silencing (Grandinetti et al., 2009). In pancreatic ductal adenocarcinoma cells (PDAC), SIN3B is also associated with the cellular senescence caused by the activated KRAS oncogene, but targeted SIN3B inactivation can delay the PDAC progression, and deletion of SIN3B reduced the KRAS-induced IL-1α expression, which produced protective inflammatory responses (Rielland et al., 2014).
In prostate adenocarcinoma, SIN3B protects against tumor local invasive, and inactivation of SIN3B or PTEN leads to acceleration of tumor progression (Bainor et al., 2017). Chromosone-associated SIN3B was thus proved important in PTEN-induced cell senescence and thereto prevented progression to invasive prostate adenocarcinoma. In human hepatocarcinoma cells (HCC), SIN3B was upregulated in the hexamethylene bisacetamide (HMBA)-induced differentiation (Tang et al., 2007). But the expression levels of MAD1, the member of MXD family, reduced in HCC tissues (Nam et al., 2008). However, the regulation of transcription by SIN3B may depend on the molecular and cellular conditions (Bansal et al., 2016). The roles of SIN3B in metastasis especially of HCC have not been well defined.
Integrins, the important adhesion molecules on the cell membrane, can induce cell migration and tumor metastasis. The integrin αV subunit is reported to be involved in the process of vascular endothelial cell migration and the tumor angiogenesis (Varner et al., 1995; Kim et al., 2000). The integrin αV complexed with β3 subunit and forms subtype αVβ3, which is associated with tumor stemness and anchorage-independent progression (Desgrosellier et al., 2009).
Cerebroside sulfotransferase (CST) is the rate-limiting enzyme for the synthesis of sulfatide in humans. CST recognizes galactocerebroside (GalCer) and transfers the sulfate groups from 3′-phospho-adenosine-5′-phosphosulfate to the 3′-hydroxyl group of the galactose of GalCer to form sulfatide (Handa et al., 1974; Fukuda et al., 2001). Sulfatide is highly expressed in the kidney, brain, myelin and other nerve tissues (Trick et al., 1999; Frank, 2000). We previously observed that sulfatide also highly expressed in hepatocellular carcinoma and promoted tumor metastasis through integrin αVβ3 by increasing the expression of integrin αV subunit (Wu et al., 2004, 2013). However, the details of regulation mechanism are still unknown. In this study, we identified SIN3B as a gene activator to enhance integrin αV gene transcription through reducing interaction with HDAC2 and MAD1.
Results
SIN3B is overexpressed in HCC
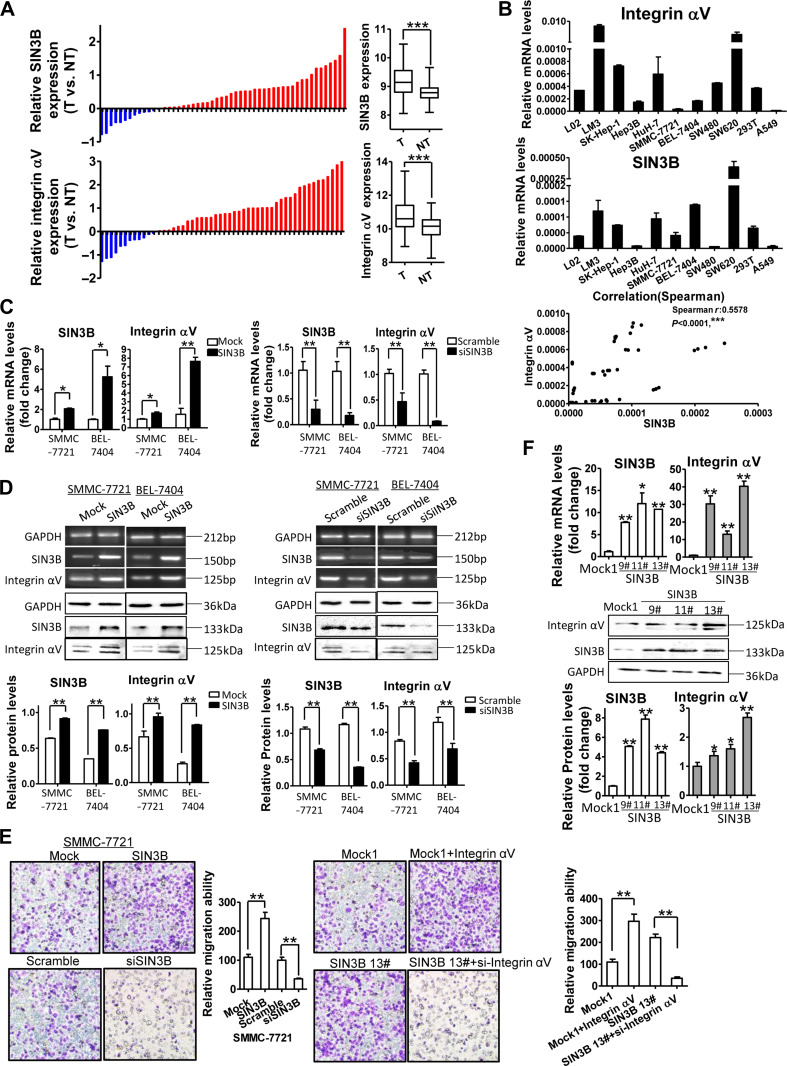
To investigate the role of SIN3B in the development or progression of HCC, we analyzed the expression of SIN3B in 331 cases of HCC from the TCGA database (IlluminaHiSeq). Among these cases, SIN3B expression was analyzed in both HCC and adjacent non-tumor tissues of 52 patients and observed that the average expression level of SIN3B in HCC tissues was significantly higher than that in adjacent non-tumor tissues. High expression of SIN3B was found in hepatocellular carcinoma tissues in 39 cases and 13 cases of adjacent tissues (Figure 1A, upper). Concomitantly, the expression of integrin αV subunit gene ITGAV was highly expressed in 40 cases of hepatocellular carcinoma tissues and 12 cases of adjacent non-tumor tissues. The average expression level of integrin αV in HCC tissues was significantly higher than that in adjacent non-tumor tissues (Figure 1A, low). The integrin αV expression was closely associated with patient’s overall survival rate in 331 cases with HCC.
Figure 1.
