Abstract
Pseudomonas aeruginosa, a gram-negative bacterium, is one of the most common pathogens colonizing the lungs of cystic fibrosis patients. P. aeruginosa secrete extracellular vesicles (EVs) that contain lipopolysaccharide (LPS), and other virulence factors that modulate the host’s innate immune response leading to an increased local pro-inflammatory response and reduced pathogen clearance, resulting in chronic infection and ultimately poor patient outcomes. Lung macrophages (LMs) are the first line of defense in the airway innate immune response to pathogens. Proper host response to bacterial infection requires communication between antigen presenting cells (APC) and T cells, ultimately leading to pathogen clearance. Here, we investigate whether EVs secreted from P. aeruginosa alter Major Histocompatibility Complex (MHC) antigen expression in lung macrophages, thereby potentially contributing to decreased pathogen clearance.
Primary lung macrophages from human subjects were collected via bronchoalveolar lavage (BAL) and exposed to EVs isolated from P. aeruginosa in vitro. Gene expression was measured with the NanoString nCounter gene expression assay. DNA methylation was measured with the EPIC array platform to assess changes in methylation. P. aeruginosa EVs suppress the expression of eleven different MHC-associated molecules in lung macrophages. Additionally, we show reduced DNA methylation in a regulatory region of gene CFB, complement factor B, as the possible driving mechanism of widespread MHC gene suppression.
Our results demonstrate MHC molecule downregulation by P. aeruginosa-derived EVs in lung macrophages, which is consistent with an immune evasion strategy employed by a prokaryote in a host-pathogen interaction, potentially leading to decreased pulmonary bacterial clearance.
Keywords: Lung Macrophage, Extracellular vesicles, Pseudomonas aeruginosa, Major histocompatibility complex Class II, Innate immunity
Summary sentence:
P. aeruginosa EVs suppress the expression of multiple MHC-associated molecules in lung macrophages.
Introduction
Cystic fibrosis (CF) is the most common fatal genetic disease that occurs in Caucasians (1), with nearly 30,000 individuals diagnosed in the US and an estimated 70,000 individuals afflicted world-wide (2). CF manifests predominantly as a pulmonary disease but also affects gastrointestinal, endocrine/exocrine and reproductive function. CF is caused by a mutation in a gene that encodes the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride and bicarbonate ion transport channel that maintains osmotic balance across multiple epithelial surfaces in the body (3, 4). Ultimately, 90% of deaths in CF patients are attributed to pulmonary dysfunction directly associated with chronic infection, underscoring the absolute importance of studying the pathogenesis of CF-relevant microbes (5).
Pseudomonas aeruginosa is an opportunistic pathogen that commonly colonizes the lung in CF patients, resulting in an accelerated decline of pulmonary function (6). P. aeruginosa is the most dominant pathogen of the lung microbiota of adult cystic fibrosis patients and also commonly infects pediatric CF patients (7). Chronic infection with gram-negative pathogens of CF, including P. aeruginosa and B. cepacia, is associated with a rapid decline in pulmonary function, more frequent exacerbations, and higher rates of mortality (8). P. aeruginosa resides primarily in the mucus overlying lung epithelial cells and secretes extracellular vesicles (EVs) that diffuse through the mucus and fuse with airway cells, thus delivering virulence factors and other toxins into the host cell cytoplasm that modify the innate immune response (9–14).
Extracellular vesicles (EVs) are lipid bilayer vesicles secreted by most eukaryotic and many prokaryotic cells, and thought to be intimately involved in trans-kingdom crosstalk (13, 15–17). EVs can range in size from 20 nM to 1000 nM depending on the producing cell and its environment (18). Gram-negative bacterial EVs contain lipopolysaccharide (LPS), a known virulence factor, on their surface (19). Moreover, it has been suggested that microbial EVs have important and underappreciated roles in intra-kingdom and inter-kingdom communication by regulating gene expression in target cells directly and indirectly via host immune receptor signaling (18).
Antigen presentation is crucial for immune responses. Major histocompatibility complex (MHC) molecules present peptides to other immune cells in order to mount an adaptive immune response (20). The host response to bacterial infection requires communication between antigen presenting cells (APC) and T cells. This communication is relayed through MHC class II antigen presentation to helper T-cells followed by adaptive T-cell or B-cell responses (21). The interaction between T-cells and MHC class II, along with the surrounding milieu, is essential for defining the phenotype and success of the inflammatory response to infection.
Macrophages are a key APC of the lung and possess remarkable immune plasticity, with the ability to sense and adapt to the local microenvironment (22, 23). Lung macrophages play pivotal roles in bacterial recognition and elimination as well as in polarization of innate and adaptive immunity. Depending on the local setting, macrophages sometimes play a role in anti-inflammatory responses, tissue repair, and homeostasis, whereas at other times, they promote inflammatory and phagocytic processes through the complex production of cytokines and cellular interaction (24, 25).
Numerous pathogens employ a strategy of host immune escape during infection, including MHC molecule modulation (26–29). EVs from the periodontal gram-negative pathogen, Porphyromonas gingivalis have been shown to inhibit surface expression of HLA-DR molecules in human umbilical cord vascular endothelial cells (30). Additionally, Cif protein, a bacterial virulence factor secreted in EVs by P. aeruginosa has been shown to inhibit MHC class I antigen presentation in airway epithelial cells (10).
The goal of this study is to assess whether EVs isolated from one of the most prevalent cystic fibrosis pathogens, P. aeruginosa, can alter expression of MHC-related molecules in lung macrophages thereby potentially modulating the macrophages’ ability to mount a proper immune response to the infection.
Materials and Methods
This study was approved by the Dartmouth-Hitchcock Institutional Review Board (#22781). All subjects were healthy non-smokers. A total of seven subjects were used in the gene expression portion of this study (3 males and 4 females), with a mean age of 27.14 (±2.9) years. Lung macrophages from an additional four healthy subjects were used for the protein validation portion of the study (three females and one male), with a mean age 28.25 (±3.4) years.
Bronchoalveolar Lavage and Macrophage Isolation.
Subjects underwent flexible bronchoscopy following local anesthesia with lidocaine to the posterior pharynx and distal trachea/carina with or without intravenous sedation per patient preference. A bronchoscope was inserted trans-orally and advanced through the vocal cords. BAL fluid was obtained from tertiary airways via rinses with 20 ml of sterile saline followed by 10 ml of air and repeated for a total of 5 times per airway. Lung macrophages were isolated as previously described (31, 32).
EV generation and isolation
A clinically-derived mucoid strain of P. aeruginosa (DH1137- gift of Dr. D. Hogan, Geisel School of Medicine at Dartmouth) was grown overnight (20 hours) in LB media. Whole cells were pelleted five times (3500 × g 20 minutes) then filtered (0.22 uM), leaving conditioned media containing the EVs. Media was adjusted to pH 4.0. Isolation of EVs was performed by mixing 600 microliters of Resin Slurry E (Norgen Bio-Tek Corp., Thorold, ON, Canada) with conditioned media, multiple inversions with a 10 minute equilibration. Resin was pelleted with centrifugation @ 800 × g for 10 minutes (Sorvall Legend X1R Benchtop centrifuge), media discarded and the tube was inverted to drain. To recover EVs, 500 microliters of 1 × PBS (pH 6.0) added to resin, tube was vortexed and allowed a 10 minute equilibration, and subsequently centrifuged @ 400 × g for 10 minutes to pellet the resin. PBS containing EVs was centrifuged two additional times (400 × g 10 minutes) to remove any residual resin. EVs in 1 × PBS were aliquoted and stored at −80°C until use.
Extracellular Vesicle Characterization
Negative staining transmission electron microscopy (TEM) was done as previously described (33). Briefly, a 10 microliter aliquot of EVs in 1 × PBS was placed on top of carbon-coated formvar support film on 300 mesh Cu grids (Electron Microscopy Science) for 1 minute, with excess liquid removed by blotting with filter paper. Sample was immediately stained with 1% uranyl acetate, blotted and air-dried in a humidity-controlled chamber. The samples were examined using a FEI (Thermo-Fisher) Tecnai 12, transmission electron microscope with an accelerating voltage of 120KV and images recorded using Digital Micrograph software via a Gatan, Rio9 digital camera system. Cryo-EV samples were prepared on freshly glow discharged C-flat copper grids (Electron Microscopy Sciences), plunged into liquid nitrogen cooled, liquified ethane and stored in liquid nitrogen using a FEI (Thermo-Fisher) Vitrobot, set to 2 second and 4 second blotting times. Cryo-TEM was carried out on a Phillips CM120 Cryo-TEM at 120KV accelerating voltage. Images were collected under low-dose conditions using Digital Micrograph software and a Gatan, Orius 4K digital camera system.
