Abstract
Bacterial infections cause a wide range of host immune disorders, resulting in local and systemic tissue damage. Antibiotics are pharmacological interventions for treating bacterial infections, but increased antimicrobial resistance and the delayed development of new antibiotics have led to a major global health threat, the so-called “superbugs”. Bacterial infections consist of two processes: pathogen invasion and host immune responses. Developing nanotherapeutics to target these two pathways may be effective for eliminating bacteria and restoring host homeostasis, thus possibly finding new treatments for bacterial infections. This review offers new approaches for developing nanotherapeutics based on the pathogenesis of infectious diseases. We have discussed how nanoparticles target infectious microenvironments (IMEs) and how they target phagocytes to deliver antibiotics to eliminate intracellular pathogens. We also review a new concept—host-directed therapy for bacterial infections, such as targeting immune cells for the delivery of anti-inflammatory agents and vaccine developments using bacterial membrane-derived nanovesicles. This review demonstrates the translational potential of nanomedicine for improving infectious disease treatments.
Introduction
Bacteria are prokaryotic microorganisms that invade the body, causing infectious diseases. Infectious diseases are a severe burden to global health.[1] Staphylococcus aureus (S. aureus, a gram-positive bacterium) and Pseudomonas aeruginosa (a gram-negative bacterium) are the most common pathogens. Infectious diseases involve interactions between bacteria and host cells, resulting in inflammatory responses to eliminate bacteria.[2] However, uncontrolled and excessive inflammation could lead to pathogenesis within a wide range of host disorders, such as meningitis,[3] pneumonia,[4] sepsis,[5] tuberculosis,[6] cholera[7] and gastritis.[8] Bacterial infection is also the leading cause of death in hospitals during surgical operations and in immunosuppressed patients.[9]
Antibiotics have significantly decreased the incidence of diseases caused by bacterial pathogens,[10] but antibiotics are increasingly less effective at killing bacteria due to antimicrobial resistance[11] and the delayed development of new antibiotics.[12] The rapid rise of antimicrobial resistance is strongly correlated with the overdose and misuse of antibiotics.[13] For example, multidrug-resistant (MDR) bacteria, as defined by the WHO (World Health Organization), are a serious problem because they no longer respond to standard antibiotic treatments, making them so-called superbugs.[14] The molecular mechanisms for developing drug resistance include pathways that are intrinsic (natural) and adaptive (acquired) to bacteria.[15] For intrinsic resistance, bacteria do not have target sites for drug delivery. In addition, low membrane permeability or MDR efflux pumps limit intracellular delivery.[16] For acquired mechanisms of drug resistance, bacteria produce enzymes that inactivate or degrade drugs inside bacteria, thus reducing the efficacy of antibiotics.[15b] Bacteria also mutate to express the ribosomal protection protein Tet (O)[17] and modify ribosomes [18] for the mitigation of antibiotics. In addition, bacteria may mutate the original drug target sites, and thus it is necessary to develop new antibiotics.[19]
Surgical operations are conventional methods for removing infectious tissues quickly.[20] If bacteria spread throughout the body and access to surgery is limited, antibiotic administration is a primary option. However, antimicrobial resistance to antibiotics is a health threat.[12] In addition, the development of antimicrobial resistance is strongly associated with the inappropriate use of antibiotics and a less targeted delivery. With the limitations of traditional antibacterial methods, recent advances in nanotechnology [21] may offer the opportunities to combat “superbugs”.
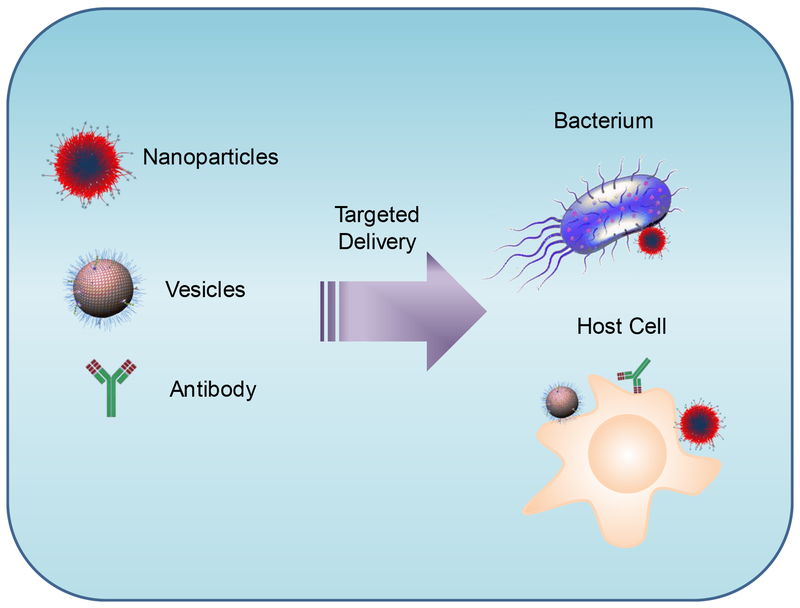
The process of bacterial infection is that bacteria invade the host tissues and activate the host immune responses. Therefore, infections include two components, invaded bacteria and host cells. To eliminate bacteria effectively and restore the host’s homeostasis, we may develop new nanoparticle drug delivery systems to target these two pathways (Scheme 1). While there are several reviews regarding antimicrobial therapies,[21e, 21g, 21i, 22] this review may offer a new approach to developing therapeutics for infectious diseases based on the pathogenesis of infectious diseases and the rational design of nanomaterials. We discuss infection microenvironments, a new target for antibiotic delivery. Antimicrobial resistance is also related to the biofilms formed by bacteria, and thus we review the current advances in nanoparticle design for preventing biofilm growth. Finally, we review a new concept of host-directed therapies by targeting infectious tissues, and we discuss vaccine developments.
Scheme 1:
The current concepts underlying antimicrobial therapies that target invading pathogens (bacteria) and infected host cells through the rational design of nanomaterials (polymeric NPs and nanovesicles) and antibodies.
1. Targeting Infectious Microenvironments (IMEs)
Immune cells (for example, macrophages) sense lipopolysaccharides on gram-negative bacteria to activate inflammation pathways, which produce cytokines (TNF-α and IL-1β).[23] Subsequently, blood vessels upregulate cellular adhesion proteins to recruit blood leukocytes to the locations where bacteria are present. Finally, leukocytes eliminate bacteria from the host.[24] During this process, the bacteria and the host form an interesting microenvironment involving the pathogens, immune cells and blood vasculature. In addition, infectious lesions possess a low pH, unique bacterial enzymes and an activated blood vasculature. Nanoparticle targeting of tumor microenvironments was proposed several decades ago. Many nanoparticles have been designed to target tumor microenvironments, and the results have demonstrated increased favorable outcomes in cancer treatments.[25] Unlike tumor microenvironments, IMEs include pathogens and host cells, and the formation of IMEs is acute and temporary and is strongly dependent on innate immune responses. Targeting IMEs may offer insights into developing new delivery systems to deliver antibiotics efficiently to infectious diseases.
1.1. Directly Targeting Bacteria
The direct delivery of antibiotics to bacteria would be a novel strategy for reducing antimicrobial resistance because of the low dose of drug administration, but most antibiotics lack a targeting ability. Recent advances in nanotechnology allow investigators to load drugs inside nanoparticles, and simultaneously, nanoparticles can be conjugated with targeting ligands or antibodies to recognize diseased tissues.[21c, 21d]
Hussain et al. proposed the in vivo screening of S. aureus in a lung infection model to identify a targeting ligand that specifically recognizes S. aureus so they could enhance antibiotic delivery to infection sites.[26] In their studies, they found that the cyclic 9 amino acid peptide CARGGLKSC (CARG) could increase vancomycin delivery after the peptide was conjugated to porous silicon nanoparticles loaded with vancomycin, thus significantly increasing mouse survival during lung bacterial infection and reducing the systemic toxicity of vancomycin. It is interesting to observe that the CARG peptides specifically bound to S. aureus rather than to Pseudomonas aeruginosa (P. aeruginosa) in vitro, and this result is consistent with in vivo studies in S. aureus-infected mouse lungs and skin when compared with the results in healthy mice and P. aeruginosa -infected mouse models. These studies demonstrate the potential of directly targeting pathogens in situ using nanoparticles to improve the bioavailability of antibiotics in infectious sites, thus possibly preventing the antimicrobial resistance caused by overdose administrations of antibiotics.
