Abstract
Colorectal cancer (CRC) is one of the most common cancers and a leading cause of mortality worldwide. Small extracellular vesicles (sEVs) are nano‐sized extracellular vesicles containing a variety of bioactive molecules, such as nucleic acids, proteins, lipids, and metabolites. Recent evidence from CRC has revealed that sEVs contribute to tumorigenesis, progression, and drug resistance, and serve as a tool for “liquid biopsy” and a drug delivery system for therapy. In this review, we summarize information about the roles of sEVs in the proliferation, invasion, migration, epithelial‐mesenchymal transition, formation of the premetastatic niche, and drug resistance to elucidate the mechanisms governing sEVs in CRC and to identify novel targets for therapy and prognostic and diagnostic biomarkers.
Keywords: Cancer Metastasis, Cancer Therapy and Diagnosis, Colorectal Cancer, Extracellular Vesicles, Small Extracellular Vesicles
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide and patients with metastasis usually have a poor prognosis. Small extracellular vesicles (sEVs) have been identified to play a significant role in intercellular communication. sEVs released from different cells exert huge effects on CRC cell proliferation and metastasis. They could also serve as biomarkers for CRC diagnosis and prognosis and be a potential drug delivery system to treat CRC patients.
Abbreviations
- BMDCs
bone marrow‐derived dendritic cells
- BMSCs
bone marrow‐derived mesenchymal stem/stromal cells
- CAF
cancer‐associated fibroblast
- CCL
CC chemokine ligand
- ceRNAs
competing endogenous RNAs
- CRC
colorectal cancer
- CSC
cancer stem cell
- CTCs
circulating tumor cells
- DCs
dendritic cells
- EGFR
epidermal growth factor receptor
- EMT
epithelial‐mesenchymal transition
- ESCRT
endosomal sorting complex required for transport
- ESEs
early‐sorting endosomes
- EVs
extracellular vesicles
- HSP
heat shock protein
- ILVs
intraluminal vesicles
- ITG
integrin
- ITGA2
integrin α2
- KLF
Krüppel‐like factor
- LARC
locally advanced rectal cancer
- LN
lymph node
- miRNAs
microRNAs
- MVBs
multivesicular bodies
- ncRNAs
noncoding RNAs
- NETs
neutrophil extracellular traps
- PD‐L1
programmed cell death ligand‐1
- PKM
pyruvate kinase
- PMN
premetastatic niche
- PTEN
phosphatase and tensin homolog deleted on chromosome ten
- Rab
Rasrelated proteins in brain
- sEVs
small extracellular vesicles
- TAMs
tumor‐associated macrophages
- TGF‐β
transforming growth factor‐β
- TLR
Toll‐like receptor
- TME
tumor microenvironment
- TSG101
tumor susceptibility 101
1. BACKGROUND
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, while the death rate ranks second, with approximately 900 000 deaths recorded annually. 1 CRC is generally asymptomatic at the early stage, which highlights the importance of early detection and diagnosis. 2 Although screening approaches have been implemented throughout the world, unfortunately, the early diagnosis rate of CRC only reaches 30‐40%, which is far from our goal. 3 Although therapies, including the application of laparoscopy, chemotherapy, radiotherapy, and target agents, have rapidly developed in recent years, the prognosis of patients with CRC generally remains poor, 4 with a 5‐year survival rate of only 10% for patients with advanced stage tumors or with metastasis. 5 Hence, the development of suitable and measurable biomarkers for an early diagnosis and predicting the prognosis has attracted increasing attention.
Extracellular vesicles (EVs) used to be identified as two main subtypes based on the mechanisms of biogenesis: endosome‐origin small extracellular vesicles (sEVs) and plasma membrane‐derived ectosomes (microvesicles/microparticles), 6 with a diameter fluctuating from 50 to 1000 nm and from 40 to 160 nm, respectively. 7 Over the past few decades, EVs have spawned great interest of their role in the progression of various cancers and their clinical potential. In view of the large number of researches of CRC focus in recent years, we will mainly summarize a specific subset of EVs referring to “exosomes” in publications. However, with an increasing understanding of EVs, it is unambiguous that the ultimate origin of vesicles cannot be discriminated yet. And it is unavoidable that most methods applied to isolate exosomes so far will coisolate heterogeneous EVs from diverse origin. As a result, based on the nomenclature illustrated in the Minimal Information for Studies on Extracellular Vesicles 2018 (MISEV2018) published by the International Society for Extracellular Vesicles (ISEV), 8 we will apply the terminology “small extracellular vesicles” (sEVs) (diameter <200 nm or <100 nm) in place of “exosomes” (Tables 1 and 2).
TABLE 1.
Main types of extracellular vesicles
TABLE 2.
MISEV2018‐recommended nomenclature 8
| Characteristics | Recommended nomenclature | |
|---|---|---|
| Physical characteristics | Size |
Small: diameter <200 nm or <100 nm Large and/or medium: >200 nm |
| Density | Low; middle; high | |
| Biochemical composition | eg, CD63+/CD81+ EVs, PD‐L1+ EVs, etc | |
| Conditions or cell of origin | eg, Apoptotic EVs, hypoxic EVs, etc | |
Although sEVs were initially underestimated as “cellular debris” and a system to dispose of cellular components when they were first discovered in 1983 by two independent groups, now they are considered to play a significant role in intercellular communication. 7 , 9 , 10 Based on emerging evidence, biomolecules loaded in sEVs are shuttled to recipient cancer cells or stromal cells, thus modulating their activity by regulating signaling pathways. sEVs are involved in tumor proliferation, metastasis, angiogenesis, immune regulation, and even drug resistance. 11 Additionally, cancer cell‐derived sEVs carry a number of cancer‐specific biomolecules, such as proteins, microRNAs (miRNAs), and lncRNAs, which might serve as biomarkers for the early detection of CRC. 12 , 13 In this review, we summarize the biogenesis of sEVs, their roles in CRC progression (Table 3), and their promising clinical applications (Table 4).
TABLE 3.
The role of the substances in CRC sEVs
| Cargo | Parent cell | Recipient cell | Pathway and target | Biofunction | Reference |
|---|---|---|---|---|---|
| MiRNAs | |||||
| miR‐21 | CAFs | CRC cells | / | Promote proliferation, invasion, and metastasis | 159 |
| miR‐92a‐3p | CAFs | CRC cells | FBXW7 and MOAP1/Wnt/β‐catenin | Promote invasion and metastasis | 29 |
| miR‐486‐5p | CRC cells | CRC cells | PLAGL2/IGF2/β‐catenin | Promote proliferation, invasion, and metastasis | 30 |
| miR‐193a | CRC cells | CRC cells | / | Promote proliferation, invasion, and metastasis | 160 |
| miR‐16‐5p | BMSCs | CRC cells | ITGA2 | Inhibit proliferation, invasion, metastasis, and promote apoptosis | 33 |
| miR‐128‐3p | CRC cells | CRC cells | Bmi1 | EMT | 113 |
| miR‐106b‐3p | CRC cells | CRC cells | DCL‐1 | Promote invasion, metastasis, and EMT | 44 |
|
miR‐25‐3p miR‐130b‐3p miR‐425‐5p |
CRC cells | TAMs | PTEN/PI3K/Akt/STAT6 | EMT | 45 |
| miR‐25‐3p | CRC cells | Endothelial cells | KLF/ZO‐1 and occluding and Claudin5 | PMN and metastasis | 45 |
| miR‐200s | CRC cells | Endotheliocytes | / | EMT | 42 , 43 |
| miR‐21 | CRC cells | TAMs | TLR7/IL‐6 | PMN | 47 |
| miR‐1229 | CRC cells | Endothelial cells | HIPK2/MEF2C/VEGF | Angiogenesis | 59 |
| miR‐146a‐5p | CSCs | CD8+ T cells | / | Promote immunosuppressive microenvironment | 80 |
| miR‐1246 | GOF mutp53 cancer cells | Macrophages | TGF‐β | Inhibit macrophage polarization | 67 |
| miR‐203 | CRC cells | Monocytes | / | Promote metastasis | 161 |
|
miR‐21‐5p miR‐155‐5p |
M2 TAMs | CRC cells | BRG1 | Promote migration, invasion, and metastasis | 87 |
| LncRNAs | |||||
| LncRNA 91H | CRC cells | CRC cells | HNRNPK | Promote invasion and metastasis | 37 |
| LncRNA UCA1 | CRC cells | CRC cells | miR‐143/MYO6 | Promote proliferation, apoptosis, and metastasis | 38 |
| LncRNA APC1 | CRC cells | Endothelial cells | p38‐MAPK | Angiogenesis | 32 |
| LncRPPH1 | CRC cells | Macrophages | / | Promote proliferation and metastasis | 86 |
| CircRNAs | |||||
| CircIFT80 | CRC cells | CRC cells | miR‐1236‐3p/HOXB7 | Promote proliferation, metastasis, and EMT | 46 |
| CircFMN2 | CRC cells | CRC cells | miR‐1182/hTERT | Promote proliferation | 39 |
| CircLONP2 | CRC cells | CRC cells | miR‐17 | Promote invasion and metastasis | 40 |
| Others | |||||
| Wnt4 | CRC cells | CRC cells | Wnt/β‐catenin | Promote proliferation and metastasis | 28 |
| CCL2 | CRC cells | TAMs | / | PMN | 69 |
| Integrins (ITGs) | CRC cells | CAFs | / | PMN | 74 |
| PD‐L1 | CRC cells | T cells | / | Promote proliferation and drug resistance | 78 |
| CXCL1, CXCL2 | CRCSC | Neutrophils | IL‐1β | Immune regulation | 94 |
| HSP | CRC cells | NK cells | Granzyme B | Initiate apoptosis | 99 |
TABLE 4.
