Abstract
Background
Ephrin type-A receptors (EPHA) are members of family of receptor tyrosine kinases and are related to tumor immunogenicity and immune microenvironment, however, the association between EPHA mutation (EPHAmut) and efficacy of immune checkpoint inhibitors (ICIs) has not been investigated in non-small cell lung cancer (NSCLC).
Methods
Multiple cohorts were used to assess the immunotherapeutic predictive performance of EPHAmut, including one discovery cohort (n=79) and two public validation cohort (cohort 1: NSCLC, n=165; cohort 2: pan-cancer, n=1662). The Cancer Genome Atlas cohort was used for prognostic analysis and mechanism exploration.
Results
In the discovery cohort, patients with EPHAmut had superior disease control rate (72.2% vs 36.1%, p=0.01) and progression-free survival (PFS) (HR 0.38; 95% CI 0.21 to 0.68; p<0.001) compared with those with wide-type EPHA (EPHAwt) in NSCLC. The association between EPHAmut and immunotherapy outcomes in NSCLC was consistently observed in the validation cohorts by multivariable models (cohort 1, PFS HR 0.59; 95% CI 0.37 to 0.96; p=0.03; cohort 2, overall survival (OS) HR 0.63; 95% CI 0.41 to 0.98; p=0.04). Further pooled estimates of the discovery and validation cohorts showed that patients with EPHAmut exhibited a significantly longer PFS and OS in lung adenocarcinoma (LUAD) while not squamous cell lung cancer (LUSC). Consistently, mechanism analysis revealed that patients with EPHAmut was associated with increased T cell signatures and downregulated transforming growth factor-β signaling compared with patients with EPHAwt in LUAD while not LUSC.
Conclusions
Our results demonstrated that EPHAmut is an independent classifier that could stratify patients with LUAD for ICIs therapy. Further prospective studies are warranted.
Trial registration number
NCC2016JZ-03, NCC2018-092.
Keywords: immunotherapy, lung neoplasms
Introduction
Recent clinical trials have revealed a promising survival advantage of immune checkpoint inhibitors (ICIs) in treating patients with non-small cell lung cancer (NSCLC).1–3 However, quite a portion of patients failed to benefit from ICIs either as single-agents or through combination administration.4–6 The biomarkers associated with the response to immunotherapy are not fully understood, and there is an urgent to identify more biomarkers to determine the responsiveness to ICIs.
Emerging predictors for immunotherapy in NSCLC have shown good performance, such as programmed death-ligand 1 (PD-L1) expression4 and tumor mutational burden (TMB),6 7 which have been validated in prospectively in random controlled trials. However, they are still imperfect, such as the utilization of TMB is sometimes limited by the uncertainty of calculating algorithm and the lack of uniform standard to determine the cut-off value.8 Moreover, even with all the established biomarkers, such as PD-L1, TMB and gene expression profile score, there are still a majority of patients with NSCLC showing unresponsiveness to ICIs. Therefore, the identification of more convenient and reliable biomarkers beyond TMB and PD-L1 expression for the prediction of ICIs benefits are needed for clinical practice.
Ephrin receptor tyrosine kinase (Eph RTKs) superfamily is the largest family of tyrosine kinases, which are the key regulators of cell–cell communication both in normal development and disease.9 10 Ephrin type-A receptors (EPHA) are members of the EPH family of RTKs with nine EPHA receptors (EPHA1-8, EPHA10). All Eph receptors have a highly conserved overall structure and similar structural features.10 11 EPHA were reported to be associated with lung cancer biology in previous studies. EPHA3-7 are most frequently mutated genes in NSCLC (in 5%–15% of the tumors)12–14 (cbioportal.org). Most EPHA3 mutations are loss-of-function missense mutations that inhibit the activating of Eph RTKs by disrupting ephrin binding, kinase activity or cell surface localization, and inactivating of EPHA3 promoted the lung cancer cells proliferation.15 Previous studies have shown that high EPHA4 expression in tumors was associated with improved outcome and EPHA4 can inhibit lung cancer cell migration and invasion, suggesting a tumor suppressor role.16 Additionally, increased EPHA4, A5 and A7 expression were identified as predictors for favorable survival.17 The EPHA2 G391R mutant promoted lung cancer cell invasiveness and growth.18 EPHA2 promotes epithelial-mesenchymal transition (EMT) through activation of Wnt/β-catenin signaling.19 Frequent EPHA5 and EPHA7 mutations in NSCLC that could be pathologically relevant were also identified, but remained to be characterized.20
As for immunomodulating properties, plenty of molecular research concerning tumor immunogenicity and immune microenvironment support the possible association between EPHA and immunotherapeutic efficacy. Considering tumor immunogenicity, EphA2/ephrin-A1 signaling in the lung was involved in T-cell maturation and chronic inflammation.21 The CD4+ T cell clone isolated from patient with melanoma, whose tumor tissues overexpressed EPHA3, could recognize the EphA3 epitope and elicit immunoreactivity against melanoma cell lines selectively.22 Moreover, EphA10 antibody administration would facilitate cytotoxic CD8+ T cells to attack malignant breast cancer cells.23 Previous studies have also demonstrated that tumor cell-intrinsic EPHA2 regulated T cell infiltration and the sensitivity to immunotherapy in pancreatic ductal adenocarcinoma through EPHA2/transforming growth factor (TGF)-β/recombinant mothers against decapentaplegic (SMAD) axis.24 25 In addition, EPHA2-knockout tumors increased the mRNA expression of CD8A and exhibited a higher signature of interferon (IFN) response and inflammatory pathways.24 As for the immune microenvironment, Eph receptors and ephrin ligands were related to the immune cell development, activation and migration.10 EPHA1 and EPHA4 expressed in T cells and facilitated migration of T cell subsets through activation of Src kinases.21 EPHA1 and EPHA3 are expressed in T cells and mediate T cell chemotaxis in vitro.26 27 In addition, EPHA2 induces significant immunoreactivity in CD8+ T cells via major histocompatibility complex I-restricted presentation against renal cell carcinoma and glioma cell lines in vitro.28 29 In addition, EPHA1-4, EPHA7, EPHA10 have reported to be involved in the activation and proliferation of immune cells including T cells, B cells and dendritic cells and antibody production.10 Based on these observations, we hypothesized that EPHA mutation might influence the status of immunogenicity and immune microenvironment and be associated with clinical benefit of ICIs, which remains unexplored in patients with NSCLC.
