Abstract
Human reticulocyte 15-lipoxygenase-1 (h15-LOX-1 or ALOX15) and platelet 12-lipoxygenase (h12-LOX or ALOX12) catalysis of docosahexaenoic acid (DHA) and the maresin precursor, 14S-hydroperoxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14S-HpDHA), were investigated to determine their product profiles and relative rates in the biosynthesis of the key maresin intermediate, 13S,14S-epoxy-4Z,7Z,9E,11E,16Z,19Z-docosahexaenoic acid (13S,14S-epoxy-DHA). Both enzymes converted DHA to 14S-HpDHA, with h12-LOX having a 39-fold greater kcat/KM value (14.0 ± 0.8 s−1 μM−1) than that of h15-LOX-1 (0.36 ± 0.08 s−1 μM−1) and a 1.8-fold greater 14S-HpDHA product selectivity, 81 and 46%, respectively. However, h12-LOX was markedly less effective at producing 13S,14S-epoxy-DHA from 14S-HpDHA than h15-LOX-1, with a 4.6-fold smaller kcat/KM value, 0.0024 ± 0.0002 and 0.11 ± 0.006 s−1 μM−1, respectively. This is the first evidence of h15-LOX-1 to catalyze this reaction and reveals a novel in vitro pathway for maresin biosynthesis. In addition, epoxidation of 14S-HpDHA is negatively regulated through allosteric oxylipin binding to h15-LOX-1 and h12-LOX. For h15-LOX-1, 14S-HpDHA (Kd = 6.0 μM), 12S-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (12S-HETE) (Kd = 3.5 μM), and 14S-hydroxy-7Z,10Z,12E,16Z,19Z-docosapentaenoic acid (14S-HDPAω-3) (Kd = 4.0 μM) were shown to decrease 13S,14S-epoxy-DHA production. h12-LOX was also shown to be allosterically regulated by 14S-HpDHA (Kd = 3.5 μM) and 14S-HDPAω-3 (Kd = 4.0 μM); however, 12S-HETE showed no effect, indicating for the first time an allosteric response by h12-LOX. Finally, 14S-HpDHA inhibited platelet aggregation at a submicrololar concentration, which may have implications in the benefits of diets rich in DHA. These in vitro biosynthetic pathways may help guide in vivo maresin biosynthetic investigations and possibly direct therapeutic interventions.
Acute inflammation is a beneficial response to injury and infection, but it can develop into chronic inflammation, which leads to disease if not properly regulated.1 Specifically, resident immune cells initiate a proinflammatory response, which is amplified by neutrophils, eosinophils, and M1-polarized macrophages by producing bioactive molecules, such as leukotrienes and prostaglandins.2 This proinflammatory response eventually transitions to the proresolution phase of inflammation, with M2-polarized macrophages producing potent molecules, such as maresins, resolvins, lipoxins, and neuroprotectins, otherwise known as specialized proresolving mediators (SPMs).3,4 This transition from inflammation to resolution is now recognized as being critical for correcting chronic inflammatory diseases, such as cardiovascular disease, Alzheimer’s disease, atherosclerosis, and cancer,5 to name a few. Most inflammatory therapeutics have so far focused on inhibiting the formation of proinflammatory molecules, such as the nonsteroidal anti-inflammatory drugs (NSAIDS); however, these molecules have limited effect on chronic inflammatory diseases. The therapeutic challenge of chronic inflammation is inhibiting the proinflammatory processes while simultaneously stimulating the proresolution processes. This goal is complicated by the fact that the biocatalysts that produce the proinflammatory molecules, such as lipoxygenase (LOX) and cyclooxygenase (COX) isozymes, are the same biocatalysts that produce the proresolution molecules.3,6 Therefore, to develop the most effective chronic inflammatory therapeutics, it is imperative that the biosynthetic pathways for SPMs are characterized in order to determine the specific roles of LOX and COX in their production.
Maresins are a class of SPMs made from docosahexaenoic acid (DHA) and were first discovered in macrophage exudates and subsequently characterized by total synthesis.7–9 They are extremely potent oxylipins that decrease the inflammatory response as well as promote healing and homeostasis.3,10,11 Maresins have picomolar to nanomolar potency in reducing polymorphonuclear cell infiltration, increasing macrophage phagocytosis of cellular debris, and increasing tissue regeneration in a planaria model.7,12,13 They also inhibit TRPV1 currents in neurons and reduce neuropathic pain in mice.14,15 The first maresin discovered was maresin 1 (MaR1, 7R,14S-dihydroxy-4Z,8E,10E,12Z,16Z,19Z-docosahexaenoic acid), followed by maresin 2 (MaR2, 13R,14S-dihydroxy-4Z,7Z,9E,11Z,16Z,19Z-docosahexaenoic acid), and then the MaR-peptide derivatives, maresin conjugates in tissue regeneration (MCTR).14,16 These maresins are proposed to be partially synthesized by LOX isozymes; however, the specific LOX isozymes involved are not well-characterized.
There are six human LOX isozymes, whose primary reactivity is to abstract a hydrogen atom from a bis-allylic methylene on a polyunsaturated fatty acid and insert molecular oxygen, generating a hydroperoxide product. The naming of specific LOX isozymes is dependent on the carbon of the substrate that becomes oxidized. The oxygenation registry starts from the ω-end of the molecule, because the fatty acid substrate enters into the active site methyl-end first (Figure 1). For example, human platelet 12-lipoxygenase (h12-LOX or ALOX12) produces the ω-9 product, 12S-HpETE, from arachidonic acid (AA), while human reticulocyte 15-lipoxygenase-1 (h15-LOX-1 or ALOX15) produces primarily the ω-6 product, 15S-HpETE. In comparison, with DHA as the substrate, the ω-9 product of h12-LOX is primarily 14S-HpDHA, while the ω-6 product of h15-LOX-1 is primarily 17S-HpDHA.
Figure 1.
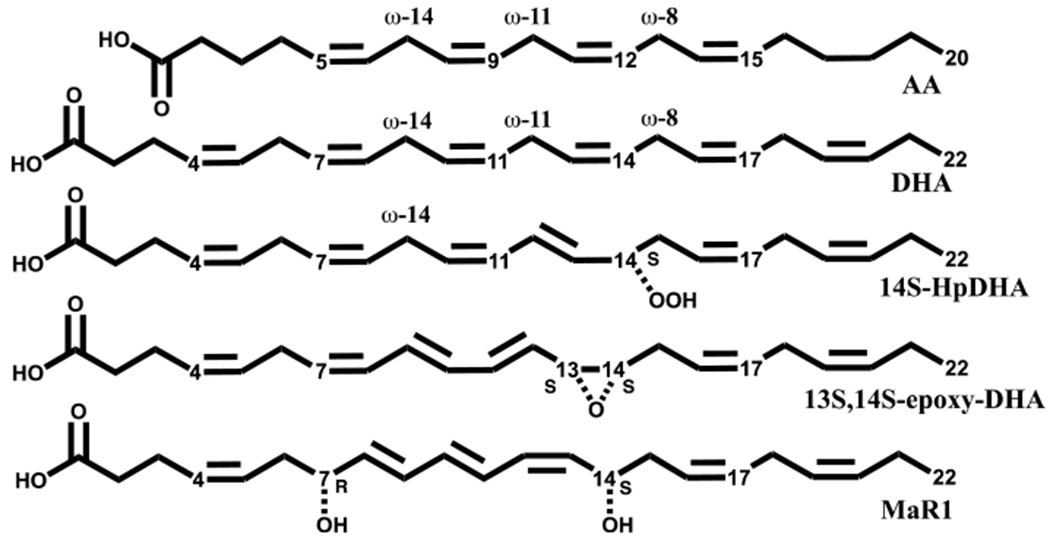
Structures of AA, DHA, 14S-HpDHA, 13S,14S-epoxy-DHA, and MaR1.
In addition to oxygenation reactions, LOX isozymes can also perform dehydration reactions, which are similar to the oxygenation reaction, but the product is an epoxide rather than a hydroperoxide. Specifically, LOX isozymes react with their own hydrogen peroxide oxylipin product and abstract a hydrogen atom from the adjacent bis-allylic methylene carbon, generating a septa-dienyl radical, similar to the oxygenation reaction. However, instead of oxygen attacking the septa-dienyl radical intermediate, the hydroperoxide moiety dehydrates to form an epoxide. The most characterized example of this mechanism is the reaction of h5-LOX with 5S-HpETE, to generate the 5,6-epoxide, leukotriene A4 (LTA4).17
The biosynthetic pathway for maresin biosynthesis utilizes both of these LOX reactions and is proposed to involve h12-LOX. First, h12-LOX is proposed to abstract a hydrogen atom from the ω-11 carbon, and then, the ω-9 carbon is oxygenated, generating 14S-HpDHA. h12-LOX then abstracts a hydrogen atom from the ω-14 carbon of 14S-HpDHA, which then dehydrates to form the critical maresin intermediate, 13S,14S-epoxy-4Z,7Z,9E,11E,16Z,19Z-docosahexaenoic acid (13S,14S-epoxy-DHA).18 This epoxide is then enzymatically hydrolyzed to form the final maresin product (Figure 1).5
While h12-LOX is the proposed biocatalyst for maresin production due to its known AA reactivity, it is theoretically possible that h15-LOX-1 is also capable of performing the same set of enzymatic reactions. While h15-LOX-1 generates primarily the ω-6 product (i.e., 15-HpETE or 17S-HpDHA), it can also generate the ω-9 product in appreciable amounts (i.e., 12-HpETE or 14S-HpDHA).19 These two hypothetical biosynthetic maresin pathways have biological ramifications, because protein levels of h12-LOX and h15-LOX-1 depend on the inflammatory stimuli and the cell type involved in the inflammatory response. For example, isolated macrophages have been shown to make MaR1 and MaR2; however, h12-LOX has no change in induction during the polarization of macrophages into the proresolving M2 phenotype, while h15-LOX-1 has a 300-fold increase in induction.5,20 Alternatively, h12-LOX is highly expressed in platelets and has been shown to produce 13S,14S-epoxy-DHA in a proinflammation state, but a hydrolase from neutrophils was required to make appreciable levels of MaR1 in neutrophil-platelet aggregates.21 Given these two possible biosynthetic pathways for producing maresins, the current work investigated the reactivities and kinetics of h12-LOX and h15-LOX-1 with the goal of determining their relative in vitro biosynthetic properties. By understanding which LOX enzyme is involved in the in vitro biosynthesis of maresins, these results may be extrapolated to their in vivo biosynthetic pathways, which could help direct inhibitor investigations and predict the cellular consequences of specific LOX inhibition.
MATERIALS AND METHODS
Chemicals.
