Abstract
Background and Aims
Artificial intelligence (AI)-based applications have transformed several industries and are widely used in various consumer products and services. In medicine, AI is primarily being used for image classification and natural language processing and has great potential to affect image-based specialties such as radiology, pathology, and gastroenterology (GE). This document reviews the reported applications of AI in GE, focusing on endoscopic image analysis.
Methods
The MEDLINE database was searched through May 2020 for relevant articles by using key words such as machine learning, deep learning, artificial intelligence, computer-aided diagnosis, convolutional neural networks, GI endoscopy, and endoscopic image analysis. References and citations of the retrieved articles were also evaluated to identify pertinent studies. The manuscript was drafted by 2 authors and reviewed in person by members of the American Society for Gastrointestinal Endoscopy Technology Committee and subsequently by the American Society for Gastrointestinal Endoscopy Governing Board.
Results
Deep learning techniques such as convolutional neural networks have been used in several areas of GI endoscopy, including colorectal polyp detection and classification, analysis of endoscopic images for diagnosis of Helicobacter pylori infection, detection and depth assessment of early gastric cancer, dysplasia in Barrett’s esophagus, and detection of various abnormalities in wireless capsule endoscopy images.
Conclusions
The implementation of AI technologies across multiple GI endoscopic applications has the potential to transform clinical practice favorably and improve the efficiency and accuracy of current diagnostic methods.
Abbreviations: ADR, adenoma detection rate; AI, artificial intelligence; AMR, adenoma miss rate; ANN, artificial neural network; BE, Barrett’s esophagus; CAD, computer-aided diagnosis; CADe, CAD studies for colon polyp detection; CADx, CAD studies for colon polyp classification; CI, confidence interval; CNN, convolutional neural network; CRC, colorectal cancer; DL, deep learning; GI, gastroenterology; HDWL, high-definition white light; HD-WLE, high-definition white light endoscopy; ML, machine learning; NBI, narrow-band imaging; NPV, negative predictive value; PIVI, preservation and Incorporation of Valuable Endoscopic Innovations; SVM, support vector machine; VLE, volumetric laser endomicroscopy; WCE, wireless capsule endoscopy; WL, white light
The American Society for Gastrointestinal Endoscopy (ASGE) Technology Committee provides reviews of existing, new, or emerging endoscopic technologies that have an impact on the practice of GI endoscopy. Evidence-based methods are used, with a MEDLINE literature search to identify pertinent clinical studies on the topic and a MAUDE (Food and Drug Administration Center for Devices and Radiological Health) database search to identify the reported adverse events of a given technology. Both are supplemented by accessing the “related articles” feature of PubMed and by scrutinizing pertinent references cited by the identified studies. Controlled clinical trials are emphasized, but in many cases data from randomized controlled trials are lacking. In such cases, large case series, preliminary clinical studies, and expert opinions are used. Technical data are gathered from traditional and web-based publications, proprietary publications, and informal communications with pertinent vendors. Reports on emerging technology are drafted by 1 or 2 members of the ASGE Technology Committee, reviewed and edited by the committee as a whole, and approved by the Governing Board of the ASGE. When financial guidance is indicated, the most recent coding data and list prices at the time of publication are provided. For this review, the MEDLINE database was searched through May 2020 for relevant articles by using relevant key words such as “machine learning,” “deep learning,” “artificial intelligence,” “computer-aided diagnosis,” “convolutional neural networks,” “gastrointestinal endoscopy,” and “endoscopic image analysis,” among others. Technology reports are scientific reviews provided solely for educational and informational purposes. Technology reports are not rules and should not be construed as establishing a legal standard of care or as encouraging, advocating, requiring, or discouraging any particular treatment or payment for such treatment.
Introduction
In recent years, a proliferation of artificial intelligence (AI)-based applications has rapidly transformed our work and home environments and our interactions with devices. AI is a broad descriptor that refers to the development and application of computer algorithms that can perform tasks that usually require human intelligence.1 Machine learning (ML) refers to AI in which the algorithm, based on the input raw data, analyzes features in a separate dataset without specifically being programmed and delivers a specified classification output (Figure 1, Figure 2, Figure 3).2,3 Examples of prevalent ML-based applications and devices include digital personal assistants on smartphones and speakers; predictive analytics that provide shopping or movie recommendations based on previous purchases or downloads or that show user-specific content on social networks; automated reading and analysis of postal addresses; automated investing based on analysis of large amounts of financial data; and autonomous vehicles.
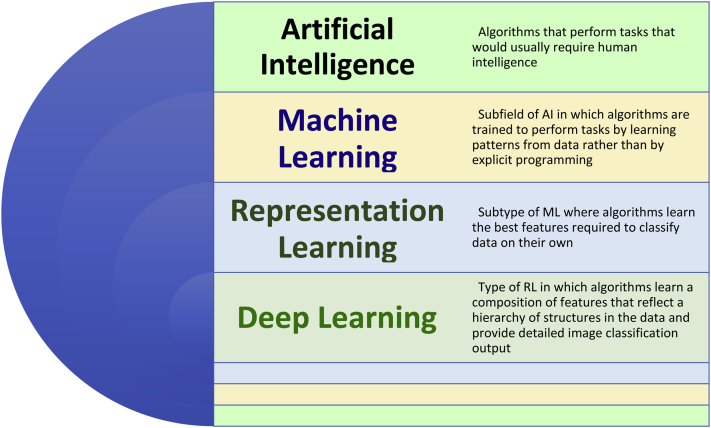
Figure 1.
Diagram representation of hierarchy of artificial intelligence domains (adapted from Goodfellow et al8 with permission). Abbreviations: AI, artificial intelligence; ML, machine learning; RL, representation learning; DL, deep learning.
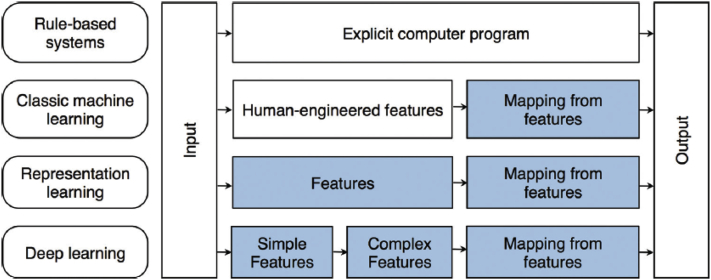
Figure 2.
Flowchart and descriptions of various types of learning and differentiation between conventional machine learning and deep learning (adapted from Chartrand et al6 with permission).
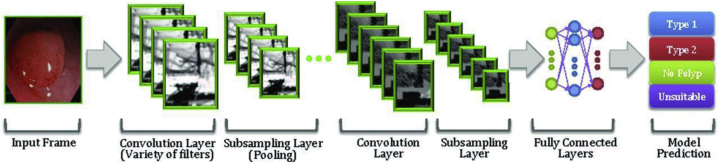
Figure 3.
An example of convolutional neural network for colorectal polyps (adapted from Byrne et al33 with permission).
One of the more common tasks to which ML has been applied is image discrimination and classification, which has many applications within medicine. In conventional ML, a training set of images with the desired categories is used to repeatedly train the system to improve performance and reduce errors. After multiple training sequences, the system performance is evaluated on an independent test set of images. Support vector machine (SVM) algorithms and artificial neural networks (ANN) are 2 commonly used conventional ML techniques.3, 4, 5 The major disadvantage of these conventional, handcrafted systems is the engineering and effort needed to design each system for a specific task. Deep learning (DL) is a transformative ML technique that overcomes many of these limitations. In contrast to SVM and ANN approaches, DL uses a back-propagation algorithm consisting of multiple layers, which enables the system itself to change the parameters in each layer based on the representations in the previous layers (representation learning) and to provide the output more efficiently. One of the major advantages of this system is transfer learning, in which a pretrained model that has learned natural image features on one task can be applied to a new task, even with a limited training dataset for the new task.6 This avoids the need to design a system de novo for each task. For example, a model that was developed to classify photographs of animals can subsequently be applied to the classification of flower types even without a large training dataset of flower images.
Convolutional neural network (CNN) is the most prominent DL technique currently in use, especially for image and pattern recognition. Other DL techniques include recurrent neural networks, which are applied for natural language processing and understanding and for development of predictive models. Several open-source software platforms that offer pretrained CNNs are available (eg, Convolutional Architecture for Fast Feature Embedding [Caffe, Berkeley AI Research, University of California, Berkeley, Calif, USA]).7 A more detailed description of the technical aspects of these techniques is beyond the scope of this document; for additional information, more comprehensive reviews in this area are available.2,6,8,9 A glossary of commonly AI-related terms and basic definitions is also included in Table 1.
