Abstract
Liquid–liquid phase separation has drawn attention as many neurodegeneration or cancer-associated proteins are able to form liquid membraneless compartments (condensates) by liquid–liquid phase separation. Furthermore, there is rapidly growing evidence that disease-associated mutation or post-translational modification of these proteins causes aberrant location, composition or physical properties of the condensates. It is ambiguous whether aberrant condensates are always causative in disease mechanisms, however they are likely promising potential targets for therapeutics. The conceptual framework of liquid–liquid phase separation provides opportunities for novel therapeutic approaches. This review summarises how the extensive recent advances in understanding control of nucleation, growth and composition of condensates by protein post-translational modification has revealed many possibilities for intervention by conventional small molecule enzyme inhibitors. This includes the first proof-of-concept examples. However, understanding membraneless organelle formation as a physical chemistry process also highlights possible physicochemical mechanisms of intervention. There is huge demand for innovation in drug development, especially for challenging diseases of old age including neurodegeneration and cancer. The conceptual framework of liquid–liquid phase separation provides a new paradigm for thinking about modulating protein function and is very different from enzyme lock-and-key or structured binding site concepts and presents new opportunities for innovation.
Keywords: cancer, condensates, drug discovery and design, liquid–liquid phase separation, neurodegeneration, therapeutics
Introduction
Liquid–liquid phase separation (LLPS) has emerged as the mechanism underlying the formation of many membraneless organelles (MLOs). This ranges from nuclear domains including nucleoli [1,2] and heterochromatin [3,4], to the formation of stress granules in the cytoplasm [5–7] and formation of signalling and adhesion bodies at the cell membrane [8,9]. Compartments formed by LLPS are termed biomolecular condensates [10,11]. They undergo constant internal rearrangement (reflecting their liquid state), they exist in equilibrium with the surrounding environment (a dilute solvent phase), their formation is reversible and they are distinct from aggregates and protein crystals or polymers [12].
Interest in LLPS has exploded, in part, through the involvement of many condensate-forming proteins in disease, most prominently neurodegenerative disease and cancer [13–15]. Condensates are typically formed by two classes of protein: Firstly, proteins with extensive intrinsically disordered regions (IDRs) termed intrinsically disordered proteins (IDPs) [10,12,16] and secondly, proteins with multiple copies of interaction domains (MCIDPs) [9,17]. High valency transient interactions between either the folded interaction domains of MCIDPs or specialised unstructured regions of particular amino acid composition (called ‘stickers’) in IDPs result in LLPS [10,18]. These proteins are termed the condensate scaffold and are required to form the condensate, while recruitment of client proteins (which cannot themselves undergo LLPS) generates the final condensate composition [19]. However, despite their clear links with key non-communicable diseases, IDPs and MCIDPs are not conventional drug targets.
The conventional view of the druggable genome is the set of protein-coding genes whose product is typical of those that can be modulated by an orally administered small molecule [20], estimated at 10–15% [20–23] of the genome (3000–5000 proteins). This does not typically include IDPs and MCIDPs. A similar proportion of the genome are disease-associated (3961 have entries in OMIM[24]). However, druggability does not correlate well with having a role in disease [20,25]—disease associated IDPs are one example. Our rapidly expanding knowledge of LLPS regulation highlights many druggable proteins which may be valuable targets for conventional drugs. Would it be possible to apply our new understanding of LLPS to make the scaffolds, conventionally undruggable proteins, into new drug targets?
Why target liquid–liquid phase separation?
A phenotype common in neurodegenerative diseases is aggregation of nuclear or cytoplasmic proteins with IDRs—this includes TAU in Alzheimer's disease and TDP-43 and FUS in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Mutations in these proteins are also associated with more severe pathology [13,26,27]. Neurodegeneration repeat expansion disorders also generate abnormal IDPs including poly-Q in Huntington's disease and poly-FR and poly-PR in ALS/FTD [28–30]. There is evidence for LLPS of all of these proteins, for example TAU canonically stabilises neuron microtubules but can also undergo LLPS [31–33] and concentrate tubulin in the condensate leading to microtubule nucleation [34]. Condensates may also have aberrant properties (different material properties, composition or location of formation) or may more rapidly transition from to an irreversible aggregate when the proteins have disease-associated mutations [7,32,35–37]. Unfortunately, the mechanism of pathogenesis in these diseases is not always clear, for example in ALS only a small proportion of cases involve TDP-43/FUS mutations and, while the mutations are likely causative, it is not completely clear if or how aberrant LLPS is involved [38]. For example, phase-separation deficient TFP43 is retains its splicing activity [39]. Nonetheless, the clear association of LLPS with disease makes these proteins targets of interest.
There is growing evidence for LLPS roles in cancer. Several IDPs associated with neurodegeneration are also oncogenes: EWSR1 (whose fusion with the transcription factor FLI1 causes Ewing's sarcoma) and FUS (whose fusion with the transcription factor CHOP causes myxoid liposarcoma). It is likely, but not proven, that aberrant LLPS of the IDR/transcription factor fusion in the nucleus leads to the activation of tumour-specific enhancers [40]. Another cancer-LLPS link is the tumour suppressor SPOP which interacts with proto-oncogenic substrates and undergoes LLPS, leading to substrate ubiquitination and degradation. Cancer-causing mutations in SPOP prevent LLPS [41]. Molecular detail of these mechanisms are still emerging, but sit as part of wider evidence for likely LLPS-cancer links. Perturbation of any of the membraneless nuclear compartments, whether large (the nucleolus or heterochromatin) or small (transcription factories, DNA damage foci and, in particular, superenhancers [42]) could alter gene expression and contribute to cancer [43–46]. More generally, mechanisms involving LLPS in cell signalling [9,17] can also underlie signalling which, when defective, may contribute to oncogenesis [13]. However overall, the relative importance of defective LLPS as a mechanism in oncogenesis and the diversity of LLPS regulation in different cancers remains to be seen.
Relevance of LLPS is also not limited to non-communicable diseases. Upon viral infection, viroplasm/viral factories often form in the cytoplasm. These MLOs have the liquid properties of condensates for vesicular stomatitis virus and rabies virus [47,48]. Bacteria and fungi can undergo a phase transition of the entire cytoplasm, thought to be protective when stressed or dormant [49,50], and formation of the bacterial nucleoid may be by LLPS [51,52]. Finally, eukaryotic parasites have their own set of vital MLOs with diverse functions which may be formed by LLPS, from the specialised RNA polymerase I transcription factory required for antigenic variation in trypanosomes [53] to the cytoplasmic messenger ribonucleoprotein particles required for expression repression in the female gametes of malaria parasites [54]. These may also be novel targets for intervention.