Oxylipin mass spectrometry standards, maresin 1 (7R,14S-dihydroxy-4Z,8E,10E,12Z,16Z,19Z-docosahexaenoic acid), 7epi-maresin 1 (7-epi-MaR), 7S,14S-dihydroxy-4Z,8E,10E,12Z,16Z,19Z docosahexaenoic acid, d5-maresin 1, and 7S/R-HDHA, 7S/R-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid were purchased from Cayman Chemical. DHA, docosapentaenoic acid (DPAω-3), and AA were purchased from Nu Chek Prep, Inc. at +99% purity. Reagent and HPLC grade chemicals were used (Fisher Scientific, Pittsburgh, PA, USA).
Expression and Purification of h15-LOX-1, h15-LOX-2, h12-LOX, and h5-LOX.
Recombinant expression and purification of wild-type h15-LOX-1 (Uniprot entry P16050), h12-LOX (Uniprot entry P18054), h5-LOX (Uniprot entry P09917), and h15-LOX-2 (Uniprot entry O15296) were performed as previously described.17,22,23
The purities of the enzymes were assessed with a sodium dodecyl sulfate polyacrylamide gel to be >95%. The metal content was assessed on a Finnigan inductively coupled plasma mass spectrometer (ICP-MS), using an iron standard solution and curve along with cobalt–EDTA as an internal standard.
Synthesis and Purification of Oxylipins.
14S-HpDHA, 14S-HDPAω-3, and 12S-HETE were synthesized in 200 mL of 25 mM HEPES buffer at pH 7.5 for h15-LOX-1 and pH 8.0 for h12-LOX. The absorbance increase at 234 nm was monitored until it reached completion and quenched with 0.4% (v/v) glacial acetic acid. The solution was extracted 3 times with 100 mL of dichloromethane and evaporated to dryness. For the reduction of 12S-hydroperoxy-5Z,8Z,10E,14Z-eicosatetraenoic acid (12S-HpETE) to 12S-HETE as well as other hydroperoxides, trimethylphosphite was added in molar excess to extract. The HETE products were then purified on HPLC with a reverse phase Higgins Haisil Semi preparative C18 column (5 μm, 250 × 10 mm) and an isocratic 55:45 mixture of 99.9% acetonitrile with 0.1% acetic acid and 99.9% water with 0.1% acetic acid. However, the docosanoid products were purified more effectively with a normal phase Phenomonex silica column (5 μm, 250 × 10 mm) and an isocratic mixture of 99% hexane, 1% isopropanol, and 0.1% trifluoroacetic acid. 7(S)-Hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid (7S-HDHA) was synthesized by reaction of DHA (25–50 μM) with h5-LOX. The reaction was carried out for 2 h in 800 mL of 25 mM HEPES, pH 7.5, containing 50 mM NaCl, 100 μM EDTA, and 200 μM ATP. The reaction was quenched with 0.5% glacial acetic acid, extracted three times with 300 mL of dichloromethane, and evaporated to dryness under N2. The enzymatically produced hydroperoxide was reduced with the addition of trimethylphosphite. The product was purified isocratically on reverse phase as were HETE products. The purity was checked by LC-MS/MS to be greater than 95%.
Product Analysis of LOX Enzymes Reacting with DHA, 14S-HpDHA, and 14S-HDHA.
h15-LOX-1 (0.125 μM) and h12-LOX (0.300 μM) were reacted in 2 mL of 25 mM HEPES (pH 7.5 for h15-LOX-1, pH 8.0 for h12-LOX) at room temperature and ambient oxygen at 250 μM. For fatty acids, 10 μM AA or DHA was reacted for 1 min, while turnover was monitored by absorbance at 234 nm. For the oxylipin, 1 μM 14S-HpDHA or 14S-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid (14S-HDHA) were reacted for 20 min with h15-LOX-1 or 1 h with h12-LOX while being monitored at 270 nm (N = 3). The reaction was quenched with 0.4% glacial acetic acid, and d5-maresin-1 was added as an extraction/ionization standard. After quenching, the reaction was extracted three times with 2 mL of DCM. This reaction was done in parallel with a no enzyme control for background subtraction. In addition, hydroperoxide determination was established by split samples, in which one was reduced with trimethyl phosphite, while the other was left unreduced. A shift in retention time indicated peroxide reduction to the alcohols. The samples were blown down under N2 to dryness, reconstituted in 50 μL of acetonitrile and 50 μL of water with 0.1% formic acid, and 90 μL was injected for LC-MS/MS analysis. Structural characterization was performed on a Sciex Excion LC, using a C18 column (Phenomenex Kinetex, 4 μm, 150 × 2.0 mm). Mobile phase A consisted of water with 0.1% (v/v) formic acid, and mobile phase B was acetonitrile with 0.1% formic acid. The flow rate was 0.400 mL/min with initial conditions (15% B) maintained for 0.75 min. Mobile phase B was held at 15% over 1 min and then ramped to 30% over 0.75 min, to 47% over 2 min, to 54% over 1.5 min, to 60% over 4.5 min, to 70% over 4.5 min, to 80% over 1 min, to 100% over 1 min, held at 100% for 2 min, and finally returned to 15% to equilibrate for 2 min.24 The chromatography system was coupled to a Sciex PDA and x500B qTOF. Analytes were ionized through electrospray ionization with a −4.0 kV spray voltage and 50, 50, and 20 PSI for ion source gases 1 and 2 and the curtain gas, respectively. The CAD gas was set to 7, while the probe temperature was 550 °C, respectively. DP was −60 V, and CE was set to −10 V with a 5 V spread. MS2 acquisition was performed using SWATH, and m/z ratios of ±0.5:343.2 (HDHAs) and 359.2 (diHDHAs) were used. All analyses were performed in negative ionization mode at the normal resolution setting. Matching retention times, UV spectra, and fragmentation patterns to known standards with at least six common fragments identified the products. In the cases in which MS standards were not available, structures were deduced from comparison to known and theoretical fragments, while double bond geometry was determined through UV spectra. In the case of 7S,14S-dihydroxy-4Z,8E,10Z,12E,16Z,19Z-docosahexaenoic acid (7S,14S-EZE-diHDHA), its stereochemistry was determined by comparing enzymatically generated sample to known standards (Figure S1).
Scheme 1. Proposed Biosynthetic Scheme of Maresins with Lipoxygenase Kinetic Parameter kcat/KM Represented as Biosynthetic Flux (B.F.)a.
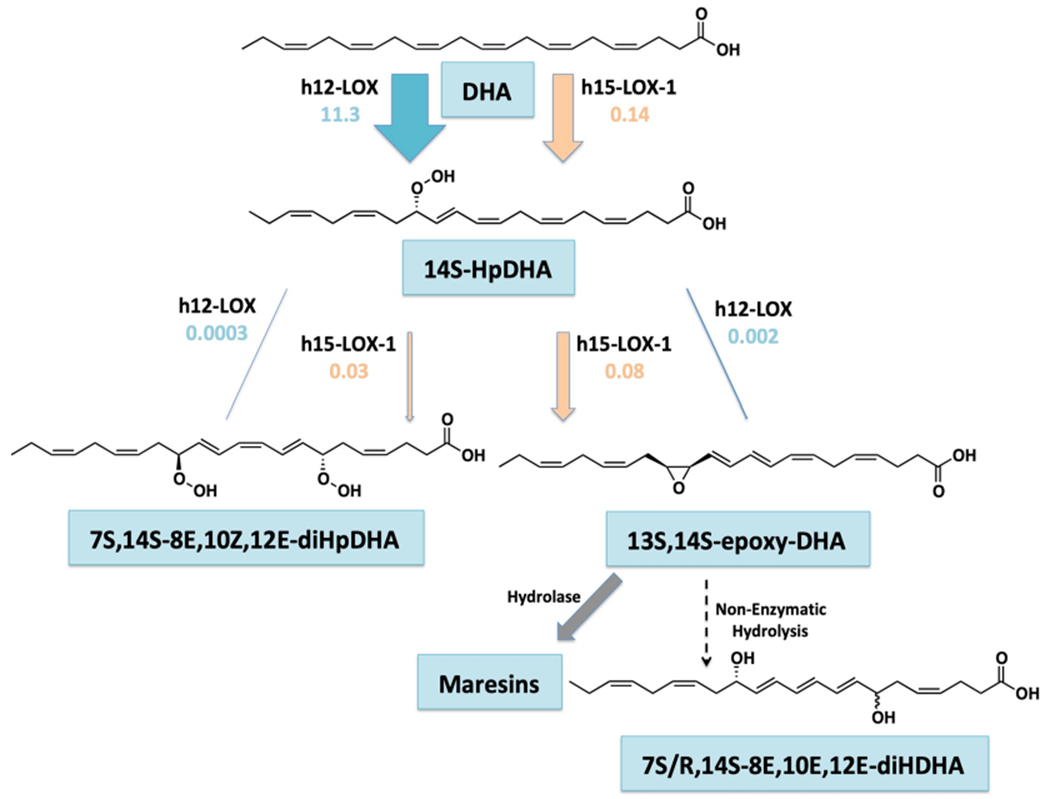
aB.F. = kcat/KM * % product. The size of the arrows are proportional to their biosynthetic flux.
Chiral Chromatography and Characterization of 7S,14S-EZE-diHDHA.
7S,14S-EZE-diHDHA was synthesized in 300 mL of 25 mM HEPES buffer (pH = 7.5), using 20 μM 7S-HDHA and h15-LOX-1 or using h5-LOX in 15 mL of buffer containing 10 μM 14S-HDHA and ~6 μM ammonium-sulfate-precipitated h5-LOX. Purified 7S,14S-EZE-diHDHA was analyzed via LC-MS/MS using a Chirapak AD-RH 2.1 × 150 mm, 5 μM chiral column coupled to a Thermo Electron LTQ. Mobile phase solvent A consisted of 99.9% acetonitrile and 0.1% formic acid, and solvent B consisted of 99.9% water and 0.1% formic acid. Separation was achieved over 60 min using an isocratic flow of 35:65 A/B for 0–30 min followed by a gradient from 35:65 A/B to 75:25 A/B from 30 to 60 min. The chromatography system was coupled to a Thermo Electron LTQ LC-MS/MS for mass analysis. Negative ionization and normal resolution settings were used. MS2 data was obtained with a mass list containing the m/z ratio of ±0.5:359.4 (diHDHAs). MS/MS fragments used to identify 7,14-diHDHA included: 341, 297, 221, 177, 141, and 123. Retention times and fragmentations were compared to MaR1 and 7-epi-MaR standards purchased from Cayman Chemicals (Ann Arbor, MI).
Steady-State Kinetics of h15-LOX-1, h15-LOX-2, and h12-LOX.