Table 1.
| Term | Definition/Description |
|---|---|
| Artificial intelligence (AI) | Branch of computer science that develops machines to perform tasks that would usually require human intelligence |
| Machine learning (ML) | Subfield of AI in which algorithms are trained to perform tasks by learning patterns from data rather than by explicit programming |
| Representation learning (RL) | Subtype of ML in which algorithms learn the best features required to classify data on their own |
| Deep learning (DL) | Type of RL in which algorithms learn a composition of features that reflect a hierarchy of structures in the data and provide detailed image classification output |
| Deep reinforcement learning (DRL) | Technique combining DL and sequential learning to achieve a specific goal over several steps in a dynamic environment |
| Training dataset | Dataset used to select the ideal parameters of a model after iterative adjustments |
| Validation dataset | A (usually) distinct dataset used to test and adjust the parameters of a model |
| Neural networks | Model of layers consisting of connected nodes broadly similar to neurons in a biological nervous system |
| Support vector machine (SVM) | Classification technique that enables identification of an optimal separation plane between categories by receiving data inputs in a testing dataset and providing outputs that can be used in a separate validation dataset |
| Recurrent neural networks | DL architecture for tasks involving sequential inputs such as speech or language and used for speech recognition and natural language processing and understanding (eg, predictive text suggestions for next words in a sequence) |
| Convolutional neural networks (CNN) | DL architecture that adaptively learns hierarchies of features through back-propagation and is used for detection and recognition tasks in images (eg, face recognition) |
| Computer-aided detection/diagnosis | Use of a computer algorithm to provide detection or a diagnosis of a specified object/region of interest |
| Transfer learning | Ability of a trained CNN model to perform a separate task by using a relatively small dataset for the new task |
As with several other areas such as consumer products and finance, AI is expected to be a disruptive technology in some medical specialties, particularly those that require analysis and interpretation of large datasets and images (eg, radiology, pathology, and dermatology).3,4,10 For example, AI is being evaluated in radiology to triage radiographs based on potential pathology to determine the order of reading by the radiologist and to calculate tumor volumes on CT scans in patients with hepatocellular carcinoma.1 A wide range of potential applications for ML and DL exists in gastroenterology, especially in the realm of GI endoscopy, which also involves acquisition and analysis of large datasets of images.11 Although computer-aided analysis and detection, which involve the use of algorithms to analyze endoscopic images and detect or diagnose specific conditions, have been areas of research for many years, the advent of DL is likely to be a transformative process in this field. Several early reports have described the application of DL and other forms of AI to varied clinical problems within GI endoscopy.
This document reviews the currently reported applications of AI in GI endoscopy, including colorectal polyp detection, classification, and real-time histologic assessment. Furthermore, the document reviews the use of AI in the analysis of wireless capsule endoscopy (WCE) images and videos, localization and diagnosis of esophageal and gastric pathology on EGD, and image analysis of endoscopic ultrasound images (Table 2). The document does not cover the application of AI techniques (eg, natural language processing) for mining and/or analysis of endoscopic or medical databases or for using demographic and clinicopathologic variables to create predictive models.
Table 2.
Reported applications of computer-aided diagnosis and artificial intelligence in various endoscopic procedures
| Procedure | Application |
|---|---|
| Colonoscopy | Detection of polyps (real time and on still images and video)∗ |
| Classification of polyps (neoplastic vs hyperplastic)∗ | |
| Detection of malignancy within polyps (depth of invasion on endocytoscopic images)∗ | |
| Presence of inflammation on endocytoscopic images∗ | |
| Wireless capsule endoscopy (WCE) | Lesion detection and classification (bleeding, ulcers, polyps)∗ |
| Assessment of intestinal motility | |
| Celiac disease (assessment of villous atrophy, intestinal motility) | |
| Improve efficiency of image review | |
| Deletion of duplicate images and uninformative image frames (eg, images with debris)∗ | |
| Upper endoscopy | Identify anatomical location∗ |
| Diagnosis of Helicobacter pylori infection status∗ | |
| Gastric cancer detection and assessing depth of invasion∗ | |
| Esophageal squamous dysplasia | |
| Detection and delineation of early dysplasia in Barrett’s esophagus∗ | |
| Real-time image segmentation in volumetric laser endomicroscopy (VLE) in Barrett’s esophagus∗ | |
| Endoscopic ultrasound (EUS) | Differentiation of pancreatic cancer from chronic pancreatitis and normal pancreas |
| Differentiation of autoimmune pancreatitis from chronic pancreatitis | |
| EUS elastography |
Applications in which use of deep learning has been reported.
Applications in endoscopy
Colorectal polyps: detection, classification, and cancer prediction
AI has been primarily evaluated in 3 clinical scenarios for neoplastic disorders of the colon: polyp detection, polyp characterization (adenomatous vs nonadenomatous), and prediction of invasive cancer within a polypoid lesion. Published computer-aided diagnosis (CAD) studies for colon polyp detection (CADe) and classification (CADx) are subject to a number of limitations. Higher-quality images may be chosen for CAD, leading to selection bias. Some advanced imaging technologies used in the literature are not widely available for clinical use. One study to date has included sessile serrated lesions in a CADe model but not in CADx, a limitation because these polyps are important precursors in up to 30% of colon cancers.12 CAD has been evaluated in most studies by using archived still images or video segments of real procedures. Although multiple systems have the processing speed to be considered “real-time capable,” to date only 2 studies have been performed during real-time colonoscopy.12,13 To be clinically useful, AI platforms in colonoscopy will need rapid image analysis with real-time information that assists the endoscopist in accurately determining the presence and/or type of polyp present.
Polyp detection
The rate of missed polyps during colonoscopy is as high as 25%.14 The subtle appearance of some polyps, quality of the bowel preparation and colonoscopist mucosal inspection technique, inherent ability, and fatigue may all contribute to missing these polyps.15,16 Improved detection of neoplastic polyps may result in a greater reduction in interval colon cancers. The incorporation of AI may reduce polyp miss rates, particularly among those endoscopists with lower adenoma detection rates (ADRs). Initial CADe studies used traditional handcrafted algorithms for image analysis17,18; however, several recent publications have reported on the use of DL for polyp detection.12,19, 20, 21
A small study assessed a computer-aided polyp detection model by using 24 archived colonoscopy videos containing 31 polyps.17 Polyp location was marked by an expert endoscopist and was used as the criterion standard. The polyp detection sensitivity and specificity for the CADe system were 70.4% and 72.4%, respectively. The model performed best for identification of small flat lesions (Paris 0-II), which may be difficult to detect endoscopically. A similar study used a different CADe model to evaluate video and still images of 25 unique polyps; the model demonstrated a sensitivity of 88% for polyp detection. Although the study used archived images, this algorithm was capable of providing real-time (0.3 second latency) analysis and reporting.18
Significant improvements have been realized in computer-aided polyp detection with the incorporation of DL technologies (Video 1, available online at www.VideoGIE.org). A single-center study designed and trained a CNN using 8641 labeled images containing 4088 unique polyps from screening colonoscopies of more than 2000 patients.19 On an independent validation set of 1330 images, the CNN system detected polyps with an accuracy of 96.4% and a false-positive rate of 7%. The investigators also tested the model on 9 colonoscopy videos in which a total of 28 polyps were detected and removed and then compared the computer-assisted image analysis with the analysis of 3 expert colonoscopists (ADRs ≥50%). The 3 experts identified 36 polyps while reviewing unaltered videos and 45 polyps while reviewing CNN-overlaid videos. When expert review with CNN overlay was used as the criterion standard, the sensitivity and specificity of the CNN alone for polyp detection in these videos were 93% and 93%, respectively (P < .00001). False positives generated by the CNN tended to occur in the settings of near-field collapsed mucosa, debris, suction marks, narrow-band imaging (NBI), and polypectomy sites. The fast processing speed (10 milliseconds per frame), the ability to identify polyps during examination with standard high-definition white light endoscopy (HD-WLE), and the ability to run the software on standard consumer-quality desktop computers suggest that the technology could be practical in a “real world” endoscopy environment.
Wang et al12 reported the first prospective randomized controlled trial demonstrating an improvement in ADR using CADe technology. Patients were randomized in a nonblinded fashion to undergo routine diagnostic colonoscopy (n = 536) or colonoscopy with the assistance of real-time computer-aided polyp detection (n = 522). The DL-based CNN system provided simultaneous visual and audio notification of polyp detection. The AI system significantly increased ADR (29.1% vs 20.3%; P < .001), mean number of adenomas per patient (0.53 vs 0.31; P < .001), and overall polyp detection rate (45% vs 29%, P < .001). The improved ADR was ascribed to a higher number of diminutive adenomas identified (185 vs 102; P < .001) because there was no statistically significant difference in detection of larger adenomas (77 vs 58; P = .075). This study supports the use of CADe as an aid to endoscopists with low ADR (20% baseline); however, the benefit of an automated polyp detection system must be validated for endoscopists with greater expertise. A small number of false positive cases were reported in the CADe group (n = 39), equivalent to 0.075 per colonoscopy. The false positives were ascribed to intraluminal bubbles, retained fecal material, wrinkled mucosa, and local inflammation. Withdrawal time was slightly increased while using the CADe system (6.9 minutes vs 6.3 minutes) because of the additional time for biopsy sampling of additional polyps detected. In addition, the CADe system increased detection of diminutive hyperplastic polyps almost 2-fold (114 vs 52; P < .001). It is likely that endoscopists, with the help of a virtual chromoendoscopy or a CADx system, could render a high-confidence optical diagnosis of diminutive hyperplastic rectosigmoid polyps supporting a detect, diagnose, and leave in situ strategy, which would result in workload and cost reductions.22
Wang et al21 performed another CADe study that aimed to assess the ability of AI to improve colon polyp detection, measured as a reduction in the adenoma miss rate (AMR). This was a single-center, open-label, prospective, tandem colonoscopy study of patients randomly assigned to undergo CADe colonoscopy (n = 184) or routine colonoscopy (n = 185), followed immediately by the endoscopist performing the other procedure. Overall, AMR was significantly lower in the CADe colonoscopy arm (13.89% vs 40.00%; P < .0001). AMR was found to be significantly lower for both diminutive (<5 mm) and small adenomas (5-9 mm) in the CADe colonoscopy group. Moreover, a post hoc video analysis attempted to measure the AMR for only “visible” polyps because this represents the maximal possibility that CADe could help to decrease the miss rate. When comparing CADe to standard high-definition white light endoscopy (HDWL) colonoscopy, only 1.59% of visible adenomas were missed by CADe colonoscopy, whereas 24.21% of visible polyps were missed in the routine colonoscopy group (P < .001).
