Abstract
Atherosclerosis is a chronic inflammatory disease associated with the development of plaques that can be converted into an acute clinical event by thrombosis or plaque rupture. Mesenchymal stem cells (MSCs) exhibit therapeutic effects for the treatment of various diseases, including atherosclerosis. In this study, we show that microRNA-145 (miR-145) is associated with atherosclerosis by microRNA sequencing and bioinformatics analysis. MSC-derived miR-145-rich exosomes could efficiently deliver miR-145 from MSCs to human umbilical vein endothelial cells (HUVECs). Treatment of miR-145-rich exosomes could downregulate JAM-A, inhibit migration in vitro, and reduce atherosclerotic plaque in vivo. Our study suggests that MSC-derived miR-145-rich exosomes have great potential for atherosclerosis prevention.
Keywords: MSC, exosomes, HUVEC, microRNA, atherosclerosis
Graphical Abstract
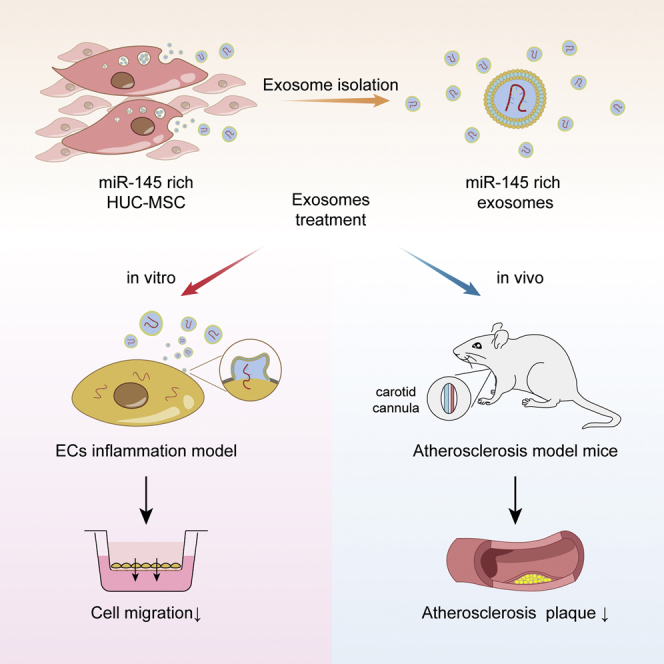
Introduction
Atherosclerosis is a disease of the large arteries and the primary cause of atherosclerotic stroke. In recent years, several genetic and environmental factors related to atherosclerosis have been identified, revealing that atherosclerosis is a chronic inflammatory disease. With the development of plaques, atherosclerosis can process to atherosclerotic stroke through thrombosis and plaque rupture. Atherosclerotic stroke is the main cause of physical disability in modern society; thus, it is necessary to develop new strategies to prevent atherosclerotic stroke.1
Junction adhesion molecule A (JAM-A, also known as F11R) is overexpressed in patients with atherosclerosis, thereby promoting atherosclerotic plaque formation.2,3 The mechanism involves combination between JAM-A and its ligand lymphocyte function-associated antigen-1 (LFA-1), which loosens tight junctions and promotes the migration of endothelial cells (ECs), resulting in increased endothelial permeability.4,5 As a tight junction-associated protein, JAM-A plays an important role in the barrier function of ECs and regulates the migration of ECs.6, 7, 8, 9 Importantly, EC migration can also lead to angiogenesis in atherosclerosis plaques, and plaques with increased angiogenesis exhibit lower stabilization.10 Accordingly, the inhibition of JAM-A may have therapeutic potential in the treatment of atherosclerosis.
Exosomes are extracellular vesicles ranging in size from 50 to 150 nm. These vesicles have been shown to have therapeutic effects in various diseases. The contents of exosomes, including microRNAs, are released to recipient cells, resulting in regulation of biological function.11 MicroRNAs inhibit the expression of specific proteins but are unstable in circulation because they are easily degraded by serum RNase. Notably, exosomes can protect microRNAs from RNases, allowing microRNAs to remain stable in circulation and to be transferred to recipient cells. Human umbilical cord mesenchymal stem cells (HUC-MSCs) produce large numbers of exosomes,12, 13, 14 and such MSC-derived exosomes are expected to be therapeutic in various diseases.15, 16, 17 However, the roles of microRNAs from MSC-derived exosomes in atherosclerosis have not yet been evaluated.
In this study, we used microRNA sequencing to identify differential expressed microRNAs in serum-derived exosomes from patients with atherosclerotic stroke compared with controls. We further evaluated the mechanisms through which the identified microRNAs could modulate EC migration and endothelial barrier function in an MSC and human umbilical vein EC (HUVEC) coculture system.
Results
Differentially expressed exosomal microRNAs between the atherosclerotic stroke and control groups
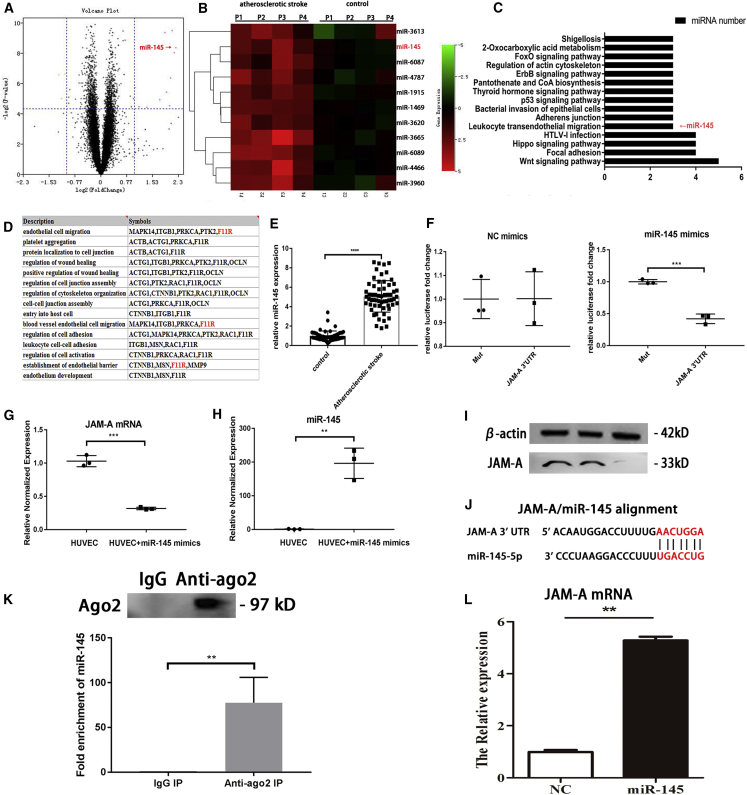
In order to identify the differential expressed exosomal microRNAs in atherosclerotic stroke, microRNA sequencing was conducted using serum-derived exosomes from 16 patients who experienced atherosclerotic stroke and from 16 healthy individuals. In total, 4,603 exosomal microRNAs were evaluated (Figure 1A), and a set of differentially expressed microRNAs was identified (fold change [FC] > 2). The top 10 differentially expressed exosomal microRNAs are listed in the heatmap in Figure 1B. Functional enrichment of these microRNAs (Figure 1C) showed that microRNA-145 (miR-145) was involved in the leukocyte transendothelial migration pathways, which is related to atherosclerosis. According to previous studies and the TargetScanHuman database, F11R (JAM-A) is a target of miR-145. Moreover, functional enrichment analysis and previous studies demonstrated that JAM-A was involved in EC migration and endothelial barrier function, which are associated with atherosclerosis (Figure 1D). Compared with the other differential expressed microRNAs, miR-145 and its targeting gene JAM-A are related with both leukocyte transendothelial migration pathway and endothelial barrier function, which convinced us miR-145 may play an important role in the early stage of atherosclerosis.