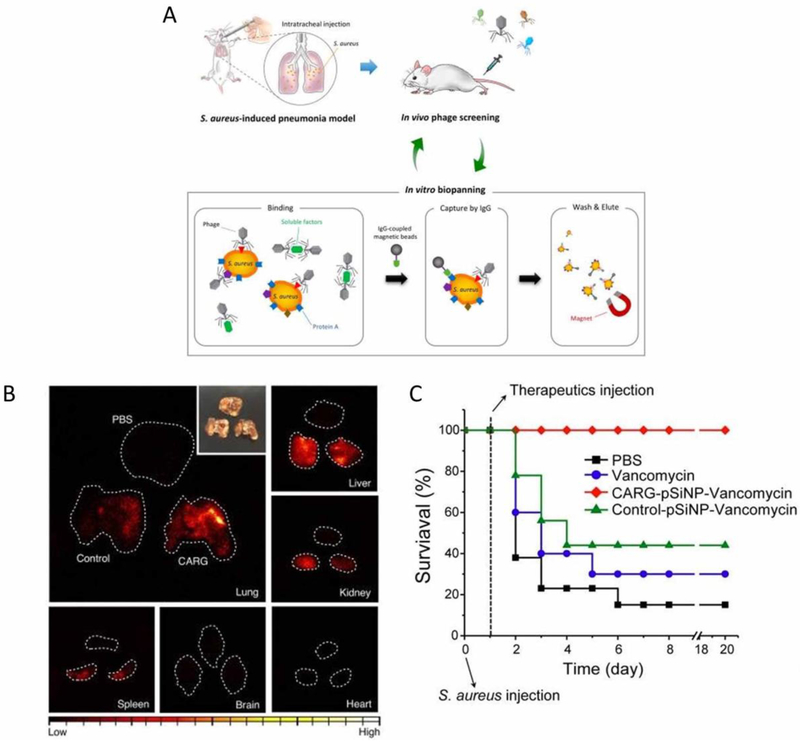
In those studies, a peptide library in a S. aureus pulmonary infection mouse model was screened, and the peptide that specifically binds to S. aureus-infected tissues was identified, namely, CARG (Figure 1A). To target bacteria in vivo, CARG peptides were conjugated to porous silicon (pSiNPs) loaded with vancomycin, and the pSiNPs were linked with PEG molecules for long circulation. To demonstrate the targeting of CARG-conjugated pSiNPs (CARG-pSiNPs) in vivo, CARG-pSiNPs and pSiNPs were intravenously injected into S. aureus-infected mice. Time-gated luminescence imaging for long-lived photoluminescence signals of pSiNPs (Figure 1B) showed a higher luminescence of CARG-pSiNPs compared to the control NPs (pSiNPs) and CARG-pSiNPs in healthy lungs. These results indicate that CARG peptides can enhance the delivery of pSiNPs to S. aureus-infected tissues, thus significantly delivering vancomycin to the locations of the bacteria. Ultimately, the authors evaluated the therapeutic efficacy of CARG-pSiNPs in an S. aureus-infected lung mouse model. Figure 1C shows the survival studies performed after the mice were given S. aureus intratracheally to cause severe pneumonia, followed by varied treatments. Ninety percent of the mice were dead after Day 6 without treatment. When the mice were treated with free vancomycin and vancomycin-loaded nontargeted pSiNPs, their survival increased, but 60% of the mice still died after 5 days. Surprisingly, the mice did not die within 20 days when they were treated with CARG-pSiNPs loaded with vancomycin. These results demonstrate that CARG-pSiNP can increase the delivery of vancomycin, thus preventing mouse deaths due to bacterial infections.
Figure 1.
(A) Schematic representation of peptide library screening with an in vivo phage display in an S. aureus-induced pneumonia model. A lung infection was generated through the intratracheal inoculation of S. aureus into the lungs. At 48-72 h after the bacterial inoculation, a CX7C-peptide library was screened by in vivo phage display. After three in vivo rounds, the enriched phage pool was subjected to in vitro biopanning on cultured S. aureus. The in vitro phage pool was further screened in vivo in infected animals. The graph shows the enrichment obtained over three rounds of in vivo biopanning. (B) Time-gated luminescence images of pSiNPs in major organs harvested from mice after 1 h of circulation (λe = 500 nm). The nanoparticles were injected intravenously into infected mice 24 h after S. aureus infection. The white dashed line designates the outer boundary of each organ. The control nanoparticles were pSiNPs grafted with polyethylene glycol only (no targeting peptide). Inset: a photograph of the lung tissues. (C) The mouse survival rate (n = 9) after the mice were intratracheally inoculated with 5 × 107 colony-forming units (CFU) of S. aureus. The mice received the following therapeutics intravenously 24 h after the bacterial inoculation: free vancomycin, CARG-pSiNP-vancomycin and control-pSiNP-vancomycin. The vancomycin was adjusted to have the same dose for each formulation (3 ± 0.2 mg kg−1). Copyright 2018 Springer Nature.
1.2. Targeting Activated Endothelium in IMEs
The direct targeting of bacteria via a ligand is a promising approach to delivering antibiotics to pathogens effectively, but this task may present various challenges. Bacterial infections are usually localized outside the blood vasculature,[27] and thus, the intravenous administration of drug carriers requires that they overcome vessel barrier to reach the location of the bacteria. In particular, bacteria invade the airway to cause lung infections. The lung is comprised of millions of tiny air sacs (called alveoli), and each alveolus is surrounded by blood capillaries to form an interface between the blood circulation and the airspace. In an alveolus, the epithelial cells are exposed to the airway and the endothelial cells form blood vessels close to the epithelial cells. Hussain [26] used a lung infection mouse model, and bacteria were administered via the airway. pSiNPs should overcome the air-blood vessel barrier to bind S. aureus bacteria, thus delivering vancomycin to the bacteria. The molecular mechanism for the vascular permeation of NPs is not clear, but it will be interesting to investigate in the future.
When considering a unique feature of infectious microenvironments (IMEs), including pathogen invasion and the host immune responses, a new concept was proposed to codeliver an antibiotic and anti-inflammation agent to the site of infection to diminish the bacteria and the inflammation response simultaneously.[28] IMEs have unique features, including a low pH,[29] bacterial enzymes,[30] and activated blood vessels.[23] The endothelium lining the lumen of the blood vasculature is quickly upregulated to express cell adhesion molecules when bacterial infections occur, such as intercellular adhesion molecule-1 (ICAM-1). Based on this observation, Zhang et al. designed a novel polymeric nanoparticle that was coated with ICAM-1 antibody to target the inflamed vasculature. The nanoparticles were disassembled in response to the low pH and the bacterial enzymes in IMEs to trigger the drug release. An antibiotic and an anti-inflammatory agent were loaded inside a polymeric micellar core via hydrophobic interaction. Last, the anti-ICAM-1 antibody was coated onto the surface of polymeric micelles. The expectation is that the multifunctional polymeric micelles may simultaneously eliminate the bacteria and mitigate the inflammation responses caused by the bacterial invasion.
Figure 2 shows a principle concept for the design of multifunctional nanoparticles targeted to IMEs. A multifunctional block copolymer with pH/enzyme-sensitive moieties and biotin linkers can form nanoparticles. An anti-ICAM-1 antibody can be linked to the surface of the polymeric micelles (so-called CIP+TPCA-1-NPs-anti-ICAM-1), as shown in Figure 2A. In an infectious lesion, the activated endothelium highly expresses ICAM-1 molecules, and the vascular permeability is increased. After intravenous injection, antibody-coated nanoparticles can recognize and bind to the inflamed vasculature to facilitate the deposition of NPs at the site of infection. Subsequently, the nanoparticles release the loaded drugs in response to the acidity and bacterial enzymes present in the IMEs (Figure 2B).