SIN3B promotes integrin αV expression in HCC. (A) SIN3B and integrin αV mRNA expressions were analyzed between HCC tissues (T) and adjacent non-tumor liver tissues (TN) in 52 patients with HCC from TCGA database. The levels of SIN3B and integrin αV mRNA were summarized on the right. (B) Integrin αV & SIN3B mRNAs were determined by qRT-PCR in 11 cell lines, and the Spearman correlation between integrin αV and SIN3B expressions was analyzed (bottom). (C) Integrin αV and SIN3B expressions in HCC cells with SIN3B overexpression or knockdown were determined by qRT-PCR. (D) Integrin αV and SIN3B expressions in SIN3B overexpression or knockdown HCC cells were determined by RT-PCR and western blot. Data of western blot were quantitatively analyzed (low panel). (E) Representative micrographs of transmembrane migration of HCC with SIN3B and integrin αV overexpression or knockdown and quantitative analysis (left). SIN3B stable transfectants (Mock clone 1 & SIN3B clone #13) were assayed for transmembrane migration after integrin αV overexpression or knockdown (right). Original magnification, 20×. (F) SIN3B and integrin αV expressions in SIN3B stable transfectants (#9, #11, #13 clones) were detected by qPCR (upper) and western blot analysis (low). Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Further analysis indicated that overexpression of both integrin αV and SIN3B was noted in 33 cases of HCC tissues and 6 cases of adjacent tissues. Overexpression of integrin αV and low expression of SIN3B were noted in 7 cases of HCC tissues, while overexpression of SIN3B and low expression of integrin αV were noted in 6 cases. Chi-square test results suggested a correlation between SIN3B and integrin αV subunit expressions (P < 0.05).
To further confirm the correlation between integrin αV and SIN3B expressions, we investigated the expression of integrin αV and SIN3B in 11 various cell lines (Figure 1B). The results showed that the expression levels of integrin αV subunit and SIN3B were significantly higher in hepatocellular carcinoma cells (LM3, SK-Hep-1, and HuH-7) than in non-tumor hepatocyte LO2 cells. Moreover, integrin αV and SIN3B expressions in high metastatic potential SW620 cells were significantly higher than those in low metastatic potential SW480 cells. The Spearman rank correlation analysis also indicated a significant correlation between integrin αV and SIN3B levels (R = 0.5578, P < 0.0001) (Figure 1B, bottom).
SIN3B promotes integrin αV gene transcription
In order to further investigate the role of SIN3B, a protein generally considered synergistic repressor factor, in the correlation with integrin αV expression in hepatocellular carcinoma cells, we transfected SIN3B cDNA into SMMC-7721 and BEL-7404 HCC cells, and interestingly noted that SIN3B overexpression robustly promoted the expression of integrin αV gene and protein (Figure 1C and D; Supplementary Figure S1A). While, silence of SIN3B by RNAi resulted in a decreased expression level of integrin αV subunit (Figure 1C and D). Further, the migration rate of SMMC-7721 cells was significantly increased after overexpression of SIN3B, but significantly decreased by the SIN3B knockdown (Figure 1E, left). However, knockdown of integrin αV subunit in the stably SIN3B-transfected cells (Figure 1F) completely abolished the migration promotion by SIN3B (Figure 1E, right).
SIN3B promotion of integrin αV subunit requires sulfatide
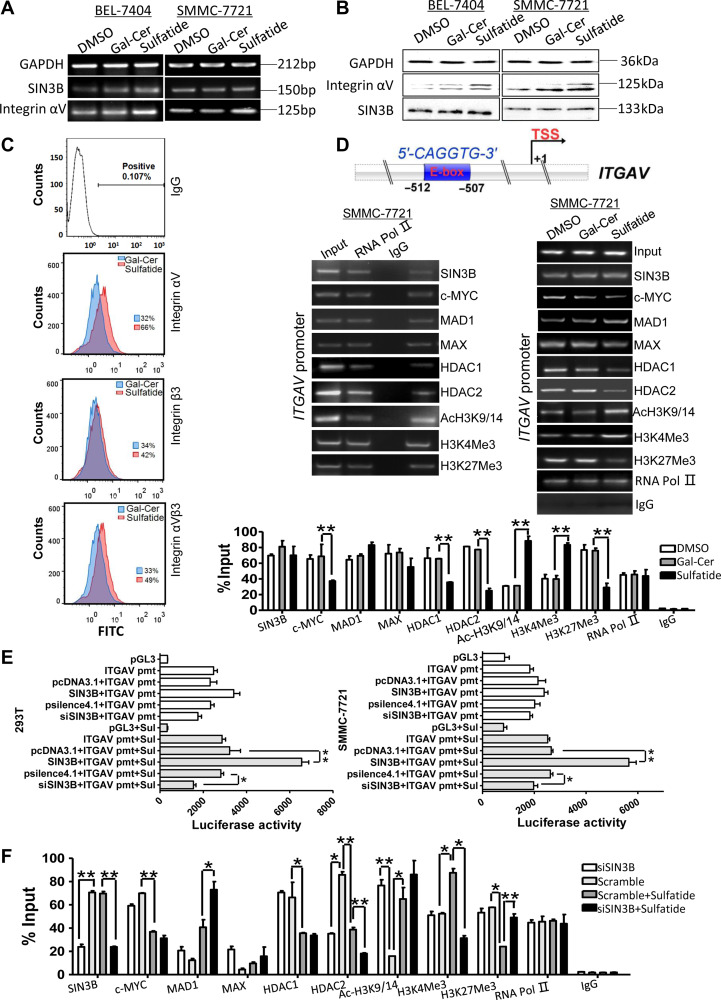
After sulfatide treatment, the expression of integrin αV was significantly increased in SMMC-7721 and BEL-7404 cells (Figure 2A and B; Supplementary Figure S1B and C) and the surface integrin αV and αVβ3 significantly increased as indicated in flow cytometry analysis data (Figure 2C). However, the expression level of SIN3B was not changed (Figure 2A and B).
Figure 2.
SIN3B promotes the transcriptional activity of ITGAV promoter in the presence of sulfatide. (A) Integrin αV and SIN3B mRNAs analysis by RT-PCR in BEL-7404 and SMMC-7721 cells treated with sulfatide 24 h. (B) Integrin αV and SIN3B proteins determination by western blot in both BEL-7404 and SMMC-7721 cells treated with sulfatide 48 h. (C) Flow cytometry analysis of cell surface integrin expression after sulfatide treatment. Normal mouse IgG antibody was as negative control. (D) Schematic diagram of E-box element in ITGAV promoter. ChIP analysis was performed with indicated antibodies. RNA Pol II as the positive control, and normal rabbit IgG as negative control. The promoter fragment of ITGAV subunit gene was detected by PCR (middle) and qPCR (bottom). (E) The activity of ITGAV gene promoter was determined using a luciferase assay system in HEK-293T cells and SMMC-7721 cells. pmt, promoter; Sul, sulfatide. (F) ChIP analysis of ITGAV gene promoter was performed in SIN3B knockdown cells, in the presence or absence of sulfatide, with indicated antibodies. The ITGAV promoter fragment was detected by qPCR. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01.