Unfortunately, IDRs linked with LLPS are a classic example of a canonically undruggable protein domain [55–61], while the protein–protein interaction domains of MCIDs (for example SUMO) are challenging targets [62,63]. IDRs are common in human proteins, half of all human proteins are 20% IDR and 10% of human proteins are more than 50% IDR [64]. Targeting enzymes which control LLPS is therefore the most plausible approach. Furthermore, disease-associated LLPS is a powerful phenotypic readout for high throughput screening [65,66]. However, direct targeting of IDPs/MCIDPs with drugs would be extremely valuable.
Targeting liquid–liquid phase separations
Targeting LLPS certainly poses challenges, however the conceptual framework of LLPS provides concrete examples and predictions for drug development. This includes both (1) ‘conventional’ drugs which target globular proteins involved in LLPS regulation by signalling or protein post-translational modifications (PTMs) and (2) ‘unconventional’ drugs which directly target the interaction domains leading to LLPS or the physical chemistry of the LLPS system.
Despite many major recent advances, it is important to note that LLPS provides a reductionist model for MLO formation. Cells deviate from physical chemistry models as they are not in equilibrium and scaffolds are often subject to PTMs [32,67–70] and energy-dependent rearrangement by helicases/chaperones [71]. Condensate scaffold are also not uniform polymers and condensates can also contain a huge number of different proteins, unlike typical physical chemistry models. The precise nature of some MLOs is also a subject of debate (whether they are liquid, gel or glass-like) and whether this reflects different phases or a continuum of viscosity.
The conventional: enzymatic inhibitors
Scaffold proteins are subject to PTMs which regulate their LLPS [72,73]. Chemical modification of the scaffold changes its physicochemical properties—if this alters it such that LLPS no longer occurs under the current cellular conditions then the condensate will dissolve or vice versa. As condensates are an enrichment of scaffold in equilibrium with the surroundings the PTM enzyme could be positioned elsewhere in the cell [74]. Many specific examples of PTM modulating LLPS are known [72,73]: Serine phosphorylation and arginine methylation of FUS both reduce LLPS [67–69] and phosphomimetic mutation of TDP-43 reduces LLPS [70]. In contrast, phosphorylation of TAU [32] and FMRP and CAPRIN1 [75] promotes LLPS and poly-SUMOylation promotes LLPS in SUMO/SIM condensate formation [19]. Aberrant PTMs are associated with disease, making the responsible kinases, phosphatases, methyltransferases, demethylases, SUMO ligases, etc. good therapeutic targets [13,72] (Figure 1A). However, identification of the correct PTM enzyme targets and finding specific small molecule activators and inhibitors will be challenging—diseases, such as ALS, can involve perturbation of many PTM enzymes [76,77]. It is also not clear that PTM interventions will be efficacious until they can be tested in disease models. For example TDP-43 is hyperphosphorylated in aggregates in ALS patients [78,79] despite phosphorylation being implicated in reducing TDP-43 LLPS [70]—evidently phosphorylation has failed to prevent aggregation in late pathology.
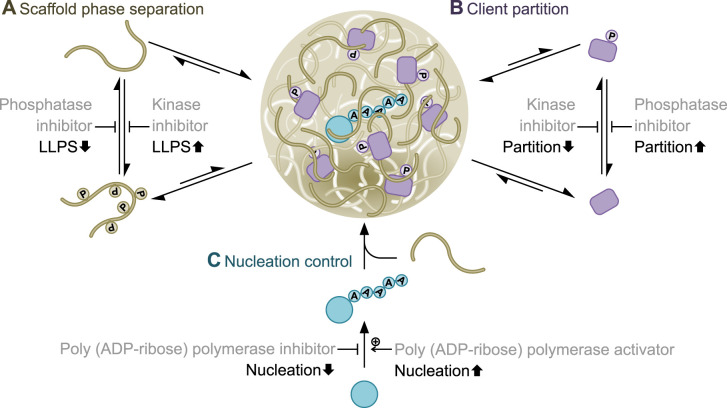
Figure 1. Possible protein liquid–liquid phase separation intervention via post-translational modifications.
Cartoon representation of a hypothetical LLPS system, modelled loosely on FUS and TDP-43 [7,67–69,81,89–91,100]. There are several potential points of intervention. (A) Phosphorylation (P) of the scaffold (coloured tan) reduces phase separation, therefore a kinase inhibitor would reduce LLPS and a phosphatase inhibitor promote LLPS. (B) Phosphorylation of a client (coloured purple) promotes partition to the condensate, therefore a kinase inhibitor would reduce client partition to the condensate and a phosphatase inhibitor would do the inverse. (C) Polyadenylation (A) of a key regulatory protein (coloured cyan) nucleates this condensate, therefore a poly(A) polymerase would promote nucleation while an inhibitor would do the inverse.
Client proteins can partition into condensates and the degree to which they do so depends on the nature of their interactions with the scaffolds and is quantified with a partition coefficient [9,19,80]. PTMs of either the scaffold or client can change the partition coefficient, for example changes to client SUMOylation altering partition to SUMO/SIM condensates [19] and changes to scaffold phosphorylation altering partition of RNA polymerase II to FUS, EWSR1 or TAF15 hydrogels [81]. As control of condensate composition is linked with condensate biochemical function, achieving the correct composition is important [82]. It is therefore also likely there are disease-related PTM enzymes which alter client partition into condensates which would also be conventional targets for small molecule drugs (Figure 1B). However, as above, identifying the relevant PTM enzymes and identifying specific small molecule inhibitors will be challenging.
The kinetics of condensate nucleation are unfavourable, meaning that under conditions where LLPS could occur it may not [12,73]. Mechanisms of nucleation is a complex topic, with only recent advances allowing direct analysis of nucleation [83,84]. Cellular structures implicated in nucleation include membranes, cytoskeleton structures, nucleic acid structures, and specific proteins [71,73,74,80,85–88] but perhaps the best example of a specific molecular mechanism is polyADP ribose (PAR) nucleating TDP-43 and FUS condensates [7,89–91]. PAR polymerases are a conventional target for small molecules and inhibitors reduce TDP-43-associated pathology in in vitro neurons, a very promising result demonstrating control of nucleation as a therapeutic target [90]. Another specific molecular example is Nephrin phosphorylation leading to the Nephrin signalling MLO [92]. Together, this points to PTMs often controlling nucleation, and the enzymes responsible would be conventional targets for small molecule drugs [90,91] (Figure 1C). However, nuanced intervention will require understanding of both when and where condensates should form for their normal function.
Finally, whether LLPS occurs is dependent on the concentration of the scaffold. Cells have many ways of regulating protein or nucleic acid level in different compartments, from simply how much has been synthesised, to organelle import/export. This is a more multi-faceted topic, but there are clear further possibilities for targeting by conventional small molecule drugs.