Reactions of h15-LOX-1 and h15-LOX-2 were performed in a 1 cm2 quartz cuvette containing 2 mL of RT, 25 mM HEPES buffer (pH 7.5), while h12-LOX reactions had the same conditions but at pH 8.0. DHA and AA concentrations were varied from 0 to 20 μM, and product formation was monitored by measuring absorbance at 234 nm using ε234 = 25 000 M−1 cm−1 for conjugated diene formation in buffer. Triplicate reactions were initiated by the addition of h15-LOX-1 (25 nM), h15-LOX-2 (220 nM), or h12-LOX (18 pM) and were monitored on a PerkinElmer Lambda 45 UV–Vis spectrophotometer. The stock concentration of 14S-HpDHA was determined by the absorbance at 234 nm in methanol using ε234 = 27 000 M−1 cm−1. 14S-HpDHA concentrations ranged from 1 to 200 μM for h15-LOX-1 and 5 to 200 μM for h12-LOX. Enzyme concentrations for oxylipin turnover were 200 nM for h15-LOX-1 and 1 μM for h12-LOX. The amount of h12-LOX is high due to its low reactivity with 14S-HpDHA. Product formation was measured by the change in absorbance at 270 nm for 7S,14S-EZE-diHDHA, 13S,14S-epoxy-DHA, and the nonenzymatic hydrolysis product, 7R/S,14S-dihydroxy-4Z,8E,10E,12E,16Z,19Z-docosahexaenoic acid (7R/S,14S-EEE-diHDHA) using the extinction coefficient for a conjugated triene in buffer, ε270 = 37 000 M−1 cm−1. 14S-Hydroxy,20-hydroxy-4Z,8Z,10Z,12E,16Z,19E-docosahexaenoic acid (14S,20-diHDHA) turnover was not included in kinetic parameter calculations because of nonoverlapping absorption spectra at 245 nm. Concentrations of the purified secondary products were assessed in methanol with an extinction coefficient of ε270 = 40 000 M−1 cm−1. KaleidaGraph (Synergy) was used to fit initial rates and the second-order derivatives (kcat/KM) to the Michaelis–Menten equation for the calculation of kinetic parameters.
Allosteric Modulation of Epoxidation of h15-LOX-1 and h12-LOX.
The allosteric effects were investigated using a constant 14S-HpDHA concentration (1 μM) and varying 12S-HETE or 14S-HDPAω-3 (1–100 μM). h15-LOX-1 (0.125 μM) or h12-LOX (0.3 μM) were used and reacted for 20 min or 1 h, respectively. 14S-HpDHA was also varied alone (1–50 μM), and product percentages were calculated (N = 3). Products were reduced and analyzed via LC-MS as described above (vide supra).
Effect of 14S-HpDHA on Human Platelet Aggregation and Lipidomics.
The University of Michigan Institutional Review Board approved all research involving human volunteers. Platelets were isolated from whole blood through serial centrifugation and resuspended in Tyrode’s buffer as previously published.25 For aggregation assays, 250 μL of washed human platelets (3 × 108/mL) were incubated with 1–10 μM 14S-HpDHA, 7S,14S-EZE-diHDHA, maresin-1, or control oxylipin 12S-hydroxy-8Z,10E,14Z-eicosatrienoic acid (12S-HETrE) for 10 min at 37 °C in a glass cuvette. The oxylipin-treated platelets were stimulated with 0.25 μg/mL collagen while stirring at 1100 rpm in a Chrono-log model 700 aggregometer, and aggregation was analyzed via a decrease in light transmittance. For lipidomics, 1 × 109 platelets resuspended in 1 mL of Tyrode’s buffer were incubated with 10 μM DHA or 17.5 μM 14(S)-HpDHA for 10 min and stimulated with collagen. Supernatants and pellets from washed platelets were extracted by a C18 cartridge using published methods and 20 ng of d5-MaR1 as an internal standard.21 The products were resuspended in 50:50 acetonitrile and 0.1% formic acid in water and analyzed by LC-MS. The m/z transition used for 14S-HDHA was 343.2 → 205.2, 11S-HDHA was 343.2 → 165.2, and 7,14-diDHA isomers was 359.2 → 177.2.
Molecular Docking.
A homology model of h15-LOX-1 was built using the substrate-mimetic inhibitor-bound porcine 12-LOX structure (pdb id 3rde). PRIME homology modeling software (Schrodinger Inc.) was used to build the model. During homology modeling, the metal ion (Fe3+) was retained, with a hydroxide ion that coordinated to the metal ion and the cocrystallized inhibitor. The subsequent model was subjected to a protein-preparation step using Protein Preparation Wizard (Schrodinger Inc.). During this step, hydrogen atoms were optimized to make better hydrogen bonding interactions, protonation states of titratable residues were assigned, and the model was energy-minimized with restraints, such that heavy atoms did not move beyond 0.3 Å. The three-dimensional (3D) structure of 14S-HpDHA was prepared from the SMILES string using LigPrep software (Schrodinger Inc.). Both the neutral and charged carboxylic acid forms of the substrate were generated, but neither protonation state of the substrate successfully docked to the active site using the standard rigid receptor docking using Glide (Schrodinger Inc.). Therefore, we used InducedFit docking to predict conformational changes necessary to accommodate this ligand. The metal ion, hydroxide ion, and metal ion coordinating residues (His360, His365, His540, His544, and Ile662) were kept fixed during the InducedFit docking; all other active site residue side chains as well as the substrate were sampled extensively. Subsequent to docking, the substrate-bound protein structures were subjected to molecular-mechanics based energy minimization (MM-GBSA) to rank them.
RESULTS
h15-LOX-1 Steady-State Kinetics and Product Distribution with DHA.
The efficiency of h15-LOX-1 in converting DHA into the peroxide MaR1 intermediate, 14S-HpDHA, was examined in order to determine its relevancy to maresin biosynthesis. First, the product profile for this reaction revealed dual product specificity with almost equal amounts of 14S-HpDHA (40%) and 17S-hydroperoxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid (17S-HpDHA) (46%) being produced, with a small amount of 11S-hydroperoxy-4Z,7Z,9E,13Z,15Z,19Z-docosahexaenoic acid (11S-HpDHA) (12%) (Table 1), consistent with previous work.19 This is indicative of the ability of h15-LOX-1 to produce both the ω-6 and ω-9 oxylipins, comparable to its production of 12S-HETE and 15S-HETE from AA. However, the increased percentage of the ω-9 oxylipin with DHA (40% 14S-HpDHA versus 10% 12S-HETE) supports the hypothesis that the increased chain length and desaturation of DHA allows for a deeper active site insertion of DHA. The production of 11S-HpDHA supports this hypothesis as well and indicates that DHA inserts into the active site even deeper than for 14S-HpDHA production, producing the ω-12 oxylipin product (11S-HpDHA).
Table 1.
Product Profile of h12-LOX and h15-LOX-1 Reacting with 10 μM DHA for 1 min
| enzyme + DHA | 11S-HpDHA (ω-12) | 14S-HpDHA (ω-9) | 17S-HpDHA (ω-6) |
|---|---|---|---|
| h15-LOX-1 | 12 ± 1% | 40 ± 1% | 46 ± 1% |
| h12-LOX | 19 ± 3% | 81 ± 3% | <1.0% |
With respect to kinetics, the rate of substrate capture26 (kcat/KM) for the reaction of h15-LOX-1 with DHA was 0.36 ± 0.1 s−1 μM−1, only 6 times slower than that of AA, its preferred substrate, indicating DHA is a competent substrate (Table 2). The lower kcat/KM is explained by the kcat value being half and the KM value being almost 3-fold greater for DHA than that for AA.
Table 2.
Steady-State Kinetics of h12-LOX and h15-LOX-1 with AA and DHAa
| enzyme + substrate | kcat (s−1) | KM (μM) | kcat/KM (s−1 μM−1) | relative kcat/KMb |
|---|---|---|---|---|
| h15-LOX-1 | ||||
| AA | 5.3 ± 0.4 | 2.7 ± 0.3 | 2.0 ± 0.2 | 1.0 |
| DHA | 2.4 ± 0.6 | 6.7 ± 3 | 0.36 ± 0.1 | 0.18 |
| h12-LOX | ||||
| AA | 18 ± 0.8 | 0.95 ± 0.1 | 19 ± 2 | 9.5 |
| DHA | 13 ± 0.2 | 0.90 ± 0.06 | 14 ± 0.8 | 7.0 |
The h15-LOX-1 with AA data from were obtained previously.22
The relative kcat/KM for h15-LOX-1 and AA is set to 1.
h12-LOX Steady-State Kinetics and Product Distribution with DHA.
The reaction of h12-LOX with DHA also revealed the production of 14S-HpDHA; however, for h12-LOX, it was the major product at 81%, with only 19% of the 11S-HpDHA product being made (Table 1). This result is in agreement with the literature, indicating that h12-LOX primarily produces the ω-9 oxylipin, 14S-HpDHA.19 However, the production of the ω-12 oxylipin from DHA, 11S-HpDHA, contrasts the fact that the ω-12 product of AA (9S-HpETE) is not produced with h12-LOX and AA.27 This result again supports the hypothesis that the longer length and higher degree of unsaturation of DHA allows for a deeper insertion into the active site, as was observed for 15-LOX-1 (vide supra).
For kinetics, the kcat/KM for this reaction was determined to be 14 ± 0.8 s−1 μM−1, which is comparable to that with AA.28 The kcat was also similar to that of AA (kcat = 13 ± 0.2 s−1), indicating similar rates of substrate capture (kcat/KM) and product release (kcat) between DHA and AA for h12-LOX (Table 2). When comparing the kinetics and product profiles of h15-LOX-1 and h12-LOX, the data revealed that h12-LOX is approximately 79-fold more efficient at producing the maresin intermediate, 14S-HpDHA, than h15-LOX-1 (39-fold from kcat/KM times 2-fold from product profile percentages equals a biosynthetic flux (B.F.) of 79-fold) (Table 3). This result is consistent with the biological role of h12-LOX considering that its primary reactivity is abstraction from the ω-11 carbon, with oxygen insertion at the ω-9 position (14S-HpDHA for DHA and 12-HpETE for AA).
Table 3.
Biosynthetic Flux (B.F.) of 14S-HpDHA of h15-LOX-1 and h12-LOX Reactions with DHAa
| enzyme + DHA | 14S-HpDHA biosynthetic flux | fold difference |
|---|---|---|
| h15-LOX-1 | 0.14 | 1.0 |
| h12-LOX | 11 | 79 |
B.F. = kcat/KM * % product.
h15-LOX-1 and h15-LOX-2 Steady-State Kinetics and Product Distribution with 14S-HpDHA and 14S-HDHA.
The kinetics and product profile of h15-LOX-1 reacting with 14S-HpDHA were examined to characterize the efficiency of h15-LOX-1 in synthesizing the key 13,14-epoxide intermediate in the biosynthesis of maresins. The major product formed was the dehydration product, 7R/S,14S-dihydroxy-4Z,8E,10E,12E,16Z,19Z-docosahexaenoic acid (7S/R,14S-EEE-diHDHA, 71%) (Table 4), which is formed by the nucleophilic attack of 13S,14S-epoxy-docosa-4Z,7Z,9E,11E,16Z-hexaenoic acid (13S,14S-epoxy-DHA) by water. The characterization of this epoxide degradation product was achieved by its reduced nature in nonreducing conditions (dialcohol instead of dihydroperoxide), its sole production at 10 μM molecular oxygen compared to atmospheric conditions (~250 μM molecular oxygen) (Figure S2), its MS fragments (m/z = 341, 323, 297, 279, 221, 177), and its similar UV absorbance shoulder heights at ~262 and ~282 nm (Figure S2).29 This molecule’s characterization is consistent with well-known literature results of h12-LOX epoxidation of 14S-HpDHA and its subsequent nonenzymatic hydrolysis.16,18 Oddly, the minor hydrolysis product, 13R/S,14S-diHDHA, was not seen, which could be due to sterics and reaction conditions. This production of 13S,14S-epoxy-DHA is consistent with a hydrogen abstraction from C9 (ω-14) by h15-LOX-1 and a subsequent dehydration to form the epoxide. The abstraction of the hydrogen atom from the ω-14 carbon is similar to the mechanism already observed in the production of 11S-HpDHA by h15-LOX-1 and DHA.