Nanoparticle Tracking Analysis (NTA) with the Nanosight NS300 instrument (Malvern, Worcestershire, UK) was used to determine mean EV particle size and concentrations as previously described (34). For NTA, EV samples were diluted in PBS and introduced into the sample chamber via syringe pump. The following script was used for EV measurements: PRIME, DELAY 120, CAPTURE 30, REPEAT 3. Other acquisition settings included: camera level of 14; camera shutter speed 13 ms; camera gain 360; laser - blue 488. NTA post acquisition settings were optimized and kept constant between samples. Software used was NTA version 3.2 Dev. Build 3.2.16.
Experimental Design and Gene Expression
Right upper lobe (RUL) lung macrophages were used in this study, as several studies have shown that the RUL is the predominant lung lobe affected in CF patients (26, 35) and our previous work has shown regional differences in the phenotype of lung macrophages (36, 37). Cells were plated in 6-well tissue culture dishes seeded at 1 × 106 cells per well. Cells were allowed to adhere for 60 minutes followed by incubation in the presence or absence (negative control) of P.a. EVs at a ratio of 100:1 (EVs:cell) per Cecil et al. (38), for up to 48 hours. We have used the 48 hour time point to simulate chronic infection. Control experiments were performed as above, in the presence or absence of Pseudomonas-derived LPS only (1000 ng/ml). RNA was isolated via RNeasy mini kit (Qiagen, Germantown, MD) as outlined in manufacturer’s instructions, and quantitated on a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA). The digital multiplexed NanoString nCounter human v3 Cancer/Immune RNA expression assay (NanoString Technologies, Seattle,WA) was performed according to manufacturer’s instructions with total RNA. Briefly, 200 ng total RNA was assayed by overnight hybridization (65 °C) with nCounter gene-specific reporter and capture probes. Following a 20 hour hybridization, reporter and capture probes are washed away using the automated nCounter sample prep station and probe/target complexes are aligned and immobilized in the nCounter Cartridge. Cartridges are then placed in the nCounter digital analyzer for data collection. nSolver Analysis software (NanoString) (V3.0) was used for data analysis including background correction by subtracting the mean of the six negative controls included on the NanoString platform and normalization using the average geometric mean of the thirty housekeeping genes included in the assay.
Immunoblotting
Total protein was obtained by lysing cells in 200 μl/well 2xLaemmli buffer with 5% β-mercaptoethanol with 1% protease inhibitors and boiling 97°C, 5 minutes. Lysates were run on 7.5% and 10% gels and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were washed in Tris buffered saline with 0.1% Tween 20, cut, and incubated overnight at 4°C in 5% bovine serum albumin containing 1:5000 primary unless otherwise noted. Primary antibodies: monoclonal mouse anti-β Actin (Abcam, cat#ab8226); monoclonal rabbit anti-CD14 (Cell Signaling Technology, cat#56082S); 1:2000 monoclonal mouse anti-CD74 (Santa Cruz Biotechnology, cat#sc-20062); monoclonal rabbit anti-HLA-DRA (Cell Signaling Technology, cat#97971T); monoclonal rabbit anti-CD9 (Cell Signaling Technology, cat#13174S). Membranes were then washed four times in TBST and incubated at room temperature for 1 hour in BSA-TBST containing 1:5000 secondary. Secondary antibodies: mouse IgGκ binding protein (Santa Cruz Biotechnology, cat#sc-516102); polyclonal goat anti-rabbit IgG (Abcam, cat#ab7090). Membranes were then washed, exposed to Chemiluminescent Substrate (Thermo Fisher), and imaged using X-ray film.
Mass Spectrometry
Sample was lysed in 8M urea, 50mM Tris HCl (pH 8.0) containing 1X Roche Complete Protease Inhibitor. Lysates were quantified by Qubit fluorometry (Life Technologies). A 20μg aliquot of each sample was digested in solution using the following protocol:sample was diluted in 25mM ammonium bicarbonate and proteins were reduced with 10mM dithiothreitol at 60°C followed by alkylation with 50mM iodoacetamide at RT.
Proteins were digested with 1μg trypsin (Promega) at 37°C for 18h. The digest was quenched with formic acid and peptides cleaned using solid phase extraction (SPE) using the Empore C18 plate (3M). A 2μg aliquot was analyzed by nano LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to aThermoFisher Fusion Lumos mass spectrometer. Peptides were loaded on a trapping column and eluted over a 75μm analytical column at 350nL/min; both columns were packed with Luna C18 resin (Phenomenex) with a 60minute gradient. The mass spectrometer was operated in data dependent mode, with MS and MS/MS performed in the Orbitrap at 60,000 FWHM resolution and 50,000 FWHM resolution, respectively. A 3s cycle time was employed for all steps. Data were searched using a local copy of Mascot with the following parameters: Enzyme: Trypsin/Database: Uniprot-Pseudomonas aeruginosa strain ATCC 15692 (forward and reverse appended with common contaminants)/Fixed modification: Carbamidomethyl (C)/Variable modifications: Oxidation (M), Acetyl (Protein N-term), Deamidation (NQ), Pyro-Glu (N-term Q).
Mass values: Monoisotopic/Peptide Mass Tolerance: 10 ppm/Fragment Mass Tolerance: 0.02 Da/Max Missed Cleavages: 2. Mascot DAT files were parsed into the Scaffold software for validation, filtering and to create a nonredundant list per sample. Data were filtered 1% protein and peptide level false discovery rate (FDR) and requiring at least two unique peptides per protein.
DNA Methylation Array.
Epigenome-wide DNA methylation profiling was performed via the Infinium Methylation EPIC Bead Chips (Illumina Inc., San Diego, CA) for the determination of methylation levels of more than 850,000 CpG sites as previously described (39). Briefly, DNA was extracted from bronchoalveolar lavage-derived cells via Qiagen (Germantown, MD) DNeasy Blood and Tissue Kit. DNA was quantitated on a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA). Bisulfite conversion of DNA was carried out with the Zymo EZ DNA methylation kit (Zymo Research, Irvine, CA) and EPIC array hybridization and scanning were performed at the University of Southern California Molecular Genomics Core.
DNA methylation array data processing.
Raw intensity data files (IDATs) from the MethylationEPIC BeadChips were processed by the minfi R/Bioconductor analysis pipeline (version 1.30) (40) with annotation file version ilm10b4.hg19. Probes known to cross-hybridize, probes associated with known SNPs, non CpGs and sex chromosomes, as well as those failing to meet a detection P-value of 0.05 in ≥ 20% samples, were excluded. Location of CpGs were determined by the method of Zhou et al.(41). Genomic contexts and CpG relation to CpG islands were provided in the Illumina EPIC annotation file. The “promoter” transcriptional context was defined as either having a “TSS200” or “TSS1500” annotation, or both in the column UCSC_RefGene_Group (TSS, transcription start site). CpGs in the “promoter” context were any with either “Promoter_Associated” or “Promoter_Associated_Cell_type_specific” in the Regulatory_Feautre_Group annotation. Likewise, the “gene body” transcriptional context was defined as having a “Body” in the UCSF RefGene Group annotation. The “enhancer” context was defined as having a FANTOM5 enhancer record. DNA methylation array data has been deposited into Gene Expression Omnibus under accession no. GSE142801. Please contact corresponding author (David.A.Armstrong@hitchcock.org) for a token to access this data.
Statistical analysis
All gene expression data were presented as means ± SD. Graph Pad Prism (v7.0) analysis was used for paired sample t-tests. Differential gene expression analysis of Nanostring data was conducted using edgeR (v3.26.8) and limma (v3.40.6) packages in R (v3.6.1)(42, 43). The normalized data from Nanostring were scaled to counts per million (cpm). 541 genes with greater than 10 cpm in at least 2 samples were used for downstream analysis. Likelihood ratio tests were conducted after fitting the gene expression data to a negative binomial generalized linear model. 310 genes were determined as differentially expressed based on its likelihood ratio test FDR value threshold of 0.05.
Results
Characterization of Extracellular Vesicles
Negative stain TEM was used to visualize isolated Pseudomonas aeruginosa EVs (Fig.1A). EVs were heterogeneous in size with a range of 30 – 600 nm in diameter. EVs show characteristic concave appearance common in negative staining. Additionally, cryo-EM was used to further visually confirm EV size range (Fig.1B), and nanoparticle tracking analysis was used for particle counting, sizing and visualization. Several batches of P.a. EVs were isolated and used over the course of the study. A representative particle trace curve from one batch of P.a. EVs analyzed via the NanoSight NS300 nanoparticle tracking instrument is shown in Fig.1C. Mean particle diameter was 191.6 nM (±94.4). Typical EV particle yield from a starting volume of 35 milliliters of P.a. conditioned media was 4.25 × 109 total particles, corresponding to 1.2 × 108 particles/ml of media. A representative video of the P.a. EV particles passing through the 488 nM laser light of the NanoSight NS300 instrument is presented in the Supplemental files as Supplemental Video 1.
