Abstract
Nrf2 (NFE2L2 – nuclear factor (erythroid-derived 2)-like 2) is a transcription factor, which is repressed by interaction with a redox-sensitive protein Keap1 (Kelch-like ECH-associated protein 1). Deregulation of Nrf2 transcriptional activity has been described in the pathogenesis of multiple diseases, and the Nrf2/Keap1 axis has emerged as a crucial modulator of cellular homeostasis. Whereas the significance of Nrf2 in the modulation of biological processes has been well established and broadly discussed in detail, the focus on Keap1 rarely goes beyond the regulation of Nrf2 activity and redox sensing. However, recent studies and scrutinized analysis of available data point to Keap1 as an intriguing and potent regulator of cellular function. This review aims to shed more light on Keap1 structure, interactome, regulation and non-canonical functions, thereby enhancing its significance in cell biology. We also intend to highlight the impact of balance between Keap1 and Nrf2 in the maintenance of cellular homeostasis.
Keywords: Keap1, Nrf2, interacting proteins, non-canonical functions
Graphical Abstract
Introduction
The fundamental role of defence pathways is to integrate adaptive cellular responses to stress events. These highly coordinated strategies aim to counteract internal disturbances, minimise acute damage, reset the homeostasis of the cell and sustain its survival. The Nrf2/Keap1 axis has been recognised as a central node for a cross-talk of cellular defence and survival pathways. Nrf2 (nuclear factor (erythroid-derived 2)-like 2) is a transcription factor, which transactivates expression of over 1000 protective genes [1,2], which comprise more than 1% of the human genome [3]. Its transcriptional activity is mainly regulated by a redox-sensitive repressor Keap1 (Kelch-like ECH-associated protein 1) [4]. Many excellent reviews deeply describe the exact mechanisms of Nrf2 regulation and its protective effects on the cellular homeostasis (exhaustively reviewed in [5]). Recent research has been broadly focused on the manipulation of Nrf2-Keap1 interaction, which enables modulation of Nrf2 activity in the therapeutic approaches [6]. The role of Keap1, however, has been attributed mainly to Nrf2 repressor function and redox sensor. Still, thanks to its extraordinary structure and reactivity, Keap1 undergoes numerous posttranslational modifications, has a very rich interactome, and, thus, is involved in various cellular processes. We aim to underline this atypical biological significance of Keap1 in this review.
Keap1 résumé
Keap1, encoded by the KEAP1 gene, was discovered in 1999. A two-hybrid screening assay identified it as a protein repressing Nrf2 transcriptional activity, sensitive to electrophiles [4]. It is also an intracellular sensor of oxidants [7]. KEAP1 evolved as a product of gene duplication. In fish, two isoforms of keap1 gene, keap1a and keap1b, are distinguished. During evolution, they separated, and keap1b was coevolving with vertebrate homologues [8,9]. Keap1 is a well-conserved protein, sharing around 95% of sequence homology between species (data from NCBI) and, importantly, possessing the same function throughout the phylogenic tree among the vertebrates [10]. The Ensembl database reports that there are 10 splicing variants of human Keap1 gene, out of which 2 alternatively spliced transcript variants, possessing 6 exons and 5 introns, encode the same protein isoform, which is the full-length 624 amino acid long. Six other transcript variants encode potential isoforms (116, 129, 172, 212, 213 and 279 amino acid long), which were computationally mapped (uniprot.org). The last 2 transcript variants encode no protein. Recently 444 amino acid long Keap1 isoform, derived from the alternative splicing variant lacking the fourth and fifth exons, has been described in human highly-metastatic hepatoma cells and other cell lines. This isoform is missing the double glycine repeat (DGR, Kelch) domain, which is responsible for interactions with Nrf2 [11].
Keap1 is expressed widely in various cell types and tissues [4]. It is mostly located in the perinuclear region of the cytoplasm, but also the nucleus, endoplasmic reticulum and inclusion bodies [12–14]. Its absolute amount ranges between 50,000 to 300,000 molecules per cell, which corresponds to ~1 μM concentration [15]. That makes it a moderately abundant protein relative to another well-known repressor and a component of E3 ubiquitin ligase hDM2 (up to 200 000 copies) [16] or transcription factors (up to 100 000 copies) [17]. It could partially explain how it preferentially is activated by Michael addition, its usual mode of redox activation [18]. Human Keap1 contains 27 cysteines (Tab.1), which comprise 4.33% of all amino acids, whereas the average content of cysteines in human proteins is 2.26% [19]. Since the median length of coding sequences of human proteins is 375 amino acids [20], the average cysteine content counts for 8 per protein, which means that Keap1 contains over two times more cysteines than the average protein in human. This fact is of particular significance since the cysteine proteome plays a fundamental role in many biological functions (please refer to [21] for more details). A large part of Keap1 cysteines is highly reactive due to surrounding positively-charged, basic amino-acids [22]. The electrophile compounds activating Nrf2 modify directly critical Keap1 cysteine residues by alkylation and oxidoreduction reactions [23]. They can be divided into four classes, depending on Keap1 cysteine preferences [24]. It was shown that the oxidants, such as H2O2 and HClO, might induce Keap1 transient oxidation, but its mechanism remains elusive [25]. Interestingly, the study carried out it a zebrafish model showed that H2O2 is capable of Nrf2 target gene (gstp1) induction in an Nrf2-dependent manner in larvae but not in embryonic cells. Therefore, it was proposed that factors additional to Nrf2 and Keap1, absent at the embryonic stage, are required for sensing of H2O2 [23]. Keap1 is a metalloprotein binding zinc, which modulates the reactivity of Keap1 critical cysteines, thus influencing Nrf2-dependent response [26]. Coordination of zinc by Keap1 may explain how it can react with H2O2 preferentially versus most other protein cysteines under physiological conditions [27]. The breaking of the O-O bond in H2O2 cannot be significantly achieved under physiological conditions by cysteine alone, even in its thiolate (S−) form because OH− is an extremely poor leaving group. Protonation of the leaving OH− or the proximity of a metal, such as Zn2+, to which the OH− could bind, facilitates the oxidation of cysteine. It is well established that Keap1 represents a sensor of environmental stress [9], creates an adaptive interface between the exposome and genome [21], and is a target for pharmacological modification as it has been comprehensively reviewed in [6,7].
Table. 1.
Keap1 is highly rich in cysteines. Comparison between human, mouse and rat.
| Human | Mouse | Rat | |
|---|---|---|---|
| gene | KEAP1 | Keap1 | Keap1 |
| aliases | iNrf2, KLHL19 | iNrf2, mKIAA0132 | iNrf2 |
| chromosome | 19 | 9 | 8 |
| cysteines | 27 | 25 | 25 |
| amino acids | 624 | 624 | 624 |
Keap1 structure
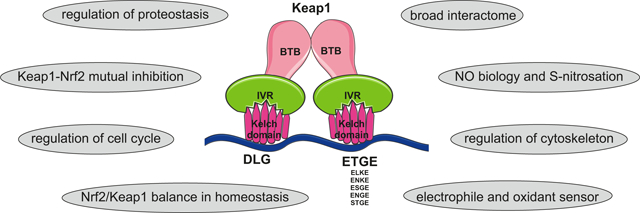
Keap1 is a member of the BTB-kelch protein family, which comprises about 50 members classified as Kelch-like (KLHL1–42, Keap1 is classified as KLHL19) or Kelch, and possessing BTB domain 1–14 (KBTBD1–14). All of these proteins contain two canonical domains in their structure: BTB (Broad-Complex, Tramtrack and Bric a brac) and Kelch domain [28,29]. Five distinct regions can be distinguished in the structure of Keap1: (i) the N-terminal region (NTR), position 1–49 aa, (ii) BTB (also named poxvirus and zinc finger (POZ)) domain, position 50–179 aa, (iii) intervening region (IVR, also named BACK domain), position 180–314 aa, (iv) double-glycine repeat (DGR, Kelch) domain, positions 327–611 aa, and (v) C-terminal region (CTR), position 612–624 aa [7], as depicted in Fig. 1A.
Fig. 1. Keap1 structure.
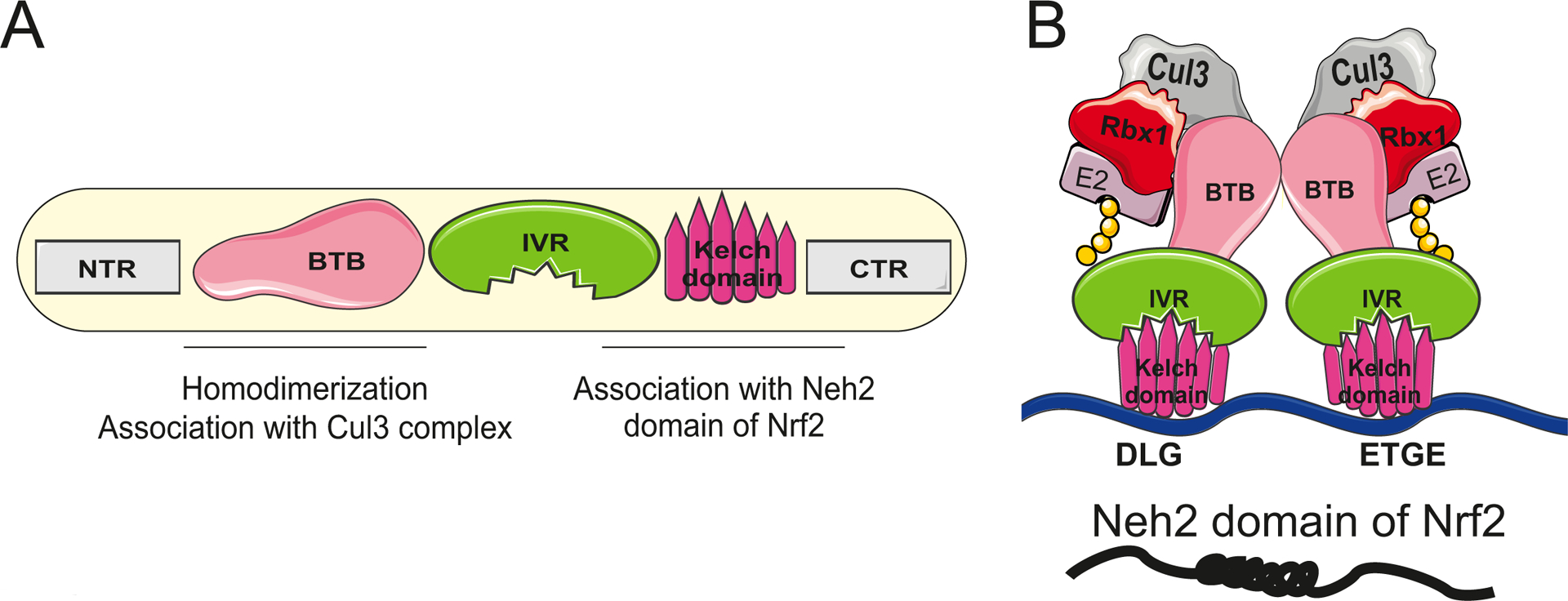
(A) The organisation of Keap1 domains. NTR – N terminal region. BTB - Broad-Complex, Tramtrack and Bric a brac. IVR- intervening region. DGR - double-glycine repeat region. CTR – C terminal region; (B) Schematic illustration of the Keap1-Cul3-E3 ligase complex interacting with the Neh2 domain of Nrf2 via the DLG and ETGE motifs. Cul3 – cullin3; Rbx1 – RING box protein 1; E2 – Ubiquitin ligase with ubiquitin (yellow circles).
The BTB domain is an evolutionarily conserved motif with the multifunctional role, including mediation of Keap1 homodimerisation, its interactions with the Cullin-3-Rbx-1-E3 ligase complex, and sensing electrophilic compounds, including the most potent activators of Nrf2, the cyclic cyanoenones, and nitric oxide through a highly reactive cysteine residue (C151) [9,30–34]. The major determinants of the high reactivity of this cysteine are basic amino acids, K131, R135, and K150, and H154, localised in the spatial proximity to C151 [9,25,35]. Keap1 homodimerisation via BTB domain is required for the interaction with Nrf2 [36], which is achieved directly by Kelch domain of Keap1 and Neh2 domain of Nrf2 through its DLG and ETGE motifs (Fig. 1B) [37].
The IVR region (position 209–314 aa) is cysteine-rich. Two critical cysteine residues C273 and C288 are required for the repression of Nrf2 nuclear accumulation [38,39] and responsible for sensing of alkenals [9]. The IVR region contains the most reactive residues of Keap1 (C257, C272, C288, and C297), which are the direct redox sensors [18]. They are a target for pharmacological activation of this pathway as it is reviewed in [6,7].