Table 4.
h15-LOX-1 and h12-LOX Product Profiles with 1 μM 14S-HpDHA Incubated for 20 min (pH = 7.5) and 60 min (pH = 8.0), Respectively, with Ambient O2 Concentrations
| enzyme + 14S-HpDHA | 13S,14S-epoxy-DHAa | 7S,14S-EZE-diHpDHA | 14S,20-diHpDHA |
|---|---|---|---|
| h15-LOX-1 | 71 ± 6% | 29 ± 6% | <1.0% |
| h12-LOX | 76 ± 11% | 12 ± 2% | 13 ± 9% |
7S/R,14S-EEE-diHDHA is the detected product from nonenzymatic hydrolysis of 13S,14S-epoxy-DHA.
The production of the minor product, 7S,14S-dihydroperoxy-4Z,8E,10Z,12E,16Z,19Z-docosahexaenoic acid (7S,14S-EZE-diHpDHA) (29%), is less easily explained (Table 4). This minor product was characterized by its shift in HPLC retention time when the dihydroperoxide was reduced to the dialcohol. In addition, characterization was determined by MS fragments (m/z = 341, 323, 297, 279, 221, 177), its UV spectra at 270 nm, which revealed appropriate shoulders for an E,Z,E conjugation between the hydroxyls, and the molecule’s alignment with the 7S,14S isomer by chiral chromatography as described in methods (Figure S1 and S2).30 In total, this characterizes 7S,14S-EZE-diHpDHA as an oxygenation product and is best explained by a reverse entry of the substrate, carboxylic acid first, into the active site. The reverse entry binding mode is well-characterized in the catalytic mechanism of h5-LOX, converting AA into 5S-HpETE.31–33 With the carboxylate group entering the active site first, it is reasonable to assume a deprotonated carboxylate would retard entry into the hydrophobic cavity. This hypothesis is supported by the 7-fold loss in 7S,14S-EZE-diHpDHA production at increased pH (Figure S3), which appears to be due to the deprotonation of the carboxylic acid. In support of this conclusion, pH levels have been shown to alter lipoxygenase product formation paralleling pH plot titrations of fatty acids along with unusually high pKas.34–36
Regarding the kinetics of h15-LOX-1 and 14S-HpDHA, its efficiency and turnover were found to be comparable to those of DHA, with kcat/KM being 0.11 ± 0.006 s−1 μM−1 and kcat being 2.0 ± 0.04 s−1 μM−1 (Table 5a). This similarity in rates is significant, since the hydrogen atom abstraction is from C9 (ω-14) on 14S-HpDHA, which is not the canonical abstraction mechanism seen when DHA is the substrate. In addition, the kinetic parameters of 14S-HDHA were markedly reduced relative to 14S-HpDHA, which is consistent with a dehydration reaction being the primary reaction mechanism for 14S-HpDHA. The lack of the hydroperoxide moiety in 14S-HDHA prohibits dehydration, and thus, only the oxygenation product is observed, 7S-hydroxy,14S-hydroperoxy-EZE-HDHA, with 14S-HDHA as the substrate.
Table 5.
Kinetics of DHA, 14S-HpDHA, and 14S-HDHA with h15-LOX-1 and h12-LOX
| h15-LOX-1 + substrate with |
kcat (s−1) |
KM (μM) |
kcat/KM (s−1
μM−1) |
relative kcat/KMa |
fold differencea |
|---|---|---|---|---|---|
| DHA | 2.4 ± 0.6 | 6.7 ± 3 | 0.36 ± 0.08 | 1.0 | 1.0 |
| 14S-HpDHA | 2.0 ± 0.04 | 18 ± 1 | 0.11 ± 0.006 | 0.31 | −3.2 |
| 14S-HDHA | 0.57 ± 0.01 | 18 ± 4 | 0.03 ± 0.002 | 0.080 | −13 |
| h12-LOX + substrate with |
kcat (s−1) |
KM (μM) |
kcat/KM (s−1
μM−1) |
relative kcat/KMa |
fold differencea |
| DHA | 13 ± 0.2 | 0.90 ± 0.06 | 14 ± 0.8 | 1.0 | 1.0 |
| 14S-HpDHA | 0.22 ± 0.02 | 88 ± 16 | 0.0024 ± 0.0002 | 0.00017 | −5880 |
| 14S-HDHA | 0.17 ± 0.03 | 210 ± 70 | 0.00080 ± 0.00003 | 0.00006 | −16 700 |
Relative and fold difference of kcat/KM of enzyme with DHA set to 1.
At various 14S-HpDHA concentrations, h15-LOX-2 was observed not to react with 14S-HpDHA under our conditions. This is understandable, since h15-LOX-2 strictly abstracts from the ω-8 methylene carbon, which is not present in 14S-HpDHA and thus explains the lack of catalysis.
h12-LOX Steady-State Kinetics and Product Distribution with 14S-HpDHA and 14S-HDHA.
In comparison, the kinetics and product profile were also investigated with h12-LOX and 14S-HpDHA. With respect to its product profile, the major product was the dehydration product, 13S,14S-epoxy-DHA, which was observed as the nonenzymatic hydrolysis product (7S/R,14S-EEE-diHDHA, 76%). This is identical to the major product of h15-LOX-1, with matching retention times on reverse phase chromatography, fragmentation patterns, and UV spectrum peak heights at 270 nm. Two minor products were also observed, the oxygenation product previously seen with h15-LOX-1, 7S,14S-EZE-diHpDHA (12%), and a new product, 14S,20-dihydroperoxy-4Z,8Z,10Z,12E,16Z,19E-docosahexaenoic acid (14S,20-diHpDHA) (13%) (Figure S2 and Table 4). The characterization of 14S,20-diHpDHA was determined by its MS fragmentation (m/z = 341, 323, 301, 297, 279, 205),37 and its absorption of 245 nm light, indicative of two unconjugated dienes (Figure S2).14,20,34 The production of 14S,20-diHpDHA is most likely due to a methyl-end first entry into the active site, however, with a restricted entry depth, thus allowing for C20 oxygenation. In support of these conclusions, the reduced oxylipin 14S-HDHA, which chemically cannot form the epoxide product, only produced the oxygenation product, 7S,14S-EZE-diHpDHA, similar to that seen for h15-LOX-1 (vide supra). It should be noted that there are no commercially available standards for these two minor products; however, 7S,14S-EZE-diHpDHA was stereochemically characterized (Figure S1). 14S,20-DiHpDHA was not stereochemically characterized due to no available standards.
The kinetics of h12-LOX with 14S-HpDHA were dramatically reduced, relative to AA. The kcat/KM was found to be 0.0024 ± 0.0002 s−1 μM−1, and the kcat was found to be 0.22 ± 0.02 s−1 μM−1 (Table 5b), which corresponds to a 5800-fold and 60-fold reduction, respectively, and indicates that 14S-HpDHA is a poor substrate, relative to DHA for h12-LOX. 14S,20-DiHpDHA was not included in the kinetic parameter calculations, because it absorbs at 245 nm and thus has a nonoverlapping spectrum. The kinetic rates of the alcohol, 14S-HDHA, were also examined and found to be about a third as efficient as 14S-HpDHA, consistent with a loss of the epoxide generating pathway (Table 5b).
When comparing the biosynthesis of 13S,14S-epoxy-DHA by both h15-LOX-1 and h12-LOX, it is observed that the kcat value of h15-LOX-1 is almost 10-fold greater than that of h12-LOX, and its kcat/KM is over 45-fold greater. When taking into account the percent production of 13S,14S-epoxy-DHA, the biosynthetic flux of h15-LOX-1 is over 40 times greater than that of h12-LOX (h15-LOX-1 kcat/KM * % = 0.078 and h12-LOX kcat/KM * % = 0.0018) (Table 6), indicating that h15-LOX-1 is a better catalyst in making the maresin intermediate, 13S,14S-epoxy-DHA, than h12-LOX and suggests that h15-LOX-1 could contribute significantly to maresin formation in vivo (vida infra).
Table 6.
Biosynthetic Flux (B.F.) of 13S,14S-Epoxy-DHA of h15-LOX-1 and h12-LOX Reactions with 14S-HpDHAa
| enzyme + 14S-HpDHA | 13S,14S-epoxy-DHA biosynthetic flux | fold difference |
|---|---|---|
| h15-LOX-1 | 0.078 | 43 |
| h12-LOX | 0.0018 | 1.0 |
B.F. = kcat/KM * % product.
h15-LOX-1 Allosteric Regulation of 14S-HpDHA Epoxidation.
It was observed that when h15-LOX-1 reacted with 14S-HpDHA at low concentration (1 μM), dehydration and the subsequent epoxide formation was the favored mechanism, resulting in the formation of the water-induced degradation product, 7R/S,14S-EEE-diHDHA (71 ± 11%). However, it was observed that at a high concentration of 14S-HpDHA (50 μM), oxygenation was favored, producing mostly 7S,14S-EZE-diHDHA (76 ± 0.5%) (Figure 2a). This was a substantial change in the reaction mechanism, with an approximately 80% increase in oxygenation products, indicating a possible allosteric interaction, as previously seen for h15-LOX-1 reacting with AA and linoleic acid (LA).22 In fact, when this response was titrated and fit to a hyperbolic curve, it resulted in a Kd value of 6.0 ± 3 μM, comparable in magnitude to previous allosteric regulators of h15-LOX-1.22
Figure 2.
