Abstract
Dysfunction of granulosa cells (GCs) leading to follicle atresia has been extensively studied as a major cause of premature ovarian insufficiency (POI), but the regulatory role of long non-coding RNAs (lncRNAs) in this process is still poorly understood. Here, we show that the lncRNA LINC02690 or GCAT1 (granulosa cell-associated transcript 1) is downregulated in GCs from patients with biochemical POI (bPOI), and we show a significant correlation between downregulated GCAT1 and serum levels of follicle-stimulating hormone and anti-Müllerian hormone. Downregulation of GCAT1 inhibited G1/S cell cycle progression and thus inhibited the proliferation of GCs. Mechanistically, we show that GCAT1 competes with cyclin-dependent kinase inhibitor 1B (CDKN1B) mRNA for polypyrimidine tract-binding protein 1 (PTBP1) binding, and thus decreased GCAT1 might promote PTBP1 binding to CDKN1B mRNA and thereby initiate CDKN1B protein (p27) translation. Together, our results suggest that downregulation of GCAT1 under conditions of bPOI inhibits the proliferation of GCs through PTBP1-dependent p27 regulation, thus suggesting a novel form of lncRNA-mediated epigenetic regulation of GC function that contributes to the pathogenesis of POI.
Graphical Abstract
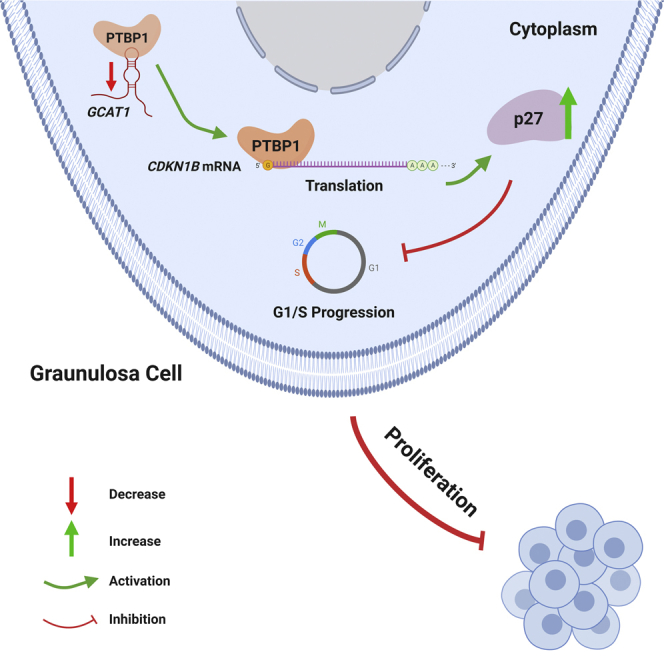
The role of lncRNAs in the development of premature ovarian insufficiency (POI) is poorly described. Qin et al. revealed that lncRNA GCAT1 partakes POI by modulating the proliferation of granulosa cells in a manner of lncRNA-protein-mRNA network, suggesting the potential of GCAT1 as novel target for POI diagnosis and therapy.
Introduction
Premature ovarian insufficiency (POI) is characterized by the occurrence of menopause before the age of 40 years resulting from the premature depletion of the ovarian primordial follicle pool. This condition is characterized by infertility, elevated serum follicle-stimulating hormone (FSH), and decreased estradiol (E2) levels. Patients with biochemical POI (bPOI) present with increased serum levels of FSH (>10 IU/L) and reduced fertility but still with regular menstruation, which is considered as the early stage of POI. POI affects 1%∼5% of all women globally and has serious impacts on reproductive and psychological health.1,2 In 20%∼25% of cases, POI is associated with genetic abnormalities, but the causes in the majority of patients remain to be elucidated.3
Granulosa cells (GCs) are follicular somatic cells that provide essential nutrients and secrete the steroids needed for the progression of folliculogenesis.4,5 The proliferation and differentiation of GCs are critical for follicle maturation and ovulation, and aberrant apoptosis and degeneration of GCs results in follicular atresia.6, 7, 8 Given that dysfunctional GCs have been implicated in POI,9,10 identifying the regulatory network of functional GCs will provide important insights into the understanding of POI pathogenesis.
In the human genome, the majority of transcripts are non-coding RNAs (ncRNAs), and less than 1.5% encode proteins.11 Defined by their length, long non-coding RNAs (lncRNAs), which are classified as longer than 200 nucleotides, account for > 80% of ncRNAs,12 and the depletion, overexpression, or mutation of lncRNAs has been shown to be associated with various cellular dysfunctions.13,14 Notably, functional lncRNAs have been shown to regulate gene expression and biological behavior in GCs, and therefore they are involved in female reproductive diseases such as polycystic ovary syndrome (PCOS), endometriosis, and POI.15, 16, 17 Nevertheless, due to the low levels of expression and poor conservation of lncRNAs compared with protein-coding RNAs, the roles and causative mechanisms of lncRNAs in POI remain unclear.
In the present study, we demonstrate that downregulation of lncRNA LINC02690, also called GC-associated transcript 1 (GCAT1), facilitated the binding of cyclin-dependent kinase inhibitor 1B (CDKN1B) mRNA to polypyrimidine tract-binding protein 1 (PTBP1), thereby initiating CDKN1B protein (p27) translation. Silencing of GCAT1 resulted in cell cycle arrest in G1 phase, and this inhibited the proliferation of GCs. These findings suggest a previously unrecognized lncRNA-protein-mRNA regulatory network that is involved in GC function and suggest a novel mechanism for POI pathogenesis.