Figure 1.
MicroRNA sequencing and function enrichment; miR-145 was chosen for next experiments
(A) Volcano plot of microRNA sequencing. (B) Heatmap of the top 10 DEGs. (C) Functional enrichment of top 10 differentially expressed microRNAs. (D) Functional enrichment of miR-145 target genes. (E) PCR was used to evaluate the expression of miR-145 in serum-derived exosomes from patients with atherosclerotic stroke (n = 60) and healthy controls (n = 60). Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. ∗∗∗∗p < 0.0001. (F) Relative luciferase expression. Unpaired t tests were used to test differences between the two groups. ∗∗p = 0.002. (G) qRT-PCR, effects of miR-145 on JAM-A mRNA in HUVECs; all data are shown as means ± SDs. n = 3. ∗∗∗p = 0.0001. (H) qRT-PCR, miR-145 expression after miR-145 mimics treatment. ∗∗p = 0.0017. (I) Western blot, lane 1, HUVEC; lane 2, HUVEC+NC mimcs; lane 3, HUVEC+miR-145 mimics. (J) JAM-A and miR-145 alignment. (K) Ago2 miR-145 RIP assay. n = 3. ∗p = 0.0093. (L) miR-145 JAM-A mRNA pulldown assay. n = 3. ∗∗p = 0.0055.
To verify that serum-derived exosomal miR-145 was differentially expressed in patients who experienced atherosclerotic stroke, the serum exosomal miR-145 expression was measured from 60 patients who experienced atherosclerotic stroke and 60 healthy individuals (Figure 1E). The results showed that exosomal miR-145 was differentially expressed between the two groups, suggesting that serum exosomal miR-145 may have potential applications as a biomarker of atherosclerotic stroke.
Dual-luciferase assays showed that JAM-A was a target of miR-145. After treatment with negative control mimics, there were no significant differences in activity of the JAM-A 3′ untranslated region (UTR) compared with that in the Mut group. However, after treatment with miR-145 mimics, JAM-A 3′ UTR activity was inhibited. This result verified that JAM-A was a target of miR-145 (Figure 1F). After treatment of HUVECs with miR-145 mimics, JAM-A mRNA levels were decreased compared with that in untreated HUVECs (Figure 1G). Argonaute-2 miR-145 RIP assay suggested that argonaute-2 can combine miR-145. miR-145 JAM-A mRNA pulldown assay showed that JAM-A mRNA can be combined with miR-145. This information demonstrated that miR-145 can suppress JAM-A expression at post-transcription level by binding to JxAM-A mRNA.
Isolation and identification of HUC-MSCs
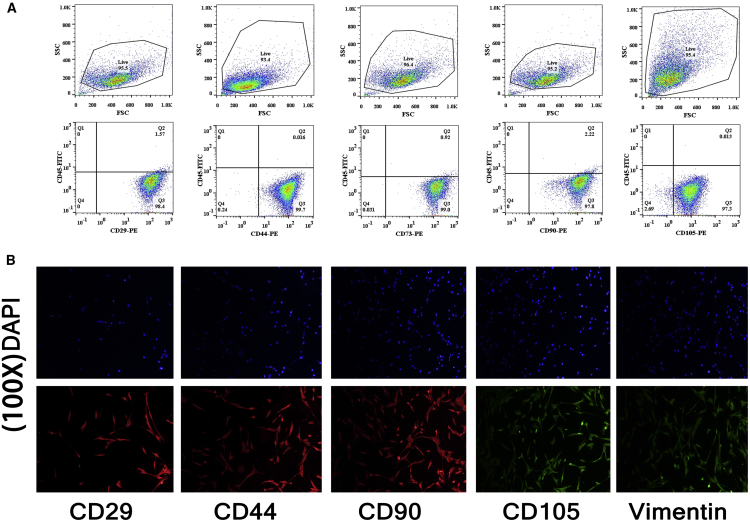
We selected HUC-MSCs as exosome producers. MSCs were isolated from the HUCs of healthy normal deliveries at the Affiliated Hospital of Qingdao University. After isolation, MSCs were identified by flow cytometry and immunofluorescence. CD29, CD44, CD73, CD90, and CD105 were used as positive markers in flow cytometry, and CD45 was used as the negative control (Figure 2A). The MSC markers CD29, CD44, CD90, CD105, and vimentin were detected by immunofluorescence analysis (Figure 2B). The results showed that the positive rate was above 95%, suggesting that the isolated cells were pure HUC-MSCs.
Figure 2.
Identification of MSCs
(A) Flow cytometry analysis of CD29, CD44, CD73, CD90, and CD105 in MSCs. (B) Immunofluorescence of MSC markers CD29, CD44, CD90, CD105, and vimentin; the images were 100 times magnified.
Isolation and identification of MSC-derived exosomes
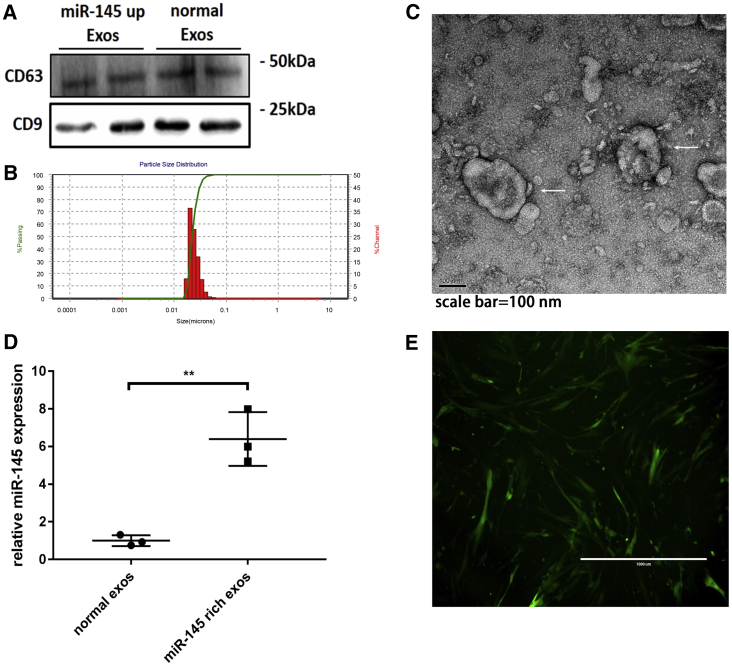
MSC-derived exosomes were isolated from MSC culture medium, and miR-145-rich exosomes were isolated from MSCs transfected with an hsa-miR-145-overexpressing lentivirus. After isolation, transmission electron microscopy, nanoparticle tracking analysis (NTA), and western blotting were used to identify exosomes. The exosome markers CD9 and CD63 were detected by western blotting (Figure 3A). NTA identified MSC-derived exosomes with a diameter of 50–150 nm (Figure 3B). Exosome structures were directly observed by electron microscopy (Figure 3C). These results supported that we successfully isolated exosomes from the cell culture medium. Additionally, PCR showed that the expression of miR-145 in miR-145-rich exosomes was increased by approximately 6.399-fold compared with that in normal exosomes (Figure 3D).