The DGR domain contains six repeats of Kelch motif, which form a six-bladed β-propeller structure. Each blade of this structure (I–VI) is built of four-stranded antiparallel ß-sheets (A–D), which enable Keap1 interactions with diverse proteins [40]. The sequences of Kelch repeats differ between particular KLHL1–42, providing the substrate selectivity [28]. However, some regions are conserved: DGR, present at the end βB strand along with individual tyrosine (βC) and tryptophan (βD) residues, arbitrates hydrophobic packing between blades. Thus, the Kelch domain is also referred to as DGR or DC (DGR and CTR) domain [41]. Those similar features enable the maintenance of overall “kelch” shape. The vast majority of kelch domains (just as in the case of Keap1) form six-bladed beta-propellers [29].
The X-ray structures of Kelch [40,42,43] and BTB domains [44] have provided essential insights in their detailed structure. The spatial organisation of all domains has been revealed by cryogenic electron microscopy. Keap1 resembles a cherry bob, where BTB domains form a stem, whereas IVR and Kelch domains are closely located and form the spherical “cherry” part (for the structural details, please refer to [45]).
Regulation of Keap1 expression, level and activity
The level and activity of Keap1 are regulated via several mechanisms: (i) transcriptional regulation, (ii) epigenetic modifications, (iii) miRNAs, (iv) somatic mutations, (v) post-translational modifications, and (vi) degradation (Fig. 2).
Fig. 2. The modes of Keap1 regulation.
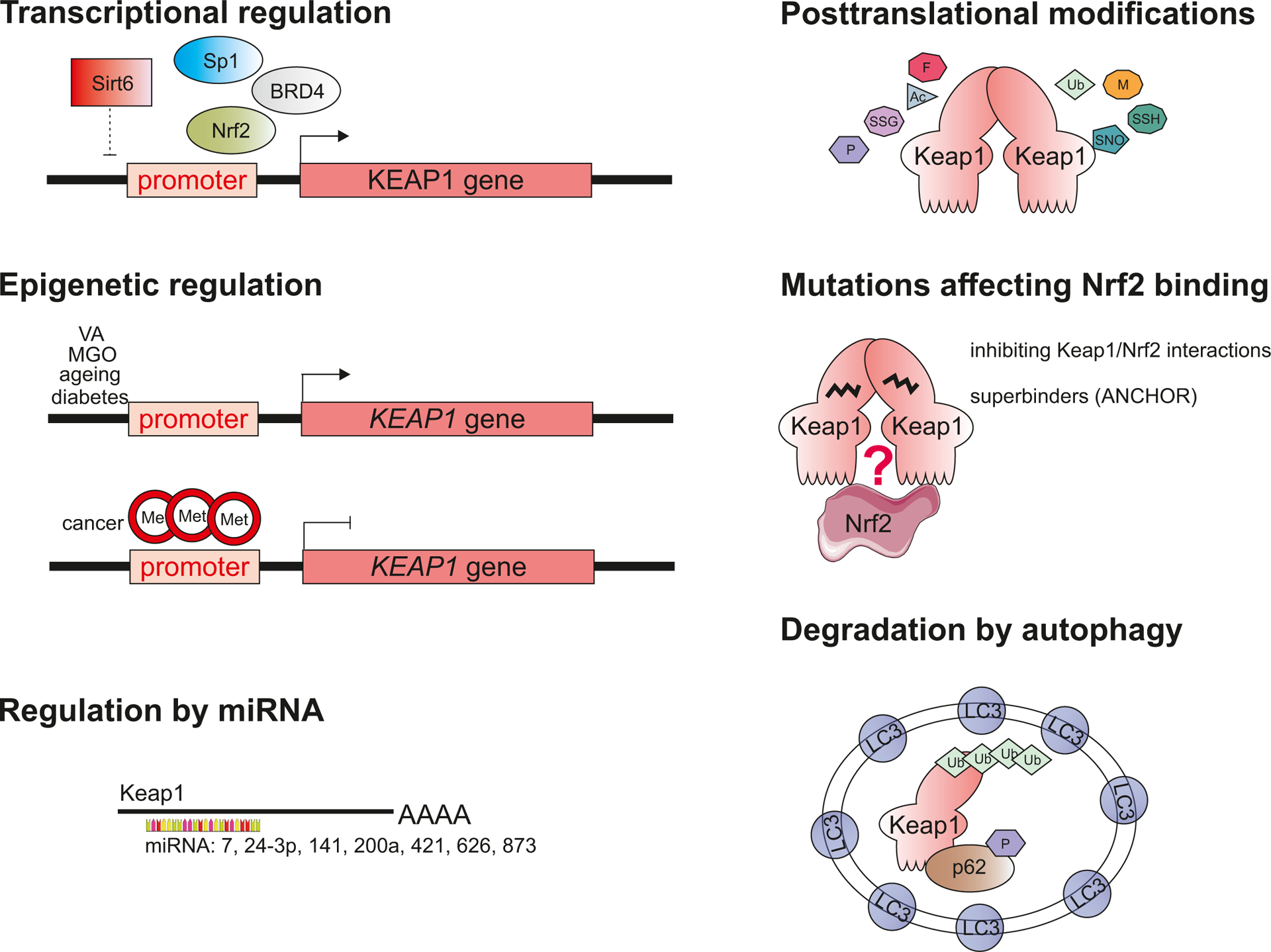
The level and activity of Keap1 are regulated via several mechanisms: transcription factors, epigenetic modifications, miRNAs, somatic mutations, post-translational modifications, and degradation. VA – valproic acid, MGO – methylglyoxal.
KEAP1 is a transcriptional target of BRD4 [46,47], SP1 [48,49], Nrf2 [50] and possibly AP2 [49]. Also, the presence of putative binding sites for transcription factors: hepatic nuclear factor (HNF-1), signal transducer and activator of transcription-6 (STAT-6), CCAAT/enhancer-binding protein (C/EBP), OCT1, aryl hydrocarbon receptor (Ahr)/Arnt heterodimers, and hypoxia-inducible factor (HIF), has been identified in murine Keap1 gene [50]. On the contrary, sirtuin 6 (Sirt6) deacetylase was shown to decrease Keap1 transcript in cardiomyocytes. The molecular mechanism of this suppression remains, however, unknown [51].
The level of Keap1 transcript is affected by differential methylation of its promoter. Keap1 increases with age in the human lens due to demethylation of its promoter, which results in higher gene expression [52]. Similar regulation is observed in diabetic conditions and in response to several compounds like methylglyoxal (MGO) [53] or valproic acid [54,55]. On the other hand, hypermethylation of the promoter is observed in many types of cancers, including lung, colon, prostate and gliomas. It also correlates with disease progression [48,49,56–59]. The promoter hypermethylation perturbs Keap1 transcription and significantly decreases its protein level [49,59]. Keap1 level can also be negatively regulated by miR-7 [60], miR-24–3p [61], miR-141 [62,63], miR-200a [64,65], miR-421 [66], miR-626 [67], and mir-873 [68].
Numerous mutations characterise the coding region of KEAP1 gene in cancer cells [69,70]. They are associated with poor prognosis as they promote the proliferation of cancer cells and cause resistance to chemotherapy in different types of cancers [reviewed in [71,72]]. Interestingly, they are spanned uniformly across the whole gene and rarely cause protein truncation. Instead, the mutations result in altered ability to bind and regulate Nrf2 degradation, usually abrogating and decreasing its binding capacity. However, there are some mutations within Keap1 which may facilitate Keap1 and Nrf2 interactions - called ‘superbinders’ (R320Q, R470C, G423V, D422N, G186R, S243C and V155F). However, despite strong binding and enhanced ubiquitination, Keap1 mutant disables Nrf2 degradation, leading to increased Nrf2 transcriptional activity [73]. This group of mutants is also called ‘ANCHOR’ (Additionally NRF2-Complexed HypomORph). Interestingly, the anchor relates only to the increased interaction with Nrf2, but not with other Keap1-interacting proteins, for which the relation is comparable to the wild-type. The ANCHOR mutations stabilise Keap1 and phase-separate in a sequestosome (SQSTM1, p62)-dependent manner [74].
Due to its reactive nature, Keap1 undergoes many post-translational modifications, including S-nitrosation, ubiquitination, alkylation, carbonylation, glycosylation, S-glutathionylation, oxidation, phosphorylation, succination, and sulfhydrylation. They serve as an alternative way of non-canonical Nrf2-activation (summarised in Tab. 2). Curiously, two of them, ubiquitination and S-nitrosation, not only modify the Keap1 residues but are also directly dependent on Keap1 [30,75]. In regard to ubiquitination, Keap1 is an adaptor protein for E3 ligase complex [30–33], which ubiquitinates cellular proteins, including Nrf2 [32], IKKβ [76,77], PGAM5 [78], Miro2 [79], MCM3, SLK, MAD2L1 [80], and possibly also Myo9b [81]. Keap1 itself also undergoes ubiquitination, which predisposes it to proteasomal-independent degradation and enables Nrf2 activation [82]. In accordance, the deubiquitinating enzyme USP15 negatively regulates Nrf2 through deubiquitination of Keap1 [83]. Concerning the S-nitrosation-Keap1 cross-talk, Keap1 may undergo S-nitrosation (SNO) at C151 and C273 residues [84–86], and recently it has been recognised as a crucial regulator of this post-translational modification [75] (further described in more detail). Keap1 also evolved as a sensor of nitric oxide [9]. Another type of feedback regulation is observed in case of MGO, which modifies Keap1 to form MICA (methylimidazole crosslink between proximal cysteine and arginine residues) and functionally inactivates it [87]. At the same time, treatment of cells with MGO induces Keap1 promoter DNA demethylation leading to overexpression of Keap1 mRNA and protein [88]. There are several post-translational modifications (malonylation, palmitoylation and acetylation), which have been reported, however, the exact impact on Keap1 was not fully elucidated.
Table 2.
Post-translational modifications of Keap1.
| Modification | Residue | Increase in Nrf2 activity | Reference |
|---|---|---|---|
| Ubiquitination* | K39, K61, K84, K97, K108, K287, K312, K615 | Yes (essential for Keap1 degradation) | [82,89] |
| S-nitrosation* | C151, C273 | Yes | [9,84–86] |
| Alkylation (itaconate) | C151, C257, C273, C288, C297 | Yes (modifies and lowers Keap1 level) | [90] |
| Carbonylation | C273, C288 | Yes | [91] |
| Glycosylation | S104 | No (crucial in mediating Nrf2 ubiquitination) | [92] |
| S-glutathionylation | C434 | Yes | [93,94] |
| MICA (methylimidazole crosslink between proximal cysteine and arginine residues) | MICA crosslink between C151 and R135 | Yes | [87] |
| Nitro fatty acids | C38, C226, C257. C273, C288, C489 | Yes | [95] |
| Oxidation | disulphide between C226 and C613; intramolecular between C151 | Yes | [25,26] |
| Phosphorylation | Y206, Y263, Y334, | Yes (enabling Keap1 nuclear export) | [96] |
| Succination | C151, C288 | Yes | [97–99] |
| Sulfhydrylation | C151 | Yes | [100] |
| Malonylation | K131 | n.d. | [101] |
| Acetylation | K131 | n.d. | [102] |
| Palmitoylation | n.d. | n.d. | [103] |
- the modifications that are both mediated by Keap1 and exerted on Keap1 protein itself.
Degradation of Keap1 occurs via p62-dependent autophagy [104], which is facilitated upon its preceding ubiquitination [82]. In basal conditions, the half-life of Keap1 is ~13h, whereas electrophiles, such as tert-butylhydroquinone (tBHQ) or 1,2-naphthoquinone (1,2-NQ) can accelerate its degradation. The half-life of modified Keap1 is shortened to 3.4h and 7.1h by tBHQ and 1,2-NQ, respectively [104].
Keap1 interactome
Nrf2 is a hallmark interacting protein for Keap1. Many methods were implemented to study this interaction and to establish the minimal interacting fragment, including surface plasmon resonance [105], fluorescence polarisation assay [106], FRET [107], and ITC [108]. They revealed that Nrf2 binds to Keap1 in stoichiometry 1:2. Neh2 domain of Nrf2 is recruited to Keap1 by ETGE and DLG motifs [109]. Both motifs interact with the same residues of Keap1 dimer (DGR/Kelch), however with different binding affinities, probably due to different acidic residue composition. The ETGE motif has 100 times higher affinity than DLG [43,110]. This interaction can be mimicked by 16-aa long fragment, containing Nrf2 ETGE motif [107,111], or even 9 aa- (LDEETGEFL) or 7 aa-long (DEETGEF) sequences [105,106]). A minimal Nrf2 recognition sequence is of 6–7 amino acids [112].
