Abstract
The importance of the microbiome for bovine udder health is not well explored and most of the knowledge originates from research on mastitis. Better understanding of the microbial diversity inside the healthy udder of lactating cows might help to reduce mastitis, use of antibiotics and improve animal welfare. In this study, we investigated the microbial diversity of over 400 quarter milk samples from 60 cows sampled from two farms and on two different occasions during the same lactation period. Microbiota analysis was performed using amplicon sequencing of the 16S rRNA gene and over 1000 isolates were identified using MALDI-TOF MS. We detected a high abundance of two bacterial families, Corynebacteriaceae and Staphylococcaceae, which accounted for almost 50% of the udder microbiota of healthy cows and were detected in all the cow udders and in more than 98% of quarter milk samples. A strong negative correlation between these bacterial families was detected indicating a possible competition. The overall composition of the udder microbiota was highly diverse and significantly different between cows and between quarter milk samples from the same cow. Furthermore, we introduced a novel definition of a dysbiotic quarter at individual cow level, by analyzing the milk microbiota, and a high frequency of dysbiotic quarter samples were detected distributed among the farms and the samples. These results emphasize the importance of deepening the studies of the bovine udder microbiome to elucidate its role in udder health.
Subject terms: Microbial communities, Microbial ecology
Introduction
There is still an ongoing discussion as to whether the mammary internal epithelial surface of the bovine udder harbours its own microbiota and whether this microbiota has a role in influencing udder health1,2. The source of microorganisms in the milk found deep inside the udder may originate from external contamination of the teat area followed by migration into the remote alveoli. Alternatively, translocation may occur from other parts of the body, such as the gut, via the blood vessels3. It has been hypothesized that translocation of bacteria from the rumen to the udder (and into the milk) may be important for the development of the intestinal microbiota of the suckling calf4.
The healthy bovine udder is a large organ comprised of four milk-producing quarters which each open to the outside of the body through a teat canal. Milk production takes place in the alveoli and collects first in the gland cistern and then in the teat cistern. In healthy udders, the quarters produce copious amounts of milk with low numbers of somatic cells and the numbers of microorganisms are sparse. Bovine mastitis causes considerable animal suffering and enormous economic losses on a global scale5. Mastitis leads to an increase in the numbers of somatic cells especially leukocytes in the milk, which may herald the infection before other clinical signs appear and also large numbers of the infecting organisms as well as a reduced milk yield. During mastitis, an imbalanced microbiota is detected in the infected quarter milk samples and this is caused by the presence of the pathogenic microorganism, the immune response of the cow and the interactions between the pathogen and the resilient microbiota6,7.
More recent studies have used molecular techniques to study the microbiology of milk from the udder and have uncovered an extraordinary diversity of bacteria. However, Taponen et al.2 summarized several previous microbiota studies and showed that the results are so variable that an identification of a generic udder microbiome is not at present possible. They also explained how a variety of factors would affect the results, especially the method of sampling. The studies reviewed by Taponen et al.2 were carried out using a variety of analysis methods and sampling techniques and, in addition, milk was obtained from different breeds of cow. Milk sampling methodology and microbiological analysis was recently studied by Dahlberg et al.8. They concluded that the contribution of external contamination of samples was a considerable problem when the bacterial load of the milk was low and this, in addition, made the removal of kit contaminations a challenge.
The presence of a healthy microbiota in diverse human body sites has been linked to a protective role against dysbiosis-related diseases such as inflammatory bowel disease or mastitis9,10.In cattle, there is evidence that mastitis changes the microbiome composition of the udder and that the functionality of the microbiome differs11,12. Furthermore, the microbiome balance within the quarter seems to be very fragile and is subject to several shaping factors from the host and from the environment (reviewed in13). More findings are needed to elucidate the role of the udder microbiome in the lactating cows and to identify its potential role against mastitis. Therefore, the aim of this work was to decipher the microbial composition at quarter level of cows regularly milked and with no sign of clinical mastitis in order to increase the knowledge of the potential microbial role. We performed a cross-sectional study starting with milk from 60 Norwegian Red cows selected from two farms and sampled at early and late lactation. Microbial composition was assessed using amplicon sequencing of the 16S rRNA gene and, in addition, we identified over 1000 isolates to confirm the presence of viable bacteria in the samples.
Results
Overall composition of the udder microbiota across farms and sampling periods
Quarter milk samples were collected from 60 cows in the first sampling period. Between the two sampling periods, eleven cows were removed from the study. All the cows were sampled during a single lactation period and the average days in milking (DIM) was 38 and 44 for farm A (“Centre for livestock production” at the Norwegian University of Life Sciences) and farm K (“Kalnes Upper Secondary School” farm), respectively in the first period and 213 and 222 during the second period. The range of parity of the cows was similar between the two farms (Table S1).
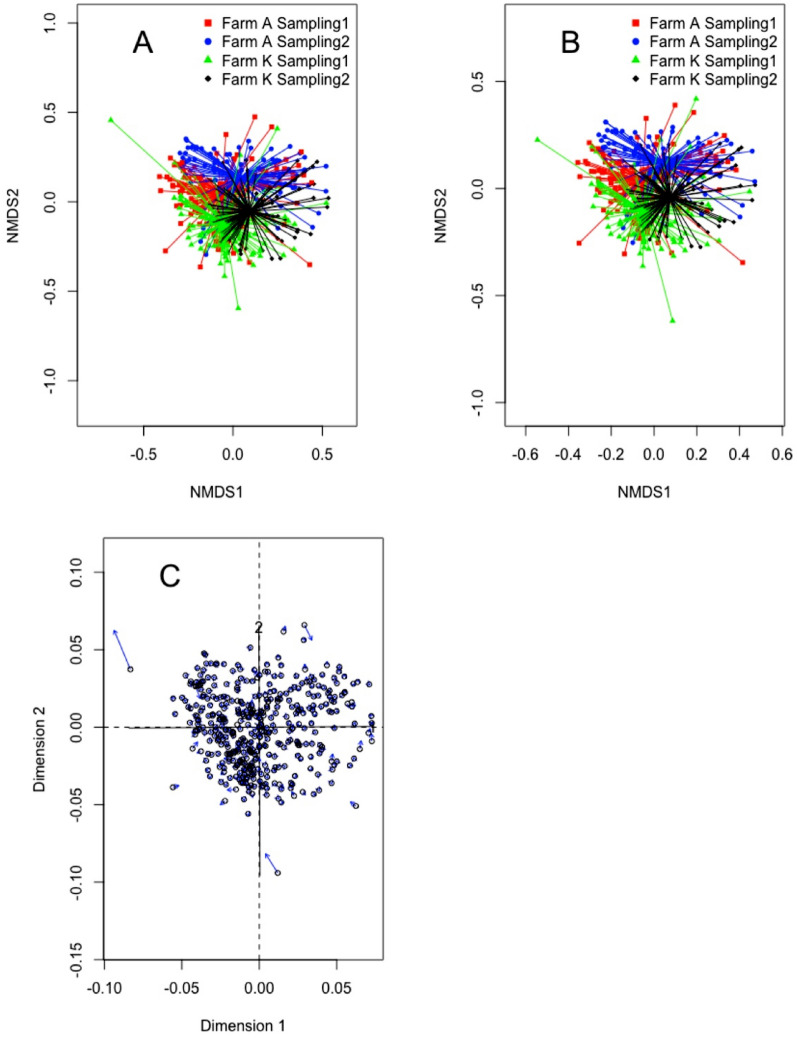
To explore the composition of the udder microbiota, amplicon sequencing of the 16S rRNA gene was performed for all the samples. From the original number of samples, a total of 403 quarter milk samples passed the quality filtering steps and were included in the study. The number of high-quality sequences obtained were 4,832,201 with an average of 12,549 sequences per milk sample (median 5988, min 109, max 102,760). A total of 10,010 sequence variants (SV) were detected and of these, 8759 were positively assigned to family level and kept for further analysis. Species richness estimation (Chao1) had an average of 89 SV per samples (median 61) and was significantly different (p < 0.05) between farm A (median 69.5) and farm K (median 58) but did not show significant difference between the two sampling periods. No significant differences were found with respect to sampling period and farms, regarding species diversity, which was calculated using Shannon index. Between-samples diversity (beta diversity) was calculated using non-metric multidimensional scaling using as input the Bray–Curtis and Jaccard distance matrix (Fig. 1). Both plots showed highly similar grouping between samples from the different farms and different sampling periods. This was also confirmed by Procrustes analysis (Fig. 1C). Multivariate homogeneity of group dispersion was not significant between the sampling periods or between the farms (p > 0.05) while PERMANOVA was significant (p < 0.001) between the two farms, between the two sampling occasions and the interaction between sampling and farms using both the Bray–Curtis and the Jaccard distance matrix.