Figure 3.
Identification of exosomes
(A) Exosome markers CD63 and CD9 were evaluated by western blotting. (B) NTA to determine the diameter of exosomes. (C) Exosomes under a transmission electron microscope. (D) Expression of miR-145 in miR-145-rich exosomes. The mean value of the fold change of normal exosomal miR-145 was set as 1. Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. n = 3. ∗∗p = 0.003. (E) MSCs were transfected with miR-145-overexpressing lentivirus.
Exosomes served as carriers of microRNAs from MSCs to ECs
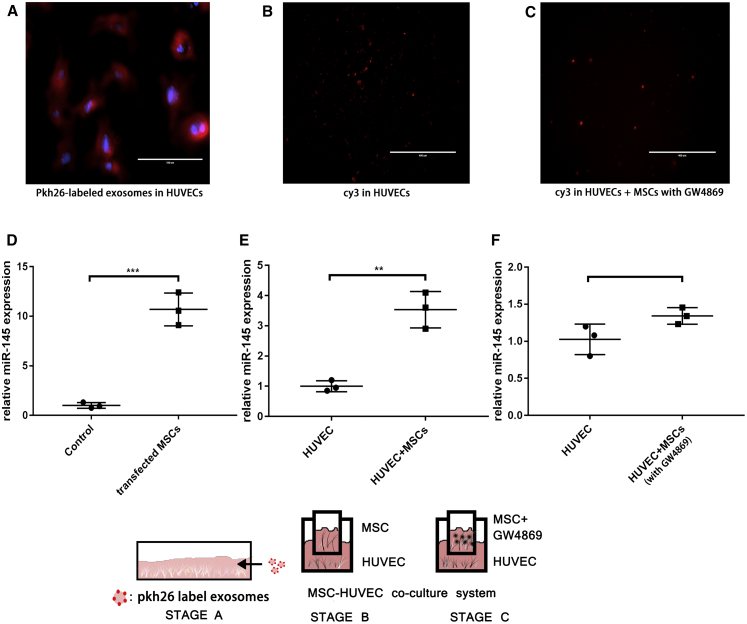
First, to investigate whether the MSC-derived exosomes could be taken up by HUVECs, Pkh26 dye (a fluorescent dye that marks the membranous structure and shows red fluorescence under excitation at 551 nm) was used to mark exosomes. After treatment with Pkh26-stained exosomes, HUVECs were observed under a microscope, and Pkh26 could be seen clearly (Figure 4A). This result suggested that MSC-derived exosomes could be absorbed by HUVECs.
Figure 4.
MSC-derived exosomes transferred miR-145 from MSCs to HUVECs
(A) Stage A. Exosomes were marked with pkh26. HUVECs were treated with pkh26-labeled exosomes. Scale bar, 100 μm. Red, pkh26; blue, DAPI. (B) Stage B. Coculture of MSCs and HUVECs. Scale bar, 400 μm. (C) Stage C. GW4869 blocked exosome secretion, and MSCs and HUVECs were cocultured. Scale bar, 400 μm. (D) miR-145 in transfected MSCs was measured by PCR. The mean value of the fold change in the control group was set as 1. Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. n = 3. ∗∗∗p = 0.0006. (E) Expression of miR-145 in HUVECs in stage B. Mean value of the fold change in the control group was set as 1. Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. n = 3. ∗∗p = 0.0022. (F) Expression of miR-145 in HUVECs in stage C. Mean value of the fold change in the control group was set as 1. Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. n = 3.
Next, we showed that the MSCs could transport microRNAs into HUVECs by secreting exosomes. Cy3 is a fluorescent dye that shows red fluorescence under excitation at 570 nm. We used Cy3-miR-145 mimics to transfect MSCs (Figure 4D) and then performed coculture of HUVECs and MSCs with MSCs in the upper layer and HUVECs in the lower layer. After coculture, Cy3 was clearly detected in HUVECs (Figure 4B), and the expression of miR-145 was also increased (Figure 4E). This result suggested that MSC-derived exosomes could serve as microRNA carriers to transport miR-145 to HUVECs.
To further investigate whether microRNA transportation from MSCs to HUVECs was an exosome-dependent procedure, the exosome secretion inhibitor GW4869 was used to block exosome secretion from transfected MSCs. The results showed that GW4869 blocked miR-145 delivery, and weak fluorescence was observed in HUVECs (Figure 4C); no differences in miR-145 expression were observed compared with that in the control (Figure 4F). These results suggested that microRNA transportation from MSCs to HUVECs was an exosome-dependent process.
miR-145-rich exosomes regulated HUVEC migration
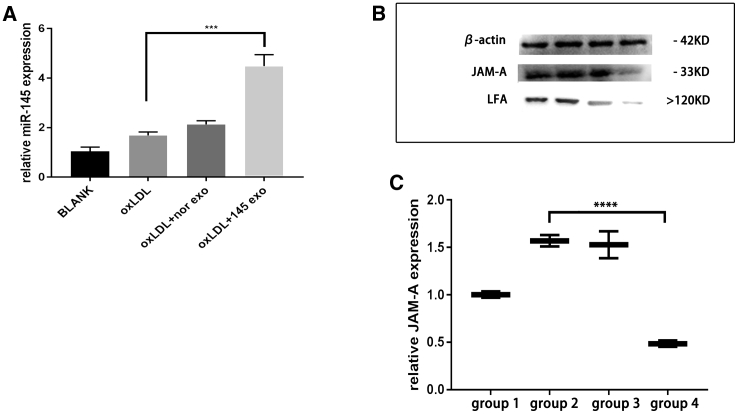
To study the effects of miR-145-rich exosomes in inflammatory HUVECs, we treated HUVECs with or without oxidised low-density lipoprotein (oxLDL), normal exosomes, and miR-145-overexpressing exosomes. qRT-PCR analyses showed that miR-145 levels were increased after treatment with miR-145-rich exosomes (Figure 5A). Additionally, the expression of JAM-A protein was decreased after treatment with miR-145-rich exosomes (Figure 5B and 5C), as was the expression of LFA-1, the ligand of JAM-A. This result suggested that MSC-derived miR-145-rich exosomes could transfer miR-145 to HUVECs and inhibit JAM-A expression.
Figure 5.
miR-145-rich exosomes inhibit JAM-A expression of HUVEC
(A) HUVECs were treated with miR-145-rich exosomes, and PCR was used to measure miR-145 expression in HUVECs. Mean value of the fold change in the control group was set as 1. Unpaired t tests were used to test differences between the two groups. All data are shown as means ± SDs. n = 3. ∗∗∗p = 0.0006. (B) Effects of miR-145-rich exosomes on JAM-A expression in HUVECs. Lane 1, blank control; lane 2, HUVECs treated with oxLDL; lane 3, HUVECs treated with oxLDL plus normal exosomes; lane 4, HUVECs treated with oxLDL plus miR-145-rich exosomes. (C) JAM-A expression in HUVECs. Group 1, blank control; group 2, HUVECs treated with oxLDL; group 3, HUVECs treated with oxLDL plus normal exosomes; group 4, HUVECs treated with oxLDL plus miR-145-rich exosomes. All data are shown as means ± SDs. n = 3. ∗∗∗∗p < 0.0001.