We herein investigated the association between EPHA mutation and the clinical efficacy of anti-PD-(L)1 treatment in NSCLCs on the basis of our discovery cohort and multiple validation cohorts. The potential mechanism was subsequently explored based on RNA expression and whole genome sequencing (WES) data in The Cancer Genome Atlas (TCGA) database.
Materials and methods
Patients and data sources
The detailed information regarding the data being analyzed in this study is summarized in online supplemental table S1, and the flow diagram of this study is depicted as figure 1. In brief, we included patients treated with anti-PD-(L)1 antibodies at National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital and Chinese Academy of Medical Sciences and Peking Union Medical College and Sun Yat-sen University Cancer Center from December 2016 to December 2018 (named China cohort) as a discovery set, all patients were treated as part of clinical trials. Eligible patients for this study were determined mainly based on the following criteria: (i) >18 years old; (ii) Eastern Cooperative Oncology Group performance status: 0–1; (iii) have advanced or recurrent NSCLC; (iv) failure after first-line platinum-based doublets chemotherapy; (v) radiologically evaluable according to Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1. CT or MRI scans were reviewed by the investigators. The comprehensive genomic profiling of 79 patients with NSCLC were implemented by WES. PD-L1 scoring was available in 49 out of 79 patients (online supplemental methods).
Figure 1.
Flow diagram of the study. BLCA, bladder cancer; DCB, durable clinical benefit; EPHA, ephrin type-A receptors; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinomas; NSCLC, non-small cell lung cancer; ORR, objective response rate; PFS, progression-free survival; SKCM, skin cutaneous melanoma.
jitc-2020-001315supp001.pdf (987.2KB, pdf)
jitc-2020-001315supp002.pdf (119.3KB, pdf)
Multiple independent public cohorts were used to validate the association between EPHA mutation and immunotherapy efficacy. The first validation cohort (validation cohort 1, n=165) was a pooled cohort consisting three public datasets with patients with NSCLC treated with ICIs with available WES data, including the datasets of 75 patients treated with anti-PD-1 plus anticytotoxic T-lymphocyte antigen 4 (anti-CTLA-4) (Hellmann cohort),12 56 patients treated with anti-PD-(L)1 with or without anti-CTLA-4 (Miao cohort)30 and 34 patients treated with anti-PD-1 (Rizvi 34 cohort).31 The second validation cohort (validation cohort 2) consisted 1662 patients with a variety of cancer types who had received at least one dose of ICI therapy (Memorial Sloan Kettering Cancer Center [MSKCC] cohort), including 350 patients with NSCLC.7 The tumor tissues were subjected to MSKCC 468-gene panel (earlier versions included 341 or 410 genes) target sequencing. We also obtained the WES data of 2599 solid tumors in TCGA along with the corresponding mRNA expression data of 2541 solid tumors across six tumor types from cBioPortal (www.cbioportal.org) to study the mechanism underlying the association between EPHA mutation and immunotherapy (online supplemental methods).
Study assessment
In China cohort, baseline tumor assessments were performed within 1–28 days prior to the initiation of the anti-PD-(L)1 treatment, with the subsequent assessments being performed every 6–8 weeks until objective disease progression. The objective response rate (ORR) was defined as the percentage of patients with confirmed complete response (CR) or partial response (PR) by RECIST V.1.1. The disease control rate (DCR) was defined as the percentage of patients with confirmed complete response (CR), PR or stable disease (SD) by RECIST V.1.1. Durable clinical benefit (DCB) was defined as the percentage of patients who achieved CR, PR or SD lasted >6 months; all other patients were considered to have no durable benefit (NDB). Progression-free survival (PFS) was defined as the time from the beginning of ICI treatment to the date of PD or death from any cause. Patients who had not progressed were censored at the date of their last scan.
In the validation cohorts, tumor response was evaluated according to RECIST V.1.1 in Hellmann cohort,12 Miao cohort30 and MSKCC cohort.7 Objective response to anti-PD-1 treatment was assessed by investigator-assessed immune-related response criteria (irRC) in Rizvi 34 cohort.31 The definitions of DCB, NCB and PFS were consistent with those in the discovery cohort.
EPHA mutation
The detailed profiles of EPHA (EPHA1-8, EPHA10) mutation in each cohort are listed in the online supplemental figure S1. The non-synonymous mutations including TRUNC (Frameshift del, Frameshift ins, nonsense, nonstop, splice region, splice site), INFRAME (Inframe del and Inframe ins) and MISSENSE mutations of at least one EPHA subtype were defined as EPHA mutation (EPHAmut) in this study.
Statistical analyses
Continuous variables were compared by Mann-Whitney U test and categorical variables were compared by χ2 test or Fisher’s exact test. Survival was estimated by Kaplan-Meier curves, with the p value determined by a log-rank test. HR was determined through the univariable and multivariable Cox regression. Variables with p<0.1 in the univariable regression and those which has been reported associated with the effect of immunotherapy in NSCLC were also included into multivariable Cox regression. Random-effect models were used to pool the effect sizes. The poor results of categorical variables were presented as relative risk (RR) and 95% CIs. Q-test and I2 statistics were used to assess the heterogeneity. A result of p>0.1 and I2 <50% indicated no significance between-study heterogeneity.
We used propensity-score matching with a ratio of 1:1 to analyze the association between EPHA status and PFS and OS in patients with NSCLC from TCGA. Propensity score was estimated by age, sex, smoking history, tumor stage, pathology, history of other malignancy and neoadjuvant therapy. False discovery rate (FDR) was used to estimate the significance of differences between the mRNA expression levels. All reported p values were two-tailed, FDR < 0.05 is considered statistically significant.
All analyses were performed using SPSS V.24.0 (SPSS, Chicago, Illinois, USA), and R V.3.5.2. Graphs in the present study were drawn by GraphPad Prism 8 and R V.3.5.2.