The clinical application of CRC sEVs components
| Cargo | Parent cell | Source of sEVs | Biomarker potential | Reference |
|---|---|---|---|---|
| MiRNAs | ||||
| miR‐181a‐5p | Hypoxic tumor cells | Plasma | Prognosis | 141 |
| miR‐486‐5p | Hypoxic tumor cells | Plasma | Prognosis | 141 |
| miR‐30d‐5p | Hypoxic tumor cells | Plasma | Prognosis | 141 |
| miR‐150‐5p | CRC cells | Serum | Diagnosis and prognosis | 143 |
| miR‐99b‐5p | CRC cells | Serum | Diagnosis and prognosis | 143 |
| miR‐27a | CRC cells | Plasma | Diagnosis and prognosis | 144 |
| miR‐130a | CRC cells | Plasma | Diagnosis and prognosis | 144 |
| miR‐92b | CRC cells | Plasma | Diagnosis | 162 |
| miR‐122 | CRC cells | Serum | Diagnosis and prognosis | 163 |
| miR‐424‐5p | CRC cells | Serum | Diagnosis | 164 |
| LncRNAs | ||||
| LNCV6_116109/LNCV6_98390/LNCV6_38772/LNCV_108266/LNCV6_84003/LNCV6_98602 | CRC cells | Plasma | Diagnosis (stages I‐II) | 3 |
| HOTTIP | CRC cells | Serum | Prognosis | 142 |
| LINC02418 | CRC cells | Serum | Diagnosis | 165 |
| CircRNAs | ||||
| hsa‐circ‐0004771 | CRC cells | Serum | Diagnosis | 166 |
| circ‐PNN | CRC cells | Serum | Diagnosis | 167 |
2. sEVs: BIOGENESIS AND CHARACTERIZATION
In this part, we only describe the biogenesis of exosomes because the process of exosomes is most well illustrated. It primarily occurs in several steps, as described below. (a) The formation of early‐sorting endosomes (ESEs): The invagination of the cellular plasma membrane leads to the formation of ESEs, which contain proteins from the cell surface and extracellular milieu. (b) The formation of multivesicular bodies (MVBs): After ESEs mature into late‐sorting endosomes, the inward budding of the endosomal membrane results in the formation of MVBs. (c) The generation of exosomes: MVBs, which are characterized by the presence of intraluminal vesicles (ILVs), either fuse with the cellular plasma membrane to release the ILVs as exosomes containing specific biomolecules into the extracellular space or fuse with lysosomes or autophagosomes to be degraded 7 (Figure 1).
FIGURE 1.
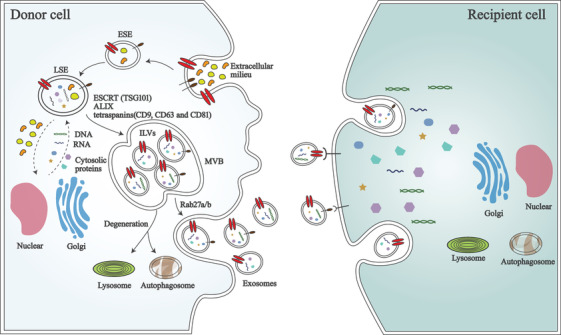
The main procedure of exosomes biogenesis and release. Cellular plasma membrane invaginates to form early‐sorting endosomes (ESEs), then ESEs mature into late‐sorting endosomes (LSEs), and inward budding of endosomal membrane results in multivesicular bodies (MVBs). Tumor susceptibility 101 (TSG101), ALG‐2 interacting protein X (ALIX), and tetraspanins (CD9, CD63, and CD81) are indispensable parts in endosomal sorting complex required for transport (ESCRT)‐dependent way in the process of MVBs biogenesis. MVBs fuse with cellular plasma membrane to release intraluminal vesicles (ILVs) as exosomes. Rab27a is associated with membrane fusion and endosomal size, whereas Rab27b is connected with exosomes redistribution. After secretion, exosomes uptaken by recipient cells could be mediated by endocytosis, fusion with the plasma membrane, or ligand/receptor interaction
Many researchers have attempted to explore the mechanisms of exosomes’ formation. The endosomal sorting complex required for transport (ESCRT) consists of four main complexes (ESCRT‐0, I, II, and III) and is known to be the most important machinery responsible for delivering specific molecules into ILVs of the MVBs and for protein recycling. ESCRT‐0, which is activated by phosphatidylinositol 3‐phosphate, recruits ESCRT‐I to form the ESCRT‐I complex by interacting with tyrosine kinase substrate prosaposin domains and ESCRT‐I subunit tumor susceptibility 101 (TSG101). The complex is necessary for sorting cargo and transferring ubiquitinated transmembrane proteins into the MVBs. ESCRT‐II is involved in the initiation of inward budding processes. ESCRT‐III is regulated by complexes I and II, and plays key roles in cargo sorting and concentration, vesicle scission, and protein recycling. Additionally, ALG‐2 interacting protein X, an accessory protein of ESCRT, functions in the ESCRT pathway to regulate the process of exosomes biogenesis. 14
In addition to the ESCRT‐dependent pathway, an ESCRT‐independent mechanism also exists, and some proteins, such as the Rasrelated proteins in brain (Rab) family (Rab27a and Rab27b), tetraspanins (CD9, CD63, and CD81), and sphingomyelinase, have been shown to engage in membrane fusion, endosomal vesicle trafficking, and vesicle release. Among these proteins, Rab27a is associated with membrane fusion and the endosomal size, whereas Rab27b knockdown redistributes multivesicular endosomes to the perinuclear region. However, researchers should focus on elucidating the precise mechanism of the ESCRT‐independent pathway, due to the insufficient understanding of the mechanism. 15
The composition of double‐layered and cup‐shaped sEVs is heterogeneous and reflects their cellular origin and physiological and pathological states. Because sEVs are synthesized by most cell types, such as T lymphocytes, B cells, and dendritic cells (DCs), sEVs contain proteins, lipids, mRNAs, noncoding RNAs (ncRNAs), and other molecules with a density ranging from 1.10 to 1.20 g/mL. 14 The contents of cancer‐derived sEVs are innately heterogeneous and may exert powerful effects on recipient cells. 16
sEVs have been isolated from bodily fluids, for example, plasma, plural effusion, breast milk, saliva, tears, and urine. Numerous sEVs isolation and enrichment techniques have been developed; however, nonvesicular molecular structures inevitably contaminate the isolated sEVs. The principal conventional methods used for isolation and enrichment are “standard” ultracentrifugation, gradient centrifugation, polymer‐based precipitation, size‐exclusion chromatography, and immunoaffinity chromatography, with the advantage of high throughput. On the other hand, recently developed methods, such as microfluidic filtering, contact‐free sorting, and immunoaffinity enrichment, have increased the enrichment efficiency and specificity, but at a lower throughput. Overall, the advantages and disadvantages of isolation methods exist objectively in both conventional and novel techniques, and thus further studies are needed to develop methods for more efficient isolation of sEVs. 14 To date, a consensus “gold‐standard” isolation method has not been identified, and the method basically depends on the downstream applications and specific scientific questions.
Although “sEVs‐specific” markers are not currently available, investigators have reported at least three positive protein markers in a semiquantitative manner, such as Western blotting or flow cytometry, to generally characterize sEVs. Positive proteins include at least one transmembrane/lipid‐bound protein (CD9 and CD63) and one cytosolic protein (TSG101). In addition, the levels of proteins that are not expected to be enriched, such as calnexin, should also be determined. Moreover, the characterization of single vesicles in a mixture is recommended to provide an indication of the heterogeneity. At least two different but complementary techniques should be utilized for characterization, such as electron microscopy (transmission) or atomic force microscopy. 17 , 18 Furthermore, several sEVs and EV databases provide lists of the contents that have been identified, and researchers could compare the components they have isolated with the components listed in the databases EVpedia, Vesiclepedia, Exocart, exoRBase, and EVmiRNA. 19 , 20 , 21 , 22 , 23
3. CRC sEVs AND METASTASIS
Patients with CRC tend to experience a high death rate due to metastasis or tumor recurrence. 24 Based on emerging evidence, sEVs critically mediate CRC metastasis by transporting biomolecules between different cell types and play a pivotal role in the intricate process of tumor biofunction. Therefore, it is imperative to determine how sEVs exert their effects on the metastasis of CRC.