By analyzing the sequence of the integrin αV promoter, we found that there was an E-box sequence at the position between −512 and −507 upstream (Figure 2D) of the transcription initiation site. By ChIP experiments, it was found that SIN3B, MAD1, MAX, HDAC1 and HDAC2 were recruited to the integrin αV promoter (Figure 2D, left). However, sulfatide treatment significantly reduced the recruitment of HDAC1 and HDAC2, but not SIN3B. H3K4Me3 and acH3K9/14 occupancy on integrin αV gene promoter was significantly increased, while H3K27Me3 significantly reduced after sulfatide treatment (Figure 2D, right).
Unexpectedly, in both HEK-293T and SMMC-7721 cells, the pGL3 basic-ITGAV promoter reporter did not respond to the co-transfection with SIN3B expression. However, after both of these cells were treated, respectively, with exogenous sulfatide, SIN3B co-transfection robustly enhanced the transcriptional activity of the integrin αV promoter (Figure 2E), which was also significantly decreased by siSIN3B. HDAC2 occupancy on integrin αV promoter, interestingly, decreased significantly after SIN3B knockdown (Figure 2F) as well as sulfatide treatment, although occupancy of HDAC1, H3K4me3, and H3K27me3 on integrin αV promoter remained similar in SIN3B knockdown cells as scramble control. After sulfatide treatment, the occupancy of HDAC1, HDAC2, H3K27me3 and c-MYC on the promoter was significantly reduced. MAX and MAD1 occupancy was enhanced in SIN3B knockdown cells, especially in the presence of sulfatide.
SIN3B exists in sulfatide-protein complex
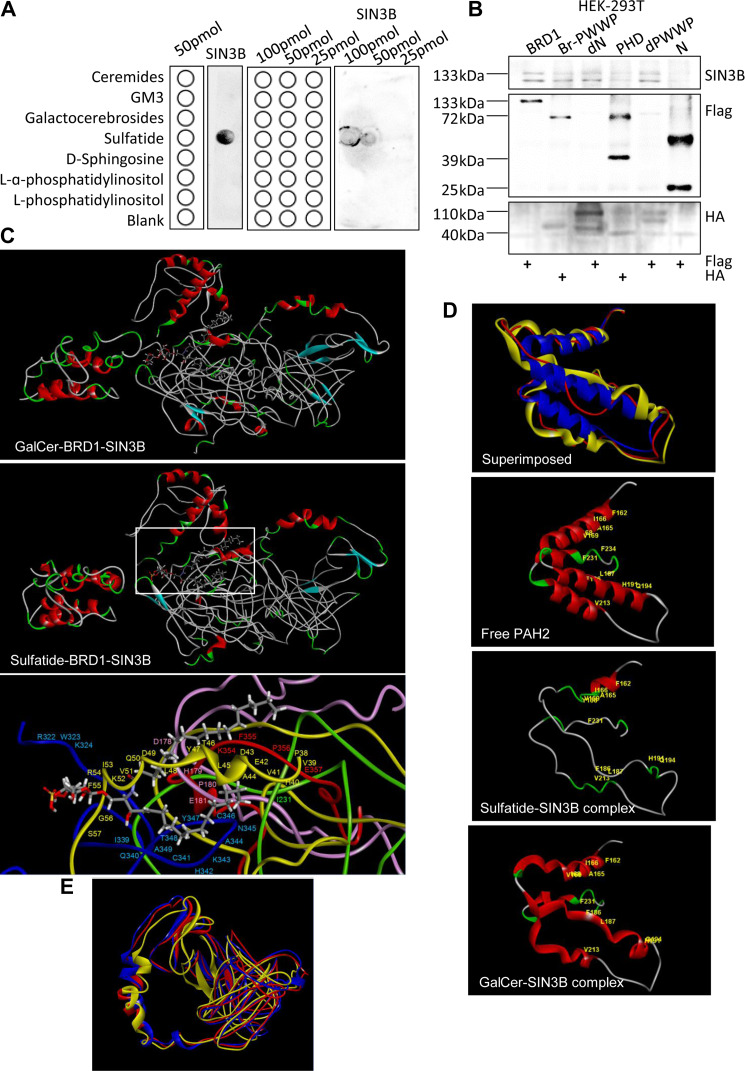
To understand how sulfatide affected SIN3B regulation, we screened possible binding proteins by coating sulfatide on magnetic beads and incubating with SMMC-7721 hepatoma cell lysates. After elution and separation by SDS-PAGE, a band near 130 kDa was noted in comparison with control group. After analysis by mass spectrometry, SIN3B was identified among the proteins (Cai et al., 2018). Further through fat blot assay using the PVDF membrane immobilized with ceramide, ganglioside GM3, galactocerebrosides, sulfatide, D-sphingosine, L-α-phosphatidylinositol and L-phosphatidylinositol, SIN3B was confirmed to bind sulfatide specifically (Figure 3A). The immunoprecipitation experiments of SIN3B and its mutant constructs showed that SIN3B was also associated with the bromo domain of BRD1 (Figure 3B).
Figure 3.
SIN3B interaction with sulfatide. (A) Fat blot analysis of immobilized ceremides, GM3, galactocerebrosides, sulfatide, D-sphingosine, L-α-phosphatidylinositol, and L-phosphatidylinositol interaction with SIN3B. The lipid concentrations were 100, 50, and 25 pmol. The schematics of lipid strip templates were shown on the left and the fat blot spotting buffer was as blank. (B) The interaction between SIN3B and BRD1 deletion mutants was examined by immunoprecipitation with HA or Flag antibody and blotting with the SIN3B antibody. (C) Molecular modeling of the sulfatide–SIN3B–BRD1 complex. Red, α-helix. Blue, β-sheet. Green, β-turn. Gray, random coil. Core complex and the amino acid residues on the active binding sites showed in white rectangle were amplified at bottom. Green, BRD1 PHD1 domain. Blue, BRD1 PHD2 domain. Yellow, SIN3B PAH1 domain. Pink, SIN3B PAH2 domain. Red, SIN3B PAH3 domain. (D) Molecular modeling of the superimposed SIN3B PAH2 domain. Red, PAH2 domain in the sulfatide–SIN3B complex. Blue, free SIN3B PAH2 domain. Yellow, PAH2 domain in virtual GalCer–SIN3B complex. The amino acid residues of secondary structure were highlighted in yellow. (E) Molecular modeling of SIN3B HID domain. Red, PAH2 domain in sulfatide–SIN3B complex. Blue, free SIN3B PAH2 domain. Yellow, PAH2 domain in virtual GalCer–SIN3B complex.