Results
Association between EPHA mutations and the clinical benefit to anti-PD-(L)1 therapy in the discovery cohort
The discovery cohort included 79 patients with stage IV NSCLC who were treated with anti-PD-(L)1 treatment and had baseline tissue samples sequenced by WES (online supplemental table S4). The median age was 55 (IQR, 47–61) years and 54 patients (68%) were male. Nearly half of the patients had a history of smoking. The major histology was adenocarcinoma (47%). Most patients (74/79, 94%) received PD-1 inhibitor monotherapy and the rest received PD-L1 inhibitor monotherapy (5/79, 6%). The median follow-up time was 21 months.
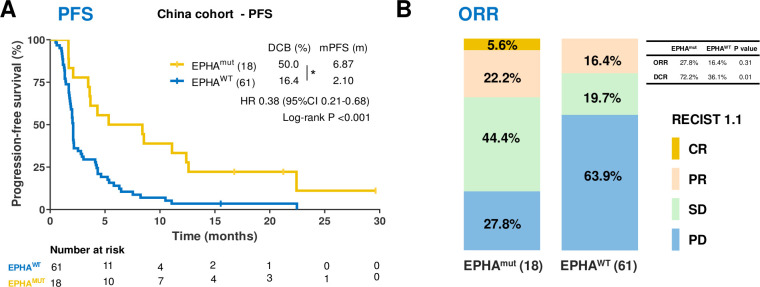
In the discovery cohort, 22.8% patients with NSCLC harbored EPHA mutations, and most EPHA mutations were missense mutations (92.1%) (online supplemental figure S1). Patients harboring EPHA mutation (EPHAmut) had superior PFS (median PFS 6.87 months vs 2.10 months, HR 0. 38; 95% CI 0.21 to 0.68; p<0.001; figure 2A), a higher DCB (50% vs 16.4%; Fisher’s exact test, p=0.009; figure 2A), higher DCR (72.2% vs 36.1%; Fisher’s exact test, p=0.01; figure 2B) and numerically higher ORR (27.8% vs 16.4%; Fisher exact test, p=0.31; figure 2B) compared with those with wild-type EPHA (EPHAwt).
Figure 2.
The association between EPHA mutation and clinical response to immune checkpoint inhibitors in China cohort. (A) Kaplan-Meier survival curves of progression-free survival (PFS) comparing the EPHAmut group and EPHAwt group in China cohort. (B) The ratio of patients with complete response (CR), partial response (PR), stable disease (SD) and progression disease (PD) treated with anti-PD-(L)1 antibody in EPHA mutation and EPHA wide-type group. *P<0.05 by Fisher’s exact test.
In the univariable analyses, besides EPHA mutation, several other indexed such as smoking history, sex and TMB (≥median vs <median) were also associated with the immunotherapeutic PFS with the HRs (95% CI) of 0.48 (0.30 to 0.76), 0.48 (0.29 to 0.79) and 0.46 (0.28 to 0.73), respectively (online supplemental table S5). In the multivariable Cox proportional hazards regression model adjusted by the smoking status, age, sex, PD-L1 expression and TMB (≥median vs <median), the association between EPHA mutation and PFS remained significant (HR 0.36; 95% CI 0.19 to 0.71; p=0.003; online supplemental table S5). Several factors which have been reported to be associated with the immunotherapeutic effectiveness, such as sex, age, histology, smoking and so on were also included simultaneously in the multivariable Cox regression model to exclude the potential confounding effects.5 32–34 These results indicated that EPHA mutations are associated with better clinical benefits of anti-PD-(L)1 therapy independent of PD-L1 expression and TMB.
Association between EPHA mutation and clinical benefits of ICIs in the validation cohorts
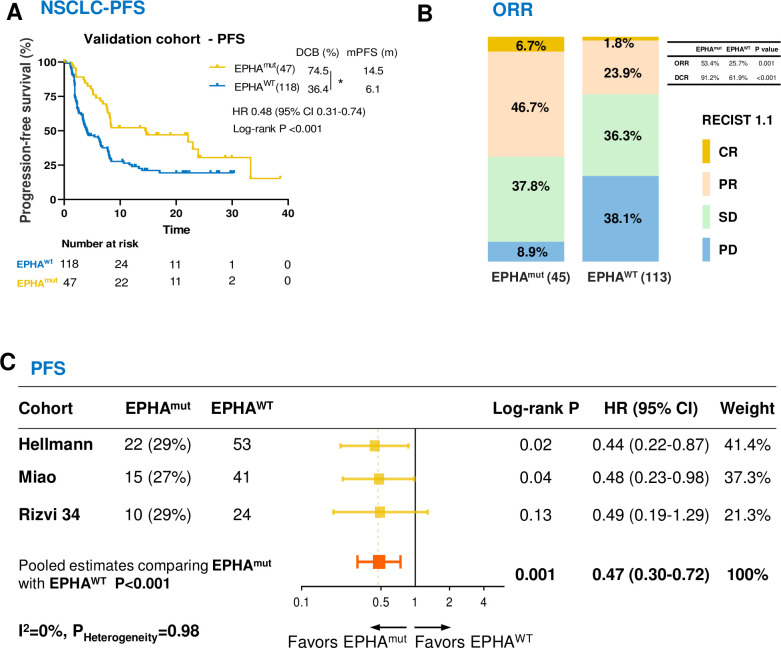
In validation cohort 1 (online supplemental table S6), EPHAmut was prevailed in 28.5% patients with NSCLC and it was associated with significantly longer PFS of ICIs (HR 0.48; 95% CI 0.31 to 0.74; p<0.001; figure 3A) and better DCB (74.5% vs 36.4%; Fisher’s exact test, p=0.02; figure 3A), DCR (91.2% vs 61.9%; Fisher’s exact test, p<0.001; figure 3B) and ORR (53.4% vs 25.7%; Fisher’s exact test, p=0.001; figure 3B), which was consistent with the discovery cohort. The trend of prolonged PFS in EPHAmut patients was consistently observed across all three datasets included in the first validation cohort (figure 3C), and the pooled analyses showed a significantly longer PFS (HR 0.47; 95% CI 0.30 to 0.72; p<0.001; figure 3C), ORR (RR 2.12; 95% CI 1.39 to 3.22; p<0.001; online supplemental figure S2) and DCB (RR 2.04; 95% CI 1.52 to 2.74; p<0.001; online supplemental figure S2) in patients with EPHAmut versus EPHAwt. Statistical analyses for study heterogeneity did not present significant in all pooled estimates (p>0.10, I2 <50%), indicating the consistency of the association between EPHAmut and favorable benefit to ICIs across the three datasets.