Results
Downregulation of lncRNA GCAT1 in GCs is clinically related with POI
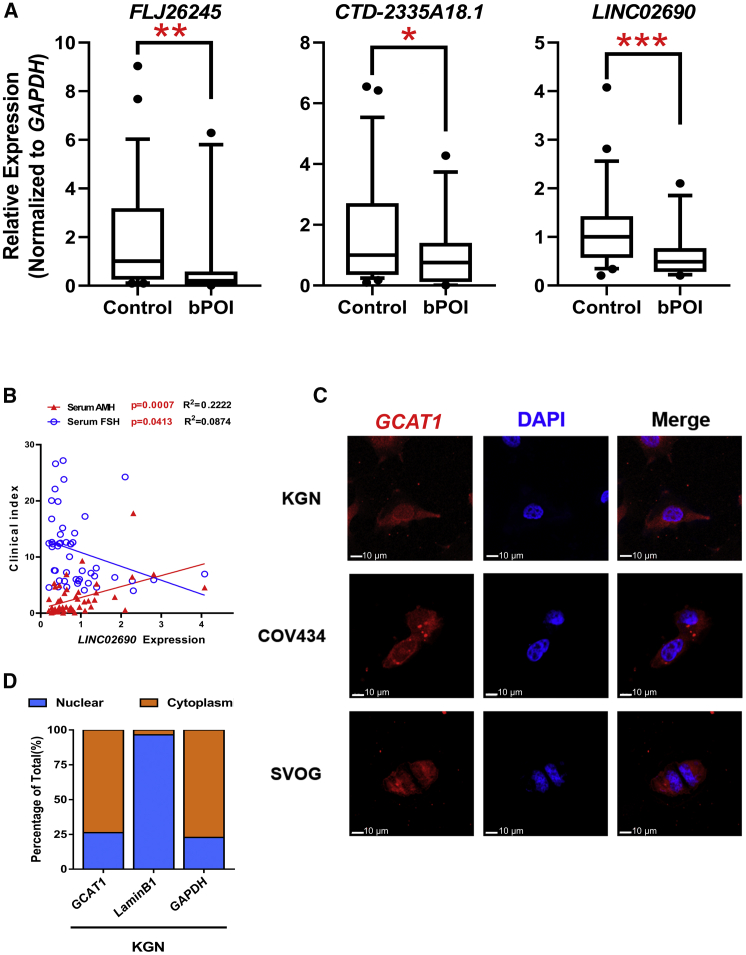
Using the Arraystar human lncRNA expression microarray v3.0, we previously17 explored the global expression profile of lncRNAs in GCs of bPOI patients (n = 10) and controls (n = 10) (GenBank: GSE135697). After initial screening by fold change (>2), length (<2,000 nt), raw signal intensity (>100), and without overlapping with coding transcripts, the top seven most differentially expressed lncRNAs (FLJ26245, RP11-626G11.1, AC139099.6, CTD-2335A18.1, C8orf69, LINC02690, and RP11-773D16.1) were filtered out. Among the seven lncRNA candidates, as confirmed by qRT-PCR, FLJ26245, CTD-2335A18.1, and LINC02690 were downregulated in independent GC samples from 24 patients with bPOI and 24 age-matched controls (Figures 1A and S1A). The Pearson correlation analysis showed that only LINC02690 was significantly correlated with serum levels of basic FSH (p = 0.0413, R2 = 0.08744, n = 48) and anti-Müllerian hormone (AMH) (p = 0.0007, R2 = 0.2222, n = 48) (Figures 1B and S1B), both of which are commonly used indicators of ovarian reserve. Therefore, we focused on LINC02690 for further investigation, considering its potential pathological and clinical value, and referred to it as GCAT1.
Figure 1.
Reduced expression of lncRNA GCAT1 in GCs is clinically relevant in POI
(A) The expression levels of three differentially expressed lncRNAs were validated by qRT-PCR in GCs from an independent cohort of patients with bPOI (n = 24) and controls (n = 24). Ct values were normalized to GAPDH. Data are presented as the median ± interquartile range. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by two-tailed Mann-Whitney U test. (B) The correlation between the expression level of GCAT1 in GCs and the serum concentration of AMH and FSH was analyzed by Pearson correlation analysis. (C) The subcellular localization of GCAT1 was detected by RNA FISH assay in KGN, COV434, and SVOG cells. (D) Relative GCAT1 expression levels in the cytoplasmic and nuclear fractions of KGN cells by qRT-PCR. Lamin B1 was used as the nuclear control, and GAPDH was used as the cytoplasmic control.
In the human genome, GCAT1 resides on chromosome 11p11.2 as part of an intergenic fragment between the genes CHST1 and SLC35C1. The coding potential calculator18 and coding-potential assessment tool19 confirmed that GCAT1 is a non-coding RNA due to its negligible protein-coding potential (Figures S2A and S2B). The single-cell RNA sequencing (RNA-seq)20 analysis showed that GCAT1 was selectively expressed in human GCs of secondary and antral follicles during folliculogenesis (Figure S2C). GCAT1 was found to be localized in both the nucleus and cytoplasm of GCs, with the majority in the cytoplasmic fraction, as shown by fluorescence in situ hybridization (FISH) and cellular fractionation followed by qRT-PCR (Figures 1C and 1D).
Silencing of GCAT1 inhibits GC proliferation
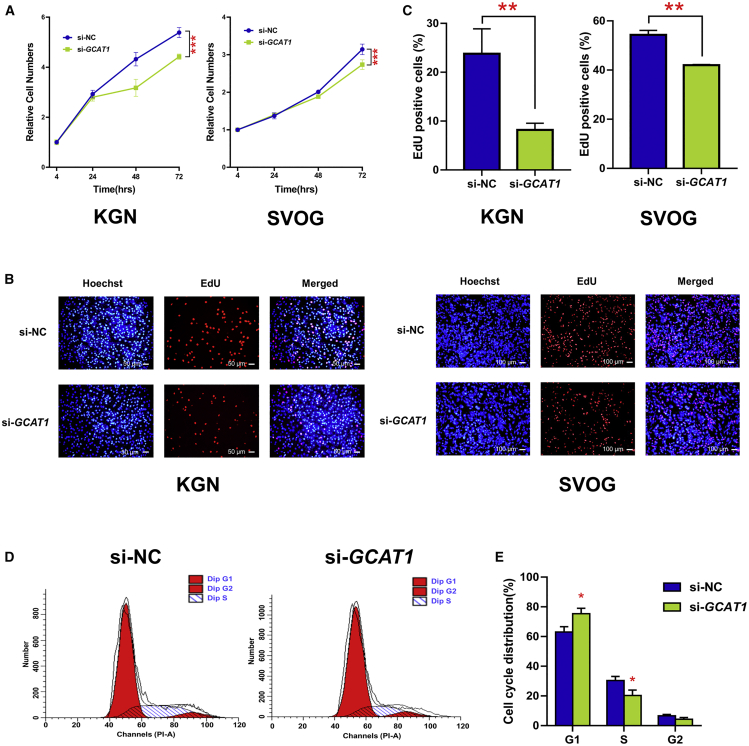
To investigate the role of GCAT1 in the biological function of GCs, cell counting kit 8 (CCK8) assays were performed to evaluate cell viability after silencing GCAT1 in KGN and SVOG cell lines using specific small interfering RNA (siRNA) (Figure S3A). As shown in Figure 2A, silencing of GCAT1 led to a significant decrease in cell number. 5-ethynyl-2′-deoxyuridine (EdU) staining showed that the reduced cell number was the result of inhibited proliferation of GCs (Figures 2B and 2C), and flow cytometry analysis showed an increased percentage of G1-phase cells and a decreased percentage of S-phase cells after silencing of GCAT1 (Figures 2D and 2E). Taken together, these results indicated that silencing GCAT1 inhibits GC proliferation by preventing the G1/S transition of the cell cycle.
