Abstract
Background
The pathophysiology of schizophrenia, a major psychiatric disorder, remains elusive. In this study, the role of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor (RXR) families, belonging to the ligand-activated nuclear receptor superfamily, in schizophrenia, was analyzed.
Methods
The PPAR/RXR family genes were screened by exploiting molecular inversion probe (MIP)-based targeted next-generation sequencing (NGS) using the samples of 1,200 Japanese patients with schizophrenia. The results were compared with the whole-genome sequencing databases of the Japanese cohort (ToMMo) and the gnomAD. To reveal the relationship between PPAR/RXR dysfunction and schizophrenia, Ppara KO mice and fenofibrate (a clinically used PPARα agonist)-administered mice were assessed by performing behavioral, histological, and RNA-seq analyses.
Findings
Our findings indicate that c.209–2delA, His117Gln, Arg141Cys, and Arg226Trp of the PPARA gene are risk variants for schizophrenia. The c.209–2delA variant generated a premature termination codon. The three missense variants significantly decreased the activity of PPARα as a transcription factor in vitro. The Ppara KO mice exhibited schizophrenia-relevant phenotypes, including behavioral deficits and impaired synaptogenesis in the cerebral cortex. Oral administration of fenofibrate alleviated spine pathology induced by phencyclidine, an N-methyl-d-aspartate (NMDA) receptor antagonist. Furthermore, pre-treatment with fenofibrate suppressed the sensitivity of mice to another NMDA receptor antagonist, MK-801. RNA-seq analysis revealed that PPARα regulates the expression of synaptogenesis signaling pathway-related genes.
Interpretation
The findings of this study indicate that the mechanisms underlying schizophrenia pathogenesis involve PPARα-regulated transcriptional machinery and modulation of synapse physiology. Hence, PPARα can serve as a novel therapeutic target for schizophrenia.
Keywords: Molecular inversion probes (MIP), Peroxisome proliferator-activated receptor α (PPARα), Schizophrenia, Synaptogenesis, Therapeutic drug, Phencyclidine (PCP)
Research in Context.
Evidence before this study
Our recent study has revealed that dietary deprivation of polyunsaturated fatty acid (PUFA) during gestational and early postnatal stages in mice elicited schizophrenia-like behavioral phenotypes in its offspring at adulthood. Additionally, we have revealed that the nuclear receptor RXR (retinoid X receptor)/PPAR (peroxisome proliferator-activated receptor) system acts as an upstream mechanism linking PUFA deficiency and the behavioral phenotypes via controlling expressions of schizophrenia-relevant genes. Nevertheless, whether PPAR/RXR is involved in the pathophysiology of schizophrenia remains an open question.
Added value of this study
We showed that PPARA variants identified in schizophrenia significantly decreased activities of PPARα as a transcription factor when compared to that of the wild-type in vitro. Newly generated Ppara KO mice exhibited a deficit in the sensorimotor gating function of the brain, and the schizophrenia-related histological abnormalities. And RNA-seq analysis revealed that PPARα regulates the expression of synaptogenesis signaling pathway-related genes. Moreover, a treatment of mice with the PPARα agonist fenofibrate alleviated spine pathology induced by phencyclidine (PCP), a schizophrenia-mimetic drug, and reduced a sensitivity to MK-801, another hallucinogenic drug.
Implications of all the available evidence
This study indicates that the mechanisms underlying schizophrenia pathogenesis involve PPARα-regulated transcriptional machinery and modulation of synapse physiology. In addition, PPARα can serve as a novel therapeutic target for schizophrenia.
Alt-text: Unlabelled box
1. Introduction
Peroxisome proliferator-activated receptor (PPAR) and retinoid X receptor (RXR) belong to the nuclear receptor superfamily. They function as transcription factors [1] by binding to the ligands, such as fatty acids and their derivatives [2, 3]. PPARs and RXRs share a basic molecular architecture: a DNA-binding domain (DBD), a C-terminal ligand-binding domain (LBD), and two transcription activation function motifs (N-terminal ligand-independent AF-1 motif and C-terminal ligand-dependent AF-2 motif). PPARs function as a transcription factor by forming heterodimers with RXRs. The PPAR/RXR heterodimers regulate target gene expression by binding to the cis-elements called PPAR response elements (PPREs) in the promoter regions of target genes [4], [5], [6].
The PPAR/RXR complexes, which are reported to regulate lipid metabolism in the liver, are therapeutic targets for metabolic disorders, such as hyperlipidemia. These genes are also expressed in the brain [7], [8], [9] and have the following known function: 1) RXRγ accelerates remyelination [10], [11], [12], [13], 2) PPARγ mediates neuroprotection [14], [15], [16], [17], [18], 3) PPARα and PPARγ modulate synaptic plasticity [19], and 4) PPAR and RXR stimulate the proliferation and differentiation of neural cells [20], [21], [22], [23].
Multiple human studies have suggested that PPAR and RXR are implicated in mental disorders, including schizophrenia. Reduced expression of RXRA and PPARA was observed in the hair follicle cells from individuals with schizophrenia [24]. Although common variants in the PPARA, RXRA, or other PPAR/RXR genes did not show any genetic association with schizophrenia in genome-wide association studies (GWAS), the variants in PPARA and RXRA genes showed methylation quantitative trait loci (meQTL) effect in the prefrontal cortex [25]. In addition, in the postmortem brain and erythrocyte membranes of the patients with schizophrenia, the composition of polyunsaturated fatty acids (PUFAs), known as endogenous ligands, for PPAR/RXR, has been changed [26, 27]. PUFAs in the erythrocyte membranes are associated with the clinical symptoms in individuals at ultra-high risk for psychosis [28]. Furthermore, n-3 PUFA supplementation has been reported to prevent the transition to psychosis in individuals at ultra-high risk for psychosis [29, 30].
Some animal studies have indicated the potential role of PPAR and RXR genes in the pathogenesis of mental disorders. We have previously shown that the gestational and early postnatal dietary deprivation of PUFAs elicited schizophrenia-like molecular, cellular, and behavioral phenotypes in mouse offspring at adulthood [24]. These prodromal phenotypes of schizophrenia resulting from the dysregulated transcription of neurodevelopment-related genes are accompanied by reduced expression of Ppara (encoding the PPARα subtype) and Rxra (encoding the RXRα subtype) genes in the mouse cortex [24]. Moreover, Ppara-deficient mice have shown repetitive behavior and cognitive inflexibility [31], and altered sleep profiles when subjected to dietary restrictions [32]. Furthermore, the role of Ppara in hippocampal synaptogenesis also suggests its potential role in neuronal function [19, 33].
In the present study, we further tested the role of RXR/PPAR pathway in schizophrenia pathogenesis. To this end, we first resequenced the RXR/PPAR genes in 1200 Japanese individuals with schizophrenia to identify loss-of-function (LoF) variants. We identified multiple variants in PPARA and further validated them by using molecular biology approaches. To corroborate the findings in human samples, we prepared Ppara KO mice and examined the phenotypes. Finally, pharmacological manipulation of PPARα was also explored as a potential therapeutic strategy for schizophrenia using model animals.
2. Methods
2.1. Study approval
All the animal experimental protocols were approved by the Ethics Committee of RIKEN (permission number: W2019–2–044). The animal experiments were carried out in accordance with the guidelines of the National Institutes of Health for the care and use of laboratory animals. Human subject studies were approved by the Ethics Committee of RIKEN (Wako 1:15–3, Wako 3 2020–09) and experiments were conducted according to the principles of the Declaration of Helsinki.
2.2. Human DNA subjects
In total, 1200 Japanese patients with schizophrenia (657 males with a mean age of 49.1 ± 14.0 years; 543 females with a mean age of 51.1 ± 14.4 years), were used for sequencing the RXR and PPAR genes. All patients were diagnosed based on the Diagnosis and Statistical Manual of Mental Disorders IV (DSM-IV) criteria, and at least two experienced psychiatrists confirmed the diagnosis. Written informed consent (IC) was obtained from all participants after explaining our study protocols and purposes. All the subjects were recruited from the Honshu area of Japan (the main island of Japan). The patients were recruited through referrals from attending physicians.
2.3. Targeted next-generation sequencing (NGS) using molecular inversion probes (MIPs)
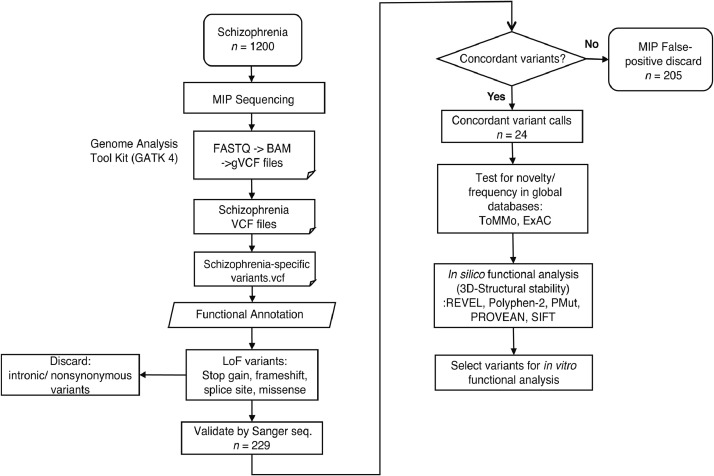
Genomic DNA was isolated from the blood samples collected from human subjects using standard methods. One hundred sixteen MIPs were designed for six genes (RXRA, RXRB, RXRG, PPARA, PPARD, and PPARG) using MIPgen [34] (http://shendurelab.github.io/MIPGEN/), which targeted the coding exons and flanking exon-intron boundaries of the genes (GRCh37 build) and covered all the known transcripts [35]. The variants identified by NGS-based sequencing were further validated using Sanger sequencing (Fig. 1, Fig. S1, Table S9). The detailed protocols are described in the Supplementary information.
Fig. 1.
Workflow for the detection of mutations using molecular inversion probes (MIPs).
2.4. Graphical three-dimensional models
Three-dimensional models of the missense variants, based on the crystal structures of the PPARγ/RXRα heterodimer (pdb: 3e00), were generated using SWISS-MODEL (http://swissmodel.expasy.org/). The detailed protocols are described in the Supplementary information.
2.5. RT-PCR analysis of hair follicle samples
The hair follicle samples were collected from ethnic Japanese participants (patient 1, woman, age 34 years; control, 3 women, mean age 45.7 ± 1.2 years). Schizophrenia was diagnosed by at least two experienced psychiatrists using the DSM-IV criteria. Total RNA isolation from the scalp hair follicle and cDNA synthesis were performed as described previously [36, 37]. PCR amplification was performed using the following primers: 5ʹ-ACTCGAGGCCGGCGATCTAG-3ʹ and 5ʹ ACTCGAGGCCGGCGATCTAG-3ʹ.