Repici et al20 performed the third multicenter, randomized trial of AI for polyp detection in real-time colonoscopy for indications of screening, surveillance, or fecal immunochemical test positivity. Participants (n = 685) were randomized in a 1:1 ratio to CADe (GI-Genius, Medtronic, Dublin, Ireland) with HDWL colonoscopy or HDWL colonoscopy alone. The CADe system improved ADR to 54.8% (187 of 341) from 40.4% (139 of 344) in the control group (relative risk, 1.30; 95% confidence interval [CI], 1.14-1.45). Adenomas detected per colonoscopy were also higher in the CADe group (mean 1.07 ± 1.54) than in the control group (mean 0.71 ± 1.20) (incidence rate ratio 1.46; 95% CI, 1.15-1.86). The improved ADR was seen in polyps <5 mm size and those 5 to 9 mm diameter without increasing withdrawal time.
Polyp classification
Alternative strategies for managing diminutive colon polyps have been proposed, including “resect and discard” or “leave in situ” paradigms.23, 24, 25 These strategies involve interrogation of the polyp using an enhanced imaging technique; the polyp is then resected and discarded if it appears adenomatous, or left in situ if it appears hyperplastic and is located in the rectosigmoid colon. However, attaining the necessary accuracy thresholds to implement these approaches has been challenging outside of expert centers.24,26 CADx may provide a support tool for endoscopists that allows more widespread attainment of the recommended accuracy thresholds.27 Potential benefits include improved cost effectiveness, shorter procedure time, and fewer adverse events resulting from unnecessary polypectomies.
A summary of published reports on AI for polyp classification is presented in Table 3. Early studies on polyp classification published in 2010 and 2011 evaluated the ability of CADx to discriminate adenomatous from hyperplastic polyps when using magnification chromoendoscopy28 or magnification NBI.29, 30, 31 These studies used traditional (non-DL) AI techniques and achieved accuracy rates for polyp classification of 85% to 98.5%. However, these studies were limited in that the image analysis software lacked real-time polyp characterization capability, required manual segmentation of the polyp margins, and analyzed images that were captured using magnification technologies that are both operator dependent and not routinely available in clinical practice.
Table 3.
Summary of reported studies on computer-aided diagnosis or detection of colorectal polyps
| Study | Design | Real time or delayed? | Lesion number (learning/validation) | Type of computer aided design | Imaging technology | Lesion size and type | Sensitivity/Specificity/Negative predictive value accuracy for neoplasia | Accuracy for surveillance interval |
|---|---|---|---|---|---|---|---|---|
| Takemura 201028 | Retrospective | Image analysis ex vivo. Not real time capable. | 72 polyps/134 polyps | Automated classification | Magnifying chromoendoscopy (Kudo pit pattern) | NR | NR/NR/NR/98.5% | NS |
| No SA | ||||||||
| Tischendorf 201029 | Post hoc analysis of prospective data | Image analysis ex vivo. Not real time capable. | 209 polyps/NS | Automated classification with SVM | Magnifying NBI | 8.1 mm avg (2-40 mm) | 90%/70%/NR | NS |
| SA excluded | 85.3% | |||||||
| Gross 201131 | Post hoc analysis of prospective data | Image analysis ex vivo. Not real time capable. | 434 polyps/NS | Automated classification with SVM | Magnifying NBI | 2-10 mm (SA; n = 2) | 95%/90.3/NR/93.1% | NS |
| Takemura 201232 | Retrospective | Image analysis ex vivo | NR/371 polyps | Automated classification with SVM | Magnifying NBI | NR | 97.8%/97.9%/NR/97.8% | NS |
| No SA | ||||||||
| Kominami 201630 | Prospective | Real time analysis of ex vivo images | NR/118 polyps | Automated classification with SVM | Magnifying NBI | ≤5 mm: 88 >5 mm: 30 |
For ≤5 mm:93%/93.3%/93%/93.2% | 92.7% |
| SA excluded | ||||||||
| Chen 201834 | Prospective validation | Image analysis ex vivo. | 2157/284 polyps | Automated classification with CNN | Magnifying NBI | SA excluded | 96.3%/78.1%/91.5%/90.1% | NS |
| Real time capability. | ||||||||
| Byrne 201933 | Prospective validation | Ex vivo video images. Real time capability (50 ms delay) | Test set: 125 videos | Automated classification with CNN | Near focus NBI | SA excluded | 98%/83%/97% | NS |
| 94% | ||||||||
| Jin 202042 | Prospective validation | Image analysis ex vivo | 2150/300 | Automated classification with CNN | NBI | ≤5 mm:300 | 83.3%/91.7%/NR/86.7% | NS |
| SA excluded | ||||||||
| Mori 201537 | Retrospective | Ex vivo of still images | NR/176 polyps | Automated classification (type NS) | Endocytoscopy | ≤10 mm:176 | 92%/79.5%/NR/89.2% | NR |
| SA excluded | ||||||||
| Mori 201635 | Retrospective | Ex vivo of still images. Real time capability. | 6051/205 polyps | Automated classification with SVM | Endocytoscopy | ≤5 mm: 139 | 89%/88%/76%/89% | 96% |
| 6-10 mm: 66 | ||||||||
| No SA | ||||||||
| Misawa 201636 | Prospective | Ex vivo of still images | 979/100 | Automated classification with SVM | Endocytoscopy with NBI | Mean 8.6 ± 10.3 mm | 84.5%/97.6%/82%/90% | NR |
| No SA | ||||||||
| Mori 201813 | Prospective | Real time colonoscopy | NS/475 polyps | Automated classification with SVM | Endocytoscopy with NBI and MB | ≤5 mm: 475 | Rectosigmoid: NR/NR/96.4%/98.1% | NR |
| No SA |
Abbreviations: CNN, convolutional neural network; MB, methylene blue; NBI, narrow-band imaging (Olympus Corporation, Center Valley, Penn, USA); NR, not reported; NS, not specified or studied; SA, serrated adenoma (includes SSA and traditional SA); SVM, support vector machine.
More recent studies have used AI technology with immediate polyp classification capability,13,30,32, 33, 34, 35, 36, 37, 38, 39, 40, 41 although only one of the studies has been evaluated in real time during an in vivo rather than recorded colonoscopy.13 These AI polyp classification studies used enhanced imaging technologies beyond HD-WLE, such as NBI, magnification NBI, endocytoscopy, confocal endomicroscopy, or laser-induced autofluorescence. In a prospective single-operator trial of 41 patients, 118 colorectal lesions were evaluated with magnifying NBI and real-time CADx using an SVM-based technique before resection.30 The diagnostic accuracy of CADx for diminutive polyp classification was 93.2%, with the pathologic diagnosis of the resected polyp serving as the criterion standard. Notably, the recommended surveillance colonoscopy interval based on real-time CAD histology prediction was concordant with pathology in 92.7% of the subset of diminutive polyps (n = 88), exceeding the Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) initiative threshold of ≥90% for the “resect and discard” strategy.23
Applying DL technology to image recognition of polyps has led to higher accuracy and faster image processing times (Video 2, available online at www.giejournal.org).33 Four studies have used CNNs to classify diminutive colon polyps as adenomatous or hyperplastic after inspection with conventional NBI,29,42 magnifying NBI,30 or near-focus NBI,32 using histology as the criterion standard. These studies trained the CNN using still images30,42 or video.29,32 On validation sets of 106 to 300 polyps, these CNNs identified adenomatous polyps in near real time (50 millisecond delay in one study) with a diagnostic accuracy of 88.5% to 94% and a negative predictive value (NPV) of 91.5% to 97%. The level of performance of the CNN in these studies met the “leave in situ” minimum threshold of a 90% NPV proposed by the American Society for Gastrointestinal Endoscopy PIVI initiative.23 Jin et al42 demonstrated that the use of CADx improved the overall accuracy of optical polyp diagnosis from 82.5% to 88.5% (P < .05). AI assistance was most beneficial for novices with limited training in using enhanced imaging techniques for polyp characterization. For the novice group of endoscopists (n = 7), the ability to correctly differentiate adenomatous from hyperplastic diminutive polyps improved with CADx from 73.8% accuracy to levels comparable to experts at 85.6% (P < .05). In contrast, colonoscopy experts (n = 4) with variable experience with NBI and formally trained experts in NBI (n = 11) demonstrated a smaller improvement with the addition of CADx, from 83.8% to 89.0% and 87.6% to 90.0%, respectively. Limitations of the study included the selection of only high-quality images for study inclusion and exclusion of sessile serrated polyps and lymphoid aggregates from the polyp population.
In a prospective study of 791 consecutive patients who underwent colonoscopy with endocytoscopes using NBI or methylene blue staining, CADx was able to characterize diminutive rectosigmoid polyps in real time with performance levels necessary to follow the “diagnose and leave in situ strategy” for nonneoplastic polyps.13 A total of 466 diminutive (including 250 rectosigmoid) polyps from 325 patients were identified. The DL system distinguished rectosigmoid adenomas from hyperplastic polyps in real time with an accuracy of 94% and an NPV of 96%. However, CAD was not useful in distinguishing neoplastic from nonneoplastic polyps proximal to the sigmoid colon (NPV 60.0%).
