Abstract
A typical approach to the joint analysis of two high-dimensional datasets is to decompose each data matrix into three parts: a low-rank common matrix that captures the shared information across datasets, a low-rank distinctive matrix that characterizes the individual information within a single dataset, and an additive noise matrix. Existing decomposition methods often focus on the orthogonality between the common and distinctive matrices, but inadequately consider the more necessary orthogonal relationship between the two distinctive matrices. The latter guarantees that no more shared information is extractable from the distinctive matrices. We propose decomposition-based canonical correlation analysis (D-CCA), a novel decomposition method that defines the common and distinctive matrices from the space of random variables rather than the conventionally used Euclidean space, with a careful construction of the orthogonal relationship between distinctive matrices. D-CCA represents a natural generalization of the traditional canonical correlation analysis. The proposed estimators of common and distinctive matrices are shown to be consistent and have reasonably better performance than some state-of-the-art methods in both simulated data and the real data analysis of breast cancer data obtained from The Cancer Genome Atlas.
Keywords: approximate factor model, canonical variable, common structure, distinctive structure, soft thresholding
1. Introduction
Many large biomedical studies have collected high-dimensional genetic and/or imaging data and associated data (e.g., clinical data) from increasingly large cohorts to delineate the complex genetic and environmental contributors to many diseases, such as cancer and Alzheimer’s disease. For example, The Cancer Genome Atlas (TCGA; Koboldt et al., 2012) project collected human tumor specimens and derived different types of large-scale genomic data such as mRNA expression and DNA methylation to enhance the understanding of cancer biology and therapy. The Human Connectome Project (Van Essen et al., 2013) acquired imaging datasets from multiple modalities (HARDI, R-fMRI, T-fMRI, MEG) across a large cohort to build a “network map” (connectome) of the anatomical and functional connectivity within the healthy human brain. These cross-platform datasets share some common information, but individually contain distinctive patterns. Disentangling the underlying common and distinctive patterns is critically important for facilitating the integrative and discriminative analysis of these cross-platform datasets (van der Kloet et al., 2016; Smilde et al., 2017).
Throughout this paper, we focus on disentangling the common and distinctive patterns of two high-dimensional datasets written as matrices for k = 1, 2 on a common set of n objects, where each of the pk rows corresponds to a mean-zero variable. A popular approach to such an analysis is to decompose each data matrix into three parts:
| (1) |
where Ck’s are low-rank “common” matrices that capture the shared structure between datasets, Dk’s are low-rank “distinctive” matrices that capture the individual structure within each dataset, and Ek’s are additive noise matrices. Model (1) has been widely used in genomics (Lock et al., 2013; O’Connell and Lock, 2016), metabolomics (Kuligowski et al., 2015), and neuroscience (Yu et al., 2017), among other areas of research. Ideally, the common and distinctive matrices should provide different “views” for each individual dataset, while borrowing information from the other. A fundamental question for model (1) is how to decompose Yk’s into the common and distinctive matrices within each dataset and across datasets.
Most decomposition methods for model (1) are based on the Euclidean space (, ·) endowed with the dot product. Such methods include JIVE (Lock et al., 2013), angle-based JIVE (AJIVE; Feng et al., 2018), OnPLS (Trygg, 2002; Löfstedt and Trygg, 2011), COBE (Zhou et al., 2016), and DISCO-SCA (Schouteden et al., 2014). A common characteristic among all these methods is to enforce the row-space orthogonality between the common and distinctive matrices within each dataset, that is, for k =1, 2. With the exception of OnPLS, these methods impose additional orthogonality across the datasets, that is, for all k and ℓ. A potential issue associated with these methods is that they inadequately consider the more desired orthogonality between the distinctive matrices D1 and D2, which guarantees that no common structure is retained therein. Specifically, the first four methods do not impose any orthogonality constraint between D1 and D2. Although DISCO-SCA and a modified JIVE (O’Connell and Lock (2016); denoted as R.JIVE) have considered the row-space orthogonality between the distinctive matrices, it may be incompatible with their orthogonal condition that for all k, ℓ = 1, 2 even as p1 = p2 = 1.
Rather than the conventionally used Euclidean space (, ·), the aim of this paper is to develop a new decomposition method for model (1) based on the inner product space (, cov), which is the vector space composed of all zero-mean and finite-variance real-valued random variables and endowed with the covariance operator as the inner product. Specifically, model (1) is a sample-matrix version of the prototype given by
| (2) |
The Euclidean space (, ·) is hence not an appropriate space for defining the common matrices Ck’s and the distinctive matrices Dk’s, because two uncorrelated non-constant random variables will almost never have zero sample correlation, i.e., the orthogonality in (, ·). The matrices defined by the aforementioned methods based on (, ·) are, in fact, estimators of the counterparts defined through model (2) on (, cov). Instead, for model (2), we introduce a common-space constraint for the common vectors , an orthogonal-space constraint for the distinctive vectors , and a parsimonious-representation constraint for the signal vectors xk := yk – ek, k = 1, 2 as follows:
| (3) |
| (4) |
| (5) |
where is the vector space spanned by entries of any random vector υ = (υ1, …, υp)⊤, and ⊥ denotes the orthogonality between two subspaces and/or random variables in (, cov). The orthogonal relationship between distinctive matrices D1 and D2 is now described by (4).
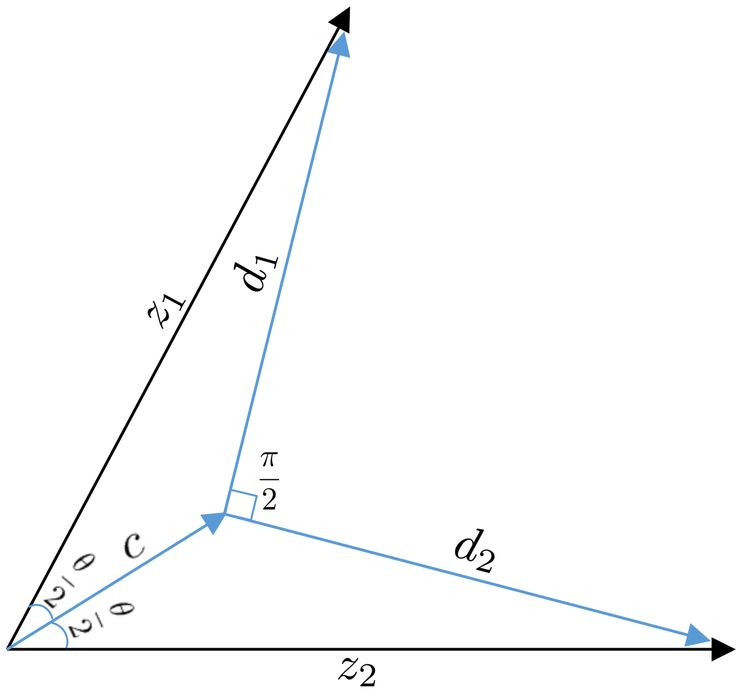
To illustrate the advantage of our proposed constraints over those imposed by the six existing methods mentioned above, we consider a toy example based on model (2) with p1 = p2 = 1. Suppose z1 and z2 are two standardized signal random variables with the same distribution and corr(z1, z2) ϵ (0, 1), i.e., their angle on (, cov), denoted as θ, in (0, π/2) (see Figure 1). We want to decompose them as zk = ck + dk for k = 1, 2. The constraints of JIVE, AJIVE, OnPLS, and COBE translated into space (, cov) do not guarantee d1 ⊥ d2, i.e., corr(d1, d2) = 0. DISCO-SCA and R.JIVE impose d1 ⊥ d2 and cj ⊥ dk for all j, k = 1, 2. Restrict as in our (5) to avoid the signal space being represented by a higher dimensional space. Then their orthogonal constraints result in either (i) d1 = d2 = 0 or (ii) that only one of d1 and d2 is a zero constant, since a two-dimensional space does not tolerate three nonzero orthogonal elements. Scenario (i) indicates z1 = c1 ≠ z2 = c2 and fails to reveal the distinctive patterns of z1 and z2. Scenario (ii) implies unequal distributions of d1 and d2, which contradicts the symmetry of z1 and z2 about 0.5(z1 + z2). However, our proposed constraints and developed method will achieve the desirable decomposition shown in Figure 1, where d1 ⊥ d2, c1 = c2 = c ∝ 0.5(z1 + z2), and moreover, ∥c∥ indicates the extent of 1/θ or corr(z1, z2).
Figure 1:
The geometry of D-CCA for two standardized random variables.
Motivated by the toy example above, we introduce a novel method, decomposition-based canonical correlation analysis (D-CCA), which generalizes the classical canonical correlation analysis (CCA; Hotelling, 1936) by further separating common vectors and distinctive vectors between signal vectors subject to constraints (3)-(5). In contrast, classical CCA only seeks the association between two random vectors by sequentially determining the mutually orthogonal pairs of canonical variables that have maximal correlations between the vector spaces respectively spanned by entries of the two random vectors. Another related but different method, the sparse CCA (Chen et al., 2013; Gao et al., 2015, 2017), focuses on the sparse linear combinations of original variables for representing canonical variables with improved interpretability, which is neither required nor pursued by our D-CCA.