Figure 3.
Association between EPHA mutations and survival in patients with non-small cell lung cancer (NSCLC) treated with immune checkpoint inhibitors in the validation cohort 1. (A) Kaplan-Meier survival curves comparing progression-free survival (PFS) between the EPHAmut group and EPHAwt group in validation cohort. (B) The ratio of patients with complete response (CR), partial response (PR), stable disease (SD) and progression disease (PD) treated with anti-PD-(L)1 antibody in EPHA mutation and EPHA wide-type group. (C) Pooled estimates of PFS. The squares in yellow represent study-specific HRs. The squares in orange indicate the pooled HRs. Horizontal lines indicate the 95% CIs. The p values for heterogeneity and the values of I2 are from the pooled analysis of study-specific HRs. *P<0.05 by Fisher’s exact test.
In the multivariable Cox proportional hazards regression model adjusted by PD-L1 expression and TMB (top 20% vs the rest) and other confounding factors, EPHAmut remained an independent predictor for superior PFS (HR 0.59; 95% CI 0.37 to 0.96; p=0.03; online supplemental table S7).
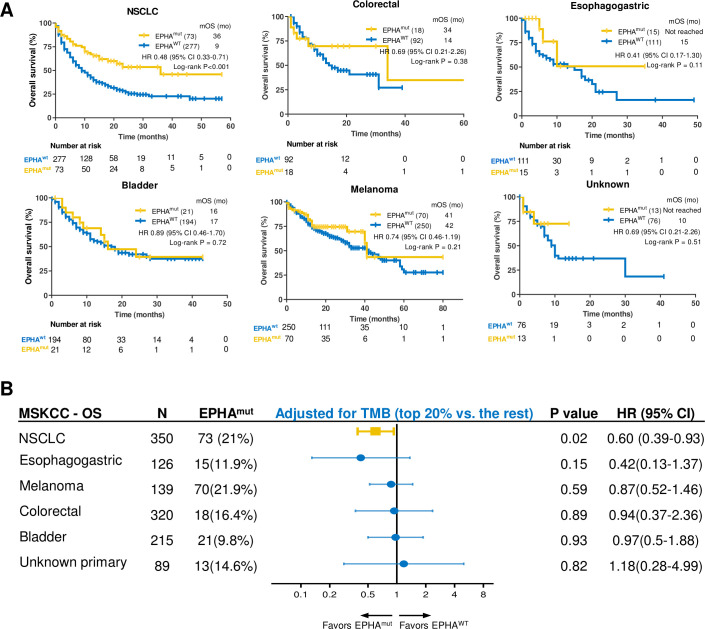
Association between EPHA mutation and ICIs efficacy in validation cohort 2
In the validation cohort 2, which comprised 1662 patients with >10 types of tumors, we further validated the association between EPHAmut and significantly longer OS in NSCLC (HR 0.48; 95% CI 0.33 to 0.71; p<0.001; figure 4A). However, no statistically significant association was observed between OS and EPHA status in other tumors (figure 4A). After adjusted for TMB (top 20% vs the rest), the association between EPHAmut and OS remained significant in NSCLC (HR 0.60; 95% CI 0.39 to 0.93; p=0.02; figure 4B). EPHAmut remained an independent predictor for ICIs in NSCLC in the multivariate Cox regression (HR 0.63; 95% CI 0.41 to 0.98; p=0.04; online supplemental table S8), as adjusted by histology, age, sex, treatment and TMB.
Figure 4.
Association between EPHA mutation and immunotherapeutic overall survival (OS) in validation cohort 2. (A) Kaplan-Meier curves of OS between EPHA mutation and EPHA wide-type group in non-small cell lung cancer (NSCLC), melanoma, bladder cancer, colorectal cancer, esophagogastric cancer, cancer of unknown primary. (B) Subgroup analysis of OS in multitype of tumors in MSKCC cohort adjusted by tumor mutational burden (TMB) (top 20% vs the rest).
In addition, all the EPHA subtypes seemed to play uniform roles in predicting the clinical survival benefit with the HRs <1.0 in the pool analysis of the discovery and validation cohorts (online supplemental figure S3). Taken together, these data indicated that EPHA mutation might be predictive of the clinical benefit of ICIs independently in NSCLC.
Association between EPHA mutations and the clinical benefit to anti-PD-(L)1 therapy in lung adenocarcinoma and lung squamous cell carcinoma
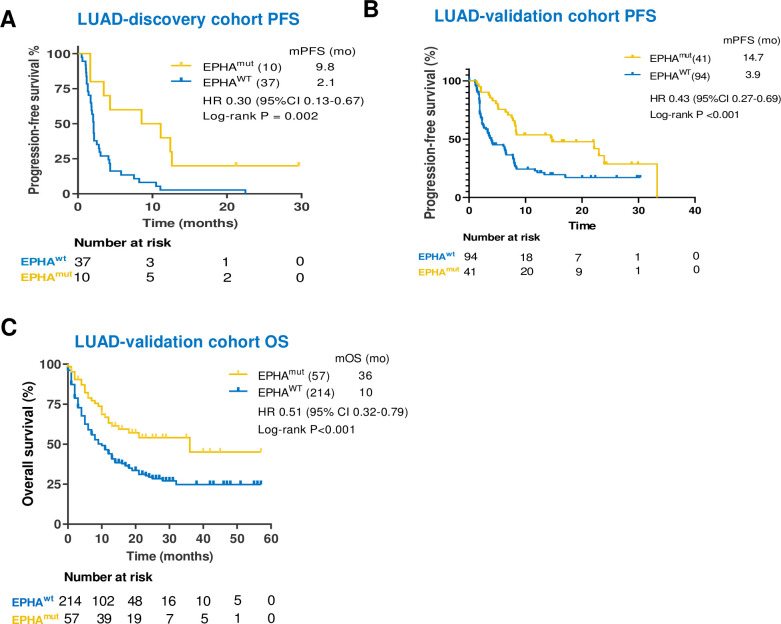
We further explored whether the association between EPHAmut and clinical benefit of immunotherapy may vary with the histological subtypes. In the discovery cohort, EPHAmut was associated with significantly longer PFS (median PFS 9.82 vs 2.10 months; HR 0.30; 95% CI 0.13 to 0.67; p=0.002; figure 5A) than EPHAwt in lung adenocarcinoma (LUAD), which was consistently observed in both validation cohorts (figure 5B, C). The pooled analysis further revealed that EPHAmut was associated with significantly longer PFS (discovery cohort plus validation cohort 1; HR 0.38; 95% CI 0.25 to 0.58; p<0.001; online supplemental figure S4) and OS (validation cohort 2; HR 0.51; 95% CI 0.33 to 0.79; p=0.001; online supplemental figure S4) in LUAD.