Fig 1. Characterization of extracellular vesicles from Pseudomonas aeruginosa (P.a.).
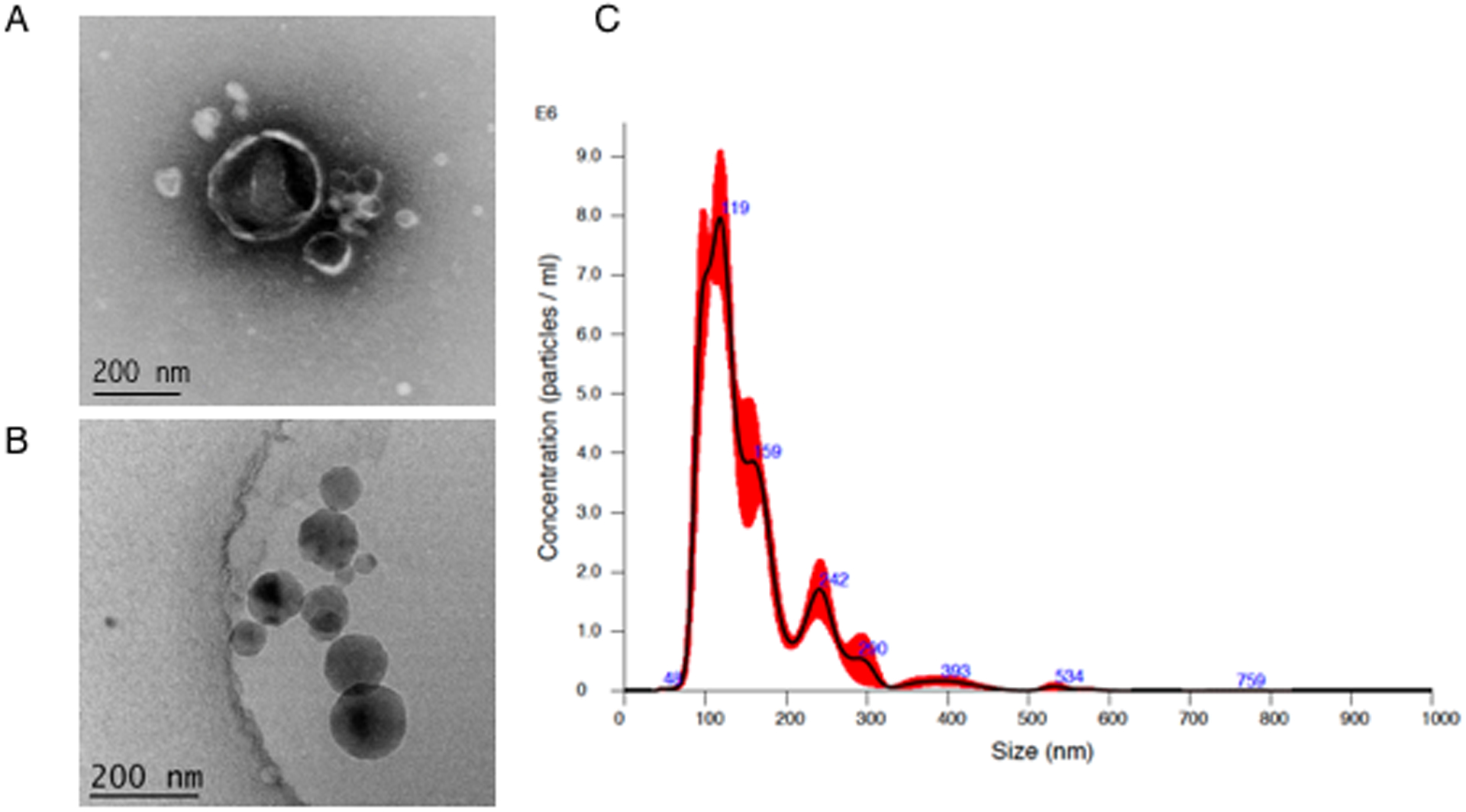
EVs from P.a. were isolated as described in methods. For visualization, EVs were negatively stained for TEM (A) or flash frozen for cryo-electron microscopy (B). Nanoparticle tracking analysis via NanoSight NS300 was used for EV particle counting and sizing (C).
A proteomics analysis was used to further characterize the P. aeruginosa EVs used in this study. Mass spectrometry was performed by MS Bio-Works, Ann Arbor, MI. A total of 26 proteins were detected by mass spectrometry, including numerous outer membrane proteins such as Elastase, LasA, OmpF, OmpH, OsmE, PagL, and PA3309, as well as extracellular and cytosolic P. aeruginosa proteins. A complete list of the proteins identified, their accession numbers, associated genes and compartmental locations are shown in Table I.
Table 1.
26 Proteins Identified by Mass Spectrometry from Pseudomonas aeruginosa EVs
| Protein | Accession Number |
Molecular Weight (kD) |
Gene | Protein Compartmental Location |
|---|---|---|---|---|
| Elastase | P14756|ELAS_PSEAE | 54 | lasB | Extracellular |
| B-type flagellin | P72151|FLICB_PSEAE | 49 | fliC | Extracellular |
| Chitin-binding protein | Q9I589|CBPD_PSEAE | 42 | cbpD | Extracellular |
| Outer membrane porin | P13794|PORF_PSEAE | 38 | oprF | Outer Membrane Vesicle |
| PhoP/Q outer membrane protein H1 | G3XD11|G3XD11_PSEAE | 22 | oprH | Outer Membrane Vesicle |
| Protease Las A | P14789|LASA_PSEAE | 46 | lasA | Extracellular |
| Elongation Factor Tu | P09591|EFTU_PSEAE | 43 | tufA | Cytosol |
| FigM | Q9HYP5|Q9HYP5_PSEAE | 11 | flgM | |
| Lipid A deacylase | Q9HVD1|PAGL_PSEAE | 18 | pagL | Outer Membrane Vesicle |
| Thioredoxin | Q9X2T1|THIO_PSEAE | 12 | trxA | Cytosol |
| Osmotically inducible lipoprotein OsmE | Q9HUT7|Q9HUT7_PSEAE | 13 | osmE | Outer Membrane |
| Uncharacterized protein | Q9HWS1|Q9HWS1_PSEAE | 16 | PA4107 | |
| Uncharacterized protein | Q9HZG8|Q9HZG8_PSEAE | 12 | PA3040 | |
| Chaperone protein DnaK | Q9HV43|DNAK_PSEAE | 68 | dnaK | Cytosol |
| Arginine/ornithine binding protein | G3XD47|G3XD47_PSEAE | 28 | aotJ | Extracellular |
| B-type flagellar hook-associated protein | Q9K3C5|FLID2_PSEAE | 49 | fliD | Extracellular |
| Probable peroxidase | Q9HY81|Q9HY81_PSEAE | 22 | PA3529 | |
| DNA-binding protein HU | P05384IDBHB_PSEAE | 9 | hupB | Cytosol |
| Usp domain-containing protein | Q9HYT5|Q9HYT5_PSEAE | 16 | PA3309 | Outer Membrane Vesicle |
| OmpA-like domain-containing protein | Q9I5A7|Q9I5A7_PSEAE | 25 | PA0833 | |
| Lysyl endopeptidase | Q9HWK6|LYSC_PSEAE | 48 | prpL | Extracellular |
| Uncharacterized protein | Q9I4N7|Q9I4N7_PSEAE | 13 | PA1093 | |
| Uncharacterized protein | Q9I384|Q9I384_PSEAE | 10 | PA1641 | |
| 4-hydroxyphenylpyruvate dioxygenase | Q9I576|HPPD_PSEAE | 40 | hpd | |
| Alkyl hydroperoxide reductase C | Q9I6Z3|Q9I6Z3_PSEAE | 21 | ahpC | Cytosol |
| Probable cold-shock protein | Q9I4H8|Q9I4H8_PSEAE | 8 | PA1159 | Cytosol |
Cytokine/chemokine and MHC gene expression
Gene expression of numerous cytokines and chemokines were altered in lung macrophages following 48-hour treatment with P.a. EVs (Table II). A number of pro-inflammatory molecules such as: IL1β, IL8, CXCL1, were notably higher, while two anti-inflammatory genes IL6 and IL10 were also higher. CCL18 and CCL23, T and B cell chemo-attractants were conspicuously lower.
Table II.