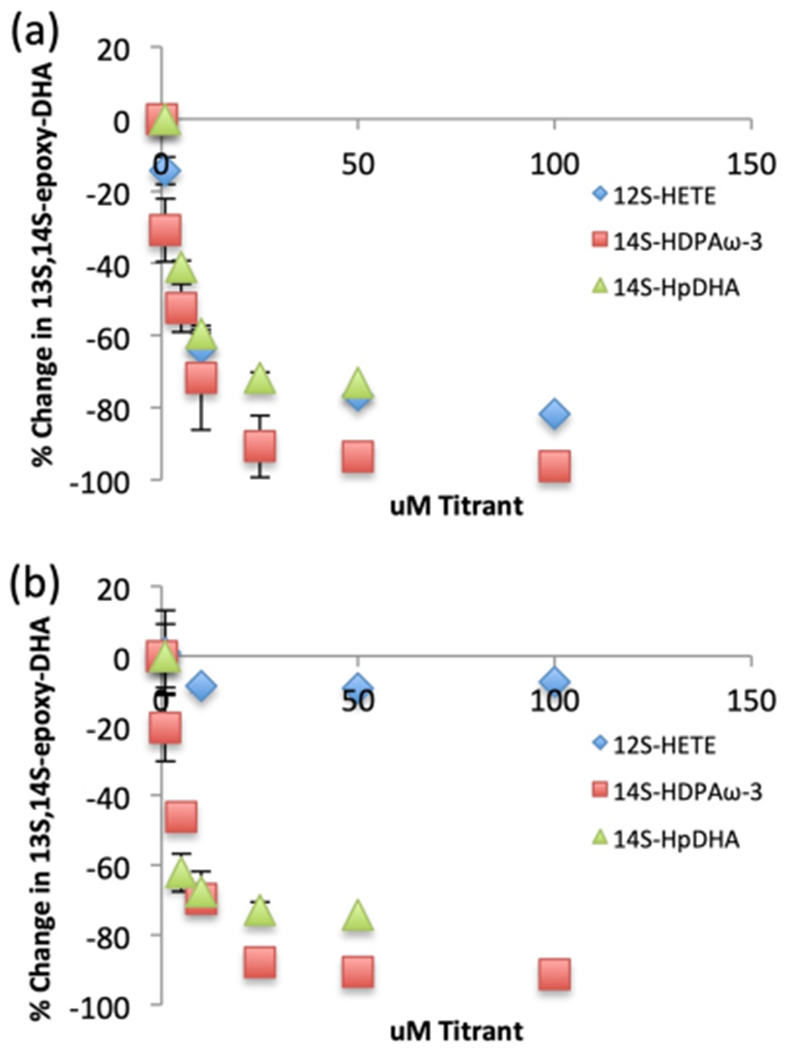
Percent of 13S,14S-epoxy-DHA (dehydration) vs 7S,14S-EZE-diHDHA/14S,20-diHpDHA (oxygenation). (a) h15-LOX-1 reaction with 1 μM 14S-HpDHA and increasing 14S-HpDHA (green triangles), increasing 14S-HDPAω-3 (red squares), and increasing 12S-HETE (blue diamonds) concentration. (b) h12-LOX reaction with 1 μM 14S-HpDHA and increasing 14S-HpDHA (green triangles), increasing 14S-HDPAω-3 (red squares), and increasing 12S-HETE (blue diamonds) concentration.
To investigate this hypothesis further, a previously reported allosteric modulator 12S-HETE and a similar LOX product to 14S-HDHA, 14S-hydroxy-7Z,10Z,12E,16Z,19Z-docosapentaenoic acid (14S-HDPAω-3), were titrated into the reaction mixture, and the epoxidation/oxygenation product percentages were recorded. With a constant concentration of 14S-HpDHA (1 μM) and a titration of 12S-HETE from 0 to 100 μM, h15-LOX-1 produced dramatically less epoxidation product, changing from 73 to 20% and resulting in a Kd of 3.5 ± 0.7 μM (Figure 2a). Repeating the same experiment with 14S-HDPAω-3 as the titrant, the epoxidation product was practically eliminated, decreasing from 71 to 2.6%, with a Kd of 4.0 ± 0.6 μM (Figure 2a). These data suggest that these three oxylipins, 14S-HpDHA, 14S-HDPAω-3, and 12S-HETE, bind to an allosteric site in h15-LOX-1 and regulate the product profile of 14S-HpDHA as the substrate. This result is significant, since 12S-HETE is found at higher concentrations in patients with essential hypertension, which could potentially affect maresin formation by allostery in vivo (vide infra).38 We are currently investigating this hypothesis further in human cell lines.
h12-LOX Allosteric Regulation of 14S-HpDHA Epoxidation.
Considering the allosteric effect observed for h15-LOX-1, the same experiments were performed to determine if an allosteric site was also present in h12-LOX and if it regulated the production of the maresin epoxide intermediate. A titration of h12-LOX with 14S-HpDHA (1 to 50 μM) decreased the epoxidation product from 76 to 19%, similar to the change seen for h15-LOX-1. In addition, the data were fit to a hyperbolic saturation curve, yielding a Kd of 3.5 ± 2 μM (Figure 2b). This data was supported with the titration of h12-LOX with 14S-HDPAω-3, which manifested a decrease in epoxidation from 72.0 to 5.0%, with a Kd of 4.6 ± 0.7 μM (Figure 2b). In contrast, 12S-HETE demonstrated no allosteric effect with h12-LOX (Figure 2b). These data indicate for the first time that h12-LOX also manifests an allosteric effect; however, it is more selective than that observed with h15-LOX-1, responding to only 14S-HpDHA and 14S-HDPAω-3 but not 12S-HETE.
Oxylipin Docking to h15-LOX-1 and h12-LOX.
To investigate the structural basis for the in vitro kinetic results indicating that h15-LOX is markedly more efficient in reacting with 14S-HpDHA than h12-LOX, computational docking was used to predict how 14S-HpDHA binds to both LOX isozymes. Initially, 14S-HpDHA was docked into h15-LOX-1 in the canonical manner with the methyl-end entering first (Figure 3a). The hydrophobic tail of 14S-HpDHA was positioned deep in the hydrophobic pocket created by residues Phe352, Ile413, Phe414, Ile417, and Met418. The carboxylate group makes a hydrogen bond with Arg402 and the hydroperoxide group on C14 makes a hydrogen bond with Glu356, thus positioning the reactive C9 4.3 Å away from the oxygen atom of the hydroxide ion.
Figure 3.
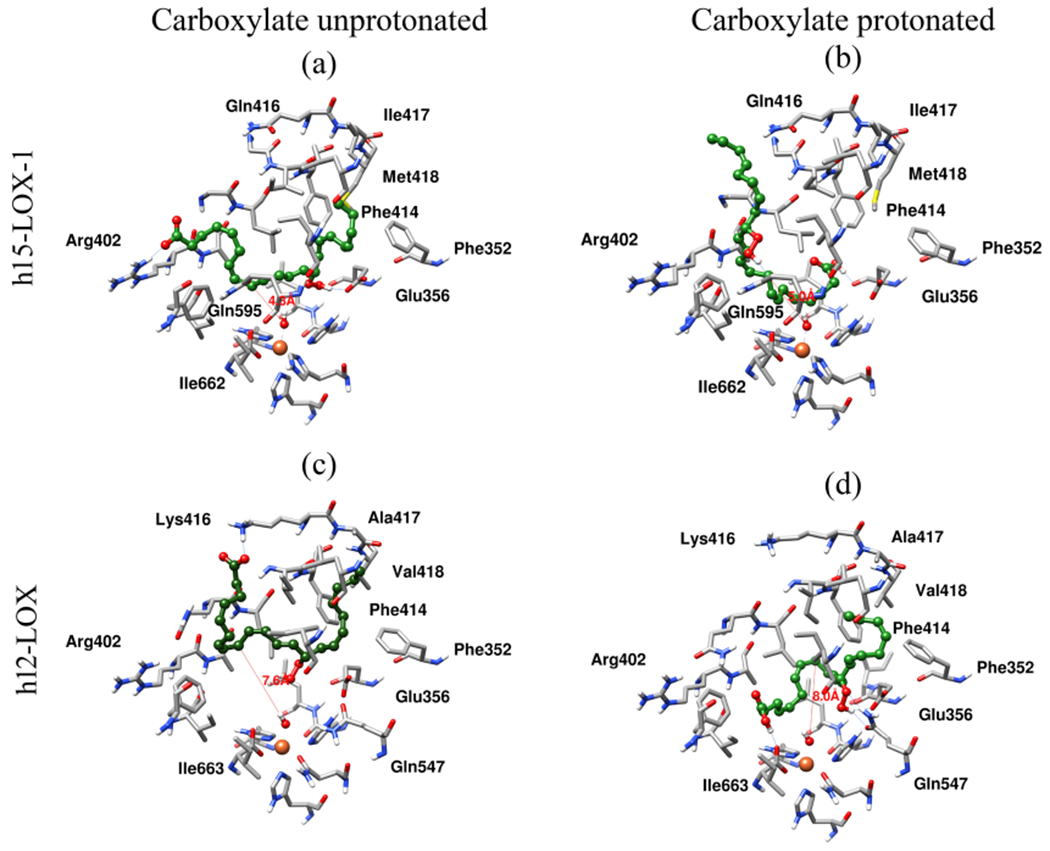
Docking pose of (a) h15-LOX-1 and 14S-HpDHA, methyl-end-bound first and (b) carboxylic-acid-end-bound first, (c) h12-LOX and 14S-HpDHA methyl-end-bound first, and (d) 14S-HpDHA, carboxylic-acid-end-bound first. Atoms and bonds of protein are shown in tube representation, and 14S-HpDHA is shown in ball-and-stick representation. Carbon atoms of protein and 14S-HpDHA are shown in gray and green colors, respectively. Nitrogen, oxygen, sulfur, and hydrogen atoms are shown in blue, red, yellow, and white colors. Hydrogen bonds between 14S-HpDHA and protein are shown with a blue color line, and the distance between the hydroxide oxygen atom to C9 of 14S-HpDHA is shown with a red color line.
In addition to dehydration, the production of a minor product, 7S,14S-EZE-diHpDHA, indicates an oxygenation reaction on C7. The close proximity of the oxygenated carbon to the carboxylate as well as the decreased production of this minor product at higher pH, implies that 14S-HpDHA may bind in the reverse orientation to generate 7S,14S-EZE-diHpDHA. Therefore, 14S-HpDHA was docked in a protonated form, in which case the predicted binding mode placed the carboxylic acid deep in the pocket, i.e., completely “flipped” relative to unprotonated 14S-HpDHA (Figure 3b), with the hydrophobic tail of 14S-HpDHA pointing toward the substrate entry opening. The proton of the carboxylic acid of 14S-HpDHA makes a hydrogen bond with the backbone carbonyl oxygen of Glu356, and the hydrogen atom of the hydroperoxide moiety makes a hydrogen bond with residue Gln595. In this binding pose, the reactive C9 is 5.0 Å from the oxygen of the hydroxide ion, which is comparable to the distance seen for the methyl-end first docking pose.
Computational docking also provides an explanation for the lower ability of h12-LOX to catalyze the analogous reaction to produce 7S,14S-EZE-diHpDHA from 14S-HpDHA. For the unprotonated, charged substrate, docking predicted a methyl-end first docking pose (Figure 3c), with the carboxylate group of 14S-HpDHA making a hydrogen bond with the side chain of Lys416 and the hydroperoxide hydrogen atom forming a hydrogen bond with the oxygen atom on the hydroxide ion liganded to the iron. This docking pose positions the reactive C9 far away from the oxygen atom of hydroxide ion (7.9 Å), which is not favorable for catalysis. Protonated 14S-HpDHA was predicted to bind much deeper in the cavity (Figure 3d), though not in a “flipped” orientation. The hydrogen atom of the carboxylic acid group makes a hydrogen bond with the C-terminal Ile663 residue, and the hydroperoxide oxygen and hydrogen atoms form hydrogen bonds with the side chains of Glu356 and Gln547, respectively. The reactive C9 is again 8.0 Å away from the hydroxide oxygen atom. In summary, both binding poses to h12-LOX place C9 significantly further from the catalytic site than in the poses predicted for h15-LOX-1, consistent with the lowered activity of h12-LOX relative to h15-LOX-1. We speculate that low but measurable activity with h12-LOX could be explained by conformational fluctuations that transiently bring C9 closer to the catalytic center.