Figure 1.
Non-metric multidimensional scaling (NMDS) plot of bovine quarter milk samples. (A) NMDS using Bray–Curtis dissimilarity matrix and (B) NMDS constructed using the Jaccard index dissimilarity matrix. (C) Procrustes analysis combining the NMDS results for Bray–Curtis and Jaccard index dissimilarity matrixes.
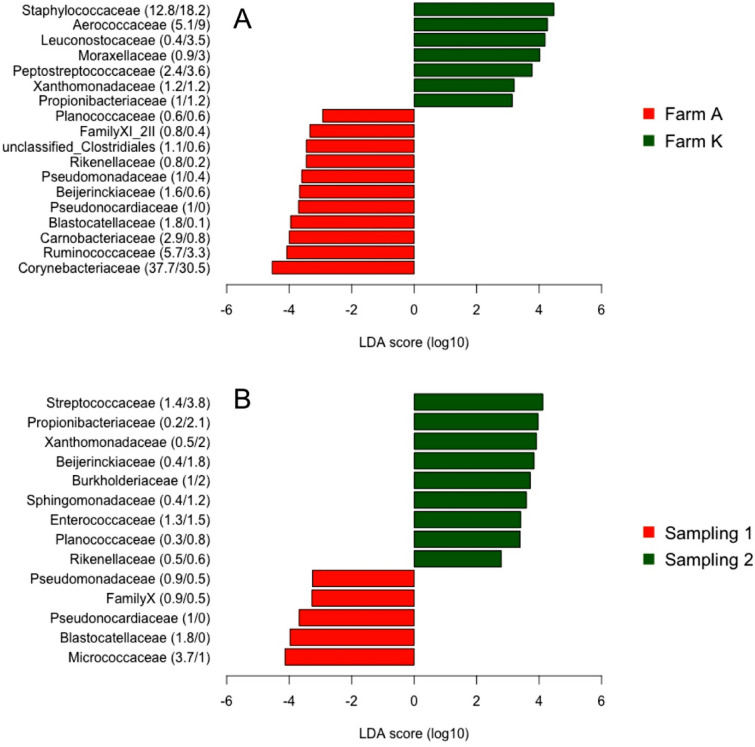
Taxonomical evaluation of the bacterial SVs identified 75 orders and 186 families. Twenty-seven families had a relative abundance greater than 0.5%. Corynebacteriaceae dominated the microbiota and accounted for more than 1/3 of the reads (34.2%) followed by Staphylococcaceae, Aerococcaceae and Ruminococcaceae (Table 1). These four families were detected in more than 80% of the quarter milk samples from both farms. Linear discriminant analysis of the relative abundances identified 51 families which were significantly different between the farms. Forty-one families were significantly enriched in quarter milk samples from farm A and 18 families from samples obtained from the farm K. In particular, the two most abundant families in the experiment (Corynebacteriaceae and Staphylococcaceae) were those that differed the most between farms (Fig. 2). In particular, Corynebacteriaceae were more abundant in farm A (38.3%, vs. 31.1% in farm K), while Staphylococcaceae showed a greater abundance in samples from farm K (18.5%, vs. 13.1%). Sequence variants assigned to the Corynebacteriaceae family were found in almost all the quarter milk samples (99 and 98% of the samples from farm A and K, respectively), while Staphylococcaceae were present in 98% of the quarter milk samples from farm K and in 93.6% of the quarter from farm A. However, these two families were detected in all the 60 cows in both sampling periods. Among the Corynebacteriaceae sequences, Corynebacterium bovis was the dominant taxa detected. The sequences assigned to this species accounted for 47.3% of all the Corynebacteriaceae sequences and 74.9% of the quarter milk samples were positive for C. bovis. Among the most abundant SVs assigned to the family Staphylococcaceae, only one SV was identified as Staph. aureus (SV_53, Table S3) and accounted for the 1.2% of all the sequences assigned to this family. Most of the Staphylococcaceae sequences were mostly identified, by sequence search, to the minor mastitis-related species of non-aureus staphylococci (NAS) group. This group included Staph. epidermidis, Staph. haemolyticus, Staph. simulans, Staph. chromogenes and Staph. xylosus.
Table 1.
Descriptive results of the microbiota summarized at family level (> 0.5%) detected in 403 quarter milk samples of 60 Norwegian Red cows.