Figure 2.
The design of an IME-responsive and biofunctional nanoparticle (NP), and the targeted delivery of nanotherapeutics at a site of infection. (A) A drug-loaded polymeric micelle is self-assembled from pH/enzyme-responsive amphiphilic block copolymers and drugs, followed by an antibody coating to target the infection sites. The poly (ß-amino ester) (PAE) segment is pH-sensitive and enzyme-responsive. The 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)] (PEGylated DSPE) on the pendants of PAE is used for drug loading. Biotins on the surface of a micelle are used for biofunctionalization. (B) Drug-loaded NPs-anti-ICAM-1 specifically target activated endothelial cells at a site of infection after intravenous (i.v.) injection. Drug-loaded NPs-anti-ICAM-1 bind to the activated endothelial cells in IMEs, crossing the blood vessel and releasing drugs triggered by the local infectious cues. Copyright 2018 Wiley.
To support this concept, the authors established an acute lung bacterial infection mouse model because the lung has a unique interface with the blood vessels and airspace, as shown in Figure 3A. When bacteria invade the lung, host inflammation responses activate the endothelium in the bloodstream to express ICAM-1 and other adhesion molecules for leukocyte recruitment and to increase vascular permeability. Using this lung infection model (Figure 3B), the authors addressed whether the given NPs can target IMEs and subsequently release payloads in response to local cues induced by P. aeruginosa. In this study, NPs and the antibiotic ciprofloxacin (CIP) were chemically conjugated with the fluorescent dyes FITC and CY5, respectively. After P. aeruginosa was intratracheally applied to the mouse lung, fluorescently labeled NPs loaded with CIP were administered by i.v. Lung bronchoalveolar lavage fluid (BALF) was collected to determine how the NPs targeted the infection sites and if the drug release was dependent on infectious clues. The results showed that both the drug (CY5-CIP) and the nanoparticles (FITC-NPs) decreased in the lung tissues, but they increased in the BALF over time (Figure 3C–F), indicating that both the drug and micellar NPs were transported from the circulation to the lung airspace. In addition, the anti-ICAM-1 coating of the NPs dramatically increased the transport of NPs when compared to nontargeted IgG2b-coated NPs (Figure 3C–F). When the fluorescence ratios of CY-CIP and FITC-NPs (Figure 3G) were analyzed from Figure 3D and Figure 3F, the NPs that were found to be coated with IgG2b showed unchangeable fluorescent ratios, suggesting that the drug and the NPs were transported together from the bloodstream to the lung airway. However, the anti-ICAM-1 coating on the NPs showed that the ratios decreased, implying that the NPs were bound to the inflamed vasculature first, and the local cues (acids and bacterial enzymes) promoted drug (CIP) release and diffused into the lung. The analysis of in vitro drug release dynamics (Figure 3H) is consistent with the result from the acute lung infection model (Figure 3C–F).
Figure 3.
(A) A major component of the lung (alveolus), which forms the interface of the airway and bloodstream. P. aeruginosa invades the lung via the airway and causes infectious diseases. Anti-ICAM-1-coated NPs bind theinflamed vasculature of the lung, and the low pH of the infection environment triggers the drug release from the pH/enzyme-responsive NPs. (B) Experimental timeline of the animal studies. Fluorescence intensity of FITC-labeled NPs (C) and CY5-labeled CIP (E) of lung tissues after the BALFs were removed. Fluorescence intensity of FITC; FITC-labeled NPs (D) and CY5-labeled CIP (F) in BALF. G) Fluorescence intensity ratios of CY5 to FITC in BALF. (H) Drug release profiles of CIP from antibody-coated NPs under different conditions. (I) A survival study in the peritonitis-induced mouse sepsis model following the i.p. injection of a lethal dose of P. aeruginosa. The mice were treated with drug formulations including PBS, free CIP, free CIP+TPCA-1, CIP+TPCA-1-NPs-IgG2b, and CIP+TPCA-1-NPs-anti-ICAM-1 4 h after bacterial injection. All the data are expressed as the means ± s.d. (n = 3). *p < 0.05. Copyright 2018 Wiley.
The authors also examined their concept in a sepsis model. Sepsis is a severe disease caused by a dysregulated host inflammation response to bacterial infections, and it has a high mortality rate.[5, 31] Despite studies on potential therapeutic targets in the past, effective treatments for sepsis are lacking. The authors treated mice with high doses of P. aeruginosa and studied their survival after treating them with anti-ICAM-1-NPs loaded with CIP (ciprofloxacin) and TPCA-1((2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide) or several control formulations. The result showed that 90% of the mice survived in the CIP/TPCA-1-anti-ICAM-1-NPs treatment group compared to 50% in the control antibody-coated NP group and 40% in the free drug group (Figure 3I). This study not only shows the development of a new NP-based drug delivery system, but it may also shift the current state of nanomedicine to a biology-driven nanotherapeutics design to improve infectious disease therapies.
2. Targeting Host Cells for Bacterial Clearance
Antimicrobial resistance is associated with bacterial survival inside host cells[32] because the cells have low permeability to antibiotic entry. For example, S. aureus is internalized inside host phagocytes (neutrophils and macrophages) after the intravenous invasion of bacteria. Usually, bacteria are eliminated by phagocytes, but the incomplete clearance of S. aureus within blood phagocytes causes the infected cells to act as “Trojan horses” to spread the bacteria. This role causes chronic and recurrent infections[33] [34] including osteomyelitis, [35] pulmonary infections[36] and endocarditis. [37] Thus, targeting intracellular bacteria may be a key to clinical success. In addition, chasing phagocytes is a focal point for the targeted delivery of antibiotics to infectious sites.
2.1. Targeting Macrophages
Lehar et al. developed antibody-antibiotic conjugates (AACs) consisting of an anti-S. aureus antibody (THIOMAB) to target S. aureus, and the bacteria were eliminated after being taken up by the host cells.[38] Figure 4A shows the design of the antibody-antibiotic conjugates. The AACs bind to the bacterial membrane, and subsequently, the complexes are taken up by phagocytic cells. Intracellular proteases cleave a protease-sensitive peptide linker inside lysosomes to release the antibiotic (dmDNA31) (Figure 4B) for bacterial killing. In the USA300 MRSA S. aureus-infection mouse model, a single administration of AACs showed higher potency than vancomycin when administered twice per day, suggesting that it targeted intracellular MRSA. This approach to S. aureus is a potential approach to treating chronic infections (Figure 4C).
Figure 4.
(A) Model of AAC (not drawn to scale). (B) Mechanism of AAC action. (C) Wild-type mice (n = 5 per group) were treated with saline, anti-β-WTA antibody used in the AAC (monoclonal antibody (mAb)), vancomycin (twice daily), or anti-MRSA AAC (a single dose of 50 mg kg−1) starting 24 h after infection. Copyright 2015 Springer Nature.
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a pathogen that invades host cells. Yeom et al. reported the conjugation of AuNPs with a DNA aptamer (AuNP-Apt) to deliver C-terminally hexahistidine-tagged A3-APO (AuNP-AptHis-A3-APOHis) in host cells. Gold nanoparticle-DNA aptamer conjugates mediated the delivery of antimicrobial peptides inside the cells to eliminate intracellular S. Typhimurium effectively.[39] AuNP-AptHis-A3-APOHis can kill intracellular bacterial in S. Typhimurium-infected HeLa cells. After S. Typhimurium was given to mice, several treatments were performed daily for five days. The results showed that the AuNP-AptHis-A3-APOHis completely protected the mice from death by bacterial infection.