Together, these opportunities from conventional drug targets highlight the value of LLPS as a phenotype for screening existing drug libraries. This is subject to the many advantages and various challenges of efficacy and specificity, perhaps best understood for kinase drugs [93], and any hits would be amenable to conventional analyses of mode of action, analyses of specificity and efficacy, optimisation of leads by medicinal chemistry paradigms—overall potentially highly productive.
The unconventional: physicochemical mechanisms
The possibility of directly targeting LLPS has gathered great interest [94], particularly through our pre-print describing small molecules which modulate LLPS of ALS-associated stress granule proteins [66]. Perhaps because of a tendency to view ligandability as a strict pre-requisite for druggability [95–98], efforts to target canonically undruggable proteins tend to focus on finding binding sites, whether they are in non-enzymatic structured domains [62], cryptic binding sites [99] or transient binding sites in IDPs [57,61]. However, IDPs and MCIDPs in a condensate are suspected to have large scale conformational unwinding which enhances the capacity to undergo high valency interactions required for LLPS [9,19,100,101]. Recent evidence points to a balance of sticker (sequence sections which tend to cross-interact) and spacer (sections which do not) leading to LLPS without tertiary structures forming [100,102–105], although some studies indicate transient tertiary structures do form [106]. Is it possible to have drug interaction with an IDP without requiring a binding pocket? LLPS suggests it is possible through a physicochemical mechanism of action and this novel approach opens up new concepts for traditionally undruggable cellular targets.
The effects of the aliphatic alcohol 1,6-hexanediol are perhaps the best evidence that physicochemical disruption of LLPS is possible. 1,6-hexanediol was originally used to analyse FG rich IDRs in the nuclear pore complex [107–109] and was later found to disrupt many MLOs [45,110,111]. However, high concentrations (0.1–1%, high mM) are required and it has many aberrant effects [112]. 1,6-hexanediol, like many compounds including alcohols and dimethylsulfoxide (DMSO), alters hydrogen bonding and therefore alters hydrophobic interactions which, in turn, alters LLPS [107–109]. General solvent effects like these are too broad to be of use as a therapeutic. However it is notable that cells maintain high adenosine triphosphate (ATP) concentrations and, like 1,6-hexanediol, ATP reduces stress granule protein LLPS in vitro [113] and in cells [114].
There are several other more therapeutically plausible approaches to directly modulate LLPS. First is through polyphasic linkage—the interplay between protein phase and protein–ligand interaction. If protein–ligand binding is preferred when the protein is in a particular phase (for example, preferring the solution over the solid phase) then more ligand binding shifts the system equilibrium to prefer the corresponding protein phase, or vice versa. Examples of polyphasic linkage are known for several solid to solution phases. The classic example is haemoglobin crystallisation dependent on the oxygen ligand concentration [115–117]. In neurodegenerative disease, huntingtin aggregation is dependent on the profilin ligand concentration, via a soluble oligomer intermediate [118]. These concepts should also apply to liquid phases, indeed LLPS of the ALS-associated protein UBQLN2 shows polyphasic linkage with its ligand ubiquitin [119]. Therefore small molecules which preferentially bind a scaffold when either soluble or when in a liquid phase will modulate LLPS and may have therapeutic potential.
Second is through partition of molecules into a condensate altering the condensate properties. This is well characterised for partition of client proteins: The partitioned protein itself confers new properties to the condensate while simultaneously displacing other proteins which partitioned to the condensate through similar client-scaffold interactions [19]. Furthermore, high concentrations of a client protein can destabilise a condensate, likely through entropic effects [10,19,82,120]. This is arguably a form of polyphasic linkage as client binding results in an effective preference for the soluble phase of the scaffold. Small molecules, like proteins, can partition into a dilute or condensate phase. The best evidence comes from non-cellular systems: partition of neutral hydrophobic and aliphatic small molecules into various synthetic polymer condensates [121–123], partition of enzyme substrates into condensates where the enzymes are clients of the condensate [124], partitioning of dye molecules into condensate models of proto-cells [125,126] and partition of small ions out of polyelectrolyte coacervate condensates [127]. The partition of a client depends on several contributing factors to the free energy of the solute (the client) in the different coexisting liquid phases likely dominated by hydrophobic interactions, charge and hydrogen bonding depending on the properties of the scaffold [10,128], in practice this will be dominated by the properties of stickers for IDP scaffolds. A small molecule which partitions to a condensate, even without strong binding, may therefore have therapeutic value.
Thirdly, more complex interactions also seem plausible, in particular interaction with the condensate surface. There is evidence for accumulation of small dye molecules to condensate interfaces and a terpolymer has been used to stabilise a protocell coacervate [126]. Here, part of the molecule partitioned into the condensate and part out in a surfactant-like manner [126] which, like surfactants, may stabilise condensates at low concentrations and destabilise them at higher concentrations.
The synergistic
It is widely suspected that condensates provide a specialised environment for biochemistry through molecular crowding, scaffold and client biochemistry and local solvent environment [128]. They can promote enzymatic activity inefficient in the cytosol [129], concentrate proteins to promote their interaction [34,80] or sequester proteins to prevent activity [130]. There are further strong predictions from biomolecular chemistry [18,128] and a rapidly growing number of in vitro models including promoting actin nucleation [131], enhanced ribozyme activity [132] and enhanced enzyme activity [124,125,133–135]. If disease-associated enzymes are condensate clients then a synergistic approach to drug optimisation could be taken—linking a small molecule inhibitor with a molecule that partitions to the condensate to confer enhanced efficacy and/or specificity.
Is a physicochemical mechanism plausible?
Existing evidence indicates a physicochemical mechanism is plausible, however existing evidence primarily involves the effect of proteins rather than small molecules on LLPS. Providing evidence for a physicochemical mode of action of a small molecule using normal drug development paradigms is challenging as it does not necessarily require strong small molecule binding to the target. Evidence for target engagement will therefore also pose challenges. However, there is early evidence small molecules can modulate LLPS from synthetic polymers. Methylene blue partitions to a polyelectrolyte coacervate through hydrophobic interactions [123] and reduces the effective strength of π–π and cation-π interactions, modulating whether LLPS or aggregation occurs and changing condensate properties [136]. Planar heterocyclic small molecules including mitoxatrone, similar to methylene blue, have been identified as preventing stress granule protein LLPS [65,66]. It was suggested they act through disrupting RNA base stacking by intercalation [65] however can act on RNA-free in vitro LLPS [66] perhaps reducing cation-π interaction required for FUS-like protein LLPS [100] (Figure 2). Our work also identified lipoamide as a stress granule LLPS modulating compound, with physicochemical effects in vitro and specific effects on stress granules in cells [66]. This suggests that small molecules that reach high concentration by partition to specific condensates could indeed alter LLPS.
