Abstract
In light of recent findings on the small proportion of variance in body mass index (BMI) explained by shared environment, and growing interests in the role of genetic susceptibility, we assessed the relative contribution of socioeconomic status (SES) and genome-wide polygenic score for BMI to explaining variation in BMI. Our final analytic sample included 4,918 white and 1,546 black individuals from the US National Longitudinal Study of Adolescent to Adult Health Wave IV (2007–2008) who had complete measures on BMI, demographics, SES, genetic data, and health behaviors. We used ordinary least-squares regression to assess variation in log(BMI) as a function of the aforementioned predictors, independently and mutually adjusted. All analyses were stratified by race/ethnicity in the main analysis, and further by sex. The age-adjusted variation in log(BMI) was 0.055 among whites and 0.066 among blacks. The contribution of SES and polygenic score ranged from less than1% to 6% and from 2% to 8%, respectively, and majority of the variation (87%–96%) in log(BMI) remained unexplained. Differential distribution of socioeconomic resources, stressors, and buffers may interact to produce systematically larger variation in vulnerable populations. More understanding of the contribution of biological, genetic, and environmental factors, as well as stochastic elements, in diverse phenotypic variance is needed in population health sciences.
Keywords: body mass index, genetic, obesity, polygenic score, socioeconomic status, variation
Abbreviations
- Add Health
National Longitudinal Study of Adolescent to Adult Health
- BMI
body mass index
- SE
standard error
- SES
socioeconomic status
- SNP
single nucleotide polymorphism
Globally, approximately 2 billion people are estimated to be overweight, with one-third of them being obese (1). Between 1980 and 2008, the prevalence of obesity increased from 28.8% to 36.9%, and the mean body mass index (BMI) increased by 0.4 per decade for men (2). During the same period, the prevalence of obesity increased from 29.8% to 38.0%, and the mean BMI increased by 0.5 per decade for women (2). Evidence also suggests increasing variation in BMI distribution within and across countries (3). Given the size of the population affected by overweight and obesity, as well as the well-known health impairments associated with overweight and obesity (4, 5), researchers have focused on identifying various modifiable risk factors ranging from global food supply system (6), societal norms (7), socioeconomic and physical environments (8, 9), and individual health behaviors (10).
In these studies that focus on population-level differences in BMI and obesity, the importance of risk factors is determined on the basis of the statistical significance and the magnitude of average summary measures. However, factors that explain a large proportion of population-level differences may not necessarily provide a basis for an effective prediction rule for individuals if they account for only a small proportion of individual-level differences (11, 12). For instance, despite prior findings establishing a strong association between socioeconomic status (SES; i.e., income and educational attainment) and mean BMI (13, 14), in a recent study in which variation in women’s BMI across 58 low- and middle-income countries was assessed, only 2% of the interindividual variance was explained by basic demographic and socioeconomic factors, including place of residence, education, household wealth, and marital status (15).
At the same time, recent genetic studies have identified many common genetic variants associated with BMI (16–18). In a large genome-wide association study for BMI, researchers identified 97 BMI-associated loci accounting for approximately 2.7% of BMI variation (18). Similar results have been found in the Japanese population (19). Although individual variants do not account for a large proportion of the phenotype, a polygenic score summarizing genome-wide genotype data allows quantification of the cumulative effect of all relevant single nucleotide polymorphisms (SNPs) in explaining disease or trait liability (20, 21). In a recent study deriving a new polygenic predictor comprising 2.1 million BMI-associated common variants, 23.4% of the heritability in BMI was estimated to be explained among UK Biobank adult participants (20). In other studies, 40% to 70% of interindividual variance in BMI has been attributed to genetic factors (22, 23).
It is also suggested that the observed effect of the polygenic score reflects not only the direct genetic effect but also indirect effects mediated via household environment (24). Study findings on gene-environment interactions suggest the role of genetic susceptibility may be accentuated in “obesogenic environments”—defined broadly as social deprivation (25) and education (26) or more specific behaviors like physical inactivity (27) and energy-dense food consumption (28, 29)—although it is unlikely that any one particular aspect of the environment would have a predominant effect over other environmental factors (30). Despite continued interest in the interactive role of socioeconomic factors and genetic variants in relation to BMI, no study, to our knowledge, has systematically examined the joint contribution of SES and genetic factors in explaining variation in BMI.
In this study, data from the National Longitudinal Study of Adolescent to Adult Health (Add Health) were used to assess the relative contribution of SES, lifestyle, and genetic factors to explaining variations in BMI. The consistency in variance explained by these factors was assessed across different subgroups defined by race/ethnicity and sex.