In order to further understand the interaction between SIN3B and sulfatide, we constructed the 3D conformation of the complex by using Discovery Studio 3.1. Sulfatide formed a more compact and stable complex with SIN3B than GalCer–SIN3B complex since the total energy for SIN3B-sulfatide complex was 12259.36111 kcal/mol, while the total energy for SIN3B–GalCer complex required 20152.25534 kcal/mol. The interaction of SIN3B molecule with sulfatide occurs mainly through the three tandem PAH domains (Figure 3C). Based on the analysis of the active binding sites by Discovery Studio 3.1, we calculated the amino acid residues in the SIN3B complex involved with sulfatide by covalent or noncovalent binding. The two fatty acid side chains of the sulfatide molecule were filled into the hydrophobic cavity formed by the SIN3B PAH1 domain (38th proline–57th amino acid residues) and the BRD1 PHD2 domain (339th isoleucine–349th alanine residues). The 180th proline of the SIN3B PAH2 domain and the 355th phenylalanine and 356th proline of the PAH3 domain were all involved to form the sulfatide-SIN3B core complex (Figure 3C, bottom).
Sulfatide affects SIN3B interaction with binding proteins
By remodeling, we observed that the 3D conformation change in the second PAH2 domain of SIN3B. In free SIN3B, the PAH2 domain was dense, since peptides (180th proline–195th glutamine, 210th glutamic acid–220th leucine, 225th glutamic acid–234th phenylalanine) form α-helices. In the complex with sulfatide, the 3D conformation of the PAH2 domain became loose and the α-helices turned to a flexible and relaxed β-turn and random coil. In the GalCer–SIN3B complex, the PAH2 domain still retained the α-helix structures (Figure 3D). The HID domain of SIN3B, although in sulfatide-SIN3B complex, was not robustly changed in conformation (Figure 3E).
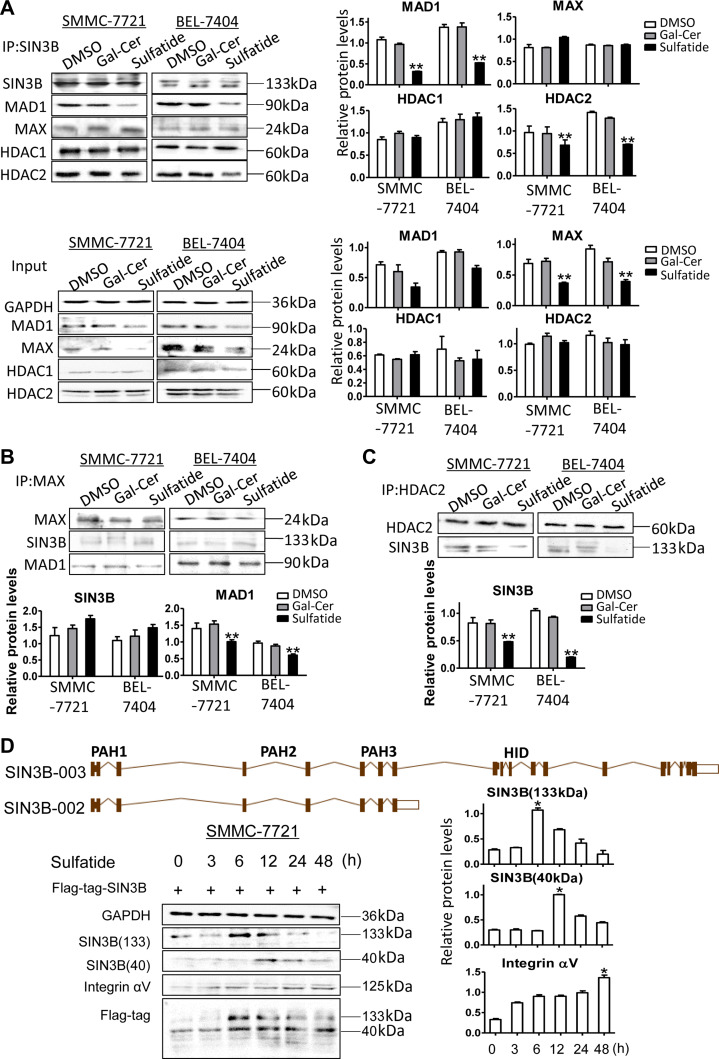
As the PAH2 domain of SIN3B is the binding site for other proteins such as MAX (Spronk, et al., 2000), we further investigated whether sulfatide-induced conformational changes would affect its interaction with other proteins. By Co-IP experiments, we found that MAD1 binding to SIN3B was reduced significantly by sulfatide as well as its expression level (Figure 4A). However, MAX in the complex of SIN3B did not change although its expression also reduced (Figure 4A and B). Sulfatide also significantly reduced the binding of SIN3B to HDAC2 (Figure 4A and C), although the binding to HDAC1 was not different. The binding affinity of SIN3B to HDAC2 did not reduce when SIN3B was knocked down, unless in the presence of high level of sulfatide (Supplementary Figure S1D).
Figure 4.
Sulfatide affects the binding of SIN3B to MAD1 and HDAC2. (A–C) The expression measurement of SIN3B, MAX, MAD1, HDAC1, and HDAC2 in whole-cell lysates was performed by western blot and quantitative analysis. Co-Immunoprecipitated (IP) complex with the corresponding antibody (SIN3B, MAX, MAD1, HDAC1, or HDAC2) was detected by western blot and quantitative analysis, respectively. (D) Ideograph of SIN3B-003 and SIN3B-002 transcript variants (upper). Integrin αV, SIN3B-003 (133 kDa) and SIN3B-002 (40 kDa) were detected at different treatment intervals by western blot (left) and quantitative analysis (right). Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01.