Adoption of AI systems in the form of a clinical decision support device could lead to more widespread use of the “leave in situ” and “resect and discard” strategies for management of diminutive colorectal polyps. Mori et al43 recently reported the first AI system that enables polyp detection followed by immediate polyp characterization in a real-time fashion by use of an endocytoscope (CF-H290ECI; Olympus Corp, Tokyo, Japan). The same group quantified the cost reduction from using an AI system to aid in the optical diagnosis of colorectal polyps.44 A diagnose and leave in situ strategy for diminutive rectosigmoid polyps supported by the AI prediction (not removed when predicted to be nonneoplastic) compared with a strategy of resecting all polyps yielded an average colonoscopy cost savings of 10.9% and gross annual reduction in reimbursement of $85.2 million in the United States.
AI may serve as the arbitrator between the endoscopist and pathologist when there exists discordant histologic characterization of diminutive colon polyps. In a series of 644 lesions ≤3 mm with a high-confidence optical diagnosis of adenoma, discrepancy between endoscopic and pathologic diagnoses occurred in 186 (28.9%) lesions.22 This included a pathologic diagnosis of hyperplastic polyp, sessile serrated polyp, and normal mucosa in 85 (13.2%), 2 (0.3%), and 99 (15.4%), respectively. Among these discordant results, Shahidi et al22 used a real-time AI clinical decision support solution, which agreed with the endoscopic diagnosis in 168 (90.3%) lesions. This raises the question of the validity of using a pathologic analysis as the criterion for characterizing colorectal lesions ≤3 mm when high-confidence optical evaluation identifies an adenoma and supports the use of AI to help decide the final pathologic diagnosis and resultant surveillance colonoscopy interval.
Detecting malignancy in colorectal polyps
Accurate optical diagnosis of T1 colorectal cancer (CRC) and the level of submucosal invasion help determine the optimal treatment approach for colorectal neoplasms. Lesions suspected to be T1 CRC confined to the SM1 layer (<1000 m) can be considered for endoscopic resection by en bloc techniques with either endoscopic mucosal resection (lesion diameter ≤2 cm) or endoscopic submucosal dissection.45,46 Deep submucosal invasion (1000 m or more) requires surgery because of the higher risk of lymph node metastasis.47 Current endoscopic assessment of depth of invasion consists of HD-WLE with morphologic examination (eg, Paris classification), NBI (selectively magnifying or near focus), and EUS.48 These advanced imaging techniques are not routinely used to assess colorectal polyps in Western countries; thus, AI may provide useful guidance for endoscopists in this setting.
A Japanese study evaluated an endocytoscopy-based CAD system to differentiate invasive cancer from nonmalignant adenomatous polyps; 5543 endocytoscopy images (2506 nonneoplasms, 2667 adenomas, and 370 invasive cancers) from 238 lesions (100 nonneoplasms, 112 adenomas, and 26 invasive cancers) were randomly selected from the database for ML.49 Sessile serrated lesions were excluded. An SVM classified these training set images and subsequently 200 validation set images (100 adenomas and 100 invasive cancers) to determine the characteristics for the diagnosis of invasive cancer. The algorithm achieved an accuracy of 94.1% (95% CI, 89.7-97.0) for identifying invasive malignancy. Ito et al50 developed an endoscopic CNN to distinguish depth of invasion for malignant colon polyps. The study included 190 images from 41 lesions (cTis = 14, cT1a = 14, and cT1b = 13). They used AlexNet and Caffe for machine learning, with the resulting CNN demonstrating a diagnostic sensitivity, specificity, and accuracy for deep invasion (cT1b) of 67.5%, 89.0%, and 81.2%, respectively.
Colonoscopy in inflammatory bowel disease
A CAD system evaluated the persistence of histologic inflammation in endocytoscopic images obtained during colonoscopy in patients with ulcerative colitis with an accuracy of 91% (83%-95%).51 A second study demonstrated the ability of a deep neural network CAD system to accurately identify patients with ulcerative colitis in endoscopic (90.1% accuracy [95% CI, 89.2%-90.9%]) and histologic remission (92.9% accuracy [95% CI, 92.1%-93.7%]) based on computer analysis of endoscopic mucosal appearance.52 No studies to date have evaluated the use of CAD for dysplasia detection and grading in the setting of surveillance colonoscopy for patients with chronic colitis.
Improving quality and training in colonoscopy
AMRs during colonoscopy are partly attributable to incomplete visual inspection of the colonic mucosal surface area. AI is being developed to provide objective and immediate feedback to the endoscopist to enhance visual inspection of colonic mucosa, potentially leading to improvements in both polyp detection and colon cancer prevention. A proof-of-concept study used an AI model to evaluate mucosal surface area inspected and several other quality metrics that contribute to adequacy of visual inspection during colonoscopy, including bowel preparation scores, adequacy of colonic distention, and clarity of the endoscopic view.53 A technically more-mature product with validation of performance is awaited. A second study used a deep CNN to assess colon bowel preparations; 5476 images from 2000 colonoscopy patients were used to train the CNN, and 592 images were used for validation using the Boston Bowel Preparation Scale as the measure of bowel preparation quality.54 Twenty previously recorded videos (30-second clips) were used to assess the real-time value of the CNN, with a reported accuracy of 89% compared with a low score among 5 expert endoscopists.
Analysis of wireless capsule endoscopy images
WCE is an established diagnostic tool for the evaluation of various small-bowel abnormalities such as bleeding, mucosal pathology, and small-bowel polyps.55 However, the review and analysis of large amounts of graphic data (up to 8 hours of video and approximately 60,000 images in a typical examination) remain major challenges. In a blinded study of 17 gastroenterologists with varied WCE experience who were shown WCE clips with variable or no pathology, the overall detection rate for any pathology was less than 50%.56 The detection rates for angioectasias, ulcers/erosions, masses/polyps, and blood were 69%, 38%, 46%, and 17%, respectively. Therefore, effective CAD systems that assist physician diagnosis are an unmet, yet critical, need.
The software that currently accompanies commercial WCE systems is capable of performing both curation functions (eg, removal of uninformative image frames such as those that contain debris or fluid) to enhance reader efficiency and rudimentary CAD functions (eg, using color to locate frames with blood). Conventional handcrafted CAD systems designed to detect one or more specific abnormalities such as bleeding, ulcers, polyps, intestinal motility, celiac disease, and Crohn’s disease have been reported but are not widely applicable. A detailed review of these WCE CAD systems is beyond the scope of this article, and the reader is referred to comprehensive reviews on this topic.57, 58, 59 As noted previously, a major limitation of conventional CAD systems is that each is designed to be specific to an image feature, and thus their performance is difficult to replicate in other datasets.57 Another challenge in designing image analysis software for WCE is that the resolution of images/video captured is of relatively lower quality compared with those acquired with high-definition endoscopes.
To overcome some of the limitations of the aforementioned CAD systems, there have been recent efforts to use DL techniques such as CNN to analyze WCE images.57,60,61 Given the large number of images collected with a relatively standard technique, WCE examinations provide an opportunity to create large, annotated databases, which are critical to developing robust CNN algorithms.62 A proof-of-principle study evaluated the ability of a CNN system to label a 120,000 image WCE dataset (100,000 image training set and 20,000 image validation set).57 The CNN system correctly classified nonpathologic image features such as intestinal wall, bubbles, turbid material, wrinkles, and clear blobs with an accuracy of 96%. In another report using a dataset of 10,000 images (2850 frames with bleeding and 7150 normal frames), an 8-layer CNN model had a precision value of 99.90% for the detection of bleeding,60 compared with 99.87%63 and 98.31%64 reported previously on this dataset when using conventional CAD systems. Similar DL systems have also been reported to detect polyps,65 angioectasias,66 small intestinal ulcers and erosions,67,68 and hookworms in WCE images.69
The most comprehensive and promising WCE study to date created a database of 113,426,569 small-bowel WCE images from 6970 patients at 77 medical centers. The CNN-based model was trained using 158,235 small bowel capsule endoscopy images from 1970 patients70 to identify and categorize small-bowel pathology that included inflammation, ulcers, polyps, lymphangiectasia, bleeding, vascular disease, protruding lesions, lymphatic follicular hyperplasia, diverticula, and parasitic disease. The validation dataset included 5000 small-bowel WCE videos interpreted by the CNN and 20 gastroenterologists (250 videos per GI). If there was discordance between the conventional analysis and CNN model, the gastroenterologists re-evaluated the video to confirm the final interpretation, which served as the criterion standard. The CNN-based algorithm was superior to the gastroenterologists in identifying abnormalities in both the per-patient analysis (sensitivity of 99.8% vs 74.57%; P < .0001) and per-lesion analysis (sensitivity of 99.90% vs 76.89%; P < .0001). Furthermore, the mean reading time per patient for the CNN model of 5.9 ± 2.23 minutes was much shorter compared with 96.6 ± 22.53 minutes for conventional reading by gastroenterologists (P < .001).
These studies suggest that DL techniques have the potential to serve as important tools to help gastroenterologists analyze small bowel capsule endoscopy images more efficiently and more accurately; however, currently no studies have used CNN to assess the impact on patient outcomes.