METHODS
Data and Sample
Data were taken from the latest wave of Add Health, an ongoing, nationally representative survey of adolescents in the United States (31). In 1994, Add Health administered in-school questionnaires to students selected through a stratified random sample of all US high schools (n = 90,118), and a subsample (the core sample) participated in home-based interviews between 1994 and 1995 (n = 20,745). During the 2007–2008 Wave IV data collection, saliva and capillary whole blood samples were collected from respondents. Of 15,701 individuals interviewed, 15,159 consented to genotyping, and 12,254 agreed to genetic data archiving. DNA extraction and genotyping were conducted on this sample using 2 genotyping platforms, Illumina Omni1 and Omni2.5 (Illumina, Inc., San Diego, California). Of the 9,129 cases in the genetic file, 8,592 individuals had valid information pertaining to the Add Health study design. We restricted our analysis to participants with relatively homogenous European American and African American genetic ancestry (n = 7,097). Imputation was conducted on SNPs “called” in greater than 98% of the samples with minor allele frequency greater than 1% using the Michigan Imputation Server (32). For the European-descent subsample, imputation analysis used the Haplotype Reference Consortium reference panel (33). For the African American subsample, imputation analysis used the 1,000 Genomes, version 3, reference panel (34). To eliminate confounding with pregnancy, we omitted 248 women respondents who were pregnant at the time of their Wave IV interviews. After a listwise deletion of individuals with missing data on covariates (n = 385 cases), we achieved an analytic sample of 6,464 cases with complete data on polygenic score for BMI and all covariates (n = 4,918 white and n = 1,546 black individuals). Data on all variables came from Wave IV, except for race/ethnicity and sex identifications, which were drawn from Wave I.
Outcome
BMI was based on measured height and weight and was calculated by dividing weight in kilograms by height in meters squared. Given that BMI was not normally distributed, we used log-transformed BMI for all analyses.
Socioeconomic Status
Individuals’ SES was measured via 3 proxy indicators. Personal income was self-reported and categorized into quintiles. Completed education was measured via a binary variable equal to 1 for respondents who reported completing a 4-year college degree (0 otherwise). Homeownership status was also a binary variable equal to 1 for those who own their home and 0 otherwise.
Polygenic Score for BMI
The genome-wide polygenic score for BMI was created from a large-scale genome-wide association study discovery sample that included Europeans only (18). Population stratification, or the nonrandom patterning of alleles across a population of different ancestry, is a potential confounder in genome-wide association studies (35). Therefore, to address this problem, apart from conducting our analyses separately within white individuals and black individuals, we also included the top 10 principle components of the variance-covariance matrix of the genetic data (36), because the first principal components estimated from genome-wide SNP data are thought to reflect genome-wide patterning of allele frequencies by shared ancestry (37). Importantly, within each of our 2 ancestral groups, we used principal components that were calculated within both groups as ancestry specific. The polygenic scores were created on all unrelated individuals (using only 1 member of a related group) and then projected onto the small number of remaining related individuals to preserve power and avoid inflated statistics (38, 39).
Other Covariates
We considered several other demographic and health behavior covariates. Sex was assessed via respondent self-reports, as was age, which we categorized into the following: 24–26 years (referent), 27–29 years, and 30–33 years. Respondents’ partnership status was defined using household roster data, and we used categories for “married/cohabiting” and “single.” Last, our analysis included binary measures of past-week physical activity (equal to 1 for those engaging in any bouts of physical activity), current daily smoking (equal to 1 for current smokers), and 2 measures of past-week fast-food and sugar-sweetened beverage consumption with categories for 0 times, 1 or 2 times, 3 or 4 times, and 5 or more times.
Statistical Analysis
We used ordinary least-squares regression to assess variation in log(BMI) as a function of the aforementioned predictors. All analyses were stratified by race/ethnicity (white vs black individuals). Although the use of European-derived polygenic score prohibits direct comparison of genetic distributions between different ancestry groups, within-ancestral groups analyses are feasible (40). The measurement error resulting from our use of a European-derived polygenic score to predict BMI in black individuals will produce attenuated estimates (41), but we can still test if this attenuation is modest enough in our black sample to demonstrate the same trends we observed in the white sample. The age-adjusted model (model 1) was used as a baseline for comparing the changes in log(BMI) variations in subsequent models, where demographic variables (i.e., sex, relationship status), SES (i.e., education, income, home ownership), polygenic score, and other health behaviors (i.e., smoking, physical activity, fast-food consumption, and sugar-sweetened beverage consumption) were included 1 at a time (models 2–5, respectively). We also assessed the joint contribution of SES and polygenic score in model 6, and all factors together in model 7. In these models, the resulting β coefficients can be interpreted as the change in BMI by 100 × (coefficient) for an increase in 1 unit in the independent variable. The proportion of variance in log(BMI) explained by covariate adjustment was computed by taking the difference in residual variance between the adjusted model and the base model, and converting to percentage: 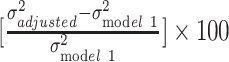 . We performed additional analysis further stratifying the study population by sex. All analyses were carried out in Stata 14 (StataCorp LLC, College Station, Texas).