2.6. Construction of plasmids
The Human Fetal Brain Marathon-Ready cDNA (Clontech, Mountain View, CA, USA) was subjected to PCR amplification to obtain the human PPARA cDNA. The PCR amplification was performed using the following primer sets: forward, 5ʹ-CAGGTACCACCACCATGGTGGACACGGAAAGCCC-3ʹ; reverse, 5ʹ-CTGGATCCTCAGTACATGTCCCTGTAGATCTCC-3ʹ. The amplified cDNA was cloned into the mammalian expression vector pcDNA3 (Invitrogen) in which the expression of the inserted cDNA is driven by the cytomegalovirus (CMV) promoter. Conventional site-directed mutagenesis was performed to generate the five PPARα mutant (Cys122Ser, Arg128Thr, His117Gln, Arg141Cys, and Arg226Trp) constructs (Table S1). The sequences of the generated plasmids were validated using Sanger sequencing.
2.7. Cell culture and transfection
CV-1 and COS-7, cell lines derived from the kidney of a male adult African green monkey, were cultured in Dulbecco's modified Eagle's medium (DMEM) (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS) (SAFC Bioscience, Brooklyn, Australia), 100 U/mL penicillin, and 100 U/mL streptomycin at 37 °C and 5% CO2. The CV-1 and COS-7 cells were transfected using Viafect (Promega, Madison, WI, USA), following the manufacturer's instructions. For the experiments involving ligand treatment, the cells were cultured in phenol red-free DMEM (Wako) supplemented with 10% charcoal-stripped FBS (Biological Industries, Cromwell, CT, USA) for 24 h before and after transfection.
2.8. Luciferase assay
The CV-1 cells were plated on 24-well cell culture plates at a concentration of 5 × 104 cells/well for 24 h. The cells were then transiently co-transfected with 0.2 µg of Cignal PPAR Reporter construct (QIAGEN, Hilden, Germany) and 0.4 µg of pcDNA3 WT PPARα or PPARα variant (His117Gln, Arg141Cys, Arg226Trp, Cys122Ser, Arg128Thr) constructs or empty expression plasmid. The Cignal PPAR Reporter comprised an inducible PPAR-responsive firefly luciferase reporter and a constitutively-expressed Renilla luciferase construct (40:1). At 24 h post-transfection, the cells were lysed using Passive Lysis Buffer (Promega). The firefly luciferase activity relative to the Renilla luciferase activity was examined to determine the PPARα activity using the dual-luciferase reporter assay system (Promega) and Lumat luminometer (Berthold Technologies, Bad Wildbad, Germany). All the luciferase assays were performed at least in triplicate.
2.9. Immunocytochemistry
The COS-7 cells were cultured on the 8-well chamber plates at a cell density of 2.5 × 104 cells/well for 24 h. The COS-7 cells were co-transfected with 0.2 µg of pAcGFP1-C1 (Clontech) and 0.4 µg of pcDNA3-PPARα-WT or variant expression constructs with 50 µM WY14643 for 24 h. The cells were fixed with 4% paraformaldehyde in 0.01 M phosphate-buffered saline (PBS) for 15 min and washed with PBS. The fixed cells were permeabilized with 0.2% Triton X-100 and were blocked with 1% Blocking Reagent (Roche Diagnostics, Mannheim, Germany). The cells were then incubated with the primary antibody against PPARα (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature. The cells were washed with PBS and were incubated with the Alexa Fluor 594-conjugated secondary antibody. The cells were then washed with PBS and incubated with 4ʹ,6-diamidino-2-phenylindole (DAPI, Roche) for nucleus staining. Next, the cells mounted with PermaFluor Mountant (Thermo Fisher Scientific, Waltham, MA, USA), and the images were captured under an FV1000-D IX81 confocal microscope (Olympus, Tokyo, Japan). The images were analyzed using the ImageJ software (https://imagej.nih.gov/ij/).
2.10. Analysis of subnuclear localization
The fluorescence intensity of nuclei or cytoplasm was calculated using ImageJ software. PPARα signals were evaluated within the outline of the nuclei and cytoplasm in areas stained with DAPI and GFP, respectively. Then, we calculated the ratio of the fluorescence intensity (cytoplasm/nuclei).
2.11. Animals
The inbred C57BL/6CrlCrlj (B6J) and closed colony Slc:ddY (ddY) mice were purchased from Charles River Laboratories, Japan (Kanagawa, Japan) and Japan SLC, Inc (Shizuoka, Japan), respectively. The mice were housed in groups of four or five in standard cages in a temperature- and humidity-controlled room with a 12 h light-dark cycle (lights on at 8:00). Animals had free access to standard lab chow and water. No special environmental enrichment was provided in the homecage. All behavioral tests were performed using male animals between 10:00 and 18:00.
2.12. Generation of Ppara KO mouse
The Ppara-deficient mice were generated using the CRISPR/Cas9 nickase method [38], [39], [40], [41], [42]. For more details, see Supplementary information.
2.13. Behavioral analyses of Ppara KO mouse
Ppara KO and control (wild-type) littermate mice aged 2–3 months were assessed in the behavioral tests. The timeline and n per experimental factor are shown in Fig. S2.
Prepulseinhibition (PPI) test: A startle reflex measurement system (O'Hara & Co., Ltd., Tokyo, Japan) was used to measure the ASR and PPI of the startle response [43, 44], following a previously described protocol [45]. A test session was comprised of 70 trials. In each trial, a paired prepulse (0, 70, 74, 78, 82, and 86 dB[A])-pulse (115 dB[A]) stimulus or no prepulse-pulse paired stimulus was administered. The percentage PPI was calculated as follows: {[(ASR amplitude of trial without prepulse) – (ASR amplitude of trial with prepulse)]/(ASR amplitude of trial without prepulse)} × 100. PPI background noise was 65 dB. The inter-stimulus interval was 10 to 20 s (average 15 s). The time between the start of the prepulse and the start of the pulse was 100 ms. The duration of the prepulse was 20 ms. The duration of the pulse was 40 ms. The number of each trial type was 10. We performed three independent experiments and analyzed the data at once.
Marble-buryingtest: The marble-burying test was performed following a previously reported protocol [46]. Transparent plastic cages (196 × 306 × 166 mm; W × D × H) were filled with white paper bedding material (Paperclean, Japan SLC) to a depth of 4 cm. The mice were placed individually in the test cages for 30 min (habituation trial) and then returned to their home cage. Next, 20 glass marbles were evenly set on the surface of the bedding materials in the cage. The mice were then returned to the test cage for 30 min test trial. The number of marbles that were buried over two-thirds of their surface area was counted. The other protocols for behavioral tests are described in the Supplementary information.
2.14. Spine analysis
The mice were anesthetized with isoflurane and were perfused transcardially with 4% paraformaldehyde in PBS. The brains were excised and cut into coronal sections to a thickness of 200 μm using a vibratome. Lipophilic dye (DiI, Invitrogen) was coated onto the tungsten particles (1.7 μm diameter, Bio-Rad, Hercules, CA, USA). The DiI-coated particles were delivered to the slices using a Helios Gene Gun system (Bio-Rad). A polycarbonate filter (pore size: 8.0 μm) (Beckton Dickinson, Franklin Lakes, NJ, USA) was inserted between the gun and the preparation to remove clusters of large particles. The density of labeling was regulated by varying the gas pressure (95–105 psi helium) [47]. The labeled structures were imaged under a confocal microscope (FV1000-D IX81, Olympus). Four to five sections/mouse were used to count the pyramidal neurons between 5.14–5.78 mm anterior to the interaural line. Pyramidal cells were identified in their morphology and randomly selected from layer V in the medial prefrontal cortex (mPFC). The images were captured in 0.45 μm steps and stacked for 3D reconstruction using ImageJ (https://imagej.nih.gov/ij/) and Spiso3D (mathematical and automated software calculating geometrical parameters of spines) [48]. All protrusions along each apical dendrite were counted from secondary dendrites toward the apical tuft at approximately 50–100 μm [49]. The spines were morphologically classified into the following two groups: immature (filopodia + thin) and mature (stubby + mushroom + branched) spines [50]. The analysis was performed in a blind manner.
2.15. RNA-seq analysis
Comparative transcriptome analysis between the following groups was performed using RNA-seq: Ppara KO mice (n = 6) versus WT littermates (n = 6) or corn oil-treated mice (n = 6) versus fenofibrate-treated mice (n = 6). RNA-seq analysis was performed as described previously [37, 51]. For more details, see Supplementary information.
2.16. PPARα agonist administration to PCP-administered mice
Six-week-old mice were administered PCP (10 mg/kg body weight/day, i.p.) or vehicle (saline, i.p.) once daily for 2 weeks [49]. Two mouse strains (B6J and ddY) were tested. However, PCP did not show any robust effect in B6J, including on spine morphology. Since the ddY mice manifested clearer and more reproducible effects of PCP administration, we used ddY mice in the current study. PCP-administered ddY mice were treated with fenofibrate (100 mg/kg/day, p.o.: Sigma-Aldrich, St. Louis, MO) or the vehicle (corn oil, p.o.: Sigma-Aldrich) once daily for 4 weeks. The spines were analyzed at the age of 12 weeks.
2.17. MK-801-induced locomotor hyperactivity test to fenofibrate-treated mice
B6J mice were chronically treated with vehicle (corn oil, p.o.) or fenofibrate (100 mg/kg/day, p.o.) once daily for 2 weeks (at 7–9 weeks of age). At 24 h post-last administration, MK-801-induced locomotor hyperactivity tests were performed. For the Methods of MK-801-induced locomotor hyperactivity test, see Supplementary information.
2.18. Statistical analysis
The data of in vitro assays were analyzed using a Fisher's exact test, χ2 test, or a one-way analysis of variance (ANOVA), followed by Dunnett's test. The data of mouse experiments were analyzed using the two-tailed Student's t-test, the two-tailed Mann-Whitney U test, one-way ANOVA followed by post hoc Tukey's test, or two-way repeated-measures ANOVA followed by post hoc Fisher's least significant difference (LSD) test or Bonferroni's comparison test. The data points with more than two standard deviations (SD) of mean were defined as outliers and were omitted from the analysis. The differences were considered statistically significant when the p-values were less than 0.05. All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA).
2.19. Role of the funding source
The funders had no role in the study design, data collection, data analysis, interpretation, or writing of the report.
3. Results
3.1. Identification of variants in the RXR and PPAR genes
The role of the PPAR or RXR genes in schizophrenia was analyzed by examining the genetic variants in the exons and flanking intronic regions of the six known RXR/PPAR genes (RXRA, RXRB, RXRG, PPARA, PPARD, and PPARG). The genomic DNA samples obtained from 1200 patients with schizophrenia in the Japanese population were subjected to MIP-based NGS (Fig. S1). Two hundred and twenty-nine variants were identified by NGS-based sequencing. Of these, 23 non-synonymous variants and one intronic splice acceptor site variant in these genes were validated by Sanger sequencing (Table 1, Fig. 1). On the other hand, 49 non-synonymous variants were identified in the Tohoku Medical Megabank Organization (ToMMo) 3.5KJPN database (https://ijgvd.megabank.tohoku.ac.jp/) [52], [53], [54], [55], [56] (Table 1), which is a whole-genome reference panel of 3554 individuals from a Japanese population and used as a control in this study.