Figure 2.
Silencing of GCAT1 inhibited GC proliferation and G1/S cell cycle progression
(A) CCK8 assay showing the viability of KGN and SVOG cells after GCAT1 silencing. Results are expressed as the mean ± SD (n = 3). ∗∗∗p < 0.001 by two-tailed Student’s t test. (B and C) EdU staining assay showing the proliferation of KGN and SVOG cells after GCAT1 silencing. Results are expressed as the mean ± SD (n = 3). ∗∗p < 0.01 by two-tailed Student’s t test. (D and E) Flow cytometry analysis of the cell cycle distribution of KGN cells after GCAT1 silencing. Results are expressed as the mean ± SD (n = 3). ∗p < 0.05 by two-tailed Student’s t test.
GCAT1 interacts with the PTBP1 and heterogeneous nuclear ribonucleoprotein K (hnRNPK) proteins
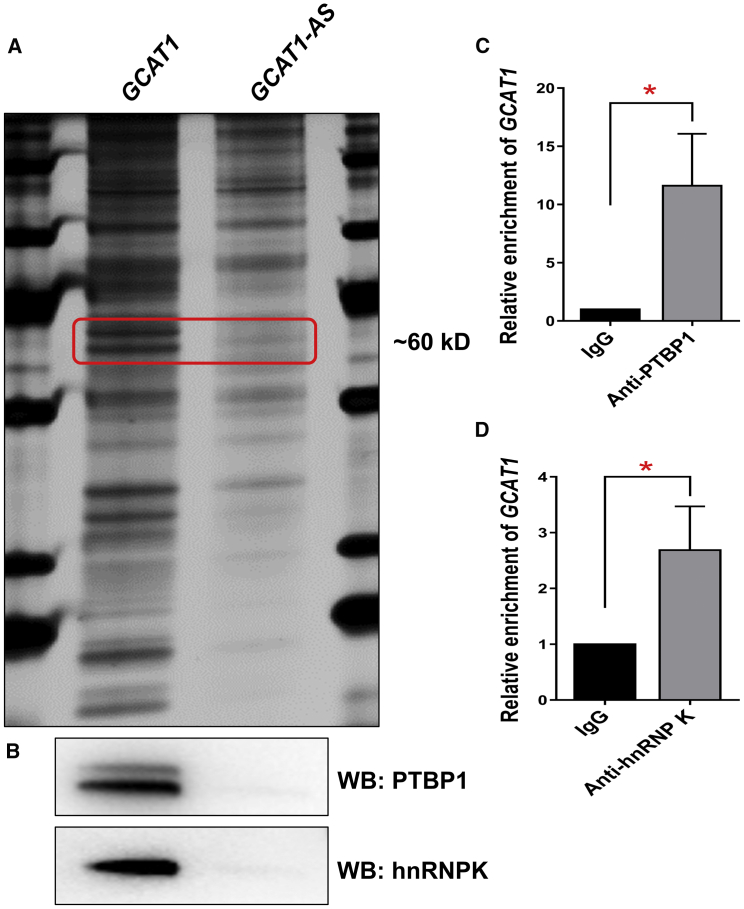
To identify the proteins that bind to GCAT1, we performed an RNA pull-down assay. All proteins pulled down from KGN cells by GCAT1 and its antisense control GCAT1-AS were separated by SDS-PAGE and visualized by silver staining (Figure 3A). The specific bands present in the GCAT1 probe lane were cut out, digested with trypsin, and analyzed by mass spectrometry. According to their unique peptides, the upper band was identified as PTBP1 (Figure S3B), and the lower band as hnRNPK proteins (Figure S3C). To further substantiate these interactions, western blot was performed, and the results showed that PTBP1 and hnRNPK were detected in the GCAT1 pull-down but not in the GCAT1-AS pull-down (Figure 3B). Consistent with these results, RNA-binding protein immunoprecipitation (RIP) confirmed the enrichment of GCAT1 by PTBP1 and hnRNPK antibodies compared with isotype immunoglobulin G (IgG) control in KGN cells (Figures 3C and 3D). Taken together, these findings demonstrate that GCAT1 directly interacts with PTBP1 and hnRNPK.
Figure 3.
GCAT1 interacts with the PTBP1 and hnRNPK proteins
(A) Detection of GCAT1-binding proteins by RNA pull-down assays. Two specific bands of ~60 kDa (red box) were pulled down by GCAT1 and subsequently identified by mass spectrometry. (B) Western blot showing the specific association between GCAT1 and PTBP1 and hnRNPK in the samples obtained from the RNA pull-down. The antisense transcript of GCAT1 was used as the negative control. (C and D) Confirmation of the interaction between GCAT1 and PTBP1 and hnRNPK by RIP using the PTBP1 antibody and hnRNPK antibody in KGN cells. Results are expressed as the mean ± SD (n = 3). ∗p < 0.05 by two-tailed Student’s t test.