. We performed additional analysis further stratifying the study population by sex. All analyses were carried out in Stata 14 (StataCorp LLC, College Station, Texas).
Ethics statement
The study was approved by the Colorado Multiple Institutional Review Board (COMIRB-16-0361).
RESULTS
Our final analytic sample (n = 6,464) was well balanced in terms of sex (52% women) and reflective of the US population of young adults in terms of educational attainment (31% with a 4-year college degree) (Table 1). A majority of the sample (>80%) was 27–33 years old. In terms of SES, the average personal annual income in the top quintile group was approximately $67,000 and in the bottom quintile group was just under $4,000. Approximately one-fourth of the sample were self-identified daily smokers, and only 14% were classified as being physically active. The mean BMI was 29.11 (standard error (SE), 0.09) for the overall sample and it varied by race/ethnicity (blacks: 30.68, SE, 0.22; whites: 28.62, SE, 0.11) and by sex (men: 28.9, SE, 0.12; women: 29.32, SE, 0.14) (Figure 1). Log(BMI) was used for the remaining analyses and presentations.
Table 1.
Descriptive Summary of Sample Characteristics by Selected Covariates (n = 6,464), US National Longitudinal Study of Adolescent to Adult Health Wave IV, 2007–2008
| Characteristic | No. | Mean (SE) | Proportion (SE) |
|---|---|---|---|
| Body mass indexa | 6,464 | 29.11 (0.09) | |
| Black race/ethnicity | 1,546 | 0.24 (0.01) | |
| Women | 3,384 | 0.52 (0.01) | |
| Age category, years | |||
| 24–26 | 1,060 | 0.16 (0.01) | |
| 27–29 | 3,335 | 0.52 (0.01) | |
| 30–33 | 2,069 | 0.32 (0.01) | |
| Partnered | 3,842 | 0.59 (0.01) | |
| Personal annual income quintileb, USD | |||
| 0–10,000 | 1,320 | 3,838.07 (100.29) | |
| 10,001–24,000 | 1,328 | 17,758.85 (107.52) | |
| 24,001–34,000 | 1,233 | 28,947.22 (80.11) | |
| 34,001–48,500 | 1,291 | 40,253.85 (110.85) | |
| 48,501–100,000 | 1,292 | 67,166.29 (450.88) | |
| Four-year college degree | 1,989 | 0.31 (0.01) | |
| Homeowner | 2,741 | 0.42 (0.01) | |
| Daily smoker | 1,702 | 0.26 (0.01) | |
| Physically active | 927 | 0.14 (0.00) | |
| Weekly fast-food consumption | |||
| 0 | 1,495 | 0.23 (0.01) | |
| 1–2 | 2,721 | 0.42 (0.01) | |
| 3–4 | 1,198 | 0.19 (0.01) | |
| ≥5 | 1,050 | 0.16 (0.01) | |
| Weekly sugar-sweetened beverageconsumption | |||
| 0 | 865 | 0.13 (0.00) | |
| 1–2 | 657 | 0.11 (0.00) | |
| 3–4 | 604 | 0.09 (0.00) | |
| ≥5 | 4,338 | 0.67 (0.01) |
Abbreviation: SE, standard error.
aCalculated as weight (kg)/height(m)2.
bMeans and SEs represent within-quintile estimates.
Figure 1.