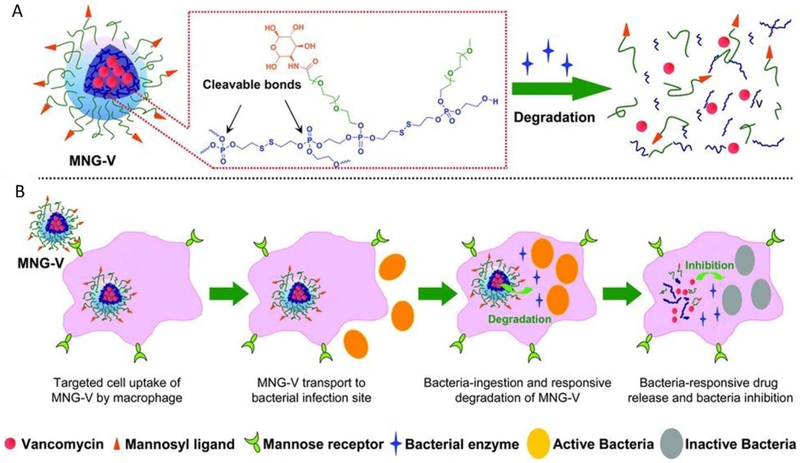
Macrophages are major phagocytes that remove bacteria, but sometimes, the bacterial clearance is not completed to cause chronic infections. Xiong et al. studied a mannosylated nanogel to deliver antibiotics to macrophages to inhibit bacterial infections.[40] Figure 5 shows the principle of the concept in material design. The nanogel contains a polyphosphoester core and mannosyl ligand-conjugated poly(ethylene glycol) on the shell. Macrophages are highly expressed mannose receptors, so the mannosylation of a nanogel can mediate the binding of the nanogel to macrophages, and the nanogel is disassembled by bacterial phosphatase and phospholipase to release antibiotics. In a zebra fish model infected by a methicillin-resistant strain of S. aureus (MRSA), the fluorescent images showed that fluorescently labeled-nanogel were internalized by macrophages. In addition, vancomycin-loaded nanogel significantly increased the survival rate of infected zebrafish.
Figure 5.
(A) Schematic illustration of vancomycin-loaded mannosylated nanogels (MNG-V) and bacteria-responsive drug release and (B) targeted uptake of MNG-V, transport, degradation, drug release and bacteria inhibition. Copyright 2012 Wiley.
2.2. Targeting Neutrophils
Neutrophils are primary phagocytes that chase bacteria for containment and clearance.[41] Targeting neutrophils using nanoparticles in situ may be a means to treat infectious diseases. Recently, Wang’s group demonstrated that albumin (a serum protein) formed nanoparticles that can specifically target activated neutrophils during inflammation according to intravital microscopy.[42] These studies allowed for the development of nanoparticle-based delivery to treat infectious diseases. P. aeruginosa is a common pathogen that causes acute pneumonia, and therefore it is necessary to target infectious lungs. Dafeng et al. loaded cefoperazone acid, a broad spectrum antibiotic, inside albumin nanoparticles to examine targeting neutrophils for the delivery of antibiotics to eradicate P. aeruginosa from the lung.[21b] These studies show that albumin NPs can selectively bind to activated neutrophils in the bloodstream and are internalized, and subsequently, neutrophils transport albumin NPs to bacterial locations. The NPs release cefoperazone acid to kill P. aeruginosa. The hijacking of activated neutrophils to deliver antibiotics may offer a novel strategy for the increased bioaction of antibiotics, thus possibly preventing antimicrobial resistance.
3. Targeting Biofilms
When bacteria invade the body, phagocytes usually actively eliminate them to maintain homeostasis. However, intracellular pathogens can manipulate the host cells for survival. This incomplete clearance of bacteria may cause bacterial colonization to form biofilms.[43] Biofilms can cause chronic infections and develop antimicrobial resistance.[14] In addition, biofilms start on the surface of medical instruments and become irreversible and aggregated to form stable biofilm organisms.[44] Biofilms show significant resistance to treatment compared with planktonic bacteria.[45] Bacterial colonization presents clinical challenges including chronic or recurrent infections, impaired wound healing and sepsis due to the limited permeability of antibiotics to bacterial biofilms.[46] Advances in nanotechnology enable physicians to tackle the clinical challenges posed by biofilms because nanoparticles may increase drug stability and solubility, enhance tissue penetration and deliver high concentrations of therapeutics. Herein, we show some examples of eradicating biofilms using nanoparticle-drug systems (Table 1).
Table 1.
Nanoparticle-drug systems for eradicating biofilms.
| Bacterial biofilm | Therapeutics | Materials | Ref. |
|---|---|---|---|
| Staphylococcal | Triclosan | Mixed-shell polymeric micelles | [49] |
| P. aeruginosa | Nitric oxide and Gentamicin | Polymer nanoparticles | [50] |
| Staphylococcus epidermidis | Methicillin | mPEG-b-PDLLA diblock copolymer nanoparticles | [51] |
| P. aeruginosa | Ciprofloxacin (CPX) | Poly-L-lysine (PL)-coated PLGA nanoparticles | [53] |
| P. aeruginosa and S. aureus | -- | DNase I-coated Polymethylmethacrylate (PMMA) film | [54] |
| S. aureus | -- | Lytic peptides-modified polyethylene terephthalate (PET) film | [55] |
3.1. Enhanced Permeability to Biofilms
Liu et al. reported on mixed-shell polymeric micelles (MSPM) consisting of two copolymers, poly(ethyleneglycol)-b-poly(ε-caprolactone) (PEG-b-PCL) as a shell and pH-responsive poly(ε-caprolactone)-b-poly(β-amino ester) (PCL-b-PAE) as a core, to carry triclosan for the eradication of biofilms (Figure 6).[47] PEG molecules may assist MSPM in penetrating biofilms, and PAE blocks at pH 5.0 promoted the disassembly of MSPMs to release triclosan. Confocal images showed that MSPMs diffused within the Staphylococcal biofilms at pH 5.0, rather than at pH 7.4. The anti-bacterial efficacy of triclosan-loaded MSPMs increased by 8 times compared with triclosan in biofilms formed by S. aureus Xen36 and S. aureus ATCC12600GFP.
Figure 6.
Schematic representation of the hypothesis and findings that drug-loaded MSPMs composed of PEG and pH-responsive PAE enhance bacterial killing. (A) Nonencapsulated antimicrobials penetrate biofilms to a limited extent and kill only the bacteria on the outside of the biofilm. Its penetration is limited by adsorption onto bacterial cell surfaces and matrix components. (B) Antimicrobials encapsulated in a SSPM nanocarrier with stealth properties will show better penetration into a biofilm than nonencapsulated ones and thus kill bacteria in deeper layers of the biofilm, providing sufficient antimicrobial release. Due to the stealth properties of the SSPM nanocarriers, there will be no targeting to bacterial cell surfaces, and, as a consequence, there will be little enzymatic degradation of micelles and antimicrobial release. (C) Antimicrobials encapsulated in a MSPM nanocarrier with stealth properties will show full penetration in a biofilm due to their stealth properties and become positively charged in the low pH vicinity of bacteria to target themselves to the bacterial cell surface and expose their micelle core (see panel D). The micelle core is subsequently hydrolyzed by bacterial lipases to release its antimicrobial content. (D) Summary of the surface adaptability of MSPMs under the influence of pH changes and lipase degradation. Copyright 2016 American Chemical Society.
Nguyen et al. reported the use of polymer nanoparticles to codeliver gentamicin and a nitric oxide (NO) donor to eradicate both biofilms and planktonic bacteria.[48] In vitro staining studies showed that simultaneous treatments of both gentamicin and NO dramatically decreased the biofilm biomass compared to untreated groups or treatment alone, indicating that there is a synergistic effect from gentamicin and NO in eradicating biofilms. In another study, mPEG-b-PDLLA di-block copolymer nanoparticles were used to deliver superparamagnetic iron oxide nanoparticles (SPIONs) and antibiotic methicillin.[49] An external magnetic field enables iron oxide nanoparticles to penetrate inside Staphylococcus biofilms. Confocal microscopy images showed that methicillin-loaded iron oxide-encapsulating polymer vesicles (IOP) completely destroyed the biofilms, but methicillin alone only killed the bacteria present on the surface of the biofilms.