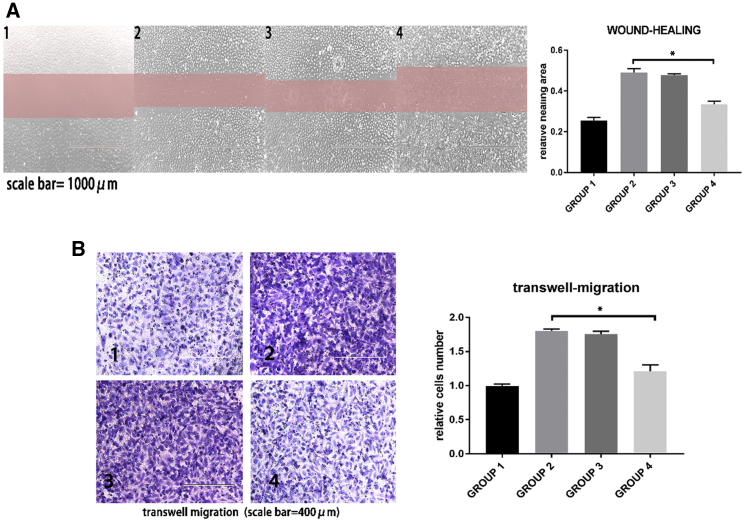
Wound-healing assays (Figure 6A) and transwell migration assays (Figure 6B) were performed to observe HUVEC migration. Following treatment with miR-145-rich exosomes, HUVECs showed slower migration, suggesting that miR-145-rich exosomes could suppress the migration of HUVECs under an inflammatory environment.
Figure 6.
HUVEC migration was evaluated by wound-healing assay and transwell migration assay
Group 1, normal HUVECs; group 2, HUVECs treated with oxLDL; group 3, HUVECs treated with oxLDL + normal exosomes; group 4, HUVECs treated with oxLDL + miR-145-rich exosomes. (A) Wound-healing assay. Compared with group 2, group 4 showed lower healing area. n = 3. ∗p = 0.0110. (B) Transwell migration assay. Compared with group 2, group 4 showed fewer migrating cells. n = 3. ∗p = 0.0131.
miR-145-rich exosomes inhibited HUVEC migration by targeting JAM-A
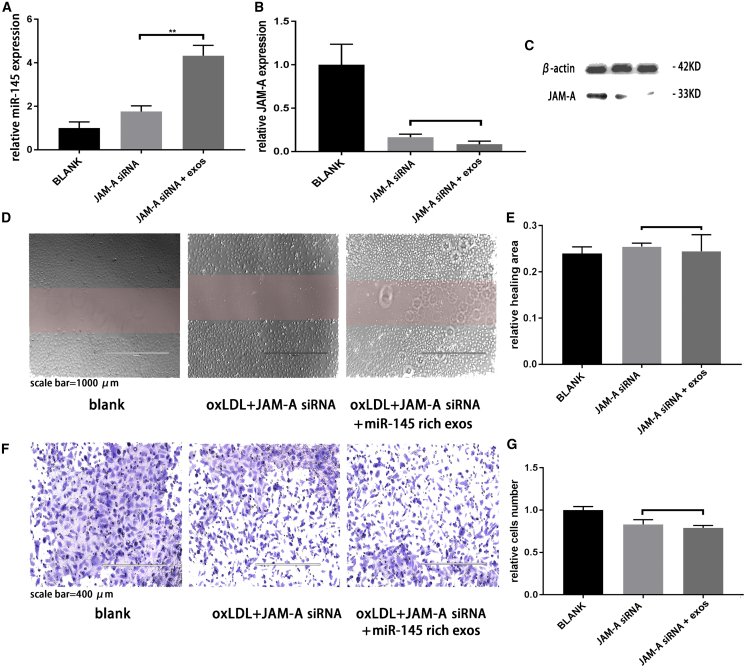
To confirm that JAM-A was targeted by miR-145-rich exosomes to inhibit migration, we performed JAM-A silencing by small interfering RNA (siRNA) in HUVECs and then treated the cells with miR-145-rich exosomes and oxLDL. miR-145 was measured by qRT-PCR (Figure 7A), and JAM-A protein was detected by western blotting (Figure 7B and 7C). Wound-healing assays (Figure 7D and 7E) and transwell migration assays (Figure 7F and 7G) were used to observe HUVEC migration. The results showed that treatment with miR-145-rich exosomes could not reduce the migration of JAM-A-silencing HUVECs under inflammatory conditions. Thus, miR-145-rich exosomes reduced HUVEC migration by inhibiting JAM-A expression.
Figure 7.
On the base of JAM-A silencing, miR-145-rich exosomes have no effect on HUVEC migration
(A) Expression of miR-145 in HUVECs was measured by PCR. n = 3. ∗p = 0.0012. (B and C) Analysis of JAM-A protein expression by western blotting. (D) Wound-healing assay. (E) Quantification of wound-healing assay data. (F) Transwell migration assay. (G) Quantification of transwell migration assay data.
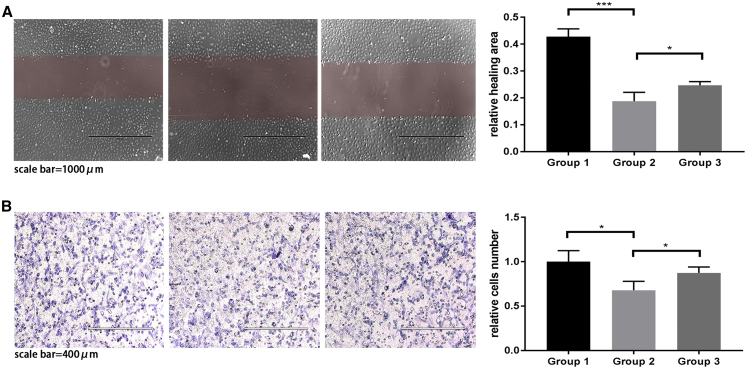
To further demonstrate that miR-145 lowers HUVEC migration, a rescue experiment was also performed. miR-145 mimics and JAM-A plasmid are transfected into HUVECs (Figure 8). On the base of miR-145 mimic treatment, JAM-A plasmid can increase HUVEC migration. This result demonstrates that miR-145 can inhibit HUVEC migration.
Figure 8.