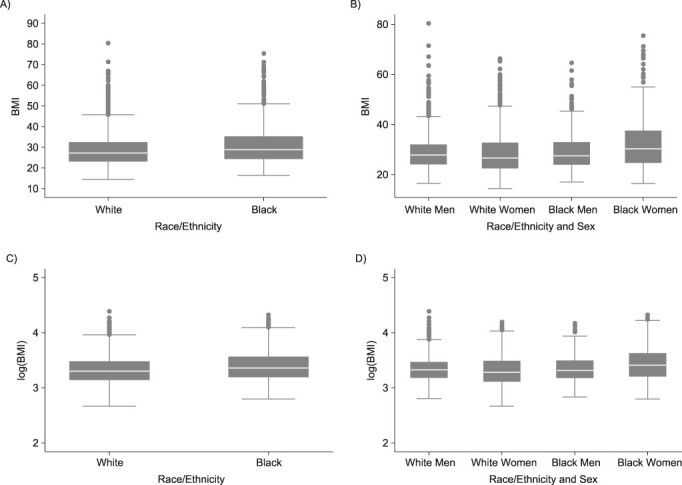
Box plots showing the distribution of (A) body mass index (BMI) by race/ethnicity, (B) BMI by race/ethnicity and sex, (C) log(BMI) by race/ethnicity, and (D) log(BMI) by race/ethnicity and sex (n = 6,172), US National Longitudinal Study of Adolescent to Adult Health Wave IV, 2007–2008.
In the fully adjusted model for whites, women had, on average, lower log(BMI) than men (β = −0.03; SE, 0.01), and completion of a 4-year college degree was associated with statistically significantly lower log(BMI) (β = −0.07; SE, 0.01) (Web Table 1, available at https://doi.org/10.1093/aje/kwaa058). The polygenic score was also statistically significantly associated with mean log(BMI) (β = 0.06; SE, 0.00). Of the total variation in age-adjusted log(BMI) among whites (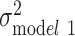 = 0.055), demographic and SES variables explained 0.21% and 2.51%, respectively (Figure 2). Polygenic score alone explained 8.39% of the variation in log(BMI), whereas health behaviors explained less than 1%. SES and polygenic score jointly explained almost 10% of the variation in log(BMI), and all the variables taken together explained up to 11.16% of the variation in log(BMI) among white individuals. However, the majority of the variation (88.84%) remained unexplained (Figure 3).
= 0.055), demographic and SES variables explained 0.21% and 2.51%, respectively (Figure 2). Polygenic score alone explained 8.39% of the variation in log(BMI), whereas health behaviors explained less than 1%. SES and polygenic score jointly explained almost 10% of the variation in log(BMI), and all the variables taken together explained up to 11.16% of the variation in log(BMI) among white individuals. However, the majority of the variation (88.84%) remained unexplained (Figure 3).
Figure 2.
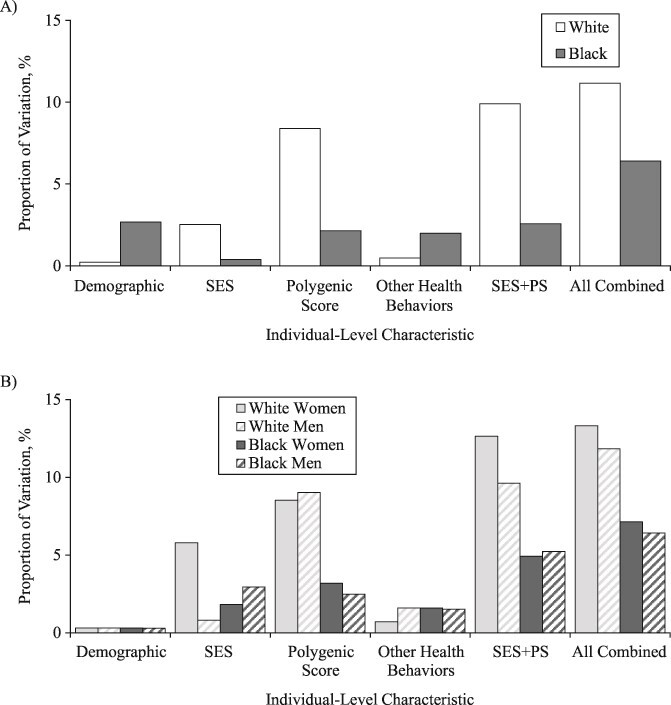
Proportion of variation in log(BMI) explained by demographic, socioeconomic status (SES), polygenic score (PS), and health behaviors, separately and jointly, stratified by (A) race/ethnicity and (B) race/ethnicity and sex, US National Longitudinal Study of Adolescent to Adult Health Wave IV, 2007–2008. BMI, body mass index.
Figure 3.