| % Read over total | Number SV | % Quarter positive in farm A | % Quarter positive in farm B | % Positive cows | Average % of reads in farm A | Average % of reads in farm K | |
|---|---|---|---|---|---|---|---|
| Corynebacteriaceae | 34.2 | 742 | 99 | 98 | 100 | 38.3 | 31.1 |
| Staphylococcaceae | 15.5 | 254 | 93.6 | 98 | 100 | 13.1 | 18.5 |
| Aerococcaceae | 7 | 261 | 83.8 | 89.4 | 100 | 5.2 | 9.2 |
| Ruminococcaceae | 4.5 | 1518 | 83.8 | 85.9 | 100 | 6 | 3.4 |
| Peptostreptococcaceae | 3 | 107 | 76.5 | 79.9 | 100 | 2.5 | 3.7 |
| Streptococcaceae | 2.6 | 155 | 52.5 | 53.8 | 95 | 2.3 | 2.9 |
| Micrococcaceae | 2.4 | 118 | 68.6 | 57.3 | 98.3 | 2 | 3.1 |
| Lachnospiraceae | 2.4 | 739 | 75 | 73.4 | 100 | 2.7 | 2.3 |
| Moraxellaceae | 2 | 209 | 67.2 | 76.9 | 100 | 1 | 3.1 |
| Leuconostocaceae | 2 | 48 | 15.2 | 70.4 | 78.3 | 0.4 | 3.6 |
| Carnobacteriaceae | 1.8 | 135 | 71.1 | 59.3 | 100 | 2.9 | 0.8 |
| Burkholderiaceae | 1.4 | 237 | 66.2 | 52.8 | 96.7 | 1.3 | 1.7 |
| Enterococcaceae | 1.4 | 87 | 29.9 | 22.6 | 78.3 | 1.3 | 1.5 |
| Xanthomonadaceae | 1.2 | 59 | 49.5 | 28.6 | 85 | 1.2 | 1.2 |
| Beijerinckiaceae | 1.1 | 76 | 48.5 | 24.1 | 88.3 | 1.6 | 0.6 |
| Propionibacteriaceae | 1.1 | 62 | 47.1 | 53.8 | 95 | 1 | 1.2 |
| Blastocatellaceae | 1 | 19 | 17.2 | 5 | 30 | 1.9 | 0.1 |
| Unclassified Clostridiales | 0.9 | 503 | 68.6 | 54.3 | 98.3 | 1.2 | 0.6 |
| Sphingomonadaceae | 0.8 | 91 | 46.1 | 42.2 | 95 | 0.9 | 0.7 |
| Family X | 0.7 | 4 | 1.5 | 0 | 5 | 0 | 0 |
| Family XI | 0.7 | 6 | 67.6 | 58.8 | 98.3 | 1 | 0.5 |
| Family XI_2 | 0.7 | 89 | 28.9 | 27.6 | 85 | 0.2 | 0.1 |
| Pseudomonadaceae | 0.7 | 150 | 52.9 | 30.2 | 96.7 | 1.1 | 0.4 |
| Planococcaceae | 0.6 | 72 | 43.1 | 32.7 | 88.3 | 0.6 | 0.6 |
| Family XI_2II | 0.6 | 182 | 60.3 | 46.7 | 96.7 | 0.8 | 0.4 |
| Rikenellaceae | 0.5 | 389 | 50 | 35.2 | 96.7 | 0.9 | 0.3 |
| Pseudonocardiaceae | 0.5 | 26 | 21.1 | 1.5 | 48.3 | 1 | 0 |
Figure 2.
Linear discriminant analysis effect size (LEfSe) scores computed between the relative abundances of the most abundant families (> 0.5%) between (A) the farms and (B) the sampling periods. Values after taxa names are the percentage of abundance detected for each group.
Interestingly, Leuconostocaceae which accounted for only 2% of the total microbiota, were found mostly in farm K. This family was found in 70% of the quarters from farm K but only in 15% of the quarters from farm A. A temporal shift of the udder microbiota could be identified between the two sampling periods. In total, 21 families were significantly more abundant during the second sampling (late Aug-Sep) and 13 were more abundant during the first sampling (Jan-Mar). In particular, Streptococcaceae and Micrococcaceae were the two families that differed the most between the two sampling periods.
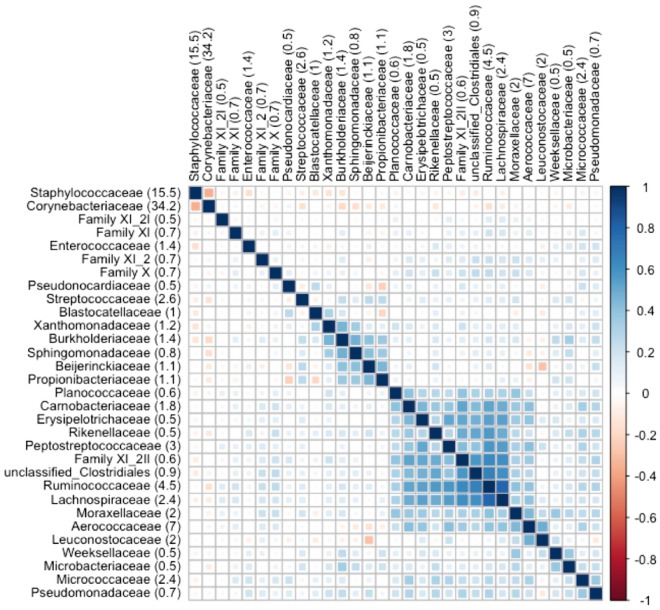
Correlation analysis of the most abundant families were performed to highlight how the presence and abundance of the most abundant families were related to each other. The strongest negative correlation was found between the Corynebacteriaceae and Staphylococcaceae families (Fig. 3). Positive correlation was found between the families of Lachnospiraceae, Ruminococcaceae, unclassified Clostriales, Family XI_2 and Peptostreptococcaceae. All these families are part of the Clostridiales order and accounted for 11.4% of the total reads in the study.
Figure 3.
Correlation analysis between the most abundant families found in the all the milk samples. Values after taxa names are the percentage of abundance detected over the entire experiment.
Individual cow microbiota
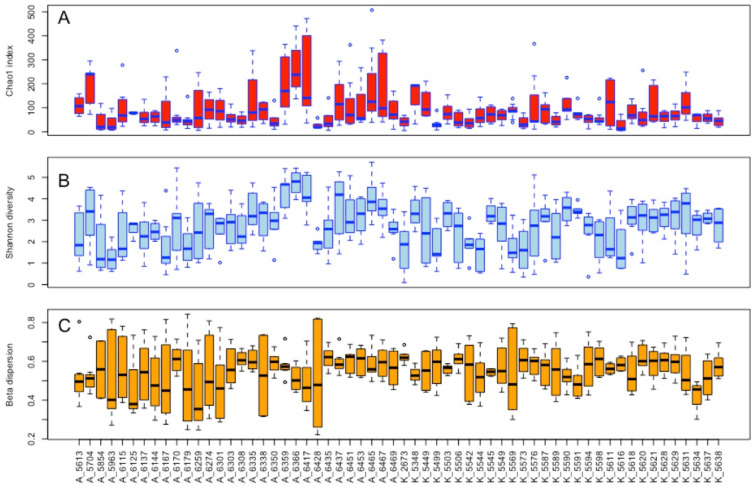
At single cow level, both the alpha diversity indexes (Chao1 estimate and Shannon diversity) were significantly different (p < 0.001) indicating that each cow contained a variable bacterial richness and diversity. Beta dispersion analysis of the microbiota within each individual cow indicated that phylogenetic and taxonomic assessments of the reads also revealed a diverse microbial population between quarters from the same cow (Fig. 4). However, the distribution of beta dispersion was larger between cows than within cows (Figure S1).
Figure 4.
Boxplot of the alpha diversity indexes (A: chao1 estimate, B: Shannon diversity) grouped per individual cows. (C) Multivariate homogeneity of group dispersions obtained using the Bray–Curtis dissimilarity matrix and grouped per cow.