Figure 2. Physicochemical intervention of protein liquid–liquid phase separation?.
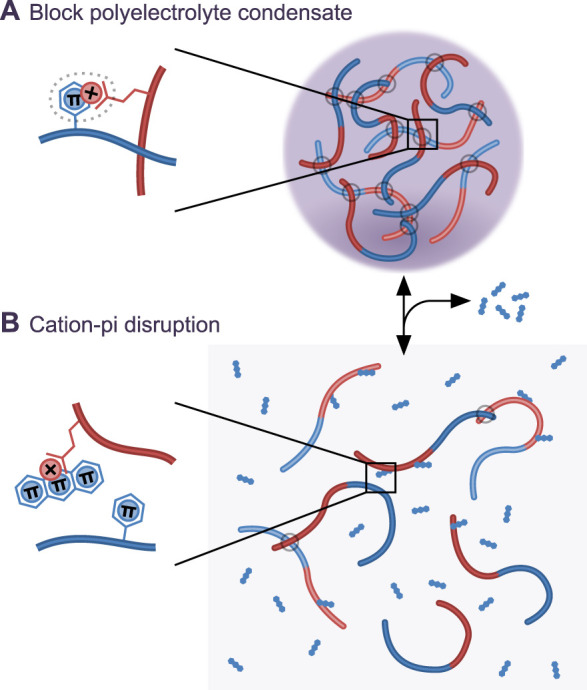
(A) Cartoon representation of a model block polyelectrolyte condensate whose formation involves cation-π interactions inspired by synthetic polymer [136] and FUS-like [100] LLPS. (B) A compound which partitions to the condensate and reduces effective strength of cation-π interactions hypothetically leads to condensate disruption.
Could this approach be efficacious? A small molecule needs to reach the target, have sufficient specificity and have a beneficial effect. Conventional small molecule therapeutics are guided by Lipinski's rule of five [137] and similar [138,139] to bias towards good oral bioavailability. Will small molecules with a physicochemical mechanism tend to meet these guidelines? And would they be amenable to medicinal chemistry to better meet these criteria? This also assumes that condensate modulation is beneficial to patients—the precise pathomechanisms are unclear in many of these diseases and aberrant condensate formation/dissolution may be a phenotype rather than a cause.
Specificity is an important concern. It is not yet clear that different condensates may be targeted specifically by partition of a small molecule to only the target condensate. 1,6-hexanediol affects many condensates [45,110–112] and sets an unfortunate precedent. On the other hand, coexisting immiscible condensates are common, perhaps best exemplified by the nucleolar sub-compartments [2]. Immiscibility points to different physicochemical scaffold properties giving specific partition of clients and presumably allowing specific partition of a small molecule [74]. Growing knowledge of the molecular grammar of the stickers involved in IDP scaffold LLPS [100,102–105] may even allow guided physicochemical drug design, analogous to structure-based drug design, although perhaps not in the near future. Broader concerns are sensitivity of condensates in target cells (e.g. cancer cells or neurons) compared with off-target cells. Aberrant condensates, such in Ewing's sarcoma [40], may be only present in the target cell, but form by LLPS using scaffolds (in this case, EWSR1) used in all cells for other physiological condensates. Therapeutic possibility here hinges on the side effects of disrupting normal EWSR1 function in balance with the benefits of disrupting a nuclear EWSR1 fusion protein.
The necessary pre-requisite for any high throughput screen is that small molecules with activity exist in the screened library, and this library is normally filtered to avoid small molecules with undesirable physicochemical properties (e.g. aggregation [140]) and small molecules which often interfere with assays (termed PAINs [141]). For a physicochemical mechanism, arguably small molecules viewed as having poor physicochemical properties should be included—small molecules which can form micelles, colloids or other aggregates might be highly effective against the physical chemistry of LLPS and be a true hit, while they would interfere with in vitro assays for recombinant enzyme inhibition. Arguably, it may also be desirable to avoid widely successful chemical fragments or privileged scaffolds to avoid interaction globular proteins which would be an off-target effect. Screening will also require novel approaches. Perhaps partition to in vitro condensates or phenotypic screens for effects on condensates in cells.
Conclusions
Many MLOs formed by LLPS are associated with disease through a combination of condensates forming in aberrant locations, with aberrant composition or aberrant physical properties. This suggests many possibilities for therapeutics. PTM enzymes are canonical drug targets and PTM often controls LLPS, condensate composition and condensate nucleation. Furthermore, presence of condensates is a simple phenotypic readout to screen for hits. However, in many neurodegenerative diseases and cancers the pathomechanism is not concrete, is LLPS actually a good target or is aberrant LLPS simply a consequence of pathology? More basic research into these disease pathomechanisms are needed to confer confidence in LLPS as a target.
More speculatively, direct targeting of LLPS may be a novel therapeutic approach. Most drug targets are enzymes, ion channels, G-protein coupled receptors, kinases, nuclear receptors and transporters. However, well over half of the genome, including IDPs, does not fall into these classes [25,142,143]. The physical chemistry of LLPS for condensate formation provides a new conceptual framework for how small molecules may interact with IDPs—one which does not necessarily involve well-defined binding sites and instead physical chemistry mechanisms. However, finding these molecules may need careful library design and re-evaluation of what defines a small molecule as drug-like. It is also not yet unambiguous if physicochemical mechanisms will be efficacious and specific.
Summary
Many roles of liquid–liquid phase separation have emerged in neurodegeneration and cancer.
Aberrant location, composition or physical properties of condensates formed by liquid–liquid phase separation are associated with disease.
Recent discoveries show how protein post-translational modification can regulate condensate nucleation, growth and composition.
There is great potential for small molecule inhibitors of post-translational modification to modulate liquid–liquid phase separation.
More speculatively, direct physicochemical action of small molecules on liquid–liquid phase separation may have therapeutic potential.
Abbreviations
- IDP
intrinsically disordered protein
- IDR
intrinsically disordered [protein] region
- LLPS
liquid–liquid phase separation
- MCIDP
multiple copies of interaction domain protein
- MLO
membraneless organelle
- PTM
post-translational modification
Competing Interests
R.J.W. is a Scientific Advisor for Dewpoint Therapeutics and has a consultancy agreement.
Funding
R.J.W. is supported by a Wellcome Trust Sir Henry Dale Fellowship [211075/Z/18/Z].