SIN3B with complete HID domain regulates integrin αV
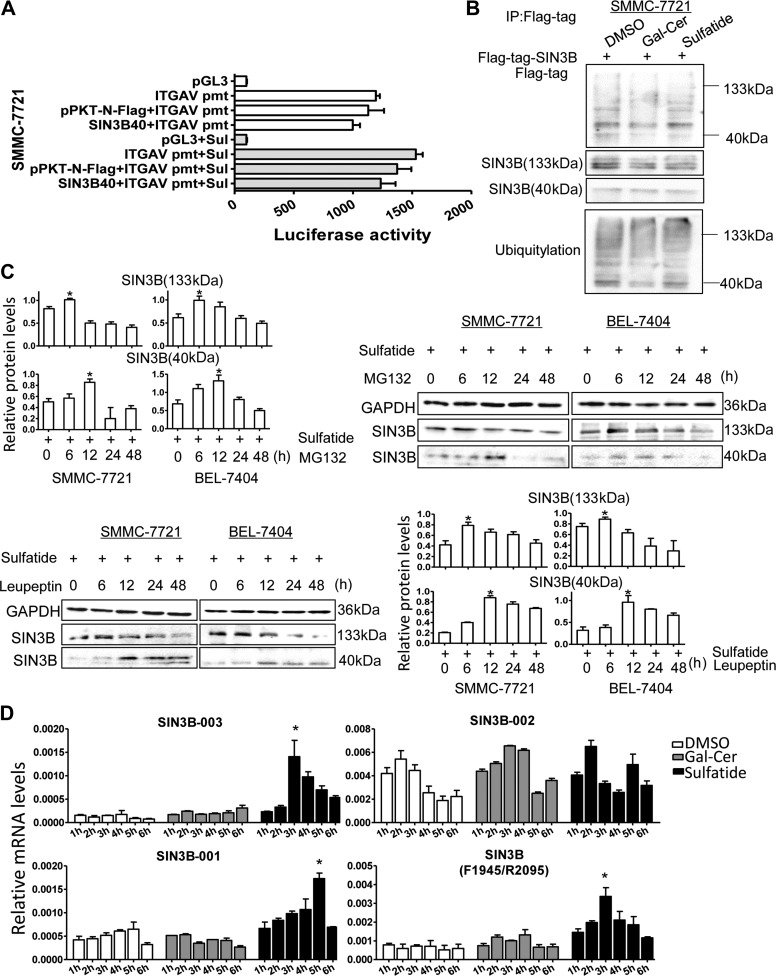
For SIN3B gene, there were 12 transcript variants. The SIN3B-002 transcript variant encodes protein with molecular weight 39507 Da because it lacks HID domain compared to full-length SIN3B (Figure 4D, top). While variant SIN3B-003 encodes the full length of SIN3B 1162 amino acids with molecular weight 133 kDa and SIN3B-001 encodes 1130 amino acids. After sulfatide treatment, 133 kDa SIN3B protein level reached its peak at 6 h, whereas 40 kDa SIN3B expression level peaked at 12 h after sulfatide treatment. The expression level of integrin αV subunit increased after 24–48 h treatment (Figure 4D, bottom). Then the SIN3B-002 cDNA was constructed into the plasmid pPKT-N-Flag-SIN3B-40, which was further co-transferred with integrin αV gene promoter reporter system. Interestingly the subtype SIN3B-002 failed to enhance the promoter activity even under sulfatide treatment (Figure 5A), but SIN3B-003 subtype with complete protein structure significantly stimulated the promoter activity (Figure 2E). We further investigated the total ubiquitination level of the SIN3B proteins, and observed the total ubiquitination level of the SIN3B protein did not change significantly after sulfatide treatment (Figure 5B). And the expression of SIN3B subtypes also did not increase at 24 and 48 h after administration of proteasome inhibitor MG132 (Figure 5C), but 40 kDa SIN3B robustly increased after 12 h treatment with leupeptin. We further detected the expression level of SIN3B transcriptional variants (Figure 5D) after sulfatide treatment by qPCR with specific primers according to exons and intron sequences in SIN3B-003 and SIN3B-002 transcripts (Supplementary Figure S2). In HCC cells, the expression level of the SIN3B-003 transcript was higher than that in the control group and reached the peak at 3 h after sulfatide treatment. While the SIN3B-001 transcript reached the peak after nearly 5 h. The results of SIN3B universal primers (F1945/R2095) were close to SIN3B-003 and peaked at 3 h after treatment, but the overall expression level was higher than that of SIN3B-003. While the SIN3B-002 transcript variant was not significantly elevated in sulfatide treatment compared with the control group (Figure 5D).
Figure 5.
Expression measurement of SIN3B subtypes in HCC. (A) The luciferase activity was determined in SMMC-7721 cells co-transfected with pPKT-N-Flag-SIN3B-40 and pGL3-ITGAV promoter. (B) Total ubiquitylation in sulfatide-treated HCC cells was analyzed by western blot. (C) SIN3B (133 kDa) and SIN3B (40 kDa) were detected by western blot, after the treatment with sulfatide and the proteasome inhibitor MG132 (10 mM) (upper) or the proteolytic enzyme inhibitor leupeptin (10 mM) (low), and quantitative analysis. (D) The transcript variants were detected in sulfatide-treated cells at different time intervals by qPCR. Data are representative of at least three independent experiments. *P < 0.05. **P < 0.01.
Discussion
Histones can be methylated or acetylated by the recruited methyltransferase or acetyltransferase for gene opening or closing. After the histones are acetylated, the DNA and histone octamers are released from the compact nucleosomes, allowing the transcription factors and the co-transcription factors to bind specifically to the exposed DNA binding site, and activate gene transcription (Sterner and Berger, 2000). The changes in histone acetylation levels, mediated by histone acetyltransferase and histone deacetylase, control dynamic regulation of gene expression (Brown et al., 2000; Joshi et al., 2013). We previously have found that the transcription of integrin αV subunit gene was associated with acetylation regulation (Wu et al., 2013; Cai et al., 2018).
In this study, we noted that the expression level of SIN3B was significantly correlated with the expression of integrin αV subunit in hepatocellular carcinoma and SIN3B expression was much higher in HCC than adjacent tissues from TCGA liver cancer database. In 11 various cancer cells, SIN3B expression was also closely correlated with integrin αV subunit. Overexpression of SIN3B in HCC cells by transfection significantly promoted either RNA or protein expression of integrin αV subunit. However, SIN3B is generally considered as a corepressor of gene transcription. Interestingly in this study, we did note that SIN3B promoted integrin αV transcription, but not suppressed it. In order to understand the regulation mechanism, we observed the regulatory effect of SIN3B on the promoter activity of integrin αV gene by dual luciferase activity assay. Unexpectedly, we found that SIN3B alone did not significantly enhance the transcriptional activity of the ectopic integrin αV gene promoter, unless in the presence of sulfatide. Previous studies revealed that sulfatide was elevated in HCC (Wu et al., 2004; Dong et al., 2014), but its role was not understood. Overexpression of the key enzyme CST for sulfatide synthesis in HCC cells resulted in acceleration of metastasis via integrin αVβ3 by the promotion of integrin αV expression (Zhang et al., 2010; Wu et al., 2013). However, the detailed mechanism of sulfatide regulation of integrin αV expression has not been well understood. This study presented the evidence that SIN3B promoted integrin αV expression especially when sulfatide was significantly elevated. Interestingly, SIN3B was identified in the complex with sulfatide and fat blot assay also indicated that SIN3B interacted with sulfatide specifically. Simulation reconstruction of SIN3B 3D conformation suggested that the PAH2 domain became loose after sulfatide binding since its α-helices turned to β-turn and random coil. The PAH2 domain is the MXD binding site for SIN3B protein. Reduction of MAD1 was indeed found in the SIN3B complex and this might be caused by the PAH2 domain conformation alteration. HDAC2 was also reduced in SIN3B complex with sulfatide although the HID domain was not affected. Less recruitment of HDAC2 might be due to the reduction of MAD1 binding or the PAH2 domain alteration. Spronk et al. (2000) has reported the aqueous solution structure of the complex of the PAH2 domain of SIN3B with the MAD1 N-terminal peptide fragment, When the SIN3B PAH2 domain binds to the MAD1 SID domain, the secondary structure of PAH2 is more stable. The PAH2-SID structure provides a structural basis for the PAH2 domain, to bind pairing α-helix selectively.