Top 20 Cytokines/Chemokines Altered in Lung Macrophages* With Pseudomonas aeruginosa EV-Treatment
| Gene | Normalized Read Counts** (+/− SD) | |
|---|---|---|
| Untreated | EV-treated (48 Hr) | |
| IL1α | 170 (80) | 3661 (960) |
| IL1β | 370 (248) | 27130 (11310) |
| IL6 | 3.3 (4.3) | 1810 (1230) |
| IL8 | 1502 (1793) | 102843 (43186) |
| IL10 | 1.6 (0.6) | 69 (44) |
| IL19 | 1.0 (0.1) | 679 (386) |
| TGFβ1 | 338 (33) | 388 (74) |
| TNF | 37 (21) | 357 (153) |
| CCL2 (MCP-1) | 48 (29) | 3861 (1882) |
| CCL3 (MIP-1α) | 3653 (452) | 10822 (6354) |
| CCL4 (MIP-1β) | 183 (246) | 4136 (2493) |
| CCL7 (MCP-3) | 4.6 (4.2) | 226.5 (144.3) |
| CCL18 (MIP-4) | 14051.3 (10181.1) | 5355.1 (4644.1) |
| CCL20 (MIP-3α) | 152.8 (221.1) | 8081.9 (4137.4) |
| CCL23 (MIP-3) | 270.6 (195.8) | 71.5 (34.2) |
| CXCL1 (GROα) | 83.6 (138.9) | 31247.4 (18633.5) |
| CXCL2 (GROβ) | 146.0 (92.1) | 5344.8 (4184.8) |
| CXCL3 (GROγ) | 290.9 (276.0) | 16089.7 (8477.7) |
| CXCL5 (ENA-78) | 1765.1 (2118.8) | 40783.8 (6306.1) |
| CXCL16 (SRPSOX) | 1325.7 (1792.7) | 2589.1 (541.1) |
n=7 subjects
NanoString PanCancer/Immunology gene expression assay
Interestingly, P.a. EVs had a considerable suppressive effect on major histocompatibility molecule gene expression. Multiple MHC Class II genes, in all subjects, were down-regulated after 48 hours of P.a. EV treatment (Fig.2), including HLA-DRA, HLA-DPA1, HLA-DPB1 (P < 0.0001), HLA-DRB3, HLA-DMB (P < 0.001), and HLA-DRB4 (P < 0.01). Additionally, gene expression of MHC Class II-related molecules CD74 (HLA-DR antigen-associated invariant chain), CTSS (Cathepsin S) (P < 0.0001) and CD9 (Tetraspanin-29) (P < 0.001) were suppressed following 48-hour P.a. EV treatment. Furthermore, two MHC Class I molecules, HLA-A and HLA-B (P < 0.001), were suppressed as well under our infection model experimental conditions (Supplemental Fig.1). A full list of the 310 differentially expressed genes following 48 hour P.a. EV-treatment is presented in Supplemental Table I.
Fig 2. MHC Class II and Class II-related gene expression is suppressed in lung macrophages following P.a. EV-treatment.
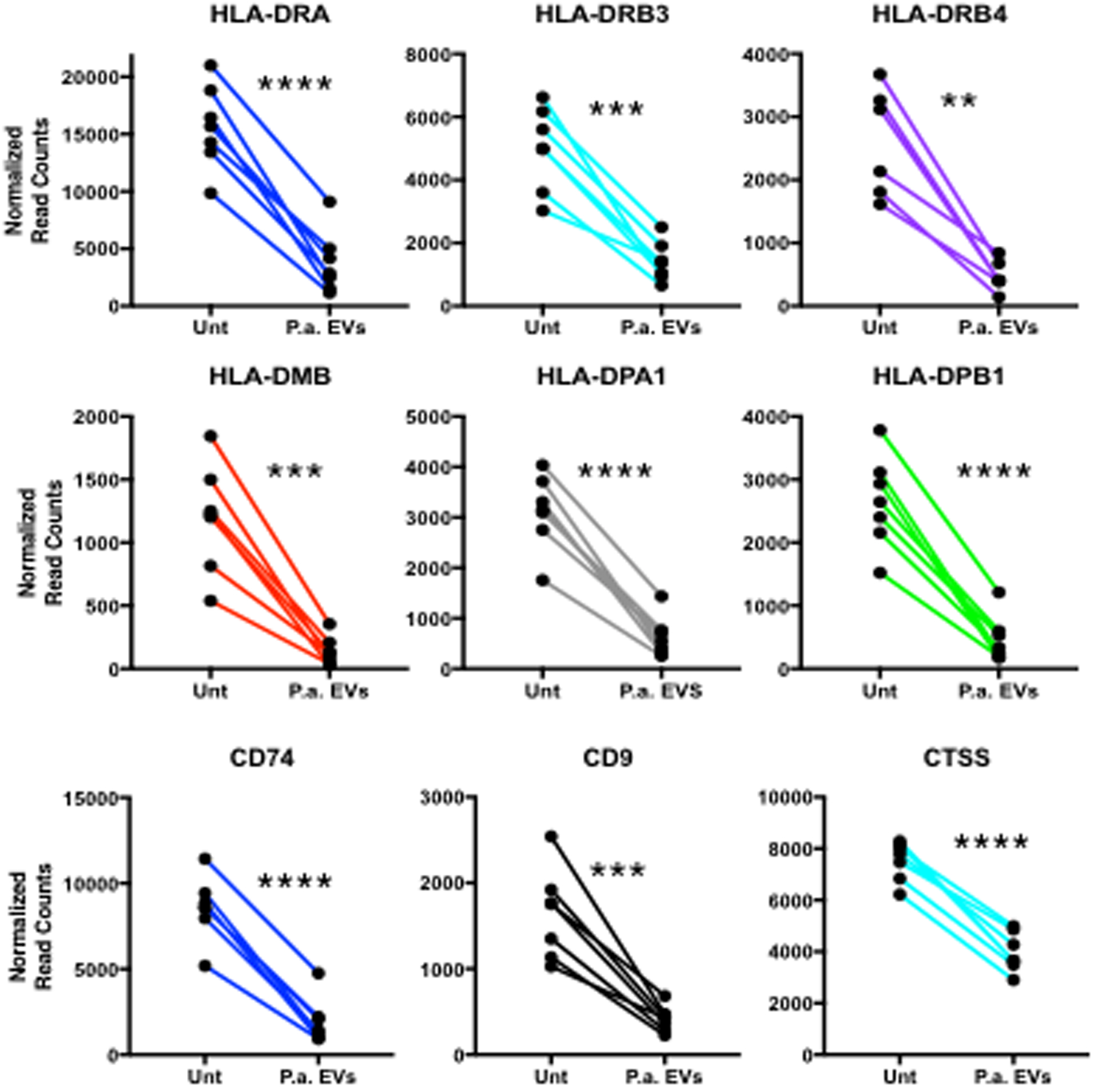
HLA-Class II subunits -DRA and -DRB, along with paralogues -DP and enzyme -DM show decreased gene expression in all subjects (n=7). Additionally, gene expression is suppressed for class II-related molecules CD74, CD9 and cathepsin L, 48 hours post-EV-treatment in lung macrophages.
** P < 0.01, *** P < 0.001, **** P < 0.0001.
To examine the potential contribution of LPS alone, which is contained on the surface of EVs, we conducted additional experiments using Pseudomonas-derived LPS in vitro. Interestingly, the effects of P.a. EVs on lung macrophages noted above were very similar to those effects seen in in vitro experiments with Pseudomonas-derived LPS alone shown in Table III. Alterations of cytokine / chemokine gene expression as well as and decreases MHC-related molecules occurred with 48-hour LPS treatment (1000ng/ml), indicating that LPS contained either on or within P.a. EVs was responsible for much, if not all, of these gene expression changes, therefore establishing the relevance of testing an infection model with EVs as a diffusible virulence agent.
Table III.
Gene Expression Changes in Lung Macrophages With P. aeruginosa EV-Treatment vs LPS Only*
| Gene | Normalized Read Counts | |||
|---|---|---|---|---|
| P. aeruginosa EVs | LPS Only | |||
| Untreated | EV-treated | Untreated | LPS-treated | |
| CCL3 | 362 | 10822 | 262 | 11336 |
| CCL3L1 | 436 | 13778 | 238 | 15346 |
| CCL4 | 183 | 4136 | 81 | 4608 |
| CCL18 | 14051 | 5355 | 6762 | 3409 |
| CD14 | 788 | 7891 | 720 | 6187 |
| CD44 | 2426 | 5581 | 3444 | 5774 |
| CD74 | 8574 | 1947 | 6127 | 1240 |
| CD81 | 6882 | 3512 | 6359 | 3368 |
| CTSS | 7537 | 3974 | 6103 | 3260 |
| CXCL2 | 146 | 5345 | 60 | 3764 |
| CXCL3 | 291 | 16090 | 74 | 13847 |
| CXCL5 | 1765 | 40784 | 708 | 39122 |
| HLA-A | 4564 | 2287 | 3204 | 2086 |
| HLA-B | 4015 | 2351 | 4153 | 2823 |
| HLA-DRA | 15646 | 3758 | 11216 | 2430 |
| HLA-DRB3 | 5002 | 1413 | 3820 | 925 |
| IL1β | 370 | 27129 | 176 | 22714 |
| IL1RN | 229 | 1086 | 463 | 2502 |
| IL8 | 1502 | 102843 | 766 | 67858 |
| LAMP1 | 2631 | 1727 | 3707 | 2095 |
| LGALS3 | 5035 | 1840 | 4480 | 1890 |
| MARCO | 1912 | 652 | 880 | 152 |
| MRC1 | 1046 | 1652 | 669 | 2276 |
| S100A8 | 407 | 2492 | 342 | 2638 |
| SH2D1B | 765 | 18478 | 347 | 17515 |
P. aeruginosa-derived LPS (1000ng/ml) 48 Hours
n=2
MHC-related protein levels in lung macrophages
As an independent validation of the gene expression results, we next measured protein levels in lung macrophages untreated or exposed to P.a. EVs for 24 and 48 hours. Lung macrophages from an additional four subjects were used in this portion of the study with representative results presented in Fig.3. Total cellular protein of CD74 and CD9 were partially reduced at 24 hours of EV-treatment and further reduced at the 48 hour time point. The MHC molecule HLA-DRA was increased at 24 hours but modestly decreased by 48 hours. However, it’s not particularly surprising to observe only modest decreases in MHC- DRA and -DRB protein at 48 hours given its half-life of 20–50 hours on human cells (44, 45) and that others have noted cell surface stabilization of MHC molecules with TLR ligand engagement (46). As additional confirmation we show both gene expression and protein analysis for CD14, a molecule that was significantly upregulated in our infection model (Supplemental Fig 2).