The “low-rank plus noise” model yk = xk + ek for each single k can be naturally formulated by a factor model as yk = Bkfk + ek, where the latent factor is an orthonormal basis of with Bk being the coefficient matrix. In factor model analysis (Bai and Ng, 2008), xk = Bkfk is called the “common component”, and ek the “idiosyncratic error”. These two terms should not be confused with our considered common vectors and distinctive vectors that are solely based on signals excluding noises . For general dynamic factor models (Forni et al., 2000), Hallin and Liška (2011) proposed a joint decomposition method, which divides each dataset into strongly common, weakly common, weakly idiosyncratic, and strongly idiosyncratic components (also see Forni et al. (2017) and Barigozzi et al. (2018)). Applying their method to our considered scenarios with no temporal dependence, and additionally assuming no correlations between signals , and noises , then for each yk, xk is the sum of strongly common and weakly common components, ek is the strongly idiosyncratic component, and no weakly idiosyncratic component exists. One may treat their strongly common and weakly common components as the common vector ck and the distinctive vector dk, respectively, but the desired orthogonality (4) is still not imposed. Especially when , xk is entirely a weakly common component, and thus the orthogonality (4) fails for the toy example shown in Figure 1. See Remark S.l in the supplementary material for more detailed discussions.
Our major contributions of this paper are as follows. The proposed D-CCA method appropriately decomposes each paired canonical variables of signal vectors x1 and x2 into a common variable and two orthogonal distinctive variables, and then collects all of them to form the common vector ck and the distinctive vector dk for each xk. The common matrix Ck and the distinctive matrix Dk are defined with columns as n realizations of ck and dk, respectively. Three challenging issues that arise in estimating the low-rank matrices defined by D-CCA are high dimensionality, the corruption of signal random vectors by unobserved noises, and the unknown signal covariance and cross-covariance matrices that are needed in CCA. To address these issues, we study the considered “low-rank plus noise” model under the framework of approximate factor models (Wang and Fan, 2017), and develop a novel estimation approach by integrating the S-POET method for spiked covariance matrix estimation (Wang and Fan, 2017) and the construction of principal vectors (Björck and Golub, 1973). Under some mild conditions, we systematically investigate the consistency and convergence rates of the proposed matrix estimators under a high-dimensional setting with min(p1, p2) > κ0n for a positive constant κ0.
The rest of this paper is organized as follows. Section 2 introduces the D-CCA method that appropriately defines the common and distinctive matrices from the inner product space (, cov). A soft-thresholding approach is then proposed for estimating the matrices defined by D-CCA. Section 3 is devoted to the theoretical results of the proposed matrix estimators under a high-dimensional setting. The performance of D-CCA and the associated estimation approach is compared to that of the aforementioned state-of-the-art methods through simulations in Section 4 and through the analysis of TCGA breast cancer data in Section 5. Possible future extensions of D-CCA are discussed in Section 6. All technical proofs are provided in the supplementary material.
Here, we introduce some notation. For a real matrix M = (Mij)1≤i≤p,1≤j≤n, the ℓ-th largest singular value and the ℓ-th largest eigenvalue (if p = n) are respectively denoted by σℓ(M) and λℓ(M), the spectral norm ∥M∥2 = σ1(M), the Frobenius norm , and the matrix norm . We use M[s:t,u:v], M[s:t,:] and M[:,u:v] to represent the submatrices (Mij)s≤i≤t,u≤j≤v, (Mij)s≤i≤t,1≤j≤n and (Mij)1≤i≤p,u≤j≤v of the p×n matrix M, respectively. Denote the Moore-Penrose pseudoinverse of matrix M by M†. Define 0p×n to be the p×n zero matrix and Ip×p to be the p×p identity matrix. Denote diag(M1, …, Mm) to be a block diagonal matrix with M1, …, Mm as its main diagonal blocks. For signal vectors xk’s, denote Σk = cov(xk), Σ12 = cov(x1, x2), rk = rank(Σk), rmin = min(r1, r2), rmax = max(r1, r2) and r12 = rank(Σ12). For a subspace B of a vector space A, denote its orthogonal complement in A by A \ B. We write a ∝ b if a is proportional to b, i.e., a = κb for some constant κ. Throughout the paper, our asymptotic arguments are by default under n → ∞. We reserve {c, cℓ}, {ck} and {Ck} for the common variables, common vectors and common matrices, respectively, and use other notation for constants, e.g., κ0.
2. The D-CCA Method
Suppose the columns of matrices Yk, Xk and Ek are, respectively, n independent and identically distributed (i.i.d.) copies of mean-zero random vectors yk, xk and ek for k = 1, 2. We consider the “low-rank plus noise” model for the observable random vector yk as follows:
| (6) |
where is a real deterministic matrix, is a mean-zero random vector of rk latent factors such that cov(fk) = Irk×rk and cov(fk, ek) = 0rk×pk, and rk is a fixed number independent of {n, p1, p2}. Write the model in a sample-matrix form by
| (7) |
where the columns of Fk are assumed to be i.i.d. copies of fk. We assume that the model given in (6) and (7) is an approximate factor model (Wang and Fan, 2017) that allows for correlations among entries of ek in contrast with the strict factor model (Ross, 1976) and has be a spiked covariance matrix for which the top rk eigenvalues are significantly larger than the rest (i.e., signals are stronger than noises). Detailed conditions for consistent estimation will be given later in Assumption 1. Although approximate factor models are often used in econometric literature (Chamberlain and Rothschild, 1983; Bai and Ng, 2002; Stock and Watson, 2002; Bai, 2003) with temporal dependence on {, } across t’s, we assume independence across the n samples as in Wang and Fan (2017) since no temporal dependence is quite natural in our motivating TCGA datasets and considered in the six competing methods mentioned in Section 1.
2.1. Definition of common and distinctive matrices
We define the common and distinctive matrices of two datasets based on the inner product space (, cov). The low-rank structure of xk in (6) indicates that the dimension of is rk.
One natural way to construct the decomposition of Xk = Ck + Dk for k = 1, 2 is to decompose the signal vectors as
| (8) |
subject to the constraints (3)-(5) with space dimensions L12 ≤ rmin and Lk ≤ rk, where βkℓ, and are real deterministic vectors, and random variables , and are, respectively, the orthogonal basis of , and . The desirable contraints (3)-(5) are now equivalent to
| (9) |
We call the common variables of x1 and x2, and the distinctive variables of xk. The columns of common matrix Ck are defined as the i.i.d. copies of ck, and those of distinctive matrix Dk are the ones of dk. The space represents the common structure of x1 and x2, or datasets X1 and X2, and the spaces correspond to their distinctive structures.
To achieve a decomposition of form (8), our D-CCA method adopts a two-step optimization strategy given in (10) and (11) below. The first step uses the classical CCA to recursively find the most correlated variables between signal spaces as follows: For ℓ = 1, …, r12,
| (10) |
where . Variables are called the ℓ-th pair of canonical variables, and their correlation is the ℓ-th canonical correlation of x1 and x2. Augment with any (rk – r12) standardized variables to be zk = (zk1, …, zkrk)⊤ such that is an orthonormal basis of . A detailed procedure to obtain a solution of will be presented later after Theorem 2. An important property of these augmented canonical variables is the bi-orthogonality shown in the following theorem.
Theorem 1 (Bi-orthogonality). The covariance matrix of z1 and z2 is
where Λ12 is a r12×r12 nonsingular diagonal matrix.
Theorem 1 implies that all correlations between and are confined between their subspaces and , and moreover, span({z1ℓ, z2ℓ}) ⊥ span({z1m, z2m}) holds for 1 ≤ ℓ ≠ m ≤ r12. We hence only need to investigate the correlations within each subspace span({z1ℓ, z2ℓ}) for 1 ≤ ℓ ≤ r12. The second step of our D-CCA defines the common variables by
| (11) |
with the constraints
| (12) (13) (14) |
Constraints (12) and (13) are actually the special case of (8) and (9) for two standardized random variables. Constraint (14) indicates that cℓ explains more variances of z1ℓ and z2ℓ when their correlation pℓ increases. Although ρℓ, here referring to the ℓ-th canonical correlation of , is always positive for 1 ≤ ℓ ≤ r12, we include ρℓ = 0 to enable (11) as a general optimization problem for any two standardized variables with nonnegative correlation. The unique solution of (11) is given by
| (15) |
where θℓ = arccos(ρℓ) is the angle between z1ℓ and z2ℓ in (, cov). More desirable than constraint (14), it easily follows from (15) that var(cℓ) is a continuous and strictly monotonic increasing function for ρℓ ϵ [0, 1]. We defer the detailed derivation of (15) to the supplementary material (Proposition S.2 and its proof). This solution is geometrically illustrated in Figure 1 with ℓ omitted in the subscriptions. Simply let dkℓ = zkℓ for r12 + 1 ≤ ℓ ≤ rk. The two-step optimization strategy arrives at the following decomposition of form (8): For k = 1, 2,
| (16) |
| (17) |
Constraints (3)-(5) or equivalently (9) are satisfied due to the bi-orthogonality in Theorem 1 and the constraints in (12) and (13).