Table 1.
Non-synonymous polymorphisms identified in the RXR/PPAR genes from 1200 schizophrenia and ToMMo 3.5KJPN samples.
| Gene | Nucleotide change | Amino acid change | dbSNP ID | MAFa (alternation alleles/total alleles) | |
|---|---|---|---|---|---|
| Schizophrenia | RXRA | c.138G>C | p.Gln46His | rs1588288848 (New) | 0.00042 (1/2400) |
| c.178G>A | p.Gly60Ser | rs773076567 | 0.00042 (1/2400) | ||
| c.188C>T | p.Pro63Leu | rs531899705 | 0.00083 (2/2400) | ||
| c.665C>T | p.Ser222Leu | rs139536056 | 0.00042 (1/2400) | ||
| c.1094C>T | p.Thr365Met | rs746817210 | 0.00042 (1/2400) | ||
| RXRB | c.292C>G | p.Pro98Ala | rs759432300 | 0.00042 (1/2400) | |
| c.310C>T | p.Pro104Ser | rs201700806 | 0.00042 (1/2400) | ||
| c.581C>T | p.Pro194Leu | rs1338024369 (New) | 0.00042 (1/2400) | ||
| RXRG | c.185C>T | p.Pro62Leu | rs781134922 (New) | 0.00083 (2/2400) | |
| c.254C>T | p.Ala85Val | rs145797395 | 0.00042 (1/2400) | ||
| c.584G>A | p.Arg195His | rs765348774 | 0.00042 (1/2400) | ||
| PPARA | c.209–2delA | – | rs1601773792 (New) | 0.00042 (1/2400) | |
| c.351C>A | p.His117Gln | rs765089560 (New) | 0.00042 (1/2400) | ||
| c.418G>A | p.Asp140Asn | rs113379388 | 0.00460 (11/2400) | ||
| c.421C>T | p.Arg141Cys | rs766370613 | 0.00042 (1/2400) | ||
| c.676C>T | p.Arg226Trp | rs1254453296 (New) | 0.00042 (1/2400) | ||
| c.680T>C | p.Val227Ala | rs1800234 | 0.05309 (121/2400) | ||
| c.1184G>A | p.Gly395Glu | rs200858936 | 0.00629 (15/2400) | ||
| PPARD | c.89A>G | p.Asn30Ser | rs149040923 | 0.00042 (1/2400) | |
| c.623C>T | p.Thr208Met | rs766827287 | 0.00042 (1/2400) | ||
| c.1144G>A | p.Glu382Lys | rs777270771 | 0.00042 (1/2400) | ||
| PPARG | c.34C>G | p.Pro12Ala | rs1801282 | 0.03270 (76/2400) | |
| c.550A>G | p.Lys184Glu | rs1575106051 (New) | 0.00042 (1/2400) | ||
| c.1111G>A | p.Glu371Lys | rs141683496 | 0.00042 (1/2400) | ||
| ToMMo 3.5KJPNb | RXRA | c.22C>T | p.Pro8Leu | rs1175967955 | 0.00014 (1/6916) |
| c.68C>T | p.Thr23Met | rs747209166 | 0.00014 (1/6898) | ||
| c.138G>C | p.Gln46His | rs1588288848 (New) | 0.00029 (2/6900) | ||
| c.178G>A | p.Gly60Ser | rs773076567 | 0.00073 (5/68,92) | ||
| c.242C>T | p.Thr81Ile | -c | 0.00015 (1/6520) | ||
| c.635G>T | p.Gly212Val | – | 0.00015 (1/6892) | ||
| c.639G>C | p.Lys213Asn | – | 0.00015 (1/6886) | ||
| c.724G>A | p.Val242Met | rs1383908613 | 0.00014 (1/6978) | ||
| c.1094C>T | p.Thr365Met | rs746817210 | 0.00014 (1/7022) | ||
| RXRB | c.44C>T | p.Pro182Ser | – | 0.00030 (2/6696) | |
| c.884C>T | p.Ala295Val | rs143679412 | 0.00014 (1/6938) | ||
| c.907A>G | p.Arg303Gly | – | 0.00014 (1/6944) | ||
| c.403G>A | p.Arg468Gln | rs760368861 | 0.00015 (1/6860) | ||
| RXRG | c.74C>T | p.Thr25Ile | rs939696922 | 0.00014 (1/7078) | |
| c.112C>T | p.Pro38Ser | – | 0.00014 (1/7084) | ||
| c.154C>T | p.Ala85Val | rs145797395 | 0.00084 (6/7102) | ||
| c.283C>A | p.Pro95Thr | – | 0.00028 (2/7094) | ||
| c.395G>A | p.Gly132Glu | – | 0.00014 (1/7106) | ||
| c.454G>A | p.Gly152Arg | – | 0.00014 (1/7108) | ||
| c.1004G>A | p.Arg335Gln | rs757609630 | 0.00028 (2/7090) | ||
| c.1157A>G | p.Asn386Ser | rs1339580376 | 0.00014 (1/7088) | ||
| c.1280G>A | p.Arg427His | rs901274500 | 0.00014 (1/7048) | ||
| c.1340C>G | p.Pro447Arg | – | 0.00014 (1/6986) | ||
| PPARA | c.29C>T | p.Pro10Leu | rs1401227237 | 0.00014 (1/6968) | |
| c.65C>G | p.Pro22Arg | rs1193512118 | 0.00044 (3/6824) | ||
| c.113C>T | p.Ser38Leu | rs575811356 | 0.00015 (1/6832) | ||
| c.116A>G | p.Gln39Arg | – | 0.00015 (1/6844) | ||
| c.418G>A | p.Asp140Asn | rs113379388 | 0.00501 (35/6988) | ||
| c.484C>G | p.Leu162Val | rs1800206 | 0.00014 (1/7032) | ||
| c.508G>A | p.Ala170Thr | rs939270657 | 0.00014 (1/7054) | ||
| c.680T>C | p.Val227Ala | rs1800234 | 0.05570 (395/7092) | ||
| c.1021C>T | p.Arg341Cys | - | 0.00014 (1/7096) | ||
| c.1184G>A | p.Gly395Glu | rs200858936 | 0.00619 (44/7106) | ||
| PPARD | c.89A>G | p.Asn30Ser | rs149040923 | 0.00088 (6/6812) | |
| c.623C>T | p.Thr208Met | rs766827287 | 0.00028 (2/7064) | ||
| c.941G>A | p.Arg314His | rs767428713 | 0.00014 (1/7056) | ||
| c.1256G>A | p.Arg419Gln | rs200904269 | 0.00015 (1/6782) | ||
| PPARG | c.34C>G | p.Pro12Ala | rs1801282 | 0.03054 (217/7106) | |
| c.43G>T | p.Asp15Tyr | – | 0.00042 (3/7106) | ||
| c.199A>G | p.Thr67Ala | rs778975987 | 0.00014 (1/7104) | ||
| c.205G>A | p.Asp69Asn | – | 0.00014 (1/7108) | ||
| c.284A>G | p.Tyr95Cys | rs1477623791 | 0.00028 (2/7106) | ||
| c.302G>A | p.Glu101Lys | – | 0.00014 (1/7104) | ||
| c.676T>G | p.Ser226Ala | – | 0.00014 (1/7080) | ||
| c.743A>G | p.Asp248Gly | – | 0.00014 (1/7100) | ||
| c.826G>A | p.Val276Ile | rs147996578 | 0.00014 (1/7100) | ||
| c.947G>A | p.Arg316His | rs28936407 | 0.00014 (1/7098) | ||
| c.1147A>G | p.Ser383Gly | – | 0.00014 (1/7090) | ||
| c.1242A>G | p.Ile414Met | – | 0.00014 (1/7096) |
We evaluated the missense variants by using the following variant annotation tools: REVEL [57], PolyPhen-2 [58], PMut [59], PROVEAN [60], and SIFT [61]. We defined the variants as harmful if all the tools predicted them “deleterious” (Table S2). Four common variants [minor allele frequency (MAF) of ≧ 0.1%] were detected in both the schizophrenia sample and the ToMMo database, but they were not harmful. In addition, the allele frequencies of those four variants were not different between the schizophrenia sample and the ToMMo dataset (Table S3). Regarding rare variants (MAF of <0.1%), we compared the number of harmful alleles per gene between the patients with schizophrenia and the ToMMo samples. As a result, the total number of harmful alleles in PPARA was significantly higher in the schizophrenia sample than in the ToMMo samples (Table 2). The total number of harmful alleles in other genes did not differ between the two groups (Table 2). The harmful missense variants (His117Gln, Arg141Cys, Arg226Trp) and the splice acceptor site variant (c.209–2del) in PPARA were detected exclusively in patients with schizophrenia but not in the ToMMo cohort. In addition, the His117Gln, Arg141Cys, or c.209–2del variant was not detected in the exomes of 125,748 individuals from the Genome Aggregation Database (gnomAD) v2.1.1 (https://gnomad.broadinstitute.org/) [62]. The allele frequency of the Arg226Trp was higher in the schizophrenia sample than in the gnomAD database (Table S4), although only one allele was found in each sample group. Therefore, we decided to focus on the PPARA gene and its variants (those exclusively detected in patients with schizophrenia and the Arg226Trp) in further study. The clinical characteristics of patients harboring these variants are described in Supplementary Material.
Table 2.
Number of harmful alleles per gene in schizophrenia and ToMMo 3.5KJPN.
| Gene | n | Harmful | Not harmful | p-value(Fisher's exact test) | |
|---|---|---|---|---|---|
| RXRA | Schizophrenia | 6 | 1 | 5 | 0.5211 |
| ToMMo 3.5KJPN a | 14 | 1 | 13 | ||
| RXRB | Schizophrenia | 3 | 0 | 3 | 1.0000 |
| ToMMo 3.5KJPN | 5 | 0 | 5 | ||
| RXRG | Schizophrenia | 4 | 1 | 3 | 1.0000 |
| ToMMo 3.5KJPN | 17 | 3 | 14 | ||
| PPARA | Schizophrenia | 3 | 3 | 0 | 0.0455 |
| ToMMo 3.5KJPN | 9 | 2 | 7 | ||
| PPARD | Schizophrenia | 3 | 0 | 3 | 1.0000 |
| ToMMo 3.5KJPN | 10 | 0 | 10 | ||
| PPARG | Schizophrenia | 2 | 1 | 1 | 0.1250 |
| ToMMo 3.5KJPN | 14 | 0 | 14 |
ToMMo 3.5KJPN: https://ijgvd.megabank.tohoku.ac.jp/
Bold character means significant difference (p < 0.05).