Ex Vivo Incubations of Human Platelets and DHA and 14S-HpDHA.
Platelets contain large amounts of h12-LOX, which produce significant quantities of oxylipins. Considering that h12-LOX produced a mixture of 14S-HpDHA (81%) and 11S-HpDHA (19%) when reacted with DHA in vitro, DHA was administered to human platelets to determine if the ex vivo metabolites were of comparable ratios. Consistently, platelets produced the same oxylipins; however, their ratio was slightly different, with 14S-HDHA manifesting 94 ± 5% and 11S-HDHA manifesting 6% ± 1% (Figure 4a). These two oxylipins were reduced approximately 70% with the addition of the selective h12-LOX inhibitor, ML355,39,40 indicating both products are produced by h12-LOX (data not shown). The observed percentage of 11S-HDHA in platelets is less than that observed in vitro and could represent a cellular allosteric effect favoring the maresin pathway by making the important intermediate, 14S-HpDHA. In addition, 11S-HDHA could be preferentially metabolized, thus reducing its relative ratio to 14S-HDHA.
Figure 4.
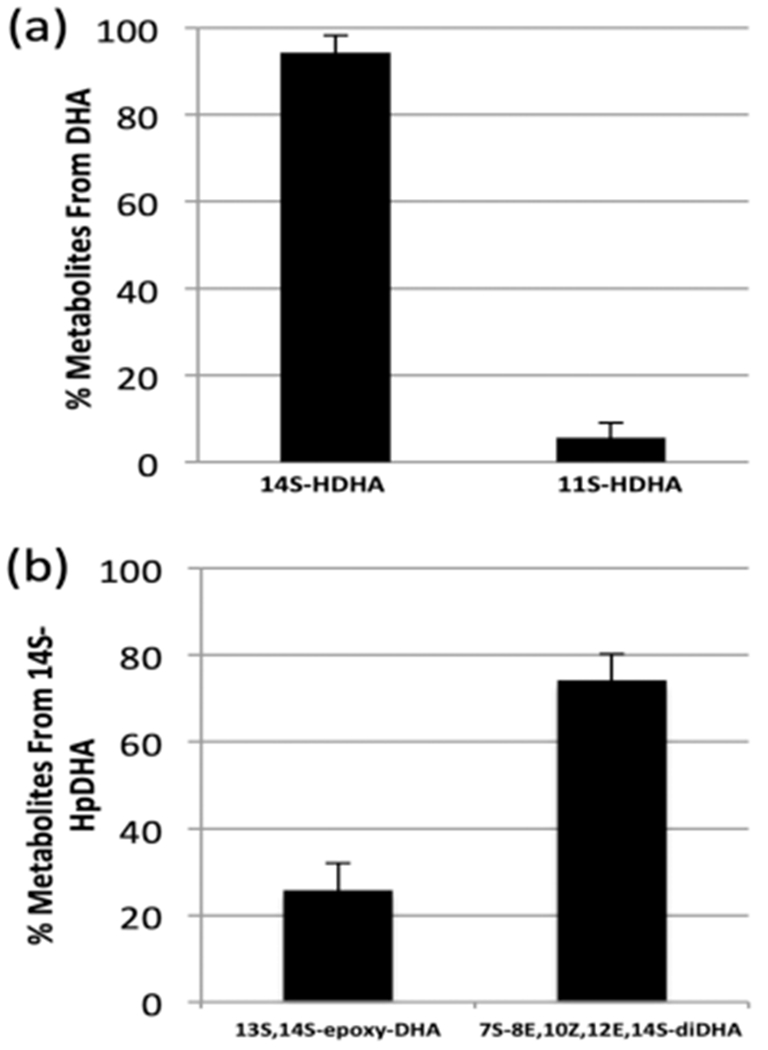
Percent of metabolites of human platelets (N = 3, 3 ×108 platelets/mL) reacting with (a) 10 μM DHA and (b) 17.5 μM 14S-HpDHA.
The in vitro kinetic data of h12-LOX also indicated that 14S-HpDHA was a poor substrate; however, this may not be the case ex vivo with human platelets. Therefore, human platelets were incubated with 14S-HpDHA (17.5 μM) to determine the quantity of oxylipins produced and the cellular product profile. The primary metabolite of 14S-HpDHA was the oxygenation product, 7S,14S-EZE-diHDHA (34.3 ng/1 × 109 platelets, 76 ± 6%), while the minor metabolite was the epoxidation product, 7R/S,14S-EEE-diHDHA (24 ± 6%, 11.8 ng/1 × 109 platelets) (Figure 4b). This percentage of products is similar to the percentage of epoxidation and oxygenation when using recombinant h12-LOX at a 20 μM 14S-HpDHA concentration (vide supra), highlighting the fact that at higher substrate concentrations, more of the oxygenation products are favored in platelets as well. With respect to the overall turnover in platelets, the amount of oxylipin products observed with 14S-HpDHA as the substrate is considerably less than that seen when AA is the substrate. With 17.5 μM AA added, 6000 ng of 12S-HpETE is produced per 1 × 109 platelets, while only 34 ng of 7S,14S-EZE-diHDHA and 12 ng of 7R/S,14S-EEE-diHDHA are produced per 1 × 109 platelets. These data show that less than 1% of 14S-HpDHA is catalyzed relative to AA, indicating that 14S-HpDHA is a poor substrate for h12-LOX ex vivo, consistent with its poor in vitro activity.
14S-HpDHA Inhibits Platelet Aggregation.
The ability of platelet-derived oxylipins to activate or inhibit platelet function depends at least in part on the metabolizing enzyme and starting PUFA.41 Therefore, isolated human platelets were treated with 14S-HpDHA for 10 min and then stimulated with collagen (0.25 μg/mL) in an aggregometer to determine the effect of 14S-HpDHA on platelet activation (Figure 5). Platelets treated with ≥1 μM of 14S-HpDHA had over a 90% reduction in aggregation compared to vehicle control, indicating that 14S-HpDHA is significantly more potent relative to the other h12-LOX-derived antiplatelet oxylipins, 7S,14S-EZE-diHDHA, MaR-1, and 12S-HETrE, which all required 5–10 μM to completely inhibit platelet aggregation.41 The novel and high potency of 14S-HpDHA demonstrate its potential value as an antiaggregation therapeutic, which we are currently investigating in more detail.
Figure 5.
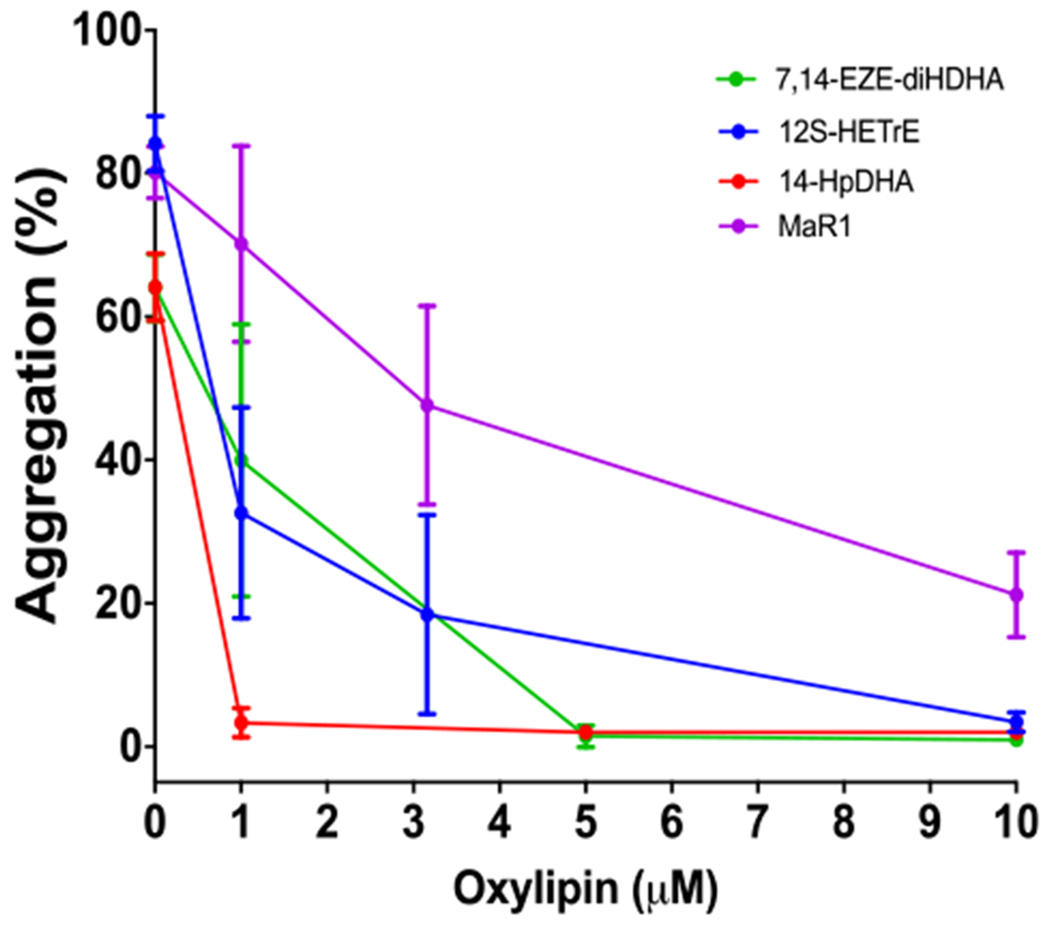
14S-HpDHA inhibits platelet aggregation. Human platelets were incubated with 14HpDHA, 12S-HETrE, 7S,14S-EZE-diHDHA, and MaR1 at concentrations ranging from 1 to 10 μM and then stimulated with collagen (0.25 μg/mL) in a Chrono-log aggregometer. Data represent the mean ± SEM of the maximum aggregation of three to five independent experiments. Statistical analysis was performed using one-way ANOVA with Dunnett’s test comparing the aggregation of platelets treated with each concentration of oxylipin to the aggregation of vehicle-treated platelets. **P < 0.01, ***P < 0.001.
DISCUSSION
Maresins are critical bioactive molecules that turn off inflammation and promote healing by reducing polymorphonuclear cell infiltration, increasing macrophage phagocytosis of cellular debris and tissue regeneration.1,12 It has been proposed that h12-LOX is the primary LOX isozyme in the in vivo maresin biosynthesis.20 In an attempt to understand the in vivo biosynthesis of MaR1, in vitro investigations have been performed to determine potential routes of biosynthesis.