Fig 3. Protein levels of MHC-related molecules decrease in lung macrophages exposed to Pseudomonas aeruginosa EVs for up to 48 hours.
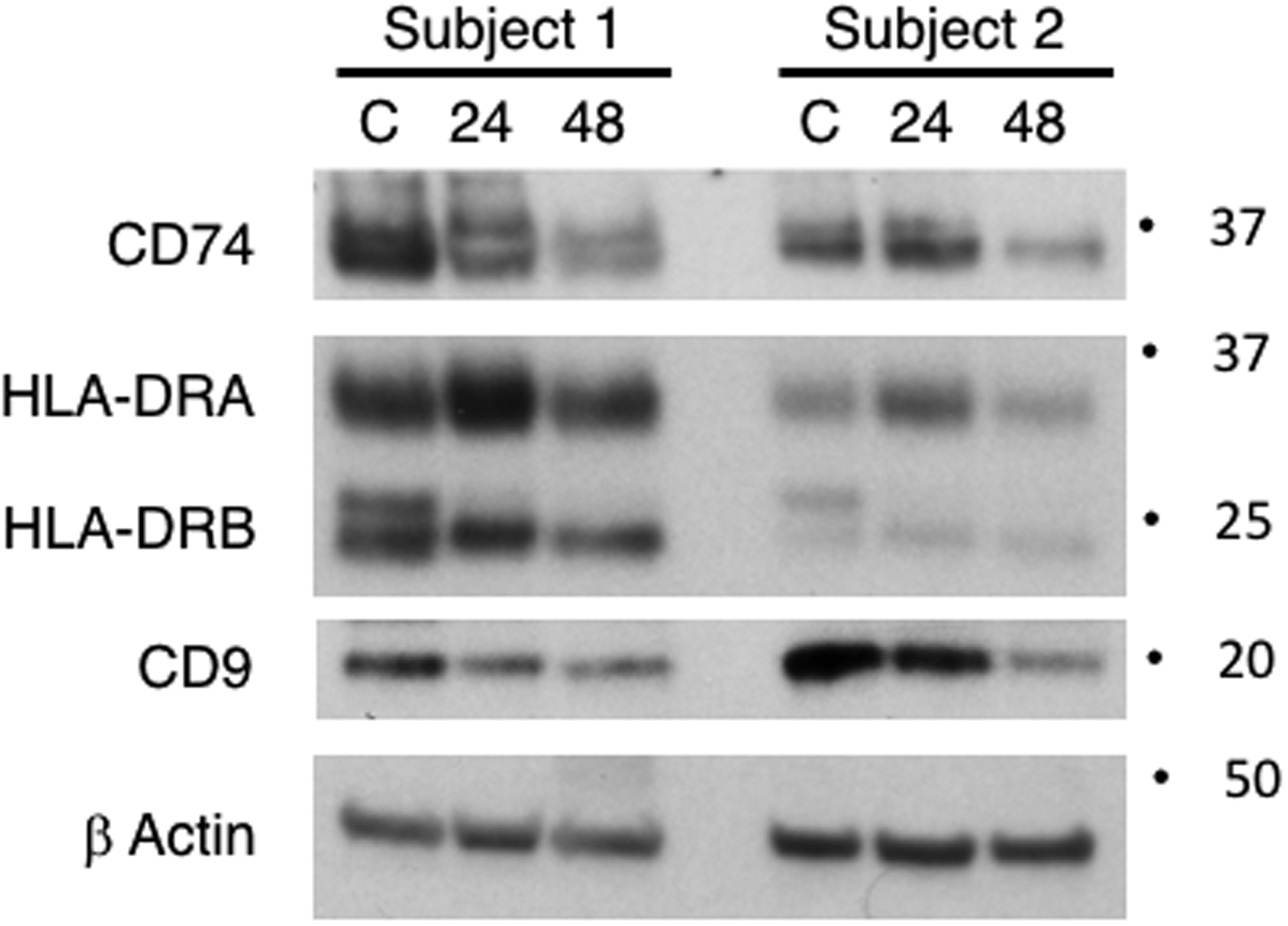
MHC class II-related proteins CD74 and CD9 decrease after 48 hours, however show some variability at the 24 hour time point. HLA-DR subunits were modestly diminished at 48 hours post P.a. EV-treatment. TLR engagement has been reported to partly stabilize HLA-DR surface molecules, potentially leading to a modest depletion at the protein level. A complement of four additional subjects were used for cellular protein studies (see methods section). Responses were similar in 3 of 4 subjects.
To explore the possible cellular mechanism of widespread reduction of MHC-related molecules in our infection model, we leveraged genome-wide DNA methylation data from the Illumina EPIC DNA methylation array and focused on regulatory regions associated with MHC molecules. P.a. EVs result in altered DNA methylation in lung macrophage MHC genes. We observed significant hypomethylation associated with P. a. EV treatment at three CpG sites in exon 8 of complement factor B gene, a MHC class III gene. Complement factor B (CFB) gene body hypomethylation related with P.a. EV treatment ranged from 5.1% – 18.8% (Table IV). These differentially methylated CpGs in the gene body of CFB are situated between MHC class I and II regions (Supplemental Fig 3). We next tested the correlation between their methylation level and expression of MHC genes. We observed a strong correlation between DNA methylation of the CFB gene body CpGs and gene expression of both HLA-DRA (Fig.4 A–C) and HLA-DRB3 (Fig.4 D–F). A detailed correlation matrix heat map of all CFB CpGs on the EPIC array and all MHC genes on the NanoString Cancer/Immune gene expression panel is shown in Fig.5, with the exon 8 containing CpGs highlighted.
Table IV.
Chromosomal Location of HLA genes and Complement Factor B (CFB) Hypomethylated CpGs
| Gene Name/EPIC cg ID | Class | Gene/CpG Location | Hypomethylation Change (%) |
|---|---|---|---|
| HLA-A | I | chr6: 29,881,755 – 30,002,076 | |
| HLA-B | I | chr6: 31,259,328 – 31,347,305 | |
| CFB | III | chr6: 31,945,650 – 31,952,084 | |
| CFB (cg12124018) | III | chr6: 31,948,906 | −15.8 |
| CFB (cg21606287) | III | chr6: 31,948,928 | −18.8 |
| CFB (cg01143314) | III | chr6: 31,948,934 | − 5.1 |
| HLA-DRA | II | chr6: 32,414,554 – 32,419,764 | |
| HLA-DRB3 | II | chr6: 32,449,765 – 32,462,852 | |
| HLA-DMB | II | chr6: 33,025,327 – 33,043,825 |
Fig 4. CFB Exon 8 CpGs methylation shows strong correlation to HLA-DRA and HLA-DRB3 gene expression.