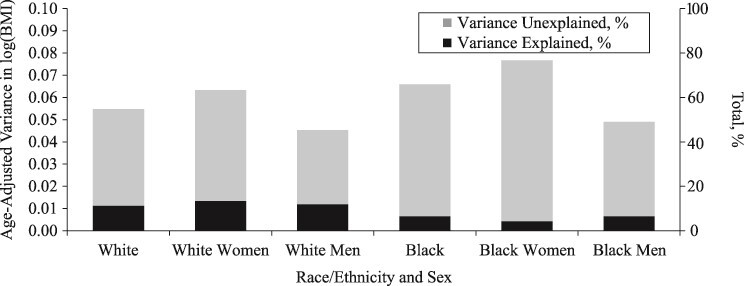
Age-adjusted variance in log(BMI) and the total percentage unexplained by demographic, socioeconomic status, polygenic score, and health behaviors, stratified by race/ethnicity and by sex, US National Longitudinal Study of Adolescent to Adult Health Wave IV, 2007–2008. BMI, body mass index.
Among black individuals, sex, relationship status, polygenic score, and smoking were all statistically significantly associated with log(BMI) in the fully adjusted model (P < 0.01) (Web Table 2). The age-adjusted variation in log(BMI) was larger among blacks than whites (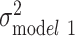 =0.066). SES factors and polygenic score each independently explained 0.38% and 2.14% of the variation in log(BMI), respectively, and were additive when considered jointly (2.57% of the variation explained in model 6) (Figure 2). All the variables taken together explained only 6% of the variance among black individuals, leaving most of the variation in log(BMI) unexplained (
=0.066). SES factors and polygenic score each independently explained 0.38% and 2.14% of the variation in log(BMI), respectively, and were additive when considered jointly (2.57% of the variation explained in model 6) (Figure 2). All the variables taken together explained only 6% of the variance among black individuals, leaving most of the variation in log(BMI) unexplained (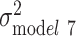 = 0.062) (Figure 3).
= 0.062) (Figure 3).
When stratified by race/ethnicity and sex, we consistently found a significant association between polygenic score and log(BMI) but not for SES and log(BMI) (Web Tables 3–6). Although we did not conduct a formal interaction test between sex and polygenic score, the largely overlapping 95% confidence interval around the β coefficient for the polygenic score for women versus men (within the same racial/ethnic group) indicated no statistically significant difference in the association between the polygenic score and log(BMI) by sex. For instance, among blacks, there was no apparent sex difference in the association between polygenic score and log(BMI) (black women: β = 0.05; SE, 0.01; 95% confidence interval: 0.03, 0.07; black men: β = 0.03; SE, 0.01; 95% confidence interval: 0.01, 0.05). The total age-adjusted variation in log(BMI) ranged from 0.045 for white men to 0.077 for black women (Figure 3). In terms of the variance explained, the contribution of SES was largest for white women (5.79%) and smallest for white men (0.81%), and the contribution of polygenic score ranged from 2.49% for black men to 9.04% for white men (Figure 2). All variables taken together explained 6%–7% of variation in log(BMI) for black men and black women, and up to 13% for white women.
DISCUSSION
Five salient findings emerged from our analysis. First, the amount of age-adjusted variation in log(BMI) was not uniform across different subgroups defined by race/ethnicity and sex; log(BMI) was approximately 70% larger for black women than for white men. Second, the contribution of SES factors in explaining variation in log(BMI) was the greatest for white women (5.8%), the least for white men (<1%), and approximately 2%–3% for black individuals. Third, the proportion of variance explained by the polygenic score was approximately 8% for whites and 2% for blacks. Fourth, the total amount of variance explained by all factors taken together was not necessarily larger for subgroups with larger log(BMI) variance. Fifth, the majority of variation in log(BMI) (87%–96%) remained unexplained by the demographic characteristics, SES, polygenic score, and health behaviors considered in our analysis.
The results concerning black individuals should be interpreted with caution because the BMI polygenic score we used was created from a genome-wide association study discovery sample that included Europeans only (18). The European-derived polygenic score is not portable across different ancestral groups, given that allelic distributions of SNPs associated with BMI vary by population due to population drift (41). However, with the caveat that measurement error will attenuate estimates, within-ancestral groups analyses are still possible (40). As expected, the estimated association between polygenic score and log(BMI) among blacks was approximately two-thirds as large as the estimated association in the white participants. Moreover, polygenic score explained 8.39% of variation in log(BMI) among whites compared with 2.14% among blacks. Hence, we can conclude that although the attenuation is due to noise and population drift, we still observe the same trends within ancestries.