Rescue experiment
Group 1, normal HUVECs; group 2, HUVECs treated with miR-145 mimics; group 3, HUVECs treated with miR-145 mimics + JAM-A plasmid. (A) Wound-healing assay. Compared with group 1, group 2 showed lower healing area. n = 3. ∗∗∗p = 0.0007. Compared with group 2, group 3 showed higher healing area. n = 3. ∗p = 0.0477. (B) Transwell migration assay. Compared with group 1, group 2 showed fewer migrating HUVECs. n = 3. ∗p = 0.0251. Compared with group 2, group 3 showed more migrating HUVECs. n = 3. ∗p = 0.0483.
miR-145-rich exosomes reduced atherosclerotic plaques in vivo
To investigate the effects of miR-145-rich exosomes in vivo, we used carotid artery cannula and a high-fat diet to establish atherosclerosis model mice (Figure 9B). We found that exosome treatment had no influence on mouse body weight. Additionally, miR-145-rich exosomes reduced plaque formation in atherosclerosis model mice, and H&E staining showed that atherosclerotic plaques were reduced after treatment with miR-145-rich exosomes (Figure 9A). Immunohistochemistry analysis and western blot also demonstrated that JAM-A expression was reduced in plaques after treatment with miR-145-rich exosomes (Figure 9C). Overall, these results showed that exosomal miR-145 inhibited JAM-A expression and reduced the formation of atherosclerotic plaques in vivo.
Figure 9.
In vivo experiments
(A) H&E staining of plaques following treatment with miR-145-rich exosomes. The chart shows lumen/artery ratio. After miR-145-rich exosome treatment, the lumen area increased, suggesting that the stenosis level decreased. ∗∗p = 0.0098. (B) Immunohistochemistry was used to evaluate JAM-A expression in plaques. (C) Western blot showed that JAM-A expression was inhibited after miR-145-rich exosomes treatment, the relative JAM-A expression is shown in the following chart. After miR-145-rich exosomes treatment, JAM-A expression decreased. ∗∗p = 0.0057.
Discussion
In this study, we evaluated the potential application of exosomal miR-145 and its target gene JAM-A in the treatment of atherosclerosis using RNA sequencing and bioinformatics analysis. Notably, our findings showed that MSC-derived exosomes could transfer miR-145 into HUVECs.
Various methods of isolating EVs have been developed, including ultracentrifugation,18 buoyant density ultracentrifugation,19 tangential flow filtration, and microfluidic filtration.20,21 Commercial kits we used to isolate exosomes are available to precipitate EV from cell culture medium; the steps are simple and time saving, but the exosomes may be coisolated with other components, and it cannot be applied in large-scale isolation. The function of EVs varies depending on their origin cells,22,23 MSC-derived exosomes conserved most advantages of MSCs, such as low immunogenicity and renewal capability.24 The interaction of exosomes with target cells is a complicated process that is mostly mediated by endocytosis-related processes, including macropinocytosis, clathrin-mediated endocytosis, phagocytosis, and lipid raft-mediating internalization.25, 26, 27 Integrins and tetraspanins play important roles in binding of exosomes and target cells. After binding, the exosomes can be absorbed by the cell membrane, and exosome contents are then released directly into cells.28 In particular, exosomes secreted by brain ECs can cross the blood-brain barrier in a process mediated by several receptors, including insulin receptor, transferrin receptor, LDL receptor-related protein, and TMEM30A on the exosome membrane.29 Taken together, these results suggest that exosomes can serve as containers across different biological barriers, transfer drugs or biological molecules to target cells, and regulate cell-cell communication.
In this study, we found significant differences in the expression of serum-derived exosomal miR-145 from patients with atherosclerosis compared with that in healthy controls. Bioinformatics analysis predicted that miR-145 and its target gene JAM-A were associated with endothelial barrier function and EC migration. Then, we isolated MSCs from the HUC, which served as a producer of exosomes, and overexpressed miR-145 in MSC-derived exosomes. We found that MSC-derived miR-145-rich exosomes could inhibit EC migration, suggesting that these exosomes may regulate HUVEC function. Finally, in vivo experiments showed that MSC-derived miR-145-rich exosomes could inhibit JAM-A expression in plaque area and reduced atherosclerosis plaque sizes. Accordingly, miR-145-containing MSC-derived exosomes may have therapeutic effects in the treatment of atherosclerosis.
Endothelial injury is the initial stage of atherosclerosis. After injury, ECs overexpress inflammatory factor and promote plaque formation. In a study by Schmitt et al.,3 researchers found that under inflammatory conditions, JAM-A expression was increased in ECs. Overexpression of endothelial JAM-A may promote atherosclerosis via binding of JAM-A and LFA-1, which attracts monocytes to adhere to ECs, or by loosening tight junctions between ECs, which enhances EC migration and increases the permeability of the endothelium.3 Moreover, an inhibitory polypeptide of JAM-A was found to inhibit the interaction between ECs and monocytes under an inflammatory environment by blocking the C2 domain of JAM-A, further supporting the role of JAM-A in mediating atherosclerosis.9
The endothelium and its intercellular tight junctional complexes represent a barrier between tissue and blood and can produce functional molecules that regulate macrophage chemotaxis.30 Numerous studies have demonstrated a variety of changes in the metabolic and synthetic activities of ECs in response to defined flow stimulation, including the expression of prostacyclin, nitric oxide, cytokines, growth factors, extracellular matrix components, and vasoactive mediators.31, 32, 33 Additionally, the endothelium can regulate smooth muscle cell migration and proliferation.34 Cholesterol-engorged macrophages, called foam cells, play key roles in early lesions of atherosclerosis. The precursors of macrophages are monocytes, a type of leukocyte. The entry of monocytes into the artery wall is mediated by adhesion and chemotactic molecules secreted by ECs. oxLDL-treated ECs will bind monocytes rather than neutrophils. Additionally, adhesion-associated molecules on the endothelial surface, such as selectins, mediate monocyte rolling and adhesion to the endothelium.35,36 Thus, ECs play key roles in atherosclerosis, and EC-targeting therapy during early atherosclerosis may inhibit plaque formation or reduce plaque size.
Exosomal microRNAs can regulate various biological functions and play important roles in many types of diseases. Importantly, because of their stability in blood, exosomal microRNAs may have applications as disease biomarkers.37 Klein-Scory and colleagues38 found that exosome-contained cellular RNA may provide information regarding early stage pancreatic cancer progression. Moreover, miR-17-92 in MSC-derived exosomes can enhance neurological recovery by promoting oligodendrogenesis, neurogenesis, and axonal outgrowth.39 Patel et al.40 found that expression of the long noncoding RNA MALAT1 in adipose-derived stem cell-derived exosomes has tremendous therapeutic potential for traumatic brain injury by modulating inflammation. Taken together, these studies suggested that exosomal microRNAs may have potential diagnostic or therapeutic value for various diseases. MicroRNAs can have more than one target gene, and one target gene can be suppressed by various microRNAs. In addition to miR-145, our RNA sequencing results also identified another 182 differentially expressed microRNAs between exosomes from patients with atherosclerosis and those from control serum. Another study identified 223 overexpressed microRNAs in THP-1 macrophages treated with lipopolysaccharide.41 Thus, further analyses of these DEGs may provide insights into the complicated mechanisms of atherosclerosis. In summary, our results have shown that miR-145 modified exosomes are efficient tools for gene drug delivery. miR-145-rich exosomes can inhibit the migration of HUVECs via targeting JAM-A in vitro, and administration of miR-145-rich exosomes can inhibit atherosclerosis plaque formation in vivo. In consideration of the poor prognosis of atherosclerotic stroke, our study suggests that the treatment with miR-145-rich exosomes can provide a new therapeutic strategy for atherosclerosis. However, our research only studies the migration of ECs via JAM-A in vitro. Previous research also reported that miR-145 could also affect the angiogenesis. More specific mechanisms of exosomal miR-145 still need to be revealed in further study.