EGD
Anatomical location and quality assessment
DL algorithms have been designed to identify and label standard anatomical structures during EGD as an important early step in accurately diagnosing various disease states of the upper GI tract. In addition, CNNs have been developed to evaluate whether images/video frames acquired by the endoscopist are informative and to improve quality of the examination by assessing for blind spots and determining the proportion of mucosal surface area examined. A CNN algorithm used a development dataset of 27,335 images and independent validation set of 17,081 images to broadly classify the anatomical location of images obtained on upper endoscopy into larynx, esophagus, stomach (upper, middle, or lower regions), or duodenum.71 The demonstrated accuracy was 97%.
A single-center study developed a real-time quality improvement DL system termed WISENSE, which identifies blind spots during EGD and creates automated photodocumentation.72 The system was developed by combining a CNN algorithm with deep reinforcement learning, a newer DL technique designed to solve dynamic decision problems. After development, testing, and validation, the algorithm was applied in a single-center randomized controlled trial of 324 patients undergoing EGD performed by experienced endoscopists. Use of WISENSE reduced blind spot detection from 22.46% to 5.86% (P < .001), increased inspection time, and improved completeness of photodocumentation. The lesser curve of the middle upper body of the stomach was the anatomic site where the algorithm aided most with blind spots. In a prospective, single-blind randomized controlled trial (n = 437) from the same institution, the additional benefit of using an AI algorithm for blind spot detection was evaluated among patients undergoing unsedated ultrathin transoral endoscopy, unsedated conventional EGD, and sedated conventional EGD.73 The blind spot rate of the AI-assisted group was lower than that of the control group among all procedures; the conventional sedated EGD combined with the AI algorithm had a lower overall blind spot rate (3.42%) than ultrathin and unsedated endoscopy (21.77% and 31.23%, respectively; P < .05).
Diagnosis of Helicobacter pylori infection
Gastric cancer is prevalent worldwide, and H pylori infection is a leading cause. Although not routinely performed in Western countries, endoscopic diagnosis of H pylori infection based on mucosal assessment is an important component of gastric cancer screening in Asia. This process is time consuming because it requires the evaluation of multiple (∼50-60) images and is associated with a steep learning curve. AI may be a useful tool to improve physician diagnostic performance for the diagnosis of H pylori infection based on pattern recognition in endoscopic images.
A 22-layer CNN was applied on a training dataset of 32,208 white light (WL) gastric images (1750 patients) from upper endoscopy and a prospective validation set of 11,481 images (397 patients) and compared to a blinded assessment by 33 gastroenterologists with a broad range of experience.74 The CNN was noted to have a diagnostic accuracy for H pylori detection similar to expert endoscopists, but 12.1% higher than beginner endoscopists. The authors reported an accuracy, sensitivity, and specificity of 87.7% (95% CI, 84-90.7), 88.9% (95% CI, 79.3-95.1), and 87.4% (95% CI, 83.3-90.8), respectively, when the CNN was given the anatomic location of the images. Another study evaluated the role of a similar CNN architecture in H pylori diagnosis on screening endoscopic images of the lesser curve of the stomach obtained during transnasal endoscopy, using a more limited dataset of 179 images (149-image developmental set and 30-image validation set); the sensitivity and specificity were both 86.7% for the algorithm.75 A CNN algorithm based on gastric mucosal HD-WLE appearance was applied to a study population (n = 1959 patients; 8.3 ± 3.3 images per patient; 56% H pylori prevalence rate) undergoing EGD and gastric biopsy. By using archived endoscopic images, the CNN achieved an H pylori diagnostic accuracy of 93.8% (95% CI, 91.2-95.8).76
One of the challenges in improving the accuracy of endoscopic diagnosis is the differentiation between gastric mucosal changes due to active H pylori infection versus eradicated infection. Using a CNN model on gastric endoscopic images (n = 98,564), Shichijo et al77 categorized patients as negative, positive, and eradicated with an accuracy of 80%, 48%, and 84%, respectively. Given that the patterns of H pylori gastritis are different in Western (antral involvement) and Eastern countries (corpus involvement), these algorithms are likely to be specific to the population in which they were studied.74 Furthermore, given the accuracy and widespread availability of breath testing and stool antigen testing for active H pylori infection in Western countries, the potential clinical utility of this DL application remains uncertain.
Diagnosis of gastric cancer and premalignant gastric lesions
Several studies have evaluated the role of CNN algorithms to improve the detection of gastric cancer and premalignant conditions such as chronic atrophic gastritis and gastric polyps. One report described the development and evaluation of a CNN for the diagnosis of gastric cancer from endoscopic images.78 The training set comprised 13,584 images of gastric cancer; the algorithm was validated on an independent set of 2296 images of gastric cancer and normal areas of the stomach derived from 77 lesions in 69 patients. In 47 seconds, the CNN was able to identify 71 of 77 lesions accurately (sensitivity 92.2%), but 161 benign lesions were misclassified as cancer (positive predictive value of 31%). The 6 missed cancers were all well-differentiated cancers that were superficially depressed. Gastritis associated with mucosal surface irregularity or change in color tone was noted in nearly half of false-positive lesions. In another study, a CNN algorithm was designed to evaluate the depth of invasion of gastric cancer based on preoperative WLE images (developmental dataset 790 images, testing set 203 images) among a group of patients who underwent surgical or endoscopic resection.79 Compared with human endoscopists, the CNN system was able to differentiate early gastric cancer from deeper submucosal invasion with a higher accuracy (by 17.25%; 95% CI, 11.63%-22.59%) and specificity (by 32.21%; 95% CI, 26.78-37.44). Other CNN systems have been developed for detection of gastric polyps80 and chronic atrophic gastritis from images of the proximal stomach81 and distal stomach.82
An algorithm to detect gastric and esophageal cancer was developed based on a dataset of 1,036,496 images from 84,424 individuals and validated on both an external retrospective dataset (28,663 cancer and 783,876 control images) and prospective dataset (4317 cancer and 62,433 control images).83 This system, named the Gastrointestinal Artificial Intelligence Diagnostic System by the investigators and provided to participating institutions through a cloud-based AI platform, was tested in a multicenter, case-control study of 1102 cancer and 3.430 control images from 175 randomly selected patients and compared to the performance of human endoscopists. Diagnostic accuracy of the system was >90% in all datasets; sensitivity was similar to that of expert endoscopists (0.942 [95% CI, 0.924-0.957] vs 0.945 [95% CI, 0.927-0.959]; P > .05) and superior to trainee endoscopists (0.722; 95% CI, 0.691-0.752; P < .001). The authors propose that this system could be used to help nonexpert endoscopists improve detection of GI cancers.
Evaluation of esophageal cancer and dysplasia
CAD studies based on image analysis features for the endoscopic diagnosis of esophageal squamous cell neoplasia60 and Barrett’s neoplasia61,62 have used DL algorithms to enhance the detection of esophageal dysplasia and cancer. A CNN algorithm was developed to detect esophageal cancer (squamous and adenocarcinoma) on stored endoscopic WL and NBI images, using 8428 training images from 384 patients and a test set of 1111 images from 49 patients and 50 controls.84 The majority of the cancers in the test set were mucosal (T1a, 82%) and squamous cell histology (84%). The algorithm had a comprehensive sensitivity of 98% when evaluating both WL and NBI images; the sensitivity when evaluating only WL or NBI images was 81% and 89%, respectively. The positive predictive value was 40%, with the majority of false positives resulting from shadows or normal anatomic impressions on the esophageal lumen.
Endoscopic detection of dysplasia in Barrett’s esophagus (BE) is challenging. De Groof et al85 developed a DL CAD system trained on 1704 high-resolution WLE images from 669 patients with nondysplastic BE or early neoplasia, the latter defined as high-grade dysplasia or early esophageal adenocarcinoma (stage T1). They subsequently validated the CAD system on 3 independent datasets totaling 377 images. For the largest dataset, the CAD system correctly classified images as containing neoplastic BE (113 of 129 images) or nondysplastic BE (149 of 168 images) with primary outcome measures of accuracy, sensitivity, and specificity of 89%, 90%, and 88%, respectively. The CAD system also achieved greater accuracy (88% vs 73%) than any of the 53 general endoscopists. The system also correctly identified the optimal site for biopsy of the dysplastic BE in 92% to 97%, a performance that was similar to that of expert endoscopists. A real-time application of this CAD system during live EGD was reported in a small pilot study of 20 patients with nondysplastic BE (n = 10) and confirmed dysplastic BE (n = 10). The CAD system accurately identified Barrett’s neoplasia at a given level in the esophagus with 90% accuracy compared to expert assessment and histology.86 A second group developed a CNN program that detected early esophageal neoplasia in real time with a high level of accuracy.87 The CNN analyzed 458 test images (225 dysplasia and 233 nondysplastic) and correctly detected early neoplasia with a sensitivity of 96.4%, specificity of 94.2%, and accuracy of 95.4%.