3.2. Splice acceptor site variant in the PPARA gene results in exon-skipping
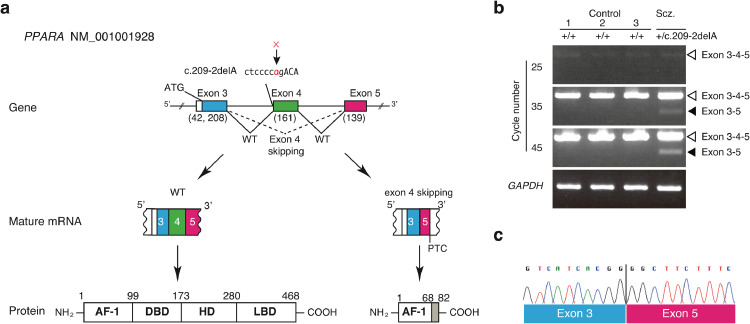
The PPARA c.209–2delA variant seemed highly deleterious as the deletion leads to the disruption of the consensus sequence of the splice acceptor site at intron 3 (between exon 3 and 4). To test this idea, the transcripts from hair follicle cells of the patient with schizophrenia and three control subjects were analyzed with reverse transcription-polymerase chain reaction. The specific primers amplified the region from exon 3 to exon 5 (Fig. 2a and b). Products with the size expected from the wild-type allele (358 bp) were detected in all the samples. In contrast, a shorter product was also detected exclusively in the patient sample (Fig. 2b). Direct sequencing of this fragment revealed that exon 4 was skipped in the mature transcripts of the c.209–2delA variant (Fig. 2c), which resulted in the introduction of a premature stop codon (PTC). The truncated protein was predicted to terminate within the AF-1 domain and to lack the DBD and LBD (Fig. 2a). The band derived from the mutant allele was weaker than that derived from the WT allele (Fig. 2b), which suggested that nonsense-mediated mRNA decay (NMD) eliminated the mutant transcript [63], [64], [65].
Fig. 2.
A splicing mutation in the PPARA gene
(a) (left) Normal splicing between adjacent exons. (right) Aberrant splicing and skipping of exon due to the deletion c.209–2delA. The deletion elicits a loss of universal splicing acceptor consensus sequence from the transcript and consequently results in exon 4 skipping.
(b) The products of reverse transcription-polymerase chain reaction (RT-PCR) obtained from total RNA of the hair follicle cells. The smaller fragment was derived from exon 4-excluded transcripts. cDNAs from three Japanese subjects without the same variant were analyzed as healthy controls.
(c) The small fragment was directly sequenced.
3.3. Decreased transcription factor activity of missense PPARα variants
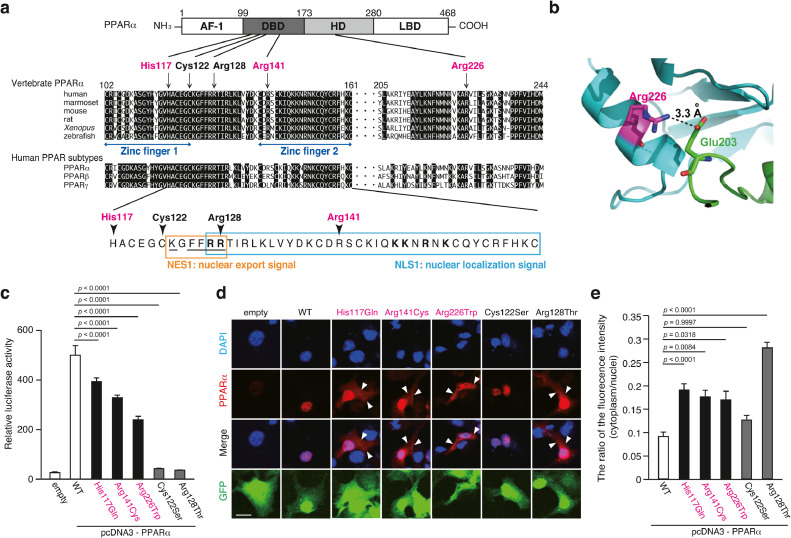
The PPARα His117, Arg141, and Arg226 residues are highly conserved among vertebrates and human PPAR subtypes (Fig. 3a). The mutations His117Gln and Arg141Cys may affect the ability of PPARα to bind to DNA as the His117 and Arg141 residues are located in the zinc finger motif of the DBD (Fig. 3a, Fig. S3a-c). The Arg226 residue was predicted to contribute to maintaining the orientation of DBD and LBD in the heterodimer by forming a salt bridge between the Glu203 residue in RXRα and consequently promoting intermolecular interaction between PPARα and RXRα (Fig. 3b). The Trp226 residue may disrupt this salt bridge as the positive charge of Arg-residue is lost, which may lead to structural destabilization of the PPARα/RXR heterodimer. These variants were expected to affect transcriptional factor activities.
Fig. 3.
Three missense variants in the PPARA gene
(a) Schematic representation of the PPARα protein. (upper) Amino acid sequence alignment among vertebrates and human subtypes of PPARα. Magenta characters and arrows indicate the position of the missense mutations found in patients with schizophrenia. Black characters and arrows indicate the position of the functional mutations reported in the previous studies [66, 67]. (lower) Amino acid sequences of NLS1 (blue square) and NES1 (orange square) of human PPARα. Bold letters in NLS1 designate a potential bipartite basic motif similar to the consensus binding signal for importin α. The underline in NES1 designates a potential KXFF(K/R)R motif similar to the consensus binding signal for CRT (calreticulin) and a putative hydrophobic NES motif for CRM1.
(b) A salt bridge between Arg226 in PPARα and Glu203 in RXRα.
(c) Activity of PPARα mutants as a transcription factor in the CV-1 cells.
The values represent the mean ± standard error (n = 3/group). The data were analyzed using one-way ANOVA (p < 0.001), followed by Dunnett's multiple comparison tests. These results were replicated in three independent experiments (data not shown).
(d) Intracellular localization of PPARα in the COS-7 cells was examined using the anti-PPARα antibody under a confocal microscope. Scale bar = 10 µm.
(e) The ratio of PPARα fluorescent intensity in the cytoplasm to that in the nuclei. The values represent the mean ± standard error (n > 97 group). The data were analyzed using a one-way ANOVA (p < 0.001), followed by Dunnett's multiple comparison tests.
The effect of missense variants on the transcription factor activity of PPARα was evaluated using the luciferase reporter assay. The CV-1cells transfected with a reporter construct that harbors a luciferase expression cassette under the control of multiple PPREs were used for the assay. The Cys122Ser and Arg128Thr variants, which are deficient in the transcription factor function [66, 67], were used as controls for the assay. The overexpression of WT PPARα significantly increased the reporter activity (Fig. 3c). The reporter activity in the cells transfected with the His117Gln, Arg141Cys, or Arg226Trp variant was significantly lower than that in the cells transfected with WT PPARα (Fig. 3c). A similar trend was obtained when the HEK293 cells were transfected with these variants (Fig. S4a).
3.4. Enhanced extranuclear localization of PPARα variants
PPARα is reported to shuttle between the nucleus and cytoplasm [68]. The PPARα protein harbors two nuclear localization signals (NLSs; NLS1 and NLS2) and two nuclear export signals (NESs; NES1 and NES2) [68] (Fig. 3a). The Arg141 residue is located within NLS1, whereas the His117 residue is located near NES1. The Arg226 residue is distant from NLS or NES (Fig. 3a). Therefore, the His117Gln and Arg141Cys mutations may affect the nuclear localization of PPARα and its DNA-binding ability.
To test this hypothesis, the COS-7 cells, which do not exhibit endogenous PPARα expression, were co-transfected with the PPARα (WT, His117Gln, Arg141Cys, Arg226Trp, Cys122Ser, or Arg128Thr)-expressing constructs and a green fluorescent protein (GFP)-expressing reporter construct. Similar the luciferase assay, the Cys122Ser and Arg128Thr variants were used as controls. The intracellular localization of PPARα in the transfected cells was examined using immunocytochemistry (Fig. 3d). The His117Gln, Arg141Cys, and Arg226Trp PPARα variants exhibited enhanced extranuclear localization when compared with WT PPARα. However, the localization of the Cys122Ser variant was not significantly different from that of WT (Fig. 3d and e). A similar trend was observed when the HEK293 (a human embryonic kidney cell line) cells were transfected with these PPARα constructs (Fig. S4b). Since PPARα is a transcriptional factor, decreased transcription factor activity of the PPARα mutants may be explained, in part, by enhanced extranuclear localization.
3.5. Distribution of Ppara mRNA in the mouse brain during neurodevelopment
To explore the biological importance of the PPARA gene in the brain, we analyzed, using in situ hybridization, the expression pattern of Ppara in the mouse brain at multiple neurodevelopmental stages, critical periods associated with the susceptibility to schizophrenia [69], and adult stage (Fig. S5a and b). The expression of Ppara peaked at embryonic day 16.5 (E16.5) when the transcript expression was detected in the brain with a relatively high expression level around the ventricular zones. The expression of Ppara declined postnatally. At the adult stage, moderate expression of Ppara was observed in the hippocampus, mPFC, cerebellar granule cells, and piriform cortex with faint but ubiquitous expression in the other regions. These results indicate that PPARA/Ppara may have an important role in neurodevelopment.
3.6. Generation and phenotype analysis of Ppara KO mice
The effects of Ppara depletion in mouse was evaluated using a Ppara KO mouse model, which was established using the clustered regularly spaced palindrome repeat (CRISPR)/Cas9n technology in the genetic background of inbred B6J mice (Fig. S6a-c). The KO mice were healthy without any visible abnormalities in growth or morphology (Fig. S6d). Additionally, the blood biochemical profiles were within the normal range (Table S5).
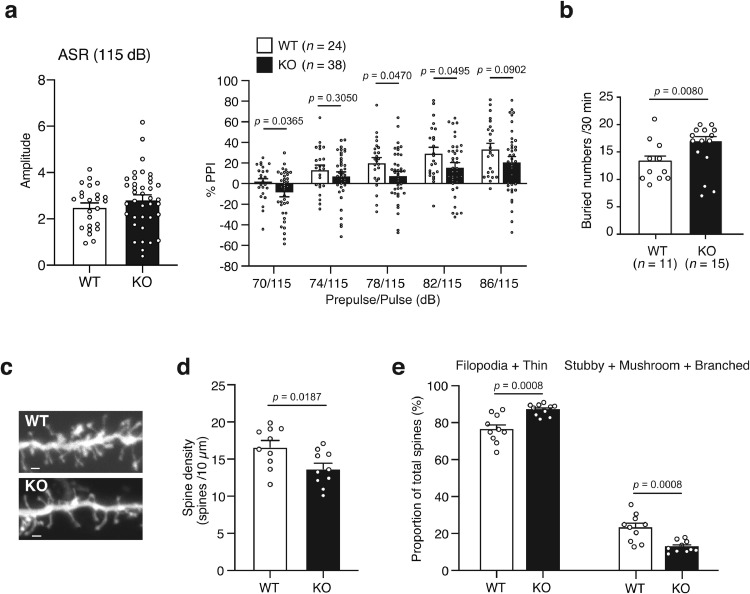
The Ppara KO mice were assessed through various behavioral tests related to schizophrenia: PPI test, psychostimulant-induced locomotor hyperactivity test, and sociability and cognitive tasks including those previously conducted to evaluate the phenotypes of Ppara KO mice [31] or Ppara expression-decreased mice [24], marble-burying, tail suspension, and the open field tests (Fig. S2). The PPI test was performed to evaluate the sensorimotor gating function, which is impaired in patients with schizophrenia and other neuropsychiatric diseases. The average startle response was not significant among Ppara KO and control mice (Fig. 4a). A two-way (prepulse x genotype) repeated-measures ANOVA demonstrated a significant difference [genotype effect: p = 0.0475, prepulse effect: p < 0.0001, interaction (genotype x prepulse): p = 0.6080] between the two groups. The Fisher's LSD post hoc test showed significant and trend differences in Ppara KO mice (70 dB, p = 0.0365; 78 dB, p = 0.0470; 82 dB, p = 0.0495; 86 dB, p = 0.0902) than controls (Fig. 4a). In the marble-burying test, the number of buried marbles was significantly higher in the KO mice than in the control mice (Fig. 4b), suggesting a higher anxiety level in KO mice. The two genotypes did not exhibit significant differences in other behavioral tests, including open field test, novel object recognition test, Y-maze test, three-chamber social interaction test, home cage activity test, tail suspension test, forced swim test, administration of MK-801-induced locomotor hyperactivity, and sucrose preference tests (Table S6).