In addition to population stratification that can bias estimation of genotype-phenotype associations, dynastic effects and assortative mating can also induce correlations through confounding between genotypes and phenotypes. Dynastic effects refer to bias that arises when inherited SNPs operate indirectly on offspring phenotype via their effects on the parents’ phenotype (24), for instance, obesity-inducing environments in this study. Lack of data on maternal and paternal genotype precludes assessment of the presence of dynastic effects in the current analysis (24, 42). Last, assortative mating refers to the nonrandom pairing of spouses on the basis of phenotypic characteristics and social homogamy (e.g., SES) (43). Assortative mating affects the distribution of allele frequencies at the population level and may lead to bias in genotype-phenotype associations (43, 44). These are potential biases and limitations of the polygenic scores used in these analyses, which are created from summary statistics of genotype-phenotype associations in large samples of unrelated individuals.
There are other data-related limitations. First, our sample was predominantly young adults aged 24–33 years. Prior studies have reported heterogeneity in some of the SNPs associated with obesity phenotype by age (45) as well as heterogeneity in the association between BMI and genetic risk score depending on the demographic characteristics of the cohort (46). Second, the majority of covariates were self-reported measures and their ability to explain variation in BMI could be biased by measurement error. Third, given the cross-sectional nature of our analysis, we do not claim any causality between predictor variables and BMI. Rather, we focused on quantifying the relative contribution of SES, polygenic score, and other relevant covariates in explaining BMI variation. Last, our models assumed linear relationship between the covariates, but specifying nonlinear and interactive relationships between social factors may improve the model fit (47). However, in a recent study comparing machine-learning algorithms of flexible functional forms and nonlinear associations of SES with conventional ordinary least-squares models in predicting women’s height, only a marginal difference was found (48).
The differential variation in BMI across subgroups defined by race/ethnicity and sex aligns with recent studies’ findings of disparate rate of weight gains across different segments of the population (3, 49, 50), with non-Hispanic black women and women with less than a high school education having the highest mean and greatest dispersion (as measured by standard deviation) in BMI in the United States (49). Monitoring the systematic differences in variability across populations, or changes in within-population variation over time, in health outcomes is useful to identify the underlying vulnerabilities and capacities (51, 52). Differential distribution of income and health resources, as well as stressors and buffers, interact to produce systematically patterned variation in population health and well-being (51). In fact, for certain physiological traits like blood pressure, racial differences in variation is speculated to emerge earlier than differences in means (53). We found BMI to be most variable among black women, arguably the most vulnerable subgroup in our sample. That variation in black women was larger than the variation in either women or blacks alone further supports that multiple social identities at the micro level likely interact with macro-level structural factors to produce disparate health outcomes (54).
An additional contribution of our study was in quantifying the extent to which SES, polygenic score, and other factors with known relevance to body weight explain the variation in BMI and how these associations vary across different subgroups. The small proportion in the variance explained by SES was comparable to that reported in a prior study in which 0.1%–6.4% of women’s BMI was explained by SES across different countries (15). A larger fraction of variance explained by SES among white women in our study can be partially explained by the relatively stronger average association observed between education and income with BMI in this subgroup. Compared with SES, the polygenic score consistently explained more variation in BMI for both white individuals and black individuals. Importantly, when considered simultaneously, SES and polygenic score had an additive, not interactive, contribution to explaining variation in BMI.
The proportion of unexplained variation in BMI was not driven by the magnitude of variation, meaning that subgroups with larger variance did not have more or fewer systematic components. The small proportion in variance explained by demographic, SES, polygenic score, and health behaviors does not mean the role of these predictors on BMI is not important (55). Yet, quantifying the residual variance and the proportion explained, in addition to the commonly reported tests of significance in average associations, can provide a more comprehensive basis for making decisions about the practical importance of the determinants for population health and health disparities by examining the agreement between the magnitude of their theoretical and empirical importance (56, 57). There are other measures, such as population attributable fraction, that can also aid quantification of genetic and environmental contribution to disease. Factors that explain more variation in health outcomes for specific groups would inform targeted policies and interventions. To this point, deepening our understanding of the contributions of biological, genetic, and environmental factors, as well as stochastic elements, in diverse phenotypic variance is needed in population health sciences.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Division of Health Policy and Management, College of Health Sciences, Korea University, Seoul, South Korea (Rockli Kim); Department of Public Health Sciences, Graduate School, Korea University, Seoul, South Korea (Rockli Kim); Harvard Center for Population and Development Studies, Boston, Massachusetts (Rockli Kim and S. V. Subramanian); Department of Sociology, University of Colorado, Denver, Colorado (Adam M. Lippert); Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts (Robbee Wedow); Department of Epidemiology, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (Robbee Wedow and Marcia P. Jimenez); Analytic Translational Genetics Unit, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts (Robbee Wedow); Department of Sociology, Harvard University, Cambridge, Massachusetts (Robbee Wedow); and Department of Social and Behavioral Sciences, Harvard T. H. Chan School of Public Health, Boston, Massachusetts (S. V. Subramanian).