The Nrf2/Keap1 complex adopts two different conformations, sequentially alternating in a cycle in basal conditions [3]. The open conformation is formed by newly synthesised Nrf2, which binds to one Keap1 molecule of the ‘cherry bob’ Keap1 dimer via high-affinity ETGE motif. This does not allow for Nrf2 ubiquitination and protects Nrf2 from proteasomal degradation. In the next step, low-affinity DLG motif of Nrf2 binds to the second member of Keap1 dimer. Thus, the complex adopts a closed conformational state, which predisposes Nrf2 for Keap1/Cul3/E3 ligase-dependent polyubiquitination and subsequent proteasomal degradation. The free Keap1 enters the next cycle then [3]. It is postulated that the inducers of Nrf2 activity block Nrf2/Keap1 complex in the closed conformation, but Nrf2 ubiquitination is not feasible then, and Nrf2 remains trapped by Keap1. Therefore, transactivation of Nrf2 target genes in achieved by de novo synthesised Nrf2, rather than through the liberation of Nrf2 from Keap1 complex [3].
The majority of Keap1-binding partners contain an evolutionally conserved ETGE motif, which is responsible for their interactions (Fig. 3) [113,114]. Direct interaction of Keap1 with other proteins via the non-canonical motifs, such as ELKE/ENKE, GLNLG, ARM domain, was also found (Tab. 3). Binding of the ETGE-containing proteins to Keap1 can displace Nrf2, preventing its degradation and enabling its translocation to the nucleus and the expression of cytoprotective genes. The interactions of Keap1 with other proteins and interaction motifs were discovered in the pull-down assay or in high-throughput studies and further confirmed by mutations and exchange of amino acids. Some of the interactors were also denoted in the Spotlite database [115]. The Keap1 interactors p62, DPP3 and Bcl-xL were also determined in cDNA screenings based on ARE-driven transcriptional activity. Another, later neither described nor verified in context of Nrf2, were D-site of albumin promoter binding protein (DBP), kinesin family member 26B (KIF26B), cAMP-responsive element-binding protein-regulated transcription coactivator 1 (TORC1), myeloid cell leukaemia sequence 1 (MCL1), and splicing factor, arginine/serine-rich 10 (SFRS10) [116]. Keap1 interactors verified by biochemical methods are presented in Table 3. All the depicted proteins, similarly to Nrf2, interact with Keap1 via its Kelch domains. The compounds disrupting the Keap1/Nrf2 interaction (e.x. sulforaphane, tBHQ or H2O2) were shown to have no effect or to increase interactions of Keap1 with its binding partners (Tab. 4). A significant part of Keap1-interacting proteins is characterised by the presence of disordered regions, which contain many acidic amino acids [117]. Kelch domain contains multiple positively-charged residues. Thus, such an interaction is favoured [118]. Additionally, previously reported Keap1-interacting proteins, like myosin VIIa [119], cortactin [120,121], RhoGAP1 [122,123], contain SH3 domains. Apart from interaction with eukaryotic proteins, Keap1 was shown to interact with Marburg virus proteins [124]. Additionally, there are also small molecules that bind to the Kelch domain [125]. Mass spectrometry-identified proteins interacting with Keap1 are listed in [80,113,126,127].
Fig. 3. Alignment of the confirmed Keap1 interaction motifs.
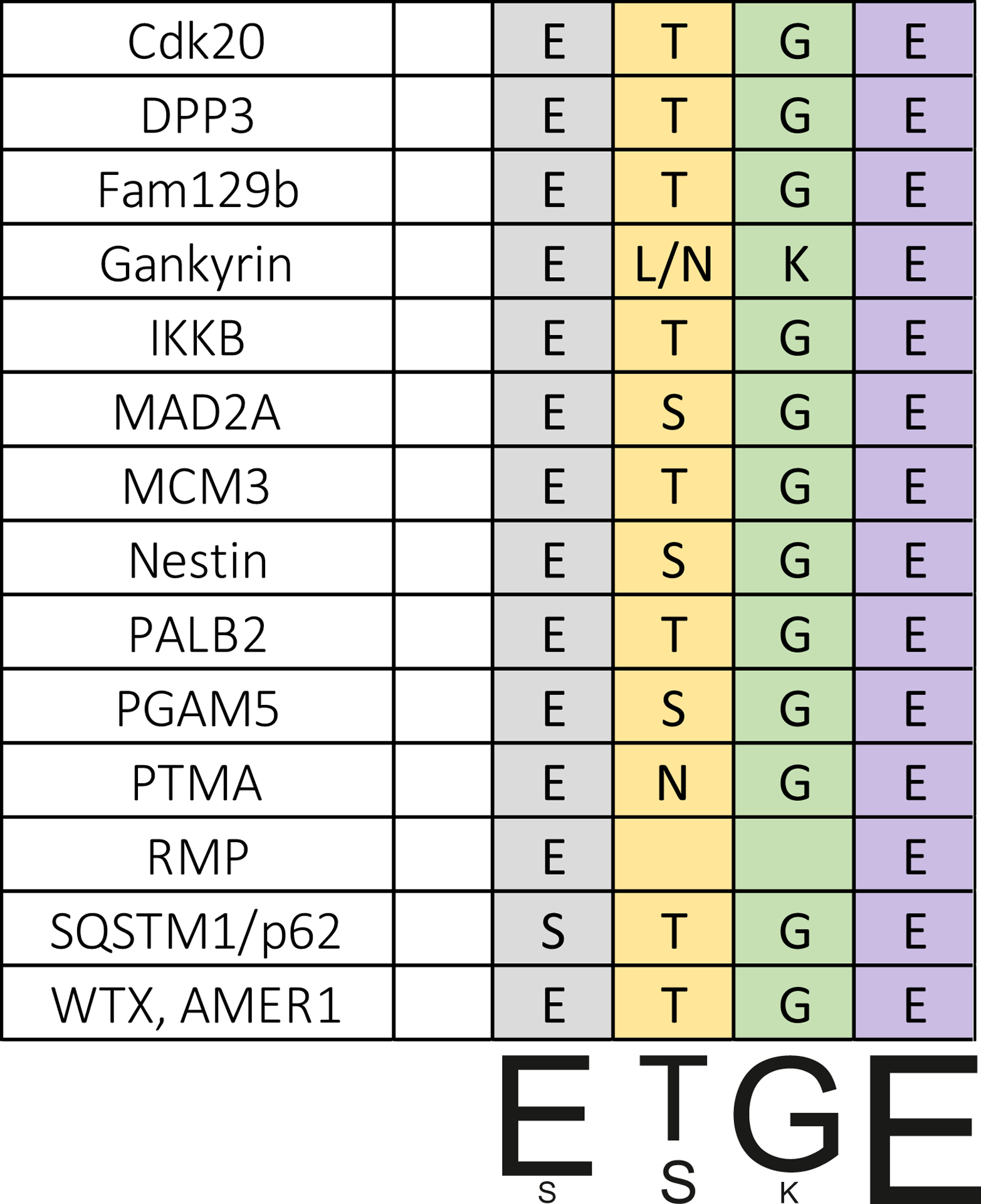
Table 3. Proteins interacting with Keap1, verified by biochemical methods.
The location of interacting amino acids was shown for human (h) proteins. Presence of the interacting motif was also verified in mouse (m) and rat (r) proteins based on the sequence search. X - unspecified amino acid.
| Protein name and symbol | Function | Motif interacting with Keap1 | Reference |
|---|---|---|---|
| Actin | Cytoskeleton | n.d. | [128] |
| BPTF/FAC1 | Chromatin regulation | n.d. | [129] |
| Cdk20 | Cell cycle | 25ETGE28, h m r | [130] |
| DPP3 | Protein degradation | 480ETGE483, h m r | [113,116,131] |
| EGFR | Growth factor signalling | n.d. | [96] |
| Fam129b | Apoptosis suppression, signalling |
708DLG710, h 718ETGE721,h m r |
[132] |
| Gankyrin | Proteasome subunit |
21ELKE24, h m r
201ENKE204, h m r |
[133] |
| GAPDH | Metabolism | n.d. | [75] |
| HBXIP | Amino-acid sensing, viral response | 110GLNLG115, h | [134] |
| HSP90 | Protein folding | n.d. | [135] |
| IKKB | Regulation of NFkB signaling | 36ETGE39, h | [76,77] |
| KPNA6 (importin α7) | Nuclear import of Keap1 | ARM domain | [136] |
| MAD2A | Cell division | 92ESGE96, h m | [137] |
| MCM3 | DNA replication | 387ESGE390, h m r | [80,138] |
| Nestin | Intermediate filament | 1414ESGE1417, h m r DLG – predicted | [127] |
| NOS3 | Production of nitric oxide | n.d. | [75] |
| p65 | Inflammation | n.d. | [139] |
| PALB2 | DNA repair | 91ETGE94, h m r | [140,141] |
| PGAM5 | Necrosis | 79ESGE82, h m r | [78,79,142,143] |
| PIDD | Apoptosis | n.d. | [126] |
| PTMA | Chromatin remodeling | 43ENGE46, h m r | [13,144,145] |
| RelA-associated inhibitor (iASPP) | p53 regulator | 239DLT241, h | [146] |
| RMP | Gene transcription regulation | EXXE, h m | [147] |
| p62 | Autophagy | 349STGE352,h m r (phosphorylated) | [148,149] |
| WTX, AMER1 | Regulation of Wnt signaling pathway | 288ETGE291, h | [73,150] |
Table 4.
The effect of compounds activating Nrf2, tBHQ, sulforaphane and H2O2, on the interaction of Keap1 with its binding partners.
Keap1 and NO-based regulation
Nitric oxide and reactive nitrogen species (RNS) induce Nrf2 nuclear translocation. It is achieved due to the S-nitrosation [9,84–86] or oxidative modification [25,26] of Keap1. However, the NO-related functions of Keap1 go beyond its post-translational modifications, aimed at the release of Nrf2.
A sensor of nitric oxide in Keap1 is cysteine C151 located in the BTB domain, which interestingly emerged in coincidence with the expansion of NOS family of proteins. NO sensing capacity of C151 is dependent on the presence of basic amino acids K131, R135 and K150, which spatially are present in the direct vicinity of C151. Perceiving of NO is accomplished by S-nitrosation of this cysteine residue [9]. However, Keap1 is not only a sensor of NO and RNS. Recently, it has also been recognised as a crucial modulator of S-nitrosation [75]. S-nitrosation it is an oxidative chemical reaction that generates a nitroso group on a cysteine thiol. Throughout years, the mechanism of formation of the nitrosothiol in the biological systems was elusive and thought to be non-enzymatic. Especially that such a reaction would require the formation of an intermediate NO+ [152], and the kinetics of such mechanisms raises constraints in in vivo conditions [153,154]. Keap1 directly interacts with nitric oxide synthase (NOS) and transnitrosating protein GAPDH (glyceraldehyde 3-phospho dehydrogenase) to govern S-nitrosation in mammalian cells. Depriving the cells of any protein of this complex inhibits protein S-nitrosation [75]. Keap1/GAPDH/NOS enzymatic complex resembles the bacterial one, which comprises of SNO synthase (Hcp), transnitrosylase GAPDH and NarGHI, as NO source [155]. It suggests that Keap1 can be a mammalian SNO synthase. It could be speculated that NO sensing by C151 and the requirement of positively charged amino acids for this process could account for such enzymatic activity of Keap1. Importantly, Keap1-related S-nitrosation of NOX4 has a fundamental significance in endothelial cells, protecting them from oxidative damage and apoptosis in the absence of Nrf2. Moreover, S-nitrosation determines EC fate, balancing between premature senescence and cellular death [75].
Keap1 in cytoskeleton regulation
Just upon discovery of Keap1 as Nrf2-interactor [4], the similarity of Keap1 to Drosophila Kelch protein was raised. Kelch is required to organise the ovarian ring canal cytoskeleton through F-actin binding and cross-linking [156]. Keap1 was shown to colocalise with a variety of both integrin and cadherin-based adhesion assemblies [119,157]. Subsequently, due to the resemblance to Drosophila Kelch protein, the interaction between Keap1 and actin was investigated. Keap1 colocalises and interacts with actin via its DGR domain. Disruption of actin leads to the release of Nrf2 from Keap1 interaction and its nuclear translocation [128]. Keap1 is present in focal adhesions and adherent junctions, but not in stress fibres [158]. It stabilises F-actin and restricts focal adhesion turnout. Additionally, its overexpression weakens the migratory capacity of cells and triggers thick stress fibres formation due to RhoA overactivity. The latter is caused by enhanced Myo9b degradation [81]. Furthermore, Keap1 regulates actin organisation, through associations with actin-regulatory proteins, such as myosin VIIa [119]. More complex regulation was reported in case of cortactin, which subcellular localisation is regulated by Keap1 binding [121]. Finally, Keap1 leads to the overabundance of RhoGAP1, the protein regulating Cdc42 activity. It impairs podosome formation and disrupts actin rearrangements, thus preventing endothelial cell migration and angiogenesis [123]. Keap1 may also interact with the other components of the cytoskeleton, tubulin [159], intermediate filament lamin [131] and nestin [127]. However, the functional significance of these interactions remains unknown.