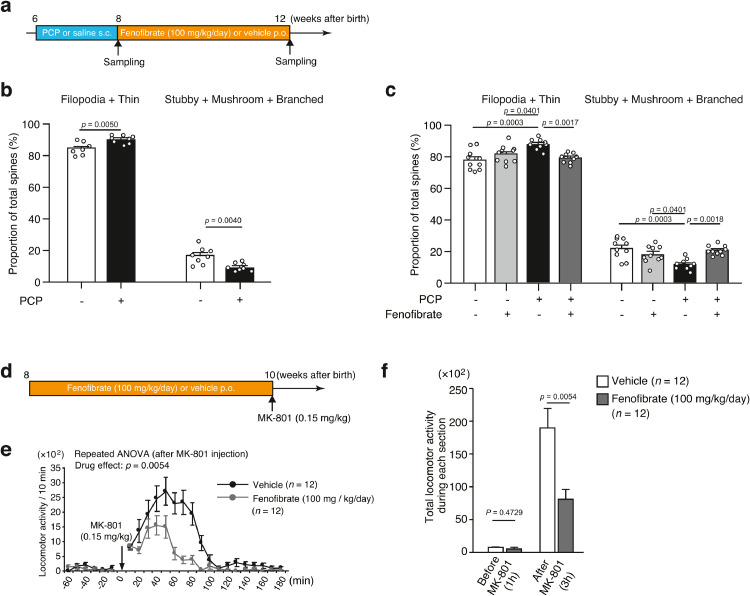
Fig. 4.
Phenotypes of Ppara knockout (KO) mice
(a) Prepulse inhibition (PPI) test. The values represent the mean ± standard error. The average of startle response was analyzed by using unpaired t-test. The PPI data were analyzed using a two-way repeated-measures ANOVA (genotype effect, p = 0.0475), followed by Fisher's LSD test.
(b) The marble-burying test. The values represent the mean ± standard error. The data were analyzed using unpaired t-tests.
(c) Typical images of dendritic spines in Ppara KO mice. Scale bar = 1 µm.
(d) Average spine density in wild-type (WT) (4 mice, ten cells) and Ppara KO (4 mice, 10 cells) mice. The values represent the mean ± standard error. The data were analyzed using the unpaired t-test.
(e) Morphological classification of spines in WT (4 mice, ten cells) and Ppara KO (4 mice, ten cells) mice. Spines were classified as immature (filopodia + thin) spines and mature (stubby + mushroom + branched) spines. The values represent the mean ± standard error. The data were analyzed using the unpaired t-test.
The postmortem brains of patients with schizophrenia and the brains of schizophrenia model animals exhibited decreased spine density and/or altered spine size [70], [71], [72]. To investigate the effects of Ppara LoF on spine density, spine density was calculated from randomly labeled apical dendrites in the mPFC (Fig. 4c), which exhibit Ppara expression (Fig. S5a). Spine density was significantly lower in the KO mice than in the control mice (Fig. 4d). Additionally, the KO mice exhibited a decreased percentage of mature spines (stubby + mushroom + branched) and an increased percentage of immature spines (filipodia + thin) (Fig. 4e). We also investigated the spine morphology of heterozygous Ppara mice. The heterozygous mice showed decreased mature spines and increased immature spines, but there were no changes in spine density (Fig. S7a and b).
3.7. Ppara regulates synaptogenesis signaling pathway-related genes
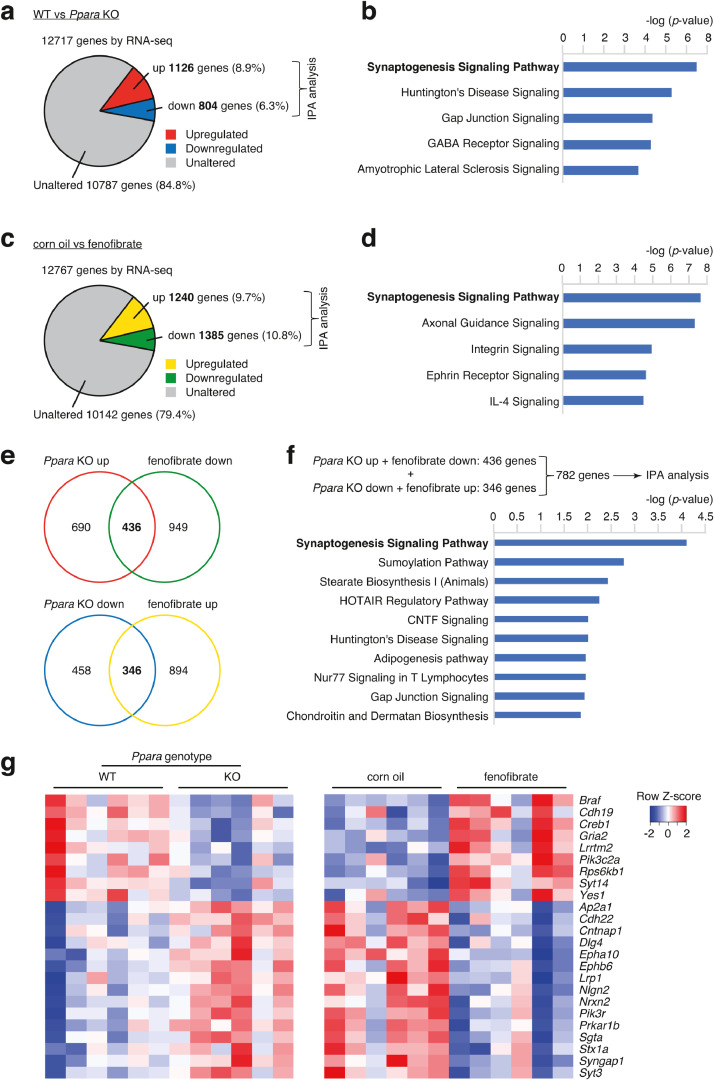
To identify the downstream genes of PPARα, gene expression patterns were compared by RNA-seq analyses between Ppara KO and WT mice, and between PPARα-agonist- and vehicle-treated mice. First, the mPFC of Ppara KO and control mice were examined by RNA-seq analysis. Of the 12,717 mapped genes, 1930 genes were differentially expressed (p < 0.05, log2 |fold change| > 0.1 or < −0.1), including 804 downregulated genes (6.3%) and 1126 upregulated genes (11.3%), between control and Ppara KO mice (Fig. 5a). Ingenuity pathway analysis (IPA) revealed that the differentially expressed genes were significantly enriched in several pathways, including the synaptogenesis signaling pathway (Fig. 5b).
Fig. 5.
RNA-seq analyses of Ppara KO mice and fenofibrate-administered mice.
(a) Pie chart showing the distribution of the number of genes that are differentially expressed between WT and Ppara KO mice.
(b) The top five canonical pathways in which the differentially expressed genes (between WT and Ppara KO mice) are enriched were identified using IPA.
(c) Pie chart showing the distribution of the number of genes that are differentially expressed between corn oil- and fenofibrate-administered mice.
(d) The top five canonical pathways in which the differentially expressed genes (between corn oil-and fenofibrate-administered mice) are enriched were identified using IPA.
(e) Venn-diagram (upper panel) showing shared genes among upregulated genes in Ppara KO mice and genes downregulated in fenofibrate-administered mice. Venn-diagram (lower panel) showing shared genes among genes downregulated in Ppara KO mice and genes upregulated in fenofibrate-administered mice.
(f) Top five canonical pathways shared among Ppara KO mice and fenofibrate-administered mice.
(g) Heatmap showing genes differentially expressed in the synaptogenesis signaling pathway.
Fenofibrate, a PPARα agonist, is reported to penetrate the blood-brain barrier [73]. The effect of fenofibrate on the gene expression profile of the brain was examined. The inbred ddY mice (aged 8 weeks) were administered a vehicle (corn oil) or fenofibrate (100 mg/kg body weight/day, p.o. for 4 weeks). The mPFC of vehicle and fenofibrate groups was subjected to RNA-seq. Of the 12,767 mapped genes, 2625 were differentially expressed (p < 0.05, log2 |fold change| >0.1 or <−0.1), including 1385 downregulated (10.8%) and 1240 upregulated genes (9.73%), between the vehicle and fenofibrate groups (Fig. 5c). IPA revealed that the differentially expressed genes were enriched in the synaptogenesis signaling pathway (Fig. 5d).
There were overlaps between the genes upregulated in the Ppara KO mice and those downregulated in the fenofibrate-administered mice. There were also overlaps between genes downregulated in the Ppara KO mice and those upregulated in fenofibrate-administered mice (Fig. 5e). The genes that fluctuate in the opposite direction between Pparα KO mice and PPARα activated mice might be candidates for the downstream factors of PPARα. IPA revealed that these genes were enriched in the canonical pathways, including synaptogenesis signaling, SUMOylation, and stearate biosynthetic pathways (Fig. 5f and g and Fig. S8a and b). Among the genes enriched in the synaptogenesis signaling pathway, those that harbor potential PPREs within 2000 bp upstream of the transcription start site were identified using the web tool, PPRE SEARCH (http://www.classicrus.com/PPRE/) (Table S7). All the genes had PPREs in the promoter regions. This indicates that these genes are potential candidates directly regulated by PPARα.