3.1. sEVs and cell proliferation, invasion, and migration in CRC
Some signaling pathways are engaged in the process of CRC formation, of which the canonical Wnt‐β‐catenin pathway is ubiquitously active in CRC development, and the most prevalent genetic events are mutations that disrupt the Wnt signaling cascade. 25 Simultaneously, different Wnts behave in a similar manner in terms of the biochemical signaling pathways or effects on target cells, and β‐catenin is crucial effector of the pathway. 26 EVs recruiting mutant β‐catenin might activate the Wnt signaling pathway to stimulate proliferation and migration of recipient cells 27 ; meanwhile, hypoxic CRC cell‐derived sEVs simultaneously shuttle Wnt4 to normoxic cells to promote invasion and migration by stimulating the Wnt signaling pathway. 28 Overexpression of cancer‐associated fibroblast (CAF)‐derived sEVs‐miR‐92a‐3p downregulates the expression of its target genes FBXW7 and MOAP1 to inhibit the ubiquitination and degradation of β‐catenin, resulting in the invasion and migration of CRC cells by activating the Wnt‐β‐catenin signaling pathway. 29 According to Liu et al, sEVs‐miR‐486‐5p, an oncogene, directly binds to PLAGL2 to stimulate CRC cell growth and migration by inducing the expression of genes involved in the IGF2‐β‐catenin signaling pathway. 30 The tumor‐suppressor gene APC ordinarily negatively regulates the canonical Wnt signaling pathway; nevertheless, in another pathway, APC downregulates PPARα binding to the lncRNA‐APC1 promoter to increase lncRNA‐APC1 expression, which suppresses CRC cell proliferation and metastasis by inhibiting sEVs production. 31 , 32
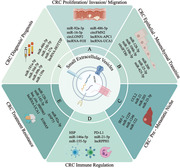
Additionally, numerous sEVs biomolecules are involved in a special signaling axis or signaling targets to alter tumor metastasis. Notably, miR‐16‐5p contained in sEVs derived from bone marrow‐derived mesenchymal stem/stromal cells (BMSCs) functions as a tumor suppressor that inhibits the proliferation, invasion, and migration and simultaneously promotes the apoptosis of CRC cells by reducing the expression of integrin α2 (ITGA2). 33 The levels of sEVs‐lncRNAs and circRNAs are associated with the regulation of metastasis and proliferation in CRC. 34 , 35 , 36 High sEVs‐lncRNA 91H levels promote invasion and migration by interacting with and modifying the expression of the RNA‐binding protein HNRNPK. 37 The oncogenic lncRNA UCA1 increases the proliferation, apoptosis, and metastasis of CRC cells both in vitro and in vivo. Furthermore, UCA1 regulates MYO6 expression by directly sponging miR‐143. 38 CircFMN2, which is abundant in serum sEVs, sponges miR‐1182 to eventually increase the expression of hTERT and substantially accelerate CRC cell proliferation. 39 Nevertheless, circLONP2, which is mainly located in the nucleus, has a vastly different function from conventional sEVs circRNAs. It serves as a vital metastasis‐initiating factor that promotes CRC invasion and metastasis in distant organs by promoting the maturation and packing of miR‐17 into cell‐derived sEVs. 40 As described above, these studies definitely illustrate that ncRNAs packed in sEVs exert important effects on recipient CRC cells and regulate their proliferation, migration, invasion, and metastasis (Figure 2A).
FIGURE 2.
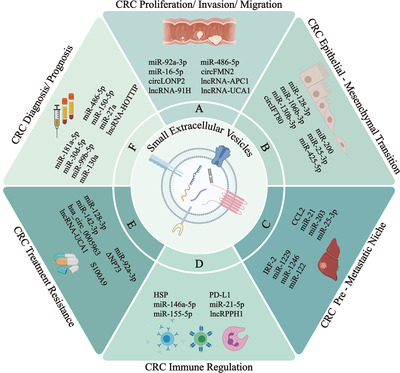
sEVs play a significant role in colorectal cancer proliferation/invasion/migration (A), epithelial‐mesenchymal transition (B), premetastatic niche (C), immune regulation (D), treatment resistance (E), and diagnosis/prognosis (F)
3.2. sEVs and the epithelial‐mesenchymal transition (EMT) of CRC
EMT is characterized by a loss of epithelial properties (ie, low levels of E‐cadherin and β‐catenin) and a gain of mesenchymal properties (ie, high levels of N‐cadherin and vimentin), and plays a predominant role in metastasis and drug resistance, and therefore is associated with a poor prognosis. 41 Accumulating evidence has revealed the association between sEVs and EMT.
Notably, some miRNAs suppress the EMT in CRC. For instance, sEVs containing a high level of miR‐128‐3p appear to selectively transfer miR‐128‐3p to oxaliplatin‐resistant CRC cells, resulting in the inhibition of the oxaliplatin‐induced EMT. 41 The miR‐200 family (miR‐200a, 200b, 200c, 141, 429) contained in the sEVs of CRC cells is transferred to blood and lymph endotheliocytes to further repress the EMT of endothelial cells. 42 , 43 In contrast, many miRNAs function to promote the EMT. High expression levels of miR‐106b‐3p, 44 miR‐25‐3p, miR‐130b‐3p, and miR‐425‐5p 45 promote the EMT through various pathways, and miR‐25‐3p facilitates the EMT by activating the canonical PTEN‐PI3K‐Akt‐STAT6 signaling axis. Circular RNAs contained in sEVs might function as competing endogenous RNAs (ceRNAs) to regulate the EMT in patients with mCRC. For example, sEVs‐circIFT80 functions as a ceRNA of miR‐1236‐3p to further regulate HOXB7 expression, which subsequently promotes the EMT of CRC 46 (Figure 2B).
3.3. sEVs and the premetastatic niche (PMN) of CRC
The survival rate of patients with CRC substantially decreases to approximately 12% when distant organ metastasis occurs, increasing the importance of identifying critical components involved in the process of tumor metastasis and exploring new strategies to prevent tumor metastasis. 47 , 48 Based on Paget's “seed‐and‐soil” theory, a novel concept of the PMN was proposed to elucidate mechanisms of tumor metastasis from primary sites to secondary or remote organ sites. 49 The PMN is a microenvironment that primarily consists of tumor‐derived cytokines, growth factors, sEVs, immune cells, and host stromal cells, and prepares a suitable site for the dissemination and growth of circulating tumor cells (CTCs). 50 , 51 Liu and Cao proposed six characteristics to define the PMN that promote metastasis and enable colonization, including vascular leakiness and angiogenesis, lymphangiogenesis, inflammation, immunosuppression, reprogramming, and organotropism. 52 As components of the PMN, tumor‐derived sEVs were recently proposed to regulate the formation of the PMN in specific organ sites. 53 , 54 Therefore, we desire to obtain insights into how sEVs regulate the formation of the PMN in CRC (Figure 2C).
3.3.1. Vascular leakiness and angiogenesis
The vascular endothelial cell layer is connected by adherens and tight junctions, which provides a physical barrier to cells and body fluids. Tumor‐derived sEVs impair the functions of EC junctions to promote vascular permeability for further CTC entry into specific sites of distant organs. 49 , 55 , 56 CRC‐derived sEVs‐miR‐25‐3p contributes to the induction of vascular leakiness and angiogenesis by downregulating the expression of the Krüppel‐like factor (KLF) family (zinc finger‐containing transcription factors), followed by the inhibition of the downstream endothelial cell targets ZO‐1, occludin, Claudin5, and VEGFR2. Consequently, sEVs‐miR‐25‐3p was capable of impairing the junctions of the endothelial cell layer, which increased the formation of the PMN and CRC metastasis in liver and lung in vivo. 57 , 58 Moreover, CRC cells delivered miR‐1229 into vascular endothelial cells to induce VEGF expression and subsequently promote angiogenesis59 (Figure 3A).
FIGURE 3.
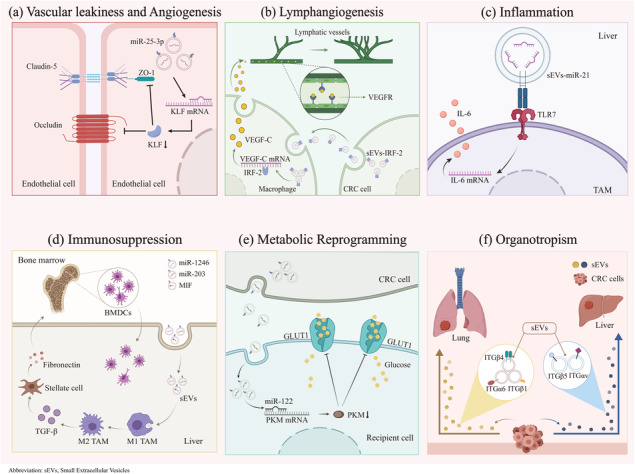
The mechanisms of sEVs in the formation of premetastatic niche in colorectal cancer. A, Vascular leakiness and angiogenesis: sEVs‐miR‐25‐3p induces vascular leakiness and angiogenesis by downregulating KLF family, ZO‐1, occludin, Claidin5, and VEGFR2. B, Lymphangiogenesis: sEVs‐IRF‐2 stimulates VEGF‐C secretion by sentinel lymph node (LN) macrophages to promote lymphangiogenesis. C, Inflammation: sEVs‐miR‐21 polarizes liver macrophages via miR‐21‐TLR7‐IL6 axis, which induces chronical inflammation. D, Immunosuppression: sEVs‐miR‐1246, miR‐203, and MIF polarize macrophages to increase TGF‐β expression, and then activate hepatic stellate cells to secrete fibronectin, which recruits bone marrow‐derived dendritic cells (BMDCs) to promote immunosuppressive microenvironment. E, Reprogramming: sEVs‐miR‐122 inhibits PKM expression to reduce GLUT1 and glucose uptake to reprogram the metabolism. F, Organotropism: sEVs‐ITGα6/ITGβ4/ITGβ1 are enriched in lung tropic sEVs, while sEVs‐ITGβ5/ITGαv were primarily liver tropic
3.3.2. Lymphangiogenesis
Lymphatic vessels probably serve as a primary point of access for the lymphatic dissemination of tumor cells, 60 and lymphangiogenesis precedes CTC arrival at distant organ sites. 61 , 62 Therefore, lymphangiogenesis is actively involved in the formation of the PMN. According to clinical data, tumor‐derived VEGFs promote premetastatic lymphangiogenesis in regional lymph nodes (LNs). 63 CRC‐derived sEVs‐IRF‐2 (interferon regulatory factor 2) has been postulated to stimulate VEGF‐C secretion by sentinel LN macrophages, resulting in lymphangiogenesis and metastasis 64 (Figure 3B).
3.3.3. Inflammation
Chronic inflammation is a critical driver of tumor progression and metastasis, and thus the local inflammatory microenvironment is an important factor contributing to the formation of the PMN. As components of the microenvironment, sEVs are involved in regulating inflammation by facilitating the infiltration of inflammatory cells. 52 Notably, inappropriate activation of Toll‐like receptor 4 (TLR4) or other signaling pathways in immune cells may lead to unexpected inflammation. 65 CRC‐derived sEVs‐miR‐21 is capable of polarizing liver macrophages into an IL‐6‐secreting phenotype by binding to TLR7 in tumor‐associated macrophages (TAMs), which contributes to the creation of an inflammatory PMN. 47 Thus, the miR‐21‐TLR7‐IL6 axis would be a potential therapeutic target for patients with CRC that has metastasized to the liver (Figure 3C).