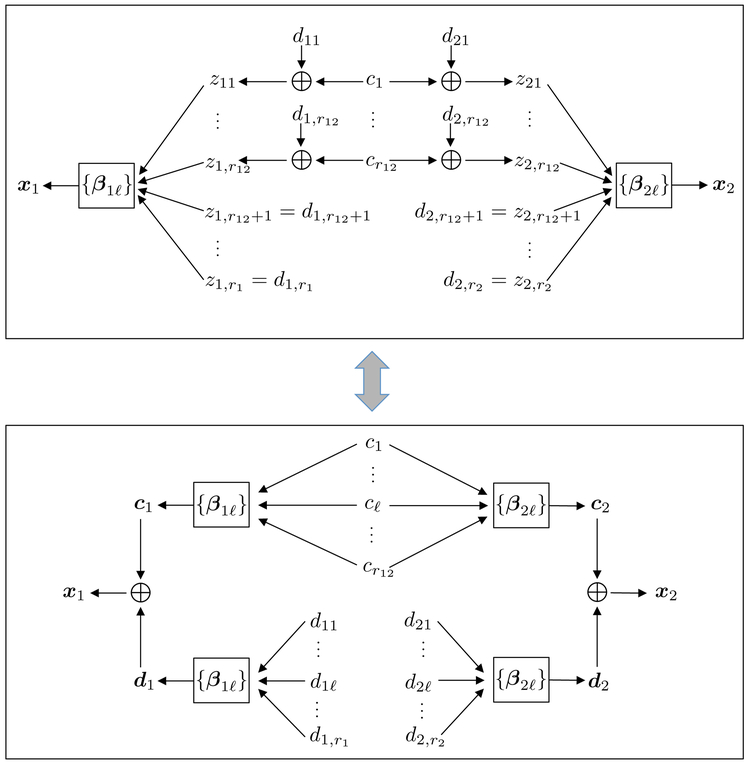
The workflow of D-CCA can be interpreted from the perspective of blind source separation (Comon and Jutten, 2010). Jointly for k = 1, 2, D-CCA first uses CCA to recover the input sources and the mixing channel that generate the output signal vector xk. Then by the constrained (11), D-CCA discovers the common components and the distinctive components , k = 1, 2 of the two sets of input sources , k = 1, 2. Finally, D-CCA separately passes and through the mixing channel to form the common vector ck and the distinctive vector dk of each k-th output signal vector xk. Figure 2 illustrates such interpretation of the D-CCA decomposition structure.
Figure 2:
The decomposition structure of D-CCA.
The solution to the CCA problem in (10) may not be unique even when ignoring a simultaneous sign change, but all solutions yield the same ck and dk as shown in the following theorem.
Theorem 2 (Uniqueness). All solutions to the problem in (10) for canonical variables give the same ck and dk defined in (16).
We now present a procedure to obtain the augmented canonical variables {z1, z2}. For k = 1, 2, let a singular value decomposition (SVD) of Σk be , where Λk = diag(σ1 (Σk), …, σrk(Σk)) and Vk is a pk×rk matrix with orthonormal columns. Let , then we have . Define
The rank of Θ is also r12. Denote a full SVD of Θ by , where Uθ1 and Uθ2 are two orthogonal matrices, and Λθ is a r1 × r2 rectangular diagonal matrix for which the main diagonal is (σ1(Θ), …, σr12 (Θ), 01×(rmin–r12)). We then define
| (18) |
which satisfies cov(zk) = Irk×rk and corr(z1, z2) = Λθ. Note that σℓ(Θ) = ρℓ for ℓ ≤ r12 are the canonical correlations between x1 and x2.
Now look back to that is defined in (16). Plugging (17) and (15) for βkℓ and cℓ in the formula together with , given in (18), we obtain
| (19) |
where AC = diag(a1, …, ar12) and for ℓ ≤ r12. Replacing random vector by its sample matrix in the rightmost of (19) yields
| (20) |
This equation is useful to our design of estimators for Ck and Dk=Xk–Ck in the next subsection.
2.2. Estimation of D-CCA matrices
In this subsection, we discuss the estimation of the matrices defined by D-CCA under model (1) for two high-dimensional datasets. For simplicity, we write the proposed estimators with true ranks r1, r2 and r12. In practice, we can replace those unknown true ranks by the estimated ranks given in Subsection 2.3 with a theoretical guarantee provided in Section 3.
Recall that Yk = Xk + Ek with k = 1, 2. Our first task is to obtain a good initial estimator, denoted by , of Xk. Under the approximate factor model given in (6) and (7), our construction of is inspired by the S-POET method (Wang and Fan, 2017) for spiked covariance matrix estimation. Let the full SVD of Yk be
| (21) |
where Uk1 and Uk2 are two orthogonal matrices and Λyk is a rectangular diagonal matrix with the singular values in decreasing order on its main diagonal. The matrix is then obtained via soft-thresholding the singular values of Yk by
| (22) |
with and . Let . Under Assumption 1 that will be given later, it can be shown that with probability tending to 1 (see the proof of Theorem 3).
We next use to develop estimators for Ck in (20) and Dk=Xk–Ck. Define the estimators of Σk and Σ12 as and , respectively. Then, based on and , we obtain estimators , , and in the same way as their true counterparts Vk, Λk, Uθk and Λθ with a r1×r2 matrix . Define and . We have
From Theorem 1 in Björck and Golub (1973), it follows that and for ℓ ≤ rmin are the principal vectors of the row spaces of and , and moreover, . Let with for and otherwise . Define estimators of Ck, Dk and Xk by
| (23) |
| (24) |
and
| (25) |
Here, we substitute for as the estimator of Xk. The latter can be written as
| (26) |
with
| (27) |
Note that . When , we have . But when , redundantly keeps the nonzero approximated samples of the zero common variable of z1ℓ and z2ℓ for .
Similar to the decomposition of given in (16) that is built on the inner product space (, cov), the decomposition of in (26) is constructed by an analogy of (12) and (15) on the space with the inner product ⟨u, υ⟩ = u⊤υ/n for any . We thus have the appealing property , which corresponds to the orthogonal relationship between the distinctive structures given in (4).
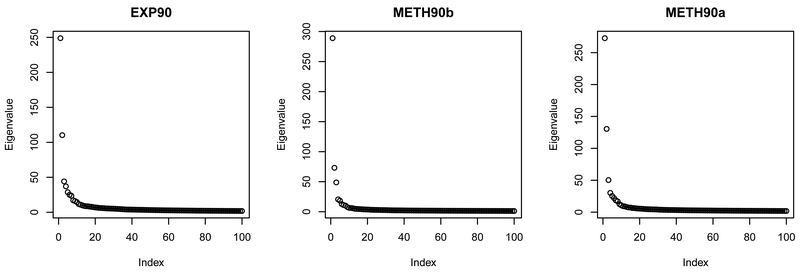
Throughout our estimation construction, the key idea is to develop a good estimator of . Thus, the S-POET method (Wang and Fan, 2017) may be replaced by any other good approach, but with possibly different assumptions. For example, given the cleaned signal data Xk’s, Chen et al. (2013) and Gao et al. (2015, 2017) showed that sparse CCA algorithms can consistently estimate the canonical coefficient matrix Γk for by imposing certain sparsity on Γk’s and that all eigenvalues of cov(xk) are bounded from above and below by positive constants. These two conditions are not assumed for our proposed method. In particular, their bounded eigenvalue condition contradicts our low-rank structure of signal xk that introduces the spiked covariance matrix cov(yk). The sparse CCA algorithms need the cleaned signal data Xk’s available beforehand. Alternatively, they may be directly applicable to the observable data Yk’s by assuming zero Ek’s, if the bounded eigenvalue condition holds for cov(yk). For the TCGA datasets in our real-data application, the scree plots given later in Figure 6 favorably suggest our spiked eigenvalue assumption. Moreover, the approximate factor model with spiked covariance structure has been widely used in various fields such as signal processing (Nadakuditi and Silverstein, 2010) and machine learning (Huang, 2017), and fits the low-rank plus noise structure considered in the six competing methods mentioned in Section 1. Our paper hence focuses on this spiked covariance model and leaves the extension to sparse CCA models for future research.
Figure 6:
The scree plot of the sample covariance matrix for each TCGA dataset.
2.3. Rank selection
In practice, matrix ranks r1, r2 and r12 are usually unknown and need to be determined. There is a rich literature on determining rk, k ϵ {1, 2}, which is the number of latent factors for the high-dimensional approximate factor model. Examples of consistent estimators include but are not limited to Bai and Ng (2002), Onatski (2010), and Ahn and Horenstein (2013). Several heuristic approaches for selecting r12, the number of nonzero canonical correlations for the high-dimensional CCA, have been proposed by Song et al. (2016). In this paper, we apply the edge distribution (ED) method of Onatski (2010) to determine rk for k = 1, 2 by
| (28) |
where is the ℓ-th eigenvalue of . The upper bound is chosen as with mk = min(n, pk) which is recommended by Ahn and Horenstein (2013), and parameter δ is calibrated as in Section IV of Onatski (2010). It is believed that r12 > 0 if two variables from different cleaned datasets have a significant nonzero correlation detected by, e.g., the normal approximation test of DiCiccio and Romano (2017). Otherwise, it is unnecessary to conduct the proposed matrix decomposition. We select the nonzero r12 by using the minimum description length information-theoretic criterion (MDL-IC) proposed by Song et al. (2016):
| (29) |
where sℓ is the ℓ-th singular value of with U12 and U22 defined in (21). The ranks r1, r2, and r12 determined by (28) and (29) perform well in our numerical studies.