3.8. PPARα agonist is a potential therapeutic drug for schizophrenia
To examine the therapeutic effect of the pharmacological activation of PPARα on schizophrenia, the ability of fenofibrate to mitigate dendritic spine loss in mice treated with phencyclidine (PCP), an NMDA receptor antagonist [49], was evaluated. The ddY mice (aged 6 weeks) were divided into the following four groups: (1) saline/vehicle, (2) saline/fenofibrate (100 mg/kg body weight/day, p.o. for 4 weeks), (3) PCP (10 mg/kg body weight/day, i.p. for 2 weeks)/vehicle, and (4) PCP/fenofibrate. The PCP-administered mice were administered a vehicle (corn oil) or fenofibrate (Fig. 6a). The repeated administration of PCP significantly decreased dendritic spine density in the mPFC (Fig. S9a and b) and resulted in decreased levels of mature spines (stubby + mushroom + branched) and increased levels of immature spines (filipodia + thin) (Fig. 6b). The chronic treatment of fenofibrate significantly mitigated the PCP-induced decrease in the number of mature spines (Fig. 6c). Spine density did not significantly differ among the groups (Fig. S9c and d).
Fig. 6.
Effect of the PPARα agonist fenofibrate.
(a) Schedule of treatment with fenofibrate.
(b) Morphological classification of spines in saline-administered mice (4 mice, 8 cells) and phencyclidine (PCP)-administered mice (3 mice, 8 cells). The values represent the mean ± standard error. The data were analyzed using unpaired t-tests.
(c) Morphological classification of spines in saline/vehicle-administered mice (4 mice, 10 cells), saline/fenofibrate-administered mice (6 mice, 10 cells), PCP/vehicle-administered mice (4 mice, 10 cells), and PCP/fenofibrate-administered mice (5 mice, 10 cells). The values represent the mean ± standard error. The data were analyzed using one-way ANOVA (p < 0.001), followed by Tukey's test.
(d) Experimental design for the MK-801-induced locomotor hyperactivity test following chronic administration of fenofibrate.
(e) Locomotor activity before and after a single injection of MK-801 (0.15 mg/kg body weight). The values represent the mean ± standard error. The data were analyzed using two-way repeated-measures ANOVA (drug effect, p < 0.001).
(f) Cumulative locomotor activity before and after MK-801 injection. The values represent the mean ± standard error. The data were analyzed using unpaired t-tests.
Next, the effect of fenofibrate on the behavior of mice was evaluated. The B6J mice were treated with fenofibrate (100 mg/kg body weight/day, p.o. for 2 weeks) (Fig. 6d), and then injected with MK-801 (0.15 mg/kg, s.c.), an NMDA receptor blocker, 24 h after the last administration of fenofibrate. The pre-treatment of fenofibrate significantly suppressed the hyper-locomotive responses evoked by MK-801. The data were analyzed using two-way (drug x time) repeated-measures ANOVA (drug effect: p = 0.0054, time effect: p < 0.0001, interaction: p < 0.0001), followed by analyses of the total locomotor activity during each section using an unpaired t-test (p = 0.0054) (Fig. 6e and f).
The severe side effects of fenofibrate include rhabdomyolysis and pancreatitis. However, the analysis of blood biochemical parameters in this study revealed that the levels of creatine phosphokinase (CPK), amylase, and other biochemical markers were within the normal range in the fenofibrate-administered mice under the assay conditions (8-week-old B6J mice, 100 mg/kg body weight/day, p.o. for 2 weeks) (Table S8).
4. Discussion
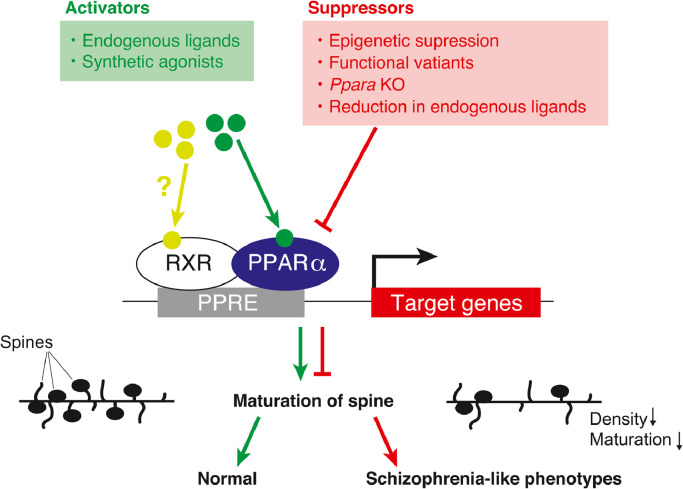
In the current study, we present evidence showing the causal relationship between the dysfunction of PPARα and schizophrenia. The rare PPARA variants identified in patients with schizophrenia (His117Gln, Arg141Cys, c.209–2del, and Arg226Trp) exhibited functional deficits, which may contribute to increased susceptibility to schizophrenia. The behavioral and histological defects in Ppara-deficient mice were similar to those in patients with schizophrenia. PPARα agonists, such as fenofibrate, may have a potential therapeutic benefit for patients with schizophrenia (Fig. 7).
Fig. 7.
Schematic illustration of the proposed role of PPARα in the pathogenesis of schizophrenia.
Accumulating evidence suggests that PPARα/Pparα plays an important role in neurodevelopment. Ppara is highly expressed in the mouse brain during the developmental period (Fig. S5a and b). The orthologs of mammalian PPARα regulate the formation of neurons and glial cells in the zebrafish [74]. In vitro studies showed that PPARα is expressed in neural stem cells, and that some PPARα target genes modulate cell proliferation [22, 75]. Notably, deficits in the neurodevelopment have been established as an important risk factor for schizophrenia [76, 77]. Collectively, these observations lead to an attractive hypothesis that the dysfunction of PPARα may contribute to the pathophysiology of schizophrenia through controlling neurodevelopment.
The results of RNA-seq analysis present multiple possibilities on the roles of PPARα in the pathogenesis of schizophrenia. First, the PPARα regulates the expression of synaptogenesis signaling pathway-related genes, including cyclic AMP response element-binding 1 (Creb1). The synaptogenesis signaling pathway-related genes harbor potential PPREs in the promoter region (Table S7). Of them, Creb1 was reported to be regulated by PPARα in the hippocampus [19, 78]. Accordingly, PPARα may be involved in dendritic spine dynamics by regulating synaptogenesis-related genes (e.g., CREB1).
Second, Sumo1 (encoding small ubiquitin-related modifier 1) expression was decreased in the mPFC of Ppara KO mice (Fig. S8a). Sumo1 is involved in the maturation of dendritic spines [79]. The Ppara KO mice showed impaired spine maturation (Fig. 4e). Therefore, PPARα can modulate spine maturation partly via the SUMOylation pathway. Importantly, Rubio et al. [80] suggested a potential role of PPARα in the pathophysiology of schizophrenia by regulating SUMOylation.
Third, the expression of Slc27a4, encoding long-chain fatty acid transport protein 4 (FATP4), was upregulated in the mPFC of the Ppara KO mice (Fig. S8b). The upregulation of FATP4 may affect the fatty acid composition of the brain. In fact, the altered fatty acid composition in the postmortem brains of patients with schizophrenia is reported [26, 27]; therefore, PPARα can be involved in the pathophysiology of schizophrenia via control of fatty acid transport.
In the present study, the Ppara KO mice showed a significantly lowered PPI in the PPI test (at the prepulse levels of 70, 78 and 82) and excessive buried marbles in the marble-burying test. Furthermore, previous studies showed that the Ppara KO mice exhibited repetitive behavior and cognitive inflexibility [31] and altered sleep behavior during fasting [32]. These results indicate that PPARA deficiency may lead to vulnerability to schizophrenia and other mental conditions such as autism.
Antipsychotics exert their therapeutic efficacy mainly through dopamine D2 receptor antagonism. However, D2 receptor antagonists are less effective against negative and cognitive symptoms of schizophrenia and are often associated with severe side effects. Thus, it has been long desired to identify the therapeutic agents that show efficacy in patients with schizophrenia through other molecular mechanisms. In the present study, we revealed that pharmacological activation of PPARα rescued the endophenotypes of schizophrenia in the mouse model.
Fenofibrate ameliorated the changes in dendritic spine morphology in the PCP-administered mice and also prevented the MK-801-induced locomotor hyperactivity. These effects were possibly mediated by the regulation of downstream genes of PPARα, such as Creb1 and Sumo1, although it is necessary to investigate the mechanism experimentally in the future. In addition, evaluating the effect of fenofibrate on other endophenotypes of schizophrenia will clarify how PPARα agonists can exert their potential efficacy on schizophrenia. It is worth examining whether PPARα agonists such as fenofibrate are effective in established genetically-manipulated animal models (e.g., Disc1 KO mouse).
It has been reported that intervention with PUFAs, one of the endogenous PPARα ligands, is useful for the prevention or treatment of schizophrenia [81, 82]. Maternal treatment with resveratrol, a PPARα agonist derived from natural products, also prevents the behavior associated with schizophrenia [83]. Taken together with our current observations, these findings indicate that PPARα may become a promising molecular target for the treatment of schizophrenia. There is a need to design PPARα agonists that can penetrate the blood-brain barrier more efficiently in order to achieve better treatment of schizophrenia.
The PPAR family proteins heterodimerize with the RXR family proteins to exert their function as a transcription factor. The RXR agonist, bexarotene, improves the positive and negative symptoms of schizophrenia (NCT00535574) [24, 84]. Therefore, the combinatorial treatment with RXR and PPAR ligands may be useful for treating patients, especially with the decreased expression of PPARA and/or RXRA. Since a subset of patients with schizophrenia exhibited downregulated PPARA and RXRA expression in the hair follicle cells [24].
There are several limitations to this study. First, the mutations in PPARA were too rare to explain the pathophysiology of schizophrenia as a whole. However, it should be noted that many factors can influence the activity of PPARα. It is necessary to examine the relationship between functional dysregulation of PPARα and schizophrenia in the future. Second, a high dose of fenofibrate is known to cause side effects such as rhabdomyolysis and pancreatitis [85], [86], [87], [88]. Therefore, it may be preferable to develop safer PPARα agonists when considering clinical applications. Third, the effect of MK-801 was alleviated by fenofibrate in ddY mice, although Ppara KO on the B6J background did not alter MK-801-induced locomotor hyperactivity. This discrepancy is probably attributed to the genetic background and should be further scrutinized.
In summary, the findings of this study indicate that the PPARα-RXR pathway could be a novel molecular target for treating schizophrenia, and will accelerate future clinical trials on schizophrenia.
Contributors
YW, MM, TO, SB and TY designed the study. YW, TO, SB, SM, KI, YI, HO, AW, YH, YN, TT, TS and TK performed the experiments. MM, TT and MI collected clinical information. YW, TO, SB, YI, HO, AW, YH, YN, TT, TM and MI acquired the data, YW, MM, TO, SB, YI, HO, AW, TK and TY analyzed the data, and YW, MM, TO, SB and TY wrote the manuscript. All authors read and approved the final version of the manuscript, and ensure it is the case.
Data sharing
The raw data obtained in this study are available on DDBJ database (http://www.ddbj.nig.ac.jp/index-e.html) with accession numbers of DRA010532 and DRA010537 with bioproject accession number of PRJDB10250 and PRJDB10264.