3.3.4. Immunosuppression
The recruitment of immune cells to establish an immunosuppressive microenvironment is a hallmark of PMN formation. In particular, bone marrow‐derived dendritic cells (BMDCs) are one of the main effector cells that suppress the antitumor response and allow primary tumor cells to overcome the immune defenses. 66 sEVs containing large amount of miR‐1246 that are derived from TP53 mutant CRC cells, miR‐203‐enriched sEVs and macrophage migration inhibitory factor‐containing sEVs originating from CRC cells are taken up by liver macrophages, resulting in the polarization of macrophages to M2 TAMs to increase transforming growth factor‐β (TGF‐β) levels, 47 , 67 subsequently inducing neighboring hepatic stellate cells to secrete fibronectin, and ultimately contributing to the recruitment of BMDCs that promote PMN formation. 68 Interestingly, a classical Chinese medicine, Dahuang Zhechong pill, reduces the expression of sEVs‐CC chemokine ligand‐2 (CCL2) to repress TAM recruitment and inhibit the polarization of M1 cells to a M2 phenotype, eventually ameliorating the formation of the PMN 69 (Figure 3D).
3.3.5. Reprogramming
Stromal, metabolic, and epigenetic reprogramming are engaged in PMN‐induced tumor metastasis. As shown in recent studies, sEVs are associated with recruiting and reprogramming host stromal cells, such as fibroblasts and epithelial cells, into the PMN to modify the PMN. 70 , 71 sEVs‐miR‐122 taken up by recipient cells specifically inhibits pyruvate kinase (PKM) expression to reduce glucose transporter 1 levels and glucose uptake, which reprogrammed the metabolism of PMN to facilitate metastasis72 (Figure 3E). sEVs‐contained miRNAs may epigenetically exhaust the expression of phosphatase and tensin homolog deleted on chromosome ten (PTEN) to induce the secretion of chemokine CCL2, subsequently recruiting tumor‐promoting myeloid cells to promote PMN formation. 73 Regrettably, few studies have examined how sEVs facilitate the reprogramming of PMN in CRC; therefore, further investigations assessing how remodeling occurs are urgently needed.
3.3.6. Organotropism
The concept of organotropism is described as the ability of certain tumors to metastasize to specific organs. Tumor‐derived sEVs express particular integrins (ITGs) that interact with extracellular matrix molecules (laminin and fibronectin) to initiate the formation of PMN in a tissue‐specific manner. ITGα6/ITGβ4/ITGβ1 are enriched in lung‐tropic sEVs, while in parallel, ITGβ5/ITGαv are primarily enriched in liver‐tropic sEVs 4 9 (Figure 3F). In a CRC model, integrin beta‐like 1 activated CAFs in remote organs through the TNFAIP3‐NF‐κB signaling pathway, and subsequently the stimulated CAF secretion of the proinflammatory cytokines IL‐6 and IL‐8 to promote formation of the PMN. 74
In summary, all these studies have undoubtedly provided a foundation of further investigations of the role of sEVs in PMN formation and propose a promising strategy to prevent metastasis of CRC.
4. CRC sEVs AND IMMUNE REGULATION
Cancer‐derived sEVs are mechanistically engaged in impairing an effective immune response by modulating the maturation and antitumor activity of immune cells, 75 , 76 which promotes the establishment of an immunosuppressive microenvironment for cancer cells. 77 Programmed cell death ligand‐1 (PD‐L1) expressed on the surface of cancer cells binds its receptor PD‐1 on effector T cells, thus attenuating their activity in antitumor immunity; not surprisingly, PD‐L1 is also present on the surface of sEVs. In a Rab27a knockout CRC model, PD‐L1+ sEVs appeared to inhibit T‐cell activity to promote tumor growth and resist to immune checkpoint protein inhibitors. 78
CD8+ T cells represent the chief antitumor effector cells in the tumor microenvironment (TME). However, the dysfunction of CD8+ T cells impairs the antitumor effect. 79 Cheng and colleagues revealed that sEVs‐miR‐146a‐5p, a major miRNA expressed in cancer stem cells (CSCs) that depends on Rab27a activation, decreased the number of tumor‐infiltrating CD8+ T cells, thus promoting the formation of an immunosuppressive cancer microenvironment in CRC. 80 CRC sEVs induced a shift in the phenotype T cells to tumor‐supporting Treg‐like cells by activating TGF‐β‐Smad signaling and inactivating SAPK signaling. 81 In addition to impairing T‐cell function, Huber et al proposed that CRC cells release numerous sEVs containing Fas‐ligand positive and tumor necrosis factor‐related apoptosis‐inducing ligands to promote CD8+ T‐cell apoptosis, thus creating an immunosuppressive microenvironment. 82
Generally, M2 TAMs, which are known as alternatively activated macrophages, are required to promote tumorigenesis by secreting pro‐tumor factors, such as inflammatory cytokines, chemokines, and angiogenic factors. 83 , 84 sEVs are capable of inducing macrophages to differentiate into the pro‐tumor M2 phenotype. 85 LncRPPH1‐loaded sEVs, which are present at high levels in serum, are transferred into macrophages to mediate macrophage M2 polarization, which in turn promotes the proliferation and metastasis of CRC cells. 86 Furthermore, Lan et al have reported that M2 macrophage‐derived sEVs containing miR‐21‐5p and miR‐155‐5p downregulate the expression of BRG1 by directly binding to the BRG1 coding sequence in CRC cells, thus resulting in the metastasis of CRC. 87
DCs are potent antigen‐presenting cells that activate T cells to induce an antitumor response. However, EVs derived from cancer cells are able to block DC activity through various signaling pathways. 88 One study reported an obvious upregulation of TNF, TGF‐β, and IL‐6 in DCs cocultured with EVs, which subsequently decreased their phagocytic activity, suppressed the proliferation of T cells, and impaired the cytolytic potential of T cells by downregulating intracellular granzyme B, perforin, and IFN‐γ. 89 sEVs‐heat shock protein (HSP) derived from B lymphoma cells more efficiently induces both the phenotypic and functional maturation of DCs, and simultaneously stimulates the antitumor activity of CD8+ T cells. 90
Although neutrophils are characterized by neutral staining, the role of neutrophils in cancers is by no means neutral. Neutrophils possess both tumor‐promoting and tumor‐suppressing functions, depending on numerous factors, 91 including polarization (N1 or N2 phenotype) and location relative to the tumor (intratumor, peritumor, or stromal). 92 , 93 CSC sEVs transported to bone marrow not only increase neutrophil survival but also reprogram these cells to the pro‐tumor phenotype that secretes IL‐1β, mainly by activating the NF‐κB signaling pathway. 94 Recently, the role of neutrophil extracellular traps (NETs), the extracellular mesh‐like structures containing DNA and cytosolic granular proteins, released from neutrophils in cancer have attracted increasing attention. 95 Specifically, sEVs derived from KRAS mutant CRC induce neutrophil accumulation and the formation of NETs to promote CRC growth and metastasis, and the effect was abolished by an anti‐IL‐8 treatment. 96 , 97 , 98 This evidence revealed roles for sEVs in the survival, migration, phenotype transition, and NET release of neutrophils.
The studies described above reveal the functions of sEVs in inhibiting antitumor immunity; however, the exact role of sEVs is still a topic of debate. Limited studies have been conducted to elucidate the antitumor effect of sEVs. HSP on the surface of sEVs from CRC cells induces the migration and cytolytic activity of natural killer (NK) cells following the release of granzyme B to initiate tumor apoptosis. 99 The other pathway of HSP is to inhibit tumor growth by converting immunosuppressive regulatory T cells to Th17 cells via IL‐6. 100 Taken together, further research is indeed needed to understand the mechanisms by which sEVs regulate the immune response in CRC, and to identify potential prognostic factors and antitumor therapies (Figure 2D).
5. CRC sEVs AND TREATMENT RESISTANCE
Important achievements in mCRC treatment have been reported, such as the application of conventional therapeutics based on 5‐fluorouracil with oxaliplatin or irinotecan, monoclonal antibodies such as panitumumab and cetuximab, 24 , 101 , 102 , 103 and pre‐ and postoperative radiotherapy. However, a large number of patients exhibit different levels of treatment resistance, which directly results in reduced survival rate. 104 Therefore, the mechanisms of primary or acquired therapy resistance must be clarified to improve the survival rate of patients with mCRC. Many researchers have analyzed sEVs released by stromal or cancer cells and their potential pivotal roles in treatment resistance 105 (Figure 2E).