Figure 5.
Patients with EPHA mutation showed a favorable clinical benefit in lung adenocarcinoma (LUAD) when treated with immune checkpoint inhibitors. (A–F) Kaplan-Meier survival curves comparing progression-free survival (PFS) or overall survival (OS) between the EPHA mutation group and EPHA wide-type group in China cohort and two validation cohorts.
After adjusted by TMB, PD-L1 and other confounding factors in the multivariate Cox regression, EPHAmut remained an independent predictor for PFS and OS efficacy of ICIs in patients with LUAD based on either the discovery cohort (PFS HR 0.34; 95% CI 0.12 to 0.95; p=0.04; online supplemental table S9) or the validation cohorts (validation cohort 1, PFS HR 0.53; 95% CI 0.31 to 0.90; p=0.02; online supplemental table S10); validation cohort 2, OS HR 0.61; 95% CI 0.38 to 1.00; p=0.05; online supplemental table S11). No difference of PFS and OS was observed in patients with lung squamous cell carcinomas (LUSC) with EPHAmut versus EPHAwt (online supplemental figure S4).
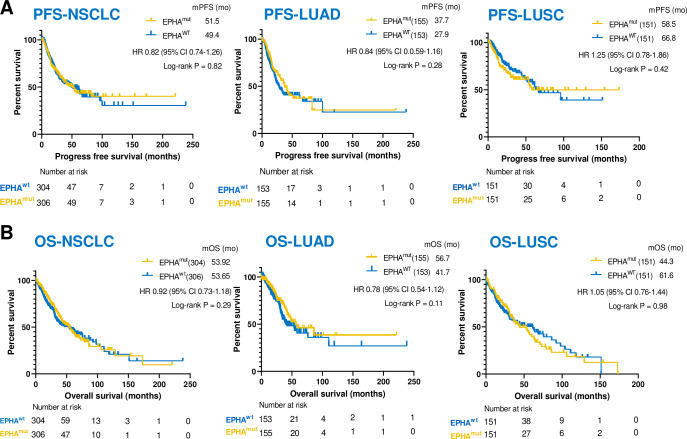
EPHA mutation was not a prognostic factor
To assess to potential prognostic value of EPHA mutation, survival analyses were further performed according to EPHA mutational status in the TCGA database. No significant difference was found in PFS or OS between EPHAmut and EPHAwt subsets in patients with NSCLC, LUAD and LUSC with standard treatment (figure 6), suggesting that EPHA mutation was not a prognostic factor.
Figure 6.
The association between EPHA mutation and progression-free survival (PFS) and overall survival (OS) in The Cancer Genome Atlas (TCGA) by propensity score matching. (A) Kaplan-Meier curves of OS between EPHA mutation and EPHA wide-type group among the patients with lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and non-small cell lung cancer (NSCLC) in TCGA database. (B) Kaplan-Meier curves of PFS between EPHA mutation and EPHA wide-type group among the patients with LUAD, LUSC and NSCLC in TCGA database.
Potential mechanisms associated with EPHA mutations in predicting the efficacy of ICIs
Considering the previously identified correlations of PD-L1 expression and TMB with the clinical benefits of ICIs, the associations of EPHA mutations with PD-L1 expression, TMB as well as neoantigen burden were analyzed to investigate the possible mechanism. We first aimed to ascertain whether co-occurrence takes place between EPHA mutations with robust predictors, including PD-L1 expression, higher TMB and predicted neoantigens. As shown, EPHAmut was associated with significantly higher TMB in both the discovery cohort (median: 317 muts vs 105 muts, Mann-Whitney U test, p<0.001; online supplemental figure S5A) and validation cohort 1 (median: 332 muts vs 123 muts, Mann-Whitney U test, p<0.001; online supplemental figure S5C), along with significantly elevated predicted neoantigens (validation cohort 1, median: 476 vs 148, Mann-Whitney U test, p<0.001; online supplemental figure S5E), suggesting the co-occurrence between the EPHAmut and elevated TMB and predicted neoantigens. However, no association was observed between EPHAmut and PD-L1 expression (≥1% vs <1%) (discovery cohort: Fisher’s exact test, p=1.00; validation cohort: Fisher’s exact test, p=0.48; online supplemental figure S5B, S5D). PD-L1 expression was relatively balanced between EPHAmut group and EPHAwt group in the discovery and validation cohorts.
We further investigated other driver mutations co-mutated with EPHA. The co-occurrence of EPHA and EGFR, STK11, ALK, ROS1 seldom occurred (online supplemental figure S6A). The incidence rates of EPHA co-mutated with KRAS and TP53 were 2.1% and 14.9% in the discovery cohort and 15.5% and 19.3% in the validation cohort in LUAD, respectively (online supplemental figure S6A). However, few co-mutations of EPHA with KRAS and TP53 existed in LUSC. We further investigated the effect of co-mutation in predicting survival of immunotherapy in LUAD. The association between EPHA mutation and survival was not influenced by KRAS mutation or TP53 mutation in LUAD as shown in online supplemental figure S7B-7C, suggesting that EPHA mutation was an independent predictor of immunotherapy.
To further explore the underlying mechanism of the predictive values of EPHA mutations to ICI efficacy, 47 immune-related signatures and 43 TGF-β signaling genes based on the RNA and WES data from TCGA database (online supplemental tables S2 and S3) were analyzed. Gene Set Enrichment Analysis (GSEA) in NSCLCs revealed a prominent enrichment of signatures related to the downregulation of TGF-β signaling, while no difference of IFN-γ signaling was observed (data not shown). Further analyses in separate populations of LUAD or LUSC illustrated that LUAD rather than LUSC subset presented the prominent enrichments of signatures related to IFN-γ signaling upregulation and TGF-β signaling downregulation in EPHAmut versus EPHAwt groups (FDR adjusted p=0.03 for both; figure 7A, B).