Supplementary material
Supplementary material is available at EBioMedicine online.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
We are grateful to the Support Unit for Bio-Material Analysis and Animal Resources Development, Research Resources Division, RIKEN Center for Brain Science, for animal care and sequencing service. This work was supported by the Strategic Research Program for Brain Sciences from Japan Agency for Medical Research and Development (AMED) under Grant Nos. JP19dm0107129 (to M.M.) and JP19dm0107083 (to T.Y.), by JSPS KAKENHI under Grant No. JP18K07578 (to M.M.) and JP20K07934 (to T.O.), and by the Grant-in-Aid for Scientific Research on Innovative Areas from the MEXT under Grant No. JP18H05435 (to T.Y.). In addition, this study was supported by grants from the SENSHIN Medical Research Foundation (to M.M.), and by RIKEN Junior Research Associate Program (to Y.W.).
Footnotes
Funding: This work was supported by the Strategic Research Program for Brain Sciences from AMED (Japan Agency for Medical Research and Development) under Grant Numbers JP19dm0107129 (to M.M.) and JP19dm0107083 (to T.Y.), by JSPS KAKENHI under Grant Number JP18K07578 (to M.M.) and JP 20K07934 (to T.O.), and by the Grant-in-Aid for Scientific Research on Innovative Areas from the MEXT under Grant Number JP18H05435 (to T.Y.). In addition, this study was supported by grants from the SENSHIN Medical Research Foundation (to M.M.), and by RIKEN Junior Research Associate Program (to Y.W.).
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.103130.
Contributor Information
Motoko Maekawa, Email: motoko.maekawa@riken.jp.
Takeo Yoshikawa, Email: takeo.yoshikawa@riken.jp.
Appendix. Supplementary materials
References
- 1.Nuclear Receptors Nomenclature C. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97(2):161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 2.de Urquiza A.M., Liu S., Sjoberg M., Zetterstrom R.H., Griffiths W., Sjovall J. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290(5499):2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 3.Grygiel-Gorniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications-a review. Nutr J. 2014;13:17. doi: 10.1186/1475-2891-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf D.J., Evans R.M. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T.P. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kersten S. Integrated physiology and systems biology of PPARalpha. Mol Metab. 2014;3(4):354–371. doi: 10.1016/j.molmet.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moreno S., Farioli-Vecchioli S., Ceru M.P. Immunolocalization of peroxisome proliferator-activated receptors and retinoid X receptors in the adult rat CNS. Neuroscience. 2004;123(1):131–145. doi: 10.1016/j.neuroscience.2003.08.064. [DOI] [PubMed] [Google Scholar]
- 8.Cimini A., Benedetti E., Cristiano L., Sebastiani P., D'Amico M.A., D'Angelo B. Expression of peroxisome proliferator-activated receptors (PPARs) and retinoic acid receptors (RXRs) in rat cortical neurons. Neuroscience. 2005;130(2):325–337. doi: 10.1016/j.neuroscience.2004.09.043. [DOI] [PubMed] [Google Scholar]
- 9.Cimini A., Cristiano L., Benedetti E., D'Angelo B., Ceru M.P. PPARs expression in adult mouse neural stem cells: modulation of PPARs during astroglial differentiaton of NSC. PPAR Res. 2007;2007:48242. doi: 10.1155/2007/48242. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Huang J.K., Jarjour A.A., Nait Oumesmar B., Kerninon C., Williams A., Krezel W. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat Neurosci. 2011;14(1):45–53. doi: 10.1038/nn.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanafy K.A., Sloane J.A. Regulation of remyelination in multiple sclerosis. FEBS Lett. 2011;585(23):3821–3828. doi: 10.1016/j.febslet.2011.03.048. [DOI] [PubMed] [Google Scholar]
- 12.Chandraratna RAN R.J., Nowak E.C. Treatment with retinoid X receptor agonist IRX4204 ameliorates experimental autoimmune encephalomyelitis. Am J Transl Res. 2016;8:1016–1026. [PMC free article] [PubMed] [Google Scholar]
- 13.Merk D. Chances and challenges of retinoid X receptor gamma targeting for regenerative multiple sclerosis treatment. Future Med Chem. 2015;7(18):2411–2413. doi: 10.4155/fmc.15.163. [DOI] [PubMed] [Google Scholar]
- 14.Benani A., Kremarik-Bouillaud P., Bianchi A., Netter P., Minn A., Dauca M. Evidence for the presence of both peroxisome proliferator-activated receptors alpha and beta in the rat spinal cord. J Chem Neuroanat. 2003;25(1):29–38. doi: 10.1016/s0891-0618(02)00093-5. [DOI] [PubMed] [Google Scholar]
- 15.Cimini A., Cristiano L., Colafarina S., Benedetti E., Di Loreto S., Festuccia C. PPARgamma-dependent effects of conjugated linoleic acid on the human glioblastoma cell line (ADF) Int J Cancer. 2005;117(6):923–933. doi: 10.1002/ijc.21272. [DOI] [PubMed] [Google Scholar]
- 16.Bernardo A., Levi G., Minghetti L. Role of the peroxisome proliferator-activated receptor-gamma (PPAR-gamma) and its natural ligand 15-deoxy-Delta12, 14-prostaglandin J2 in the regulation of microglial functions. Eur J Neurosci. 2000;12(7):2215–2223. doi: 10.1046/j.1460-9568.2000.00110.x. [DOI] [PubMed] [Google Scholar]
- 17.Genovese T., Mazzon E., Di Paola R., Cannavo G., Muia C., Bramanti P. Role of endogenous ligands for the peroxisome proliferators activated receptors alpha in the secondary damage in experimental spinal cord trauma. Exp Neurol. 2005;194(1):267–278. doi: 10.1016/j.expneurol.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Esmaeili M.A., Yadav S., Gupta R.K., Waggoner G.R., Deloach A., Calingasan N.Y. Preferential PPAR-alpha activation reduces neuroinflammation, and blocks neurodegeneration in vivo. Hum Mol Genet. 2016;25(2):317–327. doi: 10.1093/hmg/ddv477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel D., Roy A., Kundu M., Jana M., Luan C.H., Gonzalez F.J. Aspirin binds to PPARalpha to stimulate hippocampal plasticity and protect memory. Proc Natl Acad Sci U S A. 2018;115(31):E7408–E7E17. doi: 10.1073/pnas.1802021115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J., Ge H., Liu W., Zhu H., Chen Y., Zhang X. M2 microglia promotes neurogenesis and oligodendrogenesis from neural stem/progenitor cells via the PPARgamma signaling pathway. Oncotarget. 2017;8(12):19855–19865. doi: 10.18632/oncotarget.15774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 22.Cimini A., Ceru M.P. Emerging roles of peroxisome proliferator-activated receptors (PPARs) in the regulation of neural stem cells proliferation and differentiation. Stem Cell Rev. 2008;4(4):293–303. doi: 10.1007/s12015-008-9024-2. [DOI] [PubMed] [Google Scholar]
- 23.van Neerven S., Kampmann E., Mey J. RAR/RXR and PPAR/RXR signaling in neurological and psychiatric diseases. Prog Neurobiol. 2008;85(4):433–451. doi: 10.1016/j.pneurobio.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa M., Watanabe A., Iwayama Y., Kimura T., Hamazaki K., Balan S. Polyunsaturated fatty acid deficiency during neurodevelopment in mice models the prodromal state of schizophrenia through epigenetic changes in nuclear receptor genes. Transl Psychiatry. 2017;7(9):e1229. doi: 10.1038/tp.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaffe A.E., Gao Y., Deep-Soboslay A., Tao R., Hyde T.M., Weinberger D.R. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19(1):40–47. doi: 10.1038/nn.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peet M., Laugharne J., Rangarajan N., Horrobin D., Reynolds G. Depleted red cell membrane essential fatty acids in drug-treated schizophrenic patients. J Psychiatr Res. 1995;29(3):227–232. doi: 10.1016/0022-3956(95)00001-l. [DOI] [PubMed] [Google Scholar]
- 27.Arvindakshan M., Ghate M., Ranjekar P.K., Evans D.R., Mahadik S.P. Supplementation with a combination of omega-3 fatty acids and antioxidants (vitamins E and C) improves the outcome of schizophrenia. Schizophr Res. 2003;62(3):195–204. doi: 10.1016/s0920-9964(02)00284-0. [DOI] [PubMed] [Google Scholar]
- 28.Berger M., Nelson B., Markulev C., Yuen H.P., Schafer M.R., Mossaheb N. Relationship between polyunsaturated fatty acids and psychopathology in the NEURAPRO clinical trial. Front Psychiatry. 2019;10:393. doi: 10.3389/fpsyt.2019.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amminger G.P., Schafer M.R., Papageorgiou K., Klier C.M., Cotton S.M., Harrigan S.M. Long-chain omega-3 fatty acids for indicated prevention of psychotic disorders: a randomized, placebo-controlled trial. Arch Gen Psychiatry. 2010;67(2):146–154. doi: 10.1001/archgenpsychiatry.2009.192. [DOI] [PubMed] [Google Scholar]
- 30.Cadenhead K., Addington J., Cannon T., Cornblatt B., Mathalon D., McGlashan T. Omega-3 fatty acid versus placebo in a clinical high-risk sample from the north american prodrome longitudinal studies (NAPLS) consortium. Schizophr Bull. 2017;43(Suppl1):S16. [Google Scholar]
- 31.D'Agostino G., Cristiano C., Lyons D.J., Citraro R., Russo E., Avagliano C. Peroxisome proliferator-activated receptor alpha plays a crucial role in behavioral repetition and cognitive flexibility in mice. Mol Metab. 2015;4(7):528–536. doi: 10.1016/j.molmet.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo Y., Chikahisa S., Shiuchi T., Shimizu N., Tanioka D., Uguisu H. Sleep profile during fasting in PPAR-alpha knockout mice. Physiol Behav. 2020;214 doi: 10.1016/j.physbeh.2019.112760. [DOI] [PubMed] [Google Scholar]