5.1. sEVs and drug resistance
The conventional mechanisms of drug resistance have been elucidated by numerous researchers over the past few decades, including mutations in p53, overexpression of ATP‐binding cassette efflux transporters, alterations in cellular drug influx/efflux, disruption of apoptotic pathways, and single‐nucleotide polymorphisms in platinum or fluoropyrimidine targets, among others. 104 , 106
In addition to the mechanisms mentioned above, scholars’ interest has shifted to the exploration of cell‐derived sEVs. Two major monoclonal antibodies, cetuximab and panitumumab, directly bind to the extracellular domain of the epidermal growth factor receptor (EGFR) and admittedly improve the survival of patients with mCRC expressing wild type RAS, including improvements in overall survival, progression‐free survival, and response rate, both as single agents and in combination with chemotherapy. 107 , 108 EGFR is an essential component of the pathway regulating cell proliferation by activating several principal downstream pathways, the RAS‐RAF‐MAPK, PI3K‐PTEN‐AKT, and JAK‐STAT pathways, and mutations in any protein involved in these pathways, such as KRAS, BRAF, and PIK3CA, may lead to resistance to anti‐EGFR therapy. 109 sEVs derived from cetuximab‐resistant cells have been shown to “infect” cetuximab‐sensitive cells and transform them to cetuximab‐resistant cells by reducing the expression of PTEN and increasing the levels of phosphorylated AKT. 110 Although the specific role of sEVs was not clearly determined in this previous study, another study revealed that sEVs‐lncRNA UCA1 is essential to confer cetuximab resistance in CRC cells by participating in RNA‐RNA interactions. 111 Overexpression of UCA1 contributes to reducing the expression of miRNA targets, which induces the deregulation of certain signaling pathways. 112 Nevertheless, overexpression of sEVs‐transmitted miR‐128‐3p was recently shown to increase the chemosensitivity of oxaliplatin‐resistant cells by inhibiting the expression of the drug transporter MRP5 to reduce oxaliplatin efflux. 113
CSCs are characterized by the surface markers CD133 and CD44, 114 self‐renewal, and the ability to differentiate into various cell types, which are inherently chemoresistant and enriched in recurrent cancer. 115 , 116 , 117 , 118 Therefore, several researchers have elaborated the association of sEVs with the formation of CSCs and resistance to chemotherapy. sEVs derived from CAFs consist of different types of RNAs, and the direct transfer of sEVs‐miR‐92a‐3p to CRC cells primes stem cells and induces chemoresistance by activating the Wnt‐β‐catenin pathway and inhibiting FBXW7 and MOAP1. 29 , 118 , 119 sEVs derived from myeloid‐derived suppressor cells maintain the stemness of CRC cells by delivering S100A9 to bind and activate NADPH oxidase, which activates NF‐κB and STAT3. 115 sEVs from BMSCs carry miR‐142‐3p, which increases the population of CSCs by downregulating the protein Numb (inhibitor of the Notch signaling pathway), thus activating the Notch signaling pathway. 120 However, miR‐142‐3p may also inhibit the stemness of CRC cells by targeting CD133, Lgr5, and ABCG2. Therefore, the role and mechanisms of miR‐142‐3p require further study. 121
Cytotoxic drugs are very important treatments for patients with mCRC and are often administered in combination with other anticancer drugs. The sEVs‐circular RNA hsa_circ_0005963 delivered from oxaliplatin‐resistant cells is transferred to chemosensitive cells, increasing glycolysis to induce drug resistance by regulating the miR‐122‐PKM2 axis. 122 Moreover, ΔNP73, an isoform of homolog of p53, is transferred by sEVs to confer resistance to oxaliplatin, thus promoting CRC cell proliferation. 123
Overall, drug resistance exists, and an effective method is not available to address this problem. Additionally, sEVs represent a specific focus area for researchers to elucidate the mechanisms of drug resistance and improve treatments for patients with mCRC.
5.2. sEVs and radiotherapy resistance
Pre‐ and postoperative radiotherapy prolonged disease‐free survival in several randomized trials, 124 , 125 , 126 of which preoperative radiotherapy achieved better effects. 127 However, similar to resistance to chemotherapy, radioresistance occurs and the response rate is only approximately 20%. 128 A resistance mechanism related to CAF‐derived sEVs has been identified, as CAF‐derived sEVs stimulate the TGF‐β signaling pathway to program CRC cells to the CSC phenotype characterized by clonogenicity and radioresistance. 129 Studies on this topic are extremely limited; accordingly, the underlying mechanisms of radiotherapy resistance must be elucidated to provide better therapeutic strategies in the future.
6. CRC sEVs AND CLINICAL THERAPY
As described above, drug resistance is quite common in CRC. With the characteristics of high biocompatibility, low immunogenicity, and efficient delivery, sEVs have become a potential drug delivery system. 130 , 131 Early in 2008, 40 patients with advanced CRC were enrolled in a phase I clinical trial and were treated with ascites‐derived sEVs in conjunction with the granulocyte‐macrophage colony‐stimulating factor, but therapeutic responses were rarely detected, with the exception of a few patients in the stable state. 132 Recently, the University of Louisville designed a randomized phase I clinical trial to investigate the ability of plant sEVs to deliver curcumin to normal and malignant colon tissues (NCT01294072) that was based on the biocompatibility of sEVs to increase the stability, solubility, and bioactivity of curcumin in cancer cells and immune cells in colon cancer.
Some researchers have postulated that sEVs loaded with drugs, coated with certain high‐density antibodies, and functionalized with targeting ligands would display an improved tumor‐targeting ability and inhibit cell proliferation, which might represent an innovative delivery system for drugs targeting specific cancers. 133 Deregulated miRNA expression is associated with the chemotherapy resistance pathway, and thus the inhibition of oncogenic miRNAs represents a potential therapeutic strategy. 134 According to Liang et al, the co‐delivery of 5‐fluorouracil and a miR‐21 inhibitor by engineered sEVs reverses anticancer drug resistance in CRC. 135 As shown in another study, miR‐128‐3p is crucial regulator that increases intracellular oxaliplatin accumulation to increase the chemosensitivity of CRC. 113
7. CRC sEVs AND FUTURE PERSPECTIVES IN THE CLINIC
Early detection of CRC would contribute remarkably to improving the prognosis and survival of patients. With the increasing application of colonoscopy in the past few decades, the detection rate has increased. However, the invasive process is likely to create complications, such as pain, intestinal perforation, or bleeding, in patients. 136 , 137 Currently, “liquid biopsy,” an easy, noninvasive, reproducible, economic, and acceptable strategy compared to traditional biopsy, has been shown to be potential complement to surgical biopsy in the diagnostic screen and monitoring of the patient's condition. The assays of the molecules and cells present in the “liquid biopsy” include circulating tumor DNAs, CTCs, tumor‐educated platelets, circulating free RNAs, circulating miRNAs, and circulating sEVs. 138 Among these markers, sEVs are characterized by stability (the constituents are protected from degradation by the phospholipid bilayer membrane) and ready availability (abundant in the blood), which make them relatively promising biomarkers in clinical practice 139 (Figure 2F).
With the assistance of bioinformatics, a group systematically analyzed the diagnostic value of CRC‐associated sEVs long RNAs, and revealed that the combination of KRTAP5‐4, MAGEA3, and BCAR4 exhibited promise in distinguishing colorectal adenoma and cancer from the healthy tissue, suggesting a possible strategy for detecting early‐stage CRC. 140 Another study provided additional evidence that sEVs‐lncRNAs (LNCV6_116109, LNCV6_98390, LNCV6_38772, LNCV_108266, LNCV6_84003, and LNCV6_98602) are upregulated in CRC, particularly in patients with stage I‐II tumors, and these molecules might also represent early diagnostic biomarkers for CRC. 3
Bjørnetrø and colleagues identified sEVs‐miRNAs originating from hypoxic tumor cells as circulating biomarkers to predict high‐risk locally advanced rectal cancer (LARC), of which miR‐181a‐5p, miR‐486‐5p, and miR‐30d‐5p were associated with LN metastasis, organ‐invasive primary tumors, and metastasis progression, respectively. 141 The lncRNA HOTTIP not only identifies patients with CRC but also potentially functions as a valuable surrogate biomarker for presurgical risk stratification, as HOTTIP expression significantly predicted the survival time after surgery. 142 A clinical trial (NCT03874559) aims to characterize the levels of sEVs biomarkers in patients with LARC undergoing neoadjuvant chemoradiation therapy to compare the rates of sEVs biomarker expression before, during, and after chemoradiation therapy, which may identify specific prognostic biomarkers.
Interestingly, some biomolecules possess both diagnostic and prognostic properties. Circulating sEVs‐miR‐150‐5p, miR‐99b‐5p, miR‐27a, and miR‐130a levels might serve as novel diagnostic and prognostic biomarkers for patients with CRC, as sEVs‐miR‐150‐5p and miR‐99b‐5p are downregulated, whereas miR‐27a and miR‐130a are upregulated. 143 , 144
Several clinical trials (NCT03874559; NCT04227886; NCT04394572; NCT04523389) aim to characterize sEVs‐contained biomarkers to help identify specific diagnostic and/or prognostic biomarkers for CRC. NCT03874559 and NCT04227886 both aim to characterize the levels of sEVs biomarkers in patients with LARC undergoing neoadjuvant chemoradiation therapy to compare the rates of sEVs biomarker expression before, during, and after chemoradiation therapy, which may identify specific prognostic biomarkers. To step further, NCT04394572 specifically screens sEVs protein markers for CRC diagnostic and/or prognostic, while NCT04523389 wants to figure out specific sEVs miRNAs for early CRC prognosis. Though clinical trials are still under progress, once completed, they could provide an insight into sEVs‐related biomarkers to help early diagnosis and prognosis for CRC patients.
8. DISCUSSION
CRC is one of the most common and prevalent malignant cancers, with high morbidity and mortality rates being reported worldwide. The patient's condition is exacerbated as a result of a delayed diagnosis, therapy resistance, and poor prognosis. Characterized by lipid bilayer membrane and carrying a number of biomolecules, sEVs are now publicly recognized to serve as messengers involved in intercellular communication in the TME and to participate in the progression of CRC. As is summarized above, we mainly discuss how sEVs‐related molecules action to recipient cells that contributes to CRC progression, metastasis, and other potential clinical applications. Studies aiming to elucidate the detailed mechanisms would help identify potential targets to block tumor progression.