Volumetric laser endomicroscopy (VLE) is a wide-field advanced imaging technology increasingly being used for evaluation of dysplasia in BE. VLE requires the user to view and analyze a large dataset of images (1200 images in cross-sectional frame over 90 seconds from a 6-cm segment of the esophagus) in real time. To address this challenge, a CAD system was developed that used 2 specific image features (loss of layering and increased subsurface signal intensity) that are part of clinical algorithms for dysplasia identification.88 In a dataset of VLE images with histologic correlation, the sensitivity and specificity of the algorithm were 90% and 93%, respectively, compared with 83% and 71% for experts. An AI-based, real-time image segmentation system added a third VLE image feature (hyporeflective structures representing dysplastic BE glands) to the 2 previously established image features predictive of dysplasia. The VLE regions of interest were marked by the AI algorithm with color overlays to facilitate study interpretation.89
Analysis of EUS images
CAD based on digital image analysis has been reported in the EUS evaluation of pancreatic masses, especially to differentiate pancreatic cancer (PC) from chronic pancreatitis (CP). However, there are limited studies in this area and no reports on the use of DL in the analysis of EUS images. In a single-center Chinese study, representative EUS images from 262 PC patients and 126 CP patients were used to develop an SVM algorithm based on pattern classification.90 On a random sample of images from the same dataset, the model had a sensitivity and specificity of 96.2% and 93.4%, respectively, for the identification of PC. In a similar study from a different institution in China, the authors developed an SVM algorithm to evaluate EUS images from 153 PC, 43 CP, and 20 normal control patients.91 Using a similar methodology of dividing the images into training and validation sets, the authors tested the performance characteristics of the algorithm after 50 trials and reported a sensitivity and specificity of 94% and 99%, respectively. Two earlier, smaller studies evaluated the role of digital image analysis using a handcrafted ANN model based on texture analysis or grayscale variations to differentiate PC from normal pancreas or CP, achieving similar results.92,93
CAD of EUS images when using ML has also been used in the differentiation of autoimmune pancreatitis from CP.94,95 Zhu et al94 evaluated the role of a novel image descriptor (local ternary pattern variance) as an additional tool to refine standard textural and feature analyses and then constructed an SVM algorithm based on those parameters. ML by the algorithm was conducted by using 200 randomized learning trials on a set of EUS images from patients with presumed autoimmune pancreatitis (n = 81) based on the HiSORT criteria96 and CP (n = 100). The sensitivity and specificity of the algorithm were reported at 84% and 93%, respectively.
EUS elastography has been used to characterize pancreatic masses and to differentiate PC from CP and benign from malignant lymph nodes.97 In a study of 68 patients with normal pancreas (n = 22), CP (n = 11), PC (n = 32), and pancreatic neuroendocrine tumors (n = 3), investigators calculated hue histograms derived from dynamic (video) sequences during EUS elastography.95 They subsequently built an ANN algorithm that used a multilayer perceptron model to evaluate these dynamic sequences to differentiate PC from CP. The model achieved an accuracy of 95% in differentiating benign versus malignant pancreatic masses and 90% for mass-forming pancreatitis versus PC. The authors further evaluated this technique in a prospective, multicenter study (13 academic centers in Europe) of 258 patients (211 PC and 47 CP patients).98 In this study, 3 videos of 10-second duration were collected during EUS elastography; processing and analyses were performed in a blinded manner at the primary institution. Patients either had a positive cytologic or surgical pathologic diagnosis or had clinical follow-up for 6 months. The algorithm had a sensitivity and specificity of 88% and 83%, respectively, with a positive predictive value of 96% and an NPV of 57%. The authors concluded that future real-time CAD systems based on these techniques may support real-time decision- making in the evaluation of pancreatic masses.
Areas for future research
Applications for AI in GI have been the subject of research for the past 2 decades, and these potentially transformative technologies are now poised to generate clinically useful and viable tools. The most promising applications appear to be real-time colonic polyp detection and classification. DL technologies for polyp detection with demonstrated real-time capability must be prospectively assessed during actual colonoscopies to confirm performance. Similarly, AI must be further evaluated for real-time lesion characterization (eg, neoplastic vs nonneoplastic, deep vs superficial submucosal invasion). The effect of AI on relevant clinical endpoints such as ADR, withdrawal times, PIVI endpoints, and cost effectiveness remains uncertain. However, AI-based applications that will have the potential to analyze the endoscopic image and recognize landmarks to enable direct measurement of quality metrics and to supplement the clinical documentation of the procedure appear likely.
Development and incorporation of these technologies into the GI endoscopy practice should also be guided by unmet needs and particularly focused on addressing areas of potential to enhance human performance through the use of AI. These could include applications in which a large number of images have to be analyzed or careful endoscopic evaluation of a wide surface area (eg, gastric cancer or CRC screening) needs to be performed, in which AI has the potential to increase the efficiency of human performance. Although investigators have typically sought to design CAD algorithms with high sensitivity and specificity, in practice highly sensitive but less specific CAD could be used as a “red flag” technology to improve detection of early lesions with confirmation by advanced imaging modalities or histology.
One of the critical needs to develop and test DL systems is the availability of large, reliably curated image and video datasets, as few such databases exist. Although this may require effort and cooperation among stakeholders, their availability would accelerate AI research in endoscopy. The evolution of current endoscopes and image processors into “smart” devices that are capable of analyzing and processing endoscopic data has potential. Critical appraisal of the improvement in patient outcomes, cost-effectiveness, safety, and the changes in clinical practice required to incorporate and implement these tools is required. Adopting these technologies will be associated with some cost burden; a corresponding reimbursement for their use will undoubtedly affect the rate of incorporation into clinical practice. Attention to the pitfalls and successes of the incorporation of AI in other fields (eg, radiology) may yield valuable lessons for its integration in GI endoscopy.
Summary
Rapid developments in computing power in the past few years have led to widespread use of AI in many aspects of human-machine interaction, including medical fields requiring analysis of large amounts of data. Although there has been active research in image analysis and CAD for many years, the recent availability of DL techniques such as CNN has facilitated the development of tools that promise to become an integral aid to physician diagnosis in the near future. These techniques are being explored in various aspects of GI endoscopy such as automated detection and classification of colorectal polyps, WCE interpretation, diagnosis of esophageal neoplasia, and pancreatic EUS, with the intent of developing real-time tools that inform physician diagnosis and decision-making. The widespread application of DL technologies across multiple aspects of GI endoscopy has the potential to transform clinical endoscopic practice positively.
Footnotes
Disclosure: Dr Pannala is a consultant for HCL Technologies and has received travel compensation from Boston Scientific Corporation. Dr Krishnan is a consultant for Olympus Medical. Dr Melson received an investigator-initiated grant from Boston Scientific and has stock options with Virgo Imaging. Dr Schulman is a consultant for Boston Scientific, Apollo Endosurgery, and MicroTech and has research funding from GI Dynamics. Dr Sullivan is a consultant and performs contracted research for Allurion Technologies, Aspire Bariatrics, Baronova, Obalon Therapeutics; is a consultant, performs contracted research, and has stock in Elira; is a consultant for USGI Medical, GI Dynamics, Phenomix Sciences Nitinotes, Spatz FGIA, and Endotools; and performs contracted research for Finch Therapeutics and ReBiotix. Dr Trikudanathan is a speaker and has received honorarium and travel from Boston Scientific, and is on the advisory board for Abbvie. Dr Trindade is a consultant for Olympus Corporation of the Americas and PENTAX of America, Inc., has received food and beverage from Boston Scientific, and has a research grant from NinePoint Medical, Inc. Dr Watson is a consultant and speaker for Apollo Endosurgery and Boston Scientific and a consultant for Medtronic and Neptune Medical Inc. Dr Lichtenstein is a consultant for Allergan Inc. and Augmenix; a speaker for Aries Pharmaceutical; a consultant and speaker for Gyrus ACMI, Inc., and Olympus Corporation of the Americas; and has received a tuition payment from Erbe USA Inc. All other authors disclosed no financial relationships.
Supplementary data
Colonoscopy with CADe (Skout, Iterative Scopes, Cambridge, Mass) real-time identification of a 3 mm ascending colon polyp as denoted by a bounding box.
The artificial intelligence system (combined CADe) automatically detects colonic polyps which are noted by the oval bounding box. The individual polyps are then interrogated with the endocytoscope and the AI system (CADx) characterizes the polyp as neoplastic or non-neoplastic with a probability level.
References
- 1.Tang A., Tam R., Cadrin-Chenevert A. Canadian Association of Radiologists white paper on artificial intelligence in radiology. Can Assoc Radiol J. 2018;69:120–135. doi: 10.1016/j.carj.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 2.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 3.Obermeyer Z., Emanuel E.J. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang F., Jiang Y., Zhi H. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2:230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel J.L., Goyal R.K. Applications of artificial neural networks in medical science. Curr Clin Pharmacol. 2007;2:217–226. doi: 10.2174/157488407781668811. [DOI] [PubMed] [Google Scholar]
- 6.Chartrand G., Cheng P.M., Vorontsov E. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 7.Jia Y, Shelhamer E, Donahue J, et al. Caffe: convolutional architecture for fast feature embedding. MM ’14: Proceedings of the 22nd ACM International Conference on Multimedia 2014:675-678.