A.M.L. was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health under award 2P2CHD066613. M.P.J. was supported by the Ruth L. Kirschstein National Research Service Award, Individual Pre-Doctoral Fellowship (no. 5F31HL134300-02).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Conflict of interest: none declared.
REFERENCES
- 1. Seidell JC, Halberstadt J. The global burden of obesity and the challenges of prevention. Ann Nutr Metab. 2015;66(Suppl. 2):7–12. [DOI] [PubMed] [Google Scholar]
- 2. Finucane MM, Stevens GA, Cowan MJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9·1 million participants. Lancet. 2011;377(9765):557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Razak F, Corsi DJ, Subramanian S. Change in the body mass index distribution for women: analysis of surveys from 37 low-and middle-income countries. PLoS Med. 2013;10(1):e1001367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Visscher TL, Seidell JC. The public health impact of obesity. Annu Rev Public Health. 2001;22(1):355–375. [DOI] [PubMed] [Google Scholar]
- 5. Lim SS, Vos T, Flaxman AD, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380(9859):2224–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swinburn BA, Sacks G, Hall KD, et al. The global obesity pandemic: shaped by global drivers and local environments. Lancet. 2011;378(9793):804–814. [DOI] [PubMed] [Google Scholar]
- 7. Burke MA, Heiland FW. Evolving societal norms of obesity: what is the appropriate response? JAMA. 2018;319(3):221–222. [DOI] [PubMed] [Google Scholar]
- 8. Sallis JF, Conway TL, Cain KL, et al. Neighborhood built environment and socioeconomic status in relation to physical activity, sedentary behavior, and weight status of adolescents. Prev Med. 2018;110:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glass TA, McAtee MJ. Behavioral science at the crossroads in public health: extending horizons, envisioning the future. Soc Sci Med. 2006;62(7):1650–1671. [DOI] [PubMed] [Google Scholar]
- 10. Brown RE, Sharma AM, Ardern CI, et al. Secular differences in the association between caloric intake, macronutrient intake, and physical activity with obesity. Obes Res Clin Pract. 2016;10(3):243–255. [DOI] [PubMed] [Google Scholar]
- 11. Rose G. Sick individuals and sick populations. Int J Epidemiol. 2001;30(3):427–432. [DOI] [PubMed] [Google Scholar]
- 12. Merlo J, Mulinari S, Wemrell M, et al. The tyranny of the averages and the indiscriminate use of risk factors in public health: the case of coronary heart disease. SSM Popul Health. 2017;3:684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McLaren L. Socioeconomic status and obesity. Epidemiol Rev. 2007;29(1):29–48. [DOI] [PubMed] [Google Scholar]
- 14. Kinge JM, Strand BH, Vollset SE, et al. Educational inequalities in obesity and gross domestic product: evidence from 70 countries. J Epidemiol Community Health. 2015;69(12):1141–1146. [DOI] [PubMed] [Google Scholar]
- 15. Kim R, Kawachi I, Coull BA, et al. Contribution of socioeconomic factors to the variation in body-mass index in 58 low-income and middle-income countries: an econometric analysis of multilevel data. Lancet Glob Health. 2018;6(7):e777–e786. [DOI] [PubMed] [Google Scholar]
- 16. Yengo L, Sidorenko J, Kemper KE, et al. Meta-analysis of genome-wide association studies for height and body mass index in∼ 700000 individuals of European ancestry. Hum Mol Genet. 2018;27(20):3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wen W, Zheng W, Okada Y, et al. Meta-analysis of genome-wide association studies in east Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23(20):5492–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akiyama M, Okada Y, Kanai M, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49(10):1458–1467. [DOI] [PubMed] [Google Scholar]
- 20. Khera AV, Chaffin M, Wade KH, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587–596.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis CM, Vassos E. Prospects for using risk scores in polygenic medicine. Genome Med. 2017;9(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Visscher PM, Brown MA, McCarthy MI, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kong A, Thorleifsson G, Frigge ML, et al. The nature of nurture: effects of parental genotypes. Science. 2018;359(6374):424–428. [DOI] [PubMed] [Google Scholar]