Open Access
Open access for this article was enabled by the participation of University of Oxford in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
References
- 1.Brangwynne C.P., Mitchison T.J. and Hyman A.A. (2011) Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. U.S.A. 108, 4334–4339 10.1073/pnas.1017150108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M. et al. (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L. et al. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X. and Karpen G.H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aulas A. and Vande Velde C. (2015) Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front. Cell Neurosci. 9, 423 10.3389/fncel.2015.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J. et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y. et al. (2015) A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- 8.Beutel O., Maraspini R., Pombo-García K., Martin-Lemaitre C. and Honigmann A. (2019) Phase separation of Zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936.e11 10.1016/j.cell.2019.10.011 [DOI] [PubMed] [Google Scholar]
- 9.Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L. et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banani S.F., Lee H.O., Hyman A.A. and Rosen M.K. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin Y. and Brangwynne C.P. (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- 12.Hyman A.A., Weber C.A. and Jülicher F. (2014) Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 10.1146/annurev-cellbio-100913-013325 [DOI] [PubMed] [Google Scholar]
- 13.Alberti S. and Dormann D. (2019) Liquid–liquid phase separation in disease. Annu. Rev. Genet. 53, 171–194 10.1146/annurev-genet-112618-043527 [DOI] [PubMed] [Google Scholar]
- 14.Elbaum-Garfinkle S. (2019) Matter over mind: liquid phase separation and neurodegeneration. J. Biol. Chem. 294, 7160–7168 10.1074/jbc.REV118.001188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spannl S., Tereshchenko M., Mastromarco G.J., Ihn S.J. and Lee H.O. (2019) Biomolecular condensates in neurodegeneration and cancer. Traffic 20, 890–911 10.1111/tra.12704 [DOI] [PubMed] [Google Scholar]
- 16.Posey A.E., Holehouse A.S. and Pappu R.V. (2018) Phase separation of intrinsically disordered proteins. Methods Enzymol. 611, 1–30 10.1016/bs.mie.2018.09.035 [DOI] [PubMed] [Google Scholar]
- 17.Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J. et al. (2016) Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 10.1126/science.aad9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holehouse A.S. and Pappu R.V. (2018) Functional implications of intracellular phase transitions. Biochemistry 57, 2415–2423 10.1021/acs.biochem.7b01136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R. et al. (2016) Compositional control of phase-separated cellular bodies. Cell 166, 651–663 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins A.L. and Groom C.R. (2002) The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 10.1038/nrd892 [DOI] [PubMed] [Google Scholar]
- 21.Kumar R.D., Chang L.-W., Ellis M.J. and Bose R. (2013) Prioritizing potentially druggable mutations with dGene: an annotation tool for cancer genome sequencing data. PLoS ONE 8, e67980 10.1371/journal.pone.0067980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russ A.P. and Lampel S. (2005) The druggable genome: an update. Drug Discov. Today 10, 1607–1610 10.1016/S1359-6446(05)03666-4 [DOI] [PubMed] [Google Scholar]
- 23.Finan C., Gaulton A., Kruger F.A., Lumbers R.T., Shah T., Engmann J. et al. (2017) The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, eaag1166 10.1126/scitranslmed.aag1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amberger J.S., Bocchini C.A., Scott A.F. and Hamosh A.O.M.I.M. (2019) Org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 47, D1038–D1043 10.1093/nar/gky1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oprea T.I., Bologa C.G., Brunak S., Campbell A., Gan G.N., Gaulton A. et al. (2018) Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug Discov. 17, 317–332 10.1038/nrd.2018.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan Y., Du A., Gu J., Duan G., Wang C., Gui X. et al. (2019) PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 29, 233–247 10.1038/s41422-019-0141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan L., Cookson M.R., Petrucelli L. and La Spada A.R. (2018) Converging pathways in neurodegeneration, from genetics to mechanisms. Nat. Neurosci. 21, 1300–1309 10.1038/s41593-018-0237-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ash P.E.A., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.-L., Dejesus-Hernandez M. et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 10.1016/j.neuron.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K. et al. (2015) RAN translation in Huntington disease. Neuron 88, 667–677 10.1016/j.neuron.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori K., Weng S.-M., Arzberger T., May S., Rentzsch K., Kremmer E. et al. (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 10.1126/science.1232927 [DOI] [PubMed] [Google Scholar]
- 31.Ambadipudi S., Biernat J., Riedel D., Mandelkow E. and Zweckstetter M. (2017) Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 8, 275 10.1038/s41467-017-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S. et al. (2018) Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 10.15252/embj.201798049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X., Lin Y., Eschmann N.A., Zhou H., Rauch J.N., Hernandez I. et al. (2017) RNA stores tau reversibly in complex coacervates. PLoS Biol. 15, e2002183 10.1371/journal.pbio.2002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T. et al. (2017) Local nucleation of microtubule bundles through tubulin concentration into a condensed Tau phase. Cell Rep. 20, 2304–2312 10.1016/j.celrep.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeynaems S., Bogaert E., Kovacs D., Konijnenberg A., Timmerman E., Volkov A. et al. (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 10.1016/j.molcel.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conicella A.E., Zerze G.H., Mittal J. and Fawzi N.L. (2016) ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peskett T.R., Rau F., O'Driscoll J., Patani R., Lowe A.R. and Saibil H.R. (2018) A liquid to solid phase transition underlying pathological huntingtin Exon1 aggregation. Mol. Cell 70, 588–601.e6 10.1016/j.molcel.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor J.P., Brown R.H. and Cleveland D.W. (2016) Decoding ALS: from genes to mechanism. Nature 539, 197–206 10.1038/nature20413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt H.B., Barreau A. and Rohatgi R. (2019) Phase separation-deficient TDP43 remains functional in splicing. Nat. Commun. 10, 1–14 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boulay G., Sandoval G.J., Riggi N., Iyer S., Buisson R., Naigles B. et al. (2017) Cancer-Specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 10.1016/j.cell.2017.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouchard J.J., Otero J.H., Scott D.C., Szulc E., Martin E.W., Sabri N. et al. (2018) Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36.e8 10.1016/j.molcel.2018.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sur I. and Taipale J. (2016) The role of enhancers in cancer. Nat. Rev. Cancer 16, 483–493 10.1038/nrc.2016.62 [DOI] [PubMed] [Google Scholar]