Keap1 in proteostasis
The role of Nrf2 in proteostasis is mostly attributed to the transcriptional regulation of the ubiquitin-proteasome system and autophagy [reviewed in [160,161]]. Keap1 is an essential component of Rbx1-E3-ubiquitin complex, which marks various proteins for degradation, including Nrf2 [32], but also IKKβ [76,77], PGAM5 [78], Miro2 [79], MCM3, SLK, MAD2L1 [80], and possibly Myo9b [81]. Additionally, Keap1 interacts with DPP3, which participates in the hydrolysis of peptides formed during proteasomal degradation [113,116,131]. However, there is no data on how Keap1 affects DPP3 enzymatic activity. Keap1 was reported to assist in ubiquitin aggregate clearance through autophagy, where it interacts with p62 and LC3 [162]. In response to selective autophagy, it relocalises to inclusion bodies following interaction with p62 [12]. Keap1 colocalises with p62 in puncta to foster its degradation [148,162–164]. The interaction between Keap1 and p62 occurs following phosphorylation of STGE motif of p62, which can be executed by mTORC1 [165] or TAK1 [166]. Thus, an increase in the negative charge of this sequence enables interaction with positively charged residues of the Kelch domain of Keap1. The binding affinity of this pair is similar to the one observed for the DLG motif of Nrf2 and Kelch [149]. The interaction between p62 and Keap1 is enhanced when p62 undergoes phase separation (and appears as puncta) driven by cytoplasmatic DAXX (death-associated protein 6) [167]. Importantly, mutation in the KIR domain of p62, which abrogates the interaction of Keap1 with p62, has been associated with amyotrophic lateral sclerosis (ALS) [168]. High-throughput data also indicate for Keap1 interaction with essential autophagy regulator Atg5 [169]. Coimmunoprecipitation revealed direct interaction of Keap1 with proteasomal subunits PSMD2, PSMD4 and segregase Vcp/p97 [74], the latter, interestingly, had been previously reported to regulate Nrf2 stability negatively [154].
Keap1 at the mitochondrial interface
There are some contradictory data on the localisation of Keap1 in mitochondria, depending on the experimental setup and method used [14]. However, there is a growing body of evidence indicating the presence of Keap1 in the proximity of mitochondria, where it interacts with mitochondrial protein PGAM5 [78,79,143]. This interaction tethers Nrf2 at the mitochondria, thus abrogating its transcriptional activity [143]. However, a decrease either in Nrf2 or PGAM5 causes impairment of retrograde mitochondrial signalling, due to hyperactivity of Keap1-Cul3 complex and enhanced degradation of Miro2 [79]. Moreover, during an elevated generation of oxidants, Keap1/PGAM5 mediates caspase-independent cell death, called ‘oxeiptosis’ [142]. Conversely, Keap1 is crucial for the maintenance of proper mitochondrial homeostasis, as p62 recruits it to mitochondria, and together with Rbx1, mediates mitochondrial ubiquitination, thus, preventing the formation of dysfunctional megamitochondria and mitigating nonalcoholic fatty liver disease [170].
Cell cycle progression
Apart from the cytoplasmatic localisation, Keap1 is also present in the nucleus [131,136]. The Kelch domain-derived interaction with importin α7/karyopherin α6 (KPNA6) is required for the nuclear import of Keap1, which is crucial in the fine-tuning of the antioxidant response [136]. Similarly, p65 interacts with Keap1 and promotes its nuclear translocation [139]. These nucleus-related functions of Keap1 were investigated mainly in the context of Nrf2 activation. However, Keap1 may also be a crucial regulator of cell cycle progression, independently of Nrf2. Recently, Keap1 was shown to negatively regulate endothelial cell proliferation and induce senescence in those cells [75,123]. Moreover, Keap1 knockout mice fail to execute the S-phase of the cell cycle during liver regeneration properly [171]. This impairment was associated with deregulation among cell-cycle mediators and mitogenic pathways. Keap1 knockout mice were also characterised by better genome integrity and stability, possibly due to increased expression of Rad51 [171]. BRCA1 and PALB2 are essential regulators of DNA damage response, also through the recruitment of Rad51. Notably, Keap1, through sequestration of PALB2, impedes the cell-cycle dependent BRCA1-PALB2 interaction [141]. Furthermore, more in-depth studies revealed that Keap1 interacting motif in PALB2 is central to PALB2-BRCA1 complex formation and facilitation of homologous recombination in G1 cells. Keap1 recruits CRL3 ubiquitination complex to PALB2. The ubiquitin moiety on lysine 20 of PALB2 suppresses its interaction with BRCA1. Finally, inactivation of Keap1 leads to the formation of stable BRCA1-PALB2-BRCA2 complex in both G1 and S phases [141].
MCM3 (DNA replication licensing factor, mini-chromosome maintenance protein), involved in DNA replication, interacts similarly to MCM7 with Keap1-Cul3 complex in cell-cycle dependent manner. However, this interaction does not lead to MCM degradation. Instead, it regulates MCM complex dynamics [80]. Keap1, together with MCM3 and MCM-BP, were shown to form a ternary complex. The interaction with MCMBP was also reported in high throughput study [172]. Interestingly, Keap1 may interact with Cdk20 [130], Chd6 [131], Cdc34 [173], Sass6 and many other [174]. Keap1 forms also complex with ProTα [13,144,145], which regulates the compaction state of chromatin. However, this interaction was not studied concerning proliferation and cell cycle progression.
Keap1 repression and the significance of Nrf2/Keap1 balance
The unique structure of Keap1 and the resulting diversity of interacting proteins may suggest that the interplay between Keap1 and its interactome should be tightly controlled. The function of Keap1 as an Nrf2 repressor has been very well established; the issue of Keap1 repression, however, has been beyond the research mainstream until now. Recent papers have recognised that the presence of unrestrained Keap1 is detrimental for cellular homeostasis [75,79,123] and emphasised the role of Nrf2 as the repressor of Keap1 [75,123], which implicates that Nrf2 and Keap1 mutually inhibit each other. Nrf2-depleted cells exhibited substantial deregulation of mitochondrial homeostasis due to the Keap1-mediated degradation of Miro2 [79]. The function of endothelial cells (ECs) devoid of Nrf2 is aggravated due to the overabundance of unrestrained Keap1, which stabilises RhoGAP1 and potentiates protein S-nitrosation, inhibiting angiogenesis and evoking premature senescence of ECs [75,123]. The role of Nrf2 as Keap1 repressor is further supported by the phenotype of Nrf2 transcriptional knockout mice, in which the sequence coding the DNA binding domain has been disrupted by LacZ, resulting in the formation of fusion protein N-terminal Nrf2-β-gal. These animals develop well and do not exhibit abnormal phenotype unless challenged by detrimental factors. It is likely achieved by the presence of the Neh2 domain in these animals, capable of Keap1 binding [123]. The clinical significance of Nrf2/Keap1 imbalance can be implicated in age-related diseases [reviewed in [175]]. Keap1 is reported to increase during ageing [176] or in the models of premature senescence [75]. However, the exact role of Keap1, in contrast to that of Nrf2, in ageing has not been addressed yet.
Keap1 inactivation, thus defined by Nrf2 hyperactivity, is characterised by multiple disorders, including hyperkeratosis of the digestive tract [177], bone hypoplasia [178], abnormal kidney development and function [179,180], and pancreatic atrophy [181]. Nrf2 serial activation was also associated with hyperproliferation of keratinocytes and pathogenesis of psoriasis [182]. Additionally, such cells (keratinocytes with Nrf2 overactivity) are featured by sebaceous gland enlargement and seborrhea, together with thickening and hyperkeratosis of hair follicle infundibula [183]. Furthermore, when mice were expressing a constitutively active Nrf2 (caNrf2) mutant in hepatocytes, liver regeneration was impaired due to delayed proliferation and enhanced apoptosis [184]. Finally, recently it was demonstrated in adult flies, that while mild Nrf2 activation extended lifespan, a high Nrf2 expression level led to developmental lethality or, after inducible activation, it altered mitochondrial bioenergetics, led to the onset of Diabetes Type 1 hallmarks and ageing acceleration [185]. The adverse effects of Nrf2 hyperactivity often result from the overabundance of its target genes or through metabolic reprogramming [186]. Accordingly, Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis [187,188] and consequently on exogenously delivered non-essential amino-acids (NEAA) [189]. Detrimental effects of Nrf2 hyperactivity have also been described in 4 cases of human mutations in NFE2L2, which disable the binding of Nrf2 to Keap1. The patients exhibited developmental delay, failure to thrive, immunodeficiency, leukoencephalopathy, and hypohomocysteinaemia [190]. Remarkably, as described above, the lack of either Nrf2 [75,79,123] or Keap1 [177] results in detrimental effects for the cells and organisms. Dysfunction of cells devoid of Nrf2, exhibiting impaired angiogenic potential and migration, senescence onset and increased S-nitrosation of proteins in case of endothelial cells and aberrant mitochondrial trafficking in case of retinal pigment epithelial cells, can be fully rescued by concomitant deletion of Keap1 [75,79,123]. In accordance, detrimental effects of Keap1 knockout, hyperkeratosis and constriction of oesophagus accounting for juvenile lethality of these mice, are reversed in Nrf2/Keap1 double knock out animals [177]. These results evidence that Nrf2/Keap1 balance is crucial to maintain cellular homeostasis.
The physiological and clinical relevance of Nrf2 hyperactivity models, Keap1-null [177] and caNrf2 [184] mice, might be questionable, taking into consideration that human Keap1 deficiency has not been reported so far and in caNrf2 mice Neh2-deficient caNrf2 is driven by a powerful CMV promoter [184]. To address this issue, we compared the fold change of Nrf2 target gene expression in these mice, patients with mutations within Neh2 domain of Nrf2 [190] and human tumours with loss of NFE2L2 exon 2 [191]. The expression of target genes in caNrf2 mouse is usually increased by ~10-fold up to ~100-fold [184], in Keap1-null mouse by ~10–30-fold up to ~400-fold [180], in Keap1 floxed mouse, which revealed the hypomorphism of floxed Keap1 allele and milder phenotype than Keap1-null animals, by ~10-fold [192]. Expression of Nrf2 target genes in human cells isolated from patients with Neh2 mutations was increased usually by ~2-fold up to ~60-fold [190], while in human tumour samples deficient in exon 2 of NFE2L2 by ~2-fold up to ~30-fold [191]. These limitations might be relevant, taking into account ‘the hormetic characteristics’ of Nrf2 activity [193], which also supports the significance of Nrf2/Keap1 balance.
Concluding remarks
The function of Keap1 as an Nrf2 repressor has been established exceptionally well and in detail. However, Keap1 is not only a repressor of Nrf2. The non-canonical functions of Keap1 should be explored further, for example, in the context of age-related dysfunctions, when an imbalance between Nrf2 and Keap1 occurs. Moreover, these atypical functions of Keap1 and the role of Nrf2 as a Keap1 repressing protein should be taken into consideration during data interpretation.
Highlights.