3. Theoretical Properties of D-CCA Estimators
In this section, we establish asymptotic results for the high-dimensional D-CCA matrix estimators proposed in Subsection 2.2.
Assumption 1. We assume the following conditions for model given in (6) and (7).
-
(I)
Let λk1 > … > λk,rk > λk,rk+1 ≥ … ≥ λk,pk > 0 be the eigenvalues of cov(yk). There exist positive constants κ1, κ2 and δ0 such that κ1 ≤ λkℓ ≤ κ2 for ℓ > rk and minℓ≤rk(λkℓ – λk,ℓ+1)/λkℓ ≥ δ0.
-
(II)
Assume pk > κ0n with a constant κ0 > 0. When n → ∞, assume λk,rk → ∞, pk/(nλkℓ) is upper bounded for ℓ ≤ rk, λk1/λk,rk is bounded from above and below, and with γk2 given in (V) below.
-
(III)
The columns of are i.i.d. copies of , where is the full SVD of cov(yk) with . The entries of are independent with , , and the sub-Gaussian norm with a constant K > 0 for all i ≤ pk.
-
(IV)
The matrix is a diagonal matrix, and with a constant M > 0 holds for all i ≤ pk and ℓ ≤ rk.
-
(V)
Denote ek = (ek1, …, ek,pk)⊤ and fk = (fk1, …, fk,rk)⊤. Assume ∥ cov(ek)∥∞ < s0 with a constant s0 > 0. For all i ≤ pk and ℓ ≤ rk, there exist positive constants γk1, γk2, bk1 and bk2 such that for t > 0, and .
Assumption 1 follows assumptions 2.1-2.3 and 4.1-4.2 of Wang and Fan (2017) which guarantee desirable performance of the initial signal estimators ’s defined in (22). The diverging leading eigenvalues of cov(yk) assumed in conditions (I) and (II), together with the approximate sparsity constraint ∥ cov(ek)∥∞ < s0 in condition (V), indicate the necessity of sufficiently strong signals for soft-thresholding. Although Wang and Fan (2017) considered p > n, it is not difficult to relax it to pk > κ0n, as given in our condition (II). A random variable is said to be sub-Gaussian if its sub-Gaussian norm is bounded (Vershynin, 2012). Condition (III) imposes the sub-Gaussianity on all entries of with a uniform bound. Simply letting can lead to a diagonal matrix that is required by condition (IV). In condition (V), the approximately sparse constraint is imposed on cov(ek) rather than Ek. See Wang and Fan (2017) and also Fan et al. (2013) for more detailed discussions of the above assumption.
We consider the relative errors of the proposed matrix estimators in the spectral norm and also in the Frobenius norm. For convenience, we use ∥ · ∥(·) as general notation for one of these two matrix norms. Define αCκ,(·) = ∥Ck∥(·)/∥Xk∥(·) and αDκ,(·) = ∥Dk∥(·)/∥Xk∥(·).
Theorem 3. For k = 1, 2, assume , , and defined in Subsection 2.2 are constructed with true rk and r12. Suppose that r12 ≥ 1 and Assumption 1 hold. Define and
Then, we have the following relative error bounds of the matrix estimators
and the error bound of canonical correlation estimators
Provided that matrix ranks r1, r2 and r12 are correctly selected, Theorem 3 shows the consistency of the proposed matrix estimators in the relative errors that are the norms of estimation errors divided by the norms of true matrices, with associated convergence rates. The ratios αCκ,(·) and αDκ,(·) in the convergence rates of and can be removed if the relative errors are instead scaled by the norms of the signal matrices.
Although the ED estimators of r1 and r2 given in (28) are consistent under some mild conditions (Onatski, 2010), the consistency of the MDL-IC estimator in (29) for r12 is still unclear. However, the following corollary indicates the robustness of our proposed matrix estimators given in (23) and (25) when r12 is misspecified but r1 and r2 are appropriately selected.
Corollary 1. For k = 1, 2, assume , and defined in Subsection 2.2 are constructed with the unknown rk replaced by an estimator satisfying . Define with , and for . Suppose that r12 ≥ 1 and Assumption 1 hold. Then, with Δ and δθ defined in Theorem 3, we have
Corollary 1 provides an acceptable range, [, rmin], for the choice of r12 when r1 and r2 are consistently estimated, which can theoretically lead to the same convergence rates (up to a constant factor) as those in Theorem 3. Note that the distinctive matrices ’s are independent of r12.
4. Simulation Studies
We consider the following three simulation setups to evaluate the finite sample performance of the proposed D-CCA estimators comparing with the six competing methods mentioned in Section 1 and also the decomposition of Hallin and Liška (2011) (denoted as GDFM).
Setup 1: Let , with r1 = 3, r12 = 1, and λℓ(Σ1) = 500 – 200(ℓ – 1) for ℓ ≤ 3. Set for each k = 1, 2. Randomly generate V1 with orthonormal columns, which is the same for all replications. Let . Generate that are independent of . Vary dimension p1 from 100 to 1,500, the first canonical angle θ1 = arccos(ρ1) from 0° to 75° with ρ1 = corr(z11, z21), and the noise variance from 0.01 to 16.
Setup 2: Use the same settings for x1 and as in Setup 1. For x2, fix p2 = 300, and set r2 = 5 and λℓ(Σ2) = 500 – 100(ℓ – 1) for ℓ ≤ 5. Simulate with and a randomly generated V2 that is the same for all replications. Let r12 = 1. Vary p1, θ1 and according to Setup 1.
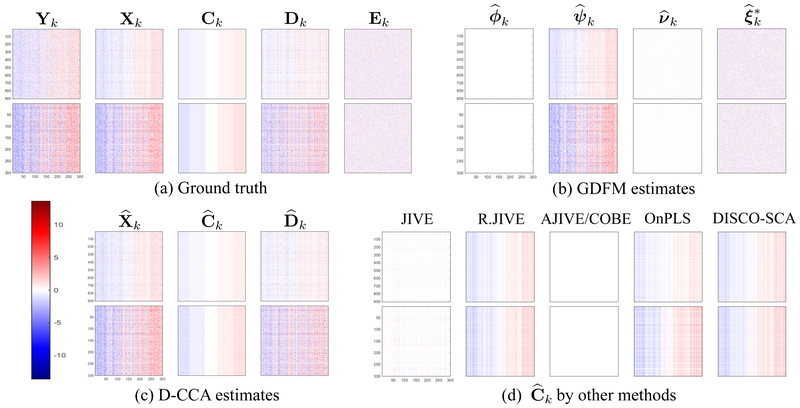
Setup 3 is for visual purposes: Fix p1 = 3p2 = 900, θ1 = 45°, and . Generate two independent variables υ1 and υ2 such that and υ2 ~ N(0, 1). Let and . Set and randomly generate for k = 1, 2. The other settings are the same as those in Setup 2.
We fixed the sample size n = 300 and conducted 1,000 replications for Setups 1 and 2. Setup 3 is only used for the purpose of visually comparing D-CCA with the seven other methods. Setup 3 is similar to Setup 2, but it has the common variable of the first pair of canonical variables following a discrete uniform distribution instead of a Gaussian distribution. We ran a single replication of Setup 3 for the visual comparison in Figure 5. To determine the ranks r1, r2, and r12, we respectively used the ED method given in (28) and the MDL-IC method in (29). Additional simulations with AR(1) matrices for are given in the supplementary material (Section S.2).
Figure 5:
Color maps for a single replication of Setup 3.
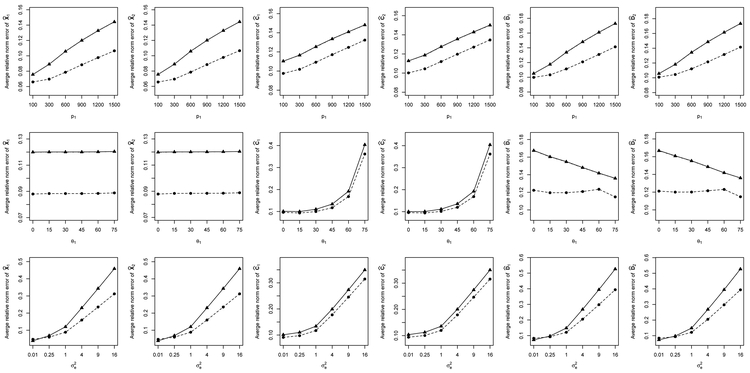
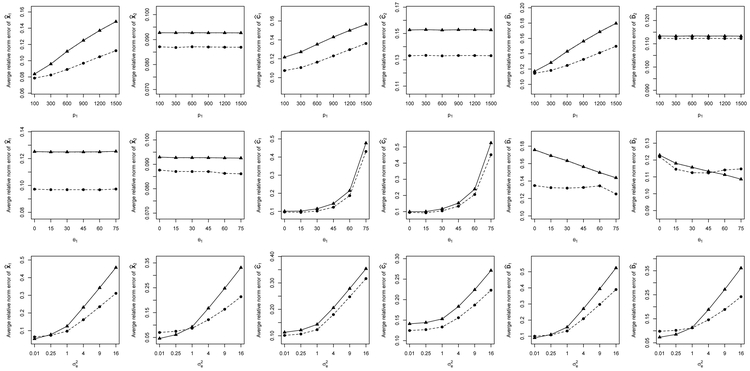
The results obtained by D-CCA for Setups 1 and 2 are summarized in Figures 3 and 4 and Table 1. The first rows of the two figures show the average relative errors (AREs) for θ1 = 45°, and varying p1; the second rows are for p1 = 900, and varying θ1; and the third rows are for p1 = 900, θ1 = 45° and varying . Both figures reveal that the curves based on the estimated ranks almost overlap with those based on the true ranks. The ranks are selected with very high accuracy (>99.7%).