Silencing GCAT1 increases p27 expression by regulating PTBP1-mediated translation
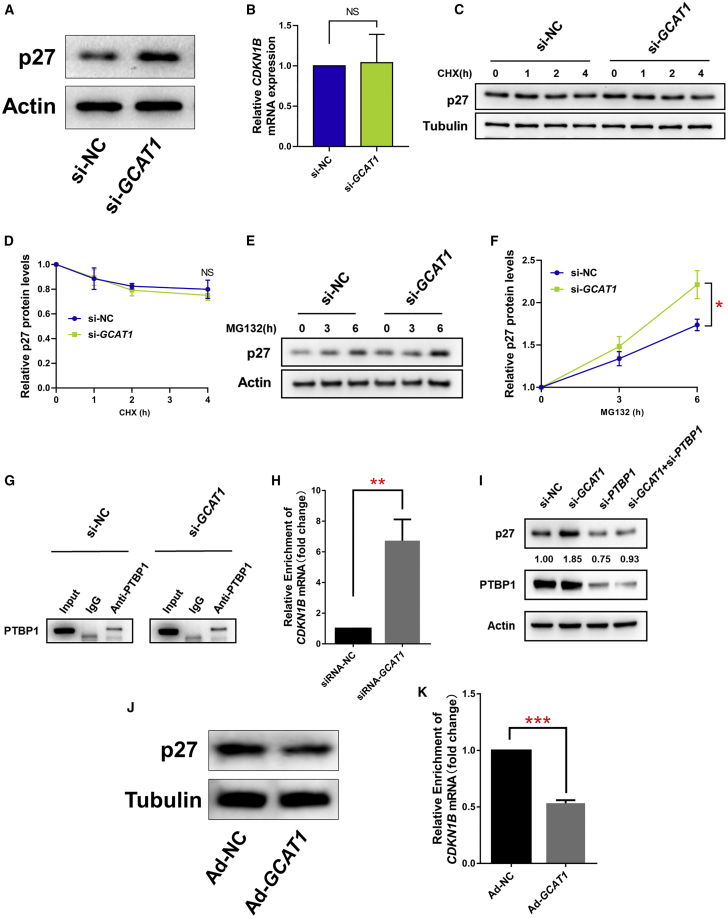
The observation that GCAT1 could directly bind to PTBP1 was particularly interesting because PTBP1 is known to bind to an internal ribosome entry segment (IRES) element in the 5′ untranslated region of CDKN1B mRNA and to promote the translation of p27, which is a well-known G1-related CDK inhibitor. We therefore further explored whether GCAT1 could modulate the expression of p27 as expected. Interestingly, silencing of GCAT1 significantly increased p27 protein levels but did not affect its mRNA level (Figures 4A and 4B), implying that GCAT1 regulates p27 expression at the post-transcriptional level. To determine how GCAT1 regulates p27 expression, we used cycloheximide (CHX) to inhibit translational activity, and the increased p27 protein after GCAT1 silencing was completely reversed (Figures 4C and 4D). In addition, MG132 was used to block the proteasome-degradation pathway, but no effect on the increased level of p27 protein was observed after GCAT1 silencing (Figures 4E and 4F). Together, these results suggest that GCAT1 regulates p27 protein levels by modulating its translation rather than through the proteasome-degradation pathway.
Figure 4.
Silencing of GCAT1 increased p27 expression by regulating PTBP1-mediated translation
(A) p27 protein levels were analyzed by western blot after GCAT1 silencing in KGN cells. The results represent three independent experiments. (B) The mRNA levels of CDKN1B were analyzed by qRT-PCR after GCAT1 silencing in KGN cells. Values of qRT-PCR were obtained from triplicate experiments and expressed as the mean ± SD (n = 3). Two-tailed Student’s t test. (C and D) Western blot showing the p27 protein levels in KGN cells after GCAT1 silencing and exposure to CHX. The results represent three independent experiments. (E and F) Western blot showing the p27 protein levels in KGN cells after GCAT1 silencing and exposure to MG132. The data represent three independent experiments. (G and H) RIP assays showed the enrichment of CDKN1B mRNA immunoprecipitated by PTBP1 antibody after silencing of GCAT1. Results are expressed as the mean ± SD (n = 3). ∗p < 0.05 by two-tailed Student’s t test. (I) Western blot showing p27 protein levels after GCAT1 silencing, PTBP1 silencing, and co-silencing in KGN cells. The data shown represent three independent experiments. (J) P27 protein levels were analyzed by western blot after GCAT1 overexpression in KGN cells. The results represent three independent experiments. (K) RIP assays showed the enrichment of CDKN1B mRNA immunoprecipitated by PTBP1 antibody after overexpression of GCAT1. Results are expressed as the mean ± SD (n = 3). ∗∗∗p < 0.001 by two-tailed Student’s t test.
Based on the binding of GCAT1 to PTBP1, we hypothesized that GCAT1 regulates p27 translation by competing with CDKN1B mRNA for PTBP1 binding. To validate this hypothesis, we first examined whether GCAT1 regulates the binding of PTBP1 to CDKN1B mRNA. As shown in Figures 4G and 4H, silencing of GCAT1 significantly increased the amount of CDKN1B mRNA immunoprecipitated by the PTBP1 antibody. More importantly, the elevated p27 protein levels accompanying GCAT1 silencing could be reversed by co-silencing of PTBP1 (Figure 4I), which implied that PTBP1 is required for GCAT1-mediated regulation of p27 protein levels. Furthermore, GCAT1 overexpression (Figure S3D) significantly reduced the protein levels of p27 as well as the binding between PTBP1 and CDKN1B mRNA (Figures 4J and 4K). Taken together, our results suggest that GCAT1 competes with CDKN1B mRNA for binding to PTBP1; therefore, the downregulation of GCAT1 increases p27 translation.