- 33.Roy A., Kundu M., Jana M., Mishra R.K., Yung Y., Luan C.H. Identification and characterization of PPARalpha ligands in the hippocampus. Nat Chem Biol. 2016;12(12):1075–1083. doi: 10.1038/nchembio.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle E.A., O'Roak B.J., Martin B.K., Kumar A., Shendure J. MIPgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics. 2014;30(18):2670–2672. doi: 10.1093/bioinformatics/btu353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maekawa M., Yamada K., Toyoshima M., Ohnishi T., Iwayama Y., Shimamoto C. Utility of scalp hair follicles as a novel source of biomarker genes for psychiatric illnesses. Biol Psychiatry. 2015;78(2):116–125. doi: 10.1016/j.biopsych.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 37.Ide M., Ohnishi T., Toyoshima M., Balan S., Maekawa M., Shimamoto-Mitsuyama C. Excess hydrogen sulfide and polysulfides production underlies a schizophrenia pathophysiology. EMBO Mol Med. 2019;11(12):e10695. doi: 10.15252/emmm.201910695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11(4):399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 41.Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimamoto-Mitsuyama C., Ohnishi T., Balan S., Ohba H., Watanabe A., Maekawa M. Evaluation of the role of fatty acid-binding protein 7 in controlling schizophrenia-relevant phenotypes using newly established knockout mice. Schizophr Res. 2019 doi: 10.1016/j.schres.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Shoji H., Miyakawa T. Relationships between the acoustic startle response and prepulse inhibition in C57BL/6J mice: a large-scale meta-analytic study. Mol Brain. 2018;11(1):42. doi: 10.1186/s13041-018-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita M., Sakakibara Y., Hall F.S., Numachi Y., Yoshida S., Kobayashi H. Impaired cliff avoidance reaction in dopamine transporter knockout mice. Psychopharmacology (Berl) 2013;227(4):741–749. doi: 10.1007/s00213-013-3009-9. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe A., Toyota T., Owada Y., Hayashi T., Iwayama Y., Matsumata M. Fabp7 maps to a quantitative trait locus for a schizophrenia endophenotype. PLoS Biol. 2007;5(11):e297. doi: 10.1371/journal.pbio.0050297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morimura N., Yasuda H., Yamaguchi K., Katayama K.I., Hatayama M., Tomioka N.H. Autism-like behaviours and enhanced memory formation and synaptic plasticity in Lrfn2/SALM1-deficient mice. Nat Commun. 2017;8:15800. doi: 10.1038/ncomms15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mataga N., Mizuguchi Y., Hensch T.K. Experience-dependent pruning of dendritic spines in visual cortex by tissue plasminogen activator. Neuron. 2004;44(6):1031–1041. doi: 10.1016/j.neuron.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 48.Mukai H., Hatanaka Y., Mitsuhashi K., Hojo Y., Komatsuzaki Y., Sato R. Automated analysis of spines from confocal laser microscopy images: application to the discrimination of androgen and estrogen effects on spinogenesis. Cereb Cortex. 2011;21(12):2704–2711. doi: 10.1093/cercor/bhr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirai Y., Fujita Y., Hashimoto R., Ohi K., Yamamori H., Yasuda Y. Dietary intake of sulforaphane-rich broccoli sprout extracts during juvenile and adolescence can prevent phencyclidine-induced cognitive deficits at adulthood. PLoS ONE. 2015;10(6) doi: 10.1371/journal.pone.0127244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Risher W.C., Ustunkaya T., Singh Alvarado J., Eroglu C. Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0107591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohnishi T., Balan S., Toyoshima M., Maekawa M., Ohba H., Watanabe A. Investigation of betaine as a novel psychotherapeutic for schizophrenia. EBioMedicine. 2019;45:432–446. doi: 10.1016/j.ebiom.2019.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishigaki K., Akiyama M., Kanai M., Takahashi A., Kawakami E., Sugishita H. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020 doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takata R., Takahashi A., Fujita M., Momozawa Y., Saunders E.J., Yamada H. 12 new susceptibility loci for prostate cancer identified by genome-wide association study in Japanese population. Nat Commun. 2019;10(1):4422. doi: 10.1038/s41467-019-12267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagasaki M., Yasuda J., Katsuoka F., Nariai N., Kojima K., Kawai Y. Rare variant discovery by deep whole-genome sequencing of 1,070 Japanese individuals. Nat Commun. 2015;6:8018. doi: 10.1038/ncomms9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaguchi-Kabata Y., Nariai N., Kawai Y., Sato Y., Kojima K., Tateno M. iJGVD: an integrative Japanese genome variation database based on whole-genome sequencing. Hum Genome Var. 2015;2:15050. doi: 10.1038/hgv.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tadaka S., Katsuoka F., Ueki M., Kojima K., Makino S., Saito S. 3.5KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 2019;6:28. doi: 10.1038/s41439-019-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez-Ferrando V., Gazzo A., de la Cruz X., Orozco M., Gelpi J.L. PMut: a web-based tool for the annotation of pathological variants on proteins, 2017 update. Nucleic Acids Res. 2017;45(W1):W222–W2W8. doi: 10.1093/nar/gkx313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maquat L.E. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118(Pt 9):1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 64.Behm-Ansmant I., Kashima I., Rehwinkel J., Sauliere J., Wittkopp N., Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581(15):2845–2853. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 65.Kurosaki T., Maquat L.E. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. 2016;129(3):461–467. doi: 10.1242/jcs.181008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juge-Aubry C.E., Gorla-Bajszczak A., Pernin A., Lemberger T., Wahli W., Burger A.G. Peroxisome proliferator-activated receptor mediates cross-talk with thyroid hormone receptor by competition for retinoid X receptor. Possible role of a leucine zipper-like heptad repeat. J Biol Chem. 1995;270(30):18117–18122. doi: 10.1074/jbc.270.30.18117. [DOI] [PubMed] [Google Scholar]
- 67.Auclair M., Vigouroux C., Boccara F., Capel E., Vigeral C., Guerci B. Peroxisome proliferator-activated receptor-gamma mutations responsible for lipodystrophy with severe hypertension activate the cellular renin-angiotensin system. Arterioscler Thromb Vasc Biol. 2013;33(4):829–838. doi: 10.1161/ATVBAHA.112.300962. [DOI] [PubMed] [Google Scholar]
- 68.Umemoto T., Fujiki Y. Ligand-dependent nucleo-cytoplasmic shuttling of peroxisome proliferator-activated receptors, PPARalpha and PPARgamma. Genes Cells. 2012;17(7):576–596. doi: 10.1111/j.1365-2443.2012.01607.x. [DOI] [PubMed] [Google Scholar]
- 69.Birnbaum R., Weinberger D.R. Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci. 2017;18(12):727–740. doi: 10.1038/nrn.2017.125. [DOI] [PubMed] [Google Scholar]
- 70.Glausier J.R., Lewis D.A. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konopaske G.T., Lange N., Coyle J.T., Benes F.M. Prefrontal cortical dendritic spine pathology in schizophrenia and bipolar disorder. JAMA Psychiatry. 2014;71(12):1323–1331. doi: 10.1001/jamapsychiatry.2014.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Broadbelt K., Byne W., Jones L.B. Evidence for a decrease in basilar dendrites of pyramidal cells in schizophrenic medial prefrontal cortex. Schizophr. Res. 2002;58(1):75–81. doi: 10.1016/s0920-9964(02)00201-3. [DOI] [PubMed] [Google Scholar]
- 73.Mysiorek C., Culot M., Dehouck L., Derudas B., Staels B., Bordet R. Peroxisome-proliferator-activated receptor-alpha activation protects brain capillary endothelial cells from oxygen-glucose deprivation-induced hyperpermeability in the blood-brain barrier. Curr Neurovasc Res. 2009;6(3):181–193. doi: 10.2174/156720209788970081. [DOI] [PubMed] [Google Scholar]
- 74.Hsieh Y.C., Chiang M.C., Huang Y.C., Yeh T.H., Shih H.Y., Liu H.F. Pparalpha deficiency inhibits the proliferation of neuronal and glial precursors in the zebrafish central nervous system. Dev Dyn. 2018;247(12):1264–1275. doi: 10.1002/dvdy.24683. [DOI] [PubMed] [Google Scholar]
- 75.Cristiano L., Cimini A., Moreno S., Ragnelli A.M., Paola Ceru M. Peroxisome proliferator-activated receptors (PPARs) and related transcription factors in differentiating astrocyte cultures. Neuroscience. 2005;131(3):577–587. doi: 10.1016/j.neuroscience.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Iritani S. What happens in the brain of schizophrenia patients?: an investigation from the viewpoint of neuropathology. Nagoya J Med Sci. 2013;75(1–2):11–28. [PMC free article] [PubMed] [Google Scholar]
- 77.Pino O., Guilera G., Gomez-Benito J., Najas-Garcia A., Rufian S., Rojo E. Neurodevelopment or neurodegeneration: review of theories of schizophrenia. Actas Esp Psiquiatr. 2014;42(4):185–195. [PubMed] [Google Scholar]
- 78.Roy A., Jana M., Corbett G.T., Ramaswamy S., Kordower J.H., Gonzalez F.J. Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep. 2013;4(4):724–737. doi: 10.1016/j.celrep.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khayachi A., Gwizdek C., Poupon G., Alcor D., Chafai M., Casse F. Sumoylation regulates FMRP-mediated dendritic spine elimination and maturation. Nat Commun. 2018;9(1):757. doi: 10.1038/s41467-018-03222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rubio M.D., Wood K., Haroutunian V., Meador-Woodruff J.H. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology. 2013;38(10):1910–1920. doi: 10.1038/npp.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pawelczyk T., Grancow-Grabka M., Trafalska E., Szemraj J., Zurner N., Pawelczyk A. An increase in plasma brain derived neurotrophic factor levels is related to n-3 polyunsaturated fatty acid efficacy in first episode schizophrenia: secondary outcome analysis of the OFFER randomized clinical trial. Psychopharmacology (Berl) 2019;236(9):2811–2822. doi: 10.1007/s00213-019-05258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider M., Levant B., Reichel M., Gulbins E., Kornhuber J., Muller C.P. Lipids in psychiatric disorders and preventive medicine. Neurosci Biobehav Rev. 2017;76(Pt B):336–362. doi: 10.1016/j.neubiorev.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 83.Ferreira F.R., de Moura N.S.B., Hassib L., Pombo T.R. Resveratrol ameliorates the effect of maternal immune activation associated with schizophrenia in adulthood offspring. Neurosci Lett. 2020;734 doi: 10.1016/j.neulet.2020.135100. [DOI] [PubMed] [Google Scholar]
- 84.Lerner V., Miodownik C., Gibel A., Sirota P., Bush I., Elliot H. The retinoid X receptor agonist bexarotene relieves positive symptoms of schizophrenia: a 6-week, randomized, double-blind, placebo-controlled multicenter trial. J Clin Psychiatry. 2013;74(12):1224–1232. doi: 10.4088/JCP.12m08160. [DOI] [PubMed] [Google Scholar]
- 85.Danis R., Akbulut S., Ozmen S., Arikan S. Rhabdomyolysis-induced acute renal failure following fenofibrate therapy: a case report and literature review. Case Rep Med. 2010;2010 doi: 10.1155/2010/537818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Enger C., Gately R., Ming E.E., Niemcryk S.J., Williams L., McAfee A.T. Pharmacoepidemiology safety study of fibrate and statin concomitant therapy. Am J Cardiol. 2010;106(11):1594–1601. doi: 10.1016/j.amjcard.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 87.Preiss D., Tikkanen M.J., Welsh P., Ford I., Lovato L.C., Elam M.B. Lipid-modifying therapies and risk of pancreatitis: a meta-analysis. JAMA. 2012;308(8):804–811. doi: 10.1001/jama.2012.8439. [DOI] [PubMed] [Google Scholar]
- 88.Wang D., Wang Y. Fenofibrate monotherapy-induced rhabdomyolysis in a patient with hypothyroidism: a rare case report and literature review. Medicine (Baltimore) 2018;97(14):e0318. doi: 10.1097/MD.0000000000010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.