3.2. Targeting Extracellular DNA to Prevent Biofilm Formation
Extracellular DNA (eDNA) supports the adhesion of bacteria and their aggregation, resulting in biofilm formation. Studies have shown that blocking eDNA production or removing eDNA during biofilm formation decreased bacterial adhesion.[50] Due to the important role of eDNA in biofilm formation, DNase I was used to target biofilms. The codelivery of DNase and antibiotics reportedly enhanced the eradication of biofilms to improve the diffusion of the antibiotic into biofilms.[51] Poly-L-lysine (PL)-coated PLGA nanoparticles were loaded with the fluoroquinolone antibiotic ciprofloxacin (CPX) used to treat P. aeruginosa infections. At a single dose, ciprofloxacin-PLGA NPs inhibited P. aeruginosa growth and reduced the biofilm mass by more than 80%. When NPs were given several times, DNAse I-loaded NPs (PLGA–PL–CPX–DNase I) completely prevented new biofilms.
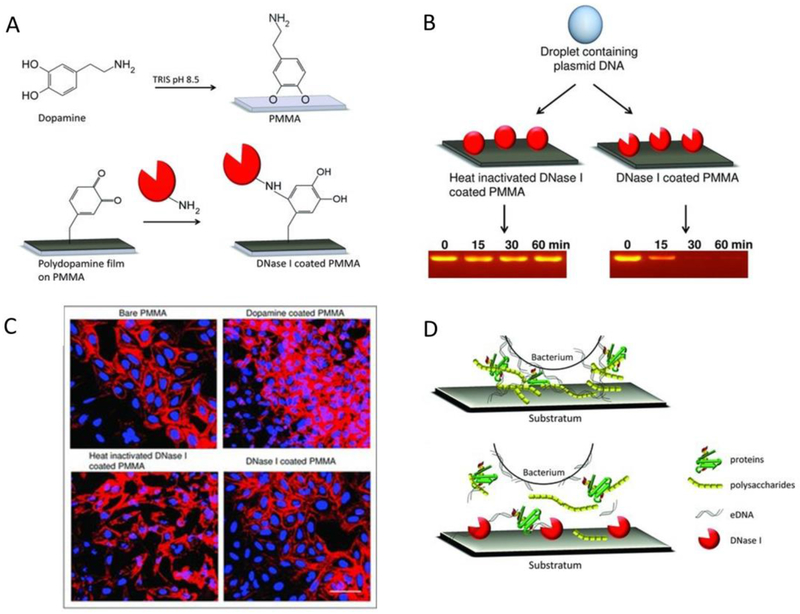
Swartjes et al. reported that DNase I-coated polymethylmethacrylate (PMMA) film could reduce bacterial adhesion and inhibit biofilms (Figure 7A to D).[52] The results showed that bacterial growth on the DNase I-coated PMMA surface was reduced by 99% for P. aeruginosa PAO1 and 95% for S. aureus ATCC 12600 in 60 min, compared to the control groups (Figure 7B). Confocal images demonstrated that P. aeruginosa PAO1 and S. aureus ATCC 12600 grew to form a thickness of 10 and 18 μm, but after a coating with DNase I-coated-PMMA, the thicknesses were only 0.2 and 3 μm, respectively (Figure 7C). These studies show the proof of concept indicating that conjugating lytic peptides or incorporated DNase I to nanoparticles may be a promising strategy for the reduction of biofilms.
Figure 7.
(A) Adhesion of dopamine to PMMA by dip coating and subsequent DNase I coupling to a polydopamine film. (B) Agarose gel showing the degradation of plasmid DNA in a droplet on PMMA coated with DNase I with and without heat inactivation. Plasmid DNA is hydrolyzed within 30 min when placed on PMMA coated with active DNAse I. (C) CLSM images of U2OS cell adhesion and proliferation on different substrata. The scale bar represents 100 μm. (D) eDNA acting as a bridge between a bacterial cell surface and various biopolymers in EPS, such as proteins and polysaccharides, play an important role in bacterial adhesion (upper). The disruption of the EPS by DNase I prevents bacterial adhesion to the substratum surface (below). Copyright 2013 Wiley.
3.3. Targeting Biofilms on Medical Instruments
Medical instruments are sources of biofilms, and removing them from instrument surfaces is a tough task. Pretreatments for medical instruments are needed to develop effective methods to inhibit biofilm formation. Traba et al. developed responsive antibacterial surfaces by coating polyethylene terephthalate (PET) with lytic peptides because they demonstrated good antibacterial properties.[53] A surface coating using this material showed excellent antibacterial and anti-biofilm activity compared with an unmodified surface. In addition, the antibacterial activities were dependent on adding the peptides to the PET. In long-term studies, catheters coated with PET loaded with peptides did not form biofilm even after 7 days, but in control catheters without coating, bacteria grew quickly, within 24 h.
4. Host-directed Therapy for Bacterial Infections
Despite the increase in antibiotics R&D, antimicrobial resistance is challenging in nature because bacteria mutate rapidly to diminish the efficacy of antibiotics and new drugs lag behind the emergency of antimicrobial resistance. There is an urgent need to develop alternative approaches to treating infectious diseases. Host-directed therapy is an emerging method to target infected host cells for increased host defense and eliminating bacteria with the aim of controlling and clearing infection.[54] Host-directed therapies include a wide range of therapeutic agents, such as small molecules, biologics (monoclonal antibodies), cytokines, nucleic acids and cellular therapy. For small molecule-based therapies, nonsteroidal anti-inflammatory drugs (NSAIDs) have been used for pneumonia. For example, when ibuprofen was used in patients with sepsis, they showed some improvement.[55] For biologic-based therapies, the neutralization of pro-inflammatory cytokines (such as anti-IL-1b and anti-TNF-α is a way to control inflammation responses induced by bacterial infections.[56] Current therapies lack tissue targeting, thus it is necessary to develop drug carriers to improve their efficacy.
4.1. Targeting Inflamed Vasculature using Neutrophil-membrane Nanovesicles
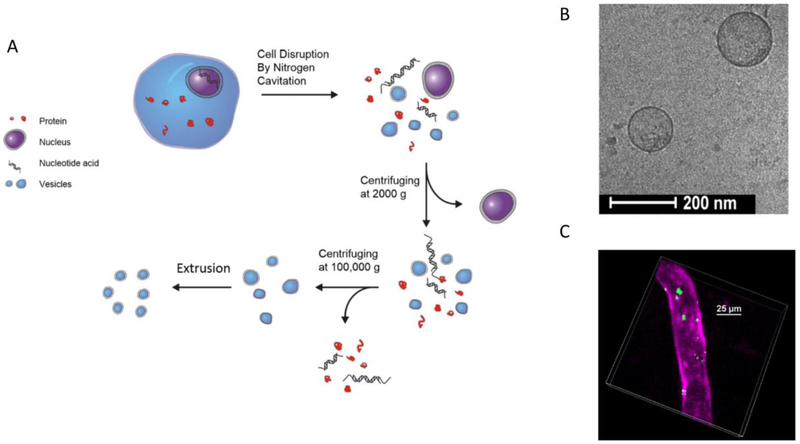
Bacterial infections usually cause pneumonia, which is attributed to acute lung injury (ALI) or the development of acute respiratory distress syndrome (ARDS). ALI/ARDS is associated with leaky pulmonary edema, hypoxia, leukocyte infiltration and inflamed vasculature.[57] Improved endothelial integrity is key to the clinical success of ALI/ARDS. With inspiration from the binding of integrins on neutrophils to ICAM-1 on endothelial cells, Jin et al. prepared nanovesicles made of the plasma membranes of differentiated HL-60 (a human neutrophil-like cell line) via nitrogen cavitation[58], and they successfully showed the enhanced delivery of anti-inflammatory drugs, TPCA-1 and piceatannol to the inflammatory loci in ALI/ARDS animal models induced by LPS.[59] In their studies, Jin et al. developed nitrogen cavitation to disrupt neutrophils physically, and subsequently, the cell membrane formed nanovesicles (Figure 8A). The cryo-TEM image showed that the nanovesicles had a shell structure with a size of 200 nm in diameter and a shell thickness of 3-4 nm (Figure 8B), equivalent to that of a cell membrane. The in vivo imaging of live mouse vasculature showed that the nanovesicles were able to bind specifically to inflamed endothelium (Figure 8C). Furthermore, the animal studies showed that neutrophil membrane nanovesicles decreased the cytokines (TNF-α, IL-1β and IL-6) in blood and increased survival when mice were challenged with a lethal dose of LPS. The studies show that the targeting of inflamed or infected vasculature may be a strategy to strengthen the endothelial integrity for improved therapies in infectious diseases.