GCAT1 regulates GC proliferation and G1/S transition via p27
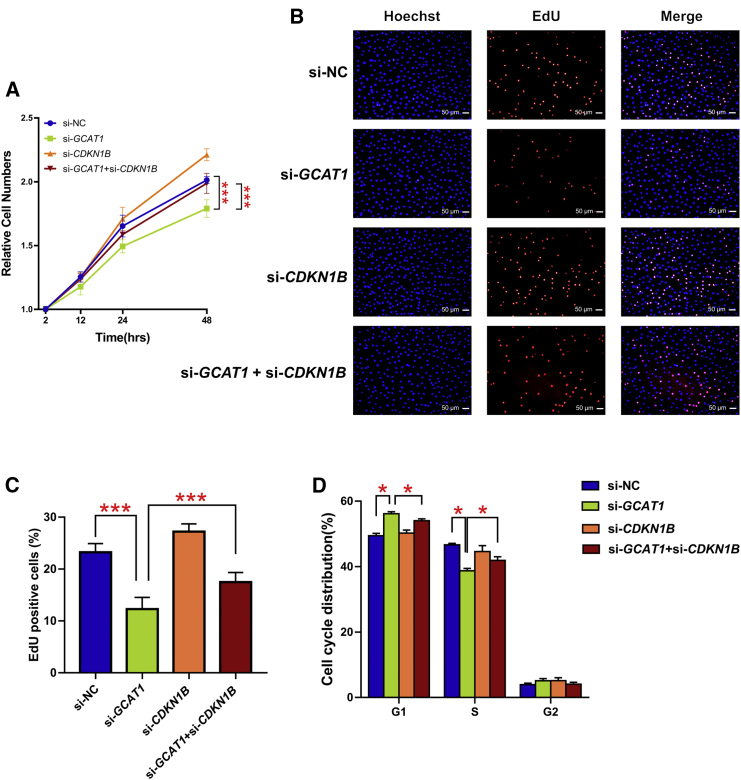
To determine whether the influence of GCAT1 on GC proliferation and G1/S cell cycle arrest was dependent on p27, GCAT1 and CDKN1B were separately or simultaneously silenced in KGN cells. As shown in Figures 5A–5C, silencing of GCAT1 significantly inhibited cell proliferation, and this was partially rescued by the simultaneous silencing of CDKN1B. Similarly, CDKN1B silencing also markedly reversed the effect of GCAT1 on G1/S arrest (Figure 5D). Collectively, these results suggest that the effect of GCAT1 deficiency on GC proliferation and G1/S arrest is mediated by p27.
Figure 5.
GCAT1 regulated GC proliferation and G1/S transition via p27
(A) The CCK8 assay showed the cell viability of KGN cells after GCAT1 silencing, CDKN1B silencing, and co-silencing. Results are expressed as the mean ± SD (n = 3). ∗∗∗p < 0.001 by two-tailed Student’s t test. (B and C) EdU staining assay showing the proliferation ability of KGN cells after GCAT1 silencing, CDKN1B silencing, and co-silencing. Results are expressed as the mean ± SD (n = 3). ∗∗∗p < 0.001 by two-tailed Student’s t test. (D) The flow cytometry cell cycle distribution of KGN cells after GCAT1 silencing, CDKN1B silencing, and co-silencing. Results are expressed as the mean ± SD (n = 3). ∗p < 0.05 by two-tailed Student’s t test.
Discussion
Here, we found that lncRNA GCAT1 was significantly downregulated in GCs from patients with bPOI and that silencing of GCAT1 inhibited G1/S cell cycle progression and the proliferation of GCs by promoting p27 translation by competing with CDKN1B mRNA for PTBP1 binding. These findings suggest a novel epigenetic mechanism consisting of lncRNA-protein-mRNA interactions in the pathogenesis of POI.
In mammals, communication between the oocyte and its neighboring GCs is critical for follicular development,8 and the proliferation and differentiation of GCs determines the follicle’s fate.21 Loss-of-function mutations within genes responsible for GC proliferation and differentiation—such as FOXL2,22 BMP15,23 and WT124—have been found to be causative for POI. Furthermore, we previously found that lncRNA HCP5 participates in the pathogenesis of POI by transcriptionally regulating MSH5 and the DNA damage repair process via YB1.17 Therefore, elucidating the lncRNA-mediated regulatory network for GC function is essential for a comprehensive understanding of POI pathogenesis. In the present study, we first showed the downregulation of lncRNA GCAT1 in GCs from patients with bPOI. Subsequent in vitro experiments showed that silencing of GCAT1 significantly inhibited GC proliferation by arresting the cell cycle in G1 phase. Similar findings have been reported in PCOS and endometriosis. For example, lncRNA LINC-01572:28 was previously found to inhibit GC proliferation by reducing p27 degradation in patients with PCOS,15 and knockdown of lncRNA MALAT1 could inhibit GC proliferation by upregulating p21 in endometriosis patients.16 Our results provide new molecular evidence that dysregulated lncRNAs are responsible for the aberrant proliferation of GCs.
Our study identified p27 as the effector through which GCAT1 regulates GC proliferation. As a member of the cyclin-dependent kinase inhibitor family, p27 is well known as an inhibitor of cell cycle progression. P27−/− female mice are infertile due to a disrupted estrus cycle and impaired luteal cell differentiation,25 and previous evidence suggests that p27 modulates ovarian development by suppressing ovarian follicle activation and promoting follicular atresia.26 In mice, the expression of p27 is predominantly observed in the oocytes and pre-GCs of primordial follicles. During follicle growth, the high level of p27 persists in oocytes, whereas significantly reduced expression of p27 is observed in GCs. It thus appears that p27 maintains the meiotic arrest of oocytes, while decreased p27 levels in GCs promote their proliferation and subsequent follicle development and ovulation.27 In this study, we found that silencing of GCAT1 increased p27 expression by fine-tuning p27 translation, which inhibited cell cycle progression and the proliferation of GCs, thus leading to follicular atresia and ovarian insufficiency.
The expression of p27 can be regulated at the transcriptional, post-transcriptional, and translational levels,28, 29, 30 and it has been shown that the RNA binding protein PTBP1 can bind to the IRES of CDKN1B mRNA, thereby initiating the translation of p27.30 In this study, we showed that the increased p27 expression after GCAT1 silencing is PTBP1-dependent and that GCAT1 competes with CDKN1B mRNA for binding to PTBP1, thereby regulating p27 translation in GCs. As a member of the heterogenous nuclear ribonucleoprotein family, PTBP1 is an RNA-binding protein responsible for RNA splicing, mRNA stability, and translation initiation.31, 32, 33 Interestingly, lncRNA TRMP and lncRNA OVAAL have been shown to compete with CDKN1B mRNA for PTBP1 binding, resulting in the suppression of p27 protein in non-small-cell lung cancer and colon cancer.34,35 Our results show that PTBP1-associated GCAT1 is a crucial regulator of p27 protein in GCs, further confirming that PTBP1 serves as an important binding partner of lncRNAs in mediating p27 translation. Collectively, our findings demonstrate the regulatory role of the dynamic interaction between GCAT1 and PTBP1 in human GC cell cycle progression and proliferation. As for the other GCAT1-binding protein hnRNPK, this might provide an alternative pathway through which GCAT1 regulates GC function, and this is worthy of future investigation.