Figure 7.
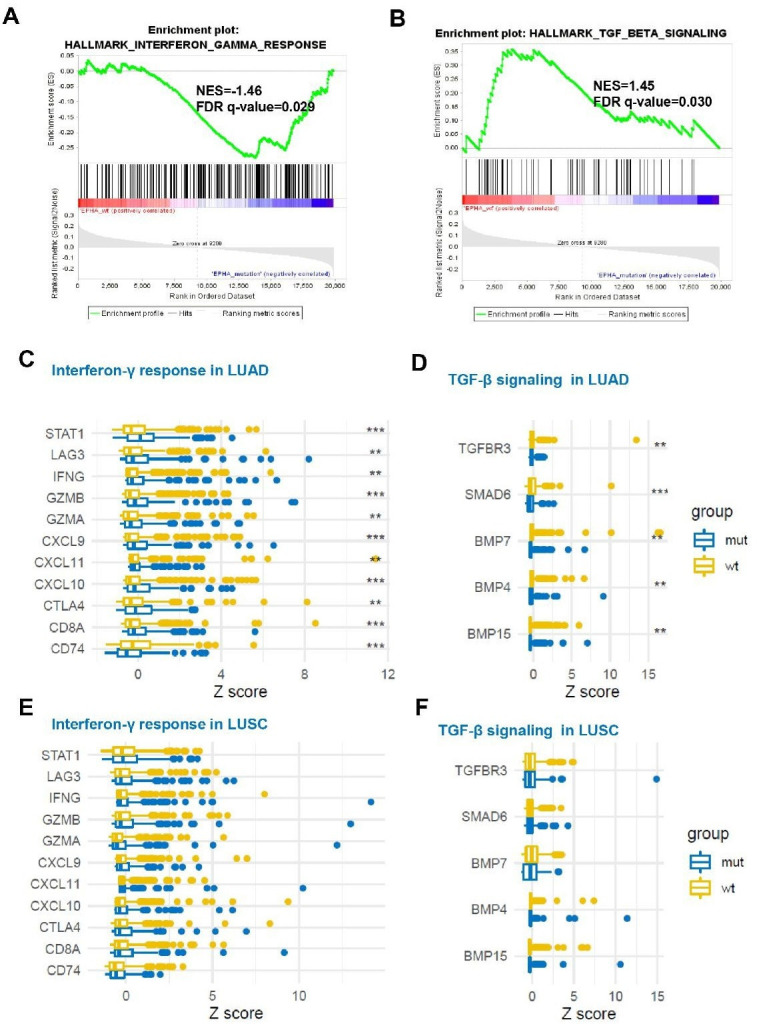
Associations between EPHA status and immune response-related genes and transforming growth factor (TGF)-β signaling-related genes mRNA expression. (A) The enrichment in TGF-β signaling by Gene Set Enrichment Analysis (GSEA) between EPHA mutation and EPHA wild-type groups in lung adenocarcinoma (LUAD). (B) The enrichment in interferon-γ response signaling by GSEA between EPHA mutation and EPHA wild-type groups in LUAD. (C) Box plot comparing the expression of immune-related genes between patients with EPHA mutation and EPHA wild-type in LUAD. (D) Box plot comparing the expression of TGF-β signaling-related genes between patients with EPHA mutation and EPHA wild-type in LUAD. (E) Box plot comparing the expression of immune-related genes between patients with EPHA mutation and EPHA wild-type in lung squamous cell carcinoma (LUSC). (F). Box plot comparing the expression of TGF-β signaling-related genes between patients with EPHA mutation and EPHA wild-type in LUSC ** False discovery rate (FDR)<0.05; ***False discovery rate (FDR)<0.01.
Furthermore, compared with EPHAwt patients, the mRNA expression levels of ten immune-related genes and five TGF-β-related genes were significantly increased and decreased, respectively, in patients with EPHAmut (FDR adjusted p<0.05; figure 7C, D; online supplemental tables S12 and S13) in the LUAD subset, while such pattern was not observed in the LUSCs (figure 7E, F; online supplemental table S12).
No significant differences of TGF-β signaling or T cell gene signature between EPHAmut and EPHAwt were observed in the other types of tumor (online supplemental tables S12 and S13). GSEA results showed no enrichment in the IFN-γ or TGF-β signaling in patients with EPHAwt in bladder, esophageal carcinoma, skin cutaneous melanoma or head and neck carcinoma cohorts based on TCGA datasets (data not shown).
Collectively, the above results showed that the superior ICI benefits in NSCLCs with EPHAmut might mainly be mainly attributed to LUAD. In terms of mechanism, the downregulation of TGF-β signaling and the increased T cell signatures mediated by EPHAmut might be on account of the different susceptibility for ICIs between LUAD and other tumors.
Discussion
In this study, we provided strong evidences that EPHAmut was associated with superior efficacy and survival benefits of ICIs independent of PD-L1 expression and TMB status in patient with LUAD. EPHAmut was not associated with the OS of NSCLCs with standard treatment in TCGA database, suggesting the predictive, but not prognostic impact of EPHAmut. To our knowledge, our study is the first to propose that the mutation of EPHA might be a predictor favorable for the ICIs delivery.
As derived from multiple discovery and the validation cohorts, EPHAmut consistently correlated well with a better clinical benefit in patients with NSCLC, especially LUAD. However, no significant differences were observed in PFS or OS in LUSC when stratified by EPHA mutation status. Moreover, the frequency of EPHA mutation was the relative balance between LUSC and LUAD (30% vs 21.3% in discovery cohort; 22.7% vs 30% in validation cohort). In addition, the activated IFN-γ signaling was only observed in LUAD but not in LUSC from EPHAmut versus EPHAwt subgroup, and thus it remained undetermined whether EPHAmut could be applied as a predictor of ICIs in LUSC. Collectively, the superior clinical benefit of ICIs in NSCLC with EPHAmut might be largely contributed from its effects in LUAD.