- 43.Rubio K., Dobersch S. and Barreto G. (2019) Functional interactions between scaffold proteins, noncoding RNAs, and genome loci induce liquid-liquid phase separation as organizing principle for 3-dimensional nuclear architecture: implications in cancer. FASEB J. 33, 5814–5822 10.1096/fj.201802715R [DOI] [PubMed] [Google Scholar]
- 44.Boeynaems S., Tompa P. and Van Den Bosch L. (2018) Phasing in on the cell cycle. Cell Div. 13, 1 10.1186/s13008-018-0034-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K. et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K. and Sharp P.A. (2017) A phase separation model for transcriptional control. Cell 169, 13–23 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinrich B.S., Maliga Z., Stein D.A., Hyman A.A. and Whelan S.P.J. (2018) Phase transitions drive the formation of vesicular stomatitis virus replication compartments. mBio 9 e02290-17 10.1128/mBio.02290-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudrière-Gesbert C., Gaudin Y. et al. (2017) Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 8, 58 10.1038/s41467-017-00102-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munder M.C., Midtvedt D., Franzmann T., Nüske E., Otto O., Herbig M. et al. (2016) A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 5, e09347 10.7554/eLife.09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parry B.R., Surovtsev I.V., Cabeen M.T., O'Hern C.S., Dufresne E.R. and Jacobs-Wagner C. (2014) The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194 10.1016/j.cell.2013.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunha S., Woldringh C.L. and Odijk T. (2001) Polymer-mediated compaction and internal dynamics of isolated escherichia coli nucleoids. J. Struct. Biol. 136, 53–66 10.1006/jsbi.2001.4420 [DOI] [PubMed] [Google Scholar]
- 52.de Vries R. (2010) DNA condensation in bacteria: interplay between macromolecular crowding and nucleoid proteins. Biochimie 92, 1715–1721 10.1016/j.biochi.2010.06.024 [DOI] [PubMed] [Google Scholar]
- 53.Navarro M. and Gull K. (2001) A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414, 759–763 10.1038/414759a [DOI] [PubMed] [Google Scholar]
- 54.Mair G.R., Braks J.A.M., Garver L.S., Dimopoulos G., Hall N., Wiegant J.C.A.G. et al. (2006) Translational repression is essential for Plasmodium sexual development and mediated by a DDX6-type RNA helicase. Science 313, 667–669 10.1126/science.1125129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ambadipudi S. and Zweckstetter M. (2016) Targeting intrinsically disordered proteins in rational drug discovery. Expert Opin. Drug Discov. 11, 65–77 10.1517/17460441.2016.1107041 [DOI] [PubMed] [Google Scholar]
- 56.Cheng Y., LeGall T., Oldfield C.J., Mueller J.P., Van Y.-Y.J., Romero P. et al. (2006) Rational drug design via intrinsically disordered protein. Trends Biotechnol. 24, 435–442 10.1016/j.tibtech.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 57.Chong B., Li M., Li T., Yu M., Zhang Y. and Liu Z. (2018) Conservation of potentially druggable cavities in intrinsically disordered proteins. ACS Omega 3, 15643–15652 10.1021/acsomega.8b02092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joshi P. and Vendruscolo M. (2015) Druggability of intrinsically disordered proteins. Adv. Exp. Med. Biol. 870, 383–400 10.1007/978-3-319-20164-1_13 [DOI] [PubMed] [Google Scholar]
- 59.Neira J.L., Bintz J., Arruebo M., Rizzuti B., Bonacci T., Vega S. et al. (2017) Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Sci. Rep. 7, 1–15 10.1038/srep39732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu C., Niu X., Jin F., Liu Z., Jin C. and Lai L. (2016) Structure-based inhibitor design for the intrinsically disordered protein c-Myc. Sci. Rep. 6, 1–11 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y., Cao H. and Liu Z. (2015) Binding cavities and druggability of intrinsically disordered proteins. Protein Sci. 24, 688–705 10.1002/pro.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makley L.N. and Gestwicki J.E. (2013) Expanding the number of ‘Druggable’ targets: non-Enzymes and protein-Protein interactions. Chem. Biol. Drug Des. 81, 22–32 10.1111/cbdd.12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voet A.R.D., Ito A., Hirohama M., Matsuoka S., Tochio N., Kigawa T. et al. (2014) Discovery of small molecule inhibitors targeting the SUMO–SIM interaction using a protein interface consensus approach. MedChemComm. 5, 783–786 10.1039/C3MD00391D [DOI] [Google Scholar]
- 64.van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K. et al. (2014) Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 10.1021/cr400525m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang M.Y., Markmiller S., Vu A.Q., Javaherian A., Dowdle W.E., Jolivet P. et al. (2019) Small-Molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819.e11 10.1016/j.neuron.2019.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wheeler R.J., Lee H.O., Poser I., Pal A., Doeleman T., Kishigami S. et al. (2019) Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv 721001 10.1101/721001 [DOI] [Google Scholar]
- 67.Hofweber M., Hutten S., Bourgeois B., Spreitzer E., Niedner-Boblenz A., Schifferer M. et al. (2018) Phase separation of FUS Is suppressed by Its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 10.1016/j.cell.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 68.Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H. et al. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q. et al. (2018) FUS phase separation Is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 10.1016/j.cell.2018.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang A., Conicella A.E., Schmidt H.B., Martin E.W., Rhoads S.N., Reeb A.N. et al. (2018) A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 10.15252/embj.201797452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Protter D.S.W. and Parker R. (2016) Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hofweber M. and Dormann D. (2019) Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Snead W.T. and Gladfelter A.S. (2019) The control centers of biomolecular phase separation: how membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell 76, 295–305 10.1016/j.molcel.2019.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wheeler R.J. and Hyman A.A. (2018) Controlling compartmentalization by non-membrane-bound organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170193 10.1098/rstb.2017.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim T.H., Tsang B., Vernon R.M., Sonenberg N., Kay L.E. and Forman-Kay J.D. (2019) Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 10.1126/science.aax4240 [DOI] [PubMed] [Google Scholar]