Materials and methods
Ethics statement
All patients enrolled in this study provided informed consent for participation. The study was carried out according to the principles of the Declaration of Helsinki and was approved by the ethics committee of our institution. All animal procedures were done in accordance with the ethics committee.
Sequencing of exosomal microRNAs from patients with atherosclerosis
Thirty-two patients were enrolled in the microRNA sequencing, including 16 patients who experienced atherosclerotic stroke and 16 healthy individuals. All patients were recruited from the Affiliated Hospital of Qingdao University and were of Chinese Han ancestry. There were no significant differences in age between the patient group and healthy group. Sixteen patients were randomly divided into four pools, and unpaired t tests were used to examine differentially expressed genes (DEGs) with a cutoff of p ≤0.05 and fold change >2. Volcano plots and heatmaps were produced using R software.
Bioinformatics analysis
Functional enrichment of the top 10 differentially expressed exosomal microRNAs was performed using DIANA-miRPath.42 The target genes of specific microRNAs were predicted using the TargetScanHuman database.43 Functional enrichment of proteins was performed using Metascape.44
Dual-luciferase assays
The reporter genes were designed by Takara (Shiga, Japan). Lipofectamine 3000 transfection reagent (Thermo Scientific, USA) was used to transfect reporter genes into HUC-MSCs. After cotransfection of miR-145 mimics and reporter plasmid, the HUVECs were cultured in fetal bovine serum (FBS)-free medium in 48-well plates for 8 h. The medium was then replaced with complete culture medium. At 72 h after transfection, cellular protein was harvested using passive lysis buffer and used for measurement of luciferase activity with a dual luciferase assay kit (Promega, Madison, WI, USA). Relative luciferase activity was calculated using raw data from firefly luciferase normalized to Renilla luciferase activity.
Isolation of HUC-MSCs from the HUC
HUCs were obtained postpartum from full-term healthy infants delivered via normal vaginal delivery at Qingdao University Affiliated Hospital after obtaining informed consent according to institutional guidelines under an approved protocol. HUC tissues were cut into 3- to 5-cm-long pieces. The surface of the cord was rinsed with sterile phosphate-buffered saline (PBS) to remove as much blood as possible. Vessels were removed to retain the Wharton’s jelly, which was then cut into 1-mm pieces and cultured in complete culture medium (90% Dulbecco’s modified Eagle’s medium [DMEM]/F12 and 10% FBS, supplemented with 1% penicillin-streptomycin). When cells were 80% confluent, they were digested with trypsin and resuspended in culture medium. Cells were diluted at a ratio of 1:2, replated, and cultured at 37°C in an incubator with an atmosphere containing 5% CO2.
Cytometry and immunofluorescence analyses for identification of HUC-MSCs
The positive markers CD44, CD90, CD105, CD29, and CD73 and the negative marker CD45 were detected by flow cytometry. CD44, CD90, CD105, CD29, and vimentin were observed by immunofluorescence. All antibodies were purchased from Abcam (Cambridge, UK).
Cell culture
HUVECs were used as model ECs. HUVECs were cultured in 90% DMEM containing 10% FBS and 1% penicillin-streptomycin. HUC-MSCs were cultured in 90% DMEM/F12 containing 10% FBS and 1% penicillin-streptomycin. All cells were cultured in 5% CO2 at 37°C in a humidified atmosphere. DMEM, FBS, and penicillin-streptomycin were purchased from BI (BI, USA), and trypsin was purchased from Thermo Scientific. Culture flasks (T25) were purchased from Corning (Corning, NY, USA).
Transfection
MSCs were transfected with 10 nM Cy3-labeled miR-145 mimic (Takara) using Lipo3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To obtain miR-145-overexpressing exosomes, MSCs were transfected with a miR-145-overexpressing lentivirus (Genechem) at a multiplicity of infection of 20. MSCs were cultured in serum-free medium for 6 h after transfection, and AFP fluorescence was observed after 3 days.
Isolation and identification MSC-derived exosomes
MSCs were cultured to 3rd passage before the experiment. For the miR-145-rich exosomes, we used the miR-145 overexpression lentivirus to transfect 3rd-passage MSCs; after 1 passage culturing, transfected MSCs were expanded to obtain the culturing medium to isolate miR-145-rich exosomes. For normal exosomes, 3rd-passage MSCs were expanded directly to obtain the culturing medium to isolate normal exosomes. All MSCs are cultured in T75 culture flasks (Corning, NY, USA). The culture medium was harvested every passage operation. To guarantee that all isolated exosomes are MSC derived, all completed culture mediums were made with exosome-depleted FBS (BI, USA).
The total exosome isolation reagent was purchased from Thermo Scientific. The harvested cell culture medium was centrifuged at 2,000 × g for 30 min to remove cells and debris. Then, 1 volume of cell-free culture medium was fixed with 0.5 volumes of reagent and incubated at 4°C overnight. Samples were centrifuged at 10,000 × g for 1 h at 4°C, supernatants were discarded, and exosome pellets were resuspended in PBS. Isolated exosomes were identified by electron microscopy observation, western blotting of exosomal markers CD9 and CD63, and NanoSight analysis.
In vivo and in vitro exosome treatment
In vitro, 2 μg of exosomes on the basis of protein measurement were added to 105 recipient cells. In vivo, 80 μg of exosomes were injected into recipient mice weekly via tail-vein injection from the 9th week to 16th week. The concentration of miR-145 in miR-145-rich exosome solution is about 0.932 × 103 copies/μL.
Electron microscopy
For observation under electron microscopy, exosomes were fixed with paraformaldehyde and loaded on Formvar- and carbon-coated copper grids. Grids were then placed in 2% gelatin, incubated at 37°C for 20 min, rinsed with PBS, and blocked with 1% cold water fish-skin gelatin. Exosomes were viewed under a Talos F200C transmission electron microscope (Thermo Fisher).
Western blotting
JAM-A expression was examined by western blotting. Proteins were isolated from HUVECs, then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% milk (Solarbio) in TBST (Solarbio) for 2 h at room temperature and incubated overnight at 4°C with primary antibodies (1:1,000 dilution; all antibodies were purchased from Abcam). The membranes were then incubated for 1 h at room temperature with a 1:2,000 dilution of secondary antibody (Abcam). Protein bands on the blots were detected using chemiluminescence horseradish peroxidase (HRP) substrate (Millipore) with a fusionX imaging system (Vilber). All antibodies were purchased from Abcam. Gray levels were calculated using ImageJ software and normalized to the level of β-actin expression. The relative expression in the blank control was set as 1.
qRT-PCR
miR-145 levels in exosomes and HUVECs were examined by qRT-PCR. Total RNA was extracted from exosomes or HUVECs, and RNA was then reverse transcribed into cDNA. In brief, the samples were incubated for 1 h at 37°C, and the reaction was terminated at 85°C. For qPCR, denaturation was performed at 95°C for 10 s, followed by amplification at 95°C for 5 s and 60°C for 20 s in 40 cycles. Mir-X microRNA first-strand synthesis and TB green qRT-PCR kits were purchased from Takara. miR-145-5p primers were designed by Takara. qRT-PCR was carried out using a Roche LightCycler 480 II. Data were normalized to U6 expression. The mean value in the control group was set as 1. All data are shown as means ± standard deviations (SDs).