In this review we emphasize the non-canonical functions of Keap1
We highlight the impact of balance between Keap1 and Nrf2 in the maintenance of cellular homeostasis
Acknowledgements
This work was supported by the National Science Centre grant SONATA BIS No. 2016/22/E/NZ3/00405 (AGP). DK acknowledges financial support from the National Science Centre Poland (NCN) under the ETIUDA doctoral scholarship based on the decision number DEC-2019/32/T/NZ3/00326. HJF is supported by grant ES023864 from the United States National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y, An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements, Biochem. Biophys. Res. Commun 236 (1997) 313–322. 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- [2].Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S, Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis, Nucleic Acids Research. 38 (2010) 5718 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Baird L, Llères D, Swift S, Dinkova-Kostova AT, Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex, Proc Natl Acad Sci U S A. 110 (2013) 15259–15264. 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M, Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain, Genes Dev. 13 (1999) 76–86. 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yamamoto M, Kensler TW, Motohashi H, The KEAP1-NRF2 System: a Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis, Physiol. Rev 98 (2018) 1169–1203. 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cuadrado A, Manda G, Hassan A, Alcaraz MJ, Barbas C, Daiber A, Ghezzi P, León R, López MG, Oliva B, Pajares M, Rojo AI, Robledinos-Antón N, Valverde AM, Guney E, Schmidt HHHW, Transcription Factor NRF2 as a Therapeutic Target for Chronic Diseases: A Systems Medicine Approach, Pharmacol. Rev 70 (2018) 348–383. 10.1124/pr.117.014753. [DOI] [PubMed] [Google Scholar]
- [7].Dinkova-Kostova AT, Kostov RV, Canning P, Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants, Arch Biochem Biophys. 617 (2017) 84–93. 10.1016/j.abb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li L, Kobayashi M, Kaneko H, Nakajima-Takagi Y, Nakayama Y, Yamamoto M, Molecular evolution of Keap1. Two Keap1 molecules with distinctive intervening region structures are conserved among fish, J. Biol. Chem 283 (2008) 3248–3255. 10.1074/jbc.M708702200. [DOI] [PubMed] [Google Scholar]
- [9].McMahon M, Lamont DJ, Beattie KA, Hayes JD, Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals, Proc. Natl. Acad. Sci. U.S.A 107 (2010) 18838–18843. 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M, Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system, Genes Cells. 7 (2002) 807–820. 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- [11].Qiu L, Wang M, Zhu Y, Xiang Y, Zhang Y, A Naturally-Occurring Dominant-Negative Inhibitor of Keap1 Competitively against Its Negative Regulation of Nrf2, Int J Mol Sci 19 (2018). 10.3390/ijms19082150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T, p62/SQSTM1 Is a Target Gene for Transcription Factor NRF2 and Creates a Positive Feedback Loop by Inducing Antioxidant Response Element-driven Gene Transcription, J. Biol. Chem 285 (2010) 22576–22591. 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB, Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes, Mol. Cell. Biol 25 (2005) 1089–1099. 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Watai Y, Kobayashi A, Nagase H, Mizukami M, McEvoy J, Singer JD, Itoh K, Yamamoto M, Subcellular localization and cytoplasmic complex status of endogenous Keap1, Genes Cells. 12 (2007) 1163–1178. 10.1111/j.1365-2443.2007.01118.x. [DOI] [PubMed] [Google Scholar]
- [15].Iso T, Suzuki T, Baird L, Yamamoto M, Absolute Amounts and Status of the Nrf2-Keap1-Cul3 Complex within Cells, Mol Cell Biol. 36 (2016) 3100–3112. 10.1128/MCB.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang YV, Wade M, Wong E, Li Y-C, Rodewald LW, Wahl GM, Quantitative analyses reveal the importance of regulated Hdmx degradation for P53 activation, PNAS. 104 (2007) 12365–12370. 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Biggin MD, Animal Transcription Networks as Highly Connected, Quantitative Continua, Developmental Cell. 21 (2011) 611–626. 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- [18].Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P, Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants, Proc. Natl. Acad. Sci. U.S.A 99 (2002) 11908–11913. 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Miseta A, Csutora P, Relationship between the occurrence of cysteine in proteins and the complexity of organisms, Mol. Biol. Evol 17 (2000) 1232–1239. 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- [20].Brocchieri L, Karlin S, Protein length in eukaryotic and prokaryotic proteomes, Nucleic Acids Res. 33 (2005) 3390–3400. 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Go Y-M, Chandler JD, Jones DP, The cysteine proteome, Free Radic. Biol. Med 84 (2015) 227–245. 10.1016/j.freeradbiomed.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P, Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups, Proc. Natl. Acad. Sci. U.S.A 98 (2001) 3404–3409. 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kobayashi M, Li L, Iwamoto N, Nakajima-Takagi Y, Kaneko H, Nakayama Y, Eguchi M, Wada Y, Kumagai Y, Yamamoto M, The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds, Mol. Cell. Biol 29 (2009) 493–502. 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Saito R, Suzuki T, Hiramoto K, Asami S, Naganuma E, Suda H, Iso T, Yamamoto H, Morita M, Baird L, Furusawa Y, Negishi T, Ichinose M, Yamamoto M, Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response, Mol. Cell. Biol 36 (2016) 271–284. 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fourquet S, Guerois R, Biard D, Toledano MB, Activation of NRF2 by Nitrosative Agents and H2O2 Involves KEAP1 Disulfide Formation, J. Biol. Chem 285 (2010) 8463–8471. 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N, Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein, Biochemistry. 44 (2005) 6889–6899. 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- [27].Forman HJ, Davies MJ, Krämer AC, Miotto G, Zaccarin M, Zhang H, Ursini F, Protein cysteine oxidation in redox signaling: caveats on sulfenic acid detection and quantification, Arch Biochem Biophys. 617 (2017) 26–37. 10.1016/j.abb.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dhanoa BS, Cogliati T, Satish AG, Bruford EA, Friedman JS, Update on the Kelch-like (KLHL) gene family, Hum Genomics. 7 (2013) 13 10.1186/1479-7364-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Prag S, Adams JC, Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals, BMC Bioinformatics. 4 (2003) 42 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA, The Keap1-BTB Protein Is an Adaptor That Bridges Nrf2 to a Cul3-Based E3 Ligase: Oxidative Stress Sensing by a Cul3-Keap1 Ligase, Molecular and Cellular Biology. 24 (2004) 8477–8486. 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Furukawa M, Xiong Y, BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase, Mol. Cell. Biol 25 (2005) 162–171. 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang DD, Lo S-C, Cross JV, Templeton DJ, Hannink M, Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex, Mol. Cell. Biol 24 (2004) 10941–10953. 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M, Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2, Mol. Cell. Biol 24 (2004) 7130–7139. 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dayalan Naidu S, Muramatsu A, Saito R, Asami S, Honda T, Hosoya T, Itoh K, Yamamoto M, Suzuki T, Dinkova-Kostova AT, C151 in KEAP1 is the main cysteine sensor for the cyanoenone class of NRF2 activators, irrespective of molecular size or shape, Sci Rep. 8 (2018) 8037 10.1038/s41598-018-26269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dinkova-Kostova AT, The Role of Sulfhydryl Reactivity of Small Molecules for the Activation of the KEAP1/NRF2 Pathway and the Heat Shock Response, Scientifica. (2012). 10.6064/2012/606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Suzuki T, Maher J, Yamamoto M, Select heterozygous Keap1 mutations have a dominant-negative effect on wild-type Keap1 in vivo, Cancer Res. 71 (2011) 1700–1709. 10.1158/0008-5472.CAN-10-2939. [DOI] [PubMed] [Google Scholar]
- [37].Zipper LM, Mulcahy RT, The Keap1 BTB/POZ Dimerization Function Is Required to Sequester Nrf2 in Cytoplasm, J. Biol. Chem 277 (2002) 36544–36552. 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- [38].Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang M-I, Kobayashi A, Yamamoto M, Kensler TW, Talalay P, Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers, Proc. Natl. Acad. Sci. U.S.A 101 (2004) 2040–2045. 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang DD, Hannink M, Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress, Mol. Cell. Biol 23 (2003) 8137–8151. 10.1128/mcb.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li X, Zhang D, Hannink M, Beamer LJ, Crystal structure of the Kelch domain of human Keap1, J. Biol. Chem 279 (2004) 54750–54758. 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- [41].Adams J, Kelso R, Cooley L, The kelch repeat superfamily of proteins: propellers of cell function, Trends Cell Biol. 10 (2000) 17–24. 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- [42].Padmanabhan B, Scharlock M, Tong KI, Nakamura Y, Kang M-I, Kobayashi A, Matsumoto T, Tanaka A, Yamamoto M, Yokoyama S, Purification, crystallization and preliminary X-ray diffraction analysis of the Kelch-like motif region of mouse Keap1, Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 61 (2005) 153–155. 10.1107/S1744309104032506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Padmanabhan B, Tong KI, Kobayashi A, Yamamoto M, Yokoyama S, Structural insights into the similar modes of Nrf2 transcription factor recognition by the cytoplasmic repressor Keap1, J Synchrotron Radiat. 15 (2008) 273–276. 10.1107/S090904950705114X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cleasby A, Yon J, Day PJ, Richardson C, Tickle IJ, Williams PA, Callahan JF, Carr R, Concha N, Kerns JK, Qi H, Sweitzer T, Ward P, Davies TG, Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO, PLoS ONE. 9 (2014) e98896 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Canning P, Sorrell FJ, Bullock AN, Structural basis of Keap1 interactions with Nrf2, Free Radic Biol Med. 88 (2015) 101–107. 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Huang M, Zhu L, Garcia JS, Li MX, Gentles AJ, Mitchell BS, Brd4 regulates the expression of essential autophagy genes and Keap1 in AML cells, Oncotarget. 9 (2018) 11665–11676. 10.18632/oncotarget.24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hussong M, Börno ST, Kerick M, Wunderlich A, Franz A, Sültmann H, Timmermann B, Lehrach H, Hirsch-Kauffmann M, Schweiger MR, The bromodomain protein BRD4 regulates the KEAP1/NRF2-dependent oxidative stress response, Cell Death Dis. 5 (2014) e1195 10.1038/cddis.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Guo D, Wu B, Yan J, Li X, Sun H, Zhou D, A possible gene silencing mechanism: hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells, Biochem. Biophys. Res. Commun 428 (2012) 80–85. 10.1016/j.bbrc.2012.10.010. [DOI] [PubMed] [Google Scholar]
- [49].Muscarella LA, Barbano R, D’Angelo V, Copetti M, Coco M, Balsamo T, la Torre A, Notarangelo A, Troiano M, Parisi S, Icolaro N, Catapano D, Valori VM, Pellegrini F, Merla G, Carella M, Fazio VM, Parrella P, Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome, Epigenetics. 6 (2011) 317–325. 10.4161/epi.6.3.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lee O-H, Jain AK, Papusha V, Jaiswal AK, An Auto-regulatory Loop between Stress Sensors INrf2 and Nrf2 Controls Their Cellular Abundance, J. Biol. Chem 282 (2007) 36412–36420. 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- [51].Kanwal A, Pillai VB, Samant S, Gupta M, Gupta MP, The nuclear and mitochondrial sirtuins, Sirt6 and Sirt3, regulate each other’s activity and protect the heart from developing obesity-mediated diabetic cardiomyopathy, FASEB J. 33 (2019) 10872–10888. 10.1096/fj.201900767R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gao Y, Yan Y, Huang T, Human age-related cataracts: epigenetic suppression of the nuclear factor erythroid 2-related factor 2-mediated antioxidant system, Mol Med Rep. 11 (2015) 1442–1447. 10.3892/mmr.2014.2849. [DOI] [PubMed] [Google Scholar]