Relative abundance distribution of the quarter microbiota showed that there was a high microbial diversity between cows, between quarters of the same cow and between sampling times of the milk from the same cow (Figure S2). In the case of several cows, one or more quarters showed a microbiota markedly different from the other quarters and was defined here as dysbiotic. Here, we introduced a new concept of a dysbiotic quarter. We used a clear cut off from the relative abundance results of the microbiota and defined a quarter as dysbiotic when one of the taxa, at family level, increases to over 60% of the reads in one quarter milk sample (within each individual and in each sampling) while remaining less than 20% in all the other quarters (this was calculated only on cows where 3 or 4 quarter milk samples were available). In this study, we identified 36 dysbiotic quarters out of 106 analysis (where 3 or 4 quarter milk microbiota was available) and detected that Staphylococcaceae (Table S4) was the most common family (20) followed by Streptococcaceae (5) and Enterococcaceae (4). We also detected good correlation between the enriched family in the dysbiotic quarter and the species identified in the same quarter by culturing method and maldi-TOF analysis. In most of these cases of a dysbiotic quarter, the SV composition of the Corynebacteriaceae was also changed (examples in Fig. 5, Figure S2). This was, however, not the case on occasions where a dysbiotic milk sample was detected (examples in Fig. 5 C and D). The shift in Corynebacteriaceae composition was detected in at least 15% of the cows (Figure S2). Dysbiotic quarters presented an increased level of potential mastitis-pathogens such as Streptococcaceae, Aerococcaceae, Enterococcaceae and other families which were present in high abundance in the dysbiotic quarter but not in the other quarters (Figure S2). Examples of these dysbiotic microbiota are presented in Fig. 5. Two of the quarters of cow 34 (Fig. 5C,D) presented an increased abundance of Staphylococcaceae in quarter left front (LF) from the first sampling while the quarter right rear (RR) from sampling 2 was dominated by Streptococcaceae. The Corynebacteriaceae population was dominated by 2 SVs (assigned to C. bovis) in all the quarters except for the quarter containing Streptococcaceae. In this quarter these 2 SVs were not detected and the most abundant Corynebacteriaceae was assigned to C. kroppenstedtii. A further example of dysbiotic udder microbiota with impact on the Corynebacterium population was cow 1, where the presence of Staphylococcaceae in the quarter RR in sampling 1 was associated with a completely different Corynebacteriaceae population within the same quarter. The same pattern was found in cow 41 where the Corynebacteriaceae population was different in the dysbiotic quarter, concurrent with increased abundance of Saccharopolyspora sp. in quarter RR from the first sampling period.
Figure 5.
Relative abundance of the quarter microbiota of cow sampled in 2 different occasions (A, C and E) and relative abundance of the sequence variants assigned to the family Corynebacteriaceae for the same cows (B, D and F). Values after the family and Corynebacteriaceae taxa indicate the percentage abundance over the entire value of the plot. Black lines indicate the relative abundance of the Corynebacteriaceae over the total microbiota.
Culturing and identification of isolates
Each quarter milk sample was further analyzed by culturing methods using 3 different agar plates (TSA blood, M17 and MRS). The TSA blood agar plate was incubated both in anaerobic and anaerobic condition. Over 2000 isolates were collected and of these only 1150 were classified at species level by maldi-TOF (score > 2). We obtained the majority of the isolates from the TSA blood agar (513 in aerobic condition and 366 in anaerobic condition), while 219 and 52 were obtained from M17 and MRS, respectively (Table S2). Seventy-five different species were detected. The most frequently isolated species was Aerococcus viridans (25%), however at genus level Staphylococcus was the most abundant genus (45%) and 15 species were identified. Staphylococcus epidermidis (22% of the total isolates), Staph. chromogenes (11%) and Staph. haemolyticus (4%) were the most abundant Staphylococcus identified. Nine species of Corynebacterium were identified, and these accounted for 5.5% of the total number of isolates. Corynebacterium amycolatum and C. bovis, were the 2 most abundant species within this genus. The majority of the Corynebacterium isolates (95%) were obtained from TSA blood agar under aerobic conditions.
In addition to isolation of bacteria, growth on TSA blood agar was also used to estimate the number of bacteria present and each quarter milk sample was then divided in different groups based on the number of colonies. We used the definition “A” from Dohoo et al.14 as valid for our study and identified quarters with possible intramammary infection if the number of colonies was > 1 per 0.01 mL (> 10 per 0.1 mL in our case). The total number of infected quarters was 173 (42%). By grouping the quarter milk samples in infected and not infected, the microbiota composition was not significantly different (adonis p value > 0.05), indicating poor correlation between the two types of analysis. However, the results from sequencing were confirmed by maldi-TOF species identification as the presence of species detected by maldi-TOF can be found in the microbiota at quarter level (Figure S2, Table S4). For example, in cow 11, the presence of several species of Enterococcus, Staphylococcus and Streptococcus was detected by maldi-TOF and a corresponding increase in the relative abundance of the family Enterococcaceae, Staphylococcaceae and Streptococcaceae was detected in some of the quarter milk samples (Table 2). In cow 34, the presence of Strep. uberis and Staph. epidermidis is highly correlated with the increase in Staphylococcaceae and Streptococcaceae in the microbiota of 2 of the quarter milk samples. For cow 41, only 2 quarter milk samples were infected and the maldi-TOF results identified the isolates as Staphylococcus epidermidis (in the same quarters). However, the quarter milk samples with dysbiotic microbiota (sampling 1 RR), showed increased abundance in a taxon included in the “other” (low abundance microbiota). Blast results of the SV with the highest abundance suggested the presence of Saccharopolyspora rectivirgula (99.51% identity).
Table 2.
Quarter milk samples with identified dysbiotic microbiota for 3 selected cows (11, 34, 41) and results from the maldi-TOF identification of the isolates for the same milk samples. The microbiota composition of these selected cows is presented in Fig. 5.
| Cow ID | Farm | Sampling period | Quarter | Species identified | Dysbiotic milk quarter | Taxonomical classification of enriched family (> 60%) |
|---|---|---|---|---|---|---|
| Cow 11 | K | 1 | LF | Corynebacterium amycolatum/Enterococcus gallinarum/Staphylococcus epidermidis | x | Enterococcaceae |
| LR | Aerococcus viridans/Staphylococcus warneri | x | Staphylococcaceae | |||
| RF | Aerococcus viridans/Staphylococcus epidermidis | |||||
| RR | Aerococcus viridans/Bacillus pumilus/Enterococcus malodoratus/Lactobacillus plantarum/Staphylococcus epidermidis/Staphylococcus xylosus | |||||
| 2 | LF | Citrobacter freundii/Enterococcus gallinarum/Staphylococcus equorum | ||||
| LR | ||||||
| RF | Aerococcus viridans/Aeromonas hydrophila/Enterococcus gallinarum/Lactococcus raffinolactis/Streptococcus parauberis | |||||
| RR | Aerococcus viridans/Corynebacterium bovis | |||||
| Cow 34 | A | 1 | LF | Staphylococcus epidermidis | x | Staphylococcaceae |
| LR | ||||||
| RF | Aerococcus viridans | |||||
| RR | Aerococcus viridans/Streptococcus uberis | x | Streptococcaceae | |||
| 2 | LF | Aerococcus viridans | ||||
| LR | ||||||
| RF | Aerococcus viridans/Pantoea agglomerans | |||||
| RR | Streptococcus uberis | |||||
| Cow 41 | A | 1 | LF | |||
| LR | Staphylococcus epidermidis | |||||
| RF | ||||||
| RR | ||||||
| 2 | LF | |||||
| LR | Staphylococcus epidermidis | |||||
| RF | ||||||
| RR | 0 | x | Pseudonocardiaceae |
Discussion
The growing interest in understanding the complex bovine udder microbiome has resulted in several published studies in recent years. These studies aimed to uncover how this microbiome influences the udder health and possibly has an important role during mastitis. In this study, we contribute to this knowledge by exploring the milk microbiota with a cross-sectional study of the milk microbiota as elucidated from over 400 quarter milk samples obtained from 60 lactating cows.