RNA interference
JAM-A silencing was achieved by JAM-A siRNA. The JAM-A siRNA was purchased from Takara.
Wound-healing assay
Cells were seeded in 6-well plates in complete culture medium. Wounds were created using 200 μL sterile pipette tips when the cells reached 90% confluence, and the plates were then washed with PBS. Closing of the wound was observed after 24 h. All experiments were performed in the presence of 5 μg/mL mitomycin C to inhibit cell proliferation. ImageJ software was used to analyze the results. Relative healing area was calculated as follows: relative healing area = (initial area − final area)/initial area. Data are shown as means ± SDs.
Transwell migration assay
Transwell chambers (8 μm) were purchased from Corning. HUVECs were seeded in the upper chambers and cultured with serum-free medium. Complete medium containing 10% serum was added to the lower chamber. After culture for 12 h, cells in the upper chamber were removed, and invasive cells embedded in the membrane of the transwells were fixed with 4% paraformaldehyde, stained with crystal violet for 15 min, and observed under a microscope. ImageJ software was used to analyze the results. The mean cell number in the control group was set as 1 for determination of the relative cell number. Data are shown as means ± SDs.
Atherosclerosis model mice
All mice were apoE−/− female mice (in C57BL/6J background, Vital River, Beijing, China) and were housed in the animal facility of the Affiliated Hospital of Qingdao University on a 12-h light-dark cycle. All mice were fed a high-fat diet (15% fat and 0.25% cholesterol; KEAOXIELI, Beijing, China), except mice in the blank control group. When mice were 8 weeks old, mice were anesthetized by intraperitoneal injection of pentobarbital sodium (4 mg/100 g) before the surgery. The right common carotid artery was inserted with a silicone ring to induce the formation of carotid atherosclerotic plaques. From the 9th to 16th week, exosomes were given every week. After the 16th week, mice were euthanized by using pentobarbital sodium (15 mg/100 g). After euthanasia, the right common carotid artery tissues were immediately frozen in liquid nitrogen and then embedded in paraffin for subsequent experiments. The plaque size was examined by H&E staining, and JAM-A expression was detected by immunohistochemistry.
H&E staining
Microscope slides containing rehydrated tissue sections were immersed in water for 30 s with agitation by hand. Slides were then dipped into a Coplin jar containing Mayer’s hematoxylin and agitated for 30 s. Next, slides were rinsed in water for 1 min and stained with 1% eosin Y solution for 10–30 s with agitation. Sections were dehydrated with two changes of 95% alcohol and two changes of 100% alcohol for 30 s each. The alcohol was extracted with two changes of xylene, and one drop of mounting medium was added, followed by coverslipping. The lumen area ratio was calculated to evaluate the level of stenosis; the formula is as follows: lumen area ratio = lumen area/artery area. Smaller lumen area ratio means higher level of stenosis.
Immunohistochemistry
Immunohistochemistry for JAM-A was performed. First, sections were permeabilized by microwaving in 10 mM citrate buffer (pH 6.0) for 15 min. Immunostaining was carried out by using the avidin-biotinylated enzyme complex method with anti-JAM-A antibodies (1 μg/mL). After incubation with the appropriate biotin-conjugated secondary antibodies and streptavidin solution, color development was performed using 3,3-diaminobenzidine tetrahydrochloride as a chromogen. Sections were counterstained using Gill-2 hematoxylin (Thermo Fisher). After staining, sections were dehydrated by ethanol and xylene. All antibodies were purchased from Abcam.
Statistical analysis
PCR, western blot, and migration assay data are shown as means ± SDs. Student’s t tests, analysis of variance, and chi-square tests were used to compare differences between groups. For western blotting, the mean of the normalized gray level in the control group was defined as a relative FC of 1. For transwell migration assays, the mean cell number in the control group was defined as a relative cell number of 1. For qRT-PCR, the relative FC was calculated as follows: FCU6 = 2(CU6 − CT); relative FC = (FCu61 + FCu62 +……+ FCu6n)/n. All data were normalized to the FC of the control group.
Acknowledgments
We gratefully acknowledge the support of many individuals who made this study possible and the contributions of plasma donors. This work was funded by the National Natural Science Foundation (81971111).
Author contributions
W.Y. conducted experiments, statistical analysis, and drafted the manuscript. R.Y., X.Z., J.W., and Z.Z. contributed to the experiment design. X.P. and A.M. contributed to the experiment design and manuscript revision.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.037.
Contributor Information
Xudong Pan, Email: drpan055@163.com.
Aijun Ma, Email: drmaj@126.com.
Supplemental information
References
- 1.Adeoye O., Hornung R., Khatri P., Kleindorfer D. Recombinant tissue-type plasminogen activator use for ischemic stroke in the United States: a doubling of treatment rates over the course of 5 years. Stroke. 2011;42:1952–1955. doi: 10.1161/STROKEAHA.110.612358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y.J., Bai D.N., Du J.X., Jin L., Ma J., Yang J.L., Cai W.B., Feng Y., Xing C.Y., Yuan L.J., Duan Y.Y. Ultrasound-guided imaging of junctional adhesion molecule-A-targeted microbubbles identifies vulnerable plaque in rabbits. Biomaterials. 2016;94:20–30. doi: 10.1016/j.biomaterials.2016.03.049. [DOI] [PubMed] [Google Scholar]
- 3.Schmitt M.M.N., Megens R.T.A., Zernecke A., Bidzhekov K., van den Akker N.M., Rademakers T., van Zandvoort M.A., Hackeng T.M., Koenen R.R., Weber C. Endothelial junctional adhesion molecule-a guides monocytes into flow-dependent predilection sites of atherosclerosis. Circulation. 2014;129:66–76. doi: 10.1161/CIRCULATIONAHA.113.004149. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt M.M.N., Fraemohs L., Hackeng T.M., Weber C., Koenen R.R. Atherogenic mononuclear cell recruitment is facilitated by oxidized lipoprotein-induced endothelial junctional adhesion molecule-A redistribution. Atherosclerosis. 2014;234:254–264. doi: 10.1016/j.atherosclerosis.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Luissint A.C., Williams H.C., Kim W., Flemming S., Azcutia V., Hilgarth R.S., Leary M.N.O., Denning T.L., Nusrat A., Parkos C.A. Macrophage-dependent neutrophil recruitment is impaired under conditions of increased intestinal permeability in JAM-A-deficient mice. Mucosal Immunol. 2019;12:668–678. doi: 10.1038/s41385-019-0143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mamdouh Z., Mikhailov A., Muller W.A. Transcellular migration of leukocytes is mediated by the endothelial lateral border recycling compartment. J. Exp. Med. 2009;206:2795–2808. doi: 10.1084/jem.20082745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nava P., Capaldo C.T., Koch S., Kolegraff K., Rankin C.R., Farkas A.E., Feasel M.E., Li L., Addis C., Parkos C.A., Nusrat A. JAM-A regulates epithelial proliferation through Akt/β-catenin signalling. EMBO Rep. 2011;12:314–320. doi: 10.1038/embor.