- [53].Periyasamy P, Ayaki M, Bidasee K, Shinohara T, Methylglyoxal Activates Chronic ER Stress Mediated Epigenetic Loss of Nrf2/Keap1 Gene Regulation in Diabetic Cataractous Lenses, Invest. Ophthalmol. Vis. Sci 54 (2013) 1148–1148. [Google Scholar]
- [54].Palsamy P, Bidasee KR, Shinohara T, Valproic acid suppresses Nrf2/Keap1 dependent antioxidant protection through induction of endoplasmic reticulum stress and Keap1 promoter DNA demethylation in human lens epithelial cells, Exp Eye Res. 121 (2014) 26–34. 10.1016/j.exer.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Veronezi GMB, Felisbino MB, Gatti MSV, Mello MLS, de C Vidal B, DNA Methylation Changes in Valproic Acid-Treated HeLa Cells as Assessed by Image Analysis, Immunofluorescence and Vibrational Microspectroscopy, PLOS ONE. 12 (2017) e0170740 10.1371/journal.pone.0170740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fabrizio FP, Sparaneo A, Centra F, Trombetta D, Storlazzi CT, Graziano P, Maiello E, Fazio VM, Muscarella LA, Methylation Density Pattern of KEAP1 Gene in Lung Cancer Cell Lines Detected by Quantitative Methylation Specific PCR and Pyrosequencing, Int J Mol Sci 20 (2019). 10.3390/ijms20112697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang R, An J, Ji F, Jiao H, Sun H, Zhou D, Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues, Biochem. Biophys. Res. Commun 373 (2008) 151–154. 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- [58].Yoo NJ, Kim HR, Kim YR, An CH, Lee SH, Somatic mutations of the KEAP1 gene in common solid cancers, Histopathology. 60 (2012) 943–952. 10.1111/j.1365-2559.2012.04178.x. [DOI] [PubMed] [Google Scholar]
- [59].Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova GS, Biswal S, Loss of Keap1 Function in Prostate Cancer Cells Causes Chemo- and Radio-resistance and Promotes Tumor Growth, Mol Cancer Ther. 9 (2010) 336 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kabaria S, Choi DC, Chaudhuri AD, Jain MR, Li H, Junn E, MicroRNA-7 activates Nrf2 pathway by targeting Keap1 expression, Free Radic. Biol. Med 89 (2015) 548–556. 10.1016/j.freeradbiomed.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xiao X, Lu Z, Lin V, May A, Shaw DH, Wang Z, Che B, Tran K, Du H, Shaw PX, MicroRNA miR-24–3p Reduces Apoptosis and Regulates Keap1-Nrf2 Pathway in Mouse Cardiomyocytes Responding to Ischemia/Reperfusion Injury, Oxid Med Cell Longev. 2018 (2018) 7042105 10.1155/2018/7042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cheng L-B, Li K, Yi N, Li X, Wang F, Xue B, Pan Y, Yao J, Jiang Q, Wu Z, miRNA-141 attenuates UV-induced oxidative stress via activating Keap1-Nrf2 signaling in human retinal pigment epithelium cells and retinal ganglion cells, Oncotarget. 8 (2017) 13186–13194. 10.18632/oncotarget.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, Zheng F, Lin X, MiR-141 Activates Nrf2-Dependent Antioxidant Pathway via Down-Regulating the Expression of Keap1 Conferring the Resistance of Hepatocellular Carcinoma Cells to 5-Fluorouracil, CPB. 35 (2015) 2333–2348. 10.1159/000374036. [DOI] [PubMed] [Google Scholar]
- [64].Eades G, Yang M, Yao Y, Zhang Y, Zhou Q, miR-200a Regulates Nrf2 Activation by Targeting Keap1 mRNA in Breast Cancer Cells, J. Biol. Chem 286 (2011) 40725–40733. 10.1074/jbc.M111.275495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yang J-J, Tao H, Hu W, Liu L-P, Shi K-H, Deng Z-Y, Li J, MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis, Cellular Signalling. 26 (2014) 2381–2389. 10.1016/j.cellsig.2014.07.016. [DOI] [PubMed] [Google Scholar]
- [66].Duan F-G, Wang M-F, Cao Y-B, null Dan Li, Li R-Z, Fan X-X, Khan I, Lai H-L, Zhang Y-Z, Hsiao WW-L, Yao X-J, Wu Q-B, Liu L, Tang Y-J, Leung EL-H, MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3’UTR and predicts poor survival in non-small cell lung cancer, Cell Death Dis. 10 (2019) 821 10.1038/s41419-019-2031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu X, Tang Y, Cheng L, Yao J, Jiang Q, Li K, Zhen Y, Targeting Keap1 by miR-626 protects retinal pigment epithelium cells from oxidative injury by activating Nrf2 signaling, Free Radical Biology and Medicine. 143 (2019) 387–396. 10.1016/j.freeradbiomed.2019.08.024. [DOI] [PubMed] [Google Scholar]
- [68].Wang J, Ishfaq M, Xu L, Xia C, Chen C, Li J, METTL3/m6A/miRNA-873–5p Attenuated Oxidative Stress and Apoptosis in Colistin-Induced Kidney Injury by Modulating Keap1/Nrf2 Pathway, Front. Pharmacol 10 (2019). 10.3389/fphar.2019.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov. 2 (2012) 401–404. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci Signal. 6 (2013) pl1 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fabrizio FP, Sparaneo A, Trombetta D, Muscarella LA, Epigenetic versus Genetic Deregulation of the KEAP1/NRF2 Axis in Solid Tumors: Focus on Methylation and Noncoding RNAs, Oxidative Medicine and Cellular Longevity. (2018). 10.1155/2018/2492063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Taguchi K, Yamamoto M, The KEAP1-NRF2 System in Cancer, Front Oncol. 7 (2017). 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hast BE, Cloer EW, Goldfarb D, Li H, Siesser PF, Yan F, Walter V, Zheng N, Hayes DN, Major MB, Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination, Cancer Res. 74 (2014) 808–817. 10.1158/0008-5472.CAN-13-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cloer EW, Siesser PF, Cousins EM, Goldfarb D, Mowrey DD, Harrison JS, Weir SJ, Dokholyan NV, Major MB, p62-dependent phase separation of patient-derived KEAP1 mutations and NRF2, Molecular and Cellular Biology. (2018) MCB.00644–17 10.1128/MCB.00644-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kopacz A, Kloska D, Proniewski B, Cysewski D, Personnic N, Piechota-Polanczyk A, Kaczara P, Zakrzewska A, Forman HJ, Dulak J, Jozkowicz A, Grochot-Przeczek A, Keap1 controls protein S-nitrosation and apoptosis-senescence switch in endothelial cells, Redox Biology. (2019) 101304 10.1016/j.redox.2019.101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kim J-E, You D-J, Lee C, Ahn C, Seong JY, Hwang J-I, Suppression of NF-kappaB signaling by KEAP1 regulation of IKKbeta activity through autophagic degradation and inhibition of phosphorylation, Cell. Signal 22 (2010) 1645–1654. 10.1016/j.cellsig.2010.06.004. [DOI] [PubMed] [Google Scholar]
- [77].Lee D-F, Kuo H-P, Liu M, Chou C-K, Xia W, Du Y, Shen J, Chen C-T, Huo L, Hsu M-C, Li C-W, Ding Q, Liao T-L, Lai C-C, Lin A-C, Chang Y-H, Tsai S-F, Li L-Y, Hung M-C, KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta, Mol. Cell 36 (2009) 131–140. 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lo S-C, Hannink M, PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex, J. Biol. Chem 281 (2006) 37893–37903. 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- [79].O’Mealey GB, Plafker KS, Berry WL, Janknecht R, Chan JY, Plafker SM, A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking, J. Cell. Sci 130 (2017) 3467–3480. 10.1242/jcs.203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mulvaney KM, Matson JP, Siesser PF, Tamir TY, Goldfarb D, Jacobs TM, Cloer EW, Harrison JS, Vaziri C, Cook JG, Major MB, Identification and Characterization of MCM3 as a Kelch-like ECH-associated Protein 1 (KEAP1) Substrate, J. Biol. Chem 291 (2016) 23719–23733. 10.1074/jbc.M116.729418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wu B, Yang S, Sun H, Sun T, Ji F, Wang Y, Xu L, Zhou D, Keap1 Inhibits Metastatic Properties of NSCLC Cells by Stabilizing Architectures of F-Actin and Focal Adhesions, Mol Cancer Res. 16 (2018) 508–516. 10.1158/1541-7786.MCR-17-0544. [DOI] [PubMed] [Google Scholar]
- [82].Zhang DD, Lo S-C, Sun Z, Habib GM, Lieberman MW, Hannink M, Ubiquitination of Keap1, a BTB-Kelch Substrate Adaptor Protein for Cul3, Targets Keap1 for Degradation by a Proteasome-independent Pathway, J. Biol. Chem 280 (2005) 30091–30099. 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- [83].Villeneuve NF, Tian W, Wu T, Sun Z, Lau A, Chapman E, Fang D, Zhang DD, USP15 negatively regulates Nrf2 through deubiquitination of Keap1, Mol. Cell 51 (2013) 68–79. 10.1016/j.molcel.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Buckley BJ, Li S, Whorton AR, KEAP1 MODIFICATION AND NUCLEAR ACCUMULATION IN RESPONSE TO S-NITROSOCYSTEINE, Free Radic Biol Med. 44 (2008) 692–698. 10.1016/j.freeradbiomed.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Ratnayake S, Dias IHK, Lattman E, Griffiths HR, Stabilising cysteinyl thiol oxidation and nitrosation for proteomic analysis, Journal of Proteomics. 92 (2013) 160–170. 10.1016/j.jprot.2013.06.019. [DOI] [PubMed] [Google Scholar]
- [86].Um H-C, Jang J-H, Kim D-H, Lee C, Surh Y-J, Nitric oxide activates Nrf2 through S-nitrosylation of Keap1 in PC12 cells, Nitric Oxide. 25 (2011) 161–168. 10.1016/j.niox.2011.06.001. [DOI] [PubMed] [Google Scholar]
- [87].Bollong MJ, Lee G, Coukos JS, Yun H, Zambaldo C, Chang JW, Chin EN, Ahmad I, Chatterjee AK, Lairson LL, Schultz PG, Moellering RE, A metabolite-derived protein modification integrates glycolysis with KEAP1–NRF2 signalling, Nature. 562 (2018) 600 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Palsamy P, Bidasee KR, Ayaki M, Augusteyn RC, Chan JY, Shinohara T, Methylglyoxal induces endoplasmic reticulum stress and DNA demethylation in the Keap1 promoter of human lens epithelial cells and age-related cataracts, Free Radic. Biol. Med 72 (2014) 134–148. 10.1016/j.freeradbiomed.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Emanuele MJ, Elia AEH, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen Y-N, Rush J, Hsu PW-C, Yen H-CS, Elledge SJ, Global identification of modular cullin-RING ligase substrates, Cell. 147 (2011) 459–474. 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, Runtsch MC, King MS, McGouran JF, Fischer R, Kessler BM, McGettrick AF, Hughes MM, Carroll RG, Booty LM, Knatko EV, Meakin PJ, Ashford MLJ, Modis LK, Brunori G, Sévin DC, Fallon PG, Caldwell ST, Kunji ERS, Chouchani ET, Frezza C, Dinkova-Kostova AT, Hartley RC, Murphy MP, O’Neill LA, Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1, Nature. 556 (2018) 113–117. 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Curtis JM, Hahn WS, Long EK, Burrill JS, Arriaga EA, Bernlohr DA, Protein Carbonylation and Metabolic Control Systems, Trends Endocrinol Metab. 23 (2012) 399–406. 10.1016/j.tem.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Chen P, Smith TJ, Wu J, Siesser PF, Bisnett BJ, Khan F, Hogue M, Soderblom E, Tang F, Marks JR, Major MB, Swarts BM, Boyce M, Chi J, Glycosylation of KEAP1 links nutrient sensing to redox stress signaling, EMBO J. 36 (2017) 2233–2250. 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Carvalho AN, Marques C, Guedes RC, Castro-Caldas M, Rodrigues E, van Horssen J, Gama MJ, S-Glutathionylation of Keap1: a new role for glutathione S-transferase pi in neuronal protection, FEBS Lett. 590 (2016) 1455–1466. 10.1002/1873-3468.12177. [DOI] [PubMed] [Google Scholar]
- [94].Wang L, Qu G, Gao Y, Su L, Ye Q, Jiang F, Zhao B, Miao J, A small molecule targeting glutathione activates Nrf2 and inhibits cancer cell growth through promoting Keap-1 S-glutathionylation and inducing apoptosis, RSC Adv. 8 (2018) 792–804. 10.1039/C7RA11935F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kansanen E, Bonacci G, Schopfer FJ, Kuosmanen SM, Tong KI, Leinonen H, Woodcock SR, Yamamoto M, Carlberg C, Ylä-Herttuala S, Freeman BA, Levonen A-L, Electrophilic Nitro-fatty Acids Activate NRF2 by a KEAP1 Cysteine 151-independent Mechanism, J. Biol. Chem 286 (2011) 14019–14027. 10.1074/jbc.M110.190710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Huo L, Li C-W, Huang T-H, Lam YC, Xia W, Tu C, Chang W-C, Hsu JL, Lee D-F, Nie L, Yamaguchi H, Wang Y, Lang J, Li L-Y, Chen C-H, Mishra L, Hung M-C, Activation of Keap1/Nrf2 signaling pathway by nuclear epidermal growth factor receptor in cancer cells, Am J Transl Res. 6 (2014) 649–663. [PMC free article] [PubMed] [Google Scholar]
- [97].Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M, Fischer R, Carmeliet P, Maxwell PH, Pugh CW, Frizzell N, Soga T, Kessler BM, El-Bahrawy M, Ratcliffe PJ, Pollard PJ, Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling, Cancer Cell. 20 (2011) 524–537. 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Burgener A-V, Bantug GR, Meyer BJ, Higgins R, Ghosh A, Bignucolo O, Ma EH, Loeliger J, Unterstab G, Geigges M, Steiner R, Enamorado M, Ivanek R, Hunziker D, Schmidt A, Müller-Durovic B, Grählert J, Epple R, Dimeloe S, Lötscher J, Sauder U, Ebnöther M, Burger B, Heijnen I, Martínez-Cano S, Cantoni N, Brücker R, Kahlert CR, Sancho D, Jones RG, Navarini A, Recher M, Hess C, SDHA gain-of-function engages inflammatory mitochondrial retrograde signaling via KEAP1-Nrf2, Nat. Immunol 20 (2019) 1311–1321. 10.1038/s41590-019-0482-2. [DOI] [PubMed] [Google Scholar]