Sampling of udder milk for microbiome analysis is difficult and challenging15,16. In order to accomplish a more representative overview of the milk microbiota that colonize the upper interior part of the udder, the sampling method used in this study was different to those previously used to study milk microbiota2,6. The sampling was performed after the cow was regularly milked with the intent to remove microbial contaminations from the teat apex and to avoid the sampling of milk present in the cistern which might contain bacteria able to enter the udder between milking. The complete removal of contaminations from the teat apex or the environment cannot be ensured by the results obtained in this study, and additional experiments are needed to compare microbiota from milk samples obtained pre- and post-milking. However, although some taxa associated with the environment, such as Bacillaceae and Pseudomonadaceae, were detected in the dataset their abundance was much lower (< 1%) than that found in milk collected from bulk farm tanks which included organisms present in cows as well as milk equipment and the farm environment17–19. Interestingly, by using this sampling regime, we obtained a microbiota that was highly dominated by two families (Corynebacteriaceae and Staphylococcaceae). Several other taxa commonly associated with mastitis and as part of the milk microbiota were also detected. These included Aerococcaceae, families within the order Clostridiales, Streptococcaceae and Enterococcaceae. The sampling regime used in the study resulted in a similar or lower bacterial richness in our samples compared to previous studies20–22. However, comparisons with other studies cannot be conclusive due to different sampling procedures, filters and bioinformatics pipelines used between the studies. In particular, the method used for SV inferences increases the richness detected of microbiota studies compared to open-reference OTU clustering methods23.
The most common taxa found in the quarter milk samples are frequently reported in previous milk microbiota studies2,20. The family Corynebacteriaceae, which includes only the genus Corynebacterium, has been previously described as part of the skin-associated microbiome in humans and it is also found in other bovine body sites, such as the teat canal and uterus24–27. Therefore, it was not unexpected to find a high abundance and frequency of Corynebacteriaceae in quarter milk samples. This was similar to previous reports of milk microbiota2,6. Surprisingly, this taxon was found in almost all the quarter milk samples from two different farms and was detected in all the 60 cows. This suggests that Corynebacterium is part of the udder core microbiota revealed in this study. The role of Corynebacterium within the udder is still unknown and this might include a protective role against mastitis pathogens. Competition for niche adaptation between Corynebacterium and other taxa and inhibition of pathogen bacteria has been previously reported in microbiome studies28,29. Of particular interest was the interaction between Corynebacterium and Staphylococcus. Here, we found that the two main taxa of this study were negatively correlated with regards to their abundance. It is not possible to know if this negative correlation arose from the increase in abundance of one of the two taxa in the quarter or the decrease of one of them because of the lack of information of the absolute number of bacteria. However, competition between Corynebacterium and Staphylococcus has been shown previously in different environments and the negative correlation found in this study is further confirmation28,30,31. Another indication of a possible protective role of Corynebacterium against dysbiosis can be drawn from the analysis of the composition of the Corynebacterium population itself. The Corynebacterium population was different in dysbiotic quarters and this was shown in several cows. In most of these cases, this dysbiosis was caused by the presence of mastitis pathogens, such as Streptococcaceae and Enterococcaceae, which might outcompete and replace the dominant Corynebacterium SVs found in the other quarters of the same cow.
Several species of Corynebacterium inhabit the udder microbiome and Hoque et al.11 identified 12 of them using a metagenomics approach. In this study, the most common Corynebacterium SVs were assigned to Corynebacterium bovis, but several other species were detected by amplicon sequencing and MALDI-TOF identification of isolates. Corynebacterium bovis was previously described as part of the teat canal microbiota and Hiitio et al.12 also detected this species in the upper part of the gland. Corynebacterium bovis is classified as a minor mastitis pathogen and usually targeted during routine testing by mastitis laboratories. In our study, we found that C. bovis was present in a large number of quarter milk samples and several isolates were also obtained by culturing. By using standard culturing methods, Goncalves et al.32 detected C. bovis in only 15.8% of composite milk samples and using sequencing methods Taponen et al.2 detected that C. bovis covered between 50 and 73% of all Corynebacterium reads in milk samples that were PCR positive for C. bovis. The presence of C. bovis in milk has also been linked to an increase number of somatic cells in the cow’s milk and to the changed milk composition in subclinical mastitis32.
The genus Staphylococcus contains one of the most frequent and well-known mastitis pathogens, Staph. aureus. The samples analyzed in this study were obtained from cows with no signs of clinical mastitis and this pathogen was infrequently found in our samples and was not the most abundant Staphylococcus species isolated. On the other hand, the non-aureus staphylococci (NAS), which included several species of skin-related and the minor mastitis pathogens, were the most abundant group within the Staphylococcaceae family. Similar to Corynebacterium, NAS are commonly found in milk microbiota and in other microbiomes such as the bovine teat canal and skin12,33.
High correlations between the incidence of several families of the order Clostridiales were detected. These families accounted for more than 10% of the total microbiota in this study and are known to inhabit the cow rumen34,35. Their high positive correlation might indicate that their presence in the milk microbiota was connected. Rumen-related families within the Clostridiales order are often detected in milk microbiota studies. Vasquez et al.5 found that members of the Clostridiales such as Ruminococcaceae and Lachnospiraceae were among the dominant taxa in milk microbiota. However, the presence of these families in milk might originate from contamination during sampling procedure from the farm environment, the teat canal or from the translocation of cow microbiota through the hypothesized endogenous entero-mammary pathway3,36. Another aspect might be that families of the order Clostridiales are able to colonize the interior of the udder due to favorable anaerobic conditions and presence of nutrients. None of the over 1000 isolates obtained were classified within the order Clostridiales. This could be due to their fastidious growth requirements as this order includes obligative anaerobe spore-forming genera.
In this study, we collected milk samples from each quarter and a considerable difference in microbiota was found for samples collected from the same cow. This was detected in several individuals. Despite factors pertaining to the individual cow, such as immunity and genetics, that are known to influence the udder microbiota37, our results indicated that within-individual microbial variation is frequent. This is supported by the fact that certain taxa were often detected in only one quarter, rather than in all four quarters. Examples of these are found in this study where, in particular, the presence of Streptococcaceae and Enterococcaceae was detected in some quarters of the same cow but not in all four.
The milk microbiota structure was found to be influenced by farm and sampling period and several of the most abundant taxa were found to be influenced by these factors. The reasons for this variation in composition of quarter microbiota between the two farms studied may be mainly due to different farming practices, animal variability and genetics. Farming practices, such as bedding material, milking systems, housing system have previously shown to have an impact on the milk microbiota38,39. Whether or not the farming practices used in both farms in this study (e. g. cows regularly milked in an automatic milking system, open barn environment) influenced the microbiota inside the udder cannot be fully confirmed. However, since hygienic conditions have been found to be important for the transmission of mastitis pathogens within the herd and it can therefore be hypothesized that taxa able to colonize the udder can be spread within the herds, thus increasing the differences in diversity between farms40.
The variation in microbiota between sampling periods can be attributed both to the season when the sampling occurred and to the days of lactation of the animals. Interestingly, Streptococcaceae were significantly more abundant in the second sampling which was performed between late Aug-Sep. Species belonging to the Streptococcaceae family, such as Strep. dysgalactiae, Strep. uberis and Strep. agalactiae are common mastitis pathogens. Previous studies showed that cows have a greater risk of Streptococcus mastitis during the summer months compared to the winter months41,42. Therefore, it is not unexpected that this taxon was more abundant in the second sampling period compared to the first one. The presence of Streptococcus in milk samples was confirmed by the identification of the isolates.