Interestingly, we showed that GCAT1 expression in GCs is significantly correlated with serum levels of AMH and FSH, both of which are the most commonly used clinic indicators for ovarian reserve. To our knowledge, this is the first lncRNA involved in POI pathogenesis that is related to ovarian reserve markers. In addition, p27 has been widely accepted as a prognostic and therapeutic marker in human cancers.36 Our results improve our understanding of the role of lncRNA-mediated p27 expression in human GCs and suggest the potential value of p27 as a preventative and therapeutic target for POI. Therefore, GCAT1 might be a promising diagnostic marker for ovarian reserve, and its interaction with p27 might hold therapeutic relevance to patients with POI.
In conclusion, our findings suggest a previously unrecognized mechanism through which lncRNA-induced inhibition of GC proliferation acts as a potential inducer of POI, and we show that downregulation of GCAT1 leads to the disruption of GC proliferation and to cell cycle arrest followed by follicle atresia and eventual ovarian insufficiency. The lncRNA-protein-mRNA regulatory network for GC function suggests a novel mechanism for POI pathogenesis. In addition, strategies aimed at inhibiting p27 expression in GCs may provide a translational method to recover GC proliferation and prevent GCAT1-related POI.
Materials and methods
Participants and study approval
A total of 24 bPOI patients receiving in vitro fertilization or intracytoplasmic sperm injection and embryo transfer at the Center for Reproductive Medicine, Shandong University (Jinan, China) were recruited. The inclusion criteria of bPOI were (1) basal serum FSH >10 IU/L, (2) <40 years of age, and (3) regular menstruation (23–35 days). Women with chromosomal abnormalities or a history of ovarian surgery, chemotherapy, or radiotherapy were excluded. As controls, 24 age-matched women with regular menstrual cycles and normal serum FSH levels (<10 IU/L) who sought infertility treatment due to tubal obstruction or male factors were enrolled. The clinical characteristics of all participants are shown in Table 1. Ovarian GC samples were collected on the day of oocyte retrieval and processed according to standard procedures.37 All experiments were conducted with the approval of the Institutional Review Board of Reproductive Medicine of Shandong University. Written informed consent was obtained from all participants.
Table 1.
Clinical characteristics of patients with bPOI and controls
| Variables | Control (n = 24) | bPOI (n = 24) | p value |
|---|---|---|---|
| Baseline characteristics | |||
| age (y) | 29.33 ± 3.48 | 31.04 ± 3.57 | 0.100a |
| BMI (kg/m2) | 21.71 (19.83, 22.61) | 21.43 (19.32, 26.51) | 0.813b |
| basal FSH (IU/L) | 5.86 (4.82, 7.09) | 13.32 (12.34, 20.01) | <0.0001b |
| basal LH (IU/L) | 5.55 (3.88, 5.84) | 5.40 (4.06, 8.62) | 0.359b |
| basal E2 (pg/mL) | 29.75 (23.00, 42.77) | 30.45 (12.90, 41.55) | 0.578b |
| AMH (ng/mL) | 3.71 (2.40, 5.29) | 0.48 (0.31, 0.94) | <0.0001b |
Data are presented as mean ± SD or median (inter-quartile range [IQR]) based on distribution. BMI, body mass index.
Student’s t test.
Mann-Whitney U test.
Cell lines and cell culture
The human granulosa-like tumor cell line KGN38 was the kind gift of the RIKEN BioResource Center in Japan (Tsukuba, Japan). Another human granulosa-like tumor cell line, COV434, was obtained from Prof. Ying Xu of Nanjing University in China (Nanjing, China). The SVOG luteinized GC line was the kind gift of Prof. Peter C.K. Leung of the University of British Columbia in Canada to the Chinese University of Hong Kong-Shandong University Joint Laboratory on Reproductive Genetics (Hong Kong, China). The KGN, SVOG, and COV434 cells were cultured in DMEM/F-12 medium (HyClone, USA) and DMEM/high-glucose medium (HyClone, USA) supplemented with 10% fetal bovine serum (Biological Industries, Israel) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere of 5% CO2 at 37°C.
RNA isolation and qRT-PCR analysis
Total RNA from GCs and the cell lines was isolated using TRIzol reagent (Invitrogen) according to standard protocols. Approximately 700 ng of RNA were reverse transcribed using the PrimeScript RT reagent kit (Takara, Kyoto, Japan). For GCAT1 detection, reverse transcription was carried out using the reverse transcriptase M-MLV (RNase H−) kit (Takara) and specific reverse transcription primers. qRT-PCR labeled by SYBR green master mix (Takara) was performed on a LightCycler 480 system (Roche, USA). The primers for qRT-PCR are listed in Table S1.
FISH
A mix of probes targeting lncRNA GCAT1 were synthesized and labeled with Cy3 modification (RiboBio, Guangzhou, China). The hybridization experiment was performed in KGN, SVOG, and COV434 cells using the fluorescent in situ hybridization kit (RiboBio) according to the manufacturer’s instructions and visualized on a spinning disc confocal microscope (ANDOR, UK).
Cell fractionation
KGN cells were fractionated using the PARIS kit (Invitrogen) according to the manufacturer’s instructions. RNA was isolated from both nuclear and cytoplasmic fractions and then analyzed by qRT-PCR.
Cell transfection
KGN and SVOG cells were transfected with 100 nM siRNAs using lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. All specific siRNAs against GCAT1, PTBP1, and CDKN1B as well as the negative control siRNA were designed and produced by Genepharma Technology (GenePharma, Shanghai, China). The sequences of the siRNAs are listed in Table S2. Adenoviruses for overexpressing GCAT1 (Ad-GCAT1) were generated and purchased from Hanbio (Shanghai, China). The empty virus expressing only GFP served as the negative control (Ad-NC).
Cell viability
KGN and SVOG cells were transfected and seeded onto 96-well plates at an initial density of 6,000 cells per well. Cell viability was assessed by the absorbance at 450 nm using the CCK8 (Beyotime, Shanghai, China) according to the manufacturer’s instructions.
EdU proliferation assays
Forty-eight hours after transfection, KGN and SVOG cells were seeded onto 96-well plates until ∼80% confluency and then incubated with 50 μM EdU diluted in complete medium for 2 hours. The EdU staining was performed using the cell-light EdU DNA cell proliferation kit (RiboBio) according to the manufacturer’s instructions, and the images were acquired on a fluorescence microscope (Olympus, Japan).