- [99].Ooi A, Wong J-C, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BWH, Tan M-H, Zhang Z, Yang XJ, Zhou M, Gardie B, Molinié V, Richard S, Tan PH, Teh BT, Furge KA, An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma, Cancer Cell. 20 (2011) 511–523. 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- [100].Xie L, Gu Y, Wen M, Zhao S, Wang W, Ma Y, Meng G, Han Y, Wang Y, Liu G, Moore PK, Wang X, Wang H, Zhang Z, Yu Y, Ferro A, Huang Z, Ji Y, Hydrogen Sulfide Induces Keap1 S-sulfhydration and Suppresses Diabetes-Accelerated Atherosclerosis via Nrf2 Activation, Diabetes. 65 (2016) 3171–3184. 10.2337/db16-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H, Cheng Z, Park J, Wan X, Liu X, Yue WW, Wanders RJA, Locasale JW, Lombard DB, de Boer VCJ, Zhao Y, Proteomic and Biochemical Studies of Lysine Malonylation Suggest Its Malonic Aciduria-associated Regulatory Role in Mitochondrial Function and Fatty Acid Oxidation, Mol. Cell Proteomics 14 (2015) 3056–3071. 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Weinert BT, Schölz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C, Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation, Cell Rep. 4 (2013) 842–851. 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- [103].Won SJ, Martin BR, Temporal profiling establishes a dynamic S-palmitoylation cycle, ACS Chem Biol. 13 (2018) 1560–1568. 10.1021/acschembio.8b00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M, Keap1 degradation by autophagy for the maintenance of redox homeostasis, Proc. Natl. Acad. Sci. U.S.A 109 (2012) 13561–13566. 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen Y, Inoyama D, Kong A-NT, Beamer LJ, Hu L, Kinetic analyses of Keap1-Nrf2 interaction and determination of the minimal Nrf2 peptide sequence required for Keap1 binding using surface plasmon resonance, Chem Biol Drug Des. 78 (2011) 1014–1021. 10.1111/j.1747-0285.2011.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Inoyama D, Chen Y, Huang X, Beamer LJ, Kong A-NT, Hu L, Optimization of fluorescently labeled Nrf2 peptide probes and the development of a fluorescence polarization assay for the discovery of inhibitors of Keap1-Nrf2 interaction, J Biomol Screen. 17 (2012) 435–447. 10.1177/1087057111430124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Schaap M, Hancock R, Wilderspin A, Wells G, Development of a steady-state FRET-based assay to identify inhibitors of the Keap1-Nrf2 protein-protein interaction, Protein Sci. 22 (2013) 1812–1819. 10.1002/pro.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M, Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model, Mol. Cell. Biol 26 (2006) 2887–2900. 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M, Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response, Mol. Cell. Biol 27 (2007) 7511–7521. 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Tong KI, Kobayashi A, Katsuoka F, Yamamoto M, Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism, Biol. Chem 387 (2006) 1311–1320. 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- [111].Lo S-C, Li X, Henzl MT, Beamer LJ, Hannink M, Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling, EMBO J. 25 (2006) 3605–3617. 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hancock R, Bertrand HC, Tsujita T, Naz S, El-Bakry A, Laoruchupong J, Hayes JD, Wells G, Peptide inhibitors of the Keap1-Nrf2 protein-protein interaction, Free Radic. Biol. Med 52 (2012) 444–451. 10.1016/j.freeradbiomed.2011.10.486. [DOI] [PubMed] [Google Scholar]
- [113].Hast BE, Goldfarb D, Mulvaney KM, Hast MA, Siesser PF, Yan F, Hayes DN, Major MB, Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination, Cancer Res. 73 (2013) 2199–2210. 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Karttunen M, Choy W-Y, Cino EA, Prediction of Binding Energy of Keap1 Interaction Motifs in the Nrf2 Antioxidant Pathway and Design of Potential High-Affinity Peptides, J Phys Chem B. 122 (2018) 5851–5859. 10.1021/acs.jpcb.8b03295. [DOI] [PubMed] [Google Scholar]
- [115].Goldfarb D, Hast B, Wang W, Major MB, An Improved Algorithm and Web Application for Predicting Co-Complexed Proteins from Affinity Purification - Mass Spectrometry Data, J Proteome Res. 13 (2014) 5944–5955. 10.1021/pr5008416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Liu Y, Kern JT, Walker JR, Johnson JA, Schultz PG, Luesch H, A genomic screen for activators of the antioxidant response element, Proc. Natl. Acad. Sci. U.S.A 104 (2007) 5205–5210. 10.1073/pnas.0700898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wright PE, Dyson HJ, Intrinsically disordered proteins in cellular signalling and regulation, Nat. Rev. Mol. Cell Biol 16 (2015) 18–29. 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Do TN, Choy W-Y, Karttunen M, Binding of Disordered Peptides to Kelch: Insights from Enhanced Sampling Simulations, J. Chem. Theory Comput 12 (2016) 395–404. 10.1021/acs.jctc.5b00868. [DOI] [PubMed] [Google Scholar]
- [119].Velichkova M, Guttman J, Warren C, Eng L, Kline K, Vogl AW, Hasson T, A human homologue of Drosophila kelch associates with myosin-VIIa in specialized adhesion junctions, Cell Motil. Cytoskeleton 51 (2002) 147–164. 10.1002/cm.10025. [DOI] [PubMed] [Google Scholar]
- [120].Du Y, Weed SA, Xiong W-C, Marshall TD, Parsons JT, Identification of a Novel Cortactin SH3 Domain-Binding Protein and Its Localization to Growth Cones of Cultured Neurons, Mol Cell Biol. 18 (1998) 5838–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ito A, Shimazu T, Maeda S, Shah AA, Tsunoda T, Iemura S-I, Natsume T, Suzuki T, Motohashi H, Yamamoto M, Yoshida M, The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1, Sci Signal. 8 (2015) ra120 10.1126/scisignal.aad0667. [DOI] [PubMed] [Google Scholar]
- [122].Barfod ET, Zheng Y, Kuang WJ, Hart MJ, Evans T, Cerione RA, Ashkenazi A, Cloning and expression of a human CDC42 GTPase-activating protein reveals a functional SH3-binding domain., J. Biol. Chem 268 (1993) 26059–26062. [PubMed] [Google Scholar]
- [123].Kloska D, Kopacz A, Cysewski D, Aepfelbacher M, Dulak J, Jozkowicz A, Grochot-Przeczek A, Nrf2 Sequesters Keap1 Preventing Podosome Disassembly: A Quintessential Duet Moonlights in Endothelium, Antioxid. Redox Signal 30 (2019) 1709–1730. 10.1089/ars.2018.7505. [DOI] [PubMed] [Google Scholar]
- [124].Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, Speller LN, Leung DW, Geisbert TW, Amarasinghe GK, Basler CF, The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway, Cell Rep. 6 (2014) 1017–1025. 10.1016/j.celrep.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Marcotte D, Zeng W, Hus J-C, McKenzie A, Hession C, Jin P, Bergeron C, Lugovskoy A, Enyedy I, Cuervo H, Wang D, Atmanene C, Roecklin D, Vecchi M, Vivat V, Kraemer J, Winkler D, Hong V, Chao J, Lukashev M, Silvian L, Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism, Bioorg. Med. Chem 21 (2013) 4011–4019. 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- [126].Ji L, Zhang R, Chen J, Xue Q, Moghal N, Tsao M-S, PIDD interaction with KEAP1 as a new mutation-independent mechanism to promote NRF2 stabilization and chemoresistance in NSCLC, Sci Rep. 9 (2019) 12437 10.1038/s41598-019-48763-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang J, Lu Q, Cai J, Wang Y, Lai X, Qiu Y, Huang Y, Ke Q, Zhang Y, Guan Y, Wu H, Wang Y, Liu X, Shi Y, Zhang K, Wang M, Peng Xiang A, Nestin regulates cellular redox homeostasis in lung cancer through the Keap1-Nrf2 feedback loop, Nat Commun. 10 (2019) 5043 10.1038/s41467-019-12925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kang M-I, Kobayashi A, Wakabayashi N, Kim S-G, Yamamoto M, Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes, PNAS. 101 (2004) 2046–2051. 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Strachan GD, Morgan KL, Otis LL, Caltagarone J, Gittis A, Bowser R, Jordan-Sciutto KL, Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein, Biochemistry. 43 (2004) 12113–12122. 10.1021/bi0494166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wang Q, Ma J, Lu Y, Zhang S, Huang J, Chen J, Bei J-X, Yang K, Wu G, Huang K, Chen J, Xu S, CDK20 interacts with KEAP1 to activate NRF2 and promotes radiochemoresistance in lung cancer cells, Oncogene. 36 (2017) 5321–5330. 10.1038/onc.2017.161. [DOI] [PubMed] [Google Scholar]
- [131].Lu K, Alcivar AL, Ma J, Foo TK, Zywea S, Mahdi A, Huo Y, Kensler TW, Gatza ML, Xia B, NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 interaction, Cancer Res. 77 (2017) 2881–2892. 10.1158/0008-5472.CAN-16-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Cheng K-C, Lin R-J, Cheng J-Y, Wang S-H, Yu J-C, Wu J-C, Liang Y-J, Hsu H-M, Yu J, Yu AL, FAM129B, an antioxidative protein, reduces chemosensitivity by competing with Nrf2 for Keap1 binding, EBioMedicine. 45 (2019) 25–38. 10.1016/j.ebiom.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Yang C, Tan Y-X, Yang G-Z, Zhang J, Pan Y-F, Liu C, Fu J, Chen Y, Ding Z-W, Dong L-W, Wang H-Y, Gankyrin has an antioxidative role through the feedback regulation of Nrf2 in hepatocellular carcinoma, J. Exp. Med 213 (2016) 859–875. 10.1084/jem.20151208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zhou X-L, Zhu C-Y, Wu Z-G, Guo X, Zou W, The oncoprotein HBXIP competitively binds KEAP1 to activate NRF2 and enhance breast cancer cell growth and metastasis, Oncogene. 38 (2019) 4028–4046. 10.1038/s41388-019-0698-5. [DOI] [PubMed] [Google Scholar]
- [135].Prince TL, Kijima T, Tatokoro M, Lee S, Tsutsumi S, Yim K, Rivas C, Alarcon S, Schwartz H, Khamit-Kush K, Scroggins BT, Beebe K, Trepel JB, Neckers L, Client Proteins and Small Molecule Inhibitors Display Distinct Binding Preferences for Constitutive and Stress-Induced HSP90 Isoforms and Their Conformationally Restricted Mutants, PLoS One. 10 (2015). 10.1371/journal.pone.0141786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Sun Z, Wu T, Zhao F, Lau A, Birch CM, Zhang DD, KPNA6 (Importin {alpha}7)-mediated nuclear import of Keap1 represses the Nrf2-dependent antioxidant response, Mol. Cell. Biol 31 (2011) 1800–1811. 10.1128/MCB.05036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Karttunen M, Choy W-Y, Cino EA, Prediction of Binding Energy of Keap1 Interaction Motifs in the Nrf2 Antioxidant Pathway and Design of Potential High-Affinity Peptides, J. Phys. Chem. B 122 (2018) 5851–5859. 10.1021/acs.jpcb.8b03295. [DOI] [PubMed] [Google Scholar]
- [138].Tamberg N, Tahk S, Koit S, Kristjuhan K, Kasvandik S, Kristjuhan A, Ilves I, Keap1-MCM3 interaction is a potential coordinator of molecular machineries of antioxidant response and genomic DNA replication in metazoa, Sci Rep. 8 (2018) 1–18. 10.1038/s41598-018-30562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Yu M, Li H, Liu Q, Liu F, Tang L, Li C, Yuan Y, Zhan Y, Xu W, Li W, Chen H, Ge C, Wang J, Yang X, Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway, Cell. Signal 23 (2011) 883–892. 10.1016/j.cellsig.2011.01.014. [DOI] [PubMed] [Google Scholar]
- [140].Ma J, Cai H, Wu T, Sobhian B, Huo Y, Alcivar A, Mehta M, Cheung KL, Ganesan S, Kong A-NT, Zhang DD, Xia B, PALB2 Interacts with KEAP1 To Promote NRF2 Nuclear Accumulation and Function, Mol Cell Biol. 32 (2012) 1506–1517. 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, Xia B, Peter M, Durocher D, A mechanism for the suppression of homologous recombination in G1 cells, Nature. 528 (2015) 422–426. 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Holze C, Michaudel C, Mackowiak C, Haas DA, Benda C, Hubel P, Pennemann FL, Schnepf D, Wettmarshausen J, Braun M, Leung DW, Amarasinghe GK, Perocchi F, Staeheli P, Ryffel B, Pichlmair A, Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway, Nat. Immunol 19 (2018) 130–140. 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lo S-C, Hannink M, PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria, Exp. Cell Res 314 (2008) 1789–1803. 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Khan H, Cino EA, Brickenden A, Fan J, Yang D, Choy W-Y, Fuzzy complex formation between the intrinsically disordered prothymosin α and the Kelch domain of Keap1 involved in the oxidative stress response, J. Mol. Biol 425 (2013) 1011–1027. 10.1016/j.jmb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- [145].Padmanabhan B, Nakamura Y, Yokoyama S, Structural analysis of the complex of Keap1 with a prothymosin α peptide, Acta Crystallogr Sect F Struct Biol Cryst Commun. 64 (2008) 233–238. 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ge W, Zhao K, Wang X, Li H, Yu M, He M, Xue X, Zhu Y, Zhang C, Cheng Y, Jiang S, Hu Y, iASPP Is an Antioxidative Factor and Drives Cancer Growth and Drug Resistance by Competing with Nrf2 for Keap1 Binding, Cancer Cell. 32 (2017) 561–573.e6. 10.1016/j.ccell.2017.09.008. [DOI] [PubMed] [Google Scholar]