In the current work, h12-LOX has been shown to be much more efficient in producing the maresin intermediate, 14S-HpDHA, than h15-LOX-1. This result is expected, since hydrogen atom abstraction from the ω-11 carbon with oxygenation at the ω-9 carbon is the canonical biosynthetic mechanism for h12-LOX (i.e., 12S-HpETE production from AA and 14S-HpDHA from DHA). For h15-LOX-1, the canonical mechanism is hydrogen atom abstraction from the ω-8 carbon, with oxygenation at the ω-6 carbon (i.e., 15S-HpETE production from AA and 17S-HpDHA from DHA), and indeed, 17S-HpDHA is the major product from DHA (46%). However, h15-LOX-1 also makes a significant amount of the noncanonical ω-9 product, 14S-HpDHA (40%), which is substantially more than that seen with AA (10% 12S-HpETE). This shift in reactivity to the oxygenation of the ω-9 carbon is most likely due to the change in the nature of the substrate, with DHA having increased length and unsaturation relative to AA. This hypothesis is supported by the fact that h15-LOX-1 also produces 12% 11S-HpDHA, which implies an even deeper fatty acid insertion into the active site for DHA and a subsequent hydrogen atom abstraction from the ω-14 carbon. This expanded reactivity of h15-LOX-1 led us to consider h15-LOX-1 as a possible partner in the next step of maresin biosynthesis, the dehydration of 14S-HpDHA to form 13S,14S-epoxy-DHA, which requires a similar ω-14 abstraction.
Utilizing 14S-HpDHA as the substrate, the present work confirmed that 13S,14S-epoxy-DHA, the maresin intermediate, was the major product from the reaction with h12-LOX.18 In this reaction, a hydrogen atom is abstracted from the ω-14 carbon, with dehydration of the hydroperoxide on the ω-9 carbon to form the ω-8,9 epoxide. In addition to this expected result, the data indicates that h15-LOX-1 also produces 13S,14S epoxy-DHA from 14S-HpDHA as the major product, similar to the epoxidation reactions of h15-LOX-1 seen before.42,43 This result demonstrates that both h12-LOX and h15-LOX-1 can abstract a hydrogen atom from the ω-14 carbon of 14S-HpDHA, enabling dehydration of the hydroperoxide to form the epoxide. For both h12-LOX and 15-LOX-1, this is a noncanonical reaction, albeit not entirely unexpected, given that 11S-HpDHA can be produced as a minor product of the DHA reaction with both h12-LOX (19%) and h15-LOX-1 (12%), which requires abstraction of a hydrogen atom from the ω-14 carbon. It should be noted that the ability of both h12-LOX and 15-LOX-1 to abstract from the ω-14 carbon is unique to the C22:6 oxylipins. Neither h12-LOX nor 15-LOX-1 react with AA to produce the corresponding 9S-HpETE, even though the ω-14 carbon of AA is also an activated methylene carbon in the middle of a 1,4-pentadiene moiety. Clearly, the longer length and higher degree of unsaturation of DHA allows for an expanded reactivity for both isozymes.
Although both h12-LOX and 15-LOX-1 react with 14S-HpDHA and produce a majority of 13S,14S-epoxy-DHA, their rates are not comparable. h15-LOX-1 produces 13S,14S-epoxy-DHA with a kcat/KM value over 40 times faster than that for h12-LOX, implicating h15-LOX-1 as a possible source of maresins in vivo. This is supported by the observation that M2-polarized macrophages and platelet-neutrophil aggregates produce substantial amounts of maresins.20,21 When M2-polarized macrophages are challenged by the h15-LOX-1 inhibitor BLX-3887, the production of maresin and 14S-HDHA are decreased by 70 and 45%, respectively.44 This proresolution polarization of macrophages also coincides with very high h15-LOX-1 expression compared to h12-LOX,12 and therefore, it is possible that the maresins in this cell type are synthesized primarily by h15-LOX-1, similar to our in vitro data. It has also been proposed that h12-LOX could be involved in the biosynthesis of maresins, since high levels of h12-LOX are found in platelets, along with neutrophil hydrolase expression, which could allow for maresin transcellular biosynthesis during aggregation. However, our observation of a low turnover of 14S-HpDHA by platelets to the epoxide products ex vivo indicates this pathway could be less important in vivo. In addition, the low reactivity of h12-LOX with 14S-HpDHA coincides with a nonproductive binding in the active site, as seen by our docking model. A possible scheme is presented for maresin formation, indicating that both h12-LOX and h15-LOX-1 could be involved in maresin biosynthesis (Scheme 1). It should be noted that these biosynthetic flux values are approximations of the possible rates in vivo, since there are many variables in the cell that could affect these rates, such as small molecules, oxygen levels, and proteins (vide infra). In addition, different optimal pH buffers were utilized for each of the LOX isozymes to achieve more accurate kinetic values.
The role of an allosteric site for both h12-LOX and h15-LOX-1 was investigated in order to determine if the generation of the maresin intermediate, 13S,14S-epoxy-DHA, was affected in vitro. In fact, the data indicates that increased 14S-HpDHA concentration did decrease 13S,14S-epoxy-DHA production and increased oxygenation products, thus suggesting a negative product feedback effect for maresin formation. The molecular mechanism for this effect could be due to the allosteric effector changing the binding site in a way that increases the frequency of the reverse entry of 14S-HpDHA into the active site (i.e., carboxylic acid-end first), e.g., by increasing the size of the active site and thereby lowering the energetic cost of burying the polar carboxylic acid in the active site. This effect was previously seen for h5-LOX catalysis31 and could explain the disparities between the in vitro and in vivo ratios of 11- and 14-oxylipins, which we are currently investigating. Another possibility is that the hydrogen bond to Gln595 allows for preferential epoxidation, which is lost due to the allosteric effect. We are currently investigating these hypotheses with active site mutagenesis.
CONCLUSIONS
In summary, we have demonstrated that while h12-LOX is a viable biocatalyst for the production of the key maresin intermediate, 13S,14S-epoxy-DHA, h15-LOX-1 maybe a more efficient pathway to maresin biosynthesis. In addition, we have identified possible allosteric regulation of maresin biosynthesis for both h12-LOX and h15-LOX-1 by oxylipins. Given that 14S-HpDHA has micromolar potency for affecting maresin formation, it is biologically feasible that maresin formation is negatively regulated by oxylipin concentrations in the cell. This fact, coupled with the high potency of maresins and the slow rate in which h12-LOX and 15-LOX-1 make the necessary epoxide intermediate, 13S,14S-epoxy-DHA, suggest that there may be multiple biosynthetic pathways to help control inflammation resolution with maresins. Finally, 14S-HpDHA was found to be a highly potent antiplatelet metabolite of DHA, which could imply additional dietary benefits for fish oil consumption. These new insights into the biochemistry of maresin biosynthesis will allow for a better understanding of the biological consequences of LOX inhibitors as well as provide a novel target for therapeutic investigations in accelerating resolution of acute inflammation.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge Prof. Kelley Moremen for helpful discussions and expressing h12-LOX in Chinese hamster ovary cells, whose protein was comparable to that expressed in SF9 insect cells.
Funding
NIH R01 GM105671 (M.H. and T.R.H.); NIH R01 HL11405 (M.H. and T.R.H.); NIH R35 GM131835 (M.H. and T.R.H.); and NIH K99HL136784 (B.E.T.).
ABBREVIATIONS
- AA
arachidonic acid
- DHA
docosahexaenoic acid
- SPM
specialized proresolving mediator
- 12S-HETE
12S-hydroxy-5Z,8Z,10E,14Z-eicosatetraenoic acid
- 12S-HETrE
12S-hydroxy-8Z,10E,14Z-eicosatrienoic acid
- 15S-HETE
15S-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid
- 7S-HDHA
7S-hydroxy-4Z,8E,10Z,13Z,16Z,19Z-docosahexaenoic acid
- 14S-HpDHA
14S-hydroperoxy-4Z,7Z,10Z,12E,16Z,19Z-docosa-hexaenoic acid
- 14S-HDHA
14S-hydroxy-4Z,7Z,10Z,12E,16Z,19Z-docosahexaenoic acid
- 17S-HpDHA
17S-hydroperoxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid
- 17S-HDHA
17S-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoic acid
- 11S-HpDHA
11S-hydroperoxy-4Z,7Z,9E,13Z,15Z,19Z-docosahexaenoic acid
- 11S-HDHA
11S-hydroxy-4Z,7Z,9E,13Z,15Z,19Z-docosahexaenoic acid
- 14S-HDPAω-3
14S-hydroxy-7Z,10Z,12E,16Z,19Z-docosapentaenoic acid
- 13S, 14S-epoxy-DHA
13S,14S-epoxy-4Z,7Z,9E,11E,16Z,19Z-docosahexaenoic acid
- maresin 1 (MaR1)
7R,14S-dihydroxy-4Z,8E,10E,12Z,16Z,19Z- docosahexaenoic acid
- 7epi-maresin 1 (7-epi-MaR)
7S,14S-dihydroxy-4Z,8E,10E,12Z,16Z,19Z-docosahexaenoic acid
- maresin 2 (MaR2)
13R,14S-dihydroxy-4Z,7Z,9E,11Z,16Z,19Z-docosahexaenoic acid
- 7S, 14S-EZE-diHDHA
7S,14S-dihydroxy-4Z,8E,10Z,12E,16Z,19Z-docosahexaenoic acid
- 7R/S, 14S-EEE-diHDHA
7R/S,14S-dihydroxy-4Z,8E,10E,12E,16Z,19Z-docosahexaenoic acid
- 7S, 14S-EZE-diHpDHA
7S,14S-dihydroperoxy-4Z,8E,10Z,12E,16Z,19Z-docosahexaenoic acid
- 14S, 20-diHpDHA
14S-hydroperoxy, 20-hydroperoxy-4Z,8Z,10Z,12E,16Z,19E-docosahexaenoic acid
- 14S, 20-diHD-HA
14S-hydroxy, 20-hydroxy-4Z,8Z,10Z,12E,16Z,19E-docosahexaenoic acid
- h15-LOX-1
human reticulocyte 15-lipoxygenase-1 (ALOX15)
- h12-LOX
human platelet 12-lipoxygenase (ALOX12)
- h5-LOX
human 5-lipoxygenase (ALOX5)
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00233.
MS characterization of oxylipin standards and pH dependence on product profiles (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00233
The authors declare the following competing financial interest(s): MPJ is a consultant to and shareholder of Schrodinger LLC, which licenses the software used in this work.
Contributor Information
Cody Freedman, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, California 95064, United States.
Adrianne Tran, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, California 95064, United States.
Benjamin E. Tourdot, Department of Pharmacology, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States
Chakrapani Kalyanaraman, Department of Pharmaceutical Chemistry, School of Pharmacy, University of California San Francisco, San Francisco, California 94158, United States.