LUAD and LUSC are distinct in disease pathology, smoking associations, metastatic trends, molecular mechanisms and patient outcomes.35 36 The differences of their molecular characteristics might contribute to the diverse immunogenic features and consequently varied response to immunotherapy. Recently, KRAS mutations were identified to represent the generation of neoantigens that reflect an improved immunogenicity, subsequently bringing superior efficacy to ICIs in NSCLC.37 In the present study, the more enrichment of KRAS mutations in EPHAmut tumors compared with EPHAwt may be one potential explanation for the distinct performance for ICIs efficacy in LUAD and LUSC (online supplemental figure S6A). The co-occurring mutations in EPHA and KRAS seemed to be associated with the optimal PFS in LUAD in present study, however, the co-mutation between EPHA and KRAS need to be validated in larger population. Moreover, EPHAmut was associated with increased T cell signatures and downregulated TGF-β only in LUAD but not in LUSC or other tumors, indicating the different transduction signaling mediated by EPHAmut across histologies, which partly explained the higher sensitivity for ICIs treatment of EPHAmut tumors in LUAD.
One critical obstacle impeding the extensive utility of PD-L1 expression and TMB is the determination of feasible cut-off values. EPHA mutation, as a dichotomous indicator, could avoid the dilemma of cut-off selection, which provided an objective and convenient approach for stratifying patients beneficial of ICIs delivery. In addition, EPHA mutations could be easily detected in peripheral blood compared with other established biomarkers such as PD-L1 expression and TMB, which will bring new insights of the invasive biomarker exploration for immunotherapy. Moreover, our results revealed a possibility of personalized immunotherapy approach combined with EPHA inhibitor in NSCLC, for the optimization of ICI treatment in clinical practice in further.38 However, this needs to be further studied.
Limitations of this study included potential statistical bias due to the retrospective profile and limited sample sizes, which, however, was minimized by the consistent results arising from multiple cohorts analyses. Second, lacking hotspot and difficult to verify the function of each EPHA mutation and indeed influenced the precision of biomarker detection, our attempt to recruit functional EPHA mutations into our EPHA mutation pattern was handicapped by the limited information available regarding the functions of different mutations. Those mutations are associated with amino acid substitutions scattered throughout the receptor and lack of hotspots, as illustrated in TCGA and COSMIC databases. The functional mutations need to be further investigated by molecular studies in cell line and xenograft model. However, the lack of hotspot mutation, which conversely decreased the risk of subjective discrimination of deleterious mutations. Third, several patients’ PD-L1 expression were missing, which may weaken the statistical effect, while there was no association between PD-L1 expression and EPHA status, and PD-L1 expression was relative balance between EPHAmut group and EPHAwt group in discovery and validation cohorts, additionally, PD-L1 expression was adjusted by multivariable Cox proportional hazards regression model as the dummy variables, minimizing the impact of PD-L1 expression insufficiency. In addition, the preliminary interpretation of mechanism underlying the association between EPHA mutation and clinical benefit needed to be further investigated by basic research. Lastly, further prospective studies are warranted.
Conclusions
Our results demonstrated that EPHAmut as an independent classifier could stratify patients with LUAD for appropriate administration of anti-PD-(L)1 therapy, and correspondingly provide a feasible and convenient approach for better clinical practice. Further prospective studies were warranted.
Acknowledgments
The authors would like to thank the staff members of the TCGA Research Network, the cBioportal data portal as well as all the authors for making their valuable research data public.
Footnotes
HB, JD, CL, WX and WF contributed equally.
Contributors: Conception and design: JW, ZW, CL, GW, SC and LZ. Collection and assembly of data: HB, SC, JD, WF, YX, GW, RW, JS, JX, XW and KF. Data analysis and interpretation: HB, JD, CL, WX, YX, GW and WF. Manuscript writing: all authors. Final approval of manuscript: all authors.
Funding: This work was supported by the National Key Research and Development Project (2019YFC1315700, to JW; 2019YFC1315704, to ZW), the National Natural Sciences Foundation Key Program (81630071, to JW); CAMS Innovation Fund for Medical Sciences (CIFMS 2016-I2M-3-008, to JW; 2017-I2M-1-005, to ZW); Aiyou Foundation (KY201701, to JW), Ministry of Education Innovation Team Development Project (IRT-17R10, to JW), CAMS key lab of translational research on lung cancer (2018PT31035, to JW), the National Natural Sciences Foundation (81871889, to ZW) and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018RC320009, to ZW).
Competing interests: CL, YX, GW and SC is the employee of Burning Rock Biotech. WX and ZZ is the employee of 3D Medicines.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the ethics committees of all participating centers, and all patients provided written informed consent.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The validation cohorts (including Rizvi 34 cohort, Hellmann cohort, Miao cohort and MSKCC cohort) used in this study were publicly available as described in the 'Materials and methods' section. The China cohort is available from the corresponding author on reasonable request.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Blumenthal GM, Zhang L, Zhang H, et al. . Milestone analyses of immune checkpoint inhibitors, targeted therapy, and conventional therapy in metastatic non-small cell lung cancer trials: a meta-analysis. JAMA Oncol 2017;3:e171029. 10.1001/jamaoncol.2017.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gettinger S, Horn L, Jackman D, et al. . Five-Year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209-003 study. J Clin Oncol 2018;36:1675–84. 10.1200/JCO.2017.77.0412 [DOI] [PubMed] [Google Scholar]
- 3.Garon EB, Hellmann MD, Rizvi NA, et al. . Five-Year overall survival for patients with advanced Non‒Small-Cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol 2019;37:2518–27. 10.1200/JCO.19.00934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reck M, Rodríguez-Abreu D, Robinson AG, et al. . Pembrolizumab versus chemotherapy for PD-L1–Positive Non–Small-Cell lung cancer. N Engl J Med Overseas Ed 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 5.Mok TSK, Wu Y-L, Kudaba I, et al. . Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819–30. 10.1016/S0140-6736(18)32409-7 [DOI] [PubMed] [Google Scholar]