- [147].Wan Z-H, Jiang T-Y, Shi Y-Y, Pan Y-F, Lin Y-K, Ma Y-H, Yang C, Feng X-F, Huang L-F, Kong X-N, Ding Z-W, Tan Y-X, Dong L-W, Wang H-Y, RPB5-mediating protein promotes cholangiocarcinoma tumorigenesis and drug resistance by competing with NRF2 for KEAP1 binding, Hepatology. (2019). 10.1002/hep.30962. [DOI] [PubMed] [Google Scholar]
- [148].Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee M-S, Tanaka K, Komatsu M, Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells, The Journal of Cell Biology. 193 (2011) 275–284. 10.1083/jcb.201102031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou Y-S, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M, The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1, Nat. Cell Biol 12 (2010) 213–223. 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- [150].Camp ND, James RG, Dawson DW, Yan F, Davison JM, Houck SA, Tang X, Zheng N, Major MB, Moon RT, Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein, J. Biol. Chem 287 (2012) 6539–6550. 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Lo S-C, Hannink M, PGAM5, a Bcl-XL-interacting Protein, Is a Novel Substrate for the Redox-regulated Keap1-dependent Ubiquitin Ligase Complex, J. Biol. Chem 281 (2006) 37893–37903. 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- [152].Kovacs I, Lindermayr C, Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation, Front Plant Sci. 4 (2013). 10.3389/fpls.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Nedospasov A, Rafikov R, Beda N, Nudler E, An autocatalytic mechanism of protein nitrosylation, Proc Natl Acad Sci U S A. 97 (2000) 13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Seth D, Hess DT, Hausladen A, Wang L, Wang Y-J, Stamler JS, A Multiplex Enzymatic Machinery for Cellular Protein S-nitrosylation, Mol. Cell 69 (2018) 451–464.e6. 10.1016/j.molcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Seth D, Hess DT, Hausladen A, Wang L, Wang Y-J, Stamler JS, A Multiplex Enzymatic Machinery for Cellular Protein S-nitrosylation, Mol. Cell 69 (2018) 451–464.e6. 10.1016/j.molcel.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Xue F, Cooley L, kelch encodes a component of intercellular bridges in Drosophila egg chambers, Cell. 72 (1993) 681–693. 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- [157].Velichkova M, Hasson T, Keap1 in adhesion complexes, Cell Motil. Cytoskeleton 56 (2003) 109–119. 10.1002/cm.10138. [DOI] [PubMed] [Google Scholar]
- [158].Velichkova M, Hasson T, Keap1 Regulates the Oxidation-Sensitive Shuttling of Nrf2 into and out of the Nucleus via a Crm1-Dependent Nuclear Export Mechanism, Molecular and Cellular Biology. 25 (2005) 4501–4513. 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M, A human interactome in three quantitative dimensions organized by stoichiometries and abundances, Cell. 163 (2015) 712–723. 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- [160].Cui T, Lai Y, Janicki JS, Wang X, Nuclear factor erythroid-2 related factor 2 (Nrf2)-mediated protein quality control in cardiomyocytes, Front Biosci (Landmark Ed). 21 (2016) 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Pajares M, Cuadrado A, Rojo AI, Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases, Redox Biol. 11 (2017) 543–553. 10.1016/j.redox.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Fan W, Tang Z, Chen D, Moughon D, Ding X, Chen S, Zhu M, Zhong Q, Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy, Autophagy. 6 (2010) 614–621. 10.4161/auto.6.5.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Duleh S, Wang X, Komirenko A, Margeta M, Activation of the Keap1/Nrf2 stress response pathway in autophagic vacuolar myopathies, Acta Neuropathol Commun. 4 (2016). 10.1186/s40478-016-0384-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lerner C, Bitto A, Pulliam D, Nacarelli T, Konigsberg M, Van Remmen H, Torres C, Sell C, Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts, Aging Cell. 12 (2013) 966–977. 10.1111/acel.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Ichimura Y, Waguri S, Sou Y-S, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee M-S, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M, Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy, Mol. Cell 51 (2013) 618–631. 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- [166].Hashimoto K, Simmons AN, Kajino-Sakamoto R, Tsuji Y, Ninomiya-Tsuji J, TAK1 Regulates the Nrf2 Antioxidant System Through Modulating p62/SQSTM1, Antioxid. Redox Signal 25 (2016) 953–964. 10.1089/ars.2016.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Yang Y, Willis TL, Button RW, Strang CJ, Fu Y, Wen X, Grayson PRC, Evans T, Sipthorpe RJ, Roberts SL, Hu B, Zhang J, Lu B, Luo S, Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response, Nat Commun. 10 (2019) 3759 10.1038/s41467-019-11671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Goode A, Rea S, Sultana M, Shaw B, Searle MS, Layfield R, ALS-FTLD associated mutations of SQSTM1 impact on Keap1-Nrf2 signalling, Mol Cell Neurosci. 76 (2016) 52–58. 10.1016/j.mcn.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Rolland T, Taşan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis A-R, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere J-C, Vega K, Walsh J, Cusick ME, Xia Y, Barabási A-L, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M, A proteome-scale map of the human interactome network, Cell. 159 (2014) 1212–1226. 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, Yanagawa T, Okamoto K, Iijima M, Sesaki H, Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease, Cell Metab. 28 (2018) 588–604.e5. 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Hu M, Zou Y, Nambiar SM, Lee J, Yang Y, Dai G, Keap1 modulates the redox cycle and hepatocyte cell cycle in regenerating liver, Cell Cycle. 13 (2014) 2349–2358. 10.4161/cc.29298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Taipale M, Tucker G, Peng J, Krykbaeva I, Lin Z-Y, Larsen B, Choi H, Berger B, Gingras A-C, Lindquist S, A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways, Cell. 158 (2014) 434–448. 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Liu X, Zhang Y, Hu Z, Li Q, Yang L, Xu G, The Catalytically Inactive Mutation of the Ubiquitin-Conjugating Enzyme CDC34 Affects its Stability and Cell Proliferation, Protein J. 37 (2018) 132–143. 10.1007/s10930-018-9766-x. [DOI] [PubMed] [Google Scholar]
- [174].Sowa ME, Bennett EJ, Gygi SP, Harper JW, Defining the human deubiquitinating enzyme interaction landscape, Cell. 138 (2009) 389–403. 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Kloska D, Kopacz A, Piechota-Polanczyk A, Nowak W, Dulak J, Jozkowicz A, Grochot-Przeczek A, Nrf2 in aging - Focus on the cardiovascular system, Vascul. Pharmacol. (2018). 10.1016/j.vph.2018.08.009. [DOI] [PubMed] [Google Scholar]
- [176].Wang R, Liu L, Liu H, Wu K, Liu Y, Bai L, Wang Q, Qi B, Qi B, Zhang L, Reduced NRF2 expression suppresses endothelial progenitor cell function and induces senescence during aging, Aging (Albany NY). 11 (2019) 7021–7035. 10.18632/aging.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M, Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation, Nat. Genet 35 (2003) 238–245. 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- [178].Yoshida E, Suzuki T, Morita M, Taguchi K, Tsuchida K, Motohashi H, Doita M, Yamamoto M, Hyperactivation of Nrf2 leads to hypoplasia of bone in vivo, Genes Cells. 23 (2018) 386–392. 10.1111/gtc.12579. [DOI] [PubMed] [Google Scholar]
- [179].Noel S, Arend LJ, Bandapalle S, Reddy SP, Rabb H, Kidney epithelium specific deletion of kelch-like ECH-associated protein 1 (Keap1) causes hydronephrosis in mice, BMC Nephrol. 17 (2016) 110 10.1186/s12882-016-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Suzuki T, Seki S, Hiramoto K, Naganuma E, Kobayashi EH, Yamaoka A, Baird L, Takahashi N, Sato H, Yamamoto M, Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus, Nat Commun. 8 (2017) 14577 10.1038/ncomms14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Hamada S, Shimosegawa T, Taguchi K, Nabeshima T, Yamamoto M, Masamune A, Simultaneous K-ras activation and Keap1 deletion cause atrophy of pancreatic parenchyma, Am. J. Physiol. Gastrointest. Liver Physiol 314 (2018) G65–G74. 10.1152/ajpgi.00228.2017. [DOI] [PubMed] [Google Scholar]
- [182].Yang L, Fan X, Wang G, 016 Hyperactivation of Nrf2 contributes to keratinocyte hyperplasia in psoriasis by promoting Keratin 6, 16 and 17 expressions, J Invest Dermatol. 137 (2017) S3 10.1016/j.jid.2017.02.029. [DOI] [PubMed] [Google Scholar]
- [183].Schäfer M, Willrodt A-H, Kurinna S, Link AS, Farwanah H, Geusau A, Gruber F, Sorg O, Huebner AJ, Roop DR, Sandhoff K, Saurat J-H, Tschachler E, Schneider MR, Langbein L, Bloch W, Beer H-D, Werner S, Activation of Nrf2 in keratinocytes causes chloracne (MADISH)-like skin disease in mice, EMBO Molecular Medicine. 6 (2014) 442–457. 10.1002/emmm.201303281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Köhler UA, Kurinna S, Schwitter D, Marti A, Schäfer M, Hellerbrand C, Speicher T, Werner S, Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis, Hepatology. 60 (2014) 670–678. 10.1002/hep.26964. [DOI] [PubMed] [Google Scholar]
- [185].Tsakiri EN, Gumeni S, Iliaki KK, Benaki D, Vougas K, Sykiotis GP, Gorgoulis VG, Mikros E, Scorrano L, Trougakos IP, Hyperactivation of Nrf2 increases stress tolerance at the cost of aging acceleration due to metabolic deregulation, Aging Cell. 18 (2019) e12845 10.1111/acel.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Fu J, Xiong Z, Huang C, Li J, Yang W, Han Y, Paiboonrungruan C, Major MB, Chen K-N, Kang X, Chen X, Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus, J. Biol. Chem 294 (2019) 327–340. 10.1074/jbc.RA118.005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sánchez-Rivera FJ, Subbaraj L, Martinez B, Bronson RT, Prigge JR, Schmidt EE, Thomas CJ, Goparaju C, Davies A, Dolgalev I, Heguy A, Allaj V, Poirier JT, Moreira AL, Rudin CM, Pass HI, Vander Heiden MG, Jacks T, Papagiannakopoulos T, Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis, Nat. Med 23 (2017) 1362–1368. 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BS, Alvarez SW, Wu WL, Karakousi TR, Zavitsanou AM, Ubriaco J, Muir A, Karagiannis D, Morris PJ, Thomas CJ, Possemato R, Vander Heiden MG, Papagiannakopoulos T, Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer, ELife. 6 (n.d.). 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].LeBoeuf SE, Wu WL, Karakousi TR, Karadal B, Jackson SR, Davidson SM, Wong K-K, Koralov SB, Sayin VI, Papagiannakopoulos T, Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids, Cell Metabolism. 0 (2019). 10.1016/j.cmet.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Huppke P, Weissbach S, Church JA, Schnur R, Krusen M, Dreha-Kulaczewski S, Kühn-Velten WN, Wolf A, Huppke B, Millan F, Begtrup A, Almusafri F, Thiele H, Altmüller J, Nürnberg P, Müller M, Gärtner J, Activating de novo mutations in NFE2L2 encoding NRF2 cause a multisystem disorder, Nat Commun 8 (2017). 10.1038/s41467-017-00932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS, Hartmaier RJ, Chmielecki J, Seshagiri S, Gentleman R, Stokoe D, Recurrent Loss of NFE2L2 Exon 2 Is a Mechanism for Nrf2 Pathway Activation in Human Cancers, Cell Rep. 16 (2016) 2605–2617. 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- [192].Taguchi K, Maher JM, Suzuki T, Kawatani Y, Motohashi H, Yamamoto M, Genetic analysis of cytoprotective functions supported by graded expression of Keap1, Mol. Cell. Biol 30 (2010) 3016–3026. 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Maher J, Yamamoto M, The rise of antioxidant signaling--the evolution and hormetic actions of Nrf2, Toxicol. Appl. Pharmacol 244 (2010) 4–15. 10.1016/j.taap.2010.01.011. [DOI] [PubMed] [Google Scholar]