Although numerous researchers have attempted to determine the mechanism by which sEVs modulate the pathogenesis of CRC, several challenges and obstacles in EVs field cannot be ignored. As proposed, there are various kinds of isolation methods for EVs isolation; however, due to limitations of isolation and characterization methods, it is still unrealistic to propose specific markers to classify each type of EVs. Besides, when function to recipient cells of EVs mentioned in research articles, we should be aware of that some non‐EVs components often coisolate with EVs, such as albumin, soluble immune active cytokines, or even platelets from plasma, which contaminates the EVs and might give wrong information of EVs function. Moreover, these contaminants may also influence the accuracy of sEVs‐based diagnosis. Therefore, when specific sEVs function is mentioned, it should be interpreted with caution. And it is important to choose and develop isolation techniques to isolate EVs with least soluble factor, which helps apply sEVs detection into clinical applications. 145
Furthermore, there is still a debate whether plasma or serum is a better source of sEVs for clinical applications. Though plasma sEVs are deemed to contain more sEVs biomarkers and better for diagnosis than serum sEVs, 146 plasma contains platelets that are able to release several types of membrane vesicles, 147 which will confound the measurement of circulating miRNA biomarkers in plasma, 148 , 149 and even more nonvesicle miRNAs. 150 Nevertheless, compared to plasma, sEVs originated miRNAs are more highly expressed in serum sEVs samples. Therefore, serum seems more preferable for cancer‐associated sEVs biomarker studies. 150
In clinical application, it is important to give a specific dose of certain medical substance. Though there are many methods to quantify EVs such as quantification of particle number by nanoparticle tracking analysis, or measurement of particulate components such as proteins, lipids, and nucleic acids are relative suitable proxy for EVs quantification, it is still imperfect and need further improvement. Some work has tried to give solutions to this issue, and they build their own quantification platforms based on antibodies, immunoassay, and imaging flow cytometry, which are able to selectively detect tumor‐derived EVs. 151 , 152 , 153 And in their work, compared to healthy donors, cancerous samples are able to release more EVs, and gene knockdown of organ‐tropic membrane proteins could decrease organ‐tropic EVs to specific organ. 154 This might indicate that increasing of EVs secretion promotes cancer progression. Besides the quantification of sEVs, how sEVs contents are regulated also spawn our attention. Sumoylated sEVs‐hnRNPA2B1, 155 ubiquitinated target proteins 156 seem to be vital for molecules to be sorted into sEVs, which might associate with cancer metastasis. Moreover, a stoichiometric analysis of sEVs‐miRNA raised the awareness that the majority of individual sEVs do not carry sufficient miRNAs, with less than one copy of miRNA per sEVs. 157 Therefore, the presence of heterogeneity of distinct subset of sEVs warrants the significance to establish specific markers of specific subset of sEVs, and further methods to isolate interested subsets of sEVs are needed.
Additionally, although some molecules were described to be latent biomarkers, more meaningful and specific biomolecules involved in CRC must be discovered to increase the accuracy of diagnosis and prognosis. In addition, more clinical trials are urgently needed to validate the application of sEVs as biomarkers for clinical screening and monitoring the patient's condition. The advances in applying sEVs in patients with CRC are still limited but have rapidly attracted attention, and sEVs are predicted to be successfully employed in clinical therapy and the diagnosis of early‐stage CRC in the future.
AUTHOR CONTRIBUTIONS
Dawei Li and Ping Wei conceived and critically reviewed the structure of manuscript. Xuefeng He drafted and revised the manuscript. Xinyang Zhong and Zijuan Hu collected literatures and completed tables. Senlin Zhao researched on the background of the study. All the authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
ACKNOWLEDGMENTS
The figures were created with BioRender.com and modified by Huaijun Chen (Department of Neurosurgery, The Second Affiliated Hospital of Zhejiang University School of Medicine), and we really appreciate the contribution.
He X, Zhong X, Hu Z, Zhao S, Wei P, Li D. An insight into small extracellular vesicles: Their role in colorectal cancer progression and potential clinical applications. Clin Transl Med. 2020;10:e249 10.1002/ctm2.249
Contributor Information
Ping Wei, Email: weiping@fudan.edu.cn.
Dawei Li, Email: li_dawei@fudan.edu.cn.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467‐1480. [DOI] [PubMed] [Google Scholar]
- 3. Hu D, Zhan Y, Zhu K, et al. Plasma exosomal long non‐coding RNAs serve as biomarkers for early detection of colorectal cancer. Cell Physiol Biochem. 2018;51(6):2704‐2715. [DOI] [PubMed] [Google Scholar]
- 4. van der Stok EP, Spaander MCW, Grunhagen DJ, Verhoef C, Kuipers EJ. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol. 2017;14(5):297‐315. [DOI] [PubMed] [Google Scholar]
- 5. Li Q, Wei P, Wu J, et al. The FOXC1/FBP1 signaling axis promotes colorectal cancer proliferation by enhancing the Warburg effect. Oncogene. 2018;38(4):483‐496. [DOI] [PubMed] [Google Scholar]
- 6. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364‐372. [DOI] [PubMed] [Google Scholar]
- 7. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harding C, Stahl P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun. 1983;113(2):650‐658. [DOI] [PubMed] [Google Scholar]
- 10. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967‐978. [DOI] [PubMed] [Google Scholar]
- 11. Guo W, Gao Y, Li N, et al. Exosomes: new players in cancer (Review). Oncol Rep. 2017;38(2):665‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toiyama Y, Okugawa Y, Fleshman J, Richard Boland C, Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta Rev Cancer. 2018;1870(2):274‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belov L, Matic KJ, Hallal S, Best OG, Mulligan SP, Christopherson RI. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J Extracell Vesicles. 2016;5:25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tschuschke M, Kocherova I, Bryja A, et al. Inclusion biogenesis, methods of isolation and clinical application of human cellular exosomes. J Clin Med. 2020;9(2):436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xie F, Zhou X, Fang M, et al. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv Sci (Weinh). 2019;6(24):1901779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278‐294. [DOI] [PubMed] [Google Scholar]
- 17. Lotvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Witwer KW, Soekmadji C, Hill AF, et al. Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. J Extracell Vesicles. 2017;6(1):1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu T, Zhang Q, Zhang J, et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89‐D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim DK, Kang B, Kim OY, et al. EVpedia: an integrated database of high‐throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013;2 10.3402/jev.v2i0.20384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue):D1241‐D1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106‐D112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145‐164. [DOI] [PubMed] [Google Scholar]
- 25. Nusse R, Clevers H. Wnt/beta‐catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985‐999. [DOI] [PubMed] [Google Scholar]
- 26. Farooqi AA, de la Roche M, Djamgoz MBA, Siddik ZH. Overview of the oncogenic signaling pathways in colorectal cancer: mechanistic insights. Semin Cancer Biol. 2019;58:65‐79. [DOI] [PubMed] [Google Scholar]
- 27. Kalra H, Gangoda L, Fonseka P, et al. Extracellular vesicles containing oncogenic mutant beta‐catenin activate Wnt signalling pathway in the recipient cells. J Extracell Vesicles. 2019;8(1):1690217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang Z, Yang M, Li Y, Yang F, Feng Y. Exosomes derived from hypoxic colorectal cancer cells transfer Wnt4 to normoxic cells to elicit a prometastatic phenotype. Int J Biol Sci. 2018;14(14):2094‐2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu JL, Wang W, Lan XL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial‐mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X, Chen X, Zeng K, et al. DNA‐methylation‐mediated silencing of miR‐486‐5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β‐catenin signal pathways. Cell Death Dis. 2018;9(10):1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morin PJ. Colorectal cancer: the APC‐lncRNA link. J Clin Invest. 2019;129(2):503‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang FW, Cao CH, Han K, et al. APC‐activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129(2):727‐743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y, Shen L, Li F, Yang J, Wan X, Ouyang M. microRNA‐16‐5p‐containing exosomes derived from bone marrow‐derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J Cell Physiol. 2019;234(11):21380‐21394. [DOI] [PubMed] [Google Scholar]
- 34. Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quinn JJ, Chang HY. Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47‐62. [DOI] [PubMed] [Google Scholar]
- 36. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao T, Liu X, He B, et al. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luan Y, Li X, Luan Y, et al. Circulating lncRNA UCA1 promotes malignancy of colorectal cancer via the miR‐143/MYO6 axis. Mol Ther Nucleic Acids. 2019;19:790‐803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR‐1182/hTERT signaling pathways. Clin Sci (Lond). 2019;133(24):2463‐2479. [DOI] [PubMed] [Google Scholar]
- 40. Han K, Wang FW, Cao CH, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA‐17. Mol Cancer. 2020;19(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212‐226. [DOI] [PubMed] [Google Scholar]
- 42. Senfter D, Holzner S, Kalipciyan M, et al. Loss of miR‐200 family in 5‐fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum Mol Genet. 2015;24(13):3689‐3698 [DOI] [PubMed] [Google Scholar]
- 43. Holzner S, Senfter D, Stadler S, et al. Colorectal cancer cell‐derived microRNA200 modulates the resistance of adjacent blood endothelial barriers in vitro. Oncol Rep. 2016;36(5):3065‐3071. [DOI] [PubMed] [Google Scholar]
- 44. Liu H, Liu Y, Sun P, et al. Colorectal cancer‐derived exosomal miR‐106b‐3p promotes metastasis by down‐regulating DLC‐1 expression. Clin Sci (Lond). 2020;134(4):419‐434. [DOI] [PubMed] [Google Scholar]
- 45. Wang D, Wang X, Si M, et al. Exosome‐encapsulated miRNAs contribute to CXCL12/CXCR4‐induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36‐52. [DOI] [PubMed] [Google Scholar]