Figure 8:
(A) The preparation of cell membrane-derived nanovesicles via nitrogen cavitation and a series of centrifugations. After purification, the intracellular components are removed and purified nanovesicles are obtained. (B) Cryo-TEM image of HL-60 cell membrane-formed nanovesicles. (C) The intravital image shows a cremaster venule from a live mouse following the i.v. injection of DiO fluorescently labeled nanovesicles (green) and Alex-Fluor-647 anti-CD31 (pink) to label the blood vessel. Copyright 2016 Elsevier.
4.2. Neutralizing cytokines using macrophage nanovesicles
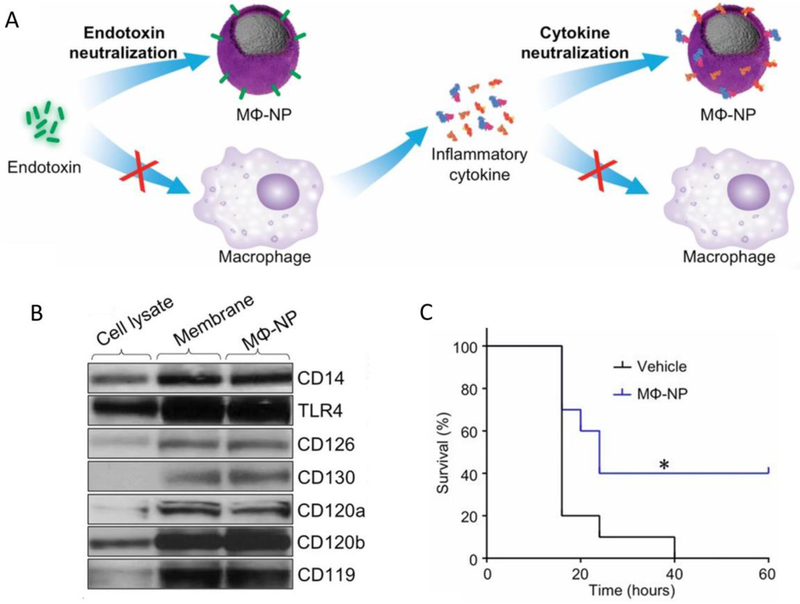
Sepsis is a life-threatening bacterial infection that is associated with a series of inflammatory responses.[5] Macrophages play an important role in neutralizing endotoxins and sequestering proinflammatory cytokines to inhibit sepsis.[60] Based on this knowledge, Thamphiwatana et al. developed a macrophage cell membrane coated with polymeric nanoparticles (MΦ-NPs) that can bind endotoxins and cytokines, thus downregulating the inflammatory cascades for treating sepsis (Figure 9A).[61] Key cell membrane proteins for LPS binding and cytokine binding receptors were confirmed by Western blot, suggesting that MΦ-NPs inherit the biological characteristics of macrophages (Figure 9B). In vitro experiments showed that 1 mg of MΦ-NPs could remove 62.5 ng of the endotoxin, 105.1 pg of IL-6, 4.3 pg of TNF-α, and 6.5 pg of IFN-γ. In vivo experiments demonstrated that MΦ-NPs decreased the TNF-α and IL-6 in mouse plasma and improved mouse survival after the administration of a lethal dose of LPS. In the E. coli-induced sepsis model, a single dose of MΦ-NPs significantly increased the survival rate and decreased the proinflammatory cytokines (Figure 9C).
Figure 9:
(A) MΦ-NPs that can bind and neutralize endotoxins and proinflammatory cytokines, thus inhibiting inflammation cascades to manage sepsis. (B) The Western blot results show membrane receptors on the macrophage cell lysate, macrophage membrane vesicles, and MΦ-NPs. (C) In the sepsis model, under a lethal dose of E. coli (1 × 107 cfu), the mouse survival rates after treatments with the vehicles and MΦ-NPs, respectively (300 mg/kg) (n = 10). Copyright 2017 National Academy of Sciences.
4.3. Vaccine Development Based on Bacterial Membrane Nanovesicles
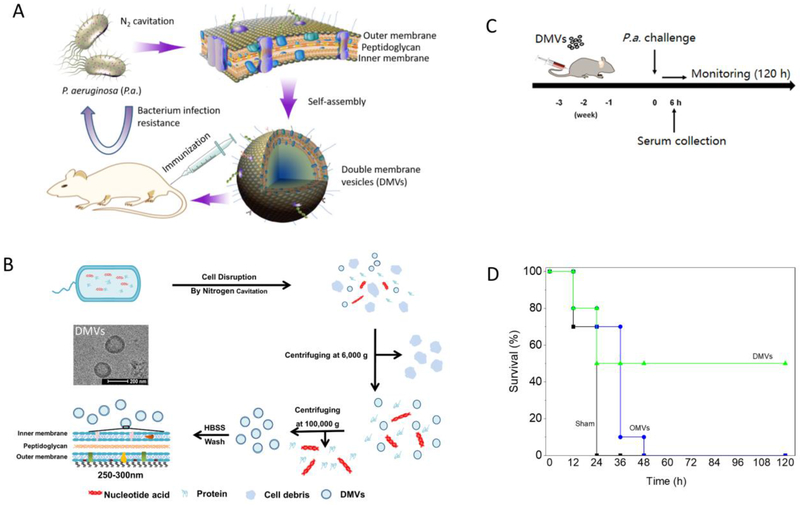
Due to multidrug resistance, alternative therapies for infectious diseases are needed, and developing vaccines is a promising strategy. Wang et al. proposed a strategy for developing a vaccine based on the generation of whole membranes from pathogenic bacteria because they possess antigens from the parent bacteria. The authors produced double-layered membrane vesicles (DMVs) derived from the P. aeruginosa membrane, and they immunized mice with the DMVs to boost host protection from bacterial infections (Figure 10A).[62] First, they prepared nanovesicles using nitrogen cavitation and characterized their structure, size, surface charge and the proteomic profiles of the nanovesicles (Figure 10B). Second, they showed that DMVs evoked the innate and adaptive immune responses of dendritic cells and T cells in vitro. They further showed that immunization with P. aeruginosa-derived DMVs can protect against P. aeruginosa-induced infection (Figure 10C and D), and they showed prolonged survival after immunization with DMVs compared to other treatments. Their study suggests a new concept for developing vaccines through the production of pathogen membrane-derived nanovesicles to strengthen host immunity, and this idea may be applied to a wide range of bacterial strains.
Figure 10:
Concept of using bacterial membrane-derived DMVs for vaccine development. (A) Schematic demonstration of DMVs for vaccine development. The hypothesis is that the bacterial membrane possesses surface proteins and the LPS of the parent bacteria, and thus the vesicles derived from the bacterial membrane can generate host immunity to bacterial infections. (B) Illustration of DMV preparation. Cultured P aeruginosa bacteria were subject to nitrogen cavitation followed by differential centrifugation to obtain pure DMVs. The structure of DMVs was confirmed by TEM. (C) Experimental design for vaccination using DMVs in the bacterial infection mouse model. (D) The mouse survival after the mice were challenged by a lethal dose of P. aeruginosa (1010 CFU each, n=10). OMVs (outer membrane vesicles as control). Copyright 2018 Elsevier.