Cell cycle analysis
KGN cells were cultured in 6-cm2 dishes. Forty-eight hours after transfection, cells were harvested and fixed in 70% pre-chilled ethanol at −20°C overnight followed by staining with 7-Aminoactinomycin D (7-AAD) (Multi Sciences, Hangzhou, China). The cells were then subjected to flow cytometry cell cycle distribution analysis.
RNA pull-down assay
To synthesize sense or antisense full-length GCAT1 RNA, the pcDNA3.1 plasmid (Invitrogen) containing the GCAT1 cDNA template was linearized with specific restriction enzymes and transcribed in vitro using the MEGAscript T7 transcription kit (Invitrogen). Cultured KGN cell lysates were incubated with magnetic bead-conjugated sense or antisense GCAT1 probes, and the co-precipitated proteins were isolated and subjected to SDS-PAGE analysis. The protein bands were further visualized by silver staining. The specific bands pulled down by GCAT1 were cut out followed by mass spectrometry or western blot.
Western blot
Cultured cells were washed twice with ice-cold PBS and then lysed in SDS lysis buffer (Beyotime) supplemented with protease inhibitor cocktail (Cell Signaling Technology, Boston, MA, USA). Cell lysates were collected immediately according to the manufacturer’s instructions and subjected to bicinchoninic acid (BCA) protein assay (Invitrogen) for concentration determination. Equal amounts of proteins in each lane were separated by SDS-PAGE and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% (w/v) skim milk and then incubated with specific primary antibodies overnight followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (Proteintech, Chicago, IL, USA). An enhanced chemiluminescence (ECL) chemiluminescence kit (Millipore) was used to detect immunoreactive protein bands using a ChemiDoc MP imaging system (Bio-Rad, USA). The details for the antibodies are provided in Table S3.
RIP
The RIP assay was performed using the EZ-Magna RIP kit (Millipore) according to the manufacturer’s instructions. For each RIP assay, about 1 × 107 KGN cells were lysed in RIP lysis buffer supplemented with RNase inhibitor and protease inhibitor cocktail and then subjected to immunoprecipitation at 4°C overnight with PTBP1 or hnRNPK antibodies. A homologous IgG was used as the isotype control. After six washes with RIP wash buffer, the co-precipitated RNA was extracted using TRIzol reagent according to standard protocols. The purified RNA was then reverse transcribed and subjected to qRT-PCR analysis.
CHX chase assay
The stability of p27 protein was determined by CHX chase assay. 24 h after transfection, KGN cells were treated with 100 μg/mL CHX for 0, 1, 2, or 4 h to block translation; DMSO was used as the control reagent. The cell lysates were analyzed by western blot.
MG132 exposure
KGN cells were transfected with indicated siRNA. After 24 h, cells were exposed to 10 μM MG132 for 0, 3, or 8 h to block the proteasome pathways; DMSO was used as the control reagent. The cell lysates were analyzed by western blot.
Statistics
Statistical analysis was performed using SPSS 23.0 (IBM, USA) and GraphPad Prism 8 (GraphPad Software, USA) to assess differences between experimental groups. Unless otherwise noted, statistical significance was analyzed by a two-tailed Student’s t test. The Mann-Whitney U-test was used for independent samples when the population could not be assumed to be normally distributed. Pearson correlation and linear regression analyses were used to determine the association between GCAT1 and clinical characteristics. Differences were considered statistically significant at p <0.05.
Acknowledgments
This work was supported by the National Key Research & Developmental Program of China (2017YFC1001100), the Science Foundation for Distinguished Young Scholars of Shandong (JQ201720), and the National Natural Science Foundation of China (81771541 and 81671413). The authors thank all participants. The authors thank BioRender.com for providing help in creating the schematic graph.
Author contributions
Y.Q. and S.Z. designed and supervised the study. D.L. and X.W. conducted the experiment. Y.D. and X.Z. contributed to acquisition of results. D.L. and X.W. wrote the manuscript. G.L., W.-Y.C., and P.C.K.L. provided the SVOG cell line. All of the authors approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.041.
Supplemental information
References
- 1.Coulam C.B., Adamson S.C., Annegers J.F. Incidence of premature ovarian failure. Obstet. Gynecol. 1986;67:604–606. [PubMed] [Google Scholar]
- 2.Webber L., Davies M., Anderson R., Bartlett J., Braat D., Cartwright B., Cifkova R., de Muinck Keizer-Schrama S., Hogervorst E., European Society for Human Reproduction and Embryology (ESHRE) Guideline Group on POI ESHRE Guideline: management of women with premature ovarian insufficiency. Hum. Reprod. 2016;31:926–937. doi: 10.1093/humrep/dew027. [DOI] [PubMed] [Google Scholar]
- 3.Qin Y., Jiao X., Simpson J.L., Chen Z.J. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dumesic D.A., Meldrum D.R., Katz-Jaffe M.G., Krisher R.L., Schoolcraft W.B. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil. Steril. 2015;103:303–316. doi: 10.1016/j.fertnstert.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 5.Li R., Albertini D.F. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe L.A., Egbert J.R. Regulation of Mammalian Oocyte Meiosis by Intercellular Communication Within the Ovarian Follicle. Annu. Rev. Physiol. 2017;79:237–260. doi: 10.1146/annurev-physiol-022516-034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Hayek S., Clarke H.J. Control of Oocyte Growth and Development by Intercellular Communication Within the Follicular Niche. Results Probl. Cell Differ. 2016;58:191–224. doi: 10.1007/978-3-319-31973-5_8. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda F., Inoue N., Manabe N., Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012;58:44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 9.Gao F., Zhang J., Wang X., Yang J., Chen D., Huff V., Liu Y.X. Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Hum. Mol. Genet. 2014;23:333–341. doi: 10.1093/hmg/ddt423. [DOI] [PubMed] [Google Scholar]