- 46. Feng W, Gong H, Wang Y, et al. circIFT80 functions as a ceRNA of miR‐1236‐3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375‐387. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47. Shao Y, Chen T, Zheng X, et al. Colorectal cancer‐derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39(11):1368‐1379. [DOI] [PubMed] [Google Scholar]
- 48. Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 is critical for premetastatic niche formation and metastasis in colorectal cancer. Cancer Res. 2017;77(13):3655‐3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoshino A, Costa‐Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sceneay J, Smyth MJ, Moller A. The pre‐metastatic niche: finding common ground. Cancer Metastasis Rev. 2013;32(3‐4):449‐464. [DOI] [PubMed] [Google Scholar]
- 51. Guo Y, Ji X, Liu J, et al. Effects of exosomes on pre‐metastatic niche formation in tumors. Mol Cancer. 2019;18(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu Y, Cao X. Characteristics and significance of the pre‐metastatic niche. Cancer Cell. 2016;30(5):668‐681. [DOI] [PubMed] [Google Scholar]
- 53. Han W, Duan Z. Roles of exosomes in liver metastases: novel diagnosis and treatment choices. J Cell Physiol. 2019;234(12):21588‐21600. [DOI] [PubMed] [Google Scholar]
- 54. Balacescu O, Sur D, Cainap C, et al. The impact of miRNA in colorectal cancer progression and its liver metastases. Int J Mol Sci. 2018;19(12):3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garcia‐Roman J, Zentella‐Dehesa A. Vascular permeability changes involved in tumor metastasis. Cancer Lett. 2013;335(2):259‐269. [DOI] [PubMed] [Google Scholar]
- 56. Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med. 2012;18(6):883‐891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zeng Z, Li Y, Pan Y, et al. Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schillaci O, Fontana S, Monteleone F, et al. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non‐metastatic cells and increase endothelial permeability: their emerging role in tumor heterogeneity. Sci Rep. 2017;7(1):4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu HY, Yu CH, Zhang HH, et al. Exosomal miR‐1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470‐477. [DOI] [PubMed] [Google Scholar]
- 60. Karnezis T, Shayan R, Caesar C, et al. VEGF‐D promotes tumor metastasis by regulating prostaglandins produced by the collecting lymphatic endothelium. Cancer Cell. 2012;21(2):181‐195. [DOI] [PubMed] [Google Scholar]
- 61. Hirakawa S, Brown LF, Kodama S, Paavonen K, Alitalo K, Detmar M. VEGF‐C‐induced lymphangiogenesis in sentinel lymph nodes promotes tumor metastasis to distant sites. Blood. 2007;109(3):1010‐1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hirakawa S, Kodama S, Kunstfeld R, Kajiya K, Brown LF, Detmar M. VEGF‐A induces tumor and sentinel lymph node lymphangiogenesis and promotes lymphatic metastasis. J Exp Med. 2005;201(7):1089‐1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wakisaka N, Hasegawa Y, Yoshimoto S, et al. Primary tumor‐secreted lymphangiogenic factors induce pre‐metastatic lymphvascular niche formation at sentinel lymph nodes in oral squamous cell carcinoma. PLoS One. 2015;10(12):e0144056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun B, Zhou Y, Fang Y, Li Z, Gu X, Xiang J. Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer. 2019;145(6):1648‐1659. [DOI] [PubMed] [Google Scholar]
- 65. Cao X. Self‐regulation and cross‐regulation of pattern‐recognition receptor signalling in health and disease. Nat Rev Immunol. 2016;16(1):35‐50. [DOI] [PubMed] [Google Scholar]
- 66. Liu Y, Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med (Berl). 2016;94(5):509‐522. [DOI] [PubMed] [Google Scholar]
- 67. Cooks T, Pateras IS, Jenkins LM, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR‐1246. Nat Commun. 2018;9(1):771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Costa‐Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre‐metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816‐826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen C, Yao X, Xu Y, et al. Dahuang Zhechong Pill suppresses colorectal cancer liver metastasis via ameliorating exosomal CCL2 primed pre‐metastatic niche. J Ethnopharmacol. 2019;238:111878. [DOI] [PubMed] [Google Scholar]
- 70. Wortzel I, Dror S, Kenific CM, Lyden D. Exosome‐mediated metastasis: communication from a distance. Dev Cell. 2019;49(3):347‐360. [DOI] [PubMed] [Google Scholar]
- 71. Shu S, Yang Y, Allen CL, et al. Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre‐metastatic microenvironment. Sci Rep. 2018;8(1):12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fong MY, Zhou W, Liu L, et al. Breast‐cancer‐secreted miR‐122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17(2):183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang L, Zhang S, Yao J, et al. Microenvironment‐induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527(7576):100‐104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ji Q, Zhou L, Sui H, et al. Primary tumors release ITGBL1‐rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast‐niche formation. Nat Commun. 2020;11(1):1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. 2017;189(3):259‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor‐derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor‐reactive activated CD8+ T lymphocytes. J Immunol. 2009;183(6):3720‐3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor‐released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67(7):2912‐2915. [DOI] [PubMed] [Google Scholar]
- 78. Poggio M, Hu T, Pai CC, et al. Suppression of exosomal PD‐L1 induces systemic anti‐tumor immunity and memory. Cell. 2019;177(2):414‐427.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet. 2019;56(1):29‐31. [DOI] [PubMed] [Google Scholar]
- 80. Cheng WC, Liao TT, Lin CC, et al. RAB27B‐activated secretion of stem‐like tumor exosomes delivers the biomarker microRNA‐146a‐5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer. 2019;145(8):2209‐2224. [DOI] [PubMed] [Google Scholar]
- 81. Yamada N, Kuranaga Y, Kumazaki M, Shinohara H, Taniguchi K, Akao Y. Colorectal cancer cell‐derived extracellular vesicles induce phenotypic alteration of T cells into tumor‐growth supporting cells with transforming growth factor‐β1‐mediated suppression. Oncotarget. 2016;7(19):27033‐27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Huber V, Fais S, Iero M, et al. Human colorectal cancer cells induce T‐cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology. 2005;128(7):1796‐1804. [DOI] [PubMed] [Google Scholar]
- 83. Zhou D, Huang C, Lin Z, et al. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26(2):192‐197. [DOI] [PubMed] [Google Scholar]
- 84. Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013;34(5):216‐223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Baig MS, Roy A, Rajpoot S, et al. Tumor‐derived exosomes in the regulation of macrophage polarization. Inflamm Res. 2020;69(5):435‐451. [DOI] [PubMed] [Google Scholar]
- 86. Liang ZX, Liu HS, Wang FW, et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes‐mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lan J, Sun L, Xu F, et al. M2 macrophage‐derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79(1):146‐158. [DOI] [PubMed] [Google Scholar]
- 88. Michielsen AJ, Hogan AE, Marry J, et al. Tumour tissue microenvironment can inhibit dendritic cell maturation in colorectal cancer. PLoS One. 2011;6(11):e27944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Valenti R, Huber V, Filipazzi P, et al. Human tumor‐released microvesicles promote the differentiation of myeloid cells with transforming growth factor‐beta‐mediated suppressive activity on T lymphocytes. Cancer Res. 2006;66(18):9290‐9298. [DOI] [PubMed] [Google Scholar]
- 90. Chen W, Wang J, Shao C, et al. Efficient induction of antitumor T cell immunity by exosomes derived from heat‐shocked lymphoma cells. Eur J Immunol. 2006;36(6):1598‐1607. [DOI] [PubMed] [Google Scholar]
- 91. Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16(7):431‐446. [DOI] [PubMed] [Google Scholar]
- 92. Shaul ME, Fridlender ZG. Tumour‐associated neutrophils in patients with cancer. Nat Rev Clin Oncol. 2019;16(10):601‐620. [DOI] [PubMed] [Google Scholar]
- 93. Nemeth T, Sperandio M, Mocsai A. Neutrophils as emerging therapeutic targets. Nat Rev Drug Discov. 2020;19(4):253‐752. [DOI] [PubMed] [Google Scholar]
- 94. Hwang WL, Lan HY, Cheng WC, Huang SC, Yang MH. Tumor stem‐like cell‐derived exosomal RNAs prime neutrophils for facilitating tumorigenesis of colon cancer. J Hematol Oncol. 2019;12(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18(2):134‐147. [DOI] [PubMed] [Google Scholar]
- 96. Tohme S, Yazdani HO, Al‐Khafaji AB, et al. Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Res. 2016;76(6):1367‐1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Najmeh S, Cools‐Lartigue J, Rayes RF, et al. Neutrophil extracellular traps sequester circulating tumor cells via beta1‐integrin mediated interactions. Int J Cancer. 2017;140(10):2321‐2330. [DOI] [PubMed] [Google Scholar]
- 98. Hisada Y, Mackman N. Cancer‐associated pathways and biomarkers of venous thrombosis. Blood. 2017;130(13):1499‐1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gastpar R, Gehrmann M, Bausero MA, et al. Heat shock protein 70 surface‐positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65(12):5238‐5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Guo D, Chen Y, Wang S, et al. Exosomes from heat‐stressed tumour cells inhibit tumour growth by converting regulatory T cells to Th17 cells via IL‐6. Immunology. 2018;154(1):132‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wood‐1994‐Chemotherapy‐for‐colorectal‐cancer. https://pubmed.ncbi.nlm.nih.gov/10778957/
- 102. Showalter SL, Showalter TN, Witkiewicz A, et al. Evaluating the drug‐target relationship between thymidylate synthase expression and tumor response to 5‐fluorouracil. Is it time to move forward. Cancer Biol Ther. 2008;7(7):986‐994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yaffee P, Osipov A, Tan C, Tuli R, Hendifar A. Review of systemic therapies for locally advanced and metastatic rectal cancer. J Gastrointest Oncol. 2015;6(2):185‐200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Li XL, Zhou J, Chen ZR, Chng WJ. P53 mutations in colorectal cancer ‐ molecular pathogenesis and pharmacological reactivation. World J Gastroenterol. 2015;21(1):84‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mannavola F, Salerno T, Passarelli A, Tucci M, Interno V, Silvestris F. Revisiting the role of exosomes in colorectal cancer. Where Are We Now? Front Oncol. 2019;9:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Dylla SJ, Beviglia L, Park IK, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3(6):e2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Van Cutsem E, Cervantes A, Nordlinger B, Arnold D, Group EGW. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Ann Oncol. 2014;25(Suppl 3):iii1‐iii9. [DOI] [PubMed] [Google Scholar]