The term dysbiosis can be defined as an alteration of the normal microbiota due to loss of diversity, reduction of commensal microbes and usually presence of pathogens which outcompete the existing microbiota. Several studies showed that disruption of diversity in udder microbiota is linked to a change in the health status of the udder and, in case of mastitis, the microbiota is dominated by the pathogen11,12. An important challenge for defining dysbiosis is the definition of a “normal” or “baseline” microbiota. Here, we decided to compare the microbial composition within the same individual and sampling to identify the “normal” microbiota and the presence of dysbiosis in only one quarter. By using this approach, we were therefore not able to identify dysbiosis that might occur in more than one quarter. However, we identified a high prevalence of dysbiosis in the bovine udder and a good correlation between the detected dysbiosis and isolation of bacterial species (within the same taxonomy) identified by maldi-TOF.
While the data presented here further support that a microbiota is present in the udder and might play a protective role, a few limitations of this study need to be considered. The somatic cell counts (SCC), used as indicator for the udder health, were not recorded at the time of sampling and indications of the health status of the quarters was obtained with culturing methods, which are biased by the conditions used for culturing and might therefore not reflect the true status. However, samples were collected from lactating cows, milked during regular farm practices with no sign of clinical mastitis during the collection days. Most microbiota studies suffer from the lack of absolute number of bacteria and results are presented in relative abundance where each feature is constrained to each other. Changes in abundance of one taxon will therefore impact the relative abundance of the others. The microbiota study presented here was performed using short-read sequencing, a method which achieves a lower resolution during taxonomical assignation of the reads. Furthermore, milk from cows with no sign of clinical mastitis harvest a low abundance microbial biomass and in case of an introduction of one taxon into the udder, this might have a big impact on the abundance of the resident microbiota of the udder. The cow breed has previously shown to be an important factor shaping the milk microbiota22. In this study, we selected 60 cows of Norwegian Red breed and therefore our results need to take into consideration possible breed-to-breed diversity when compared to other studies where more popular breeds are used (e. g. Holstein or Friesian). The microbiota of milk from the udder of Norwegian Red cows has not previously been reported.
Conclusion
The understanding of the complexity of the udder microbiome is enriched with the increased number of studies and some additional conclusions can be drawn from this work. The milk collected from mammary quarters contained a complex microbiota which was composed primarily by the two taxa, Corynebacteriaceae and Staphylococcaceae. These two taxa were detected in all the cows and are thus identified as part of the core microbiota with potential implications for the health of the udder. Evidence of interactions between Corynebacteriaceae and Staphylococcaceae for niche adaptation are also shown by the negative correlations of their abundances. Disruption of “normal” microbiota (dysbiosis) was detected frequently in quarters showing, in most of the cases, the presence of mastitis-related pathogens. This occurred in cows used for regular milk production and our results provide evidences that the microbiome balance within the udder is dynamic and very fragile and subject to several shaping factors both from the host and from the environment. This study is an additional step to decipher the potential role of the udder microbiome in cow health.
Materials and methods
Study animals
Sixty Norwegian Red cows selected from two dairy herds in the south-eastern part of Norway were included in this study. Thirty cows were sampled from the “Centre for livestock production” at the Norwegian University of Life Sciences (farm A) and thirty cows were selected from the “Kalnes Upper Secondary School” farm (farm K). Both farms operate under the regulations of the Norwegian Food Safety Authority regarding food production and animal care. The farm owners provided their permission for the sampling and for the use of their information in this study. Information about farm management is presented in Table S1. Samples were collected without the use of invasive procedures. Metadata from each individual cow were obtained from the Norwegian Cattle Health Recording System43. Cows were considered healthy and included in the study if (1) no sign of clinical mastitis were detected by the personnel, (2) there was no record of mastitis (reported by the farmers or the veterinarians) in the Norwegian Cattle Health Recording System 2 weeks before the sampling and (3) the cows were not under any antimicrobial treatment.
Sample collection
Milk samples were collected from each quarter in two occasions (January–March 2018 and late August–September 2018) corresponding to the early and late lactation period. At each day of sampling, 5 cows were selected thus obtaining 20 quarter milk samples which were used for analysis. Only 2 samplings per week were performed. On the day of sampling, collection of the milk was performed towards the end of the regular milking routine by trained personnel. After removal of the milking apparatus, milk (50 mL) was collected following the “Procedure for Collecting Milk Samples” of the National Mastitis Council (NMC, www.nmconline.org). The sample tubes were immediately stored in ice and transported to the laboratory within 2 h from the last sampling and immediately prepared for analysis. A total of 430 samples were successfully collected during the experiment. Samples were not included in the analysis if the cow showed signs of mastitis or the cow was no longer part of the herd during the second sampling.
Microbiological analysis and DNA extraction
Milk samples for microbiota analysis (40 mL) were centrifuged in an Heraeus Multifuge X3R centrifuge (ThermoFischer Scientific, Massachusetts, United States), using a FIS-6X100Y Fiberlite Rotor (ThermoFischer Scientific) at 8000 rpm for 10 min. Sterile foam coated Critical Swab cotton swabs (VWR, Pennsylvania, United States) were used to remove the cream layer. The supernatant was discarded, and the milk pellet resuspended and collected in 1.5 mL Eppendorf tubes that were immediately frozen at − 20 °C awaiting further processing. DNA extraction was performed using the DNeasy PowerFood Microbial Kit (Qiagen, Düsseldorf, Germany) starting from step 2 in the detailed protocol DNeasy Powerfood Microbial Kit Handbook. For increased efficiency in lysis of difficult species an additional incubation for 10 min at 65 °C on a heat block was performed at step 4 in the protocol, before proceeding with the rest of the protocol from step 5. Elution of DNA was performed in 50 µl of elution buffer before samples were placed in storage at − 20 °C.
For growth and identification of isolates, 100 µl raw milk were plated on de Man, Rogosa, and Sharpe agar (MRS; Difco Laboratories, Detroit, MI), M17 agar (Merck, Darmstadt, Germany) and TSA blood agar plates (ThermoFischer Scientific, Massachusetts, United States). Plates with MRS and M17 were incubated at 30 °C under anaerobic conditions for two days, while blood plates were incubated at 37 °C under both aerobic and anaerobic conditions for 24 h. Airtight containers and AnaeroGen 3.5L sachets (ThermoFischer Scientific) were used to achieve anaerobic conditions. After incubation, up to 10 phenotypically distinct bacterial colonies (representing the most phenotypic diversity on plate and not the proportion of the total isolates on the plate) were streaked out on new plates and re-incubated. All strains were collected and stored at -80 °C in cryomedia with 5% DMSO and 15% glycerol. TSA blood agar plate were also used to group the milk quarter milk samples based on the number of colonies present. The plates were grouped in 0 colonies, 1 < 10, 10 < 100, 100 < 1000 and > 1000 colonies. We considered the definition “A” from Dohoo et al.14 as valid for our study and identified quarters with intramammary infection if the number of colonies was > 1 per 0.01 mL (> 10 per 0.1 mL in our case) in one of the two conditions used for TSA blood agar (aerobic, anaerobic).