We simulated the spatial structure of SIN3B complex with sulfatide by Discovery Studio 3.1 and compared it with the free SIN3B. It was found that the fatty acid side chains of sulfatide were linked to the amino acid residues of the PAH1 domain of SIN3B, formed by hydrophobic lacunar binding. Wherein the 38th proline, 39th and 41th valine, 45th leucine, 47th tyrosine, 48th leucine, 51th valine, 53th isoleucine, and 55th phenylalanine hydrophobic amino acid residues provided active binding sites for sulfatide on the PAH1 domain. The binding of SIN3B with sulfatide resulted in the release of four α-helices in the PAH2 domain of SIN3B. In addition to the 162th phenylalanine, 165th alanine, 166th isoleucine, 168th tyrosine, and 169th valine in α1-helix, the 186th phenylalanine, 187th leucine, 191th leucine, and 194th tyrosine in α2-helix, the 213th valine in α3-helix, and the 231th phenylalanine and 234th phenylalanine in α4-helix all changed position, making SIN3B and MAD1 not form a stable complex. Sulfatide also reduced the binding of SIN3B with HDAC2. Although we did not note that sulfatide caused SIN3B HID domain conformation change in the simulation, the reduction of SIN3B binding to MAD1 may lead to a decrease in HDAC2 binding to SIN3B and collapse of deacetylation complex on the promoter. It is also possible that the conformation of four α-helices in the PAH2 domain has an important effect on the binding of SIN3B HID domain to HDAC2. Furthermore, we investigated the regulation of integrin αV subunit by the SIN3B subtype lacking the HID domain by luciferase activity assay and found that the regulation of SIN3B promoting integrin αV gene expression in hepatocellular carcinoma depended on the HID domain.
In summary, sulfatide changed the spatial conformation of the PAH2 domain of SIN3B after the interaction, reducing its binding capacity to MAD1 and HDAC2. These effects of sulfatide reduced the recruitment of HDAC2 deacetylation complexes on the integrin αV gene promoter and limited the deacetylation, resulting in the integrin αV subunit promoter histone remained at a higher level of acetylation, and facilitated the transcription of the integrin αV subunit gene.
Materials and methods
Case analysis from TCGA database
The human hepatocellular carcinoma gene expression (IlluminaHiSeq) data were analyzed from TCGA (The Cancer Genome Atlas, https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/). There were 331 HCC cases with clinical follow-up data from the database and 52 cases with gene expression array data in both liver cancer and adjacent non-tumor tissue samples.
Cell culture and treatment
Human hepatocellular carcinoma cell lines SMMC-7721 and BEL-7404 were from the Shanghai Institute of Biochemistry & Cellular Biology, Chinese Academy of Science, Shanghai, China. HCC-derived cell lines LM3, Hep3B and HuH-7 were from Liver Cancer Institute of Zhongshan Hospital, Fudan University, Shanghai, China. Human colorectal cancer cells SW480 and SW620 were from Fudan University Shanghai Cancer Center. Hepatoma cells SK-Hep-1, human alveolar adenocarcinoma cell A549, and human embryonic kidney cells HEK293T were obtained from the American Type Culture Collection and cultured according to the recommended conditions. For the treatment, cells were incubated with GalCer, sulfatide (2 μmol/L, 24 h for RNA analysis, 48 h for protein assays) (Sigma-Aldrich).
Plasmids and transfection
The SIN3B cDNA was from GeneCopoeia Inc. Rockville, MD 20850, USA. Then the SIN3B cDNA and its variant cDNA were obtained by polymerase chain reaction with specific primers (Table 1) and cloned into the HindIII and XbaI sites of the plasmids pcDNA3.1B and pPKT-N-Flag. Specific interference sequences (Table 1) targeting SIN3B were constructed into the BamHI and HindIII sites of pSilencer 4.1 plasmid. The expression plasmid of integrin αV subunit gene was constructed as in our previous study (Wu et al., 2013; Wang et al., 2016). BRD1 and its mutant constructs were generous gifts from Prof. Atsushi Lwama, Chiba University, Japan (Mishima et al., 2011).
Table 1.
Primer sequences used for PCR.
| Primer | Sequence |
|---|---|
| hGAPDH s79 | 5′-CGGATTTGGTCGTATTGGG-3′ |
| hGAPDH a291 | 5′-CGCTCCTGGAAGATGGTGAT-3′ |
| hβ-actin F1409 | 5′-CCGAGGACTTTGATTGCA-3′ |
| hβ-actin R1508 | 5′-GTGGGGTGGCTTTTAGGA-3′ |
| ITGAV F372 | 5′-GACAGTCCTGCCGAGTA-3′ |
| ITGAV R497 | 5′-CTGGGTGGTGTTTGCT-3′ |
| ITGAV promoter F | 5′-AGCTCCTGAGCCTGGGT-3′ |
| ITGAV promoter R | 5′-CAACAGTCGCACGGAAGT-3′ |
| SIN3B F1945 | 5′-GAGCCGCACCTCATCTTT-3′ |
| SIN3B R2095 | 5′-AGAGGCTGGGCACGAACT-3′ |
| SIN3B EX8 1 | 5′-GTATGAAAACTTCCTCCGCTG-3′ |
| SIN3B-003 AS 3 | 5′-ACGAACCCTGGAGAAGTGTC-3′ |
| SIN3B-002 AS 2 | 5′-GGTGGTGTATTGCTCCTTA-3′ |
| SIN3B-001 AS 4 | 5′-CGTTCAGTACCTCCTTGCAGA-3′ |
| SIN3B F35 HindIII | 5′-AAAAAAGCTTATGGCGCACGCTGG-3′ |
| SIN3B R3424 XbaI | 5′-CCCCTCTAGAGGGCGAGGCCGGGC-3′ |
| SIN3B top | 5′-GATCCTTTATCGCATCTATGGCAGTTCAAGAGATCGCCATAGATGCGATAAAAGA-3′ |
| SIN3B bot | 5′-AGCTTCTTTTATCGCATCTATGGCGATCTCTTGAATCGCCATAGATGCGATAAAG-3′ |
Polymerase chain reaction
The total RNAs of cells were isolated with Trizol reagent (invitrogen, Thermo Fisher Scientific) according to the manufacturer’s protocol. Reverse transcription reactions were performed using reverse transcriptase M-MLV (TaKaRa). Real-time PCR was performed with the CFX-connect qPCR detection system (Bio-Rad) using the Bestar® SybrGreen qPCR Mastermix (DBI® Bioscience) according to the manufacture’s instruction. The specific primers are listed in Table 1.