- 8.Goodfellow I., Bengio Y., Courville A. MIT Press; Cambridge, MA: 2016. Deep learning. [Google Scholar]
- 9.François-Lavet V., Henderson P., Islam R. An introduction to deep reinforcement learning. Foundation and Trends in Machine Learning. 2018;11:219–354. [Google Scholar]
- 10.Shameer K., Johnson K.W., Glicksberg B.S. Machine learning in cardiovascular medicine: are we there yet? Heart. 2018;104:1156–1164. doi: 10.1136/heartjnl-2017-311198. [DOI] [PubMed] [Google Scholar]
- 11.Berzin T.M., Topol E.J. Adding artificial intelligence to gastrointestinal endoscopy. Lancet. 2020;395:485. doi: 10.1016/S0140-6736(20)30294-4. [DOI] [PubMed] [Google Scholar]
- 12.Wang P., Berzin T.M., Glissen Brown J.R. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori Y., Kudo S.E., Misawa M. Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 14.Corley D.A., Levin T.R., Doubeni C.A. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:2541. doi: 10.1056/NEJMc1405329. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S., Thosani N., Ladabaum U. Adenoma miss rates associated with a 3-minute versus 6-minute colonoscopy withdrawal time: a prospective, randomized trial. Gastrointest Endosc. 2017;85:1273–1280. doi: 10.1016/j.gie.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 16.Leufkens A.M., van Oijen M.G., Vleggaar F.P. Factors influencing the miss rate of polyps in a back-to-back colonoscopy study. Endoscopy. 2012;44:470–475. doi: 10.1055/s-0031-1291666. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Esparrach G., Bernal J., Lopez-Ceron M. Exploring the clinical potential of an automatic colonic polyp detection method based on the creation of energy maps. Endoscopy. 2016;48:837–842. doi: 10.1055/s-0042-108434. [DOI] [PubMed] [Google Scholar]
- 18.Tajbakhsh N., Gurudu S.R., Liang J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imaging. 2016;35:630–644. doi: 10.1109/TMI.2015.2487997. [DOI] [PubMed] [Google Scholar]
- 19.Urban G., Tripathi P., Alkayali T. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology. 2018;155:1069–1078.e8. doi: 10.1053/j.gastro.2018.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repici A., Badalamenti M., Maselli R. Efficacy of real-time computer-aided detection of colorectal neoplasia in a randomized trial. Gastroenterology. 2020;159:512–520.e7. doi: 10.1053/j.gastro.2020.04.062. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Liu P, Glissen Brown JR, et al. Lower adenoma miss rate of computer-aided detection-assisted colonoscopy vs routine white-light colonoscopy in a prospective tandem study. Gastroenterology. Epub 2020 Jun 17. [DOI] [PubMed]
- 22.Shahidi N., Rex D.K., Kaltenbach T. Use of endoscopic impression, artificial intelligence, and pathologist interpretation to resolve discrepancies between endoscopy and pathology analyses of diminutive colorectal polyps. Gastroenterology. 2020;158:783–785.e1. doi: 10.1053/j.gastro.2019.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Rex D.K., Kahi C., O'Brien M. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011;73:419–422. doi: 10.1016/j.gie.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 24.Ladabaum U., Fioritto A., Mitani A. Real-time optical biopsy of colon polyps with narrow-band imaging in community practice does not yet meet key thresholds for clinical decisions. Gastroenterology. 2013;144:81–91. doi: 10.1053/j.gastro.2012.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuiper T., van den Broek F.J., van Eeden S. New classification for probe-based confocal laser endomicroscopy in the colon. Endoscopy. 2011;43:1076–1081. doi: 10.1055/s-0030-1256767. [DOI] [PubMed] [Google Scholar]
- 26.Patel S.G., Schoenfeld P., Kim H.M. Real-time characterization of diminutive colorectal polyp histology using narrow-band imaging: implications for the resect and discard strategy. Gastroenterology. 2016;150:406–418. doi: 10.1053/j.gastro.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu Dayyeh B.K., Thosani N., Konda V. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015;81:502.e1–502.e16. doi: 10.1016/j.gie.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Takemura Y., Yoshida S., Tanaka S. Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest Endosc. 2010;72:1047–1051. doi: 10.1016/j.gie.2010.07.037. [DOI] [PubMed] [Google Scholar]
- 29.Tischendorf J.J., Gross S., Winograd R. Computer-aided classification of colorectal polyps based on vascular patterns: a pilot study. Endoscopy. 2010;42:203–207. doi: 10.1055/s-0029-1243861. [DOI] [PubMed] [Google Scholar]
- 30.Kominami Y., Yoshida S., Tanaka S. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643–649. doi: 10.1016/j.gie.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Gross S., Trautwein C., Behrens A. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354–1359. doi: 10.1016/j.gie.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Takemura Y., Yoshida S., Tanaka S. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video) Gastrointest Endosc. 2012;75:179–185. doi: 10.1016/j.gie.2011.08.051. [DOI] [PubMed] [Google Scholar]
- 33.Byrne M.F., Chapados N., Soudan F. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P.J., Lin M.C., Lai M.J. Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Mori Y., Kudo S.E., Chiu P.W. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110–1118. doi: 10.1055/s-0042-113609. [DOI] [PubMed] [Google Scholar]
- 36.Misawa M., Kudo S.E., Mori Y. Characterization of colorectal lesions using a computer-aided diagnostic system for narrow-band imaging endocytoscopy. Gastroenterology. 2016;150:1531–1532.e3. doi: 10.1053/j.gastro.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Mori Y., Kudo S.E., Wakamura K. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos) Gastrointest Endosc. 2015;81:621–629. doi: 10.1016/j.gie.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Kuiper T., Alderlieste Y.A., Tytgat K.M. Automatic optical diagnosis of small colorectal lesions by laser-induced autofluorescence. Endoscopy. 2015;47:56–62. doi: 10.1055/s-0034-1378112. [DOI] [PubMed] [Google Scholar]
- 39.Aihara H., Saito S., Inomata H. Computer-aided diagnosis of neoplastic colorectal lesions using 'real-time' numerical color analysis during autofluorescence endoscopy. Eur J Gastroenterol Hepatol. 2013;25:488–494. doi: 10.1097/MEG.0b013e32835c6d9a. [DOI] [PubMed] [Google Scholar]
- 40.Inomata H., Tamai N., Aihara H. Efficacy of a novel auto-fluorescence imaging system with computer-assisted color analysis for assessment of colorectal lesions. World J Gastroenterol. 2013;19:7146–7153. doi: 10.3748/wjg.v19.i41.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andre B., Vercauteren T., Buchner A.M. Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol. 2012;18:5560–5569. doi: 10.3748/wjg.v18.i39.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin E.H., Lee D., Bae J.H. Improved accuracy in optical diagnosis of colorectal polyps using convolutional neural networks with visual explanations. Gastroenterology. 2020;158:2169–2179.e8. doi: 10.1053/j.gastro.2020.02.036. [DOI] [PubMed] [Google Scholar]
- 43.Mori Y., Kudo S.E., Misawa M. Simultaneous detection and characterization of diminutive polyps with the use of artificial intelligence during colonoscopy. VideoGIE. 2019;4:7–10. doi: 10.1016/j.vgie.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori Y, Kudo SE, East JE, et al. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: an add-on analysis of a clinical trial (with video). Gastrointest Endosc. Epub 2020 Mar 30. [DOI] [PubMed]
- 45.Ikematsu H., Yoda Y., Matsuda T. Long-term outcomes after resection for submucosal invasive colorectal cancers. Gastroenterology. 2013;144:551–559. doi: 10.1053/j.gastro.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Yoda Y., Ikematsu H., Matsuda T. A large-scale multicenter study of long-term outcomes after endoscopic resection for submucosal invasive colorectal cancer. Endoscopy. 2013;45:718–724. doi: 10.1055/s-0033-1344234. [DOI] [PubMed] [Google Scholar]
- 47.Ferlitsch M., Moss A., Hassan C. Colorectal polypectomy and endoscopic mucosal resection (EMR): European Society of Gastrointestinal Endoscopy (ESGE) Clinical Guideline. Endoscopy. 2017;49:270–297. doi: 10.1055/s-0043-102569. [DOI] [PubMed] [Google Scholar]
- 48.Backes Y., Moss A., Reitsma J.B. Narrow band imaging, magnifying chromoendoscopy, and gross morphological features for the optical diagnosis of T1 colorectal cancer and deep submucosal invasion: a systematic review and meta-analysis. Am J Gastroenterol. 2017;112:54–64. doi: 10.1038/ajg.2016.403. [DOI] [PubMed] [Google Scholar]
- 49.Takeda K., Kudo S.E., Mori Y. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798–802. doi: 10.1055/s-0043-105486. [DOI] [PubMed] [Google Scholar]
- 50.Ito N., Kawahira H., Nakashima H. Endoscopic diagnostic support system for cT1b colorectal cancer using deep learning. Oncology. 2019;96:44–50. doi: 10.1159/000491636. [DOI] [PubMed] [Google Scholar]
- 51.Maeda Y., Kudo S.E., Mori Y. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video) Gastrointest Endosc. 2019;89:408–415. doi: 10.1016/j.gie.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 52.Takenaka K, Ohtsuka K, Fujii T, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. Epub 2020 Feb 12. [DOI] [PubMed]
- 53.Thakkar S., Carleton N.M., Rao B. Use of artificial intelligence-based analytics from live colonoscopies to optimize the quality of the colonoscopy examination in real time: proof of concept. Gastroenterology. 2020;158:1219–1221.e2. doi: 10.1053/j.gastro.2019.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J., Wu L., Wan X. A novel artificial intelligence system for the assessment of bowel preparation (with video) Gastrointest Endosc. 2020;91:428–435.e2. doi: 10.1016/j.gie.2019.11.026. [DOI] [PubMed] [Google Scholar]