- 76.Guo W., Vandoorne T., Steyaert J., Staats K.A. and Van Den Bosch L. (2020) The multifaceted role of kinases in amyotrophic lateral sclerosis: genetic, pathological and therapeutic implications. Brain, awaa022 10.1093/brain/awaa022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hu J.-H., Zhang H., Wagey R., Krieger C. and Pelech S.L. (2003) Protein kinase and protein phosphatase expression in amyotrophic lateral sclerosis spinal cord. J. Neurochem. 85, 432–442 10.1046/j.1471-4159.2003.01670.x [DOI] [PubMed] [Google Scholar]
- 78.Kametani F., Obi T., Shishido T., Akatsu H., Murayama S., Saito Y. et al. (2016) Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 23281 10.1038/srep23281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neumann M., Kwong L.K., Lee E.B., Kremmer E., Flatley A., Xu Y. et al. (2009) Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. (Berl) 117, 137–149 10.1007/s00401-008-0477-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woodruff J.B., Gomes B.F., Widlund P.O., Mahamid J., Honigmann A. and Hyman A.A. (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077.e10 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 81.Kwon I., Kato M., Xiang S., Wu L., Theodoropoulos P., Mirzaei H. et al. (2013) Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 10.1016/j.cell.2013.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ditlev J.A., Case L.B. and Rosen M.K. (2018) Who's In and who's Out—Compositional control of biomolecular condensates. J. Mol. Biol. 430, 4666–4684 10.1016/j.jmb.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan T., Kandola T.S., Wu J., Venkatesan S., Ketter E., Lange J.J. et al. (2018) Quantifying nucleation in vivo reveals the physical basis of prion-like phase behavior. Mol. Cell 71, 155–168.e7 10.1016/j.molcel.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Posey A.E. and Pappu R.V. (2018) A first glimpse of nucleation of phase transitions in living cells. Mol. Cell 71, 1–3 10.1016/j.molcel.2018.06.028 [DOI] [PubMed] [Google Scholar]
- 85.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J. et al. (2009) Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 86.Kim S., Kalappurakkal J.M., Mayor S. and Rosen M.K. (2019) Phosphorylation of nephrin induces phase separated domains that move through actomyosin contraction. Mol. Biol. Cell 30, 2996–3012 10.1091/mbc.E18-12-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang J.T., Smith J., Chen B.-C., Schmidt H., Rasoloson D., Paix A. et al. (2014) Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife 3, e04591 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garcia-Jove Navarro M., Kashida S., Chouaib R., Souquere S., Pierron G., Weil D. et al. (2019) RNA is a critical element for the sizing and the composition of phase-separated RNA–protein condensates. Nat. Commun. 10, 1–13 10.1038/s41467-019-11241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M. et al. (2015) Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGurk L., Mojsilovic-Petrovic J., Van Deerlin V.M., Shorter J., Kalb R.G., Lee V.M. et al. (2018) Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 6, 84 10.1186/s40478-018-0586-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGurk L., Gomes E., Guo L., Mojsilovic-Petrovic J., Tran V., Kalb R.G. et al. (2018) Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 10.1016/j.molcel.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Banjade S. and Rosen M.K. (2014) Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3, e04123 10.7554/eLife.04123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.-A. et al. (2017) The target landscape of clinical kinase drugs. Science 358, eaan4368 10.1126/science.aan4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mullard A. (2019) Biomolecular condensates pique drug discovery curiosity. Nat. Rev. Drug Discov. 18, 324–326 10.1038/d41573-019-00069-w [DOI] [PubMed] [Google Scholar]
- 95.Edfeldt F.N.B., Folmer R.H.A. and Breeze A.L. (2011) Fragment screening to predict druggability (ligandability) and lead discovery success. Drug Discov. Today 16, 284–287 10.1016/j.drudis.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 96.Sakharkar M.K., Rajamanickam K., Babu C.S., Madan J., Chandra R. and Yang J. (2019) Preclinical: Drug Target Identification and Validation in Human In Encyclopedia of Bioinformatics and Computational Biology (Ranganathan S., Gribskov M., Nakai K. and Schönbach C., eds), pp. 1093–1098, Academic Press, Oxford [Google Scholar]
- 97.Vukovic S. and Huggins D.J. (2018) Quantitative metrics for drug–target ligandability. Drug Discov. Today 23, 1258–1266 10.1016/j.drudis.2018.02.015 [DOI] [PubMed] [Google Scholar]
- 98.Yuan Y., Pei J. and Lai L. (2013) Binding site detection and druggability prediction of protein targets for structure-based drug design. Curr. Pharm. Des. 19, 2326–2333 10.2174/1381612811319120019 [DOI] [PubMed] [Google Scholar]
- 99.Vajda S., Beglov D., Wakefield A.E., Egbert M. and Whitty A. (2018) Cryptic binding sites on proteins: definition, detection, and druggability. Curr. Opin. Chem. Biol. 44, 1–8 10.1016/j.cbpa.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J., Choi J.-M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M. et al. (2018) A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Majumdar A., Dogra P., Maity S. and Mukhopadhyay S. (2019) Liquid–Liquid phase separation Is driven by large-scale conformational unwinding and fluctuations of intrinsically disordered protein molecules. J. Phys. Chem. Lett. 10, 3929–3936 10.1021/acs.jpclett.9b01731 [DOI] [PubMed] [Google Scholar]
- 102.Yang Y., Jones H.B., Dao T.P. and Castañeda C.A. (2019) Single amino acid substitutions in stickers, but not spacers, substantially alter UBQLN2 phase transitions and dense phase material properties. J. Phys. Chem. B 123, 3618–3629 10.1021/acs.jpcb.9b01024 [DOI] [PubMed] [Google Scholar]
- 103.Martin E.W., Holehouse A.S., Peran I., Farag M., Incicco J.J., Bremer A. et al. (2020) Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 10.1126/science.aaw8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A. et al. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choi J.-M., Holehouse A.S. and Pappu R.V. (2020) Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 10.1146/annurev-biophys-121219-081629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peran I. and Mittag T. (2020) Molecular structure in biomolecular condensates. Curr. Opin. Struct. Biol. 60, 17–26 10.1016/j.sbi.2019.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patel S.S., Belmont B.J., Sante J.M. and Rexach M.F. (2007) Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- 108.Ribbeck K. and Görlich D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shulga N. and Goldfarb D.S. (2003) Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol. Cell Biol 23, 534–542 10.1128/MCB.23.2.534-542.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Updike D.L., Hachey S.J., Kreher J. and Strome S. (2011) P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939–948 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I. et al. (2015) Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wheeler J.R., Matheny T., Jain S., Abrisch R. and Parker R. (2016) Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y. et al. (2017) ATP as a biological hydrotrope. Science 356, 753–756 10.1126/science.aaf6846 [DOI] [PubMed] [Google Scholar]