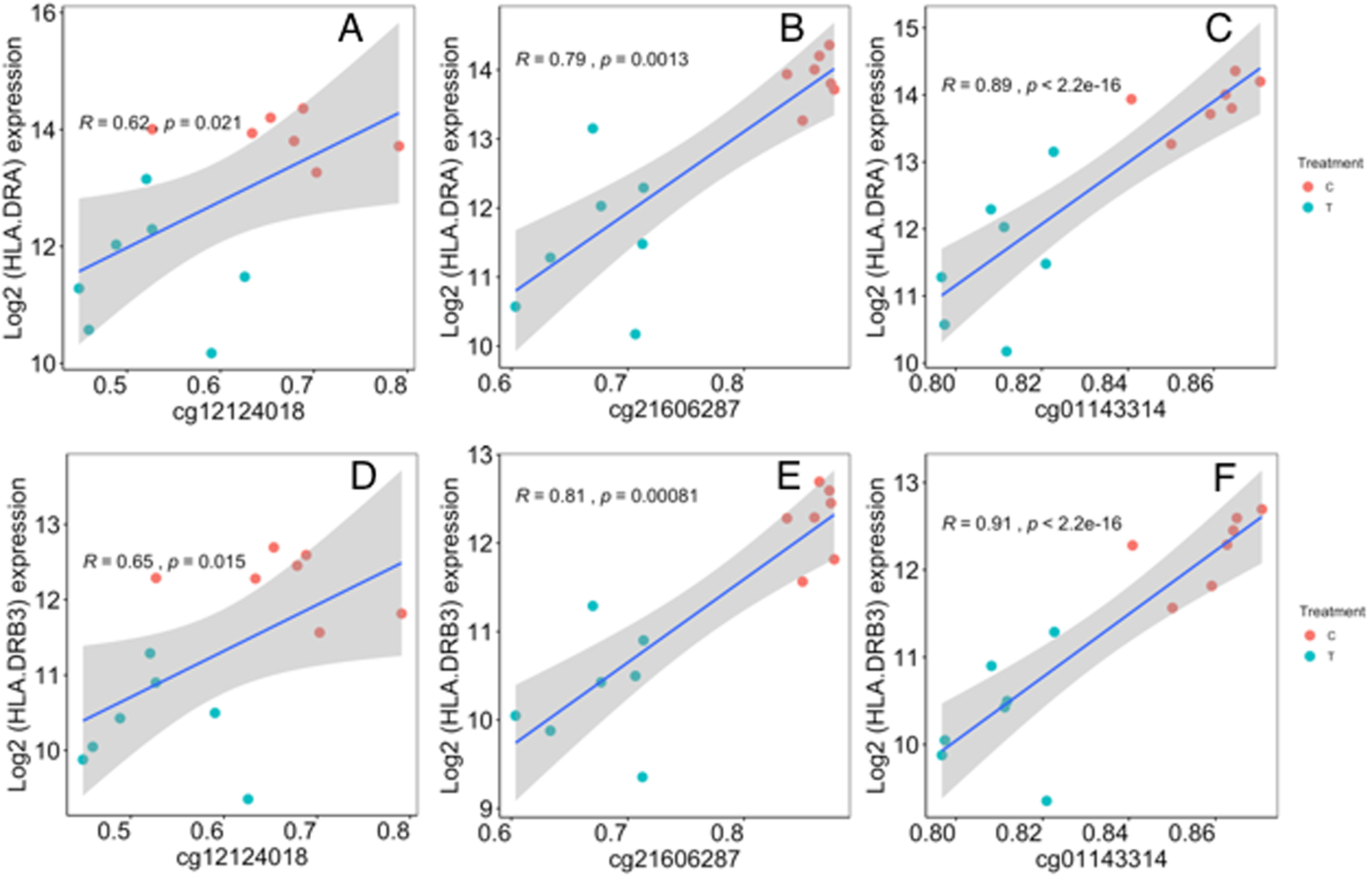
Correlation coefficients and linear regression p values were determined for CpGs in exon 8 (cg12124018, cg21606287, cg01143314) associated with gene expression of HLA-DRA (Figure 6A-C) and HLA-DRB3 (Figure 6D-F). Correlations were calculated using the Spearman rank correlation method. Linear regression lines and its 95% confidence intervals is indicated by the blue lines and gray bands, respectively.
Figure 5. Correlation plot of CFB-associated CpG methylation and MHC- related gene expression.
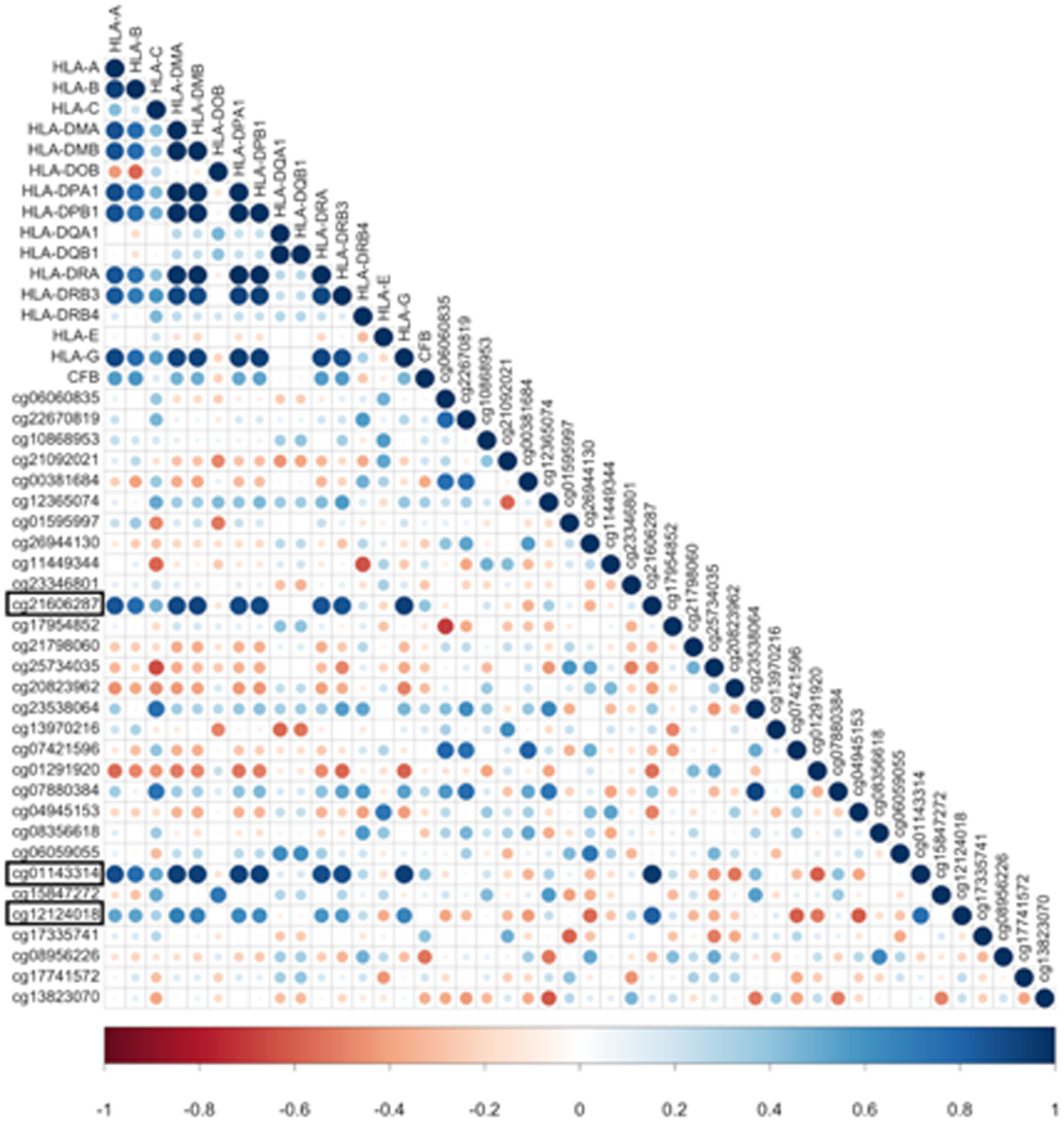
Strong correlation (R > 0.55, Spearman rank correlation method) exists between methylation of CFB CpGs (cg21606287, cg01143314, cg12124018) and gene expression of multiple MHC-related genes. Hypomethylated CFB CpGs in exon 8 are highlighted in black rectangle. Size of the circles indicates the absolute value of corresponding correlation coefficients. Color indicates the direction of correlation from strong negative correlation (dark red, correlation coefficient=−1) to strong positive correlation (dark blue, correlation coefficient= 1).
Discussion
The goal of this study was to determine if EVs - lipid bilayer vesicles secreted from P. aeruginosa, could alter lung macrophage gene expression, specifically at MHC genes associated with antigen presentation and pathogen clearance. We have used human lung macrophages and P.a. EVs as an in vitro model for lung infection, simulating how microbial EVs may diffuse some distance from their cell of origin, potentially affecting innate immune cells not in direct physical contact with individual microbes at sites of bacterial colonization in the lung. Our results demonstrate that when macrophages are exposed to P.a. EVs, MHC-related genes and protein levels are suppressed. Additionally, we observed hypomethylation of a DNA regulatory region situated directly between MHC class I and class II genes within complement factor B (CFB) gene body. Hypomethylation of three closely situated CpGs within the CFB gene showed strong correlation with decreased MHC gene expression, hence the potential driving mechanism of MHC Class II mRNA suppression.