- 55.Wang A., Banerjee S., Barth B.A. Wireless capsule endoscopy. Gastrointest Endosc. 2013;78:805–815. doi: 10.1016/j.gie.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 56.Zheng Y., Hawkins L., Wolff J. Detection of lesions during capsule endoscopy: physician performance is disappointing. Am J Gastroenterol. 2012;107:554–560. doi: 10.1038/ajg.2011.461. [DOI] [PubMed] [Google Scholar]
- 57.Segui S., Drozdzal M., Pascual G. Generic feature learning for wireless capsule endoscopy analysis. Comput Biol Med. 2016;79:163–172. doi: 10.1016/j.compbiomed.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 58.Iakovidis D.K., Koulaouzidis A. Software for enhanced video capsule endoscopy: challenges for essential progress. Nat Rev Gastroenterol Hepatol. 2015;12:172–186. doi: 10.1038/nrgastro.2015.13. [DOI] [PubMed] [Google Scholar]
- 59.Liedlgruber M., Uhl A. Computer-aided decision support systems for endoscopy in the gastrointestinal tract: a review. IEEE Rev Biomed Eng. 2011;4:73–88. doi: 10.1109/RBME.2011.2175445. [DOI] [PubMed] [Google Scholar]
- 60.Jia X., Meng M.Q. A deep convolutional neural network for bleeding detection in wireless capsule endoscopy images. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:639–642. doi: 10.1109/EMBC.2016.7590783. [DOI] [PubMed] [Google Scholar]
- 61.Malagelada C., Drozdzal M., Segui S. Classification of functional bowel disorders by objective physiological criteria based on endoluminal image analysis. Am J Physiol Gastrointest Liver Physiol. 2015;309:G413–G419. doi: 10.1152/ajpgi.00193.2015. [DOI] [PubMed] [Google Scholar]
- 62.Leenhardt R., Li C., Le Mouel J.P. CAD-CAP: a 25,000-image database serving the development of artificial intelligence for capsule endoscopy. Endosc Int Open. 2020;8:E415–E420. doi: 10.1055/a-1035-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan Y., Li B., Meng M.Q. Bleeding frame and region detection in the wireless capsule endoscopy video. IEEE J Biomed Health Inform. 2016;20:624–630. doi: 10.1109/JBHI.2015.2399502. [DOI] [PubMed] [Google Scholar]
- 64.Fu Y., Zhang W., Mandal M. Computer-aided bleeding detection in WCE video. IEEE J Biomed Health Inform. 2014;18:636–642. doi: 10.1109/JBHI.2013.2257819. [DOI] [PubMed] [Google Scholar]
- 65.Yuan Y., Meng M.Q. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys. 2017;44:1379–1389. doi: 10.1002/mp.12147. [DOI] [PubMed] [Google Scholar]
- 66.Leenhardt R., Vasseur P., Li C. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189–194. doi: 10.1016/j.gie.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 67.Aoki T., Yamada A., Aoyama K. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357–363.e2. doi: 10.1016/j.gie.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 68.Fan S., Xu L., Fan Y. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol. 2018;63:165001. doi: 10.1088/1361-6560/aad51c. [DOI] [PubMed] [Google Scholar]
- 69.He J.Y., Wu X., Jiang Y.G. Hookworm detection in wireless capsule endoscopy images with deep learning. IEEE Trans Image Process. 2018;27:2379–2392. doi: 10.1109/TIP.2018.2801119. [DOI] [PubMed] [Google Scholar]
- 70.Ding Z., Shi H., Zhang H. Gastroenterologist-level identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology. 2019;157:1044–1054.e5. doi: 10.1053/j.gastro.2019.06.025. [DOI] [PubMed] [Google Scholar]
- 71.Takiyama H., Ozawa T., Ishihara S. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci Rep. 2018;8:7497. doi: 10.1038/s41598-018-25842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu L., Zhang J., Zhou W. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut. 2019;68:2161–2169. doi: 10.1136/gutjnl-2018-317366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen D., Wu L., Li Y. Comparing blind spots of unsedated ultrafine, sedated, and unsedated conventional gastroscopy with and without artificial intelligence: a prospective, single-blind, 3-parallel-group, randomized, single-center trial. Gastrointest Endosc. 2020;91:332–339.e3. doi: 10.1016/j.gie.2019.09.016. [DOI] [PubMed] [Google Scholar]
- 74.Shichijo S., Nomura S., Aoyama K. Application of convolutional neural networks in the diagnosis of helicobacter pylori infection based on endoscopic images. EBioMedicine. 2017;25:106–111. doi: 10.1016/j.ebiom.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Itoh T., Kawahira H., Nakashima H. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open. 2018;6:E139–E144. doi: 10.1055/s-0043-120830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng W., Zhang X., Kim J.J. High accuracy of convolutional neural network for evaluation of Helicobacter pylori infection based on endoscopic images: preliminary experience. Clin Transl Gastroenterol. 2019;10:e00109. doi: 10.14309/ctg.0000000000000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shichijo S., Endo Y., Aoyama K. Application of convolutional neural networks for evaluating Helicobacter pylori infection status on the basis of endoscopic images. Scand J Gastroenterol. 2019;54:158–163. doi: 10.1080/00365521.2019.1577486. [DOI] [PubMed] [Google Scholar]
- 78.Hirasawa T., Aoyama K., Tanimoto T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653–660. doi: 10.1007/s10120-018-0793-2. [DOI] [PubMed] [Google Scholar]
- 79.Zhu Y., Wang Q.C., Xu M.D. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806–815.e1. doi: 10.1016/j.gie.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 80.Zhang X., Chen F., Yu T. Real-time gastric polyp detection using convolutional neural networks. PloS One. 2019;14 doi: 10.1371/journal.pone.0214133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guimaraes P., Keller A., Fehlmann T. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4–6. doi: 10.1136/gutjnl-2019-319347. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y., Li F., Yuan F. Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Dig Liver Dis. 2020;52:566–572. doi: 10.1016/j.dld.2019.12.146. [DOI] [PubMed] [Google Scholar]
- 83.Luo H., Xu G., Li C. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645–1654. doi: 10.1016/S1470-2045(19)30637-0. [DOI] [PubMed] [Google Scholar]
- 84.Horie Y., Yoshio T., Aoyama K. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25–32. doi: 10.1016/j.gie.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 85.de Groof A.J., Struyvenberg M.R., van der Putten J. Deep-learning system detects neoplasia in patients with Barrett's esophagus with higher accuracy than endoscopists in a multistep training and validation study with benchmarking. Gastroenterology. 2020;158:915–929.e4. doi: 10.1053/j.gastro.2019.11.030. [DOI] [PubMed] [Google Scholar]
- 86.de Groof A.J., Struyvenberg M.R., Fockens K.N. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video) Gastrointest Endosc. 2020;91:1242–1250. doi: 10.1016/j.gie.2019.12.048. [DOI] [PubMed] [Google Scholar]
- 87.Hashimoto R., Requa J., Dao T. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video) Gastrointest Endosc. 2020;91:1264–1271.e1. doi: 10.1016/j.gie.2019.12.049. [DOI] [PubMed] [Google Scholar]
- 88.Swager A.F., van der Sommen F., Klomp S.R. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839–846. doi: 10.1016/j.gie.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 89.Trindade A.J., McKinley M.J., Fan C. Endoscopic surveillance of Barrett's esophagus using volumetric laser endomicroscopy with artificial intelligence image enhancement. Gastroenterology. 2019;157:303–305. doi: 10.1053/j.gastro.2019.04.048. [DOI] [PubMed] [Google Scholar]
- 90.Zhu M., Xu C., Yu J. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. PloS One. 2013;8:e63820. doi: 10.1371/journal.pone.0063820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang M.M., Yang H., Jin Z.D. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest Endosc. 2010;72:978–985. doi: 10.1016/j.gie.2010.06.042. [DOI] [PubMed] [Google Scholar]
- 92.Das A., Nguyen C.C., Li F. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest Endosc. 2008;67:861–867. doi: 10.1016/j.gie.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 93.Norton I.D., Zheng Y., Wiersema M.S. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625–629. doi: 10.1067/mge.2001.118644. [DOI] [PubMed] [Google Scholar]
- 94.Zhu J., Wang L., Chu Y. A new descriptor for computer-aided diagnosis of EUS imaging to distinguish autoimmune pancreatitis from chronic pancreatitis. Gastrointest Endosc. 2015;82:831–836.e1. doi: 10.1016/j.gie.2015.02.043. [DOI] [PubMed] [Google Scholar]
- 95.Saftoiu A., Vilmann P., Gorunescu F. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086–1094. doi: 10.1016/j.gie.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 96.Chari S.T., Smyrk T.C., Levy M.J. Diagnosis of autoimmune pancreatitis: the Mayo Clinic experience. Clin Gastroenterol Hepatol. 2006;4:1010–1016. doi: 10.1016/j.cgh.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 97.Giovannini M. Endoscopic ultrasound elastography. Pancreatology. 2011;11(Suppl 2):34–39. doi: 10.1159/000323496. [DOI] [PubMed] [Google Scholar]
- 98.Saftoiu A., Vilmann P., Gorunescu F. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84–90.e1. doi: 10.1016/j.cgh.2011.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Colonoscopy with CADe (Skout, Iterative Scopes, Cambridge, Mass) real-time identification of a 3 mm ascending colon polyp as denoted by a bounding box.
The artificial intelligence system (combined CADe) automatically detects colonic polyps which are noted by the oval bounding box. The individual polyps are then interrogated with the endocytoscope and the AI system (CADx) characterizes the polyp as neoplastic or non-neoplastic with a probability level.