Figure 3:
Average relative errors of D-CCA estimates under Setup 1 in spectral norm (○) and Frobenius norm (△) using true r1, r2 and r12, and those in spectral norm (●) and Frobenius norm (▴) using , and .
Figure 4:
Average relative errors of D-CCA estimates under Setup 2 in spectral norm (○) and Frobenius norm (▵) using true r1, r2 and r12, and those in spectral norm (●) and Frobenius norm (▴) using , and .
Table 1:
Averages (standard errors) of D-CCA estimates for the first canonical angle/correlation.
| (p1, ) | θ1 = 0°/ρ1 = 1 | θ1 = 45°/ρ1 = 0.707 | θ1 = 60°/ρ1 = 0.5 | θ1 = 75°/ρ1 = 0.259 |
|---|---|---|---|---|
| Setup 1 | ||||
| (100, 1) | 3.59°(0.21°)/0.998(0.000) | 44.7°(2.38°)/0.710(0.029) | 59.3°(2.88°)/0.509(0.043) | 73.5°(3.06°)/0.284(0.051) |
| (600, 1) | 3.61°(0.21°)/0.998(0.000) | 44.7°(2.39°)/0.710(0.029) | 59.4°(2.89°)/0.509(0.043) | 73.5°(3.07°)/0.284(0.051) |
| (900, 1) | 3.61°(0.21°)/0.998(0.000) | 44.7°(2.39°)/0.710(0.029) | 59.4°(2.90°)/0.509(0.043) | 73.5°(3.09°)/0.283(0.052) |
| (l500, 1) | 3.61°(0.21°)/0.998(0.000) | 44.7°(2.39°)/0.710(0.029) | 59.3°(2.89°)/0.509(0.043) | 73.5°(3.08°)/0.284(0.051) |
| (900, 0.01) | 0.36°(0.02°)/1.000(0.000) | 44.6°(2.38°)/0.711(0.025) | 59.3°(2.89°)/0.508(0.038) | 73.5°(3.08°)/0.280(0.046) |
| (900, 1) | 3.61°(0.21°)/0.998(0.000) | 44.7°(2.39°)/0.709(0.026) | 59.4°(2.90°)/0.507(0.038) | 73.5°(3.09°)/0.280(0.046) |
| (900, 9) | 11.0°(0.66°)/0.992(0.001) | 45.6°(2.43°)/0.705(0.026) | 59.9°(2.92°)/0.504(0.039) | 73.7°(3.08°)/0.279(0.046) |
| (900, 16) | 14.9°(0.91°)/0.966(0.004) | 46.4°(2.47°)/0.688(0.028) | 60.4°(2.93°)/0.492(0.040) | 73.9°(3.06°)/0.273(0.047) |
| Setup 2 | ||||
| (100, 1) | 3.58°(0.21°)/0.998(0.000) | 44.5°(2.36°)/0.712(0.029) | 59.0°(2.83°)/0.514(0.042) | 72.7°(2.90°)/0.296(0.048) |
| (600, 1) | 3.59°(0.21°)/0.998(0.000) | 44.5°(2.36°)/0.712(0.029) | 59.0°(2.83°)/0.514(0.042) | 72.7°(2.89°)/0.297(0.048) |
| (900, 1) | 3.60°(0.21°)/0.998(0.000) | 44.5°(2.37°)/0.712(0.029) | 59.0°(2.84°)/0.514(0.043) | 72.7°(2.90°)/0.296(0.048) |
| (1500, 1) | 3.60°(0.21°)/0.998(0.000) | 44.5°(2.36°)/0.712(0.029) | 59.0°(2.82°)/0.514(0.042) | 72.7°(2.89°)/0.296(0.048) |
| (900, 0.01) | 0.36°(0.02°)/1.000(0.000) | 44.4°(2.35°)/0.714(0.029) | 59.0°(2.82°)/0.515(0.042) | 72.7°(2.89°)/0.297(0.048) |
| (900, 1) | 3.60°(0.21°)/0.998(0.000) | 44.5°(2.37°)/0.712(0.029) | 59.0°(2.84°)/0.514(0.043) | 72.7°(2.90°)/0.296(0.048) |
| (900, 9) | 10.9°(0.64°)/0.982(0.002) | 45.4°(2.41°)/0.701(0.030) | 59.6°(2.87°)/0.506(0.043) | 73.0°(2.93°)/0.292(0.049) |
| (900,16) | 14.6°(0.87°)/0.967(0.004) | 46.3°(2.45°)/0.691(0.031) | 60.0°(2.89°)/0.499(0.044) | 73.2°(2.93°)/0.289(0.049) |
Consider Figure 3 of Setup 1 as an example. We have nearly identical plots for the two datasets that are generated from the same distribution. From the first row, where all considered cases have almost the same set of average ratios {αCk,(·), αDk,(·)}, all the AREs become bigger as the dimension p1 increases. For the second row, the increasing canonical angle θ1 results in a change in the average ratios αCk,2 from 0.997 down to 0.18 and in αck,F from 0.74 down to 0.14; αDk,2 is stable around 0.78 for the first 5 values of θ1 and then increases to 0.87 at θ1 = 75°; and αDk,F changes from 0.67 to 0.93. Meanwhile, this leads to increasing AREs of and decreasing AREs of , but does not affect the AREs of . The third row shows that all the AREs increase as the noise variance becomes bigger. Note that increasing is equivalent to decreasing the eigenvalues of Σk by scaling to 1. These results agree with the influence of p1, α and λ1(Σk) on the convergence rates given in Theorem 3.
For Setup 2, with similar arguments, we find a similar pattern of estimation performance for D-CCA, as shown in the second and third rows and the plots of the first dataset in the first row of Figure 4. For the first row of Figure 4, the considered cases of the second dataset have a fixed dimension p2 and stable ratios {αC2,(·), αD2,(·)}. The corresponding AREs are still acceptable and interestingly are not much impacted by the change in the dimension p1 of the first dataset. From Table 1, we see that the estimated canonical angles and correlations perform well for Setups 1 and 2 even in the presence of strong noise levels.
The comparison of D-CCA and the seven other methods is shown in Tables 2 and 3, and Figure 5. First consider these methods other than GDFM (Hallin and Liška, 2011). Table 2 reports the results for Setups 1 and 2 when we set p1 = 900, θ1 = 45° (i.e., ρ1 = 0.707), and . All methods except OnPLS have comparably good performance for the estimation of signal matrices. As expected, D-CCA outperforms all the six competing methods in terms of estimating the common and distinctive matrices. In particular, AJIVE and COBE are unable to discover the common matrices. Figure 5 visually shows a similar comparison based on a single replication of Setup 3. The signal, common, and distinctive matrices are recovered well by the D-CCA method. In contrast, the common matrix estimators estimated from the six state-of-the-art methods significantly differ from the ground truth. AJIVE and COBE still yield zero matrices as the estimators of the common matrices, which appears not reasonable when the first canonical correlation ρ1 has a high value of 0.707. Table 3 shows the proportion of significant nonzero correlations among the p1×p2 pairs of variables between d1 and d2 that were detected by the normal approximation test (DiCiccio and Romano, 2017) using each method’s estimates of D1 and D2. The procedure of Benjamini and Hochberg (1995) was applied to the multiple tests to control false discovery rate at 0.05. Results are omitted for AJIVE and COBE with , and also for D-CCA and R.JIVE due to zero correlation estimates by . All the other methods have a large amount of significant nonzero correlations retained between their distinctive structures.