In cystic fibrosis, lung macrophages have been shown to exhibit decreased phagocytosis and defective bacterial clearance (21, 22, 47). This phenotype is likely influenced by both intrinsic (i.e. CFTR expression) (48) and extrinsic factors (i.e. polymicrobial environment) (49, 50). A number of recent studies have examined how virulence and other pathogen-derived factors as well as whole extracellular vesicles can affect host cells in a pathogen-host interaction. Milillo et al. (51) showed that RNA from Brucella abortus contributes to inhibit MHC-II-restricted antigen presentation in macrophages. Additionally, Bomberger et al. (10) showed that P. aeruginosa virulence factor CFTR inhibitory factor (Cif) inhibits TAP function and MHC class I antigen presentation in epithelial cells. Also, Park et al. were one of the first studies to show that outer membrane vesicles or EVs from P. aeruginosa could cause pulmonary inflammation in vivo in the absence of whole bacteria (52). Finally, Cecil et al. (38) demonstrated that EVs from oral pathogens: P. gingivalis, T. denticola and T. forsythia have significant immunomodulatory effects on monocytes and macrophages.
In this report we demonstrate for the first time that EVs from a common CF pathogen, P. aeruginosa, suppress expression of MHC-related molecules in human lung macrophages. Nine different MHC class II-related genes/molecules including: HLA-DRA, -DRBs, -DMB, -DPs (chromosome 6), CD74 (chr5), CD9 (chr1) and CTSS (chr12) are diminished with P.a. EV-exposure. HLA-DRA and -DRB are the heterodimeric subunits of the MHC II complex that bind processed antigenic fragments within the phagolysosome (27). HLA-DMB is the enzyme that removes CLIP from the HLA-DR cleft so that the antigenic fragments can bind (27). HLA-DP are class II beta chain paralogues. CD74, also called Ii, is the class II invariant chain that initially blocks the MHC II cleft in the antigen-processing compartment or phagolysosome (27). CD9 regulates MHC II trafficking within the cell (53). CTSS is cathepsin S, a lysosomal cysteine protease that degrades CD74 and pathogen peptides within the phagolysosome (27). All of these molecules are intimately involved in MHC class II antigen processing and presentation to CD4+ T cells as a bridge between innate and adaptive immunity during a pathogen infection.
Additionally, we asked if the MHC-related effects seen may be attributed to LPS alone or some other yet to be identified virulence factor within EVs. Interestingly, experiments with LPS alone demonstrated that lipopolysaccharide itself was responsible for much, if not all, of these gene expression changes in lung macrophages in our infection model.
The response we are observing has some of the hallmarks of endotoxin tolerance, however, other aspects, specifically: IL1β and MHC molecules, are not consistent with tolerance. Studies on the mechanism of endotoxin tolerance have long history but are still in constant progress. However, the exact mechanism of endotoxin tolerance is still elusive and many questions remain to be answered (54). In our infection model it may be that human lung macrophages contain cell specific subsets of tolerizeable genes and non-tolerizeable genes with different behaviors after endotoxin stimulation. What we are observing may be a combination of an active host response (i.e. endotoxin tolerance) and an active pathogen-derived process to circumvent the host’s immune response. Future studies should be directed toward answering this question.
Furthermore, we have identified via genome-wide DNA methylation analysis, 3 specific CpGs on chromosome 6 within the complement factor B (CFB) gene, situated directly between MHC class I and MHC class II regions that may serve as a DNA regulatory region for MHC molecule gene expression. The CpGs of CFB are located approximately 500 kb from the class II HLA-DRA gene and 700 kb from the class I HLA-B gene. Hypomethylation (range 5.1 – 18.8%) of these CpG sites is strongly correlated with gene expression of multiple MHC genes.
Over 200 MHC molecules are clustered in a 3 MB region of chromosome 6 in humans. Multiple DNA regulatory regions (i.e. enhancers, silencers, DHS regions) may be involved in MHC molecule co-regulation (55). One type of regulatory element, enhancers, do not always act upon the closest gene promoter but can bypass neighboring genes to regulate genes located more distantly along a chromosome (56). And in some cases, individual enhancers have been found to regulate multiple genes (57). One of the most extreme examples is the location of a DNA regulatory element, in this case an enhancer, is located 1 Mb upstream from the Sonic Hedgehog SHH gene, within an intron of the unrelated LMBR1 gene (58, 59). We believe CFB gene methylation could be another example of a distal DNA regulatory element located in a neighboring gene (gene A i.e. CFB), where hypomethylation in the gene CFB results in transcriptional repression of gene B (i.e. MHC genes).
In addition to the MHC genes on chromosome 6, other MHC-related genes such as cathepsin S and CD74 are located on alternate chromosomes (chromosomes 1 & 5 respectively). This suggests that P.a. EVs possess some factor(s) that specifically target MHC molecule suppression regardless of their chromosomal location as an immune evasion strategy. The exact mechanism of gene regulation across multiple differing chromosomes remains to be determined and should be the focus of future investigations.
This study has a number of strengths, however, this work also has a number of limitations. We cannot rule out that virulence factors or other yet unidentified molecules within P. aeruginosa EVs beyond LPS, may be contributing to the effects we are seeing in our infection model. Additionally, it’s not currently known if the response seen is an active host response (i.e. tolerance), an operative pathogen process to circumvent host defenses or some combination of both.
In conclusion, in our infection model, we have demonstrated the immunosuppressive potential of P. aeruginosa EVs on lung macrophages. These effects appear to be largely attributable to LPS contained on or within EVs and may be contingent upon the diffusible nature of EVs. In the polymicrobial community of a CF lung infection, the local milieu is significantly more complicated than presented in our model, but, based on the observations demonstrated here, we suggest that the down regulation of MHC molecules by P.a. EVs may be contributing, along with other extrinsic factors from P.a. and additional pathogens, to decreased antigen presentation and pathogen clearance in lung infection.
Supplementary Material
Funding:
This work is supported by the NIH R01HL122372 (to A.A.) and NIH R01CA216265 (to B.C.C). Subject enrollment and clinical sample collection were achieved with the assistance of the CF Translational Research Core at Dartmouth, which is jointly funded by the NIH (P30DK117469 to Dean Madden) and CFF (STANTO19R0 to Bruce Stanton). The Electron Microscopy Core Facility at the University of Massachusetts Medical School is funded in part by Award Number (S10 OD025113-01) from the NIH, National Center for Research Resources.
“The funders (Madden and Stanton) had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.”