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw S.K., Ma S., Kim M.B., Rao R.M., Hartman C.U., Froio R.M., Yang L., Jones T., Liu Y., Nusrat A. Coordinated redistribution of leukocyte LFA-1 and endothelial cell ICAM-1 accompany neutrophil transmigration. J. Exp. Med. 2004;200:1571–1580. doi: 10.1084/jem.20040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sladojevic N., Stamatovic S.M., Keep R.F., Grailer J.J., Sarma J.V., Ward P.A., Andjelkovic A.V. Inhibition of junctional adhesion molecule-A/LFA interaction attenuates leukocyte trafficking and inflammation in brain ischemia/reperfusion injury. Neurobiol. Dis. 2014;67:57–70. doi: 10.1016/j.nbd.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra R., O’Neil G.L., Harding I.C., Cheng M.J., Mensah S.A., Ebong E.E. Glycocalyx in Atherosclerosis-Relevant Endothelium Function and as a Therapeutic Target. Curr. Atheroscler. Rep. 2017;19:63. doi: 10.1007/s11883-017-0691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abels E.R., Breakefield X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai R.C., Arslan F., Lee M.M., Sze N.S., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. (Amst.) 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Yeo R.W., Lai R.C., Zhang B., Tan S.S., Yin Y., Teh B.J., Lim S.K. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 2013;65:336–341. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Sarmah D., Agrawal V., Rane P., Bhute S., Watanabe M., Kalia K., Ghosh Z., Dave K.R., Yavagal D.R., Bhattacharya P. Mesenchymal Stem Cell Therapy in Ischemic Stroke: A Meta-analysis of Preclinical Studies. Clin. Pharmacol. Ther. 2018;103:990–998. doi: 10.1002/cpt.927. [DOI] [PubMed] [Google Scholar]
- 16.Vu Q., Xie K., Eckert M., Zhao W., Cramer S.C. Meta-analysis of preclinical studies of mesenchymal stromal cells for ischemic stroke. Neurology. 2014;82:1277–1286. doi: 10.1212/WNL.0000000000000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yong K.W., Choi J.R., Mohammadi M., Mitha A.P., Sanati-Nezhad A., Sen A. Mesenchymal Stem Cell Therapy for Ischemic Tissues. Stem Cells Int. 2018;2018:8179075. doi: 10.1155/2018/8179075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 19.Lobb R.J., Becker M., Wen S.W., Wong C.S.F., Wiegmans A.P., Leimgruber A., Möller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan C.Y., Lai R.C., Wong W., Dan Y.Y., Lim S.K., Ho H.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res. Ther. 2014;5:76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies R.T., Kim J., Jang S.C., Choi E.J., Gho Y.S., Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 22.Choi D.S., Kim D.K., Kim Y.K., Gho Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 23.Jong A.Y., Wu C.H., Li J., Sun J., Fabbri M., Wayne A.S., Seeger R.C. Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells. J. Extracell. Vesicles. 2017;6:1294368. doi: 10.1080/20013078.2017.1294368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding M., Shen Y., Wang P., Xie Z., Xu S., Zhu Z., Wang Y., Lyu Y., Wang D., Xu L. Exosomes Isolated From Human Umbilical Cord Mesenchymal Stem Cells Alleviate Neuroinflammation and Reduce Amyloid-Beta Deposition by Modulating Microglial Activation in Alzheimer’s Disease. Neurochem. Res. 2018;43:2165–2177. doi: 10.1007/s11064-018-2641-5. [DOI] [PubMed] [Google Scholar]
- 25.Tian T., Zhu Y.L., Zhou Y.Y., Liang G.F., Wang Y.Y., Hu F.H., Xiao Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014;289:22258–22267. doi: 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svensson K.J., Christianson H.C., Wittrup A., Bourseau-Guilmain E., Lindqvist E., Svensson L.M., Mörgelin M., Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013;288:17713–17724. doi: 10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzner D., Schnaars M., van Rossum D., Krishnamoorthy G., Dibaj P., Bakhti M., Regen T., Hanisch U.K., Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 28.Rana S., Zöller M. Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem. Soc. Trans. 2011;39:559–562. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 29.Haqqani A.S., Delaney C.E., Tremblay T.L., Sodja C., Sandhu J.K., Stanimirovic D.B. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10:4. doi: 10.1186/2045-8118-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusis A.J. Atherosclerosis. Nature. 2000;407:233–241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimbrone M.A., Jr., Nagel T., Topper J.N. Biomechanical activation: an emerging paradigm in endothelial adhesion biology. J. Clin. Invest. 1997;99:1809–1813. doi: 10.1172/JCI119346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnick N., Gimbrone M.A., Jr. Hemodynamic forces are complex regulators of endothelial gene expression. FASEB J. 1995;9:874–882. doi: 10.1096/fasebj.9.10.7615157. [DOI] [PubMed] [Google Scholar]
- 33.Topper J.N., Gimbrone M.A., Jr. Blood flow and vascular gene expression: fluid shear stress as a modulator of endothelial phenotype. Mol. Med. Today. 1999;5:40–46. doi: 10.1016/s1357-4310(98)01372-0. [DOI] [PubMed] [Google Scholar]
- 34.Gimbrone M.A., Jr. Vascular endothelium, hemodynamic forces, and atherogenesis. Am. J. Pathol. 1999;155:1–5. doi: 10.1016/S0002-9440(10)65090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong Z.M., Chapman S.M., Brown A.A., Frenette P.S., Hynes R.O., Wagner D.D. The combined role of P- and E-selectins in atherosclerosis. J. Clin. Invest. 1998;102:145–152. doi: 10.1172/JCI3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins R.G., Velji R., Guevara N.V., Hicks M.J., Chan L., Beaudet A.L. P-Selectin or intercellular adhesion molecule (ICAM)-1 deficiency substantially protects against atherosclerosis in apolipoprotein E-deficient mice. J. Exp. Med. 2000;191:189–194. doi: 10.1084/jem.191.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y., Shen Z., Yu X.Y. Transport of microRNAs via exosomes. Nat. Rev. Cardiol. 2015;12:198. doi: 10.1038/nrcardio.2014.207-c1. [DOI] [PubMed] [Google Scholar]
- 38.Klein-Scory S., Tehrani M.M., Eilert-Micus C., Adamczyk K.A., Wojtalewicz N., Schnölzer M., Hahn S.A., Schmiegel W., Schwarte-Waldhoff I. New insights in the composition of extracellular vesicles from pancreatic cancer cells: implications for biomarkers and functions. Proteome Sci. 2014;12:50. doi: 10.1186/s12953-014-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xin H., Katakowski M., Wang F., Qian J.Y., Liu X.S., Ali M.M., Buller B., Zhang Z.G., Chopp M. MicroRNA cluster miR-17-92 Cluster in Exosomes Enhance Neuroplasticity and Functional Recovery After Stroke in Rats. Stroke. 2017;48:747–753. doi: 10.1161/STROKEAHA.116.015204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel N.A., Moss L.D., Lee J.Y., Tajiri N., Acosta S., Hudson C., Parag S., Cooper D.R., Borlongan C.V., Bickford P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflammation. 2018;15:204. doi: 10.1186/s12974-018-1240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortega F.J., Mercader J.M., Moreno-Navarrete J.M., Rovira O., Guerra E., Esteve E., Xifra G., Martínez C., Ricart W., Rieusset J. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 42.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.