Table 2:
Averages (standard errors) of norm ratios when p1 = 900, θ1 = 45° and .
| Ratio | Method | Spectral norm | Frobenius norm | Spectral norm | Frobenius norm |
|---|---|---|---|---|---|
| Setup 1 |
Setup 2 |
||||
| k = 1 / k = 2 | k = 1 / k = 2 | k = 1 / k = 2 | k = 1 / k = 2 | ||
| D-CCA | 0.088(0.010)/0.088(0.010) | 0.120(0.006)/0.120(0.006) | 0.097(0.012)/0.087(0.017) | 0.125(0.007)/0.093(0.006) | |
| JIVE | 0.108(0.005)/0.109(0.005) | 0.141(0.004)/0.141(0.004) | 0.116(0.005)/0.067(0.004) | 0.145(0.004)/0.090(0.002) | |
| R.JIVE | 0.109(0.018)/0.089(0.015) | 0.139(0.013)/0.140(0.009) | 0.108(0.018)/0.102(0.026) | 0.139(0.012)/0.105(0.011) | |
| AJIVE | 0.080(0.004)/0.081(0.004) | 0.116(0.003)/0.116(0.004) | 0.081(0.004)/0.051(0.002) | 0.116(0.003)/0.082(0.002) | |
| OnPLS | 0.390(0.111)/0.399(0.112) | 0.315(0.076)/0.321(0.077) | 0.397(0.111)/0.550(0.116) | 0.320(0.077)/0.331(0.064) | |
| DISCO-SCA | 0.083(0.003)/0.083(0.004) | 0.154(0.004)/0.154(0.005) | 0.084(0.004)/0.053(0.002) | 0.174(0.005)/0.093(0.002) | |
| COBE | 0.080(0.004)/0.081(0.004) | 0.116(0.003)/0.116(0.004) | 0.081(0.004)/0.051(0.002) | 0.116(0.003)/0.082(0.002) | |
| D-CCA | 0.117(0.028)/0.120(0.027) | 0.134(0.028)/0.136(0.027) | 0.123(0.028)/0.133(0.036) | 0.143(0.029)/0.153(0.038) | |
| JIVE | 0.996(0.009)/0.996(0.008) | 1.024(0.014)/1.024(0.013) | 0.998(0.009)/0.990(0.015) | 1.037(0.025)/1.013(0.019) | |
| R.JIVE | 1.000(0.043)/0.576(0.032) | 1.003(0.043)/0.588(0.031) | 1.003(0.049)/0.576(0.041) | 1.006(0.052)/0.589(0.042) | |
| AJIVE | 1(0)/1(0) | 1(0)/1(0) | 1(0)/1(0) | 1(0)/1(0) | |
| OnPLS | 0.787(0.112)/0.777(0.113) | 0.817(0.143)/0.805(0.142) | 0.779(0.105)/0.796(0.071) | 0.804(0.117)/0.815(0.098) | |
| DISCO-SCA | 1.023(0.065)/1.023(0.066) | 1.057(0.087)/1.058(0.089) | 0.772(0.183)/1.052(0.112) | 0.826(0.227)/1.190(0.237) | |
| COBE | 1(0)/1(0) | 1(0)/1(0) | 1(0)/1(0) | 1(0)/1(0) | |
| D-CCA | 0.121(0.016)/0.122(0.016) | 0.148(0.010)/0.149(0.009) | 0.133(0.018)/0.112(0.019) | 0.156(0.011)/0.113(0.009) | |
| JIVE | 0.703(0.040)/0.703(0.040) | 0.541(0.023)/0.541(0.023) | 0.704(0.040)/0.599(0.036) | 0.546(0.024)/0.371(0.016) | |
| R.JIVE | 0.689(0.040)/0.405(0.032) | 0.535(0.019)/0.337(0.020) | 0.690(0.041)/0.350(0.031) | 0.536(0.022)/0.238(0.017) | |
| AJIVE | 0.706(0.040)/0.706(0.040) | 0.538(0.022)/0.539(0.022) | 0.705(0.040)/0.605(0.035) | 0.538(0.022)/0.369(0.015) | |
| OnPLS | 0.655(0.093)/0.654(0.095) | 0.574(0.064)/0.576(0.066) | 0.656(0.094)/0.658(0.113) | 0.574(0.063)/0.476(0.057) | |
| DISCO-SCA | 0.704(0.049)/0.704(0.049) | 0.558(0.041)/0.559(0.041) | 0.532(0.114)/0.628(0.067) | 0.462(0.092)/0.432(0.078) | |
| COBE | 0.706(0.040)/0.706(0.040) | 0.538(0.022)/0.539(0.022) | 0.705(0.040)/0.605(0.035) | 0.538(0.022)/0.369(0.015) | |
| GDFM | 0.080(0.004)/0.081(0.004) | 0.116(0.003)/0.116(0.004) | 0.081(0.004)/0.052(0.002) | 0.116(0.003)/0.082(0.002) | |
| GDFM | 0.083(0.003)/0.083(0.004) | 0.154(0.004)/0.154(0.005) | 0.084(0.004)/0.053(0.002) | 0.174(0.005)/0.093(0.002) | |
| GDFM | 0.080(0.003)/0.080(0.004) | 0.099(0.003)/0.099(0.003) | 0.082(0.003)/0.047(0.002) | 0.128(0.004)/0.044(0.001) | |
Table 3:
The proportions of significant nonzero correlations between d1 and d2 for simulation setups (with p1 = 900, θ1=45° and ) and TCGA datasets. Averages (standard errors) are shown for Setups 1 and 2. Significant correlations are detected by the normal approximation test (DiCiccio and Romano, 2017) using and , with false discovery rate controlled at 0.05.
| Method | Setup 1 | Setup 2 | Setup 3 | EXP90/METH90b | EXP90/METH90a |
|---|---|---|---|---|---|
| D-CCA | |||||
| JIVE | 69.9%(2.5%) | 60.8%(3.0%) | 98.7% | 85.0% | 58.2% |
| R.JIVE | |||||
| AJIVE | |||||
| OnPLS | 56.6%(11.1%) | 32.7%(8.2%) | 52.5% | 72.9% | 68.6% |
| DISCO-SCA | 50.3%(4.6%) | 25.2%(6.7%) | 25.1% | 67.8% | 64.2% |
| COBE | |||||
| GDFM | 70.3%(2.4%) | 61.5%(2.7%) | 98.6% | 100% | 100% |
| GDFM | 73.8%(1.8%) | 64.8%(2.3%) | 97.0% | 85.8% | 87.0% |
Now consider the GDFM method (Hallin and Liška, 2011). We set the sample temporal cross-covariances to be zero in GDFM estimation for our simulated data and TCGA datasets that have no temporal dependence. GDFM decomposes each data matrix by with each component’s name shown in Table 6. By Remark S.1 (in the supplementary material), theoretically for our simulated i.i.d. data with no correlations between signals and noises, the weakly idiosyncratic matrix νk is zero, and the joint common matrix and the marginal common matrix χk are both equal to the signal matrix Xk. Moreover, the strongly common matrix ϕk is zero, when , i.e., the first canonical correlation ρ1 between x1 and x2 is smaller than 1. The above theoretical results are evidenced by our simulations. In Table 2, the relative errors of estimators and to signal Xk are as comparably small as those of by our D-CCA and the other five well performed methods. The similarly small norm ratios of to numerically support νk = 0. The squares of these quantities are much smaller, and especially in the Frobenius norm are equivalent to matrix-variation ratios. The strongly common matrix estimate is zero for the setups, with ρ1 = 0.707 < 1, considered in the table. These numerical evidences are more clearly seen in Figure 5(b) under a similar setup.
Table 6:
Ranks, variation ratios (), and SWISS scores of GFDM matrix estimates for TCGA datasets.
| Matrix Estimate | EXP90 / METH90b | EXP90 / METH90a | ||
|---|---|---|---|---|
| Rank (VR) | SWISS | Rank (VR) | SWISS | |
| (joint common) | 4 / 4 | 0.373 / 0.569 | 4 / 4 | 0.378 / 0.850 |
| (marginal common) | 3 (0.986) / 3 (0.986) | 0.364 / 0.566 | 3 (0.990) / 3 (0.974) | 0.372 / 0.851 |
| (strongly common) | 2 (0.755) / 2 (0.626) | 0.288 / 0.348 | 2 (0.770) / 2 (0.372) | 0.302 / 0.613 |
| (weakly common) | 1 (0.231) / 1 (0.360) | 0.764 / 0.991 | 1 (0.220) / 1 (0.602) | 0.811 / 0.996 |
| (weakly idiosyncratic) | 1 (0.014)/ 1 (0.014) | 0.987 / 0.760 | 1 (0.010) / 1 (0.026) | 0.997 / 0.812 |
| 2 (0.245) / 2 (0.374) | 0.777 / 0.982 | 2 (0.230) / 2 (0.628) | 0.819 / 0.988 | |
| (strongly idiosyncratic) | 656 (2.070) / 656 (1.196) | 0.985 / 0.987 | 656 (2.151) / 656 (1.764) | 0.977 / 0.989 |
| (marginal idiosyncratic) | 657 (2.084) / 657 (1.211) | 0.985 / 0.983 | 657 (2.160) / 657 (1.790) | 0.977 / 0.987 |
5. Analysis of TCGA Breast Cancer Data
In this section, we apply the proposed D-CCA method to analyze genomic datasets produced from TCGA breast cancer tumor samples. We investigate the ability to separate tumor subtypes for matrices obtained from D-CCA in comparison to those obtained from the six competing methods as well as GDFM that are mentioned in Section 1. We consider the mRNA expression data and DNA methylation data for a common set of 660 samples. The two datasets are publicly available at https://tcga-data.nci.nih.gov/docs/publications and have been respectively preprocessed by Ciriello et al. (2015) and Koboldt et al. (2012). The 660 samples were classified by Ciriello et al. (2015) into 4 subtypes using the PAM50 model (Parker et al., 2009) based on mRNA expression data. Specifically, the samples consist of 112 basal-like, 55 HER2-enriched, 331 luminal A, and 162 luminal B tumors.