- 6.Reck M, Schenker M, Lee KH, et al. . Nivolumab plus ipilimumab versus chemotherapy as first-line treatment in advanced non-small-cell lung cancer with high tumour mutational burden: patient-reported outcomes results from the randomised, open-label, phase III CheckMate 227 trial. Eur J Cancer 2019;116:137–47. 10.1016/j.ejca.2019.05.008 [DOI] [PubMed] [Google Scholar]
- 7.Samstein RM, Lee C-H, Shoushtari AN, et al. . Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Büttner R, Gosney JR, Skov BG, et al. . Programmed Death-Ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. J Clin Oncol 2017;35:3867–76. 10.1200/JCO.2017.74.7642 [DOI] [PubMed] [Google Scholar]
- 9.Ieguchi K, Maru Y. Roles of EphA1/A2 and Ephrin-A1 in cancer. Cancer Sci 2019;110:841–8. 10.1111/cas.13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling TK, Lamb TJ. Emerging roles for Eph receptors and ephrin ligands in immunity. Front Immunol 2019;10:1473. 10.3389/fimmu.2019.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisabeth EM, Falivelli G, Pasquale EB. Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol 2013;5. 10.1101/cshperspect.a009159. [Epub ahead of print: 01 Sep 2013]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellmann MD, Nathanson T, Rizvi H, et al. . Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018;33:843–52. 10.1016/j.ccell.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. . Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109–21. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 14.Campbell JD, Alexandrov A, Kim J, et al. . Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–16. 10.1038/ng.3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang G, Song W, Amato K, et al. . Effects of cancer-associated EphA3 mutations on lung cancer. J Natl Cancer Inst 2012;104:1183–98. 10.1093/jnci/djs297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saintigny P, Peng S, Zhang L, et al. . Global evaluation of Eph receptors and ephrins in lung adenocarcinomas identifies EphA4 as an inhibitor of cell migration and invasion. Mol Cancer Ther 2012;11:2021–32. 10.1158/1535-7163.MCT-12-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaginis C, Tsoukalas N, Bournakis E, et al. . Ephrin (Eph) receptor A1, A4, A5 and A7 expression in human non-small cell lung carcinoma: associations with clinicopathological parameters, tumor proliferative capacity and patients' survival. BMC Clin Pathol 2014;14:8. 10.1186/1472-6890-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J-J, Xie D. The roles and therapeutic potentials of Ephs and ephrins in lung cancer. Exp Cell Res 2013;319:152–9. 10.1016/j.yexcr.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Xiao D, Li G, et al. . Epha2 promotes epithelial-mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer cells. Oncogene 2014;33:2737–47. 10.1038/onc.2013.238 [DOI] [PubMed] [Google Scholar]
- 20.Ding L, Getz G, Wheeler DA, et al. . Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069–75. 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulthard MG, Morgan M, Woodruff TM, et al. . Eph/Ephrin signaling in injury and inflammation. Am J Pathol 2012;181:1493–503. 10.1016/j.ajpath.2012.06.043 [DOI] [PubMed] [Google Scholar]
- 22.Chiari R, Hames G, Stroobant V, et al. . Identification of a tumor-specific shared antigen derived from an Eph receptor and presented to CD4 T cells on HLA class II molecules. Cancer Res 2000;60:4855–63. [PubMed] [Google Scholar]
- 23.Taki S, Kamada H, Inoue M, et al. . A novel bispecific antibody against human CD3 and ephrin receptor A10 for breast cancer therapy. PLoS One 2015;10:e0144712. 10.1371/journal.pone.0144712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markosyan N, Li J, Sun YH, et al. . Tumor cell-intrinsic EphA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J Clin Invest 2019;129:3594–609. 10.1172/JCI127755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conejo-Garcia JR. Breaking barriers for T cells by targeting the EPHA2/TGF-β/COX-2 axis in pancreatic cancer. J Clin Invest 2019;129:3521–3. 10.1172/JCI130316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aasheim H-C, Delabie J, Finne EF. Ephrin-A1 binding to CD4+ T lymphocytes stimulates migration and induces tyrosine phosphorylation of Pyk2. Blood 2005;105:2869–76. 10.1182/blood-2004-08-2981 [DOI] [PubMed] [Google Scholar]
- 27.Holen HL, Nustad K, Aasheim H-C. Activation of EphA receptors on CD4+CD45RO+ memory cells stimulates migration. J Leukoc Biol 2010;87:1059–68. 10.1189/jlb.0709497 [DOI] [PubMed] [Google Scholar]
- 28.Tatsumi T, Herrem CJ, Olson WC, et al. . Disease stage variation in CD4+ and CD8+ T-cell reactivity to the receptor tyrosine kinase EphA2 in patients with renal cell carcinoma. Cancer Res 2003;63:4481–9. [PubMed] [Google Scholar]
- 29.Zhang JG, Eguchi J, Kruse CA, et al. . Antigenic profiling of glioma cells to generate allogeneic vaccines or dendritic cell-based therapeutics. Clin Cancer Res 2007;13:566–75. 10.1158/1078-0432.CCR-06-1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao D, Margolis CA, Vokes NI, et al. . Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018;50:1271–81. 10.1038/s41588-018-0200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizvi NA, Hellmann MD, Snyder A, et al. . Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbst RS, Baas P, Kim D-W, et al. . Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 33.Nosaki K, Saka H, Hosomi Y, et al. . Safety and efficacy of pembrolizumab monotherapy in elderly patients with PD-L1-positive advanced non-small-cell lung cancer: pooled analysis from the KEYNOTE-010, KEYNOTE-024, and KEYNOTE-042 studies. Lung Cancer 2019;135:188–95. 10.1016/j.lungcan.2019.07.004 [DOI] [PubMed] [Google Scholar]
- 34.Reck M, Rodríguez-Abreu D, Robinson AG, et al. . Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 35.Coudray N, Ocampo PS, Sakellaropoulos T, et al. . Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559–67. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, Hua X, Zhu B, et al. . Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med 2016;13:e1002162. 10.1371/journal.pmed.1002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Zheng S, Jin R, et al. . The superior efficacy of anti-PD-1/PD-L1 immunotherapy in KRAS-mutant non-small cell lung cancer that correlates with an inflammatory phenotype and increased immunogenicity. Cancer Lett 2020;470:95–105. 10.1016/j.canlet.2019.10.027 [DOI] [PubMed] [Google Scholar]
- 38.Torres-Adorno AM, Vitrac H, Qi Y, et al. . Eicosapentaenoic acid in combination with EphA2 inhibition shows efficacy in preclinical models of triple-negative breast cancer by disrupting cellular cholesterol efflux. Oncogene 2019;38:2135–50. 10.1038/s41388-018-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizvi H, Sanchez-Vega F, La K, et al. . Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633–41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001315supp001.pdf (987.2KB, pdf)
jitc-2020-001315supp002.pdf (119.3KB, pdf)