Western blot analysis and immunoprecipitation
The proteins obtained were separated on an 8% SDS-PAGE and transferred to a PVDF membrane. The blot was probed with the anti-integrin αV, SIN3B, c-MYC, MAD1, MAX, HDAC1, HDAC2 and BRD1 antibodies (Santa Cruz Biotechnology and Bioworld Technology, Inc.), respectively, followed by a horseradish peroxidase-conjugated secondary antibody. The density of developed protein bands was analyzed using TotalLab v2.01 (Nonlinear Dynamics Ltd). Immunoprecipitation assays were performed as previously described (Hu et al., 2008). Briefly, cells were lysed in RIPA lysis buffer with protease inhibitors, and then incubated with corresponding antibody. The protein-antibody complexes were then collected with pre-washed agarose-protein G beads (CTB, Pointbio) at 4°C overnight, and were analyzed by standard immunoblot procedures.
Migration assays
Cell migration was performed using Millipore Millicell (8.0 μm) (Wu et al., 2010). Briefly, 0.1 ml of 5 × 105 cells/ml suspension was seeded in the upper chamber Millicell with 8.0 μm polyethylene terephthalate membrane (Millipore). Then 0.5 ml DMEM medium containing 10% fetal bovine serum was added to the lower chamber in a 24-well plate. After incubating at 37°C with 5% CO2 for 6 h, the cells in the upper Millicell were removed with a cotton bud gently. The microporous membrane was fixed in 4% paraformaldehyde for 15 min and stained with 1% crystal violet for 10 min. Then the cells on the downside surface of the membrane of the Millicell were counted.
Flow cytometry analysis
SMMC-7721 cells treated with sulfatide for 48 h were detached by 5 mM EDTA, washed with 1 mM EDTA in 0.84% NaCl, and suspended in DMEM with 10% FBS to the final concentration of 1 × 107 cells/ml. Cells were incubated with 5 μg/ml FITC-labeled integrin αV or β3 antibody for 1 h at 4°C, and the FITC-mouse IgG antibody as the control. After cells were washed in PBS, cell surface immunofluorescence of all samples was analyzed by a flow cytometer FACS scan (Cytomics™ FC 500, Beckman Coulter).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed with the EZ-ChIP kit (Merck Millipore) according to the manufacturer’s instructions using anti-SIN3B, c-MYC, MAD1, MAX, H3K4Me3 and H3K27Me3 antibodies. The occupancy of the integrin αV gene promoter of purified DNAs was detected by PCR and quantitative PCR. The primer sequences are given in Table 1.
Luciferase assays
The activities of firefly luciferase and renilla luciferase were measured with the dual luciferase reporter assay system (Promega) 48 h after transfection.
Mass spectrometry
Sulfatide was coated and coupled with epoxy magnetic particles based on the previous report (Sun et al., 2009). Then the particles were incubated with 500 μl SMMC-7721 cell lysate overnight at 4°C. After washing and SDS-PAGE, the specific bands at 133 kDa for sulfatide compared to GalCer in Coomassie Brilliant Blue stained gel were excised for MS analysis as our previous report (Cai, et al., 2018).
Fat blot assay
The assay was according to the lipid-binding analysis method by Munnik and Wierzchowiecka (2013). Ceremides, ganglioside GM3, galactocerebrosides, sulfatide, D-sphingosine, L-α-phosphatidylinositol and L-phosphatidylinositol (Sigma-Aldrich) were suspended in resuspension buffer (CHCl3/MeOH/H2O=20:9:1, v/v), and diluted to gradient concentration by spotting buffer (CHCl3/MeOH/50 mM HCl/Ponceau S=250:500:200:2, v/v). The lipids were spotted on PVDF membrane strips and dried at room temperature in the dark. The membranes were blocked in fat blot buffer (50 mM Tris–HCl, 150 mM NaCl, pH 7.5) containing 3% fatty acid-free BSA for 1 h at room temperature, and incubated with SMMC-7721 cell lysates (protein 5 μg/ml) in blocking buffer-0.1% Tween overnight at 4°C. After three washes with fat blot buffer-0.1% Tween, the membranes were incubated with anti-SIN3B antibody and goat anti-rabbit IgG HRP secondary antibody (Jackson ImmunoReseach Lab, Inc.). The blots were visualized using Immunobilon Western (Millipore).
Molecular modeling
The molecular models were created using the Build Module of Discovery Studio (DS) version 3.1 Client (BIOVIA). The SIN3B model was created with the crystal structure of the PAH1 domain (PDB ID: 2CZY), PAH2 domain (PDB ID: 2F05), PAH3 domain (PDB ID: 2RMR) and HID domain (PPDB ID: 1ZS8).The BRD1 model was created with the PHD1 domain (PDB ID: 2KU3), PHD2 domain (PDB ID: 2LQ6), bromo domain (PDB ID: 3RCW) and PWWP domain (PDB ID: 3LYI). Two complex models were immersed in an implicit solvent model using generalized born with implicit membrane(GBIM) and minimized with Steepest Descendent and Conjugate Gradient algorithms to reach the minimum convergence (i.e. 0.1 kcal/mol/Å) after 500 iterations. The long-range electrostatic interactions were treated by spherical cutoff method, and hydrogen bonds were constrained with the SHAKE algorithm. All energy minimizations and molecular dynamics simulation were performed using Chemistry at Harvard Macromolecular Mechanics 27 (CHARMm27) force field parameters. The trajectories collected after the production steps were analyzed, and the most energetically favorable conformation was chosen.
Quantitative analysis
Data are presented as mean ± SD. Statistical differences were measured using one-way ANOVA and Student’s t-test. Chi-square test and Spearman rank correlation analysis were used for the analysis of ratio difference. Statistical analysis was performed using SPSS software v22 (IBM) and P-values <0.05 were considered significant.
Supplementary Material
Funding
This work was supported by the National Natural Science Foundation of China (31570800 and 31400689).
Conflict of interest
none declared.
Author contributions
Q.C. did the experiments and acquired data; P.Z., Y.L., C.K., H.X., B.Q., and R.W. did partial experiments; Q.C., P.Z., Y.D., and C.K. interpreted the data; Q.C. drafted the manuscript; X.Z.W. designed the study and revised the manuscript.