- 25. Young AI, Wauthier F, Donnelly P. Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat Commun. 2016;7:12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu S, Walter S, Marden J, et al. Genetic vulnerability to diabetes and obesity: does education offset the risk? Soc Sci Med. 2015;127:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li S, Zhao JH, Luan J, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7(8):e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi Q, Chu AY, Kang JH, et al. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ. 2014;348:g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qi Q, Downer MK, Kilpeläinen TO, et al. Dietary intake, FTO genetic variants, and adiposity: a combined analysis of over 16,000 children and adolescents. Diabetes. 2015;64(7):2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tyrrell J, Wood AR, Ames RM, et al. Gene–obesogenic environment interactions in the UK biobank study. Int J Epidemiol. 2017;46(2):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris KM, Halpern CT, Haberstick BC, et al. The national longitudinal study of adolescent health (add health) sibling pairs data. Twin Res Hum Genet. 2013;16(1):391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McVean G, Altshuler D, Durbin R, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamer D, Sirota L. Beware the chopsticks gene. Mol Psychiatry. 2000;5(1):11–13. [DOI] [PubMed] [Google Scholar]
- 36. Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 37. Price AL, Zaitlen NA, Reich D, et al. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braudt DB, Harris KM. Polygenic Scores (PGSs) in the National Longitudinal Study of Adolescent to Adult Health (Add Health) – Release 1. Chapel Hill, NC: Carolina Population Center, University of North Carolina at Chapel Hill; 2018. [Google Scholar]
- 39. Lee JJ, Wedow R, Okbay A, et al. Gene discovery and polygenic prediction from a 1.1-million-person GWAS of educational attainment. Nat Genet. 2018;50(8):1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belsky DW, Domingue BW, Wedow R, et al. Genetic analysis of social-class mobility in five longitudinal studies. Proc Natl Acad Sci. 2018;115(31):E7275–E7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martin AR, Gignoux CR, Walters RK, et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 2017;100(4):635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brumpton B, Sanderson E, Hartwig FP, et al. Within-family studies for Mendelian randomization: avoiding dynastic, assortative mating, and population stratification biases. bioRxiv. 2019;602516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yengo L, Robinson MR, Keller MC, et al. Imprint of assortative mating on the human genome. Nat Hum Behav. 2018;2(12):948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hartwig FP, Davies NM, Davey SG. Bias in Mendelian randomization due to assortative mating. Genet Epidemiol. 2018;42(7):608–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winkler TW, Justice AE, Graff M, et al. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11(10):e1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walter S, Mejía-Guevara I, Estrada K, et al. Association of a genetic risk score with body mass index across different birth cohorts. JAMA. 2016;316(1):63–69. [DOI] [PubMed] [Google Scholar]
- 47. Seligman B, Tuljapurkar S, Rehkopf D. Machine learning approaches to the social determinants of health in the health and retirement study. SSM Popul Health. 2018;4:95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Daoud A, Kim R, Subramanian S. Predicting women's height from their socioeconomic status: a machine learning approach. Soc Sci Med. 2019;238:112486. [DOI] [PubMed] [Google Scholar]
- 49. Krishna A, Razak F, Lebel A, et al. Trends in group inequalities and interindividual inequalities in BMI in the United States, 1993–2012. Am J Clin Nutr. 2015;101(3):598–605. [DOI] [PubMed] [Google Scholar]
- 50. Kim R, Kawachi I, Coull BA, et al. Patterning of individual heterogeneity in body mass index: evidence from 57 low-and middle-income countries. Eur J Epidemiol. 2018;33(8):741–750. [DOI] [PubMed] [Google Scholar]
- 51. Galea S, Ahern J, Karpati A. A model of underlying socioeconomic vulnerability in human populations: evidence from variability in population health and implications for public health. Soc Sci Med. 2005;60(11):2417–2430. [DOI] [PubMed] [Google Scholar]
- 52. Lewontin R, Levins R. Schmalhausen's law. Capital Nat Social. 2000;11(4):103–108. [Google Scholar]
- 53. Himmelstein DU, Levins R, Woolhandler S. Beyond our means: patterns of variability of physiological traits. Int J Health Serv. 1990;20(1):115–124. [DOI] [PubMed] [Google Scholar]
- 54. Bowleg L. The problem with the phrase women and minorities: intersectionality—an important theoretical framework for public health. Am J Public Health. 2012;102(7):1267–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosenthal R, Rubin DB. A note on percent variance explained as a measure of the importance of effects. J Appl Soc Psychol. 1979;9(5):395–396. [Google Scholar]
- 56. Good R, Fletcher HJ. Reporting explained variance. J Res Sci Teach. 1981;18(1):1–7. [Google Scholar]
- 57. O'grady KE. Measures of explained variance: cautions and limitations. Psychol Bull. 1982;92(3):766–777. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