Library preparation and sequencing
DNA concentration was measured for all samples using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc, Wilmington, DE, USA). The V3 and V4 region of the 16S rRNA was amplified using the primers Uni340F (CCTACGGGRBGCASCAG) and Bac806R (GGACTACYVGGGTATCTAAT). The PCR reaction contained 1× of the Q5 Hot Start High-Fidelity 2× Master Mix (New England Biolabs, Massachusetts, United States), 1× of EvaGreen Dye 20× in water (Biotium, California, United States), 0.5 µM of each primers and three microliter of DNA in a final volume of 20 µL. Amplification was performed in a LightCycler 480 Instrument II (Roche, Basel, Switzerland) using initial denaturation at 98 °C for 30 s, followed by 35 cycles of denaturation at 98 °C for 15 s, annealing at 53 °C for 30 s and elongation at 72 °C for 20 s. The final elongation was performed at 72 °C for 10 min. The PCR product was then purified using 0.7× of Agencourt AMPure XP beads (Beckman Coulter, Inc, Brea, CA, USA) according to the manufacturer's instruction. A second PCR was performed using the purified PCR product (4 µL) to incorporate the primers with adapters and the P5 and P7 Nextera indexes (Illumina, San Diego, CA, USA). The PCR conditions were the same as above except that annealing temperature was 55 °C and a total of 10 cycles were used. Libraries were cleaned and normalized using the SequalPrep Normalization Plate (96) Kit (ThermoFischer Scientific) and pooled together. The final library concentration was then measured using Qubit 2 with the dsDNA HS kit (ThermoFischer Scientific) and quantitated using the KAPA Library Quantification kit (Illumina) before being sequenced on an Illumina MiSeq platform (Illumina) using the 2× 300 bp V3 kit (Illumina). Sequence data that support the findings of this study have been deposited in the EBI data with accession number: PRJEB35792.
Sequence data analysis
Sequence data quality were evaluated using FastQC version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were quality filtered and trimmed using the Dada2 package44 using truncating of forward reads set to 265 bases and truncating of reverse reads set to 220 bases. The error model in Dada2 was created using 1 million random filtered reads. Sequence variants (SV) was inferred using the DADA2 algorithm44 and removal of chimeras was performed using the function “removeBimeraDenovo” in the Dada2 R package. Sequence variants shorter than 375 base pairs were removed from the final table. Taxonomy was assigned using the Decipher R package45 against the SILVA SSU database46. Samples with less than 2000 reads and SV with less than 10 sequences were removed from the table. Normalization of the SV table was performed using the metagenomeSeq R package using the cumNorm function47. Individual SV sequences were also queried against the BLAST non-redundant (nr) database for taxonomical identification of the species or group of species. The sequences with the highest % identity and lowest e-value where used to assign the name of the species (or group of species if similar results were obtained) to the SV. This was done for SV assigned to the genus Corynebacterium and Staphylococcus (Table S3). The most abundant sequence variants assigned to the genus Staphylococcus were classified as non-aureus staphylococcus (NAS) if they obtained the highest identity with one of the main NAS species identified in mastitis cases (Staph. chromogenes, Staph. simulans, Staph. epidermidis, Staph. haemolyticus and Staph. xylosus). Negative and positive controls were included for each sequencing run. Negative controls were included during (1) sampling by using a sterile 50 mL falcon tubes with 20 mL ultraclean PCR grade water Sigma W45002 (Merck, Germany) (2) DNA extraction using only extraction kit reagents and plastic were also included (3) library preparation using PCR grade water. The positive controls were used during library preparation and for each round of sequencing and consisted of a serial dilution of the ATCC MSA 2002, 20 Strain Even Mix Whole Cell Material (ATCC, Virginia, United States). Positive and negative controls sequence data were used to filter potential contamination from the data. The R package “decontam” was used to remove SV which were identified as contamination during each sequence run. In addition, SVs present in positive controls but not included in the list of bacteria of the ATCC MSA2002 were also removed from the final dataset. In total 4592 SV were removed from the experiment and of these 72.3% were not identified at family level (Table S5) and accounted for 10.6% of the total reads.
Statistical analysis
All statistical analyses were performed using the R software48. Chao1 estimate and Shannon diversity were chosen to evaluate the alpha diversity and were calculated using the R package Vegan49. Pairwise comparison of the alpha diversity indexes between group levels was performed using the Wilcoxon rank-sum test. Multivariate homogeneity of group dispersions was calculated using the function “betadisper” available in the R package Vegan. Permutational analysis using dissimilarity matrix (“adonis” function from the R package Vegan) was used to test differences in the composition of the community between groups of samples (n. of permutation 999). Bray–Curtis and Jaccard dissimilarity matrixes were selected as input for ordination analysis using non-metric multidimensional scaling (NMDS) and the results of these two ordinations were compared using Procrustes analysis50. Linear discriminant analysis effect size51 was used to identify families which were significantly more abundant among groups of samples. Correlation analysis between the 30 most abundant families was performed using Pearson correlation coefficient for all individual taxa based on their relative abundance.
Isolates identification
The MALDI-TOF Biotyper system (Bruker, Bremen, Germany) was used for identification of the isolates at species level. Fresh cultures were prepared from the frozen stock by inoculation in the same media of isolation. A single colony was collected and applied to ground steel Maldi target plates (Bruker) using sterilized wooden toothpicks and then 1 µL 70% formic acid was added for on-plate direct extraction before applying 1 µL HCCA matrix (Bruker) and identification. Automated data analysis of the raw spectra was performed using the MALDI BioTyper 1.1 software (Bruker) with default settings to create a list of the most significant peaks of the spectrums. Each spectrum was automatically compared with the MALDI Biotyper database using the parameter of the pattern-matching algorithms. Identification of the bacterial species was considered only for results > 2.
Supplementary information
Acknowledgements
This work received financial contributions from the Norwegian Foundation for Research Levy on Agricultural Products (FFL) and the Norwegian Agricultural Agreement Research Fund (JA) (Grant No. 267623), the Faculty of Chemistry, Biotechnology and Food Science at the Norwegian University of Life Sciences and TINE SA.
Author contributions
D.P.: planning of experiment, data and statistical analysis, writing of the paper. R.M.: performed the sampling, DNA extraction, sequencing and Maldi-TOF identification of isolates. A.B. analysis of the cow data and participate in the writing and editing of the paper. J.N. plan the experiment and contributed to writing and editing of the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77054-6.