Abbreviations:
- BAL
Bronchoalveolar lavage
- CF
Cystic fibrosis
- CFB
complement factor B
- EV
Extracellular vesicle
- LM
Lung Macrophage
- LPS
Lipopolysaccharide
- MHC
major histocompatibility complex
- NTA
Nanoparticle tracking analysis
References
- 1.O’Sullivan BP, and Freedman SD. 2009. Cystic fibrosis. Lancet 373: 1891–1904. [DOI] [PubMed] [Google Scholar]
- 2.Foundation CF 2018. Cystic Fibrosis Foundation Patient Registry In 2017 Annual Data Report, Bethesda, MD [Google Scholar]
- 3.Elborn JS, and Shale DJ. 1990. Cystic fibrosis. 2. Lung injury in cystic fibrosis. Thorax 45: 970–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elborn JS 2016. Cystic fibrosis. Lancet 388: 2519–2531. [DOI] [PubMed] [Google Scholar]
- 5.Gilligan PH 1991. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev 4: 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malhotra S, Hayes D Jr., and Wozniak DJ. 2019. Cystic Fibrosis and Pseudomonas aeruginosa: the Host-Microbe Interface. Clin Microbiol Rev 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briard B, Mislin GLA, Latge JP, and Beauvais A. 2019. Interactions between Aspergillus fumigatus and Pulmonary Bacteria: Current State of the Field, New Data, and Future Perspective. J Fungi (Basel) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ, Ranganathan SC, Waters VJ, and Ratjen FA. 2014. Antibiotic management of lung infections in cystic fibrosis. II. Nontuberculous mycobacteria, anaerobic bacteria, and fungi. Ann Am Thorac Soc 11: 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeppen K, Barnaby R, Jackson AA, Gerber SA, Hogan DA, and Stanton BA. 2019. Tobramycin reduces key virulence determinants in the proteome of Pseudomonas aeruginosa outer membrane vesicles. PLoS One 14: e0211290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bomberger JM, Ely KH, Bangia N, Ye S, Green KA, Green WR, Enelow RI, and Stanton BA. 2014. Pseudomonas aeruginosa Cif protein enhances the ubiquitination and proteasomal degradation of the transporter associated with antigen processing (TAP) and reduces major histocompatibility complex (MHC) class I antigen presentation. J Biol Chem 289: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O’Toole GA, and Stanton BA. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 5: e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis TN, and Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, and Stanton BA. 2016. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog 12: e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renelli M, Matias V, Lo RY, and Beveridge TJ. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150: 2161–2169. [DOI] [PubMed] [Google Scholar]
- 15.Buzas EI, Gyorgy B, Nagy G, Falus A, and Gay S. 2014. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 10: 356–364. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, Comstock LE, Gandhi R, and Weiner HL. 2016. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 19: 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Properzi F, Logozzi M, and Fais S. 2013. Exosomes: the future of biomarkers in medicine. Biomark Med 7: 769–778. [DOI] [PubMed] [Google Scholar]
- 18.Tsatsaronis JA, Franch-Arroyo S, Resch U, and Charpentier E. 2018. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol 26: 401–410. [DOI] [PubMed] [Google Scholar]
- 19.Cecil JD, Sirisaengtaksin N, O’Brien-Simpson NM, and Krachler AM. 2019. Outer Membrane Vesicle-Host Cell Interactions. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Jiang X, Bao J, Wang Y, Liu H, and Tang L. 2018. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front Immunol 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruscia EM, and Bonfield TL. 2016. Innate and Adaptive Immunity in Cystic Fibrosis. Clin Chest Med 37: 17–29. [DOI] [PubMed] [Google Scholar]
- 22.Bruscia EM, and Bonfield TL. 2016. Cystic Fibrosis Lung Immunity: The Role of the Macrophage. J Innate Immun 8: 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing Z, Afkhami S, Bavananthasivam J, Fritz DK, D’Agostino MR, Vaseghi-Shanjani M, Yao Y, and Jeyanathan M. 2020. Innate immune memory of tissue-resident macrophages and trained innate immunity: Re-vamping vaccine concept and strategies. J Leukoc Biol. [DOI] [PubMed] [Google Scholar]
- 24.Hirayama D, Iida T, and Nakase H. 2017. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi G, Cavazza A, Spagnolo P, Bellafiore S, Kuhn E, Carassai P, Caramanico L, Montanari G, Cappiello G, Andreani A, Bono F, and Nannini N. 2017. The role of macrophages in interstitial lung diseases: Number 3 in the Series “Pathology for the clinician” Edited by Peter Dorfmuller and Alberto Cavazza. Eur Respir Rev 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra S, Hayes D Jr., and Wozniak DJ. 2019. Mucoid Pseudomonas aeruginosa and regional inflammation in the cystic fibrosis lung. J Cyst Fibros. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche PA, and Furuta K. 2015. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol 15: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tortorella D, Gewurz BE, Furman MH, Schust DJ, and Ploegh HL. 2000. Viral subversion of the immune system. Annu Rev Immunol 18: 861–926. [DOI] [PubMed] [Google Scholar]
- 29.Antonia AL, Gibbs KD, Trahair ED, Pittman KJ, Martin AT, Schott BH, Smith JS, Rajagopal S, Thompson JW, Reinhardt RL, and Ko DC. 2019. Pathogen Evasion of Chemokine Response Through Suppression of CXCL10. Front Cell Infect Microbiol 9: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srisatjaluk R, Kotwal GJ, Hunt LA, and Justus DE. 2002. Modulation of gamma interferon-induced major histocompatibility complex class II gene expression by Porphyromonas gingivalis membrane vesicles. Infect Immun 70: 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bessich JL, Nymon AB, Moulton LA, Dorman D, and Ashare A. 2013. Low levels of insulin-like growth factor-1 contribute to alveolar macrophage dysfunction in cystic fibrosis. J Immunol 191: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Armstrong DA, Salas LA, Hazlett HF, Nymon AB, Dessaint JA, Aridgides DS, Mellinger DL, Liu X, Christensen BC, and Ashare A. 2018. Genome-wide DNA methylation profiling shows a distinct epigenetic signature associated with lung macrophages in cystic fibrosis. Clin Epigenetics 10: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung MK, and Mun JY. 2018. Sample Preparation and Imaging of Exosomes by Transmission Electron Microscopy. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oosthuyzen W, Sime NE, Ivy JR, Turtle EJ, Street JM, Pound J, Bath LE, Webb DJ, Gregory CD, Bailey MA, and Dear JW. 2013. Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol 591: 5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nemec SF, Bankier AA, and Eisenberg RL. 2013. Upper lobe-predominant diseases of the lung. AJR Am J Roentgenol 200: W222–237. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong DA, Chen Y, Dessaint JA, Aridgides DS, Channon JY, Mellinger DL, Christensen BC, and Ashare A. 2019. DNA Methylation Changes in Regional Lung Macrophages Are Associated with Metabolic Differences. Immunohorizons 3: 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong DA, Nymon AB, Ringelberg CS, Lesseur C, Hazlett HF, Howard L, Marsit CJ, and Ashare A. 2017. Pulmonary microRNA profiling: implications in upper lobe predominant lung disease. Clin Epigenetics 9: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cecil JD, O’Brien-Simpson NM, Lenzo JC, Holden JA, Singleton W, Perez-Gonzalez A, Mansell A, and Reynolds EC. 2017. Outer Membrane Vesicles Prime and Activate Macrophage Inflammasomes and Cytokine Secretion In Vitro and In Vivo. Front Immunol 8: 1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kling T, Wenger A, Beck S, and Caren H. 2017. Validation of the MethylationEPIC BeadChip for fresh-frozen and formalin-fixed paraffin-embedded tumours. Clin Epigenetics 9: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, and Irizarry RA. 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W, Laird PW, and Shen H. 2017. Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res 45: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, McCarthy DJ, and Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanzavecchia A, Reid PA, and Watts C. 1992. Irreversible association of peptides with class II MHC molecules in living cells. Nature 357: 249–252. [DOI] [PubMed] [Google Scholar]
- 45.Muller KP, Schumacher J, and Kyewski BA. 1993. Half-life of antigen/major histocompatibility complex class II complexes in vivo: intra- and interorgan variations. Eur J Immunol 23: 3203–3207. [DOI] [PubMed] [Google Scholar]
- 46.De Gassart A, De Angelis Rigotti F, and Gatti E. 2013. MHC-II ubiquitination. Methods Mol Biol 960: 517–527. [DOI] [PubMed] [Google Scholar]
- 47.Barnaby R, Koeppen K, Nymon A, Hampton TH, Berwin B, Ashare A, and Stanton BA. 2018. Lumacaftor (VX-809) restores the ability of CF macrophages to phagocytose and kill Pseudomonas aeruginosa. Am J Physiol Lung Cell Mol Physiol 314: L432–L438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorio C, Buffelli M, Angiari C, Ettorre M, Johansson J, Vezzalini M, Viviani L, Ricciardi M, Verze G, Assael BM, and Melotti P. 2011. Defective CFTR expression and function are detectable in blood monocytes: development of a new blood test for cystic fibrosis. PLoS One 6: e22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baishya J, and Wakeman CA. 2019. Selective pressures during chronic infection drive microbial competition and cooperation. NPJ Biofilms Microbiomes 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruscia EM, Zhang PX, Ferreira E, Caputo C, Emerson JW, Tuck D, Krause DS, and Egan ME. 2009. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator−/− mice. Am J Respir Cell Mol Biol 40: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milillo MA, Trotta A, Serafino A, Marin Franco JL, Marinho FV, Alcain J, Genoula M, Balboa L, Costa Oliveira S, Giambartolomei GH, and Barrionuevo P. 2019. Bacterial RNA Contributes to the Down-Modulation of MHC-II Expression on Monocytes/Macrophages Diminishing CD4(+) T Cell Responses. Front Immunol 10: 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park KS, Lee J, Jang SC, Kim SR, Jang MH, Lotvall J, Kim YK, and Gho YS. 2013. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am J Respir Cell Mol Biol 49: 637–645. [DOI] [PubMed] [Google Scholar]
- 53.Rocha-Perugini V, Martinez Del Hoyo G, Gonzalez-Granado JM, Ramirez-Huesca M, Zorita V, Rubinstein E, Boucheix C, and Sanchez-Madrid F. 2017. CD9 Regulates Major Histocompatibility Complex Class II Trafficking in Monocyte-Derived Dendritic Cells. Mol Cell Biol 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu D, Cao S, Zhou Y, and Xiong Y. 2019. Recent advances in endotoxin tolerance. J Cell Biochem 120: 56–70. [DOI] [PubMed] [Google Scholar]
- 55.Kolovos P, Knoch TA, Grosveld FG, Cook PR, and Papantonis A. 2012. Enhancers and silencers: an integrated and simple model for their function. Epigenetics Chromatin 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, and Bejerano G. 2013. Enhancers: five essential questions. Nat Rev Genet 14: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, and Locksley RM. 2001. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol 2: 842–847. [DOI] [PubMed] [Google Scholar]
- 58.Stadhouders R, van den Heuvel A, Kolovos P, Jorna R, Leslie K, Grosveld F, and Soler E. 2012. Transcription regulation by distal enhancers: who’s in the loop? Transcription 3: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sagai T, Hosoya M, Mizushina Y, Tamura M, and Shiroishi T. 2005. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132: 797–803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