To quantify the extent of subtype separation, we adopt the standardized within-class sum of squares (SWISS; Cabanski et al., 2010)
for matrix A = (Aij)p×n, where is the average of the j-th sample’s subtype on the i-th row and . is the average of the i-th row’s elements. The SWISS score represents the variation within the subtypes as a proportion of the total variation. A lower score indicates better subtype separation. For the mRNA expression data, we filtered out the subset consisting of the 1,195 variably expressed genes with marginal SWISS≤0.9 from the original 20,533 genes, and denote this subset as EXP90. The 2,083 variably methylated probes of the DNA methylation data, originally with 21,986 probes, are included in the analysis. We denote the 881 probes with marginal SWISS≤0.9 as METH90b and the remaining 1,202 probes as METH90a. We conducted the analysis for the pair of EXP90 and METH90b as well as the pair of EXP90 and METH90a.
The ranks and proportions of explained signal variation for the matrix estimators obtained by D-CCA and the six competing methods (except GDFM) are given in Table 4, and their SWISS scores are shown in Table 5. We see in Table 4 that D-CCA, AJIVE and COBE give much lower ranks for the estimated signal matrices than the other methods. Particularly for the EXP90 dataset, the rank of obtained by the remaining four methods is inconsistent for the two pairs. As shown in the scree plots of Figure 6, the ranks of signal matrices selected by D-CCA and AJIVE look reasonable because the few most leading principal components of the observed data are captured for denoising, while the signal matrix ranks for the METH90b and METH90a datasets seem to be underestimated by COBE. Using D-CCA, the estimated canonical correlations and angles of signal vectors are (0.934, 0.431) and (20.9°, 64.4°) between the EXP90 and METH90b datasets, and are (0.610, 0.275) and (52.4°, 74.0°) between the EXP90 and METH90a datasets.
Table 4:
Ranks (and proportions of explained signal variation, i.e., ) of matrix estimates for TCGA datasets.
| Matrix | Method | EXP90 / METH90b | EXP90 / METH90a |
|---|---|---|---|
| D-CCA | 2 / 3 | 2 / 3 | |
| JIVE | 35 / 18 | 41 / 29 | |
| R.JIVE | 40 / 27 | 44 / 49 | |
| AJIVE | 2 / 3 | 2 / 3 | |
| OnPLS | 13 / 10 | 12 / 10 | |
| DISCO-SCA | 13 / 13 | 17 / 17 | |
| COBE | 2 / 1 | 2 / 2 | |
| D-CCA | 2 (0.472) / 2 (0.301) | 2 (0.120) / 2 (0.062) | |
| JIVE | 1 (0.068) / 1 (0.086) | 3 (0.236) / 3 (0.167) | |
| R.JIVE | 1 (0.212) / 1 (0.505) | 3 (0.274) / 3 (0.602) | |
| AJIVE | 0 / 0 | 0 / 0 | |
| OnPLS | 3 (0.516) / 3 (0.510) | 2 (0.455) / 2 (0.166) | |
| DISCO-SCA | 6 (0.732) / 6 (0.571) | 8 (0.745) / 8 (0.363) | |
| COBE | 0 / 0 | 0 / 0 | |
| D-CCA | 2 (0.223) / 3 (0.506) | 2 (0.564) / 3 (0.797) | |
| JIVE | 34(0.932) / 17 (0.914) | 38 (0.764) / 26 (0.833) | |
| R.JIVE | 39 (0.788) / 26 (0.495) | 41 (0.726) / 46 (0.398) | |
| AJIVE | 2 (1) / 3 (1) | 2 (1) / 3 (1) | |
| OnPLS | 10 (0.484) / 7 (0.490) | 10 (0.545) / 8 (0.834) | |
| DISCO-SCA | 7 (0.268) / 7 (0.429) | 9 (0.255) / 9 (0.637) | |
| COBE | 2 (1) / 1 (1) | 2 (1) / 2 (1) |
Table 5:
SWISS scores for TCGA breast cancer subtypes. Lower scores indicate better subtype separation.
| Matrix | Method | EXP90 / METH90b | EXP90 / METH90a |
|---|---|---|---|
| Yk | For all | 0.773 / 0.814 | 0.773 / 0.952 |
| D-CCA | 0.313 / 0.623 | 0.313 / 0.925 | |
| JIVE | 0.632 / 0.698 | 0.643 / 0.920 | |
| R.JIVE | 0.642 / 0.689 | 0.647 / 0.931 | |
| AJIVE | 0.314 / 0.623 | 0.314 / 0.925 | |
| OnPLS | 0.523 / 0.669 | 0.515 / 0.905 | |
| DISCO-SCA | 0.526 / 0.663 | 0.553 / 0.904 | |
| COBE | 0.314 / 0.545 | 0.314 / 0.926 | |
| D-CCA | 0.240 / 0.269 | 0.528 / 0.606 | |
| JIVE | 0.831 / 0.831 | 0.639 / 0.736 | |
| R.JIVE | 0.373 / 0.373 | 0.564 / 0.885 | |
| AJIVE | NA / NA | NA / NA | |
| OnPLS | 0.398 / 0.312 | 0.419 / 0.494 | |
| DISCO-SCA | 0.447 / 0.400 | 0.470 / 0.717 | |
| COBE | NA / NA | NA / NA | |
| D-CCA | 0.623 / 0.940 | 0.320 / 0.979 | |
| JIVE | 0.691 /0.741 | 0.830 / 0.963 | |
| R.JIVE | 0.833 / 0.997 | 0.874 / 0.998 | |
| AJIVE | 0.314 / 0.623 | 0.314 / 0.925 | |
| OnPLS | 0.878 / 0.978 | 0.871 / 0.989 | |
| DISCO-SCA | 0.935 / 0.992 | 0.944 / 0.995 | |
| COBE | 0.314 / 0.545 | 0.314 / 0.926 |
From Table 5, for the pair of EXP90 and METH90b datasets, the matrix obtained by all the seven methods gains an improved SWISS score compared to the noisy data matrix Yk. Other than AJIVE and COBE with , a clear pattern of increasing SWISS scores, from to and then to , can be seen for the remaining methods except JIVE. This indicates that an enhanced ability to separate the tumor samples by subtype can be expected when integrating two datasets that can exhibit such a distinction to a moderate extent. Also note that the estimated common matrices of our D-CCA have the lowest SWISS scores. While considering the pair of EXP90 and METH90a datasets, for all the seven methods we find a big gap between the SWISS scores of the two estimated signal matrices, and that the denoised matrix of the METH90a dataset still has nearly no discriminative power with SWISS close to 1. The ability on subtype separation seems more likely to be a distinctive feature of EXP90 dataset comparing to METH90a dataset. The estimated distinctive matrix of EXP90 is thus expected to have a lower SWISS score than its estimated common matrix. However, only D-CCA meets this point, except that AJIVE and COBE yield zero common matrices. The failure of the six competing methods may be caused by their inappropriate decomposition constructions, which are mentioned in Section 1. In particular, from Table 3, we see that a lot of significant nonzero correlations exist among all gene-probe pairs based on the estimated distinctive matrices, respectively, obtained by JIVE, OnPLS and DISCO-SCA.
The GDFM method (Hallin and Liška, 2011) was also applied to the TCGA datasets. Table 6 summarizes the results of GDFM matrix estimates. As estimators of signal matrix Xk, matrix has comparable rank and SWISS score as those of our D-CCA estimator given in Tables 4 and 5. Besides, , i.e., νk = 0, is numerically suggested by the remarkably small variation ratios of to that are likely just induced by estimation errors. With very large ranks and uninformative SWISS scores, both and appear to be noises. One may let , (or ), and (or ) for GDFM. Inspecting Table 6 reveals that the discussion given in the preceding paragraph also holds even when we include GDFM.
6. Discussion
In this paper, we study a typical model for the joint analysis of two high-dimensional datasets. We develop a novel and promising decomposition-based CCA method, D-CCA, to appropriately define the common and distinctive matrices. In particular, the conventionally underemphasized orthogonal relationship between the distinctive matrices is now well designed on the space of random variables. A soft-thresholding-based approach is then proposed for estimating these D-CCA-defined matrices with a theoretical guarantee and satisfactory numerical performance. The proposed D-CCA outperforms some state-of-the-art methods in both simulated and real data analyses.