References
- 1.Rainard P. Mammary microbiota of dairy ruminants: fact or fiction? Vet. Res. 2017 doi: 10.1186/s13567-017-0429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taponen S, et al. Bovine milk microbiome: a more complex issue than expected. Vet. Res. 2019 doi: 10.1186/s13567-019-0662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young W, Hine BC, Wallace OAM, Callaghan M, Bibiloni R. Transfer of intestinal bacterial components to mammary secretions in the cow. Peerj. 2015 doi: 10.7717/peerj.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Addis MF, et al. The bovine milk microbiota: insights and perspectives from-omics studies. Mol. Biosyst. 2016;12:2359–2372. doi: 10.1039/c6mb00217j. [DOI] [PubMed] [Google Scholar]
- 5.Halasa T, Huijps K, Østerås O, Hogeveen H. Economic effects of bovine mastitis and mastitis management: a review. Vet. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- 6.Vasquez AK, et al. The microbiome of Escherichia coli and culture-negative nonsevere clinical mastitis: characterization and associations with linear score and milk production. J. Dairy Sci. 2019;102:578–594. doi: 10.3168/jds.2018-15062. [DOI] [PubMed] [Google Scholar]
- 7.Oikonomou G, et al. Microbiota of cow's milk; distinguishing healthy, sub-clinically and clinically diseased quarters. PLoS ONE. 2014 doi: 10.1371/journal.pone.0085904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlberg J, et al. Microbiota data from low biomass milk samples is markedly affected by laboratory and reagent contamination. PLoS ONE. 2019 doi: 10.1371/journal.pone.0218257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 10.Patel SH, et al. Culture independent assessment of human milk microbial community in lactational mastitis. Sci. Rep. 2017 doi: 10.1038/s41598-017-08451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoque MNA, et al. Metagenomic deep sequencing reveals association of microbiome signature with functional biases in bovine mastitis. Sci. Rep. 2019 doi: 10.1038/s41598-019-49468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falentin H, et al. Bovine teat microbiome analysis revealed reduced alpha diversity and significant changes in taxonomic profiles in quarters with a history of mastitis. Front. Microbiol. 2016 doi: 10.3380/Fmicb.2016.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derakhshani H, et al. Invited review: microbiota of the bovine udder: contributing factors and potential implications for udder health and mastitis susceptibility. J. Dairy Sci. 2018;101:10605–10625. doi: 10.3168/jds.2018-14860. [DOI] [PubMed] [Google Scholar]
- 14.Dohoo IR, et al. Diagnosing intramammary infections: evaluation of definitions based on a single milk sample. J. Dairy Sci. 2011;94:250–261. doi: 10.3168/jds.2010-3559. [DOI] [PubMed] [Google Scholar]
- 15.Hiitio H, et al. The effect of sampling technique on PCR-based bacteriological results of bovine milk samples. J. Dairy Sci. 2016;99:6532–6541. doi: 10.3168/jds.2015-10811. [DOI] [PubMed] [Google Scholar]
- 16.Lima SF, Bicalho MLD, Bicalho RC. Evaluation of milk sample fractions for characterization of milk microbiota from healthy and clinical mastitis cows. PLoS ONE. 2018 doi: 10.1371/journal.pone.0193671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skeie SB, Haland M, Thorsen IM, Narvhus J, Porcellato D. Bulk tank raw milk microbiota differs within and between farms: a moving goalpost challenging quality control. J. Dairy Sci. 2019;102:1959–1971. doi: 10.3168/jds.2017-14083. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues MX, Lima SF, Canniatti-Brazaca SG, Bicalho RC. The microbiome of bulk tank milk: characterization and associations with somatic cell count and bacterial count. J. Dairy Sci. 2017;100:2536–2552. doi: 10.3168/jds.2016-11540. [DOI] [PubMed] [Google Scholar]
- 19.Li N, et al. Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci. Rep. 2018 doi: 10.1038/s41598-018-20862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metzger SA, et al. A cohort study of the milk microbiota of healthy and inflamed bovine mammary glands from dryoff through 150 days in milk. Front. Vet. Sci. 2018 doi: 10.3389/fvets.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derakhshani H, Plaizier JC, De Buck J, Barkema HW, Khafipour E. Composition of the teat canal and intramammary microbiota of dairy cows subjected to antimicrobial dry cow therapy and internal teat sealant. J. Dairy Sci. 2018;101:10191–10205. doi: 10.3168/jds.2018-14858. [DOI] [PubMed] [Google Scholar]
- 22.Cremonesi P, et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE. 2018 doi: 10.1371/journal.pone.0205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nearing JT, Douglas GM, Comeau AM, Langille MGI. Denoising the Denoisers: an independent evaluation of microbiome sequence error-correction approaches. Peerj. 2018;6:4. doi: 10.7717/peerj.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dimitriu PA, et al. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. Mbio. 2019 doi: 10.1128/mBio.00839-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callewaert C, et al. Characterization of Staphylococcus and Corynebacterium clusters in the human axillary region. PLoS ONE. 2013 doi: 10.1371/journal.pone.0070538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braem G, et al. Culture-independent exploration of the teat apex microbiota of dairy cows reveals a wide bacterial species diversity. Vet. Microbiol. 2012;157:383–390. doi: 10.1016/j.vetmic.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 27.Ballas P, et al. Corynebacterium endometrii sp. nov., isolated from the uterus of a cow with endometritis. Int. J. Syst. Evol. Microbiol. 2019 doi: 10.1099/ijsem.0.003728. [DOI] [PubMed] [Google Scholar]
- 28.Hardy BL, et al. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. Mbio. 2019 doi: 10.1128/mBio.02491-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. Mbio. 2016;7:e01725–e11715. doi: 10.1128/mBio.01725-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks BW, Barnum DA. The susceptibility of bovine udder quarters colonized with Corynebacterium bovis to experimental infection with Staphylococcus aureus or Streptococcus agalactiae. Can. J. Comp. Med. 1984;48:146–150. [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goncalves JL, et al. Effects of bovine subclinical mastitis caused by Corynebacterium spp. on somatic cell count, milk yield and composition by comparing contralateral quarters. Vet. J. 2016;209:87–92. doi: 10.1016/j.tvjl.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 34.Stewart RD, et al. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 2019;37:953. doi: 10.1038/s41587-019-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henderson G, et al. Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. Peerj. 2019 doi: 10.7717/peerj.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Addis MF, et al. Evaluation of milk cathelicidin for detection of bovine mastitis. J. Dairy Sci. 2016;99:8250–8258. doi: 10.3168/jds.2016-11407. [DOI] [PubMed] [Google Scholar]
- 37.Catozzi C, et al. Short communication: intra- and inter-individual milk microbiota variability in healthy and infected water buffalo udder quarters. J. Dairy Sci. 2019;102:7476–7482. doi: 10.3168/jds.2019-16352. [DOI] [PubMed] [Google Scholar]
- 38.Doyle CJ, Gleeson D, O'Toole PW, Cotter PD. Impacts of seasonal housing and teat preparation on raw milk microbiota: a high-throughput sequencing study. Appl. Environ. Microb. 2017 doi: 10.1128/AEM.02694-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzger SA, et al. Influence of sampling technique and bedding type on the milk microbiota: results of a pilot study. J. Dairy Sci. 2018;101:6346–6356. doi: 10.3168/jds.2017-14212. [DOI] [PubMed] [Google Scholar]
- 40.Ruegg PL. A 100-year review: mastitis detection, management, and prevention. J. Dairy Sci. 2017;100:10381–10397. doi: 10.3168/jds.2017-13023. [DOI] [PubMed] [Google Scholar]
- 41.Pantoja JCF, Hulland C, Ruegg PL. Somatic cell count status across the dry period as a risk factor for the development of clinical mastitis in the subsequent lactation. J. Dairy Sci. 2009;92:139–148. doi: 10.3168/jds.2008-1477. [DOI] [PubMed] [Google Scholar]
- 42.Pantoja JCF, Hulland C, Ruegg PL. Dynamics of somatic cell counts and intramammary infections across the dry period. Prev. Vet. Med. 2009;90:43–54. doi: 10.1016/j.prevetmed.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Østerås O, et al. Results and evaluation of thirty years of health recordings in the Norwegian dairy cattle population. J. Dairy Sci. 2007;90:4483–4497. doi: 10.3168/jds.2007-0030. [DOI] [PubMed] [Google Scholar]
- 44.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581. doi: 10.1038/NMETH.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murali A, Bhargava A, Wright ES. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 2018 doi: 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200. doi: 10.1038/NMETH.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 49.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/J.1654-1103.2003.Tb02228.X. [DOI] [Google Scholar]
- 50.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. doi: 10.1007/Bf02291478. [DOI] [Google Scholar]
- 51.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011 doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.