Gao et al. also reported developments in bacterial outer membrane vesicle (OMV)-based vaccines.[63] They developed E. coli (Escherichia coli) OMVs coated onto gold nanoparticles (BM-AuNPs), with a size of less than 50 nm, and the gold nanoparticles enhanced the stability of the OMVs in buffers. Antigen-presenting dendritic cells in lymph nodes were increased in immunized mice after treating with BM-AuNPs compared with OMVs. BM-AuNPs also elevated the IFN-γ and IL-17, but not the IL-4, indicating that the generation of Th1 and Th17 promoted T cell responses against bacterial infection. These two studies provide the foundation for developing the next generation of nanoparticle-based vaccines to treat infectious diseases.
5. Conclusion and Perspectives
In this review, we focused on discussing new approaches to the design of new nanotherapeutics to solve the current challenges associated with bacterial infections. The specific question in infectious diseases is antimicrobial resistance, because bacteria mutate rapidly and developments in new antibiotics lag behind the pace of microbial resistance. The targeted delivery of therapeutics may be a promising strategy to overcome antimicrobial resistance. Recent advances in nanotechnology and nanoscience offer opportunities to deliver therapeutics specifically to diseased tissues because multifunctional nanoparticle delivery systems can load various drugs. This review is focused on how to design nanomaterials rationally to treat infectious diseases based on an understanding of the pathogenesis of diseases and their microenvironments.
Bacterial infections include two major components: pathogenic bacteria and the host immune response. In this review, we have discussed two strategies for treating infectious diseases, namely targeting bacteria through the delivery of antibiotics and targeting host cells to mitigate inflammation responses and to strengthen immunity via vaccines. For the targeted delivery of antibiotics, we have discussed a new concept, targeting infectious microenvironments (IMEs). IMEs are totally different from well-studied tumor microenvironments[64] because IMEs are acute and dynamic, and they are involved with pathogens and host immune responses. We show two examples in which inorganic NPs and polymeric micelles were designed to improve antibiotic delivery in infectious lesions. Targeting IMEs for the treatment of infectious diseases is promising since this strategy may repurpose clinical drugs (small molecules) for improved antimicrobial resistance. It is also necessary to develop in vivo imaging tools[65] (intravital microscopy) in real time to investigate how NPs interact with IMEs and how NPs respond to local cues from IMEs for controlled drug release.
Phagocytes (neutrophils and macrophages) chase bacteria for clearance. Targeting these phagocytes offers new opportunities to treat infectious diseases effectively. We have shown that targeting macrophages can deliver antibiotics into the host cells. Another example is the hijacking of neutrophils using albumin NPs to deliver antibiotics inside infectious lesions.
In the second section, we discussed the targeting of host cells for infectious disease therapies, as the so-called host-directed therapy. This is also a new research area. We are focused on reviewing cell membrane-derived nanovesicles to construct drug delivery systems. We have demonstrated that neutrophil membrane-derived nanovesicles can specifically target inflamed vasculatures to strengthen the lung vascular integrity for preventing lung pneumonia. In this vein, bacterial membrane-formed nanovesicles provide protection from bacterial infections after nanovesicles are used for mouse immunization.
In summary, this review presents the current literature on new therapeutic developments for treating infectious diseases, and it reorganizes these studies based on the pathogenesis of infectious diseases. We believe that this review may be a starting point for material engineers and clinical scientists to think about how to design the next generation of nanotherapeutics to overcome antimicrobial resistance in infectious diseases.
Acknowledgements
This work was supported by the National Institute of Health via grant RO1GM116823 awarded to Z. W.
References
- [1].a) Wang S, Gao J, Wang Z, Wiley Interdiscip Rev Nanomed Nanobiotechnol 2018, e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schwechheimer C, Kuehn MJ, Nat Rev Microbiol 2015, 13, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wilson JW, Schurr MJ, LeBlanc CL, Ramamurthy R, Buchanan KL, Nickerson CA, Postgrad Med J 2002, 78, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heckenberg SG, Brouwer MC, van de Beek D, Handb Clin Neurol 2014, 121, 1361. [DOI] [PubMed] [Google Scholar]
- [4].van der Poll T, Opal SM, Lancet 2009, 374, 1543. [DOI] [PubMed] [Google Scholar]
- [5].Angus DC, van der Poll T, N Engl J Med 2013, 369, 2063. [DOI] [PubMed] [Google Scholar]
- [6].Maartens G, Wilkinson RJ, Lancet 2007, 370, 2030. [DOI] [PubMed] [Google Scholar]
- [7].Nelson EJ, Harris JB, Morris JG Jr., Calderwood SB, Camilli A, Nat Rev Microbiol 2009, 7, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Humphries RM, Linscott AJ, Clin Microbiol Rev 2015, 28, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Dijkshoorn L, Nemec A, Seifert H, Nat Rev Microbiol 2007, 5, 939. [DOI] [PubMed] [Google Scholar]; b) Argyres MI, New England Journal of Medicine 2010, 363, 1483. [DOI] [PubMed] [Google Scholar]
- [10].Aminov RI, Frontiers in Microbiology 2010, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roope LSJ, Smith RD, Pouwels KB, Buchanan J, Abel L, Eibich P, Butler CC, Tan PS, Walker AS, Robotham JV, Wordsworth S, Science 2019, 364. [DOI] [PubMed] [Google Scholar]
- [12].Tyers M, Wright GD, Nat Rev Microbiol 2019, 17, 141. [DOI] [PubMed] [Google Scholar]
- [13].a) Ventola CL, P T 2015, 40, 277. [PMC free article] [PubMed] [Google Scholar]; b) Ventola CL, P T 2015, 40, 344. [PMC free article] [PubMed] [Google Scholar]
- [14].Fair RJ, Tor Y, Perspect Medicin Chem 2014, 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Alekshun MN, Levy SB, Cell 2007, 128, 1037. [DOI] [PubMed] [Google Scholar]; b) Shi X, Zhang CY, Gao J, Wang Z, Wiley Interdiscip Rev Nanomed Nanobiotechnol 2019, e1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Piddock LJV, Nature Reviews Microbiology 2006, 4, 629. [DOI] [PubMed] [Google Scholar]
- [17].Li W, Atkinson GC, Thakor NS, Allas U, Lu CC, Chan KY, Tenson T, Schulten K, Wilson KS, Hauryliuk V, Frank J, Nature Communications 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weisblum B, Antimicrobial Agents and Chemotherapy 1995, 39, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stapleton PD, Taylor PW, Sci Prog 2002, 85, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin J, Nishino K, Roberts MC, Tolmasky M, Aminov RI, Zhang L, Front Microbiol 2015, 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Wang Z, Tiruppathi C, Minshall RD, Malik AB, ACS Nano 2009, 3, 4110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chu D, Gao J, Wang Z, ACS Nano 2015, 9, 11800. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang Z, Tiruppathi C, Cho J, Minshall RD, Malik AB, IUBMB Life 2011, 63, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Torchilin VP, Nat Rev Drug Discov 2014, 13, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Xin Q, Shah H, Nawaz A, Xie W, Akram MZ, Batool A, Tian L, Jan SU, Boddula R, Guo B, Liu Q, Gong JR, Adv Mater 2018, e1804838. [DOI] [PubMed] [Google Scholar]; f) Rajchakit U, Sarojini V, Bioconjug Chem 2017, 28, 2673. [DOI] [PubMed] [Google Scholar]; g) Ding X, Wang A, Tong W, Xu FJ, Small 2019, 15, e1900999. [DOI] [PubMed] [Google Scholar]; h) Lakshminarayanan R, Ye E, Young DJ, Li Z, Loh XJ, Adv Healthc Mater 2018, 7, e1701400. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Hutnick MA, Pokorski JK, Mol Pharm 2018, 15, 2910. [DOI] [PubMed] [Google Scholar]; j) Dhanasekar M, Jenefer V, Nambiar RB, Babu SG, Selvam SP, Neppolian B, Bhat SV, Materials Research Bulletin 2018, 97, 238. [Google Scholar]