References
- Bainor A.J., Deng F.M., Wang Y., et al. (2017). Chromatin-associated protein SIN3B prevents prostate cancer progression by inducing senescence. Cancer Res. 77, 5339–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal N., David G., Farias E., et al. (2016). Emerging roles of epigenetic regulator Sin3 in cancer. Adv. Cancer Res. 130, 113–135. [DOI] [PubMed] [Google Scholar]
- Brown C.E., Lechner T., Howe L., et al. (2000). The many HATs of transcription coactivators. Trends Biochem. Sci. 25, 15–19. [DOI] [PubMed] [Google Scholar]
- Cai Q.Q., Dong Y.W., Qi B., et al. (2018). BRD1-mediated acetylation promotes integrin αV gene expression via interaction with sulfatide. Mol. Cancer Res. 16, 610–622. [DOI] [PubMed] [Google Scholar]
- David G., Grandinetti K.B., Finnerty P.M., et al. (2008). Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc. Natl Acad. Sci. USA 105, 4168–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Barnes L.A., Shields D.J., et al. (2009). An integrin αvβ3-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat. Med. 15, 1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.W., Wang R., Cai Q.Q., et al. (2014). Sulfatide epigenetically regulates miR-223 and promotes the migration of human hepatocellular carcinoma cells. J. Hepatol. 60, 792–801. [DOI] [PubMed] [Google Scholar]
- Frank M. (2000). MAL, a proteolipid in glycosphingolipid enriched domains: functional implications in myelin and beyond. Prog. Neurobiol. 60, 531–544. [DOI] [PubMed] [Google Scholar]
- Fukuda M., Hiraoka N., Akama T.O., et al. (2001). Carbohydrate-modifying sulfotransferases: structure, function, and pathophysiology. J. Biol. Chem. 276, 47747–47750. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanz P., Quintanilla A., Lafita M.C., et al. (2014). Sin3b interacts with Myc and decreases Myc levels. J. Biol. Chem. 289, 22221–22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandinetti K.B., Jelinic P., DiMauro T., et al. (2009). Sin3B expression is required for cellular senescence and is up-regulated upon oncogenic stress. Cancer Res. 69, 6430–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzenda A., Lomberk G., Zhang J.S., et al. (2009). Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa S., Yamato K., Ishizuka I., et al. (1974). Biosynthesis of seminolipid: sulfation in vivo and in vitro. J. Biochem. 75, 77–83. [DOI] [PubMed] [Google Scholar]
- Hu P., Shi B., Geng F., et al. (2008). E-cadherin core fucosylation regulates nuclear β-catenin accumulation in lung cancer cells. Glycoconj. J. 25, 843–850. [DOI] [PubMed] [Google Scholar]
- Hurlin P.J., and Huang J. (2006). The MAX-interacting transcription factor network. Semin. Cancer Biol. 16, 265–274. [DOI] [PubMed] [Google Scholar]
- Joshi P., Greco T.M., Guise A.J., et al. (2013). The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 9, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Harris M., and Varner J.A. (2000). Regulation of integrin αvβ3-mediated endothelial cell migration and angiogenesis by integrin α5β1 and protein kinase A. J. Biol. Chem. 275, 33920–33928. [DOI] [PubMed] [Google Scholar]
- Laherty C.D., Yang W.M., Sun J.M., et al. (1997). Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89, 349–356. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Miyagi S., Saraya A., et al. (2011). The Hbo1-Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 118, 2443–2453. [DOI] [PubMed] [Google Scholar]
- Munnik T., and Wierzchowiecka M. (2013). Lipid-binding analysis using a fat blot assay. Methods Mol. Biol. 1009, 253–259. [DOI] [PubMed] [Google Scholar]
- Nam C.W., Park N.H., Park B.R., et al. (2008). Mitotic checkpoint gene MAD1 in hepatocellular carcinoma is associated with tumor recurrence after surgical resection. J. Surg. Oncol. 97, 567–571. [DOI] [PubMed] [Google Scholar]
- Rielland M., Cantor D.J., Graveline R., et al. (2014). Senescence-associated SIN3B promotes inflammation and pancreatic cancer progression. J. Clin. Invest. 124, 2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spronk C.A., Tessari M., Kaan A.M., et al. (2000). The Mad1-Sin3B interaction involves a novel helical fold. Nat. Struct. Biol. 7, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Sterner D.E., and Berger S.L. (2000). Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Yang G., Sun S., et al. (2009). The hydroxyl-functionalized magnetic particles for purification of glycan-binding proteins. Curr. Pharm. Biotechnol. 10, 753–760. [DOI] [PubMed] [Google Scholar]
- Tang J., Niu J.W., Xu D.H., et al. (2007). Alteration of nuclear matrix-intermediate filament system and differential expression of nuclear matrix proteins during human hepatocarcinoma cell differentiation. World J. Gastroenterol. 13, 2791–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick D., Decker J., Groene H.J., et al. (1999). Regional expression of sulfatides in rat kidney: immunohistochemical staining by use of monospecific polyclonal antibodies. Histochem. Cell Biol. 111, 143–151. [DOI] [PubMed] [Google Scholar]
- Varner J.A., Brooks P.C., and Cheresh D.A. (1995). REVIEW: the integrin αVβ3: angiogenesis and apoptosis. Cell Adhes. Commun. 3, 367–374. [DOI] [PubMed] [Google Scholar]
- Wang R., Qi B., Dong Y.W., et al. (2016). Sulfatide interacts with and activates integrin αVβ3 in human hepatocellular carcinoma cells. Oncotarget 7, 36563–36576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng X., Cheng X., Wu X., et al. (2014). Sin3B mediates collagen type I gene repression by interferon γ in vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 447, 263–270. [DOI] [PubMed] [Google Scholar]
- Wu W., Dong Y.W., Shi P.C., et al. (2013). Regulation of integrin αV subunit expression by sulfatide in hepatocellular carcinoma cells. J. Lipid Res. 54, 936–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.H., Shi B.Z., Zhao Q.L., et al. (2010). Fucosylated glycan inhibition of human hepatocellular carcinoma cell migration through binding to chemokine receptors. Glycobiology 20, 215–223. [DOI] [PubMed] [Google Scholar]
- Wu X.Z., Honke K., Zhang Y.L., et al. (2004). Lactosylsulfatide expression in hepatocellular carcinoma cells enhances cell adhesion to vitronectin and intrahepatic metastasis in nude mice. Int. J. Cancer 110, 504–510. [DOI] [PubMed] [Google Scholar]
- Zhang C.Y., Hu P., Fu D., et al. (2010). 3′-Sulfo-Lex is important for regulation of integrin subunit αV. Biochemistry 49, 7811–7820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.