Steve Perry, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, California 95064, United States.
Michael Holinstat, Department of Pharmacology, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States.
Matthew P. Jacobson, Department of Pharmaceutical Chemistry, School of Pharmacy, University of California San Francisco, San Francisco, California 94158, United States.
Theodore R. Holman, Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, California 95064, United States.
REFERENCES
- (1).Tabas I, and Glass CK (2013) Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science 339, 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, Li Y, Wang X, and Zhao L (2018) Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 9, 7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Serhan CN, Chiang N, and Van Dyke TE (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 8, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wongrakpanich S, Wongrakpanich A, Melhado K, and Rangaswami J (2018) A Comprehensive Review of Non-Steroidal Anti-Inflammatory Drug Use in The Elderly. Aging Dis 9, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sugimoto MA, Sousa LP, Pinho V, Perretti M, and Teixeira MM (2016) Resolution of Inflammation: What Controls Its Onset? Front. Immunol 7, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wisastra R, and Dekker FJ (2014) Inflammation, Cancer and Oxidative Lipoxygenase Activity are Intimately Linked. Cancers 6, 1500–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Serhan CN, Yang R, Martinod K, Kasuga K, Pillai PS, Porter TF, Oh SF, and Spite M (2009) Maresins: novel macrophage mediators with potent antiinflammatory and proresolving actions. J. Exp. Med 206, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rodriguez AR, and Spur BW (2015) First total synthesis of the macrophage derived anti-inflammatory and pro-resolving lipid mediator Maresin 2. Tetrahedron Lett 56, 256–259. [Google Scholar]
- (9).Rodriguez AR, and Spur BW (2012) Total synthesis of the macrophage derived anti-inflammatory lipid mediator Maresin 1. Tetrahedron Lett 53, 4169–4172. [Google Scholar]
- (10).Wang CW, Colas RA, Dalli J, Arnardottir HH, Nguyen D, Hasturk H, Chiang N, Van Dyke TE, and Serhan CN (2016) Maresin 1 Biosynthesis and Proresolving Anti-infective Functions with Human-Localized Aggressive Periodontitis Leukocytes. Infect. Immun 84, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tang S, Wan M, Huang W, Stanton RC, and Xu Y (2018) Maresins: Specialized Proresolving Lipid Mediators and Their Potential Role in Inflammatory-Related Diseases. Mediators Inflammation 2018, 2380319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Wuest SJ, Crucet M, Gemperle C, Loretz C, and Hersberger M (2012) Expression and regulation of 12/15-lipoxygenases in human primary macrophages. Atherosclerosis 225, 121–127. [DOI] [PubMed] [Google Scholar]
- (13).Serhan CN, Dalli J, Karamnov S, Choi A, Park CK, Xu ZZ, Ji RR, Zhu M, and Petasis NA (2012) Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB J. 26, 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dalli J, Vlasakov I, Riley IR, Rodriguez AR, Spur BW, Petasis NA, Chiang N, and Serhan CN (2016) Maresin conjugates in tissue regeneration biosynthesis enzymes in human macrophages. Proc. Natl. Acad. Sci. U. S. A 113, 12232–12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jo YY, Lee JY, and Park CK (2016) Resolvin E1 Inhibits Substance P-Induced Potentiation of TRPV1 in Primary Sensory Neurons. Mediators Inflammation 2016, 5259321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Deng B, Wang CW, Arnardottir HH, Li Y, Cheng CY, Dalli J, and Serhan CN (2014) Maresin biosynthesis and identification of maresin 2, a new anti-inflammatory and pro-resolving mediator from human macrophages. PLoS One 9, No e102362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Smyrniotis CJ, Barbour SR, Xia Z, Hixon MS, and Holman TR (2014) ATP Allosterically Activates the Human 5-Lipoxygenase Molecular Mechanism of Arachidonic Acid and 5(S)-Hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic Acid. Biochemistry 53, 4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dalli J, Zhu M, Vlasenko NA, Deng B, Haeggstrom JZ, Petasis NA, and Serhan CN (2013) The novel 13S,14S-epoxymaresin is converted by human macrophages to maresin 1 (MaR1), inhibits leukotriene A(4) hydrolase (LTA(4)H), and shifts macrophage phenotype. FASEB J. 27, 2573–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kutzner L, Goloshchapova K, Heydeck D, Stehling S, Kuhn H, and Schebb NH (2017) Mammalian ALOX15 orthologs exhibit pronounced dual positional specificity with docosahexaenoic acid. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1862, 666–675. [DOI] [PubMed] [Google Scholar]
- (20).Dalli J, and Serhan CN (2012) Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood 120, E60–E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Abdulnour REE, Dalli J, Colby JK, Krishnamoorthy N, Timmons JY, Tan SH, Colas RA, Petasis NA, Serhan CN, and Levy BD (2014) Maresin 1 biosynthesis during platelet-neutrophil interactions is organ-protective. Proc. Natl. Acad. Sci. U. S. A 111, 16526–16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wecksler AT, Kenyon V, Deschamps JD, and Holman TR (2008) Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry 47, 7364–7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Wecksler AT, Kenyon V, Garcia NK, Deschamps JD, van der Donk WA, and Holman TR (2009) Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry 48, 8721–8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yang J, Schmelzer K, Georgi K, and Hammock BD (2009) Quantitative profiling method for oxylipin metabolome by liquid chromatography electrospray ionization tandem mass spectrometry. Anal. Chem 81 , 8085–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Green AR, Freedman C, Tena J, Tourdot BE, Liu B, Holinstat M, and Holman TR (2018) 5 S,15 S-Dihydroperoxyeicosatetraenoic Acid (5,15-diHpETE) as a Lipoxin Intermediate: Reactivity and Kinetics with Human Leukocyte 5-Lipoxygenase, Platelet 12-Lipoxygenase, and Reticulocyte 15-Lipoxygenase-1. Biochemistry 57, 6726–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Northrop DB (1998) On the meaning of Km and V/K in enzyme kinetics. J. Chem. Educ 75, 1153–1157. [Google Scholar]
- (27).Wecksler AT, Garcia NK, and Holman TR (2009) Substrate specificity effects of lipoxygenase products and inhibitors on soybean lipoxygenase-1. Bioorg. Med. Chem 17, 6534–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ikei KN, Yeung J, Apopa PL, Ceja J, Vesci J, Holman TR, and Holinstat M (2012) Investigations of human platelet-type 12-lipoxygenase: role of lipoxygenase products in platelet activation. J. Lipid Res 53, 2546–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Balas L, Guichardant M, Durand T, and Lagarde M (2014) Confusion between protectin D1 (PD1) and its isomer protectin DX (PDX). An overview on the dihydroxy-docosatrienes described to date. Biochimie 99, 1–7. [DOI] [PubMed] [Google Scholar]
- (30).Chen P, Fenet B, Michaud S, Tomczyk N, Vericel E, Lagarde M, and Guichardant M (2009) Full characterization of PDX, a neuroprotectin/protectin D1 isomer, which inhibits blood platelet aggregation. FEBS Lett 583, 3478–3484. [DOI] [PubMed] [Google Scholar]
- (31).Browner MF, Gillmor SA, and Fletterick R (1998) Burying a charge. Nat. Struct. Biol 5, 179–179. [Google Scholar]
- (32).Newcomer ME, and Brash AR (2015) The structural basis for specificity in lipoxygenase catalysis. Protein Sci 24, 298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Coffa G, Imber AN, Maguire BC, Laxmikanthan G, Schneider C, Gaffney BJ, and Brash AR (2005) On the relationships of substrate orientation, hydrogen abstraction, and product stereochemistry in single and double dioxygenations by soybean lipoxygenase-1 and its Ala542Gly mutant. J. Biol. Chem 280, 38756–38766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dobson EP, Barrow CJ, Kralovec JA, and Adcock JL (2013) Controlled formation of mono- and dihydroxy-resolvins from EPA and DHA using soybean 15-lipoxygenase. J. Lipid Res 54, 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Gardner HW (1989) Soybean Lipoxygenase-1 Enzymatically Forms Both (9S)-and (13S)-Hydroperoxides From Linoleic Acid by a pH-Dependent Mechanism. Biochim. Biophys. Acta, Lipids Lipid Metab 1001, 274–281. [DOI] [PubMed] [Google Scholar]
- (36).Glickman MH, and Klinman JP (1995) Nature of Rate-Limiting Steps in the Soybean Lipoxygenase-1 Reaction. Biochemistry 34, 14077–14092. [DOI] [PubMed] [Google Scholar]
- (37).Yokokura Y, Isobe Y, Matsueda S, Iwamoto R, Goto T, Yoshioka T, Urabe D, Inoue M, Arai H, and Arita M (2014) Identification of 14,20-dihydroxy-docosahexaenoic acid as a novel anti-inflammatory metabolite. J. Biochem 156, 315–321. [DOI] [PubMed] [Google Scholar]
- (38).Gonzalez-Nunez D, Claria J, Rivera F, and Poch E (2001) Increased levels of 12(S)-HETE in patients with essential hypertension. Hypertension 37, 334–338. [DOI] [PubMed] [Google Scholar]
- (39).Luci DK, Jameson JB, Yasgar A, Diaz G, Joshi N, Kantz A, Markham K, Perry S, Kuhn N, Yeung J, Kerns EH, Schultz L, Holinstat M, Nadler JL, Taylor-Fishwick DA, Jadhav A, Simeonov A, Holman TR, and Maloney DJ (2014) Synthesis and Structure-Activity Relationship Studies of 4-((2-Hydroxy-3-methoxybenzyl)amino)benzenesulfonamide Derivatives as Potent and Selective Inhibitors of 12-Lipoxygenase. J. Med. Chem 57, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Adili R, Tourdot BE, Mast K, Yeung J, Freedman JC, Green A, Luci DK, Jadhav A, Simeonov A, Maloney DJ, Holman TR, and Holinstat M (2017) First Selective 12-LOX Inhibitor, ML355, Impairs Thrombus Formation and Vessel Occlusion In Vivo With Minimal Effects on Hemostasis. Arterioscler., Thromb, Vasc. Biol 37, 1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Yeung J, Hawley M, and Holinstat M (2017) The expansive role of oxylipins on platelet biology. J. Mol. Med 95, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Pistorius K, Souza PR, De Matteis R, Austin-Williams S, Primdahl KG, Vik A, Mazzacuva F, Colas RA, Marques RM, Hansen TV., and Dalli J (2018) PDn-3 DPA Pathway Regulates Human Monocyte Differentiation and Macrophage Function. Cell Chem. Biol 25, 749–760 e749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ivanov I, Kuhn H, and Heydeck D (2015) Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15). Gene 573, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Werner M, Jordan PM, Romp E, Czapka A, Rao ZG, Kretzer C, Koeberle A, Garscha U, Pace S, Claesson HE, Serhan CN, Werz O, and Gerstmeier J (2019) Targeting biosynthetic networks of the proinflammatory and proresolving lipid metabolome. FASEB J. 33, 6140–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