There are many possible further studies beyond the current proposed D-CCA. The first is to generalize the D-CCA for three or more datasets. We may assume that at least two datasets have mutually orthogonal distinctive structures. An immediate idea starts from substituting the multiset CCA (Kettenring, 1971) for the two-set CCA in D-CCA. However, the challenge is that the iteratively obtained sets of canonical variables are not guaranteed to have the bi-orthogonality given in Theorem 1. Hence, we cannot follow the proposed D-CCA to simply break down the decomposition problem to each set of canonical variables, and need a more sophisticated design to meet the desirable constraint. Another direction is to incorporate the nonlinear relationship between the two datasets. The D-CCA only considers the linear relationship by using the traditional CCA based on Pearson’s correlation. It is worth trying the kernel CCA (Fukumizu et al., 2007) or the distance correlation (Székely et al., 2007) to capture the nonlinear dependence. Inspired by the time series analysis of Hallin and Lišska (2011) and Barigozzi et al. (2018), we also expect to generalize D-CCA to general dynamic factor models with comparisons to their methods. These interesting and challenging studies are under investigation and will be reported in future work.
Supplementary Material
Acknowledgments
Dr. Zhu’s work was partially supported by NIH grants MH086633 and MH116527, NSF grants SES-1357666 and DMS-1407655, a grant from the Cancer Prevention Research Institute of Texas, and the endowed Bao-Shan Jing Professorship in Diagnostic Imaging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or any other funding agency.
References
- Ahn SC and Horenstein AR (2013), “Eigenvalue ratio test for the number of factors,” Econometrica, 81, 1203–1227. [Google Scholar]
- Bai J (2003), “Inferential theory for factor models of large dimensions,” Econometrica, 71, 135–171. [Google Scholar]
- Bai J and Ng S (2002), “Determining the number of factors in approximate factor models,” Econometrica, 70, 191–221. [Google Scholar]
- Bai J and Ng S (2008), “Large dimensional factor analysis,” Foundations and Trends in Econometrics, 3, 89–163. [Google Scholar]
- Barigozzi M, Hallin M, and Soccorsi S (2018), “Identification of global and local shocks in international financial markets via general dynamic factor models,” Journal of Financial Econometrics, DOI: 10.1093/jjfinec/nby006. [DOI] [Google Scholar]
- Benjamini Y and Hochberg Y (1995), “Controlling the false discovery rate: a practical and powerful approach to multiple testing,” Journal of the Royal Statistical Society, Series B, 57, 289–300. [Google Scholar]
- Björck A and Golub GH (1973), “Numerical methods for computing angles between linear subspaces,” Mathematics of Computation, 27, 579–594. [Google Scholar]
- Cabanski CR, Qi Y, Yin X, Bair E, Hayward MC, Fan C, Li J, Wilkerson MD, Marron JS, Perou CM, and Hayes DN (2010), “SWISS MADE: Standardized within class sum of squares to evaluate methodologies and dataset elements,” PLoS ONE, 5, e9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain G and Rothschild M (1983), “Arbitrage, factor structure, and mean-variance analysis on large asset markets,” Econometrica, 51, 1281–1304. [Google Scholar]
- Chen M, Gao C, Ren Z, and Zhou HH (2013), “Sparse CCA via precision adjusted iterative thresholding,” arXiv preprint arXiv:1311.6186. [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. (2015), “Comprehensive molecular portraits of invasive lobular breast cancer,” Cell, 163, 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comon P and Jutten C (2010), Handbook of Blind Source Separation: Independent Component Analysis and Applications, Academic Press. [Google Scholar]
- DiCiccio CJ and Romano JP (2017), “Robust permutation tests for correlation and regression coefficients,” Journal of the American Statistical Association, 112, 1211–1220. [Google Scholar]
- Fan J, Liao Y, and Mincheva M (2013), “Large covariance estimation by thresholding principal orthogonal complements,” Journal of the Royal Statistical Society: Series B, 75, 603–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Jiang M, Hannig J, and Marron J (2018), “Angle-based joint and individual variation explained,” Journal of Multivariate Analysis, 166, 241–265. [Google Scholar]
- Forni M, Hallin M, Lippi M, and Reichlin L (2000), “The generalized dynamic-factor model: Identification and estimation,” Review of Economics and statistics, 82, 540–554. [Google Scholar]
- Forni M, Hallin M, Lippi M, and Zaffaroni P (2017), “Dynamic factor models with infinite-dimensional factor space: Asymptotic analysis,” Journal of Econometrics, 199, 74–92. [Google Scholar]
- Fukumizu K, Bach FR, and Gretton A (2007), “Statistical consistency of kernel canonical correlation analysis,” Journal of Machine Learning Research, 8, 361–383. [Google Scholar]
- Gao C, Ma Z, Ren Z, and Zhou HH (2015), “Minimax estimation in sparse canonical correlation analysis,” The Annals of Statistics, 43, 2168–2197. [Google Scholar]
- Gao C, Ma Z, and Zhou HH (2017), “Sparse CCA: Adaptive estimation and computational barriers,” The Annals of Statistics, 45, 2074–2101. [Google Scholar]
- Hallin M and Liška R (2011), “Dynamic factors in the presence of blocks,” Journal of Econometrics, 163, 29–41. [Google Scholar]
- Hotelling H (1936), “Relations between two sets of variates,” Biometrika, 28, 321–377. [Google Scholar]
- Huang H (2017), “Asymptotic behavior of support vector machine for spiked population model,” Journal of Machine Learning Research, 18, 1–21. [Google Scholar]
- Kettenring JR (1971), “Canonical analysis of several sets of variables,” Biometrika, 58, 433–451. [Google Scholar]
- Koboldt D, Fulton R, McLellan M, Schmidt H, Kalicki-Veizer J, McMichael J, Fulton L, Dooling D, Ding L, et al. (2012), “Comprehensive molecular portraits of human breast tumours,” Nature, 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuligowski J, Pérez-Guaita D, Sànchez-Illana À, León-Gonzàlez Z, de la Guardia M, Vento M, Lock EF, and Quintàs G (2015), “Analysis of multi-source metabolomic data using joint and individual variation explained (JIVE),” Analyst, 140, 4521–4529. [DOI] [PubMed] [Google Scholar]
- Lock EF, Hoadley KA, Marron JS, and Nobel AB (2013), “Joint and individual variation explained (JIVE) for integrated analysis of multiple data types,” Annals of Applied Statistics, 7, 523–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfstedt T and Trygg J (2011), “OnPLS-a novel multiblock method for the modelling of predictive and orthogonal variation,” Journal of Chemometrics, 25, 441–455. [Google Scholar]
- Nadakuditi RR and Silverstein JW (2010), “Fundamental limit of sample generalized eigenvalue based detection of signals in noise using relatively few signal-bearing and noise-only samples,” IEEE Journal of Selected Topics in Signal Processing, 4, 468–480. [Google Scholar]
- O’Connell MJ and Lock EF (2016), “R.JIVE for exploration of multi-source molecular data,” Bioinformatics, 32, 2877–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onatski A (2010), “Determining the number of factors from empirical distribution of eigenvalues,” The Review of Economics and Statistics, 92, 1004–1016. [Google Scholar]
- Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, and Bernard PS (2009), “Supervised risk predictor of breast cancer based on intrinsic subtypes,” Journal of Clinical Oncology, 27, 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SA (1976), “The arbitrage theory of capital asset pricing,” Journal of Economic Theory, 13, 341–360. [Google Scholar]
- Schouteden M, Van Deun K, Wilderjans TF, and Van Mechelen I (2014), “Performing DISCO-SCA to search for distinctive and common information in linked data,” Behavior Research Methods, 46, 576–587. [DOI] [PubMed] [Google Scholar]
- Smilde AK, Mage I, Naes T, Hankemeier T, Lips MA, Kiers HAL, Acar E, and Bro R (2017), “Common and distinct components in data fusion,” Journal of Chemometrics, 31, e2900. [Google Scholar]
- Song Y, Schreier PJ, Ramirez D, and Hasija T (2016), “Canonical correlation analysis of high-dimensional data with very small sample support,” Signal Processing, 128, 449–458. [Google Scholar]
- Stock JH and Watson MW (2002), “Forecasting using principal components from a large number of predictors,” Journal of the American statistical association, 97, 1167–1179. [Google Scholar]
- Székely GJ, Rizzo ML, and Bakirov NK (2007), “Measuring and testing dependence by correlation of distances,” The Annals of Statistics, 35, 2769–2794. [Google Scholar]
- Trygg J (2002), “O2-PLS for qualitative and quantitative analysis in multivariate calibration,” Journal of Chemometrics, 16, 283–293. [Google Scholar]
- van der Kloet F, Sebastian-Leon P, Conesa A, Smilde A, and Westerhuis J (2016), “Separating common from distinctive variation,” BMC Bioinformatics, 17, S195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, and Ugurbil K (2013), “The WU-Minn human connectome project: an overview,” NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershynin R (2012), “Introduction to the non-asymptotic analysis of random matrices,” in Compressed Sensing, Cambridge University Press, Cambridge, pp. 210–268. [Google Scholar]
- Wang W and Fan J (2017), “Asymptotics of empirical eigenstructure for high dimensional spiked covariance,” The Annals of Statistics, 45, 1342–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Risk BB, Zhang K, and Marron J (2017), “Jive integration of imaging and behavioral data,” NeuroImage, 152, 38–49. [DOI] [PubMed] [Google Scholar]
- Zhou G, Cichocki A, Zhang Y, and Mandic DP (2016), “Group component analysis for multiblock data: common and individual feature extraction,” IEEE Transactions on Neural Networks and Learning Systems, 27, 2426–2439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.