- 114.Wurtz J.D. and Lee C.F. (2018) Stress granule formation via ATP depletion-triggered phase separation. New. J. Phys. 20, 045008 10.1088/1367-2630/aab549 [DOI] [Google Scholar]
- 115.Tisel W.A., Haire R.N., White J.G., Rosenberg A. and Middaugh C.R. (1980) Polyphasic linkage between protein solubility and ligand binding in the hemoglobin-polyethylene glycol system. J. Biol. Chem. 255, 8975–8978 PMID: [PubMed] [Google Scholar]
- 116.Wyman J. and Gill S.J. (1990) Binding and Linkage: Functional Chemistry of Biological Macromolecules, University Science Books, Mill Valley, California, U.S.A [Google Scholar]
- 117.Wyman J. and Gill S.J. (1980) Ligand-linked phase changes in a biological system: applications to sickle cell hemoglobin. Proc. Natl Acad. Sci. U.S.A. 77, 5239–5242 10.1073/pnas.77.9.5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Posey A.E., Ruff K.M., Harmon T.S., Crick S.L., Li A., Diamond M.I. et al. (2018) Profilin reduces aggregation and phase separation of huntingtin N-terminal fragments by preferentially binding to soluble monomers and oligomers. J. Biol. Chem. 293, 3734–3746 10.1074/jbc.RA117.000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dao T.P., Martyniak B., Canning A.J., Lei Y., Colicino E.G.. Cosgrove M.S., et al. (2019) ALS-Linked Mutations affect UBQLN2 oligomerization and phase separation in a position- and amino acid-Dependent manner. Structure 27, 937–951.e5 10.1016/j.str.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nguemaha V. and Zhou H.-X. (2018) Liquid-Liquid phase separation of patchy particles illuminates diverse effects of regulatory components on protein droplet formation. Sci. Rep. 8, 1–11 10.1038/s41598-018-25132-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Willauer H.D., Huddleston J.G. and Rogers R.D. (2002) Solute partitioning in aqueous biphasic systems composed of polyethylene glycol and salt: the partitioning of small neutral organic species. Ind. Eng. Chem. Res. 41, 1892–1904 10.1021/ie010598z [DOI] [Google Scholar]
- 122.Zhao M., Eghtesadi S.A., Dawadi M.B., Wang C., Huang S., Seymore A.E. et al. (2017) Partitioning of small molecules in hydrogen-bonding complex coacervates of poly(acrylic acid) and poly(ethylene glycol) or pluronic block copolymer. Macromolecules 50, 3818–3830 10.1021/acs.macromol.6b02815 [DOI] [Google Scholar]
- 123.Zhao M. and Zacharia N.S. (2016) Sequestration of methylene blue into polyelectrolyte complex coacervates. Macromol. Rapid Commun. 37, 1249–1255 10.1002/marc.201600244 [DOI] [PubMed] [Google Scholar]
- 124.Davis B.W., Aumiller W.M., Hashemian N., An S., Armaou A. and Keating C.D. (2015) Colocalization and sequential enzyme activity in aqueous biphasic systems: experiments and modeling. Biophys. J. 109, 2182–2194 10.1016/j.bpj.2015.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Koga S., Williams D.S., Perriman A.W. and Mann S. (2011) Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 3, 720–724 10.1038/nchem.1110 [DOI] [PubMed] [Google Scholar]
- 126.Yewdall N.A., Buddingh B.C., Altenburg W.J., Timmermans S.B.P.E., Vervoort D.F.M., Abdelmohsen L.K.E.A. et al. (2019) Physicochemical characterization of polymer-Stabilized coacervate protocells. Eur. J. Chem. Biol. 20, 2643–2652 10.1002/cbic.201900195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li L., Srivastava S., Andreev M., Marciel A.B., de Pablo J.J. and Tirrell M.V. (2018) Phase behavior and salt partitioning in polyelectrolyte complex coacervates. Macromolecules 51, 2988–2995 10.1021/acs.macromol.8b00238 [DOI] [Google Scholar]
- 128.Nakashima K.K., Vibhute M.A. and Spruijt E. (2019) Biomolecular chemistry in liquid phase separated compartments. Front. Mol. Biosci. 6, 21 10.3389/fmolb.2019.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aguilera-Gomez A. and Rabouille C. (2017) Membrane-bound organelles versus membrane-less compartments and their control of anabolic pathways in Drosophila. Dev. Biol. 428, 310–317 10.1016/j.ydbio.2017.03.029 [DOI] [PubMed] [Google Scholar]
- 130.Alberti S. (2017) The wisdom of crowds: regulating cell function through condensed states of living matter. J. Cell Sci. 130, 2789–2796 10.1242/jcs.200295 [DOI] [PubMed] [Google Scholar]
- 131.McCall P.M., Srivastava S., Perry S.L., Kovar D.R., Gardel M.L. and Tirrell M.V. (2018) Partitioning and enhanced self-Assembly of actin in polypeptide coacervates. Biophys. J. 114, 1636–1645 10.1016/j.bpj.2018.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strulson C.A., Molden R.C., Keating C.D. and Bevilacqua P.C. (2012) RNA catalysis through compartmentalization. Nat. Chem. 4, 941–946 10.1038/nchem.1466 [DOI] [PubMed] [Google Scholar]
- 133.Sokolova E., Spruijt E., Hansen M.M.K., Dubuc E., Groen J., Chokkalingam V. et al. (2013) Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc. Natl Acad. Sci. U.S.A. 110, 11692–7 10.1073/pnas.1222321110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stroberg W. and Schnell S. (2018) Do cellular condensates accelerate biochemical reactions? lessons from microdroplet chemistry. Biophys. J. 115, 3–8 10.1016/j.bpj.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kojima T. and Takayama S. (2018) Membraneless compartmentalization facilitates enzymatic cascade reactions and reduces substrate inhibition. ACS Appl. Mater Interfaces 10, 32782–32791 10.1021/acsami.8b07573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang S., Zhao M., Dawadi M.B., Cai Y., Lapitsky Y., Modarelli D.A. et al. (2018) Effect of small molecules on the phase behavior and coacervation of aqueous solutions of poly(diallyldimethylammonium chloride) and poly(sodium 4-styrene sulfonate). J. Colloid Interface Sci. 518, 216–224 10.1016/j.jcis.2018.02.029 [DOI] [PubMed] [Google Scholar]
- 137.Lipinski C.A., Lombardo F., Dominy B.W. and Feeney P.J. (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 10.1016/S0169-409X(96)00423-1 [DOI] [PubMed] [Google Scholar]
- 138.Leeson P.D. and Springthorpe B. (2007) The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 6, 881–890 10.1038/nrd2445 [DOI] [PubMed] [Google Scholar]
- 139. (2007) A decade of drug-likeness. Nat. Rev. Drug Discov. 6, 853 10.1038/nrd2460 [DOI] [Google Scholar]
- 140.Reker D., Bernardes G.J.L. and Rodrigues T. (2019) Computational advances in combating colloidal aggregation in drug discovery. Nat. Chem. 11, 402–418 10.1038/s41557-019-0234-9 [DOI] [PubMed] [Google Scholar]
- 141.Baell J.B. and Holloway G.A. (2010) New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 10.1021/jm901137j [DOI] [PubMed] [Google Scholar]
- 142.Rodgers G., Austin C., Anderson J., Pawlyk A., Colvis C., Margolis R. et al. (2018) Glimmers in illuminating the druggable genome. Nat. Rev. Drug Discov. 17, 301–302 10.1038/nrd.2017.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G. et al. (2017) A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 10.1038/nrd.2016.230 [DOI] [PMC free article] [PubMed] [Google Scholar]