- 10.De Cian M.C., Gregoire E.P., Le Rolle M., Lachambre S., Mondin M., Bell S., Guigon C.J., Chassot A.A., Chaboissier M.C. R-spondin2 signaling is required for oocyte-driven intercellular communication and follicular growth. Cell Death Differ. 2020;27:2856–2871. doi: 10.1038/s41418-020-0547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 14.Ng S.Y., Bogu G.K., Soh B.S., Stanton L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J., Xu J., Wang W., Zhao H., Liu H., Liu X., Liu J., Sun Y., Dunaif A., Du Y., Chen Z.J. Long non-coding RNA LINC-01572:28 inhibits granulosa cell growth via a decrease in p27 (Kip1) degradation in patients with polycystic ovary syndrome. EBioMedicine. 2018;36:526–538. doi: 10.1016/j.ebiom.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Liu Y.D., Chen S.L., Chen X., Ye D.S., Zhou X.Y., Zhe J., Zhang J. Down-regulation of long non-coding RNA MALAT1 inhibits granulosa cell proliferation in endometriosis by up-regulating P21 via activation of the ERK/MAPK pathway. Mol. Hum. Reprod. 2019;25:17–29. doi: 10.1093/molehr/gay045. [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Zhang X., Dang Y., Li D., Lu G., Chan W.Y., Leung P.C.K., Zhao S., Qin Y., Chen Z.J. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020;48:4480–4491. doi: 10.1093/nar/gkaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang Y.J., Yang D.C., Kong L., Hou M., Meng Y.Q., Wei L., Gao G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017;45(W1):W12–W16. doi: 10.1093/nar/gkx428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Yan Z., Qin Q., Nisenblat V., Chang H.M., Yu Y., Wang T., Lu C., Yang M., Yang S. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol. Cell. 2018;72:1021–1034.e4. doi: 10.1016/j.molcel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 21.Moley K.H., Schreiber J.R. Ovarian follicular growth, ovulation and atresia. Endocrine, paracrine and autocrine regulation. Adv. Exp. Med. Biol. 1995;377:103–119. doi: 10.1007/978-1-4899-0952-7_7. [DOI] [PubMed] [Google Scholar]
- 22.Kuo F.T., Bentsi-Barnes I.K., Barlow G.M., Pisarska M.D. Mutant Forkhead L2 (FOXL2) proteins associated with premature ovarian failure (POF) dimerize with wild-type FOXL2, leading to altered regulation of genes associated with granulosa cell differentiation. Endocrinology. 2011;152:3917–3929. doi: 10.1210/en.2010-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixit H., Rao L.K., Padmalatha V.V., Kanakavalli M., Deenadayal M., Gupta N., Chakrabarty B., Singh L. Missense mutations in the BMP15 gene are associated with ovarian failure. Hum. Genet. 2006;119:408–415. doi: 10.1007/s00439-006-0150-0. [DOI] [PubMed] [Google Scholar]
- 24.Wang H., Li G., Zhang J., Gao F., Li W., Qin Y., Chen Z.J. Novel WT1 Missense Mutations in Han Chinese Women with Premature Ovarian Failure. Sci. Rep. 2015;5:13983. doi: 10.1038/srep13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiyokawa H., Kineman R.D., Manova-Todorova K.O., Soares V.C., Hoffman E.S., Ono M., Khanam D., Hayday A.C., Frohman L.A., Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 26.Rajareddy S., Reddy P., Du C., Liu L., Jagarlamudi K., Tang W., Shen Y., Berthet C., Peng S.L., Kaldis P., Liu K. p27kip1 (cyclin-dependent kinase inhibitor 1B) controls ovarian development by suppressing follicle endowment and activation and promoting follicle atresia in mice. Mol. Endocrinol. 2007;21:2189–2202. doi: 10.1210/me.2007-0172. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Sanz J., Arluzea J., Matorras R., González-Santiago N., Bilbao J., Yeh N., Barlas A., Romin Y., Manova-Todorova K., Koff A., de la Hoz C. Increased number of multi-oocyte follicles (MOFs) in juvenile p27Kip1 mutant mice: potential role of granulosa cells. Hum. Reprod. 2013;28:1023–1030. doi: 10.1093/humrep/des436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang W., Shen J., Wu M., Arsura M., FitzGerald M., Suldan Z., Kim D.W., Hofmann C.S., Pianetti S., Romieu-Mourez R. Repression of transcription of the p27(Kip1) cyclin-dependent kinase inhibitor gene by c-Myc. Oncogene. 2001;20:1688–1702. doi: 10.1038/sj.onc.1204245. [DOI] [PubMed] [Google Scholar]
- 29.Kamura T., Hara T., Matsumoto M., Ishida N., Okumura F., Hatakeyama S., Yoshida M., Nakayama K., Nakayama K.I. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 30.Cho S., Kim J.H., Back S.H., Jang S.K. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicka K., Bushell M., Spriggs K.A., Willis A.E. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 32.Georgilis A., Klotz S., Hanley C.J., Herranz N., Weirich B., Morancho B., Leote A.C., D’Artista L., Gallage S., Seehawer M. PTBP1-Mediated Alternative Splicing Regulates the Inflammatory Secretome and the Pro-tumorigenic Effects of Senescent Cells. Cancer Cell. 2018;34:85–102.e9. doi: 10.1016/j.ccell.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui J., Placzek W.J. PTBP1 modulation of MCL1 expression regulates cellular apoptosis induced by antitubulin chemotherapeutics. Cell Death Differ. 2016;23:1681–1690. doi: 10.1038/cdd.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y., Wang C., Zhao K., Zhang G., Wang D., Mei Y. TRMP, a p53-inducible long noncoding RNA, regulates G1/S cell cycle progression by modulating IRES-dependent p27 translation. Cell Death Dis. 2018;9:886. doi: 10.1038/s41419-018-0884-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang B., Zhang Y.Y., Guo S.T., Kong L.F., Cheng Q., Liu G.Z., Thorne R.F., Zhang X.D., Jin L., Wu M. Dual functions for OVAAL in initiation of RAF/MEK/ERK prosurvival signals and evasion of p27-mediated cellular senescence. Proc. Natl. Acad. Sci. USA. 2018;115:E11661–E11670. doi: 10.1073/pnas.1805950115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu I.M., Hengst L., Slingerland J.M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 37.Xu X., Chen X., Zhang X., Liu Y., Wang Z., Wang P., Du Y., Qin Y., Chen Z.J. Impaired telomere length and telomerase activity in peripheral blood leukocytes and granulosa cells in patients with biochemical primary ovarian insufficiency. Hum. Reprod. 2017;32:201–207. doi: 10.1093/humrep/dew283. [DOI] [PubMed] [Google Scholar]
- 38.Nishi Y., Yanase T., Mu Y., Oba K., Ichino I., Saito M., Nomura M., Mukasa C., Okabe T., Goto K. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142:437–445. doi: 10.1210/endo.142.1.7862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.