- 108. Sforza V, Martinelli E, Ciardiello F, et al. Mechanisms of resistance to anti‐epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J Gastroenterol. 2016;22(28):6345‐6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bardelli A, Corso S, Bertotti A, et al. Amplification of the MET receptor drives resistance to anti‐EGFR therapies in colorectal cancer. Cancer Discov. 2013;3(6):658‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang S, Zhang Y, Qu J, et al. Exosomes promote cetuximab resistance via the PTEN/Akt pathway in colon cancer cells. Braz J Med Biol Res. 2017;51(1):e6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yang YN, Zhang R, Du JW, et al. Predictive role of UCA1‐containing exosomes in cetuximab‐resistant colorectal cancer. Cancer Cell Int. 2018;18:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA‐RNA interactions. Mol Ther Nucleic Acids. 2018;12:229‐241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liu T, Zhang X, Du L, et al. Exosome‐transmitted miR‐128‐3p increase chemosensitivity of oxaliplatin‐resistant colorectal cancer. Mol Cancer. 2019;18(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114. Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin‐like growth factor‐I receptor inhibition. Cancer Res. 2009;69(5):1951‐1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang Y, Yin K, Tian J, et al. Granulocytic myeloid‐derived suppressor cells promote the stemness of colorectal cancer cells through exosomal S100A9. Adv Sci (Weinh). 2019;6(18):1901278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhou J, Chng WJ. Identification and targeting leukemia stem cells: the path to the cure for acute myeloid leukemia. World J Stem Cells. 2014;6(4):473‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11(1):97‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hu YB, Yan C, Mu L, et al. Exosomal Wnt‐induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. 2019;38(11):1951‐1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hu Y, Yan C, Mu L, et al. Fibroblast‐derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One. 2015;10(5):e0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li H, Li F. Exosomes from BM‐MSCs increase the population of CSCs via transfer of miR‐142‐3p. Br J Cancer. 2018;119(6):744‐755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Shen WW, Zeng Z, Zhu WX, Fu GH. MiR‐142‐3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl). 2013;91(8):989‐1000. [DOI] [PubMed] [Google Scholar]
- 122. Wang X, Zhang H, Yang H, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Soldevilla B, Rodriguez M, San Millan C, et al. Tumor‐derived exosomes are enriched in DeltaNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum Mol Genet. 2014;23(2):467‐478. [DOI] [PubMed] [Google Scholar]
- 124. Sauer R, Becker H, Hohenberger W, et al. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351(17):1731‐1740. [DOI] [PubMed] [Google Scholar]
- 125. Wolmark N, Wieand HS, Hyams DM, et al. Randomized trial of postoperative adjuvant chemotherapy with or without radiotherapy for carcinoma of the rectum: National Surgical Adjuvant Breast and Bowel Project Protocol R‐02. J Natl Cancer Inst. 2000;92(5):388‐396. [DOI] [PubMed] [Google Scholar]
- 126. Roh MS, Colangelo LH, O'Connell MJ, et al. Preoperative multimodality therapy improves disease‐free survival in patients with carcinoma of the rectum: NSABP R‐03. J Clin Oncol. 2009;27(31):5124‐5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467‐1480. [DOI] [PubMed] [Google Scholar]
- 128. Krishnan S, Janjan NA, Skibber JM, et al. Phase II study of capecitabine (Xeloda) and concomitant boost radiotherapy in patients with locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2006;66(3):762‐771. [DOI] [PubMed] [Google Scholar]
- 129. Liu L, Zhang Z, Zhou L, et al. Cancer associated fibroblasts‐derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp Cell Res. 2020;391(2):111956. [DOI] [PubMed] [Google Scholar]
- 130. Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6(4):287‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Syn NL, Wang L, Chow EK, Lim CT, Goh BC. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35(7):665‐676. [DOI] [PubMed] [Google Scholar]
- 132. Dai S, Wei D, Wu Z, et al. Phase I clinical trial of autologous ascites‐derived exosomes combined with GM‐CSF for colorectal cancer. Mol Ther. 2008;16(4):782‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Li Y, Gao Y, Gong C, et al. A33 antibody‐functionalized exosomes for targeted delivery of doxorubicin against colorectal cancer. Nanomedicine. 2018;14(7):1973‐1985. [DOI] [PubMed] [Google Scholar]
- 134. Bullock MD, Silva AM, Kanlikilicer‐Unaldi P, et al. Exosomal non‐coding RNAs: diagnostic, prognostic and therapeutic applications in cancer. Noncoding RNA. 2015;1(1):53‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Liang G, Zhu Y, Ali DJ, et al. Engineered exosomes for targeted co‐delivery of miR‐21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Doubeni CA, Gabler NB, Wheeler CM, et al. Timely follow‐up of positive cancer screening results: a systematic review and recommendations from the PROSPR Consortium. CA Cancer J Clin. 2018;68(3):199‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Issa IA, Noureddine M. Colorectal cancer screening: an updated review of the available options. World J Gastroenterol. 2017;23(28):5086‐5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy‐based biomarkers of treatment response and resistance. Cancer Cell. 2020;37(4):485‐495. [DOI] [PubMed] [Google Scholar]
- 139. Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Dong L, Lin W, Qi P, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1158‐1166. [DOI] [PubMed] [Google Scholar]
- 141. Bjørnetrø T, Redalen KR, Meltzer S, et al. An experimental strategy unveiling exosomal microRNAs 486‐5p, 181a‐5p and 30d‐5p from hypoxic tumour cells as circulating indicators of high‐risk rectal cancer. J Extracell Vesicles. 2019;8(1):1567219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Oehme F, Krahl S, Gyorffy B, et al. Low level of exosomal long non‐coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019;16(10):1339‐1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhao YJ, Song X, Niu L, Tang Y, Song X, Xie L. Circulating exosomal miR‐150‐5p and miR‐99b‐5p as diagnostic biomarkers for colorectal cancer. Front Oncol. 2019;9:1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu X, Pan B, Sun L, et al. Circulating exosomal miR‐27a and miR‐130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(7):746‐754. [DOI] [PubMed] [Google Scholar]
- 145. Shu S, Yang Y, Allen CL, et al. Purity and yield of melanoma exosomes are dependent on isolation method. J Extracell Vesicles. 2020;9(1):1692401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Cao F, Gao Y, Chu Q, et al. Proteomics comparison of exosomes from serum and plasma between ultracentrifugation and polymer‐based precipitation kit methods. Electrophoresis. 2019;40(23‐24):3092‐3098. [DOI] [PubMed] [Google Scholar]
- 147. Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha‐granules. Blood. 1999;94(11):3791‐3799. [PubMed] [Google Scholar]
- 148. Mitchell AJ, Gray WD, Hayek SS, et al. Platelets confound the measurement of extracellular miRNA in archived plasma. Sci Rep. 2016;6:32651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Cheng HH, Yi HS, Kim Y, et al. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 2013;8(6):e64795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Chiam K, Mayne GC, Wang T, et al. Serum outperforms plasma in small extracellular vesicle microRNA biomarker studies of adenocarcinoma of the esophagus. World J Gastroenterol. 2020;26(20):2570‐2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wei P, Wu F, Kang B, et al. Plasma extracellular vesicles detected by single molecule array technology as a liquid biopsy for colorectal cancer. J Extracell Vesicles. 2020;9(1):1809765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Kang YT, Hadlock T, Jolly S, Nagrath S. Extracellular vesicles on demand (EVOD) chip for screening and quantification of cancer‐associated extracellular vesicles. Biosens Bioelectron. 2020;168:112535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ricklefs FL, Maire CL, Reimer R, et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019;8(1):1588555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wu AY, Sung YC, Chen YJ, et al. Multiresolution imaging using bioluminescence resonance energy transfer identifies distinct biodistribution profiles of extracellular vesicles and exomeres with redirected tropism. Adv Sci (Weinh). 2020;7(19):2001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3'‐untranslated regions. Biol Direct. 2013;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Smith VL, Jackson L, Schorey JS. Ubiquitination as a mechanism to transport soluble mycobacterial and eukaryotic proteins to exosomes. J Immunol. 2015;195(6):2722‐2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888‐14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Akers JC, Gonda D, Kim R, Carter BS, Chen CC. Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus‐like vesicles, and apoptotic bodies. J Neurooncol. 2013;113(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Bhome R, Goh RW, Bullock MD, et al. Exosomal microRNAs derived from colorectal cancer‐associated fibroblasts: role in driving cancer progression. Aging (Albany NY). 2017;9(12):2666‐2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Teng Y, Ren Y, Hu X, et al. MVP‐mediated exosomal sorting of miR‐193a promotes colon cancer progression. Nat Commun. 2017;8:14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Takano Y, Masuda T, Iinuma H, et al. Circulating exosomal microRNA‐203 is associated with metastasis possibly via inducing tumor‐associated macrophages in colorectal cancer. Oncotarget. 2017;8(45):78598‐78613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Min L, Chen L, Liu S, et al. Loss of circulating exosomal miR‐92b is a novel biomarker of colorectal cancer at early stage. Int J Med Sci. 2019;16(9):1231‐1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Sun L, Liu X, Pan B, et al. Serum exosomal miR‐122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer. 2020;11(3):630‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Dai W, Zhou J, Wang H, Zhang M, Yang X, Song W. miR‐424‐5p promotes the proliferation and metastasis of colorectal cancer by directly targeting SCN4B. Pathol Res Pract. 2020;216(1):152731. [DOI] [PubMed] [Google Scholar]
- 165. Zhao Y, Du T, Du L, et al. Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer. Cell Death Dis. 2019;10(8):568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa‐circ‐0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Xie Y, Li J, Li P, et al. RNA‐seq profiling of serum exosomal circular RNAs reveals Circ‐PNN as a potential biomarker for human colorectal cancer. Front Oncol. 2020;10:982. [DOI] [PMC free article] [PubMed] [Google Scholar]