- [22].Angsantikul P, Fang RH, Zhang L, Bioconjug Chem 2018, 29, 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dinarello CA, Cell 2010, 140, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Soehnlein O, Steffens S, Hidalgo A, Weber C, Nat Rev Immunol 2017, 17, 248. [DOI] [PubMed] [Google Scholar]
- [25].Wang AZ, Langer R, Farokhzad OC, Annu Rev Med 2012, 63, 185. [DOI] [PubMed] [Google Scholar]
- [26].Hussain S, Joo J, Kang J, Kim B, Braun GB, She ZG, Kim D, Mann AP, Molder T, Teesalu T, Carnazza S, Guglielmino S, Sailor MJ, Ruoslahti E, Nat Biomed Eng 2018, 2, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].a) Li X, Kolltveit KM, Tronstad L, Olsen I, Clin Microbiol Rev 2000, 13, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee WY, Moriarty TJ, Wong CH, Zhou H, Strieter RM, van Rooijen N, Chaconas G, Kubes P, Nat Immunol 2010, 11, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang CY, Gao J, Wang Z, Adv Mater 2018, 30, e1803618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Radovic-Moreno AF, Lu TK, Puscasu VA, Yoon CJ, Langer R, Farokhzad OC, ACS Nano 2012, 6, 4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xiong MH, Bao Y, Yang XZ, Wang YC, Sun B, Wang J, J Am Chem Soc 2012, 134, 4355. [DOI] [PubMed] [Google Scholar]
- [31].Deutschman CS, Tracey KJ, Immunity 2014, 40, 463. [DOI] [PubMed] [Google Scholar]
- [32].Thwaites GE, Gant V, Nat Rev Microbiol 2011, 9, 215. [DOI] [PubMed] [Google Scholar]
- [33].Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G, Nature Reviews Microbiology 2006, 4, 295. [DOI] [PubMed] [Google Scholar]
- [34].Foster TJ, Nature Reviews Microbiology 2005, 3, 948. [DOI] [PubMed] [Google Scholar]
- [35].Wright JA, Nair SP, Int J Med Microbiol 2010, 300, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thompson BT, Chambers RC, Liu KD, N Engl J Med 2017, 377, 1904. [DOI] [PubMed] [Google Scholar]
- [37].McDonald JR, Infect Dis Clin North Am 2009, 23, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Andlen RV, DePalatis L, Raab H, Hazenbos WL, Morisaki JH, Kim J, Park S, Darwish M, Lee BC, Hernandez H, Loyet KM, Lupardus P, Fong RN, Yan DH, Halouni CC, Luis E, Khalfin Y, Plise E, Heong JC, Lyssikatos JP, Strandh M, Koefoed K, Andersen PS, Flygare JA, Tan MW, Brown EJ, Ariathasan SM, Nature 2015, 527, 323. [DOI] [PubMed] [Google Scholar]
- [39].Yeom JH, Lee B, Kim D, Lee JK, Kim S, Bae J, Park Y, Lee K, Biomaterials 2016, 104, 43. [DOI] [PubMed] [Google Scholar]
- [40].Xiong MH, Li YJ, Bao Y, Yang XZ, Hu B, Wang J, Advanced Materials 2012, 24, 6175. [DOI] [PubMed] [Google Scholar]
- [41].a) Mayadas TN, Cullere X, Lowell CA, Annu Rev Pathol 2014, 9, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ermert D, Zychlinsky A, Urban C, Methods Mol Biol 2009, 470, 293. [DOI] [PubMed] [Google Scholar]
- [42].Wang Z, Li J, Cho J, Malik AB, Nat Nanotechnol 2014, 9, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kostakioti M, Hadjifrangiskou M, Hultgren SJ, Cold Spring Harb Perspect Med 2013, 3, a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Flemming HC, Neu TR, Wozniak DJ, J Bacteriol 2007, 189, 7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Singh S, Singh SK, Chowdhury I, Singh R, Open Microbiol J 2017, 11, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu H, Moser C, Wang HZ, Hoiby N, Song ZJ, Int J Oral Sci 2015, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu Y, Busscher HJ, Zhao B, Li Y, Zhang Z, van der Mei HC, Ren Y, Shi L, ACS Nano 2016, 10, 4779. [DOI] [PubMed] [Google Scholar]
- [48].Nguyen TK, Selvanayagam R, Ho KKK, Chen RX, Kutty SK, Rice SA, Kumar N, Barraud N, Duong HTT, Boyer C, Chemical Science 2016, 7, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Geilich BM, Gelfat I, Sridhar S, van de Ven AL, Webster TJ, Biomaterials 2017, 119, 78. [DOI] [PubMed] [Google Scholar]
- [50].Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW, Plos One 2009, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baelo A, Levato R, Julian E, Crespo A, Astola J, Gavalda J, Engel E, Mateos-Timoneda MA, Torrents E, Journal of Controlled Release 2015, 209, 150. [DOI] [PubMed] [Google Scholar]
- [52].Swartjes JJTM, Das T, Sharifi S, Subbiahdoss G, Sharma PK, Krom BP, Busscher HJ, van der Mei HC, Advanced Functional Materials 2013, 23, 2843. [Google Scholar]
- [53].Traba C, Liang JF, Journal of Controlled Release 2015, 198, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].a) Zumla A, Rao M, Wallis RS, Kaufmann SH, Rustomjee R, Mwaba P, Vilaplana C, Yeboah-Manu D, Chakaya J, Ippolito G, Azhar E, Hoelscher M, Maeurer M, c. Host-Directed Therapies Network, Lancet Infect Dis 2016, 16, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Maeurer M, Rao M, Zumla A, Curr Opin Pulm Med 2016, 22, 203. [DOI] [PubMed] [Google Scholar]; c) Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R, Nat Rev Drug Discov 2018, 17, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bernard GR, Wheeler AP, Russell JA, Schein R, Summer WR, Steinberg KP, Fulkerson WJ, Wright PE, Christman BW, Dupont WD, Higgins SB, Swindell BB, N Engl J Med 1997, 336, 912. [DOI] [PubMed] [Google Scholar]
- [56].Johnston B, Conly J, Can J Infect Dis Med Microbiol 2006, 17, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].a) Matthay MA, Ware LB, Zimmerman GA, J Clin Invest 2012, 122, 2731. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Matthay MA, Zemans RL, Annu Rev Pathol 2011, 6, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mehta D, Malik AB, Physiol Rev 2006, 86, 279. [DOI] [PubMed] [Google Scholar]
- [58].Dong X, Gao J, Zhang CY, Hayworth C, Frank M, Wang Z, ACS Nano 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].a) Gao, Chu, Wang, J Control Release 2016, 224, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gao, Wang, Wang, Biomaterials 2017, 135, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fang RH, Kroll AV, Gao W, Zhang L, Adv Mater 2018, 30, e1706759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Thamphiwatana S, Angsantikul P, Escajadillo T, Zhang Q, Olson J, Luk BT, Zhang S, Fang RH, Gao W, Nizet V, Zhang L, Proc Natl Acad Sci U S A 2017, 114, 11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wang SH, Gao J, Li M, Wang LG, Wang ZJ, Biomaterials 2018, 187, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gao W, Fang RH, Thamphiwatana S, Luk BT, Li J, Angsantikul P, Zhang Q, Hu CM, Zhang L, Nano Lett 2015, 15, 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].a) Chu D, Dong X, Shi X, Zhang C, Wang Z, Adv Mater 2018, 30, e1706245. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chu D, Dong X, Zhao Q, Gu J, Wang Z, Adv Mater 2017, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wang Z, Theranostics 2016, 6, 2431. [DOI] [PMC free article] [PubMed] [Google Scholar]